
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 12 February 2019
Sec. Plant Pathogen Interactions
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00183
Type three secretion system (T3SS) is essential for Ralstonia solanacearum to cause disease in host plants and we previously screened AroG1 as a candidate with impact on the T3SS expression. Here, we focused on two putative DAHP synthases of AroG1 and AroG2, which control the first step of the shikimate pathway, a common route for biosynthesis of aromatic amino acids (AAA), to characterize their functional roles and possible links with virulence in R. solanacearum. Deletion of aroG1/2 or aroG1, but not aroG2, significantly impaired the T3SS expression both in vitro and in planta, and the impact of AroG1 on T3SS was mediated with a well-characterized PrhA signaling cascade. Virulence of the aroG1/2 or aroG1 mutants was completely diminished or significantly impaired in tomato and tobacco plants, but not the aroG2 mutants. The aroG1/2 mutants failed to grow in limited medium, but grew slowly in planta. This significantly impaired growth was also observed in the aroG1 mutants both in planta and limited medium, but not in aroG2 mutants. Complementary aroG1 significantly restored the impaired or diminished bacterial growth, T3SS expression and virulence. Supplementary AAA or shikimic acid, an important intermediate of the shikimate pathway, significantly restored diminished growth in limited medium. The promoter activity assay showed that expression of aroG1 and aroG2 was greatly increased to 10-20-folder higher levels with deletion of the other. All these results demonstrated that both AroG1 and AroG2 are involved in the shikimate pathway and cooperatively essential for AAA biosynthesis in R. solanacearum. The AroG1 plays a major role on bacterial growth, T3SS expression and pathogenicity, while the AroG2 is capable to partially carry out the function of AroG1 in the absence of AroG1.
Like in many pathogenic bacteria of animals and plants, the syringe-like type three secretion system (T3SS) is essential for Ralstonia solanacearum to cause disease in host plants, which is a causal agent of bacterial wilt disease on more than 450 plant species worldwide (Genin et al., 2005; Genin and Denny, 2012; Jiang et al., 2017). Bacteria use this syringe-like apparatus to inject virulence factors, called type III effectors (T3Es), into host cytosol to subvert host defense and cause diseases (Cunnac et al., 2004; Tasset et al., 2010; Fujiwara et al., 2016; Popa et al., 2016). R. solanacearum is heterogeneous and currently regarded as a Ralstonia solanacearum species complex (RSSC), while the T3SS is highly conserved in RSSC strains, which is encoded by approximately 20 genes of hypersensitive response and pathogenicity (hrp) gene cluster and is globally controlled by a complex regulation network (Cunnac et al., 2004; Valls et al., 2006; Hikichi et al., 2007; Genin and Denny, 2012).
In general, the T3SS and entire T3Es (more than 100 T3Es in RSSC) are directly controlled by a master regulator HrpB, which is an AraC type of transcriptional regulator and binds directly to plant-inducible promoter (PIP) motif in the promoter regions of its target genes (Cunnac et al., 2004; Mukaihara et al., 2004, 2010). Two close paralogs of OmpR/PhoB family of two-component response regulators, HrpG and PrhG, positively regulate the hrpB expression in a parallel manner (Plener et al., 2010; Zhang et al., 2013). Expression of the hrpB and T3SS is not activated until the bacterium gets contact with host signals or some mimic signals, such as in nutrient- limited media that mimic plant apoplastic fluids (Arlat et al., 1992; Angot et al., 2006; Yoshimochi et al., 2009b). These signals are presumed to be recognized by an outer membrane protein PrhA and transferred to HrpG by a well-characterized signaling cascade of PrhA-PrhR/I-PrhJ or some novel signaling cascades (Marenda et al., 1998; Genin et al., 2005; Yoshimochi et al., 2009a; Zuluaga et al., 2013). Host signals can greatly increase the T3SS expression to about 10–20-folder higher levels than that in nutrient-limited media (Marenda et al., 1998; Yoshimochi et al., 2009b). HrpG and PrhG can respond to host signals by phosphorylation at certain residues and in turn activate the hrpB expression, while their regulation mechanism remains to be further elucidated (Yoshimochi et al., 2009b; Zhang et al., 2013). Moreover, a LysR type of transcriptional regulator PhcA negatively regulates hrpG expression, which is activated at high cell density and binds to promoters of prhI/R genes to repress their expression, and in tandem to repress the hrpB expression, while the PhcA positively regulates prhG expression (Genin et al., 2005; Yoshimochi et al., 2009a; Plener et al., 2010; Zhang et al., 2013). This results in dual regulation pathways of PhcA on hrpB expression, and R. solanacearum might switch from using the HrpG to PrhG for hrpB activation in a cell density-dependent manner (Zhang et al., 2013).
In order to further elucidate the global regulation on T3SS in R. solanacearum, we generated a popA-lacZYA fusion to monitor expression profiles of the T3SS in OE1-1, and screened several candidates with impact on expression of the T3SS by transposon mutagenesis, including the AroG1 (Rsc2660 in GMI1000) (Zhang et al., 2013), which is annotated as a putative DAHP synthase and catalyzes the formation of 3-deoxy-D- arabino-heptulosonate-7-phosphate (DAHP) by condensation of phosphoenolpyruvate (PEP) and erythrose 4-phosphate (E4P) (Ogino et al., 1982; Herrmann, 1983). This is the first step in the shikimate pathway that comprises seven steps beginning with the condensation of PEP and E4P, and ending with the formation of chorismate, and is a common route for biosynthesis of aromatic amino acids (AAA), including L-phenylalanine (Phe), L-tyrosine (Tyr) and L-Tryptophan (Trp), in bacteria, fungi, plants, and some protists (Herrmann and Weaver, 1999; Sprenger, 2006; Maeda and Dudareva, 2012). Chorismate is a common precursor for individual postchorismate pathways that lead to biosynthesis of AAA and their derivatives, such as vitamin K, ubiquinone and folic acid (Bentley, 1990; Dosselaere and Vanderleyden, 2001; Gosset et al., 2001). To date, all the enzymes, their corresponding genes and metabolic intermediates in the shikimate pathway have been well characterized in Gram-negative Escherichia coli and Gram-positive Bacillus subtilis (Pittard and Yang, 2005; Sprenger, 2006). A total of three isoenzymes of DAHP synthases of AroF, AroG and AroH, have been identified in E. coli, expression of which is inhibited by their corresponding production of AAA, while only one DAHP synthase is identified in B. subtilis, expression of which is not affected by its corresponding production of AAA (Umbarger, 1978; Herrmann, 1983; Pittard, 1996; Wu et al., 2005).
With genome searching1, only AroG, including two putative DAHP synthases of AroG1 and AroG2 (Rsc0743 in GMI1000) is annotated in RSSC, which share 58% of identities at amino acids (AA), and are greatly conserved in RSSC. As a vascular phytopathogenic bacterium, extensive proliferation in xylem vessels and its producing extracellular polysaccharide (EPS) slime have been believed to be the other main virulence factors in R. solanacearum, which severely block the sap flow in xylem vessels and causes plants rapidly stunting and wilting (Roberts et al., 1988; Denny, 1995; Vasse et al., 1995). In addition to T3SS and EPS, several molecular determinants are also involved in pathogenicity of R. solanacearum (Genin and Denny, 2012). Here, we focused on these two putative DAHP synthases of AroG1 and AroG2 to characterize their functional roles in AAA biosynthesis and possible links with virulence in R. solanacearum.
Ralstonia solanacearum strains used in this study are listed in Table 1, which are derivatives of two typical strains of OE1-1 and GMI1000. OE1-1 is virulent on both tomato and tobacco plants (Kanda et al., 2003), while GMI1000 is virulent on tomato plants but elicits HR in tobacco leaves (Salanoubat et al., 2002). R. solanacearum strains were grown at 28°C in nutrient-rich medium (B medium) or nutrient-limited medium (sucrose medium, hrp-inducing medium) (Yoshimochi et al., 2009b). E. coli DH12S and S17-1 were grown in Luria-Bertani medium at 37°C for plasmid construction and conjugational transfer, respectively.
In this study, mutants with in-frame deletion of target genes were generated with the pK18mobsacB based homologue recombination (Zhang et al., 2011). For plasmid construction, two DNA fragments (about 600-bp) flanking the target gene were PCR amplified from OE1-1 genomic DNA with respective primer-pairs, and subjected for joint PCR to generate DNA fragment, in which coding sequence (CDS) of target gene was absent. These DNA fragments were finally sub-cloned into pK18mobsacB to get pk18daroG1 and pk18daroG2, respectively. After validating sequence, these plasmids were transferred from E. coli S17-1 into R. solanacearum strains by conjugation and the aroG1 and aroG2 mutants were generated with confirmation of colony PCR. The aroG1 was further deleted from aroG2 mutants to generate aroG1/2 mutants. Primers used in this study were listed in Supplementary Table S1.
In this study, genetic complementation was performed with pUC18-mini-Tn7T-Gm based site specific chromosome integration system (Choi et al., 2005; Zhang et al., 2011). For plasmid construction, DNA fragment, containing the aroG1 and upstream region of about 600-bp (empirically harboring native promoter) was PCR amplified from OE1-1 genomic DNA, and finally sub-cloned into pUC18-mini-Tn7T-Gm to get pUCaroG1. After validating sequence, the complementary aroG1 was integrated into chromosome of R. solanacearum strains at a single attTn7 site (25-bp downstream of glmS) and confirmed by colony PCR (Zhang et al., 2011).
Mutants with promoter-exchanged aroG1, aroG2 or truncated AroG2 were generated with the above site specific chromosome integration system. For promoter exchange, upstream regions of aroG1 and aroG2 (about 600-bp to start codon, empirically harboring promoter) were PCR amplified from OE1-1 genomic DNA, and subjected for joint PCR to fuse promoter of aroG2 and aroG1 with CDS of aroG1 and aroG2, correspondingly. These DNA fragments were finally sub-cloned into pUC18-mini-Tn7T-Gm to get pUCg2p::G1 (aroG2 promoter-aroG1 CDS) and pUCg1p::G2 (aroG1 promoter-aroG2 CDS), respectively. After validating sequence, these promoter exchanged aroG1 and aroG2 were integrated into chromosomes of aroG1 or aroG1/2 mutants to generate desired mutants (Table 1).
