- 1Department of Microbiology, School of Medicine, University of Patras, Patras, Greece
- 2Division of Infectious Diseases, School of Medicine, University of Patras, Patras, Greece
- 3Department of Public Health, School of Medicine, University of Patras, Patras, Greece
Methicillin-resistant Staphylococcus aureus (MRSA) remains an important cause of nosocomial and community-associated infections due to its ability to produce toxins and evade host’s immune responses. The aim of the present study was to investigate the association of monocytes immune response in terms of cytokines produced after inoculation with different MRSA clones. Thirty-one clinical MRSA strains were selected on the basis of clonal types, accessory gene regulator (agr) groups and toxin genes carriage. Isolates were identified as S. aureus by Gram stain, catalase, coagulase production and PCR for nuc gene. The presence of mecA, lukS/lukF-PV (Panton-Valentine Leukocidin) and tst (Toxic Shock Syndrome Toxin-1) genes, as well as, the determination of agr groups was performed by PCR. Clonality was investigated by means of multi-locus sequence typing (MLST). Peripheral blood mononuclear cells were stimulated with live bacterial cells for 45 min at a ratio of 1:10. Cells were incubated for 10 h and supernatants were collected. The levels of Tumor Necrosis Factor alpha (TNFa), IL-1b, IL-8, IL-6, IL-12p40, IL-10, interferon-gamma (IFN-γ) and IL-2, were measured by Human Cytokine Multiplex Immunoassay kit. Thirteen strains were tst and 12 lukS/lukF-PV-positive. Seven strains belonged to ST80 and ST225, five to ST30 and ST239, while the remaining seven isolates were grouped together as “other.” Strains belonging to ST80 induced statistically lower levels of TNFa, IL-1b, IL-8, IL-6, IL-10, IFN-γ, and IL-2. PVL-positive strains classified into ST80 clone induced statistically lower concentrations of most cytokines as compared to PVL-positive strains belonging to other clones, tst-positive strains and toxin-negative ones. Strains of agr3 group belonging to ST80 induced statistically lower concentrations of most tested cytokines as compared to agr3 strains not-belonging to ST80, agr2 or agr1. This low induction of immune response by MRSA ST80 cannot be attributed to the presence of neither lukS/lukF-PV nor agr3.
Introduction
Staphylococcus aureus remains an important cause of infections, especially in skin and soft tissue, but can also provoke severe ones such as necrotizing pneumonia, bacteraemia and endocarditis (van Belkum et al., 2009). In Greece, S. aureus accounts for 9% of bacteraemias, while 40% of them are caused by MRSA (Kolonitsiou et al., 2017). MRSA was initially isolated from healthcare-associated infections causing outbreaks and gradually becoming endemic in the hospital setting with ST225 and ST239 being the most prominent clones (Drougka et al., 2014; Monecke et al., 2018). Since late 90s CA-MRSA arised worldwide and rapidly disseminated in the community (Witte, 2009; Drougka et al., 2014). The virulence of CA-MRSA is mainly due to the production of PVL which has a cytolytic activity on immune system’s cells (Witte, 2009; Drougka et al., 2014). In the Mediterranean area and especially in Greece such infections are mainly due to ST80 clone which during the last decade invaded the healthcare setting causing a large proportion of healthcare-associated infections (Witte, 2009; Drougka et al., 2014).
Asymptomatic nasal carriage of S. aureus is a common phenomenon, with approximately 50% of the general population being transient carriers and an additional 25% being persistent ones (von Eiff et al., 2001). Colonization is the prerequisite for invasive infection since most of infections are of endogenous origin caused by colonizing isolates (von Eiff et al., 2001). The capacity of several clones to disseminate can be partially due to their ability to evade host’s immune responses by secreting different immune evasion protein complexes, leading to persistent colonization increasing the risk of transmission (McCarthy and Lindsay, 2013). The main regulatory system of S. aureus that ensures adaptation of staphylococci to the environment acting as a quorum-sensing is the Agr system (Le and Otto, 2015). Up-regulation of virulence factors by Agr contributes to acute infection in several animal models, whereas, down-regulation by this system of microbial surface components has been implicated in enhanced biofilm formation and bacterial colonization. Among the Agr-controlled toxins are the pore-forming toxins, including the PVL (Le and Otto, 2015).
The aim of the present study was to investigate the association of monocytes’ immune response in terms of pro-inflammatory cytokines produced after inoculation with variable MRSA clones, classified into different agr groups.
Materials and Methods
Bacterial Isolates
Thirty-one clinical MRSA strains recovered from inpatients and outpatients with skin and soft tissue infections (SSTIs) or bloodstream infections (BSIs) at the University General Hospital of Patras during 2011–2014 were selected to be studied, representing the main clones identified in Greece, classified into different agr groups and carrying the genes of the superantigen Toxic Shock Syndrome Toxin-1 (TSST-1; tst) and the PVL (lukS/lukF-PV) (Table 1). The study was carried out under the Hospital Surveillance Program for multi-drug resistant bacterial colonization and infections and in accordance with the recommendations of the Ethics Committee of the University General Hospital of Patras that waived the need of inform consent from the patients with S. aureus infection that the strains were included in the study, whereas, a written informed consent was obtained, in accordance with the Declaration of Helsinki, from healthy volunteers from which PBMCs were collected. The study was approved by the Ethics Committee of the University General Hospital of Patras.
