
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 07 January 2019
Sec. Virology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.03255
This article is part of the Research Topic Innovative Approaches to Virus Diagnosis and Control View all 27 articles
The term “virosphere” describes both the space where viruses are found and the space they influence, and can extend to their impact on the environment, highlighting the complexity of the interactions involved. Studying the biology of viruses and the etiology of virus disease is crucial to the prevention of viral disease, efficient and reliable virus diagnosis, and virus control. Electron microscopy (EM) is an essential tool in the detection and analysis of virus replication. New EM methods and ongoing technical improvements offer a broad spectrum of applications, allowing in-depth investigation of viral impact on not only the host but also the environment. Indeed, using the most up-to-date electron cryomicroscopy methods, such investigations are now close to atomic resolution. In combination with bioinformatics, the transition from 2D imaging to 3D remodeling allows structural and functional analyses that extend and augment our knowledge of the astonishing diversity in virus structure and lifestyle. In combination with confocal laser scanning microscopy, EM enables live imaging of cells and tissues with high-resolution analysis. Here, we describe the pivotal role played by EM in the study of viruses, from structural analysis to the biological relevance of the viral metagenome (virome).
Since the recognition of viruses as the causative agents of disease in the last decades of the nineteenth century (reviewed in Mettenleiter, 2017), scientists have striven to elucidate their structure (Figure 1). The high resolving power of electron microscopy (EM) permits studies at nanometer scale, providing direct images of viruses for diagnosis and research. The term “Anthropocene” indicates the enormous impact of humankind on geology and ecology (Crutzen, 2002), and this holds true also for the virosphere and its impact. EM explores and validates new concepts in virology in the Anthropocene age. This is underlined by the emergence of new taxonomic groups comprising giant viruses and “virophages” (Koonin et al., 2015) and the generation of new scientific terms such as virosphere, virome, synthetic virology, or bionic viruses (Suttle, 2007; Guenther et al., 2014; Koch et al., 2016).
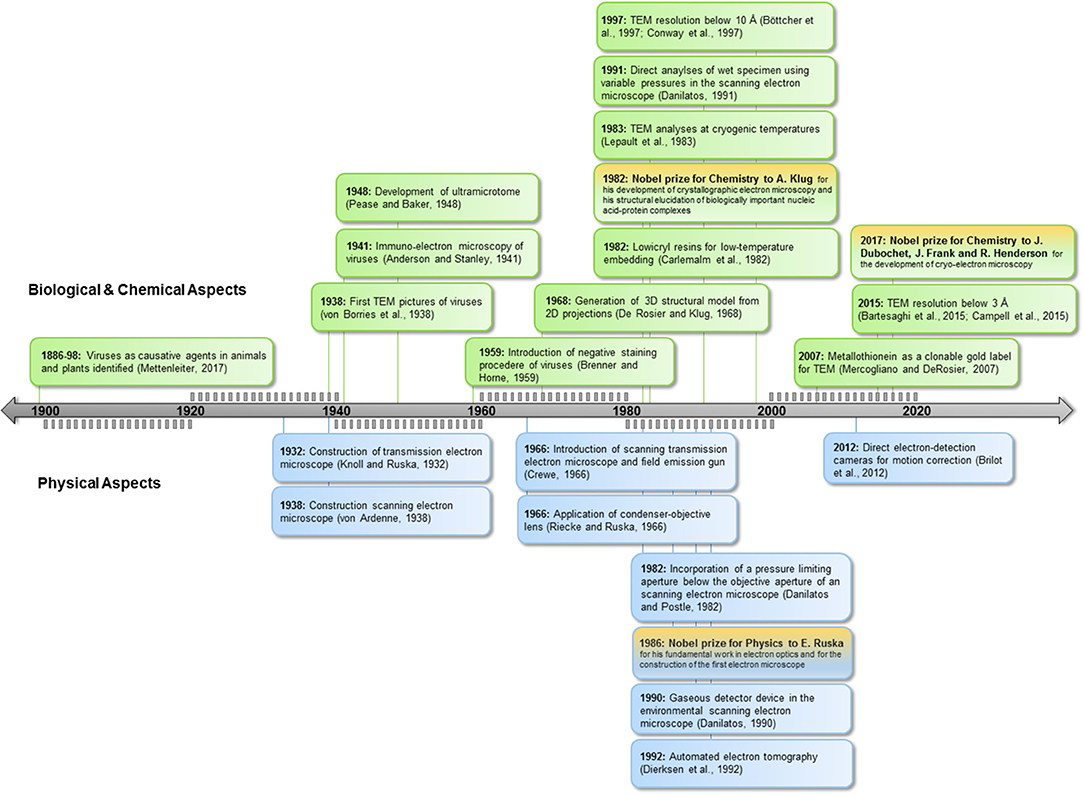
Figure 1. Milestones in EM. Below timeline: achievements in technology, above timeline: applications for biology. TEM, Transmission electron microscopy; SEM, scanning electron microcopy.
Humans act directly or indirectly as virus vectors or dead-end hosts. Climate change, global trade, and travel open virus highways, leading to high dynamics in virus spread and virus evolution. This challenges routine diagnostics based on ELISA or PCR technologies; the strength of EM lies in its ability to image the whole spectrum of interactions, including resistance and non-reactions in the case of new virus isolates or species. EM can determine functional features of viruses and underlying mechanisms of interactions relevant in nature as well as in synthetic virology (see Figures 1, 2).
