- 1Department of Respiratory Medicine, Nagasaki University Hospital, Nagasaki, Japan
- 2Department of Infectious Diseases, Nagasaki University Graduate School of Biomedical Sciences, Nagasaki, Japan
- 3Department of Laboratory Medicine, Nagasaki University Graduate School of Biomedical Sciences, Nagasaki, Japan
Candida glabrata is one of the leading causes of candidiasis and serious invasive infections in hosts with weakened immune systems. C. glabrata is a haploid budding yeast that resides in healthy hosts. Little is known about the mechanisms of C. glabrata virulence. Autophagy is a ‘self-eating’ process developed in eukaryotes to recycle molecules for adaptation to various environments. Autophagy is speculated to play a role in pathogen virulence by supplying sources of essential proteins for survival in severe host environments. Here, we investigated the effects of defective autophagy on C. glabrata virulence. Autophagy was induced by nitrogen starvation and hydrogen peroxide (H2O2) in C. glabrata. A mutant strain lacking CgAtg1, an autophagy-inducing factor, was generated and confirmed to be deficient for autophagy. The Cgatg1Δ strain was sensitive to nitrogen starvation and H2O2, died rapidly in water without any nutrients, and showed high intracellular ROS levels compared with the wild-type strain and the CgATG1-reconstituted strain in vitro. Upon infecting mouse peritoneal macrophages, the Cgatg1Δ strain showed higher mortality from phagocytosis by macrophages. Finally, in vivo experiments were performed using two mouse models of disseminated candidiasis and intra-abdominal candidiasis. The Cgatg1Δ strain showed significantly decreased CFUs in the organs of the two mouse models. These results suggest that autophagy contributes to C. glabrata virulence by conferring resistance to unstable nutrient environments and immune defense of hosts, and that Atg1 is a novel fitness factor in Candida species.
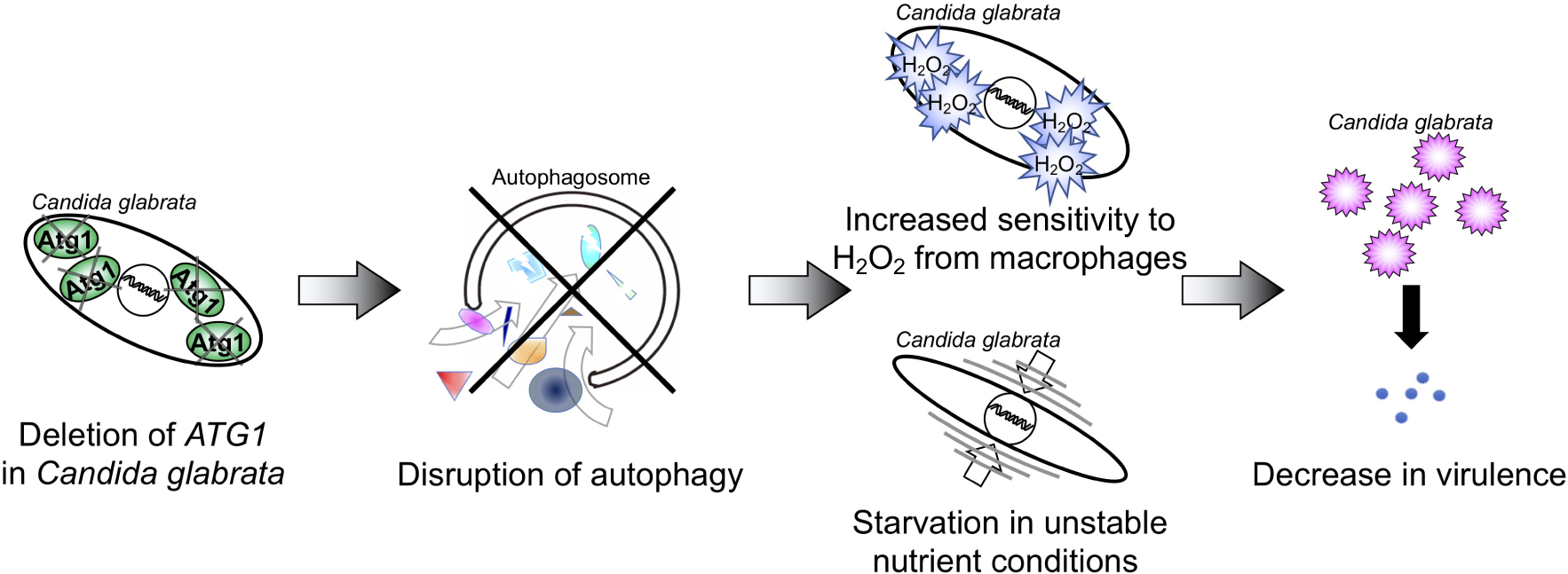
Graphical Abstract. The deletion of ATG1 disrupted autophagy function in C. glabrata, leading to increased sensitivity to hydrogen peroxide (H2O2) and nutrient starvation. These phenotypes were associated with decreased fitness and virulences in host environments.
Introduction
Autophagy is an evolutionarily conserved biological process in eukaryotes that involves the degradation of cytosolic molecules for recycling intracellular materials (Klionsky et al., 2016). Autophagy has several functions: supplying nutrients under starvation conditions, maintaining homeostasis and protein levels, clearing damaged and/or non-functional proteins, and preventing bacterial and viral infections. These functions contribute to cell longevity, development, and differentiation as well as tumor suppression (Jin et al., 2017; Chen et al., 2018; Hansen et al., 2018). In the induction of autophagy, autophagosomes, which are double-membrane cytosolic vesicles, are formed to incorporate target proteins. In the model yeast Saccharomyces cerevisiae, autophagosomes fuse with vacuoles, and proteins are degraded in the vacuoles.
Fungal pathogens also use autophagy to survive in a host environment with unstable nutrient supply (Khan et al., 2012). However, little is known about the contribution of fungal autophagy to their persistence and virulence in hosts. In addition, immune defenses such as macrophages generate ROS to kill infecting pathogens (Gonzalez-Parraga et al., 2003; Thorpe et al., 2004), but it has remained obscure whether fungal autophagy affects this oxidative stress response.
Candida species are a genus of opportunistic fungal pathogens that cause severe invasive infections in immunocompromised patients (Miceli et al., 2011). Candida glabrata is the second most common cause of candidiasis in humans (Roetzer et al., 2011). The genetic background of C. glabrata is closely related to that of S. cerevisiae. C. glabrata is a commensal yeast and capable of surviving in the host longer than other Candida species (Roetzer et al., 2011). We hypothesized that autophagy contributes to these functions in C. glabrata.
Autophagy has several subcategories. Macroautophagy covers a broad range of protein degradation processes that are mainly induced upon starvation (Suzuki, 2013). Pexophagy and mitophagy are specific to the degradation of damaged peroxisomes and mitochondria, respectively (Suzuki, 2013). Recently, pexophagy and mitophagy have been suggested to be related to C. glabrata virulence (Roetzer et al., 2010; Nagi et al., 2016). In the present study, we analyzed macroautophagy.
Macroautophagy (hereinafter simply referred to as autophagy) is induced by Atg proteins in yeasts (Yorimitsu and Klionsky, 2005). Atg1 is a component of an Atg protein complex and is essential for autophagy induction (Wang and Kundu, 2017). Candida glabrata Atg1 (CgAtg1) is also predicted to be important for autophagy, because ATG genes are highly conserved between S. cerevisiae and C. glabrata. Here, we showed that C. glabrata autophagy is induced by nitrogen starvation and H2O2. The CgATG1-deleted mutant of C. glabrata exhibited deficient adaptation to starvation and H2O2 in vitro. An ex vivo experiment using mouse peritoneal macrophages demonstrated that the Cgatg1Δ strain was phagocytosed by macrophages and showed low viability. Autophagy was revealed to be important for C. glabrata survival in two mouse models of invasive candidiasis.
Materials and Methods
Ethics Statement
Animal experiments were conducted according to the Guide for the Care and Use of Laboratory Animals (National Research Council, National Academy Press, Washington, DC, 2011) and all of the institutional regulations and guidelines for animal experimentation after pertinent review and approval by the Institutional Animal Care and Use Committee of Nagasaki University (approval number 1407281164-4).
Culture Conditions
C. glabrata was routinely cultured at 30°C in SC-trp (Dunham et al., 2015) or YPD agar [1% yeast extract, 2% peptone, 2% dextrose, and 2% Bacto agar (BD Biosciences, B242720)], unless otherwise indicated. SD-N [0.17% yeast nitrogen base without amino acids and ammonium sulfate (BD Biosciences, 233520) and 2% dextrose] was used for the nitrogen starvation condition (Budovskaya et al., 2004).
