- 1Department of ODS & Research, School of Dentistry, Meharry Medical College, Nashville, TN, United States
- 2Bioinformatics Core, School of Graduate Studies/Research & School of Medicine, Meharry Medical College, Nashville, TN, United States
- 3Department of Public Health, School of Graduate Studies & Research, Meharry Medical College, Nashville, TN, United States
- 4Department of Internal Medicine, Division of Gastroenterology & Hepatology, Meharry Medical College, Nashville, TN, United States
- 5Department of Periodontology, College of Dentistry, University of Florida, Gainesville, FL, United States
- 6Department of Biochemistry, Cancer Biology, Neuroscience & Pharmacology, Meharry Medical College, Nashville, TN, United States
- 7Department of Nutrition Metabolism & Oral Surgery, University of Texas Medical Branch at Galveston, Galveston, TX, United States
Impaired colon motility is one of the leading problems associated with inflammatory bowel disease (IBD). An expanding body of evidence supports the role of microbiome in normal gut function and in progression of IBD. The objective of this work is to determine whether diseased full thickness colon specimens, including the neuromuscular region (critical for colon motility function), contain specific oral and gut pathogens. In addition, we compared the differences in colon microbiome between Caucasians (CA) and African Americans (AA). Thirty-nine human full thickness colon (diseased colon and adjacent healthy colon) specimens were collected from Crohn's Colitis (CC) or Ulcerative Colitis (UC) patients while they underwent elective colon surgeries. We isolated and analyzed bacterial ribosomal RNA (rRNA) from colon specimens by amplicon sequencing of the 16S rRNA gene region. The microbiome proportions were quantified into Operational Taxonomic Units (OTUs) by analysis with Quantitative Insights Into Microbial ecology (QIIME) platform. Two hundred twenty-eight different bacterial species were identified by QIIME analysis. However, we could only decipher the species name of fifty-three bacteria. Our results show that proportion of non-detrimental bacteria in CC or UC colon samples were altered compared to adjacent healthy colon specimens. We further show, for the first time in full thickness colon specimens, that microbiome of CC and UC diseased specimens is dominated by putative oral pathogens belonging to the Phyla Firmicutes (Streptococcus, Staphylococcus, Peptostreptococcus), and Fusobacteria (Fusobacterium). In addition, we have identified patterns of differences in microbiome levels between CA and AA specimens with potential implications for health disparities research. Overall, our results suggest a significant association between oral and gut microbes in the modulation of colon motility in colitis patients.
Introduction
Inflammatory bowel disease (IBD) is comprised of Crohn's disease / Crohn's colitis (CC) and Ulcerative colitis (UC). The term Colitis, refers to general inflammation of the inner lining of the colon arising from numerous underlying causes including idiopathic infection, IBD (either CC or UC), ischemic colitis, allergic reactions, and/or microscopic colitis. Distally, gingivitis, and periodontal disease are chronic inflammatory gum diseases associated with orange, red, yellow, purple, and green complex bacterial infections in sub-gingival areas of oral cavity (Popova et al., 2013).
Previous studies have shown that periodontal disease (PD) is a significant risk factor and contributor to many systemic diseases, including IBD (Vavricka et al., 2013). Several factors including genetic, dietary, and environmental factors could influence the pathogenesis of microbiome (oral and gut) which in turn may increase the incidence of periodontitis and IBD (Lira-Junior and Figueredo, 2016; Agossa et al., 2017). In addition, Porphyromonas gingivalis known to cause PD altered the gut microbiota leading to increased gut epithelial permeability and endotoxemia, which causes systemic inflammation (Hajishengallis, 2015). In addition, many earlier studies have shown intestinal colonization of oral bacteria in the pathogenesis of IBD (Strauss et al., 2011; Atarashi et al., 2017).
Innumerable number of studies have shown that the gut microbiome including Phyla Proteobacteria, Firmicutes, and Bacteroidetes contribute to normal gut function (Mariat et al., 2009; Koliada et al., 2017; Walker et al., 2018; Zhao et al., 2018). Colon motility is mainly regulated by neuromuscular portion of the colon and this was shown to be impaired in colitis patients; putatively due to a reduction in neuronal nitric oxide (NO) synthase (nNOS) protein expression and/or neuronal degeneration (Bassotti et al., 2014; Gangula et al., 2017). Previous studies have analyzed the microbiome in feces and/or colon mucosal biopsy specimens of colitis patients (Gibson et al., 1991; Bibiloni et al., 2006). However, the relationship/interaction between oral and gut bacteria in the development and/or exacerbation of inflammatory disease in the colon (containing neuromuscular tissue) was under studied. In addition, data is limited on how oral bacteria interact with and influence the large intestinal flora, thereby contributing to colitis. Since motility of the colon is impaired in colitis patients and neuromuscular tissue play a role in the motility function (Geboes and Collins, 1998; Poli et al., 2001), we hypothesize that the interaction between oral and gut microbiome may play a significant role in the inflammatory processes associated with the development and progression of colitis seen in certain patient populations. Furthermore, we hypothesize that difference in microbiome may exist between CA and AA colitis patients, potentially contributing to health disparities in IBD.
Methods
Ethics Statement
The participants provided both written and verbal informed consent to Collaborative Human Tissue Networking (CHTN) Consortium to collect specimens while they underwent elective colon surgeries.
Collection of Specimens
Frozen full thickness colon specimens were obtained from Cooperative Human Tissue Networking (CHTN). Thirty-nine human full thickness colon (moderate to severe diseased colon and adjacent healthy colon) specimens were collected from CC and UC male and female patients (ages between 18 and 75 years old) while they underwent elective colon surgeries. The specimens include Ulcerative (n = 13), Crohn's (n = 13) and adjacent healthy (n = 13) specimens. Characteristics of participants included Caucasians (CA) (n = 30) and African Americans (AA) (n = 9). CC male and female patients presented with symptoms like fever, fatigue, diarrhea, blood in stool, mouth sores, abdominal cramping, and pain around the anus, reduced appetite, and weight loss. While UC male and female patients presented with additional signs like rectal pain, rectal bleeding, and inability to defecate despite urgency.
Extraction of DNA, Amplification of 16S rRNA Gene and Amplicon Sequencing
DNA extraction and microbial analysis were performed in the University of North Carolina at Chapel Hill School of Medicine Microbiome Core Facility (UNC: MC). We identified a conserved region of the 16S rRNA gene of 550 bp to amplify. This encompassed variable regions V3–V4 from the colon genomic DNA using primers 16S rRNA-F 5′-AGAGTTTGATCCTGGCTCAG-3′and 16S rRNA-R 5′-GCTGCCTCCCGTAGGAGT-3′ and overhang adapter sequences appended to the primer pair for compatibility with Illumina index and sequencing adapters. Briefly, each 16SrRNA amplicon was purified using AMPure XP reagent (Beckman Coulter, Indianapolis, IN, USA). Specifically, each sample was amplified using a limited cycle PCR program, adding Illumina sequencing adapters and optional dual-index barcodes [index 1(i7) and index 2(i5)] (Illumina, San Diego, CA, USA) to the amplicon target. The final libraries were purified using AMPure XP reagent, quantified and normalized prior to pooling. The DNA library pool was denatured with NaOH, diluted with hybridization buffer and heat denatured before loading on to the MiSeq reagent cartridge and to the MiSeq instrument (Illumina). The standard Illumina paired-end 250 base pair (PE250) protocol was used for sequencing the16S rRNA amplicons (Illumina, CA, USA).
