
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 18 December 2018
Sec. Microbial Physiology and Metabolism
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.03149
This article is part of the Research Topic Actinobacteria, a Source of Biocatalytic Tools View all 19 articles
 Yi-Rui Yin1†
Yi-Rui Yin1† Peng Sang2†
Peng Sang2† Wen-Dong Xian1
Wen-Dong Xian1 Xin Li1
Xin Li1 Jian-Yu Jiao1
Jian-Yu Jiao1 Lan Liu1
Lan Liu1 Wael N. Hozzein3,4
Wael N. Hozzein3,4 Min Xiao1*
Min Xiao1* Wen-Jun Li1,5*
Wen-Jun Li1,5*The bioconversion of lignocellulose in various industrial processes, such as biofuel production, requires the degradation of cellulose. Actinomadura amylolytica YIM 77502T is an aerobic, Gram-positive actinomycete that can efficiently degrade crystalline cellulose by extracellular cellulases. Genomic analysis of A. amylolytica identified 9 cellulase and 11 β-glucosidase genes that could potentially encode proteins that digest cellulose. Extracellular proteome characterization of A. amylolytica cell-free culture supernatant by liquid chromatography tandem mass spectrometry analysis revealed that 4 of these cellulases and 2 of these β-glucosidases functioned during cellulose hydrolysis. Thin-layer chromatography analysis revealed extracellular β-glucosidases play a major role in carboxyl methyl cellulose (CMC) degradation of products in culture supernatants. In this study, 2 of the identified secreted β-glucosidases, AaBGL1 and AaBGL2, were functionally expressed in Escherichia coli and found to have β-glucosidase activity with wide substrate specificities, including for p-nitrophenyl β-D-glucopyranoside (pNPG), p-nitrophenyl-beta-D-cellobioside (pNPC), and cellobiose. Moreover, AaBGL1 and AaBGL2 had high tolerances for glucose. After adding these β-glucosidases to commercial cellulases, the degradation rates of CMC, Avicel, birch sawdust, and corncob powder increased by 37, 42, 33, and 9%, respectively. Overall, this work identifies an alternative potential source of β-glucosidases with potential applications in commercial cellulose utilization and the bioenergy industry.
Cellulose is an ecologically friendly material, the main component of lignocellulosic materials, especially agro-industrial residues, such as wheat straw, rice straw, corn stover, bagasse, and wood chips, and comprises 35–50% of a plant’s dry weight (Khandeparker and Numan, 2008; Brinchi et al., 2013). Cellulose is a polysaccharide consisting of a backbone of β-1,4-glucose units. A series of enzymes is essential for the complete degradation of cellulose into glucose, including endo-1,4-β-glucanases (endoglucanase, EC 3.2.1.4), exo-1,4-β-glucanases (i.e., cellobiohydrolases; EC 3.2.1.91), and β-glucosidases (EC 3.2.1.21). Endoglucanases and cellobiohydrolases act on internal cellulose chains and cellulose chain ends to release smaller fragments and cellobiose, respectively, and then the cello-oligosaccharides are ultimately degraded to glucose by β-glucosidases. The accumulation of cello-oligosaccharides, such as cellobiose and cellotriose, inhibits the functioning of both endoglucanases and cellobiohydrolases during simultaneous saccharification and, therefore, β-glucosidases play a crucial role in enzymatic degradation of cellulose by relieving product inhibition of cello-oligosaccharides for cellulases (Chauve et al., 2010).
A variety of microorganisms, including filamentous fungi, bacteria, and archaea, have the ability to produce cellulases (Jayant et al., 2011; Thomas et al., 2017). The filamentous fungus Trichoderma reesei (i.e., anamorph of Hypocrea jecorina) is a cellulase overproducer widely used in commercial and industrial lignocellulose degradation and bioethanol production (Henrissat et al., 1985; Huang et al., 2014). It secretes various endoglucanases, exoglucanases and beta-glucosidases to break down and convert cellulose into glucose (Ouyang et al., 2006; Peterson and Nevalainen, 2012). However, the β-glucosidase activity of these cellulase mixtures is too low to prevent inhibition by cellobiohydrolase following cellobiose accumulation (Saloheimo et al., 2002; Zhang et al., 2010). The low β-glucosidase activity of the extracellular secretions of T. reesei limits their application in degradation of cellulosic biomass (Nieves et al., 1998; Rahman et al., 2009). Therefore, the discovery of additional β-glucosidases is necessary to enhance the efficiency of cellulose hydrolysis.
Based on analysis of sequences, enzymatic properties, and three-dimensional protein structures, β-glucosidases that hydrolyze substrates through double displacement mechanisms, which also permit enzymes to transglycosylate, are classified as members of glycoside hydrolase (GH) families 1, 3, 5, 9, 16, 30, and 116 (Thongpoo et al., 2013)1. A large number of β-glucosidases from the GH1 and GH3 families have been purified from microorganisms and characterized (Wang et al., 2005). Most bacterial β-glucosidases employed in cellulose hydrolysis belong to the GH1 family (Cantarel et al., 2009). A majority of these bacterial β-glucosidases are sensitive to glucose, with only a few being glucose tolerant (Bohlin et al., 2010; Datta, 2016), which is a major barrier to efficient utilization of cellulose (Mallerman et al., 2014). To effectively hydrolyze cellulose and accumulate high levels of monosaccharides during enzymatic hydrolysis of lignocelluloses, the GH1 β-glucosidase should have a high glucose tolerance.
In the work presented here, two secreted GH1 β-glucosidases, AaBGL1 and AaBGL2, were identified by secretome and genomic analyses of Actinomadura amylolytica YIM 77502T. AaBGL1 and AaBGL2 were then functionally expressed in Escherichia coli BL21 and the resulting recombinant proteins were purified and characterized. Both AaBGL1 and AaBGL2 can degrade cellobiose and p-nitrophenyl β-D-glucopyranoside (pNPG). AaBGL1 can also effectively degrade p-nitrophenyl β-D-cellobioside (pNPC). Furthermore, AaBGL1 and AaBGL2 are highly tolerant of glucose. After adding these β-glucosidases to commercial cellulase, the degradation rate of carboxyl methyl cellulose (CMC), Avicel, corncob powder, and birch sawdust increased. The demand for β-glucosidases that are insensitive to glucose is increasing with commercial cellulase utilization. Compared with other β-glucosidases, both AaBGL1 and AaBGL2 had high Ki values. These results suggest GH1 β-glucosidases secreted from A. amylolytica have potential applications in industrial cellulose utilization.
Actinomadura amylolytica YIM 77502T (i.e., DSM 45822T and CCTCC AA 2012024T) was stored in our laboratory (Jiao et al., 2015). The E. coli DH5α strain (Invitrogen, United States) was used for cloning and the E. coli BL21 DE3 strain (Invitrogen, United States) was utilized as the host for expression of proteins. The pET28a vector (Invitrogen, United States) was used to construct recombinant plasmids. A. amylolytica YIM 77502T was cultured on R2A medium (0.5 g/l yeast extract, 0.5 g/l peptone, 0.5 g/l glucose, 0.5 g/l soluble starch, 0.5 g/l casein acid hydrolysates, 0.5 g/l sodium pyruvate, 0.3 g/l K2HPO4, and 0.024 g/l MgSO4) at 40°C. Recombinant E. coli strains were grown at 37°C in lysogeny broth (LB) medium (1% tryptone, 0.5% yeast extract, and 1% sodium chloride; pH 7.0) containing kanamycin (50 μg/ml).
