- 1Department of Infectious Disease, Shanghai Public Health Clinical Center, Fudan University, Shanghai, China
- 2Department of Infectious Disease, Huashan Hospital, Fudan University, Shanghai, China
- 3Department of Internal Medicine, Shanghai Medical College, Fudan University, Shanghai, China
Gut microbiota dysbiosis, which has been linked to many neurological diseases, is common in HIV infection. However, its role in the pathogenesis of neurocognitive impairment is still not established. In this study, a total of 85 HIV infected subjects, naïve to antiretroviral therapy, were classified into two groups—those with HIV-associated neurological diseases (HAND) and those without, using the Montreal Cognitive Assessment (MoCA) test. Fecal samples were collected from all subjects and microbiota were analyzed by 16S rRNA amplicon sequencing. Subjects with HAND were older (P < 0.001), with lower levels of education (P = 0.002), lower CD4 T-cell counts (P = 0.032), and greater heterosexual preference (P < 0.001), than those without HAND. Gut microbiota from subjects with HAND showed significantly lower α-diversity compared to gut microbiota from subjects without HAND (Shannon index, P = 0.003). To exclude confounding bias, 25 subjects from each group, with comparable age, gender, CD4 T-cell count, educational level and sexual preference were further analyzed. The two groups showed comparable α-diversity (for SOB index, Shannon index, Simpson index, ACE index, and Chao index, all with P-value > 0.05) and β-diversity (ANOSIM statistic = 0.010, P = 0.231). There were no significant differences in microbiota composition between the two groups after the correction for a false discovery rate. Consistently, microbiota from the two groups presented similar predictive functional profiles. Gut microbiota dysbiosis is not independently associated with neurocognitive impairment in people living with HIV.
Introduction
The prognosis for HIV-infected patients has improved significantly in the past two decades, as the incidence of various opportunistic infections decreased substantially due to combined antiretroviral therapy (cART) (Antiretroviral Therapy Cohort Collaboration, 2017). However, the neurological symptoms caused by HIV infection are still not well-controlled (Fauci and Marston, 2015). In 2007, these neurological symptoms were designated as HIV-associated neurocognitive disorder (HAND) (Antinori et al., 2007). A recent study reported that almost half of people living with HIV/AIDS (PLWHA) naïve to cART, demonstrated neurocognitive impairment (Robertson et al., 2018). It has been estimated that approximately 15–55% of PLWHA suffer from HAND despite effective cART (McArthur et al., 1993; Heaton et al., 2010). HIV-associated dementia (HAD), the most severe form of HAND, has been reduced from 20 to 5% as a result of broader cART coverage, while other milder forms of HAND such as asymptomatic neurocognitive disorder (ANI) and mild neurocognitive disorder (MND) remain common (Gates et al., 2016; Saylor et al., 2016). PLWHA with HAND are less able to deal with complicated activities, resulting in poor medication compliance which leads to a shorter life expectancy and a lower quality of life (Berger and Brew, 2005; Antinori et al., 2007; Tozzi et al., 2007). However, the pathogenesis of HAND remains unknown.
Growing evidence indicates that many neurological diseases, including Parkinson’s disease, Alzheimer’s disease, and multiple sclerosis, are linked to gut microbiota dysbiosis (Hindson, 2017; Wu et al., 2017; Qian et al., 2018). Abnormalities in gut microbiota may affect brain function through several mechanisms, including the modulation of the signaling pathways of the microbiota-gut-brain axis and the regulation of both the production and absorption of neurotransmitters and neurotoxic products (e.g., kynurenine downstream products) (Vecsei et al., 2013).
In HIV-infection, leaky gut and an increased plasma level of microbiota products contribute to immune activation, which has been linked to HAND (Eden et al., 2007; Sandler and Douek, 2012; Yilmaz et al., 2013; Eden et al., 2016). Furthermore, gut microbiota dysbiosis is common in HIV-infection, especially in those with low CD4 T-cell counts (Tincati et al., 2016; Ribeiro et al., 2017; Hamad et al., 2018; Williams et al., 2018). Whether gut microbiota play a role in the pathogenesis of HAND, currently remains unclear. Herein, we studied the gut microbiome of PLWHA with and without HAND to determine whether HAND is associated with differences in gut microbiota.
Materials and Methods
Study Settings and Design
We enrolled PLWHA at the Shanghai Public Health Clinical Center (SHPCC), Shanghai, China from September 2015 to July 2016. All PLWHA older than 18 years and naïve to cART were eligible to participate. The exclusion criteria included antibiotic/probiotic administration within 4 weeks prior to enrollment; complications including cardiopulmonary diseases, hematological diseases, malignant tumor, opportunistic infections, chronic hepatitis virus B and C infection, and syphilis; history of inflammatory bowel disease; history of central nervous system diseases or mental illness prior to HIV diagnosis; as well as pregnant women. This study was approved by the Ethics Committee of SPHCC. All subjects provided written informed consent in accordance with the Declaration of Helsinki.
Enrolled PLWHA were allocated using the Montreal Cognitive Assessment (MoCA) test (Pendlebury et al., 2012). PLWHA with scores less than 26/30 points were assigned to the HAND group, while subjects that scored greater than 26/30 points were placed in the non-HAND group.
Sample Collection and DNA Extraction
At the time of recruitment, fecal and blood samples were collected from all subjects. The fecal samples were collected in disposable plastic sterile dung cups and properly handled. After collection, they were stored at -80°C until DNA extraction was performed using the QIAamp DNA Stool Mini Kit (Qiagen, Düsseldorf, Germany).
HIV RNA and CD4 T-cell counts were performed routinely at the clinical laboratory of the SPHCC, using Roche COBAS® AmpliPrep/COBAS® TaqMan® HIV-1 test, version 2.0 (CAP/CTM v2.0; Roche, Basel, Switzerland) and CytomicsTM FC 500 (Beckman Coulter, Brea, CA, United States) flow cytometry, respectively.
16S rRNA Gene Sequencing and Analysis
The targeted V3-V4 region of the bacterial 16S rRNA gene from extracted DNA, was PCR-amplified with the universal primers 341F (5′-AGA GTT TGA TCM TGG CTC AG-3′) and 805R (5′-GAC TGG AGT TCC TTG GCA CCC GAG AAT TCC A-3′) (Sakamoto et al., 2000; Chakravorty et al., 2007; Nossa et al., 2010; Haas et al., 2011). The 16S rRNA gene sequencing was performed on an Illumina MiSeq instrument (Illumina, San Diego, United States) at Sangon Biotech Co., Ltd. (Shanghai, China). All reads were demultiplexed, preprocessed, and subsequently analyzed with the Quantitative Insights into Microbial Ecology (QIIME) software package (Caporaso et al., 2010). Operational taxonomic unit (OTU) clustering was performed at a 97% similarity threshold using the QIIME pipeline. The relative abundance of the taxa at the phylum and genus levels were calculated. Alpha diversity analysis was implemented to measure the diversity of species in each sample and to calculate species diversity indices such as the SOB (the observed richness), ACE (abundance based coverage estimated), Chao, Shannon, and Simpson (Sun et al., 2016). Beta diversity was measured by unweighted and weighted UniFrac metrics and the distances were visualized by principal coordinates analysis (PCoA) (Ji et al., 2018). The OTU abundance was standardized by PICRUSt (phylogenetic investigation of communities by reconstruction of unobserved states) to conduct the 16S predictive functional analysis, that is, to remove the influence of the number of copies of the 16S marker gene in the genome of the species (Tang et al., 2018). The PICRUSt software stores the Kyoto encyclopedia of genes and genomes (KEGG) ortholog information corresponding to the greengene ID. The greengene ID obtains KEGG ortholog (KO) information corresponding to the OTU.