Processing of Sequence Reads
Data was analyzed and microbial proportions using Operational Taxonomic Units (OTUs) were determined using Quantitative Insights Into Microbial ecology (QIIME) pipeline (Caporaso et al., 2010a) in the Meharry Medical College Bioinformatics Core. Briefly, generated raw reads were preprocessed for adapter removal. Processed sequence reads were obtained as fastq files and were converted into fasta, quality and flow files using Mothur package (Schloss et al., 2009). The initial number of fasta sequences obtained were 31,09,793. First, the fasta files were cleaned of host reads by mapping on to 9 mm mouse genome. Then, the primer sequences and barcode sequences were removed, demultiplexed and quality filtered. The number of high quality sequences remaining after quality filtering was 16,64,769. The OTUs were picked by de novo strategy. The high quality sequences were clustered at 97% identity using UCLUST inbuilt in QIIME pipeline to generate 3994 OTUs and taxonomy was assigned to OTU representative sequences using UCLUST (Edgar, 2010). The picked sequences were aligned using PyNAST aligner (Caporaso et al., 2010b). The chimeric sequences and singleton OTUs were removed using ChimeraSlayer (Haas et al., 2011). We constructed a phylogenetic tree for the sequences using FastTree version 2.1.3 (data not shown) (Price et al., 2010). Next, an OTU table was constructed and taxa were summarized using the 894 OTUs obtained from QIIME pipeline. α-diversity metrics was computed using Chao1 (abundance-based richness estimator) and Shannon analysis (diversity index) and Rarefaction plots were constructed (data not shown). β-diversity metrics was computed using weighted and unweighted Principal Coordinates Analysis (PCoA) (data not shown) (Gower, 2005). A Taxonomic Summary Bar plot showing OTUs assigned to Phyla-level taxonomy per sample was subsequently constructed (Figure 1). Bar Plots showing the relative abundance of bacteria at the Phyla-level between races, diseased tissue and healthy tissue groups is shown in Figure 3. Sample-specific sequences were deposited in the MGRAST database (accession number: b3b851ba2c6d676d343739393937332e33) and was assigned an MG-RAST project ID (mgs675214) (Keegan et al., 2016). In addition, sample-specific sequences were deposited in the NCBI (BioProject: PRJNA496071).
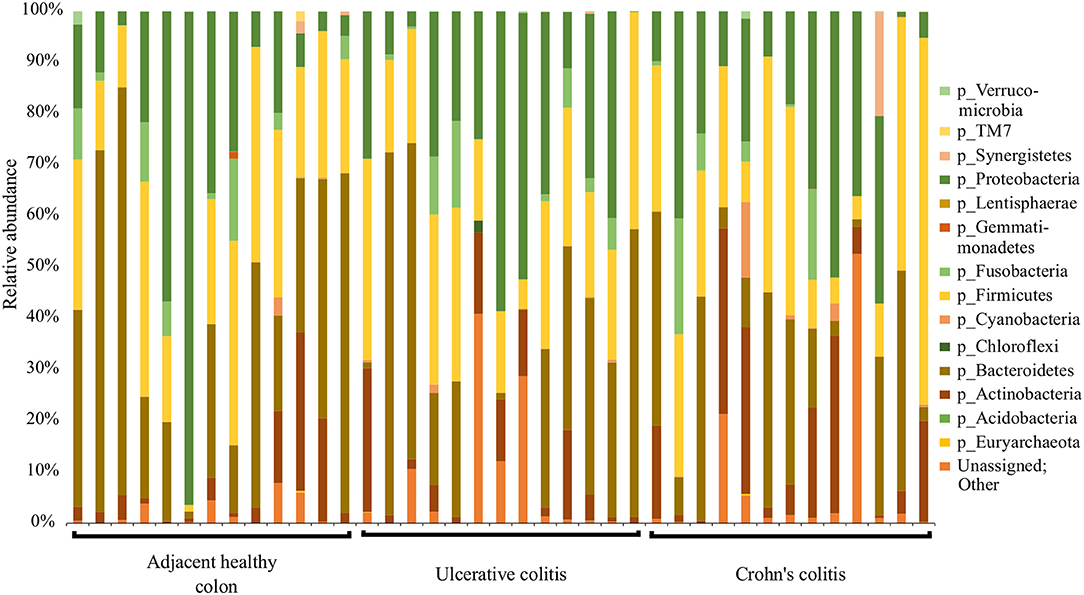
Figure 1. Summary of major bacterial taxa contributing to the communities detected in the 39 specimens at the Phylum level in the colitis and adjacent healthy specimen groups. Specimens are categorized into adjacent healthy colon (n = 13), Ulcerative colitis (UC, n = 13), and Crohn's colitis (CC, n = 13). Data represented are the relative abundances (%) of each phylum identified in each specimen.
The pathogenic and beneficial oral and gut bacteria were identified using the NCBI Genome database (https://www.ncbi.nlm.nih.gov/genome/). This analysis was performed to assess the pathogenic and healthy bacterial proportions in human full thickness colon specimens (Tables 1–3).
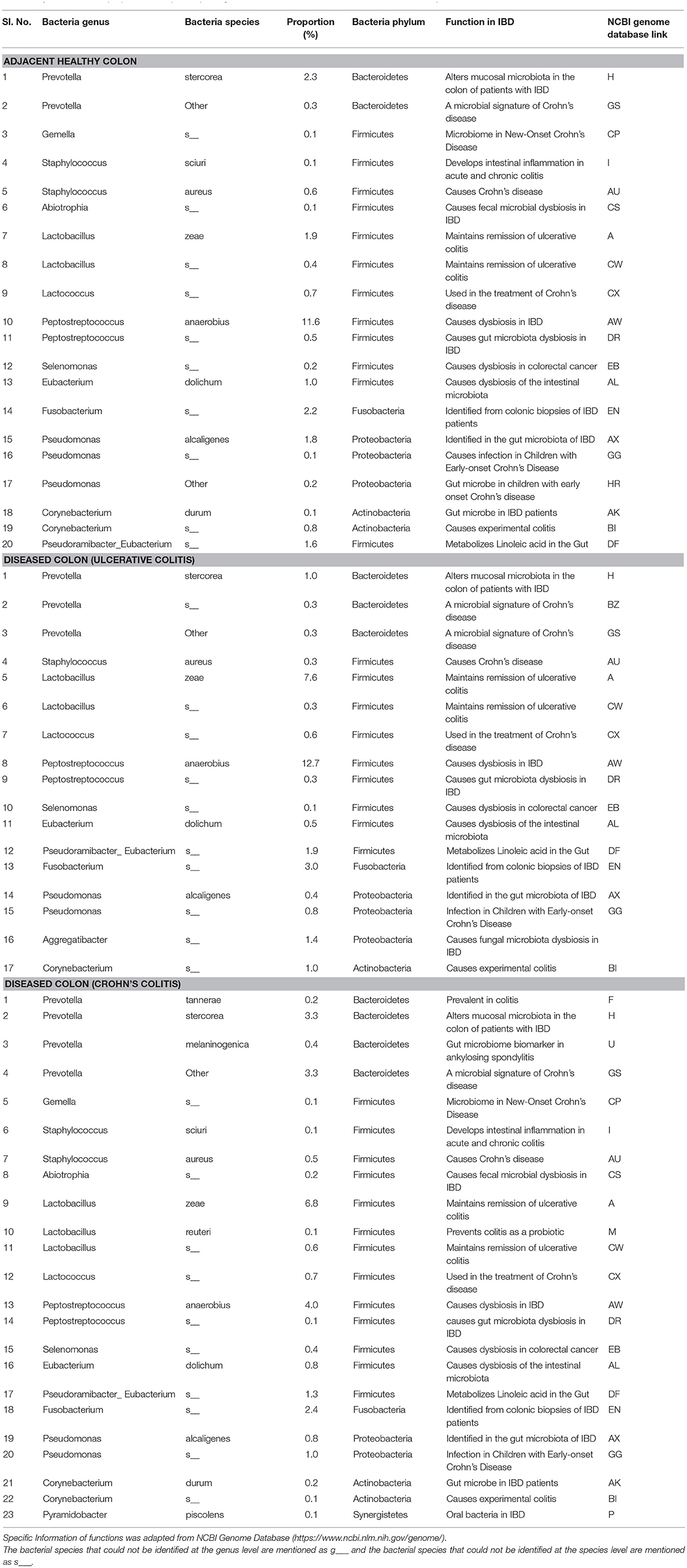
Table 1. Functions and proportions of specific pathogenic Oral bacteria colonized in full thickness colon specimens.
