
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 05 February 2019
Sec. Food Microbiology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.00135
The spoilage potential and biofilm formation of Shewanella baltica are reported to be regulated by Quorum sensing (QS) system from the phenotype point of view, but the specific mechanism is not fully understood. In the present study, the QS autoinducers were detected by UHPLC-MS/MS, cell density-dependent luxR-type genes were obtained through autoregulation experiments among a series of candidates in S. baltica OS155 (The SSO of large yellow croaker). The direct interaction between cyclo-(L-Pro-L-Phe) (PP) and LuxR01 as well as LuxR02 proteins was revealed via in vitro binding assay. Deletion of luxR-type genes (luxR01 and luxR02) impaired spoilage potential and biofilm formation of S. baltica OS155 in various degrees. Transcriptional analysis and qRT-PCR validation showed that spoilage and biofilm-related genes torS, speF, and pomA were down-regulated in luxR01 and luxR02 deletion strains. In addition, exogenous PP promoted spoilage potential and biofilm formation, which could be attenuated by luxR01 or luxR02 deletion. Our results revealed an explicit QS system employing PP as autoinducer and two orphan LuxRs as receptors which positively regulated spoilage capacity and biofilm formation via transcriptional regulation of corresponding genes in S. baltica OS155, which provides potential specific targets for seafood preservation involving QS system.
Seafood is one of the most highly perishable food products because of the chemical effects of atmospheric oxygen and the growth of spoilage microorganisms (Özogul et al., 2004). Bacterial metabolism caused by microbial growth is the major cause of seafood spoilage and results in the unacceptable characteristics including quality and safety impairment (Robson et al., 2007). However, only a portion of the original seafood microbiota, named SSOs, has strong adaptability to storage conditions, and thus prevail over the rest of the microbiota, reaching high populations and producing corresponding spoilage products (Parlapani et al., 2015). Biochemical analysis on the basis of nucleotide metabolism, production of TMA, hypoxanthine (Hx), total volatile basic nitrogen (TVB-N), and BAs have been commonly utilized to estimate fish quality. Large yellow croaker (Pseudosciaena crocea) is commercially important because of its delicious taste and good nutrition, representing one of the largest yield of single species in marine net-cage farming in China. Shewanella baltica, a commonly dominant spoilage bacteria in seafood products such as iced sea salmon (Hozbor et al., 2006), gutted sea bass (Parlapani et al., 2015), chilled fresh Mediterranean swordfish (Pantazi et al., 2008) and tropical prawns (Chinivasagam et al., 1998; Zhu et al., 2015), has received increasing attention in recent years due to its significant role in seafood spoilage, and has also been identified as the SSO of large yellow croaker stored at 4°C (Gu et al., 2013; Zhu et al., 2016).
The term QS was first put forward by Fuqua et al. (1994). QS is a cell density-dependent signaling system in various bacterial species (Fuqua et al., 1994; Bassler and Losick, 2006). These bacteria autonomously sense a threshold concentration of the autoinducers (corresponding to a critical cell density) produced by the microbial community, leading to gene expression alterations through transcriptional regulators, and thus regulate their behaviors to be beneficial to survival (Kievit and Iglewski, 2000; Smith et al., 2004). Therefore, QS-controlled physiological activities, such as bioluminescence, virulence factors secretion, biofilm formation, sporulation and so on, require the collective action of the group to be effective (Bassler and Losick, 2006; Papenfort and Bassler, 2016). Gram-positive bacteria employ oligopeptide as QS autoinducers and membrane-bound sensor histidine kinases as QS receptors, and Gram-negative bacteria generally utilize LuxIR-type proteins sensing N-acyl-homoserine lactones (AHLs) signals primarily for intraspecies communication (Whitehead et al., 2001; Waters and Bassler, 2005). Moreover, LuxS/Autoinducer-2 (AI-2) has been supposed to be universal signal in both Gram-negative and Gram-positive bacteria (Smith et al., 2004), which has been observed in several bacteria (Schauder and Bassler, 2001; Taga et al., 2001; Duan et al., 2003). Recently, a range of cyclic dipeptides (diketopiperazines, DKPs) produced by multiple Gram-negative bacteria have been reported to modulate AHLs-specific sensor system (Holden et al., 1999; Degrassi et al., 2002; Park et al., 2006), previous studies have demonstrated the positive role of DKPs as QS signals in promoting biofilm formation (Zhu et al., 2015, 2016), torA and ODC genes expression (Zhu et al., 2016) as well as the production of TVB-N, TMA, putrescine and extracellular proteases (Gu et al., 2013; Zhu et al., 2015, 2016). As a result, DKPs have been proposed to represent a novel naturally occurring QS autoinducers which can act as intra- and inter-species regulators (Shiner et al., 2005; Klose, 2006). In addition, 2698 of the 3550 luxR genes in the NCBI databases have been found as orphans which had no adjacent luxI gene in the chromosome according to a survey conducted by Hudaiberdiev et al. (2015), many of which were devoid of the AHL-binding motifs, thus may responded to exogenous signals besides AHL type molecules (Hudaiberdiev et al., 2015). These results provide strong evidence for the alleged role of DKPs in bacterial QS system, however, Campbell et al. (2009) found that the native DKPs exhibited neither antagonistic nor agonistic activities in reporter strains, and suggested that non-native DKPs influenced QS-regulated outcomes not through the direct interaction with LuxR-type proteins (Campbell et al., 2009). Therefore, questions remain on the genuine physiological function of DKPs in bacteria.
The QS systems involved in the physiological and clinical aspects of bacteria have attracted considerable attention and have been widely investigated (Ruffin et al., 2016; Wu et al., 2016). However, there is a lack of knowledge on the role of QS in food spoilage, especially in seafood. As cell–cell communication commonly exists in bacteria, QS should participate in the microbial ecology of food, and studies on the spoilage aspect of QS have been carried out in the past few years. It has been reported that bacterial spoilage of bean sprouts was affected by QS (Rasch et al., 2005), and milk spoilage by psychrotrophic bacteria Pseudomonas spp., Serratia spp., Enterobacter spp., and Hafnia alvei was also resulted from QS (Pinto et al., 2007). AHL-modulated exoenzyme activities have been reported in Serratia proteamaculans B5a isolated from cold-smoked salmon (Christensen et al., 2003), similarly, C4-HSL increased exoenzyme production in Pseudomonas psychrophila PSPF19 and promoted the spoilage of refrigerated freshwater fish (Bai and Rai Vittal, 2014). However, the molecular mechanism of how QS system regulates seafood spoilage remains to be revealed.
Bacterial biofilms are assemblages of bacterial cells with self-produced polymeric matrix or glycocalyx adherent to surface or interface (Costerton et al., 1999; Hall-Stoodley et al., 2004). QS has been reported to regulate the biofilm formation in pathogens, such as Pseudomonas aeruginosa (Davies et al., 1998), Streptococcus mutans (Li et al., 2002), Staphylococcus aureus (Yarwood et al., 2004) and Streptococcus pneumoniae (Vidal et al., 2011). Recently, AHLs and DKPs have been reported to promote the formation of biofilms in spoilage bacteria Pseudomonas psychrophila PSPF19 and S. baltica (Bai and Rai Vittal, 2014; Zhu et al., 2015, 2016). However, it remains unknown how biofilm is regulated by QS system in spoilage microorganisms.
Shewanella baltica is a facultative anaerobic, rods-shaped, and psychrotrophic Gram-negative bacterium with the optimal growth temperature around 25°C but can also grow at 0°C (Francoise, 2010). As the SSO of refrigerated shrimp and large yellow croaker, S. baltica produced a series of DKPs which could promote their spoilage capacity (Gu et al., 2013; Zhu et al., 2015, 2016), but the interaction between DKPs and typical QS receptors LuxR-type proteins (Holden et al., 1999; Degrassi et al., 2002; Park et al., 2006) has yet to be studied. In this study, we identified a series of orphan luxR-type genes by homology search within the representative strain S. baltica OS678 in NCBI database. Then we demonstrated the direct interaction between PP and LuxR-type protein (LuxR01 and LuxR02) in S. baltica. After constructing a series of orphan luxR-type gene deletion mutants (SB7301, SB7302, SB7303, SB7304, SB7305, SB7306, and SB7307), we demonstrated that the spoilage potential and biofilm formation were positively regulated by DKP-LuxR type QS system via governing the expression of torS, speF, and pomA genes. This work improved our understanding of the QS system employed in S. baltica and provided potential targets for controlling undesirable bacteria growth in foods.
Shewanella baltica 73 strain was demonstrated as the SSO of large yellow croaker (Pseudosciaene crocea) (Gu et al., 2013), which showed 99% identity with S. baltica OS155 (NCBI Taxonomy ID: 325240). Thus, the target strain in this study was uniformly named as S. baltica OS155. All bacterial strains and plasmids involved in this study are listed in Supplementary Table S1.
Escherichia coli and S. baltica strains were grown in Luria-Bertani (LB) medium at 37 and 30°C, respectively. When necessary, additives are used as follows: DAP, 50 μg/mL; Gm, 15 μg/mL; ampicillin, 100 μg/mL; IPTG, 0.1 mM.
On the basis that S. baltica OS678 is the representative strain of S. baltica species in NCBI database, we identified target genes in S. baltica OS155 by homology search with S. baltica OS678. The identification of these genes was listed in Supplementary Table S2. Among them, all of the chosen luxR-type genes contain sequence encoding a DNA-binding helix-turn-helix domain characteristic of genuine luxR genes and the conserved domains in each LuxR-type proteins were shown in Supplementary Figure S1.
The AHL standards (95% or higher purity), including C4-HSL, C6-HSL, C8-HSL, C10-HSL, C12-HSL, C14-HSL, 3-oxo-C8-HSL, 3-oxo-C10-HSL, 3-oxo-C12-HSL, and 3-oxo-C14-HSL were obtained from Sigma-Aldrich (St. Louis, MO, United States). The three cyclic dipeptides (DKPs) including PL, LL and PP were synthesized by Sangon Biotech Company (Shanghai, China). Reference standards were prepared as 1 μg/mL solution in methanol. S. baltica strains were grown in LB medium overnight (postexponential phase) at 30°C. Culture supernatants were extracted three times with equal volumes of acidified ethyl acetate (0.5% formic acid), and the extracts were evaporated under nitrogen flow (Rasch et al., 2005). Fish organs were sampled after storage at room temperature for 48 h and extracted as previously described (Buch et al., 2003). The evaporated extracts were redissolved in 1 mL methanol and stored at -20°C for further analysis using UHPLC-MS/MS. Mass spectra were recorded by means of a triple-quadrupole tandem mass spectrometer (TSQ Quantiva, Thermo Fisher Scientific, Waltham, MA, United States) coupled to a Surveyor high performance liquid chromatography system (Thermo Fisher Scientific, Waltham, MA, United States) which was equipped with a thermostated (4°C) autosampler and a 150 mm × 2.0 mm i.d., 1.8 μm Synergi Hydro RPHPLC column (Phenomenex, Aschaffenburg, Germany) kept at 30°C. The mobile phase was composed of formic acid in water (0.1%; A) and methanol (100%; B). The LC gradient was listed in Supplementary Table S3. The mass spectrometer was operated in the positive electrospray ionization mode (H-ESI) with a spray voltage of 3500 V. The temperature of the capillary was 350°C, and the sheath and auxiliary gas (nitrogen) were adjusted to 45 and 12 arbitrary units, respectively. The retention times and MS parameters used for identification and quantification of AHLs and DKPs were listed in Table 1.