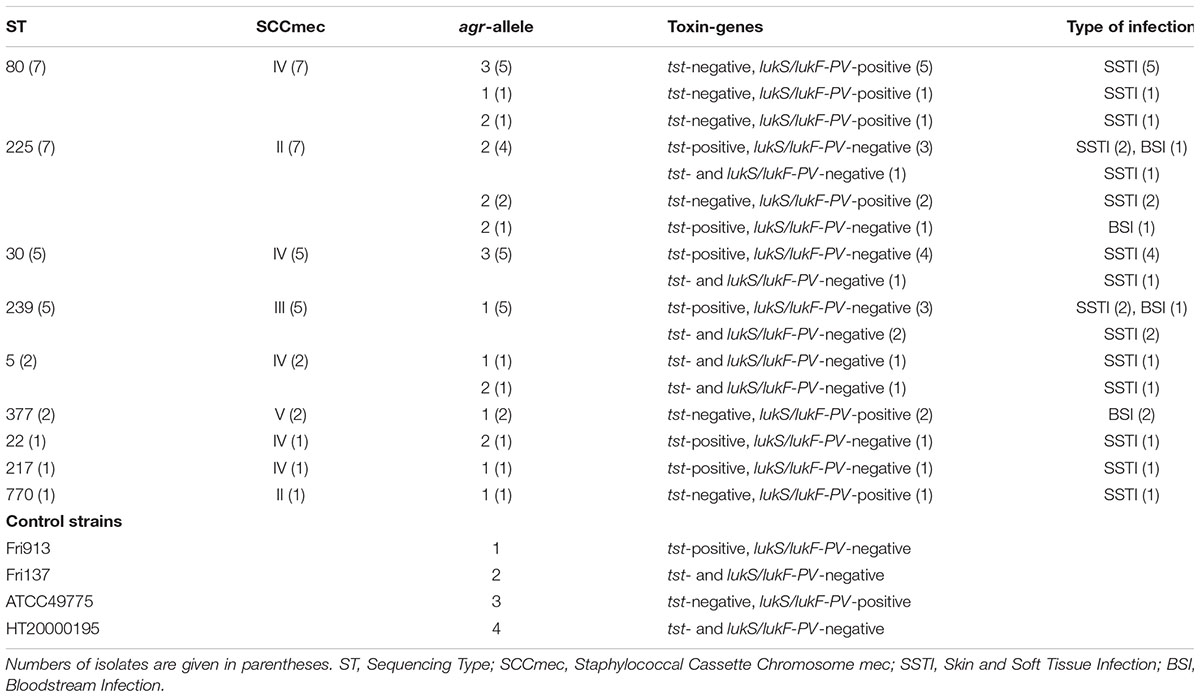
Table 1. Clinical and molecular characteristics of methicillin-resistant S. aureus strains included in the study.
S. aureus Identification
Isolates were identified as S. aureus by Gram stain, catalase and coagulase production (Slidex Staph plus test, bioMerieux S.A., Marcy l’Etoile, France) and verified by molecular methods (PCR for nuc gene) (Zhang et al., 2004).
Molecular Characterization
The presence of mecA, lukS/lukF-PV (PVL) and tst (TSST-1) genes as well as the determination of agr groups was performed by PCR (Jarraud et al., 2002). Positive control strains were: Fri 913 (agr1, tst-positive), Fri 137 (agr2), ATCC 49775 (agr3, lukS/lukF-PV-positive) and HT 20000195 (agr4) (Jarraud et al., 2002). The genes encoding the adhesins Fib (adhesin binding to fibrinogen), ClfA (adhesin binding to fibrinogen, involved in platelet aggregation and immune invasion), ClfB (adhesin binding to fibrinogen, cytokeratin 10, loricrin and involved in adhesion to epithelial cells and platelet aggregation), FnBPA (adhesin binding to fibronectin), Eno (encoding laminin binding protein) and EbpS (adhesin binding to elastin) were investigated by PCRs as described previously (Jarraud et al., 2002; Peacock et al., 2002; Tristan et al., 2003). Reference strains NCTC13552, Fri 913, ATCC119095, ATCC49775, ATCC31890, were used as controls. Clonality was investigated in all strains by MLST1. MRSA clones were defined according to their STs.
Isolation of Human Peripheral Blood Mononuclear Cells
Peripheral blood mononuclear cells were isolated from buffy coats derived from blood donation bottles of six healthy volunteers by density centrifugation on Ficoll density gradient (Biochrom AG, Berlin, Germany) as previously described (Spiliopoulou et al., 2012). Briefly, collected mononuclear cells were washed in PBS and resuspended in RPMI-1640 medium supplemented with 10% heat-inactivated fetal calf serum (Biochrom AG) and 2 mM L-glutamine (HyClone), (CM). Cells at a density of 1 × 106 cells/mL per well, were then seeded in 24-well flat bottom tissue culture plates (Sarstedt, Nümbrecht, Germany) and cultured at 37°C in a humidified, 5% CO2 atmosphere.
Cytokine Measurement
PBMC (106 cells/mL) were stimulated with live bacterial cells for 45 min at a ratio of 1:10, as in preliminary experiments, this ratio was proved to be the most efficient in cytokine production. (Spiliopoulou et al., 2012) Afterward, extracellular bacteria were lysed by lysostaphin (Sigma-Aldrich, St. Louis, MO, United States), and medium was replaced by CM supplemented with antibiotics. (Spiliopoulou et al., 2012) Cells were incubated for 10 h, since in preliminary experiment with the four control strains (Fri 913, Fri 137, ATCC 49775 and HT 20000195) in 2, 4, 6, 12, 24, and 48 h, all cytokines reached their peak between 6 and 12 h. Each experiment was carried out with mononuclear cells isolated from a single donor and performed in triplicate, with PBMCs from three donors; calculation of mean values was performed. Results for all 31 strains are based on at least three experiments from three different donors. LPS (10 ng/mL) was used as a positive control, and PBMCs without bacteria or LPS were used to assess spontaneous levels of cytokine secretion (negative control). Supernatants were collected, and the levels of TNFa, IL-1b, IL-8, IL-6, IL-12p40, IL-10, IFN-γ, and IL-2, were measured by Human Cytokine Multiplex Immunoassay kit (LINCO Research Inc., St. Charles, MO, United States) using Luminex®xMAPTM technology. A five-parameter regression formula was used to calculate the cytokine concentrations in samples from standard curves.