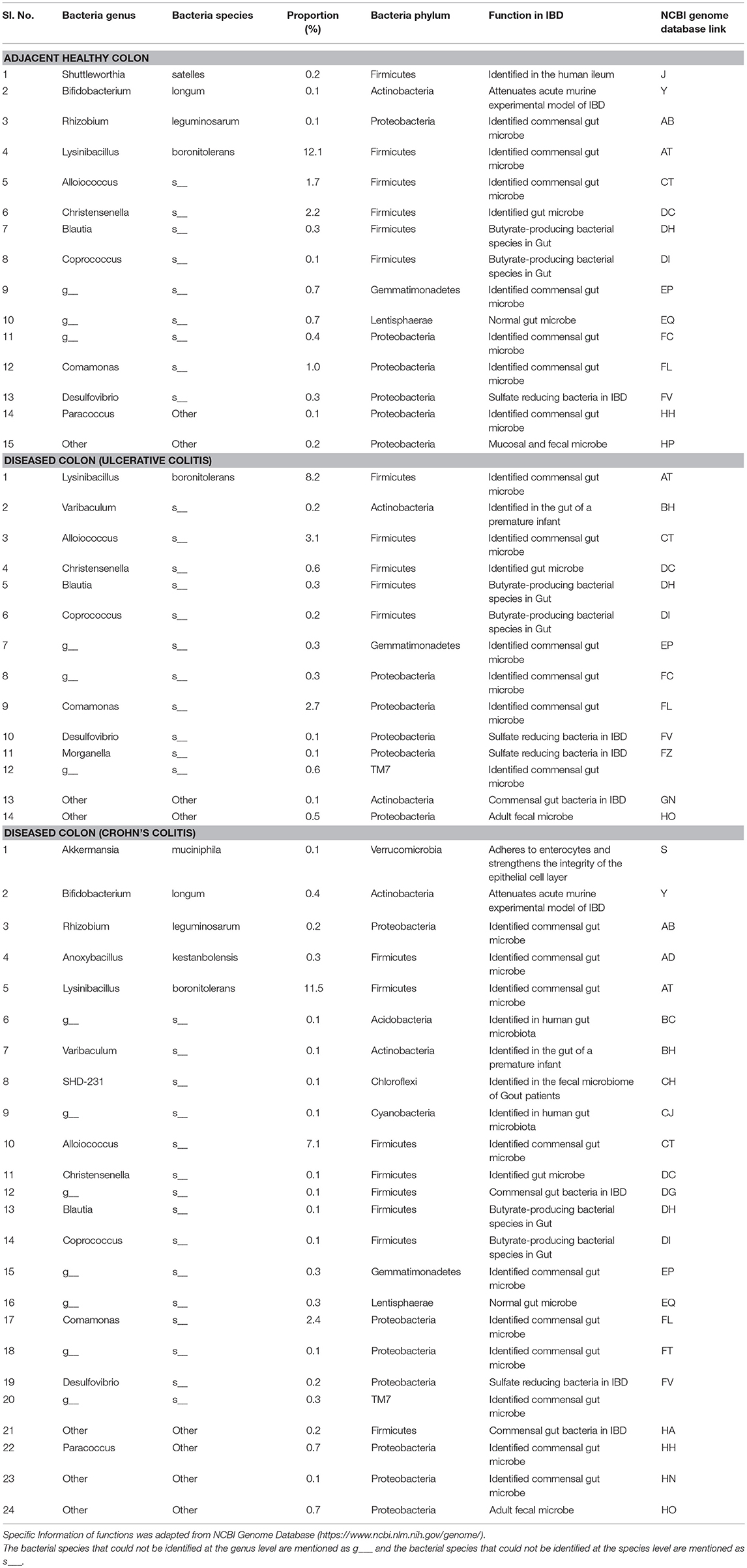
Table 2. Functions and proportions of specific beneficial Gut bacteria colonized in full thickness colon specimens.
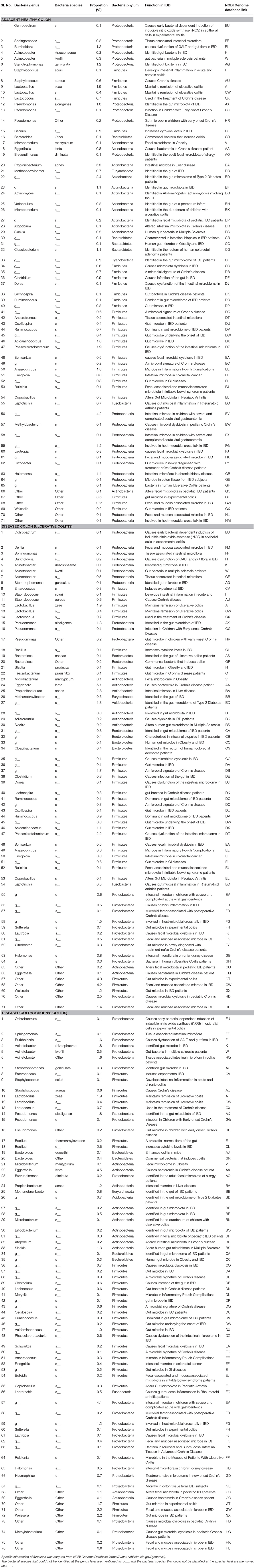
Table 3. Functions and proportions of specific pathogenic Gut bacteria colonized in full thickness colon specimens.
Statistical Analysis and Evaluation
Statistical analysis was performed between the healthy and diseased groups and based on race classification (n = 13 CC, n = 13 UC, n = 13 non-disease healthy patients, n = 30 CA and n = 9 AA). A non-parametric Mann-Whitney U Test p-value < 0.05 of bacterial 16S rRNA OTUs between the groups was considered statistically significant. IBM SPSS software package version 23 (IBM Analytics, USA) was used to conduct statistical analysis.
