- School of Microbiology, Centre for Synthetic Biology and Biotechnology, Environmental Research Institute, APC Microbiome Institute, University College Cork, Cork, Ireland
In yeasts, proteins of the Major Superfamily Transporter selectively bind and allow the uptake of sugars to permit growth on varied substrates. The genome of brewer’s yeast, Saccharomyces cerevisiae, encodes multiple hexose transporters (Hxt) to transport glucose and other MFS proteins for maltose, galactose, and other monomers. For sugar uptake, the dairy yeast, Kluyveromyces lactis, uses Rag1p for glucose, Hgt1 for glucose and galactose, and Lac12 for lactose. In the related industrial species Kluyveromyces marxianus, there are four genes encoding Lac12-like proteins but only one of them, Lac12, can transport lactose. In this study, which initiated with efforts to investigate possible functions encoded by the additional LAC12 genes in K. marxianus, a genome-wide survey of putative MFS sugar transporters was performed. Unexpectedly, it was found that the KHT and the HGT genes are present as tandem arrays of five to six copies, with the precise number varying between isolates. Heterologous expression of individual genes in S. cerevisiae and mutagenesis of single and multiple genes in K. marxianus was performed to establish possible substrates for these transporters. The focus was on the sugar galactose since it was already reported in K. lactis that this hexose was a substrate for both Lac12 and Hgt1. It emerged that three of the four copies of Lac12, four Hgt-like proteins and one Kht-like protein have some capacity to transport galactose when expressed in S. cerevisiae and inactivation of all eight genes was required to completely abolish galactose uptake in K. marxianus. Analysis of the amino acid sequence of all known yeast galactose transporters failed to identify common residues that explain the selectivity for galactose. Instead, the capacity to transport galactose has arisen three different times in K. marxianus via polymorphisms in proteins that are probably ancestral glucose transporters. Although, this is analogous to S. cerevisiae, in which Gal2 is related to glucose transporters, there are not conserved amino acid changes, either with Gal2, or among the K. marxianus galactose transporters. The data highlight how gene duplication and functional diversification has provided K. marxianus with versatile capacity to utilise sugars for growth.
Introduction
Kluyveromyces marxianus is a yeast traditionally found in fermented dairy products such as cheese and kefir (Gethins et al., 2016; Coloretti et al., 2017; Tittarelli et al., 2018). Because of its connection with these products, K. marxianus has been granted QPS (Qualified Presumption of Safety) and GRAS (Generally Regarded as Safe) status in the EU and US, respectively, designating this yeast as safe to use in food applications (Fonseca et al., 2008). K. marxianus is also found during the production of other non-dairy products such as chocolate and tequila/mezcal (Lappe-Oliveras et al., 2008; Ho et al., 2014). In order to establish and often colonise these diverse environments, the yeast possesses the capacity to access a range of sugar polymers. This involves the production of specific enzymes that degrade complex substrates and transporters that mediate the uptake of monomers into the cell. For example, Cocoa bean fermentation, an essential step in chocolate production, is generally carried out by a consortium of indigenous species that includes K. marxianus (De Vuyst and Weckx, 2016). In this environment the pectinase enzyme produced by K. marxianus degrades the pectin present in the cocoa pulp allowing the establishment of other yeast species required in the fermentation process (Schwan and Wheals, 2004). K. marxianus is also present as part of the natural flora of the Agave plant, used to produce the alcoholic beverages tequila and mezcal (Escalante-Minakata et al., 2008). Degradation of complex fructans to simple sugars by the enzyme inulinase is essential in the establishment of K. marxianus in this niche. Besides utilising various substrates and producing relevant enzymes, K. marxianus also has other interesting physiological traits, uncommon to most yeasts (Lane and Morrissey, 2010). For example, it is able to grow at high temperatures (up to 50°C), exhibits the fastest growth rate among eukaryotes and naturally produces significant amounts of flavour molecules such as esters and fusel alcohols (Morrissey et al., 2015). This species is also used for some commercial applications, for example, the production of bioethanol from cheese whey and production of the flavour molecule 2-phenylethanol (Fonseca et al., 2008; Morrissey et al., 2015; Varela et al., 2017a).
The prospect of further developing K. marxianus for cell-factory applications has motivated researchers to sequence the genome of several strains (Jeong et al., 2012; Suzuki et al., 2014; Inokuma et al., 2015; Lertwattanasakul et al., 2015; Ortiz-Merino et al., 2018). A comparison of these genomes showed that K. marxianus displays a high level of genome variation at a structural and sequence level. Divergence in DNA sequence was found to be relativity high with up to 3% divergence between some isolates. Also, different strains were found to exist in haploid, diploid, and triploid states (Ortiz-Merino et al., 2018). This high degree of variation between genomes is not surprising considering previous observations of phenotypic diversity across isolates (Lane et al., 2011). Lactose utilisation, for example, is a variable trait with some strains displaying good and poor growth on lactose (Carvalho-Silva and Spencer-Martins, 1990; Lane et al., 2011). This phenomenon is due to polymorphisms in the LAC12 gene, which encodes a lactose permease that mediates lactose uptake into the cell. Strains that display good and poor lactose consumption carry functional (LAC12-B) and non-functional (LAC12-A) alleles of the lactose transporter, respectively (Varela et al., 2017b). The sequence differences between the two alleles translate into 11 amino acid changes in the protein sequence that are sufficient to alter the function of the transporter (Ortiz-Merino et al., 2018). Unbiased analyses of genome sequences showed that isolates from dairy environments are diploid or triploid and carry the LAC12-B variant while non-dairy strains are haploid and contain the LAC12-A haplotype that encodes the non-functional lactose transporter (Ortiz-Merino et al., 2018). Although this analysis was performed in a relatively small number of genomes it raises interesting questions regarding the evolution of lactose utilisation in K. marxianus.
Information on the transport of other sugars, such as glucose and galactose, is limited in K. marxianus. Early experiments established the existence of three carbon-repressible proton-sugar transporters that mediate glucose/galactose, fructose, and lactose uptake (De Bruijne et al., 1988). Induction of the different systems was shown to depend on the carbon source used to grow the cells: in yeast cells grown on lactose, the activity of a high-affinity proton-glucose transporter was detected whereas a low-affinity glucose transporter was present in cells grown in glucose. Galactose was only taken up by cells grown in lactose and not by glucose-grown cells, which indicates that the galactose transport system requires induction by lactose or presumably galactose (Gasnier, 1987). The genes encoding these transporters were not identified in K. marxianus but both low and high-affinity glucose uptake systems were described the related yeast, Kluyveromyces lactis (Wésolowski-Louvel and Goffrini, 1992; Billard et al., 1996; Schaffrath and Breunig, 2000). In most strains, the low-affinity system is encoded by two tandemly-arrayed genes, KHT1 and KHT2 but some strains carry a single gene, RAG1, that arose from a recombination event between KHT1 and KHT2 (Weirich et al., 1997). The high affinity transport system is encoded by HGT1, which was found to exist as a single copy (Billard et al., 1996). Expression of this gene was found to be constitutive whereas expression of KHT1 and KHT2 genes was glucose inducible (Milkowski et al., 2001). Interestingly, a strain carrying null mutations for both of the uptake systems is still able to grow on glucose, which points out to the existence of another glucose transporter not yet identified (Milkowski et al., 2001). Galactose uptake was also investigated in K. lactis where the LAC12 gene was found to play a role as a low-affinity galactose transporter (Riley et al., 1987). In that study, strains mutated in LAC12 were greatly impaired in galactose uptake, which led to the idea that galactose was exclusively transported via Lac12p. Conversely, further studies on the topic showed that the high-affinity glucose carrier, HGT1, also acts as a high-affinity galactose transporter in K. lactis (Baruffini et al., 2006). Disruption of both LAC12 and HGT1 abolished galactose growth completely, ruling out the existence of other galactose transporters. It was later concluded that LAC12 and HGT1 encode a low-affinity and high-affinity galactose system, respectively (Baruffini et al., 2006).
Previously, we showed that there are four copies of the LAC12 gene K. marxianus but only one of these encodes a functional lactose transporter (Varela et al., 2017b). All the copies of LAC12 are found in sub-telomeric regions and it was not determined whether the additional copies could function in the transport of sugars other than lactose, The recent implementation of the CRISPR-Cas9 system in K. marxianus creates new opportunities to study the molecular basis of sugar uptake in this yeast (Löbs et al., 2017; Nambu-Nishida et al., 2017; Juergens et al., 2018). In this study, we used comparative genomics to identify putative sugar transporters in multiple K. marxianus strains. This revealed that there was also expansion in the number of genes that were homologous to the K. lactis KHT1/2 and HGT1 genes. Functional analysis to establish the capacity of these duplicated genes to transport galactose revealed a high degree of functional overlap. This work highlights the existence of a species-specific gene expansion and raises fascinating questions about the evolution of galactose and lactose utilisation in K. marxianus.