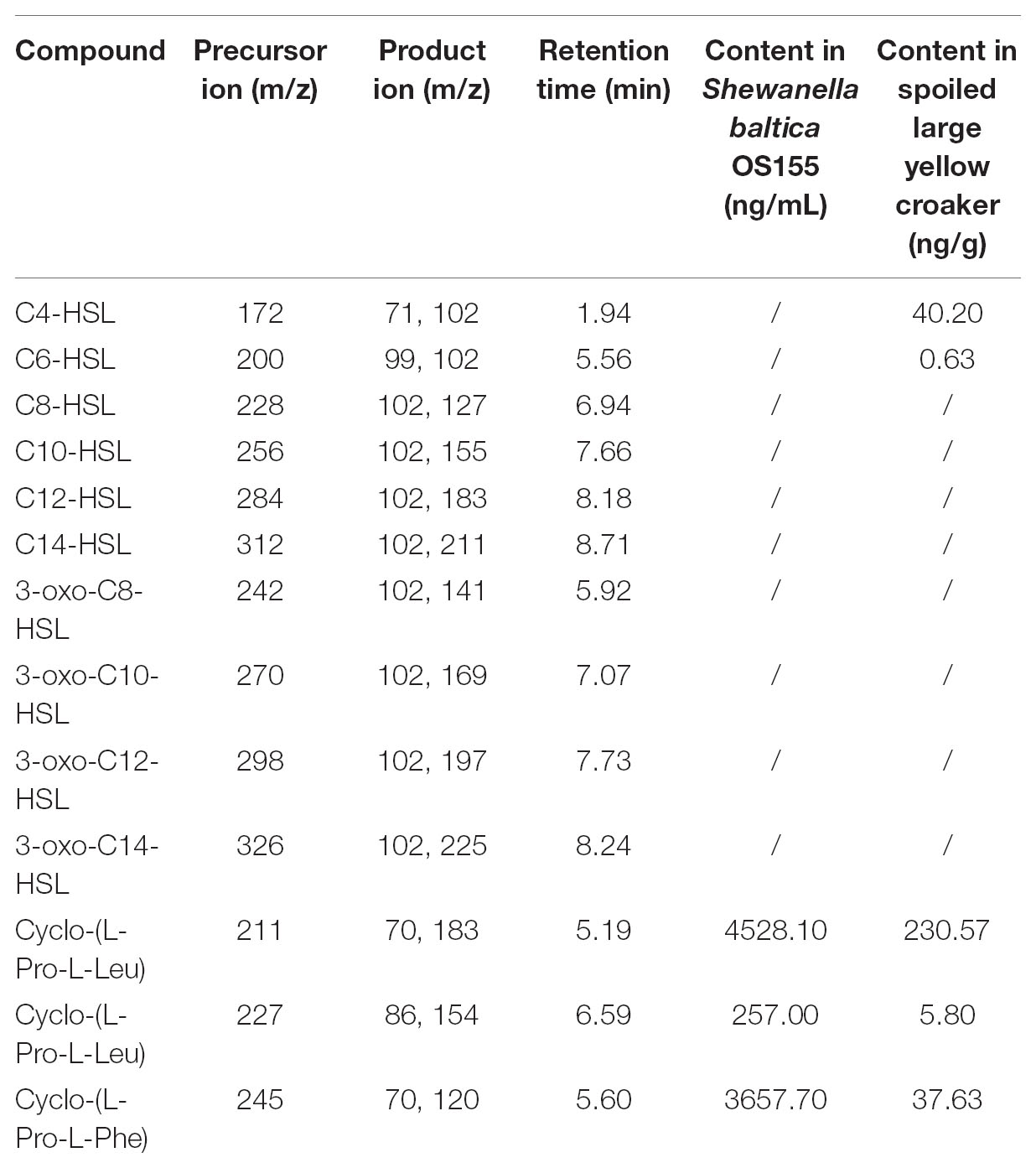
Table 1. MS conditions used for AHLs and DKPs analysis, and the identification and quantification of AHLs and DKPs in Shewanella baltica OS155 and spoiled large yellow croaker.
RNA was extracted from postexponential cultures using an RNeasy mini kit (Qiagen) according to the manufacturer’s instructions. Genomic DNA contamination was eliminated by treating total RNA with Turbo DNA free (Thermo Fisher Scientific, Waltham, MA, United States). cDNA was synthesized using random hexamers (GE Healthcare) and SuperScript II reverse transcriptase (Invitrogen, Grand Island, NY, United States). qRT-PCR was performed as described previously (Burr et al., 2006). The relative gene expression was calculated using the critical threshold (ΔΔCt) method with 16S rRNA being the internal control.
The late exponential culture supernatant of S. baltica OS155 was harvested by centrifugation and subject to a 0.22 μm filter to remove all remaining cells, then filtered supernatants were dried and resuspended in PBS buffer to a tenth of the original volume (10×). The early exponential cells of S. baltica OS155 were treated with late exponential culture extract of a final 1× concentration (Balaban and Novick, 1995). Equal volume of PBS was used as a control. The cells were further cultured for 3 h, then the expression level of provisional luxR-type genes (luxR01, luxR02, luxR03, luxR04, luxR05, and luxR06 genes) were quantified by qRT-PCR.
The provisional LuxR-type proteins-coding sequences (luxR01, luxR02, luxR03, luxR04, luxR05, and luxR06 genes) were fused with N-terminal FLAG tag (GACTACAAGGACGACGATGACAAG) and cloned into the expression vector pET-15b at the site of Nde I and BamH I using One Step Cloning Kit (Vazyme, Hangzhou, China). Target genes and primers used are listed in Supplementary Table S4. The recombinant vectors were verified by sequencing and transformed into the host E. coli BL21 (DE3) for protein expression. The empty vector pET-15b was served as a control. The recombinant strains were cultured in LB medium with ampicillin at 37°C for 4 h. The protein expression was induced with 0.1 mM IPTG overnight at 16°C (Kleber-Janke and Becker, 2000). Cells were collected and treated with B-PER Complete Bacterial Protein Extraction Reagent (Thermo Fisher Scientific, Waltham, MA, United States) for mild protein extraction according to the manufacturer’s instruction. The recombinant proteins were purified using ANTI-FLAG M2 magnetic beads (Sigma-Aldrich, St. Louis, MO, United States) based on the specific binding between M2 antibody and FLAG tag (Brizzard et al., 1994). The purified target proteins and the vector control were analyzed by SDS-PAGE and Coomassie brilliant blue R-250 staining (Bio-Rad, Hercules, CA, United States).
The purified beads-FLAG-LuxRs complexes (LuxR01, LuxR02, LuxR03, LuxR04, LuxR05, and LuxR06 proteins) were treated with a mixture containing 1 μM C4-HSL, C6-HSL, PL, LL, and PP in PBS buffer, respectively. The binding system was incubated at 4°C for 1 h with gentle rotating. The binding complexes were collected and washed with PBS buffer to remove all of the non-specifically bound molecules. The target complexes were resuspended in PBS buffer and boiled three times to release the bound molecules into the supernatant, which was collected and extracted twice with ethyl acetate. The ethyl acetate fractions were pooled, evaporated under nitrogen flow to dryness, and finally resuspended in 1 mL methanol. The empty vector pET-15b was served as a control.
In-frame deletion mutants (luxR01, luxR02, luxR03, luxR04, luxR05, and luxR06 genes deletion mutants) were constructed as reported previously (Gao et al., 2006; Jin et al., 2013). In brief, attB1 and attB2 sequences were flanking the specific sequences fused by the upstream and downstream regions of the target genes (in-frame deletion construct) for subsequent attB-attP (BP) recombination with pHGM01. Target genes and primers used are listed in Supplementary Table S4. The resulting recombination mixture was transformed into the donor E. coli WM3064, the fragment between attP1 and attP2 site within pHGM01 was replaced by the in-frame deletion construct, removing the ccdB suicide gene, thus E. coli WM3064 was allowed to grow on plates containing Gm and DAP. Integration of the recombinant vectors into the S. baltica OS155 chromosome from E. coli WM3064 was carried out by conjugation at a ratio of 2:1 (donor:recipient). PCR-verified transconjugants were grown in NaCl-less LB liquid overnight and then plated onto NaCl-less LB agar containing 10% sucrose. At last, Gm-sensitive and sucrose-resistant colonies were screened by PCR as deletion mutants, which were further verified through DNA sequencing. The constructed in-frame deletion strains for luxR01, luxR02, luxR03, luxR04, luxR05, and luxR06 genes were named SB7301, SB7302, SB7303, SB7304, SB7305, and SB7306 mutants, respectively. The constructed in-frame deletion strain for both luxR01 and luxR02 genes was named SB7307. Deletion mutants were screened by PCR amplification using the outside primers (5′-O and 3′-O) and verified by DNA sequencing (Supplementary Figure S2).
For the TMA analysis, S. baltica OS155 wild-type strain and all gene deletion mutants (SB7301, SB7302, SB7303, SB7304, SB7305, SB7306, and SB7307) were cultured in the LB broth with 10 and 100 mM TMAO, respectively. The TMA was quantified spectrophotometrically by following the colorimetric formation of the picric acid salt of TMA (Dyer, 1945). The wild-type strain was used as control and water was used as blank.
For putrescine analysis, S. baltica OS155 wild-type strain and all gene deletion mutants (SB7301, SB7302, SB7303, SB7304, SB7305, SB7306, and SB7307) were cultured in LB broth with 0.5/2% L-Ornithine monohydrochloride and 0.5/2% L-Arginine monohydrochloride supplemented with 0.005% pyridoxal-5′-phosphate. The postexponential cell culture supernatant was obtained as test samples by centrifuging at 5,000 g for 10 min at 4°C. Putrescine standard was purchased from Sigma-Aldrich (St. Louis, MO, United States), the standard solutions were prepared in 0.1 M HCL to a final concentration of 0.0, 0.1, 0.5, 1.0, 5.0, 12.5, 25, and 50 μg/ml, respectively. The extraction and pre-column derivatization of test samples or BA standards were carried out based on the methods developed by Kim et al. (2009). BAs were measured using HPLC according to the procedures reported by Ben-Gigirey et al. (1998) with little modification. The wild-type strain was used as control.
The static biofilm formation in S. baltica OS155 wild-type strain and all gene deletion mutants (SB7301, SB7302, SB7303, SB7304, SB7305, and SB7306) was quantified by microtiter plate assay as described previously with some modifications (Djordjevic et al., 2002). Overnight cultures of target strains were diluted at 1:100 ratio in a fresh sterile LB medium and then transferred into 96-well microtiter plates. After further incubation without shaking at 30°C for 60 h, LB medium was removed and the plates were washed thrice with sterile distilled water. The plates were air-dried and stained with 1% crystal violet solution in water for 20 min. The wells were washed thoroughly with sterile distilled water, after drying, 95% ethanol was used to dissolve the stained biofilm. The absorbance was measured at 595 nm using a microplate reader. Wild-type strain was used as control and pure LB liquid medium was used as blank.
The density and thickness of biofilm were analyzed by CLSM. Overnight cultures of target strains were diluted at 1:100 ratio in a fresh sterile LB medium. 2 mL dilutions were transferred into the 35 mm glass bottom culture dishes with 20 mm micro-well (Cellvis, Sunnyvale, CA, United States). After further incubation without shaking at 30°C for 60 h, the non-adhering cells were removed by washing thrice with sterile distilled water. Biofilms were fluorescent-labeled with Syto 9 dye, a green cell-permeant nucleic acid marker (Syto 9) in LIVE/DEAD BacLight Bacterial Viability kit (Invitrogen, Grand Island, NY, United States), following the manufacturer’s instructions for biofilm staining. Image acquisition was performed using a Zeiss LSM 800 Confocal Laser Scanning Microscope (Zeiss, Jena, Germany). All biofilms were scanned at 800 Hz using a 63× oil immersion objective lens with a 488-nm argon laser set at 25% intensity. A range of 500–600 nm was chosen to collect the Syto 9 emission fluorescence. Two stacks of horizontal plane images with a z-step of 1 μm were acquired per dish. Three-dimensional projections of the biofilms and the maximum thickness were acquired for three biofilm replicas per strain using IMARIS 7.1 software (Bitplane, Switzerland) (Guilbaud et al., 2015).