Actinomadura amylolytica YIM 77502T was cultured in R2A and then genomic DNA was extracted using a MasterPure Gram-positive DNA Purification kit (Epicentre MGP04100) following the standard DNA isolation procedure recommended by the manufacturer with modifications (Wu et al., 2009). Whole-genome sequencing was performed on the Illumina HiSeq 2500-PE125 platform with massively parallel sequencing Illumina technology at the Beijing Novogene Bioinformatics Technology Co., Ltd. All high-quality paired reads were assembled using the SOAP de novo2 into a number of scaffolds (Gu et al., 2013). Glimmer v3.0 (Chen et al., 2011) was used for gene prediction in assembled sequences of A. amylolytica YIM 77502T. The sequence data described here have been deposited in GenBank (Accession number: CP032402). Proteins encoded by A. amylolytica were annotated using cluster of orthologous groups (COG) protein functional classification (Tatusov et al., 2003). Carbohydrate-active enzymes (CAZymes) of A. amylolytica YIM 77502T were identified using the CAZyme Analysis Toolkit3 (Petit et al., 2015). The glycoside hydrolase families were analyzed using HMMER software based on the Pfam database4 (Finn et al., 2011).
Degradation of filter paper was assessed by inoculating strain A. amylolytica YIM 77502T into a test tube containing filter paper basic medium (NaH2PO4, KNO3, and MgSO4) after static culture at 40°C for 1 month. A. amylolytica was cultured in cellulose basic medium (NaH2PO4, KNO3, MgSO4, and Avicel 2 g/l) at 40°C with shaking at 180 rpm for 14 days. The supernatant was harvested by centrifuging for 10 min at 12,000 × g at 4°C.
The cellulose degradation activity of the above fermentation broth was measured using CMC (Sigma, United States), Avicel (Sigma, United States), and cellobiose (Sigma, United States). Fermentation broth (100 μl) was added to 400 μl PBS buffer (pH 7) containing 1% (w/v) CMC or Avicel to test cellulase activity. This mixture was incubated at different temperatures (25–90°C) for 1 h. The release of reducing sugar from CMC was determined using the 3,5-dinitrosalicylic acid assay and measuring the absorbance at 540 nm (Miller, 1959). One unit (IU) of CMCase activity was defined as the volume of fermentation broth required to release 1 μmol of reducing sugar from CMC per minute. Hydrolytic products of the fermentation broth from CMC were identified by thin-layer chromatography (TLC) using silica gel 60 plates (Merck, Darmstadt, Germany) developed with 1-butanol/acetic acid/water (2:1:1, v/v/v). Sugars were detected by heat treatment at 120°C for 10 min after spraying the plates with freshly prepared 5% (v/v) H2SO4 in ethanol (Yin et al., 2015). Glucose (G1), cellobiose (G2), cellotriose (G3), and cellotetrose (G4) were used as sugar standards. The sugar standards was purchased from Sigma, United States.
The β-glucosidase activity of the fermentation broth was measured using cellobiose. Fermentation broth (100 μl) was added to 400 μl PBS buffer (pH 7) containing 1% (w/v) cellobiose at different temperatures (25–80°C) for 30 min to test β-glucosidase activity. The release of glucose from cellobiose was determined based on the absorbance at 490 nm of the mixture using a Glucose Oxidase Assay Kit (Abnova, China). One unit (IU) of β-glucosidase activity was defined as the volume of fermentation broth required to release 2 μmol glucose from cellobiose per minute.
The above fermentation broth was transferred into 5 kD ultra-filter concentrators (Sartorius, Germany) and centrifuged for 30 min at 4°C in a swing bucket rotor, resulting in the medium become concentrated by 30-fold. Extracellular protein was precipitated overnight by ammonium sulfate precipitation. The precipitate was resuspended in 150 μL supernatant. Proteins were isolated and digested as described by Hornburg et al. (2014). Peptides were desalted on C18 StageTips as previously described and subjected to liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis (Schwanhäusser et al., 2013). The peptides were then separated using a nano high-performance liquid chromatography (HPLC; Thermo Fisher Scientific) with a C18 column. HPLC was directly coupled to a quadrupole-Orbitrap mass spectrometer via a nano electrospray ion source (Q ExactiveTM, Thermo Fisher Scientific). The mass spectrometry (MS) raw data was processed by MaxQuant (v. 1.3.8.2; Cox and Mann, 2008). MS/MS spectra were searched against the A. amylolytica YIM 77502T genomic database.
The putative β-glucosidase genes aabgl1 (1335 bp) and aabgl2 (1431 bp) from the genomic sequences of A. amylolytica were amplified via polymerase chain reaction (PCR) with the four designed primers listed in Table 1 using TransStarFastPfu Fly DNA Polymerase (TransGen Biotech, China). The PCR cycle consisted of denaturation at 98°C for 3 min, followed by 34 cycles at 98°C for 10 s, 65°C for 20 s, and 72°C for 45 s, and then a final incubation at 72°C for 5 min for the final extension. The PCR products were cloned into the pET28a plasmid, which had been digested with BamH I and Hind III, using the pEASY-Uni Seamless Cloning and Assembly Kit (TransGen Biotech, China). Recombinant plasmids with the correct size fragments were sequenced. Subsequently, the DNA from the correct recombinant plasmids were transformed into E. coli BL21 (DE3) for protein expression.
DNA and protein sequences were aligned using the BLASTx and BLASTp programs (Madden, 2002)5, respectively. Signal peptides were predicted using SignalP (Bendtsen et al., 2004)6. The primary structures of the amino acid sequences were deduced and analyzed using EXPASY tools7. Multiple alignments with protein sequences of the closely related AaBGL1 and AaBGL2 (retrieved from NCBI database) were carried out using Clustal X (Thompson et al., 1997). Phylogenetic analyses were performed using the MEGA 5 software package (Tamura et al., 2011). Trees were constructed using the maximum likelihood (ML) method with a Poisson correction model. Structural models of AaBGL1 and AaBGL2 were generated with the MODELLER package (Sali and Blundell, 1993) using the β-glucosidase BglM-G1 mutant H75R (PDB ID, 5NS7; sequence identity, 47%) from a marine metagenome and β-xylosidase (PDB ID, 1GNX; sequence identity, 52%) from Streptomyces sp. as templates.
To express the recombinant AaBGL1 and AaBGL2 proteins, the above transformants were cultured overnight in LB medium containing 100 μg/ml kanamycin at 37°C and with shaking at 220 rpm. One ml of overnight culture was inoculated into 100 ml fresh LB medium containing 100 μg/ml kanamycin and incubated at 37°C with shaking at 220 rpm. To induce expression of the recombinant β-glucosidases, 0.1 ml of 100 mM IPTG was added to the cell suspension until the cells reached mid-exponential phase (OD600≈0.6) and incubated at 25°C with shaking at 220 rpm for 6 h. Cell pellets were harvested by centrifuging at 8,000 × g at 4°C for 10 min and resuspended in 20 ml buffer A (20 mM sodium phosphate, 300 mM NaCl, and 10 mM Tris; pH 8.0).
The resuspended cells were disrupted by ultrasonication. The lysates were centrifuged at 12000 × g for 30 min at 4°C. Cell-free extracts were transferred onto a Ni-chelating affinity column (GE, United States) as the recombinant proteins possessed an N-terminal His-tag. The column was then washed with ten column volumes of buffer A and then ten column volumes of buffer A containing 20 mM imidazole (pH 8.0) and eluted with buffer A containing 300 mM imidazole (pH 8.0). The eluted proteins were desalted using disposable PD-10 Desalting Columns (GE, United States) and the desalted proteins were used for enzyme characterization. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was performed using a 10% polyacrylamide gel that was then stained with Coomassie brilliant blue dye R-250 (Liu et al., 2010). Protein concentrations were determined with Bradford Protein Assay Kit (Order NO. C503031, Sangon Biotech, China) using bovine serum albumin as the standard.
