- 1Division of Natural Sciences, Indiana Wesleyan University, Marion, IN, United States
- 2Department of Microbiology, Southern Illinois University, Carbondale, IL, United States
Lake Fryxell, situated in the McMurdo Dry Valleys of Antarctica, is an intriguing aquatic ecosystem because of its perennial ice cover, highly stratified water column, and extreme physicochemical conditions, which collectively restrict lake biodiversity to solely microbial forms. To expand our current understanding of the cultivable biodiversity of Lake Fryxell, water samples were collected from depths of 10 and 17 m, and pure cultures of eight diverse strains of aerobic, chemoorganotrophic bacteria were obtained. Despite having high 16S rRNA gene sequence similarity to mesophilic bacteria inhabiting various temperate environments, all Lake Fryxell isolates were psychrotolerant, with growth occurring at 0∘C and optimal growth from 18–24∘C for all isolates. Phylogenetic analyses showed the isolates to be members of six taxonomic groups, including the genera Brevundimonas, Arthrobacter, Sphingobium, Leifsonia, and Pseudomonas, as well as the family Microbacteriaceae (one strain could not reliably be assigned to a specific genus based on our analysis). Pseudomonas strain LFY10 stood out as a useful tool for teaching laboratory activities because of its substantial cold adaptation (visible growth is evident in 1–2 days at 4∘C), beta-hemolytic activity, and halotolerance to 8.5% (w/v) NaCl. These cold-adapted bacteria likely play a role in carbon mineralization and other nutrient cycling in Lake Fryxell, and their characterization broadens our understanding of microbial biodiversity in aquatic polar ecosystems.
Introduction
The McMurdo Dry Valleys of East Antarctica (Figure 1A) comprise a windswept polar desert that receives minimal annual precipitation (3–50 mm water equivalent per year; Fountain et al., 2009) and consists of nutrient-poor, rocky–sandy soils. The valleys contain several permanently ice-covered lakes that contain a broad diversity of microorganisms adapted to harsh physicochemical conditions, including constant freezing temperatures, nutrient limitation, and low incident light with long periods of almost total darkness during the austral winter. Lake Fryxell, located in Taylor Valley, has a water column of 18 m covered by 5–6 m of ice that prevents wind mixing and limits light penetration (Figure 1B). Beneath the ice cover, Lake Fryxell waters are fresh and supersaturated with trapped gasses, especially O2 from phytoplankton photosynthesis (Vincent, 1981; Priscu et al., 1987; Figure 1C). Deeper strata are brackish and anoxic and experience the upward diffusion of dissolved sulfide produced by sulfate-reducing bacteria (Karr et al., 2005; Sattley and Madigan, 2010; Figure 1C). Dissolved O2 and benthic sulfide coexist as opposing gradients to produce a biologically active chemocline from 9–10.5 m, wherein numbers of sulfur-oxidizing bacteria peak (Sattley and Madigan, 2006).
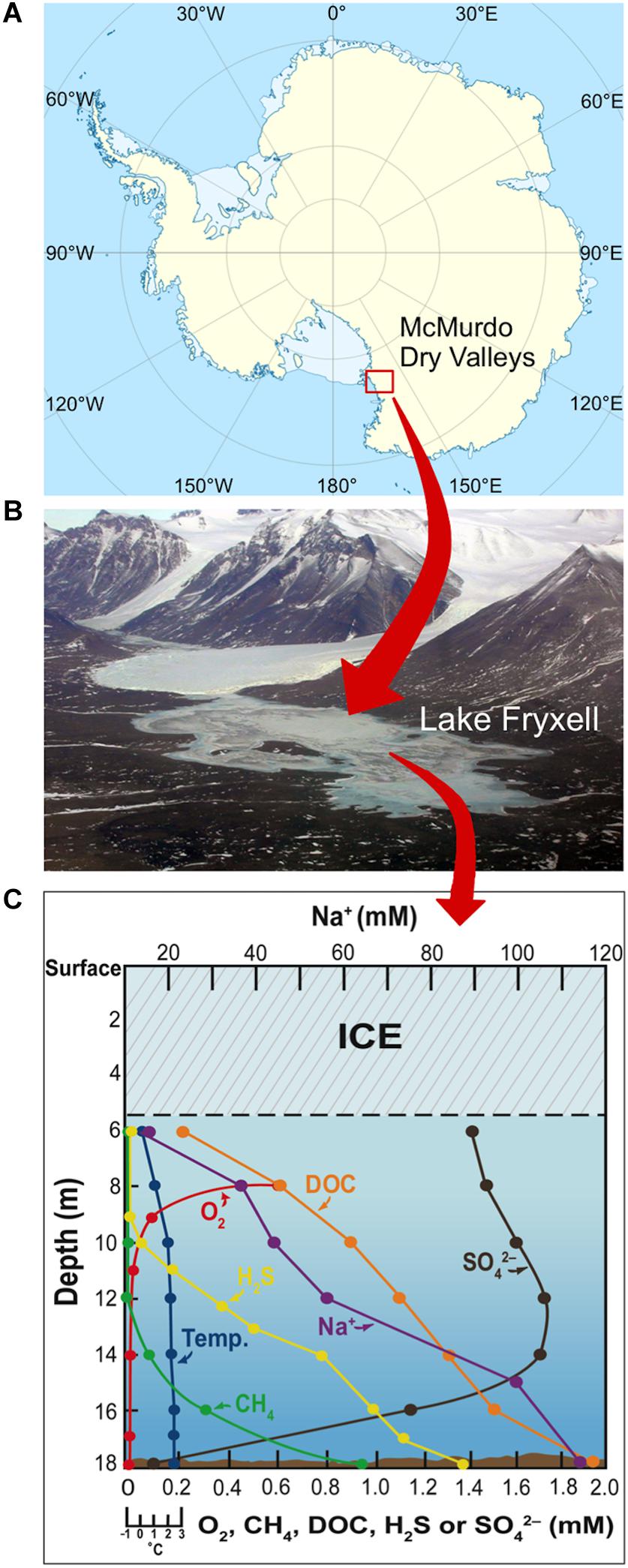
Figure 1. Geomorphology and physicochemistry of Lake Fryxell, Antarctica. (A) Site of the McMurdo Dry Valleys of East Antarctica. (B) Aerial view of Lake Fryxell in Taylor Valley. (C) Physicochemistry of the highly stratified Lake Fryxell water column (adapted from Sattley and Madigan, 2010). DOC, dissolved organic carbon.
Several studies have characterized novel bacterial isolates from Lake Fryxell microbial mats (Van Trappen et al., 2004a,b) and sediments (Sattley and Madigan, 2007, 2010), as well as the water column itself (Jung et al., 2004; Sattley and Madigan, 2006; Sattley et al., 2008). Other studies have employed culture-independent methods to examine these habitats (Brambilla et al., 2001; Karr et al., 2003, 2005, 2006; Kwon et al., 2017). Taken together, these investigations have revealed that extensive microbial diversity exists in Lake Fryxell and that only a handful of species have been isolated and characterized. Of the Lake Fryxell water column isolates, bacteria that grow phototrophically or drive microbial sulfur cycling have been the best characterized, including anoxygenic phototrophs, sulfur-oxidizing bacteria, and sulfate-reducing bacteria (Karr et al., 2003, 2005; Sattley and Madigan, 2006, 2010). By contrast, very few enrichments targeting aerobic, heterotrophic bacteria from the Lake Fryxell water column have been performed.
Although Antarctic lakes exhibit a surprising degree of microbial diversity, we have relatively few isolates from these ecosystems compared to temperate aquatic environments and know little of their genetic or biochemical potential. We have thus employed both defined and complex culture media and specific incubation conditions to enrich for cold-adapted, chemoorganotrophic bacteria from different depths of the Lake Fryxell water column. These enrichments yielded pure cultures of eight diverse strains of bacteria, and here we describe the morphology, physiology, and phylogeny of these isolates with a focus on Pseudomonas strain LFY10, a potentially useful strain for teaching laboratories in microbiology.
Materials and Methods
Sample Collection and Strain Isolation
Water samples were collected from Lake Fryxell, Antarctica (77°36.630′S, 163°08.853′E) using a peristaltic pump-driven limnological sampling device, as previously described (Sattley and Madigan, 2010; Sattley et al., 2017). Sampled portions of the water column included both the chemocline (10 m) and the anoxic benthos (17 m) of the lake. Primary enrichment cultures were then established at 0, 4, and 8°C at in situ pH (pH 7–7.4; Lee et al., 2004) in two ways. The first method was to prepare spread plates of Lake Fryxell water samples inoculated onto R2A medium (Reasoner and Geldreich, 1985). Colonies on these plates that had distinct morphological properties were then successively streaked for isolation on medium R2A at 4°C.
A second enrichment method was employed in which we added 1 ml of lake water to each of several 125-ml Erlenmeyer flasks containing 25 ml of a defined minimal medium containing glucose as sole carbon source (hereafter referred to as “ammonium-glucose medium”), as previously described (Vander Schaaf et al., 2015). The flasks were then incubated on a gently rotating orbital shaker placed inside a cooling box. After two weeks, turbid cultures were transferred to agar plates containing the same ammonium-glucose medium, and pure cultures were obtained by streaking for isolation. Axenic cultures of all bacterial strains were confirmed by microscopic observation and routinely maintained by periodic transfer into tubes of liquid R2A or the ammonium-glucose medium incubated at 4°C.
Morphology
Cell morphology was determined using a Leica Microsystems Model DM1000 phase-contrast light microscope. Capsule stains were performed using Maneval’s method (Maneval, 1941). Motility was determined by microscopic observation and visual assessment of stab inoculations into semi-solid (0.35% agar) sulfide-indole-motility (SIM) medium (Becton, Dickinson and Company, Sparks, MD, United States; Zimbro et al., 2009).
For transmission electron microscopy (TEM), samples of late logarithmic/early stationary phase cells grown at 4°C were fixed in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, post-fixed in buffered 1% osmium tetroxide containing 0.8% potassium ferricyanide, and stained in 1% aqueous uranyl acetate. The samples were then dehydrated with a graded series of acetonitrile and embedded in EMbed-812 resin. Thin sections (80 nm) were stained with 2% uranyl acetate and lead citrate and imaged using an FEI/Philips CM-10 transmission electron microscope (FEI Company, Hillsboro, Oregon, United States). Samples for scanning electron microscopy (SEM) were fixed in buffered 2.5% glutaraldehyde, post-fixed in 1% osmium tetroxide, dehydrated in a graded ethanol series, and critical-point dried. Specimens were then platinum-coated and cryosamples imaged using an FEI NOVA nanoSEM field emission scanning electron microscope.