Statistical Analysis
SPSS statistics version 23.0 (SPSS, Chicago, IL, United States) was used. Difference of cytokine production was assessed by two-tailed t-test. p < 0.05 was considered as statistically significant.
Results
Molecular and Clinical Characteristics of Isolates
All 31 strains were mecA-positive (MRSA). Twenty-six were from patients with SSTIs, whereas the remaining five were isolated from bactereamic patients. Toxin genes presence was verified in 25 strains; 13 were tst-positive and 12 lukS/lukF-PV-positive. No strain carried simultaneous both toxin genes. The adhesin genes were detected in most strains; clfA in 21 strains (68%), clfB and ebpS in 28 strains each (90%), fib and eno in 30 strains each (97%) and fnbA in 29 strains (94%). The main clones were ST80 (seven strains), ST225 (seven), ST30 (five), ST239 (five), while the remaining seven isolates, grouped together as “other STs,” belonged to ST5 (two), ST377 (two), ST22 (one), ST217 (one) and ST770 (one) (Table 1). Strains were classified into agr1 (11), agr2 and agr3 (10 strains each); no MRSA belonged to agr4 group. The presence of adhesin genes were not clonal related.
Cytokine Measurement
Strains belonging to ST80 were compared to other STs concerning cytokine production in supernatants of PBMCs (Figure 1). ST80 induced statistically lower levels of TNFa [as compared to ST30 (p 0.002), ST225 (p < 0.001), ST239 (p < 0.001) and other STs (p 0.014)], IL-1b [as compared to ST30 (p 0.007), ST225 (p 0.002) and ST239 (p < 0.001)], IL-6 [as compared to ST225 (p 0.019), ST239 (p < 0.001) and other STs (p 0.014)], IFN-γ [as compared to ST30 (p 0.001), ST225 (p < 0.001), ST239 (p 0.040) and other STs (p 0.003)], IL-2 [as compared to ST30 (p < 0.001), ST225 (p < 0.001) and other STs (p 0.003)] and IL-10 [as compared to ST30 (p 0.042), ST225 (p 0.001), ST239 (p 0.003) and other STs (p 0.032)].
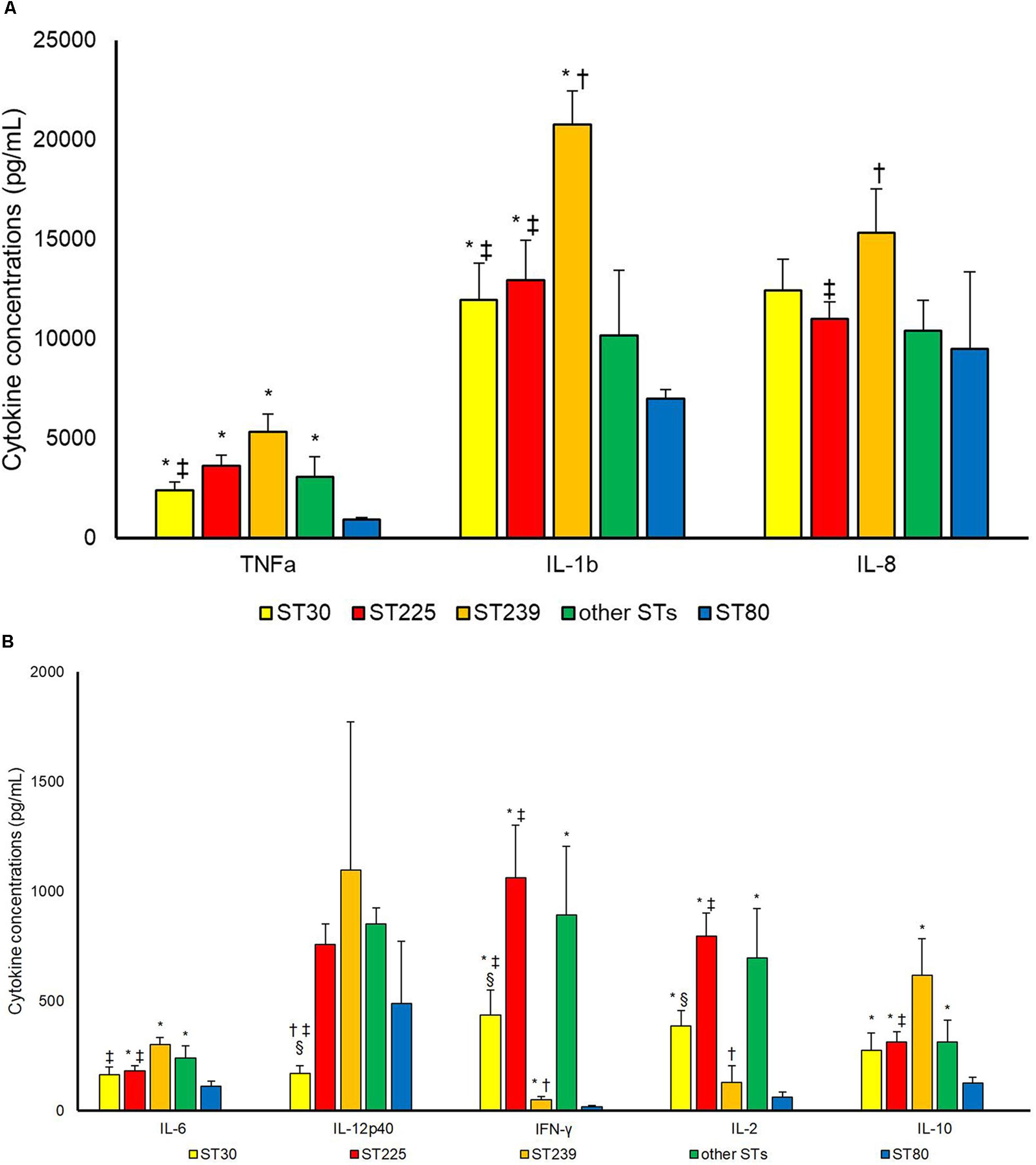
Figure 1. Cytokineconcentrations (pg/mL) (A) TNF-a, IL-1b, IL-8; (B) IL6, IL12p40, IFN-γ, IL-2, IL-10 in supernatant upon incubation of PBMCs with S. aureus cells. Differences between different Sequencing Types (STs) were assessed. ∗P < 0.05 in comparison with ST80; †P < 0.05 in comparison with other STs; ‡P < 0.05 in comparison with ST239; §P < 0.05 in comparison with ST225.
