- 1National and Local Joint Engineering Research Center of Storage, Processing and Safety Control Technology for Fresh Agricultural and Aquatic Products, Bohai University, Jinzhou, China
- 2Fresh Food Storage and Processing Technology Research Institute of Liaoning Provincial Universities, Bohai University, Jinzhou, China
- 3College of Food Science and Project Engineering, Bohai University, Jinzhou, China
- 4State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, Beijing, China
IncI-complex plasmids can be divided into seven subgroups IncI1, IncI2, IncIγ, IncB/O, IncK1, IncK2, and IncZ. In this study, a replicon-based scheme was proposed for typing IncI-complex plasmids into four separately clustering subgroups IncI2, IncI1/B/O, IncIγ/K1 and IncK2/Z, the last three of which were combined from IncI1 and IncB/O, IncIγ and IncK1, and IncK2 and IncZ, respectively. Four IncIγ/K1 plasmids p205880-NR2, p14E509-CTXM, p11011-CTXM and p61806-CTXM were fully sequenced and compared with IncIγ/K1 reference pCT, IncI2 reference R721, IncI1/B/O reference R64 and IncK2/Z reference pO26-CRL-125. These plasmids shared conserved gene organization in the replication and conjugal transfer regions, but displaying considerable sequence diversity among different subgroups. Remarkable modular differences were observed in the maintenance and transfer leading regions. p205880-NR2 contained no resistance genes or accessory modules, while the other seven plasmids acquired one or more accessory modules, which harbored mobile elements [including unit transposons, insertion sequence (IS)-based transposition units and individual IS elements] and associated resistance markers (especially including those involved in resistance to β-lactams, aminoglycosides, tetracyclins, phenicols, streptomycins, trimethoprims, sulphonamides, tunicamycins and erythromycins). Data presented here provided a deeper insight into diversification and evolution of IncI-complex plasmids.
Introduction
Plasmids of the I incompatibility complex (IncI-complex) produce I-type pili. Based on morphological and serological similarities of their pili, IncI-complex plasmids can be divided into at least seven subgroups, namely IncI1 ( = IncIα or Com1), IncI2 ( = IncIδ), IncIγ, IncB/O, IncK1, IncK2, and IncZ (Tschäpe and Tietze, 2010; Papagiannitsis et al., 2011; Rozwandowicz et al., 2017). IncI-complex plasmids are low copy-number, narrow-host-range, conjugative plasmids, varying in size from 50 to 250 kb. The backbone of an IncI-complex plasmid can be dividedinto regions of replication, maintenance, transfer leading and conjugal transfer (Sampei et al., 2010). Two protein genes (repZ and repY), one regulatory RNA gene (inc) and four functional DNA sequences (CIS, oriV, ter, and iterons) in the replication region are essential for plasmid replication and copy number control (Sampei et al., 2010). The partitioning genes parA and parB in the maintenance region are responsible for active partition of replicated DNA into daughter cells during cell division. Two gene clusters tra and pil in the conjugal transfer region encoded the thick rigid pilus as the primary pili implicated in conjugation transfer, and the thin flexible pilus increasing conjugation rate in liquid medium, respectively (Bradley, 1984). A multiple inversion system named shufflon mediates rearrangement of PilV protein by a plasmid-encoded site-specific recombinase Rci (Komano et al., 1987). This recombination event selects one of seven different pilV genes, which is responsible for the determination of recipient specificity (Komano et al., 1994).
The purpose of this study is to provide a deeper insight into genomic diversity and evolution of IncI-complex plasmids. First, a replicon-based scheme was established to divide IncI-complex plasmids into four subgroups IncI2, IncI1/B/O, IncIγ/K1, and IncK2/Z. Then, four IncIγ/K1 plasmids p14E509-CTXM, p11011-CTXM, p61806-CTXM, and p205880-NR2 were fully sequenced and compared to the reference plasmids pCT (accession number FN868832) (Cottell et al., 2011), R721 (accession number AP002527), R64 (accession number AP005147) (Sampei et al., 2010), and pO26-CRL-125 (accession number KC340959) (Venturini et al., 2013) of the above four subgroups.
Materials and Methods
Bacterial Isolates
Escherichia coli 14E509 was isolated in 2014 from a urine specimen of an infant with pneumonia in a public hospital in Henan City, China. E. coli 11011 was isolated in 2014 from a bile specimen of an elderly patient with bile duct calculus in a public hospital in Ningbo City, China. E. coli 61806 was isolated in 2015 from a whole blood specimen of an elderly patient with septic shock in a teaching hospital in Henan City, China. Klebsiella pneumoniae 205880 was isolated in 2012 from a sputum specimen of an elderly patient with pneumonia in a teaching hospital in Chongqing City, China.