Materials and Methods
Strains and Growth Conditions
The yeast strains used in this study are listed in Table 1. K. marxianus CBS6556 and NBRC1777 were obtained from the Westerdijk Fungal Biodiversity Institute and the NITE Biological Research Centre culture collections, respectively. Saccharomyces cerevisiae EBY.VW4000 was kindly provided by Dr. Eckhard Boles, Goethe University Frankfurt, Germany. K. marxianus strains were typically grown in YPD medium (10 g L-1 yeast extract, 20 g L-1 peptone, 20 g L-1 glucose). When used in transformation experiments, NBRC1777 was plated onto YPD plates containing 200 μg mL-1 of Hygromycin B (Sigma-Aldrich, St. Louis, MI, United States). K. marxianus mutants were grown on mineral media (MM) supplemented with 20 and 0.1 g L-1 glucose, or galactose (Fonseca et al., 2007). S. cerevisiae EBY.VW4000, used for heterologous expression of sugar transporters (Wieczorke et al., 1999), was maintained in synthetic complete (SC) medium (1.7 g L-1 yeast nitrogen base, 5 g L-1 ammonium plus synthetic complete drop-out lacking uracil and or L-leucine) (Formedium, Norfolk, United Kingdom) supplemented with 20 g L-1 maltose. For functional analysis the EBY.VW4000 strains expressing the different transporters were grown on SC – maltose, washed, serially diluted and spotted onto SC plates containing glucose or galactose to a final concentration of 20 or 1 g L-1, as described in the text. Yeast strains were grown at 30°C with 200 rpm agitation unless otherwise mentioned. E. coli DH5α was used for cloning purposes. The strain was maintained in LB medium (5 g L-1 yeast extract, 10 g L-1 bactopeptone, 10 g L-1 NaCl) supplemented with 100 μg mL-1 ampicillin when required.
Identification of Potential Sugar Transporters
A genome-wide identification of sugar transporters was performed in the K. marxianus CBS6556 and K. lactis CBS2359 genomes (Accession Nos. PRJNA89605 and PRJNA12377, respectively). The predicted proteomes from the two species were independently compared against the TransportDB 2.0 database to identify possible candidates (Elbourne et al., 2017). After filtering out hits with low identity (<35%) and coverage (<80%), the resulting sequences were submitted to the TMHMM server1 for the prediction of transmembrane domains. All sequences containing two or more transmembrane domains were considered as potential transporters. In order to classify these transporters, their sequences were aligned against the PFAM database (Finn et al., 2014) using HMMER v 3.0 (Finn et al., 2011). Sequences belonging to the MFS superfamily were extracted, aligned using MUSCLE 3.8 (Edgar, 2004) and then used to construct a maximum-likelihood tree using PhyML 3.1 (Guindon et al., 2010). Tree visualisation was carried out using the ETE 3 toolkit (Huerta-Cepas et al., 2016).
To compare the structure of the KHT and HGT loci across species, the publicly available Kluyveromyces genomes were aligned and visualised using progressiveMauve (Darling et al., 2010). K. marxianus genomes from different strains (CBS6556, NBRC1777, UFV-3, CBS397, CBS4857, and DMKU3) were also analysed using this software.
Heterologous Expression of Sugar Transporters
Plasmids and primers used in this study are listed in Tables 2, 3, respectively. The plasmid p426 was used to clone and express the KHT and HGT genes in S. cerevisiae. The p426 backbone, ScTEF1 promoter, ScCYC1 terminator and the KMXK_E03650 gene from K. marxianus CBS6556 were amplified by PCR with primers containing overlapping regions. PCR products were then purified and assembled using a Gibson assembly kit (New England Biolabs, Inc., Ipswich, MA, United States). The resulting plasmid, which expresses the KMXK_E03650 gene under the control of the ScTEF1 promoter, was used as a backbone for cloning the rest of the transporters. The primers p426-TEF-F and p426-CYC-R, were used to PCR amplify the p426- KMXK_E03650 plasmid from the ScTEF1 promoter to the ScCYC1 terminator (excluding the KMXK_E03650 gene). Then, purified PCR products from the rest of the CBS6556 transporters were individually combined with the purified p426 backbone and cloned by Gibson assembly. The KHT genes KMAR_50343 and KMAR_50344, absent in CBS6556, were PCR amplified from NBRC1777 and cloned following the same procedure. All the PCR reactions were performed using DNA Phusion Polymerase (New England Biolabs, Inc., Ipswich, MA, United States). Plasmids were finally introduced into S. cerevisiae EBY.VW4000 by transformation using the LiAc/SS carrier DNA/PEG method (Gietz and Schiestl, 2007). Expression of the LAC12 genes was also tested using plasmids previously described (Varela et al., 2017b). After transformations, the cells carrying the p426-KHT/HGT and LAC12 plasmids were plated onto SC media lacking uracil and leucine, respectively.
Construction of K. marxianus Mutants
The CRISPR-Cas9 system was used to disrupt the genes encoding galactose transporters in K. marxianus NBRC1777, including the LAC12, HGT genes and the KHT gene, KMXK_E03670. The pUDP002 plasmid, containing the Cas9 gene and the elements required to express the guide RNA (gRNA) molecule in K. marxianus (Juergens et al., 2018), was modified to simplify cloning of target sequences. Long overlapping oligonucleotides encoding the hammerhead ribozyme, a BsaI cloning site, the gRNA structural part and the HDV ribozyme were chemically synthesised (Integrated DNA Technologies, Coralville, IA, United States) and annealed in a thermocycler machine. In parallel, the pUDP002 plasmid was amplified by PCR using the pUDP002-F and pUDP002-R primers. The 205 bp DNA-duplex and the plasmid backbone were assembled via Gibson assembly to yield the pUCC001 plasmid (Supplementary Figure S1). Design of the gRNA sequences targeting the genes of interest was carried out using the software sgRNAcas9 (Xie et al., 2014). Complementary oligonucleotides containing the gRNA sequences and specific overhangs (5′-CGTC-3′ and 5′-AAAC-3′) were annealed, phosphorylated and cloned into pUCC001 by Golden Gate assembly, as described previously (Vyas et al., 2015; Ng and Dean, 2017). Presence of the target sequences in the pUCC001 plasmid was confirmed by colony PCR using the forward target primer and BSA-R. Plasmids were introduced into NBRC1777 by transformation using the LiAc/SS carrier DNA/PEG method and transformants checked by PCR and DNA sequencing. In order to eliminate the pUCC001 plasmid from the mutants, the strains were grown on YPD broth overnight without selection and diluted 1/10 into fresh YPD next morning. The procedure was repeated twice and then the strains were grown on YPD medium supplemented with Hygromycin B to confirm plasmid loss. To construct strains carrying mutations at additional loci, the procedure was repeated sequentially with one gene being mutated at a time. Primers used for cloning the gRNA molecules and checking transformants are listed in Table 3. The genotype of all the K. marxianus mutants constructed in this study is shown in Table 1.
Results
Extra LAC12 Copies Encode Galactose Transporters
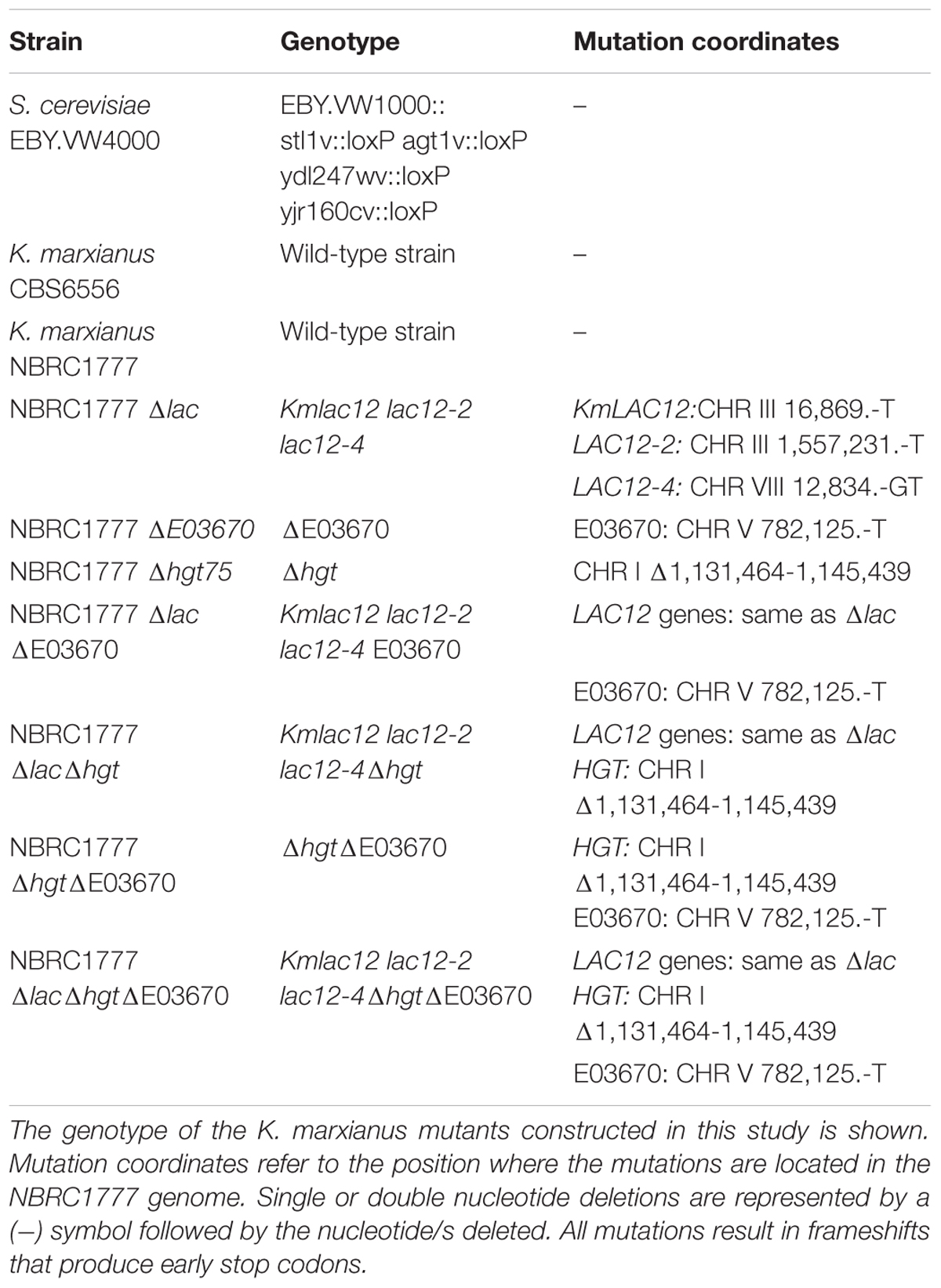
It was recently shown that K. marxianus contains four copies of the lactose permease gene with only KmLAC12 encoding a functional lactose transporter (Varela et al., 2017b). Lactose transport by Lac12p appears to be a trait that evolved in a particular dairy lineage of K. marxianus (Ortiz-Merino et al., 2018) and it was hypothesised that the proteins encoded by the LAC12 genes might have other substrates. Because the K. lactis Lac12p protein is known to transport lactose and galactose (Riley et al., 1987), it was decided to test whether the additional K. marxianus Lac12 proteins were also galactose transporters. To test this, the different LAC12 genes were expressed in S. cerevisiae EBY.VW4000 (Figure 1A), a yeast strain that has been subjected to systematic disruption of all known hexose transporters (Wieczorke et al., 1999). As previous work demonstrated that Lac12p from the Lac+ and Lac- lineages of K. marxianus were functionally different, both KmLAC12397 (lac+) and KmLAC126556 (lac-) were tested. The S. cerevisiae strains expressing KmLAC12, LAC12-2, and LAC12-4 grew in 2% galactose medium indicating that these proteins could transport galactose into the cell, though it was apparent that the CBS397 version of LAC12 promoted better growth that the equivalent gene from CBS6556. To check whether the Lac12 proteins also supported growth where high-affinity galactose transport was required, the different strains were also grown on 0.1% galactose. In this case, only the strain expressing LAC12-4 exhibited strong growth. The LAC12-3 strain did not confer growth under any condition. Since these results suggested that KmLAC12, LAC12-2, and LAC12-4 encode galactose transporters, we then tested whether inactivation of all three genes in K. marxianus would generate a strain unable to grow using galactose as a sole carbon source. This entailed the sequential construction of single, double and triple mutants using a modified version of the previously published CRISPR-Cas9 system (Juergens et al., 2018). K. marxianus strain NBRC 1777 was used as this is a haploid strain in which CRISPR-Cas9 works efficiently. The three LAC12s were targeted using this system and indel mutations, inactivating the genes by producing premature stop codons, were obtained in all cases. The triple lac12 lac12-2 lac12-4 mutant (designated K. marxianus NBRC1777 Δlac), and the wild-type strain were grown in 1 and 0.1% galactose media but no major differences in growth between wild-type and mutants were observed on plates (data not shown) or in liquid (Figure 1B). This indicates that there is an additional galactose transporter(s) in K. marxianus, an idea compatible with previous studies in K. lactis where HGT1 was reported to encode a high-affinity galactose transporter (Baruffini et al., 2006).