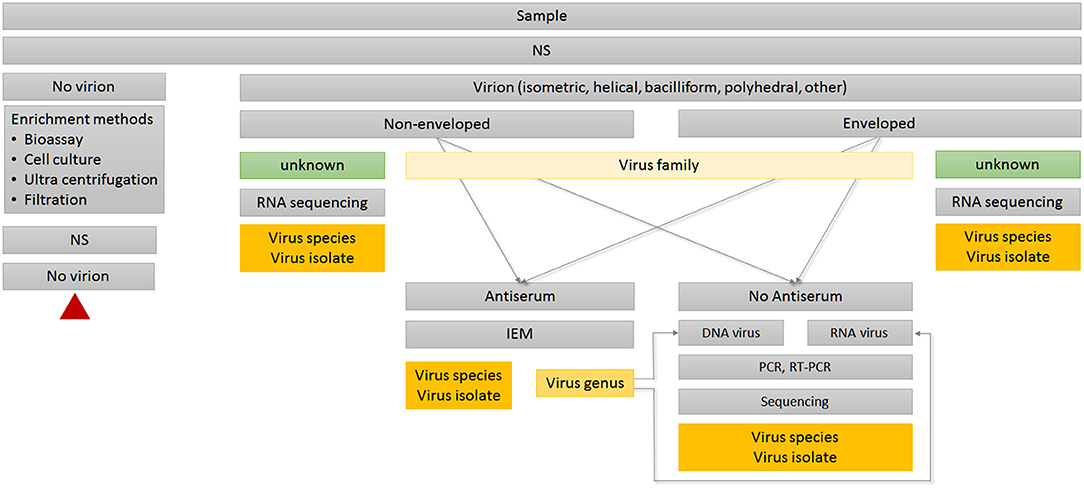
Figure 2. Decision tree for routine diagnosis using TEM. Red triangle indicates terminal node. IEM, Immuno-electron microscopy; NS, negative staining.
This review describes the versatility of EM as a universal means of virus detection, and describes its development from descriptive tool to the most powerful technique available to virologists today.
A change seen in host phenotype can be a first indication of viral presence, but further analysis is often needed to confirm a virus infection. In plants, viral symptoms can vary in mixed infections, in different environmental or growing conditions, and depending on species or cultivar.
If available, transmission electron microscopy (TEM) is a good initial step in virus diagnosis for several reasons (Huger, 1967, 1974; Koenig and Lesemann, 2001; Gentile and Gelderblom, 2014). The method of choice for direct detection—provided tissue preservation is not an issue—is tissue homogenization followed by negative staining (NS). NS has been established as a fast, robust and universal EM technique for over 60 years (Brenner and Horne, 1959) and still plays an essential role in this field (De Carlo and Harris, 2011). Such dip preparations work equally well on fresh, aged, partly degenerated, or dried specimens. NS can be applied universally to all biological tissues, organs, and cell cultures, as well as to soil and aquatic samples, e.g., from streams, irrigation, sewage outlets, lakes, oceans, etc.
The hallmark of TEM is its “open view” nature, since it provides an immediate overview of actual status, discerning amount and shape of virus(es) present, including the unexpected (Gentile and Gelderblom, 2014; Gelderblom and Madeley, 2018). As a first step in pathogen recognition, it requires only minute amounts of samples carrying high virus loads. TEM is unbiased against RNA or DNA genomes since it targets proteins, the viral capsid or ribonucleoprotein (RNP) complexes as input (King et al., 2012). In most cases, the observed morphology allows immediate preliminary classification to family level based on particle structure, size, and stability. Thus, TEM serves as a decisive tool to determine which of the available methods (e.g., bioassays, serological, or molecular biology approaches) should subsequently be used to further identify virus genus and species (Figure 2).
Immuno-electron microscopy (IEM) is based on the same serological principles as ELISA, and can be used for further virus identification during routine TEM diagnosis (Figure 2). IEM has the advantage that it works directly with raw serum, so no further purification of immunoglobulins or conjugation steps are required. Due to the small reaction volumes required, antibody consumption is low. Most TEM laboratories keep comprehensive collections of antisera specific to a broad spectrum of virus species and isolates. Antisera can be stored long term at 4°C in the presence of 0.05% sodium azide without significant loss of activity. Depending on the composition of antigens, as well as the epitopes present in the original virus purification used for immunization, polyclonal antisera can be heterogeneous in their reaction. Therefore, antisera can serve different objectives during routine diagnosis (Griffiths and Lucocq, 2014). Some antisera are suitable for capturing multiple viruses within a genus using IEM, whereas serological relationships will become visible through strength of antibody attachment in the decoration step. A homologous reaction displays the virion tightly packed with antibodies, whereas heterogonous viruses or emerging isolates will have only a weak antibody coating (Richert-Pöggeler et al., 2015). If available, monoclonal antibodies targeting single epitopes provide highest specificity and reproducibility for distinguishing different virus isolates (Griffiths and Lucocq, 2014).
Based on its ability to recognize contaminations, rapid application of TEM is essential for quality control of reference material or of virus preparations used for antibody production or directly for vaccination. Prompt production of reliable and comparable results is essential for efficient diagnostics during unexpected regional virus outbreaks or epidemics, making TEM an integral part of the protocols followed by national reference laboratories. Laue and Möller (2016) recently generated a publicly available database of EM images. Expansion of such archives will facilitate recognition of newly emerging as well as unexpected viruses. For instance, plant viruses can serve as indicators to predict contamination of irrigation water with pathogenic human viruses (Shrestha et al., 2018).
Due to the potentially serious consequences of viral outbreaks, rigorous, and universally recognized training of personnel dealing with virus identification is crucial.
Training in Diagnostic Electron Microscopy (DEM) of human and veterinary infectious diseases can be obtained in an annual External Quality Assurance (EQA) scheme prepared by the Robert Koch Institute.
Comprehensive studies of virus biology and host responses require a combination of molecular biology, biochemical analyses and EM (Ni et al., 2014; Klupp et al., 2017; Garcia-Ruiz et al., 2018). The simultaneous discovery of viruses as novel infectious entities in both the plant and animal kingdom (Figure 1) revealed that viruses follow the same basic principles, modified according to host cell and environment (Ahlquist, 2006; Richert-Pöggeler and Minarovits, 2014). Various input materials can be used for functional analysis with distinct objectives. Homogenized tissues with high virus concentrations, as well as purified virus preparations, are suitable for determination of particle size. If suitable antibodies are available, low virus titer in the original material can be enriched by pre-incubation of the grid. For unknown viruses, data gained from virion measurements and morphology provides information on genome organization and genome length as well as particle stability. This is also applicable to validation of artificially generated virus genomes, infectious full-length clones or derived virus mutants (Laufer et al., 2018). Particle length distribution discloses valuable data on the multi-component nature of a virus and the encapsidation of subgenomic or satellite sequences (Lin and Hsu, 1994; King et al., 2012; Ni et al., 2014). IEM was seminal in demonstrating the bipolar structure of some helical viruses (Torrance et al., 2006; Menzel et al., 2009).
