- 1Environment and Life Sciences Research Center, Desert Agriculture and Ecosystems Program, Kuwait Institute for Scientific Research, Kuwait City, Kuwait
- 2ARC Centre for Mine Site Restoration, Department of Environment and Agriculture, Curtin University, Perth, WA, Australia
- 3School of Biological Sciences, The University of Western Australia, Perth, WA, Australia
This research examined the general soil fungi and AM fungal communities associated with a Lonely Tree species (Vachellia pachyceras) existing in the Sabah Al-Ahmad Natural Reserve located at the Kuwait desert. The goals of the study were to describe the general fungal and AM fungal communities present in the rhizospheric, non-rhizospheric soils and roots of V. pachyceras, respectively, as well as local and non-local V. pachyceras seedlings when grown under standard nursery growing environments. Soil and root samples were analyzed for an array of characteristics including soil physicochemical composition, and culture-independent method termed PCR-cloning, intermediate variable region of rDNA, the large subunit (LSU) and internal transcribed spacer (ITS) region sequence identifications. The results reveal that the fungal phylotypes were classified in four major fungal phyla namely Ascomycota, Basidiomycota, Chytridiomycota, and Zygomycota. The largest assemblage of fungal analyses showed communities dominated by members of the phylum Ascomycota. The assays also revealed a wealth of incertae sedis fungi, mostly affiliated to uncultured fungi from diverse environmental conditions. Striking difference between rhizosphere and bulk soils communities, with more fungal diversities and Operational Taxonomic Units (OTUs) richness associated with both the field and nursery rhizosphere soils. In contrast, a less diverse fungal community was found in the bulk soil samples. The characterization of AM fungi from the root system demonstrated that the most abundant and diversified group belongs to the family Glomeraceae, with the common genus Rhizophagus (5 phylotypes) and another unclassified taxonomic group (5 phylotypes). Despite the harsh climate that prevails in the Kuwait desert, studied roots displayed the existence of considerable number of AM fungal biota. The present work thus provides a baseline of the fungal and mycorrhizal community associated with rhizosphere and non-rhizosphere soils and roots of only surviving V. pachyceras tree from the Kuwaiti desert and seedlings under nursery growing environments.
Introduction
Kuwait is located in the northeastern corner of the Arabian Desert where it constitutes a part of the northwestern coastal flat of the Arabian Gulf. This arid region severely suffered in recent years from prolonged drought (Khalaf, 1989; Brown and Porembski, 1997; El-Sheikh et al., 2010). This has ultimately steered a decline in soil and plant productivity (El-Sheikh et al., 2010), leading to further desertification. Furthermore, increases in anthropogenic activities have exacerbated these processes and under incessant risk of desertification (Wang et al., 2006). Consequently, about 90% of Kuwait’s territory (an area of approximately 18,000 km2) is occupied by the desert (Kaitharan, 2013). Such a degraded soil situation needs to be reversed to its original state to conserve key native plant species and this may include a series of interventions aimed to sustain soil stability and productivity.
In Kuwait, Acacia gerrardii (Syn Vachellia gerrardii) is considered the only native tree species existing in the desert ecosystem (Boulos and Al-Dosari, 1994). A. pachyceras O. Schwartz, synonym A. gerrardii Benth., subsequently referred to as A. gerrardii, commonly known as “Lonely Tree (LT).” A. gerrardii is considered the only native tree species existing in the Kuwait desert ecosystem (Boulos and Al-Dosari, 1994) and is only available in Sabah Al-Ahmad Natural Reserve (a 320 km2 protected area formerly acclaimed as Kuwait’s first National Park) where it is as key-stone species (Boulos and Al-Dosari, 1994; Kaitharan, 2013). It belongs to the Acacia genus, one of the largest genera of leguminous tree and shrubs that has a wide distribution throughout the world (Pedley, 1986; Sene and Sylla, 2014). Recently this iconic tree is genetically identified as V. pachyceras based on multi-locus plastid gene sequences (Suleiman et al., 2018). Many members of this genus are recognized as species that are useful for re-vegetation of water-stressed and low-nutrient soil environments (Sene et al., 2012b; Sene and Sylla, 2014). V. pachyceras was, however, damaged by the gulf war in 1990 and, the remaining plants patchiness are under the threat of extinction (Kaitharan, 2013). Conservation efforts are therefore required to make this species’ survival in Kuwait’s terrestrial ecosystem and ensuring sustainability and preserving soil biological attributes.
There is increasing evidence that trees can play a key role in ecosystem rehabilitation or restoration (Donfack et al., 1995; Vincke et al., 2010; Sene and Sylla, 2014). Many studies have supported that the presence of trees provide a number of ecological advantages from increased soil organic matter content (Hobbie et al., 2006, 2007; Peichl et al., 2006; Rivest et al., 2009; Sene et al., 2012a, 2013; Sene and Sylla, 2014), to biodiversity conservation (Akpo et al., 2003; Grouzis and Akpo, 2006; Sene et al., 2012a; Hortal et al., 2013), and improved soil microbial activity and nutrient cycling rates (Hobbie et al., 2006; Sene et al., 2013). In addition, trees in desert regions often represent “fertility islands” for many species (Rodríguez-Zaragoza et al., 2008; Kavamura et al., 2013), which in turn influence long-term vegetation dynamics and ecosystem processes. Moreover, they have a role in combating land degradation through stabilizing soil surfaces by preventing soil erosion and in facilitating plant recruitment and survivorship (El-Sheikh et al., 2010; Sene et al., 2012b, 2013; Kavamura et al., 2013; Sene and Sylla, 2014).
Microbial associations have been pointed as an important strategy to guarantee plant growth and survival (van der Heijden et al., 1998, 2008; Sene et al., 2010) and the effect is more pronounced under arid conditions such as desert areas. Soil microorganisms are particularly known to play key roles in ecosystems, and mediate many ecological processes that are central to ecosystem functioning. These processes include nutrient acquisition (Kahindi et al., 1997; Sene et al., 2010), nitrogen cycling, carbon cycling, soil formation (Rillig and Mummey, 2006; Sene and Sylla, 2014), decomposition processes (Hobbie et al., 2006, 2007; Peichl et al., 2006), and the regulation and maintenance of plant biodiversity (van der Heijden et al., 1998, 2008; Sene and Sylla, 2014). Plants adapted to harsh environments and their associated soil microorganisms within these habitats make both partners highly competitive and adaptive (Basil et al., 2004). Highly diverse groups of fungi are represented among these soil microbial communities and play fundamental physiological and ecological roles in desert ecosystems (Meiser et al., 2014; Sene and Sylla, 2014; Powell et al., 2015). Arid region soil microbes are adapted to sustain extreme environmental conditions and maintain a leading role in ecosystem processes (Pointing and Belnap, 2012). AM fungi are one of the most important associations in terrestrial ecosystems, influencing plant productivity through the acquisition of nutrients and water (Smith and Read, 2008; Allen, 2011). They can enhance plant establishment by buffering different environmental stresses and enhancing soil properties (Jeffries and Barea, 2001). The growth and development of A. nilotica is improved in association with AM fungi (Reddell, 1993). Osonubi et al. (1992) reported that inoculation of AM fungi to A. nilotica has the capacity to increase drought tolerance and plant biomass. Although numerous studies have reported the importance of mycorrhizal symbiosis for desert tree species, the symbiotic status of this unique tree, V. pachyceras (LT) has never been investigated in Kuwait. Until today, no attempt has been undertaken to identify indigenous fungal or mycorrhizal populations associated with the root system of this about to extinct LT species. Therefore, it is crucial to assess the status of native fungal and mycorrhizal propagules present in the roots of surviving plants and rhizosphere soils in the Kuwait desert before undertaking revegetation programs and introducing any inoculation technologies.
The main objective of this study was to conduct a field and seedling nursery study to investigate the root systems and rhizosphere soils of the V. pachyceras in desert and nursery conditions. This is in addition to the examination and comparison of the rhizosphere fungal and mycorrhizal communities of local and non-local V. pachyceras when grown under standard nursery seedling growing conditions and media; and to evaluate plant performance related to fungal and mycorrhizal associations. The characterization of general fungal and mycorrhizal populations and functional structures were revealed using both morphological and advanced DNA-based molecular techniques. This research is the first effort in Kuwait to assess the root mycorrhizal structure of V. pachyceras and its rhizosphere soil fungal community composition under both desert (LT) and nursery conditions (local and non-local V. pachyceras).
Materials and Methods
Study Site and Sampling
Field Sampling
The experimental site was located at the Sabah Al-Ahmed Natural Reserve, Kuwait (N 29°34.909′, E047°47.734′ around the only surviving single V. pachyceras tree, locally known as the LT). Soil samples were collected using soil corer of 3 × 30 cm from 0 to 30 cm depth for rhizospheric soil. Lateral roots were followed through the soil excavation channels created around 80–100 cm distance from the main trunk. Each replicate sample was thus a composite of 3–4 soil samples collected, which were mixed well by placing in a zip lock plastic bag and labeled as LT-S. Additional soil samples were collected 100 m away from the LT at three different points, and served as the non-rhizospheric control soil (CTL) in subsequent analyses and it is labeled as LTCS. Roots containing root nodules were also collected in triplicates from this tree roots and labeled as LT-AM. The representative soil and root samples were collected and brought to the laboratory and stored in a refrigerator until required for analysis. Roots were cut into 1–2 cm pieces and stored in 2% cetyl trimethylammonium bromide (CTAB) at -20°C for molecular characterization of AM fungal community.
Nursery Sampling
Two month old seedlings of native and non-local V. pachyceras were transplanted into one-gallon pots containing a soil mixture of agricultural soil, peat moss, potting soil and perlite (at 2:1:1:1 ratio, v/v basis) and is named hereafter as the commercial soil mix. The commercial soil mix is used conventionally in Kuwaiti nurseries for producing large-scale nursery seedlings for the restoration program at a national scale. Therefore, the current study was also intended to investigate fungal community structures in the commercial soil mix used for growing local and non-local V. pachyceras. Fifteen local and non-local V. pachyceras seedlings each were grown in one-gallon plastic pots in the nursery, and arranged in three replicate rows with five plants in each row for duration of 1 year. Steam-sterilized commercial soil mix (SAB Potting Soil-Plantaflor) was used for this study. Non-sterilized crude commercial soil mix was used as non-rhizospheric control soil (PsBp).
Seedlings were destructively sampled for plant and soil sample collection. Soil from five pots from each replicate row was pooled together and thoroughly mixed in a plastic bag to form a single composite sample per replicate in both seedling groups. Three replicate samples were used throughout the experiment. Samples were labeled as PsApLT and PsApSA for the local (Loney Tree) and non-local (Saudi Arabia) V. pachyceras seedlings, respectively. Representative soil and root samples collected as described for field soil and root samples. Several root pieces were cut into 1–2 cm pieces and stored in 2% CTAB at -20°C for molecular characterization of fungal population.
Molecular Characterization of Fungal Communities in Soil Samples
DNA Extraction and Amplification of ITS rDNA Region
Total community DNA from 0.25 g bulk soil and rhizosphere samples of V. pachyceras was extracted using a PowerSoil DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA, United States) with an addition of 0.05 g of skim milk powder in the lysis buffer. The remaining steps were performed according to the manufacturer’s instructions. Isolated soil DNAs were stored at below –20°C until PCR amplification. Amplification was performed using the fungal universal primers ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′; Gardes and Bruns, 1993) /ITS4 (5′-TCCTCCGCTTATTGATATGC-3′; White et al., 1990).
Polymerase chain amplification (PCR) was carried out in a 25-μL reaction and consisted of 1 μL soil DNA, 1 U of Taq DAN polymerase (Sigma-Aldrich), 3 mM MgCl2, 0.2 mM dNTP mix 0.2 mg/ml of BSA, and 0.3 mM concentration of each primer. The following thermocycle program was used for amplification: 94°C for 4 min followed by 35 cycles of 94, 50, and 72°C for 60 s each, and an extension period of 72°C for 10 min using a MJ Research PTC-225 Peltier Thermal Cycler. Negative control (containing no template DNA) reactions were also conducted to assess for any experimental contamination. The PCR products, 5 μl sub-samples, were observed by electrophoresis on 1 × Tris-acetate-EDTA (TAE) agarose (1% w/v) with suitable DNA size standards (Mass RulerTM, DNA Ladder Mix, Invitrogen, Canada) to ratify the size and estimate the quantity of the generated amplicons. The PCR products were visualized using ethidium bromide (0.25 μg L-1).