Exogenous PP was added at 10 μM when cells were grown to early exponential phase (about 3 h), an equal volume of DMSO was used as control. After further culture for 3 h, TMA, putrescine and biofilm formation analyses were carried out as described above.
Wild-type strain and SB7301 mutant (which is of the same status as SB7302 according to our previous results) were grown overnight and total RNA was extracted from the cell pellets. RNA samples were treated with Ribo-Zero Magnetic kit to remove ribosomal RNA. RNA transcripts were fragmented using Truseq™ RNA sample prep Kit for Library construction. The fragmented RNA sequences were reverse transcribed, random primers were used to synthesize the second cDNA strand. After adapter ligation, the second cDNA strand was digested with uracil-DNA glycosylase. The remaining fragments were PCR amplified for 15 cycles and the amplicons were selected by 2% agarose gel electrophoresis. The cDNA products were subjected to high-throughput sequencing with an Illumina HiSeq 2000 sequencer to obtain single reads of 100 bp using the forward sequencing primer.
All the statistical results were analyzed with three replicates according to a completely randomized design. The Student’s t-test was employed to find out significance difference between the means. Data were analyzed statistically by repeated measures using SPSS 17.0 procedures and p < 0.05 was considered as statistically significant.
Our previous study has isolated S. baltica OS155 from large yellow croaker and identified it as the SSO during low-temperature storage. To better investigate the QS system in S. baltica OS155, we first identified and quantified the hypothetical autoinducers produced by S. baltica OS155. Using the previously described UHPLC-MS/MS method (Wang et al., 2017), all AHLs and DKPs standards were separated within 9 min. Specifically, PL, LL, and PP were detected in the overnight culture supernatant of S. baltica OS155 in LB medium (Supplementary Figure S3A), and C4-HSL, C6-HSL, PL, LL, and PP were detected in spoiled large yellow croaker (Supplementary Figure S3B). In most situations, DKPs showed a much higher concentration than AHLs and thus were assumed to be the predominant autoinducers in both S. baltica OS155 and spoiled large yellow croaker (Table 1).
Most QS autoinducers are accumulated at high cell density, and the activated LuxR-type proteins act as transcription regulators to control the expression of relevant genes including themselves in a feedback loop. Based on this, we first introduced postexponential (high cell density) supernatants of S. baltica OS155 into their early exponential phase (low cell density) culture and analyzed the subsequent gene expression alteration. By qRT-PCR and statistical analyses, we found that luxR01, luxR02, luxR03, luxR05, and luxR06 genes were up-regulated in different degrees with statistical significance when induced by self-culture supernatant (Figure 1). This result preliminarily demonstrated luxR01, luxR02, luxR03, luxR05, and luxR06 genes as genuine luxR-type genes in QS system of S. baltica OS155.
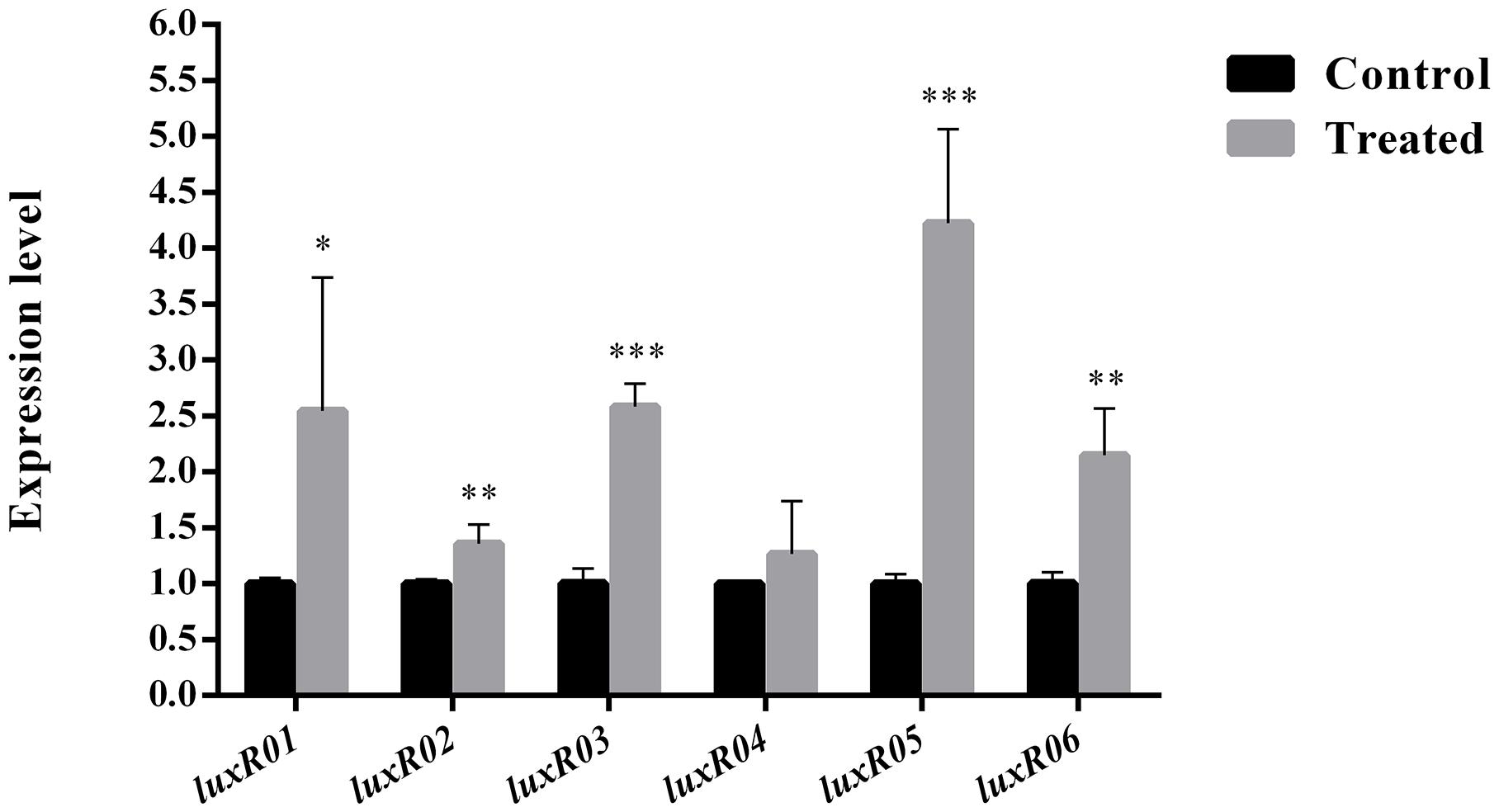
Figure 1. Identification of cell density-dependent LuxR-type proteins in Shewanella baltica OS155. Effects of post exponential supernatant extract on the transcription of provisional luxR-type genes, including luxR01, luxR02, luxR03, luxR04, luxR05 and luxR06, were quantified by qRT-PCR in S. baltica OS155. Data was presented as the mean ± standard deviation (n = 3, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
LuxR-type protein is typical QS receptor in Gram-negative bacteria whose activity is modulated by autoinducer. In light of this, we evaluated the interaction between the detected hypothetical autoinducers and LuxR-type proteins in S. baltica OS155. The purified six LuxR-type proteins were verified by SDS-PAGE and Coomassie brilliant blue staining (Supplementary Figure S4). We introduced the target proteins-bound magnetic beads into the intermixture of hypothetical QS autoinducers to enrich the specific autoinducers, which were further extracted by ethyl acetate and subject to UHPLC-MS/MS for identification. As shown in Figure 2, PP was detected in LuxR01 and LuxR02 protein binding system as compared with the pET-15b vector control, yet other LuxR-type proteins were failed to bind any hypothetical autoinducers. Our data suggested that PP is an autoinducer recognized by LuxR01 and LuxR02 proteins in S. baltica OS155.
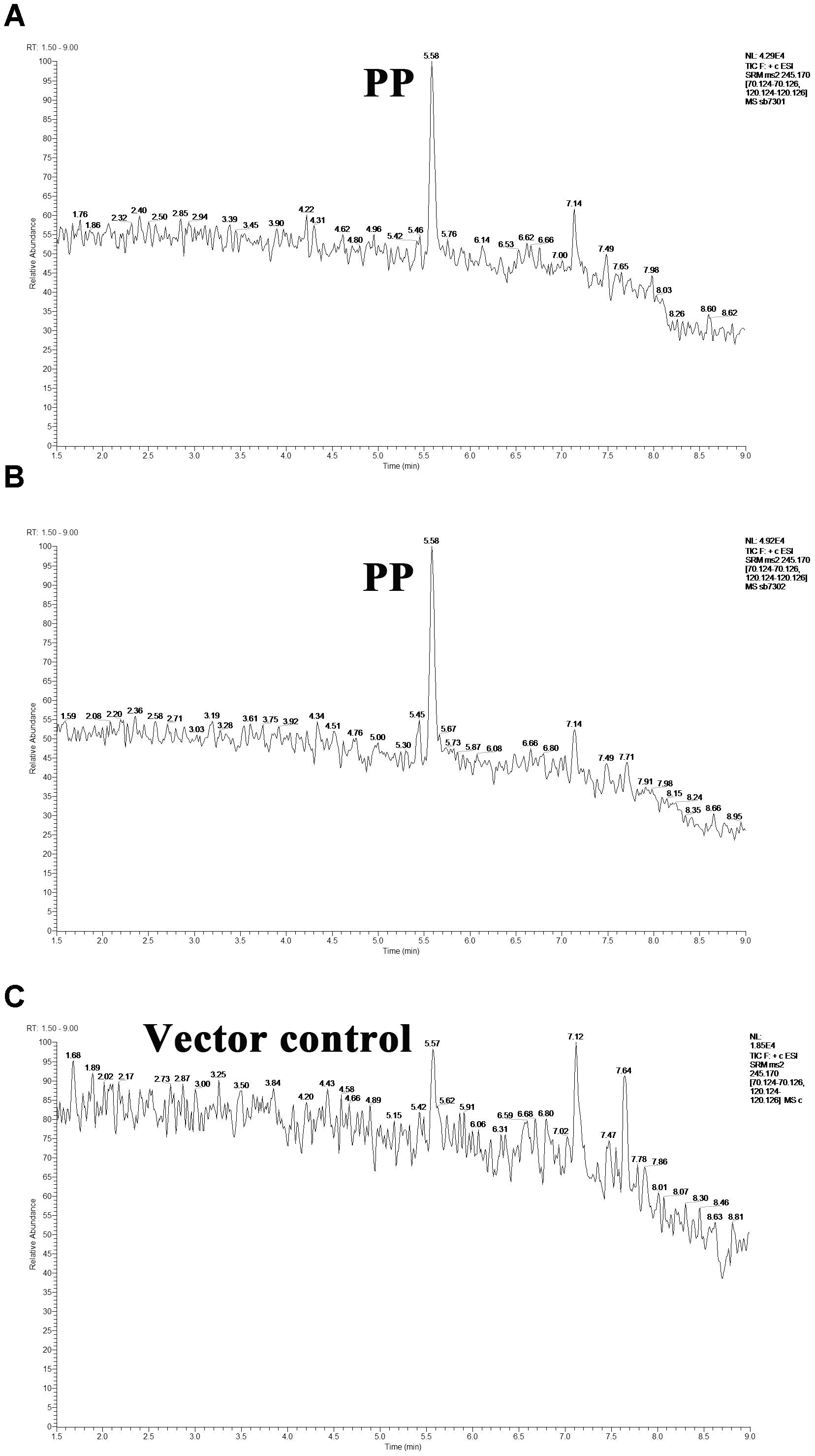
Figure 2. The direct interaction between cyclo-(L-Pro-L-Phe) (PP) molecule and LuxR01 as well as LuxR02 proteins. PP molecule captured by purified LuxR01 (A) and LuxR02 (B) in the mixture of autoinducer candidates was detected using UHPLC-MS/MS as compared with the pET-15b vector control (C). PP: cyclo-(L-Pro-L-Phe).