Genomic DNA Sequencing and Sequence Assemble
Genomic DNA was isolated from each isolate using a Qiagen Blood & Cell Culture DNA Maxi Kit (Qiagen, Hilden, Germany). The genomic DNA of strain 61806 was sequenced from a mate-pair libraries with average insert size of 5 kb (ranged from 2 to 10 kb) using a MiSeq sequencer (Illumina, CA, United States). Quality control, removing adapters and low quality reads, were performed using Trimmomatic 0.36 (Bolger et al., 2014). The filtered clean reads were then assembled using Newbler 2.6 (Nederbragt, 2014), followed by extraction of the consensus sequence with CLC Genomics Workbench 3.0 (Qiagen Bioinformatics). Gaps between contigs were filled using a combination of PCR and Sanger sequencing using an ABI 3730 Sequencer (LifeTechnologies, CA, United States), and Gapfiller V1.11 (Boetzer and Pirovano, 2012) was used for gap closure.
For all the other three isolates (14E509, 11011 and 205880), genome sequencing was performed with a sheared DNA library with average size of 15 kb (ranged from 10 to 20 kb) on a PacBio RSII sequencer (Pacific Biosciences, CA, United States), as well as a paired-end library with an average insert size of 400 bp (ranged from 150 to 600 kb) on a HiSeq sequencer (Illumina, CA, United States). The paired-end short Illumina reads were used to correct the long PacBio reads utilizing proovread (Hackl et al., 2014), and then the corrected PacBio reads were assembled de novo utilizing SMARTdenovo (available from https://github.com/ruanjue/smartdenovo).
Sequence Annotation and Genome Comparison
Open reading frames and pseudogenes were predicted using RAST 2.0 (Brettin et al., 2015) combined with BLASTP/BLASTN (Boratyn et al., 2013) searches against the UniProtKB/Swiss-Prot database (Boutet et al., 2016) and the RefSeq database (O’Leary et al., 2016). Annotation of resistance genes, mobile elements, and other features was carried out using the online databases including CARD (Jia et al., 2017), ResFinder (Zankari et al., 2012), ISfinder (Siguier et al., 2006), INTEGRALL (Moura et al., 2009), and Tn Number Registry (Roberts et al., 2008). Multiple and pairwise sequence comparisons were performed using MUSCLE 3.8.31 (Edgar, 2004) and BLASTN, respectively. Gene organization diagrams were drawn in Inkscape 0.48.11.
Phylogenetic Analysis
The nucleotide sequences of repZ coding regions of indicative plasmids were aligned using MUSCLE 3.8.31 (Edgar, 2004). Unrooted neighbor-joining trees were generated from aligned repZ sequences using MEGA7 (Kumar et al., 2016), and evolutionary distances were estimated using maximum composite likelihood method, with a bootstrap iteration of 1000.
Plasmid Conjugal Transfer
Plasmid conjugal transfer experiments were carried out with the rifampin-resistant E. coli EC600 or the sodium azide-resistant E. coli J53 being used as a recipient and each of the 14E509, 11011 and 61806 isolates as a donor. Three milliliters of overnight cultures of each of donor and recipient bacteria were mixed together, harvested and resuspended in 80 μl of Brain Heart Infusion (BHI) broth (BD Biosciences, NJ, United States). The mixture was spotted on a 1 cm2 hydrophilic nylon membrane filter with a 0.45 μm pore size (Millipore, MA, United States) that was placed on BHI agar (BD Biosciences, NJ, United States) plate and then incubated for mating at 37°C for 12 to 18 h. Bacteria were washed from filter membrane and spotted on Muller–Hinton (MH) agar (BD Biosciences, NJ, United States) plates containing 1000 μg/ml rifampin or 200 μg/ml sodium azide together with 200 μg/ml ampicillin for selecting an E. coli transconjugant that carrying blaCTX-M.
Double-Disk Synergy Test
To detect ESBL activity, the double-disk synergy test was performed as recommended by the National Committee for Clinical and Laboratory Standards Institute (CLSI) guideline (CLSI, 2015). Briefly, the bacterial strains tested were spread onto MH agar plates and four disks containing cefotaxime (30 μg), ceftazidime (30 μg), cefotaxime (30 μg) plus clavulanic acid (10 μg), and ceftazidime (30 μg) plus clavulanic acid (10 μg) were applied to each agar plate. The agar plates were incubated overnight at 37°C, and the production of ESBL was inferred when the zone of either cephalosporin was ≥5 mm larger than those without clavulanic acid.
Bacterial Antimicrobial Susceptibility Test
Bacterial antimicrobial susceptibility was tested by BioMérieux VITEK 2 and interpreted as per the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2015).