Molecular Characterization of Arbuscular Mycorrhizal Fungal Communities in Root System
PCR Amplification
A nested PCR was required to obtain sufficient amplicons for the molecular characterization of AM fungal communities from the roots of V. pachyceras tree from the field and seedlings grown in nursery under different conditions. Before isolation of genomic DNA, roots were rinsed in sterile distilled water for 48–72 h to remove excess CTAB. Next, 50 mg of roots were bead-grinded with a Tissue Lyser II (Qiagen) using the DNeasy Plant Mini Kit (Qiagen) and following the manufacturer’s protocol. Amplification was performed using the fungal universal primers LR1- (5′-GCA TAT CAA TAA GCG GAG GA-3′) and NDL22 (5′-TGG TCC GTG TTT CAA GAC G-3′) (van Tuinen et al., 1998). The polymerase chain reaction (PCR) followed the protocol of Brito et al. (2012). The reaction was carried out in a 25-μL reaction and consisted of 1 μL of total DNA, 1 U of Taq DNA polymerase (Sigma-Aldrich), 1.5 mM of MgCl2, 0.2 mM of dNTP mix, 0.5 mM of each primer and 25 μg/μl of T4 gene 32 protein (New England Biolabs Inc.). The following thermocycle program was used for amplification: 94°C for 4 min followed by 30 cycles of 94, 56, and 72°C for 60 s each, and an extension period of 72°C for 10 min using a MJ Research PTC-225 Peltier Thermal Cycler.
The nested PCR was performed on this amplicon, previously diluted at 1:50 ratio with the primers LR1 and FLR4 (Gollotte et al., 2004). PCR conditions were almost identical to those for the first reaction; however, instead of T4 gene 32 protein, BSA at 0.2 mg/ml was added to the reaction and the annealing temperature was 55oC for PCR optimization. This PCR was migrated in a 1.2% agarose gel stained with ethidium bromide and visualized under UV light. Samples where amplification failed were processed once again from the first PCR.
Cloning, Sequencing and Determination of Phylotypes and Phylogenetic Analyses
The 100 ng of PCR amplicons were cloned into a pGEM-T Easy Vector System II (Promega) using the procedure suggested by the manufacturer. The samples were stored in TTE buffer (triton X-100 1%; Tris–HCl pH 8.0 20 mM; EDTA pH 8.0 2 mM) at -20°C until use. At least 10 clones per sample were selected and transformed PCR products were sequenced using the Sanger method with two 16-capillary genetic analyzers 3130XL (Applied Biosystems).
DNA sequences were edited using BioEdit software, version 7.0.5 (1Hall, 1999) in order to resolve oligonucleotide ambiguities. The BLASTn (Basic Local Alignment Search Tool nucleotidic) search program algorithm2 was used to query the National Center for Biotechnology Information GenBank (NCBI) for highly similar sequences. When sequence similarity of at least 97% was achieved, these were considered to be in the same phylotypes (Moebius-Clune et al., 2013). Thus, one representative of each phylotype was used to continue phylogenetic analyses. Sequences were edited, aligned and queried on GenBank (NCBI) using ClustalX version 1.81 (Thompson et al., 1994) software. Operational taxonomic units (OTUs) were determined based on 97% similarity and the sequences with at least 97% similarity we considered in the same OTU, which could represent one species (Quince et al., 2008). One representative of each OTU was used to continue phylogenetic analyses. However, closely related sequences obtained were incorporated in phylogenetic analyses. Phylogenetic analyses were conducted using MEGA 6.0 (Tamura et al., 2013). Evolutionary distances were calculated as described by Jukes and Cantor (1969). Firstly, analyses were performed using the Neighbor-Joining (NJ) method (Saitou and Nei, 1987) and adopting the Kimura two-parameter method (Kimura, 1980). Secondly, maximum likelihood (ML) method based on the Kimura two-parameter model (Kimura, 1980) and 1,000 bootstrap replicates (Felsenstein, 1985) was used to compute the final tree. Finally, Bayesian inference of phylogeny was calculated using MrBayes version 3.2.2 (Ronquist et al., 2012), assuming a 4 × 4 model and non-variable substitution rates among sites – gamma rates. Analyses were constructed on two runs of four Markov chain Monte Carlo analyses where 2,000,000 generations were produced with burning fraction at 0.5 rate. These were sampled every 100 generations for 10,000 trees generated (Ronquist et al., 2012).
Statistical Analyses
Simpson’s (D), Shannon–Wiener (H) and Pielou’s evenness (E) diversity indices were calculated using the following formula; D = 1-Σ (pi)2, H = -Σpi log(pi), where pi = proportion of frequency of the ith phylotype in a sample. Phylotypes evenness was calculated as; E = H/log(S). Where H = Shannon Wiener diversity and S = phylotypes richness, i.e., total number of phylotypes. Fungal community diversities were compared and the specific levels of taxonomic resolution (rarefaction) were determined. The coverage saturation (C) was also calculated in order to verify the sufficiency of the sampling effort: C = 1 – (n1/N), where n1 is the number of phylotypes that occurred once, and N is the total number of phylotypes examined. In order to determine similarity associations between fungal communities among the different samples, a dendrogram was constructed on the basis of a similarity matrix using Morisita–Horn’s similarity coefficient (Magurran, 2004). Analyses were conducted using the “vegan” package in R software (R Development Core Team, 2010).
Results
Molecular Characterization of Fungal Communities in Soil Samples
Fungal Community Composition and Phylogenetic Diversity
The total community DNA isolated from soil samples was of high molecular weight (700–800 bp by using the primer pair ITS1F-ITS4) and sufficient purity for successful amplification of ITS rDNA fragments. The ITS rDNA fragments were obtained from all DNA samples by direct PCR amplification. The majority of fungal ITS rDNA sequences (68.34%) had high sequence similarity, up to 100%, with those of environmental fungi or known species in the NCBI database. However, 5% of the sequences in our database could not match more than 500 bp with sequences in the NCBI database; most of these sequences matched members of Fungi incertiae sedis. Eighteen clone libraries were generated from bulk and rhizosphere soil samples and 8 to 17 clones were successfully sequenced per library. Phylogenetic assignment of phylotypes was performed according to best sequence matches based on BLASTn analyses. Data obtained from BLAST analyses are summarized in Table 1. We globally obtained 48 OTUs among the 217 classifiable clone sequences (Table 1). The Ascomycota, Basidiomycota, traditional Zygomycota and traditional Chytridiomycota represented the majority of fungal sequences derived from our clone libraries (Table 1). However, a large number of sequences that matched members of Fungi incertae sedis was found. Because of the taxonomic distances between these phyla, the phylogenetic trees were inferred separately (Figures 1–3).
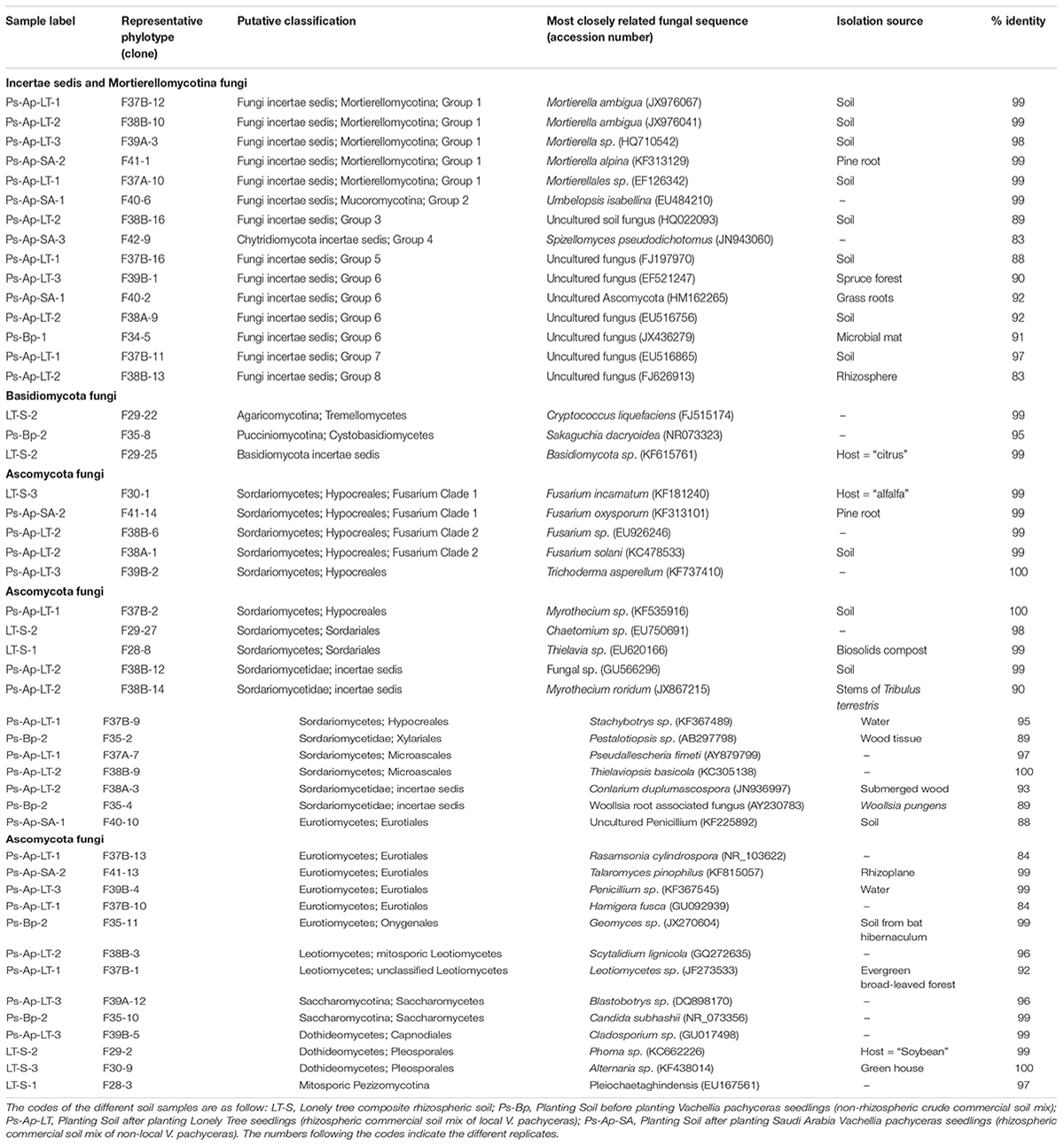
Table 1. BLAST results in Genebank database for the representing clones sequences from soil fungal communities.
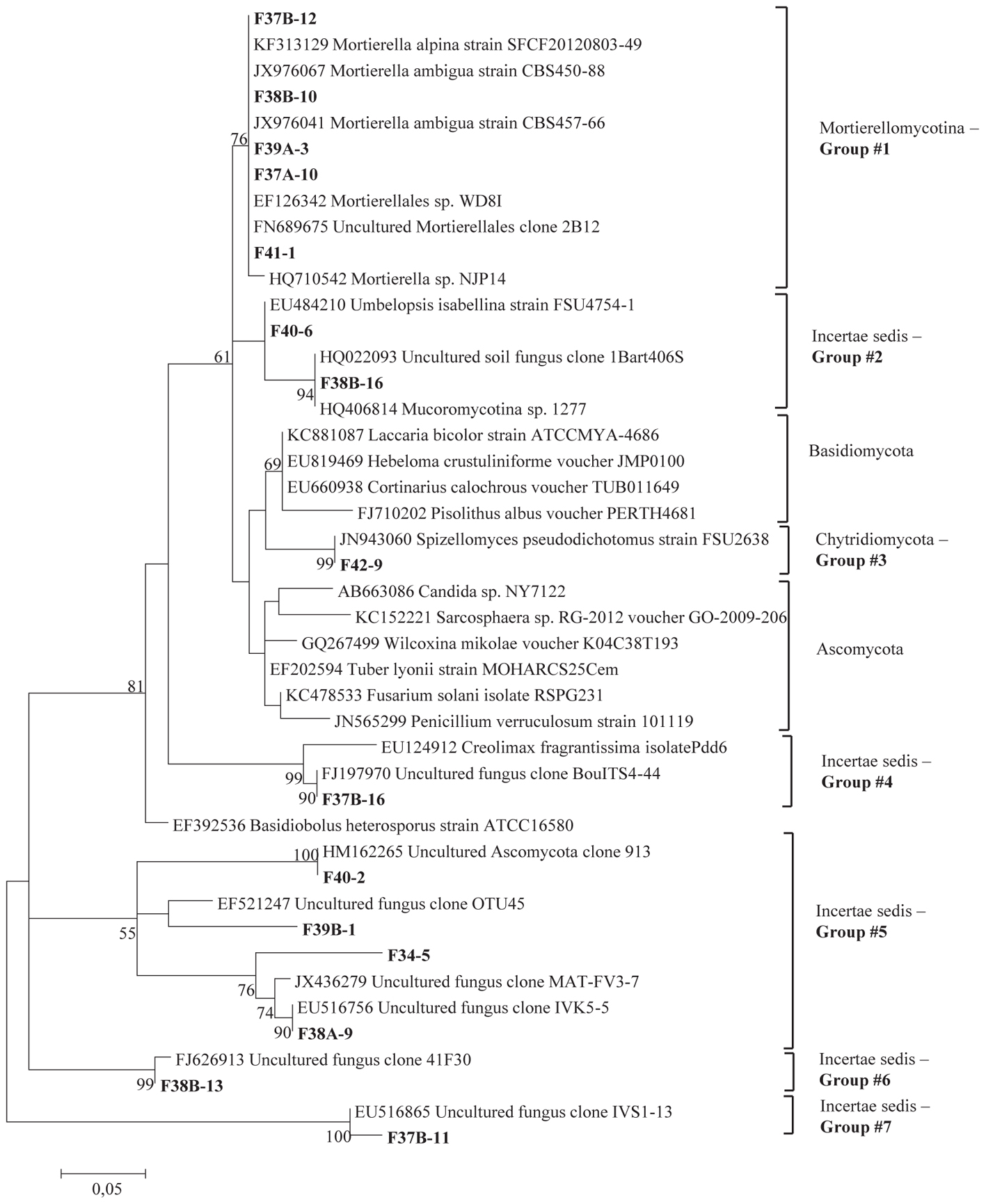
Figure 1. Phylogenetic tree showing maximum likelihood (ML) analysis of “Uncertae sedis fungi.” Bold sequences are from this study. Bootstrap percentage values (50%) generated from 1000 replicates from ML and posterior probabilities (>50%) from Bayesian analysis are shown as (ML bootstrap value/Bayesian posterior probabilities).