The unique N-terminal region of 43 AA in AroG2 is one of remarkable difference between AroG1 and AroG2, and we generated mutants with N-terminal truncated AroG2 by integrating truncated aroG2 into aroG1 or aroG1/2 mutants with the above site specific chromosome integration system. Two DNA fragments flanking deletion region (129-bp after start codon) were PCR amplified from OE1-1 genomic DNA, respectively, and subjected for joint PCR to generate desired DNA fragment, in which the region of 129-bp was absent. This DNA fragment was finally sub-cloned into pUC18-mini-Tn7T-Gm to get pUCaroG2N. After validating sequence, this truncated aroG2 was integrated into chromosome of aroG1 or aroG1/2 mutants to generate desired mutants (Table 1).
Reporter strains with aroG1-lacZYA and aroG2-lacZYA were generated with the site specific chromosome integration system. In general, promoter-less lacZYA was fused to aroG1 or aroG2 at about 54-bp after start codon, in which 6-bp of nucleotide acids were replaced with Kpn I for lacZYA insertion. The Kpn I site was generated by PCR primers and the DNA fragment containing promoter region and Kpn I site was firstly cloned into pUC18-mini-Tn7T-Gm and then promoter-less lacZYA was inserted to get pUCaroG1-lacZYA and pUCaroG2-lacZYA, respectively. After validating sequence, these reporter fusions were integrated into chromosome of aroG1 or aroG1/2 mutants to generate desired mutants (Table 1).
In this study, expression of genes, which were fused with promoter-less lacZYA, were evaluated with the β-galactosidase assay both in vitro and in planta as previously described (Zhang et al., 2013). Enzyme activity in vitro was expressed in Miller Units (Miller, 1992), and that in planta was normalized with luminescence divided by cells number. Each assay was independently repeated for at least four times, and each trial included three replications. Mean values of all experiments were averaged with SD and statistical significance was assessed using a post hoc Dunnett test following ANOVA.
In this study, tomato plants (Solanum lycopersicum cv. Moneymaker) and tobacco plants (Nicotiana tabacum CV. Bright Yellow) were grown at 25°C for 2–3 or 3–4 weeks, respectively, and subjected for virulence assay. Tomato plants were inoculated by methods of soil-soaking, which mimics natural invasion through roots, and petiole inoculation, which enables direct invasion into xylems vessels (Yao and Allen, 2007; Zhang et al., 2013). Tobacco plants were inoculated by methods of soil-soaking and leaf-infiltration, which enables direct invasion into host plants (Zhang et al., 2013). For soil-soaking, unwounded plants were inoculated by pouring bacterial suspension onto the soil to a final concentration of 1 × 107 cfu g-1 soil. For petiole inoculation, 2 μl of bacteria suspension at 108 cfu ml-1 was dropped onto the freshly cut surface of tomato petioles. For leaf-infiltration, about 50 μl of bacterial suspension at 108 cfu ml-1 was infiltrated into tobacco leaves. Each assay was repeated independently for at least four times and each trial included 12 plants. Wilt symptoms of plants were inspected as 1–4 disease index and mean values of all experiments were averaged with SD. Disease index was recorded with integer representation and the SD was extremely high. In consideration of aesthetic appearance, the SD was not presented in figures for virulence assay. Statistical significance was assessed using a post hoc Dunnett test following ANOVA.
HR test was performed in tobacco leaves. Briefly, approximately 50 μl of bacterial suspension at 108 cfu ml-1 was infiltrated into tobacco leaves with a blunt-end syringe and development of necrotic lesions in leaves was recorded periodically (Zhang et al., 2015). Each test was repeated independently at least for four times and each trial included four plants. The representative result was presented.
Bacterial growth in media, including rich and limited medium was measured with the optical density at 600 nm (OD600). For growth complementation, AAA or shikimic acid (SA) was supplemented into limited medium at a concentration of 0.1 mM and the OD600 was assayed. Growth in tomato stems and tobacco leaves was assessed as described previously (Zhang et al., 2013). Briefly, stem tissues from tomato plants and leaf disks from tobacco plants were removed daily for quantification of cells number by dilution plating. Cells density in stems and leaves was expressed in log10 cfu g-1 and log10 cfu cm-2, respectively. Each assay was repeated independently for at least four times and each trial included four plants. Mean values of all experiments were averaged with SD and statistical significance was assessed using a post hoc Dunnett test following ANOVA.
AroG catalyzes the first step in the shikimate pathway that controls AAA biosynthesis. We therefore evaluated the growth of aroG mutants in nutrient rich and limited media. The aroG2 mutant (RQ5803) exhibited identical growth as RK5050 in both media, while growth of aroG1 mutant (RQ5687) was significantly impaired in both media (Figures 1A,B). Growth of aroG1/2 mutant (RQ5806) was much less than that of RQ5687 in rich medium (Figure 1A), while it failed to grow in limited medium, which remained the OD600 at about 0.1 till 24 h post inoculation (hpi) (Figure 1B). Note that growth of aroG1 mutant and aroG1/2 mutant could eventually reach to the maximum OD600 as RK5050 at 24 hpi in rich medium (Figure 1A). Complementary aroG1 fully restored the impaired growth of RQ5687 and diminished growth of RQ5806 to that of RK5050 in limited medium (Figure 1B), indicating that both AroG1 and AroG2 are cooperatively essential for bacterial growth of R. solanacearum. The AroG1 plays a major role on growth, while the AroG2 becomes to partially function in the absence of AroG1.
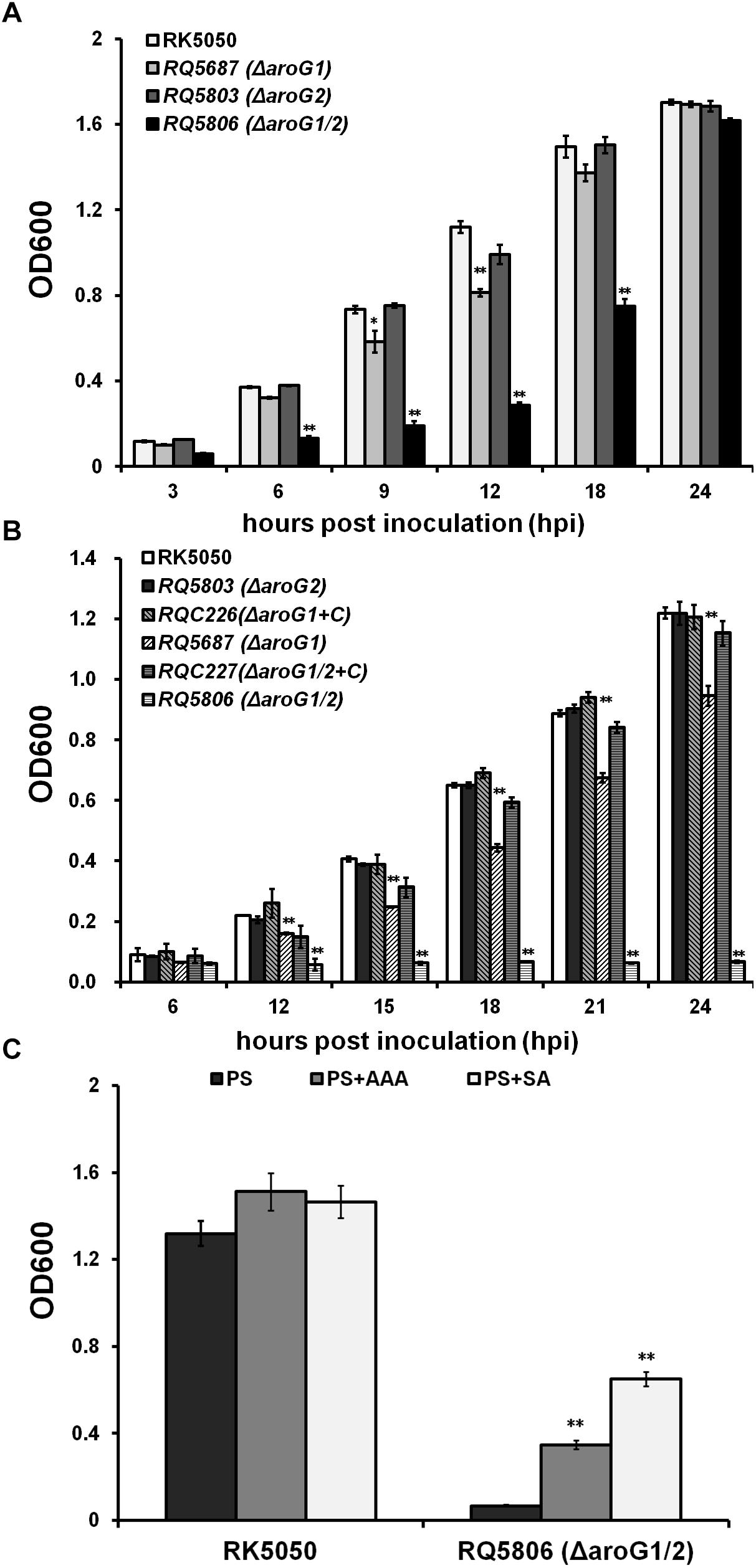
Figure 1. Growth of aroG mutants of OE1-1 in media of (A) nutrient-rich medium (B medium), (B) nutrient limited medium (sucrose medium, PS), (C) PS supplemented with AAA or SA. Each of three AAA (L-Phe, L-Tyr, and L-Trp) or SA was added into sucrose medium at a concentration of 0.1 mM. Cells suspension (OD600 = 1.0, washed twice with distilled water) was inoculated into media as a ratio of 1/100, and OD600 was measured periodically. RQC226 and RQC227 refer to aroG1 and aroG1/2 mutants complemented with aroG1 of OE1-1, respectively. Mean values of four independent trials with three replications per trial were averaged and presented with SD (error bars). Statistical significance between RK5050 (wild type) and aroG mutants was assessed using a post hoc Dunnett test following ANOVA. Significance level, ∗∗ indicates P < 0.01 (t-test).
A total of three isoenzymes of DAHP synthase, including AroF, AroG, and AroH, have been identified in some bacteria to date, while only the AroG, including AroG1 and AroG2 are annotated in genomes of RSSC, which are highly conserved in RSSC and share more than 90% identities at AAs, respectively. GMI1000 is well known to be different from OE1-1 on tobacco plants, and we ascertained whether AroG1 and AroG2 played roles on growth of GMI1000. Consistent with that of OE1-1, the aroG2 mutant (GF0033) exhibited identical growth as GMI1000 in both media, while growth of aroG1 mutant (GF0032) was significantly impaired both in rich medium (data not shown) and sucrose medium (Supplementary Figure S1). Growth of aroG1/2 mutant (GF0034) was also significantly impaired in rich medium (data not shown) but diminished in limited medium (Supplementary Figure S1). As expected, the AroG1 of OE1-1 significantly restored diminished growth of GF0034 and fully restored the impaired growth of GF0032 to that of wild- type strain in sucrose medium (Supplementary Figure S1), confirming that the involvement of AroG1 and AroG2 on bacterial growth is conserved in RSSC, and the AroG1 of OE1-1 is functionally equivalent to that of GMI1000.