When using embedded material, EM facilitates single cell analyses as well as direct comparison of adjacent cells from distinct tissues, e.g., leaf parenchyma with vasculature (Palani et al., 2009). Modified organelles, membranes as well as generated structures harboring viral replication complexes can be correlated with the infection process (Fernández de Castro et al., 2013; Tilsner et al., 2013; Gómez-Aix et al., 2015). The ultrastructural localization, function and interaction of viral proteins as well as dsRNA molecules with the host have been investigated in artificial expression systems (Kleinow et al., 2009; Tilsner et al., 2013; Kovalev et al., 2016). These methods were also applied to study systemic spread in the host including intracellular and intercellular movement with cell-to-cell movement and long distance movement. Recent studies have revealed the function of host susceptibility factors comprising membrane-acting ESCRT and SNARE proteins during replication of tombusviruses and production of budded baculovirions, respectively (Kovalev et al., 2016; Guo et al., 2017; Garcia-Ruiz et al., 2018). A process akin to autophagy was shown to be beneficial for replication of coccolithoviruses in algae (Schatz et al., 2014).
Immunogold labeling enhances imaging of antibody binding (Palani et al., 2009; Pacesa et al., 2017); furthermore, as well as facilitating localization and quantification, it enables multiple labeling on the same object by using gold particles of different size (Palani et al., 2009; Griffiths and Lucocq, 2014; Erokhina et al., 2017). In metal-tagging TEM (METTEM) viral proteins are fused with metallothionein, which, after incubation with gold salts, leads to the production of electron-dense gold nanoclusters (Fernández de Castro et al., 2014). This technology has been used in combination with immunogold labeling and for three-dimensional (3D) imaging of the interaction of viral replicase with viral RNA (Kovalev et al., 2016; Fernández de Castro et al., 2017).
Accuracy in virus quantification can be improved by using a scanning transmission electron microscopy detector (STEM, Hartel et al., 1996) in a scanning electron microscope (SEM) (Blancett et al., 2017).
Combined use of TEM and SEM improves characterization of larger objects like baculovirus occlusion bodies (Gencer et al., 2018). TEM enables high resolution of virion structure, localization, and measurement of particles and ultrastructural details within embedded occlusion body preparations. On the other hand, SEM analyses are ideal for high throughput screening of samples, quality control of preparations, and measurements for comparison of different isolates (Gencer et al., 2018).
Incorporation of pressure-limiting apertures and gaseous detection devices allows direct investigation of hydrated biological samples using SEM (Figure 1). Variable pressure can be adjusted to object sensitivity (Griffin, 2007). This technique enables high-throughput screening of material for virus-transmitting arthropod vectors. Furthermore, live imaging of developmental stages and vector interaction with the host surface is now possible.
The use of back scattered electron detectors in field emission SEM permits an enlarged field of view. Thus, large cellular volumes embedded in resin sections can be visualized at high resolution (Rizzo et al., 2016).
SEM works well in direct combination with light microscopy, and datasets for 3D reconstruction can be obtained easily (Rizzo et al., 2016; Clarke and Royle, 2018). The correlation of light and electron microscopy (CLEM) combines the advantages of both methods—the ability to simultaneously locate the target in a comparatively large volume and determine its ultrastructure (Fernández de Castro et al., 2014; Madela et al., 2014; Fonta and Humbel, 2015; Rizzo et al., 2016). With the increasing speed of developments in the field of microscopy, CLEM offers a broad spectrum of applications depending on the specific question. Good knowledge of viral replication is mandatory for designing antiviral strategies and therapies. Here, by employing fluorescent-tagged molecules, CLEM can be very helpful in finding cells of interest within layers of tissue from living samples or derived cultures (Fonta and Humbel, 2015; Hellström et al., 2015).
The introduction of direct electron detectors (DEDs) and advances in image processing have extended the resolution limit of electron cryo-microscopy (cryo-EM) into the atomic range (Kühlbrandt, 2014), allowing ab initio atomic model building. Cryo-EM is ideally suited to exploring the 3D structure of macromolecular assemblies, and elucidation of the 3D arrangement of such complexes helps understand their function in living cells. These technological developments have always involved analyses of viruses, particularly plant viruses, because their symmetrical capsids, as well as the availability of highly pure samples, greatly facilitates reconstruction. Tobacco mosaic virus (TMV)—one of the very first objects to be seen in an electron microscope (Kausche et al., 1939)—has been used to evaluate 3D reconstructions from data recorded on different DEDs (Fromm et al., 2015), illustrating improvements in resolution into the 3 Å range compared to the 4–5 Å obtained with CCD cameras (Clare and Orlova, 2010) under optimal conditions.
Encapsidation of the viral genome is an essential step of virus particle assembly and, more generally, of the viral life cycle. Cryo-EM now paves the way to elucidating mechanisms of capsid assembly and genome encapsidation, and to understanding the mechanisms that ensure only the viral genome is specifically packaged from among a background of myriad host DNAs/RNAs. Cowpea mosaic virus (CPMV)—a positive-sense, single-stranded RNA plant virus—and other members of the order Picornavirales have been investigated intensively in recent decades. Very recently, high-resolution cryo-EM structures of wild type and empty virus-like particles have been determined, implicating the C-terminal region of the small coat protein (CP) subunit as being required for virus assembly (Hesketh et al., 2015). The wild-type structure reveals the dense nature of the RNA inside the capsid shell, with an arrangement suggesting extensive base-pairing during encapsidation. The resolution was high enough to identify amino acid side-chains of the CP that interact directly with the encapsidated RNA. The circular single-stranded DNA genomes of geminiviruses—major plant pathogens in crop plants worldwide—are encapsidated in characteristic D5-symmetric twin particles formed by two incomplete icosahedra. Some years ago, the first cryo-EM structures of geminiviruses [one a mastrevirus (Zhang et al., 2001), the other a begomovirus (Böttcher et al., 2004)], revealed details of the structure of these unique particles, which have eluded crystallography until now. With recent advances in cryo-EM, high-resolution structures now reveal the fine detail of the organization of the single capsid protein in the particle, revealing the important role played by the N-terminus of the protein in different positions (Hipp et al., 2017; Hesketh et al., 2018). Together with atomic models of the capsid proteins, these new cryo-EM maps provide the first clues as to how the protein–genomic DNA interactions and assembly of these unique particles might occur.