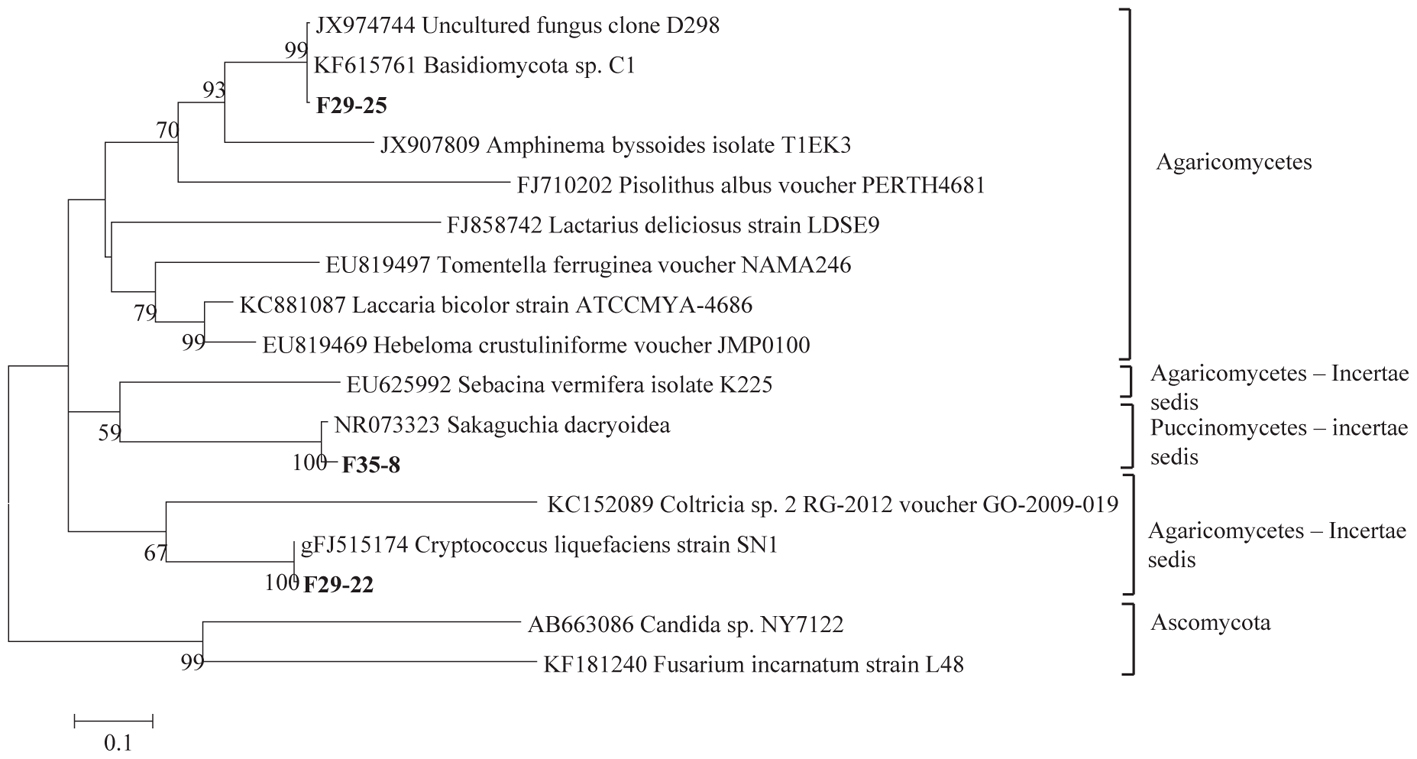
Figure 2. Phylogenetic tree showing ML analysis of Basidiomycota. Bold sequences are from this study. Bootstrap percentage values (50%) generated from 1000 replicates from ML and posterior probabilities (>50%) from Bayesian analysis are shown as (ML bootstrap value/Bayesian posterior probabilities).
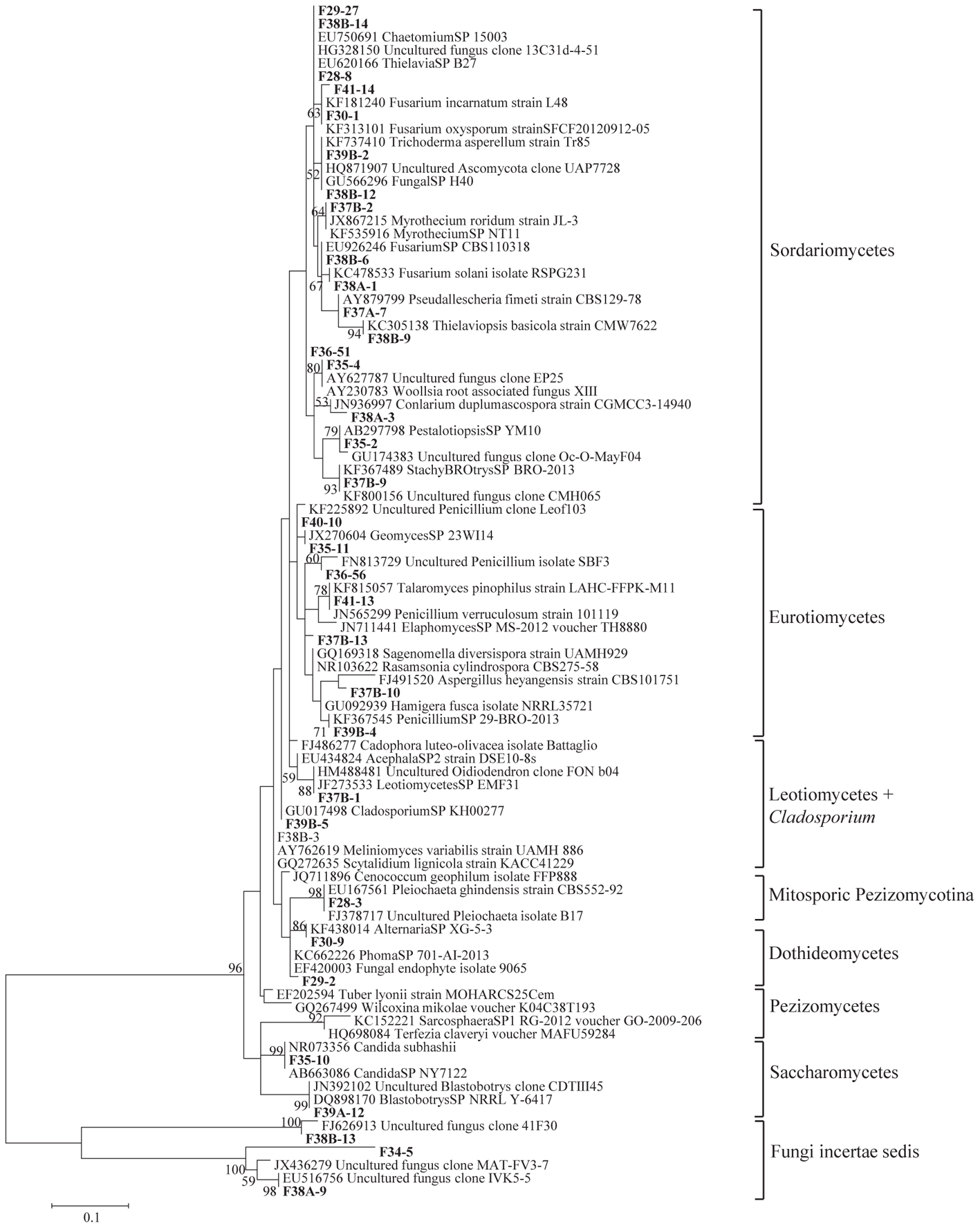
Figure 3. Phylogenetic tree showing ML analysis of Ascomycota. Bold sequence are from the study. Bootstrap percentage values (50%) generated from 1000 replicates from ML and posterior probabilities (>50%) from Bayesian analysis are shown as (ML bootstrap value/Bayesian posterior probabilities).
Sequence analysis showed that members of the phylum Ascomycota were the most common group in this study (Table 1). Twenty-three fungal sequences belonged to this group, which showed high similarities to their closest relatives. Of the Ascomycota (Figure 3), Sordariomycetes were by far the most abundant (16 OTUs), followed by the Eurotiomycetes (6 OTUs), Leotiomycetes (2 OTUs), Saccharomycetes (2 OTUs), Dothideomycetes (3 OTUs) and mitosporic Pezizomycota (1 OTU). Mortierellomycota were the second largest contributor in terms of phylogenetic diversity with 5 OTUs belonging to this sub-phylum (Figure 1). Traditional Chytridiomycota were detected but were rare in this study. Besides those fungal phyla, Basidiomycota were represented by 3 OTUs belonging to the Agaricomycetes (Figure 2). The remaining sequences, mostly affiliated to uncultured fungi from diverse environments, matched members of Fungi incertae sedis (Table 1 and Figure 1). They were divided into six groups outlined in Figure 1 and were represented by 10 OTUs. It is also remarkable to note that Sordariomycetes were widely distributed across the different soil samples, whereas other classes such as Saccharomycetes were almost exclusively found in bulk soils. The most abundant genera and species were Mortierella sp. (Mortierellomycotina) and Fusarium sp. (Sordariomycetes).
Rhizosphere Effect
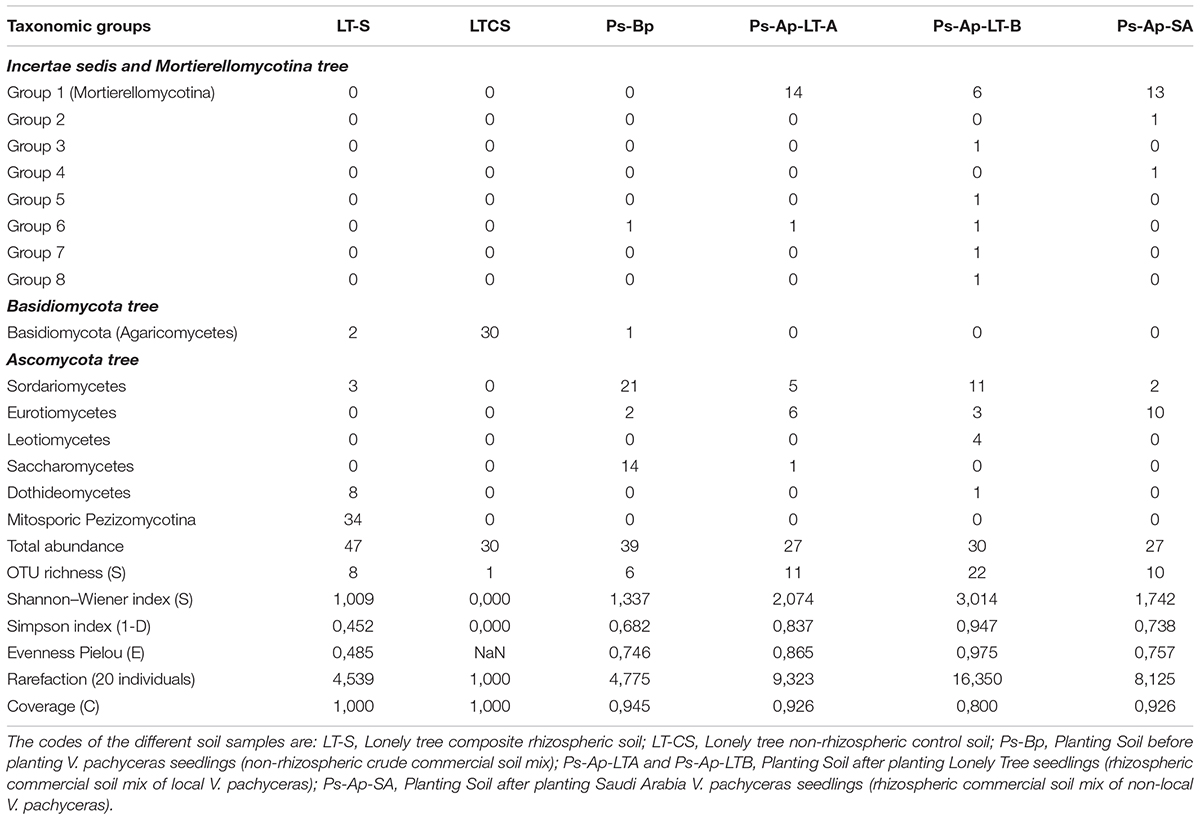
The degree to which fungal communities are influenced in the rhizospheres of plants compared to the non-rhizospheric control bulk soils was analyzed at a level of class. A differential distribution pattern of the detected fungal taxa among the different soil rhizospheres was observed at both field and nursery environments (Table 2). In more detail, the rhizosphere soils (LT-S) were clearly dominated by fungal phylotypes of the mitosporic Pezizomycotina class (72.5% of the communities) followed by members of the Dothideomycetes, which accounted for 17.02%. The remaining individual classes were relatively rare and accounted for up to 6.3% of the fungal communities. In contrast, the non-rhizosphere (bulk soil) samples (LT-CS) showed the lowest biodiversity in terms of community structure (Table 1). They were represented by fungal phylotypes of a single class; Basidiomycetes, which had clearly increased its abundance in bulk soil samples (100% of fungal communities).