AroG controls the first step for AAA biosynthesis, which are essential for bacterial growth, and the aroG1/2 mutants were auxotrophic in limited medium. We therefore evaluated whether their diminished growth was due to the deficient of AAA. Three AAA, including L-Phe, L-Tyr, and L-Trp, were supplemented into sucrose medium and OD600 was assessed. Supplementary AAA enables the aroG1/2 mutant (RQ5806) to grow in sucrose medium, even though it could just partially recover the diminished growth, which reached the maximum OD600 of approximately 0.4 at 24 hpi (Figure 1C). SA is an important intermediate in the shikimate pathway, and we evaluated whether supplementary SA could recover the diminished growth of RQ5806. Supplementary SA could also partially restore the diminished growth of RQ5806 to an OD600 of approximately 0.7 at 24 hpi (Figure 1C). All these suggested that both AroG1 and AroG2 are involved in the shikimate pathway, and hence are responsible for AAA biosynthesis in R. solanacearum.
We previously used a popA-lacZYA fusion to monitor the T3SS expression in OE1-1 and screened AroG1 as one candidate with impact on the T3SS expression (Zhang et al., 2013). We therefore generated the aroG1 and aroG2 mutants to ascertain whether they affected T3SS expression in sucrose medium (hrp-inducing medium). Consistent with that of transposon mutants, the popA expression in aroG1 mutant (RQ5687) was significantly reduced (112 versus 298 Miller Units of RK5050), while no alteration in aroG2 mutant (RQ5803), and complementary aroG1 fully restored the reduced popA expression to that of RK5050 (Figure 2A). The popA expression was further reduced in aroG1/2 mutant (RQ5806) (76 versus 298 Miller Units), and complementary aroG1 fully restored the reduced popA expression to that of RK5050 (Figure 2A). These results indicates that both AroG1 and AroG2 are cooperatively essential for T3SS expression in R. solanacearum. The AroG1 plays a major role on T3SS expression, while AroG2 becomes to function in the absence of AroG1.
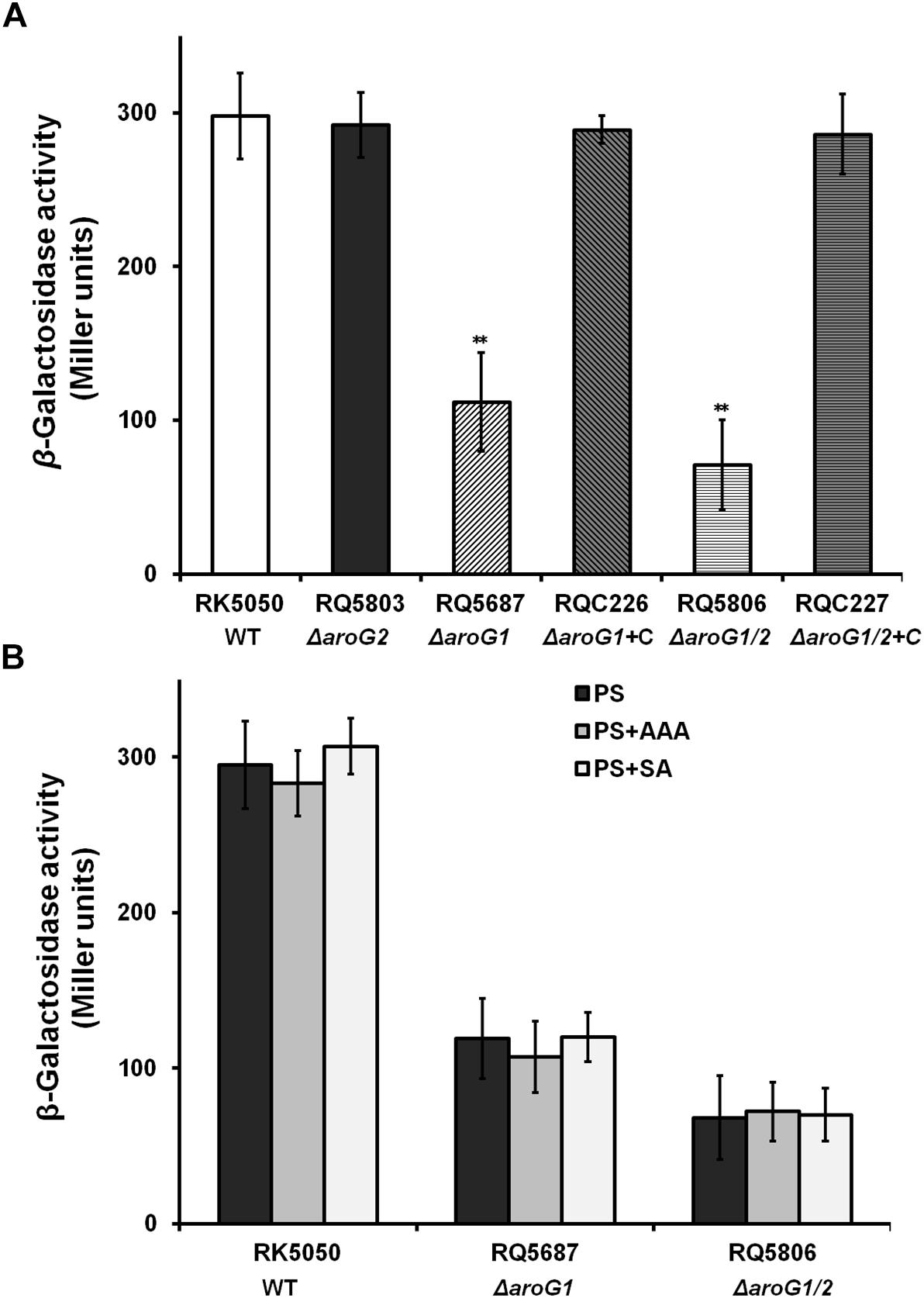
Figure 2. Expression of popA in aroG mutants of OE1-1 in (A) PS and (B) PS supplemented with AAA or SA. Cells (OD600 = 1.0) were inoculated into PS medium as a ratio of 1/100, grown to an OD600 of approximately 0.1 and subjected for β-galactosidase assay. RQC226 and RQC227 refer to aroG1 and aroG1/2 mutants complemented with aroG1, respectively. Enzymatic activities were presented in Miller units. Mean values of four independent trials with three replications per trial were averaged and presented with SD (error bars). Statistical significance between RK5050 (wild-type) and aroG mutants was assessed using a post-hoc Dunnett test following ANOVA. Significance level, ∗∗ indicates P < 0.01 (t-test).
AroG1 and AroG2 control biosynthesis of SA and AAA, and we investigated whether this reduced popA expression was due to the deficient of SA or AAA. Supplementary SA and AAA did not alter the popA expression in RQ5687 (ΔaroG1) and RQ5806 (ΔaroG1/2) (Figure 2B), indicating that impact of AroG1 and AroG2 on T3SS is not mediated with their downstream products of AAA and SA.
AroG1 and AroG2 are highly conserved in RSSC and we investigated whether they affected the T3SS expression in GMI1000, which exhibits different phenotypes from OE1-1 in tobacco plants. Consistent with that in OE1-1, the popA expression in GF0032 (GMI1000, ΔaroG1) and GF0034 (GMI1000, ΔaroG1/2) was significantly reduced, but not in GF0033 (GMI1000, ΔaroG2) (Supplementary Figure S2). As expected, the AroG1 of OE1-1 fully restored the reduced popA expression in GF0032 and GF0034 to that of wild-type strain (Supplementary Figure S2), confirming that the impact of AroG1 and AroG2 on T3SS is conserved in RSSC and the AroG1 of OE1-1 is functionally equivalent to that of GMI1000.
The T3SS of R. solanacearum is directly controlled by the master regulator HrpB, and deletion of aroG1 significantly impaired the hrpB expression in sucrose medium (Figure 3A), confirming that the impact of AroG1 on T3SS is mediated through the HrpB. Two close paralogs of HrpG and PrhG positively regulate the hrpB expression in a parallel way. Deletion of aroG1 significantly impaired the hrpG expression only in sucrose medium, but not prhG expression in either rich or sucrose medium (Figures 3A,B). PrhN positively regulates the prhG expression, while its expression was not affected with the deletion of aroG1 in either of media (Figures 3A,B). The hrpG expression is regulated by the well-characterized PrhA signaling cascade, including PrhA, PrhI/R and PrhJ. Interestingly, their expression was significantly impaired with the aroG1 deletion in sucrose medium (Figures 3A,B), indicating that the impact of AroG1 on T3SS is mediated with the well-characterized PrhA-PrhI/R-PrhJ-HrpG signaling cascade.
