- 1Key Laboratory of Agro-Ecological Processes in Subtropical Region, Institute of Subtropical Agriculture, Chinese Academy of Sciences – National Engineering Laboratory for Pollution Control and Waste Utilization in Livestock and Poultry Production – Hunan Provincial Engineering Research Center for Healthy Livestock and Poultry Production – Scientific Observing and Experimental Station of Animal Nutrition and Feed Science in South-Central, Ministry of Agriculture, Changsha, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3Hunan International Joint Laboratory of Animal Intestinal Ecology and Health, Laboratory of Animal Nutrition and Human Health, College of Life Sciences, Hunan Normal University, Changsha, China
The aim of current study was to determine variations in sow’s gut microbiota, serum immunity, and milk metabolite profile mediated by lysozyme supplementation. Twenty-four pregnant sows were assigned to a control group without supplementation and two treatments with 0.5 kg/t and 1.0 kg/t lysozyme provided in formula feed for 21 days (n = 8 per treatment). Microbiota analysis and metagenomic predictions were based on 16s RNA high-throughput sequencing. Milk metabolome was assessed by untargeted liquid chromatography tandem mass spectrometry. Serum biochemical indicators and immunoglobulins were also determined. Gut microbial diversity of sows receiving 1.0 kg/t lysozyme treatment was significantly reduced after the trial. Spirochaetes, Euryarchaeota, and Actinobacteria significantly increased while Firmicutes showed a remarkable reduction in 1.0 kg/t group compared with control. Lysozyme addition rebuilt sow’s gut microbiota to beneficial composition identified by reduced richness of Escherichia coli and increased abundance of Lactobacillus amylovorus. Accordingly, microbial metabolic functions including pyrimidine metabolism, purine metabolism, and amino acid related enzymes were significantly up-regulated in 1.0 kg/t group. Microbial metabolic phenotypes like the richness of Gram-positive bacteria and oxidative stress tolerance were also significantly reduced by lysozyme treatment. Serum alanine transaminase (ALT) activity and IgA levels were significantly down-regulated in the 1.0 kg/t group compared with control, but IgM levels showed a significantly increase in 1.0 kg/t group. Milk metabolites such as L-glutamine, creatine, and L-arginine showed significantly dose-dependent changes after treatment. Overall, lysozyme supplementation could effectively improve the composition, metabolic functions, and phenotypes of sow’s gut microbiota and it also benefit sows with better serum immunity and milk composition. This research could provide theoretical support for further application of lysozyme in promoting animal gut health and prevent pathogenic infections in livestock production.
Introduction
Enteric infections caused by pathogens, like Enterotoxigenic Escherichia coli (ETEC), have a significant negative effect on neonatal survival and animal health in swine production (Oliver and Wells, 2013; Wells et al., 2015; Huang et al., 2018). Animal infants infected by pathogenic bacteria often suffer from persistent diarrhea and serious inflammation (Huang et al., 2018; York, 2018). Prolonged inflammation of the intestinal tract leads to substantial destruction of the intestinal epithelia, resulting in malnutrition and impairing the early growth of infants (Zhao et al., 2012; Zhang et al., 2016; Patel and Kuyucak, 2017). Application of antibiotics in formula feed is well established method and can improve growth rates of piglets (Thymann et al., 2007). However, abuse of antibiotics is contributing to the high level of drug resistance in microbial communities and rising concerns regarding human health (Zhao et al., 2012; Oliver and Wells, 2013; Long et al., 2016; Oh et al., 2016). An alternative to antibiotics is lysozyme, an enzyme and natural broad-spectrum bactericide commonly found in tears, saliva, and milk, and that is a vitally important immune system activator under physiological conditions (Maga et al., 2006a, 2012; Lee et al., 2009). During bacterial infection of the intestine, mammalian Paneth cells are also able to secrete lysozyme via secretory autophagy to maintain intestinal homeostasis (Bel et al., 2017).
Breast milk contains lysozyme (<0.065 μg/mL), along with lactoferrin and secretory IgA (SIgA), which greatly aid the establishment of beneficial gut microbiota in newborns (Maga et al., 2012; Oliver and Wells, 2015). Lysozyme functions by cleaving the β-1,4-glycosidic bond between N-acetylmuramic acid and N-acetylglucosamine residues of the bacterial peptidoglycan, causing a loss of cellular membrane integrity and cell lysis (Oliver and Wells, 2013; Long et al., 2016). Lysozyme was reported to be more effective against Gram-positive bacteria (Masschalck and Michiels, 2003; Touch et al., 2003; Gao et al., 2017), and it may also indirectly affect several Gram-negative bacterial species (Maga et al., 2006a,b). Previous studies revealed that lysozyme promotes the beneficial microbe community and reduces detrimental microbes within gut microbial communities (Ko et al., 2009; Maga et al., 2012). Lysozyme could be effective against a wide range of gastrointestinal pathogens, such as Listeria monocytogenes, Clostridium perfringens, Candida spp., and Helicobacter pylori in vitro (Brundige et al., 2008; Zhang et al., 2016). Bacterial sensitivity to lysozyme is also due to the activation of innate components of the immune system, such as increased neutrophil activation during inflammation (Ragland et al., 2017; Huang et al., 2018). It has been reported that lysozyme may possess an anti-inflammatory effect via inhibiting JNK phosphorylation (Tagashira et al., 2018). Furthermore, lysozyme is capable of enhancing intestinal SIgA secretion, cause macrophage activation, and promote rapid clearance of bacterial pathogens (Lee et al., 2009; Wells et al., 2015; Ragland et al., 2017).
Recent studies reported that lysozyme sourced from chicken eggs showed significant advances in improving growth performance, intestinal morphology, gut microbiota composition, and immunity of piglets (May et al., 2012; Oliver and Wells, 2013, 2015; Oliver et al., 2014; Wells et al., 2015). For instance, weaned piglets received a hen-egg white lysozyme treatment shown better intestinal growth and development, as well as decreased ETEC counts on the intestinal mucosa and serum proinflammatory cytokines (Nyachoti et al., 2012). Moreover, lysozyme produced by transgenic animals and structurally modified lysozyme was shown to possess significant antimicrobial properties against pathogens like ETEC in piglets (Nattress and Baker, 2003; Maga et al., 2006a; Brundige et al., 2008; Tong et al., 2011; Nyachoti et al., 2012; Lu et al., 2015). Piglets that consumed lysozyme-transgenic goats’ milk (containing human lysozyme at 67% of the concentration in human breast milk) showed better intestinal morphology and fewer total coliform counts (Brundige et al., 2008).
Gut microbiota plays multiple roles in animal growth and health, including energy extraction from the diet, gut barrier function and immunity as well as growth performance (Shulman et al., 2008; Cox et al., 2014). Increasing evidence suggests that maternal diet during pregnancy modifies an offspring’s microbiota composition and intestinal development in a long-term manner (Chu et al., 2016; Cheng et al., 2018). Nutritional intervention on sows with additives results in greater neonatal survival and infant health (Oliver and Wells, 2015). However, none of these studies above provided a systematic overview of the effects of lysozyme on sow gut microbiota and its potential interactions with immune systems and milk composition. Given this, in the present study, 24 pregnant sows were assigned to a control group without supplementation and two treatments with 0.5 kg/t and 1.0 kg/t lysozyme additives provided in formula feed (n = 8, per group). After the 21-day supplementation, the effects of lysozyme on sow’s gut microbiota, milk metabolite profile, and serum biochemical indices were systematically investigated and associations among them mediated by lysozyme treatment were also revealed for the first time.
Materials and Methods
Animals and Ethics Statement
All procedures involving animals were carried out in accordance with guidelines for animal studies issued by the Animal Care and Use Committee of the Institute of Subtropical Agriculture, Chinese Academy of Sciences (Zhou et al., 2018). Modified hen-egg white lysozyme additives were obtained from Shanghai E. K. M Biotechnology Co. Ltd., Shanghai, China. A total of 24 multiparous hybrid pregnant sows (Landrace × Yorkshire) with an average parity of 4.67 ± 1.50 were selected to this study and then randomly assigned to three groups (n = 8 per treatment), including a control group (CN) without supplementation and two treatments with 0.5 kg/t (LA) and 1.0 kg/t (LB) lysozyme provided in formula feed. Lysozyme was pre-mixed in formula feeds and the daily intake of every sow is about 10 kg (For lysozyme, about 5 g in LA group and 10 g in LB group every day). The present study started 24 days before the expected date of confinement. Sows did not have diseases like diarrhea and had never received antibiotics before the study. Lysozyme supplementation continued for 21 days till prenatal fasting. Milk from all investigated sows were collected on the due date.
Sample Collection and Processing
After the 21-day supplementation, fresh feces from each individual were collected on the same day using 5 mL sterile centrifugal tubes, immediately frozen in liquid nitrogen, and stored at -80°C until DNA extraction. After the 21-day supplementation, the sows were restrained for blood sampling and approximately 5 mL blood was collected in a vacuum tube from the sow’s auricular vein and directly centrifuged at 1500 × g for 15 min. The supernatant of each sample was then divided into subsamples and stored at -20°C until analysis. On parturition day, milk from each individual was collected with 5 mL sterile centrifugal tubes and immediately frozen at -20°C until further analysis.
Microbiota Analysis Based on 16S RNA High-Throughput Sequencing
Eight fecal samples from sows in each group (n = 8 per treatment) were chosen for microbiota analysis and total bacterial DNA was extracted from approximately 0.25 g of feces using a QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The diversity and composition of the bacterial community were determined by high-throughput sequencing of the microbial 16S rRNA genes. The V4 hypervariable region of the 16S rRNA genes was PCR amplified using 515F: 5′-GTGCCAGCMGCCGCGGTAA-3′ and 806R: 5′-GGACTACHVGGGTWTCTAAT-3′ primers, Illumina adaptors, and molecular barcodes. Paired-end sequencing was performed on the Illumina HiSeq 2500 platform (Novogene, Beijing, China). Raw 16S data sequences were obtained before being screened and assembled using the QIIME (v1.9.0) (Caporaso et al., 2010) and FLASH software packages. UPARSE (v7.0.1001) (Edgar, 2013) was used to analyze the high-quality sequences and determine OTUs. Subsequently, high-quality sequences were aligned against the SILVA reference database1 and clustered into OTUs at a 97% similarity level using the UCLUST algorithm2. Each OTU was assigned to a taxonomic level with the Ribosomal Database Project Classifier program v2.203. The assembled HiSeq sequences obtained in the present study were submitted to the NCBI’s Sequence Read Archive (SRA, No. PRJNA415259) for open access.