Statistical Analysis
Data analysis was conducted using IBM SPSS version 20.0 software (IBM SPSS, Inc., Armonk, NY, United States) and GraphPad Prism version 5.0 (GraphPad Software, La Jolla, CA, United States). Continuous variables were described using mean and standard deviation (SD), while categorical variables were described by numbers and percentages. For characteristics analyses and diversity indices between groups, differences were estimated by the Student’s t-test, the Mann–Whitney U-test, or the chi-square test, where appropriate. All tests were two-sided and P < 0.05 was considered statistically significant. P-values were corrected to control the false discovery rate (FDR) < 0.05 using the Benjamini–Hochberg method.
Results
Demographic Characteristics of the PLWHA
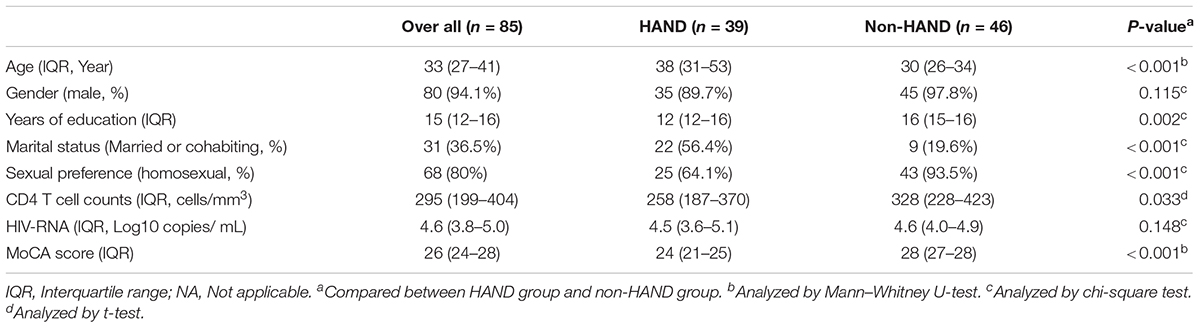
A total of 85 subjects were enrolled in this study. Among these 85 PLWHA, 39 were assigned to the HAND group and 46 were classified as non-HAND. PLWHA in the HAND group were older, with lower education levels, lower CD4 T-cell counts, and greater heterosexual preference than those in the non-HAND group (Table 1). As PLWHA in the HAND group had lower CD4 T-cell counts and greater heterosexual preference compared with those in the non-HAND group, the gut microbiota in the HAND group also showed significantly lower α-diversity when compared with that in the non-HAND group, as expected (Figure 1).
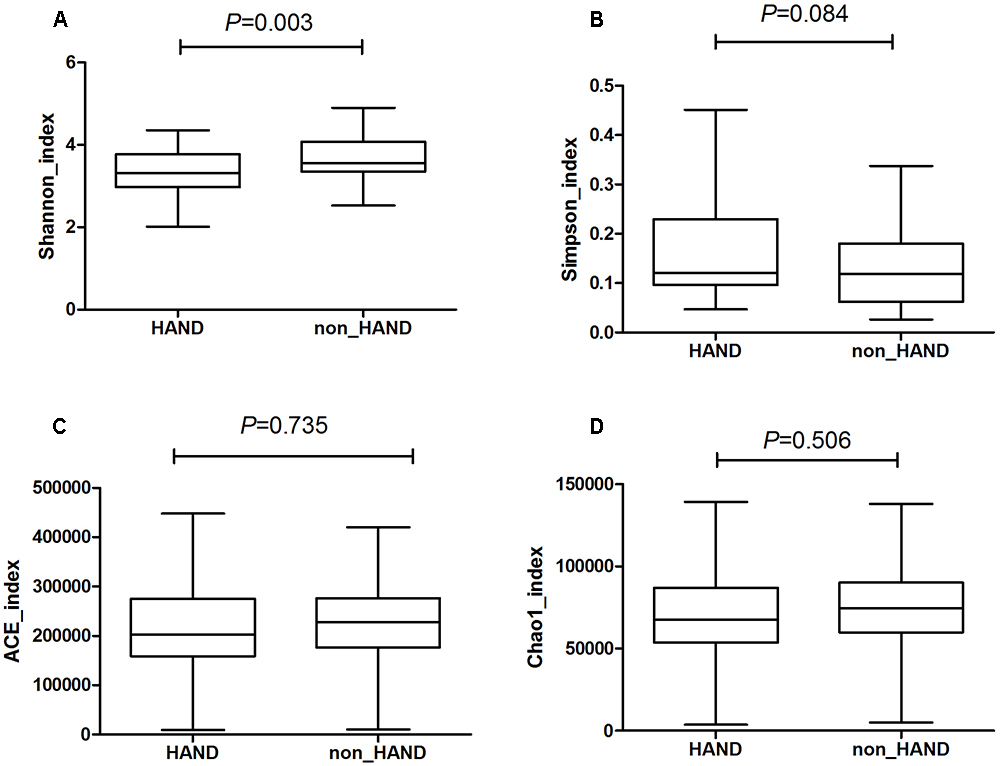
Figure 1. Significantly lower α-diversity of gut microbiota from HAND group compared with that from non-HAND group. (A) Shannon index: t-test; (B) Simpson index: Mann–Whitney test; (C) ACE index: Mann–Whitney test; (D) Chao index: t-test.