Nucleotide Sequence Accession Numbers
The complete sequence of plasmids p14E509-CTXM, p11011-CTXM, p205880-NR2, and p61806-CTXM were submitted to GenBank under accession numbers MG764547, MF344575, MF344577, and MF344576, respectively.
Results and Discussion
Replicon-Based Typing of IncI-Complex Plasmids
To better understand the evolutionary relationship of IncI-complex plasmids, a phylogenetic tree (Figure 1) was constructed from the nucleotide sequences of repZ (replication initiation) coding regions of 66 representative (arbitrarily selected) sequenced IncI-complex plasmids (Supplementary Table S1). These 66 plasmids could be divided into four separately clustering subgroups, namely IncI2, IncI1/B/O, IncIγ/K1, and IncK2/Z, the last three of which were combined from IncI1 and IncB/O, IncIγ and IncK1, and IncK2 and IncZ, respectively. As shown by pairwise comparison of repZ sequences, plasmids within each of these four subgroups shared >95% nucleotide identity, by contrast <95% sequence identity was observed between different subgroups (Supplementary Table S2).
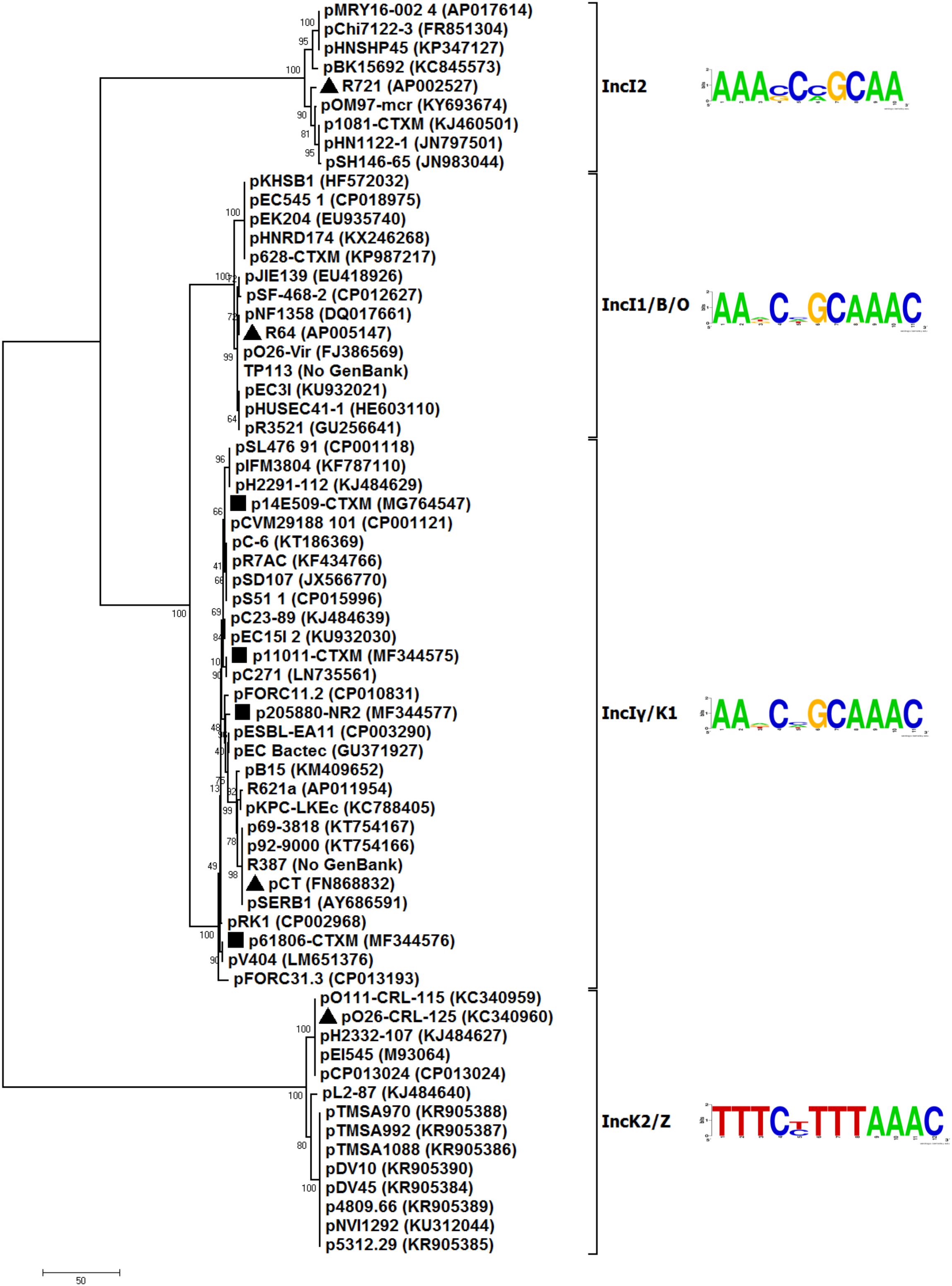
Figure 1. Neighbor-joining phylogenetic tree. The degree of support (percentage) for each cluster of associated taxa, as determined by bootstrap analysis, is shown next to each branch. The bar corresponds to the scale of sequence divergence. Triangles indicate reference plasmids for IncIγ/K1, IncI2, IncI1/B/O, and IncK2/Z subgroups, while squares denote four plasmids sequences in this study.