β-glucosidase activity was assayed using cellobiose and p-nitrophenyl-β-D-glucopyranoside (pNPG). Activity against cellobiose was determined by adding 10 μg of purified protein to a 200-μl reaction mixture containing 2% (w/v) cellobiose. After 10 min, the activity was tested using a Glucose Oxidase Assay Kit (Abnova, China). One unit (U) of β-glucosidase activity was defined as the amount of enzyme required to release 2 μmol glucose from cellobiose per minute. When using pNPG as the substrate, 10 μg protein was added to a 200-μl reaction mixture containing 2.5 mM pNPG (Sigma, St. Louis, MO, United States). After 5 min of incubation at the optimal temperature, the reaction was stopped by adding 0.6 ml of 1 M Na2CO3 (Yang et al., 2015). The pNP was measured by monitoring the absorbance at 405 nm (Harnpicharnchai et al., 2009). One unit of β-glucosidase activity was equivalent to 1 μmol of pNP released from the pNPG in 1 min.
The optimal temperatures for AaBGL1 and AaBGL2 were determined by measuring β-glucosidases activity at different temperatures (10–80°C) at the optimal pH according to the activity assay method. For thermostability analysis, purified β-glucosidases were incubated in buffer A at temperatures ranging between 10 to 80°C for 30, 60, and 120 min. The samples were rapidly cooled in an ice-water bath and residual activity was measured by the standard method.
The optimal pHs for purified recombinant AaBGL1 and AaBGL2 were investigated in pHs ranging from 2.0 to 11 in buffer (50 mM Na2HPO4-citric acid, pH 2.0–8.0; 50 mM Tris-HCl, pH 8.0–9.0; and 50 mM glycine-NaOH, pH 9.0–11.0). The stabilities of purified AaBGL1 and AaBGL2 at different pHs were assessed by incubating these enzymes in buffers with different pHs as described above at 4°C for 12 and 24 h. The amount of residual activity at the optimal temperature was then determined according to the activity assay method.
To evaluate the effects of metal ions and chemical reagents on enzymatic activity, 10 mM of various metal ions and chemical reagents (K+, Na+, Fe3+, Mg2+, Mn2+, Ca2+, Cu2+, Co2+, Zn2+, Ni2+, SDS, and EDTA) were added individually to the reaction system. The control was tested using the same process described above without any additive in the reaction mixture.
To investigate the effect of the end product glucose on catalytic activity, the reaction was carried out in the presence of glucose concentrations ranging from 0 to 3000 mM. The concentration of the initial substrate pNPG was 1 mM. For the control, the same reaction system was used, but glucose was not added.
To determine the substrate specificity of AaBGL1 and AaBGL2, cellobiose, beechwood xylan, CMC, Avicel, and different p-nitrophenyl derivatives, such as p-nitrophenyl-β-D-glucopyranoside (pNPG), p-nitrophenyl-α-D-glucopyranoside (pNP-α-G), p-nitrophenyl β-D-xylopyranoside (pNPX), and p-nitrophenyl-β-D-cellobioside (pNPC), were used as substrates to measure enzymatic activity. All these substrates were purchased from Sigma, United States. The release of pNP was determined by measuring the absorbance at 405 nm of the mixture using pNP as the standard. One unit (U) of activity was defined as the amount of enzyme released from 1 μmol pNP/min. β-glucosidase activity for cellobiose was tested using the Glucose Oxidase Assay Kit (Abnova, China). One unit (U) of β-glucosidase activity was defined as the amount of enzyme required to release 2 μmol glucose/min from cellobiose. Beechwood xylan, glucan, CMC, and Avicel were measured using the 3,5-dinitrosalicylic acid assay (Miller, 1959). One unit (U) of enzymatic activity was defined as the amount of enzyme required to release 1 μmol glucose or xylose-equivalent reducing sugars per minute. All the substrates were purchased from Sigma (St. Louis, MO, United States).
The kinetic constants (Vmax and Km) of AaBGL1 and AaBGL2 were determined using different concentrations of cellobiose (0.2–2 mg/ml) and pNPG (0.5–5.0 mg/ml) at the optimal pH and temperature for 5 min. The Vmax and Km of the β-glucosidases for cellobiose and pNPG were calculated using Lineweaver-Burk plots.
Enzymatic hydrolysis of different lignocellulosic materials, such as CMC, Avicel, birch sawdust, and corncob, was performed by adding the appropriate cellulases (Sangon Biotech, China) and/or AaBGL1 and AaBGL2 to a 0.5 ml reaction system containing 50 mM Na2HPO4-citric acid buffer (pH 6.0) and 0.5 ml of 1% (w/v) substrate at 40 and 50°C for 24 h. The dosages of cellulases and β-glucosidases (AaBGL1 and AaBGL2) were 0.2 and 0.02 U/mg, respectively, based on the dry weight of substrate (Ye et al., 2017). Pretreatment of birch sawdust and corncob with ionic liquid (1-ethyl-3-methylimidazolium acetate) was carried out according to the method described in previous studies (Cheng et al., 2012). After enzymatic hydrolysis, the reactions were terminated by boiling for 10 min. The supernatants were collected by centrifugation at 4°C at 10000 × g for 10 min. The glucose contents of the hydrolysis liquors were measured using the Glucose Oxidase Assay Kit (Abnova, China).
The complete genome of A. amylolytica YIM 77502T consisted of a single circular 6.8 Mbp chromosome, and a G+C content of 73.28%. The genome contained 6,373 coding sequences with an average length of 922 bp. The general genomic features of A. amylolytica are listed in Supplementary Table S1. Among the predicted genes, 95.8% (6,105 genes) were assigned a function and 4.2% (268 genes) had unknown functions. In addition, 86.4% of the sequence encoded genes, while 13.6% belonged to internal sequences. The functions of these genes were mainly gene transcription (590), signal transduction (373), amino acid transport and metabolism (354), carbohydrate transport and metabolism (344), lipid transport and metabolism (324), energy production and conversion (289), coenzyme transport and metabolism (284), and cell wall/membrane/envelope biogenesis (273), as shown in Supplementary Figure S1.
A CAZyme analysis was conducted to identify potential enzymes with plant cell-wall degradation ability. Through this analysis, 81 carbohydrate-binding modules (CBMs) distributed in 12 families, 63 glycoside hydrolases (GHs) in 32 families, 2 polysaccharide lyases (PLs) in 2 families, 22 carbohydrate esterases (CEs) in 5 families, 71 glycosyl transferases (GTs) in 14 families, and 12 auxiliary activities (AAs) in 4 families were found to be encoded in the genome of A. amylolytica (Figure 1A). GH1 (glycoside hydrolase family 1) and GH3 were predicted to function as β-glucosidases and GH5, GH6, GH12, GH48 and GH74 were predicted cellulases. Altogether, 11 β-glucosidase genes and 9 cellulase genes were predicted in the genome of A. amylolytica (Figure 1B).