Results
Relative Abundance Analysis
QIIME analysis showed about two hundred twenty-eight bacterial species in entire 39 specimens (Tables 1–5). However, non-ambiguous annotation at the species name resulted in fifty-three bacterial identifications. The dominant phyla across all samples (both diseased and healthy specimens) were Bacteroidetes (46.92%), followed by Firmicutes (27.8%), and Proteobacteria (24.5%). Most importantly, our results indicate that putative oral pathogens (belonging to mostly Phylum Firmicutes) dominated the microbiome of diseased specimens (Figure 1). Adjacent healthy specimens show an increased abundance of Phylum Bacteroidetes (~ 57%, containing mostly symbiotic and/or beneficial bacteria) population, which is altered in disease categories (Figure 2).
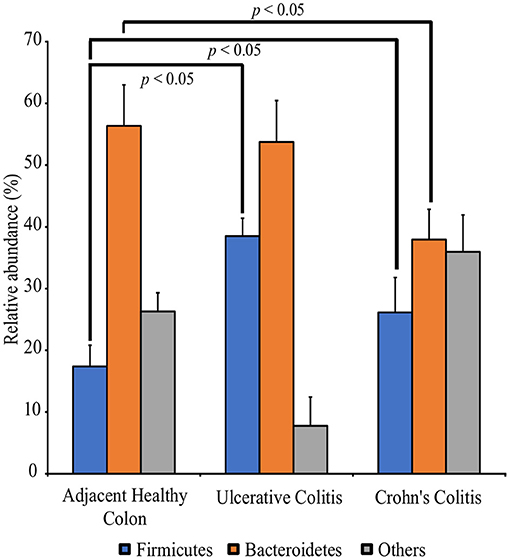
Figure 2. Perturbation of the full thickness colon microbiome in Colitis specimens as compared to adjacent healthy specimens. Specimens are categorized into adjacent healthy colon (n = 13), Ulcerative colitis (UC, n = 13) and Crohn's colitis (CC, n = 13). Data represented are the mean of relative abundances of each phylum identified in specimens belonging to each group while error bars indicate standard error. Diseased specimens demonstrate a balance between the Phyla Firmicutes and Bacteroidetes. Conversely, healthy colon specimens demonstrate a significantly higher proportion of Phyla Bacteroidetes. *p < 0.05 by Mann-Whitney U Test (n = 13 under each group).
Differential Expression of Microbiomes in the Colon of CA and AA Patients
Figure 3 show racial differences of various bacterial phyla in adjacent healthy, UC and CC full thickness colon specimens. The tissue specimens from Caucasians represented a significantly higher proportion (p < 0.05) of the oral pathogen, Fusobacterium, and gut bacteria, Parabacteroides (Bacteroidetes). CA specimens also showed significantly higher levels (p < 0.05) of Phyla Proteobacteria including Citrobacter, Hemophilus, Acinetobacter, Pseudomonas, and Stenotrophomonas as compared to AA. Whereas, the AA specimens were observed to have a significantly higher proportion (p < 0.05) of Prevotella (Bacteroidetes) and Clostridia (Firmicutes) (Figure 3; Table 4).
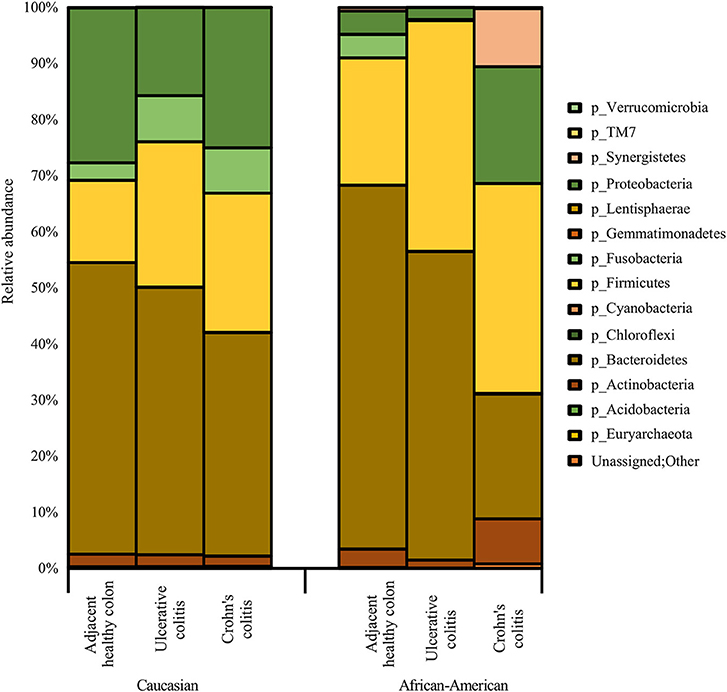
Figure 3. Summary of major bacterial taxa showing the relative abundance of oral and gut bacteria at the Phylum level in the colitis and adjacent healthy specimen groups under each race. Data represented are the mean of relative abundances of each Phyla detected in samples belonging to each group. The dominant phyla across all samples (both diseased and healthy specimens) were Bacteroidetes, followed by Firmicutes and Proteobacteria. Other major phyla observed among these specimens also include Actinobacteria, Fusobacteria, and Synergistetes. The Phylum Proteobacteria did not show any significant difference between healthy colon specimens and diseased colon specimens. A larger proportion of unassigned bacteria (0.3%) was identified in AA Crohn's Colitis patients compared to other groups.
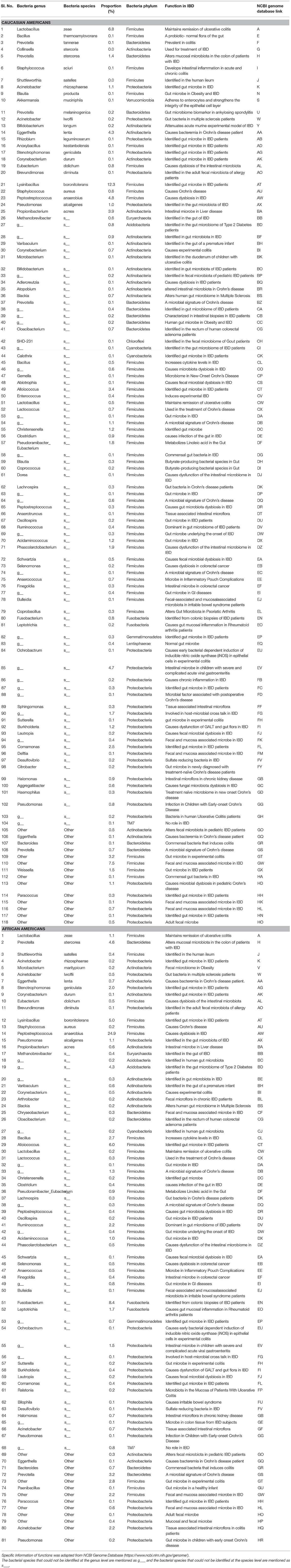
Table 4. Functions and Proportions of bacterial species identified in the full thickness human colon specimens of Caucasians and African Americans.
As depicted in Figure 3, the adjacent healthy colon specimens, UC and CC contained ~ 1%, ~ 7% and ~ 7% of sequence reads, respectively that were un-assignable to any taxon with a larger proportion of them identified in AA Colitis patients. Other major phyla observed among these specimens also include Proteobacteria (Adjacent healthy: 23.8%; UC: 26.5% and CC: 23.1%), Actinobacteria (Adjacent healthy: 6.7%; UC: 8.1% and CC: 14.1%), Fusobacteria (Adjacent healthy: 4.2%; UC: 3.6% and CC: 4.0%), and Synergistetes (Adjacent healthy: 0.2%; UC: 0.04% and CC: 1.5%). The Phylum Proteobacteria did not show any significant difference between healthy colon specimens and diseased colon specimens (Table 4).