It has been reported that QS system underpins the spoilage potential of spoilage microorganisms. On this basis, we explored the role of each LuxR-type protein in regulating spoilage-related activity, including TMA and putrescine production. We established SB7301, SB7302, SB7303, SB7304, SB7305, SB7306, and SB7307 mutans, and tested the spoilage potential of these strains. As shown in Figures 3A1,A2, all mutant strains except for SB7305 were partially decreased in TMA production compared with wide type strain. However, putrescine production was reduced only in SB7301 and SB7302 mutants (Figures 3B1,B2). In addition, the reduction of TMA and putrescine production in SB7301 and SB7302 mutants cultured in higher concentration of precursors were larger than that cultured in lower concentration of precursors (Figure 3). Consequently, it seems that LuxR01 and LuxR02 exert a greater role in spoilage potential than other LuxR-type proteins. Remarkably, double mutant SB7307 showed a more significant reduction in both TMA and putrescine production (reduction to around 40%) than SB7301 and SB7302 mutants (Supplementary Figure S5).
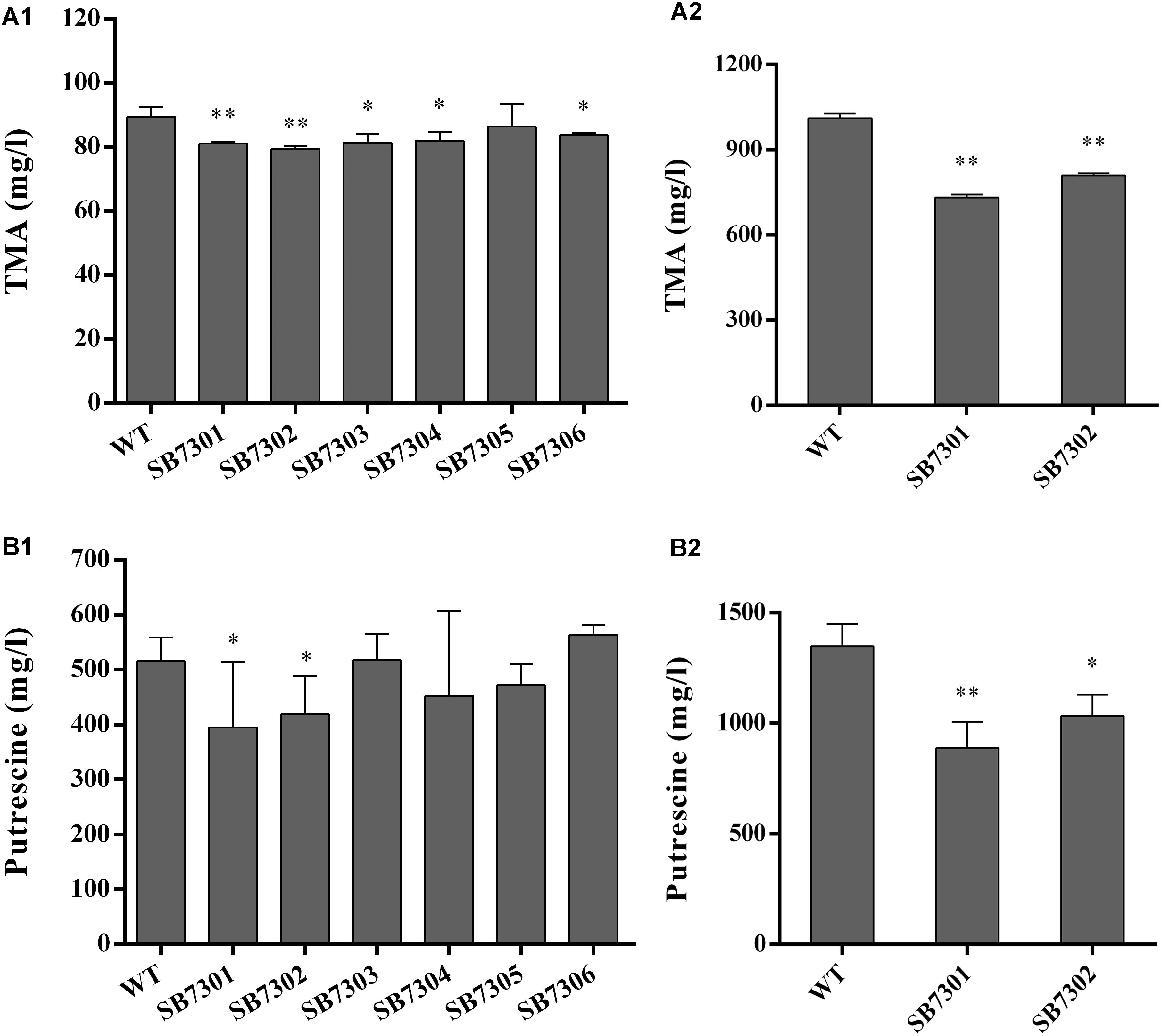
Figure 3. Spoilage potential of gene deletion mutants. The TMA and putrescine production of wild-type and each deletion mutant (SB7301, SB7302, SB7303, SB7304, SB7305, and SB7306) strain were measured at the addition of 10 mM (A1) and 100 mM (A2) TMAO, 0.5% (B1), and 2% (B2) L-Ornithine monohydrochloride and L-Arginine monohydrochloride, respectively. Data was presented as the mean ± standard deviation (n = 3, ∗p < 0.05, ∗∗p < 0.01).
It has been well studied that QS system is critical in regulating biofilm formation. Consequently, we investigated the regulatory role of each luxR-type gene in biofilm formation through microtiter plate assay. The results showed that SB7301, SB7302, and SB7303 were significantly deficient in biofilm formation, the absence of any of these genes reduced biofilm to less than half (Figure 4A). Given the excellent performance of LuxR01 and LuxR02 in PP detection and spoilage activity, we further analyzed the biofilm formation of SB7301 and SB7302 mutants by CLSM imaging. As shown in Figure 4B, the mean maximum thickness of biofilm was calculated by IMARIS software, and showed that SB7301 (P < 0.001) and SB7302 (P < 0.001) formed significantly thinner biofilm than wild-type strain, which was in accordance with the results of microtiter plate assay (Figure 4C). These results indicated that QS receptors LuxR01 and LuxR02 played important roles in spoilage potential as well as biofilm formation in S. baltica OS155.
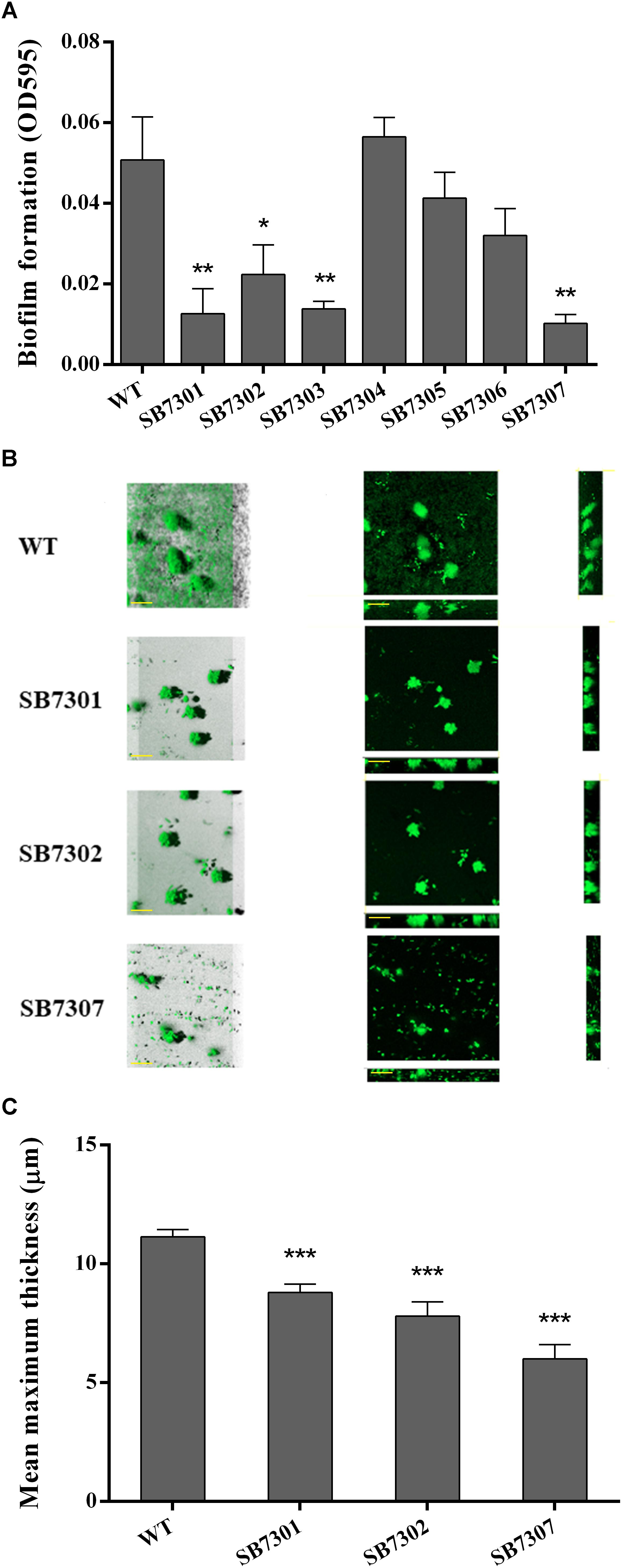
Figure 4. Biofilm formation activity of gene deletion mutants. (A) Biofilm formation of wild-type and each deletion mutant (SB7301, SB7302, SB7303, SB7304, SB7305, and SB7306) strains was quantified by microtiter plate assay. (B) Biofilm formation of wild-type, SB7301 and SB7302 mutant strains was analyzed by CLSM imaging in blend mode view (left panel) and section mode view (right panel). The scale bars represent 50 μm. (C) The mean maximum thickness of biofilms obtained from CLSM imaging. Data was presented as the mean ± standard deviation (n = 3, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
According to our previous results, PP could be recognized by cell density-dependent proteins LuxR01 and LuxR02, which were found to be responsible for spoilage activity and biofilm formation. Then we attempt to verify whether these LuxR-type proteins exert their function via interaction with PP molecule. To that end, we added exogenous PP to the culture system of wild-type strain, SB7301 and SB7302 mutants and measured their spoilage and biofilm formation activity. Both microtiter plate assay and CLSM imaging showed that PP significantly facilitated the biofilm formation in wild-type strain but not SB7301 and SB7302 mutants (Figure 5). Besides, PP partially enhanced the spoilage activity of S. baltica OS155, which could be attenuated by the deletion of luxR01 or luxR02 gene (Figure 6). These data suggested that LuxR01 and LuxR02 interacted with PP synergistically to mediate the QS-induced phenotypes such as biofilm formation and spoilage potential in different degrees.