The Ps-Ap-LT soils (grown in optimal nursery conditions) contain fungal phylotypes of at least four dominant classes: Sordariomycetes (36.7%), Leotiomycetes (13.3%), and Eurotiomycetes (10%) for the sample B and, Sordariomycetes (18.5%) and Eurotiomycetes (22.2%) for the sample A. Close relatives of Mortierellomycotina incertae sedis fungi were also common in these soil samples with 20 and 51.85% for the Ps-Ap-LT sample B and sample A, respectively. The other classes are of minor importance and accounted for up to 3.4% of the soil fungal communities. The Ps-Ap-SA treatment was dominated by fungal phylotypes of the Mortierellomycotina incertae sedis (48.15% of fungal communities), followed by members of the Eurotiomycetes (37.04%). The remaining classes were relatively rare and accounted for up to 7.4% of the soil communities. The non-rhizosphere (bulk soil) samples (Ps-Bp) were affiliated to three main classes of Ascomycota: Sordariomycetes (54%), Saccharomycetes (36%) and Eurotiomycetes (5.1%). Other phylotypes from this clone library are of minor importance, with sequences having nearest hits to members of Agaricomycetes (2.5% of fungal communities) and incertae sedis Group 6 (2.5%).
Globally taken, the rhizosphere samples LT-S, Ps-Ap-LT-A, Ps-Ap-LT-B, and Ps-Ap-SA treatments, with 8, 11, 22, and 10 OTUs, respectively, showed the highest richness of fungal communities. The field and nursery bulk soil samples disclosed 1 and 6 OTUs, respectively. Indices of Shannon (H) for diversity and Simpson (1–D) for evenness were also calculated, and data were broadly in agreement with those reported with the richness index (Table 2). Collectively, data from this study illustrate that Bulk soil samples were consistently less even than rhizosphere soils. Difference in coverage were also marked among the different samples, the nursery rhizosphere soils (Ps-Ap-LT and Ps-Ap-SA) showing lesser values of rarefaction index, suggesting that sample efforts are further needed to saturate the organismal richness in these samples.
In addition, principal component (PCA) and Cluster analyses (based on the Morisita–Horn’s similarity coefficient) were conducted to compare similarities between fungal communities of the different samples. According to these analyses, marked differences exist in the fungal community compositions of soil samples: two main clusters were distinguished (Figure 5). The first cluster included the rhizosphere soils collected from both V. pachyceras (Ps-Ap-LT-A, Ps-Ap-LT-B, and Ps-Ap-SA treatments) grown in nursery conditions and, the second cluster comprised the Lonely Tree (LT-S) and bulk soil (LT-CS and Ps-Bp) samples. However, Figure 4 showed that nursery bulk soils (Ps-Bp) were segregated from LT-S and LT-CS samples, thus suggesting that their soil fungal communities were different. Ps-Ap-LT was replicated in order to verify that the sample effort is reliable. Figure 5 clearly shows that both replicates were grouped together and roughly included in the same branch cluster.
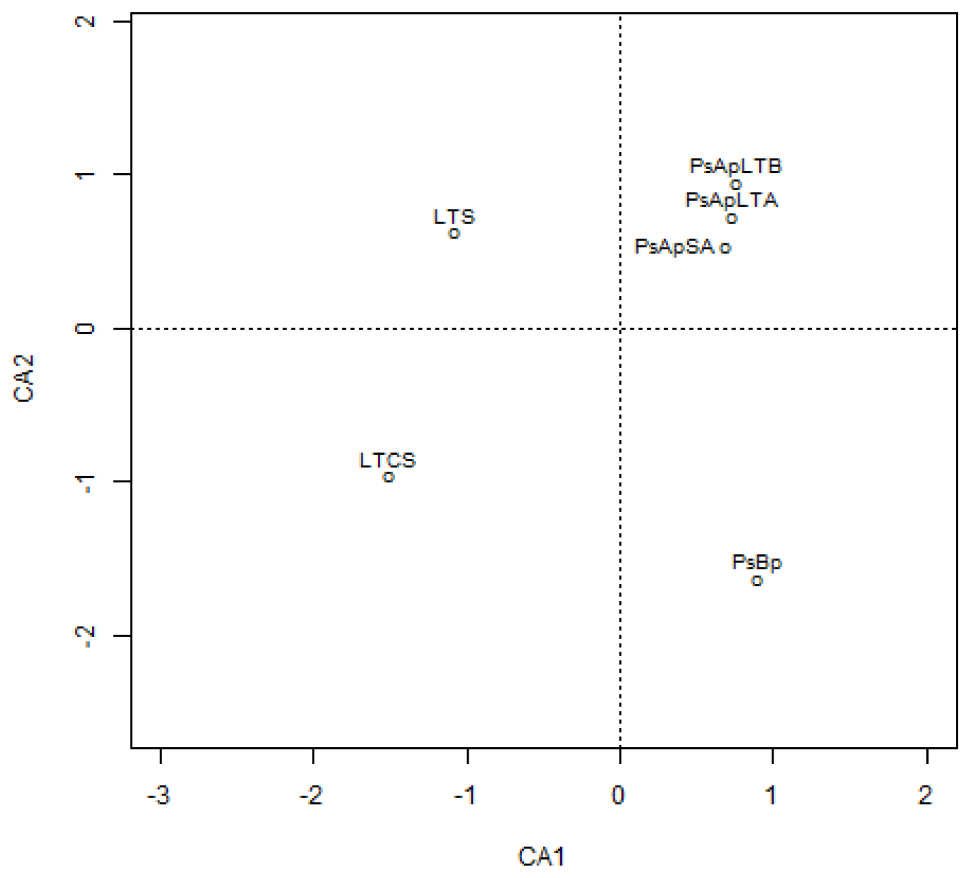
Figure 4. Correspondence analysis of the different fungal communities in soil rhizosphere of Vachellia pachyceras in diverse conditions. Soil samples are positioned along the first two DA axes, where Eigen values are 0.9919 for CA1 and 0.9536 for CA2. The codes of the different soil samples are: LT-S, Lonely tree composite rhizospheric soil; LT-CS, Lonely tree non-rhizospheric control soil; Ps-Bp, Planting Soil before planting V. pachyceras seedlings (non-rhizospheric crude commercial soil mix); Ps-Ap-LTA and Ps-Ap-LTB, Planting Soil after planting Lonely Tree seedlings (rhizospheric commercial soil mix of local V. pachyceras); Ps-Ap-SA, Planting Soil after planting Saudi Arabia V. pachyceras seedlings (rhizospheric commercial soil mix of non-local V. pachyceras).
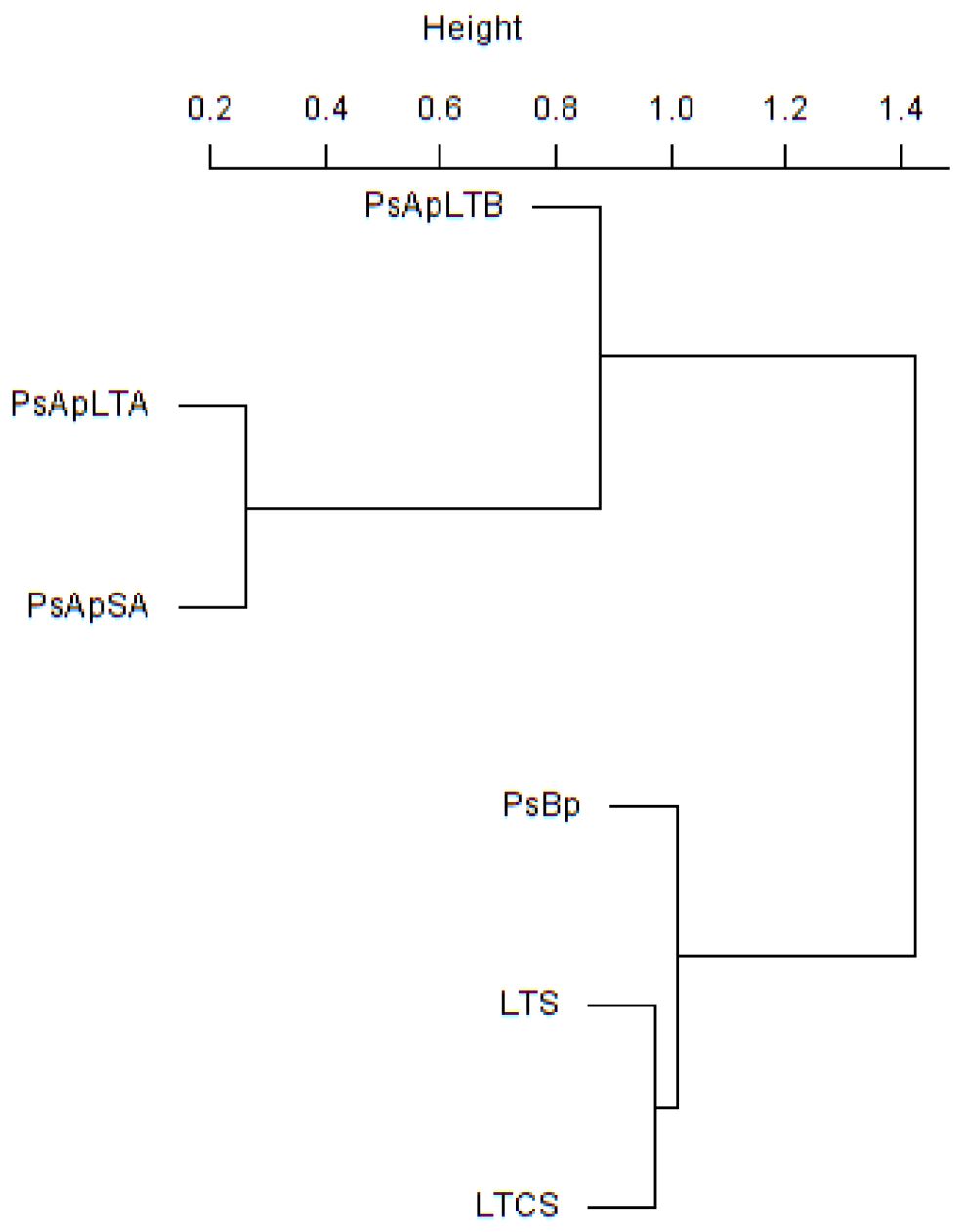
Figure 5. Cluster analysis based on Morisita-Horn similarity coefficient for different fungal communities in soil rhizosphere of V. pachyceras in diverse conditions. The codes of the different soil samples are: LT-S, Lonely tree composite rhizospheric soil; LT-CS, Lonely tree non-rhizospheric control soil; Ps-Bp, Planting Soil before planting V. pachyceras seedlings (non-rhizospheric crude commercial soil mix); Ps-Ap-LTA and Ps-Ap-LTB, Planting Soil after planting Lonely Tree seedlings (rhizospheric commercial soil mix of local V. pachyceras); Ps-Ap-SA, Planting Soil after planting Saudi Arabia V. pachyceras seedlings (rhizospheric commercial soil mix of non-local V. pachyceras).
Molecular Characterization of Arbuscular Mycorrhizal Fungal Communities From Root System
DNA extractions were performed using the Plant DNeasy kits (Qiagen, ON) from roots stored in CTAB. The expected 750–800 bp PCR fragment was obtained with the nested PCR with LR1-FLR 4. From the total DNA amplicons, 15 clone libraries were produced, and 3 to 14 clones were successfully sequenced per library (Table 3). Among all 133 clones sequenced, 10 phylotypes could be identified (Table 3).