Strain and Plasmid Construction
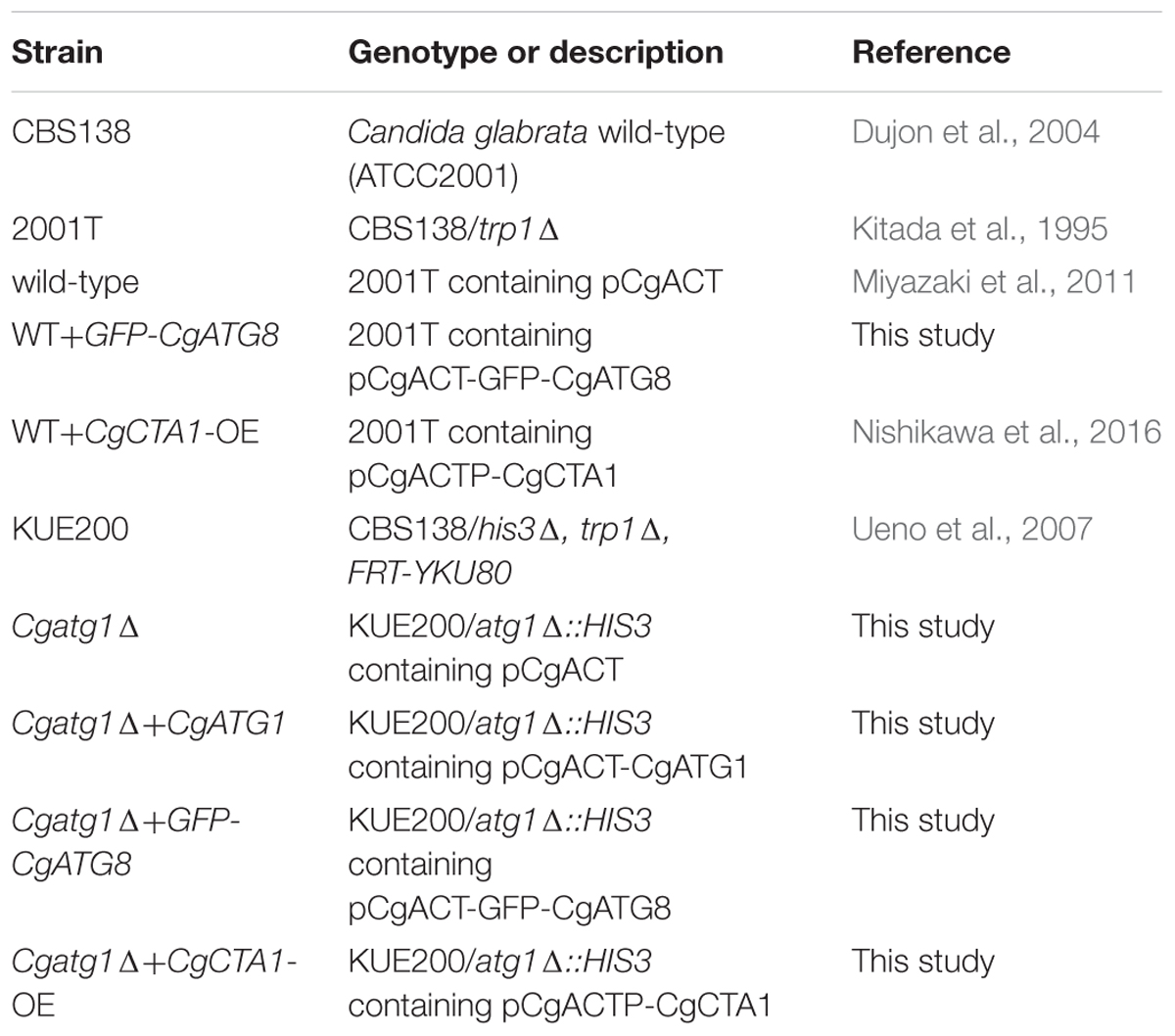
C. glabrata strains, plasmids, and primers used in this study are listed in Tables 1–3, respectively. Sequence information of C. glabrata genes was obtained from the Candida genome database1.
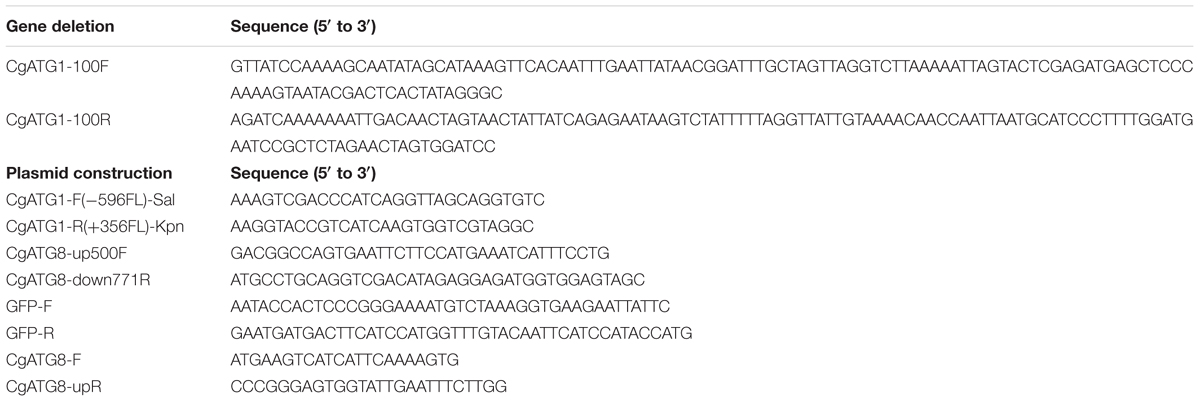
The C. glabrata atg1Δ strain was constructed using a one-step PCR-based technique as described previously (Miyazaki et al., 2010a, 2011). Briefly, an CgATG1 deletion construct was amplified from pBSK-HIS using primers tagged with 100-bp sequences homologous to the flanking regions of the CgATG1 ORF (CgATG1-100F and CgATG1-100R). C. glabrata parent strains were transformed with the deletion construct, and the resulting transformants were selected by histidine prototrophy (Miyazaki et al., 2011). Successful homologous recombination was verified by diagnostic PCR, and the absence of CgATG1 mRNA expression was confirmed by real-time qRT-PCR (data not shown). Transformation of C. glabrata was performed using the lithium acetate protocol, as described previously (Cormack and Falkow, 1999).
pCgACT-CgATG1, in which CgATG1 was expressed under the control of the CgATG1 native promoter, was constructed as follows: a 3,781-bp DNA fragment containing the CgATG1 promoter, ORF, and 3′-UTR was amplified using CgATG1-F(-596FL)-Sal and CgATG1-R(+356FL)-Kpn, digested with SalI and KpnI, and inserted into the SalI-KpnI site of pCgACT (Kitada et al., 1996). An CgATG1-reconstituted strain and its control strains were constructed by transformation of the Cgatg1Δ strain with pCgACT-CgATG1 and pCgACT, respectively. They were selected by tryptophan prototrophy and verified by qRT-PCR (data not shown).
pCgACT-GFP-CgATG8, in which N-terminally GFP-tagged CgATG8 was expressed under the control of the CgATG8 native promoter, was constructed using In-Fusion HD Cloning Plus CE (Clontech Laboratories, 638916). Briefly, a 1,600-bp DNA fragment containing the CgATG8 promoter, ORF, and 3′UTR was amplified using CgATG8-up500F and CgATG8-down771R, and inserted into the EcoRI-SalI site of pCgACT by the In-Fusion reaction to generate pCgACT-CgATG8. GFP (yEGFP1) was amplified from pYGFP1 (Cormack et al., 1997) using GFP-F and GFP-R, and inserted between the CgATG8 promoter and the ORF in pCgACT-CgATG8 by the In-Fusion reaction to generate pCgACT-GFP-CgATG8. The insertion site of the vector was produced by a PCR reaction using pCgACT-CgATG8 as the template and the primers CgATG8-F and CgATG8-upR. The C. glabrata wild-type strain 2001T and the Cgatg1Δ strain were transformed with pCgACT-GFP-CgATG8.
Growth Curve Construction
Logarithmic-phase C. glabrata cells were adjusted to 5 × 106 cells/ml and incubated in SC-trp broth at 37°C. The number of cells was counted at 2, 4, 6, 8, 24, and 48 h. Doubling times were calculated as previously described (Geber et al., 1995), except that the cells were counted using a hemocytometer instead of OD600. The averages of the doubling times were obtained from four independent experiments.
Spot Assay
Spot assays were performed as described previously (Miyazaki et al., 2010b). Briefly, logarithmic-phase cells grown in SC-trp broth were harvested and adjusted to a concentration of 2 × 107 cells/ml. Serial 10-fold dilutions were prepared, and 5 μl of each dilution was spotted onto agar plates and incubated at 30°C for 48 h, unless otherwise indicated.
Immunoblotting
Anti-GFP (Roche, 11814460001), anti-Pgk1 (OriGene EU, AP21371AF-N), anti-mouse IgG-hrp (GE Healthcare, NA931V), and anti-rabbit IgG-hrp (GE Healthcare, NA934V) antibodies were purchased. Logarithmic-phase cells were harvested and lysed using a Minute Total Protein Extraction Kit for Microbes with Thick Cell Walls (Invent Biotechnologies, YT-015) according to the accompanying instructions. Lysates were separated by SDS–PAGE and transferred to a polyvinylidene difluoride membrane (BIO-RAD, 1704156). Each protein was detected using the indicated antibodies, enhanced chemiluminescent substrate (Thermo Fisher Scientific, 34096), and a ChemiDoc Touch Imaging System (BIO-RAD, Hercules, CA, United States).