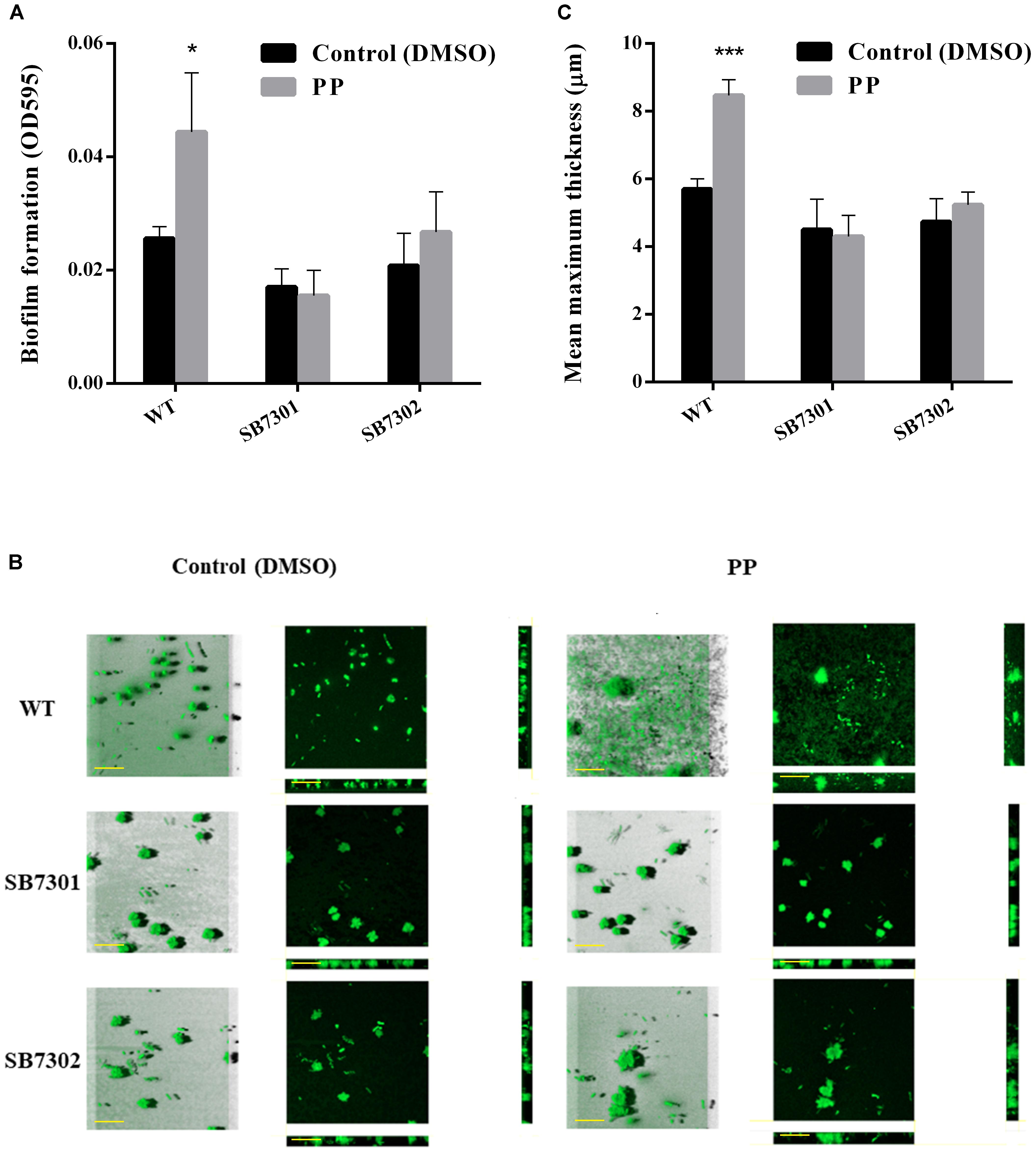
Figure 5. Cyclo-(L-Pro-L-Phe) promotes biofilm formation in a LuxR-type protein-dependent manner. (A) Biofilm formation of wild-type strain, SB7301 and SB7302 mutants with DMSO (control) and PP treatment was quantified by microtiter plate assay. (B) Biofilm formation of wild-type strain, SB7301 and SB7302 mutants with DMSO (control) and PP treatment was analyzed by CLSM imaging in blend mode view (left panel) and section mode view (right panel). The scale bars represent 50 μm. (C) The mean maximum thickness of biofilm formed in wild-type strain, SB7301 and SB7302 mutants with DMSO (control) and PP treatment was calculated by IMARIS software. Data was presented as the mean ± standard deviation (n = 3, ∗p < 0.05, ∗∗∗p < 0.001).
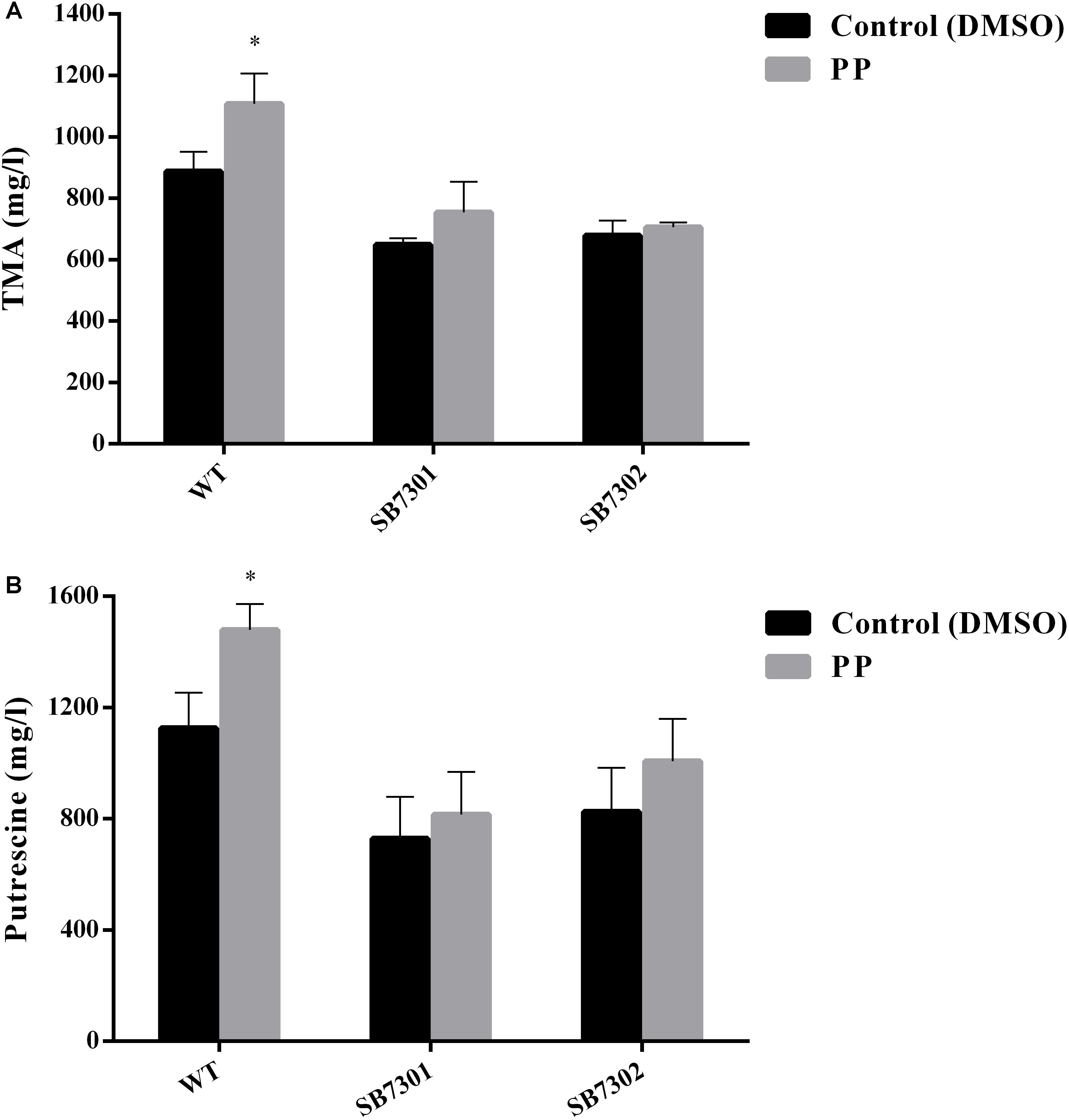
Figure 6. Cyclo-(L-Pro-L-Phe) promotes spoilage activity in a LuxR-type protein-dependent manner. The TMA (A) and putrescine (B) production of wild-type strain, SB7301 and SB7302 mutants with DMSO (control) and PP treatment were measured. Data was presented as the mean ± standard deviation (n = 3, ∗p < 0.05, ∗∗p < 0.01).
We have demonstrated that PP molecule enhanced spoilage activity partially and biofilm formation in S. baltica OS155 through the direct interaction with LuxR01 or LuxR02 proteins. To further investigate the detailed mechanism of DKP-LuxR type QS system in regulating spoilage activity and biofilm formation, high-throughput transcriptome analysis was used to identify the downstream genes regulated by LuxR01 and LuxR02 proteins. We first chose SB7301 mutant unbiasedly to be subject to transcriptome analysis. A total of 335 genes exhibited significant changes in expression level compared with wild-type strain were found, including 197 upregulated genes and 138 downregulated genes (Supplementary Data Set S1). These differentially expressed genes were further grouped according to the pathway they involved in based on the KEGG (Kyoto encyclopedia of genes and genomes) database. Results showed that these genes mainly participated in amino acid metabolism, lipid metabolism, carbohydrate metabolism, energy metabolism, signal transduction, cell motility and membrane transport (Supplementary Figure S6), which were related to spoilage potential to varying degrees. Among them, torS was responsible for TMA production (Jourlin et al., 1996), speF was responsible for putrescine production (Kashiwagi et al., 1991), and pomA was involved in biofilm formation (McCarter, 2001; Thormann et al., 2004). According to KEGG database, torS and pomA belong to two-component system, and speF belong to arginine and proline metabolism system (Supplementary Figure S7). All of these genes were significantly downregulated inSB7301 and SB7302 mutants compared with wild-type strain and was verified by qRT-PCR (Figure 7). These results revealed that DKP-LuxR type QS system is responsible for spoilage activity and biofilm formation in S. baltica OS155 via the transcriptional regulation of torS, speF, and pomA genes.