Bacterial Species Identified in a Significantly Higher Proportion in Diseased Colon Tissues
As shown in Figure 3, diseased colon specimens represented a significantly higher proportion (p < 0.05) of gut bacteria belonging to Phylum Firmicutes including Blautia producta, Faecalibacterium prausnitzii, Anoxybacillus kestanbolensis, Ruminococcus gnavus, Eubacterium dolichum, Lysinibacillus boronitolerans, and oral bacteria including Staphylococcus sciuri, Staphylococcus aureus, Streptococcus anginosus.
In contrast, healthy colon specimens were significantly dominated (p < 0.05) by oral bacteria belonging to Phylum Actinobacteria that includes; Corynebacterium kroppenstedtii, Corynebacterium durum. Additionally, healthy colon specimens were dominated by gut bacteria belonging to Phylum Actinobacteria that includes; Colinsella stercoris, Colinsella aerofaciens, Kocuria rhizophila, Eggerthella lenta, Propionibacterium granulosum, Propionibacterium acnes, Actinomyces europaeus, Rothia dentocariosa, and Phylum Bacteroidetes that includes; Bacteroides fragilis, Bacteroides eggerthii, Bacteroides caccae, Parabacteroides distasonis (Figure 3).
Alpha Diversity and Beta Diversity Analyses
Alpha diversity and beta diversity metrics were computed to analyse the diversity of bacterial species within each sample and between samples. To assess our sampling efficiency, we plotted rarefaction curves (Chao1 and Shannon) for all 39 specimens. Increased diversity (Shannon) in the diseased samples compared to control samples was observed. From the rarefaction curves, it is evident that most AA samples require additional sampling whereas Caucasian samples do not (data not shown).
Since, outliers exhibiting different microbiome profiles were observed both in the healthy and disease groups, we performed principle coordinate analysis (PCoA analysis) and hierarchial clustering to obtain a holistic view of the microbiome profile in each sample. Two dimensional PCoA plots revealed that control samples which had similar microbiome profiles as suggested by histograms and OTU heat map clustered together (data not shown).
Pathogenic Oral and Gut Flora Abundantly Colonized in Diseased Colon Specimens
The pathogenic oral bacteria identified abundantly in diseased colon specimens as compared to healthy colon specimens were Porphyromonas, Prevotella, Gemella, Staphylococcus, Streptococcus, Abiotrophia, Granulicatella, Lactobacillus, Lactococcus, Peptostreptococcus, Selenomonas, Veillonella, Parvimonas, Eubacterium, Fusobacterium, Pseudomonas, Aggregatibacter, and Corynebacterium (Table 1).
Pathogenic gut bacteria identified abundantly in diseased colon specimens as compared to healthy colon specimens include Ochrobactrum, Delftia, Sphingomonas, Burkholderia, Acinetobacter, Stenotrophomonas, Enterococcus, Granulicatella, Staphylococcus, Streptococcus, Lactobacillus, Lactococcus, Pseudomonas, Bacillus, Campylobacter, and Bacteroides (Table 3).
Discussion
Our study demonstrates significant perturbations among bacteria belonging to Phyla Bacteroidetes and Firmicutes in full-thickness diseased colon specimens containing neuromuscular compartment (Figure 2). Our studies further show that the proportion of pathogenic bacteria are higher in diseased compared to adjacent healthy colon specimens. We suggest that pathogenic bacteria belonging to these two phyla have a greater impact on colon motility function in colitis patients (Tables 1, 3). Although the incidence of IBD is increasing among African Americans (AA), the underlying causes are completely unknown (Sofia et al., 2014). Our study further highlight a significant disparity in bacterial dysbiosis among AA compared to CA colitis patients (Figure 3).
CA specimens had significantly higher levels of Fusobacterium, Parabacteroides, Citrobacter, Haemophilus, Acinetobacter, Pseudomonas, and Stenotrophomonas. Fusobacterium nucleatum is known to have a well-characterized role in the oral cavity. We have determined that Fusobacterium can be recovered from human full thickness colon specimens and this could indicate their ability to survive and proliferate inside host cells. Parabacteroides was found to be dominant in the acute phase of IBD in CA patients. Citrobacter is an epithelial cell adherent pathogen and can subvert inflammation in colitis. Pseudomonas interacts with the mucosal layer of colon and disrupts the mucosal barrier integrity leading to colitis in CA patients.
The AA specimens had significantly higher levels of Prevotella and Clostridia. Prevotella augments T-helper cells mediated colon mucosal inflammation by activating Toll-like receptor 2 leading to production of T-helper cells polarizing cytokines by antigen-presenting cells, including interleukins. In addition, Prevotella induce epithelial cells to produce interleukins and cytokines that can promote recruitment of neutrophils and mucosal T-helper cell immune responses. Prevotella can mediate inflammation of the mucosa leading to the circulation of bacteria, bacterial products and other inflammatory mediators. Prevotella could augment release of inflammatory mediators from immune cells and various stromal cells in colitis in AA patients. Clostridium can disrupt gut immune dormancy and cause infectious colitis in AA patients. Collectively, our data suggest that the presence of pathogenic bacteria in AA full thickness diseased specimens could adversely affect colon motility.
Additionally, our data in UC and CC specimens show the presence of several orange (Prevotella, Peptostreptococcus, Eubacterium, Fusobacterium, and Campylobacter), red (Porphyromonas), purple (Veillonella), and yellow (Streptococcus) complex putative oral pathogens known to cause gingivitis and periodontitis among IBD patients (Tables 1, 3). Previous studies using mucosal biopsies and feces have shown that gut microbiota in bowel diseases is characterized by an increase in certain phyla such as Proteobacteria, Firmicutes, genus Bifidobacterium, as well as a reduction in the amounts of genera Ruminococcus, Clostridia and (in some cases) Faecalibacterium (Lane et al., 2017; Nishida et al., 2018). However, none of the earlier studies using feces have shown a shift in the balance between Phyla Bacteroidetes and Firmicutes among UC or CC patients; even though this was observed in healthy individuals (Mariat et al., 2009; Koliada et al., 2017). In contrary to our results, one study using mucosal biopsies has shown a significantly decreased Firmicutes to Bacteroidetes ratio in both UC and CC compared with controls (Kabeerdoss et al., 2015). Collectively, our data suggest that the putative oral pathogens found in diseased colon specimens may modulate the proportion of non-detrimental gut bacteria, thus potentially worsening the condition of the colon in colitis patients.