Measurement of Intracellular ROS Levels
Intracellular ROS levels in C. glabrata were analyzed by measuring fluorescent DCF derived from the reaction of CM-H2DCFDA (Thermo Fisher Scientific, C6827) and ROS. Cell-permeable CM-H2DCFDA is not fluorescent; it reacts with ROS to produce fluorescent DCF. Logarithmic-phase cells were washed with PBS, resuspended in PBS with 10 μM CM-H2DCFDA, and incubated at 30°C for 1 h. The cells were washed, resuspended in SC-trp with H2O2 (Wako, 084-07441), tert-butyl hydroperoxide (Wako, 026-13451), menadione (Wako, 132-08132), or diamide (Sigma-Aldrich, D3648), and cultured at 30°C for 1 h. The cells were washed and resuspended in PBS, and fluorescence was measured with a PHERAstar FS multi-mode microplate reader (BMG LABTECH, Offenburg, Germany) at 485 nm fluorescence excitation wavelength and 520 nm emission wavelength. The cell count of each sample was determined, and relative fluorescence intensity per cell was calculated.
Lifespan Assay
Incubation of 2.5 × 106/ml cells was started in ultrapure water (Thermo Fisher Scientific, 10977-023) at 30°C. The cells were harvested at multiple time points and plated on YPD agar. The number of viable cells was determined by counting colonies after incubation for 36 h at 30°C, and the number of CFUs per ml was calculated.
Macrophage Infection Assay
To collect mouse peritoneal macrophages, peritoneal exudates were obtained from 8-week-old male BALB/c mice (Japan SLC, Shizuoka, Japan) by lavage 4 days after intraperitoneal injection of 2 ml of sterile 4% thioglycollate broth (BD Biosciences, 211716). After washing with DMEM, peritoneal macrophages were incubated in DMEM with 10% FBS and 100 μg/ml gentamycin (Nacalai Tesque, 11980-14) at 37°C under 5% CO2. Macrophages of 1.5 × 105 cells were seeded in 24-well plates and incubated in DMEM with 10% FBS for 24 h. For infection assays, 2.5 × 105 C. glabrata cells were resuspended in DMEM, added to the macrophages, and incubated. The actual C. glabrata cell number was confirmed by plating the cell suspension on YPD agar. Two hours after infection, the infected macrophages were washed thrice with PBS to remove non-phagocytosed C. glabrata cells and incubated in fresh DMEM. At multiple time points post-infection, the infected macrophages were observed under a microscope (BZ-X700, Keyence, Osaka, Japan). Then, the macrophages were lysed in water, and phagocytosed C. glabrata cells were plated on YPD agar containing streptomycin sulfate salt (Sigma-Aldrich, S1277) and penicillin G sodium salt (Sigma-Aldrich, P3032). The number of viable C. glabrata cells was determined by counting colonies after incubation for 36 h at 30°C, and the number of CFU per well was calculated.
Mouse Studies
Mouse experiments were performed as described previously (Miyazaki et al., 2010a,b). Briefly, to prepare cells for injection, cultured C. glabrata cells were resuspended in sterile saline and adjusted to 5 × 108 or 1 × 109 cells/ml. The actual C. glabrata concentration was confirmed by plating the adjusted cell suspension on YPD agar. The tail veins or abdominal cavities of 8-week-old female BALB/c mice (Japan SLC) were injected with 200 μl suspensions of each C. glabrata strain. The mice were sacrificed 7 days post-injection, and the target organs were excised. The organs were homogenized, diluted, and plated on YPD agar containing streptomycin sulfate salt and penicillin G sodium salt. Colonies were counted after incubation for 36 h at 30°C, and the number of CFU per organ was calculated.
Results
Atg1 Is Necessary for Normal Growth Rate, Survival, and Autophagy During Nitrogen Starvation and Oxidative Stress
The Cgatg1Δ strain showed slightly slower growth than the wild-type strain and the CgATG1-reconstituted strain under the SC-trp growth condition (Figure 1). Doubling times of the wild-type, Cgatg1Δ, and CgATG1-reconstituted strains in SC-trp medium were 1.173, 1.912, and 1.254 h, respectively. Spot assays revealed growth defects in the Cgatg1Δ strain under the nitrogen starvation condition and in the presence of H2O2 (Figure 2A). These phenotypes recovered to the wild-type levels by the reintroduction of intact CgATG1 into the mutant.
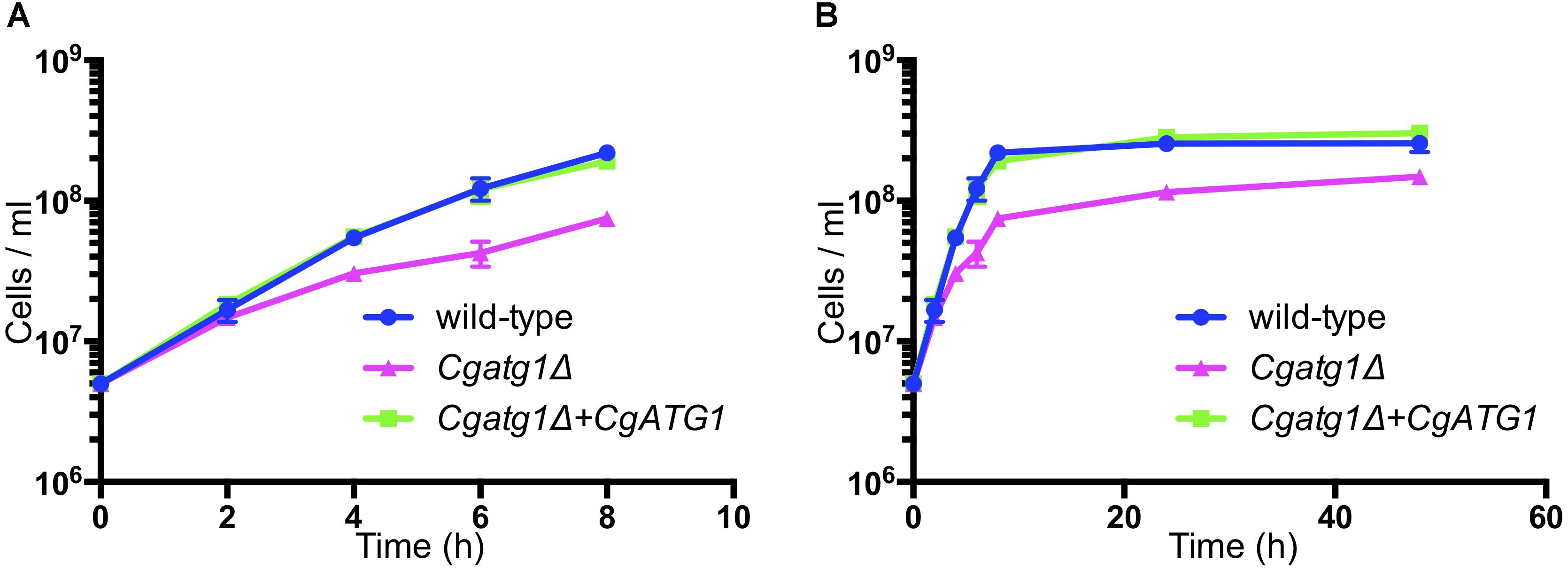
Figure 1. Growth curves of C. glabrata strains. Logarithmic-phase C. glabrata cells were grown in SC-trp broth at 37°C and counted at multiple time points. Graphs show data for the first 8 h of culture (A) to focus on the log phase and 48 h of culture (B). C. glabrata strains: wild-type (blue circles), Cgatg1Δ (magenta triangles), and Cgatg1Δ+CgATG1 (CgATG1-reconstituted Cgatg1Δ, green squares). The means ± SE of four independent experiments are shown. Doubling times: wild-type, 1.173 h; Cgatg1Δ, 1.912 h; Cgatg1Δ+CgATG1, 1.254 h.