As toxins were concerned (Figure 2), PVL-positive strains classified into ST80 induced statistically lower concentrations of TNFa (p 0.007), IL-1b (p 0.030), IL-6 (p 0.024), IFN-γ (p 0.015), IL-2 (p 0.019) and IL-10 (p 0.010) production as compared to PVL-positive strains belonging to other clones. ST80 PVL-positive clone induced also lower cytokine production as concerned to tst-positive strains [TNFa (p 0.001), IL-1b (p 0.019), IL-6 (p 0.027), IFN-γ (p 0.007), IL-2 (p 0.002) and IL-10 (p 0.043)] and toxin-negative ones [TNFa (p 0.001), IL-1b (p 0.004), IL-6 (p 0.002), IFN-γ (p 0.023), IL-2 (p 0.017) and IL-10 (p 0.002)]. No difference (p > 0.050) among tst-positive, non-ST80 PVL-positive and toxin-negative was observed.
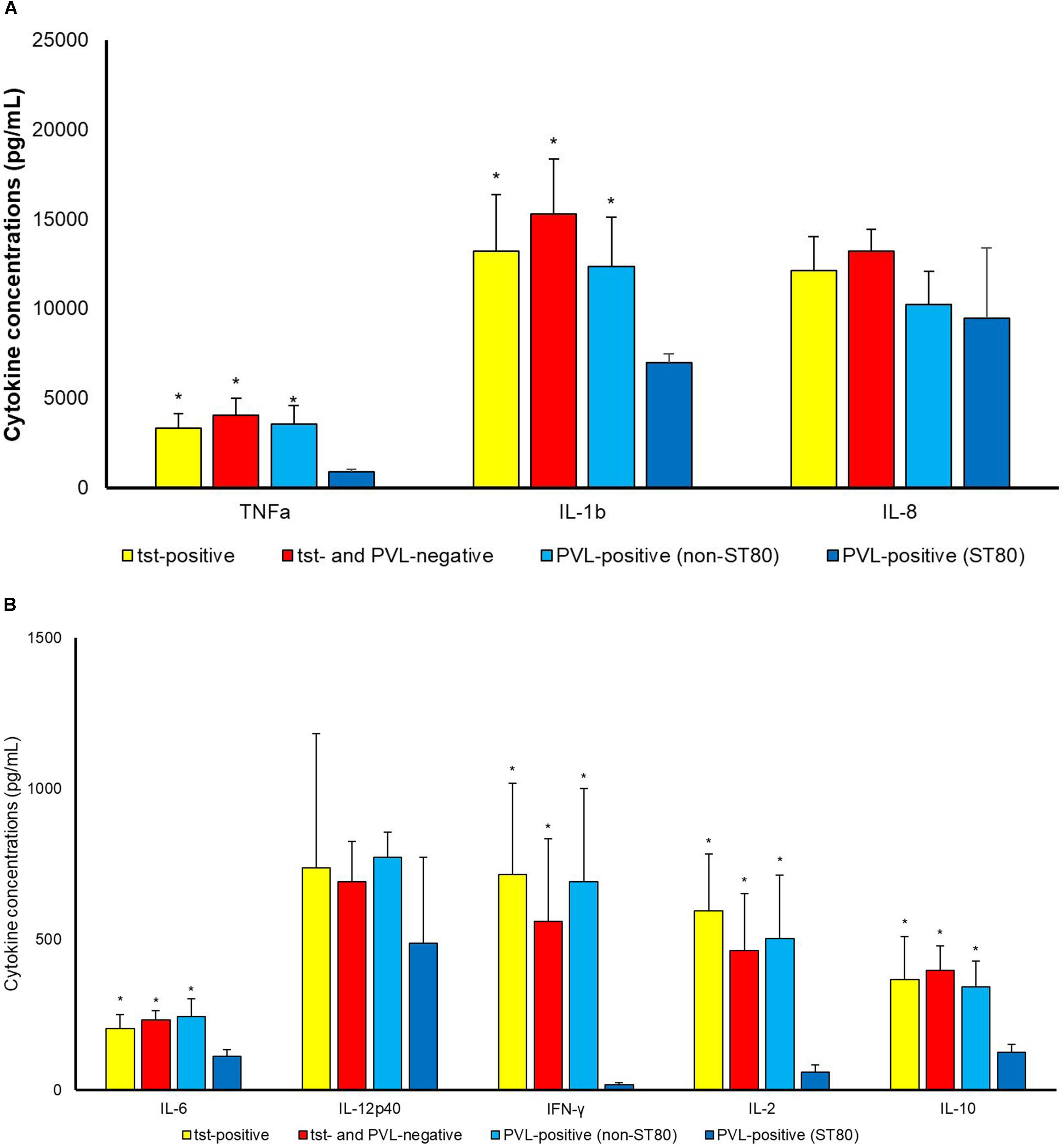
Figure 2. Cytokine concentrations (pg/mL) (A) TNF-a, IL-1b, IL-8; (B) IL6, IL12p40, IFN-γ, IL-2, IL-10 in supernatant upon incubation of PBMCs with S. aureus cells. Differences between tst-positive, tst- and PVL-negative and PVL-positive (belonging to ST80 and to STs other than ST 80) S. aureus were assessed. ∗P < 0.05 in comparison with PVL-positive belonging to ST80.