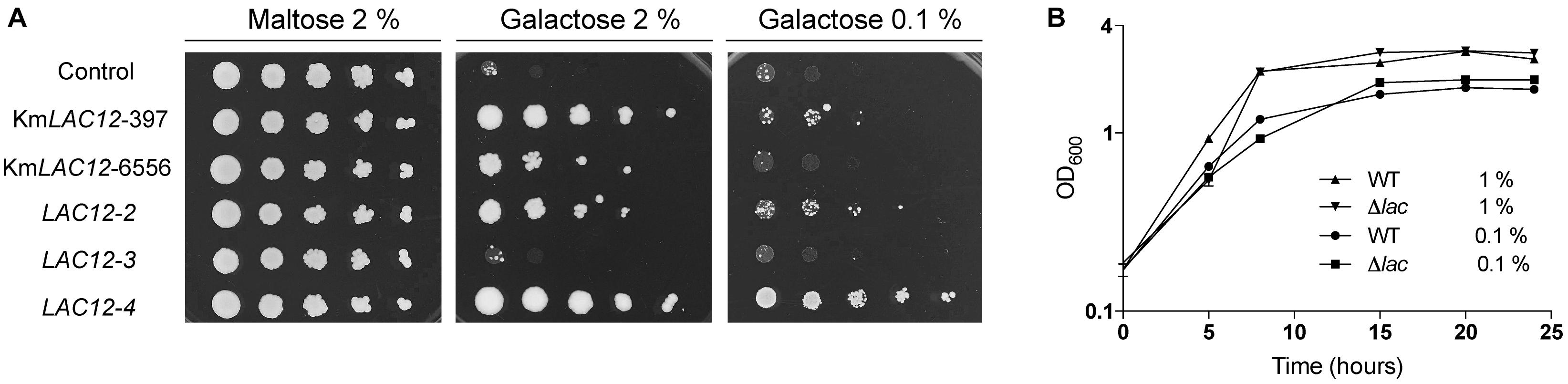
Figure 1. Role of the LAC12 genes in galactose transport. (A) Heterologous expression of the Kluyveromyces marxianus LAC12 genes in Saccharomyces cerevisiae EBY.VW4000. Yeast strains were grown on SC maltose to an OD600 of 2 then washed, diluted serially and spotted onto SC plates supplemented with maltose, galactose or glucose, as indicated. Plates were incubated for 5 days at 30°C. The LAC12-2 and LAC12-4 genes were obtained from K. marxianus CBS397, as described by Varela et al. (2017b). (B) Growth phenotype of K. marxianusΔlac mutant carrying null mutations for the KmLAC12, LAC12-2, and LAC12-4 genes. NBRC1777 and the Δlac strain were grown overnight on MM supplemented with 1 or 0.1% galactose and then transferred to fresh medium containing the same amount of sugar to an OD600 of 0.1. Growth was recorded every 6 h using a spectrophotometer. The experiment was conducted in triplicates.
The KHT and HGT Gene Family Is Expanded in K. marxianus
A genome-wide annotation of membrane transporters was performed to identify transporters of the major facilitator superfamily (MFS) that could potentially encode galactose transporters in K. marxianus CBS6556. The predicted proteomes of K. marxianus and K. lactis were compared against the TransportDB database, filtered by number of transmembrane domains, and categorised into protein families (Supplementary Table S1). Through this process, a total of 49 and 41 MFS transporters were identified in the K. marxianus and K. lactis proteomes, respectively. Interestingly, this analysis revealed that, compared to K. lactis, there has been an expansion of transporters of this superfamily in K. marxianus with five copies of the putative low-affinity glucose transporter Kht and six copies of the putative high-affinity glucose transporter Hgt1 identified. To gain a better understanding of the relationship between these sequences, the proteins were aligned, and a maximum-likelihood phylogenetic tree was constructed (Figure 2). The Hxt hexose transporter-like proteins (Hxt proteins) in S. cerevisiae, were also included in this analysis. Three of the Kht proteins (KMXK_E03650, KMXK_E03660, and KMXK_E03670) are related to the Kht2 transporter from K. lactis. In contrast, KMXK_E03680 and KMXK_E03690 are more similar to the K. lactis Kht1 protein and are related in sequence to the S. cerevisiae hexose transporter Hxt5. All the Hgt1-like transporters resemble to the K. lactis Hgt1p transporter with KMXK_A02960 being the closest in sequence to the K. lactis protein.
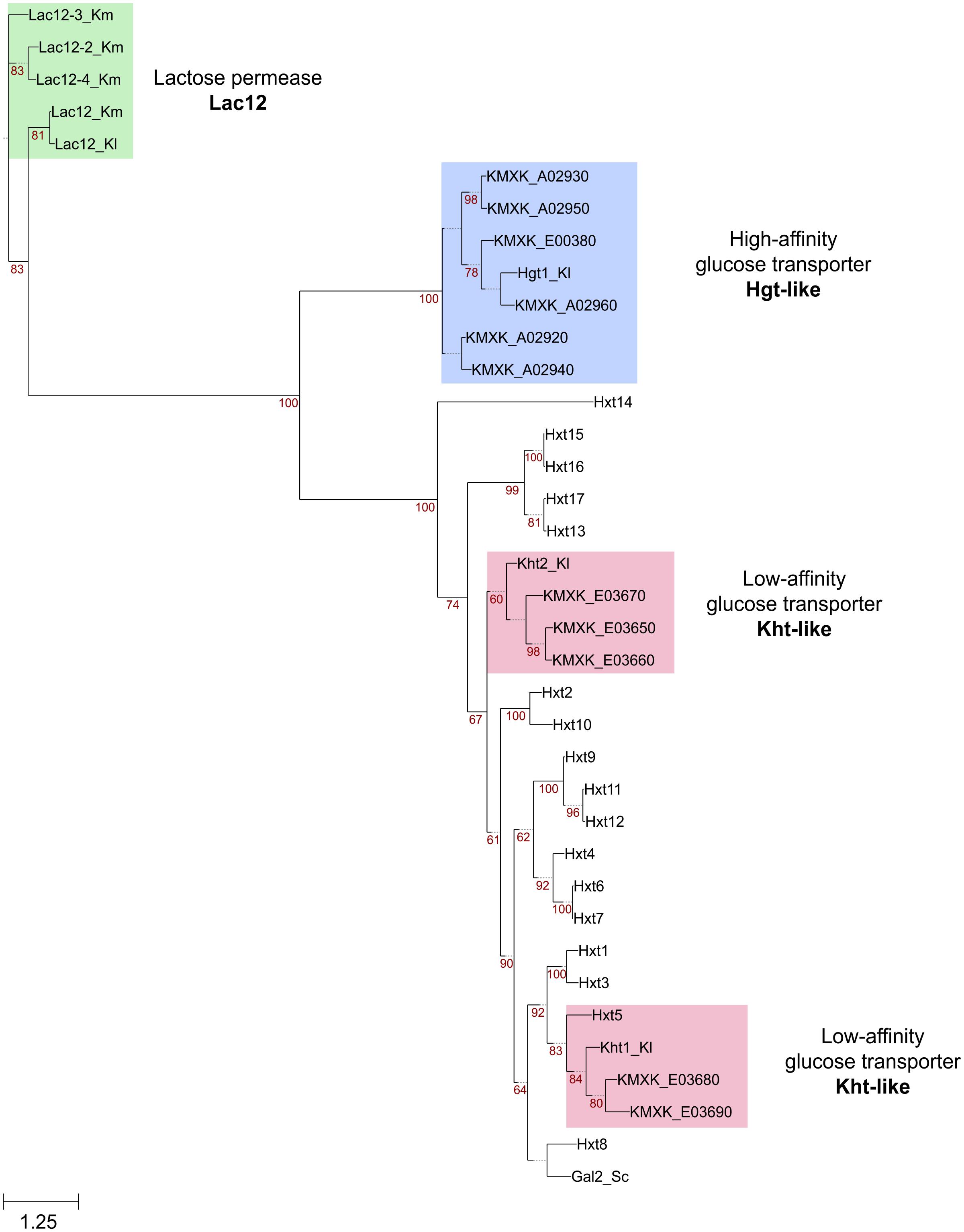
Figure 2. Phylogenetic tree of K. marxianus sugar transporters. The K. marxianus Kht, Hgt, and Lac12 protein sequences were aligned using MUSCLE and then used to construct a maximum-likelihood tree using PhyML with a bootstrapping value set to 500. The Kluyveromyces lactis Kht and Hgt sequences, and the Hxt sequences from S. cerevisiae were also used in this analysis. Branches with bootstrapping values over 60 are shown. The Kht and Hgt sequences are highlighted in red and blue boxes, respectively. Lac12 sequences are shown in green.