Putative iterons (RepZ-binding sites) were found to be located 176 to 301 bp downstream of repZ for all these 66 plasmids analyzed (Supplementary Table S1). Plasmids within each of these four subgroups shared a conserved iteron motif (Figure 1) and an identical iteron copy number (Supplementary Table S1). Iteron motifs from different subgroups dramatically differed from one another except for IncIγ/K1 and IncI1/B/O (these two had very similar iteron motifs). IncK2/Z plasmids had two copy numbers of iteron, while plasmids of all the other subgroups had three copy numbers.
Plasmid compatibility among original seven subgroups of IncI-complex had been partially validated experimentally (Praszkier et al., 1991; Rozwandowicz et al., 2017). IncIγ reference plasmid R621a was compatible with IncI1 plasmids (Bird and Pittard, 1982), and IncK2 plasmids was compatible with IncK1 reference plasmid pCT and IncB/O reference plasmid pR3521 (Rozwandowicz et al., 2017). IncZ plasmids were compatible with IncK1, IncIγ and IncI1 plasmids (Praszkier and Pittard, 2005). Plasmid compatibility among the four IncI-complex subgroups IncIγ/K1, IncI1/B/O, IncK2/Z, and IncI2 needed to be elucidated.
Overview of Sequenced IncIγ/K1 Plasmids
The four IncIγ/K1 plasmids p14E509-CTXM, p11011-CTXM, p61806-CTXM, and p205880-NR2 sequenced in the present work varied in size from about 83 kb to nearly 132 kb with variation in corresponding number of predicted ORFs (Table 1). The former three plasmids integrated various accessory modules (defined as acquired DNA regions associated with or bordered by mobile elements), which harbored resistance genes and metabolic gene clusters, and associated unit transposons, insertion sequence (IS)-based transposition units and individual IS elements. By contrast, p205880-NR2 contained no resistance genes or accessory modules (Table 1 and Supplementary Figure S1), thereby representing a prototype IncIγ/K1 plasmid.
These four IncIγ/K1 plasmids, together with pCT (IncIγ/K1 reference), R721 (IncI2 reference), R64 (IncI1/B/O reference) and pO26-CRL-125 (IncK2/Z reference), were included in a genomic comparison. Considerable modular and sequence diversity were found among the backbones of these eight plasmids (Supplementary Table S3 and Figure 2). IncIγ/K1 plasmids p61806-CTXM and pCT shared >99% nucleotide identity across >81% of their backbone sequences. IncIγ/K1 plasmids p14E509-CTXM, p11011-CTXM and p205880-NR2, and IncI1/B/O plasmid R64 shared >99% nucleotide identity across >74% of their backbone sequences. pCT and IncK2/Z plasmid pO26-CRL-125 shared >95% nucleotide identity over >70% of their backbone sequences. The backbone of IncI2 plasmid R721 displayed very low level (<3% BLAST coverage and >82% nucleotide identity) of sequence identity to the other seven plasmids.