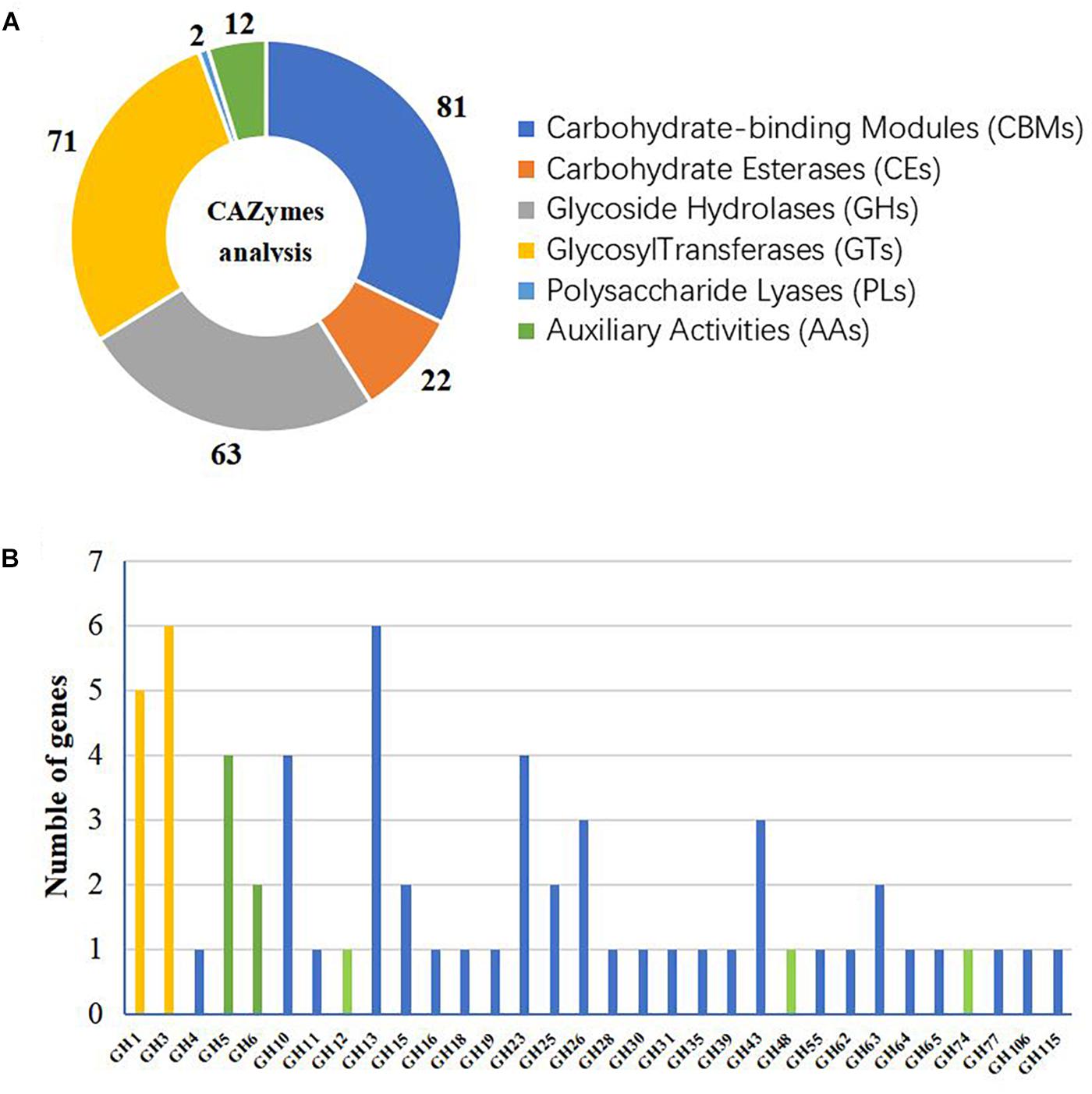
Figure 1. Glycoside hydrolase (GH) families of Actinomadura amylolytica YIM 77502T. (A) CAZyme analysis. (B) Number of GH genes. Histogram colors indicate different functions: yellow indicates predicted β-glucosidase, green indicates predicted cellulase, and blue indicates other predicted functions.
As shown in Figure 2A, filter paper was degraded by A. amylolytica YIM 77502T after culturing 1 month. TLC demonstrated the hydrolytic products of fermentation broth from CMC were mainly glucose and cellobiose (Figure 2B). Fermentation broths from cultures in microcrystalline cellulose basic medium for 2 weeks exhibited CMC and cellobiose activity under test conditions. High CMCase activity was found from 50 to 65°C, where the highest β-glucosidase activity was observed at 50°C (Figures 2C,D).
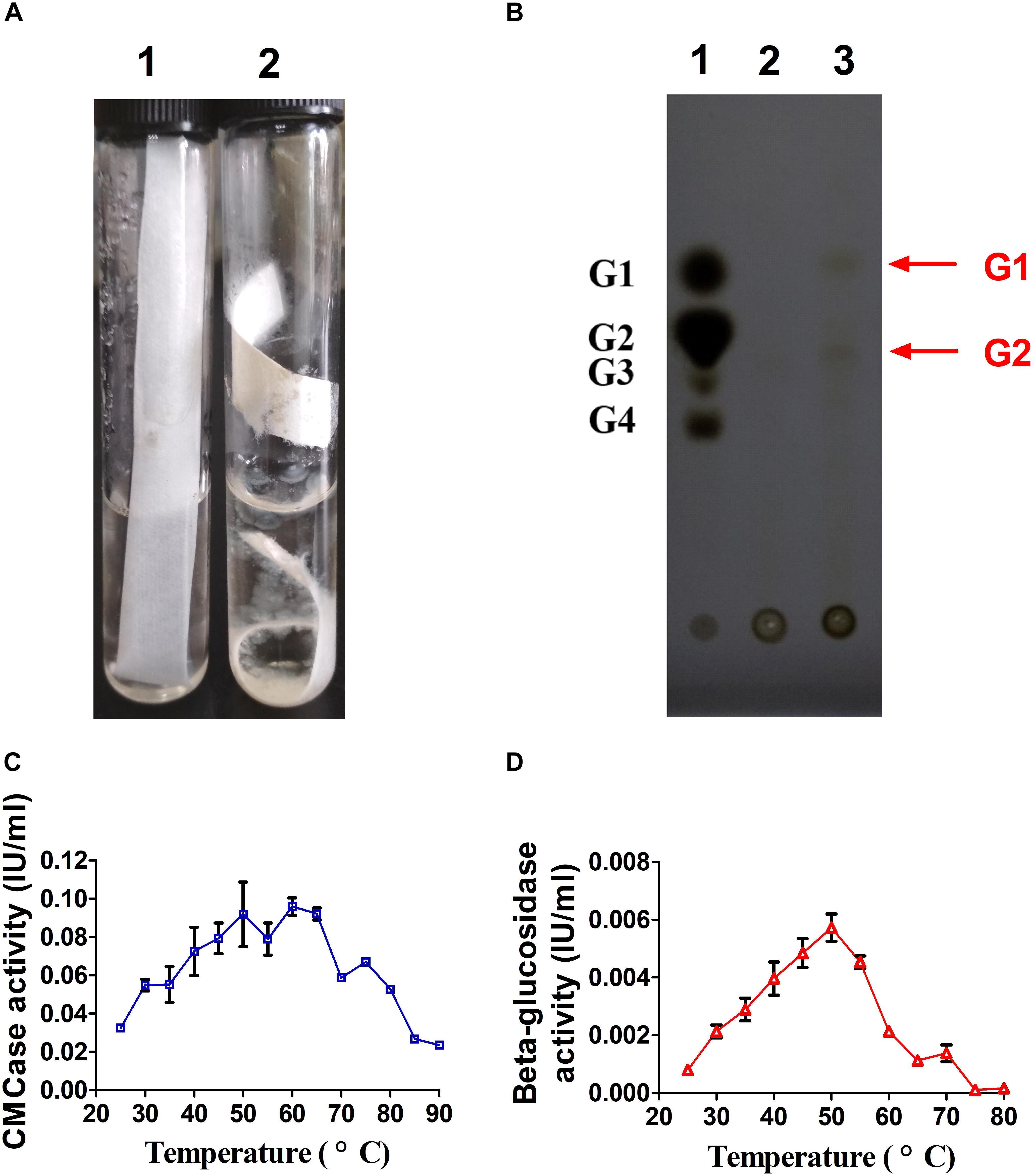
Figure 2. Cellulase activity of A. amylolytica YIM strain 77502T. (A) Hydrolysis experiment using filter paper. (B) Thin-layer chromatography plate analysis of hydrolytic products of carboxyl methyl cellulose (CMC) in fermentation broth of A. amylolytica, which was cultured with microcrystalline cellulose. (C) CMCase activity of fermentation broth. (D) β-glucosidase activity of fermentation broth.