Physiological Studies
A modified liquid R2A medium lacking soluble starch was used to determine the growth temperature range and optima and salinity tolerance of all eight isolates. To establish cardinal growth temperatures, triplicate 10-ml screw-cap tubes containing 3 ml of medium (pH 7.2) were inoculated and incubated for one week at selected temperatures within a range of −2 to 39°C. After a seven-day incubation period, growth was assessed spectrophotometrically using a Spectronic 20D+ (Spectronic Instruments, Inc.), and absorbance readings (OD540) were converted into a percentage of the maximum absorbance value. Growth rate experiments were conducted in a similar fashion, with absorbance measurements taken a total of ten times at two- to three-day intervals over a period of 19 days.
To determine salinity tolerances, triplicate cultures of each isolate were established in liquid R2A media supplemented with NaCl (0–12% [w/v], depending on the strain) and incubated at 15°C. Increments of 2% NaCl were used to determine approximate salinity ranges, and relevant concentrations in 0.25% NaCl increments were further tested to define each strain’s salinity tolerance. Cultures were incubated for up to one month, and growth was evaluated by periodic visual observations of turbidity and confirmed by microscopic observation.
To investigate carbon source utilization, culture tubes containing the defined ammonium-glucose medium were prepared as previously described (Vander Schaaf et al., 2015), with a variety of individual filter-sterilized carbon sources substituted for glucose. Duplicate tubes were inoculated and incubated at 15°C for 10 days. After incubation, growth was assessed spectrophotometrically (OD540), and absorbance readings were assigned +/− symbols to denote growth intensity and simplify presentation.
Biochemical assays for the activity of specific catabolic enzymes for cysteine, tryptophan, citrate, phenylalanine, urea, gelatin, and starch were performed for all strains as previously described (Vander Schaaf et al., 2015). Likewise, the potential for anaerobic growth was tested in all strains using anoxic media containing 10 mM (f.c.) of either dimethyl sulfoxide, sodium nitrate, or sodium fumarate as alternative electron acceptors (Vander Schaaf et al., 2015). Anoxic conditions were achieved in screw-cap tubes completely filled with medium reduced with 0.3 mM (f.c.) sodium sulfide and containing resazurin (1 mg/l) as a redox indicator. Hemolytic activity was assayed for strain LFY10 by inoculating sheep blood agar (SBA) plates and incubating at 4 and 22°C. Following incubations of five days and 48 h, respectively, colonies of fully grown cultures were visually inspected to determine hemolysis.
Phylogenetic Analyses
Small subunit (16S) rRNA genes were PCR amplified from genomic DNA isolated from mid-exponential phase cells using bacterial primers 8F (5-AGAGTTTGATCCTGGCTCAG-3) and 1525R (5-AAGGAGGTGATCCAGCC-3) and the following cycling parameters: initial denaturation at 94°C for 30 s; 30 cycles of 30 s at 94°C, 30 s at 54°C, and 60 s at 72°C; and a 10-min extension at 72°C. DNA-free controls were included in all PCR reactions, and 16S rRNA gene amplification products were confirmed to be of proper size (∼1500 bp) by comparison to a 1 Kb DNA ladder (Invitrogen, Carlsbad, CA, United States). Bacterial DNA from each strain was purified from gels using the QIAquick Gel Extraction Kit (QIAGEN Sciences, Germantown, MD, United States) according to the manufacturer’s instructions. Sequence fragments of purified 16S rRNA genes were assembled, and multiple sequence alignments were generated using UGENE version 1.11.0 (Okonechnikov et al., 2012), MUSCLE (Edgar, 2004) and MEGA 7 (Kumar et al., 2016). Positions with gaps or missing data were eliminated from the analysis. 16S rRNA sequences of all other strains used in phylogenetic analyses were obtained from the National Center for Biotechnology Information (NCBI) GenBank database.
Phylogenetic relationships were determined using BLAST (Johnson et al., 2008), the Ribosomal Database Project (Cole et al., 2009), and the List of Prokaryotic Names with Standing in Nomenclature (Parte, 2018). Methods of phylogenetic tree construction are described in their legends. Sequence identity percentages were generated using NCBI’s megaBLAST algorithm (Camacho et al., 2009).
16S rRNA gene sequences from all eight strains of Lake Fryxell bacteria were deposited into GenBank under the accession numbers shown in the phylogenetic trees. Cultures of the Lake Fryxell isolates were preserved at −80°C in growth medium containing 10% (w/v) dimethyl sulfoxide (DMSO) and are available to qualified researchers from the corresponding author upon written request.
Results
Enrichment, Isolation, and Morphology
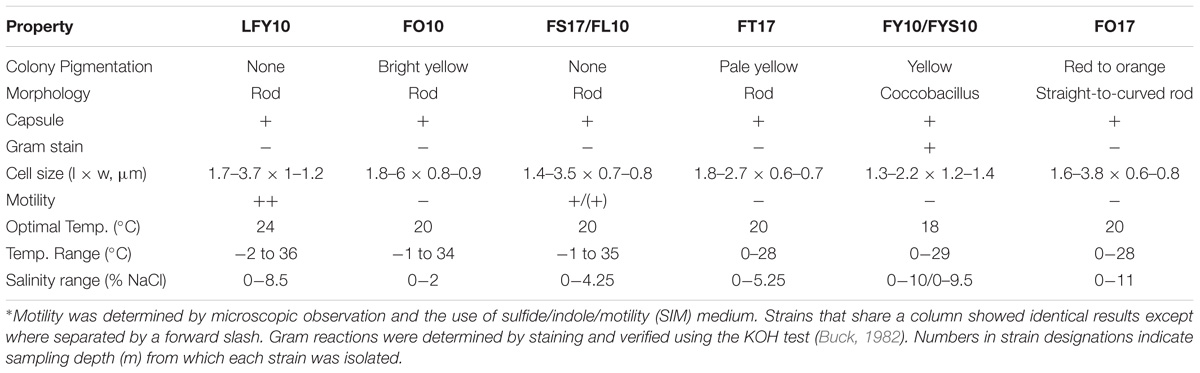
Following an initial incubation period of two (at 8°C) to five (at 0°C) weeks, depending on temperature, primary enrichment cultures of Lake Fryxell water samples spread on plates of R2A medium yielded a diversity of pigmented colonies ranging from pale yellow to reddish-orange. By contrast, liquid enrichment cultures in ammonium-glucose medium produced glossy, unpigmented colonies. Colonies from primary enrichment cultures that showed morphological variation and rapid growth were selected for further study. A total of eight strains of cold-active, aerobic, chemoorganotrophic bacteria were isolated from the Lake Fryxell water samples: five strains from a depth of 10 m and three strains from a depth of 17 m (Table 1). Numbers in strain designations denote the sampling depth (m) from which each strain was isolated.
When grown on plates of medium R2A, colonies of strains FY10 and FYS10 were yellow, while those of strain FO10 were bright yellow, strain FT17 pale yellow and translucent, strain FO17 reddish-orange, and strains FS17, FL10, and LFY10 colorless and opaque (Table 1). Most strains produced circular, shiny colonies with entire margins, but colonies of strain LFY10 had undulated margins and a mucoid appearance. Cells of strains FY10 and FYS10 were coccobacilli; cell morphologies of all other strains were rods of various sizes (Table 1 and Figure 2).
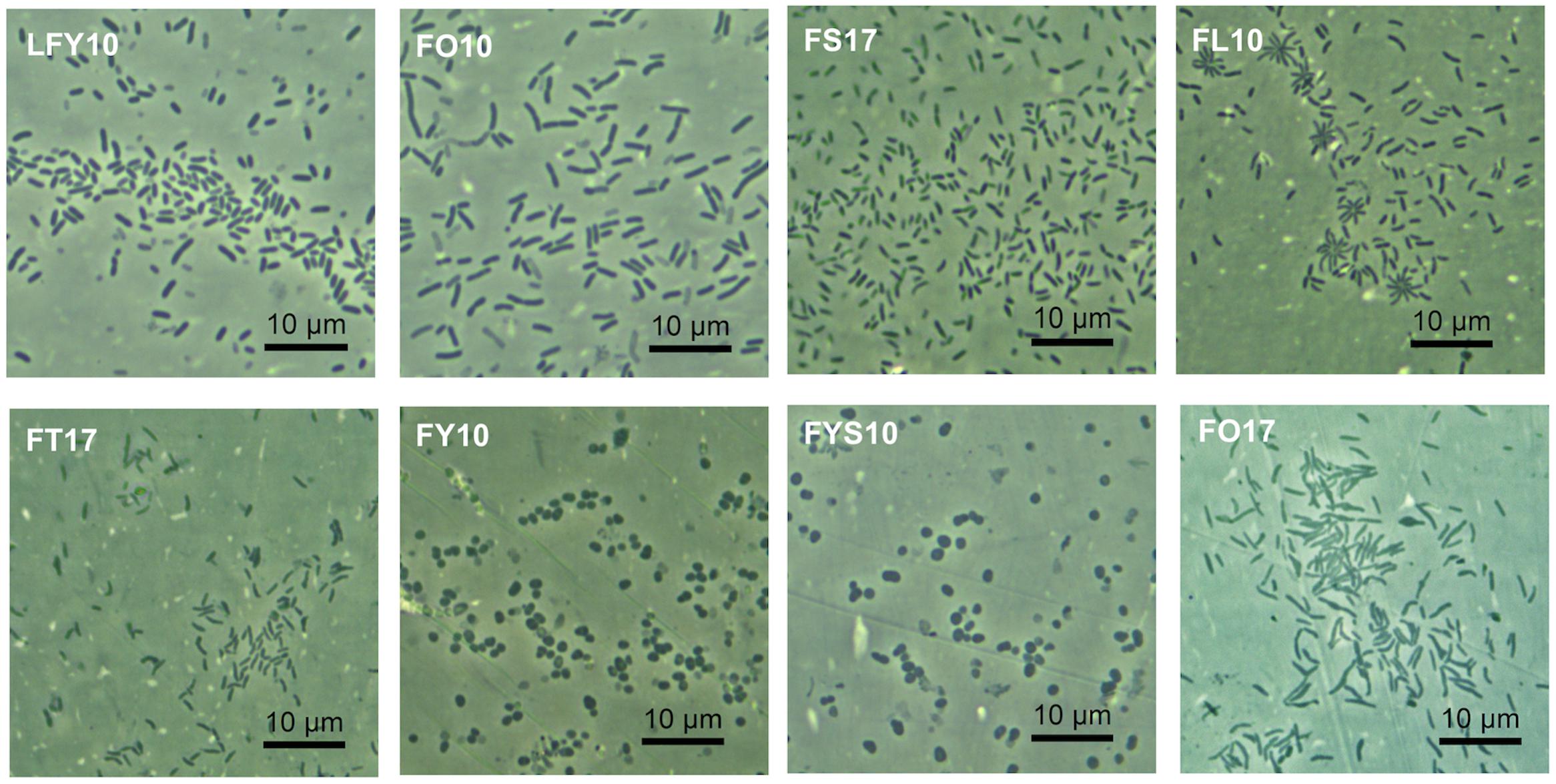
Figure 2. Cellular arrangement and morphology of Lake Fryxell isolates. Photomicrographs (1000× total magnification) show the eight bacterial Lake Fryxell isolates obtained during this study. Inset text denotes strain designation.