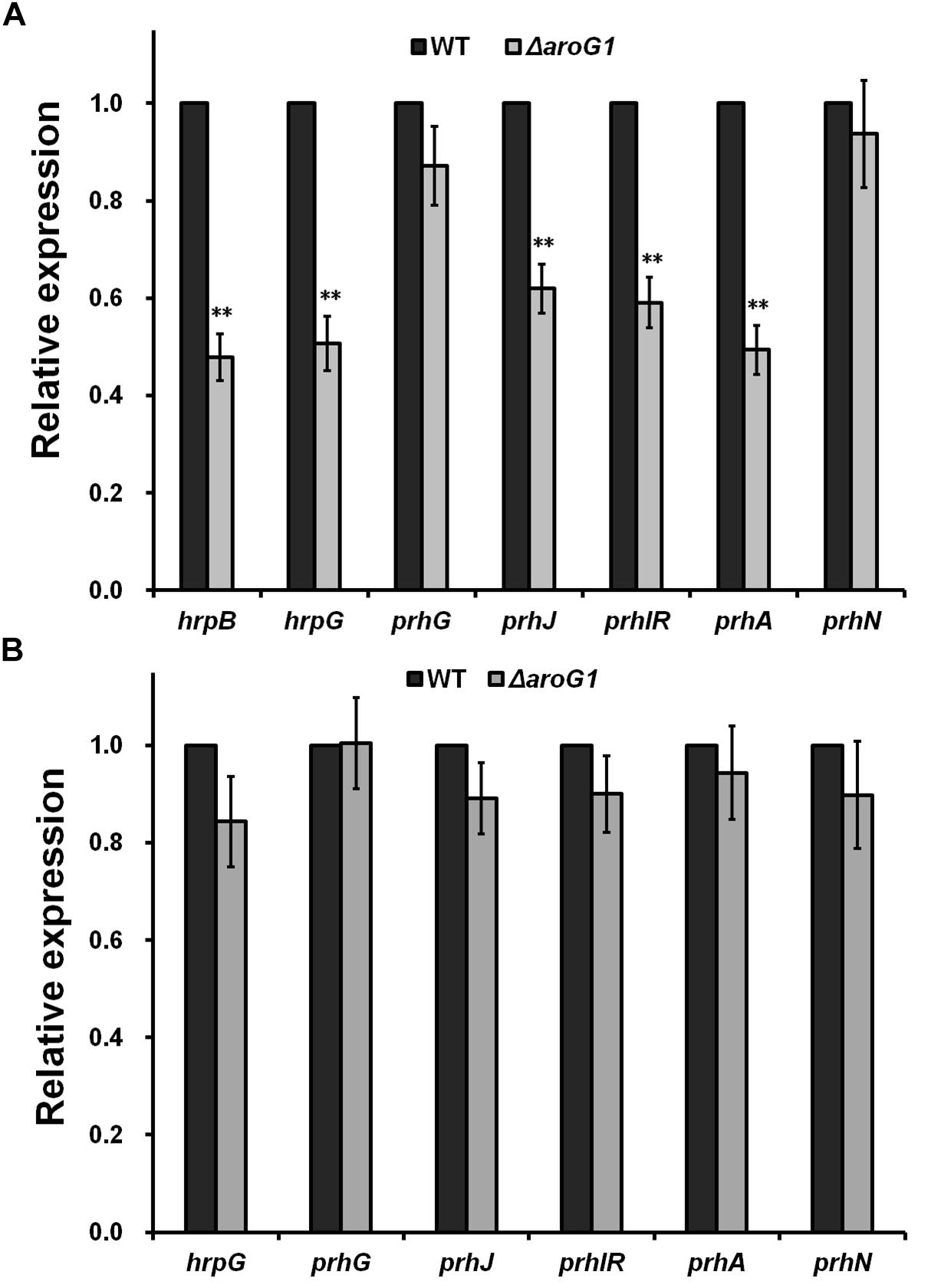
Figure 3. Relative expression of genes involved in T3SS regulation with aroG1 deletion in (A) PS, (B) B medium. Dark bars, reporter strains (the control); gray bars, aroG1 mutants corresponding to each reporter strain. Cells were grown to an OD600 of about 0.1 and subjected for β-galactosidase assay. Enzymatic activities with aroG1 deletion was divided with that of control and relative values were presented. Mean values of four independent trials with three replications per trial were averaged and presented with SD (error bars). Statistical significance between the control and aroG1 mutants was assessed using a post hoc Dunnett test following ANOVA. Significance level, ∗∗ indicates P < 0.01 (t-test).
The T3SS is essential for pathogenicity of R. solanacearum in host plants and we investigated whether AroG1 and AroG2 play roles on pathogenicity. RK5050 invaded and killed tomato plants at about 13 days post inoculation (dpi) with the soil-soaking inoculation and 7 dpi with petiole-inoculation. RQ5803 (ΔaroG2) exhibited almost identical virulence as RK5050 in tomato plants with both inoculation methods, while RQ5687 (ΔaroG1) and RQ5806 (ΔaroG1/2) completely lost pathogenicity in tomato plants with both inoculation methods, and complementary aroG1 fully restored their diminished virulence to that of RK5050 (Figures 4A,B).
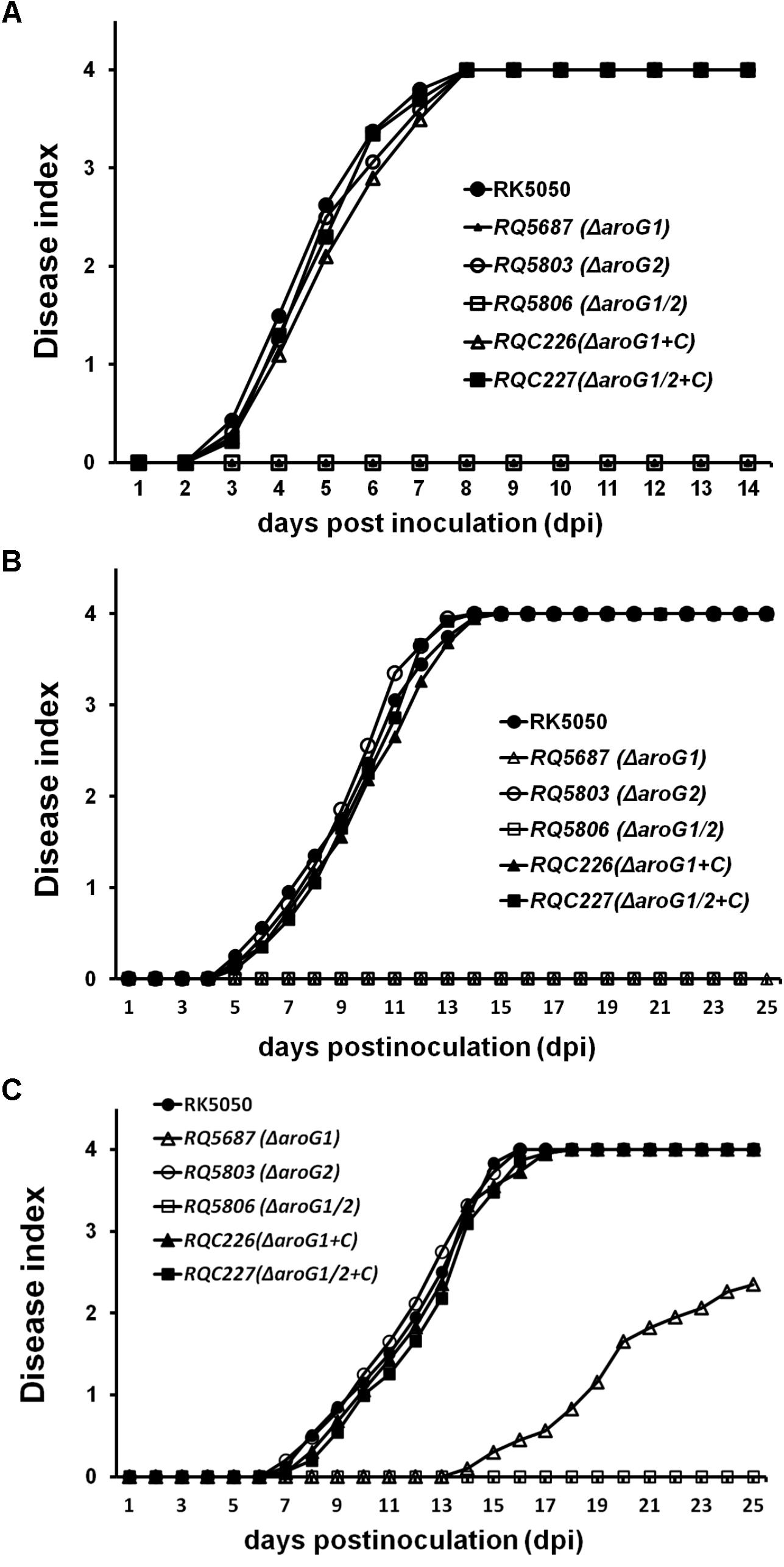
Figure 4. Pathogenicity test of aroG mutants on tomato plants (A,B) and tobacco plants (C). (A) Petiole inoculation, about 3 μl of bacterial suspension at 108 cfu ml-1 was dropped onto freshly cut surface of petioles; (B) soil-soaking inoculation, a bacterial suspension was poured into the soil of plants at a final concentration of 107 cfu g-1 of soil; (C) leaf infiltration in tobacco leaves, about 50 μl of bacterial suspension at 108 cfu ml-1 was infiltrated into tobacco leaves with a blunt-end syringe. Plants were inspected daily for wilt symptoms, and scored on a disease index scale from 0 to 4 (0, no wilting; 1, 1–25% wilting; 2, 26–50% wilting; 3, 51–75% wilting; and 4, 76–100% wilted or dead). RQC226 and RQC227 refer to aroG1 and aroG1/2 mutants complemented with aroG1, respectively. Each assay was repeated in at least four independent trials and each trial contains at least 12 plants. Mean values of all results were averaged with SD.
When challenged tobacco plants with inoculation methods of soil-soaking (data not shown) and leaf infiltration (Figure 4C), RQ5803 (ΔaroG2) exhibited identical virulence as RK5050, while RQ5806 (ΔaroG1/2) completely lost the pathogenicity in tobacco plants (Figure 4C). RQ5687 (ΔaroG1) was avirulent on tobacco plants with soil-soaking (data not shown), while it exhibited significantly less virulence than RK5050 in leaf- infiltrated tobacco plants, which killed about half tobacco plants at 25 dpi (Figure 4C). Complementary aroG1 fully restored the diminished or impaired virulence of aroG1 or aroG1/2 mutants in tomato and tobacco plants (Figures 4A–C), indicating that the AroG1 and AroG2 are cooperatively essential for pathogenicity of R. solanacearum in different host plants. The AroG1 plays essential role for pathogenicity in both tomato and tobacco plants, while the AroG2 is capable to partially function for pathogenicity in tobacco plants in the absence of AroG1.
Despite high similarity, AroG1 and AroG2 exhibited different properties on bacterial growth and pathogenicity. We firstly investigated whether this difference was due to their different promoter activities. Fusions of aroG1-lacZYA and aroG2-lacZYA were introduced into different strains, and their promoter activities were evaluated with the β-galactosidase assay. Deletion of aroG2 greatly increased the aroG1 expression to about 15-fold higher level in rich medium and 10-fold higher level in limited medium compared with those in wild-type strains (Table 2). The aroG2 expression was also greatly increased with aroG1 deletion by about 20-fold higher levels in both media, while the expression of aroG1 and aroG2 was not altered with deletion of themselves (Table 2). The expression of aroG1 and aroG2 was also greatly increased in aroG1/2 mutants, which was much less than those with deletion of single aroG1 and aroG2 (Table 2).
DAHP synthases are usually sensitive to their corresponding AAA. For instance, the AroG is Phe-sensitive in E. coli (11). In the present study, L-Phe was supplemented into rich and limited media at a concentration of 1 mM to evaluate its feedback impact on aroG expression. Expression of aroG1 and aroG2 was significantly repressed with supplementary L-Phe in both media (Table 2), confirming that AroG1 and AroG2 are also Phe-sensitive in R. solanacearum. In consideration of the fact that AroG1 and AroG2 are cooperatively essential for AAA biosynthesis, growth and pathogenicity in R. solanacearum, this bacterium can greatly initiate the expression level of aroG1 and aroG2 in the absence of the other. And hence, the highly expressed AroG1 can alone fulfill the growth and pathogenicity, and highly expressed AroG2 is capable to partially substitute the function of AroG1.