Advances in cryo-EM have revealed near-atomic structures of rod-shaped and flexible filamentous plant viruses. In contrast to the right-handed helical organization of the CPs of rod-shaped Tobamovirus (Fromm et al., 2015) and Hordeivirus (Clare et al., 2015), the particles of Potexviruses (Agirrezabala et al., 2015; DiMaio et al., 2015) and a Potyvirus (Zamora et al., 2017) are arranged in left-handed helices. Despite low sequence identity, the CPs of these flexible filamentous viruses share a common fold and a conserved RNA binding site (Valle, 2018). The CP structures also facilitated the identification of nucleoproteins from segmented negative-strand RNA viruses as structural homologs (Agirrezabala et al., 2015; Zamora et al., 2017).
Apart from deciphering key aspects of genome encapsidation and assembly of virus particles, cryo-EM may also facilitate the development of plant virus-like particles for use in biomedical and nanotechnology applications. Such virus-like particles could accommodate foreign material or can be chemically modified for coupling targets while still retaining the ability to assemble efficiently into particles (Koch et al., 2016; Meshcheriakova et al., 2017).
Besides cryo-EM of single particles, cryo-electron tomography has facilitated a major leap in our understanding of viral infection, revealing the structure and components involved in virus replication (Ertel et al., 2017). Further 3D imaging technologies, such as 3D reconstructions of serial sections and focused ion beam scanning electron microscopy (FIB/SEM) will also help explore aspects of the viral life cycle (Risco et al., 2014; Villinger et al., 2015) but are beyond the scope of this review.
The latest master species list (MSL32) assembled by the International Committee on Taxonomy of Viruses (ICTV) classifies 4853 virus species covering all host phyla. The NCBI database records 7512 sequenced viral genomes from all kingdoms. Viral abundance extrapolated from studies on the virosphere is estimated at 1031–1032 (Perales et al., 2015). However, <10% of sequences obtained from metagenomic surveys showed homology to GenBank accessions (Suttle, 2007). Comparing the sheer numbers of what is already classified with as yet uncharacterized viruses predicts the future demand for EM and its manifold applications in diagnosis, functional analysis, and high resolution characterization.
High-resolution EM in combination with generation of mutants of infectious viruses provides a powerful tool for the detection and study of structural aberrations and their impact on virus replication and evolution. The biological relevance of the coexistence of isometric and bacilliform particles, as occurs in the family Caulimoviridae, representing dsDNA viruses, or Bromoviridae comprising multipartite positive ssRNA viruses, is still unknown. In the case of filoviruses—enveloped negative sense ssRNA viruses—the three different shapes and virion lengths observed have been assigned to different numbers of encapsidated viral genomes (Booth et al., 2013). Such polyploidy, accompanied by elongated particles, has also been described for one member of the Caulimoviridae (Geijskes et al., 2004), and awaits further functional analysis.
The ubiquitous nature, high mobility and genetic versatility of viruses makes them ideal for mediating horizontal DNA transfer. As demonstrated in recent years, a combination of molecular, next-generation sequencing and EM technologies has shown that viruses can encapsidate host nucleic acids corresponding to their genome composition (Ghoshal et al., 2015). It will be interesting to see to what extent such hetero-encapsidation promotes the crossing of kingdom borders by viruses (Balique et al., 2015). High resolution EM will be essential for risk evaluation with regard to human health of artificially designed spheres or rods for use in synthetic virology or nanotechnology.
International efforts by virologists have already established platforms for direct communication and scientific exchange (Gould et al., 2012; Roenhorst et al., 2017). The networks generated coordinate dissemination of viruses and material, and define standards for establishing and maintaining virus collections as well as data archiving. Providing bioinformatic tools for database security and data management seems essential for efficient application of this new technology in virus diagnostics and control. These global networks have already proven successful in pathogen diagnoses and virus epidemiology (Romette et al., 2018). Strengthening cooperation between virologists from different fields to fully exploit technical expertise and in-depth knowledge of virus hosts will be necessary to tackle future challenges posed by the high dynamics of the virosphere.
KR-P, KF, KH, and RK contributed to writing and editing the manuscript including figures. KR-P wrote sections for introduction and outlook. KR-P, KF, and RK wrote the sections on routine diagnosis and functional virology and KH described cryo EM. All authors approved the version to be published.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Sylke Weißenberg for her expertise and help in figure design and formatting. We are grateful to Helen Rothnie for language editing.
Agirrezabala, X., Méndez-López, E., Lasso, G., Sánchez-Pina, M. A., Aranda, M., and Valle, M. (2015). The near-atomic cryoEM structure of a flexible filamentous plant virus shows homology of its coat protein with nucleoproteins of animal viruses. Elife 4:e11795. doi: 10.7554/eLife.11795
Ahlquist, P. (2006). Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat. Rev. Microbiol. 4, 371–382. doi: 10.1038/nrmicro1389
Anderson, T. F., and Stanley, W. M. (1941). A study by means of the electron microscope of the reaction between tobacco mosaic virus and its antiserum. J. Biol. Chem. 139, 339–344.