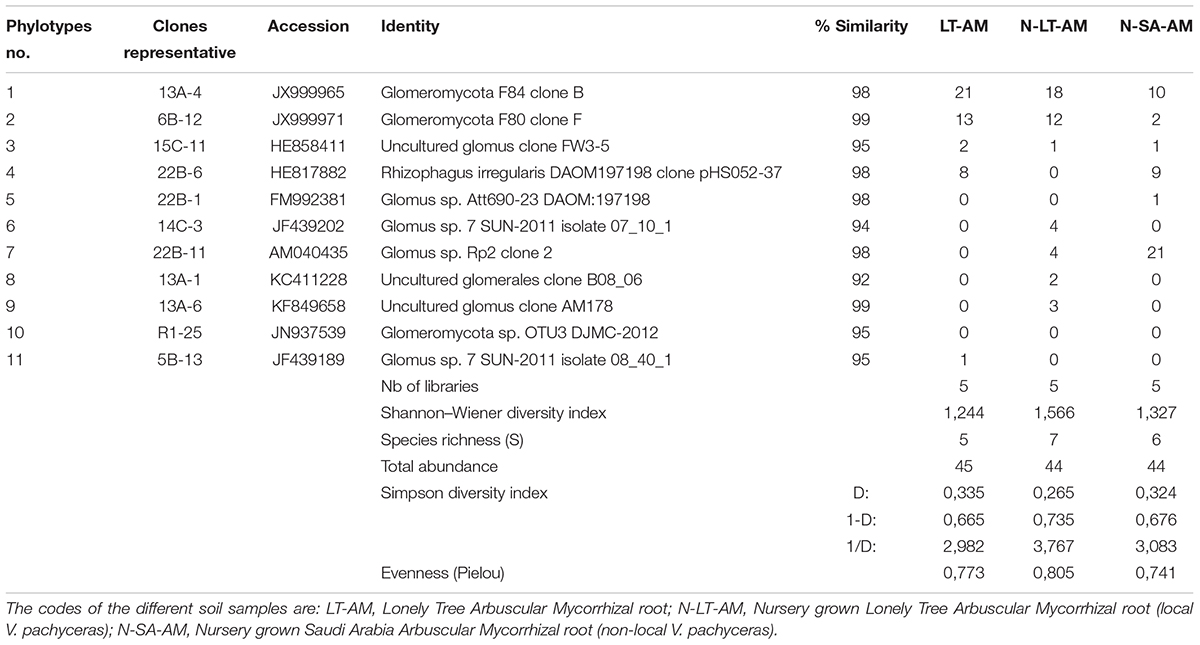
Table 3. BLAST results for the representing clone’s sequences from AMF communities in roots and statistical analysis.
Nucleotide BLAST results in GenBank database for the representing clones sequenced are shown in Table 3. First, analysis was performed excluding “Uncultured/environmental sample sequences.” If similarity percentage between the query and GenBank sequence was below 95%, nucleotide BLAST was computed again including “Uncultured/environmental sample sequences.” This was the case for phylotypes #3, 8 and 9, for which similarity with NCBI-deposited sequences was below 95%. Thus, the matching “Uncultured/environmental sample sequences” was included in further analyses. Branches that delimit phylotypes and their matching sequence were well supported, with 73 to 99% of bootstraps values and 0.67 to 1.00 of Bayesian posterior probabilities. Tree topology acquired after computing NJ and ML analyses were similar. Figure 6 present the ML tree on which is observed a different branching pattern between ML and Bayesian analyses at the family level, so it does not affect interpretation. Phylogenetic tree (Figure 6) showing molecular phylogenetic analysis obtained by maximum likelihood (ML) analysis of arbuscular mycorrhizal fungi. The most abundant and diversified main group was found within the family Glomeraceae. However, in the phylogenetic tree, on the right side it is showing two taxonomic groups. In the first group, it is assigned into two sub-groups clustering the genera Rhizophagus and Sclerocystis in which 5 different phylotypes included with the exception of phylotype 4, where a sequence of Rhizophagus irregularis DAOM 197198 was included in the branch group, no other type was included in the phylotype branches. Thus, a species could not be assigned to these groups. In the second group, only a single sub-group assigned and clustering as Glomeraceae group-1, in which another 5 phylotypes with unclassified taxonomic group included. According to cluster analysis in Figure 7, a closer relationships exist among AMF communities from Lonely Tree (LT-AM) and (N-LT-AM) than the Saudi Arabia tree (N-SA-AM). For AMF composition and frequency in the different communities (Table 3), it can be observed that the most common phylotype for all root types is the phylotype no. 1. Phylotype no. 2 is very abundant for LT-AM and N-LT-AM, and phylotype no. 7 is abundant for N-SA-AM. The highest diversity indices are found in seedlings N-LT-AM, followed by N-SA-AM and LT-AM.
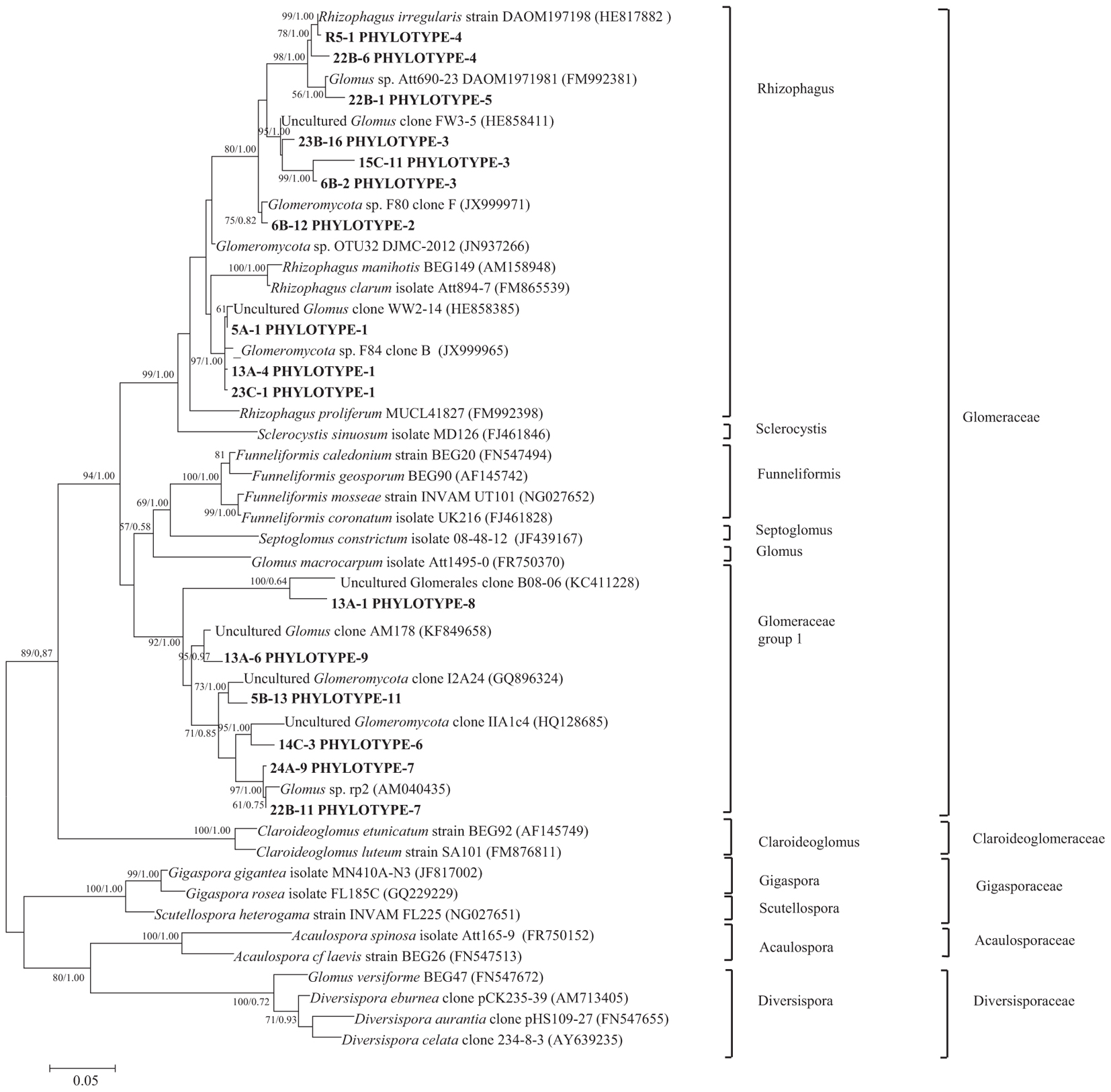
Figure 6. Phylogenetic tree showing ML analysis of arbuscular mycorrhizal fungi. Bootstrap percentage values (50%) generated from 1000 replicates from ML and posterior probabilities (>50%) from Bayesian analysis are shown as (ML bootstrap value/Bayesian posterior probabilities). Bold sequences are from this study.
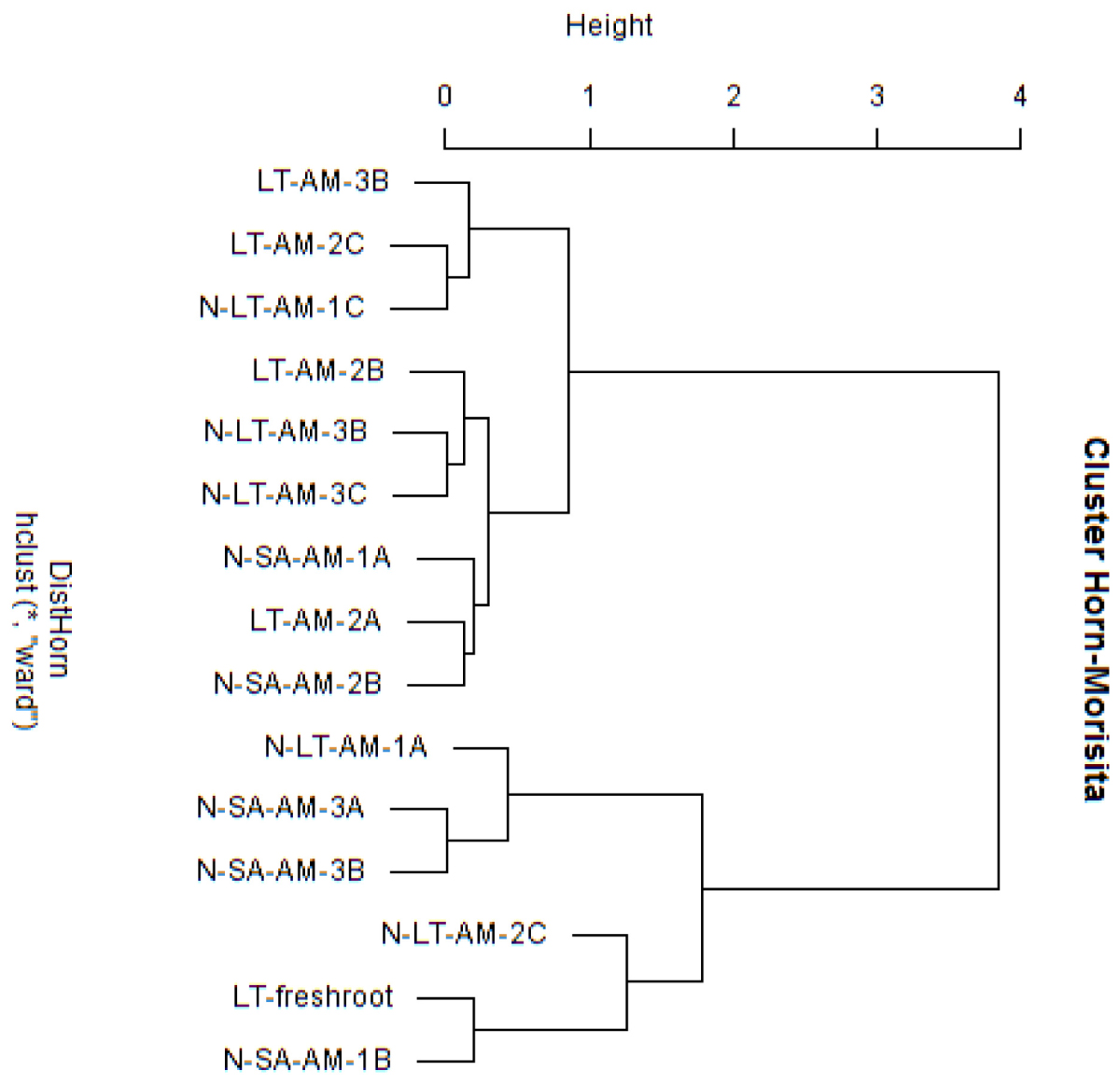
Figure 7. Cluster analysis based on Morisita-Horn similarity coefficient for different arbuscular mycorrhizal fungal communities in roots of V. pachyceras in diverse conditions. The codes of the different soil samples are: LT-AM, Lonely Tree Arbuscular Mycorrhizal root; N-LT-AM, Nursery grown Lonely Tree Arbuscular Mycorrhizal root (local V. pachyceras); N-SA-AM, Nursery grown Saudi Arabia Arbuscular Mycorrhizal root (non-local V. pachyceras). LT, Lonely Tree; The clone number were added at the end of the sample codes.