The secretome of A. amylolytica was examined by MS. Secretome analysis revealed 209 proteins produced by A. amylolytica, which were mainly involved in carbohydrate transport and metabolism (43), amino acid transport and metabolism (23), protein turnover (18), cell wall/membrane/envelope biogenesis (16), energy production and conversion (10), and signal transduction (10), as shown in Supplementary Figure S2. Among these secreted proteins, 4 cellulases belonging to GH5 and GH6, 4 xylanases belonging to GH10, and two β-glucosidases belonging to GH1 were identified among the secretion proteins (Figure 3). These two β-glucosidases were designated AaBGL1 and AaBGL2.
According to the genome sequences of A. amylolytica YIM 77502T, two putative genes encoding β-glucosidases AaBGL1A (GenBank: MH974516) and AaBGL2 (GenBank: MH974517) were amplified by PCR and introduced into the pET28a vector to construct recombinant plasmids pET28a-aabgl1 and pET28a-aabgl2. AaBGL1 and AaBGL2 consisted of 444 and 476 residues with theoretical molecular weights of 48.47 and 52.61 KDa, respectively. Signal peptides were not predicted at the N-termini of AaBGL1 and AaBGL2 based on analysis with Signal P 4.1 Server8. Sequence analysis revealed AaBGL1 and AaBGL2 contained distinct catalytic modules of glycosyl hydrolase family 1 (GH1) in the predicted enzyme proteins. The deduced amino acid sequence of AaBGL1 had the highest amino acid sequence identity (89%) to β-glucosidase (NCBI: WP 103938629.1) from Actinomadura echinospora. AaBGL2 had 84% similarity with β-glucosidases from Thermomonospora curvata (NCBI: WP 012852091.1) and Actinomadura echinospora (NCBI: WP 103937196.1). As shown in Figures 4A,B, the predicted three-dimensional model structures of AaBGL1 and AaBGL2 were very similar to other GH1 β-glucosidases with known structures The monomeric subunit of GH1 β-glucosidase adopts the expected topology of a single (α/β)8 barrel, with additional units of quite well-conserved secondary structures inserted between the α/β units. A phylogenetic analysis of the protein sequences revealed AaBGL1 and AaBGL2 also clustered with β-glucosidases (Supplementary Figures S3, S4).
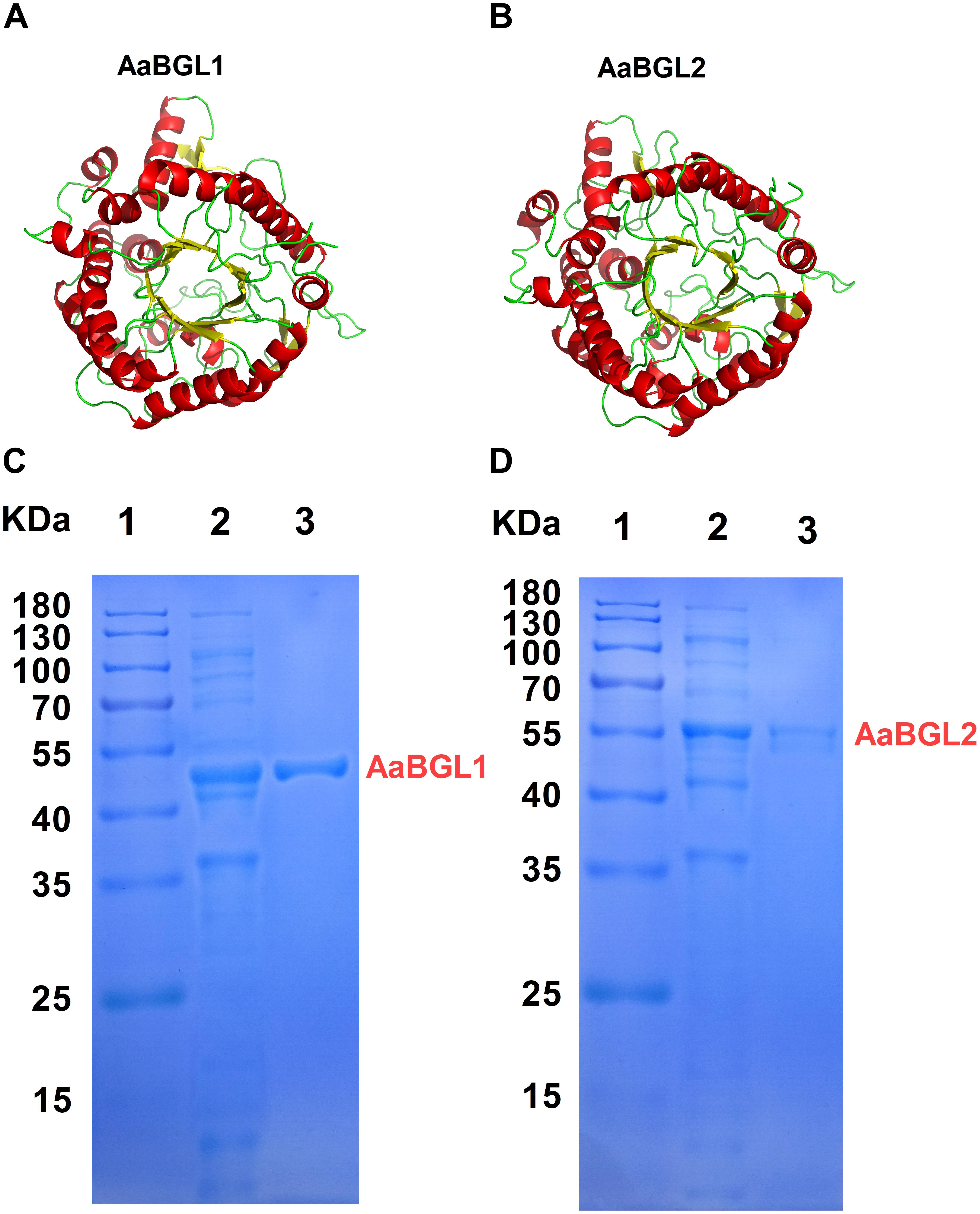
Figure 4. Three-dimensional model and purification of β-glucosidases AaBGL1 and AaBGL2. Structural models of (A) AaBGL1 and (B) AaBGL2. Sodium dodecyl sulfate polyacrylamide gel electrophoresis analysis of recombinant (C) AaBGL1 and (D) AaBGL2 produced by E. coli BL21. Lane 1, protein molecular weight marker, mass indicated on the left; lane 2, total protein in IPTG-induced E. coli BL21/pET28a-aabgl1 or pET28a-aabgl2; lane 3, purified AaBGL1 or AaBGL2.
The β-glucosidase genes were successfully expressed in E. coli BL21 (DE3) and the resulting recombinant proteins with His-tagged N-termini were purified by Ni-NTA affinity chromatography. SDS-PAGE analysis indicated the molecular masses of the recombinant β-glucosidase proteins were in good agreement with the theoretical ones (Figures 4C,D).
AaBGL1 exhibited high activity at 10–65°C at the optimal pH of 6.0. AaBGL2 exhibited the highest activity at 50°C and a pH of 6.0 (Figure 5A). Within 2 h, AaBGL1 and AaBGL2 were stable below 50°C, but their activity rapidly decreased when the temperature rose above 50°C (Supplementary Figures S5A, S6A). The thermostabilities of AaBGL1 and AaBGL2 were similar to reported β-glucosidases from fungi and bacteria, which were lost most activity when the temperature above 50°C (Santos et al., 2016; Florindo et al., 2018).