An inspection of the K. marxianus CBS6556 genome revealed that most of the additional KHT and HGT genes are present as tandem repeats, the sole exception being the HGT1 homolog KMXK_E00380. To determine whether this expansion was unique to strain CBS6556, the KHT and HGT loci were compared across a set of publicly available K. marxianus genomes and other Kluyveromyces genomes (K. lactis, K. aestuarii, K. wickerhamii, and K. dobzhanskii) (Figure 3). In all species except K. marxianus, two KHT genes and a unique HGT gene were present. The expansion observed in strain CBS6556 was seen in all five additional K. marxianus strains assessed but the copy number was found to vary. While K. marxianus strains CBS6556, DMKU3, and CBS397 carry five copies of the KHT genes, strains NBRC1777, UFV3 and CBS4857 encode six copies of the transporter (Figure 3A). For consistency, the K. marxianus CBS6556 gene nomenclature is used throughout this paper, with Supplementary Table S2 listing the names for the orthologous genes in each of the other well-annotated K. marxianus genomes. An alignment of the NBRC1777 and CBS6556 sequences showed that the 5′ and 3′ sections of the KMXK_E03660 sequence are identical to KMAR_50344 and KMAR_50345, respectively, suggesting that KMXK_E03660 arose by a recombination event between two KHT genes (Supplementary Figure S2). A similar scenario was observed at the HGT1 locus where some strains (NBRC1777, CBS6556, CBS397, and CBS4857) carry five and others (DMKU3 and UFV3) three copies of the gene (Figure 3B). A more detailed phylogenetic tree that includes all Hgt1 sequences shows a topology that is consistent with the presence of a single HGT1 gene in ancestral Kluyveromyces species. This gene duplicated twice in K. marxianus, once to give rise to KMXK 0E00380 and once to give rise to the ancestor of KMXK 0A02920, KMXK 0A02930 KMXK 0A02940, and KMXK 0A02950 (data not shown). A comparison of the NBRC1777 and DMKU3 sequences established that a reduction in copy number occurred as a result of recombination between the second and fourth HGT genes, leading to a loss of the third gene (Supplementary Figure S3).
Figure 3. Organisation of the KHT and HGT genes in K. marxianus genomes. (A) Genomic context of the KHT genes in K. marxianus strains NBRC1777, UFV3 and CBS4857 (top), CBS6556, DMKU3 and CBS397 (middle) and rest of Kluyveromyces genomes, represented by K. lactis (bottom). (B) Position of the HGT genes in the CBS6556, NBRC1777, CBS397 and CBS4857 genomes (top), DMKU3 and UFV3 (middle), and K. lactis (bottom). Sequence conservation across the different regions is represented by purple areas connecting the genes in different genomes. Sugar transporter genes are shown in green. Genes flaking these clusters are shown in blue.
Functional Analysis of the KHT and HGT Genes
The KHT1/2 and HGT1 orthologs in K. lactis were previously shown to encode glucose transporters, with Hgt1 also able to transport galactose into the cell. To functionally characterise the K. marxianus gene families, the K. marxianus CBS6556 genes were cloned and expressed in S. cerevisiae EBY.VW4000. The S. cerevisiae strains carrying the individual K. marxianus KHT and HGT genes were tested for growth on plates with the sole carbon source either glucose or galactose. Two sugar concentrations, 2 and 0.1% were used, as the lower concentration can establish whether the transporter has high affinity for the substrate (Figure 4). All the S. cerevisiae strains expressing KHT genes grew on both concentrations of glucose, indicating that all five of these proteins are high-affinity glucose transporters (Figure 4A). The only growth that was observed on galactose plates was with the strain expressing KMXK_E03670 on 2% galactose, showing that KMXK_E03670 can also function as a low-affinity galactose transporter. The pattern observed in strains expressing the HGT genes was very different (Figure 4B). Only KMXK_A02960 encoded a glucose transporter, whereas three genes, KMXK_A02960, KMXK_A02940, and KMXK_A02920, encoded proteins that could function as low-affinity galactose transporters, and the strain expressing KMXK_A02950 showed weak growth on 0.1% galactose.
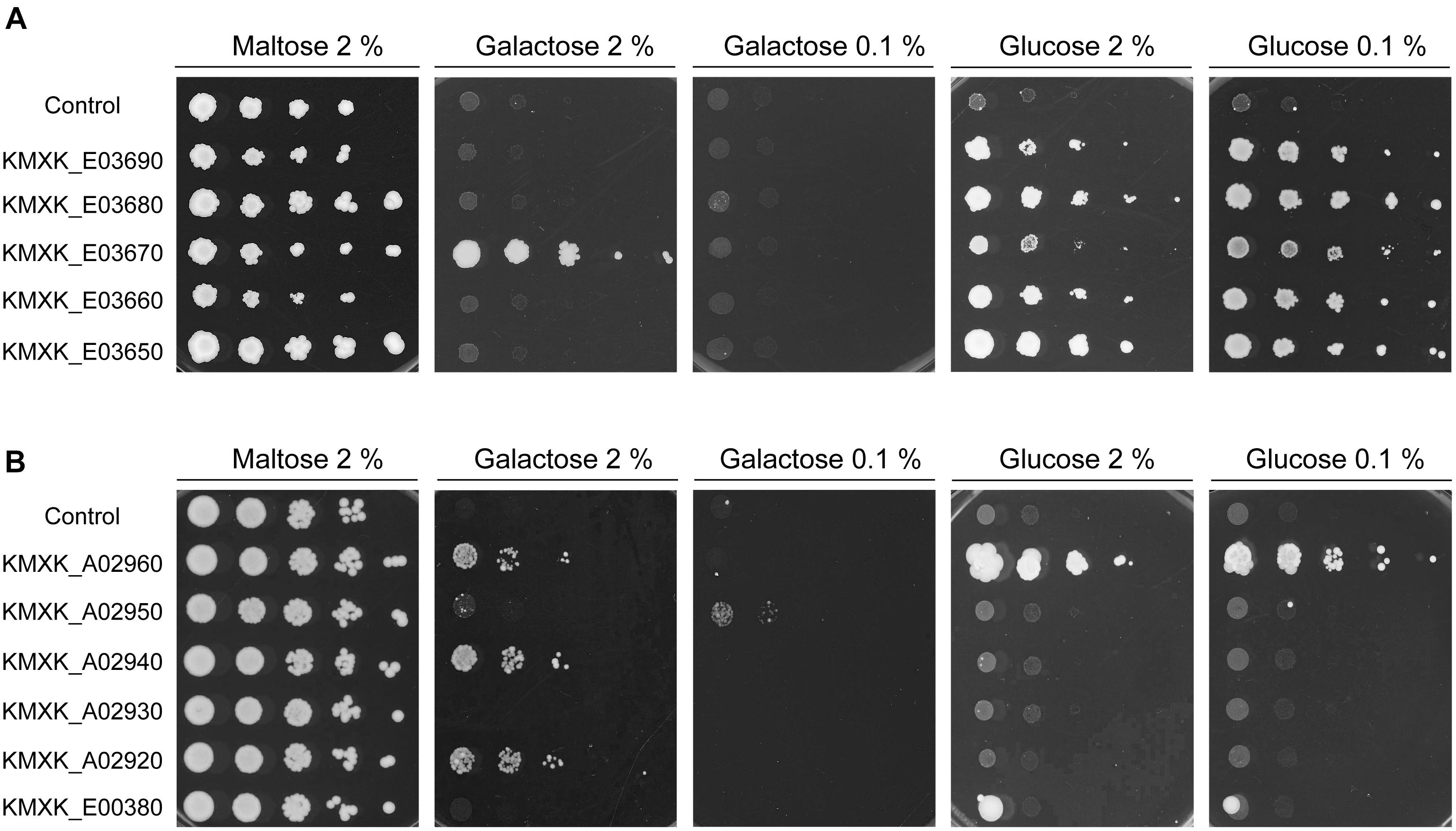
Figure 4. Functional analysis of the KHT and HGT genes in S. cerevisiae EBY.VW4000. S. cerevisiae strains expressing the KHT and HGT genes from CBS6556 were grown as previously described in Figure 1 and spotted onto SC plates containing maltose, galactose, or glucose. (A) KHT genes. (B) HGT genes.