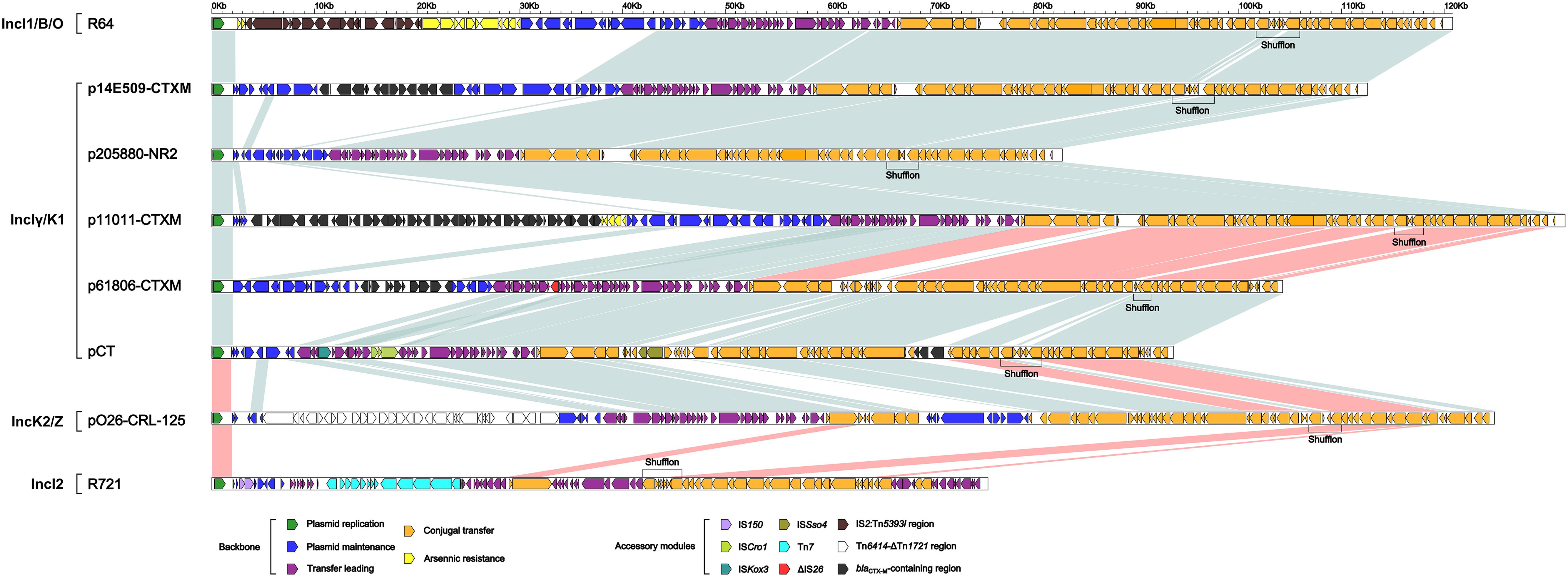
Figure 2. Linear comparison of sequenced plasmids. Genes are denoted by arrows and colored based on gene function classification. Shading regions denote regions of homology (light blue: >95% nucleotide identity; light red: very low nucleotide identity with conserved gene functions).
Each of these eight plasmids had its unique backbone genes or gene loci, especially including those in the maintenance regions. All these plasmids shared the backbone genes or gene loci inc-repY-repZ-CIS-oriV-ter (plasmid replication), parA (maintenance), and nicA, rlx, rci, pil and tra (conjugal transfer) and also the conserved gene organization in the replication and conjugal transfer regions, but with remarkable nucleotide and amino acid diversity among different subgroups.
Notably, p61806-CTXM and pCT had IncIγ/K1-type conjugal transfer regions, while p14E509-CTXM, p11011-CTXM and p205880-NR2 had IncI1/B/O-type conjugal transfer regions as observed in R64. It was speculated that homologous recombination-mediated horizontal transfer of conjugal transfer regions occurred between IncIγ/K1 and IncI1/B/O.
Accessory Modules
p14E509-CTXM, p11011-CTXM, p61806-CTXM, pCT, R721, R64 and pO26-CRL-125 carried different profiles of accessory modules (Table 1). R721 harbored two separate accessory modules, namely Tn7 (see reference Zhan et al., 2018 for gene organization) and IS150. R64 contained a single 17.2 kb accessory module, designated the IS2:Tn5393l region, which was generated from insertion of Tn5393l [a novel derivative of prototype Tn5393c (Cain and Hall, 2011)] into IS2 (Supplementary Figure S2). pO26-CRL-125 contained a 28.9 kb Tn6414-ΔTn1721 region (as its sole accessory module) with a Tn6414-oriVIncP-1α–ΔTn1721 structure (Supplementary Figure S3). Tn6414 was a novel derivative of Tn21, which was resulted from insertion of In2 into a backbone structure carrying the core transposition module tnpAR-res-tnpM together with the mer locus. Tn6414 differed from Tn21 by insertion of In13 instead of In2. In13 is atypical due to integration of two overlapping transposons Tn6029 and Tn4325 downstream of the dfrA5 single-gene cassette.
IncI-complex plasmids carry a wide range of resistance genes (Supplementary Table S4 which extended-spectrum β-lactamase (ESBL) genes are often identified. Indeed, each of the four IncIγ/K1 plasmids p14E509-CTXM, pCT, p61806-CTXM, and p11011-CTXM carried a blaCTX-M-containing region (Figure 3); besides, individual IS elements were found as additional accessory modules: ISKox3, ISCro1, and ISSso4 in pCT, and ΔIS26 in p61806-CTXM. In these four blaCTX-M-containing regions, different truncated versions of prototype ISEcp1-blaCTX-M-14 or 65-IS903D unit were connected with additional resistance regions: truncated aacC2-tmrB region in p14E509-CTXM, ΔTn6295 in p61806-CTXM, and the other three copies of truncated ISEcp1-blaCTX-M-14 or 65-IS903D unit in p11011-CTXM. Complex homologous recombination, which was probably mediated by the common region IS15DI, would be involved in assembly of these repeated blaCTX-M-containing regions. Connection of blaCTX-M-containing regions with additional resistance regions led to MDR of p14E509-CTXM and p11011-CTXM.