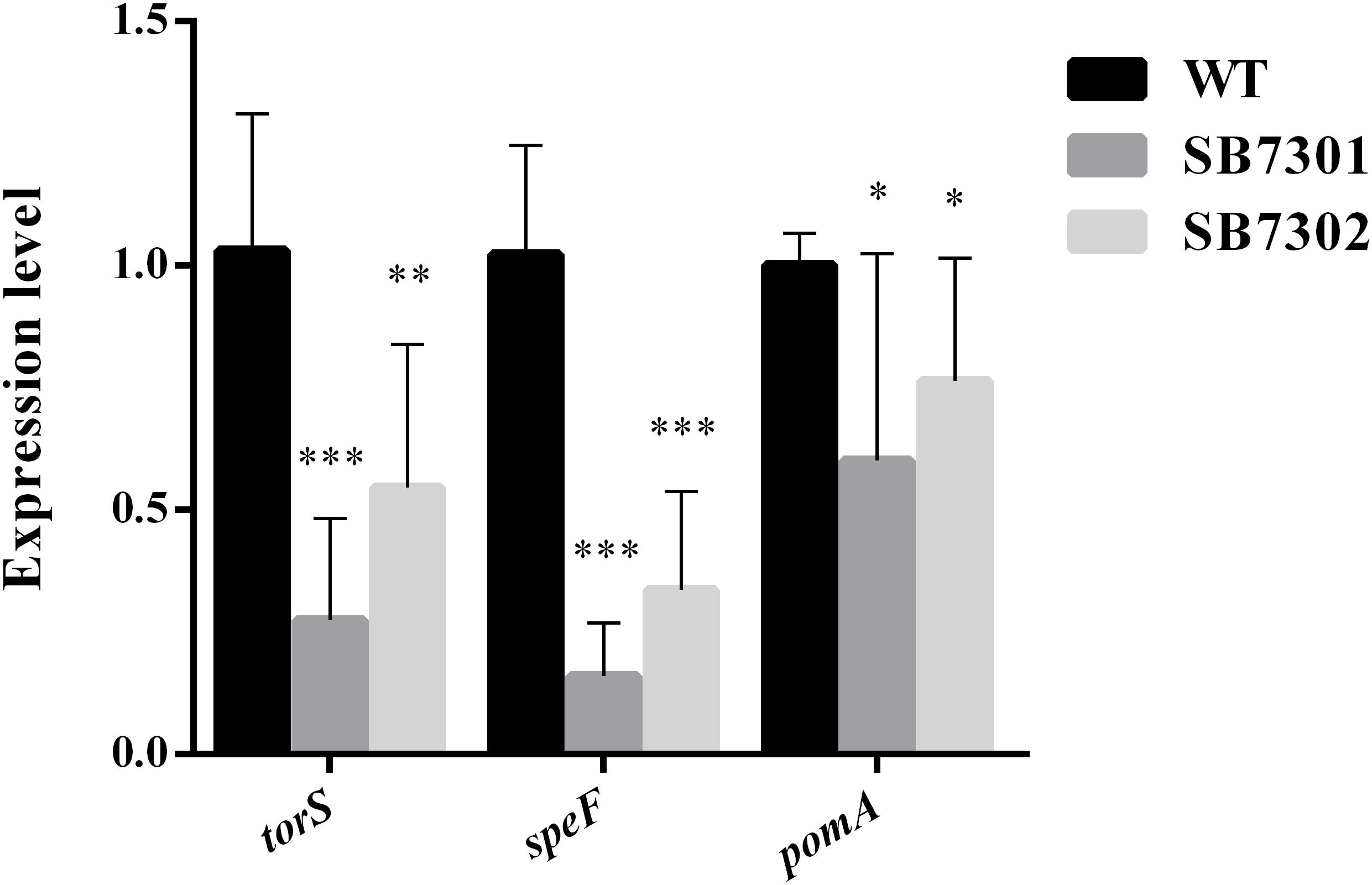
Figure 7. Transcription verification in gene deletion mutants. The expression level of torS, speF, and pomA genes in wild-type strain, SB7301 and SB7302 mutants were quantified by qRT-PCR. Data was presented as the mean ± standard deviation (n = 3, ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001).
LuxR-type receptors as cytoplasmic transcription factors together with AHLs synthesized by cognate LuxI-type synthases are commonly used in Gram-negative bacteria and have been intensively studied (Kievit and Iglewski, 2000; Reading and Sperandio, 2006; Papenfort and Bassler, 2016). In this study, three DKPs (PP, PL, and LL) were detected predominant in cell-free culture fluids of S. baltica OS155 as well as spoiled large yellow croaker, moreover, we failed to detect any AHLs in pure culture of S. baltica OS155, which was largely consistent with previous studies (Gu et al., 2013; Zhu et al., 2015, 2016). According to a recent survey presented by Hudaiberdiev et al. (2015), an unexpectedly high number of annotated LuxR proteins in NCBI databases were found to be orphans lacking luxI genes in their vicinity on the chromosome, and thus it was speculated that new signaling molecules rather than AHLs may interact with orphan LuxR proteins (Hudaiberdiev et al., 2015). On the basis of these supporting data described above, our work took the interaction between DKPs and orphan LuxR proteins as a focal point to explore QS system and concomitantly the underlying mechanism of how QS regulates the spoilage potential and biofilm formation in S. baltica OS155.
Based on the cell density-dependent principle, we demonstrated that the orphan luxR-type genes (luxR01, luxR02, luxR03, luxR05, and luxR06) were upregulated in different degrees by postexponential supernatant extract of S. baltica OS155 with statistical significance, indicating their involvement in QS system preliminarily. We found that PP could interact with orphan LuxR01 and LuxR02 proteins directly, and contrastively other LuxR-type proteins employed in our study were failed to bind any hypothetical autoinducers employed. However, their cell density-dependent expression suggests the existence of other autoinducers in S. baltica OS155. Moreover, results showed that only SB7301 and SB7302 mutants had defects in all the tested spoilage indexes simultaneously, and SB7307 mutant had a more significant reduction in spoilage potential. Since the reduction of TMA and putrescine production of SB7301 and SB7302 mutants was not so large when using 10 mM and 0.5% precursors, a higher concentration of substrates was used for further analysis. The larger reduction of TMA and putrescine production in SB7301 and SB7302 mutants cultured in higher concentration of precursors indicated that limited the reaction as well as the generation of product is substrates rather than the enzymes with low concentration of substrates. Combined with our in vitro binding assay results, we propose that LuxR01 and LuxR02 proteins were DKP-dependent QS receptors as well as key biofilm regulators, and putrescine production were also dual-regulated by LuxR01 and LuxR02. However, the specific ratio of LuxR01 and LuxR02 in regulating these phenotypes and the function of other LuxRs tested here need further study. In addition, according to the conserved domain of LuxRs (Supplementary Figure S1), LuxR02 has a response regulator domain which may receive the signal from the sensor partner in bacterial two-component systems like the canonical QS systems in Vibrio harveyi and V. cholera (Papenfort and Bassler, 2016), indicating that LuxR02 may participate in a two-component QS system with novel extracellular signal compounds and regulate corresponding gene expressions in S. baltica OS155. Further work should be done to reveal the specific roles of other orphan LuxR-type proteins and whether LuxR01 interact with LuxR02 to regulate these phenotypes. The proposed model was further confirmed via exogenous addition of synthetic PP to wild-type, SB7301 and SB7302 mutant strains. Results exhibited different degrees of increase in TMA, putrescine production and biofilm formation in wild-type strain rather than the two mutants, indicating that the interaction between PP and LuxR01 as well as LuxR02 receptors was involved in controlling spoilage potential and biofilm formation in S. baltica OS155. On the contrary, two previous reports demonstrated that exposure to exogenous PL but not PP promoted spoilage-related metabolites and biofilm formation in S. baltica (Zhu et al., 2015, 2016). This contradiction may result from the different strains we used. Up to now, there have been 10 genomes of S. baltica species in NCBI database1, the difference between these strains is listed in Supplementary Data Set S2. The strain used in our study is S. baltica OS155 which shows 98% identity and 87% query cover in genome with the representative strain S. baltica OS678. On the other hand, the concentration of DKPs they used (100 μM) was an order of magnitude larger than that of DKPs used here (10 μM), which was chosen our experiments to better mimic in vivo environment based on the quantification of PP in S. baltica OS155 (Table 1). It is possible that different DKPs have different optimum concentrations, and a higher concentration may have some side effects, e.g., the optimal concentration range for their activity will be revealed by further study. Although different strains may employ different mechanisms, all of these results indicate a role of DKPs in adjusting physiological behaviors such as spoilage potential and biofilm formation.
The reduction of trimethylamine oxide (TMAO) into TMA regulated by the torCAD operon is only induced in the presence of TMAO (Jourlin et al., 1996). The tor operon is regulated by its upstream genes, torS, torT, and torR, and disruption of each one can influence tor operon induction. The torS and torR genes encode a two-component regulatory system in which TorS is an unorthodox sensor and TorR is a response regulator (Jourlin et al., 1996). TorT interacts with TMAO upon its appearance to form the TorT-TMAO complex which further interacts with the TorS sensor, then TorS undergoes a conformational change to act as a kinase (Baraquet et al., 2006), which transphosphorylates TorR via a four-step phosphorelay (Jourlin et al., 1997). Once phosphorylated, TorR activates the expression of tor operon (Simon et al., 1995). In this study, we demonstrated that torS as a signal-transduction cascade mediator was transcriptionally regulated by LuxR-type receptors LuxR01 and LuxR02, which provides evidence connecting QS with spoilage-related gene. It is noteworthy that torS rather than torA was regulated by QS system, the indirect regulation of TorA amplified the QS signal and ensured the precise expression of TorA only at the presence of TMAO, avoiding the waste of energy when TMAO is absent.
The function of speF was well studied in E. coli, the speF-potE operon encoding an inducible ornithine decarboxylase and an ornithine/putrescine antiporter is induced at acidic pH, the ornithine decarboxylase activity was observed in the cells cultured at pH 5.2, but not at pH 7.0 (Kashiwagi et al., 1991). Since the internal pH was close to 7.0 in unstressed cells (Wilks and Slonczewski, 2007), E. coli utilized multiple mechanisms to ensure that SpeF remained inactive to reduce the consumption of amino acids around neutral pH, while constitutive ornithine decarboxylase was present (Kanjee et al., 2011). Our study showed that speF gene was significantly downregulated in SB7301 and SB7302 mutants as compared to wild-type strain. The regulation of the inducible ornithine decarboxylase but not the constitutive ornithine decarboxylase by LuxR-type proteins allowed conclusion to be drawn that QS controlled the putrescine production of S. baltica via consumption of amino acids in a well-organized and energy-saving way. However, it remains unknown whether the QS-dependent regulation of SpeF expression is in parallel with the acid stress-dependent regulation.
Flagella are supposed to play a key role in the initial steps of adhesion of bacteria to surfaces and biofilm formation (Kogure and Ikemoto, 1998; McCarter, 2001). Flagella rotation are powered by flagellar motor which is responsible for relaying the transmembrane proton or sodium ion gradient into energy (Hirota and Imae, 1983; Blair, 2003). Two cytoplasmic membrane proteins, MotA and MotB or PomA and PomB, form a complex at a molar ratio of 2: 1 as the stator of motor to conduct ions and generate the torque for rotation (Sato and Homma, 2000; McCarter, 2001; Berg, 2003; Kojima and Blair, 2004). In this study, the pomA gene was significantly downregulated in SB7301 and SB7302 mutants. PomA is homologous to MotA in E. coli and Salmonella, together with PomB to form the stator of the rotary flagellar machine. It was reported the interruption of the pomA but not motA gene yielded a non-swimming mutant and affected progression of biofilm development into pronounced three-dimensional architecture in S. oneidensis MR-1, and thus it was suggested that rotation of the polar flagellum was sodium dependent in analogy to V. parahaemolyticus where pomA/B potentially used a sodium gradient as energy source (McCarter, 2001; Thormann et al., 2004). In combination with exhibited biofilm phenotypes in SB7301 and SB7302 mutants, these results led to a model that LuxR-type proteins affected biofilm formation via regulating the stator component of flagellar motor PomA potentially using a sodium gradient. However, the specific relationship between biofilm formation and spoilage potential still remains unknown.
Overall, we propose a model for the mechanism of spoilage activity regulation in S. baltica OS155 (Figure 8). During the storage of large yellow croaker, the SSO S. baltica OS155 produces autoinducer molecule PP that are recognized by QS receptors LuxR01 and LuxR02, resulting in the modulation of TMA, putrescine production and biofilm formation via transcriptional regulation of torS, speF and pomA. This study provided evidence ascribing DKPs to QS autoinducers which can interact directly with orphan LuxR-type proteins, and improved our understanding of the mechanisms involved in seafood spoilage, offering new insights into improving seafood preservation based on the QS-dependent spoilage of SSO.
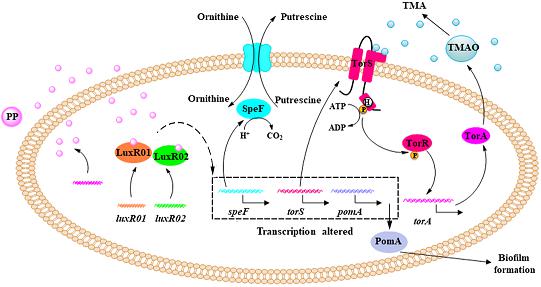
Figure 8. Scheme representing the QS system in regulating spoilage potential and biofilm formation in S. baltica OS155. At high cell density, QS signal PP reaches a threshold level and is detected by specific receptor LuxR01 and/or LuxR02 protein, which acts as transcription regulator to alter the expression of torS, speF, and pomA. Among them, TorS functions as a signal-transduction cascade mediator and transphosphorylates TorR upon the presence of TMAO, which finally activate torA expression to reduce TMAO into TMA. speF gene is responsible for the production of putrescine from ornithine. pomA gene promotes the biofilm formation by regulating swimming motility.