To definitively confirm that the K. marxianus genome encodes multiple galactose transporters, CRISPR-Cas9 was deployed to inactivate the genes encoding all of these potential transporters. The entire HGT locus was targeted for inactivation by using a single CRISPR gRNA that had target sequences in the first and last genes of the cluster and thus should lead to generation of double stranded breaks at each end. Repair of these breaks led to deletion of all functional HGT1 copies at this locus and generated the strain that lacks any intact HGT1-like gene at this locus (Supplementary Figure S4). The strain retains KMXK_E00380, but this gene is not a galactose transporter (Figure 4). A CRISPR gRNA that successfully targeted the KHT-like gene, KMXK_E03670, was also tested and by sequential mutagenesis it was possible to make a set of K. marxianus mutants that lacked various combinations of the three functional LAC12 genes (Δlac), all five HGT-like genes (Δhgt), and KMXK_E03670 (DE03670). These mutants were assessed for their ability to grow on medium where the sole carbon source was 2 or 0.1% glucose or galactose (Figure 5). None of the mutants was impaired in growth on glucose, which was not unexpected since all mutants still contain several of the Kht-like proteins, which were shown to transport glucose (Figure 4). On galactose, strains individually carrying the Δlac, ΔE03670, or Δhgt mutations did not show growth differences when compared to the wild-type strain, but some combinations of these mutations showed clear growth impairment. On 2% galactose, only growth of the strain inactivated in all putative galactose transporter genes (ΔlacΔE03670 Δhgt) was strongly impaired, corroborating the proposal that there is functional redundancy for galactose transport. On 0.1% galactose, all the strains carrying combinations of mutants were growth impaired to some extent, but the strongest phenotypes were in the triple mutant ΔlacΔE03670 Δhgt and the double mutant ΔlacΔhgt, where growth was essentially abolished. A minor and intermediate growth impairment was seen in the ΔlacΔE03670 and ΔhgtΔE03670 strains, respectively.
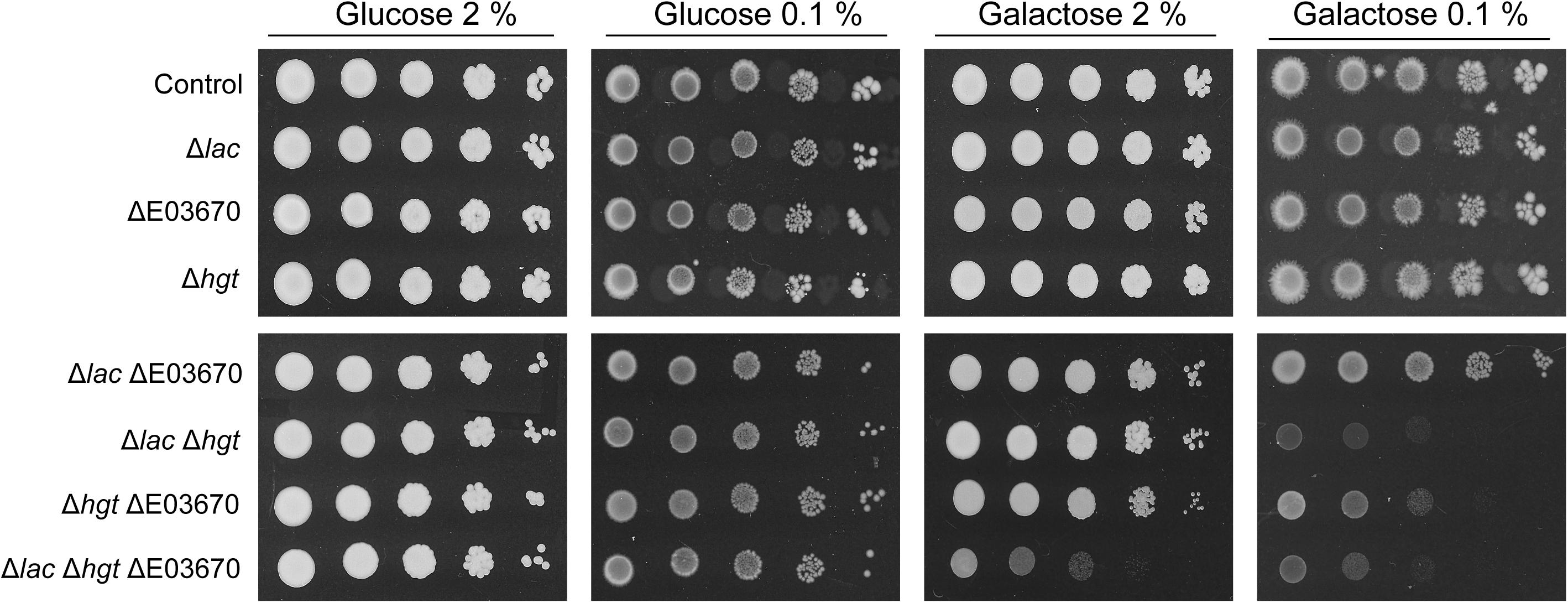
Figure 5. Phenotype of K. marxianus strains carrying mutations in galactose transport genes. The strains were grown on MM supplemented with 20 g/L to an OD600 of 2 then washed, diluted serially, and spotted onto MM plates supplemented with galactose or glucose, as indicated. The Δlac strain contains mutations in the KmLAC12, LAC12-2 and LAC12-4 genes, the ΔE03670 strain contains a single nucleotide deletion in the E03670 gene and Δhgt carries a deletion of the whole region containing the HGT genes. The precise genotype of these mutants is shown in Table 1. The wild-type strain NBRC1777 was used as a control in this experiment.
The Lac12, Hgt, and Kht protein sequences were compared to identify amino acid residues potentially involved in galactose transport. Previous studies of the Gal2 transporter from S. cerevisiae previously identified three aromatic amino acid residues that are critical for galactose recognition and transport, Tyr352, Tyr446, and Phe504 (Kasahara et al., 1997; Kasahara and Kasahara, 2000) but a full protein alignment containing all the sequences analysed in this study revealed that these positions are not exclusively present in galactose transporters (Supplementary Figure S5). This observation indicates that the relevance of these sites cannot be universally extrapolated to the K. marxianus sequences. The phylogeny of the proteins (Figure 2) illustrated that there are distinct subclades that include proteins both positive and negative for galactose transport so alignments of proteins within each subclade were performed to see whether distinguishing amino acids could be identified. Comparison of the Lac12 sequences led to the identification of 38 amino acids that differ between the sequences of Lac12, Lac12-2 and Lac12-4, positive for galactose transport and Lac12-3, negative for galactose transport (Supplementary Figure S6). A similar analysis to the Kht sequences revealed 24 amino acids that could be critical for galactose transport in these proteins (Supplementary Figure S7). The most striking finding was that there is a unique position in these proteins that differs in a consistent way between KMXK_A02960, KMXK_A02940, KMXK_A02920 and Hgt1_Kl, which allow growth on 2% galactose, and KMXK_A02930, KMXK_A02950 and KMXK_E00380, which do not (Figure 6). In galactose transporting proteins, the residue at position 435 is alanine, whereas it is threonine in the proteins that do not transport this sugar. To test whether this residue plays a critical role in sugar recognition and transport, the threonine residue in KMXK_A02930 was replaced by alanine via site-directed mutagenesis. However, heterologous expression of this variant did not confer growth on galactose, indicating that this modification alone is not critical for galactose transport (data not shown).

Figure 6. Multiple sequence alignment of the Hgt proteins. The Hgt sequences were aligned using MUSCLE 3.8. The Hgt1 sequence from K. lactis was also included in this comparison. The position where galactose and non-galactose transporters differ is marked in purple in the alignment (position 435 in KMXK_A02960). This residue is located in the twelfth transmembrane domain, according to TMHMM predictions. Sequences encoding galactose transporters are shown in blue.
Discussion
Expansion of Major Facilitator Superfamily Transporters in K. marxianus
In this study, it was shown that the phenomenon of duplicated proteins of the MFS superfamily in K. marxianus is not restricted to LAC12, but also includes the KHT and HGT gene families. In contrast to LAC12, where the duplications comprise individual genes distributed in different sub-telomeric regions, and with the exception of the HGT-like gene (KMXK_E00380), the additional KHT and HGT genes have arisen as tandem duplications. Within the Kluyveromyces genus, there are two copies of KHT, termed KHT1 and KHT2 in all species (data not shown), but the larger tandem duplications only occurred in K. marxianus, suggesting that some particular evolutionary pressure has driven this selection. It is intriguing to see that there is variability in the number of expanded genes among the seven sequenced strains we examined, demonstrating a certain amount of genome fluidity. Although the K. marxianus strains included in this study come from diverse environments and collections, all have been propagated for some time in laboratories or used for industrial processes. It is possible, therefore, that the copy number reductions, which have arisen by inter-gene homologous recombination, are a consequence of propagation in “non-natural” settings. In this regard, the phenomenon mirrors the well-known KHT1 – KHT2 recombination in K. lactis that gave rise to RAG1 (Weirich et al., 1997). Indeed, many studies of sugar transport in K. lactis only consider RAG1 (which mainly resembles KHT2) in the context of a low-affinity glucose transporter (Breunig et al., 2000). The alternative scenario, that laboratory selection led to the expansions, is not compatible with the same expansions and divergence being observed in completely independent strain lineages.