Balique, F., Lecoq, H., Raoult, D., and Colson, P. (2015). Can plant viruses cross the kingdom border and be pathogenic to humans? Viruses 7, 2074–2098. doi: 10.3390/v7042074
Bartesaghi, A., Merk, A., Banerjee, S., Matthies, D., Wu, X., Milne, J. L. S., et al. (2015). 2.2 Å resolution cryo-EM structure of β-galactosidase in complex with a cell-permeant inhibitor. Science 348, 1147–1151. doi: 10.1126/science.aab1576
Blancett, C. D., Fetterer, D. P., Koistinen, K. A., Morazzani, E. M., Monninger, M. K., Piper, A. E., et al. (2017). Accurate virus quantitation using a Scanning Transmission Electron Microscopy (STEM) detector in a scanning electron microscope. J. Virol. Methods 248, 136–144. doi: 10.1016/j.jviromet.2017.06.014
Booth, T. F., Rabb, M. J., and Beniac, D. R. (2013). How do filovirus filaments bend without breaking? Trends Microbiol. 21, 583–593. doi: 10.1016/j.tim.2013.08.001
Böttcher, B., Unseld, S., Ceulemans, H., Russell, R. B., and Jeske, H. (2004). Geminate structures of African cassava mosaic virus. J. Virol. 78, 6758–6765. doi: 10.1128/jvi.78.13.6758-6765.2004
Böttcher, B., Wynne, S. A., and Crowther, R. A. (1997). Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature 386, 88–91. doi: 10.1038/386088a0
Brenner, S., and Horne, R. W. (1959). A negative staining method for high resolution electron microscopy of viruses. Biochim. Biophys. Acta 34, 103–110. doi: 10.1016/0006-3002(59)90237-9
Brilot, A. F., Chen, J. Z., Cheng, A., Pan, J., Harrison, S. C., Potter, C. S., et al. (2012). Beam-induced motion of vitrified specimen on holey carbon film. J. Struct. Biol. 177, 630–637. doi: 10.1016/j.jsb.2012.02.003
Campbell, M. G., Veesler, D., Cheng, A., Potter, C. S., and Carragher, B. (2015). 2.8 Å resolution reconstruction of the Thermoplasma acidophilum 20S proteasome using cryo-electron microscopy. Elife 4:e06380. doi: 10.7554/eLife.06380
Carlemalm, E., Garavito, R. M., and Villiger, W. (1982). Resin development for electron microscopy and an analysis of embedding at low temperature. J. Microscopy 126, 123–143. doi: 10.1111/j.1365-2818.1982.tb00362.x
Clare, D. K., and Orlova, E. V. (2010). 4.6 Å Cryo-EM reconstruction of tobacco mosaic virus from images recorded at 300 keV on a 4k x 4k CCD camera. J. Struct. Biol. 171, 303–308. doi: 10.1016/j.jsb.2010.06.011
Clare, D. K., Pechnikova, E. V., Skurat, E. V., Makarov, V. V., Sokolova, O. S., Solovyev, A. G., et al. (2015). Novel inter-subunit contacts in barley stripe mosaic virus revealed by cryo-electron microscopy. Structure 23, 1815–1826. doi: 10.1016/j.str.2015.06.028
Clarke, N. I., and Royle, S. J. (2018). Correlating light microscopy with serial block face scanning electron microscopy to study mitotic spindle architecture. Methods Cell Biol.145, 29–43. doi: 10.1016/bs.mcb.2018.03.010
Conway, J. F., Cheng, N., Zlotnick, A., Wingfield, P. T., Stahl, S. J., and Steven, A. C. (1997). Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature 386, 91–94. doi: 10.1038/386091a0
Crewe, A. V. (1966). Scanning electron microscopes: is high resolution possible? Science 154, 729–738. doi: 10.1126/science.154.3750.729
Danilatos, G. D. (1990). Theory of gaseous detector device in the environmental scanning electron microscope. Adv. Electronics Electron. Phys. 71, 1–102. doi: 10.1016/S0065-2539(08)60388-1
Danilatos, G. D. (1991). Review and outline of environmental SEM at present. J. Microsc. 162, 391–402. doi: 10.1111/j.1365-2818.1991.tb03149.x
Danilatos, G. D., and Postle, R. (1982). The environmental scanning electron microscope and its applications. Scan. Electron. Microsc. 1982 (Pt 1), 1–16.
De Carlo, S., and Harris, J. R. (2011). Negative staining and cryo-negative staining of macromolecules and viruses for TEM. Micron 42, 117–131. doi: 10.1016/j.micron.2010.06.003
De Rosier, D. J., and Klug, A. (1968). Reconstruction of three dimensional structures from electron micrographs. Nature 217, 130–134. doi: 10.1038/217130a0
Dierksen, K., Typke, D., Hegerl, R., Koster, A. J., and Baumeister, W. (1992). Towards automatic electron tomography. Ultramicroscopy 40, 71–87. doi: 10.1016/0304-3991(92)90235-C
DiMaio, F., Chen, C. C., Yu, X., Frenz, B., Hsu, Y. H., Lin, N. S., et al. (2015). The molecular basis for flexibility in the flexible filamentous plantviruses. Nat. Struct. Mol. Biol. 22, 642–644. doi: 10.1038/nsmb.3054
Erokhina, T. N., Lazareva, E. A., Richert-Poeggeler, K. R., Sheval, E. V., Solovyev, A. G., and Morozov, S. Y. (2017). Subcellular localization and detection of tobacco mosaic virus ORF6 protein by immunoelectron microscopy. Biochemistry 82, 60–66. doi: 10.1134/s0006297917010060
Ertel, K. J., Benefield, D., Castano-Diez, D., Pennington, J. G., Horswill, M., den Boon, J. A., et al. (2017). Cryo-electron tomography reveals novel features of a viral RNA replication compartment. Elife 6:25940. doi: 10.7554/eLife.25940
Fernández de Castro, I., Fernández, J. J., Barajas, D., Nagy, P. D., and Risco, C. (2017). Three-dimensional imaging of the cellular assembly of a functional viral RNA replicase complex. J. Cell Sci. 130, 260–268. doi: 10.1242/jcs.181586