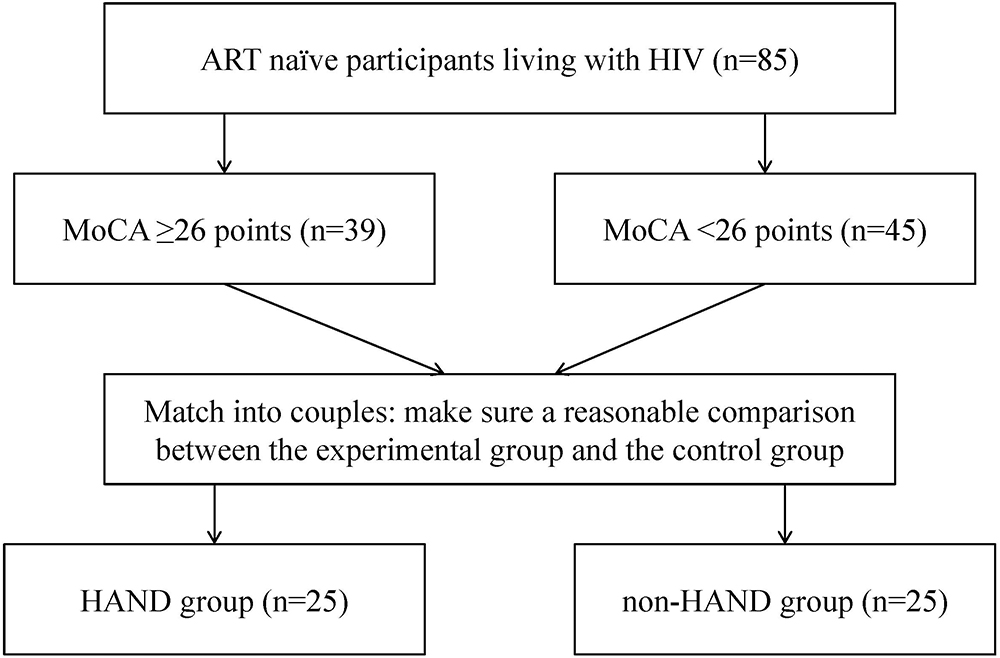
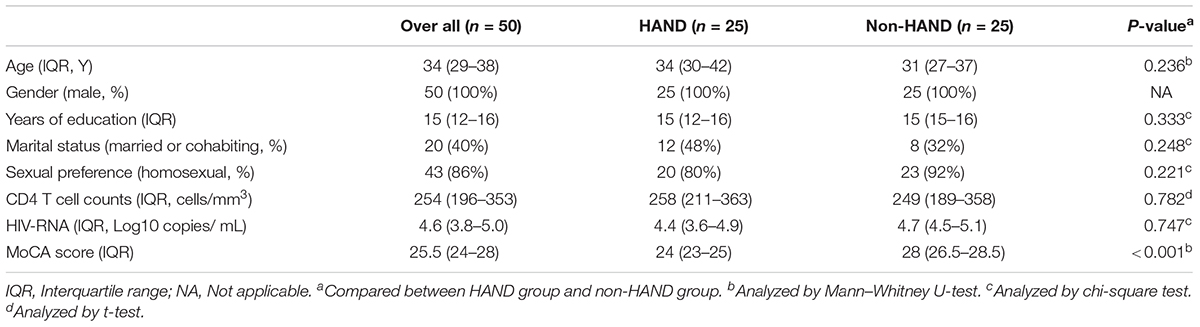
To limit confounding bias, we matched the two groups for age, gender, immunological stage, education level, and sexual preference in 50 subjects that underwent further analysis, with 25 subjects in each group. A flowchart describing the recruitment and grouping methods of all PLWHA is shown in Figure 2.
The demographic characteristics of these subgroups are shown in Table 2. All 50 PLWHA were male, aged 33.5 (28.5–38), with 15 (12–16) years of education. The majority (86%) of the group had a homosexual preference and modest immunosuppression [CD4 T-cell counts 254 (196–353) cells/mm3] (Table 2). The MoCA scores in the HAND and non-HAND groups were 24 (23–25) and 28 (26.5–28.5), respectively.
Gut Microbiota Diversity Index
We compared the α-diversity by SOB, Shannon, Simpson, ACE, and Chao indices, respectively, between the two groups. The species α-diversity of gut microbiota was not significantly different between the HAND and non-HAND groups [SOB index (median 112.0 vs. 127.0, P = 0.290), Shannon index (median 2.044 vs. 2.356, P = 0.190), Simpson index (median 0.236 vs. 0.200, P = 0.467), ACE index (median 151.2 vs. 146.4, P = 0.884), and Chao index (median 143.6 vs. 144.0, P = 0.266) Figures 3A–E].
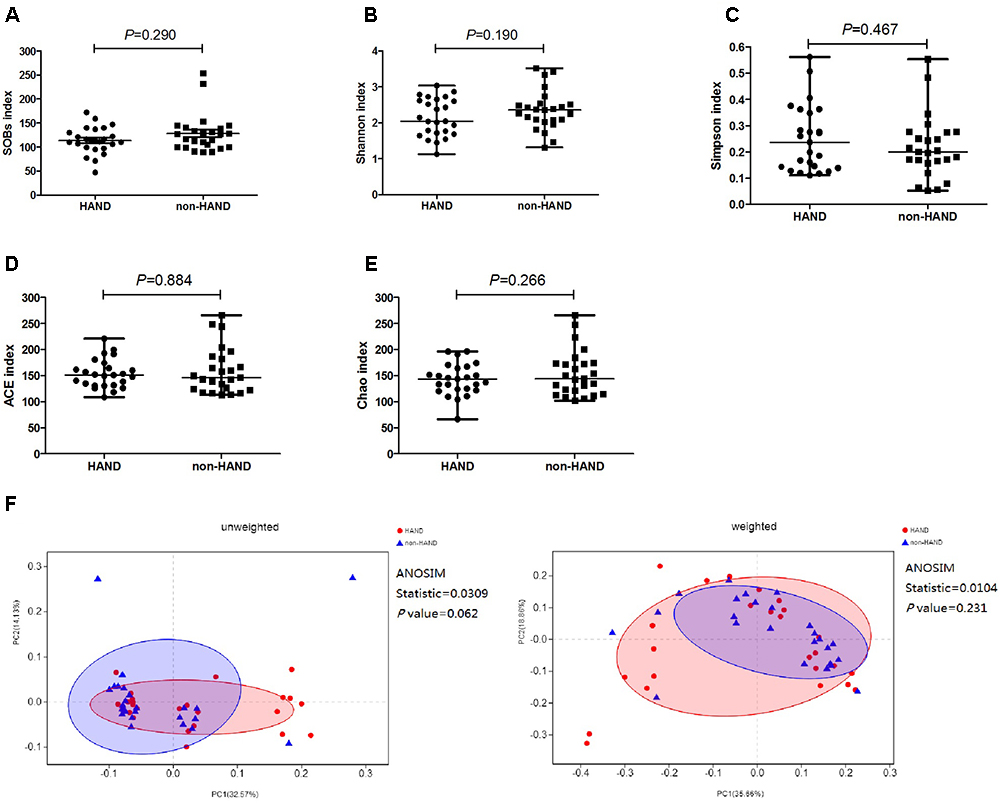
Figure 3. Differences in species α-diversity of gut microbiota between HAND and non-HAND groups in subgroup analysis. (A) SOBs index, Mann–Whitney test; (B) Shannon index, t-test; (C) Simpson index, Mann–Whitney test; (D) ACE index, Mann-Whitney test; (E) Chao index, t-test. (F) Unweighted and weighted analyses of similarities(ANOSIMs) and principal coordinates analysis(PCOA) based on the distance matrix of UniFrac dissimilarity of the fecal microbial communities in HAND group and non-HAND groups. Each symbol represented a sample, HAND group (red circle), non-HAND group (blue triangle). ANOSIM statistic showed the community variation between the compared groups, and P-values were indicated.
Furthermore, the species β-diversity of fecal microbiota was also not significantly different between the HAND and non-HAND groups (Figure 3F). We evaluated β-diversity based on the unweighted (qualitative, ANOSIM statistic = 0.031, P = 0.062) and the weighted (quantitative, ANOSIM statistic = 0.010, P = 0.231) UniFrac distance matrix of the differences between groups in the fecal microbial communities.