Discussion
Fungal Community Structures
To our knowledge, this study is the first attempt using PCR based molecular approach to reveal the diversity of general fungal communities and AM fungi associated with the only surviving nationally important tree specises – V. pachyceras in Kuwait desert. The present work provides us with a preview of general fungal communities associated with rhizosphere and non-rhizosphere soils of V. pachyceras from the Sabah Al-Ahmad Natural Reserve of the Kuwait desert and nursery growing media. Data showed that despite a hostile climate that the Kuwait desert represents, the studied area maintained surprisingly diverse fungal biodiversity, largely consisting of fungi adapted to harsh environmental conditions exists in desert. Our results are in agreement with a recent report showing unpredictably huge fungal biodiversity in the Middle Eastern desert soils (Murgia et al., 2018). For the overall community composition of this study site, the majority of fungal sequences recovered were classified into four major fungal phyla: Ascomycota, Basidiomycota, Zygomycota and Chytridiomycota, which account for 80% of the OTUs. Consistent with the previous studies (El-Said and Saleem, 2008; Bates et al., 2012; Sterflinger et al., 2012; Abed et al., 2013; Bastida et al., 2013), Ascomycota was the most abundant phylum (48%), whereas Basidiomycota accounted for a much smaller percentage of the community (10.4%). A large body of investigation has supported this dominance of Ascomycota fungi in arid and semi-arid soils. In a recent study, soil sample from Saudi Arabia and Jordan deserts revealed that the most abundant fungal phyla was Ascomycota similar to our results (Murgia et al., 2018). Abed et al. (2013) showed that in the arid desert of the Arabian Peninsula Ascomycota represents >86% of their pyrosequencing reads, while forming more than 98% of the observed isolates. Using molecular techniques, Green et al. (2008) and Bates and Garcia-Pichel (2009) reported dominance of Ascomycota fungi in soils from the Chihuahuan desert (83.3% of 989 sequences) and in Colorado Plateau (87–91% of 135 sequences), respectively. Other data from Grishkan et al. (2006) have also shown this dominance in the Negev desert (98% of 58 species). The results, however, are disclosed to those of a few scale surveys of desert soils (Connell et al., 2006; Fell et al., 2006) where Basidiomycota was the dominant phylum. These analyses and our data from the Kuwait desert indicate the variations of fungal community composition among desert lands. Molecular analyses data of this study clearly demonstrated these variations in soil fungal community composition and reflected in the non-rhizospheric control LT bulk soil, which had very low levels of fungal composition with OTUs Richness – 1 and Shannon–Weiner Index (S) – 0.0. In all other soils, such as nursery soils tested were found much higher OTUs Richness and higher Shannon–Weiner Index as these soils have relatively higher organic matter and maintained in optimal growing conditions or have vegetation effects. Chytridiomycota and Zygomycota fungi seem to be underrepresented, compared to the number of sequences in the Ascomycota. Similar results have been previously reported in Omani desert and Chihuahuan desert by Green et al. (2008).
Of the Ascomycota, the most OTU-rich fungal classes were Sordariomycetes and Eurotiomycetes. The former is the only class showing high diversity in each soil sample, indicating their high ecological plasticity. Described members of the Sordariomycetes are assumed to be cosmopolitan, and function as plant and animal pathogens, endophytes of plants, and saprobes involved in decomposition and nutrient cycling (Brunner et al., 2011; Abed et al., 2013; Qin et al., 2014). In the present study, the majority of OTUs sequences belonging to the Sordariomycetes matched previously described species and was related to the following genera: Fusarium, Myrothecium, Trichoderma, Chaetomium, Thielavia, Stachybotrys, Pestalotiopsis, Pseudallescheria, Thielaviopsis, and Conlarium. They mostly belonged to the orders Hypocreales, Sordariales, Xylariales, and Microascales. These fungi most likely play a role in organic plant material breakdown in a symbiotic or mutualistic relationship with plant species (Meiser et al., 2014; Powell et al., 2015).
The largest assemblages of fungal OTUs belonging to the Eurotiomycetes comprise members of the genera Rasamsonia, Talaromyces, Penicillium, Hamigera, and Geomyces. They belonged to the orders Euritiales and Onygenales, which include cellulolytic soil saprophytes fungi (Sugiyama et al., 1999; Berbee, 2001). Most of the OTUs in the Leotiomycetes class belonged to the genera Scytalidium and Leotiomycetes whereas most of the OTUs in the Saccharomycetes class belonged to the genera Blastobotrys and Candida. Saccharomycetes class includes genera of ascomycetous yeasts and one pathogenic on human (Candida) (Berbee, 2001).
Dothideomycetes OTUs were assembled into three genera Cladosporium, Phoma, and Alternaria. They mostly belonged to the order Pleosporales. Fungi belonging to this order (most notably Alternaria) were used before as indicative of desert settings (Bates et al., 2012; Sterflinger et al., 2012). This is mainly because they typically have darkly pigmented spores or hyphae stained with allomelanins, which may provide protection from excessive exposure to UV radiation (Bates et al., 2012). They can often be found as endophytes or epiphytes of living plants, and also as saprobes degrading cellulose, keratin and other complex carbohydrates in dead or partially digested plant matter in leaf litter (Porras-Alfaro et al., 2010; Nguyen et al., 2011; Abed et al., 2013). Nevertheless, species of the genus Aspergillus, being very abundant in Israel (Grishkan and Nevo, 2010), Saudi Arabia and Libya (Abdel-Hafez, 1981, 1982), were missing in our samples.
Basidiomycota have been reported to be diverse in soil ecosystems (Buée et al., 2009), but is not confirmed here. We showed that only 6.25% of the OTUs belong to this phylum. Most sequences matched species belonging to the Cryptococcus and Sakaguchia genera. These genera are known to comprise a number of human associated species, as either opportunists or pathogens (de Hoog et al., 2000). Dominance of yeast genera including Cryptococcus genus is also reported in Antarctica in a study by Arenz and Blachette (2011).
Most OTUs sequences in the Zygomycota and Chytridiomycota phyla matched species belonging to the Mortierella and Spizellomyces genera, reportedly common fungal groups in soils (Brown and Jumpponen, 2014; Zhang et al., 2014). Members of the former are reported to mineralize readily available dissolved organic substrates rather than breaking down soil litter polymers (Schmidt et al., 2008; Brown and Jumpponen, 2014), while the later has been found to infect spores of arbuscular mycorrhizal fungi (Ross and Ruttencutter, 1977; Daniels, 1981). OTU sequences belonging to the chytrids were also detected in soil crusts of the Sultanate of Oman in the Arabian Desert (Abed et al., 2013). However, their detection in arid deserts, although in low abundance (<3% of total OTUs) is intriguing and more research is needed in order to determine the ecological role of aquatic Chytridiomycetes fungi associated with desert lands.
Several OTUs sequences mostly affiliated to uncultured fungi from diverse environments, matched members of Fungi incertae sedis. These sequences were assembled into six groups and seem to correspond to a well-supported clad of Ascomycota, equivalent to endophytic of dark septate fungi. The detection of dark-colored fungi is a typical feature of desert soils mainly because of their ability to survive high solar radiation and temperature (Grishkan et al., 2006). Further analyses are required to explore functional attributes of these fungal species and to classify them at the genus level.
Rhizosphere Effect on Soil Fungal Communities
Microbial activity in deserts are concentrated in brief periods of high soil wetness following rainfall events and are expected to be greater in rhizosphere soils, depending on the plant type (Hollister et al., 2010; Zhang et al., 2014). We sought to investigate the rhizosphere effect of the LT V. pachyceras and the non-local Saudi Arabia V. pachyceras, on soil fungal community structures by assessing fungal diversity in soils from the same species grown in optimal nursery environments. The foremost aspect of fungal community structures that is so clear as to be unassailable in this study is the rhizosphere effect, with more fungal diversity and OTUs richness associated with both field and nursery rhizosphere soils. In contrast, a less diverse fungal community was found in the bulk soils. Such plant-dependent enrichment has received increasing support recently (Xu et al., 2012; Meiser et al., 2014; Welc et al., 2014; Zhang et al., 2014). Indeed, plant may exude a variety of carbon sources that can be consumed by fungal communities, thus creating more niches for them to occupy and promoting increased fungal richness (Weber et al., 2011). However, ideal nursery growing conditions with nutrient availability may also favor microbial population in soils. In the absence of root plants, the nutrient limitations and the combined disturbance of climatic conditions in deserts could preclude the growth and hamper the hyphal proliferation of many fungi (Meiser et al., 2014). Thus, only fungal species that might be highly specialized to such an ecological niche could be found (Bates et al., 2012). Those data and the results from fungal community structure, pointed out the complexity of the interdependency of soil microbial diversity with plant species.
Although the nursery experiment was not designed to specifically test the impacts of soil properties on the soil fungal communities, the results of this study suggest that they have an effect. Compared to the field soils, fungal communities in the nursery soils were more diverse owing to soil chemical richness and substrate. This is analogous to an edaphic condition effect and has been reported in other studies (de Castro et al., 2008; Qin et al., 2014; Welc et al., 2014). Thus, we hypothesized that by providing a nutritional resource and stable substrate, nursery sustains the growth of many more fungi than do field environments. Nevertheless, from this study, this edaphic condition effect seems to be overwhelmed by the response of fungi to the root plant presence. It is not surprising that differences in soil chemical properties may drive the observed higher fungal communities and diversity associated with nursery potting soils and LT rhizospheric soils compared to LT non-rhizospheric control soils.
Arbuscular Mycorrhizal (AM) Communities From Root System
This study is the first to use of molecular technique to reveal the AM fungal communities from the roots of V. pachyceras the only surviving native tree species in Kuwait desert and the roots of nursery grown V. pachyceras seedlings. Our attempt was to uncovering AM fungal community composition and obtained a baseline data that was previously unknown with this unique tree in Kuwait. Initial staining of roots confirmed the presence of AM fungi in the LT (V. pachyceras) and in the roots of nursery-grown V. pachyceras seedlings (Supplementary Material). This observation is further supported by the molecular characterization of AM communities associated with root systems of all the test plants of this study. Results from molecular characterization, a total of 10 phylotypes were identified, in which 5 phylotypes revealed with most abundant group from the family Glomeraceae with the presence of genera Rhizophagus formerly known as Glomus sp., suggesting specific recruitment preference of AM fungi partners by the single plants (Xu et al., 2017). However, another 5 phylotypes were also revealed with unclassified taxonomic group under Glomeraceae group 1. Furthermore, our result revealed that the diversity of AM fungi was more pronounced in the nursery-grown Vachellia seedlings than in the desert habitat of the LT roots. The identified phylotypes belonged to the Family Glomeraceae supporting the results of the examination of the spores in the soil, which morphologically resembles Glomeraceae (Supplementary Material). Nevertheless, the unclassified taxonomic group with five phylotypes revealed in this investigation suggests that further detailed analysis is required to elucidating the unclassified group fully. Similar to our findings, many studies have shown that AM fungi belonging to the genus Glomus and Rhizophagus were the most dominant in desert as well as other ecosystems, and most common AM fungi revealed throughout the world (Pande and Tarafdar, 2004; Shi et al., 2007; Al-Yahya’ei et al., 2011; Xu et al., 2017). The abundance of Glomeraceae in complex arid regions may be due to its ability to withstand harsh arid climatic conditions and resistance to high temperatures (Bever et al., 2009; Barto et al., 2011).
Even though AM fungal infection was detected morphologically under microscope from dried and thick roots typical for desert plants, DNA extraction was successful but the amplification and sequencing of such DNA samples was inadequate. Further standardization of amplification and sequencing is required for the desert AM fungal communities unique to Kuwait desert environment. Furthermore, it might be because of insufficient representation of AM fungal taxa in existing reference databases used. Nevertheless, the present research revealed a considerable diversity among the different root samples tested that has not been earlier depicted. The impact of environmental attributes on AM fungal communities that may influence shape these communities are still not adequately known (Xu et al., 2017). Data of this study showed that despite the harsh climate that prevails in the Kuwait desert, studied roots displayed the presence of AM fungal biota. However, the greatest diversity was noted in the roots of seedling from nursery, suggesting that diversity in the harsh desert conditions is lower compared to when seedlings were grown in nursery soil mix under ideal nursery conditions. It should be noted that not all sequences produced a match with a described well-known species; only the composition of the communities at the class level for the taxonomic rank could be characterized. However, it seems that soil factors may influence fungal composition and diversity more than tree species characteristics and their root system (Oehl et al., 2017). Although AM fungal spore counts are often low in arid soils and zero counts are common (Cui and Nobel, 1992; Requena et al., 1996; Titus et al., 2002). Data from this work demonstrate that a good number of AM fungal species exist in the Kuwaiti desert that need to be fully explored. However, this study established foundation work of the AM fungal communities associated with roots of V. pachyceras in Kuwait desert as well as nursery seedlings. This research delivers essential insight about ecological characteristics of V. pachyceras. Further detail information is still required about the revealed AM fungal population, and then there is possibility to improve better seedling production in nursery and survival rates of V. pachyceras seedlings when planted in desert conditions by increasing population of certain AM fungal species in roots and rhizosphere.