Phylogeny
Phylogenetic studies revealed that the Lake Fryxell isolates belonged to five distinct phylogenetic clades (Figure 3). BLAST analyses of 16S rRNA gene sequences indicated that strains FT17 and FO17 were members of the family Microbacteriaceae. Both of these strains were closely related (≥99% 16S rRNA gene sequence identity) to Leifsonia rubra, but strain FT17 also clustered tightly with species of Plantibacter and Salinibacterium. Strains FY10 and FYS10 aligned closely with Arthrobacter flavus, sharing >99% 16S rRNA gene sequence identity. Strains FS17 and FL10 showed 100% sequence identity to each other and also to three other Antarctic strains of the genus Brevundimonas (strains VS55, VW55, and VW50; Figure 3). Strain FO10 grouped within the genus Sphingobium, having an identical 16S rRNA gene sequence to Antarctic Sphingobium sp. VY55 and 99% identity to Sphingobium xenophagum (Pal et al., 2006).
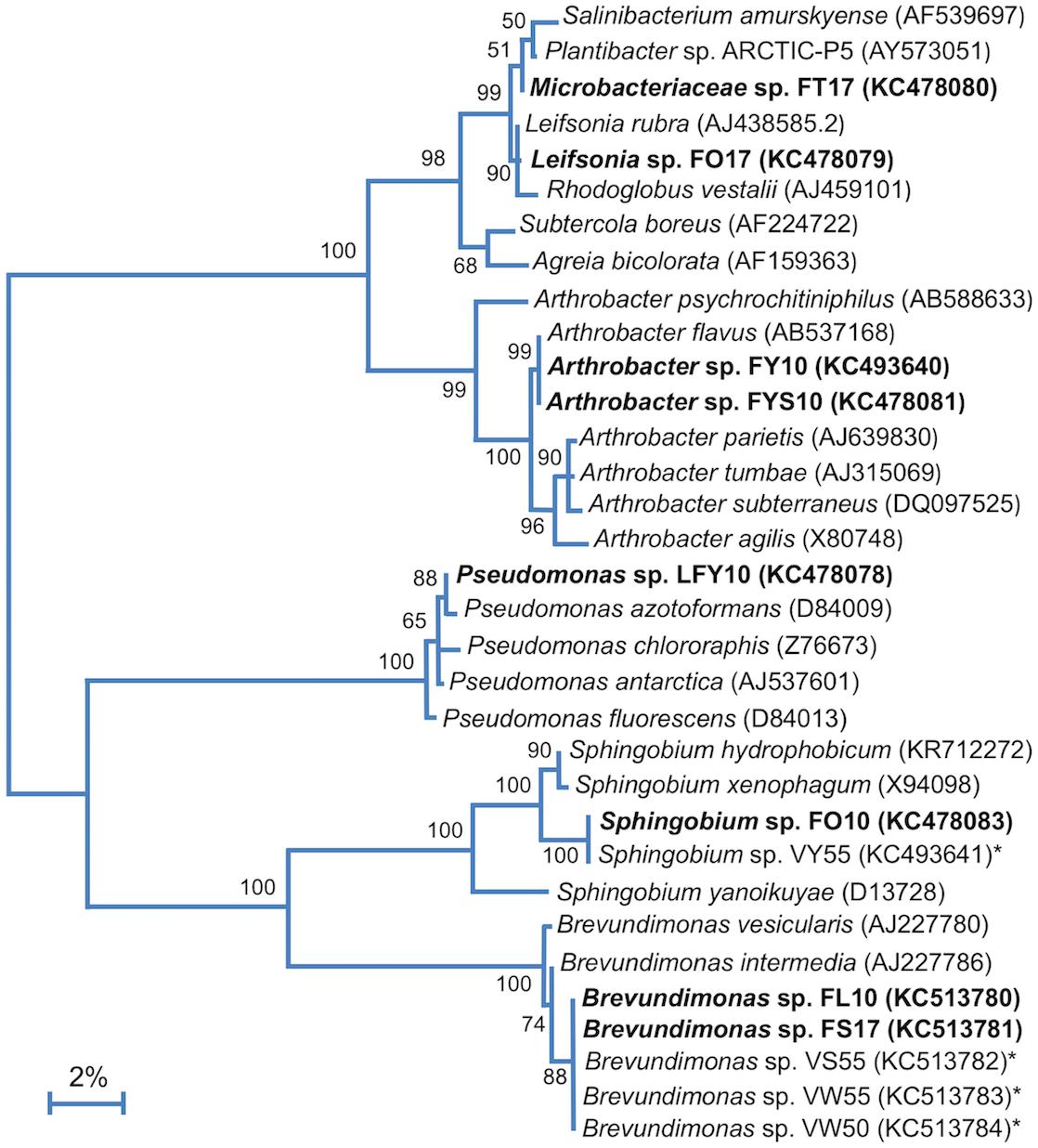
Figure 3. 16S rRNA phylogenetic analysis of the Lake Fryxell bacterial isolates (bold). 16S rRNA gene sequences from Lake Fryxell isolates and type strains of related species were aligned using Muscle (n = 1311 in final data set). The tree was constructed in Mega7 (v. 7.0.26) using the Maximum Likelihood method and the Jukes-Cantor model. 500 bootstrap replicates were obtained to determine validity (values shown at branch points indicate the percentage of replicates in which the node was observed). Asterisks (∗) indicate isolates from nearby Lake Vanda described in our previous work (Vander Schaaf et al., 2015). GenBank accession numbers are shown in parentheses. The scale bar indicates a distance equivalent to 2 nucleotide changes per 100 nucleotides.
Phylogenetic analysis of strain LFY10, a Gram-negative rod (Figure 4), showed it to group within the P. fluorescens clade of the genus Pseudomonas (Figures 3, 5). The closest well-characterized relative of strain LFY10 was Pseudomonas azotoformans (>99% identity). Other relatives of strain LFY10 included several Antarctic Pseudomonas species, including P. meridiana, P. proteolytica, and P. antarctica, all isolated from Wright Valley cyanobacterial mat samples and glacial streams (Reddy et al., 2004), as well as the more distantly related Pseudomonas sp. UC-1, isolated from South Pole ice samples (Madigan et al., 2017).
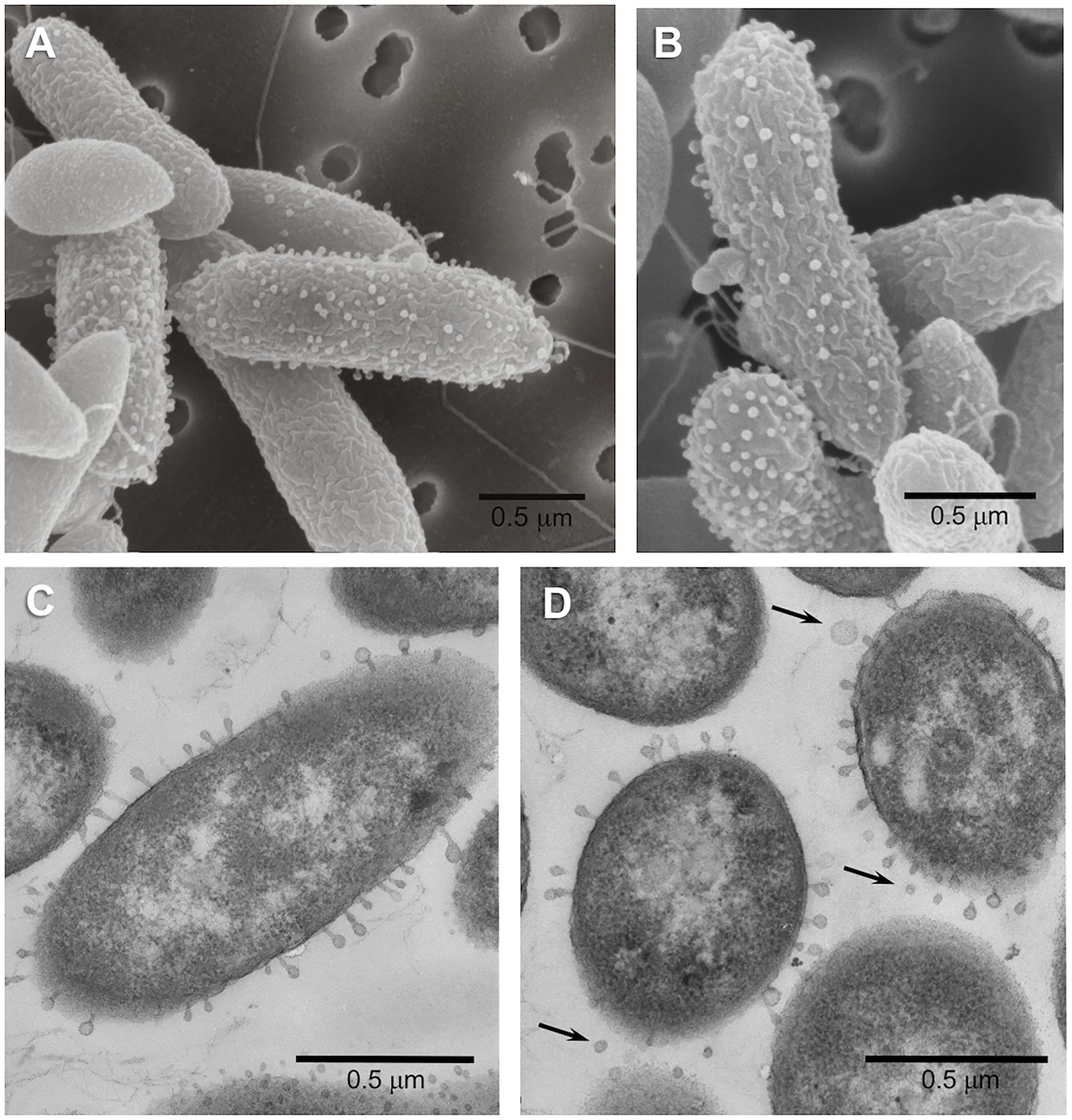
Figure 4. Scanning (A,B) and transmission (C,D) electron micrographs of cells of Pseudomonas strain LFY10 grown at 4°C. Surface blebbing can be seen in both SEM and TEM imaging and are visible only on some cells. These structures, some of which appear to be completely detached (arrows, D), are likely membrane vesicles produced by this Gram-negative bacterium. Note also the presence of polar flagella in the SEM images.