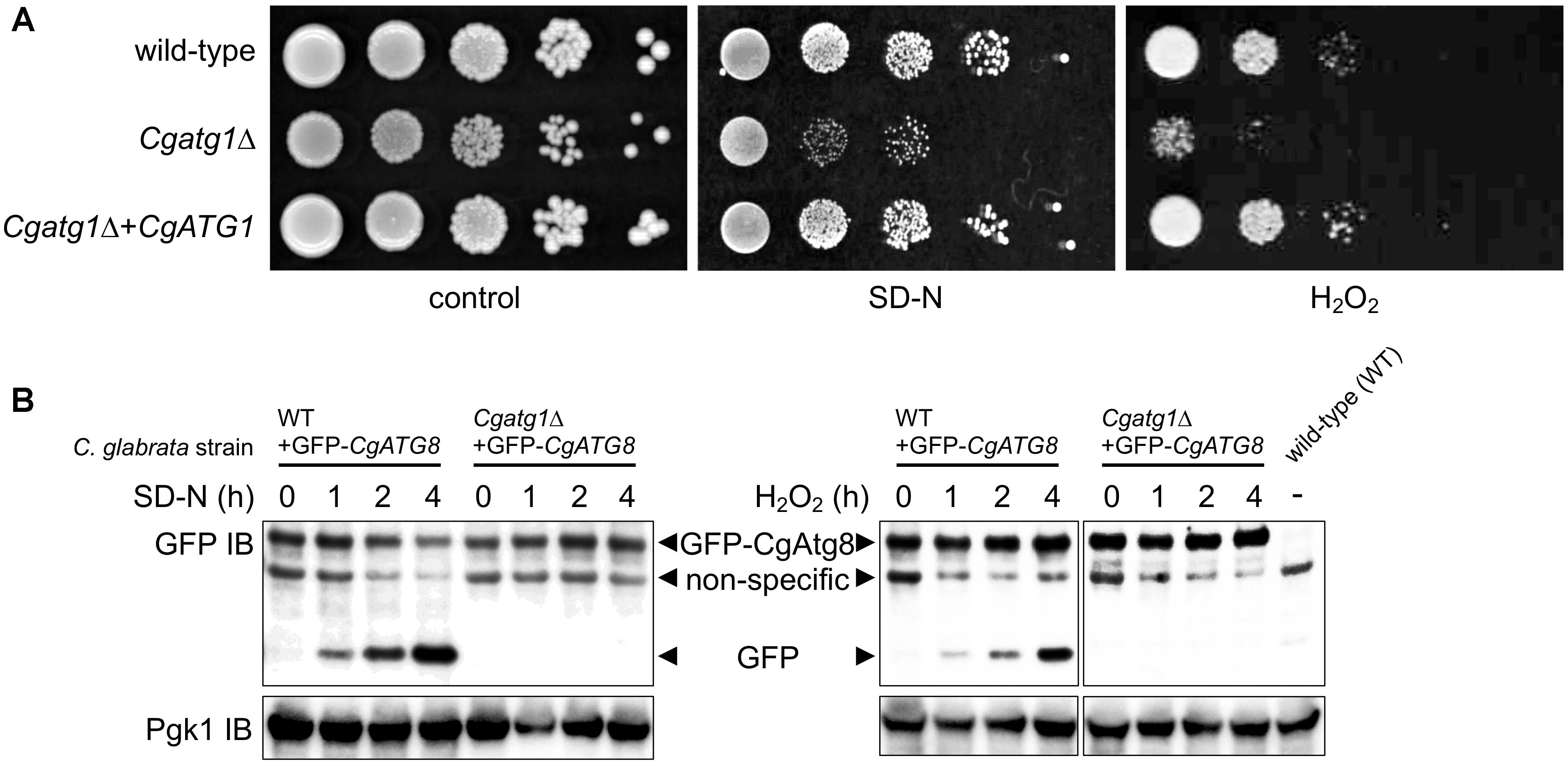
Figure 2. CgATG1-deleted C. glabrata is sensitive to nitrogen starvation and H2O2. (A) Logarithmic-phase cells were serially 10-fold diluted, spotted onto agar plates of SC-trp, SD-N, and SC-trp with 13.3 mM H2O2, and cultured at 30°C for 48 h. (B) N-terminally GFP-tagged CgAtg8 was expressed in the wild-type strain and the Cgatg1Δ strain. Cells were cultured in SD-N (left) or SC-trp with 13.3 mM H2O2 (right) for the indicated hours. Protein was extracted and immunoblotted using anti-GFP antibody and anti-Pgk1 antibody as a loading control. The protein extract of the wild-type strain without GFP-Atg8 expression was loaded as a negative control.
It remains to be established whether or not autophagy is induced by H2O2 in C. glabrata. The induction of autophagy by starvation and the necessity of CgAtg1 for autophagy also need to be confirmed, as reports of autophagy in C. glabrata are still few. In S. cerevisiae, GFP-Atg8 is known to be cleaved by autophagy, resulting in the release of GFP, which can be detected as a marker of autophagy (Shintani and Klionsky, 2004). GFP-CgAtg8 was expressed in C. glabrata from a transformed plasmid containing the CgATG8 native promoter followed by an N-terminally GFP-tagged CgATG8 ORF. GFP-Atg8 and GFP bands were specifically observed in GFP-Atg8-expressing strains, but not in the negative control strain containing an empty vector (Figure 2B). GFP bands gradually became more intense as a result of nitrogen starvation and H2O2 exposure in the wild-type strain, whereas no GFP bands were observed over the time-course in the Cgatg1Δ strain (Figure 2B), indicating that autophagy was induced in response to nitrogen starvation and H2O2 exposure in a CgAtg1-dependent manner in C. glabrata.
In addition to the spot assay, a lifespan assay was performed to examine the sensitivity of the Cgatg1Δ strain to the starvation condition using pure water. Compared to the wild-type strain, the Cgatg1Δ strain showed a rapid decline in viability within 2 h (Figure 3A). Changes in C. glabrata viability became gradual after 2 h, and the Cgatg1Δ cells died after 4 weeks, in contrast to the wild-type cells that remained viable at 4 weeks in water (Figure 3B). Overall, these results suggest that autophagy plays a role in the adaptation to starvation in C. glabrata.
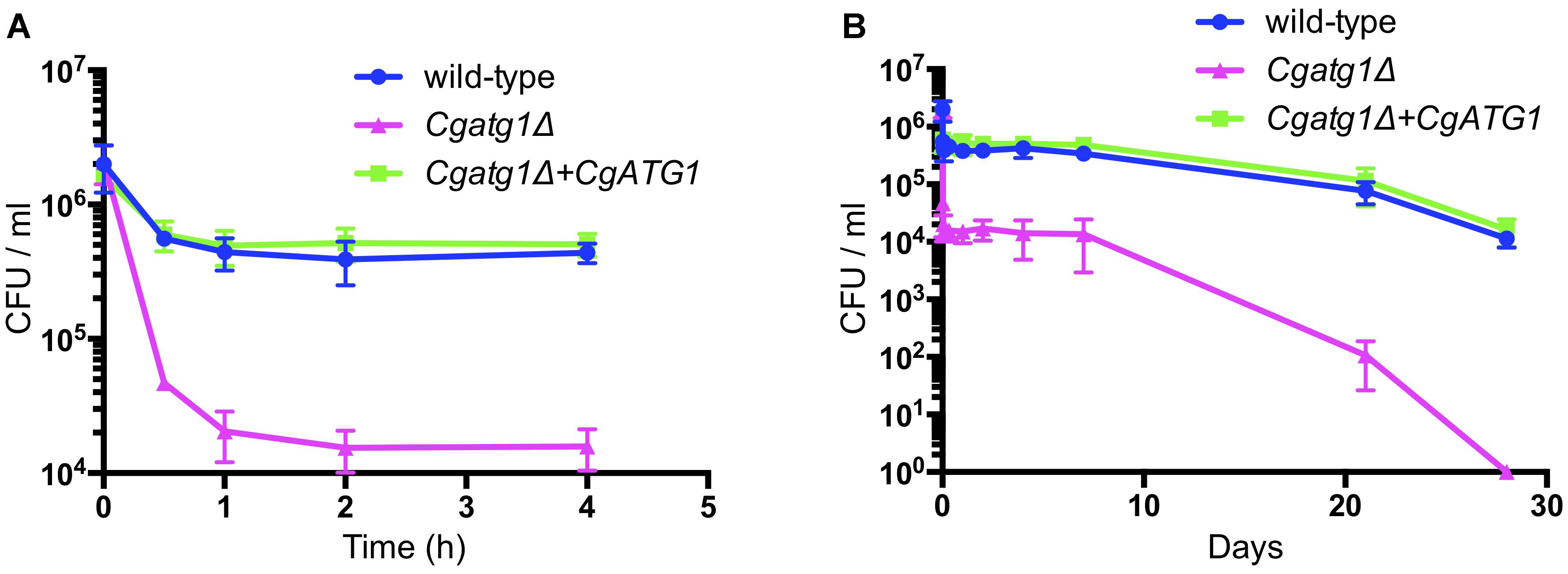
Figure 3. CgAtg1 is important for C. glabrata viability under nutrient starvation conditions. C. glabrata strains were incubated in pure water at 30°C, harvested at multiple time points, and cultured in YPD agar. Then, the number of CFU was counted. Graphs show data for the first 4 h (A) and over 28 days (B) of incubation. The means ± SE of three independent experiments are shown.