A remarkable structural difference between the AroG1 and AroG2 is a N-terminal region of 43 AA, which is unique in AroG2 (Figure 5A). We firstly investigated whether this structural difference is responsible for their functional difference. We deleted this unique N-terminal region of 43 AA in AroG2 and evaluated its contribution on growth and virulence in aroG1 and aroG1/2 mutants. Whereas this truncated AroG2-N failed to restore the diminished growth of aroG1/2 mutant (RQ5806) in limited medium (Figure 5B), or the diminished virulence of aroG1 mutant (RQ5687) in tomato plants (Figure 5C), confirming that this unique N-terminal region in AroG2 is not the cause for AroG2 to function weakly for growth and pathogenicity.
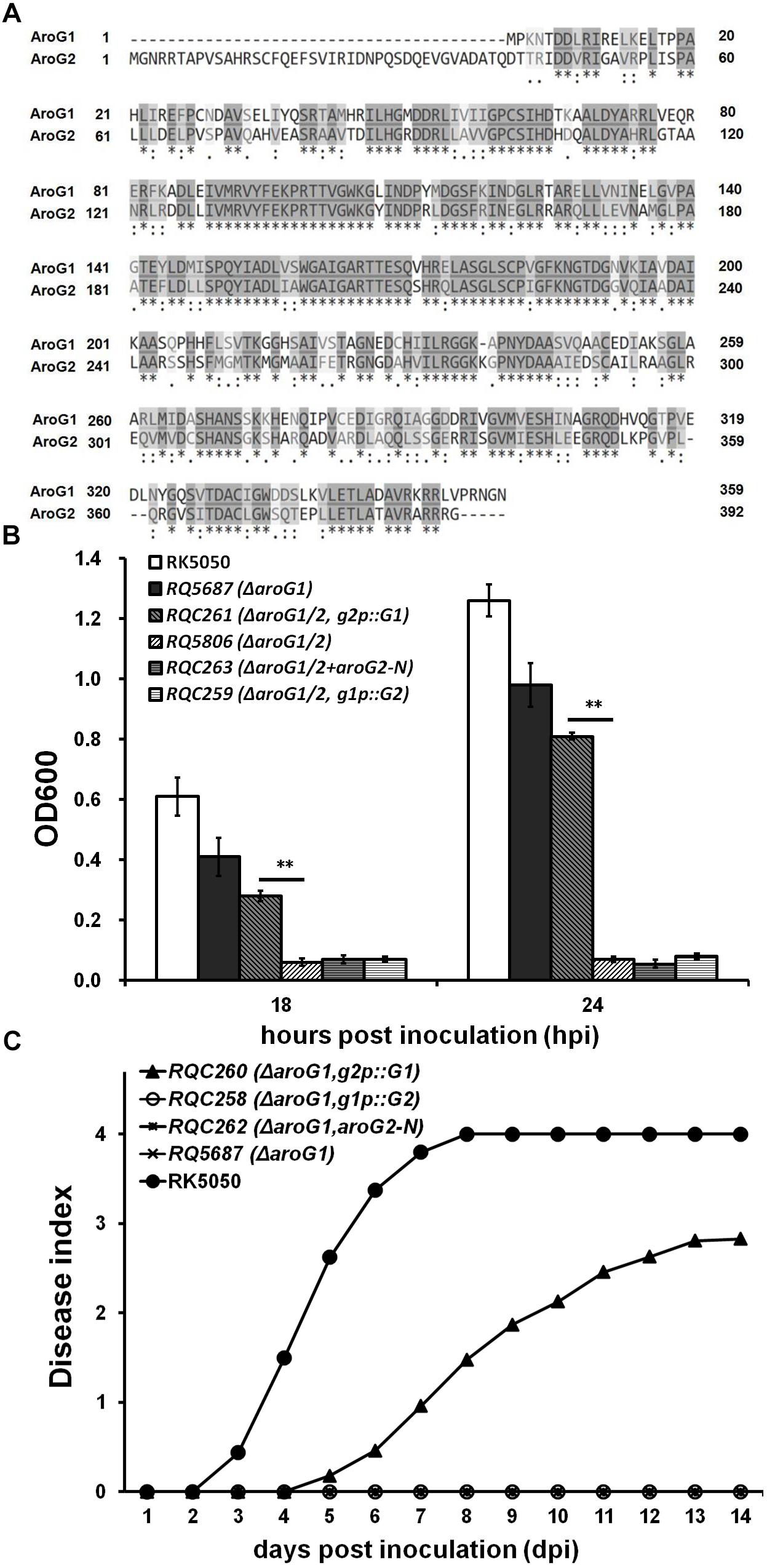
Figure 5. Functional characterization of mutated aroG1/aroG2 in different backgrounds. (A) ClustalW analysis of AroG1 and AroG2 (58% identity and 88% similarity), (B) growth assay and (C) virulence assay of aroG mutants with mutated aroG1/aroG2. The N-terminal region of 43 AA is unique in AroG2 and this region was deleted to generate the truncated AroG2-N. The g2p::G1 fused promoter of aroG2 with CDS of aroG1, and g1p::G2 fused promoter of aroG1 with CDS of aroG2. The g2p::G1, g1p::G2 and AroG2-N was integrated into chromosome of aroG1 and aroG1/2 mutants, respectively. Cells were inoculated into sucrose medium and OD600 was measured at 18 and 24 hpi for growth assay. Tomato plants were inoculated with petiole inoculation for virulence assay. Mean values of four independent trials and each trial contains at least 12 plants were averaged and presented with SD. Significance level, ∗∗ indicates P < 0.01 (t-test)
We thus transferred to investigate whether their promoters are cause for functional difference between the AroG1 and AroG2. Above promoter activity assay showed that the promoter activity of aroG2 was greatly increased with the aroG1 deletion, which was much higher than that of aroG1 with the aroG2deletion. We therefore generated the promoter-exchanged aroG1 and aroG2 and evaluated their contribution on growth and virulence in aroG1 and aroG1/2 mutants. The fusion of g2p::G1, fused promoter of aroG2 with CDS of aroG1, can partially restore the diminished growth of aroG1/2 mutant in limited medium (Figure 5B), and the diminished virulence of aroG1 mutant in tomato plants, which killed about half of petiole-inoculated tomato plants at 14 dpi (Figure 5C). Whereas the fusion of g1p::G2, fused promoter of aroG1 with CDS of aroG2, failed to restore the diminished growth of aroG1/2 mutant in limited medium, or the diminished virulence of aroG1 mutant in tomato plants (Figures 5B,C), indicating that different promoter activities between aroG1 and aroG2 are not crucial cause for their functional difference on growth and pathogenicity.
Extensive proliferation in xylem vessels is one of the main virulence determinants of R. solanacearum, and we investigated whether AroG1 and AroG2 are required for the in planta growth of R. solanacearum. The wild-type strains cause petiole-inoculated wilted and died at 3 and 7 dpi, respectively, and we quantified their growth in tomato stems at 3 and 6 dpi, respectively, which reached to approximately 108 cfu g-1 at 3 dpi, and reached to the maximum of approximately 1010 cfu g-1 at 6 dpi (Figure 6A). The aroG2 mutant was same virulent as wild-type strains in tomato plants, and it exhibited identical growth as wild-type strains in tomato stems at 3 and 6 dpi (Figure 6A). The aroG1 and aroG1/2 mutants did not cause disease in tomato plants and we quantified their growth in tomato stems till to 20 dpi. The aroG1 mutant proliferated to approximately 105 cfu g-1 at 3 dpi, increased slowly to the maximum of approximately 109 cfu g-1 at 10 dpi, and remained this density till to 20 dpi (Figure 6A). The aroG1/2 mutant exhibited significantly less proliferation than aroG1 mutant in tomato stems, which proliferated to approximately 102 cfu g-1 at 3 dpi and increased slowly to the maximum of approximately 109 cfu g-1 at 20 dpi (Figure 6A).
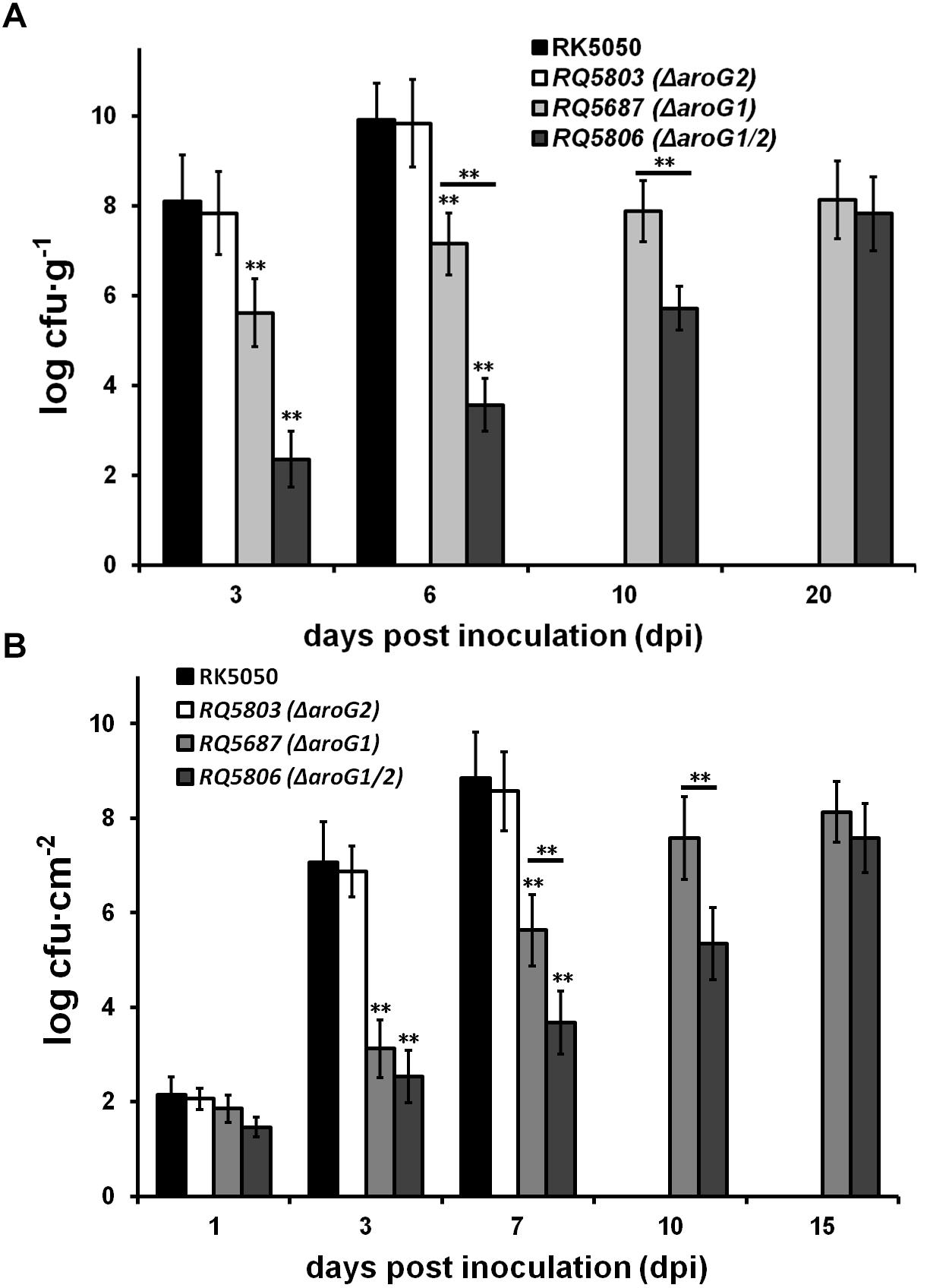
Figure 6. Growth assay of aroG mutants in planta of (A) tomato stems and (B) tobacco leaves. Stem species were removed from petiole-inoculated tomato plants, leaf disks were punched from leaf-infiltrated tobacco plants, and subjected for quantification of cells number by dilution plating, periodically. Mean values of four independent trials and each trial contains four plants were averaged and presented with SD (error bars). Statistical significance between RK5050 and aroG mutants or between aroG1 and aroG1/2 mutants was assessed using a post hoc Dunnett test following ANOVA. Significance level, ∗∗ indicates P < 0.01 (t-test).