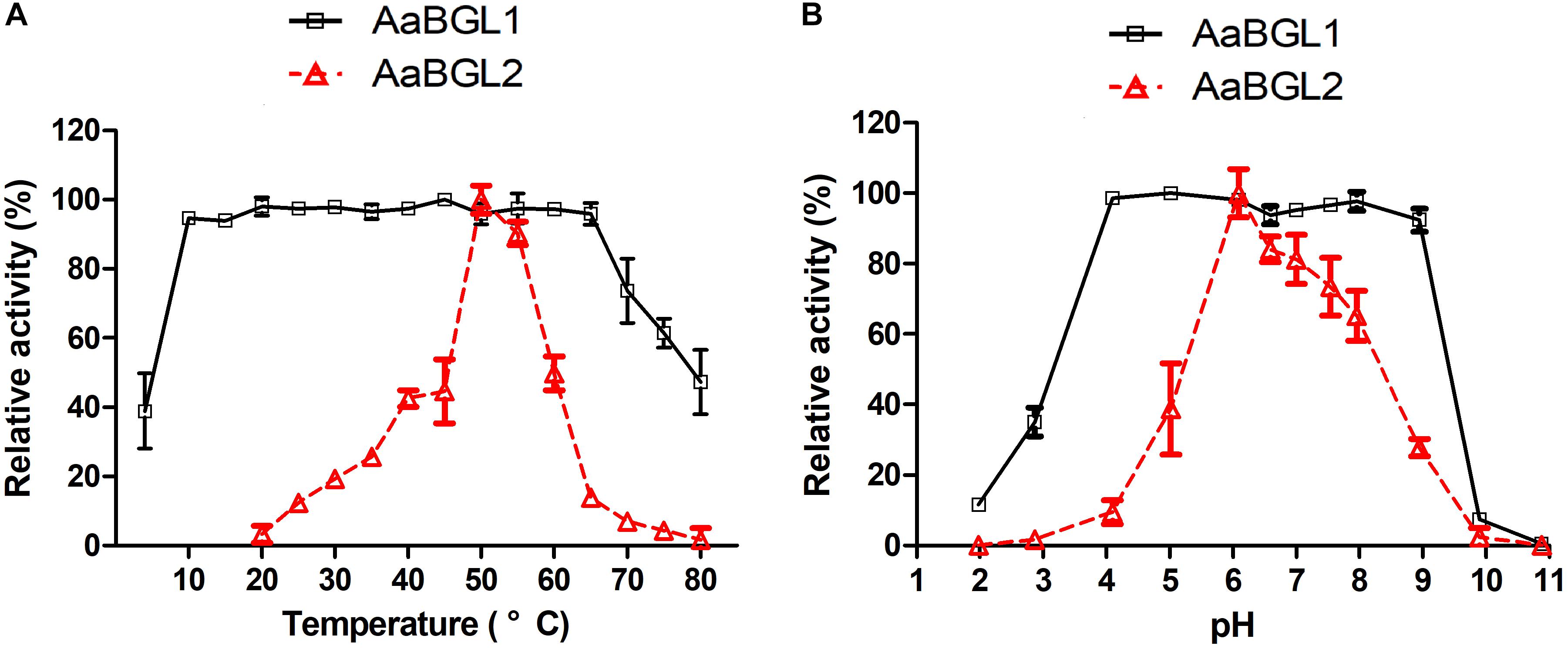
Figure 5. Effects of temperature and pH on the activity and stability of the recombinant AaBGL1 and AaBGL2. (A,B) Effect of (A) temperature and (B) pH on the activity of the recombinant β-glucosidases. The primary activity was designated 100%. Each value in the figure represents the mean ± standard deviation (n = 3). AaBGL1: 100% = 6.2 U/mg. AaBGL2 100% = 5.6 U/mg.
The effects of pH on the activity of the β-glucosidases AaBGL1 and AaBGL2 were assessed. AaBGL1 functioned in a broad optimum pH, retaining more than 95% of the catalytic activity at pHs ranging from 4.0 to 9.0. The optimum pH of AaBGL2 was 6 (Figure 5B). As shown in Supplementary Figures S5B, S6B, AaBGL1 was stable in the pH range of 4.0–9.0, retaining 100% residual activity after incubating for 12 h in pH buffer at 25°C. AaBGL2 was relatively pH stable, retaining more than 60% residual activity after incubation for 24 h at pH 4–10 at 25°C.
The influence of various metal ions and chemical reagents on the activity of AaBGL1 and AaBGL2 was also investigated and the results are shown in Supplementary Figure S7. The enzymatic activity of AaBGL1 was 102 ± 0.3% and 100 ± 1.2% by K+ and Mg2+, respectively. Na+, Ca2+, Co2+, Ni2+, and SDS ions at a concentration of 10 mM did not affect AaBGL1 activity. Fe3+ and Mn2+ slightly inhibited both AaBGL1 and AaBGL2 activity, while Cu2+, Co2+, Zn2+, Ni2+, and EDTA severely inhibited AaBGL1 activity. The enzymatic activity of AaBGL2 was not influenced by K+, Na+, Mg2+, or Ca2+; however, the of addition Cu2+, Co2+, Zn2+, Ni2+, SDS, and EDTA strongly inhibited AaBGL2 activity.
The glucose tolerances of AaBGL1 and AaBGL2 were determined using 1 mM pNPG as a substrate. The activity of AaBGL1 and AaBGL2 measured in the absence of exogenous glucose was set as 100%. As shown in Figure 6, glucose had no effect on AaBGL1 activity when the added glucose concentration was less than 1000 mM. Even when the added glucose concentration was 2000 times greater than the pNPG concentration, AaBGL1 still retained more than 40% activity, suggesting AaBGL1 is highly tolerant of glucose. AaBGL2 also retained 40% activity when the added glucose concentration was 500 times greater than the pNPG concentration (Figure 6). This suggests AaBGL2 is also a glucose-tolerant β-glucosidase.
Analysis of substrate specificity revealed AaBGL1 hydrolyzed pNPG, cellobiose, pNP-α-G, and pNPC, but not pNPX, beechwood xylan, CMC, or Avicel (Table 2). AaBGL2 exhibited activity for pNPG and cellobiose, but not other tested substrates. Based on Lineweaver-Burk plots (Supplementary Figures S8, S9), the Kcat, Km, and Vmax of AaBGL1 and AaBGL2 calculated using the pNPG and cellobiose as substrates are shown in Table 3.
To evaluate the potential use of AaBGL1 and AaBGL2 in degradation of lignocellulose, enzymatic hydrolysis of various lignocellulosic materials, including CMC, Avicel, and ionic liquid-pretreated birch sawdust and corncob powder, were performed. As shown in Figure 7, the glucose concentrations in the hydrolysis liquors were ordered Avicel > corncob > birch sawdust > CMC. After adding the β-glucosidases AaBGL1 and AaBGL2 to commercial cellulases, the degradation rates of CMC, Avicel, birch sawdust, and corncob powder increased by 37, 42, 33, and 9%, respectively. This suggests AaBGL1 and AaBGL2 cooperated with commercial cellulases in cellulose degradation.