No Differences in Gut Microbiota Composition Between the HAND and Non-HAND Groups
At the phylum level, species abundance of actinobacteria was higher in the HAND group than in the non-HAND group (4.459 vs. 2.108%; P = 0.042; Figure 4A). However, this difference disappeared after FDR correction (adjusted P = 0.541). At the genus level, lower abundances of Faecalibacterium (8.304 vs. 12.23%; P = 0.028), Catenibacterium (0.552 vs. 2.877%; P = 0.040), and Ruminococcaceae (0.7261 vs. 1.138%; P = 0.009) were detected in the HAND group in contrast to the non-HAND group (Figure 4B). After for FDR correction, the differences in the abundance of Faecalibacterium (adjusted P = 0.625), Catenibacterium (adjusted P = 0.625), and Ruminococcaceae (adjusted P = 0.625) were not significant between the aforementioned two groups.
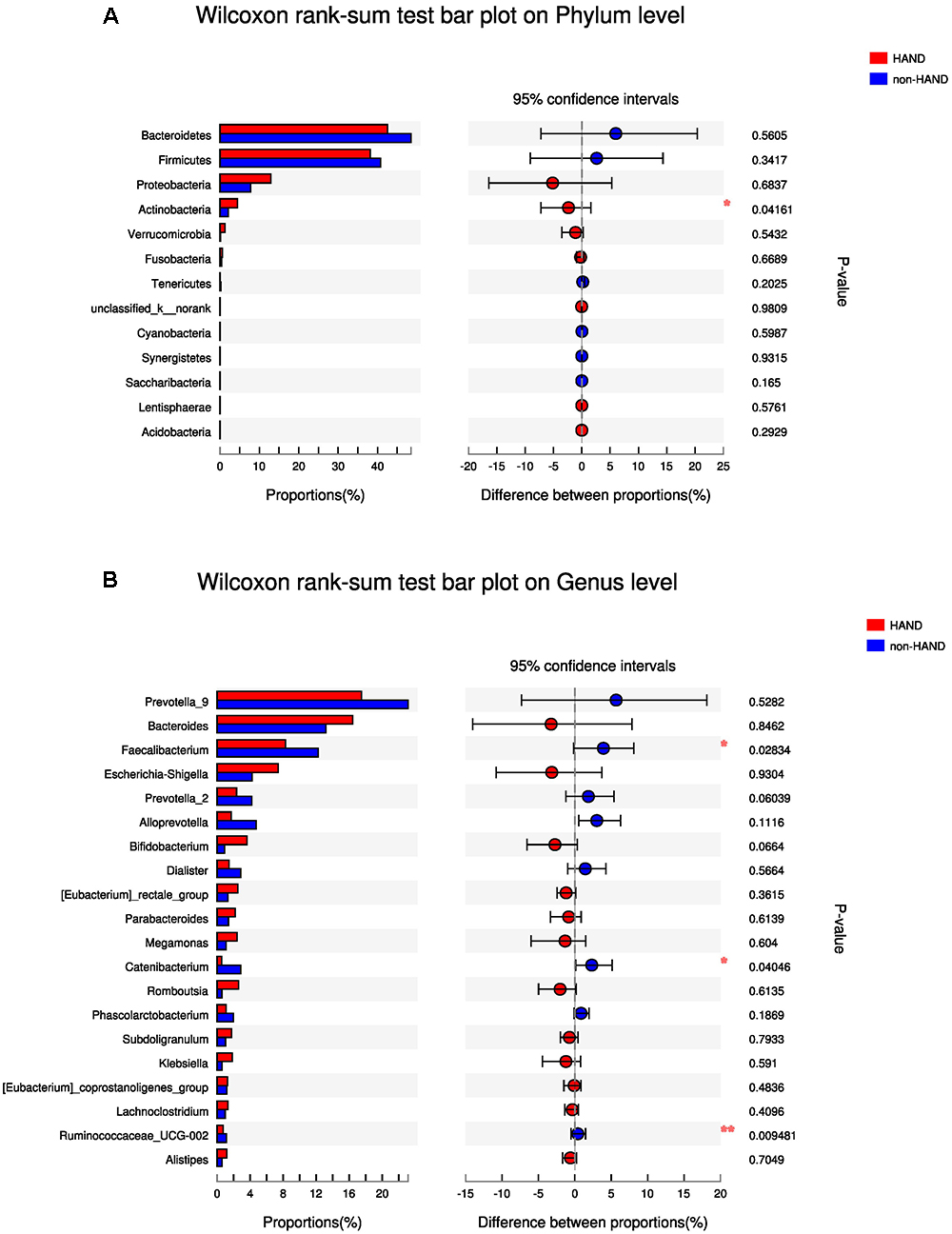
Figure 4. Difference in gut microbiota composition between the HAND group and the non-HAND group. (A) Wilcoxon rank-sum test bar plot on Phylum level. (B) Wilcoxon rank-sum test bar plot on Genus level. Each color represented a group, HAND group (red), non-HAND group (blue). ∗P < 0.05 and ∗∗P < 0.001. Confidence intervals were estimated using a percentile bootstrapping method.
Predictive Function Profile of the Gut Microbiota
Predictive functional profiling using the KEGG pathway showed that gut microbiota in the HAND group were associated with higher abundances in the cellular processes compared to the non-HAND groups (mean 344019 vs. 277003; P = 0.027). As microbiota function in cellular processes mainly include cell growth and death, motility, transport, and catabolism, we conducted a level 2 analysis of the KEGG pathways. After FDR correction, no statistical differences were detected between the two groups (Figure 5).
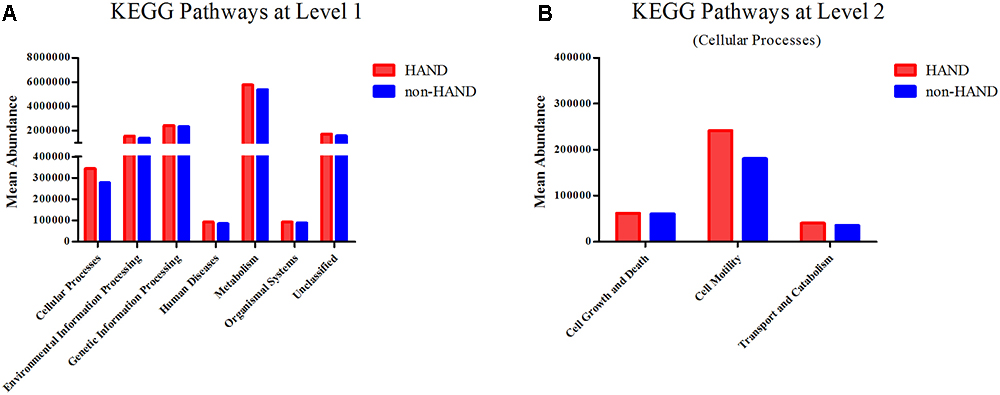
Figure 5. Functional predictions for the fecal microbiome of the HAND and non-HAND groups by KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways. (A) KEGG pathways at level 1, t-test. (B) KEGG pathways at level 2, t-test.