Analyses of the Correlation Between H2O2 Sensitivity and Intracellular ROS Levels
Intracellular ROS levels were increased by H2O2 in all the strains tested (Figure 4A). The Cgatg1Δ strain had a higher ROS level than the wild-type strain and the CgATG1-reconstituted strain regardless of the presence or absence of H2O2. High ROS levels in the Cgatg1Δ strain were also observed after exposure to other ROS-generating agents, including tert-butyl hydroperoxide (an oxidant), menadione (a superoxide-generating agent), and diamide (an inhibitor of hydroxyl radical metabolism by decreasing the glutathione pool) (Supplementary Figure S1). The elevations of ROS levels in the Cgatg1Δ strain by these three ROS-generating agents were mild (approximately 10- to 15-fold relative to the control), while H2O2 markedly elevated the ROS level in the Cgatg1Δ strain (over 60-fold relative to the control). The sensitivity of the Cgatg1Δ strain to H2O2 was eliminated by the overexpression of CgCTA1 (CaTalase A 1) that encodes a catalase involved in the metabolism of H2O2 (Figure 4B). Consistent with the spot assay, the intracellular ROS amount in the Cgatg1Δ strain decreased to the level of the wild-type strain by CgCTA1 overexpression (Figure 4C). These results suggest that autophagy affects intracellular ROS levels and resistance to H2O2 in C. glabrata. CgCTA1 overexpression partially rescued the growth rate of the Cgatg1Δ strain under the SC-trp growth condition (Figures 4D,E). Doubling times of the wild-type, CgCTA1-overexpressed wild-type, Cgatg1Δ, and CgCTA1-overexpressed Cgatg1Δ strains in SC-trp medium were 1.136, 1.032, 1.995, and 1.499 h, respectively.
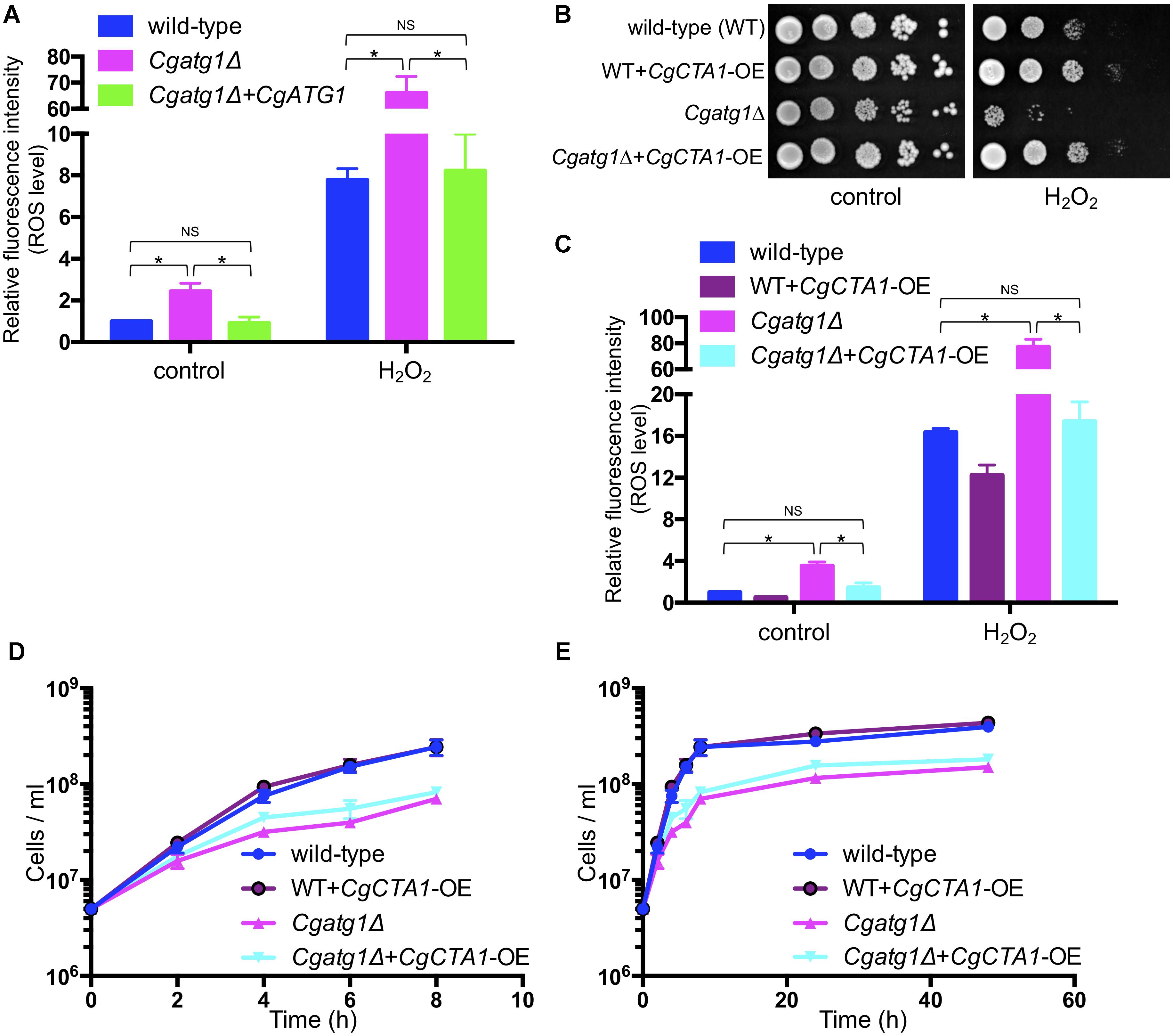
Figure 4. Analyses of intracellular ROS level with H2O2 and effect of catalase. (A) C. glabrata strains containing CM-H2DCFDA were cultured in SC-trp with/without 13.3 mM H2O2 at 30°C for 1 h, and the fluorescence produced by the reaction of CM-H2DCFDA and ROS in the cells was measured. Relative fluorescence intensity per cell was calculated, and the fluorescence intensity of the wild-type strain without H2O2 was defined as 1. The means ± SE of three independent experiments are shown. Statistical analyses were performed using two-tailed Student’s t-tests. ∗P < 0.01; NS, no significance (P > 0.05). (B–E) Spot assays (B), measurement of intracellular ROS levels (C), and growth curve construction (D,E) were performed as in Figure 2A (B), Figure 4A (C), and Figures 1A,B (D,E) to analyze the effect of CgCTA1 overexpression. C. glabrata strains: wild-type (blue), WT+CgCTA1-OE (purple), Cgatg1Δ (magenta), and Cgatg1Δ+CgCTA1-OE (cyan). (D,E) Doubling times: wild-type, 1.136 h; WT+CgCTA1-OE, 1.032 h; Cgatg1Δ, 1.995 h; Cgatg1Δ+CgCTA1-OE, 1.499 h.
CgATG1 Deletion Decreases Resistance of C. glabrata to Macrophages
Activated macrophages generate H2O2 for phagocytosis (Forman and Torres, 2001). In addition, nutrients are restricted in the engulfed state by macrophages. Autophagy in C. glabrata might act to resist the immune response. To assess this hypothesis, an ex vivo experiment was performed using macrophages collected from mouse abdominal cavities. C. glabrata strains were added to wells containing the peritoneal macrophages and co-incubated, and viable C. glabrata cells phagocytosed by the macrophages were counted. Growth of the Cgatg1Δ strain was suppressed during the 96-h co-incubation, whereas growth of the wild-type strain and the CgATG1-reconstituted strain was increased despite phagocytosis by macrophages (Figure 5A). During the 96-h period post-infection, the increase in the number of wild-type C. glabrata cells was remarkable, whereas Cgatg1Δ cells were hardly observed (Figure 5B). This suggests that autophagy is required for resistance to macrophages in C. glabrata.