Growth of these mutants was also evaluated in tobacco leaves, which was infiltrated with cells suspension at a low concentration of 104 cfu ml-1. Proliferation of the wild-type strains started from approximately 102 cfu cm-2 at 1 dpi and increased daily to approximately 109 cfu cm-2 at 7 dpi, when infiltrated leaves wilted and dried (Figure 6B). Both the aroG1 and aroG1/2 mutants proliferated slowly from approximately 102 cfu cm-2 at 1 dpi to approximately 108 cfu cm-2 at 15 dpi, when tobacco leaves became yellow (Figure 6B). Growth of the aroG1/2 mutants in tobacco leaves was significantly less slowly than that of aroG1 mutants (Figure 6B).
Expression of the T3SS can be enhanced to about 20-fold higher level in planta than that in vitro (hrp-inducing medium), and we evaluated whether AroG1 and AroG2 are required for the T3SS expression in planta. The T3SS expression in wild-type strain (RK5050) was assayed till to 6 dpi in tomato stems, when plants withered and died, which reached to the maximum at 5 dpi and decreased rapidly at 6 dpi (Figure 7A). The T3SS expression in aroG1 and aroG1/2 mutants increased slowly till to 20 dpi, which was significantly less than that of RK5050 in tomato stems (Figure 7A). Different from that in hrp-inducing medium, the aroG1 and aroG1/2 mutants exhibited almost identical T3SS expression in tomato stems (Figure 7A).
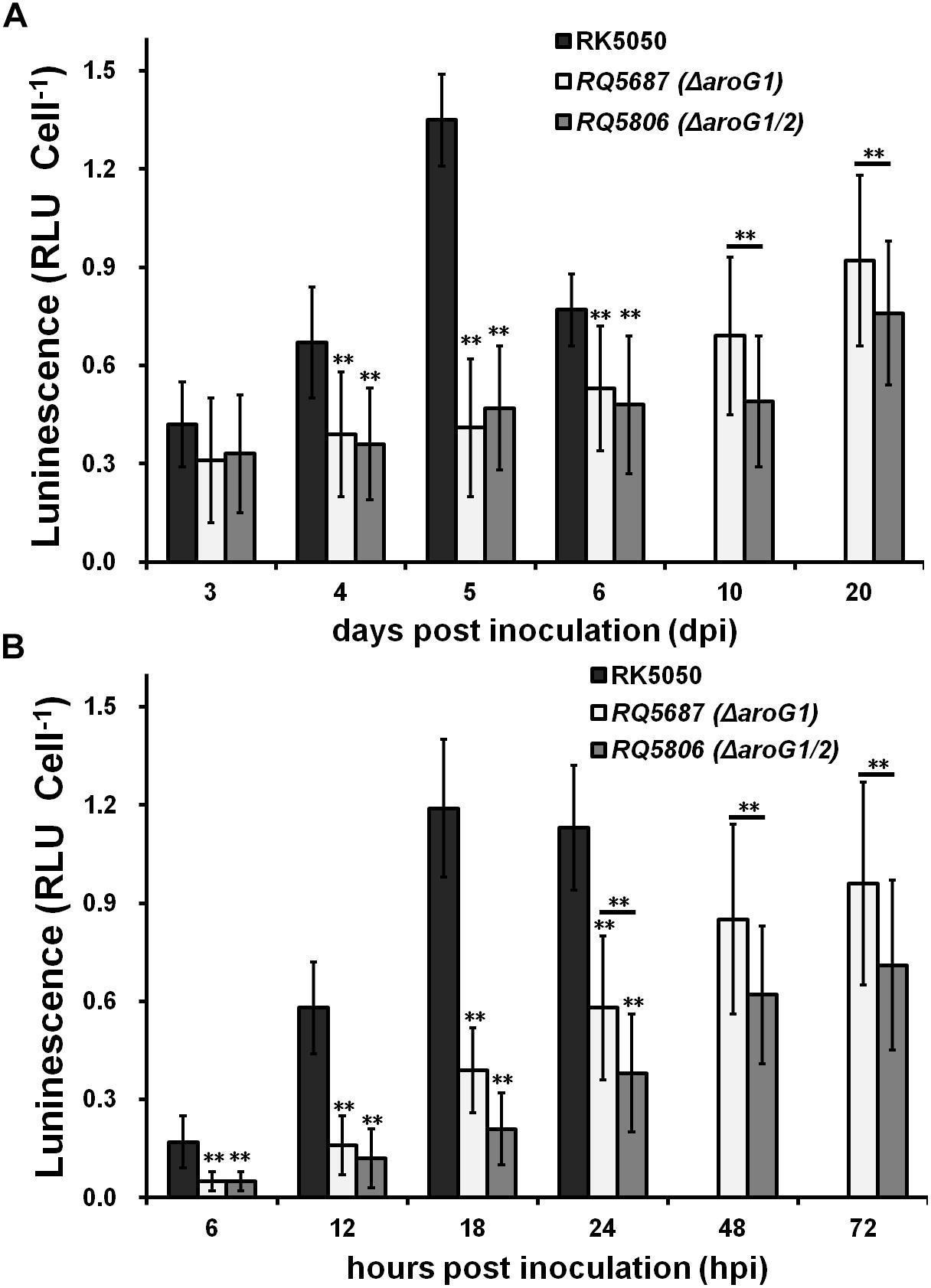
Figure 7. Expression of popA of aroG mutants in planta of (A) tomato stems and (B) tobacco leaves. Tomato plants were inoculated with petiole inoculation and stem species were removed periodically for enzyme assay with Galacto-Light Plus kit. Enzyme assay of RK5050 was performed till 6 dpi, when tomato plants became wilted and died, while that of aroG1 and aroG1/2 mutants was performed till 20 dpi since tomato plants remained healthy. Tobacco leaves were inoculated with leaf infiltration and leaf disks were punched periodically for enzyme assay. Enzyme assay of RK5050 was performed till 24 hpi, when tobacco leaves became wilted, while that of aroG1 and aroG1/2 mutants was performed till 72 hpi since tobacco leaves remained healthy. Cells number was quantified by dilution plating and luminescence was evaluated using GloMax20 luminometer (Promega). Enzymatic activity was presented with luminescence normalized with cells numbers. Mean values of four independent trials with four replications per trial were averaged and presented with SD. Significance level, ∗∗ indicates P < 0.01 (t-test).
The T3SS expression in RK5050 was assayed up to 24 hpi in tobacco leaves, when tobacco leaves became wilting, which increased slowly and reached to the maximum at about 18 hpi (Figure 7B). The T3SS expression in aroG1 and aroG1/2 mutants was assayed up to 72 hpi, which increased slowly and reached to the maximum at 72 hpi, but they were significantly less than that of RK5050 in tobacco leaves (Figure 7B). Different from that in tomato stems, the aroG1/2 mutant exhibited significantly less T3SS expression than aroG1 mutant in tobacco leaves (Figure 7B).
Deletion of aroG1 and aroG2 significantly impaired the in planta expression of T3SS, which is responsible for the HR elicitation of GMI1000 in tobacco leaves, and we investigated whether GMI1000 requires AroG1 and AroG2 for HR elicitation. Cells suspension at 108 cfu ml-1 was infiltrated into tobacco leaves with a blunt-end syringe and development of necrotic lesions was investigated periodically. It was intriguing that both aroG1 and aroG1/2 mutants exhibited identical development of necrotic lesions as GMI1000 in tobacco leaves (Supplementary Figure S3), indicating that AroG1 and AroG2 are not required for the HR elicitation of GMI1000 in tobacco leaves.
In the present study, we provided genetic evidence to demonstrate that two putative DAHP synthases of AroG1 and AroG2 are cooperatively essential for biosynthesis of AAA in R. solanacearum. Three isoenzymes of DAHP synthases, AroF, AroG and AroH, have been identified in many bacteria to control the first step in the shikimate pathway (Herrmann, 1983; Herrmann and Weaver, 1999; Sprenger, 2006). Only the AroG was annotated in genomes of RSSC strains. The aroG1/2 mutants were indeed auxotrophic in limited medium, and supplementary AAA or SA significantly restored the diminished growth of aroG1/2 mutants in limited medium, confirming that AroG1 and AroG2 are involved in the shikimate pathway and thus are responsible for AAA biosynthesis in R. solanacearum. The shikimate pathway can also lead to production of some AAA-derivatives, such as vitamin K, folic acid and ubiquinone, which are also important for bacterial growth (Dosselaere and Vanderleyden, 2001; Gosset et al., 2001). It can explain the fact that growth recovery with SA was much better than that with AAA. Supplementary SA and AAA can just partially restore diminished growth of aroG1/2 mutants in limited medium, indicating that AroG1 and AroG2 might be involved in the production of some novel compounds, which are also important for growth.