Physiology
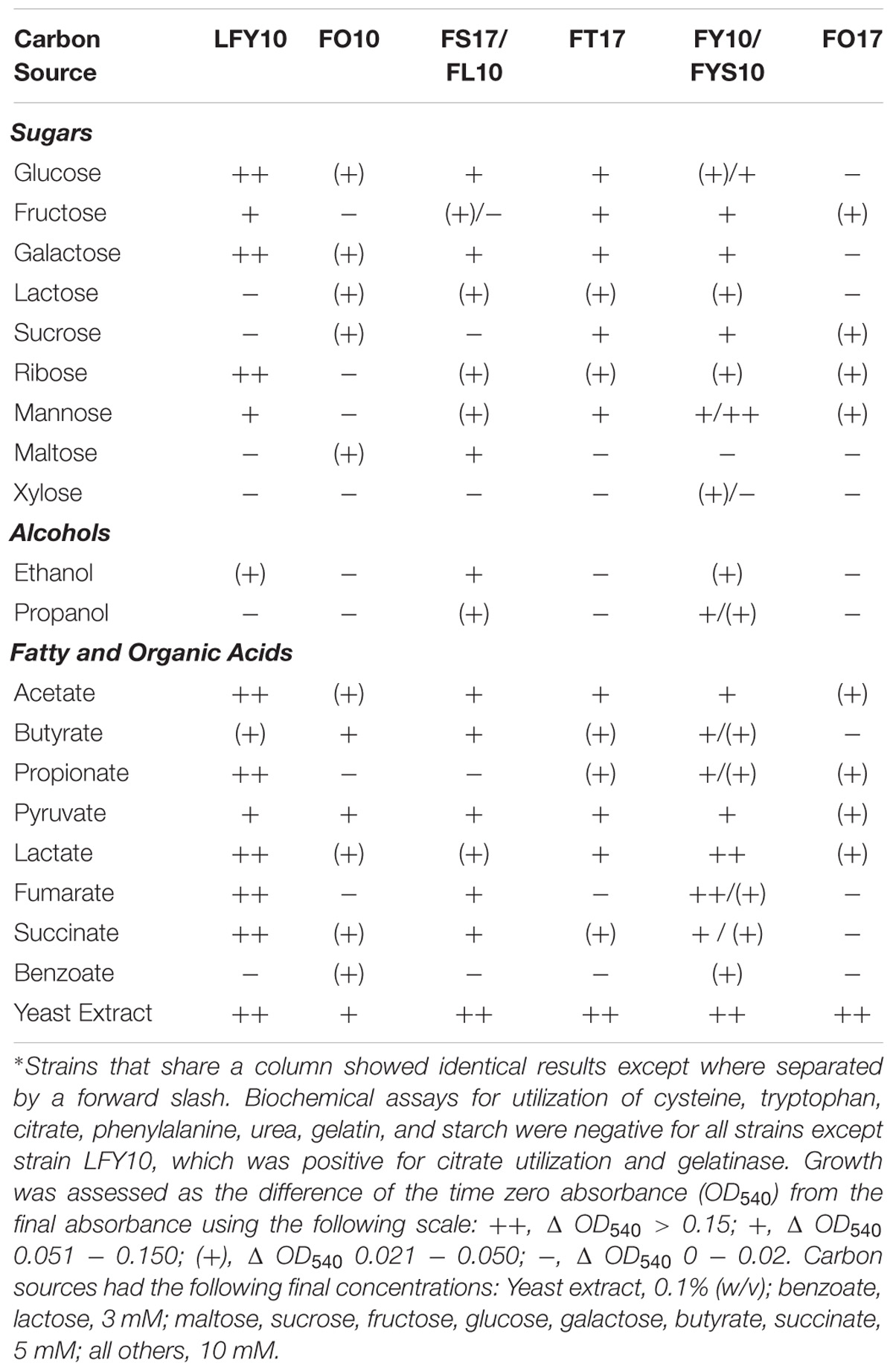
All strains showed robust growth on R2A medium. None of the isolates grew anaerobically, either by fermentation or anaerobic respiration of alternative electron acceptors (nitrate, fumarate, and DMSO). Carbon source utilization profiles varied among the Lake Fryxell isolates, but metabolic versatility was a general trend (Table 2). Yeast extract supported good growth of all Lake Fryxell isolates, and all strains were able to use acetate, pyruvate, and lactate. Strain FO17 was the most fastidious isolate, as growth of this strain was poor on any substrate tested except yeast extract.
Temperature ranges and optima for the eight Lake Fryxell strains were similar, but not identical (Table 1). The most cold-tolerant isolate, strain LFY10, also displayed the widest temperature range, with slow growth occurring down to −2°C and a maximum growth temperature of 36°C (Figure 6). Although all isolates grew at the near-freezing in situ Lake Fryxell temperatures (Figure 1), the optimum growth temperature for all strains was near 20°C; thus, all strains exhibited a psychrotolerant rather than psychrophilic phenotype (Table 1).
None of the isolates required NaCl for growth, but several strains showed significant salt tolerance. Strains LFY10, FY10, FYS10, and FO17 all grew at 8% NaCl (Table 1), and the addition of low levels (up to 1%, w/v) of NaCl stimulated their growth (data not shown). Strain FO17 showed the greatest halotolerance, as it grew in medium containing up to 11% (w/v) NaCl. Strain FO10 was the most sensitive to salt, with no growth occurring above 2% (w/v) NaCl.
Specific Features of Pseudomonas Strain LFY10
Strain LFY10 was studied in more detail because it was the most metabolically diverse of all isolates, showing the strongest growth on the greatest number of carbon sources (Table 2) and growing rapidly at 0°C (Figure 6A). Strain LFY10 was unique in biochemical tests, as it was the only isolate to show enzymatic activity for catabolism of citrate and gelatin. In addition, when incubated on blood agar plates, strain LFY10 showed beta-hemolysis at both 4 and 22°C (data not shown).
Electron micrographs of both ultrathin sections and intact cells of strain LFY10 revealed an unusual morphological feature. By TEM, small “bleb-like” structures could be seen on the surface of some but not all cells of strain LFY10 and were visible on both cross-sectioned and longitudinally sectioned cells (Figure 4). Some of these protuberances appeared to have a “head and tail” morphology reminiscent of attached bacteriophages. However, due to their inconsistent size and shape (and other reasons to be discussed below), it is more likely that these structures are membrane vesicles emerging from the outer membrane of this Gram-negative bacterium.
Discussion
Phylogenetic analyses showed the Lake Fryxell strains to lie within the families Sphingomonadaceae, Micrococcaceae, Microbacteriaceae, Caulobacteraceae, and Pseudomonadaceae. These families contain predominantly terrestrial and aquatic bacteria, along with outliers of clinical significance. Of note was the isolation of two strains of Arthrobacter (strains FY10 and FYS10), a genus primarily known for inhabiting soil. At least nine other recently described Antarctic species were assigned to this genus, including isolates from Wright Valley cyanobacterial mats (A. flavus and A. roseus), aquatic sediments (A. antarcticus and A. ardleyensis), soil (A. cryotolerans, A. gangotriensis, and A. livingstonensis), penguin guano (A. psychrochitiniphilus), and seawater (A. kerguelensis) (Reddy et al., 2000, 2002; Gupta et al., 2004; Chen et al., 2005; Wang et al., 2009; Pindi et al., 2010; Ganzert et al., 2011a). Arthrobacter strains FY10 and FYS10 showed close phylogenetic alignment (>99% identity) to Wright Valley isolate A. flavus and also had a similar phenotype. The Lake Fryxell Arthrobacter strains and A. flavus are all non-motile coccobacilli (Figure 2) with distinctive bright yellow pigmentation, and growth occurs within similar temperature and salinity ranges (Reddy et al., 2000). Interestingly, however, Arthrobacter strains FY10 and FYS10 showed considerably higher metabolic flexibility compared to A. flavus, a bacterium that utilizes only sorbitol as a carbon source (Table 2; Reddy et al., 2000).
Brevundimonas strains FL10 and FS17 had 16S rRNA gene sequences identical to three strains of Brevundimonas characterized in our previous study of Lake Vanda (Figure 3), an ice-covered lake located in Wright Valley, which lies adjacent to Lake Fryxell (Vander Schaaf et al., 2015). All five of these Antarctic strains are closely related to the temperate aquatic species Brevundimonas intermedia and B. mediterranea (Poindexter, 1964; Abraham et al., 1999; Fritz et al., 2005). Despite having identical 16S rRNA sequences, strains FL10 and FS17 exhibited differences in fructose utilization and motility. Moreover, these strains were isolated from depths in Lake Fryxell having different physicochemical conditions, with strain FL10 originating from much lower dissolved sodium and sulfide concentrations at 10 m than the brackish, sulfide-rich waters from the strain FS17 isolation depth of 17 m. Species of Brevundimonas have a widespread and diverse aquatic distribution, especially in marine habitats, and the isolation of strains FL10 and FS17 from a coastal and brackish Antarctic lake is thus consistent with this generalization (Fritz et al., 2005; Ryan and Pembroke, 2018).
Two strains from the family Microbacteriaceae were isolated from Lake Fryxell. Phylogenetic analyses of one of these isolates, designated Microbacteriaceae sp. FT17, revealed closely related species from three different genera: Leifsonia rubra; Plantibacter sp. ARCTIC-P5 (from an unpublished study of Arctic bacteria); and Salinibacterium amurskyense, a halotolerant marine bacterium from the Sea of Japan (Han et al., 2003). A marine origin seems likely for strain FT17, but it is unclear whether it represents a novel species. Another isolate of the family Microbacteriaceae, which we designated Leifsonia sp. FO17, was morphologically and phylogenetically similar to Leifsonia rubra; both bacteria produce bright red-to-orange colonies, have a curved-rod morphology, stain Gram positively, and are non-motile. Interestingly, of 17 reported Leifsonia species, six (including L. rubra) are either psychrotolerant or psychrophilic and originate from Antarctica, with Leifsonia pindariensis, isolated from a Himalayan glacier in India, as the sole exception (Reddy et al., 2003, 2008; Pindi et al., 2009; Ganzert et al., 2011b).