Metagenomic Prediction and Metabolic Phenotype Analysis
Functional metagenomes of all samples were predicted using PICRUSt v1.1.3 (Langille et al., 2013). OTUs were determined according to the instructions provided in the Genome Prediction Tutorial for PICRUSt. Metagenomes4 were predicted from the copy number normalized 16S rRNA data in PICRUSt using the predict metagenomes.py script against the functional database of KEGG Orthology. Functional categories at different levels were computed with the script categorize by function.py. Functional differences within groups were explored using LEfSe and specific analysis was performed through the Galaxy server (Mukherjee et al., 2017). Output files from the PICRUSt analysis were collected and analyzed by R software (v3.5.1) for further statistical interrogation and graphical depictions of all predicted functional datasets. BugBase5 was employed to predict organism-level microbiome phenotypes using 16S RNA datasets and mapping file according to the tutorial (Ward et al., 2017).
Serum Biochemical Indices and Immunoglobulins Analysis
Eight serum samples from each group (n = 8 per treatment) were chosen for further analysis of biochemical and immune indices. Serum parameters investigated in the present study included: total protein (TP), blood urea nitrogen (BUN), creatinine (CREA), cholesterol (CHO), triglycerides (TG), high- and low-density lipoprotein cholesterol (HDL-C, LDL-C), glucose (GLU), albumin (ALB), globulin (GLO), AST and ALT. All parameters were measured using the TBA-120FR biochemistry analyzer provided by the Biochemical Analysis Center of Hunan Normal University Hospital. IgG, IgM, and IgA were measured with enzyme-linked immunosorbent assay kits (Cusabio Biotech Co., Hubei, China) according to the manufacturer’s instructions and previous research (Lv et al., 2018).
Untargeted Metabolomic Analysis
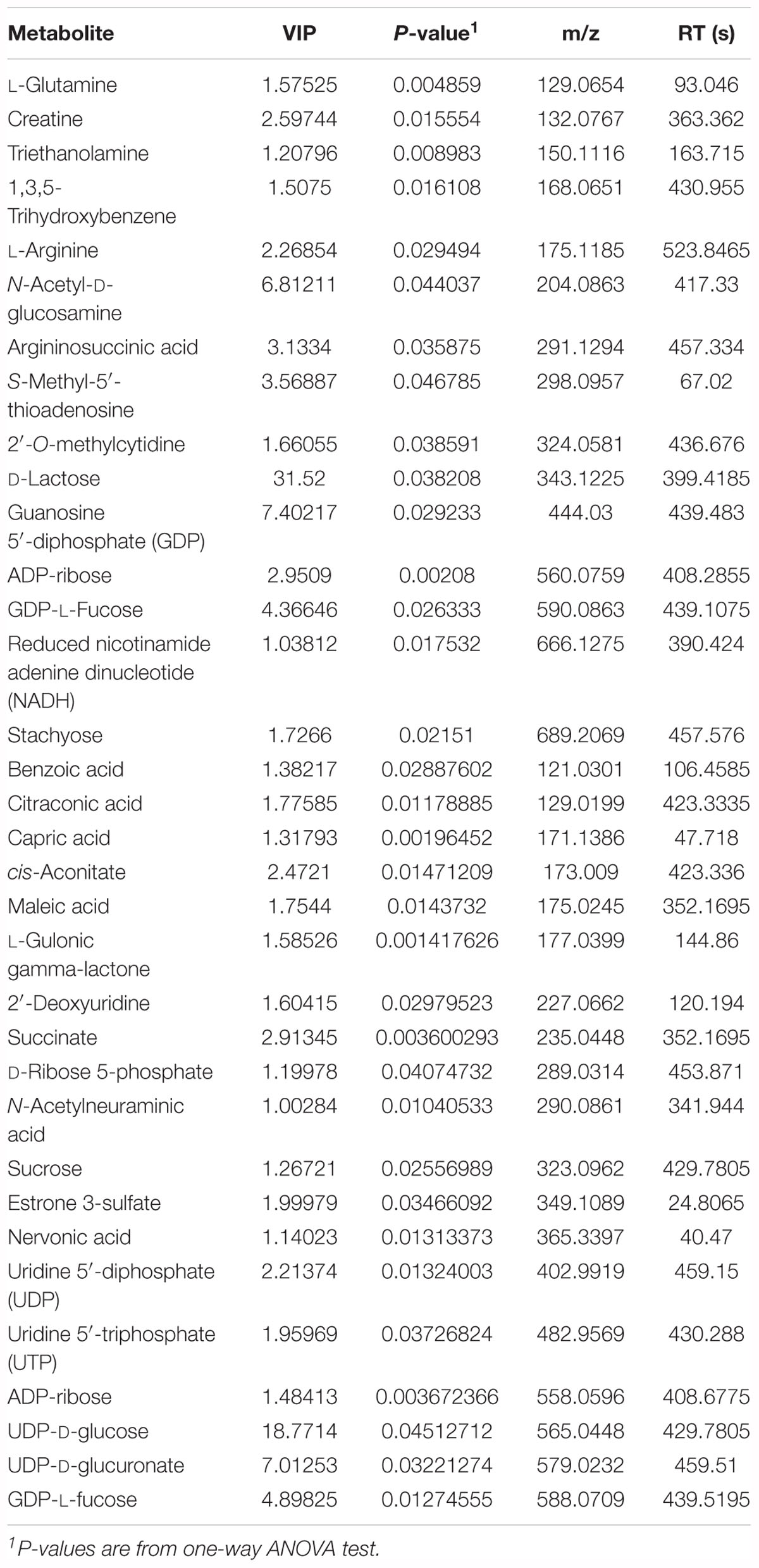
Milk from sows with nearby delivery time (within 48 h, n = 6 per treatment) were chosen for untargeted metabolomic analysis based on the liquid chromatography–tandem mass spectrometry (LC–MS/MS) platform. Milk samples were slowly thawed at 4°C first, and then 100 μl of each sample was added to 400 μl pre-cooled methanol/acetonitrile (1:1, v/v), vortex mixed, stood at -20°C for 60 min, centrifuged at 14,000 × g for 20 min at 4°C, took the supernatant and vacuum dried. For mass spectrometry, 100 μl of acetonitrile aqueous solution (acetonitrile:water = 1:1, v/v) was reconstituted, vortexed, centrifuged at 14,000 × g for 5 min at 4°C, and supernatants were taken for further analysis on LC–MS/MS platform (Bioprofile Co. Ltd., Shanghai, China). Each sample was tested by positive ion and negative ion mode using electrospray ionization (ESI). Samples were separated by ultra performance liquid chromatography (UPLC) and analyzed by mass spectrometry using a Triple-TOF 5600 mass spectrometer (AB SCIEX). The raw data was converted to .mzXML format by ProteoWizard (Adusumilli and Mallick, 2017), and then the XCMS program (Tautenhahn et al., 2012) was used for peak alignment, retention time correction, and peak area extraction. Metabolomic data were analyzed with MetaboAnalyst v4.0 (Chong et al., 2018) online version6. Key metabolites were filtered by VIP scores and rules used in our previous research (Ren et al., 2015).
Statistical Analysis
All statistical analyses were performed using SPSS 25.0 software (SPSS Inc., Chicago, IL, United States). Alpha and beta diversity were analyzed with QIIME (v1.7.0) and displayed with R software (v3.5.1) and details can be found in the legends of the corresponding figures and tables. The differences among groups were compared using one-way ANOVA and Tukey-Kramer multiple comparison tests. P-values <0.05 were used to indicate statistical significance.
Results
Lysozyme Significantly Altered the Diversity and Composition of Sow’s Gut Microbiota
Gut microbial diversity, evidenced by the Shannon index, showed a significant reduction in the 1.0 kg/t group compared to the control (p = 0.0014, Figure 1A) and no remarkable differences were found in indicators of microbial richness (ACE and Chao1, Supplementary Table 1). The principal coordinate analysis (PCoA) based on Bray–Curtis dissimilarity revealed that microbiota showed obvious segregation from the control group to lysozyme-treated groups (Figure 1B). In addition, non-metric multidimensional scaling (NMDS) plots of β-diversity weighted unifrac (Figure 1C) also confirmed the differences between control and lysozyme-treated groups [all P < 0.05 by Anosim analysis and multi-response permutation procedure (MRPP)]. Furthermore, an unweighted pair-group method with arithmetic mean (UPGMA) analysis based on weighted unifrac distances were applied and the phylogeny showed the relationships of all observed samples. The phylogeny revealed that Firmicutes, Bacteroidetes, Proteobacteria, Fibrobacteres, and Spirochaetes are the dominant bacteria in sow’s gut microbiome (Supplementary Figure 1).