Discussion
Gut microbiota dysbiosis has been linked to neurocognitive disorders in the HIV-negative population (Miklossy, 2011; Poole et al., 2013; Yang et al., 2016; Zhan et al., 2016; Noble et al., 2017). Nevertheless, an understanding of its role in the pathogenesis of HAND is still lacking to date. In the current study, we showed that gut microbiota dysbiosis is not independently associated with HAND. To the best of our knowledge, this is the first study to explore the association between gut microbiota and HAND.
We reported, along with others, that gut microbiota dysbiosis in PLWHA is common (Nowak et al., 2015; Carding et al., 2017; Desai and Landay, 2018; Lu et al., 2018; Missailidis et al., 2018). Gut microbiota from PLWHA showed significantly lower α-diversity compared with that from HIV-uninfected controls (Sun et al., 2016; Hamad et al., 2018). In this study, we found that microbiota in the HAND group also displayed significantly lower α-diversity than that in the non-HAND group, indicating associations between gut microbiota and HAND. Consistent with these results, a recent pilot study showed that supplementation with a probiotic containing Streptococcus salivarius, S. thermophilus, Bifidobacteria, Lactobacillus spp., and S. faecium improved neuropsychological performance in PLWHA (Ceccarelli et al., 2017). However, in our subgroup analysis adjusting for gender, age, education level, and CD4 T-cell count, all of which has been linked to HAND, the diversity and composition of the gut microbiota was comparable between the two groups in the current study (Wang et al., 2013). Moreover, no significant differences in the predictive function of the gut microbiota was found between the HAND and non-HAND groups. It is known that confounders such as age, gender, CD4 T-cell counts, and sexual preference also have significant effects on gut microbiota (Nowak et al., 2015; Ji et al., 2018). We and others have recently demonstrated that low CD4 T-cell counts, rather than HIV serostatus, predict the presence and recovery of gut dysbiosis in HIV-infected subjects (Guillen et al., 2018; Ji et al., 2018; Zhou et al., 2018). Furthermore, other studies found that MSM (men who have sex with men) had a significantly richer and more diverse fecal microbiota than non-MSM individuals, independent of HIV infection (Noguera-Julian et al., 2016; Kelley et al., 2017). Therefore, our results do not preclude the role of intestinal dysbiosis in the pathogenesis of HAND. However, our data do show that gut microbiota dysbiosis is not independently associated with HAND. Moreover, we do not know whether the compositions of the virome and fungal microbiome in the gut may contribute to HAND, which warrants further investigation.
Gut microbiota may contribute to neurological diseases by regulating both the production and absorption of neurotoxic products. Stimulation of the kynurenine pathway has been closely linked to the pathogenesis of HAND (Heyes et al., 2001; Smith et al., 2001; Valle et al., 2004; Vamos et al., 2009; Davies et al., 2010; Kandanearatchi and Brew, 2012; Vecsei et al., 2013; Myint and Kim, 2014). Indeed, even long-term ART in PLWHA did not normalize the overactivated kynurenine pathway (Chen et al., 2014, 2018). The levels of Lactobacilli and Proteobacteria in the gut have been correlated with kynurenine pathway activity and disease progression, respectively (Vujkovic-Cvijin et al., 2013; Zelante et al., 2013). Therefore, they might contribute to the development of HAND. However, no significant differences in these bacteria (Lactobacilli 0.044 vs. 0.046%, P = 0.710; Proteobacteria 12.94 vs. 7.79%, P = 0.684) were observed in our study.
Gut microbial dysbiosis has been associated with HIV pathogenesis (Vyboh et al., 2015; Dillon et al., 2016). Indeed, gut microbial dysbiosis is associated with many immunological parameters including T-cell activation and plasma lipopolysaccharide and soluble CD14 levels, all of which have been linked to disease progression (Dillon et al., 2014; Dinh et al., 2015; Nowak et al., 2015; Vazquez-Castellanos et al., 2015; Monaco et al., 2016; Noguera-Julian et al., 2016; Serrano-Villar et al., 2017; Vazquez-Castellanos et al., 2018). Moreover, interventions that modulate alterations in gut microbiota improved these markers in some clinical trials (Irvine et al., 2010; Gori et al., 2011; Kristoff et al., 2014). However, few studies have deciphered the association between microbiota and clinical diseases in HIV infection. Recently, altered microbiota dysbiosis has been linked to coronary heart disease, type 2 diabetes, and poor CD4 T-cell recovery (Hoel et al., 2018; Kehrmann et al., 2018; Lu et al., 2018). Unfortunately, these studies involved relatively small sample sizes and did not take into account other factors that significantly effects gut microbiota (e.g., CD4 T-cell count and sexual preference), which might bias their results. Interestingly, a landmark study recently showed no effect of experimental gut microbiota dysbiosis on disease progression in an SIV infection model, indicating that the associations we observed in PLWHA may result from the confounders (Ortiz et al., 2018). Therefore, we suggest that future microbiome studies on HIV infection should take these confounders into consideration (Noguera-Julian et al., 2016; Tincati et al., 2016; Ribeiro et al., 2017; Hamad et al., 2018; Klatt and Manuzak, 2018; Williams et al., 2018).
There are some limitations in our study that should be addressed. Firstly, the relatively small sample size might generate bias in our results. Secondly, in using MoCA as a reference for grouping, PLWHA exhibited a narrow score distribution interval, leading to a lack of HAD subjects. A recent study showed that the intestinal microbiota of people living with HAD was highly specific (Pérez-Santiago et al., 2017). Finally, all subjects we enrolled were ART naïve. Therefore, our results cannot be extended to PLWHA who have already been treated.
Conclusion
In conclusion, our research shows that gut microbiota dysbiosis is not independently associated with neurocognitive impairment in PLWHA.
Author Contributions
JC, JyS, and HL conceived and designed the study. FZ, JY, YJ, and JC acquired and analyzed the data. FZ, MS, and JjS assessed the cognitive status of the PLWHA. JW, LL, YS, and RZ were responsible for PLWHA recruitment. FZ performed the laboratory work. FZ and JY wrote the first draft of the manuscript. FZ, JC, and HL contributed to the final version of the manuscript. All authors read and approved the final manuscript.