Conclusion
In conclusion, despite harsh climatic conditions prevailing in Kuwait desert, our results demonstrate surprisingly a diverse number of general fungal and AM fungal resources exists in the studied area that remain to be fully characterized. Our study from both rhizospheric and bulk soil revealed four major fungal phyla and classified as Ascomycota, Basidiomycota, Zygomycota and Chytridiomycota. In which Ascomycota is the most abundant phylum (48%) followed by Basidiomycota, Zygomycota and Chytridiomycota. A great number of fungal ITS rDNA sequences were related to a wealth of incertae sedis fungi, suggesting further works are needed to classify them at the genus level. Similar to many other studies, we also found distinct fungal composition and diversity between the rhizospheric and non-rhizospheric soils. The most abundant AM fungal group identified was among the family Glomeraceae. Evidently, further detail taxonomic information on is still required in order to performing quantitative comparisons of relative phylotypes obtained in this research. The current research would however, assists set the basis for future research and might be helpful to determine the strategies used by this fungal microbiota in response to hot and dry weather conditions for plant fitness. Furthermore, to exploit the potential for the use of these fungal species as biofertilizer to unfavorable desert conditions, this might lead to improvement in restoration and revegetation strategies for about to extinct LT species in Kuwait. Apparently, this study is a first effort using molecular approach and advances our existing knowledge about the general fungal, and AM fungal communities related with this nationally important unique tree since the tree species is considered endangered in Kuwait.
Author Contributions
MS and AQ planned and conceptualized the experiments. MS, AQ, AM, and MS conducted the experiments and wrote the manuscript. KD, LC, PN, and NB supervised and reviewed the manuscript. All authors approved the manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the Kuwait Institute for Scientific Research (KISR) for providing encouragement throughout the experiments. We also thank Damase Khasa and his research team at Université Laval, Québec, Canada for their technical assistance, data analysis, and valuable comments and reviewing an earlier versions of this manuscript.
Footnotes
References
Abdel-Hafez, S. I. I. (1981). Halophilic fungi of desert soils in Saudi Arabia. Mycopathologia 75, 75–80. doi: 10.1007/BF00505781
Abdel-Hafez, S. I. I. (1982). Survey of the mycoflora of desert soils in Saudi Arabia. Mycopathologia 80, 3–8. doi: 10.1007/BF00437171
Abed, R. M. M., Al-Sadi, A. M., Al-Shehi, M., Al-Hinai, S., and Robinson, M. D. (2013). Diversity of free-living and lichenized fungal communities in biological soil crusts of the Sultanate of Oman and their role in improving soil properties. Soil Biol. Biochem. 57, 695–705. doi: 10.1016/j.soilbio.2012.07.023
Akpo, L. E., Banoin, M., and Grouzis, M. (2003). Effet de l’arbre sur la production et la qualité fourragère de la végétation herbacée : bilan pastoral en milieu sahélien. Rev. Méd. Vét. 154, 619–628.
Allen, M. F. (2011). Linking water and nutrients through the vadose zone: a fungal interface between the soil and plant systems. J. Arid Land 3, 155–163. doi: 10.3724/SP.J.1227.2011.00155
Al-Yahya’ei, M. N., Oehl, F., Vallino, M., Lumini, E., Redecker, D., Wiemken, A., et al. (2011). Unique arbuscular mycorrhizal fungal communities uncovered in date palm plantations and surrounding desert habitats of Southern Arabia. Mycorrhiza 21, 195–209. doi: 10.1007/s00572-010-0323-5
Arenz, B. E., and Blachette, R. A. (2011). Distribution and abundance of soil fungi in Antarctica at sites of the Peninsula. Ross Sea region and McMurdo Dry Valleys. Soil Biol. Biochem. 43, 308–315. doi: 10.1016/j.soilbio.2010.10.016
Barto, E. K., Hilker, M., Muller, F., Mohney, B. K., Weidenhamer, J. D., and Rilling, M. C. (2011). The fungal last lane: common mycorrhizal networks extend bioactive zones of allelochemicals in soils. PLoS One 6:e27195. doi: 10.1371/journal.pone.0027195
Basil, A. J., Strap, J. L., Knotek-Smith, H. M., and Crawford, D. L. (2004). Studies on the microbial populations of the rhizosphere of big sagebrush (Artemisia tridentata). J. Ind. Microbiol. Biotechnol. 31, 278–288. doi: 10.1007/s10295-004-0140-y
Bastida, F., Hernández, T., Albaladejo, J., and García, C. (2013). Phylogenetic and functional changes in the microbial community of long-term restored soils under semiarid climate. Soil Biol. Biochem. 65, 12–21. doi: 10.1016/j.soilbio.2013.04.022
Bates, S. T., and Garcia-Pichel, F. (2009). A culture-independent study of free-living fungi in biological soil crusts of the Colorado Plateau: their diversity and relative contribution to microbial biomass. Environ. Microbiol. 11, 56–67. doi: 10.1111/j.1462-2920.2008.01738.x
Bates, S. T., Nash, T. H., and Garcia-Pichel, F. (2012). Patterns of diversity for fungal assemblages of biological soil crusts from the southwestern United States. Mycologia 104, 353–361. doi: 10.3852/11-232
Berbee, M. L. (2001). The phylogeny of plant and animal pathogens in the Ascomycota. Physiol. Mol. Plant. Pathol. 59, 165–187. doi: 10.1006/pmpp.2001.0355
Bever, J. D., Richardson, S. C., Lawrence, B. M., Holmes, J., and Watson, M. (2009). Preferential allocation to beneficial symbiont with spatial structure maintains mycorrhizal mutualism. Ecol. Lett. 12, 13–21. doi: 10.1111/j.1461-0248.2008.01254.x
Boulos, L., and Al-Dosari, M. (1994). Checklist of the flora of Kuwait. J. Univ. Kuwait 21, 203–218.
Brito, I., Goss, M. J., de Carvalho, M., Chatagnier, O., and van Tuinen, D. (2012). Impact of tillage system on arbuscular mycorrhiza fungal communities in the soil under Mediterranean conditions. Soil Tillage Res. 121, 63–67. doi: 10.1016/j.still.2012.01.012
Brown, G., and Porembski, S. (1997). The maintenance of species diversity by miniature dunes in a sand-depleted Haloxylon salicornicum community in Kuwait. J. Arid Environ. 37, 461–473. doi: 10.1006/jare.1997.0286
Brown, S. P., and Jumpponen, A. (2014). Contrasting primary successional trajectories of fungi and bacteria in retreating glacier soils. Mol. Ecol. 23, 481–497. doi: 10.1111/mec.12487
Brunner, I., Plötze, M., Rieder, S., Zumsteg, A., Furrer, G., and Frey, B. (2011). Pioneering fungi from the Damma glacier forefield in the Swiss Alps can promote granite weathering. Geobiology 9, 266–279. doi: 10.1111/j.1472-4669.2011.00274.x
Buée, M., Reich, M., Murat, C., Morin, E., Nilsson, R. H., Uroz, S., et al. (2009). 454 Pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity. New Phytol. 184, 449–456. doi: 10.1111/j.1469-8137.2009.03003.x
Connell, L., Redman, R., Craig, S., and Rodriguez, R. (2006). Distribution and abundance of fungi in the soils of Taylor Valley. Antarctica. Soil Biol. Biochem. 38, 3083–3094. doi: 10.1016/j.soilbio.2006.02.016
Cui, M., and Nobel, P. S. (1992). Nutrient status, water uptake and gas exchange for three desert succulents infected with Mycorrhizal fungi. New Phytol. 122, 643–649. doi: 10.1111/j.1469-8137.1992.tb00092.x
Daniels, B. A. (1981). The influence of hyperparasites of vesicular-arbuscular mycorrhizal fungi on growth of citrus. Phytopathology 71, 212–213.
de Castro, A. P., Quirino, B. F., Pappas, G., Kurokawa, A. S., Neto, E. L., and Krüger, R. H. (2008). Diversity of soil fungal communities of Cerrado and its closely surrounding agriculture fields. Arch. Microbiol. 190:129. doi: 10.1007/s00203-008-0374-6
de Hoog, G. S., Guarro, J., and Figueras, M. J. (2000). in Atlas of Clinical Fungi, 2nd Edn, ed. G. S. De Hoog (Utrecht: Centraalbureau voor Schimmelcultures).
Donfack, P., Floret, Ch, and Pontanier, R. (1995). Secondary succession in abandoned fields of dry tropical northern Cameroon. J. Veg. Sci. 6, 499–508. doi: 10.2307/3236348
El-Said, A. H. M., and Saleem, A. (2008). Ecological and physiological studies on soil fungi at Western Region. Libya. Mycobiol. 36, 1–9. doi: 10.4489/MYCO.2008.36.1.001
El-Sheikh, M. A., Abbadi, G. A., and Bianco, P. M. (2010). Vegetation ecology of phytogenic hillocks (nabkhas) in coastal habitats of JalAz-Zor National Park. Kuwait: role of patches and edaphic factors. Flora 205, 832–840. doi: 10.1016/j.flora.2010.01.002
Fell, J. W., Scorzetti, G., Connell, L., and Craig, S. (2006). Biodiversity of micro-eukaryotes in Antarctic dry valley soils with < 5% soil moisture. Soil Biol. Biochem. 38, 3107–3119. doi: 10.1016/j.soilbio.2006.01.014
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Gardes, M., and Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x
Gollotte, A., van Tuinen, D., and Atkinson, D. (2004). Diversity of arbuscular Mycorrhizal fungi colonising roots of the grass species Agrostis capillaris and Lolium perenne in a field experiment. Mycorrhiza 14, 111–117. doi: 10.1007/s00572-003-0244-7
Green, L. E., Porras-Alfaro, A., and Sinsabaugh, R. L. (2008). Translocation of nitrogen and carbon integrates biotic crust and grass production in desert grassland. J. Ecol. 96, 1076–1085. doi: 10.1111/j.1365-2745.2008.01388.x
Grishkan, I., and Nevo, E. (2010). Spatiotemporal distribution of soil microfungi in the Makhtesh Ramon area, central negev desert. Israel. Fungal Ecol. 3, 326–337. doi: 10.1016/j.funeco.2010.01.003
Grishkan, I., Zaady, E., and Nevo, E. (2006). Soil crust microfungi along a southward rainfall gradient in desert ecosystems. Eur. J. Soil Biol. 42, 33–42. doi: 10.1016/j.ejsobi.2005.09.014
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucl. Acids Symp. Ser. 41, 95–98.
Hobbie, S. E., Ogdahl, M., Chorover, J., Chadwick, O. A., Oleksyn, J., Ztykowiak, R., et al. (2007). Tree species effects on soil organic matter dynamics: the role of soil cation composition. Ecosystems 10, 999–1018. doi: 10.1007/s10021-007-9073-4
Hobbie, S. E., Reich, P. B., Oleksyn, J., Ogdahl, M., Zytkowiak, R., Hale, C., et al. (2006). Tree species effects on decomposition and forest floor dynamics in a common garden. Ecology 87, 2288–2297. doi: 10.1890/0012-9658(2006)87[2288:TSEODA]2.0.CO;2
Hollister, E. B., Schadt, C. W., Palumbo, A. V., Ansley, R. J., and Boutton, T. W. (2010). Structural and functional diversity of soil bacterial and fungal communities following woody plant encroachment in the southern Great Plains. Soil Biol. Biochem. 42, 1816–1824. doi: 10.1016/j.soilbio.2010.06.022
Hortal, S., Bastida, F., Armas, C., Lozano, Y. M., Moreno, J. L., Garcia, C., et al. (2013). Soil microbial community under a nurse-plant species changes in composition, biomass and activity as the nurse grows. Soil Biol. Biochem. 64, 139–146. doi: 10.1016/j.soilbio.2013.04.018
Jeffries, P., and Barea, J. M. (2001). “Arbuscular mycorrhiza—a key component of sustainable plant–soil ecosystems,” in The Mycota (A Comprehensive Treatise on Fungi as Experimental Systems for Basic and Applied Research), Vol. 9, ed. B. Hock (Berlin: Springer), 95–113. doi: 10.1007/978-3-662-07334-6-6
Jukes, T. H., and Cantor, C. R. (1969). “Evolution of protein molecules,” in: Mammalian Protein Metabolism, ed. H. N. Munro (New York, NY: Academic Press), 21–132. doi: 10.1016/B978-1-4832-3211-9.50009-7
Kahindi, J. H. P., Woomer, P., George, T., de Souza Moreira, F. M., Karanja, N. K., and Giller, K. E. (1997). Agricultural intensification, soil biodiversity and ecosystem function in the tropics: the role of nitrogen-fixing bacteria. Appl. Soil Ecol. 6, 55–76. doi: 10.1016/S0929-1393(96)00151-5
Kavamura, V. N., Taketani, R. G., Lançoni, M. D., Andreote, F. D., Mendes, R., and de Melo, I. S. (2013). Water regime influences bulk soil and rhizosphere of Cereus jamacaru bacterial communities in the brazilian caatinga biome. PLoS One 8:e73606. doi: 10.1371/journal.pone.0073606
Khalaf, F. (1989). Textural characteristics and genesis of the aeolian sediments in the Kuwait desert. Sedimentology 36, 253–271. doi: 10.1111/j.1365-3091.1989.tb00606.x
Kimura, M. (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Meiser, A., Bálint, M., and Schmitt, I. (2014). Meta-analysis of deep-sequenced fungal communities indicates limited taxon sharing between studies and the presence of biogeographic patterns. New Phytol. 201, 623–635. doi: 10.1111/nph.12532
Moebius-Clune, D. J., Anderson, Z. U., and Pawlowska, T. E. (2013). Arbuscular mycorrhizal fungi associated with a single agronomic plant host across the landscape: the structure of an assemblage. Soil Biol. Biochem. 64, 181–190. doi: 10.1016/j.soilbio.2012.10.043
Murgia, M., Fiamma, M., Barac, A., Deligios, M., Mazzarello, V., Paglietti, B., et al. (2018). Biodiversity of fungi in hot desert sands. MicrobiologyOpen doi: 10.1002/mbo3.595 [Epub ahead of print].