Strains of agr3 group belonging to ST80 induced statistically lower concentrations of most tested cytokines as compared to agr3 strains not-belonging to ST80 [TNFa (p 0.005), IL-1b (p 0.018), IFN-γ (p 0.003), IL-2 (p 0.001) and IL-10 (p 0.042)], agr2 [TNFa (p < 0.001), IL-1b (p 0.008), IL-6 (p 0.019), IL-12p40 (p < 0.001), IFN-γ (p 0.001), IL-2 (p < 0.001) and IL-10 (p 0.003)] or agr1 types [TNFa (p 0.017), IL-1b (p 0.042), IL-6 (p 0.003), IL-12p40 (p < 0.001), IL-2 (p 0.002) and IL-10 (p 0.035)] (Figure 3).
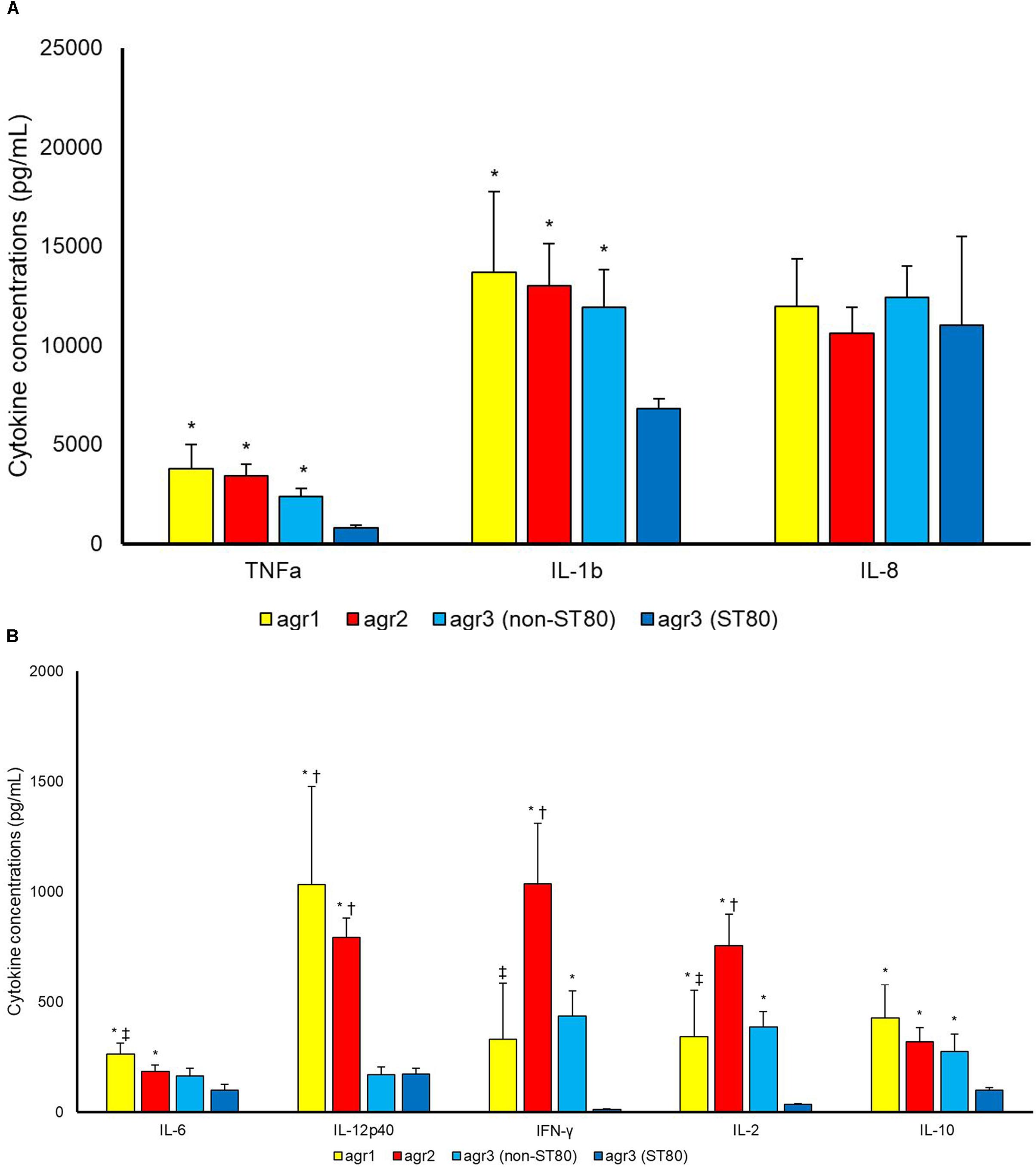
Figure 3. Cytokine concentrations (pg/mL) (A) TNF-a, IL-1b, IL-8; (B) IL6, IL12p40, IFN-γ, IL-2, IL-10 in supernatant upon incubation of PBMCs with S. aureus cells. Differences between agr1-positive, agr2-positive and agr3-positive (belonging to ST80 and to STs other than ST80) S. aureus were assessed. ∗P < 0.05 in comparison with agr3-positive belonging to ST80. †P < 0.05 in comparison with agr3-positive belonging to ST other than ST80. ‡P < 0.05 in comparison with agr2-positive.