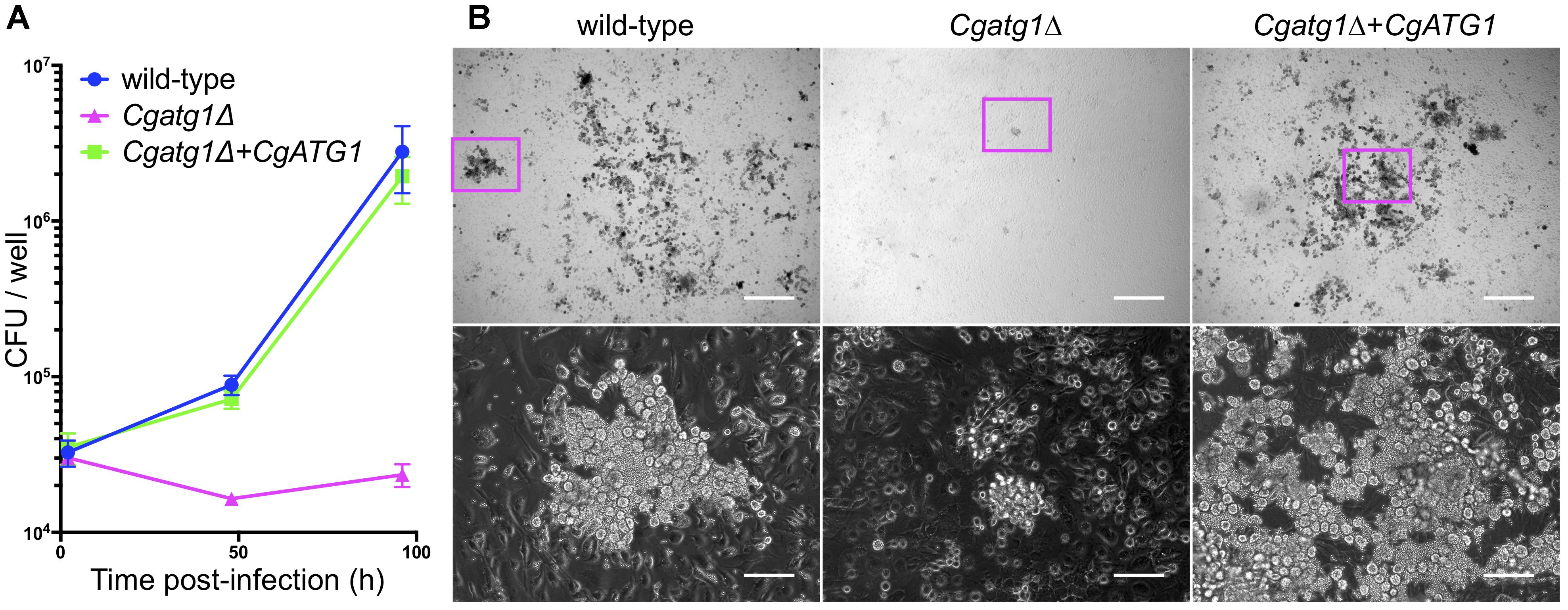
Figure 5. CgATG1-deleted C. glabrata is sensitive to phagocytosis by macrophages. Mouse peritoneal macrophages were infected with C. glabrata strains in DMEM supplemented with 10% FBS. (A) Macrophages including C. glabrata were harvested at 2, 48, and 96 h post-infection, lysed in water, and plated on YPD agar for CFU count. The means ± SE of three independent experiments are shown. (B) Microscopic images of macrophages infected with C. glabrata for 96 h. Bottom panels are enlarged images of magenta boxes in the respective upper panels. Growing C. glabrata cells were observed in the wild-type strain and the Cgatg1Δ+CgATG1 strain, but rarely in the Cgatg1Δ strain. Bar: 500 μm (upper), 100 μm (bottom).
CgAtg1 Is Important for C. glabrata Persistence in Hosts
In mouse experiments with C. glabrata, DC mouse models are used because C. glabrata frequently spreads in patients through blood flow. Macrophages mainly cause inflammation in the abdominal cavity, although they also function in the blood and subsequent organs. To observe C. glabrata intraperitoneal infection, a mouse model of IAC was also employed. Liver, spleen, and kidney are the typical organs examined in the DC mouse model. The pancreas is the predominant organ for C. glabrata infection in the IAC mouse model, while the kidney is retroperitoneal and has not yielded stable results in previous studies (Cheng et al., 2013, 2014) or in our preliminary experiments (data not shown). Decreases in the number of CFU were observed in the liver and spleen of the DC mouse model (Figure 6A) and all organs examined in the IAC mouse model (Figure 6B) when the mice were colonized by the Cgatg1Δ strain. In the kidney of the DC mouse model, the number of CFU slightly decreased in the Cgatg1Δ strain without statistical significance (Figure 6A). These results suggest that autophagy positively affects C. glabrata viability in the host.
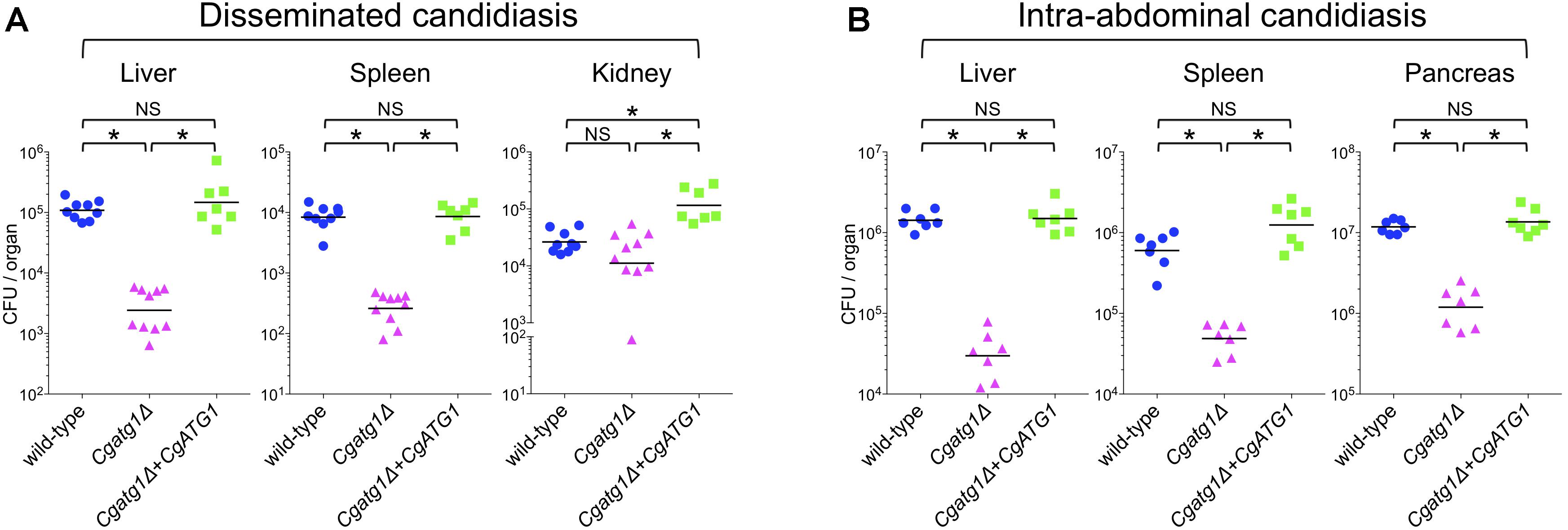
Figure 6. CgATG1 deletion decreases C. glabrata viability in mouse models of disseminated and intra-abdominal candidiasis. Groups of 7 to 10 BALB/c mice were inoculated with 1 × 108 cells intravenously (A) or 2 × 108 cells intraperitoneally (B) of each C. glabrata strain. Target organs were excised 7 days after injection. The organ homogenates were plated on YPD agar for CFU count. Numbers of CFU from each organ are indicated for individual mice in the scatter plots. Each geometric mean is shown by a bar. Statistical analyses were performed using the Wilcoxon Mann–Whitney test. ∗P < 0.01; NS, no significance (P > 0.05). Representative data of two independent experiments are shown.
Discussion
Research on autophagy as a driving force of virulence is still in its developing stages compared to fundamental studies in humans and S. cerevisiae (Klionsky et al., 2016). We examined autophagy in the pathogenic fungus C. glabrata by conducting in vitro, ex vivo, and in vivo experiments, and found that autophagy has positive effects on C. glabrata virulence.
In vitro, CgAtg1 was required for autophagy, and the Cgatg1Δ strain was sensitive to the nutrient starvation condition, suggesting that autophagy in C. glabrata is important for adaptation to starvation, which has been observed in other fungal species such as Candida albicans and Cryptococcus neoformans (Palmer et al., 2008; Yu et al., 2015). Furthermore, rapid cell death and long-term survival defects were noted in the Cgatg1Δ strain in the lifespan assay, suggesting that autophagy functions in both adaptation to sudden environmental changes and for persistence of C. glabrata in hosts.
In pathogenic fungi, autophagy has been indicated to be dispensable for C. albicans virulence, whereas it is required for the infection process of C. neoformans (Palmer et al., 2008). C. glabrata is also a commensal yeast, but its infection mechanism is different from that of C. albicans. C. albicans forms hyphae to invade across the host environment and to escape from nutrient-starved loci and macrophages (Desai, 2018). C. glabrata seems to require autophagy because it lacks the capacity to form hyphae but adheres to many different objects for a long time (Timmermans et al., 2018).
The functional mechanisms of autophagy in oxidative stress response remain to be elucidated. This study showed high intracellular ROS levels in the Cgatg1Δ strain, especially by H2O2 addition. CgCTA1 overexpression rescued the resistance of the Cgatg1Δ strain to H2O2; however, this result does not indicate that autophagy promotes the expression of CgCTA1. Rather, factors other than the expression level of CgCTA1 are thought to be the cause of the high ROS levels in the Cgatg1Δ strain, as ROS elevation was also induced by other ROS-generating agents that are not metabolized by Cta1 (Supplementary Figure S1). Indeed, the rescue of the growth rate of the Cgatg1Δ strain in SC-trp by CTA1 overexpression was only partial and not up to the level of the wild-type strain (Figures 4D,E). Cta1 is considered to reverse the defect of the Cgatg1Δ strain only for adaptation to H2O2, but not for the overall functions of autophagy. Multiple factors are involved in the ROS elevation, as intracellular ROS levels including H2O2 are changed by various stresses and regulatory factors, and autophagy affects a wide range of molecules.