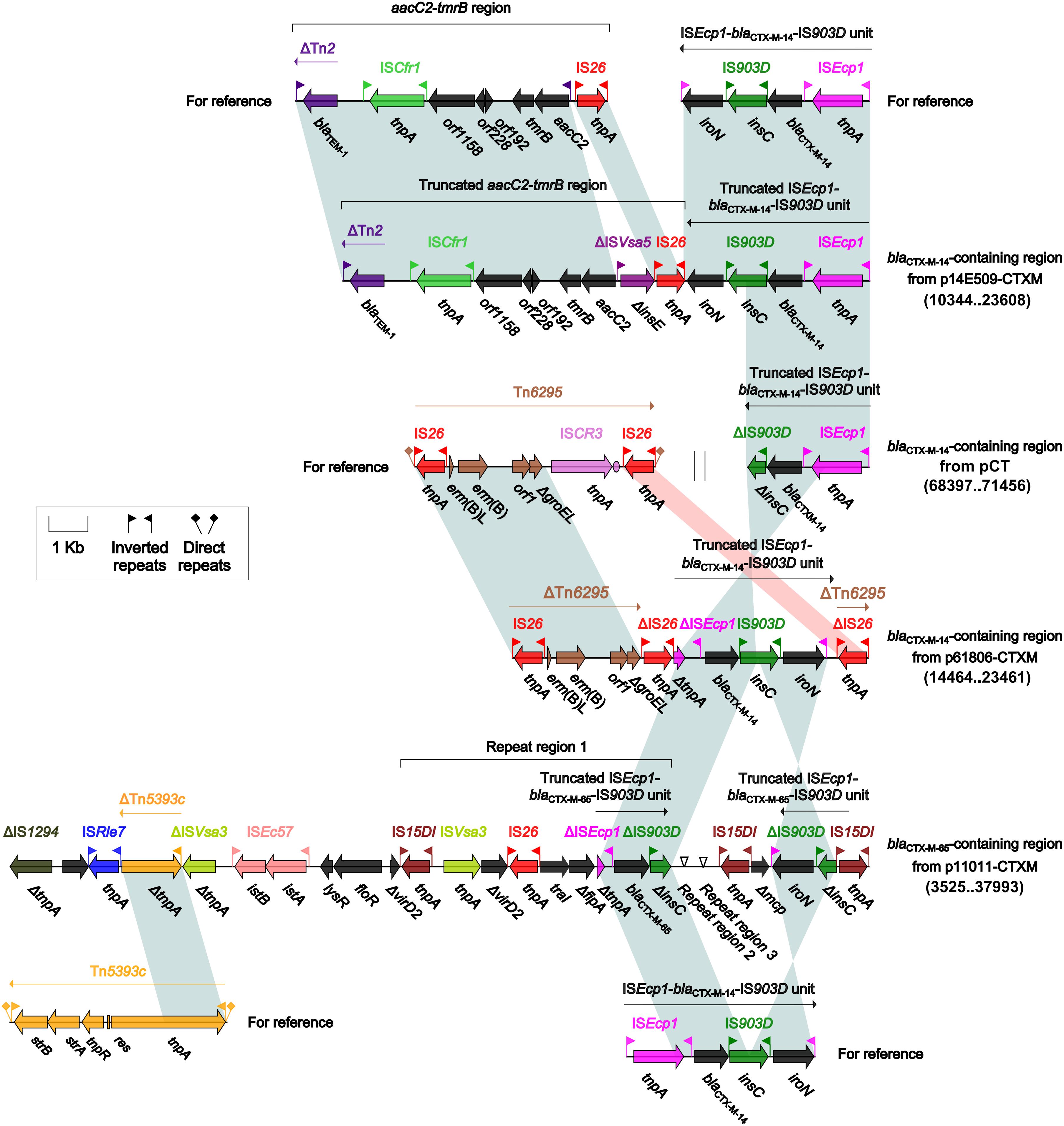
Figure 3. blaCTX-M-containingregions. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on function classification. Shading denotes regions of homology (>95% nucleotide identity). Numbers in brackets indicate nucleotide positions within corresponding plasmids. The accession numbers of the aacC2–tmrB region, the ISEcp1–blaCTX-M-14–IS903D unit, Tn6295 and Tn5393c for reference are JX101693, KP987215, KX646543, and AF262622, respectively.