Funding
This study was sponsored by the National Natural Science Foundation of China (NSFC No. 81571977), the Shanghai Municipal Commission of Health and Family Planning (No. 20164Y0015), and the Medical Science Support Program of the Shanghai Science and Technology Committee (No. 16411960400). The Major National Science and Technology Projects of China (2017ZX09304027 and 2017ZX10202101-002). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Open access publication fees are granted by the funders.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Antinori, A., Arendt, G., Becker, J. T., Brew, B. J., Byrd, D. A., Cherner, M., et al. (2007). Updated research nosology for HIV-associated neurocognitive disorders. Neurology 69, 1789–1799. doi: 10.1212/01.WNL.0000287431.88658.8b
Antiretroviral Therapy Cohort Collaboration (2017). Survival of HIV-positive patients starting antiretroviral therapy between 1996 and 2013: a collaborative analysis of cohort studies. Lancet HIV 4, e349–e356. doi: 10.1016/S2352-3018(17)30066-8
Berger, J. R., and Brew, B. (2005). An international screening tool for HIV dementia. AIDS 19, 2165–2166. doi: 10.1097/01.aids.0000194798.66670.6e
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Carding, S. R., Davis, N., and Hoyles, L. (2017). Review article: the human intestinal virome in health and disease. Aliment. Pharmacol. Ther. 46, 800–815. doi: 10.1111/apt.14280
Ceccarelli, G., Fratino, M., Selvaggi, C., Giustini, N., Serafino, S., Schietroma, I., et al. (2017). A pilot study on the effects of probiotic supplementation on neuropsychological performance and microRNA-29a-c levels in antiretroviral-treated HIV-1-infected patients. Brain Behav. 7:e00756. doi: 10.1002/brb3.756
Chakravorty, S., Helb, D., Burday, M., Connell, N., and Alland, D. (2007). A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods 69, 330–339. doi: 10.1016/j.mimet.2007.02.005
Chen, J., Shao, J., Cai, R., Shen, Y., Zhang, R., Liu, L., et al. (2014). Anti-retroviral therapy decreases but does not normalize indoleamine 2,3-dioxygenase activity in HIV-infected patients. PLoS One 9:e100446. doi: 10.1371/journal.pone.0100446
Chen, J., Xun, J., Yang, J., Ji, Y., Liu, L., Qi, T., et al. (2018). Plasma indoleamine 2,3-dioxygenase activity is associated with the size of HIV reservoir in patients receiving antiretroviral therapy. Clin. Infect. Dis. doi: 10.1093/cid/ciy676 [Epub ahead of print].
Davies, N. W., Guillemin, G., and Brew, B. J. (2010). Tryptophan, neurodegeneration and HIV-associated neurocognitive disorder. Int. J. Tryptophan. Res. 3, 121–140. doi: 10.4137/IJTR.S4321
Desai, S. N., and Landay, A. L. (2018). HIV and aging: role of the microbiome. Curr. Opin. HIV AIDS 13, 22–27. doi: 10.1097/COH.0000000000000433
Dillon, S. M., Frank, D. N., and Wilson, C. C. (2016). The gut microbiome and HIV-1 pathogenesis: a two-way street. AIDS 30, 2737–2751. doi: 10.1097/QAD.0000000000001289
Dillon, S. M., Lee, E. J., Kotter, C. V., Austin, G. L., Dong, Z., Hecht, D. K., et al. (2014). An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 7, 983–994. doi: 10.1038/mi.2013.116
Dinh, D. M., Volpe, G. E., Duffalo, C., Bhalchandra, S., Tai, A. K., Kane, A. V., et al. (2015). Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J. Infect. Dis. 211, 19–27. doi: 10.1093/infdis/jiu409
Eden, A., Marcotte, T. D., Heaton, R. K., Nilsson, S., Zetterberg, H., Fuchs, D., et al. (2016). Increased intrathecal immune activation in virally suppressed HIV-1 infected patients with neurocognitive impairment. PLoS One 11:e0157160. doi: 10.1371/journal.pone.0157160
Eden, A., Price, R. W., Spudich, S., Fuchs, D., Hagberg, L., and Gisslen, M. (2007). Immune activation of the central nervous system is still present after >4 years of effective highly active antiretroviral therapy. J. Infect. Dis. 196, 1779–1783. doi: 10.1086/523648
Fauci, A. S., and Marston, H. D. (2015). Ending the HIV-AIDS pandemic–follow the science. N. Engl. J. Med. 373, 2197–2199. doi: 10.1056/NEJMp1502020
Gates, T. M., Cysique, L. A., Siefried, K. J., Chaganti, J., Moffat, K. J., and Brew, B. J. (2016). Maraviroc-intensified combined antiretroviral therapy improves cognition in virally suppressed HIV-associated neurocognitive disorder. AIDS 30, 591–600. doi: 10.1097/QAD.0000000000000951
Gori, A., Rizzardini, G., Van’T, L. B., Amor, K. B., van Schaik, J., Torti, C., et al. (2011). Specific prebiotics modulate gut microbiota and immune activation in HAART-naive HIV-infected adults: results of the ”COPA” pilot randomized trial. Mucosal Immunol. 4, 554–563. doi: 10.1038/mi.2011.15
Guillen, Y., Noguera-Julian, M., Rivera, J., Casadella, M., Zevin, A. S., Rocafort, M., et al. (2018). Low nadir CD4+ T-cell counts predict gut dysbiosis in HIV-1 infection. Mucosal Immunol. 12, 232–246. doi: 10.1038/s41385-018-0083-7
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., Giannoukos, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi: 10.1101/gr.112730.110
Hamad, I., Abou, A. R., Ravaux, I., Mokhtari, S., Tissot-Dupont, H., Michelle, C., et al. (2018). Metabarcoding analysis of eukaryotic microbiota in the gut of HIV-infected patients. PLoS One 13:e0191913. doi: 10.1371/journal.pone.0191913
Heaton, R. K., Clifford, D. B., Franklin, D. J., Woods, S. P., Ake, C., Vaida, F., et al. (2010). HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER Study. Neurology 75, 2087–2096. doi: 10.1212/WNL.0b013e318200d727
Heyes, M. P., Ellis, R. J., Ryan, L., Childers, M. E., Grant, I., Wolfson, T., et al. (2001). Elevated cerebrospinal fluid quinolinic acid levels are associated with region-specific cerebral volume loss in HIV infection. Brain 124, 1033–1042. doi: 10.1093/brain/124.5.1033
Hindson, J. (2017). Multiple sclerosis: a possible link between multiple sclerosis and gut microbiota. Nat. Rev. Neurol. 13:705. doi: 10.1038/nrneurol.2017.142