Gene duplication followed by functional divergence is a common evolutionary mechanism. The restriction to K. marxianus and the relatively high degree of sequence similarity point to these expansions being relatively recent events. Other studies of bacterial and eukaryotic genomes found that recent gene duplications usually encode proteins that are secreted or involved in membrane functions (Kondrashov et al., 2002), which is also the case for these K. marxianus MFS proteins. In additional to duplication, which allows proteins to acquire alternative functions, tandem arrays of genes of similar sequence allow intra-locus recombination, which can give rise to recombinants that may have novel functions or to loss of genes that are functionally redundant (Mendonça et al., 2011). Such recombinations were identified in some K. marxianus strains but no novel functions were identified for the recombinant proteins. Since, however, the phenotypic space assessed was restricted to glucose and galactose transport, other functions remain a possibility.
There is a dearth of knowledge on the natural ecology of K. marxianus. It has traditionally been considered a dairy yeast but whole genome comparisons and studies of Lac12p variation showed that only a subset of K. marxianus strains are adapted to this niche (Ortiz-Merino et al., 2018). Examination of the origins of K. marxianus strains in culture collections shows that the yeast can be isolated from plants, fruits and other non-dairy niches plant sources (Suzuki et al., 2014; Zhong et al., 2016). It can be speculated that the expansion and functional diversification of MFS sugar transporters has its origins in pressures in the evolutionary history of K. marxianus. Glucose limitation was shown to trigger duplication of the hexose transporters in S. cerevisiae (Brown et al., 1998). In that study, three tandem copies of a hybrid transporter formed by recombination between the HXT6 and HXT7 genes were found in an evolved strain. Further experiments showed that this strain outcompetes the parental strain in low glucose competition experiments. It is also known that cellular stress can be a driver of gene duplication and evolution of new functions (Kondrashov, 2012). It could be speculated that duplication of MFS genes in K. marxianus arose in a period of stress or nutrient limitation, with new functions being acquired later. It remains to be determined whether these duplications play any role in the particular traits of K. marxianus, such as thermotolerance and rapid growth (Lane and Morrissey, 2010).
Functional Redundancy of Galactose Transporters in K. marxianus
The genetics of galactose transport has been well-studied in K. lactis, but it is now clear that the situation in K. marxianus is more complicated. There are also some common features. For example, in K. lactis, HGT1 encodes a protein capable of transporting both glucose and galactose (Billard et al., 1996; Baruffini et al., 2006). The syntenic arrangement, high degree of sequence similarity, and functional analysis of KMXK_A02960 indicates that these genes are orthologous. It is interesting to note that a previous screen for xylose transporters from K. marxianus reported that KMXK_A02960 transports xylose and arabinose and named the gene kmAXT1 (Knoshaug et al., 2015). That study did not report the duplicated genes and paralogs were not tested. Thus, it remains to be determined whether other Hgt1-like proteins can also transport pentose sugars. Regarding the hexose galactose, however, it was shown that two of the HGT1-like genes encode proteins capable of low-affinity transport, and one of high-affinity transport, and only the Hgt1 ortholog KMXK_A02960 has retained the capacity for glucose transport (Figure 4). It was also known from work on K. lactis that Lac12 is a low affinity galactose transporter, so it was not a surprise to find the same result in this study. The duplicated genes LAC12-2 and LAC12-4 also encode proteins that can transport galactose, but the unexpected finding was that Lac12-4 is also capable of supporting excellent growth on 0.1% galactose and is a high affinity galactose transporter (Figure 1). The KHT genes in K. lactis are thought to encode low affinity glucose transporters (Milkowski et al., 2001), though it must be considered that many of the studies were performed with a strain carrying RAG1, the recombined gene that contains the KHT1 promoter and the 5′ sequence of KHT1 along with most of KHT2. The possible role of K. lactis Kht1 and Kht2 in transporting galactose or other sugars does not seem to have received a lot of attention. In K. marxianus, there are three copies each of KHT1 (two in strain CBS6556) and KHT2, all of which encode high affinity glucose transporters. Only one of the six Kht-like proteins, KMXK_E03670, also has the capacity to transport galactose. In total, eight K. marxianus proteins with some capacity to transport galactose when heterologously expressed in S. cerevisiae were identified, indicating a remarkable functional overlap. Despite this, the proteins are not completely redundant as shown by the analysis of strains carrying different combinations of inactivated genes. Inactivation of any one of the transporters (or set of similar transporters) did not have a discernible effect but any pairwise combination did affect growth on 0.1% galactose (Figure 5). In contrast, it was necessary to inactivate all identified galactose transporters to block growth on 2% galactose, consistent with the data that low affinity transporters were more prevalent than high affinity ones. Inferences as to whether particular transporters are high or low affinity are made based on whether they support growth on 0.1% galactose but detailed kinetic studies would be required to be definitive on this point. In general, high-affinity transporters are thought to be induced and allow growth when the substrate concentration is low, whereas low-affinity transporters, which require high substrate concentrations, can allow a higher uptake rate and flux. A study examining the kinetics of sugar transport in K. marxianus CCT7735 (UFV3) found two galactose transport systems; a low-affinity system present in lactose-grown cells and a second system able to transport glucose and galactose (da Silveira et al., 2018). It is possible that these two systems are comprised of several of the transporters identified in this study. For example, the LAC12 genes could encode the low-affinity galactose transport system. This idea is supported by heterologous expression data (Figure 1) and data indicating that expression of these genes is induced by lactose and repressed by glucose (Varela et al., 2017b). The Hgt and the KMXK_E03670 Kht transporter could be part of the second transport system mentioned. However, studying the expression of these genes and the kinetics of galactose transport in K. marxianus mutants is required to understand the physiological role of these transporters. The range of substrates assessed could also be investigated since no substrates have yet been identified for some transporters, and it is also possible some of those shown here to transport glucose or galactose might actually have higher activity toward another substrate.
It is notoriously difficult to predict substrates for MFS sugar transporters and this family includes proteins able to transport various ions, sugar alcohols, and other molecules across the cell membrane (Lewinson et al., 2006). Having a set of proteins, some of which transport galactose and some of which do not offered a good opportunity to try to identify particular amino acids that might be important for distinguishing galactose from other sugars. Similar work has already been performed with S. cerevisiae Gal2, which diverged from an ancestral glucose permease. In that case, three critical amino acids were identified (Kasahara et al., 1997; Kasahara and Kasahara, 2000). From examination of the K. marxianus galactose transporters, however, it was immediately apparent that these residues are not diagnostic for galactose transporters in general (Supplementary Figure S5). In fact, there are no conserved amino acids that can be seen in all K. marxianus galactose transporters. Examination of the amino acid sequences within each sub-clade (Lac, Hgt, Kht) gave some clues as to what changes are required to enable galactose transport. This is most evident within the Hgt sub-clade where a single residue differentiates galactose and non-galactose transporters (Figure 6). However, the substitution of this residue in KMXK_A02930 (threonine to alanine substitution) was not sufficient to render it a galactose transporter (data not shown). This result is not completely unexpected considering that this position is a threonine in the galactose transporter KMXK_E03670 and is not a conserved alanine in other proteins that transport galactose (Supplementary Figure S5). Also, this residue is located in the 12th transmembrane domain, away from the pore that interacts with the substrate. Structural studies in the human transporter GLUT1, a MFS transporter that shares the topology and substrate recognition mechanism of yeast transporters, showed that the transmembrane segments 1, 2, 4, 5, 7, 8, 10, and 11 form the tunnel required for substrate transport (Kasahara et al., 2009; Sheena et al., 2011). Altogether, these findings suggest that sugar specificity in these proteins results from the combined contributions of multiple amino acids. It is apparent that different solutions can be found to create the correct environment in the substrate channel for galactose recognition and transport. Given the very close overall sequence similarity between these proteins, they will be a valuable tool for structural studies to model and identify exactly how galactose is selected for transport. Further investigations are needed to try to understand why K. marxianus has this array of transporters, for galactose and for other sugars.
Author Contributions
JV, MP, and NM performed the experimental work. JV and JM wrote the manuscript and conceived the project.
Funding
JV was supported by the YEASTCELL Marie Curie ITN project which received funding from the People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme FP7/2007-2013/ under REA Grant Agreement No. 606795. NM works on the CHASSY project, which has received funding from the European Union’s Horizon 2020 Framework Programme for Research and Innovation – Grant Agreement No. 720824.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Dr. Arun Rajkumar for critically reading the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.03330/full#supplementary-material
FIGURE S1 | Map of the CRISPR-Cas9 plasmid used in this study. The pUCC001 plasmid was obtained by introducing a double-restriction site for the type IIS restriction enzyme, BsaI (gray shading) into the pUDP002 plasmid (Juergens et al., 2018). The desired target sequence can be chemically synthesized as a pair of complementary DNA oligos containing specific overhangs compatible with pUCC001. This oligo duplex can then be cloned between the HH ribozyme and the gRNA scaffold sequences (purple and blue shading, respectively) by Golden Gate assembly.
FIGURE S2 | Recombination event leading to the formation of KMXK_E03660. (A) A scheme depicting the recombination event between the KHT genes, KMAR_50344 and KMAR_50343, is shown. The 5′ end of KMAR_50344 (gray) and the 3′ end of KMAR_50343 (purple) recombined to form KMXK_E03660 (red). (B) A DNA alignment of the KMAR_50344, KMAR_50343, and KMXK_E03660 sequences is shown. Three sections of the alignment are shown; the 5′ end of the three sequences, where KMXK_E03660 is identical to KMAR_50344 (top), the recombination point (middle) and the 5′ section of the gene where KMXK_E03660 is identical to KMAR_50343 (bottom). The different sequences are shown in colors as in the panel above. The exact recombination point is indicated with a red vertical line in position 371. Mismatches between the KMAR_50344/KMAR_50343 sequences and KMXK_E03660 are shown in bold.