Fernández de Castro, I., Sanz-Sánchez, L., and Risco, C. (2014). Metallothioneins for correlative light and electron microscopy. Methods Cell Biol. 124, 55–70. doi: 10.1016/B978-0-12-801075-4.00003-3
Fernández de Castro, I., Volonté, L., and Risco, C. (2013). Virus factories: biogenesis and structural design. Cell. Microbiol. 15, 24–34. doi: 10.1111/cmi.12029
Fonta, C. L., and Humbel, B. M. (2015). Correlative microscopy. Arch. Biochem. Biophys. 581, 98–110. doi: 10.1016/j.abb.2015.05.017
Fromm, S. A., Bharat, T. A., Jakobi, A. J., Hagen, W. J., and Sachse, C. (2015). Seeing tobacco mosaic virus through direct electron detectors. J. Struct. Biol. 189, 87–97. doi: 10.1016/j.jsb.2014.12.002
Garcia-Ruiz, H., Diaz, A., and Ahlquist, P. (2018). Intermolecular RNA recombination occurs at different frequencies in alternate forms of brome omsaic virus RNA replication compartments. Viruses 10:3. doi: 10.3390/v10030131
Geijskes, R. J., Braithwaite, K. S., Smith, G. R., Dale, J. L., and Harding, R. M. (2004). Sugarcane bacilliform virus encapsidates genome concatamers and does not appear to integrate into the Saccharum officinarum genome. Arch. Virol. 149, 791–798. doi: 10.1007/s00705-003-0260-0
Gelderblom, H. R., and Madeley, D. (2018). Rapid viral diagnosis of Orthopoxviruses by electron microscopy: optional or a must? Viruses 10:142. doi: 10.3390/v10040142
Gencer, D., Bayramoglu, Z., Nalcacioglu, R., Kleespies, R. G., Demirbag, Z., and Demir, I. (2018). Characterisation of three Alphabaculovirus isolates from the gypsy moth, Lymantria dispar dispar (Lepidoptera: Erebidae), in Turkey. Biocontr. Sci. Technol. 28, 107–121. doi: 10.1080/09583157.2017.1420752
Gentile, M., and Gelderblom, H. R. (2014). Electron microscopy in rapid viral diagnosis: an update. New Microbiol. 37, 403–422.
Ghoshal, K., Theilmann, J., Reade, R., Maghodia, A., and Rochon, D. (2015). Encapsidation of host RNAs by cucumber necrosis virus coat protein during both agroinfiltration and infection. J. Virol. 89, 10748–10761. doi: 10.1128/jvi.01466-15
Gómez-Aix, C., García-García, M., Aranda, M. A., and Sánchez-Pina, M. A. (2015). Melon necrotic spot virus Replication Occurs in Association with Altered Mitochondria. Mol. Plant-Microbe Interact. 28, 387–397. doi: 10.1094/MPMI-09-14-0274-R
Gould, E. A., de Lamballerie, X., Coutard, B., Fooks, A. R., Outlaw, M., Drosten, C., et al. (2012). The European Virus Archive: a new resource for virology research. Antiviral Res. 95, 167–171. doi: 10.1016/j.antiviral.2012.05.005
Griffin, B. J. (2007). Variable pressure and environmental scanning electron microscopy: imaging of biological samples. Methods Mol. Biol. 369, 467–495. doi: 10.1007/978-1-59745-294-6_23
Griffiths, G., and Lucocq, J. M. (2014). Antibodies for immunolabeling by light and electron microscopy: not for the faint hearted. Histochem. Cell Biol. 142, 347–360. doi: 10.1007/s00418-014-1263-5
Guenther, C. M., Kuypers, B. E., Lam, M. T., Robinson, T. M., Zhao, J., and Suh, J. (2014). Synthetic virology: engineering viruses for gene delivery. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 6, 548–558. doi: 10.1002/wnan.1287
Guo, Y., Yue, Q., Gao, J., Wang, Z., Chen, Y. R., Blissard, G. W., et al. (2017). Roles of cellular NSF protein in entry and nuclear egress of budded virions of Autographa californica Multiple Nucleopolyhedrovirus. J. Virol. 91, 1–28. doi: 10.1128/JVI.01111-17
Hartel, P., Rose, H., and Dinges, C. (1996). Conditions and reasons for incoherent imaging in STEM. Ultramicroscopy 63, 93–114. doi: 10.1016/0304-3991(96)00020-4
Hellström, K., Vihinen, H., Kallio, K., Jokitalo, E., and Ahola, T. (2015). Correlative light and electron microscopy enables viral replication studies at the ultrastructural level. Methods 90, 49–56. doi: 10.1016/j.ymeth.2015.04.019
Hesketh, E. L., Meshcheriakova, Y., Dent, K. C., Saxena, P., Thompson, R. F., Cockburn, J. J., et al. (2015). Mechanisms of assembly and genome packaging in an RNA virus revealed by high-resolution cryo-EM. Nat. Commun. 6:10113. doi: 10.1038/ncomms10113
Hesketh, E. L., Saunders, K., Fisher, C., Potze, J., Stanley, J., Lomonossoff, G. P., et al. (2018). The 3.3Å structure of a plant geminivirus using cryo-EM. Nat. Commun. 9:2369. doi: 10.1038/s41467-018-04793-6
Hipp, K., Grimm, C., Jeske, H., and Böttcher, B. (2017). Near-atomic resolution structure of a plant Geminivirus determined by electron cryomicroscopy. Structure 25, 1303–1309.e1303. doi: 10.1016/j.str.2017.06.013
Huger, A. M. (1967). “Elektronenmikroskopie bei der Diagnose von Insektenkrankheiten,” in Proc. Intern. Colloq. “Insect Pathology and Microbial Control”, ed F. A. van der Laan (North Holland Publ. Co., Amsterdam), 29–53.
Huger, A. M. (1974). Methoden und Bedeutung der Diagnose von Insektenkrankheiten/ Methods and importance of diagnosis of insect diseases. J. Plant Dis. Protect. 372–388.