In addition to the regulation of intracellular ROS, it is speculated that fungal autophagy might affect the repair of cellular damage caused by H2O2. The adaptation to H2O2 has been analyzed in pexophagy in mammals and plants (Lee et al., 2014; Zientara-Rytter and Subramani, 2016). Peroxisomes generate H2O2 and are self-oxidized. These damaged peroxisomes are degraded by pexophagy for recycling, which leads to efficient maintenance of functional and fresh peroxisomes and the cleaning of junk; this is called ‘quality control.’ Recent studies in mammals have suggested that macroautophagy is also activated by H2O2 (Filomeni et al., 2015; Yin et al., 2015; Nah et al., 2017). From previous reports and this study, autophagy is supposed to have a two-step function in the adaptation to H2O2 (Supplementary Figure S2). The first step is the metabolism of overall ROS, and the second is the quality control of proteins and organelles injured by H2O2. Further investigations are needed to elucidate the precise molecular mechanisms in C. glabrata and other species.
The Cgatg1Δ strain exhibited slightly impaired growth and high ROS levels in the SC-trp medium (Figures 1, 2A, 4A). These defects are speculated to be caused by cellular stresses generated under this control condition, whereas the wild-type strain might have adapted to the stresses through a basal level of autophagy. The high ROS levels in the Cgatg1Δ strain in the absence of any exogenous ROS agents are considered to be caused by the defect in metabolism of endogenous ROS generated by C. glabrata basal activities such as respiration. The endogenous ROS may also be a cause of the slow growth of Cgatg1Δ strain. Indeed, the doubling time of Cgatg1Δ strain was shortened to some extent by CTA1 overexpression, although it was not rescued to the level of wild-type strain (Figures 4D,E). Stimuli other than ROS such as excessive ions, high osmolarity, and high temperature were analyzed, but the Cgatg1Δ strain was not sensitive to these stresses (Supplementary Figure S3).
The function of autophagy in the adaptation to starvation is supposed to enhance C. glabrata viability in hosts, as nutrient conditions are thought to be unstable in various host environments. In addition, the effect of autophagy on ROS metabolism and resistance to H2O2 in C. glabrata was predicted to contribute to its survival against ROS-generating immune defenses, such as macrophages. An ex vivo experiment using mouse peritoneal macrophages revealed low viability of the Cgatg1Δ strain in the macrophages (Figure 5). This defect can be attributed to the decreased resistance of the Cgatg1Δ strain to both nutrient starvation and to the H2O2 encountered when phagocytosed by macrophages.
In vivo mouse experiments confirmed that autophagy is a ‘fitness factor’ in C. glabrata. Differences between the wild-type strain and the Cgatg1Δ strain are thought to originate from the basic growth rate, the resistance to insufficient nutrient environments, and the oxidative stress response to macrophages, as well as other effects of autophagy that were not addressed in this study. Both the IAC mouse model, which was employed to examine effect of peritoneal macrophages, and the DC mouse model showed attenuated viability of the Cgatg1Δ strain. These results suggest that immune defense not only by macrophages but also by other monocytes in the blood and subsequently infected organs might contribute to the decreased CFU of the Cgatg1Δ strain. However, the number of CFU from kidneys was not significantly different between the wild-type strain and the Cgatg1Δ strain in the DC mouse model. Although the exact reason is unclear, one possible explanation is that renal macrophages concentrate to the tubulointerstitium (Xu and Shinohara, 2017), a compartment of the kidney, and might not respond effectively to C. glabrata cells that usually invade the vasculature and nephrons in the kidney. Renal macrophages have a role in host defense against C. albicans (Lionakis et al., 2013; Ngo et al., 2014), but this function may be dependent on the Candida species. C. glabrata infection does not strongly attract immune cells, including neutrophils, and thus rarely causes severe inflammation (Kasper et al., 2015). Nevertheless, the results suggest that the extent of influence of autophagy in C. glabrata is substantially different among infected organs and tissues.
Although our experiments in vitro and ex vivo suggested the possibility of its contribution to virulence, obvious differences in viability and health status were not observed between the mice infected with the wild-type and Cgatg1Δ strains. From our present results in vivo, C. glabrata autophagy is concluded to be a fitness factor for longer survival in hosts.
Conclusion
Atg1 has been broadly investigated in many eukaryotes and established as a key player in autophagy induction (Wang and Kundu, 2017). However, reports of autophagy and Atg1 focusing on virulence are limited, especially in Candida species. The current study demonstrated for the first time that Atg1 is required for autophagy in C. glabrata (Graphical Abstract). Persistence in hosts is a characteristic of C. glabrata infection. The Cgatg1Δ phenotypes indicate that autophagy may play an important role in fitness and survival of this fungus in infected hosts. The detailed characteristics of CgAtg1, including functional mechanisms, modifications, and interactions with other factors, need to be elucidated in future studies.
Author Contributions
SS and TM designed the project, conducted the experiments, analyzed the data, and prepared the manuscript. All authors reviewed and approved the final version of the manuscript.
Funding
This work was partially supported by the Research Program on Emerging and Re-emerging Infectious Diseases from the Japan Agency for Medical Research and Development (AMED) (Grant No. JP18fk0108008 to TM and SK) and the Japan Society for the Promotion of Science (JSPS) KAKENHI (Grant No. JP17K10018 to SS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to the technical staff and the secretaries in our laboratories at Nagasaki University for excellent experimental support and office work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00027/full#supplementary-material
Abbreviations
ATG, autophagy-related; CFU, colony forming units; Cg, Candida glabrata; CTA1, catalase A 1; DC, disseminated candidiasis; DCF, dichlorofluorescein; DMEM, Dulbecco’s modified Eagle’s medium; ECL, enhanced chemiluminescence; FBS, fetal bovine serum; GFP, green fluorescent protein; H2O2, hydrogen peroxide; IAC, intra-abdominal candidiasis; ORF, open reading frame; PBS, phosphate-buffered saline; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; ROS, reactive oxygen species; SC-trp, synthetic complete medium lacking tryptophan; SD-N, synthetic dextrose medium lacking nitrogen; SE, standard error; YPD, yeast extract peptone dextrose.
Footnotes
References
Budovskaya, Y. V., Stephan, J. S., Reggiori, F., Klionsky, D. J., and Herman, P. K. (2004). The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J. Biol. Chem. 279, 20663–20671. doi: 10.1074/jbc.M400272200
Chen, X., He, Y., and Lu, F. (2018). Autophagy in stem cell biology: a perspective on stem cell self-renewal and differentiation. Stem Cells Int. 2018:9131397. doi: 10.1155/2018/9131397
Cheng, S., Clancy, C. J., Hartman, D. J., Hao, B., and Nguyen, M. H. (2014). Candida glabrata intra-abdominal candidiasis is characterized by persistence within the peritoneal cavity and abscesses. Infect. Immun. 82, 3015–3022. doi: 10.1128/IAI.00062-14
Cheng, S., Clancy, C. J., Xu, W., Schneider, F., Hao, B., Mitchell, A. P., et al. (2013). Profiling of Candida albicans gene expression during intra-abdominal candidiasis identifies biologic processes involved in pathogenesis. J. Infect. Dis. 208, 1529–1537. doi: 10.1093/infdis/jit335
Cormack, B. P., Bertram, G., Egerton, M., Gow, N. A., Falkow, S., and Brown, A. J. (1997). Yeast-enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans. Microbiology 143( Pt 2), 303–311. doi: 10.1099/00221287-143-2-303
Cormack, B. P., and Falkow, S. (1999). Efficient homologous and illegitimate recombination in the opportunistic yeast pathogen Candida glabrata. Genetics 151, 979–987.
Desai, J. V. (2018). Candida albicans Hyphae: from growth initiation to invasion. J. Fungi 4:E10. doi: 10.3390/jof4010010
Dujon, B., Sherman, D., Fischer, G., Durrens, P., Casaregola, S., Lafontaine, I., et al. (2004). Genome evolution in yeasts. Nature 430, 35–44. doi: 10.1038/nature02579
Dunham, M., Gartenberg, M., and Brown, G. W. (2015). Methods in Yeast Genetics and Genomics, 2015 Edition: A CSHL Course Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press.