Transferability and Antimicrobial Susceptibility
The blaCTX-M-carrying plasmids p14E509-CTXM, p11011-CTXM and p61806-CTXM could be transferred from the wild-type isolates into E. coli J53 or EC600 through conjugation, generating three transconjugants 14E509-CTXM-J53, 11011-CTXM-EC600 and 61806-CTXM-EC600, respectively. All these three transconjugants had the ESBL activity (data not shown) and, as expected, were resistant to ampicillin, piperacillin, cefazolin, cefuroxime and ceftazidime but remained susceptible to ampicillin/sulbactam, piperacillin/tazobactam, imipenem and meropenem (Supplementary Table S5).
Ethics Statement
The use of human specimens and all related experimental protocols were approved by the Committee on Human Research of the Henan Provincial People’s Hospital, Ningbo Medical Treatment Center Lihuili Hospital, the First Affiliated Hospital of Henan University, and the First Affiliated Hospital of Chongqing Medical University, and carried out in accordance with the approved guidelines. The research involving biohazards and all related procedures were approved by the Biosafety Committee of the Beijing Institute of Microbiology and Epidemiology.
Author Contributions
DsZ and ZY conceived the study and designed experimental procedures. DfZ, YZ, JF, LH, and XJ performed the experiments. ZZ, HY, WY, BG, JW, and JL analyzed the data. ZZ, YZ, and JF contributed reagents and materials. DsZ, ZY, DfZ and YZ wrote this manuscript.
Funding
This work was supported by the National Key Program (2018ZX10733402) for Infectious Diseases of China and the National Key R&D Program of China (2018YFC1200100).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00048/full#supplementary-material
FIGURE S1 | Plasmid schematic maps. Genes are denoted by arrows, and the backbone and accessory module regions are highlighted in black and gray, respectively. The innermost circle presents GC-skew [(G-C)/(G+C)], with a window size of 500 bp and a step size of 20 bp. The next-to-innermost circle presents GC content.
FIGURE S2 | The IS2:Tn5393l region from R64. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on function classification.
FIGURE S3 | The MDR region from pO26-CRL-125. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on function classification. Shading denotes regions of homology (>95% nucleotide identity). Numbers in brackets indicate nucleotide position within pO26-CRL-125. The accession numbers of Tn1721 and Tn21 for reference are X61367 and AF071413, respectively.
Footnotes
References
Bird, P. I., and Pittard, J. (1982). An unexpected incompatibility interaction between two plasmids belonging to the I compatibility complex. Plasmid 8, 211–214. doi: 10.1016/0147-619X(82)90059-2
Boetzer, M., and Pirovano, W. (2012). Toward almost closed genomes with GapFiller. Genome Biol. 13:R56. doi: 10.1186/gb-2012-13-6-r56
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boratyn, G. M., Christiam, C., Cooper, P. S., George, C., Amelia, F., Ning, M., et al. (2013). BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41, 29–33. doi: 10.1093/nar/gkt282
Boutet, E., Lieberherr, D., Tognolli, M., Schneider, M., Bansal, P., Bridge, A. J., et al. (2016). UniProtKB/Swiss-Prot, the manually annotated section of the uniprot knowledgebase: how to use the entry view. Methods Mol. Biol. 1374, 23–54. doi: 10.1007/978-1-4939-3167-5_2
Bradley, D. E. (1984). Characteristics and function of thick and thin conjugative pili determined by transfer-derepressed plasmids of incompatibility groups I1, I2, I5, B, K and Z. J. Gen. Microbiol. 130, 1489–1502. doi: 10.1099/00221287-130-6-1489
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Cain, A. K., and Hall, R. M. (2011). Transposon Tn5393e carrying the aphA1-containing transposon Tn6023 upstream of strAB does not confer resistance to streptomycin. Microb. Drug Resist. 17, 389–394. doi: 10.1089/mdr.2011.0037
CLSI (2015). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fifth Informational Supplement M100-S25. Wayne, PA: Clinical and Laboratory Standards Institute.