As previously described in our study of Lake Vanda water column isolates (Vander Schaaf et al., 2015), Sphingobium sp. FO10 was phylogenetically identical to Sphingobium sp. VY55 and closely related to Sphingomonas spp. Ant17 and BP-7 (Sakai et al., 2007). Although Sphingobium spp. VY55 and FO10 share the same optimum temperature and bright yellow pigmentation, strain FO10 showed lesser halotolerance and a slightly wider temperature range than its Lake Vanda counterpart (Vander Schaaf et al., 2015).
Phylogenetic analyses clustered Pseudomonas sp. LFY10 within the P. fluorescens group of the genus Pseudomonas (Figure 5). Under our culture conditions, colonies of strain LFY10 were opaque and did not produce the signature fluorescent yellow pigmentation exhibited by many species in the P. fluorescens group. The fluorescent pigments, called pyoverdines, specifically chelate ferric (Fe3+) iron and play a key role in Fe3+ scavenging during iron-limiting conditions (Ringel and Brüser, 2018). Although it may be that genes for pigment biosynthesis simply were not expressed under the culture conditions employed, another explanation is that strain LFY10 may not have the genetic capacity to produce these pigments. Levels of dissolved iron (Fe2+) in Lake Fryxell are approximately 0.2 μM L−1 at 10 m (Lee et al., 2004). Therefore, strain LFY10 could presumably obtain iron more efficiently by importing Fe2+ through a cation transporter than by sequestering Fe3+ through the production of fluorescent siderophores, potentially rendering genes for pyoverdine biosynthesis expendable.
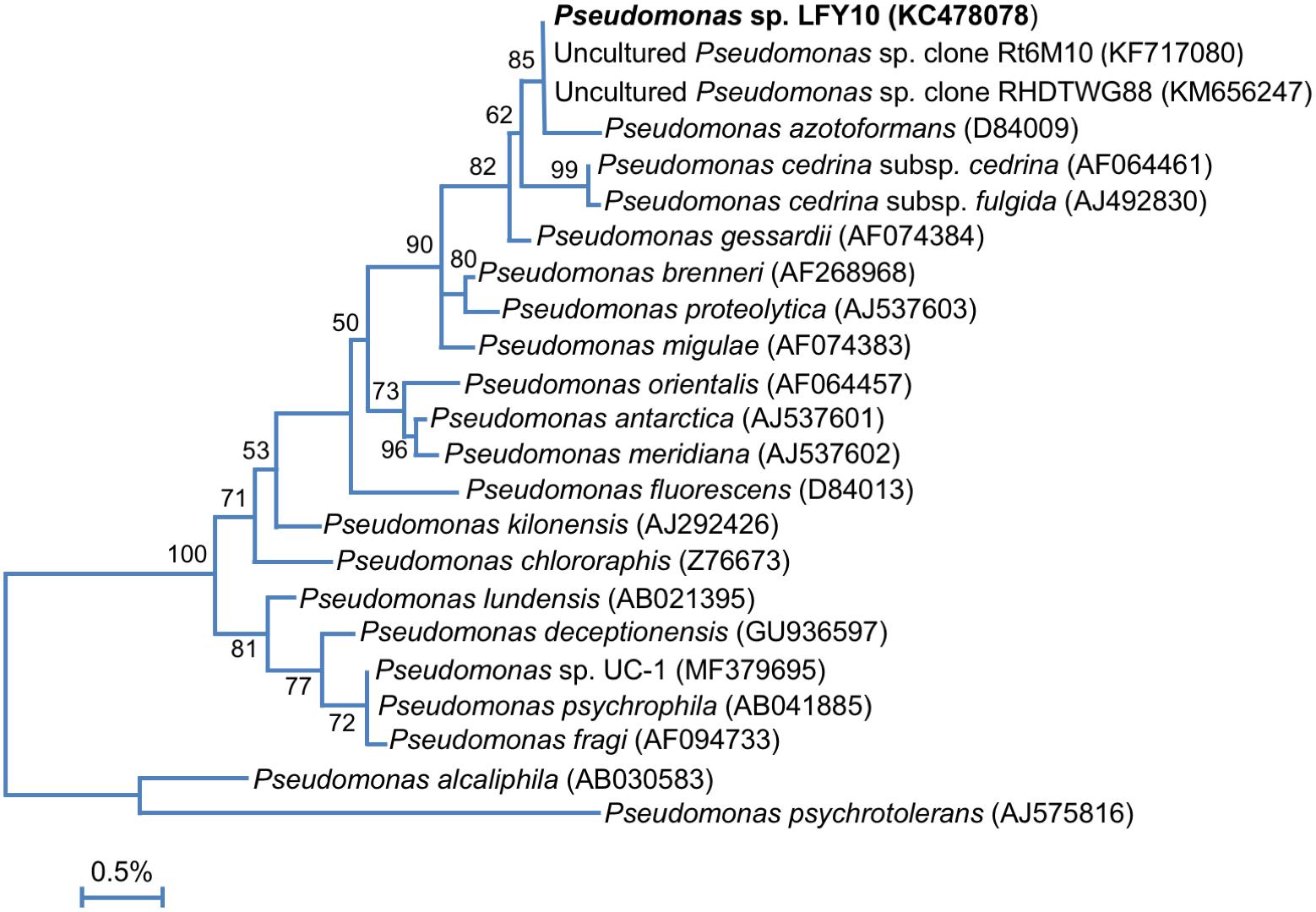
Figure 5. 16S rRNA phylogenetic analysis of strain LFY10 and related Pseudomonas strains. 16S rRNA gene sequences from strain LFY10 and other Pseudomonas species were aligned using Muscle (n = 1387 in final data set). The tree was constructed as for Figure 3. GenBank accession numbers are shown in parentheses; type strains of each species were used unless otherwise noted. The scale bar indicates a distance equivalent to 0.5 nucleotide changes per 100 nucleotides.
A megaBLASTn search revealed that two 16S rRNA gene sequences identical to that from Pseudomonas strain LFY10 had been previously identified in two dissimilar studies, one analyzing the root microbiota of Vitis vinifera from an Argentinian vineyard (Salomon et al., 2013) and the other an uncultured clone from an unpublished study in Arunachal Pradesh, India (Figure 5). Strain LFY10 also aligned closely with P. azotoformans, a mesophilic pathogen of rice and other cereals (Fang et al., 2016), as well as the aforementioned aquatic and cold-adapted Antarctic species P. antarctica, P. meridiana, and P. proteolytica (Reddy et al., 2004). With growth temperatures ranging from −2 to 36°C and an optimum growth temperature of 24°C (Table 1 and Figure 6), the psychrotolerant Arctic isolate P. psychrophila (Yumoto et al., 2001) most closely matches the growth temperature response of strain LFY10.
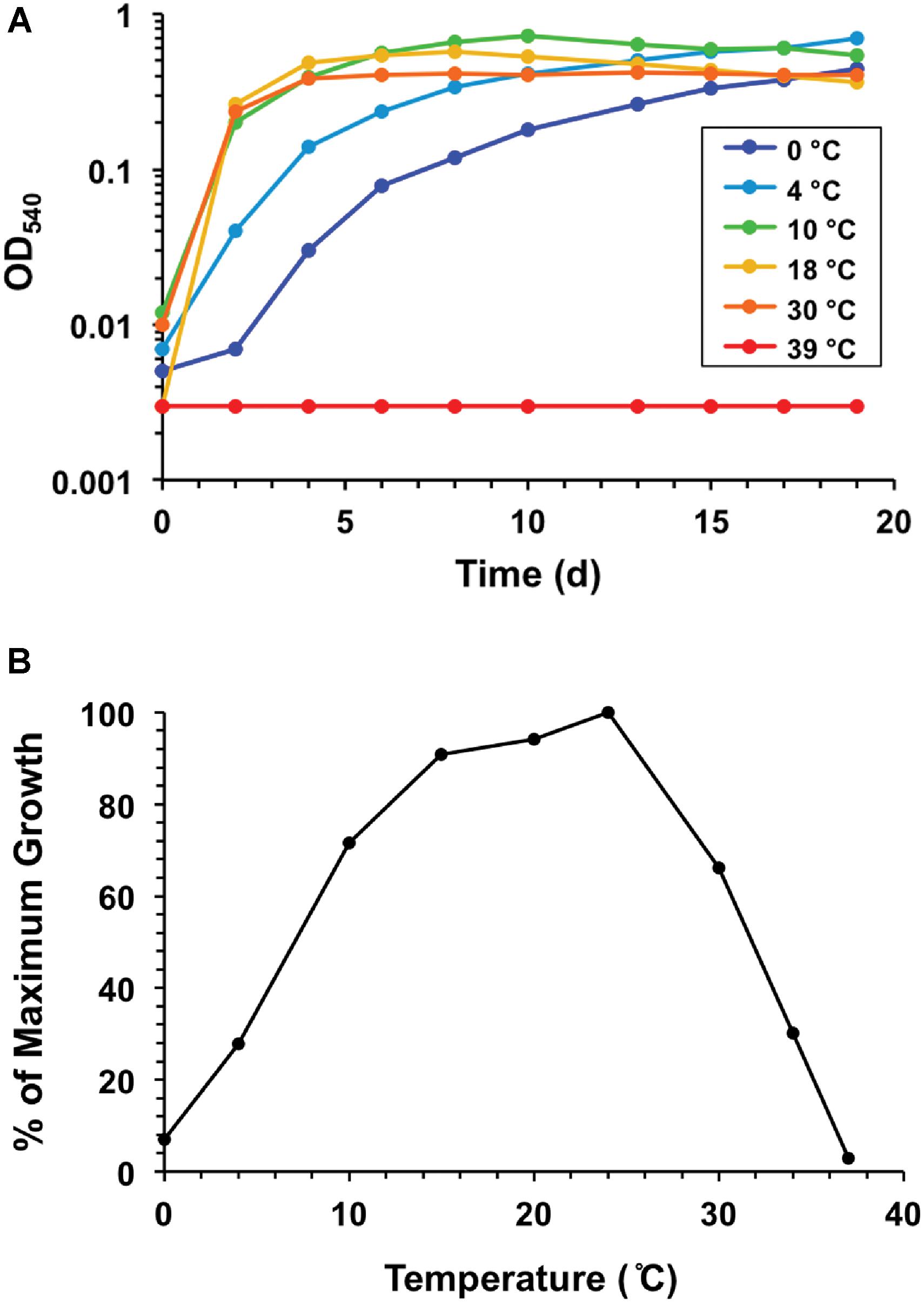
Figure 6. Growth response to temperature for strain LFY10. (A) Growth of strain LFY10 over a range of temperatures. Strong growth at low temperatures (0, 4, and 10°C) in combination with no growth at 39°C demonstrates clear cold adaptation. (B) Expression of growth rate data as percent maximum growth. The temperature supporting the highest cell density after one week of incubation was assigned a value of 100, and the percentage of maximum growth was calculated for each remaining temperature. Slow growth at −2°C was observed (data not shown). Of the eight Lake Fryxell isolates obtained in this study, strain LFY10 showed the fastest growth rate at 0°C and the widest temperature range for growth (Table 1).