Oral bacterial species like Porphyromonas, Peptostreptococcus, Eubacterium, Fusobacterium, Streptococcus salivarius, S. mitis, S. bovis, Veillonella spp., Staphylococcus aureus, S. epidermidis, and Campylobacter spp. can convert nitrate to nitrite. A large amount of bioactive NO is found in the gastrointestinal tract, generated by dietary sources and by conversion of anaerobic bacteria in the oral cavity, or by anaerobic reaction with nitrate in the colon by Escherichia coli spp. The entero-salivary nitrate conversion pathway provides a rich source of bioactive NO and nitrate-reducing bacteria, such as Veillonella. In this pathway, nitrate is obtained by the salivary gland and is then concentrated in the saliva. Various facultative anaerobic bacteria on the top of the tongue effectively reduces nitrate to nitrite. The bacteria then use the nitrate and the nitrite as electron acceptors in their respiration process. This also helps the host in the first steps of converting nitrate to NO. The salivary nitrate then reaches the systemic circulation, various enzymatic reactions occur leading to reduction to NO, and other reactive nitrogen intermediates. The oral cavity plays an important role the production of nitric oxide, and specifically, employs the nitrate-nitrite-NO pathway in the oral cavity. It is well known that oral cavity bacteria can migrate to the colon. Taken together, our data suggest that the putative oral pathogens found in diseased colon specimens may survive by exploiting the nitrate-nitrite-NO pathway to modulate the proportion of non-detrimental gut bacteria, thus potentially worsening the condition of colon in colitis patients (Figure 4).
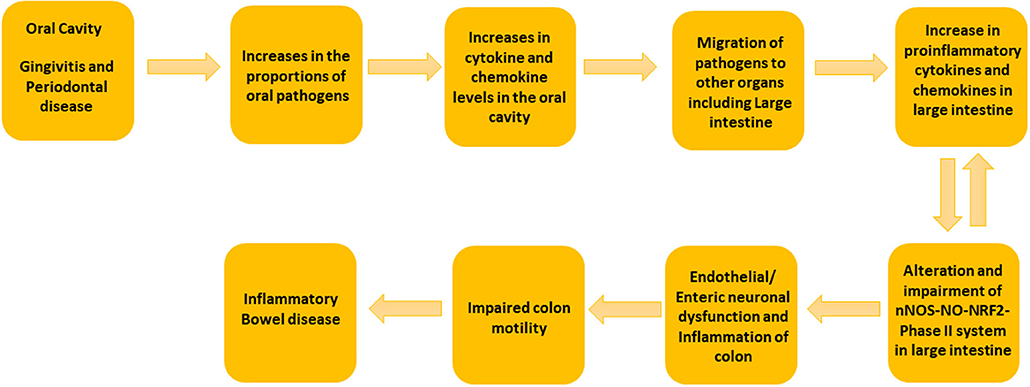
Figure 4. Schematic Representation of the suggested mechanism involved in the development of colitis by oral and gut microbiome. We propose that the increase in the concentrations of putative oral pathogens elevates the cytokine and chemokine levels in oral cavity. When putative oral pathogens travel to the gut, they can colonize locally and lead to the elevated levels of proinflammatory cytokines. This can effect on nNOS-NO-NRF2-Phase II system in the large intestine and could lead to colon dysmotility and colitis.
Previous studies suggest that enteric neurons and smooth muscle mediated gut motility is impaired in colitis patients (Snape et al., 1991; Vermillion et al., 1993). IBD associated gut inflammation affects the morphological and functional changes in the myenteric/enteric nervous system (ENS) and nitric oxide (NO) synthesis (Takahashi, 2003; Kono et al., 2004). Experimental studies have also shown that gut bacteria have a role in oxidative stress induced gut inflammation by controlling metabolic endotoxemia in obese mice (Cani et al., 2008). We have shown that polybacterial oral infection decrease the expression of nNOS and NRF2-phase II enzymes in the gut and this could lead to impaired colon motility (Gangula et al., 2015; Walker et al., 2018).
Some of the gut bacteria we have identified in the full thickness colon specimens in the present study, including Bacteroides, Prevotella, Pseudomonas, etc., have been identified in colon mucosal biopsies in earlier studies (Bibiloni et al., 2006). These bacteria evoke inflammatory responses affecting the innermost lining of colon. Many specific beneficial bacteria, including members of Bacteroides and Prevotella groups, C. coccoides, and Lactic acid bacteria were known to be decreased in colitis patients (Gibson et al., 1991). Specimens used in prior studies were colon mucosal biopsies or stool samples; but not full thickness colon specimens (Gibson et al., 1991; Bibiloni et al., 2006). Full-thickness colon consists of four layers of tissue including mucosa, submucosa, muscularis, and serosa.
Novel to this research design, full thickness colon specimens were obtained because colitis patients often experience colon motility abnormalities (Snape et al., 1991; Annese et al., 1997; Vrees et al., 2002). Several lines of evidence suggest that nitrergic neurons that releases NO via nNOS are known to play a pivotal role in colon motility (Kono et al., 2004; Winston et al., 2013). Previous studies have demonstrated that nitrergic neurons are degenerated in colitis (Onori et al., 2005; Sung et al., 2006). Recent studies from our laboratory indicate that nNOS, as well as antioxidants (NRF2 regulated-Phase II enzymes) protein expression are down-regulated in diseased colon specimens (Myers et al., 2014; Gangula et al., 2017). Furthermore, our previous studies demonstrated that polybacterial infection led to a decrease in nNOS, NRF2 and antioxidants protein expression in the colon tissues (Gangula et al., 2015). In addition, studies have shown that NO may play homeostatic role in gut inflammation (Kolios et al., 2004). Taken together, our data suggest that elevated levels of oral and gut pathogens in diseased colon full thickness specimens could contribute to impaired nNOS-NO-NRF2-Phase II system and colon motility abnormalities in IBD patients (Figure 4).
To our knowledge, our study is the first to report the presence of several microbiota of unknown function in IBD including Micrococcus luteus, Chloracidobacteria, Arthrobacter, Propionicimonas, Paludibacter, Chryseobacterium, Calothrix, and Novosphingobium (Table 5). These new microbiota members have not been identified in mucosal/fecal specimens in previous studies, suggesting that these bacteria are primarily colonized in the neuromuscular compartment. Additional studies are warranted to characterize the novel bacteria and investigate their specific role in colon motility and constipation in IBD patients.
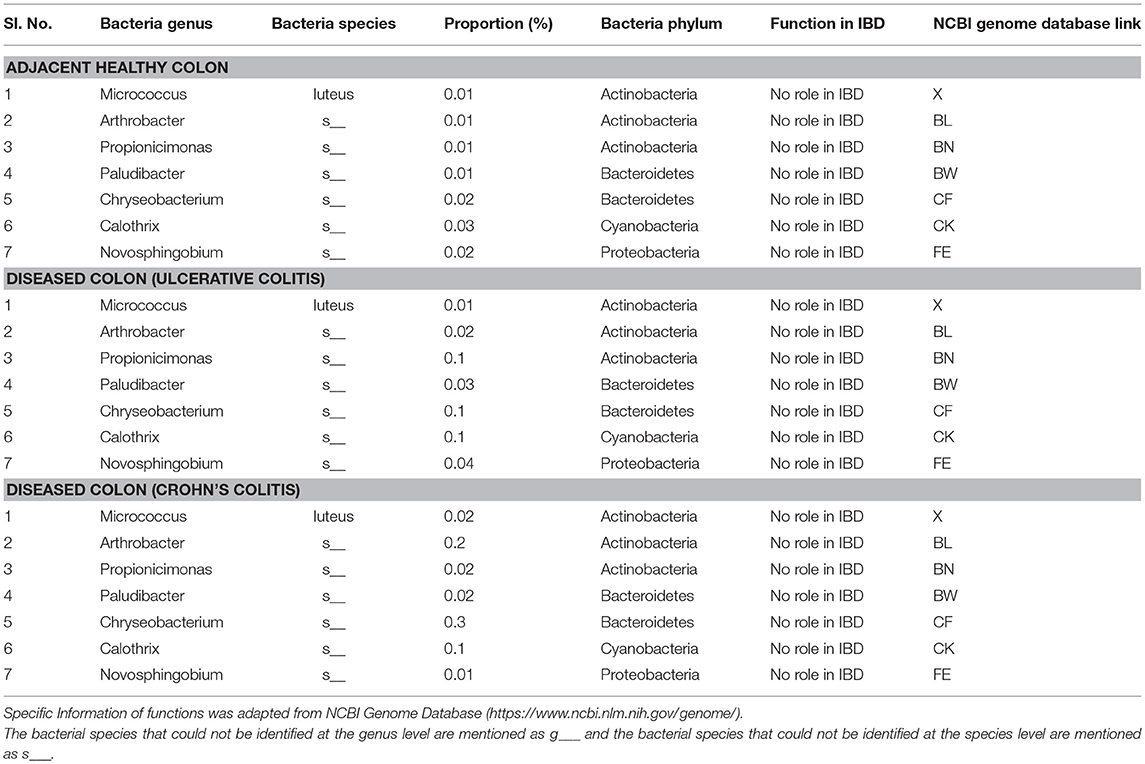
Table 5. Proportions of bacterial species of unknown function colonized in full thickness colon of colitis patients.