FIGURE S3 | Recombination event leading to the formation of KLMA_10546. (A) A scheme depicting the recombination event between KMXK_A02950 and KMXK_A02930 is shown. The 5′ end of KMXK_A02950 (gray) and the 3′ end of KMXK_A02930 (purple) recombined to form KLMA_10546 (red). (B) A DNA alignment of these sequences is shown as in Supplementary Figure S2. The exact recombination point is indicated with a red vertical line in position 230. Mismatches between the KMXK_A02950/KMXK_A02930 sequences and KLMA_10546 are shown in bold.
FIGURE S4 | Construction of an HGT null mutant in K. marxianus NBRC1777. The CRISPR-Cas9 system was used to induce mutagenesis of the HGT genes by targeting the 5′ CGCTCCAACCTGGGGTAT 3′ sequence present in all HGT genes with a single gRNA. (A) Scheme showing the repair event occurred upon targeting the HGT genes with the CRISPR-Cas9 system. Recombination between the two ends of the HGT loci is shown by gray shading. Stop codons in the recombinant HGT, obtained as consequence of the deletion, are represented by a red line. Primers used to screen the mutant colonies are shown as red arrows. (B) Screening of hgt mutant colonies. Transformants were screen by PCR using the DiagHGT primers. Lane 1, NBRC1777 wild-type. Lanes 2 and 3, NBRC1777 strains transformed with the pUCC001-HGT plasmid. DNA ladder Hyperladder I. (C) Recombination events in mutants hgt75 (top panel) and hgt77 (bottom panel). DNA sequence alignments between the wild-type and mutant sequences are shown. Blue and purple boxes represent the genes A02960 and A02920 where the recombination events took place. Coordinates in the NBRC1777 genome (chromosome I) are shown as numbers above the alignments. The protospacer sequence is shown in bold.
FIGURE S5 | Multiple sequence alignment of Lac12, Kht, Hgt, and Gal2 sequences. The protein sequence alignment was generated using MUSCLE 3.8 and visualised in Jalview 2. The S. cerevisiae Gal2 transporter was included in this analysis as the three amino acid positions critical for galactose transport have been identified in this transporter. These residues (Tyr352, Tyr446, and Phe504) are marked in red in the alignment.
FIGURE S6 | Multiple sequence alignment of Lac12 sequences. The multiple sequence alignment was generated using MUSCLE 3.8 and imported into Jalview 2 for visualisation. Sequence differences between galactose transporters (Lac12_Km, Lac12-2_Km, and Lac12-4_Km) and Lac12-3, unable to transport galactose, are marked in purple.
FIGURE S7 | Multiple sequence alignment of Kht sequences. The sequence alignment was computed as described in Supplementary Figures S6, S7. Sequence differences between the galactose transporter KMXK_E03670 and the rest of the proteins are shown in purple.
TABLE S1 | Annotation of putative MFS transporters in K. marxianus and K. lactis.
TABLE S2 | Equivalence of KHT and HGT genes in different K. marxianus genomes. Genes marked with an asterisk have arisen by recombination between two genes.
Footnotes
References
Baruffini, E., Goffrini, P., Donnini, C., and Lodi, T. (2006). Galactose transport in Kluyveromyces lactis: major role of the glucose permease Hgt1. FEMS Yeast Res. 6, 1235–1242. doi: 10.1111/j.1567-1364.2006.00107.x
Billard, P., Ménart, S., Blaisonneau, J., Bolotin-Fukuhara, M., Fukuhara, H., and Wésolowski-Louvel, M. (1996). Glucose uptake in Kluyveromyces lactis: role of the HGT1 gene in glucose transport. J. Bacteriol. 178, 5860–5866. doi: 10.1128/jb.178.20.5860-5866.1996
Breunig, K. D., Bolotin-Fukuhara, M., Bianchi, M., Bourgarel, D., Falcone, C., and Ferrero, L. L. (2000). Regulation of primary carbon metabolism in Kluyveromyces lactis. Enzyme Microb. Technol. 26, 771–780. doi: 10.1016/S0141-0229(00)00170-8
Brown, C. J., Todd, K. M., and Rosenzweig, R. F. (1998). Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Mol. Biol. Evol. 15, 931–942. doi: 10.1093/oxfordjournals.molbev.a026009
Carvalho-Silva, M., and Spencer-Martins, I. (1990). Modes of lactose uptake in the yeast species Kluyveromyces marxianus, Antonie van Leeuwenhoek. Int. J. Gen. Mol. Microbiol. 57, 77–81. doi: 10.1007/BF00403158
Coloretti, F., Chiavari, C., Luise, D., Tofalo, R., Fasoli, G., Suzzi, G., et al. (2017). Detection and identification of yeasts in natural whey starter for parmigiano reggiano cheese-making. Int. Dairy J. 66, 13–17. doi: 10.1016/J.IDAIRYJ.2016.10.013
da Silveira, F. A., Diniz, R. H. S., Sampaio, G. M. S., Brandão, R. L., da Silveira, W. B., and Castro, I. M. (2018). Sugar transport systems in Kluyveromyces marxianus CCT 7735. Antonie Van Leeuwenhoek. doi: 10.1007/s10482-018-1143-4 [Epub ahead of print].
Darling, A. E., Mau, B., and Perna, N. T. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147
De Bruijne, A. W., Schuddemat, J., Van den Broek, P. J. A., and Van Steveninck, J. (1988). Regulation of sugar transport systems of Kluyveromyces marxianus: the role of carbohydrates and their catabolism. BBA – Biomembr. 939, 569–576. doi: 10.1016/0005-2736(88)90104-6
De Vuyst, L., and Weckx, S. (2016). The cocoa bean fermentation process: from ecosystem analysis to starter culture development. J. Appl. Microbiol. 121, 5–17. doi: 10.1111/jam.13045
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Elbourne, L. D. H., Tetu, S. G., Hassan, K. A., and Paulsen, I. T. (2017). TransportDB 2.0: a database for exploring membrane transporters in sequenced genomes from all domains of life. Nucleic Acids Res. 45, D320–D324. doi: 10.1093/nar/gkw1068
Escalante-Minakata, P., Blaschek, H. P., Barba de la Rosa, A. P., Santos, L., and De León-Rodríguez, A. (2008). Identification of yeast and bacteria involved in the mezcal fermentation of Agave salmiana. Lett. Appl. Microbiol. 46, 626–630. doi: 10.1111/j.1472-765X.2008.02359.x
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. doi: 10.1093/nar/gkt1223
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Fonseca, G. G., Gombert, A. K., Heinzle, E., and Wittmann, C. (2007). Physiology of the yeast Kluyveromyces marxianus during batch and chemostat cultures with glucose as the sole carbon source. FEMS Yeast Res. 7, 422–435. doi: 10.1111/j.1567-1364.2006.00192.x
Fonseca, G. G., Heinzle, E., Wittmann, C., and Gombert, A. K. (2008). The yeast Kluyveromyces marxianus and its biotechnological potential. Appl. Microbiol. Biotechnol. 79, 339–354. doi: 10.1007/s00253-008-1458-6
Gasnier, B. (1987). Characterization of low- and high-affinity glucose transports in the yeast Kluyveromyces marxianus. Biochim. Biophys. Acta 903, 425–433. doi: 10.1016/0005-2736(87)90049-6
Gethins, L., Rea, M. C., Stanton, C., Ross, R. P., Kilcawley, K., O’Sullivan, M., et al. (2016). Acquisition of the yeast Kluyveromyces marxianus from unpasteurised milk by a kefir grain enhances kefir quality. FEMS Microbiol. Lett. 363:fnw165. doi: 10.1093/femsle/fnw165
Gietz, R. D., and Schiestl, R. H. (2007). High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34. doi: 10.1038/nprot.2007.13
Guindon, S., Dufayard, J.-F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Ho, V. T. T., Zhao, J., and Fleet, G. (2014). Yeasts are essential for cocoa bean fermentation. Int. J. Food Microbiol. 174, 72–87. doi: 10.1016/j.ijfoodmicro.2013.12.014
Huerta-Cepas, J., Serra, F., and Bork, P. (2016). ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635–1638. doi: 10.1093/molbev/msw046
Inokuma, K., Ishii, J., Hara, K. Y., Mochizuki, M., Hasunuma, T., and Kondo, A. (2015). Complete genome sequence of Kluyveromyces marxianus NBRC1777, a nonconventional thermotolerant yeast. Genome Announc. 3:e389-15. doi: 10.1128/genomeA.00389-15
Jeong, H., Lee, D.-H., Kim, S. H., Kim, H.-J., Lee, K., Song, J. Y., et al. (2012). Genome sequence of the thermotolerant yeast Kluyveromyces marxianus var. marxianus KCTC 17555. Eukaryot. Cell 11, 1584–1585. doi: 10.1128/EC.00260-12
Juergens, H., Varela, J. A., de Vries, A. R. G., Perli, T., Gast, V. J. M., Gyurchev, N. Y., et al. (2018). Genome editing in Kluyveromyces and Ogataea yeasts using a broad-host-range Cas9/gRNA co-expression plasmid. FEMS Yeast Res. 18. doi: 10.1093/femsyr/foy012
Kasahara, M., Shimoda, E., and Maeda, M. (1997). Amino acid residues responsible for galactose recognition in yeast Gal2 transporter. J. Biol. Chem. 272, 16721–16724. doi: 10.1074/JBC.272.27.16721
Kasahara, T., and Kasahara, M. (2000). Three aromatic amino acid residues critical for galactose transport in yeast Gal2 transporter. J. Biol. Chem. 275, 4422–4428. doi: 10.1074/JBC.275.6.4422
Kasahara, T., Maeda, M., Boles, E., and Kasahara, M. (2009). Identification of a key residue determining substrate affinity in the human glucose transporter GLUT1. Biochim. Biophys. Acta – Biomembr. 1788, 1051–1055. doi: 10.1016/J.BBAMEM.2009.01.014
Knoshaug, E. P., Vidgren, V., Magalhães, F., Jarvis, E. E., Franden, M. A., Zhang, M., et al. (2015). Novel transporters from Kluyveromyces marxianus and Pichia guilliermondii expressed in Saccharomyces cerevisiae enable growth on l -arabinose and d -xylose. Yeast 32, 615–628. doi: 10.1002/yea.3084
Kondrashov, F. A. (2012). Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc. R. Soc. B Biol. Sci. 279, 5048–5057. doi: 10.1098/rspb.2012.1108
Kondrashov, F. A., Rogozin, I. B., Wolf, Y. I., and Koonin, E. V. (2002). Selection in the evolution of gene duplications. Genome Biol. 3:RESEARCH0008.