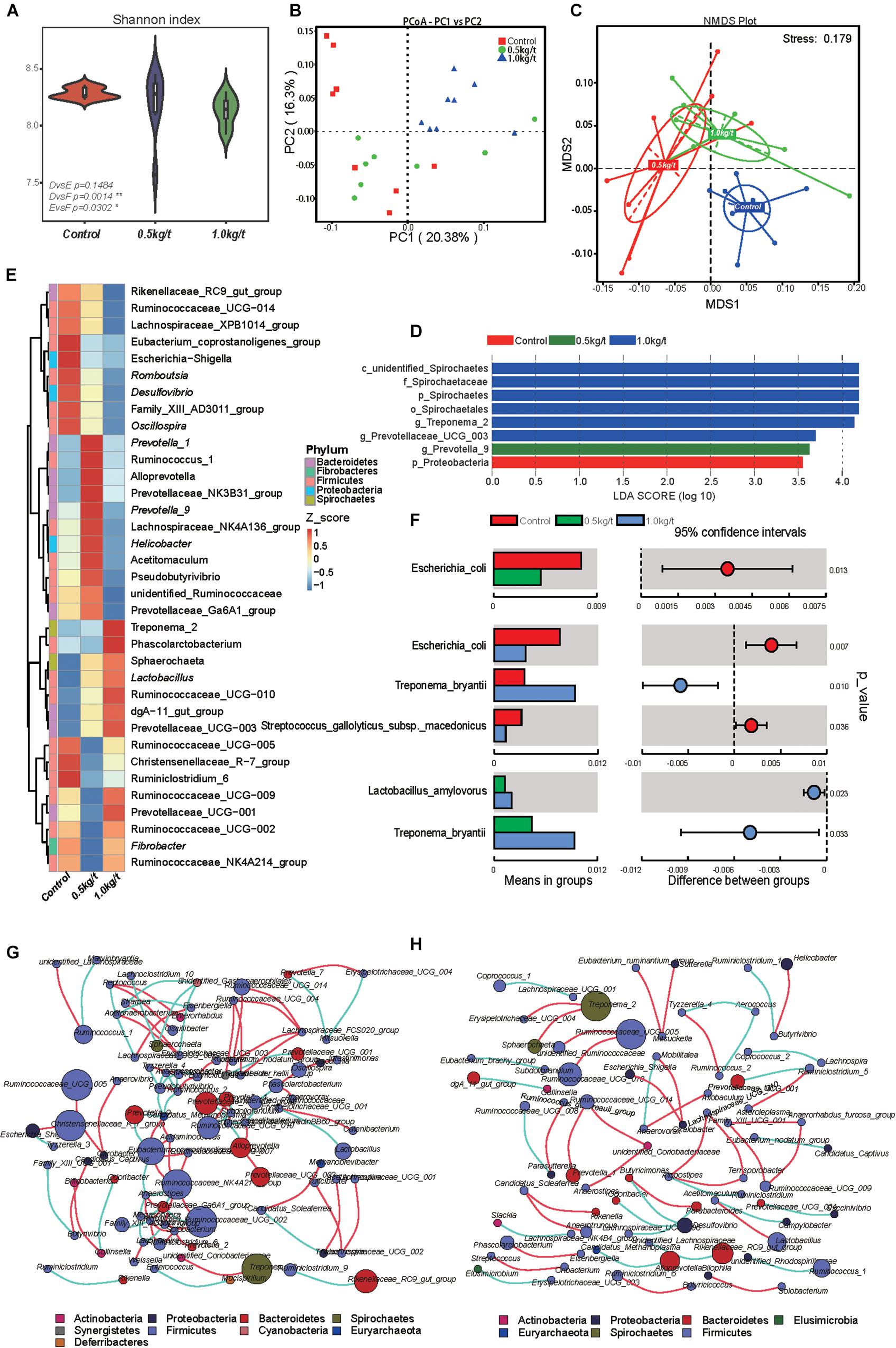
Figure 1. Effects of different lysozyme levels on sow’s gut microbiota. (A) The microbial alpha diversity (as accessed by Shannon index) based on whole OTU table. The distribution and density of samples are displayed in violin plot. Boxes represent the interquartile ranges, the inside black plots represent the median, and circles are outliers. P-values are from Wilcoxon rank sum test. (B,C) Principal coordinate analysis (PCoA) and non-metric multidimensional scaling (NMDS) analysis based on the OTU table. Significant P-values of Anosim and multi-response permutation procedure (MRPP) between groups emphasize the differences in microbial community structure. (D) LEfse analysis at different microbial taxonomic levels [linear discriminant analysis (LDA) score = 3.5]. (E) Heatmap tree shows genera significantly different among groups and their phylogenic relationships. The abundance profiles are expressed by z-scores, and genera were clustered based on Bray–Curtis distance in the clustering tree. (F) T-test bar plot of significantly differed species between groups. (G,H) Spearman’s correlation networks based on genera profile. Control group (G) and 1.0 kg/t treated group (H) showed alterations in microbial relationships.
Further, variations in the microbial composition of all groups were explored. LEfSe analysis of the bacterial community was used to filter the significantly different OTUs among groups and the results showed that there exist dramatic differences in microbial composition between the 1.0 kg/t group and the control group (Figure 1D). Spirochaetes, Euryarchaeota, and Actinobacteria significantly increased but Firmicutes showed a remarkable reduction in the 1.0 kg/t treated group compared with the control (Table 1). The heat map (according to the top 35 most different genera) shows the taxonomic distributions among each group (Figure 1E). Specifically, Escherichia coli showed a dramatically dose-response reduction in both the 0.5 kg/t and 1.0 kg/t lysozyme-treated groups (Figure 1F). Furthermore, Lactobacillus amylovorus showed a significant increase in the 0.5 kg/t group (Figure 1F).
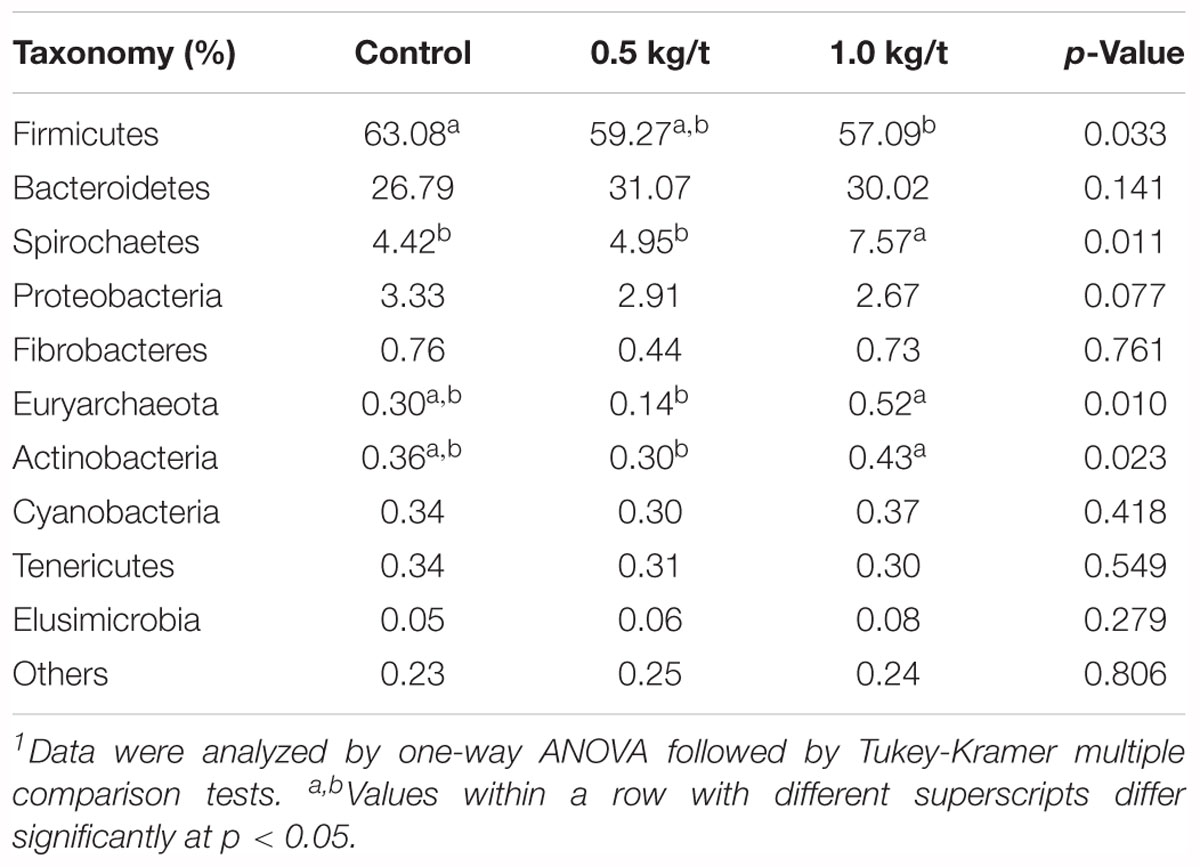
Table 1. Changes in major microbial phylum of sow’s gut microbiota after the 21-day lysozyme supplementation1.
To further determine the relationships among different microbes in the control and the 1.0 kg/t group, network analysis of gut microbiome was determined by calculating Spearman’s correlation coefficients among all genera. The results revealed that lysozyme supplementation with 1.0 kg/t rebuilt interactions among different genera (Figure 1H). Compared with the control (Figure 1G), gut microbiota shaped by 1.0 kg/t lysozyme treatments had fewer cross-linking, more positive correlations and shorter interactions indicated by lower graph densities (GD) (0.0037 and 0. 0027 in the control and 1.0 kg/t group, respectively), lower average degree (AD) (1.64 and 1.17), lower network diameters (ND) (5 and 4), lower average path lengths (APL) (1.66 and 1.17), higher modularity (MD) (0.87 and 0.94), and higher cluster coefficients (CC) (0.48 and 0.66).
Lysozyme Shifted the Metabolic Functions of Sow’s Gut Microbiota
To investigate further changes in microbial metabolic functions in gut microbiota driven by lysozyme treatment, Picrust was used to generate the metagenome based on 16S RNA sequencing results. The principal component analysis (PCA) based of KEGG annotation results revealed that the metabolic functions of sow’s gut microbiota showed obvious segregation from the control group to lysozyme-treated groups (Figure 2A). Furthermore, 80 pathways were found to significantly differ among groups at KEGG level 3, including those associated with cellular processes, environmental factors, genetic information processing, organismic systems, metabolism, and human diseases (Supplementary Table 2). LEfSe analysis of the KEGG annotation results was used to filter the significantly differed pathways among groups (Figure 2B) and results showed that there exist dramatic differences in microbial composition between the 1.0 kg/t group and the control group (Figure 2D), which is in line with variations in microbial structure (Figures 1D–F). In current study, microbial metabolism related pathways at KEGG Level 3 were specifically concerned and filtered. The heat map (according to the most different metabolism related pathways) showed the specific functional pathway distribution among each group (Figure 2C). Moreover, pyrimidine metabolism, purine metabolism, and amino acid related enzymes were significantly upregulated in the 1.0 kg/t lysozyme-treated group (Figure 2D).