Discussion
ST80 has been proven an extremely successful MRSA clone in the Mediterranean area (Drougka et al., 2014). In Greece it represents the predominant clone among CA-MRSA, accounting for more than 85% of the isolates, while since 2008 a gradual increase of healthcare-associated MRSA infections was caused by this clone, highlighting its capacity of dissemination in the community and the hospital setting (Witte, 2009; Drougka et al., 2014). This dissemination and capacity to colonize could indicate a propensity in evading host defenses.
CA-MRSA ability to colonize and infect healthy individuals and provoke severe infection is partially due to production of PVL. PVL triggers the production of IL-8 by neutrophils and of IL-1b by monocytes and macrophages (Perret et al., 2012). PVL and TSST-1 remain important for the recruitment of neutrophils having a clear proinflammatory cytokine response. (Yoong and Pier, 2012). In our study, there are conflicting results concerning PVL-positive strains. PVL-positive strains not belonging to ST80 clone had similar cytokine response as compared to PVL-negative strains, while PVL-positive strains belonging to ST80 showed a significantly lower cytokine production by PBMC as compared to the aforementioned groups.
TSST-1 is another important toxin belonging to the category of staphylococcal superantigens and is primarily responsible for toxic shock syndrome development, by triggering a massive release of proinflammatory cytokines producing an overwhelming inflammatory response (Nakagawa et al., 2005; Xu and McCormick, 2012). In a rabbit model of infective endocarditis and sepsis, comparing different lineages of S. aureus, even though no difference in lethality was observed among investigated clones, strains carrying tst gene caused lethal sepsis (King et al., 2016). In the present study, pro-inflammatory immune response of PBMC tst-positive MRSA isolates did not differ from those that did not carry tst gene.
Panton-Valentine Leukocidin, TSST-1 and other virulence factors’ production is regulated mainly by the agr system, which is also responsible for adaption of individual toxin production according to the phase of colonization and infection (Cheung et al., 2011; Le and Otto, 2015). We found that isolates carrying agr3, especially those belonging to ST80, induced lower levels of cytokine concentrations in monocyte supernatant as compared to those carrying agr1 and agr2. In murine models of S. aureus pneumonia and bacteraemia, deletion of agr resulted in better survival (Heyer et al., 2002; Bubeck Wardenburg et al., 2007), while the presence of agr2 resulted in higher mortality in MRSA bacteraemia (Cechinel et al., 2016). Our hypothesis is that ST80 agr3 induces lower virulence factor production, including PVL, resulting in lower pro-inflammatory cytokine induction, but more studies are needed to elucidate this correlation.
The onset and duration of S. aureus colonization is dependent of immune system response. A combined induction of multiple pro-inflammatory chemokines and cytokines, including IL-1β, IL-6, TNF-α, IFN-γ, IL-8, IL-12, is necessary to clear S. aureus from human nasal mucosa (Cole et al., 2016). The host’s failure to amount an elaborative combined response can lead to persistent colonization (Cole et al., 2016). As shown in the present study, strains belonging to ST80 induced significantly lower production of most of aforementioned pro-inflammatory mediators as compared to other STs (ST30, ST239, ST225), favoring colonization.
As it was previously shown, IFN-γ plays an important role in controlling staphylococcal colonization (Satorres et al., 2009). It enhances pro-inflammatory response of human mast cells in response to S. aureus by generating reactive oxygen species which leads to bacterial killing (Swindle et al., 2015). As macrophages are concerned, IFN-γ increases their phagocytic response and accelerates S. aureus killing (Greenlee-Wacker and Nauseef, 2017). Another role of IFN-γ is that confers resistance of keratinocytes to S. aureus alpha toxin induced cell death (Brauweiler et al., 2016). In the present study, ST80 induces a significantly lower IFN-γ production by monocytes as compared to ST30, ST239, and ST225, which can partially explain its ability to evade host immune response and promote chronic colonization (Prabhakara et al., 2011).
The present study has several limitations. First, we did not measure the level of tst and lukS/lukF-PV genes expression or agr activity. Secondly, mRNA expression levels of cytokines were not performed. Even though the strains included in the present study belonged to globally disseminated clones, other clones such as ST8 or ST1, which are the predominant in North America were not included (Witte, 2009). Moreover, even though the adhesin genes genetic background of the strains was determined, no adhesion activity of each MRSA clone to PBMCs was evaluated. The study is an in vitro one, so studies in murine models and in colonized or infected patients are needed to elucidate the immune response in vivo of different clones and especially ST80. We did not include isolates carrying the agr4 system since these isolates were rarely detected in our setting.
Conclusion
In conclusion, MRSA strains belonging to ST80 clone induce lower level of pro-inflammatory cytokines production by monocytes as compared to all other clones. This difference cannot be attributed neither to the presence of lukS/lukF-PV genes nor to agr3 system that are usually present in ST80. This low induction of immune response by MRSA ST80 probably leads to evasion of hosts’ immune defenses and can partially explain its silent and successful dissemination in the community and the hospital setting in Greece.