Kausche, G. A., Pfankuch, E., and Ruska, H. (1939). Die Sichtbarmachung von pflanzlichem Virus im Übermikroskop. Naturwissenschaften 27, 292–299. doi: 10.1007/bf01493353
King, A. M. Q., Lefkowitz, E., Adams, M. J., and Carstens, E. B. (2012). Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier.
Kleinow, T., Tanwir, F., Kocher, C., Krenz, B., Wege, C., and Jeske, H. (2009). Expression dynamics and ultrastructural localization of epitope-tagged Abutilon mosaic virus nuclear shuttle and movement proteins in Nicotiana benthamiana cells. Virology 391, 212–220. doi: 10.1016/j.virol.2009.06.042
Klupp, B. G., Hellberg, T., Granzow, H., Franzke, K., Dominguez Gonzalez, B., Goodchild, R. E., et al. (2017). Integrity of the linker of nucleoskeleton and cytoskeleton is required for efficient herpesvirus nuclear egress. J. Virol. 91:19. doi: 10.1128/jvi.00330-17
Knoll, M., and Ruska, E. (1932). Beitrag zur geometrischen Elektronenoptik, I und II. Annalen der Physik. 12, 607–661. doi: 10.1002/andp.19324040506
Koch, C., Eber, F. J., Azucena, C., Forste, A., Walheim, S., Schimmel, T., et al. (2016). Novel roles for well-known players: from tobacco mosaic virus pests to enzymatically active assemblies. Beilstein J. Nanotechnol. 7, 613–629. doi: 10.3762/bjnano.7.54
Koenig, R., and Lesemann, D.-E. (2001). Plant Virus Identification. Encyclopedia of Life Sciences. Chichester: John Wiley and Sons, Ltd.
Koonin, E. V., Dolja, V. V., and Krupovic, M. (2015). Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology 479–480, 2–25. doi: 10.1016/j.virol.2015.02.039
Kovalev, N., Fernández de Castro Martin, I., Pogany, J., Barajas, D., Pathak, K., Risco, C., et al. (2016). Role of viral RNA and Co-opted cellular ESCRT-I and ESCRT-III factors in formation of tombusvirus spherules harboring the tombusvirus replicase. J. Virol. 90, 3611–3626. doi: 10.1128/JVI.02775-15
Kühlbrandt, W. (2014). Biochemistry. the resolution revolution. Science 343, 1443–1444. doi: 10.1126/science.1251652
Laue, M., and Möller, L. (2016). The virusexplorer DEM - a database for diagnostic electron microscopy of viruses. Zenodo. doi: 10.5281/zenodo.221524
Laufer, M., Mohammad, H., Maiss, E., Richert-Pöggeler, K., Dall'Ara, M., Ratti, C., et al. (2018). Biological properties of Beet soil-borne mosaic virus and Beet necrotic yellow vein virus cDNA clones produced by isothermal in vitro recombination: insights for reassortant appearance. Virology 518, 25–33. doi: 10.1016/j.virol.2018.01.029
Lepault, J., Booy, F. P., and Dubochet, J. (1983). Electron microscopy of frozen hydrated biological suspensions. J. Microsc. 129, 89–102. doi: 10.1111/j.1365-2818.1983.tb04163.x
Lin, N. S., and Hsu, Y. H. (1994). A satellite RNA associated with bamboo mosaic potexvirus. Virology 202, 707–714. doi: 10.1006/viro.1994.1392
Madela, K., Banhart, S., Zimmermann, A., Piesker, J., Bannert, N., and Laue, M. (2014). “Chapter 5 - A simple procedure to analyze positions of interest in infectious cell cultures by correlative light and electron microscopy,” in Methods in Cell Biology, eds T. Müller-Reichert and P. Verkade (Amsterdam: Elsevier), 93–110. doi: 10.1016/B978-0-12-801075-4.00005-7
Menzel, W., Goetz, R., Lesemann, D. E., and Vetten, H. J. (2009). Molecular characterization of a closterovirus from carrot and its identification as a German isolate of Carrot yellow leaf virus. Arch. Virol. 154, 1343–1347. doi: 10.1007/s00705-009-0428-3
Mercogliano, C. P., and DeRosier, D. J. (2007). Concatenated metallothionein as a clonable gold label for electron microscopy. J. Struct. Biol. 160, 70–82. doi: 10.1016/j.jsb.2007.06.010
Meshcheriakova, Y., Durrant, A., Hesketh, E. L., Ranson, N. A., and Lomonossoff, G. P. (2017). Combining high-resolution cryo-electron microscopy and mutagenesis to develop cowpea mosaic virus for bionanotechnology. Biochem. Soc. Trans. 45, 1263–1269. doi: 10.1042/BST20160312
Mettenleiter, T. C. (2017). “The first virus hunters,” in Advances in Virus Research, Vol. 99, eds M. Beer and D. Höper (Burlington: Academic Press), 1–16. doi: 10.1016/bs.aivir.2017.07.005
Ni, P., Vaughan, R. C., Tragesser, B., Hoover, H., and Kao, C. C. (2014). The plant host can affect the encapsidation of brome mosaic virus (BMV) RNA: BMV virions are surprisingly heterogeneous. J. Mol. Biol. 426, 1061–1076. doi: 10.1016/j.jmb.2013.09.007
Pacesa, M., Hendrickx, R., Bieri, M., Flatt, J. W., Greber, U. F., and Hemmi, S. (2017). Small-size recombinant adenoviral hexon protein fragments for the production of virus-type specific antibodies. Virol. J. 14:158. doi: 10.1186/s12985-017-0822-5
Palani, P. V., Chiu, M., Chen, W., Wang, C. C., Lin, C. C., Hsu, C. C., et al. (2009). Subcellular localization and expression of bamboo mosaic virus satellite RNA-encoded protein. J. Gen. Virol. 90, 507–518. doi: 10.1099/vir.0.004994-0
Pease, D. C., and Baker, R. F. (1948). Sectioning techniques for electron microscopy using a conventional microtome. Proc. Soc. Exp. Biol. Med. 67, 470–474.