Nguyen, L. M., Buttner, M. P., Cruz, P., Smith, S. D., and Robleto, E. A. (2011). Effects of elevated atmospheric CO2 on rhizosphere soil microbial communities in a Mojave Desert ecosystem. J. Arid Environ. 75, 917–925. doi: 10.1016/j.jaridenv.2011.04.028
Oehl, F., Laczko, E., Oberholzer, H.-R., Jansa, J., and Egli, S. (2017). Diversity and biogeography of arbuscular mycorrhizal fungi in agricultural soils. Biol. Fert. Soils 53, 777–797. doi: 10.1007/s00374-017-1217-x
Osonubi, O., Bakare, O. N., and Mulongoy, K. (1992). Interactions between drought stress and vesicular-arbuscular mycorrhiza on the growth of Faidherbia albida (syn. Acacia albida) and Acacia nilotica in sterile and non-sterile soils. Biol. Fert. Soils 14, 159–165. doi: 10.1007/BF00346056
Pande, M., and Tarafdar, J. (2004). Arbuscular mycorrhizal fungal diversity in neem-based agroforestry systems in Rajasthan. Appl. Soil Ecol. 26, 233–241. doi: 10.1016/j.apsoil.2003.12.009
Pedley, L. (1986). “Australian Acacia: taxonomy and phytogeography,” in Australian Acacias in developing countries, Proceedings of the International Workshop at the Forestry Training Centre, ed. J. W. Turnbull (Gympie: ACIAR), 11–16.
Peichl, M., Thevathasan, N. V., Gordon, A. M., Huss, J., and Abohassan, R. A. (2006). Carbon sequestration potentials in temperate tree-based intercropping systems, Southern Ontario. Canada. Agroforest. Syst. 66, 243–257. doi: 10.1007/s10457-005-0361-8
Pointing, S. B., and Belnap, J. (2012). Microbial colonization and controls in dryland systems. Nat. Rev. Microbiol. 10:654. doi: 10.1038/nrmicro2854
Porras-Alfaro, A., Herrera, J., Natvig, D. O., Lipinski, K., and Sinsabaugh, R. L. (2010). Diversity and distribution of soil fungal communities in a semiarid grassland. Mycologia 103, 10–21. doi: 10.3852/09-297
Powell, J. T., Chatziefthimiou, A. D., Banack, S. A., Cox, P. A., and Metcalf, J. S. (2015). ). Desert crust microorganisms, their environment, and human health. J. Arid Environ. 112, 127–133. doi: 10.1016/j.jaridenv.2013.11.004
Qin, H., Wang, H., Strong, P. J., Li, Y., Xu, Q., and Wu, Q. (2014). Rapid soil fungal community response to intensive management in a bamboo forest developed from rice paddies. Soil Biol. Biochem. 68, 177–184. doi: 10.1016/j.soilbio.2013.09.031
Quince, C., Curtis, T. P., and Sloan, W. T. (2008). The rational exploration of microbial diversity. ISME J. 2, 997–1006. doi: 10.1038/ismej.2008.69
R Development Core Team (2010). R: A Language and Environment for Statistical Computing. Computer Programme. Available at: http://www.R-project.org/
Reddell, P. (1993). “Soil constraints to the growth of nitrogen-fixing trees in tropical environments,” in Symbioses in Nitrogen-Fixing Trees, eds N. S. Subba Rao and C. Rodriguez-Barrueco (New York, NY: International Science publisher), 65–84.
Requena, N., Jeffries, P., and Barea, J. M. (1996). Assessment of natural mycorrhizal potential in a desertified semiarid ecosystem. Appl. Environ. Microbiol. 62, 842–847.
Rivest, D., Cogliastro, A., Vanasse, A., and Olivier, A. (2009). Production of soybean associated with different hybrid poplar clones in a tree-based intercropping system in southwestern Québec. Canada. Agric. Ecosyst. Environ. 131, 51–60. doi: 10.1016/j.agee.2008.08.011
Rillig, M. C., and Mummey, D. L. (2006). Tansley review-mycorrhizas and soil structure. New Phytol. 171, 41–53.
Rodríguez-Zaragoza, S., González-Ruíz, T., González-Lozano, E., Lozada-Rojas, A., Mayzlish-Gati, E., and Steinberger, Y. (2008). Vertical distribution of microbial communities under the canopy of two legume bushes in the Tehuacán desert. Mexico. Eur. J. Soil Biol. 44, 373–380. doi: 10.1016/j.ejsobi.2008.05.003
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Ross, J. P., and Ruttencutter, R. (1977). Population dynamics of two versicular-arbuscular endomycorrhizal fungi and the role of hyperparasitic fungi. Phytopathology 67, 490–496.
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Schmidt, S. K., Wilson, K. L., Meyer, A. F., Schadt, C. W., Porter, T. M., and Moncalvo, J. M. (2008). “The missing fungi-new insights from culture-independent molecular studies of soil,” in Accessing Uncultivated Microorganisms: From the Environment to Organisms and Genomes and Back, ed. K. Zengler (Washington, DC: American Society for Microbiology Press), 55–56.
Sene, G., Samba-Mbaye, R., Thiao, M., Khasa, D., and Kane, A. (2012a). The abundance and diversity of legume-nodulating rhizobia and arbuscular mycorrhizal fungal communities in soil samples from deforested and man-made forest systems in a semiarid Sahel region in Senegal. Eur. J. Soil Biol. 52, 30–40. doi: 10.1016/j.ejsobi.2012.05.005
Sene, G., Thiao, M., Manga, A., Kane, A., and Samba-Mbaye, R. (2012b). Arbuscular mycorrhizal soil infectivity and spores distribution across plantations of tropical, subtropical and exotic tree species: a case study from the forest reserve of Bandia, Senegal. Afr. J. Ecol. 50, 218–232. doi: 10.1111/j.1365-2028.2011.01315.x
Sene, G., and Sylla, S. N. (2014). L’utilisation de plantations d’essences forestières au sahel: impacts sur la microflore symbiotique sous jacente et les caractéristiques physico-chimiques des sols. Press. Acad. Francophones 384.
Sene, G., Thiao, M., Samba-Mbaye, R., Khasa, D., Kane, A., Mbaye, M. S., et al. (2013). The abundance and diversity of legume-nodulating rhizobia in 28-year-old plantations of tropical, subtropical and exotic tree species: a case study from the forest reserve of Bandia. Senegal. Microbiol. Ecol. 65, 128–144. doi: 10.1007/s00248-012-0094-y
Sene, G., Thiao, M., Samba-Mbaye, R., Ndoye, F., Kane, A., Diouf, D., et al. (2010). Response of three peanut cultivars toward inoculation with two Bradyrhizobium strains and an arbuscular mycorrhizal fungus in Senegal. Afr. J. Microbiol. Res. 4, 2520–2527.
Shi, Z. Y., Zhang, L. Y., Li, X. L., Feng, G., Tian, C. Y., and Christie, P. (2007). Diversity of arbuscular mycorrhizal fungi associated with desert ephemerals in plant communities of Junggar Basin, northwest China. Appl. Soil Ecol. 35, 10–20. doi: 10.1016/j.apsoil.2006.06.002
Sterflinger, K., Tesei, D., and Zakharova, K. (2012). Fungi in hot and cold deserts with particular reference to microcolonial fungi. Fungal Ecol. 5, 453–462. doi: 10.1016/j.funeco.2011.12.007
Sugiyama, M., Ohara, A., and Mikawa, T. (1999). Molecular phylogeny of onygenalean fungi based on small subunit ribosomal DNA (SSU rDNA) sequences. Mycoscience 40, 251–258. doi: 10.1007/BF02463962
Suleiman, M. K., Quoreshi, A. M., Bhat, N. R., and Manuvel, A. J. (2018). Species identification of Vachellia pachyceras from Kuwait and its relatives Vachellia gerrardii and Vachellia tortilis based on multilocus plastid gene sequences. Edinburgh J. Bot. 75, 73–90. doi: 10.1017/S0960428617000385
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Titus, J. H., Nowak, R. S., and Smith, S. D. (2002). Soil resource heterogeneity in the mojave desert. J. Arid Environ. 52, 269–292. doi: 10.1006/jare.2002.1010
van der Heijden, M. G. A., Bardgett, R. D., and van Straalen, N. M. (2008). The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11, 296–310. doi: 10.1111/j.1461-0248.2007.01139.x
van der Heijden, M. G. A., Klironomos, J. N., and Ursic, M. (1998). Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396, 69–72.
van Tuinen, D., Jacquot, E., Zhao, B., Gollotte, A., and Gianinazzi-Pearson, V. (1998). Characterization of root colonization profiles by a microcosm community of arbuscular mycorrhizal fungi using 25S rDNA-targeted nested PCR. Mol. Ecol. 7, 879–887. doi: 10.1046/j.1365-294x.1998.00410.x
Vincke, C., Diédhiou, I., and Grouzis, M. (2010). Long-term dynamics and structure of woody vegetation in the Ferlo (Senegal). J. Arid Environ. 74, 268–276. doi: 10.1016/j.jaridenv.2009.08.006
Wang, X., Chen, F., and Dong, Z. (2006). The relative role of climatic and human factors in desertification in semiarid China. Global Environ. Chang. 16, 48–57. doi: 10.1016/j.gloenvcha.2005.06.006
Weber, C. F., Zak, D. R., Hungate, B. A., Jackson, R. B., Vilgalys, R., Evans, R. D., et al. (2011). Responses of soil cellulolytic fungal communities to elevated atmospheric CO2 are complex and variable across five ecosystems. Environ. Microbiol. 13, 2778–2793. doi: 10.1111/j.1462-2920.2011.02548.x
Welc, M., Frossard, E., Egli, S., Bünemann, E. K., and Jansa, J. (2014). Rhizosphere fungal assemblages and soil enzymatic activities in a 110-years alpine chronosequence. Soil Biol. Biochem. 74, 21–30. doi: 10.1016/j.soilbio.2014.02.014
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (Cambridge, MA: Academic Press),315–322.
Xu, L., Ravnskov, S., Larsen, J., and Nicolaisen, M. (2012). Linking fungal communities in roots, rhizosphere, and soil to the health status of Pisum sativum. FEMS Microbiol. Ecol. 82, 736–745. doi: 10.1111/j.1574-6941.2012.01445.x
Xu, X., Chen, C., Zhang, Z., Sun, Z., Chen, Y., Jiang, J., et al. (2017). The influence of environmental factors on communities of arbuscular mycorrhizal fungi associated with Chenopodium ambrosioides revealed by MiSeq sequencing investigation. Sci. Rep. 7:45134. doi: 10.1038/srep45134
Zhang, X., Xu, S., Li, C., Zhao, L., Feng, H., Yue, G., et al. (2014). The soil carbon/nitrogen ratio and moisture affect microbial community structures in alkaline permafrost-affected soils with different vegetation types on the Tibetan plateau. Res. Microbiol. 165, 128–139. doi: 10.1016/j.resmic.2014.01.002
Keywords: soil fungal communities, Lonely Tree, Vachellia pachyceras, rhizosphere soils, Kuwait desert ecosystem
Citation: Suleiman MK, Dixon K, Commander L, Nevill P, Quoreshi AM, Bhat NR, Manuvel AJ and Sivadasan MT (2019) Assessment of the Diversity of Fungal Community Composition Associated With Vachellia pachyceras and Its Rhizosphere Soil From Kuwait Desert. Front. Microbiol. 10:63. doi: 10.3389/fmicb.2019.00063
Received: 28 August 2018; Accepted: 15 January 2019;
Published: 31 January 2019.
Edited by:
Raffaella Balestrini, Institute for Sustainable Plant Protection, Italian National Research Council (IPSP-CNR), ItalyReviewed by:
Cristiana Sbrana, Istituto di Biologia e Biotecnologia Agraria (IBBA), ItalyRavindra Nath Kharwar, Banaras Hindu University, India
Copyright © 2019 Suleiman, Dixon, Commander, Nevill, Quoreshi, Bhat, Manuvel and Sivadasan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Majda K. Suleiman, bWtoYWxpbEBraXNyLmVkdS5rdw== Ali M. Quoreshi, YXF1b3Jlc2hpQGtpc3IuZWR1Lmt3
 Majda K. Suleiman
Majda K. Suleiman Kingsley Dixon2
Kingsley Dixon2 Paul Nevill
Paul Nevill Ali M. Quoreshi
Ali M. Quoreshi Narayana R. Bhat
Narayana R. Bhat Anitha J. Manuvel
Anitha J. Manuvel Mini T. Sivadasan
Mini T. Sivadasan