Author Contributions
IS, EA, and FK designed the experiments. FK, AS, and ED performed the experiments. FK, MP-O, and EJ collected the data. MP-O, FK, EJ, and IS performed the analysis. MP-O and FK wrote the manuscript. All authors contributed to manuscript revision, read and approved the submitted version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Part of this work was presented as a Poster at the 27th European Congress of Clinical Microbiology and Infectious Diseases, 22–25 April 2017, Vienna, Austria.
Abbreviations
Agr, accessory gene regulator; BSI, bloodstream infection; CA-MRSA, community-acquired methicillin-resistant S. aureus; CM, complete medium; IFN-γ, interferon-gamma; IL, interleukin; LPS, Lipopolysaccharide; MLST, Multi Locus Sequence Typing; PBMC, Peripheral blood mononuclear cell; PVL, Panton-Valentine Leukocidin; SSTI, skin and soft tissue infection; ST, sequence type; TNFa, Tumor Necrosis Factor alpha; TSST-1, Toxic Shock Syndrome Toxin-1.
Footnotes
References
Brauweiler, A. M., Goleva, E., and Leung, D. Y. (2016). Interferon-gamma protects from Staphylococcal alpha toxin-induced keratinocyte death through apolipoprotein L1. J. Invest. Dermatol. 136, 658–664. doi: 10.1016/j.jid.2015.12.006
Bubeck Wardenburg, J., Patel, R. J., and Schneewind, O. (2007). Surface proteins and exotoxins are required for the pathogenesis of Staphylococcus aureus pneumonia. Infect. Immun. 75, 1040–1044. doi: 10.1128/IAI.01313-06
Cechinel, A., Machado, D. P., Turra, E., Pereira, D., Dos Santos, R. P., Rosa, R. G., et al. (2016). Association between accessory gene regulator polymorphism and mortality among critically Ill patients receiving vancomycin for nosocomial MRSA bacteremia: a cohort study. Can. J. Infect. Dis. Med. Microbiol. 2016:8163456. doi: 10.1155/2016/8163456
Cheung, G. Y., Wang, R., Khan, B. A., Sturdevant, D. E., and Otto, M. (2011). Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect. Immun. 79, 1927–1935. doi: 10.1128/IAI.00046-11
Cole, A. L., Muthukrishnan, G., Chong, C., Beavis, A., Eade, C. R., Wood, M. P., et al. (2016). Host innate inflammatory factors and staphylococcal protein A influence the duration of human Staphylococcus aureus nasal carriage. Mucosal Immunol. 9, 1537–1548. doi: 10.1038/mi.2016.2
Drougka, E., Foka, A., Liakopoulos, A., Doudoulakakis, A., Jelastopulu, E., Chini, V., et al. (2014). A 12-year survey of methicillin-resistant Staphylococcus aureus infections in Greece: ST80-IV epidemic? Clin. Microbiol. Infect. 20, O796–O803. doi: 10.1111/1469-0691.12624
Greenlee-Wacker, M. C., and Nauseef, W. M. (2017). IFN-gamma targets macrophage-mediated immune responses toward Staphylococcus aureus. J. Leukoc. Biol. 101, 751–758. doi: 10.1189/jlb.4A1215-565RR
Heyer, G., Saba, S., Adamo, R., Rush, W., Soong, G., Cheung, A., et al. (2002). Staphylococcus aureus agr and sarA functions are required for invasive infection but not inflammatory responses in the lung. Infect. Immun. 70, 127–133. doi: 10.1128/IAI.70.1.127-133.2002
Jarraud, S., Mougel, C., Thioulouse, J., Lina, G., Meugnier, H., Forey, F., et al. (2002). Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 70, 631–641. doi: 10.1128/IAI.70.2.631-641.2002
King, J. M., Kulhankova, K., Stach, C. S., Vu, B. G., and Salgado-Pabon, W. (2016). Phenotypes and virulence among Staphylococcus aureus USA100, USA200, USA300, USA400, and USA600 clonal lineages. mSphere 1:e00071-16. doi: 10.1128/mSphere.00071-16
Kolonitsiou, F., Papadimitriou-Olivgeris, M., Spiliopoulou, A., Stamouli, V., Papakostas, V., Apostolopoulou, E., et al. (2017). Trends of bloodstream infections in a university Greek hospital during a three-year period: incidence of multidrug-resistant bacteria and seasonality in gram-negative predominance. Pol. J. Microbiol. 66, 171–180. doi: 10.5604/01.3001.0010.7834
Le, K. Y., and Otto, M. (2015). Quorum-sensing regulation in staphylococci-an overview. Front. Microbiol. 6:1174. doi: 10.3389/fmicb.2015.01174
McCarthy, A. J., and Lindsay, J. A. (2013). Staphylococcus aureus innate immune evasion is lineage-specific: a bioinfomatics study. Infect. Genet. Evol. 19, 7–14. doi: 10.1016/j.meegid.2013.06.012
Monecke, S., Slickers, P., Gawlik, D., Muller, E., Reissig, A., Ruppelt-Lorz, A., et al. (2018). Molecular typing of ST239-MRSA-III from diverse geographic locations and the evolution of the SCCmec III element during its intercontinental spread. Front. Microbiol. 9:1436. doi: 10.3389/fmicb.2018.01436