Perales, C., Moreno, E., and Domingo, E. (2015). Clonality and intracellular polyploidy in virus evolution and pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 112, 8887–8892. doi: 10.1073/pnas.1501715112
Richert-Pöggeler, K. R., and Minarovits, J. (2014). “Diversity of latent plant-virus interactions and their impact on the virosphere,” in Plant Virus-Host Interaction. Molecular Approaches and Viral Evolution, eds R. K. Gaur, T. Hohn, and P. Sharma (Elsevier Inc.), 263–275. doi: 10.1016/B978-0-12-411584-2.00014-7
Richert-Pöggeler, K. R., Turhal, A. K., Schuhmann, S., Maaß, C., Blockus, S., Zimmermann, E., et al. (2015). Carlavirus Biodiversity in Horticultural Host Plants: Efficient Virus Detection and Identification Combining Electron Microscopy and Molecular Biology Tools. Leuven, Belgium: International Society for Horticultural Science (ISHS), 37–45. doi: 10.17660/ActaHortic.2015.1072.3
Riecke, W. D., and Ruska, E. (1966). “A 100-kV transmission electron microscope with single-field condenser objective,” in Proceeding 6th International Congress on Electron Microscopy (Kyoto: Maruzen Co. Ltd.), 19–20.
Risco, C., de Castro, I. F., Sanz-Sánchez, L., Narayan, K., Grandinetti, G., and Subramaniam, S. (2014). Three-dimensional imaging of viral infections. Ann. Rev. Virol. 1, 453–473. doi: 10.1146/annurev-virology-031413-085351
Rizzo, N. W., Duncan, K. E., Bourett, T. M., and Howard, R. J. (2016). Backscattered electron SEM imaging of resin sections from plant specimens: observation of histological to subcellular structure and CLEM. J. Microsc. 263, 142–147. doi: 10.1111/jmi.12373
Roenhorst, J. W., Lacomme, C., Nisbet, C., Leichtfried, T., Menzel, W., Winter, S., et al. (2017). Euphresco project VirusCollect – fulfilling the need for a common collection of plant viruses and viroids for reference. EPPO Bull. 47, 41–47. doi: 10.1111/epp.12353
Romette, J. L., Prat, C. M., Gould, E. A., de Lamballerie, X., Charrel, R., Coutard, B., et al. (2018). The European Virus Archive goes global: a growing resource for research. Antiviral Res. 158, 127–134. doi: 10.1016/j.antiviral.2018.07.017
Schatz, D., Shemi, A., Rosenwasser, S., Sabanay, H., Wolf, S. G., Ben-Dor, S., et al. (2014). Hijacking of an autophagy-like process is critical for the life cycle of a DNA virus infecting oceanic algal blooms. New Phytol. 204, 854–863. doi: 10.1111/nph.13008
Shrestha, S., Shrestha, S., Shindo, J., Sherchand, J. B., and Haramoto, E. (2018). Virological quality of irrigation water sources and Pepper Mild Mottle Virus and Tobacco Mosaic Virus as index of pathogenic virus contamination level. Food Environ. Virol. 10, 107–120. doi: 10.1007/s12560-017-9324-2
Suttle, C. A. (2007). Marine viruses - major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812. doi: 10.1038/nrmicro1750
Tilsner, J., Linnik, O., Louveaux, M., Roberts, I. M., Chapman, S. N., and Oparka, K. J. (2013). Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. J. Cell Biol. 201, 981–995. doi: 10.1083/jcb.201304003
Torrance, L., Andreev, I. A., Gabrenaite-Verhovskaya, R., Cowan, G., Makinen, K., and Taliansky, M. E. (2006). An unusual structure at one end of potato potyvirus particles. J. Mol. Biol. 357, 1–8. doi: 10.1016/j.jmb.2005.12.021
Valle, M. (2018). “Structural homology between nucleoproteins of ssRNA viruses,” in Subcellular Biochemstry 88. Virus Protein and Nucleoprotein Complexes, eds J. Harris and D. Bhella (Singapore: Springer) 129–145. doi: 10.1007/978-981-10-8456-0_6
Villinger, C., Neusser, G., Kranz, C., Walther, P., and Mertens, T. (2015). 3D analysis of HCMV induced-nuclear membrane structures by FIB/SEM tomography: insight into an unprecedented membrane morphology. Viruses 7, 5686–5704. doi: 10.3390/v7112900
Von Ardenne, M. (1938). Das Elektronen-Rastermikroskop. Zeitschrift für Physik 109, 553–572. doi: 10.1007/BF01341584
Von Borries, B., Ruska, E., and Ruska, H. (1938). Bakterien und Virus in übermikroskopischer Aufnahme. Klin. Wochenschr. 17, 921–925. doi: 10.1007/BF01775798
Zamora, M., Méndez-López, E., Agirrezabala, X., Cuesta, R., Lavin, J. L., Sánchez-Pina, M. A., et al. (2017). Potyvirus virion structure shows conserved protein fold and RNA binding site in ssRNA viruses. Sci. Adv. 3, 1–7. doi: 10.1126/sciadv.aao2182
Keywords: electron microscopy, virus diagnosis, cryo electron microscopy, correlative microscopy, scanning electron microscopy, virus replication
Citation: Richert-Pöggeler KR, Franzke K, Hipp K and Kleespies RG (2019) Electron Microscopy Methods for Virus Diagnosis and High Resolution Analysis of Viruses. Front. Microbiol. 9:3255. doi: 10.3389/fmicb.2018.03255
Received: 30 August 2018; Accepted: 14 December 2018;
Published: 07 January 2019.
Edited by:
John P. Carr, University of Cambridge, United KingdomReviewed by:
Peter Metcalf, The University of Auckland, New ZealandCopyright © 2019 Richert-Pöggeler, Franzke, Hipp and Kleespies. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katja R. Richert-Pöggeler, a2F0amEucmljaGVydC1wb2VnZ2VsZXJAanVsaXVzLWt1ZWhuLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.