The shikimate pathway is a common route in microorganisms and plants that leads to production of AAA and derivatives (Herrmann and Weaver, 1999; Sprenger, 2006). AAA and some derivatives can be detected in apoplastic and xylem extracts of tomato plants, and R. solanacearum can metabolize these compounds inside tomato plants and facilitate it to thrive in tomato plants (Zuluaga et al., 2013). It is consistent with the fact that the aroG1/2 mutants failed to grow in limited medium, but grew slowly in host plants. Extensive proliferation in xylem vessels is one of the most important virulence determinants of R. solanacearum in host plants (Roberts et al., 1988; Denny, 1995). It was as expected that the aroG1 and aroG1/2 mutants exhibited completely diminished or significantly weakened virulence in host plants since their proliferation was significantly impaired in tomato and tobacco plants. Growth of aroG2 mutants was not altered in tomato and tobacco plants, and they exhibited identical virulence as the wild-type strain in host plants. Tobacco plants exhibit different metabolic activities on secondary metabolites, i.e., salicylic acid, from tomato plants (Bellés et al., 1999, 2006). Moreover, different host plants usually display different symptoms, depending upon infecting strains (Lin et al., 2008). It can explain the fact that the aroG1 mutants display different phenotypes on different host plants that completely lost the virulence in tomato plants, but remained weakened virulence in tobacco plants.
The T3SS is another essential virulence determinants of R. solanacearum (Vasse et al., 2000; Genin et al., 2005), which was significantly impaired in aroG1 and aroG1/2 mutants both in vitro and in planta. It is consistent with above virulence results that aroG1 and aroG1/2 mutants exhibit completely diminished or significantly weakened virulence in host plants. The T3SS expression was not altered in aroG2 mutants either in vitro or in planta. It is as expected that aroG2 mutants exhibited identical virulence as wild-type strains. The AroG2 seems not to function for bacterial growth, T3SS expression and virulence in the presence of AroG1. In consideration of facts that the aroG1/2 mutants display enhanced phenotypes on bacterial growth, T3SS expression and virulence compared to the aroG1 mutants, the AroG2 is capable of carrying out part of functions of AroG1 in the absence of AroG1. The aroG1 and aroG1/2 mutants grew slowly in planta that eventually got to the maximal densities of approximately 108 cfu g-1 in tomato stems and 108 cfu cm-2 in tobacco leaves, respectively. Note that tomato plants become wilting when R. solanacearum proliferates to a density of about 108-9 cfu g-1 in stems and tobacco leaves become withered at about 107-8 cfu cm-2 (Zhang et al., 2011, 2013). Whereas the aroG1 and aroG1/2 mutants failed to cause any wilting symptom in tomato plants even they reached to this high density at 10–20 dpi, indicating that these slow growth might enable host plants to initiate effective resistance reaction leisurely, or AroG1 and AroG2 are also required for some novel virulence determinants.
The hrpB expression was significantly impaired in aroG1 mutant, which is consistent with the fact that HrpB directly controls the entire T3SS (Cunnac et al., 2004; Tamura et al., 2005). The hrpB expression is positively regulated by two close paralogs of HrpG and PrhG in a parallel way (Zhang et al., 2013), while only the hrpG expression was significantly impaired with the aroG1 deletion in sucrose medium, suggesting that the impact of AroG1 on T3SS is mediated through the HrpG-HrpB pathway, but independent of PrhG. Expression of prhA, prhIR, and prhJ was significantly impaired in aroG1 mutants, which form the well-known PrhA signaling cascade and positively regulate the hrpG expression in tandem, confirming that the impact of AroG1 on T3SS is mediated through the well-characterized PrhA-prhI/R-PrhJ-HrpG signaling cascade. Since the T3SS assembly and secretion of T3Es require a lo of energy, T3SS expression is not activated until R. solanacearum gets contact with some signals, i.e., host signals or mimic signals in nutrient-limited medium. The aroG1/2 mutants failed to grow in limited medium, which should suffer some extreme conditions, at least the extreme starvation. R. solanacearum should stop all unessential activities, at least coordinating for energy conservation, while the T3SS expression was not completely diminished in the aroG1/2 mutants under this extreme condition, which was remained to about a quarter of the wild-type strain. The T3SS should play some roles on the survival of R. solanacearum under these extreme conditions.
DAHP synthases are usually sensitive to their corresponding AAA (McCandliss et al., 1978; Umbarger, 1978; Herrmann, 1983; Pittard, 1996). Supplementary L-Phe can significantly repress the expression of aroG1 and aroG2 in R. solanacearum. This feedback repression results in relatively low expression levels of aroG1 and aroG2 in wild-type strains (about 50 Miller Units in both rich and limited media). The aroG2 expression was greatly increased to about 20-fold higher level with aroG1 deletion. We above concluded that both AroG1 and AroG2 cooperatively function for bacterial growth, T3SS expression and virulence. The AroG2 is capable of carrying out part of functions of AroG1 in the absence of AroG1, which might be due to greatly expressed AroG2 in the absence of AroG1. The aroG1 expression was greatly enhanced to about 14-fold higher level with the aroG2 deletion, and hence, the highly expressed AroG1 can alone fulfill growth, T3SS expression and pathogenicity in the absence of AroG2. The promoter activity of aroG2 in aroG1 mutants was much higher than that of aroG1 in aroG2 mutants, while the promoter of aroG2 could just partially substitute native promoter for aroG1 to fulfill the growth and pathogenicity. On the contrary, promoter of aroG1 failed to enable AroG2 to fulfill the growth and pathogenicity, suggesting that functional difference between AroG2 and AroG1 should be due to their structural difference, but not the promoter activities. A remarkable structural difference between AroG2 and AroG1 is the unique N-terminal region of 43 AA in AroG2. Whereas the truncated AroG2 failed to restore the growth and pathogenicity in aroG1 mutants. Some AA residues have been characterized to be important for the function of DAHP isoenzymes (Jossek et al., 2001; Sprenger, 2006), indicating that certain AA residues are responsible for functional difference between AroG2 and AroG1.
Taken together, our genetic results demonstrated that two putative DAHP synthases of AroG1 and AroG2 are involved in the shikimate pathway and are responsible for the AAA biosynthesis in R. solanacearum. They cooperatively exhibit essential functions on bacterial growth, T3SS expression and pathogenicity. The AroG1 plays a major role on these phenotypes, while AroG2 is capable to partially carry out the function of AroG1 in the absence of AroG1.
YZ and KO conceived and designed the experiments. WZ and JL performed the experiments. XS, YH, KO, and YZ analyzed and discussed the results. YZ wrote and revised the manuscript.
We express thanks for funding support from the National Natural Science Foundation of China (31670082), the Chongqing Research Program of Basic Research and Frontier Technology (cstc2016jcjyA0470) to YZ and the Graduate student scientific research innovation projects in Chongqing (CYS18105) to WZ.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00183/full#supplementary-material
Angot, A., Peeters, N., Lechner, E., Vailleau, F., Baud, C., Gentzbittel, L., et al. (2006). Ralstonia solanacearum requires F-box-like domain-containing type III effectors to promote disease on several host plants. Proc. Natl. Acad. Sci. U.S.A. 103, 14620–14625. doi: 10.1073/pnas.0509393103
Arlat, M., Gough, C. L., Zischek, C., Barberis, P. A., Trigalet, A., and Boucher, C. (1992). Transcriptional organization and expression of the large hrp gene cluster of Pseudomonas solanacearum. Mol. Plant Microbe Interact. 5, 187–193. doi: 10.1094/MPMI-5-187
Bellés, J. M., Garro, R., Fayos, J., Navarro, P., Primo, J., and Conejero, V. (1999). Gentisic acid as a pathogen-inducible signal, additional to salicylic acid for activation of plant defenses in tomato. Mol. Plant Microbe Interact. 12, 227–235. doi: 10.1094/MPMI.1999.12.3.227
Bellés, J. M., Garro, R., Pallás, V., Fayos, J., Rodrigo, I., and Conejero, V. (2006). Accumulation of gentisic acid as associated with systemic infections but not with the hypersensitive response in plant-pathogen interactions. Planta 223, 500–511. doi: 10.1007/s00425-005-0109-8
Bentley, R. (1990). The shikimate pathway: a metabolic tree with many branches. Crit. Rev. Biochem. Mol. Biol. 25, 307–384. doi: 10.3109/10409239009090615
Choi, K. H., Gaynor, J. B., White, K. G., Lopez, C., Bosio, C. M., Karkhoff- Chweizer, R. R., et al. (2005). A Tn7-based broad range bacterial cloning and expression system. Nat. Methods 2, 443–448. doi: 10.1038/nmeth765
Cunnac, S., Occhialini, A., Barberis, P., Boucher, C., and Genin, S. (2004). Inventory and functional analysis of the large Hrp regulon in Ralstonia solanacearum: identification of novel effector proteins translocated to plant host cells through the type III secretion system. Mol. Microbiol. 53, 115–128. doi: 10.1111/j.1365-2958.2004.04118.x
Denny, T. P. (1995). Involvement of bacterial polysaccharides in plant pathogenesis. Annu. Rev. Phytopathol. 33, 173–197. doi: 10.1146/annurev.py.33.090195.001133
Dosselaere, F., and Vanderleyden, J. (2001). A metabolic node in action: chorismate-utilizing enzymes in microorganisms. Crit. Rev. Microbiol. 27, 75–131. doi: 10.1080/20014091096710
Fujiwara, S., Kawazoe, T., Ohnishi, K., Kitagawa, T., Popa, C., Valls, M., et al. (2016). RipAY, a plant pathogen effector protein, exhibits robust γ-Glutamyl cyclotransferase activity when stimulated by eukaryotic thioredoxins. J. Biol. Chem. 291, 6813–6830. doi: 10.1074/jbc.M115.678953
Genin, S., Brito, B., Denny, T. P., and Boucher, C. (2005). Control of the Ralstonia solanacearum type III secretion system (Hrp) genes by the global virulence regulator PhcA. FEBS Lett. 579, 2077–2081. doi: 10.1016/j.febslet.2005.02.058
Genin, S., and Denny, T. P. (2012). Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 50, 67–89. doi: 10.1146/annurev-phyto-081211-173000
Gosset, G., Bonner, C. A., and Jensen, R. A. (2001). Microbial origin of planttype 2-keto-3-deoxy-D-arabino-heptulosonate 7-phosphate synthases, exemplified by the chorismate- and Trpptophan-regulated enzyme from Xanthomonas campestris. J. Bacteriol. 183, 4061–4070. doi: 10.1128/JB.183.13.4061-4070.2001
Herrmann, K. M. (1983). “The common aromatic biosynthetic pathway,” in Amino Acids: Biosynthsis and Genetic Regulation, eds K. M. Herrmann and R. L. Somervillle (Reading, MA: Addison-Wesley), 301–322.