YW and LF designed the research. FW and CW analyzed the data. YW, FW, and CW performed the research. FW, CW, and LF wrote the manuscript. XL contributed to new reagents or analytic tools. YW developed the software necessary to perform and record experiments.
This study was financially supported by the Zhejiang Provincial Natural Science Foundation of China (Grant Number LZ15C200001), the National Natural Science Foundation of China (Grant Numbers 31571913 and 31772050), and the open funding of Beijing Advanced Innovation Center for Food Nutrition and Human Health, Beijing Technology and Business University (BTBU).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00135/full#supplementary-material
FIGURE S1. The conserved domains in provisional LuxR-type proteins of Shewanella baltica OS155. CitB: DNA-binding response regulator, NarL/FixJ family, contains REC and HTH domains. Response_reg: response regulator receiver domain, this domain receives the signal from the sensor partner in bacterial two-component systems. It is usually found N-terminal to a DNA binding effector domain; HTH_LUXR: helix_turn_helix, Lux Regulon.
FIGURE S2. PCR verification of gene deletion mutants of S. baltica OS155. Deletion mutants (SB7301, SB7302, SB7303, SB7304, SB7305, and SB7306) were screened by PCR amplification using the outside primers (5′-O and 3′-O). W, wide-type strain; D, deletion mutant.
FIGURE S3. Identification and quantification of hypothetical autoinducers in S. baltica OS155 and spoiled large yellow croaker. AHLs and DKPs in S. baltica OS155 (A) and spoiled large yellow croaker (B) were measured by UHPLC-MS/MS. C4-HSL, N-butanoylhomoserine lactone; C6-HSL, N-hexanoylhomoserine lactone; PL, cyclo-(L-Pro-L-Leu); LL, cyclo-(L-Leu-L-Leu); PP, cyclo-(L-Pro-L-Phe).
FIGURE S4. Purified recombinant LuxR-type proteins. FLAG-tagged LuxR-type proteins (LuxR01–LuxR06) were expressed in E. coli BL21 (DE3) and purified by FLAG M2 beads. Empty plasmid pET-15b was used as a control. An aliquot of the purified proteins were qualified by SDS-PAGE with 12% (left) or 15% (right) gels. ∗ M2 antibody light chain.
FIGURE S5. Spoilage potential of the double mutant. The TMA (A) and putrescine (B) production of wild-type and the double mutant SB7307 strain were measured. Data was presented as the mean ± standard deviation (n = 3, ∗∗p < 0.01, ∗∗∗p < 0.001).
FIGURE S6. Kyoto encyclopedia of genes and genomes (KEGG) enrichment of differentially expressed genes in SB7301 mutant. The 335 differentially expressed genes are involved in amino acid metabolism, lipid metabolism, carbohydrate metabolism, energy metabolism, signal transduction, cell motility and membrane transport.
FIGURE S7. The cluster analysis diagram of differentially expressed genes belonging to two-component system and arginine and proline metabolism system in SB7301 mutant. The differential expression profile of genes involved in two-component system and arginine and proline metabolism system was summarized and presented in colored mosaic matrix. SBAL_RS16580: torS, SBAL_RS21090: speF, SBAL_RS06905: pomA.
TABLE S1. Bacterial strains and plasmids used in this study.
TABLE S2. Identification of target genes involved in this study in S. baltica OS155.
TABLE S3. LC gradient.
TABLE S4. Target genes and primers used in this study.
DATA SET S1. Annotation results of 335 differentially expressed genes.
DATA SET S2. The list of 10 strains of S. baltica species.
3-oxo-C10-HSL, N-(3-oxodecanoyl)-homoserine lactone; 3-oxo-C12-HSL, N-(3-oxododecanoyl)-homoserine lactone; 3-oxo-C14-HSL, N-(3-oxotetradecanoyl)-homoserine lactone; 3-oxo-C8-HSL, N-(3-oxooctanoyl)-homoserine lactone; AHLs, N-acyl -homoserine lactones; BAs, biogenic amines; C10-HSL, N-decanoylhomoserine lactone; C12-HSL, N-dodecanoylhomoserine lactone; C14-HSL, N-tetradecanoylhomoserine lactone; C4-HSL, N-butanoylhomoserine lactone; C6-HSL, N-hexanoylhomoserine lactone; C8-HSL, N-octanoylhomoserine lactone; CLSM, confocal laser scanning microscopy; DAP, 2, 6-diaminopimelic acid; DKPs, diketopiperazines; Gm, gentamycin; IPTG, isopropyl-β-D-thiogalactoside; LL, cyclo-(L-Leu-L-Leu); PL, cyclo-(L-Pro-L-Leu); PP, cyclo-(L-Pro-L-Phe); QS, quorum sensing; SSOs, specific spoilage organisms; TMA, trimethylamine; TMAO, trimethylamine oxide.
Bai, A. J., and Rai Vittal, R. (2014). Quorum sensing regulation and inhibition of exoenzyme production and biofilm formation in the food spoilage bacteria Pseudomonas psychrophila PSPF19. Food Biotechnol. 28, 293–308. doi: 10.1080/08905436.2014.963601
Balaban, N., and Novick, R. P. (1995). Autocrine regulation of toxin synthesis by Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 92, 1619–1623. doi: 10.1073/pnas.92.5.1619
Baraquet, C., Théraulaz, L., Guiral, M., Lafitte, D., Méjean, V., and Jourlin-Castelli, C. (2006). TorT, a member of a new periplasmic binding protein family, triggers induction of the tor respiratory system upon trimethylamineN-oxide electron-acceptor binding in Escherichia coli. J. Biol. Chem. 281, 38189–38199. doi: 10.1074/jbc.M604321200
Bassler, B. L., and Losick, R. (2006). Bacterially speaking. Cell 125, 237–246. doi: 10.1016/j.cell.2006.04.001
Ben-Gigirey, B., Vieites Baptista de Sousa, J. M., Villa, T. G., and Barros-Velazquez, J. (1998). Changes in biogenic amines and microbiological analysis in Albacore (Thunnus alalunga) muscle during frozen storage. J. Food Prot. 61, 608–615. doi: 10.4315/0362-028X-61.5.608
Berg, H. C. (2003). The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72, 19–54. doi: 10.1146/annurev.biochem.72.121801.161737
Blair, D. F. (2003). Flagellar movement driven by proton translocation. FEBS Lett. 545, 86–95. doi: 10.1016/S0014-5793(03)00397-1
Brizzard, B. L., Chubet, R. G., and Vizard, D. L. (1994). Immunoaffinity purification of FLAG epitope-tagged bacterial alkaline phosphatase using a novel monoclonal antibody and peptide elution. Biotechniques 16, 730–735.
Buch, C., Sigh, J., Nielsen, J., Larsen, J. L., and Gram, L. (2003). Production of acylated homoserine lactones by different serotypes of Vibrio anguillarum, both in culture and during infection of rainbow trout. Syst. Appl. Microbiol. 26, 338–349. doi: 10.1078/072320203322497365
Burr, T., Barnard, A. M. L., Corbett, M. J., Pemberton, C. L., Simpson, N. J. L., and Salmond, G. P. C. (2006). Identification of the central quorum sensing regulator of virulence in the enteric phytopathogen, Erwinia carotovora: the VirR repressor. Mol. Microbiol. 59, 1891–1891. doi: 10.1111/j.1365-2958.2006.05052.x
Campbell, J., Lin, Q., Geske, G. D., and Blackwell, H. E. (2009). New and unexpected insights into the modulation of LuxR-type quorum sensing by cyclic dipeptides. ACS Chem. Biol. 4, 1051–1059. doi: 10.1021/cb900165y
Chinivasagam, H. N., Bremner, H. A., Wood, A. F., and Nottingham, S. M. (1998). Volatile components associated with bacterial spoilage of tropical prawns. Int. J. Food Microbiol. 42, 45–55. doi: 10.1016/S0168-1605(98)00057-9
Christensen, A. B., Riedel, K., Eberl, L., Flodgaard, L. R., Molin, S., Gram, L., et al. (2003). Quorum-sensing-directed protein expression in Serratia proteamaculans B5a. Microbiology 149, 471–483. doi: 10.1099/mic.0.25575-0
Costerton, J. W., Stewart, P. S., and Greenberg, E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322. doi: 10.1126/science.284.5418.1318
Davies, D. G., Parsek, M. R., Pearson, J. P., Iglewski, B. H., Costerton, J. W., and Greenberg, E. P. (1998). The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280, 295–298. doi: 10.1126/science.280.5361.295
Degrassi, G., Aguilar, C., Bosco, M., Zahariev, S., Pongor, S., and Venturi, V. (2002). Plant growth-promoting Pseudomonas putida WCS358 produces and secretes four cyclic dipeptides: cross-talk with quorum sensing bacterial sensors. Curr. Microbiol. 45, 250–254. doi: 10.1007/s00284-002-3704-y
Djordjevic, D., Wiedmann, M., and McLandsborough, L. A. (2002). Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl. Environ. Microbiol. 68, 2950–2958. doi: 10.1128/AEM.68.6.2950-2958.2002
Duan, K., Dammel, C., Stein, J., Rabin, H., and Surette, M. G. (2003). Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol. Microbiol. 50, 1477–1491. doi: 10.1046/j.1365-2958.2003.03803.x
Dyer, W. J. (1945). Amines in fish muscle: I. Colorimetric determination of trimethylamine. J. Fish Res. Board Can. 6, 351–358. doi: 10.1139/f42-042
Francoise, L. (2010). Occurrence and role of lactic acid bacteria in seafood products. Food Microbiol. 27, 698–709. doi: 10.1016/j.fm.2010.05.016
Fuqua, W. C., Winans, S. C., and Greenberg, E. P. (1994). Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176, 269–275. doi: 10.1128/jb.176.2.269-275.1994
Gao, W., Liu, Y., Giometti, C. S., Tollaksen, S. L., Khare, T., Wu, L., et al. (2006). Knock-out of SO1377 gene, which encodes the member of a conserved hypothetical bacterial protein family COG2268, results in alteration of iron metabolism, increased spontaneous mutation and hydrogen peroxide sensitivity in Shewanella oneidensis MR-1. BMC Genomics 7:76. doi: 10.1186/1471-2164-7-76
Gu, Q., Fu, L., Wang, Y., and Lin, J. (2013). Identification and characterization of extracellular cyclic dipeptides as quorum-sensing signal molecules from Shewanella baltica, the specific spoilage organism of Pseudosciaena crocea during 4 degrees C storage. J. Agric. Food Chem. 61, 11645–11652. doi: 10.1021/jf403918x
Guilbaud, M., Piveteau, P., Desvaux, M., Brisse, S., and Briandet, R. (2015). Exploring the diversity of Listeria monocytogenes biofilm architecture by high-throughput confocal laser scanning microscopy and the predominance of the honeycomb-like morphotype. Appl. Environ. Microbiol. 81, 1813–1819. doi: 10.1128/AEM.03173-14
Hall-Stoodley, L., Costerton, J. W., and Stoodley, P. (2004). Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108. doi: 10.1038/nrmicro821
Hirota, N., and Imae, Y. (1983). Na+-driven flagellar motors of an alkalophilic Bacillus strain YN-1. J. Biol. Chem. 258, 10577–10581.