Hoel, H., Hove-Skovsgaard, M., Hov, J. R., Gaardbo, J. C., Holm, K., Kummen, M., et al. (2018). Impact of HIV and Type 2 diabetes on gut microbiota diversity, tryptophan catabolism and endothelial dysfunction. Sci. Rep. 8:6725. doi: 10.1038/s41598-018-25168-3
Irvine, S. L., Hummelen, R., Hekmat, S., Looman, C. W., Habbema, J. D., and Reid, G. (2010). Probiotic yogurt consumption is associated with an increase of CD4 count among people living with HIV/AIDS. J. Clin. Gastroenterol. 44, e201–e205. doi: 10.1097/MCG.0b013e3181d8fba8
Ji, Y., Zhang, F., Zhang, R., Shen, Y., Liu, L., Wang, J., et al. (2018). Changes in intestinal microbiota in HIV-1-infected subjects following cART initiation: influence of CD4+ T cell count. Emerg Microbes Infect 7:113. doi: 10.1038/s41426-018-0117-y
Kandanearatchi, A., and Brew, B. J. (2012). The kynurenine pathway and quinolinic acid: pivotal roles in HIV associated neurocognitive disorders. FEBS J. 279, 1366–1374. doi: 10.1111/j.1742-4658.2012.08500.x
Kehrmann, J., Menzel, J., Saeedghalati, M., Obeid, R., Schulze, C., Holzendorf, V., et al. (2018). Gut microbiota in HIV-infected individuals linked to coronary heart disease. J. Infect. Dis. 219, 497–508. doi: 10.1093/infdis/jiy524
Kelley, C. F., Kraft, C. S., de Man, T. J., Duphare, C., Lee, H. W., Yang, J., et al. (2017). The rectal mucosa and condomless receptive anal intercourse in HIV-negative MSM: implications for HIV transmission and prevention. Mucosal Immunol. 10, 996–1007. doi: 10.1038/mi.2016.97
Klatt, N. R., and Manuzak, J. A. (2018). Microbial effects on immunity in HIV: virus, gender or sexual preference induced? EBioMedicine 31, 7–8. doi: 10.1016/j.ebiom.2018.04.008
Kristoff, J., Haret-Richter, G., Ma, D., Ribeiro, R. M., Xu, C., Cornell, E., et al. (2014). Early microbial translocation blockade reduces SIV-mediated inflammation and viral replication. J. Clin. Invest. 124, 2802–2806. doi: 10.1172/JCI75090
Lu, W., Feng, Y., Jing, F., Han, Y., Lyu, N., Liu, F., et al. (2018). Association between gut microbiota and CD4 recovery in HIV-1 infected patients. Front. Microbiol. 9:1451. doi: 10.3389/fmicb.2018.01451
McArthur, J. C., Hoover, D. R., Bacellar, H., Miller, E. N., Cohen, B. A., Becker, J. T., et al. (1993). Dementia in AIDS patients: incidence and risk factors. Multicenter AIDS Cohort Study. Neurology 43, 2245–2252. doi: 10.1212/WNL.43.11.2245
Miklossy, J. (2011). Alzheimer’s disease - a neurospirochetosis. Analysis of the evidence following Koch’s and Hill’s criteria. J. Neuroinflammation 8:90. doi: 10.1186/1742-2094-8-90
Missailidis, C., Neogi, U., Stenvinkel, P., Troseid, M., Nowak, P., and Bergman, P. (2018). The microbial metabolite trimethylamine-N-oxide in association with inflammation and microbial dysregulation in three HIV cohorts at various disease stages. AIDS 32, 1589–1598. doi: 10.1097/QAD.0000000000001813
Monaco, C. L., Gootenberg, D. B., Zhao, G., Handley, S. A., Ghebremichael, M. S., Lim, E. S., et al. (2016). Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe 19, 311–322. doi: 10.1016/j.chom.2016.02.011
Myint, A. M., and Kim, Y. K. (2014). Network beyond IDO in psychiatric disorders: revisiting neurodegeneration hypothesis. Prog. Neuropsychopharmacol. Biol. Psychiatry 48, 304–313. doi: 10.1016/j.pnpbp.2013.08.008
Noble, E. E., Hsu, T. M., and Kanoski, S. E. (2017). Gut to brain dysbiosis: mechanisms linking western diet consumption, the microbiome, and cognitive impairment. Front. Behav. Neurosci. 11:9. doi: 10.3389/fnbeh.2017.00009
Noguera-Julian, M., Rocafort, M., Guillen, Y., Rivera, J., Casadella, M., Nowak, P., et al. (2016). Gut microbiota linked to sexual preference and HIV infection. EBioMedicine 5, 135–146. doi: 10.1016/j.ebiom.2016.01.032
Nossa, C. W., Oberdorf, W. E., Yang, L., Aas, J. A., Paster, B. J., Desantis, T. Z., et al. (2010). Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World J. Gastroenterol. 16, 4135–4144. doi: 10.3748/wjg.v16.i33.4135
Nowak, P., Troseid, M., Avershina, E., Barqasho, B., Neogi, U., Holm, K., et al. (2015). Gut microbiota diversity predicts immune status in HIV-1 infection. AIDS 29, 2409–2418. doi: 10.1097/QAD.0000000000000869
Ortiz, A. M., Flynn, J. K., DiNapoli, S. R., Vujkovic-Cvijin, I., Starke, C. E., Lai, S. H., et al. (2018). Experimental microbial dysbiosis does not promote disease progression in SIV-infected macaques. Nat. Med. 24, 1313–1316. doi: 10.1038/s41591-018-0132-5
Pendlebury, S. T., Mariz, J., Bull, L., Mehta, Z., and Rothwell, P. M. (2012). MoCA, ACE-R, and MMSE versus the National Institute of Neurological Disorders and Stroke-Canadian Stroke Network vascular cognitive impairment harmonization standards neuropsychological battery after TIA and stroke. Stroke 43, 464–469. doi: 10.1161/STROKEAHA.111.633586
Pérez-Santiago, J., Gianella, S., Bharti, A., Cookson, D., Heaton, R., Grant, I., et al. (2017). “The human gut microbime and HIV-associated neurocognitive disorders,” in Proceedings of the 13th International Symposium on NeuroVirology, San Diego, CA.
Poole, S., Singhrao, S. K., Kesavalu, L., Curtis, M. A., and Crean, S. (2013). Determining the presence of periodontopathic virulence factors in short-term postmortem Alzheimer’s disease brain tissue. J. Alzheimers Dis. 36, 665–677. doi: 10.3233/JAD-121918
Qian, Y., Yang, X., Xu, S., Wu, C., Song, Y., Qin, N., et al. (2018). Alteration of the fecal microbiota in Chinese patients with Parkinson’s disease. Brain Behav. Immun. 70, 194–202. doi: 10.1016/j.bbi.2018.02.016
Ribeiro, A., Heimesaat, M. M., and Bereswill, S. (2017). Changes of the intestinal microbiome-host homeostasis in HIV-infected individuals —- a focus on the bacterial gut microbiome. Eur. J. Microbiol. Immunol. 7, 158–167. doi: 10.1556/1886.2017.00016
Robertson, K. R., Jiang, H., Kumwenda, J., Supparatpinyo, K., Marra, C. M., Berzins, B., et al. (2018). HIV associated neurocognitive impairment in diverse resource limited settings. Clin. Infect. Dis. doi: 10.1093/cid/ciy767 [Epub ahead of print].