In summary, this study have identified specific bacterial pathogens potentially associated with colon motility in IBD patients. The observations showed that some putative oral pathogens belonging to the Phyla Firmicutes (Streptococcus, Staphylococcus, Peptostreptococcus), and Fusobacteria (Fusobacterium) dominated in the microbiomes of CC and UC diseased specimens and might involve the modulation of colon motility in IBD.
Study Limitations
The limitations of the study include the smaller sample size across disease and race groups making this as a preliminary study. In spite of the limitations in sample size and the fact that some of the identified bacteria were not significantly altered in colitis specimens, we were still able to observe differences in the microbiome between CA and AA colitis patients. This could be due to amplicon sequencing of a shorter conserved region of 16S rRNA gene instead of in depth shotgun sequencing. Moreover, we did not profile the oral microbiome from oral specimens (dental plaque, etc.) in the same IBD patients from whom full thickness colon specimens were collected. Finally, host-microbiome interaction studies are needed to better discern specific roles of the oral and gut bacteria in the development of colitis. Future studies are aimed to collect oral and fecal specimens therefore a comparative experiments in regards to changes in microbiome, along with specific key proteins will be conducted from the same patient.
Author Contributions
VD, SM, KS, SP, SS, PG, and MT have contributed both for data analysis and manuscript preparation. DS, CF-D, LK, SA, and JS have contributed in manuscript preparation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors sincerely thank University of North Carolina at Chapel Hill School of Medicine- Microbiome Core Facility (UNC; MC) for performing amplicon sequencing. We thank CHTN for providing diseased and healthy specimens. We further thank dental student doctors Mr. A. Arab and J. Hodges for their initial contribution on this project. The Meharry Bioinformatics Core is funded in part by NIH grants MD007593 and MD007586. SA is supported by NIMHD R-CTR grant No. 54544MD007593 and NCI grant No. 5454CA163069. This work is supported in part by 1SC1GM121282 funded to PG. In addition, Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number SG1GM121282 (PG).
References
Agossa, K., Dendooven, A., Dubuquoy, L., Gower-Rousseau, C., Delcourt-Debruyne, E., and Capron, M. (2017). Periodontal manifestations of inflammatory bowel disease: emerging epidemiologic and biologic evidence. J. Periodontal Res. 52, 313–324. doi: 10.1111/jre.12422
Annese, V., Bassotti, G., Napolitano, G., Usai, P., Andriulli, A., and Vantrappen, G. (1997). Gastrointestinal motility disorders in patients with inactive Crohn's disease. Scand. J. Gastroenterol. 32, 1107–1117.
Atarashi, K., Suda, W., Luo, C., Kawaguchi, T., Motoo, I., Narushima, S., et al. (2017). Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science 358, 359–365. doi: 10.1126/science.aan4526
Bassotti, G., Antonelli, E., Villanacci, V., Salemme, M., Coppola, M., and Annese, V. (2014). Gastrointestinal motility disorders in inflammatory bowel diseases. World J. Gastroenterol. 20, 37–44. doi: 10.3748/wjg.v20.i1.37
Bibiloni, R., Mangold, M., Madsen, K. L., Fedorak, R. N., and Tannock, G. W. (2006). The bacteriology of biopsies differs between newly diagnosed, untreated, Crohn's disease and ulcerative colitis patients. J. Med. Microbiol. 55, 1141–1149. doi: 10.1099/jmm.0.46498-0
Cani, P. D., Bibiloni, R., Knauf, C., Waget, A., Neyrinck, A. M., Delzenne, N. M., et al. (2008). Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 57, 1470–1481. doi: 10.2337/db07-1403
Caporaso, J. G., Bittinger, K., Bushman, F. D., DeSantis, T. Z., Andersen, G. L., and Knight, R. (2010a). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinform. 26, 266–267. doi: 10.1093/bioinformatics/btp636
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010b). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Gangula, P., Ravella, K., Chukkapalli, S., Rivera, M., Srinivasan, S., Hale, A., et al. (2015). Polybacterial periodontal pathogens alter vascular and gut BH4/nNOS/NRF2-phase II enzyme expression. PLoS ONE 10:e0129885. doi: 10.1371/journal.pone.0129885.
Gangula, P. R., Smoot, D. T., Izban, M., Ballard, B., and Adunyah, S. (2017). “Expression of NO and NRF2 enzymes in Human Colitis,” in Proceedings of the RCMI Translational Science 2017 Conference (Washington, DC), 78.
Geboes, K., and Collins, S. (1998). Structural abnormalities of the nervous system in Crohn's disease and ulcerative colitis. Neurogastroenerol. Motil. 10, 189–202.