Holden, M. T., Ram, C. S., De, N. R., Stead, P., Bainton, N. J., Hill, P. J., et al. (1999). Quorum-sensing cross talk: isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other gram-negative bacteria. Mol. Microbiol. 33, 1254–1266. doi: 10.1046/j.1365-2958.1999.01577.x
Hozbor, M. C., Saiz, A. I., Yeannes, M. I., and Fritz, R. (2006). Microbiological changes and its correlation with quality indices during aerobic iced storage of sea salmon (Pseudopercis semifasciata). LWT Food Sci. Technol. 39, 99–104. doi: 10.1016/j.lwt.2004.12.008
Hudaiberdiev, S., Choudhary, K. S., Vera Alvarez, R., Gelencser, Z., Ligeti, B., Lamba, D., et al. (2015). Census of solo LuxR genes in prokaryotic genomes. Front. Cell. Infect. Microbiol. 5:20. doi: 10.3389/fcimb.2015.00020
Jin, M., Jiang, Y., Sun, L., Yin, J., Fu, H., Wu, G., et al. (2013). Unique organizational and functional features of the cytochrome c maturation system in Shewanella oneidensis. PLoS One 8:e75610. doi: 10.1371/journal.pone.0075610
Jourlin, C., Ansaldi, M., and Méjean, V. (1997). Transphosphorylation of the torr response regulator requires the three phosphorylation sites of the tors unorthodox sensor in Escherichia coli. J. Mol. Biol. 267, 770–777. doi: 10.1006/jmbi.1997.0919
Jourlin, C., Bengrine, A., Chippaux, M., and Méjean, V. (1996). An unorthodox sensor protein (tors) mediates the induction of the tor structural genes in response to trimethylamine n-oxide in Escherichia coli. Mol. Microbiol. 20, 1297–1306. doi: 10.1111/j.1365-2958.1996.tb02648.x
Kanjee, U., Gutsche, I., Ramachandran, S., and Houry, W. A. (2011). The enzymatic activities of the Escherichia coli basic aliphatic amino acid decarboxylases exhibit a ph zone of inhibition. Biochemistry 50, 9388–9398. doi: 10.1021/bi201161k
Kashiwagi, K., Suzuki, T., Suzuki, F., Furuchi, T., Kobayashi, H., and Igarashi, K. (1991). Coexistence of the genes for putrescine transport protein and ornithine decarboxylase at 16 min on Escherichia coli chromosome. J. Biol. Chem. 266, 20922–20927.
Kievit, T. R. D., and Iglewski, B. H. (2000). Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68, 4839–4849. doi: 10.1128/IAI.68.9.4839-4849.2000
Kim, M.-K., Mah, J.-H., and Hwang, H.-J. (2009). Biogenic amine formation and bacterial contribution in fish, squid and shellfish. Food Chem. 116, 87–95. doi: 10.1016/j.foodchem.2009.02.010
Kleber-Janke, T., and Becker, W. M. (2000). Use of modified BL21(DE3) Escherichia coli cells for high-level expression of recombinant peanut allergens affected by poor codon usage. Protein Exp. Purif. 19, 419–424. doi: 10.1006/prep.2000.1265
Klose, K. E. (2006). Increased chatter: cyclic dipeptides as molecules of chemical communication in Vibrio spp. J. Bacteriol. 188, 2025–2026. doi: 10.1128/JB.188.6.2025-2026.2006
Kogure, K., and Ikemoto, E. H. (1998). Attachment of vibrio alginolyticus to glass surfaces is dependent on swimming speed. J. Bacteriol. 180, 932–937.
Kojima, S., and Blair, D. F. (2004). The bacterial flagellar motor: structure and function of a complex molecular machine. Int. Rev. Cytol. 233, 93–134. doi: 10.1016/S0074-7696(04)33003-2
Li, Y. H., Tang, N., Aspiras, M. B., Lau, P. C. Y., Lee, J. H., Ellen, R. P., et al. (2002). A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184, 2699–2708. doi: 10.1128/JB.184.10.2699-2708.2002
McCarter, L. L. (2001). Polar flagellar motility of the vibrionaceae. Microbiol. Mol. Biol. Rev. 65, 445–462.
Özogul, F., Polat, A., and Özogul, Y. (2004). The effects of modified atmosphere packaging and vacuum packaging on chemical, sensory and microbiological changes of sardines (Sardina pilchardus). Food Chem. 85, 49–57. doi: 10.1128/MMBR.65.3.445-462.2001
Pantazi, D., Papavergou, A., Pournis, N., Kontominas, M. G., and Savvaidis, I. N. (2008). Shelf-life of chilled fresh Mediterranean swordfish (Xiphias gladius) stored under various packaging conditions: microbiological, biochemical and sensory attributes. Food Microbiol. 25, 136–143. doi: 10.1016/j.foodchem.2003.05.006
Papenfort, K., and Bassler, B. L. (2016). Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588. doi: 10.1038/nrmicro.2016.89
Park, D. K., Lee, K. E., Baek, C. H., Kim, I. H., Kwon, J. H., Lee, W. K., et al. (2006). Cyclo(Phe-Pro) modulates the expression of ompU in Vibrio spp. J. Bacteriol. 188, 2214–2221. doi: 10.1128/JB.188.6.2214-2221.2006
Parlapani, F. F., Haroutounian, S. A., Nychas, G. J., and Boziaris, I. S. (2015). Microbiological spoilage and volatiles production of gutted European sea bass stored under air and commercial modified atmosphere package at 2 degrees C. Food Microbiol. 50, 44–53. doi: 10.1016/j.fm.2015.03.006
Pinto, U. M., de Souza Viana, E., Martins, M. L., and Vanetti, M. C. D. (2007). Detection of acylated homoserine lactones in gram-negative proteolytic psychrotrophic bacteria isolated from cooled raw milk. Food Control 18, 1322–1327. doi: 10.1016/j.foodcont.2006.09.005
Rasch, M., Andersen, J. B., Nielsen, K. F., Flodgaard, L. R., Christensen, H., Givskov, M., et al. (2005). Involvement of bacterial quorum-sensing signals in spoilage of bean sprouts. Appl. Environ. Microbiol. 71, 3321–3330. doi: 10.1128/AEM.71.6.3321-3330.2005
Reading, N. C., and Sperandio, V. (2006). Quorum sensing: the many languages of bacteria. FEMS Microbiol. Lett. 254, 1–11. doi: 10.1111/j.1574-6968.2005.00001.x
Robson, A. A., Kelly, M. S., and Latchford, J. W. (2007). Effect of temperature on the spoilage rate of whole, unprocessed crabs: Carcinus maenas, Necora puber and Cancer pagurus. Food Microbiol. 24, 419–424. doi: 10.1016/j.fm.2006.07.016
Ruffin, M., Bilodeau, C., Maille, E., LaFayette, S. L., McKay, G. A., Trinh, N. T., et al. (2016). Quorum-sensing inhibition abrogates the deleterious impact of Pseudomonas aeruginosa on airway epithelial repair. FASEB J. 30, 3011–3025. doi: 10.1096/fj.201500166R
Sato, K., and Homma, M. (2000). Functional reconstitution of the Na+-driven polar flagellar motor component of Vibrio alginolyticus. J. Biol. Chem. 275, 5718–5722. doi: 10.1074/jbc.275.8.5718
Schauder, S., and Bassler, B. L. (2001). The languages of bacteria. Genes Dev. 15:1468. doi: 10.1101/gad.899601
Shiner, E. K., Rumbaugh, K. P., and Williams, S. C. (2005). Inter-kingdom signaling: deciphering the language of acyl homoserine lactones. FEMS Microbiol. Rev. 29, 935–947. doi: 10.1016/j.femsre.2005.03.001
Simon, G., Jourlin, C., Ansaldi, M., Pascal, M. C., Chippaux, M., and Méjean, V. (1995). Binding of the torr regulator to cis-acting direct repeats activates tor operon expression. Mol. Microbiol. 17, 971–980. doi: 10.1111/j.1365-2958.1995.mmi_17050971.x
Smith, J. L., Fratamico, P. M., and Novak, J. S. (2004). Quorum sensing: a primer for food microbiologists. J. Food Prot. 67, 1053–1070. doi: 10.4315/0362-028X-67.5.1053
Taga, M. E., Semmelhack, J. L., and Bassler, B. L. (2001). The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42, 777–793. doi: 10.1046/j.1365-2958.2001.02669.x
Thormann, K. M., Saville, R. M., Shukla, S., Pelletier, D. A., and Spormann, A. M. (2004). Initial Phases of biofilm formation in Shewanella oneidensis MR-1. J. Bacteriol. 186, 8096–8104. doi: 10.1128/JB.186.23.8096-8104.2004
Vidal, J. E., Ludewick, H. P., Kunkel, R. M., Zahner, D., and Klugman, K. P. (2011). The LuxS-dependent quorum-sensing system regulates early biofilm formation by Streptococcus pneumoniae strain D39. Infect. Immun. 79, 4050–4060. doi: 10.1128/IAI.05186-11
Wang, Y., Zhang, X., Wang, C., Fu, L., Yi, Y., and Zhang, Y. (2017). Identification and quantification of acylated homoserine lactones in Shewanella baltica, the specific spoilage organism of Pseudosciaena crocea by ultra-high-performance liquid chromatography coupled to triple quadrupole mass spectrometry. J. Agric. Food Chem. 65, 4804–4810. doi: 10.1021/acs.jafc.7b01209
Waters, C. M., and Bassler, B. L. (2005). Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346. doi: 10.1146/annurev.cellbio.21.012704.131001
Whitehead, N. A., Barnard, A. M., Slater, H., Simpson, N. J., and Salmond, G. P. (2001). Quorum-sensing in gram-negative bacteria. FEMS Microbiol. Rev. 25, 365–404. doi: 10.1111/j.1574-6976.2001.tb00583.x
Wilks, J. C., and Slonczewski, J. L. (2007). pH of the cytoplasm and periplasm of Escherichia coli: rapid measurement by green fluorescent protein fluorimetry. J. Bacteriol. 189, 5601–5607. doi: 10.1128/JB.00615-07
Wu, B., Capilato, J., Pham, M. P., Walker, J., Spur, B., Rodriguez, A., et al. (2016). Lipoxin A4 augments host defense in sepsis and reduces Pseudomonas aeruginosa virulence through quorum sensing inhibition. FASEB J. 30, 2400–2410. doi: 10.1096/fj.201500029R
Yarwood, J. M., Bartels, D. J., Volper, E. M., and Greenberg, E. P. (2004). Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 186, 1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004
Zhu, J., Zhao, A., Feng, L., and Gao, H. (2016). Quorum sensing signals affect spoilage of refrigerated large yellow croaker (Pseudosciaena crocea) by Shewanella baltica. Int. J. Food Microbiol. 217, 146–155. doi: 10.1016/j.ijfoodmicro.2015.10.020
Keywords: quorum sensing, diketopiperazines, orphan LuxR-type proteins, spoilage, biofilm formation
Citation: Wang Y, Wang F, Wang C, Li X and Fu L (2019) Positive Regulation of Spoilage Potential and Biofilm Formation in Shewanella baltica OS155 via Quorum Sensing System Composed of DKP and Orphan LuxRs. Front. Microbiol. 10:135. doi: 10.3389/fmicb.2019.00135
Received: 25 April 2018; Accepted: 21 January 2019;
Published: 05 February 2019.
Edited by:
Aldo Corsetti, Università degli Studi di Teramo, ItalyReviewed by:
Yang Zhu, Netherlands Organisation for Applied Scientific Research (TNO), NetherlandsCopyright © 2019 Wang, Wang, Wang, Li and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linglin Fu, ZnVsaW5nbGluQG1haWwuempnc3UuZWR1LmNu; ZnVsbDExMDNAaG90bWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.