Considering the physiological and phylogenetic characteristics of the bacteria isolated and described in this study, as well as the fact that Lake Fryxell is situated fewer than 10 km inland from the sea, it seems probable that at least some of these strains have a marine origin. With the possible exception of strain FO10, which shows little halotolerance, these bacteria may be derived from marine microorganisms that were deposited into Lake Fryxell, either by past flooding of Taylor Valley or aerosolization, and subsequently adapted to a planktonic lifestyle in an ice-sealed, stratified water column.
In addition to its physiological properties described above, an unexpected morphological feature of strain LFY10 was the bleb-like structures extending from the outer membrane of some but not all cells (Figure 4). Although the extensions had a phage-like morphology, closer inspection revealed that their dimensions and overall shape varied considerably. The structures had diameters ranging from 30 to 100 nm, and therefore, with such variation, it is unlikely that they were bacteriophages or particles of some other gene transfer agent, which have a defined and constant size (Solioz and Marrs, 1977; Lang et al., 2013, 2017). In addition to this, cultures of strain LFY10 grew robustly upon repeated transfer in both liquid and solid culture media—showing no universal virion-coating of cells in electron micrographs or other signs of phage infection (such as cell lysis)—and produced no detectable free viral particles.
It is likely that the observed surface structures on cells of strain LFY10 were membrane vesicles (MVs). The production of MVs is widespread among microorganisms, discovered first in Gram-negative bacteria and since identified in species from all three domains of life (Deatherage and Cookson, 2012; Anand and Chaudhuri, 2016). Pseudomonas fluorescens, a close relative to strain LFY10, produces MVs that are formed when the outer membrane of the cell wall bulges outward, fills with periplasm, and then pinches off to form a spherical vesicle (Guyard-Nicodème et al., 2007; Anand and Chaudhuri, 2016), a process that is consistent with our observations of strain LFY10. In addition, MVs vary in size from 20 to 250 nm (Beveridge, 1999; Nevot et al., 2006; Anand and Chaudhuri, 2016; Pathirana and Kaparakis-Liaskos, 2016), dimensions that correspond with the diameters of the structures extending from the cells of strain LFY10 (Figure 4).
Many Gram-negative bacteria shed MVs during the course of normal growth (Beveridge, 1999; Hagemann et al., 2014; Pathirana and Kaparakis-Liaskos, 2016). In the pathogen Pseudomonas aeruginosa, MVs play key roles in biofilm formation and pathogenesis (Park et al., 2014; Metruccio et al., 2016). However, in several cold-adapted Antarctic bacteria, including strains of Pseudomonas syringae, Shewanella vesiculosa, Pseudoalteromonas sp. M4.2, Psychrobacter fozii, and Marinobacter guineae (Frias et al., 2010; Pérez-Cruz et al., 2013, 2015; Kulkarni et al., 2015), vesicle shedding is a reaction to stress through SOS response activation and can be triggered by conditions such as extreme temperature or hyperosmolarity (Guyard-Nicodème et al., 2007; Frias et al., 2010; Metruccio et al., 2016).
Some species of cold-adapted bacteria also produce extracellular polymeric substances (EPS), which form a thick capsular layer that yields mucoid colonies (Frias et al., 2010), a feature displayed by strain LFY10. In accordance with observations of other Antarctic bacteria made by Frias et al. (2010), electron micrographs of cells of strain LFY10 showed putative MVs in various stages of formation, including several that were detached and spherical (Figure 4D). Membrane vesicles, which embed themselves in EPS, have been shown to be an integral part of this capsular layer in cold-adapted microorganisms, such as Pseudoalteromonas antarctica (Nevot et al., 2006). While the EPS in the capsular layer have been implicated in functions such as cryoprotection (Arctic marine bacterium Colwellia psychrerythraea 34H) and trace metal binding (Antarctic marine Pseudoalteromonas spp.), the role of MVs in the capsular layers is less understood (Marx et al., 2009; Ewert and Deming, 2011; Carillo et al., 2015; Casillo et al., 2018; Tribelli and López, 2018). Proposed functions for MVs in capsular layers include intracellular protein exchange, cell-cell signaling, or survival functions similar to those of the exopolymeric capsular components, but remain unclear without further study (Frias et al., 2010).
A final notable feature of Pseudomonas strain LFY10 is that it exhibits a number of ideal characteristics for use as a model cold-active bacterium in microbiology teaching labs. First, it is an excellent substitute for more commonly used psychrotolerant bacteria. For example, species of Listeria are often used to demonstrate the growth of microbes at low temperature. However, these species often take up to a week to show visible growth on tryptic soy agar (TSA) plates incubated at refrigerator temperatures (4–5°C). By contrast, strain LFY10 produces visible colonies within 24–48 h at 4°C on the same medium. In addition to growth temperature characteristics, strain LFY10 may be used to demonstrate halotolerance, as it is capable of growth in TSA containing up to 8.5% (w/v) NaCl (a demonstration commonly performed in teaching labs; data not shown) and has been shown to grow at even higher salinities in defined media (Heinzelmann et al., 2015a,b). Moreover, growth of strain LFY10 on blood agar plates may also be used to demonstrate beta-hemolysis, as colonies of this bacterium produce a characteristic clearing of red blood cells when grown on this medium (data not shown). Finally, strain LFY10 does not grow at 37°C (approximate human body temperature), and close relatives of this strain are of environmental or marine origin and are not reported to cause pathogenesis in mammals, minimizing the biosafety concerns that arise when using certain other Pseudomonas spp. (e.g., Pseudomonas aeruginosa) in a teaching lab setting.
Conclusion
In conclusion, with its permanent ice cover, seasonal swings of constant darkness or daylight, steady and near-freezing temperatures, and water column that shifts from hyperoxic to anoxic and from freshwater to brackish, the physicochemical conditions of Lake Fryxell have selected for a diversity of hardy microorganisms. Harboring exclusively microbial species, Lake Fryxell has become a favored habitat for studying microbial nutrient cycling in an ice-capped, polar environment. The collective ability of these strains to catabolize a wide range of carbon sources, including sugars, alcohols, and fatty and organic acids (Table 2), suggests that such nutritional versatility provides a fitness advantage and may catalyze dissolved organic carbon (DOC; Figure 1) turnover in the ice-sealed Lake Fryxell water column. The wide growth temperature range of the bacteria described here, including an ability to grow at 0°C, and their impressive metabolic versatility could signal their importance as key contributors to carbon turnover in this challenging aquatic ecosystem.
Data Availability Statement
The datasets generated for this study can be found in the GenBank nucleotide repository1 using either the strain names or accession numbers highlighted in Figure 3.
Author Contributions
NVS, AC, AH, CJR, CLR, EH, and WMS performed laboratory experiments. JB wrote the initial manuscript draft. WMS designed the experiments and supervised the research. MM wrote the grant proposal that funded sample collection, and WMS wrote grant proposals to support laboratory investigations. WMS and MM edited the final version of the submitted manuscript.
Funding
This research was supported in part by the Hodson Research Institute (Indiana Wesleyan University) and a Lilly Faculty Scholarship Award (IWU) to WMS. Lake Fryxell water samples were obtained through the support of National Science Foundation grant MCB0237576 to MM.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jeanice Troutman and Lynn Illeck at the Clemson University Genomics Institute for 16S rRNA gene sequencing and Chia-ping Huang and Debby Sherman at the Purdue University Life Science Microscopy Facility for the electron micrographs of Pseudomonas strain LFY10. We thank the McMurdo LTER Project for Lake Fryxell DOC and [Na+] data presented in Figure 1, Petroleum Helicopters International for field support, and the McMurdo LTER limnology team for sampling assistance. We also thank Lauren Slater for laboratory assistance and kindly acknowledge Tom Beatty and Andrew Lang for helpful discussion regarding the surface structures formed by strain LFY10.
Footnotes
References
Abraham, W.-R., Strömpl, C., Meyer, H., Lindholst, S., Moore, E. R. B., Christ, R., et al. (1999). Phylogeny and polyphasic taxonomy of Caulobacter species. Proposal of Maricaulis gen. nov. with Maricaulis maris (Poindexter) comb. nov. as the type species, and emended description of the genera Brevundimonas and Caulobacter. Int. J. Syst. Bacteriol. 49, 1053–1073. doi: 10.1099/00207713-49-3-1053
Anand, D., and Chaudhuri, A. (2016). Bacterial outer membrane vesicles: new insights and applications. Mol. Membr. Biol. 33, 125–137. doi: 10.1080/09687688.2017.1400602
Beveridge, T. J. (1999). Structures of Gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181, 4725–4733.
Brambilla, E., Hippe, H., Hagelstein, A., Tindall, B. J., and Stackebrandt, E. (2001). 16S rDNA diversity of cultured and uncultured prokaryotes of a mat sample from Lake Fryxell, McMurdo Dry Valleys, Antarctica. Extremophiles 5, 23–33. doi: 10.1007/s007920000169
Buck, J. D. (1982). Nonstaining (KOH) method for determination of Gram reactions of marine bacteria. Appl. Environ. Microbiol. 44, 992–993.