Cottell, J. L., Webber, M. A., Coldham, N. G., Taylor, D. L., Cerdeno-Tarraga, A. M., Hauser, H., et al. (2011). Complete sequence and molecular epidemiology of IncK epidemic plasmid encoding blaCTX-M-14. Emerg. Infect. Dis. 17, 645–652. doi: 10.3201/eid1704.101009
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Hackl, T., Hedrich, R., Schultz, J., and Forster, F. (2014). proovread: large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics 30, 3004–3011. doi: 10.1093/bioinformatics/btu392
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Komano, T., Kim, S. R., Yoshida, T., and Nisioka, T. (1994). DNA rearrangement of the shufflon determines recipient specificity in liquid mating of IncI1 plasmid R64. J. Mol. Biol. 243, 6–9. doi: 10.1006/jmbi.1994.1625
Komano, T., Kubo, A., and Nisioka, T. (1987). Shufflon: multi-inversion of four contiguous DNA segments of plasmid R64 creates seven different open reading frames. Nucleic Acids Res. 15, 1165–1172. doi: 10.1093/nar/15.3.1165
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Moura, A., Soares, M., Pereira, C., Leitão, N., Henriques, I., and Correia, A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098. doi: 10.1093/bioinformatics/btp105
Nederbragt, A. J. (2014). On the middle ground between open source and commercial software - the case of the Newbler program. Genome Biol. 15:113. doi: 10.1186/gb4173
O’Leary, N. A., Wright, M. W., Rodney, B. J., Stacy, C., Diana, H., Rich, M. V., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745. doi: 10.1093/nar/gkv1189
Papagiannitsis, C. C., Tzouvelekis, L. S., Kotsakis, S. D., Tzelepi, E., and Miriagou, V. (2011). Sequence of pR3521, an IncB plasmid from Escherichia coli encoding ACC-4, SCO-1, and TEM-1 β-Lactamases. Antimicrob. Agents Chemother. 55, 376–381. doi: 10.1128/AAC.00875-10
Praszkier, J., and Pittard, A. J. (2005). Control of replication in I-complex plasmids. Plasmid 53, 97–112. doi: 10.1016/j.plasmid.2004.12.005
Praszkier, J., Wei, T., Siemering, K., and Pittard, J. (1991). Comparative analysis of the replication regions of IncB, IncK, and IncZ plasmids. J. Bacteriol. 173, 2393–2397. doi: 10.1128/jb.173.7.2393-2397.1991
Roberts, A. P., Chandler, M., Courvalin, P., Guedon, G., Mullany, P., Pembroke, T., et al. (2008). Revised nomenclature for transposable genetic elements. Plasmid 60, 167–173. doi: 10.1016/j.plasmid.2008.08.001
Rozwandowicz, M., Brouwer, M. S. M., Zomer, A. L., Bossers, A., Harders, F., Mevius, D. J., et al. (2017). Plasmids of distinct IncK lineages show compatible phenotypes. Antimicrob. Agents Chemother. 61:e01954-16. doi: 10.1128/AAC.01954-16
Sampei, G., Furuya, N., Tachibana, K., Saitou, Y., Suzuki, T., Mizobuchi, K., et al. (2010). Complete genome sequence of the incompatibility group I1 plasmid R64. Plasmid 64, 92–103. doi: 10.1016/j.plasmid.2010.05.005
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, 32–36. doi: 10.1093/nar/gkj014
Tschäpe, H., and Tietze, E. (2010). Characterization of conjugative plasmids belonging to a new incompatibility group (IncZ). J. Basic Microbiol. 23, 393–401.
Venturini, C., Hassan, K. A., Roy Chowdhury, P., Paulsen, I. T., Walker, M. J., and Djordjevic, S. P. (2013). Sequences of two related multiple antibiotic resistance virulence plasmids sharing a unique IS26-related molecular signature isolated from different Escherichia coli pathotypes from different hosts. PLoS One 8:e78862. doi: 10.1371/journal.pone.0078862
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Keywords: IncI-complex plasmids, IncIγ/K1, CTX-M, multi-drug resistance, plasmids
Citation: Zhang D, Zhao Y, Feng J, Hu L, Jiang X, Zhan Z, Yang H, Yang W, Gao B, Wang J, Li J, Yin Z and Zhou D (2019) Replicon-Based Typing of IncI-Complex Plasmids, and Comparative Genomics Analysis of IncIγ/K1 Plasmids. Front. Microbiol. 10:48. doi: 10.3389/fmicb.2019.00048
Received: 28 September 2018; Accepted: 14 January 2019;
Published: 29 January 2019.
Edited by:
Charles W. Knapp, University of Strathclyde, United KingdomReviewed by:
Jian-Hua Liu, South China Agricultural University, ChinaLiang Li, Los Angeles Biomedical Research Institute, United States
Copyright © 2019 Zhang, Zhao, Feng, Hu, Jiang, Zhan, Yang, Yang, Gao, Wang, Li, Yin and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhe Yin, amVycnk5eWluQDE2My5jb20= Dongsheng Zhou, ZG9uZ3NoZW5nemhvdTE5NzdAZ21haWwuY29t
†These authors have contributed equally to this work
 Defu Zhang1,2,3,4†
Defu Zhang1,2,3,4† Wenhui Yang
Wenhui Yang Jianrong Li
Jianrong Li Zhe Yin
Zhe Yin Dongsheng Zhou
Dongsheng Zhou