Gibson, G. R., Cummings, J. H., and Macfarlane, G. T. (1991). Growth and activities of sulphate-reducing bacteria in gut contents of healthy subjects and patients with ulcerative colitis. FEMS Microbiol. Lett. 86, 103–111. doi: 10.1111/j.1574-6968.1991.tb04799.x
Gower, J. C. (2005). “Principal coordinates analysis,” in Encyclopedia of Biostatistics, eds P. Armitage and T. Colton, (John Wiley & Sons Ltd). doi: 10.1002/0470011815
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., Giannoukos, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi: 10.1101/gr.112730.110
Hajishengallis, G. (2015). Periodontitis: from microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 15, 30–44. doi: 10.1038/nri3785
Kabeerdoss, J., Jayakanthan, P., Pugazhendhi, S., and Ramakrishna, B. S. (2015). Alterations of mucosal microbiota in the colon of patients with inflammatory bowel disease revealed by real time polymerase chain reaction amplification of 16S ribosomal ribonucleic acid. Indian J Med Res. 142, 23–32. doi: 10.4103/0971-5916.162091
Keegan, K. P., Glass, E. M., and Meyer, F. (2016). MG-RAST, a metagenomics service for analysis of microbial community structure and function. Methods Mol. Biol. 1399, 207–233. doi: 10.1007/978-1-4939-3369-3_13
Koliada, A., Syzenko, G., Moseiko, V., Budovska, L., Puchkov, K., Perederiy, V., et al. (2017). Association between body mass index and Firmicutes/Bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol. 17:120. doi: 10.1186/s12866-017-1027-1
Kolios, G., Valatas, V., and Ward, S. G. (2004). Nitric oxide in inflammatory bowel disease: a universal messenger in an unsolved puzzle. Immunology 113, 427–437. doi: 10.1111/j.1365-2567.2004.01984.x
Kono, T., Chisato, N., Ebisawa, Y., Asama, T., Sugawara, M., Ayabe, T., et al. (2004). Impaired nitric oxide production of the myenteric plexus in colitis detected by a new bioimaging system. J. Surg. Res. 117, 329–336. doi: 10.1016/j.jss.2003.11.004
Lane, E. R., Zisman, T. L., and Suskind, D. L. (2017). The microbiota in inflammatory bowel disease: current and therapeutic insights. J. Inflamm. Res. 10, 63–73. doi: 10.2147/JIR.S116088
Lira-Junior, R., and Figueredo, C. M. (2016). Periodontal and inflammatory bowel diseases: is there evidence of complex pathogenic interactions? World J. Gastroenterol. 22, 7963–7972. doi: 10.3748/wjg.v22.i35.7963
Mariat, D., Firmesse, O., Levenez, F., Guimaraes, V. D., Sokol, H., Dore, J., et al. (2009). The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 9:123. doi: 10.1186/1471-2180-9-123
Myers, J. N., Schaffer, M. W., Korolkova, O. Y., Williams, A. D., Gangula, P. R., and M'Koma, A. E. (2014). Implications of the colonic deposition of free hemoglobin-α chain: a previously unknown tissue by-product in inflammatory bowel disease. Inflamm. Bowel Dis. 20, 1530–1547. doi: 10.1097/MIB.0000000000000144
Nishida, A., Inoue, R., Inatomi, O., Bamba, S., Naito, Y., and Andoh, A. (2018). Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin. J. Gastroenterol. 11, 1–10. doi: 10.1007/s12328-017-0813-5
Onori, L., Aggio, A., D'Alo, S., Muzi, P., Cifone, M. G., Mellillo, G., et al. (2005). Role of nitric oxide in the impairment of circular muscle contractility of distended, uninflamed mid-colon in TNBS-induced acute distal colitis in rats. World J. Gastroenterol. 11, 5677–5684. doi: 10.3748/wjg.v11.i36.5677
Poli, E., Lazzeretti, M., Grandi, D., Pozzoli, C., and Coruzzi, G. (2001). Morphological and functional alterations of the myenteric plexus in rats with TNBS-induced colitis. Neurochem. Res. 26, 1085–1093. doi: 10.1023/A:1012313424144
Popova, C., Dosseva-Panova, V., and Panov, V. (2013). Microbiology of periodontal diseases. a review. Biotechnol. Biotechnol. Equip. 27, 3754–3759. doi: 10.5504/BBEQ.2013.0027
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490. doi: 10.1371/journal.pone.0009490.
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Snape, W. J. Jr., Williams, R., and Hyman, P. E. (1991). Defect in colonic smooth muscle contraction in patients with ulcerative colitis. Am. J. Physiol. 261, G987–G991. doi: 10.1152/ajpgi.1991.261.6.G987
Sofia, M. A., Rubin, D. T., Hou, N., and Pekow, J. (2014). Clinical presentation and disease course of inflammatory bowel disease differs by race in a large tertiary care hospital. Dig Dis Sci. 59, 2228–2235. doi: 10.1007/s10620-014-3160-0
Strauss, J., Kaplan, G. G., Beck, P. L., Rioux, K., Panaccione, R., Devinney, R., et al. (2011). Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel Dis. 17, 1971–1978. doi: 10.1002/ibd.21606
Sung, T. S., La, J. H., Kim, T. W., and Yang, I. S. (2006). Alteration of nitrergic neuromuscular transmission as a result of acute experimental colitis in rat. J. Vet. Sci. 7, 143–150. doi: 10.4142/jvs.2006.7.2.143
Takahashi, T. (2003). Pathophysiological significance of neuronal nitric oxide synthase in the gastrointestinal tract. J. Gastroenterol. 38, 421–430. doi: 10.1007/s00535-003-1094-y
Vavricka, S. R., Manser, C. N., Hediger, S., Vogelin, M., Scharl, M., Biedermann, L., et al. (2013). Periodontitis and gingivitis in inflammatory bowel disease: a case-control study. Inflamm. Bowel. Dis. 19, 2768–2777. doi: 10.1097/01.MIB.0000438356.84263.3b
Vermillion, D. L., Huizinga, J. D., Riddell, R. H., and Collins, S. M. (1993). Altered small intestinal smooth muscle function in Crohn's disease. Gastroenterol. 104, 1692–1699.
Vrees, M. D., Pricolo, V. E., Ptenti, F. M., and Cao, W. (2002). Abnormal motility in patients with ulcerative colitis: the role of inflammatory cytokines. Arch. Surg. 137, 439–445. doi: 10.1001/archsurg.137.4.439
Walker, M. Y., Pratap, S., Southerland, J. H., Farmer-Dixon, C. M., Lakshmyya, K., and Gangula, P. R. (2018). Role of oral and gut microbiome in nitric oxide-mediated colon motility. Nitric Oxide 73, 81–88. doi: 10.1016/j.niox.2017.06.003
Winston, J. H., Li, Q., and Sarna, S. K. (2013). Paradoxical regulation of ChAT and nNOS expression in animal models of Crohn's colitis and ulcerative colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 305, G295–302. doi: 10.1152/ajpgi.00052.2013
Keywords: colitis, colon motility, nitric oxide (NO), antioxidants, oral microbiome, operational taxonomic units (OTUs), gut microbiome
Citation: Dinakaran V, Mandape SN, Shuba K, Pratap S, Sakhare SS, Tabatabai MA, Smoot DT, Farmer-Dixon CM, Kesavalu LN, Adunyah SE, Southerland JH and Gangula PR (2019) Identification of Specific Oral and Gut Pathogens in Full Thickness Colon of Colitis Patients: Implications for Colon Motility. Front. Microbiol. 9:3220. doi: 10.3389/fmicb.2018.03220
Received: 11 September 2018; Accepted: 11 December 2018;
Published: 04 January 2019.
Edited by:
Nurul-Syakima Ab Mutalib, UKM Medical Molecular Biology Institute (UMBI), MalaysiaReviewed by:
Christopher L. Hemme, University of Rhode Island, United StatesChenyang Wang, Nanjing University Medical School, China
Copyright © 2019 Dinakaran, Mandape, Shuba, Pratap, Sakhare, Tabatabai, Smoot, Farmer-Dixon, Kesavalu, Adunyah, Southerland and Gangula. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pandu R. Gangula, cGdhbmd1bGFAbW1jLmVkdQ==
†These authors have contributed equally to this work
 Vasudevan Dinakaran
Vasudevan Dinakaran Sammed N. Mandape2†
Sammed N. Mandape2† Siddharth Pratap
Siddharth Pratap Mohammad Ali Tabatabai
Mohammad Ali Tabatabai Lakshmyya N. Kesavalu
Lakshmyya N. Kesavalu Janet Hayes Southerland
Janet Hayes Southerland Pandu R. Gangula
Pandu R. Gangula