Herrmann, K. M., and Weaver, L. M. (1999). The shikimate pathway. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 473–503. doi: 10.1146/annurev.arplant.50.1.473
Hikichi, Y., Yoshimochi, T., Tsujimoto, S., Shinohara, R., Nakaho, K., Kanda, A., et al. (2007). Global regulation of pathogenicity mechanism of Ralstonia solanacearum. Plant Biotech. 24, 149–154. doi: 10.5511/plantbiotechnology.24.149
Jiang, G., Wei, Z., Xu, J., Chen, H., Zhang, Y., She, X., et al. (2017). Bacterial wilt in china: history, current status, and future perspectives. Front. Plant Sci. 8:1549. doi: 10.3389/fpls.2017.01549
Jossek, R., Bongaerts, J., and Sprenger, G. A. (2001). Characterization of a new feedback-resistant 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase of Escherichia coli. FEMS Microbiol. Lett. 202, 145–148. doi: 10.1016/S0378-1097(01)00311-1
Kanda, A., Ohnishi, S., Tomiyama, H., Hasegawa, H., Yasukohchi, M., Kiba, A., et al. (2003). Type III secretion machinery-deficient mutants of Ralstonia solanacearum lose their ability to colonize resulting in loss of pathogenicity. J. Gen. Plant Pathol. 69, 250–257. doi: 10.1007/s10327-003-0041-3
Lin, Y. M., Chou, I. C., Wang, J. F., Ho, F. I., Chu, Y. J., Huang, P. C., et al. (2008). Transposon mutagenesis reveals differential pathogenesis of Ralstonia solanacearum on tomato and Arabidopsis. Mol. Plant Microbe. Interact. 21, 1261–70. doi: 10.1094/MPMI-21-9-1261
Maeda, H., and Dudareva, N. (2012). The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 63, 73–105. doi: 10.1146/annurev-arplant-042811-105439
Marenda, M., Brito, B., Callard, D., Genin, S., Barberis, P., Boucher, C., et al. (1998). PrhA controls a novel regulatory pathway required for the specific induction of Ralstonia solanacearum hrp genes in the presence of plant cells. Mol. Microbiol. 27, 437–453. doi: 10.1046/j.1365-2958.1998.00692.x
McCandliss, R. J., Poling, M. D., and Herrmann, K. M. (1978). 3-Deoxy-d-arabino- heptulosonate 7-phosphate synthase. Purification and molecular characterization of the phenylalanine-sensitive isoenzyme from Escherichia coli. J. Biol. Chem. 253, 4259–4265.
Miller, J. H. (1992). “The lac system,” in A Short Course in Bacterial Genetics. A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria, ed. J. H. Miller (Plainview, TX: Cold Spring Harbor Laboratory Press), 43–80.
Mukaihara, T., Tamura, N., and Iwabuchi, M. (2010). Genome-wide identification of a large repertoire of Ralstonia solanacearum type III effector proteins by a new functional screen. Mol. Plant Microbe Interact. 23, 251–262. doi: 10.1094/MPMI-23-3-0251
Mukaihara, T., Tamura, N., Murata, Y., and Iwabuchi, M. (2004). Genetic screening of Hrp type III-related pathogenicity genes controlled by the HrpB transcriptional activator in Ralstonia solanacearum. Mol. Microbiol. 54, 863–875. doi: 10.1111/j.1365-2958.2004.04328.x
Ogino, T., Garner, C., Markley, J. L., and Herrmann, K. M. (1982). Biosynthesis of aromatic compounds: 13C NMR spectroscopy of whole Escherichia coli cells. Proc. Natl. Acad. Sci. U.S.A. 79, 5828–5832. doi: 10.1073/pnas.79.19.5828
Pittard, J. (1996). “Biosynthesis of the aromatic amino acids,” in Escherichia coli and Salmonella: Cellular and Molecular Biology, Vol. 1, ed. F. C. Neidhardt (Washington, DC: ASM), 458–484.
Pittard, J., and Yang, J. (2005). “Biosynthesis of phenylalanine and tyrosine. EcoSal module 3618,” in Ecosal Escherichia coli and Salmonella: Cellular and Molecular Biology, eds A. Böck, R. I. I. I. Curtiss, J. B. Kaper, F. C. Neidhardt, T. Nyström, K. E. Rudd, et al. (Washington, DC: ASM).
Plener, L., Manfredi, P., Valls, M., and Genin, S. (2010). PrhG, a transcriptional regulator responding to growth conditions, is involved in the control of the type III secretion system regulon in Ralstonia solanacearum. J. Bacteriol. 192, 1011–1019. doi: 10.1128/JB.01189-09
Popa, C., Li, L., Gil, S., Tatjer, L., Hashii, K., Tabuchi, M., et al. (2016). The effector AWR5 from the plant pathogen Ralstonia solanacearum is an inhibitor of the TOR signalling pathway. Sci. Rep. 6:27058. doi: 10.1038/srep27058
Roberts, D. P., Denny, T. P., and Schell, M. A. (1988). Cloning of the egl gene of Pseudomonas solanacearum and analysis of its role in phytopathogenicity. J. Bacteriol. 170, 1445–1451. doi: 10.1128/jb.170.4.1445-1451.1988
Salanoubat, M., Genin, S., Artiguenave, F., Gouzy, J., Mangenot, S., Arlat, M., et al. (2002). Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415, 497–502. doi: 10.1038/415497a
Sprenger, G. A. (2006). Aromatic Amino Acids. Amino Acid Biosynthesis∼Pathways, Regulation and Metabolic Engineering. Berlin: Springer.
Tamura, N., Murata, Y., and Mukaihara, T. (2005). Isolation of Ralstonia solanacearum hrpB constitutive mutants and secretion analysis of hrpB-regulated gene products that share homology with known type III effectors and enzymes. Microbiology 151, 2873–2884. doi: 10.1099/mic.0.28161-0
Tasset, C., Bernoux, M., Jauneau, A., Pouzet, C., Brière, C., Kieffer-Jacquinod, S., et al. (2010). Autoacetylation of the Ralstonia solanacearum effector PopP2 targets a lysine residue essential for RRS1-R-mediated immunity in Arabidopsis. PLoS Pathog. 6:e1001202. doi: 10.1371/journal.ppat.1001202
Umbarger, H. E. (1978). Amino acid biosynthesis and its regulation. Annu. Rev. Biochem. 47, 533–606. doi: 10.1146/annurev.bi.47.070178.002533
Valls, M., Genin, S., and Boucher, C. (2006). Integrated regulation of the type III secretion system and other virulence determinants in Ralstonia solanacearum. PLoS. Pathog. 2:e82. doi: 10.1371/journal.ppat.0020082
Vasse, J., Frey, P., and Trigalet, A. (1995). Microscopic studies of intercellular infection and protoxylem invasion of tomato roots by Pseudomonas solanacearum. Mol. Plant Microbe Interact. 8, 241–251. doi: 10.1094/MPMI-8-0241
Vasse, J., Genin, S., Frey, P., Boucher, C., and Brito, B. (2000). The hrpB and hrpG regulatory genes of Ralstonia solanacearum are required for different stages of the tomato root infection process. Mol. Plant Microbe Interact. 13, 259–267. doi: 10.1094/MPMI.2000.13.3.259
Wu, J., Sheflyan, G. Y., and Woodard, R. W. (2005). Bacillus subtilis 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase revisited: resolution of two long-standing enigmas. Biochem. J. 390, 583–590. doi: 10.1042/BJ20050294
Yao, J., and Allen, C. (2007). The plant pathogen Ralstonia solanacearum needs aerotaxis for normal bio?lm formation and interactions with its tomato host. J. Bacteriol. 189, 6415–6424. doi: 10.1128/JB.00398-07
Yoshimochi, T., Hikichi, Y., Kiba, A., and Ohnishi, K. (2009a). The global virulence regulator PhcA negatively controls the Ralstonia solanacearum hrp regulatory cascade by repressing expression of the PrhIR signaling proteins. J. Bacteriol. 191, 3424–3428. doi: 10.1128/JB.01113-08
Yoshimochi, T., Zhang, Y., Kiba, A., Hikichi, Y., and Ohnishi, K. (2009b). Expression of hrpG and activation of response regulator HrpG are controlled by distinct signal cascades in Ralstonia solanacearum. J. Gen. Plant. Pathol. 75, 196–204. doi: 10.1007/s10327-009-0157-1
Zhang, Y., Chen, L., Takehi, Y., Kiba, A., Hikichi, Y., and Ohnishi, K. (2013). Functional analysis of Ralstonia solanacearum PrhG regulating the hrp regulon in host plants. Microbiology 159, 1695–1704. doi: 10.1099/mic.0.067819-0
Zhang, Y., Kiba, A., Hikichi, Y., and Ohnishi, K. (2011). prhKLM genes of Ralstonia solanacearum encode novel activators of hrp regulon and are required for pathogenesis in tomato. FEMS. Microbiol. Lett. 317, 75–82. doi: 10.1111/j.1574-6968.2011.02213.x
Zhang, Y., Li, J., Zhang, W., Shi, H., Luo, F., Hikichi, Y., et al. (2018). A putative LysR-type transcriptional regulator PrhO positively regulates the type III secretion system and contributes to the virulence of Ralstonia solanacearum. Mol. Plant Pathol. 19, 1808–1819. doi: 10.1111/mpp.12660
Zhang, Y., Luo, F., Wu, D., Hikichi, Y., Kiba, A., Igarashi, Y., et al. (2015). PrhN, a putative marR family transcriptional regulator, is involved in positive regulation of type III secretion system and full virulence of Ralstonia solanacearum. Front. Microbiol. 6:357. doi: 10.3389/fmicb.2015.00357
Keywords: Ralstonia solanacearum, DAHP synthase, AroG, type III secretion system, pathogenesis
Citation: Zhang W, Li J, Shi X, Hikichi Y, Zhang Y and Ohnishi K (2019) Functional Characterization of Two Putative DAHP Synthases of AroG1 and AroG2 and Their Links With Type III Secretion System in Ralstonia solanacearum. Front. Microbiol. 10:183. doi: 10.3389/fmicb.2019.00183
Received: 04 October 2018; Accepted: 23 January 2019;
Published: 12 February 2019.
Edited by:
Brigitte Mauch-Mani, University of Neuchâtel, SwitzerlandReviewed by:
Carmen R. Beuzón, Universidad de Málaga, SpainCopyright © 2019 Zhang, Li, Shi, Hikichi, Zhang and Ohnishi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Zhang, YmlveW9uZ3poYW5nQHN3dS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.