Filomeni, G., De Zio, D., and Cecconi, F. (2015). Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. 22, 377–388. doi: 10.1038/cdd.2014.150
Forman, H. J., and Torres, M. (2001). Redox signaling in macrophages. Mol. Aspects Med. 22, 189–216. doi: 10.1016/S0098-2997(01)00010-3
Geber, A., Hitchcock, C. A., Swartz, J. E., Pullen, F. S., Marsden, K. E., Kwon-Chung, K. J., et al. (1995). Deletion of the Candida glabrata ERG3 and ERG11 genes: effect on cell viability, cell growth, sterol composition, and antifungal susceptibility. Antimicrob. Agents Chemother. 39, 2708–2717. doi: 10.1128/AAC.39.12.2708
Gonzalez-Parraga, P., Hernandez, J. A., and Arguelles, J. C. (2003). Role of antioxidant enzymatic defences against oxidative stress H(2)O(2) and the acquisition of oxidative tolerance in Candida albicans. Yeast 20, 1161–1169. doi: 10.1002/yea.1029
Hansen, M., Rubinsztein, D. C., and Walker, D. W. (2018). Autophagy as a promoter of longevity: insights from model organisms. Nat. Rev. Mol. Cell Biol. 19, 579–593. doi: 10.1038/s41580-018-0033-y
Jin, Y., Hong, Y., Park, C. Y., and Hong, Y. (2017). Molecular interactions of autophagy with the immune system and cancer. Int. J. Mol. Sci. 18:E1694. doi: 10.3390/ijms18081694
Kasper, L., Seider, K., and Hube, B. (2015). Intracellular survival of Candida glabrata in macrophages: immune evasion and persistence. FEMS Yeast Res. 15:fov042. doi: 10.1093/femsyr/fov042
Khan, I. A., Lu, J. P., Liu, X. H., Rehman, A., and Lin, F. C. (2012). Multifunction of autophagy-related genes in filamentous fungi. Microbiol. Res. 167, 339–345. doi: 10.1016/j.micres.2012.01.004
Kitada, K., Yamaguchi, E., and Arisawa, M. (1995). Cloning of the Candida glabrata TRP1 and HIS3 genes, and construction of their disruptant strains by sequential integrative transformation. Gene 165, 203–206. doi: 10.1016/0378-1119(95)00552-H
Kitada, K., Yamaguchi, E., and Arisawa, M. (1996). Isolation of a Candida glabrata centromere and its use in construction of plasmid vectors. Gene 175, 105–108. doi: 10.1016/0378-1119(96)00132-1
Klionsky, D. J., Abdelmohsen, K., Abe, A., Abedin, M. J., Abeliovich, H., Acevedo-Arozena, A., et al. (2016). Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222. doi: 10.1080/15548627.2015.1100356
Lee, H. N., Kim, J., and Chung, T. (2014). Degradation of plant peroxisomes by autophagy. Front. Plant Sci. 5:139. doi: 10.3389/fpls.2014.00139
Lionakis, M. S., Swamydas, M., Fischer, B. G., Plantinga, T. S., Johnson, M. D., Jaeger, M., et al. (2013). CX3CR1-dependent renal macrophage survival promotes Candida control and host survival. J. Clin. Invest. 123, 5035–5051. doi: 10.1172/jci71307
Miceli, M. H., Diaz, J. A., and Lee, S. A. (2011). Emerging opportunistic yeast infections. Lancet Infect. Dis. 11, 142–151. doi: 10.1016/S1473-3099(10)70218-8
Miyazaki, T., Inamine, T., Yamauchi, S., Nagayoshi, Y., Saijo, T., Izumikawa, K., et al. (2010a). Role of the Slt2 mitogen-activated protein kinase pathway in cell wall integrity and virulence in Candida glabrata. FEMS Yeast Res. 10, 343–352. doi: 10.1111/j.1567-1364.2010.00611.x
Miyazaki, T., Yamauchi, S., Inamine, T., Nagayoshi, Y., Saijo, T., Izumikawa, K., et al. (2010b). Roles of calcineurin and Crz1 in antifungal susceptibility and virulence of Candida glabrata. Antimicrob. Agents Chemother. 54, 1639–1643. doi: 10.1128/AAC.01364-09
Miyazaki, T., Izumikawa, K., Nagayoshi, Y., Saijo, T., Yamauchi, S., Morinaga, Y., et al. (2011). Functional characterization of the regulators of calcineurin in Candida glabrata. FEMS Yeast Res. 11, 621–630. doi: 10.1111/j.1567-1364.2011.00751.x
Nagi, M., Tanabe, K., Nakayama, H., Ueno, K., Yamagoe, S., Umeyama, T., et al. (2016). Iron-depletion promotes mitophagy to maintain mitochondrial integrity in pathogenic yeast Candida glabrata. Autophagy 12, 1259–1271. doi: 10.1080/15548627.2016.1183080
Nah, J., Yoo, S.-M., Jung, S., Jeong, E. I., Park, M., Kaang, B.-K., et al. (2017). Phosphorylated CAV1 activates autophagy through an interaction with BECN1 under oxidative stress. Cell Death Dis. 8, e2822. doi: 10.1038/cddis.2017.71
Ngo, L. Y., Kasahara, S., Kumasaka, D. K., Knoblaugh, S. E., Jhingran, A., and Hohl, T. M. (2014). Inflammatory monocytes mediate early and organ-specific innate defense during systemic candidiasis. J. Infect. Dis. 209, 109–119. doi: 10.1093/infdis/jit413
Nishikawa, H., Miyazaki, T., Nakayama, H., Minematsu, A., Yamauchi, S., Yamashita, K., et al. (2016). Roles of vacuolar H+-ATPase in the oxidative stress response of Candida glabrata. FEMS Yeast Res. 16:fow054. doi: 10.1093/femsyr/fow054
Palmer, G. E., Askew, D. S., and Williamson, P. R. (2008). The diverse roles of autophagy in medically important fungi. Autophagy 4, 982–988. doi: 10.4161/auto.7075
Roetzer, A., Gabaldon, T., and Schuller, C. (2011). From Saccharomyces cerevisiae to Candida glabrata in a few easy steps: important adaptations for an opportunistic pathogen. FEMS Microbiol. Lett. 314, 1–9. doi: 10.1111/j.1574-6968.2010.02102.x
Roetzer, A., Gratz, N., Kovarik, P., and Schuller, C. (2010). Autophagy supports Candida glabrata survival during phagocytosis. Cell Microbiol. 12, 199–216. doi: 10.1111/j.1462-5822.2009.01391.x
Shintani, T., and Klionsky, D. J. (2004). Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 279, 29889–29894. doi: 10.1074/jbc.M404399200
Suzuki, K. (2013). Selective autophagy in budding yeast. Cell Death Differ. 20, 43–48. doi: 10.1038/cdd.2012.73
Thorpe, G. W., Fong, C. S., Alic, N., Higgins, V. J., and Dawes, I. W. (2004). Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative-stress-response genes. Proc. Natl. Acad. Sci. U.S.A. 101, 6564–6569. doi: 10.1073/pnas.0305888101
Timmermans, B., De Las Penas, A., Castano, I., and Van Dijck, P. (2018). Adhesins in Candida glabrata. J. Fungi 4:E60. doi: 10.3390/jof4020060
Ueno, K., Uno, J., Nakayama, H., Sasamoto, K., Mikami, Y., and Chibana, H. (2007). Development of a highly efficient gene targeting system induced by transient repression of YKU80 expression in Candida glabrata. Eukaryot. Cell 6, 1239–1247. doi: 10.1128/EC.00414-06
Wang, B., and Kundu, M. (2017). Canonical and noncanonical functions of ULK/Atg1. Curr. Opin. Cell Biol. 45, 47–54. doi: 10.1016/j.ceb.2017.02.011
Xu, S., and Shinohara, M. L. (2017). Tissue-resident macrophages in fungal infections. Front. Immunol. 8:1798. doi: 10.3389/fimmu.2017.01798
Yin, J., Duan, J., Cui, Z., Ren, W., Li, T., and Yin, Y. (2015). Hydrogen peroxide-induced oxidative stress activates NF-κB and Nrf2/Keap1 signals and triggers autophagy in piglets. RSC Adv. 5, 15479–15486. doi: 10.1039/c4ra13557a
Yorimitsu, T., and Klionsky, D. J. (2005). Autophagy: molecular machinery for self-eating. Cell Death Differ. 12(Suppl. 2), 1542–1552. doi: 10.1038/sj.cdd.4401765
Yu, Q., Jia, C., Dong, Y., Zhang, B., Xiao, C., Chen, Y., et al. (2015). Candida albicans autophagy, no longer a bystander: its role in tolerance to ER stress-related antifungal drugs. Fungal Genet. Biol. 81, 238–249. doi: 10.1016/j.fgb.2015.02.008
Keywords: autophagy, Atg1, Candida glabrata, virulence, reactive oxygen species
Citation: Shimamura S, Miyazaki T, Tashiro M, Takazono T, Saijo T, Yamamoto K, Imamura Y, Izumikawa K, Yanagihara K, Kohno S and Mukae H (2019) Autophagy-Inducing Factor Atg1 Is Required for Virulence in the Pathogenic Fungus Candida glabrata. Front. Microbiol. 10:27. doi: 10.3389/fmicb.2019.00027
Received: 28 October 2018; Accepted: 09 January 2019;
Published: 25 January 2019.
Edited by:
Dominique Sanglard, Université de Lausanne, SwitzerlandReviewed by:
Erika Shor, Public Health Research Institute (PHRI), United StatesMiguel Cacho Teixeira, Universidade de Lisboa, Portugal
Copyright © 2019 Shimamura, Miyazaki, Tashiro, Takazono, Saijo, Yamamoto, Imamura, Izumikawa, Yanagihara, Kohno and Mukae. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Taiga Miyazaki, dGFpZ2EtbUBuYWdhc2FraS11LmFjLmpw Shintaro Shimamura, c3NoaW1hbXVAbmFnYXNha2ktdS5hYy5qcA==
 Shintaro Shimamura
Shintaro Shimamura Taiga Miyazaki
Taiga Miyazaki Masato Tashiro2
Masato Tashiro2 Takahiro Takazono
Takahiro Takazono Tomomi Saijo
Tomomi Saijo Koichi Izumikawa
Koichi Izumikawa