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Carillo, S., Casillo, A., Pieretti, G., Parrilli, E., Sannino, F., Bayer-Giraldi, M., et al. (2015). A unique capsular polysaccharide structure from the psychrophilic marine bacterium Colwellia psychrerythraea 34H that mimics antifreeze (glyco)proteins. J. Am. Chem. Soc. 137, 179–189. doi: 10.1021/ja5075954
Casillo, A., Lanzetta, R., Parrilli, M., and Corsaro, M. M. (2018). Exopolysaccharides from marine and marine extremophilic bacteria: structures, properties, ecological roles and applications. Mar. Drugs 16:E69. doi: 10.3390/md16020069
Chen, M., Xiao, X., Wang, P., Zeng, X., and Wang, F. (2005). Arthrobacter ardleyensis sp. nov., isolated from Antarctic lake sediment and deep-sea sediment. Arch. Microbiol. 183, 301–305. doi: 10.1007/s00203-005-0772-y
Cole, J. R., Wang, Q., Cardenas, E., Fish, J., Chai, B., Farris, R. J., et al. (2009). The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, 141–145. doi: 10.1093/nar/gkn879
Deatherage, B. L., and Cookson, B. T. (2012). Membrane vesicle release in Bacteria, eukaryotes, and Archaea: a conserved yet underappreciated aspect of microbial life. Infect. Immun. 80, 1948–1957. doi: 10.1128/IAI.06014-11
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Ewert, M., and Deming, J. W. (2011). Selective retention in saline ice of extracellular polysaccharides produced by the cold-adapted marine bacterium Colwellia psychrerythraea strain 34H. Ann. Glaciol. 52, 111–117. doi: 10.3189/172756411795931868
Fang, Y., Wu, L., Chen, G., and Feng, G. (2016). Complete genome sequence of Pseudomonas azotoformans S4, a potential biocontrol bacterium. J. Biotech. 227, 25–26. doi: 10.1016/j.jbiotec.2016.04.020
Fountain, A. G., Nylen, T. H., Monaghan, A., Basagic, H. J., and Bromwich, D. (2009). Snow in the Mcmurdo dry valleys, Antarctica. Int. J. Climatol. 30, 633–642. doi: 10.1002/joc.1933
Frias, A., Manresa, A., de Oliveira, E., Loìpez-Iglesias, C., and Mercadeì, E. (2010). Membrane vesicles: a common feature in the extracellular matter of cold-adapted Antarctic bacteria. Microb. Ecol. 59, 476–486. doi: 10.1007/s00248-009-9622-9
Fritz, I., Strömpl, C., Nikitin, D. I., Lysenko, A. M., and Abraham, W.-R. (2005). Brevundimonas mediterranea sp. nov., a non-stalked species from the Mediterranean Sea. Int. J. Syst. Evol. Microbiol. 55, 479–486. doi: 10.1099/ijs.0.02852-0
Ganzert, L., Bajerski, F., Mangelsdorf, K., Lipski, A., and Wagner, D. (2011a). Arthrobacter livingstonensis sp. nov. and Arthrobacter cryotolerans sp. nov., salt-tolerant and psychrotolerant species from Antarctic soil. Int. J. Syst. Evol. Microbiol. 61, 979–984. doi: 10.1099/ijs.0.021022-0
Ganzert, L., Bajerski, F., Mangelsdorf, K., Lipski, A., and Wagner, D. (2011b). Leifsonia psychrotolerans sp. nov., a psychrotolerant species of the family Microbacteriaceae from Livingston Island, Antarctica. Int. J. Syst. Evol. Microbiol. 61, 1938–1943. doi: 10.1099/ijs.0.021956-0
Gupta, P., Reddy, G. S. N., Delille, D., and Shivaji, S. (2004). Arthrobacter gangotriensis sp. nov. and Arthrobacter kerguelensis sp. nov. from Antarctica. Int. J. Syst. Evol. Microbiol. 54, 2375–2378. doi: 10.1099/ijs.0.63110-0
Guyard-Nicodème, M., Bazire, A., Heìmery, G., Meylheuc, T., Molleì, D., Orange, N., et al. (2007). Outer membrane modifications of Pseudomonas fluorescens MF37 in response to hyperosmolarity. J. Proteome Res. 7, 1218–1225. doi: 10.1021/pr070539x
Hagemann, S., Stöger, L., Kappelmann, M., Hassl, I., Ellinger, A., and Velimirov, B. (2014). DNA-bearing membrane vesicles produced by Ahrensia kielensis and Pseudoalteromonas marina. J. Basic Microbiol. 54, 1062–1072. doi: 10.1002/jobm.201300376
Han, S. K., Nedashkovskaya, O. I., Mikhailov, V. V., Kim, S. B., and Bae, K. S. (2003). Salinibacterium amurskyense gen. nov., sp. nov., a novel genus of the family Microbacteriaceae from the marine environment. Int. J. Syst. Evol Microbiol. 53, 2061–2066. doi: 10.1099/ijs.0.02627-0
Heinzelmann, S. M., Chivall, D., M’Boule, D., Sinke-Schoen, D., Villanueva, L., Sinnighe Damsté, J. S., et al. (2015a). Comparison of the effect of salinity on the D/H ratio of fatty acids of heterotrophic and photoautotrophic microorganisms. FEMS Microbiol. Lett. 362:fnv065. doi: 10.1093/femsle/fnv065
Heinzelmann, S. M., Villanueva, L., Sinke-Schoen, D., Sinnighe Damsté, J. S., Schouten, S., and van der Meer, M. T. J. (2015b). Impact of metabolism and growth phase on the hydrogen isotopic composition of microbial fatty acids. Front. Microbiol. 6:408. doi: 10.3389/fmicb.2015.00408
Johnson, M., Zaretskaya, I., Raytselis, Y., Merezhuk, Y., McGinnis, S., and Madden, T. L. (2008). NCBI BLAST: a better web interface. Nucleic Acids Res. 36, W5–W9. doi: 10.1093/nar/gkn201
Jung, D. O., Achenbach, L. A., Karr, E. A., Takaichi, S., and Madigan, M. T. (2004). A gas vesiculate planktonic strain of the purple non-sulfur bacterium Rhodoferax antarcticus isolated from Lake Fryxell, Dry Valleys, Antarctica. Arch. Microbiol. 182, 236–243. doi: 10.1007/s00203-004-0719-8
Karr, E. A., Ng, J. M., Belchik, S. M., Sattley, W. M., Madigan, M. T., and Achenbach, L. A. (2006). Biodiversity of methanogenic and other Archaea in the permanently frozen Lake Fryxell, Antarctica. Appl. Environ. Microbiol. 72, 1663–1666. doi: 10.1128/AEM.72.2.1663-1666.2006
Karr, E. A., Sattley, W. M., Jung, D. O., Madigan, M. T., and Achenbach, L. A. (2003). Remarkable diversity of phototrophic purple bacteria in a permanently frozen Antarctic lake. Appl. Environ. Microbiol. 69, 4910–4914. doi: 10.1128/AEM.69.8.4910-4914.2003
Karr, E. A., Sattley, W. M., Rice, M. R., Jung, D. O., Madigan, M. T., and Achenbach, L. A. (2005). Diversity and distribution of sulfate-reducing bacteria in permanently frozen Lake Fryxell, McMurdo Dry Valleys, Antarctica. Appl. Environ. Microbiol. 71, 6353–6359. doi: 10.1128/AEM.71.10.6353-6359.2005
Kulkarni, H. M., Swamy, Ch. V. B, and Jagannadham, M. V. (2015). The proteome of the outer membrane vesicles of an Antarctic bacterium Pseudomonas syringae Lz4W. Data Brief. 4, 406–409. doi: 10.1016/j.dib.2015.07.001
Kumar, S., Stetcher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 30, 1870–1874. doi: 10.1093/molbev/msw054
Kwon, M., Kim, M., Takacs-Vesbach, C., Lee, J., Hong, S. G., Kim, S. J., et al. (2017). Niche specialization of bacteria in permanently ice-covered lakes of the McMurdo Dry Valleys, Antarctica. Environ. Microbiol. 19, 2258–2271. doi: 10.1111/1462-2920.13721
Lang, A., Zhaxybayeva, O., and Beatty, J. T. (2013). Gene transfer agents: phage-like elements of genetic exchange. Nat. Rev. Microbiol. 10, 472–482. doi: 10.1038/nrmicro2802
Lang, A. S., Westbye, A. B., and Beatty, J. T. (2017). The distribution, evolution, and roles of gene transfer agents in prokaryotic genetic exchange. Annu. Rev. Virol. 4, 87–104. doi: 10.1146/annurev-virology-101416-041624
Lee, P. A., Mickucki, J. A., Foreman, C. M., Priscu, J. C., DiTullio, G. R., Riseman, S. F., et al. (2004). Thermodynamic constraints on microbially mediated processes in lakes of the McMurdo Dry Valleys, Antarctica. Geomicrobiol. J. 21, 221–237. doi: 10.1080/01490450490275884
Madigan, M. T., Kempher, M. L., Bender, K. S., Sullivan, P., Matthew Sattley, W., Dohnalkova, A. C., et al. (2017). Characterization of a cold-active bacterium isolated from the South Pole “Ice Tunnel”. Extremophiles 21, 891–901. doi: 10.1007/s00792-017-0950-2
Maneval, W. E. (1941). Staining bacteria and yeasts with acid dyes. Biotech. Histochem. 16, 13–19. doi: 10.3109/10520294109106189
Marx, J. G., Carpenter, S. D., and Deming, J. W. (2009). Production of cryoprotectant extracellular polysaccharide substances (EPS) by the marine psychrophilic bacterium Colwellia psychrerythraea strain 34H under extreme conditions. Can. J. Microbiol. 55, 63–72. doi: 10.1139/W08-130
Metruccio, M. M. E., Evans, D. J., Gabriel, M. M., Kadurugamuwa, J. L., and Fleiszig, S. M. J. (2016). Pseudomonas aeruginosa outer membrane vesicles triggered by human mucosal fluid and lysozyme can prime host tissue surfaces for bacterial adhesion. Front. Microbiol. 7:871. doi: 10.3389/fmicb.2016.00871
Nevot, M., Deronceleì, V., Messner, P., Guinea, J., and Mercadeì, E. (2006). Characterization of outer membrane vesicles released by the psychrotolerant bacterium Pseudoalteromonas antarctica NF3. Environ. Microbiol. 8, 1523–1533. doi: 10.1111/j.1462-2920.2006.01043.x
Okonechnikov, K., Golosova, O., and Fursov, M. (2012). Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28, 1166–1167. doi: 10.1093/bioinformatics/bts091
Pal, R., Bhasin, V. K., and Lal, R. (2006). Proposal to reclassify [Sphingomonas xenophaga Stolz et al. 2000 and [Sphingomonas taejonensis Lee et al. 2001 as Sphingobium xenophagum comb. nov. and Sphingopyxis taejonensis comb. nov., respectively. Int. J. Syst. Evol. Microbiol. 56, 667–670. doi: 10.1099/ijs.0.64161-0
Park, A. J., Surette, M. D., and Khursigara, C. M. (2014). Antimicrobial targets localize to the extracellular vesicle-associated proteome of Pseudomonas aeruginosa grown in a biofilm. Front. Microbiol. 5:464. doi: 10.3389/fmicb.2014.00464
Parte, A. C. (2018). LPSN – List of Prokaryotic names with Standing in Nomenclature (bacterio.net), 20 years on. Int. J. Sys. Evol. Microbiol. 68, 1825–1829. doi: 10.1099/ijsem.0.002786
Pathirana, R. D., and Kaparakis-Liaskos, M. (2016). Bacterial membrane vesicles: biogenesis, immune regulation and pathogenesis. Cell. Microbiol. 18, 1518–1524. doi: 10.1111/cmi.12658
Pérez-Cruz, C., Carrioìn, O., Delgado, L., Martinez, G., Loìpez-Iglesias, C., and Mercadeì, E. (2013). A new type of outer membrane vesicle produced by the Gram-negative bacterium Shewanella vesiculosa M7T: implications for DNA content. Appl. Environ. Microbiol. 79, 1874–1881. doi: 10.1128/AEM.03657-12
Pérez-Cruz, C., Delgado, L., Loìpez-Iglesias, C., and Mercadeì, E. (2015). Outer-inner membrane vesicles naturally secreted by Gram-negative pathogenic bacteria. PLoS One 10:e0116896. doi: 10.1371/journal.pone.0116896
Pindi, P. K., Kishore, K. H., Reddy, G. S. N., and Shivaji, S. (2009). Description of Leifsonia kafniensis sp. nov. and Leifsonia antarctica sp. nov. Int. J. Syst. Evol. Microbiol. 59, 1348–1352. doi: 10.1099/ijs.0.006643-0
Pindi, P. K., Manorama, R., Begum, Z., and Shivaji, S. (2010). Arthrobacter antarcticus sp. nov., isolated from an Antarctic marine sediment. Int. J. Syst. Evol. Microbiol. 60, 2263–2266. doi: 10.1099/ijs.0.012989-0
Poindexter, J. S. (1964). Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 28, 231–295.