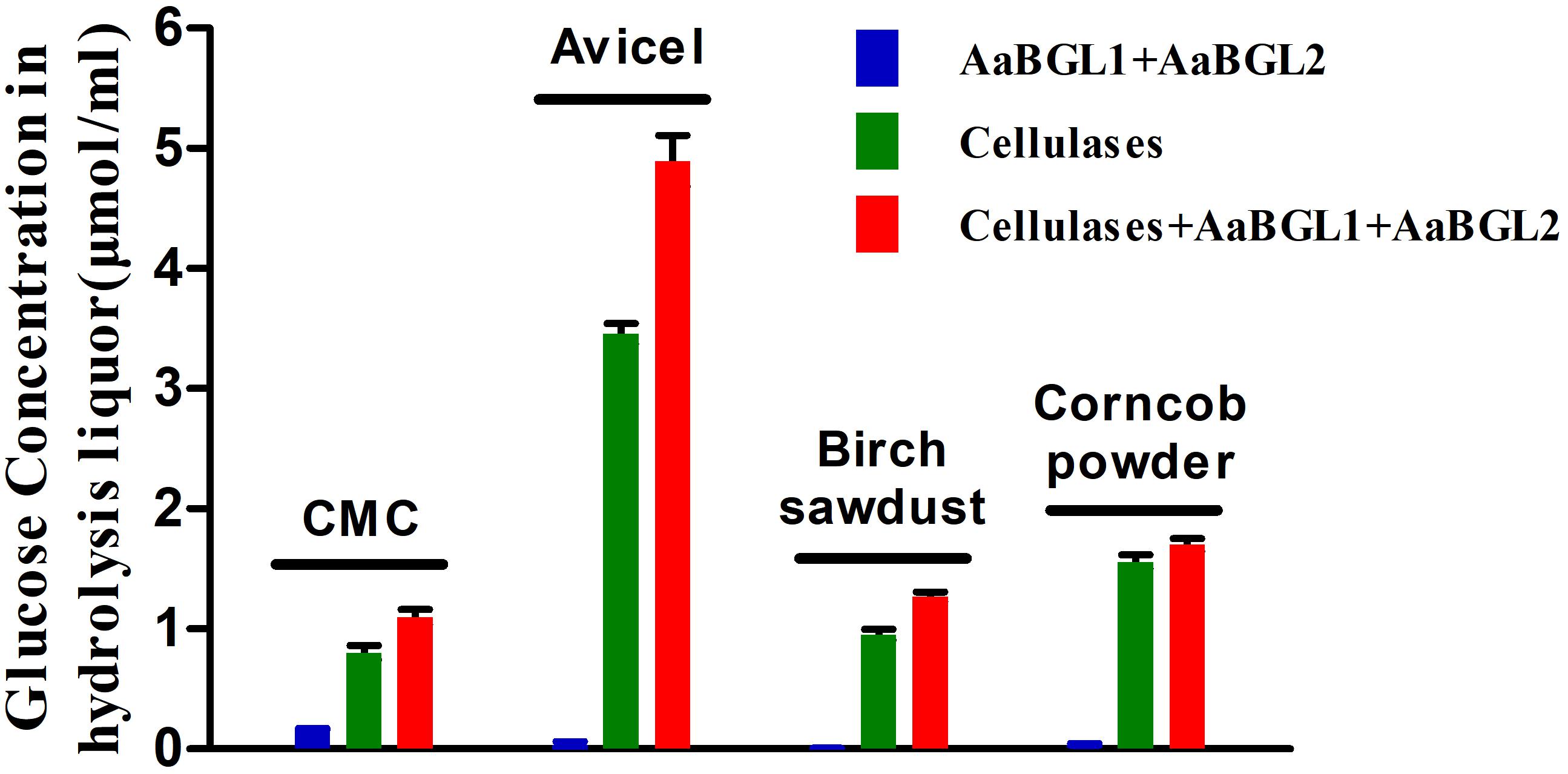
Figure 7. Cooperation of AaBGL1 and AaBGL2 with commercial cellulases in cellulosic material degradation.
An aerobic Gram-positive actinomycete, A. amylolytica YIM 77502T can digest cellulose rapidly and efficiently. A large number of glycosidase enzymes, including 9 cellulase genes and 11 β-glucosidase genes, have been predicted in the genome of A. amylolytica. Other actinomycetes, such as Thermobifida fusca (Wilson, 2010), Thermobifida halotolerans YIM 90462T (Yin et al., 2015), Thermoactinospora rubra YIM 77501T (Yin et al., 2017), and Streptomyces strains (Ventorino et al., 2016), also display highly lignocellulose-degrading activities. To date, many cellulose-degrading enzymes have been identified from cellulolytic actinomycetes (Vanee et al., 2017), indicating cellulolytic actinomycetes, such as A. amylolytica, are good potential sources of cellulose hydrolases.
In this research, 4 cellulases and 2 β-glucosidases were identified from the secretome of A. amylolytica YIM 77502T cultured with microcrystalline cellulose. These enzymes may be involved in the degradation of cellulose, but no signal peptides were found in the 2 identified β-glucosidases, AaBGL1 and AaBGL2. Broeker et al. (2018) also found 32 proteins lacking signal peptides in the secretome of Clostridium stercorarium. These 2 β-glucosidases of A. amylolytica found in the secretome may be secreted through the release of proteins from cells through cell lysis or alternative secretion pathways. The sequence similarity, tertiary structures, and ML phylogenetic trees of AaBGL1 and AaBGL2 suggest GH1 β-glucosidases are conserved. Discovery of AaBGL1 and AaBGL2 in the secretome of A. amylolytica indicates LC-MS/MS is potentially a highly useful method of discovering new proteins with specific functions.
During cellulose degradation, cellulolytic microorganisms secrete some hydrolytic enzymes, including free cellulases (endoglucanase, cellobiohydrolases, and β-glucosidases) and cellulosome, which consist of a wide variety of polysaccharide-degrading enzymes (e.g., cellulases, hemicellulases, and pectinases) (Wilson, 2009; Artzi et al., 2017). Most bacteria and fungi hydrolyze cellulose by secreting free cellulases (Cavedon et al., 1990; Ja’afaru, 2013). Meanwhile, a cellulosome is a supramolecular multienzyme complex that can efficiently degrade lignocellulose and is found in just a few bacterial species, including Clostridium (Ruminiclostridium) clariflavum and Clostridium (Ruminiclostridium) thermocellum (Bayer et al., 2004; Shiratori et al., 2009; Shinoda et al., 2018). As described above, A. amylolytica YIM 77502T secreted cellulases extracellularly to hydrolyze cellulose. A total of 344 and 38 genes in the genome of A. amylolytica were found to potentially be involved in carbohydrate transport and secretory transport of proteins, respectively (Supplementary Figure S1). Of these genes, membrane transporters are membrane proteins associated with transportation of macromolecules, such as proteins, across biological membranes (Saier et al., 2006). ATP-binding cassette transporters (ABC transporters) are members of a transport system superfamily and involved in the translocation of various substrates, such as glucose, cellobiose, and galactose, across membranes (Kemner et al., 1997; Petit et al., 2015).
Membrane transporters and ABC transporters from A. amylolytica possibly transfer cellulose-degrading enzymes and carbohydrates. Through the activity of A. amylolytica extracellular enzymes, secretome and TLC plate analyses of CMC hydrolysis products, including endoglucanase, cellobiohydrolases, and β-glucosidases, revealed hydrolysis of cellulose to oligosaccharides (cellobiose as the main product) and monosaccharide (glucose). As shown in a schematic diagram (Figure 8), cellulose-degrading enzymes are secreted into the extracellular medium and the cellulosic substrate is degraded by synergistic hydrolysis of these free glycosidolytic enzymes. The cellulose hydrolyze these products (cellobiose and glucose), which are then transported to the cytoplasm across the cell membrane through ABC transporters. The oligosaccharides are eventually hydrolyzed to monosaccharides by intracellular β-glucosidases. Finally, glucose entrance into the Embden-Meyerhof-Parnas pathway and tricarboxylic acid cycle provided material (carbon source) and energy (ATP) required for the growth and reproduction of A. amylolytica. This is the process of cellulose hydrolysis in pure culture in the laboratory simulation, but cellulose does not exist in a pure state in nature. Besides cellulose, there are also large amounts of hemicellulose, lignin, and pectin in plant biomass. Xylan, one of the main components of hemicellulose, can be hydrolyzed by xylanases. As shown in Figures 3, 4 GH10 xylanases were identified in the secretome of A. amylolytica, indicating they were also induced by cellulose. Robison reported that Trichoderma reesei Rut C-30 produces extracellular xylanases when grown on cellulose (Robison, 1984). The complete hydrolysis of lignocellulose requires the synergistic cooperation of cellulases, xylanase, pectinase, laccase, and other enzymes (Van Dyk and Pletschke, 2012). Therefore, for A. amylolytica, the collaborative expression of cellulases and xylanases may be a functional adaptation for hydrolysing plant biomass to be used as a carbon source.