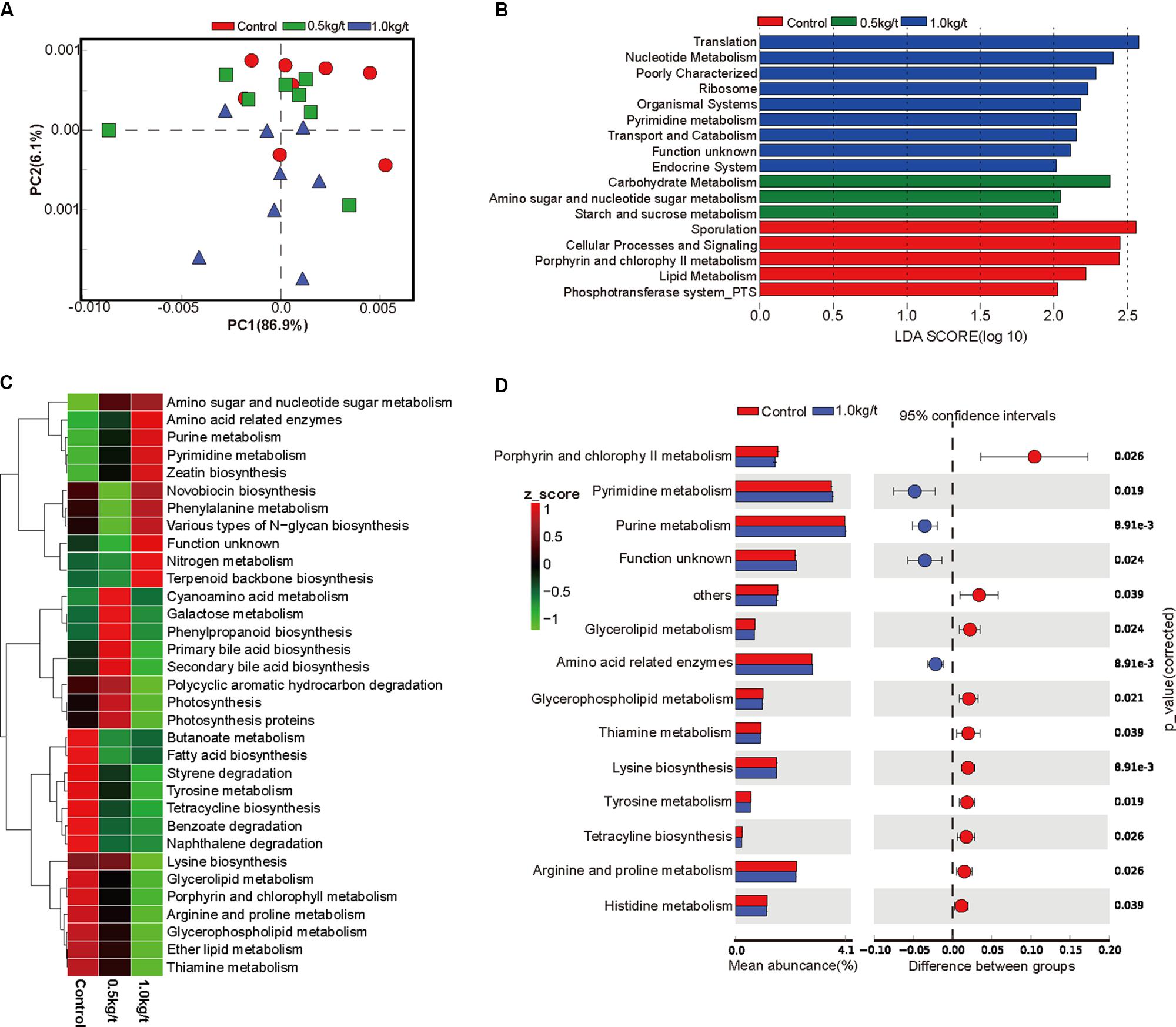
Figure 2. Lysozyme supplementation shifted the metabolic functions of sow’s gut microbiota. (A) Principal components analysis (PCA) plot of functional profiles upon lysozyme supplementation. (B) LEfse analysis at different KEGG taxonomic levels [linear discriminant analysis (LDA score = 2.0)]. (C) Heatmap tree shows metabolism related pathway significantly different among groups at KEGG level 3 and their phylogenic relationships. (D) T-test bar plot of significantly differed pathways between 1.0 kg/t treated groups and control group at KEGG level 3.
Lysozyme Mediated Dose-Dependent Changes in Gut Microbial Metabolic Phenotypes
To determine the reported impact of lysozyme on Gram-positive bacteria, organism-level coverage of functional pathways and biologically interpretable phenotypes were predicted via BugBase, an algorithm that predicts organism-level coverage of functional pathways, as well as biologically interpretable phenotypes, such as oxygen tolerance, gram staining and pathogenic potential, within complex microbiomes using either whole-genome shotgun or marker gene sequencing data. Results showed that richness of anaerobic bacteria was significantly higher in 0.5 kg/t group than 1.0 kg/t group (Figures 3A–C). Results demonstrated that the richness of Gram-positive bacteria were significantly down-regulated by lysozyme treatments, while Gram-negative bacteria showed a significant increase (Figures 3D,E). Further, mobile genetic elements and oxidative stress tolerance of gut microbiota were reduced by increased lysozyme level (Figures 3F,H). Otherwise, lysozyme supplementation significantly increased biofilm formation in the lysozyme-treated groups (Figures 3G,I).
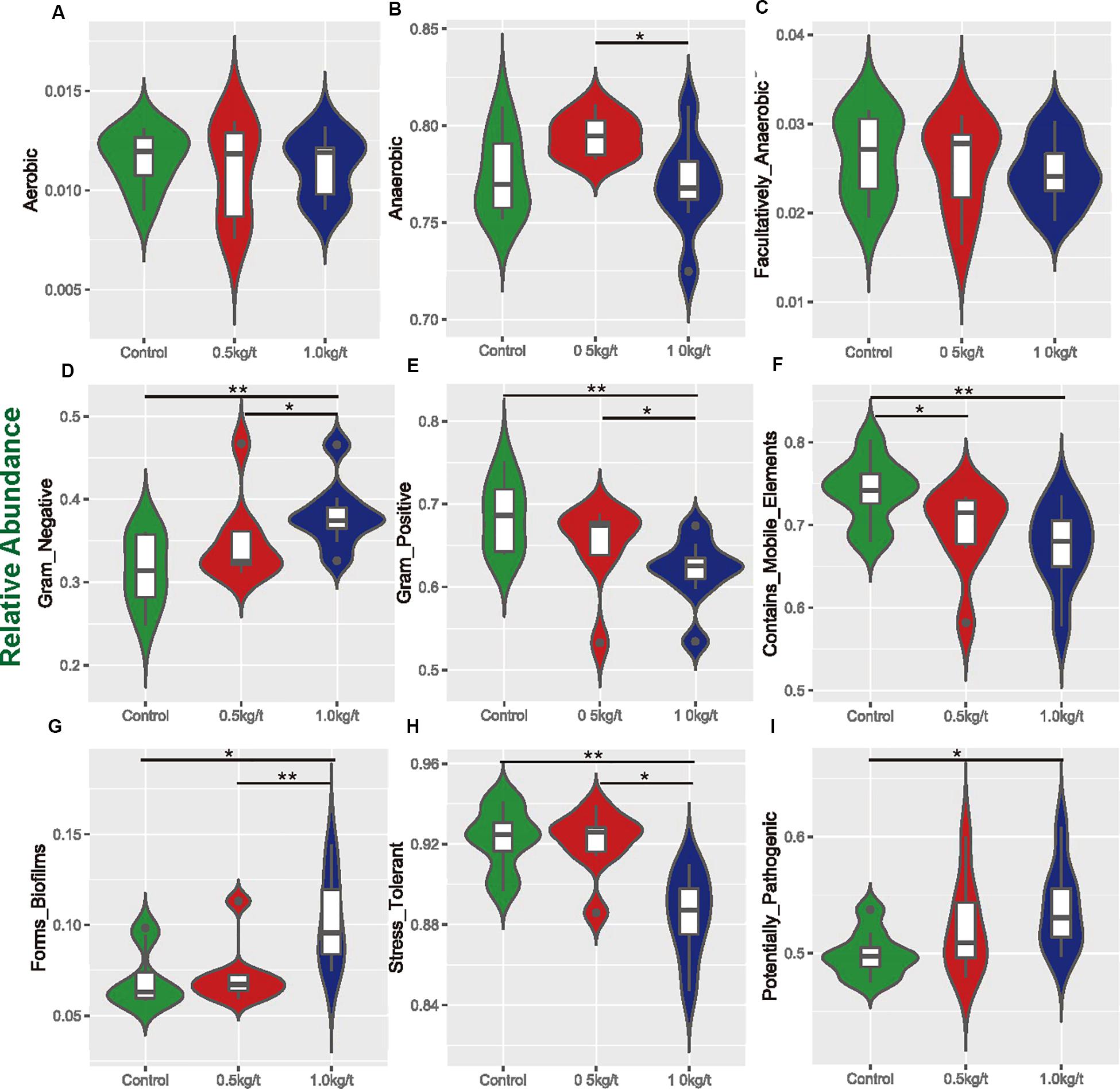
Figure 3. Variations in metabolic phenotypes of sow’s microbiota driven by lysozyme treatment. These results were generated from BugBase online (https://bugbase.cs.umn.edu/). (A–C) Oxygen utilizing. (D,E) Gram bacterial classification. (F) Mobile-element containing. (G) Biofilm forming. (H) Oxidative stress tolerant. (I) Potential pathogenic risk. Discrete phenotype relative abundances were compared using pair-wise Mann–Whitney U tests with false discovery rate correction, ∗P < 0.05, ∗∗P < 0.01.
Lysozyme Significantly Changed Sow Serum Immunity
To identify the impact of lysozyme on sow’s immunity, serum biochemical indices and immunoglobulins were determined. Results showed that serum alanine transaminase (ALT) levels were significantly down-regulated by lysozyme treatment (Figure 4B). Serum aspartate transaminase showed no difference among groups (Figures 4A,C). Dietary lysozyme supplementation had no significant impact on serum metabolite profiles (e.g., HDL-c, LDL-c, TG, and BUN, Supplementary Table 3). For immunoglobulins, serum IgM levels were significantly higher in the 1.0 kg/t group compared with the control, while IgA levels were significantly lower in the 1.0 kg/t group (Figures 4D–F).