Priscu, J. C., Nedwell, D. B., and Embley, T. M. (1987). Photosynthate distribution by microplankton in permanently ice-covered Antarctic desert lakes. Limnol. Oceanogr. 32, 260–270. doi: 10.4319/lo.1987.32.1.0260
Reasoner, D. J., and Geldreich, E. E. (1985). A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49, 1–7.
Reddy, G. S. N., Aggarwal, R. K., Matsumoto, G. I., and Shivaji, S. (2000). Arthrobacter flavus sp. nov., a psychrophilic bacterium isolated from a pond in McMurdo Dry Valley, Antarctica. Int. J. Syst. Evol. Microbiol. 50, 1553–1561. doi: 10.1099/00207713-50-4-1553
Reddy, G. S. N., Matsumoto, G. I., Schumann, P., Stackebrandt, E., and Shivaji, S. (2004). Psychrophilic pseudomonads from Antarctica: Pseudomonas antarctica sp. nov., Pseudomonas meridiana sp. nov. and Pseudomonas proteolytica sp. nov. Int. J. Syst. Evol. Microbiol. 54, 713–719. doi: 10.1099/ijs.0.02827-0
Reddy, G. S. N., Prabagaran, S. R., and Shivaji, S. (2008). Leifsonia pindariensis sp. nov., isolated from the Pindari glacier of the Indian Himalayas, and emended description of the genus Leifsonia. Int. J. Syst. Evol. Microbiol. 58, 2229–2234. doi: 10.1099/ijs.0.65715-0
Reddy, G. S. N., Prakash, J. S. S., Matsumoto, G. I., Stackebrandt, E., and Shivaji, S. (2002). Arthrobacter roseus sp. nov., a psychrophilic bacterium isolated from an Antarctic cyanobacterial mat sample. Int. J. Syst. Evol. Microbiol. 52, 1017–1021. doi: 10.1099/ijs.0.02131-0
Reddy, G. S. N., Prakash, J. S. S., Srinivas, R., Matsumoto, G. I., and Shivaji, S. (2003). Leifsonia rubra sp. nov. and Leifsonia aurea sp. nov., psychrophiles from a pond in Antarctica. Int. J. Syst. Evol. Microbiol. 53, 977–984. doi: 10.1099/ijs.0.02396-0
Ringel, M. T., and Brüser, T. (2018). The biosynthesis of pyoverdines. Microb. Cell. 5, 424–437. doi: 10.15698/mic2018.10.649
Ryan, M. P., and Pembroke, J. T. (2018). Brevundimonas spp: emerging global opportunistic pathogens. Virulence 9, 480–493. doi: 10.1080/21505594.2017.1419116
Sakai, K., Yamanaka, H., Moriyoshi, K., Oshmoto, T., and Ohe, T. (2007). Biodegradation of bisphenol A and related compounds by Sphingomonas sp. strain BP-7 isolated from seawater. Biosci. Biotech. Bioch. 71, 51–57. doi: 10.1271/bbb.60351
Salomon, M. V., Bottini, R., de Souza Filho, G. A., Cohen, A. C., Moreno, D., Gil, M., et al. (2013). Bacteria isolated from roots and rhizosphere of Vitis vinifera retard water losses, induce abscisic acid accumulation and synthesis of defense-related terpenes in in vitro cultured grapevine. Physiol. Plant. 151, 359–374. doi: 10.1111/ppl.12117
Sattley, W. M., Burchell, B. M., Conrad, S. D., and Madigan, M. T. (2017). Design, construction, and application of an inexpensive, high-resolution water sampler. Water 9:578. doi: 10.3390/w9080578
Sattley, W. M., Jung, D. O., and Madigan, M. T. (2008). Psychrosinus fermentans gen. nov., sp. nov., a lactate-fermenting bacterium from near-freezing oxycline waters of a meromictic Antarctic Lake. FEMS Microbiol. Lett. 287, 121–127. doi: 10.1111/j.1574-6968.2008.01300.x
Sattley, W. M., and Madigan, M. T. (2006). Isolation, characterization, and ecology of cold-active, chemolithotrophic, sulfur-oxidizing bacteria from perennially ice-covered Lake Fryxell, Antarctica. Appl. Environ. Microbiol. 72, 5562–5568. doi: 10.1128/AEM.00702-06
Sattley, W. M., and Madigan, M. T. (2007). Cold-active acetogenic bacteria from surficial sediments of perennially ice-covered Lake Fryxell, Antarctica. FEMS Microbiol. Lett. 272, 48–54. doi: 10.1111/j.1574-6968.2007.00737.x
Sattley, W. M., and Madigan, M. T. (2010). Temperature and nutrient induced responses of Lake Fryxell sulfate-reducing prokaryotes and description of Desulfovibrio lacusfryxellense sp. nov., a pervasive, cold-active, sulfate-reducing bacterium from Lake Fryxell, Antarctica. Extremophiles 14, 357–366. doi: 10.1007/s00792-010-0315-6
Solioz, M., and Marrs, B. (1977). The gene transfer agent of Rhodopseudomonas capsulata: purification and characterization of its nucleic acid. Arch. Biochem. Biophys. 181, 300–307. doi: 10.1016/0003-9861(77)90508-2
Tribelli, P. M., and López, N. I. (2018). Reporting key features in cold-adapted bacteria. Life 8:8. doi: 10.3390/life8010008
Van Trappen, S., Vandecandelaere, I., Mergaert, J., and Swings, J. (2004a). Algoriphagus antarcticus sp. nov., a novel psychrophile from microbial mats in Antarctic lakes. Int. J. Syst. Evol. Microbiol. 54, 1969–1973. doi: 10.1099/ijs.0.02973-0
Van Trappen, S., Vandecandelaere, I., Mergaert, J., and Swings, J. (2004b). Flavobacterium degerlachei sp. nov., Flavobacterium frigoris sp. nov. and Flavobacterium micromati sp. nov., novel psychrophilic bacteria isolated from microbial mats in Antarctic lakes. Int. J. Syst. Evol. Microbiol. 54, 85–92. doi: 10.1099/ijs.0.02857-0
Vander Schaaf, N. A., Cunningham, A. M. G., Cluff, B. P., Kraemer, C. K., Reeves, C. L., Riester, C. J., et al. (2015). Cold-active, heterotrophic bacteria from the highly oligotrophic waters of Lake Vanda, Antarctica. Microorganisms 3, 391–406. doi: 10.3390/microorganisms3030391
Vincent, W. F. (1981). Production strategies in Antarctic inland waters: phytoplankton eco-physiology in a permanently ice-covered lake. Ecology 62, 1215–1224. doi: 10.2307/1937286
Wang, F., Gai, Y., Chen, M., and Xiao, X. (2009). Arthrobacter psychrochitiniphilus sp. nov., a psychrotrophic bacterium isolated from Antarctica. Int. J. Syst. Evol. Microbiol. 59, 2759–2762. doi: 10.1099/ijs.0.008912-0
Yumoto, I., Kusano, T., Shingyo, T., Nodasaka, Y., Matsuyama, H., and Okuyama, H. (2001). Assignment of Pseudomonas sp. strain E-3 to Pseudomonas psychrophila sp. nov., a new facultatively psychrophilic bacterium. Extremophiles 5, 343–349. doi: 10.1007/s007920100199
Keywords: Antarctica, Lake Fryxell, McMurdo Dry Valleys, chemoorganotroph, psychrotolerant, halotolerant, Pseudomonas
Citation: Baker JM, Vander Schaaf NA, Cunningham AMG, Hang AC, Reeves CL, Huffman ER, Riester CJ, Madigan MT and Sattley WM (2019) Chemoorganotrophic Bacteria From Lake Fryxell, Antarctica, Including Pseudomonas Strain LFY10, a Cold-Adapted, Halotolerant Bacterium Useful in Teaching Labs. Front. Microbiol. 10:156. doi: 10.3389/fmicb.2019.00156
Received: 22 September 2018; Accepted: 22 January 2019;
Published: 06 February 2019.
Edited by:
Thulani Peter Makhalanyane, University of Pretoria, South AfricaReviewed by:
Isabelle Raymond-Bouchard, McGill University, CanadaMichael Paul Manzella, Colorado State University, United States
Copyright © 2019 Baker, Vander Schaaf, Cunningham, Hang, Reeves, Huffman, Riester, Madigan and Sattley. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: W. Matthew Sattley, bWF0dGhldy5zYXR0bGV5QGluZHdlcy5lZHU=
†These authors have contributed equally to this work
‡Present address: Jennifer M. Baker, Rackham Graduate School, University of Michigan, Ann Arbor, MI, United States Nicole A. Vander Schaaf, Van Andel Research Institute, Grand Rapids, MI, United States
 Jennifer M. Baker
Jennifer M. Baker Nicole A. Vander Schaaf1†‡
Nicole A. Vander Schaaf1†‡ Anna M. G. Cunningham
Anna M. G. Cunningham Anna C. Hang
Anna C. Hang Michael T. Madigan
Michael T. Madigan W. Matthew Sattley
W. Matthew Sattley