Nakagawa, S., Kushiya, K., Taneike, I., Imanishi, K., Uchiyama, T., and Yamamoto, T. (2005). Specific inhibitory action of anisodamine against a staphylococcal superantigenic toxin, toxic shock syndrome toxin 1 (TSST-1), leading to down-regulation of cytokine production and blocking of TSST-1 toxicity in mice. Clin. Diagn. Lab. Immunol. 12, 399–408. doi: 10.1128/CDLI.12.3.399-408.2005
Peacock, S. J., Moore, C. E., Justice, A., Kantzanou, M., Story, L., Mackie, K., et al. (2002). Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect. Immun. 70, 4987–4996. doi: 10.1128/IAI.70.9.4987-4996.2002
Perret, M., Badiou, C., Lina, G., Burbaud, S., Benito, Y., Bes, M., et al. (2012). Cross-talk between Staphylococcus aureus leukocidins-intoxicated macrophages and lung epithelial cells triggers chemokine secretion in an inflammasome-dependent manner. Cell. Microbiol. 14, 1019–1036. doi: 10.1111/j.1462-5822.2012.01772.x
Prabhakara, R., Harro, J. M., Leid, J. G., Keegan, A. D., Prior, M. L., and Shirtliff, M. E. (2011). Suppression of the inflammatory immune response prevents the development of chronic biofilm infection due to methicillin-resistant Staphylococcus aureus. Infect. Immun. 79, 5010–5018. doi: 10.1128/IAI.05571-11
Satorres, S. E., Alcaraz, L. E., Cargnelutti, E., and Di Genaro, M. S. (2009). IFN-gamma plays a detrimental role in murine defense against nasal colonization of Staphylococcus aureus. Immunol. Lett. 123, 185–188. doi: 10.1016/j.imlet.2009.03.003
Spiliopoulou, A. I., Kolonitsiou, F., Krevvata, M. I., Leontsinidis, M., Wilkinson, T. S., Mack, D., et al. (2012). Bacterial adhesion, intracellular survival and cytokine induction upon stimulation of mononuclear cells with planktonic or biofilm phase Staphylococcus epidermidis. FEMS Microbiol. Lett. 330, 56–65. doi: 10.1111/j.1574-6968.2012.02533.x
Swindle, E. J., Brown, J. M., Radinger, M., Deleo, F. R., and Metcalfe, D. D. (2015). Interferon-gamma enhances both the anti-bacterial and the pro-inflammatory response of human mast cells to Staphylococcus aureus. Immunology 146, 470–485. doi: 10.1111/imm.12524
Tristan, A., Ying, L., Bes, M., Etienne, J., Vandenesch, F., and Lina, F. (2003). Use of multiplex PCR to identify Staphylococcus aureus adhesins involved in human hematogenous infections. J. Clin. Microbiol. 41, 4465–4467. doi: 10.1128/JCM.41.9.4465-4467.2003
van Belkum, A., Melles, D. C., Nouwen, J., Van Leeuwen, W. B., Van Wamel, W., Vos, M. C., et al. (2009). Co-evolutionary aspects of human colonisation and infection by Staphylococcus aureus. Infect. Genet. Evol. 9, 32–47. doi: 10.1016/j.meegid.2008.09.012
von Eiff, C., Becker, K., Machka, K., Stammer, H., and Peters, G. (2001). Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group. N. Engl. J. Med. 344, 11–16. doi: 10.1056/NEJM200101043440102
Witte, W. (2009). Community-acquired methicillin-resistant Staphylococcus aureus: what do we need to know? Clin. Microbiol. Infect. 15(Suppl. 7), 17–25. doi: 10.1111/j.1469-0691.2009.03097.x
Xu, S. X., and McCormick, J. K. (2012). Staphylococcal superantigens in colonization and disease. Front. Cell. Infect. Microbiol. 2:52. doi: 10.3389/fcimb.2012.00052
Yoong, P., and Pier, G. B. (2012). Immune-activating properties of Panton-Valentine leukocidin improve the outcome in a model of methicillin-resistant Staphylococcus aureus pneumonia. Infect. Immun. 80, 2894–2904. doi: 10.1128/IAI.06360-11
Zhang, K., Sparling, J., Chow, B. L., Elsayed, S., Hussain, Z., Church, D. L., et al. (2004). New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J. Clin. Microbiol. 42, 4947–4955. doi: 10.1128/JCM.42.11.4947-4955.2004
Keywords: S. aureus, MLST, PVL, toxic shock syndrome toxin, accessory gene regulator (agr), methicillin-resistant S. aureus (MRSA)
Citation: Kolonitsiou F, Papadimitriou-Olivgeris M, Spiliopoulou A, Drougka E, Jelastopulu E, Anastassiou ED and Spiliopoulou I (2019) Methicillin-Resistant Staphylococcus aureus ST80 Induce Lower Cytokine Production by Monocytes as Compared to Other Sequence Types. Front. Microbiol. 9:3310. doi: 10.3389/fmicb.2018.03310
Received: 19 October 2018; Accepted: 19 December 2018;
Published: 09 January 2019.
Edited by:
Eugenia Bezirtzoglou, Democritus University of Thrace, GreeceReviewed by:
Shigeru Kamiya, Kyorin University, JapanEfthymia Petinaki, University of Thessaly, Greece
Copyright © 2019 Kolonitsiou, Papadimitriou-Olivgeris, Spiliopoulou, Drougka, Jelastopulu, Anastassiou and Spiliopoulou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matthaios Papadimitriou-Olivgeris, cGFwYWRpbWl0cmlvdW1hdEBnbWFpbC5jb20=
 Fevronia Kolonitsiou
Fevronia Kolonitsiou Matthaios Papadimitriou-Olivgeris
Matthaios Papadimitriou-Olivgeris Anastasia Spiliopoulou1
Anastasia Spiliopoulou1 Eleni Jelastopulu
Eleni Jelastopulu Iris Spiliopoulou
Iris Spiliopoulou