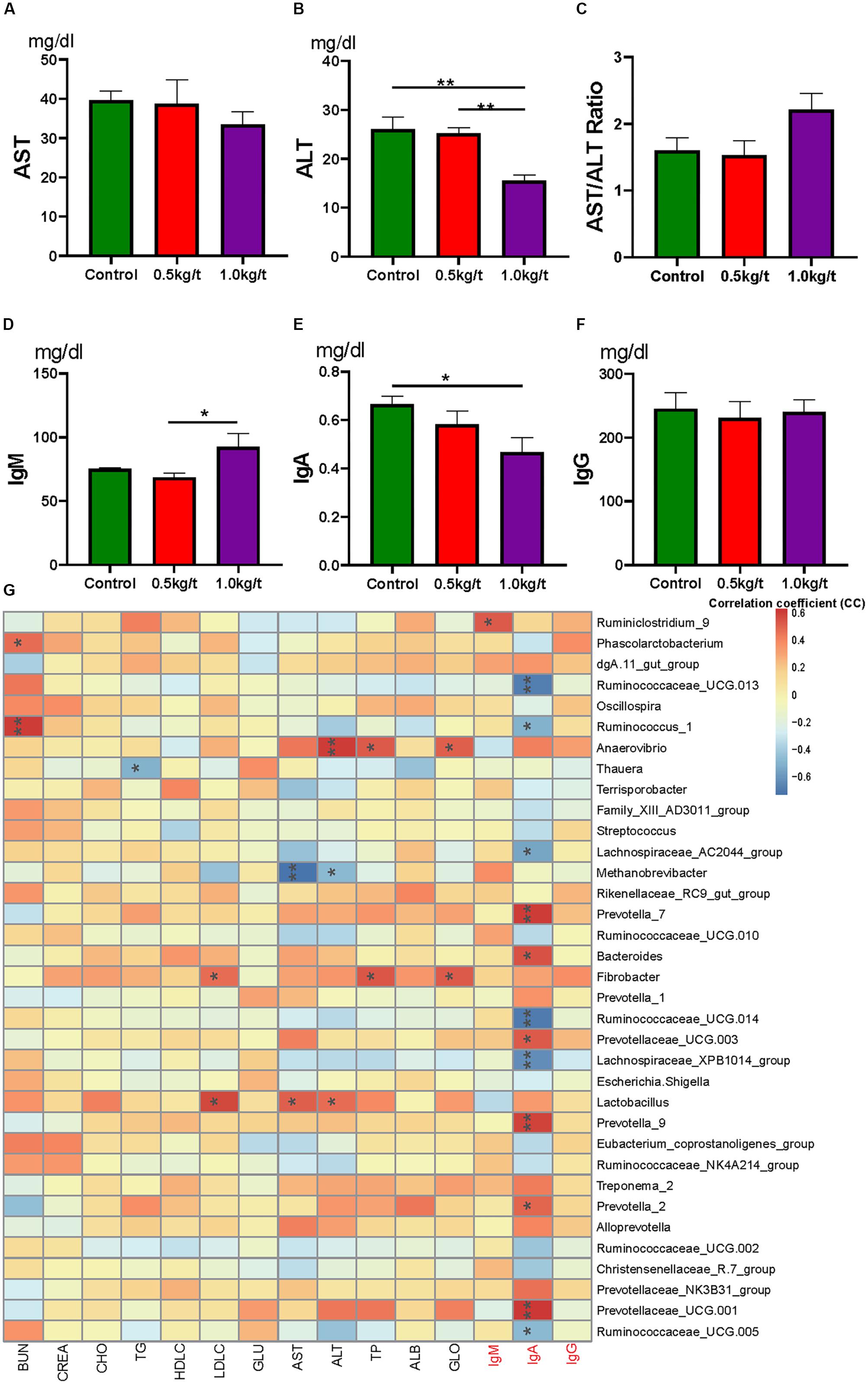
Figure 4. Lysozyme mediated changes in serum indices and correlations with gut microbiota. (A) Serum aspartate transaminase (AST), (B) serum alanine transaminase (ALT), (C) AST/ALT ratio, (D–F). Serum immunoglobulins, IgM (D), IgA (E), and IgG (F). (G) Heatmap of the Spearman r correlations between the gut microbiota significantly modified serum biochemical makers after 21-day lysozyme supplementation. ∗P < 0.05, ∗∗P < 0.01 (following the Spearman correlation analysis). BUN, blood urea nitrogen; CREA, creatinine; TG, triglycerides; GLU, glucose; HDL-C, LDL-C, high- and low-density lipoprotein cholesterol; CHO, cholesterol; AST, aspartate transaminase; ALT, alanine transaminase; TP, total protein; ALB, albumin; GLO, globulin; A/G, albumin/globulin ratio.
To further explore the relationship between immunity and the altered gut microbiome driven by lysozyme treatment, Spearman’s correlation coefficients between serum biochemical makers and immunoglobulins and major genera were calculated and visualized with heatmaps. Twelve genera, including Prevotella, Ruminococcaceae UGG, and Bacteroides showed significant correlations with IgA (Figure 4G). Ruminiclostridium 9 showed a significant positive relation with IgM. Methanobrevibacter showed a significantly negative relationship with AST, while Lactobacillus showed a significant positive correlation (Figure 4G).
Variations in Sow’s Milk Metabolite Profile Driven by Different Lysozyme Levels
To evaluate the effects of lysozyme treatment on sow’s milk, untargeted LC–MS/MS was applied to assess the metabolite profiles after the 21-day supplementation. PCA of metabolites cannot effectively distinguish the variations among groups (Figure 5A), therefore, partial least squares - discriminant analysis (PLS-DA) and sparse partial least squares - discriminant analysis (sPLS-DA) were used (Figures 5B,C). Both techniques revealed a distinct partition between lysozyme-treated groups and the control. The heat map (according to the most different metabolites) showed specific metabolite distributions within each group (Figure 5D). Twenty metabolic makers were identified by PLS-DA including uridine 5′-diphosphate (UDP), UDP-D-glucuronate, acamprosate, and triethanolamine (Figure 5E). In addition, 10 metabolites were filtered by sPLS-DA including succinate, L-glutamine, and UDP-D-glucuronate (Supplementary Figure 2). Given this, metabolites significantly differed among groups were filtered and combined via both PLS-DA and sPLS-DA (Table 2). Moreover, significant differences metabolites between the 1.0 kg/t and the control groups were also filtered via PLS-DA (Supplementary Figure 3) and metabolic makers were obtained (Supplementary Table 4). Milk metabolites such as L-glutamine, creatine and L-arginine showed significantly dose-dependent changes after treatment (Figure 5 and Table 2).
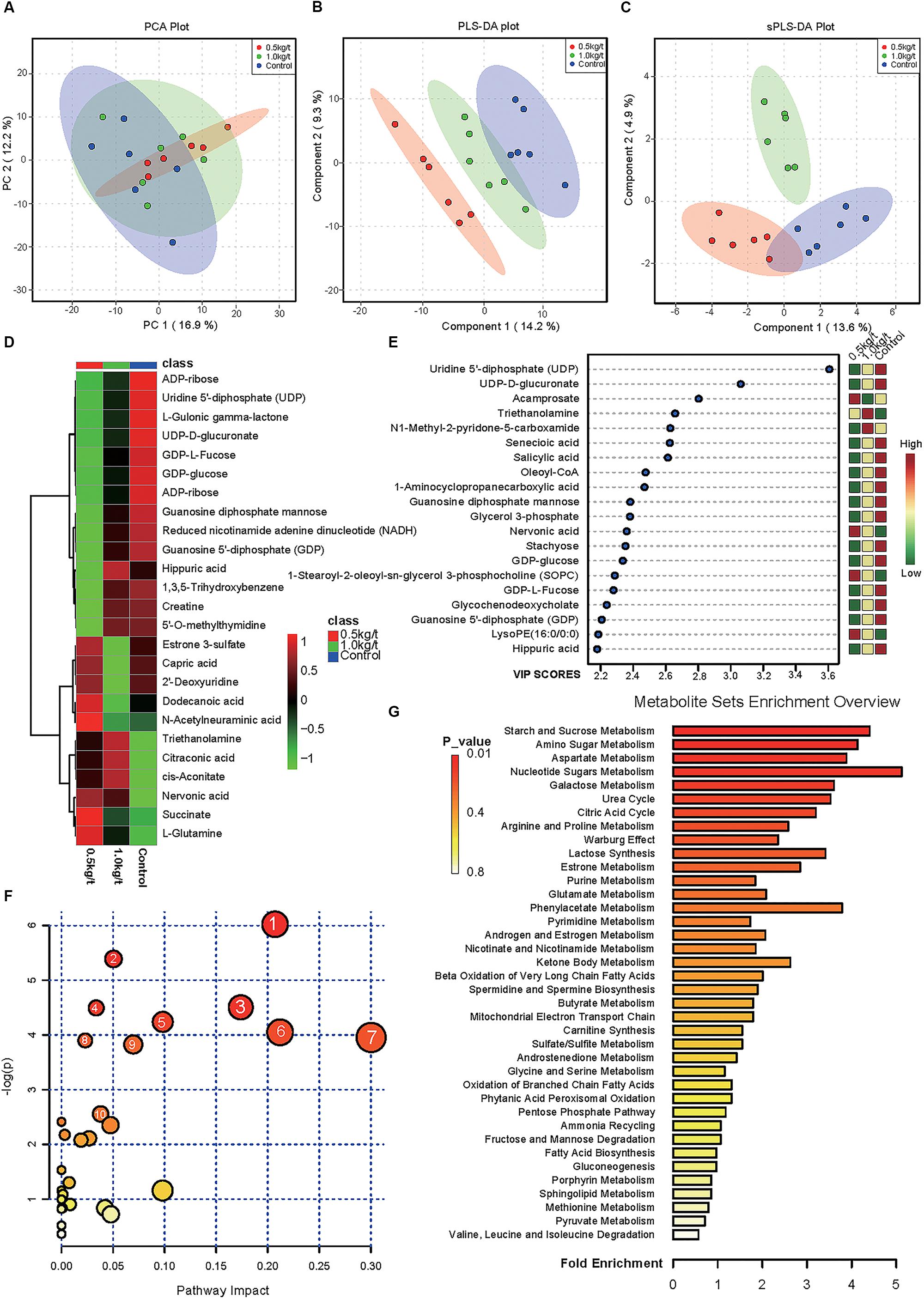
Figure 5. Changes in sow’s milk metabolite profile shaped by different lysozyme levels. (A) Principal components analysis (PCA) plot of metabolic profiles of sow’s milk upon lysozyme supplementation. (B,C) Partial least squares - discriminant analysis (PLS-DA) score plot (B) and sparse partial least squares - discriminant analysis (sPLS-DA) plot (C) of metabolic profiles and the explained variances are shown in brackets. (D) Heatmap tree shows metabolite significantly different among groups and their phylogenic relationships. The abundance profiles are expressed by z-scores. (E) Important features identified by PLS-DA. The colored boxes on the right indiate the relative concentrations of the corresponding metabolite in each group under study. (F) Bubble diagram of significantly differed pathways. The numbers are in accord with Table 3 and representing filtered pathways. (G) Bar plot for metabolite sets enrichment. The bar length represent fold enrichment and the color legend indicating p-value.