Sakamoto, M., Umeda, M., Ishikawa, I., and Benno, Y. (2000). Comparison of the oral bacterial flora in saliva from a healthy subject and two periodontitis patients by sequence analysis of 16S rDNA libraries. Microbiol. Immunol. 44, 643–652. doi: 10.1111/j.1348-0421.2000.tb02545.x
Sandler, N. G., and Douek, D. C. (2012). Microbial translocation in HIV infection: causes, consequences and treatment opportunities. Nat. Rev. Microbiol. 10, 655–666. doi: 10.1038/nrmicro2848
Saylor, D., Dickens, A. M., Sacktor, N., Haughey, N., Slusher, B., Pletnikov, M., et al. (2016). HIV-associated neurocognitive disorder–pathogenesis and prospects for treatment. Nat. Rev. Neurol. 12, 234–248. doi: 10.1038/nrneurol.2016.27
Serrano-Villar, S., Vazquez-Castellanos, J. F., Vallejo, A., Latorre, A., Sainz, T., Ferrando-Martinez, S., et al. (2017). The effects of prebiotics on microbial dysbiosis, butyrate production and immunity in HIV-infected subjects. Mucosal Immunol. 10, 1279–1293. doi: 10.1038/mi.2016.122
Smith, D. G., Guillemin, G. J., Pemberton, L., Kerr, S., Nath, A., Smythe, G. A., et al. (2001). Quinolinic acid is produced by macrophages stimulated by platelet activating factor. Nef and Tat. J. Neurovirol. 7, 56–60. doi: 10.1080/135502801300069692
Sun, Y., Ma, Y., Lin, P., Tang, Y. W., Yang, L., Shen, Y., et al. (2016). Fecal bacterial microbiome diversity in chronic HIV-infected patients in China. Emerg. Microbes Infect. 5:e31. doi: 10.1038/emi.2016.25
Tang, R., Wei, Y., Li, Y., Chen, W., Chen, H., Wang, Q., et al. (2018). Gut microbial profile is altered in primary biliary cholangitis and partially restored after UDCA therapy. Gut 67, 534–541. doi: 10.1136/gutjnl-2016-313332
Tincati, C., Douek, D. C., and Marchetti, G. (2016). Gut barrier structure, mucosal immunity and intestinal microbiota in the pathogenesis and treatment of HIV infection. AIDS Res. Ther. 13:19. doi: 10.1186/s12981-016-0103-1
Tozzi, V., Balestra, P., Bellagamba, R., Corpolongo, A., Salvatori, M. F., Visco-Comandini, U., et al. (2007). Persistence of neuropsychologic deficits despite long-term highly active antiretroviral therapy in patients with HIV-related neurocognitive impairment: prevalence and risk factors. J. Acquir. Immune Defic. Syndr. 45, 174–182. doi: 10.1097/QAI.0b013e318042e1ee
Valle, M., Price, R. W., Nilsson, A., Heyes, M., and Verotta, D. (2004). CSF quinolinic acid levels are determined by local HIV infection: cross-sectional analysis and modeling of dynamics following antiretroviral therapy. Brain 127, 1047–1060. doi: 10.1093/brain/awh130
Vamos, E., Pardutz, A., Klivenyi, P., Toldi, J., and Vecsei, L. (2009). The role of kynurenines in disorders of the central nervous system: possibilities for neuroprotection. J. Neurol. Sci. 283, 21–27. doi: 10.1016/j.jns.2009.02.326
Vazquez-Castellanos, J. F., Serrano-Villar, S., Jimenez-Hernandez, N., Soto, D. R. M., Gayo, S., Rojo, D., et al. (2018). Interplay between gut microbiota metabolism and inflammation in HIV infection. ISME J. 12, 1964–1976. doi: 10.1038/s41396-018-0151-8
Vazquez-Castellanos, J. F., Serrano-Villar, S., Latorre, A., Artacho, A., Ferrus, M. L., Madrid, N., et al. (2015). Altered metabolism of gut microbiota contributes to chronic immune activation in HIV-infected individuals. Mucosal Immunol. 8, 760–772. doi: 10.1038/mi.2014.107
Vecsei, L., Szalardy, L., Fulop, F., and Toldi, J. (2013). Kynurenines in the CNS: recent advances and new questions. Nat. Rev. Drug Discov. 12, 64–82. doi: 10.1038/nrd3793
Vujkovic-Cvijin, I., Dunham, R. M., Iwai, S., Maher, M. C., Albright, R. G., Broadhurst, M. J., et al. (2013). Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci. Transl. Med. 5:193ra91. doi: 10.1126/scitranslmed.3006438
Vyboh, K., Jenabian, M., Mehraj, V., and Routy, J. (2015). HIV and the gut microbiota, partners in crime: breaking the vicious cycle to unearth new therapeutic targets. J. Immunol. Res. 2015, 1–9. doi: 10.1155/2015/614127
Wang, Z., Zheng, Y., Liu, L., Shen, Y., Zhang, R., Wang, J., et al. (2013). High prevalence of HIV-associated neurocognitive disorder in HIV-infected patients with a baseline CD4 count ≤ 350 cells/muL in Shanghai, China. Biosci. Trends 7, 284–289.
Williams, W. B., Han, Q., and Haynes, B. F. (2018). Cross-reactivity of HIV vaccine responses and the microbiome. Curr. Opin. HIV AIDS 13, 9–14. doi: 10.1097/COH.0000000000000423
Wu, S. C., Cao, Z. S., Chang, K. M., and Juang, J. L. (2017). Intestinal microbial dysbiosis aggravates the progression of Alzheimer’s disease in Drosophila. Nat. Commun. 8:24. doi: 10.1038/s41467-017-00040-6
Yang, I., Corwin, E. J., Brennan, P. A., Jordan, S., Murphy, J. R., and Dunlop, A. (2016). The infant microbiome: implications for infant health and neurocognitive development. Nurs. Res. 65, 76–88. doi: 10.1097/NNR.0000000000000133
Yilmaz, A., Yiannoutsos, C. T., Fuchs, D., Price, R. W., Crozier, K., Hagberg, L., et al. (2013). Cerebrospinal fluid neopterin decay characteristics after initiation of antiretroviral therapy. J. Neuroinflammation 10:62. doi: 10.1186/1742-2094-10-62
Zelante, T., Iannitti, R. G., Cunha, C., De Luca, A., Giovannini, G., Pieraccini, G., et al. (2013). Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39, 372–385. doi: 10.1016/j.immuni.2013.08.003
Zhan, X., Stamova, B., Jin, L. W., DeCarli, C., Phinney, B., and Sharp, F. R. (2016). Gram-negative bacterial molecules associate with Alzheimer disease pathology. Neurology 87, 2324–2332. doi: 10.1212/WNL.0000000000003391
Keywords: HIV, cognitive, HIV-associated neurocognitive disorder, gut microbiota, predictive function
Citation: Zhang F, Yang J, Ji Y, Sun M, Shen J, Sun J, Wang J, Liu L, Shen Y, Zhang R, Chen J and Lu H (2019) Gut Microbiota Dysbiosis Is Not Independently Associated With Neurocognitive Impairment in People Living With HIV. Front. Microbiol. 9:3352. doi: 10.3389/fmicb.2018.03352
Received: 10 October 2018; Accepted: 31 December 2018;
Published: 28 January 2019.
Edited by:
Dimitris G. Hatzinikolaou, National and Kapodistrian University of Athens, GreeceReviewed by:
Maria Jose Gosalbes, Centre for Biomedical Network Research (CIBER), SpainJohn Zaunders, St Vincent’s Hospital Sydney, Australia
Copyright © 2019 Zhang, Yang, Ji, Sun, Shen, Sun, Wang, Liu, Shen, Zhang, Chen and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Chen, qtchenjun@163.com Hongzhou Lu, luhongzhou@fudan.edu.cn
†Co-first authors
 Fengdi Zhang
Fengdi Zhang Junyang Yang1†
Junyang Yang1† Jun Chen
Jun Chen Hongzhou Lu
Hongzhou Lu