Lane, M. M., Burke, N., Karreman, R., Wolfe, K. H., O’Byrne, C. P., and Morrissey, J. P. (2011). Physiological and metabolic diversity in the yeast Kluyveromyces marxianus. Antonie Van Leeuwenhoek 100, 507–519. doi: 10.1007/s10482-011-9606-x
Lane, M. M., and Morrissey, J. P. (2010). Kluyveromyces marxianus: a yeast emerging from its sister’s shadow. Fungal Biol. Rev. 24, 17–26. doi: 10.1016/j.fbr.2010.01.001
Lappe-Oliveras, P., Moreno-Terrazas, R., Arrizón-Gaviño, J., Herrera-Suárez, T., García-Mendoza, A., and Gschaedler-Mathis, A. (2008). Yeasts associated with the production of Mexican alcoholic nondistilled and distilled Agave beverages. FEMS Yeast Res. 8, 1037–1052. doi: 10.1111/j.1567-1364.2008.00430.x
Lertwattanasakul, N., Kosaka, T., Hosoyama, A., Suzuki, Y., Rodrussamee, N., Matsutani, M., et al. (2015). Genetic basis of the highly efficient yeast Kluyveromyces marxianus: complete genome sequence and transcriptome analyses. Biotechnol. Biofuels 8:47. doi: 10.1186/s13068-015-0227-x
Lewinson, O., Adler, J., Sigal, N., and Bibi, E. (2006). MicroReview Promiscuity in multidrug recognition and transport: the bacterial MFS Mdr transporters. Mol. Microbiol. 61, 277–284. doi: 10.1111/j.1365-2958.2006.05254.x
Löbs, A.-K., Engel, R., Schwartz, C., Flores, A., and Wheeldon, I. (2017). CRISPR–Cas9-enabled genetic disruptions for understanding ethanol and ethyl acetate biosynthesis in Kluyveromyces marxianus. Biotechnol. Biofuels 10:164. doi: 10.1186/s13068-017-0854-5
Mendonça, A. G., Alves, R. J., and Pereira-Leal, J. B. (2011). Loss of genetic redundancy in reductive genome evolution. PLoS Comput. Biol. 7:e1001082. doi: 10.1371/journal.pcbi.1001082
Milkowski, C., Krampe, S., and Weirich, J. (2001). Feedback regulation of glucose transporter gene transcription in Kluyveromyces lactis by glucose uptake. J. Bacteriol. 183, 5223–5229. doi: 10.1128/JB.183.18.5223
Morrissey, J. P., Etschmann, M. M. W., Schrader, J., and Billerbeck, G. M. (2015). Cell factory applications of the yeast Kluyveromyces marxianus for the biotechnological production of natural flavour and fragrance molecules. Yeast 32, 3–16. doi: 10.1002/YEA.3054
Nambu-Nishida, Y., Nishida, K., Hasunuma, T., and Kondo, A. (2017). Development of a comprehensive set of tools for genome engineering in a cold- and thermo-tolerant Kluyveromyces marxianus yeast strain. Sci. Rep. 7:8993. doi: 10.1038/s41598-017-08356-5
Ng, H., and Dean, N. (2017). Dramatic improvement of CRISPR/Cas9 editing in Candida albicans by increased single guide RNA expression. mSphere 2:e00385-16. doi: 10.1128/mSphere.00385-16
Ortiz-Merino, R. A., Varela, J. A., Coughlan, A. Y., Hoshida, H., da Silveira, W. B., Wilde, C., et al. (2018). Ploidy variation in Kluyveromyces marxianus separates dairy and non-dairy isolates. Front. Genet. 9:94. doi: 10.3389/fgene.2018.00094
Riley, M. I., Sreekrishna, K., Bhairi, S., and Dickson, R. C. (1987). Isolation and characterization of mutants of Kluyveromyces lactis defective in lactose transport. Mol. Gen. Genet. 208, 145–151. doi: 10.1007/BF00330435
Schaffrath, R., and Breunig, K. D. (2000). Genetics and molecular physiology of the yeast Kluyveromyces lactis. Fungal Genet. Biol. 30, 173–190. doi: 10.1006/fgbi.2000.1221
Schwan, R. F., and Wheals, A. E. (2004). The microbiology of cocoa fermentation and its role in chocolate quality. Crit. Rev. Food Sci. Nutr. 44, 205–221. doi: 10.1080/10408690490464104.
Sheena, A., Mohan, S. S., Haridas, N. P. A., and Anilkumar, G. (2011). Elucidation of the glucose transport pathway in glucose transporter 4 via steered molecular dynamics simulations. PLoS One 6:e25747. doi: 10.1371/journal.pone.0025747
Suzuki, T., Hoshino, T., and Matsushika, A. (2014). Draft genome sequence of Kluyveromyces marxianus strain DMB1, isolated from sugarcane bagasse hydrolysate. Genome Announc. 2:e00733-14. doi: 10.1128/genomeA.00733-14
Tittarelli, F., Varela, J. A., Gethins, L., Stanton, C., Ross, R. P., Suzzi, G., et al. (2018). Development and implementation of multilocus sequence typing to study the diversity of the yeast Kluyveromyces marxianus in Italian cheeses. Microb. Genomics doi: 10.1099/mgen.0.000153 [Epub ahead of print].
Varela, J. A., Gethins, L., Stanton, C., Ross, P., and Morrissey, J. P. (2017a). “Applications of Kluyveromyces marxianus in Biotechnology,” in Yeast Diversity in Human Welfare, eds T. Satyanarayana and G. Kunze (Singapore: Springer Singapore), 439–453. doi: 10.1007/978-981-10-2621-8_17
Varela, J. A., Montini, N., Scully, D., Van der Ploeg, R., Oreb, M., Boles, E., et al. (2017b). Polymorphisms in the LAC12 gene explain lactose utilisation variability in Kluyveromyces marxianus strains. FEMS Yeast Res. 17. doi: 10.1093/femsyr/fox021
Vyas, V. K., Barrasa, M. I., and Fink, G. R. (2015). A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Sci. Adv. 1:e1500248. doi: 10.1126/sciadv.1500248
Weirich, J., Goffrini, P., Kuger, P., Ferrero, I., and Breunig, K. D. (1997). Influence of mutations in hexose-transporter genes on glucose repression in Kluyveromyces lactis. Eur. J. Biochem. 249, 248–257. doi: 10.1111/j.1432-1033.1997.t01-1-00248.x
Wésolowski-Louvel, M., and Goffrini, P. (1992). Glucose transport in the yeast Kluyveromyces lactis. Mol. Gen. 101, 89–96. doi: 10.1007/BF00587565
Wieczorke, R., Krampe, S., Weierstall, T., Freidel, K., Hollenberg, C. P., and Boles, E. (1999). Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 464, 123–128. doi: 10.1016/S0014-5793(99)01698-1
Xie, S., Shen, B., Zhang, C., Huang, X., and Zhang, Y. (2014). sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS One 9:e100448. doi: 10.1371/journal.pone.0100448
Zhong, S., Liu, H., Zhang, H., Han, T., Jia, H., Xie, Y., et al. (2016). Effects of Kluyveromyces marxianus isolated from tibetan mushrooms on the plasma lipids, egg cholesterol level, egg quality and intestinal health of laying hens. Rev. Bras. Ciência Avícola 18, 261–268. doi: 10.1590/1806-9061-2015-0070
Keywords: sugar transport, genome evolution, gene duplication, HGT1, hexose transport, RAG1, KHT1, major facilitator superfamily
Citation: Varela JA, Puricelli M, Montini N and Morrissey JP (2019) Expansion and Diversification of MFS Transporters in Kluyveromyces marxianus. Front. Microbiol. 9:3330. doi: 10.3389/fmicb.2018.03330
Received: 01 October 2018; Accepted: 21 December 2018;
Published: 10 January 2019.
Edited by:
Jean Marie François, UMR5504 Laboratoire d’Ingénierie des Systèmes Biologiques et des Procédés (LISBP), FranceReviewed by:
Cécile Neuvéglise, Institut National de la Recherche Agronomique (INRA), FranceBoris Ugarte Stambuk, Federal University of Santa Catarina, Brazil
Copyright © 2019 Varela, Puricelli, Montini and Morrissey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John P. Morrissey, ai5tb3JyaXNzZXlAdWNjLmll
 Javier A. Varela
Javier A. Varela Martina Puricelli
Martina Puricelli Noemi Montini
Noemi Montini John P. Morrissey
John P. Morrissey