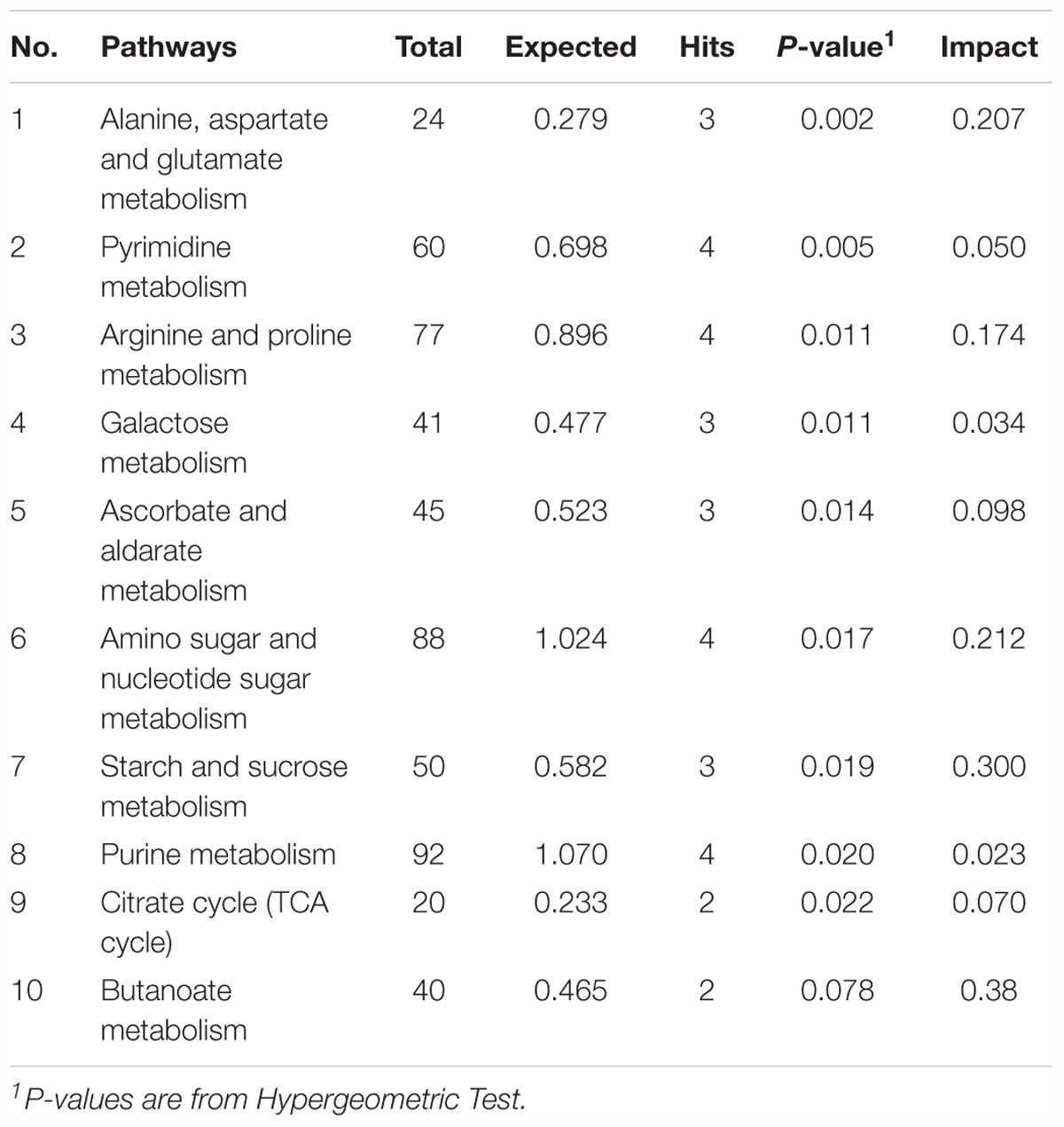
To explore the biological functions of these metabolic makers, metabolite-set enrichment analysis was performed via MetaboAnalyst v4.0 (Figure 5G). Pathway topology analysis using relative centrality revealed nine significantly different (P < 0.05) enriched pathways (Figure 5F), including alanine, aspartate, and glutamate metabolism, pyrimidine metabolism, arginine and proline metabolism, galactose metabolism, ascorbate and aldarate metabolism, amino sugar and nucleotide sugar metabolism, starch and sucrose metabolism, purine metabolism, and the citrate cycle (Table 3).
To further investigate the correlation between milk metabolic indicators and the altered gut microbiome driven by lysozyme treatment, Spearman’s correlation coefficients between significantly different metabolites and major genera were also calculated and visualized with heat map (Supplementary Figure 4A). For instance, L-glutamine showed significant positive correlations with Lactobacillus, Ruminococcus 1 and Lachnospiraceae MK4A136 group. L-Arginine showed significant negative correlations with Sphaerochaeta, Prevotella 1, Pseudobutyrivibrio, and Prevotella 9. Moreover, lysozyme-supplementation mediated associations among milk composition, serum immunity, and gut microbiota were also summarized and bacteria showed significant impact on both serum biomarkers and milk metabolic makers were filtered (Supplementary Figure 4B).
Discussion
The application of antibiotics in feeds at subtherapeutic levels could improve performance and overall health and is used extensively throughout the swine industry (Oliver and Wells, 2015). Because of the rising concerns about antibiotic resistance and human health, many countries including China (starting in 2020), America, and the European Union banned the use of antibiotics in swine production (Oliver and Wells, 2013, 2015; Oliver et al., 2014). Thus, research into alternatives is important for the future development of the livestock industry. Recent studies also revealed that maternal diets and gut microbiota could directly affect the offspring development early in life (Chu et al., 2016; Cheng et al., 2018; Wang et al., 2018).
Sows treated with probiotic combinations resulted in improved microbiota diversity in neonatal piglets (Veljovic et al., 2017). In the current study, microbial diversity evidenced by the Shannon index showed a significant reduction in the 1.0 kg/t lysozyme-treated group (p = 0.0014, Figure 1A). No differences in microbial richness were found. Previous studies reported that lysozyme could be effective against a wide range of gastrointestinal pathogens, such as Listeria monocytogenes, Clostridium perfringens, Candida spp., and Helicobacter pylori in vitro (Brundige et al., 2008; Zhang et al., 2016). Reduced microbial diversity indicating a better intestinal condition and physiological preparation for parturition (Veljovic et al., 2017; Cheng et al., 2018). Furthermore, Spirochaetes, Euryarchaeota, and Actinobacteria significantly increased but Firmicutes showed a remarkable reduction in the 1.0 kg/t group compared with the control group. As previous researches reported, ETEC is the major causative agent of diarrhea in weaned pigs, which attaches to the intestinal mucosa, leading to compromised barrier function and malabsorption of large molecules (Ko et al., 2009; Nyachoti et al., 2012; Lu D. et al., 2014). Higher concentrations of lysozyme in milk confer enteric health benefits and prevents ETEC infections in young animals (Leon-Sicairos et al., 2006; Nyachoti et al., 2012; Cooper et al., 2013; Lu D. et al., 2014). Hence, pathogenic bacteria enriched species in this study were also concerned. For instance, Escherichia coli showed a dramatically dose-response reduction in both the 0.5 kg/t and 1.0 kg/t lysozyme-treated groups (Figure 1F). Other reported pathogens which could be against by lysozyme like Helicobacter also reduced in lysozyme treated groups but it was not significant (Figure 4E). Conventional probiotics like Lactobacillus amylovorus and Treponema bryantii significantly increased in the 0.5 kg/t and 1.0 kg/t groups (Figure 1F). Lactobacillus is reported to improve the intestinal environment and activate intestinal mucosal immunity in many species, resulting in enhanced SIgA production of the innate immune system, which is essential for the prevention of fimbriae-mediated colonization and the maintenance of intestinal barrier function (Yang et al., 2014; Huang et al., 2018). In summary, lysozyme addition rebuilt sow’s gut microbiota to better composition identified by reduced richness of E. coli and increased abundance of Lactobacillus amylovorus.
For microbial metabolic functions, results showed that pyrimidine metabolism, purine metabolism, and amino acid related enzymes were significantly upregulated in the 1.0 kg/t lysozyme-treated group. Lysozyme may promote the shift to greater amino acid and nucleotide metabolism in gut microbial communities (Figure 2). These metabolic functional changes in gut microbiota are revealed for the first time. Besides, lysozyme mediated dose-dependent changes in gut microbial metabolic phenotypes. For instance, higher lysozyme levels in formula diet significantly down-regulated the richness of Gram-positive bacteria. Our findings confirmed that lysozyme has a robust antimicrobial activity against Gram-positive bacteria, and to a much lesser degree, against Gram-negative bacteria (Masschalck and Michiels, 2003). Lysozyme supplementation also significantly increased biofilm formation in the lysozyme-treated groups (Figure 3G). Formation of microbial community biofilms was reported to be closely related to drug resistance and pathogenesis (Adil et al., 2014; Lu J. et al., 2014). The mechanism of these changes in metabolic phenotypes also remain unknown. In conclusion, lysozyme supplementation also shifted the metabolic functions and phenotypes of sow’s gut microbiota.
Lysozyme is an important modulator in innate immunity and plays a crucial role in preventing intestinal inflammation (Long et al., 2016; Patel and Kuyucak, 2017; Ragland et al., 2017). Mammalian Paneth cells are able to secrete lysozyme via secretory autophagy to maintain intestinal homeostasis to against pathogenic infections (Bel et al., 2017). For serum immunity, serum IgA levels were significantly reduced by increased lysozyme levels. Meanwhile, serum IgM was significantly higher in the 1.0 kg/t group compared with the control group. Lower serum IgA indicates a lower risk of allergy in the postnatal period, which differs from mucosal SIgA (Klobasa et al., 1985; Hansen et al., 2017). IgM is an important anti-inflammatory factor and increased IgM also indicating a better immune status (Vaschetto et al., 2017). What is more, serum ALT levels were also significantly down-regulated by lysozyme treatment (p = 0.001). ALT was reported to be a liver-specific enzyme that is released into serum following acute liver injury (Robertson et al., 2016). Here, down-regulated serum ALT levels also indicate improved overall health (Robertson et al., 2016). In short, lysozyme supplementation could benefit sows with better immune status via down-regulating serum ALT and IgM and reducing serum IgA.
To determine the effect of dietary lysozyme on sow’s milk metabolites, untargeted LC–MS/MS were used to explore the metabolome of all groups. Results showed that metabolites including L-glutamine, succinate, triethanolamine, and L-arginine were significantly up-regulated by lysozyme supplementation. It should be noted that L-glutamine could enhance tight junction integrity and proliferation of intestinal porcine epithelial cells (Rhoads et al., 1997; Wang et al., 2015, 2016), which is essential for normal intestinal development. Also, succinate is a metabolite that improves glycemic control through the activation of intestinal gluconeogenesis (De Vadder et al., 2016). Our previous work revealed that ethanolamine can enhance intestinal function by altering gut microbiome and mucosal anti-stress capacity (Zhou et al., 2017, 2018). Moreover, L-arginine was also shown to play a significant role in shaping the gut microbiota and innate immunity of piglets, thus improving gut development and protecting against pathogenic infection (Li et al., 2012; Ren et al., 2014; Wu et al., 2015). Further, pathway enrichment and topology analysis of these metabolites revealed nine significantly different (P < 0.05) enriched pathways (Figure 5F) including alanine, aspartate, and glutamate metabolism and pyrimidine metabolism, arginine, and proline metabolism, which also further supports the conclusions noted above. To sum up, dietary lysozyme supplementation significantly altered the milk composition of sows, which may benefit the development of their offspring (Chu et al., 2016; Cheng et al., 2018; Wang et al., 2018).
Associations among gut microbiota, serum immunity and milk metabolites were also explored for the first time (Supplementary Figure 4B). This research proved that lysozyme has widespread influence on sow health including gut microbiota, serum immunity, and milk composition for the first time. In conclusion, lysozyme supplementation could effectively improve the composition, metabolic functions and phenotypes of sow’s gut microbiota and it also benefit sows with better immune status and milk composition. This research could provide theoretical support for further application of lysozyme in promoting animal gut health and prevent pathogenic infections in livestock production.
Author Contributions
JZ, XX, and YY designed the study. JZ, XX, KW, LZ, and YS carried out the animal trials and sample analysis. JZ, JY, and XX wrote and revised the manuscript.
Funding
The project was funded by the National Program on Key Basic Research Project (2017YFD0500503), the National Natural Science Foundation of China (31572420), and Natural Science Foundation of Hunan Province (2018JJ1028).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Shanghai E.K.M Biotechnology Company for material assistance. JZ specially thanks XX for her support and encouragement these years.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00177/full#supplementary-material
Footnotes
- ^ https://www.arb-silva.de/
- ^ https://drive5.com/usearch/manual/uclust_algo.html
- ^ https://rdp.cme.msu.edu/
- ^ http://picrust.github.io/picrust/
- ^ https://bugbase.cs.umn.edu/
- ^ http://www.metaboanalyst.ca/faces/home.xhtml
References
Adil, M., Singh, K., Verma, P. K., and Khan, A. U. (2014). Eugenol-induced suppression of biofilm-forming genes in Streptococcus mutans: an approach to inhibit biofilms. J. Glob. Antimicrob. Resist. 2, 286–292. doi: 10.1016/j.jgar.2014.05.006
Adusumilli, R., and Mallick, P. (2017). Data conversion with proteowizard msconvert. Methods Mol. Biol. 1550, 339–368. doi: 10.1007/978-1-4939-6747-6_23
Bel, S., Pendse, M., Wang, Y., Li, Y., Ruhn, K. A., Hassell, B., et al. (2017). Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science 357, 1047–1052. doi: 10.1126/science.aal4677
Brundige, D. R., Maga, E. A., Klasing, K. C., and Murray, J. D. (2008). Lysozyme transgenic goats’ milk influences gastrointestinal morphology in young pigs. J. Nutr. 138, 921–926. doi: 10.1093/jn/138.5.921
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Cheng, C., Wei, H., Xu, C., Xie, X., Jiang, S., and Peng, J. (2018). Maternal soluble fiber diet during pregnancy changes the intestinal microbiota, improves growth performance, and reduces intestinal permeability in piglets. Appl. Environ. Microbiol. 84, e1047–18. doi: 10.1128/AEM.01047-18
Chong, J., Soufan, O., Li, C., Caraus, I., Li, S., Bourque, G., et al. (2018). MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46, W486–W494. doi: 10.1093/nar/gky310
Chu, D. M., Antony, K. M., Ma, J., Prince, A. L., Showalter, L., Moller, M., et al. (2016). The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Med. 8:77. doi: 10.1186/s13073-016-0330-z
Cooper, C. A., Garas Klobas, L. C., Maga, E. A., and Murray, J. D. (2013). Consuming transgenic goats’ milk containing the antimicrobial protein lysozyme helps resolve diarrhea in young pigs. PLoS One 8:e58409. doi: 10.1371/journal.pone.0058409
Cox, L. M., Yamanishi, S., Sohn, J., Alekseyenko, A. V., Leung, J. M., Cho, I., et al. (2014). Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158, 705–721. doi: 10.1016/j.cell.2014.05.052
De Vadder, F., Kovatcheva-Datchary, P., Zitoun, C., Duchampt, A., Backhed, F., and Mithieux, G. (2016). Microbiota-produced succinate improves glucose homeostasis via intestinal gluconeogenesis. Cell Metab. 24, 151–157. doi: 10.1016/j.cmet.2016.06.013
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Gao, X., Guo, M., Zhang, Z., Shen, P., Yang, Z., and Zhang, N. (2017). Baicalin promotes the bacteriostatic activity of lysozyme on S. Aureus in mammary glands and neutrophilic granulocytes in mice. Oncotarget 8, 19894–19901. doi: 10.18632/oncotarget.15193
Hansen, I. S., Hoepel, W., Zaat, S. A. J., Baeten, D. L. P., and den Dunnen, J. (2017). Serum IgA immune complexes promote proinflammatory cytokine production by human macrophages, monocytes, and kupffer cells through FcalphaRI-TLR cross-talk. J. Immunol. 199, 4124–4131. doi: 10.4049/jimmunol.1700883
Huang, G., Li, X., Lu, D., Liu, S., Suo, X., Li, Q., et al. (2018). Lysozyme improves gut performance and protects against enterotoxigenic Escherichia coli infection in neonatal piglets. Vet. Res. 49:20. doi: 10.1186/s13567-018-0511-4
Klobasa, F., Habe, F., Werhahn, E., and Butler, J. E. (1985). Changes in the concentrations of serum IgG, IgA and IgM of sows throughout the reproductive cycle. Vet. Immunol. Immunopathol. 10, 341–353. doi: 10.1016/0165-2427(85)90023-6
Ko, K. Y., Mendoncam, A. F., Ismail, H., and Ahn, D. U. (2009). Ethylenediaminetetraacetate and lysozyme improves antimicrobial activities of ovotransferrin against Escherichia coli O157:H7. Poult. Sci. 88, 406–414. doi: 10.3382/ps.2008-00218
Langille, M. G., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Lee, M., Kovacs-Nolan, J., Yang, C., Archbold, T., Fan, M. Z., and Mine, Y. (2009). Hen egg lysozyme attenuates inflammation and modulates local gene expression in a porcine model of dextran sodium sulfate (DSS)-induced colitis. J. Agric. Food Chem. 57, 2233–2240. doi: 10.1021/jf803133b
Leon-Sicairos, N., Lopez-Soto, F., Reyes-Lopez, M., Godinez-Vargas, D., Ordaz-Pichardo, C., and de la Garza, M. (2006). Amoebicidal activity of milk, apo-lactoferrin, sIgA and lysozyme. Clin. Med. Res. 4, 106–113. doi: 10.3121/cmr.4.2.106
Li, Q., Liu, Y., Che, Z., Zhu, H., Meng, G., Hou, Y., et al. (2012). Dietary L-arginine supplementation alleviates liver injury caused by Escherichia coli LPS in weaned pigs. Innate Immun. 18, 804–814. doi: 10.1177/1753425912441955
Long, Y., Lin, S., Zhu, J., Pang, X., Fang, Z., Lin, Y., et al. (2016). Effects of dietary lysozyme levels on growth performance, intestinal morphology, non-specific immunity and mRNA expression in weanling piglets. Anim. Sci. J. 87, 411–418. doi: 10.1111/asj.12444
Lu, D., Li, Q., Wu, Z., Shang, S., Liu, S., Wen, X., et al. (2014). High-level recombinant human lysozyme expressed in milk of transgenic pigs can inhibit the growth of Escherichia coli in the duodenum and influence intestinal morphology of sucking pigs. PLoS One 9:e89130. doi: 10.1371/journal.pone.0089130
Lu, D., Liu, S., Shang, S., Wu, F., Wen, X., Li, Z., et al. (2015). Production of transgenic-cloned pigs expressing large quantities of recombinant human lysozyme in milk. PLoS One 10:e0123551. doi: 10.1371/journal.pone.0123551
Lu, J., Turnbull, L., Burke, C. M., Liu, M., Carter, D. A., Schlothauer, R. C., et al. (2014). Manuka-type honeys can eradicate biofilms produced by Staphylococcus aureus strains with different biofilm-forming abilities. PeerJ 2:e326. doi: 10.7717/peerj.326
Lv, D., Xiong, X., Yang, H., Wang, M., He, Y., Liu, Y., et al. (2018). Effect of dietary soy oil, glucose, and glutamine on growth performance, amino acid profile, blood profile, immunity, and antioxidant capacity in weaned piglets. Sci. China Life Sci. 61, 1233–1242. doi: 10.1007/s11427-018-9301-y
Maga, E. A., Cullor, J. S., Smith, W., Anderson, G. B., and Murray, J. D. (2006a). Human lysozyme expressed in the mammary gland of transgenic dairy goats can inhibit the growth of bacteria that cause mastitis and the cold-spoilage of milk. Foodborne Pathog. Dis. 3, 384–392.
Maga, E. A., Shoemaker, C. F., Rowe, J. D., Bondurant, R. H., Anderson, G. B., and Murray, J. D. (2006b). Production and processing of milk from transgenic goats expressing human lysozyme in the mammary gland. J. Dairy Sci. 89, 518–524.
Maga, E. A., Desai, P. T., Weimer, B. C., Dao, N., Kultz, D., and Murray, J. D. (2012). Consumption of lysozyme-rich milk can alter microbial fecal populations. Appl. Environ. Microbiol. 78, 6153–6160. doi: 10.1128/AEM.00956-12
Masschalck, B., and Michiels, C. W. (2003). Antimicrobial properties of lysozyme in relation to foodborne vegetative bacteria. Crit. Rev. Microbiol. 29, 191–214. doi: 10.1080/713610448
May, K. D., Wells, J. E., Maxwell, C. V., and Oliver, W. T. (2012). Granulated lysozyme as an alternative to antibiotics improves growth performance and small intestinal morphology of 10-day-old pigs. J. Anim. Sci. 90, 1118–1125. doi: 10.2527/jas.2011-4297
Mukherjee, A., Chettri, B., Langpoklakpam, J. S., Basak, P., Prasad, A., Mukherjee, A. K., et al. (2017). Bioinformatic approaches including predictive metagenomic profiling reveal characteristics of bacterial response to petroleum hydrocarbon contamination in diverse environments. Sci. Rep. 7:1108. doi: 10.1038/s41598-017-01126-3
Nattress, F. M., and Baker, L. P. (2003). Effects of treatment with lysozyme and nisin on the microflora and sensory properties of commercial pork. Int. J. Food Microbiol. 85, 259–267. doi: 10.1016/S0168-1605(02)00545-7
Nyachoti, C. M., Kiarie, E., Bhandari, S. K., Zhang, G., and Krause, D. O. (2012). Weaned pig responses to Escherichia coli K88 oral challenge when receiving a lysozyme supplement. J. Anim. Sci. 90, 252–260. doi: 10.2527/jas.2010-3596
Oh, M., Lee, J., Jeong, Y., and Kim, M. (2016). Synergistic antilisterial effects of mixtures of lysozyme and organic acids. J. Food Prot. 79, 2184–2189. doi: 10.4315/0362-028X.JFP-16-156
Oliver, W. T., and Wells, J. E. (2013). Lysozyme as an alternative to antibiotics improves growth performance and small intestinal morphology in nursery pigs. J. Anim. Sci. 91, 3129–3136. doi: 10.2527/jas.2012-5782
Oliver, W. T., and Wells, J. E. (2015). Lysozyme as an alternative to growth promoting antibiotics in swine production. J. Anim. Sci. Biotechnol. 6:35. doi: 10.1186/s40104-015-0034-z
Oliver, W. T., Wells, J. E., and Maxwell, C. V. (2014). Lysozyme as an alternative to antibiotics improves performance in nursery pigs during an indirect immune challenge. J. Anim. Sci. 92, 4927–4934. doi: 10.2527/jas.2014-8033
Patel, D., and Kuyucak, S. (2017). Computational study of aggregation mechanism in human lysozyme [D67H]. PLoS One 12:e0176886. doi: 10.1371/journal.pone.0176886
Ragland, S. A., Schaub, R. E., Hackett, K. T., Dillard, J. P., and Criss, A. K. (2017). Two lytic transglycosylases in Neisseria gonorrhoeae impart resistance to killing by lysozyme and human neutrophils. Cell Microbiol. 19:e12662. doi: 10.1111/cmi.12662
Ren, W., Chen, S., Yin, J., Duan, J., Li, T., Liu, G., et al. (2014). Dietary arginine supplementation of mice alters the microbial population and activates intestinal innate immunity. J. Nutr. 144, 988–995. doi: 10.3945/jn.114.192120
Ren, W. K., Yin, J., Gao, W., Chen, S., Duan, J. L., Liu, G., et al. (2015). Metabolomics study of metabolic variations in enterotoxigenic Escherichia coli-infected piglets. RSC Adv. 5, 59550–59555. doi: 10.1039/C5RA09513A
Rhoads, J. M., Argenzio, R. A., Chen, W., Rippe, R. A., Westwick, J. K., Cox, A. D., et al. (1997). L-glutamine stimulates intestinal cell proliferation and activates mitogen-activated protein kinases. Am. J. Physiol. 272, G943–G953. doi: 10.1152/ajpgi.1997.272.5.G943
Robertson, F. P., Bessell, P. R., Diaz-Nieto, R., Thomas, N., Rolando, N., Fuller, B., et al. (2016). High serum aspartate transaminase levels on day 3 postliver transplantation correlates with graft and patient survival and would be a valid surrogate for outcome in liver transplantation clinical trials. Transpl. Int. 29, 323–330. doi: 10.1111/tri.12723
Shulman, R. J., Eakin, M. N., Czyzewski, D. I., Jarrett, M., and Ou, C. N. (2008). Increased gastrointestinal permeability and gut inflammation in children with functional abdominal pain and irritable bowel syndrome. J. Pediatr. 153, 646–650. doi: 10.1016/j.jpeds.2008.04.062
Tagashira, A., Nishi, K., Matsumoto, S., and Sugahara, T. (2018). Anti-inflammatory effect of lysozyme from hen egg white on mouse peritoneal macrophages. Cytotechnology 70, 929–938. doi: 10.1007/s10616-017-0184-2
Tautenhahn, R., Patti, G. J., Rinehart, D., and Siuzdak, G. (2012). XCMS Online: a web-based platform to process untargeted metabolomic data. Anal. Chem. 84, 5035–5039. doi: 10.1021/ac300698c
Thymann, T., Sorensen, K. U., Hedemann, M. S., Elnif, J., Jensen, B. B., Banga-Mboko, H., et al. (2007). Antimicrobial treatment reduces intestinal microflora and improves protein digestive capacity without changes in villous structure in weanling pigs. Br. J. Nutr. 97, 1128–1137. doi: 10.1017/S0007114507691910
Tong, J., Wei, H., Liu, X., Hu, W., Bi, M., Wang, Y., et al. (2011). Production of recombinant human lysozyme in the milk of transgenic pigs. Transgenic Res. 20, 417–419. doi: 10.1007/s11248-010-9409-2
Touch, V., Hayakawa, S., Fukada, K., Aratani, Y., and Sun, Y. (2003). Preparation of antimicrobial reduced lysozyme compatible in food applications. J. Agric. Food Chem. 51, 5154–5161. doi: 10.1021/jf021005d
Vaschetto, R., Clemente, N., Pagni, A., Esposito, T., Longhini, F., Mercalli, F., et al. (2017). A double blind randomized experimental study on the use of IgM-enriched polyclonal immunoglobulins in an animal model of pneumonia developing shock. Immunobiology 222, 1074–1080. doi: 10.1016/j.imbio.2017.09.002
Veljovic, K., Dinic, M., Lukic, J., Mihajlovic, S., Tolinacki, M., Zivkovic, M., et al. (2017). Promotion of early gut colonization by probiotic intervention on microbiota diversity in pregnant sows. Front. Microbiol. 8:2028. doi: 10.3389/fmicb.2017.02028
Wang, B., Wu, Z., Ji, Y., Sun, K., Dai, Z., and Wu, G. (2016). L-Glutamine enhances tight junction integrity by activating CaMK Kinase 2-AMP-activated protein kinase signaling in intestinal porcine epithelial cells. J. Nutr. 146, 501–508. doi: 10.3945/jn.115.224857
Wang, H., Zhang, C., Wu, G., Sun, Y., Wang, B., He, B., et al. (2015). Glutamine enhances tight junction protein expression and modulates corticotropin-releasing factor signaling in the jejunum of weanling piglets. J. Nutr. 145, 25–31. doi: 10.3945/jn.114.202515
Wang, J., Zheng, J., Shi, W., Du, N., Xu, X., Zhang, Y., et al. (2018). Dysbiosis of maternal and neonatal microbiota associated with gestational diabetes mellitus. Gut 67, 1614–1625. doi: 10.1136/gutjnl-2018-315988
Ward, T., Larson, J., Meulemans, J., Hillmann, B., Lynch, J., Sidiropoulos, D., et al. (2017). BugBase predicts organism level microbiome phenotypes. bioRxiv [Preprint]. doi: 10.1101/133462
Wells, J. E., Berry, E. D., Kalchayanand, N., Rempel, L. A., Kim, M., and Oliver, W. T. (2015). Effect of lysozyme or antibiotics on faecal zoonotic pathogens in nursery pigs. J. Appl. Microbiol. 118, 1489–1497. doi: 10.1111/jam.12803
Wu, L., Liao, P., He, L., Feng, Z., Ren, W., Yin, J., et al. (2015). Dietary L-arginine supplementation protects weanling pigs from deoxynivalenol-induced toxicity. Toxins 7, 1341–1354. doi: 10.3390/toxins7041341
Yang, K. M., Jiang, Z. Y., Zheng, C. T., Wang, L., and Yang, X. F. (2014). Effect of Lactobacillus plantarum on diarrhea and intestinal barrier function of young piglets challenged with enterotoxigenic Escherichia coli K88. J. Anim. Sci. 92, 1496–1503. doi: 10.2527/jas.2013-6619
York, A. (2018). Cellular microbiology: lysozyme protects bacteria from beta-lactams. Nat. Rev. Microbiol. 16:183. doi: 10.1038/nrmicro.2018.26
Zhang, X., Jiang, A., Yu, H., Xiong, Y., Zhou, G., Qin, M., et al. (2016). Human lysozyme synergistically enhances bactericidal dynamics and lowers the resistant mutant prevention concentration for metronidazole to Helicobacter pylori by increasing cell permeability. Molecules 21:E1435. doi: 10.3390/molecules21111435
Zhao, J., Xu, J., Wang, J., Zhao, Y., Zhang, L., He, J., et al. (2012). Impacts of human lysozyme transgene on the microflora of pig feces and the surrounding soil. J. Biotechnol. 161, 437–444. doi: 10.1016/j.jbiotec.2012.05.018
Zhou, J., Xiong, X., Wang, K., Zou, L., Lv, D., and Yin, Y. (2017). Ethanolamine metabolism in the mammalian gastrointestinal tract: mechanisms, patterns, and importance. Curr. Mol. Med. 17, 92–99. doi: 10.2174/1566524017666170331161715
Keywords: milk, gut microbiota, lysozyme, metabolome, serum immunity
Citation: Zhou J, Xiong X, Yin J, Zou L, Wang K, Shao Y and Yin Y (2019) Dietary Lysozyme Alters Sow’s Gut Microbiota, Serum Immunity and Milk Metabolite Profile. Front. Microbiol. 10:177. doi: 10.3389/fmicb.2019.00177
Received: 08 February 2018; Accepted: 22 January 2019;
Published: 06 February 2019.
Edited by:
Aldo Corsetti, Università degli Studi di Teramo, ItalyReviewed by:
Shakti Singh, Los Angeles Biomedical Research Institute, United StatesFernando Ariel Genta, Fundação Oswaldo Cruz (Fiocruz), Brazil
Friedrich Götz, University of Tübingen, Germany
Copyright © 2019 Zhou, Xiong, Yin, Zou, Wang, Shao and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xia Xiong, eHhAaXNhLmFjLmNu
 Jian Zhou
Jian Zhou Xia Xiong
Xia Xiong Jia Yin3
Jia Yin3 Lijun Zou
Lijun Zou