Two β-glucosidases (AaBGL1 and AaBGL2) secreted by A. amylolytica exhibited synergistic cooperation with commercial cellulases. After adding AaBGL1 and AaBGL2, the decomposition rate of Avicel (40%), CMC (37%), and ionic liquid-pretreated birch sawdust (33%) increased by more than 30%. Meanwhile, the degradation rate of ionic liquid-pretreated corncob improved only 9%. Compared to wood, such as birch sawdust, corncob contains more hemicellulose as it is more than one-third of the dry matter of the corncob (Sun et al., 2014). This indicates degradation of substrate by the synergistic action of different kinds of cellulases is related to the purity or amount of cellulose in the substrate. It was also suggested that cellulases need to cooperate with other enzymes, such as hemicellulases (e.g., xylanase) and pectinase, when lignocellulose is degraded. The importance of the cocktail method for degradation of lignocellulosic materials also indicates that more new enzyme resources need to be found to enhance this synergistic degradation (Ibrahim, 2011).
The enzymatic properties of AaBGL1 and AaBGL2 include that, like other β-glucosidases, their activities were completely inhibited by Cu2+ (Chen et al., 2010; Wu et al., 2018). However, the activity of AaBGL1 was not affected by 10 mM SDS. A strong denaturant of proteins, SDS can inactivate most enzymes (Joo and Chang, 2010). The SDS stability of AaBGL1 suggests it is suitable for application in industrial purposes. During cellulose hydrolysis by cellulases, the hydrolysis products inhibit the activity of the enzymes. Most β-glucosidases are sensitive to the final product of glucose, which limits the use of β-glucosidase and efficient degradation of cellulose. The inhibition constants (Ki) of AaBGL1 and AaBGL2 were 1502 and 193.5 mM glucose, respectively. Meanwhile, the Ki of most fungal β-glucosidases, such as β-glucosidases from Penicillium citrinum UFV1, Chaetomium globosum, and Neurospora crassa, are between 0.1 and 10 mM (Bohlin et al., 2010; Da Costa et al., 2016). This indicates AaBGL1 and AaBGL2 are glucose-tolerant β-glucosidases. The glucose-tolerances of β-glucosidases are of great significance due to increases in glucose concentration and the conversion rates of soluble fermentable sugars from cellulose degradation.
Actinomadura amylolytica YIM 77502T exhibited CMCase and β-glucosidase activity when cultivated at 40°C using Avicel as the sole carbon source. Two GH1 β-glucosidases, designated AaBGL1 and AaBGL2, were identified in the secretome of A. amylolytica by LC-MS/MS. AaBGL1 and AaBGL2 were successfully expressed in E. coli BL21, and the recombinant proteins were purified and characterized. Both AaBGL1 and AaBGL2 were highly glucose-tolerant β-glucosidases and exhibited synergistic cooperation with commercial cellulases. Overall, the β-glucosidases studied in this work (AaBGL1 and AaBGL2) had different specificities and characteristics and could be used in different biotechnological applications, such as bioethanol production.
Y-RY, MX, and W-JL conceived the study. PS and WH were responsible for bioinformatics analysis of the genome and secretome. W-DX and XL cultured strains and collected samples. Y-RY and J-YJ separated proteins. LL and MX measured enzymatic activity. Y-RY and PS performed the data analysis and mapping. Y-RY, PS, MX, and W-JL wrote the manuscript. All authors discussed the results and commented on the manuscript. All authors read and approved the final manuscript.
This research was supported by the Infrastructure work project of the China Ministry of Science and Technology (No. 2015FY110100), Guangzhou Municipal People’s Livelihood Science and Technology Plan (No. 201803030030), and China Postdoctoral Science Foundation (Grant No. 2017M622861). The authors are grateful to the Deanship of Scientific Research, King Saud University for funding through Vice Deanship of Scientific Research Chairs. W-JL was supported by the Guangdong Province Higher Vocational Colleges & Schools Pearl River Scholar Funded Scheme (2014).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.03149/full#supplementary-material
FIGURE S1 | Clusters of orthologous groups (COG) functional classification of Actinomadura amylolytica genome-encoded proteins.
FIGURE S2 | COG functional classification of secretome of A. amylolytica.
FIGURE S3 | Phylogenetic dendrogram obtained by maximum likelihood (ML) analysis based on amino acid sequences showing the phylogenetic position of AaBGL1 with related β-glucosidases. Bootstrap values (expressed as a percentage of 1000 replications) are given at nodes.
FIGURE S4 | Phylogenetic dendrogram obtained by maximum likelihood (ML) analysis based on amino acid sequences showing the phylogenetic position of AaBGL2 with related β-glucosidases. Bootstrap values (expressed as a percentage of 1000 replications) are given at nodes.
FIGURE S5 | Effects of temperature and pH on the stability of recombinant AaBGL1. (A,B) The effect of (A) temperature and (B) pH on stability.
FIGURE S6 | Effects of temperature and pH on the stability of recombinant AaBGL2. (A,B) The effect of (A) temperature and (B) pH on stability.
FIGURE S7 | Effects of metal ions and reagents on the activity of (A) AaBGL1 and (B) AaBGL2. Statistical analysis was performed using one-way ANOVA followed by Tukey’s test for comparison of multiple treatment groups. Data marked with ∗ and ∗∗ were statistically significant different at the p < 0.05 and p < 0.001 cut-off values, respectively.
FIGURE S8 | Lineweaver-Burk plots of AaBGL1 for pPNG (A) and cellobiose (B).
FIGURE S9 | Lineweaver-Burk plots of AaBGL2 for pPNG (A) and cellobiose (B).
TABLE S1 | The general genomic features of A. amylolytica.
Artzi, L., Bayer, E. A., and Morais, S. (2017). Cellulosomes: bacterial nanomachines for dismantling plant polysaccharides. Nat. Rev. Microbiol. 15, 83–95. doi: 10.1038/nrmicro.2016.164
Bayer, E. A., Belaich, J. P., Shoham, Y., and Lamed, R. (2004). The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58, 521–554. doi: 10.1146/annurev.micro.57.030502.091022
Bendtsen, J. D., Nielsen, H., Heijne, G. V., and Brunak, S. (2004). Improved prediction of signal peptides: signalp 3.0. J. Mol. Biol. 340, 783–795. doi: 10.1016/j.jmb.2004.05.028
Bohlin, C., Olsen, S. N., Morant, M. D., Patkar, S., Borch, K., and Westh, P. (2010). A comparative study of activity and apparent inhibition of fungal beta-glucosidases. Biotechnol. Bioeng. 107, 943–952. doi: 10.1002/bit.22885
Brinchi, L., Cotana, F., Fortunati, E., and Kenny, J. M. (2013). Production of nanocrystalline cellulose from lignocellulosic biomass: technology and applications. Carbohydr. Polym. 94, 154–169. doi: 10.1016/j.carbpol.2013.01.033
Broeker, J., Mechelke, M., Baudrexl, M., Mennerich, D., Hornburg, D., Mann, M., et al. (2018). The hemicellulose-degrading enzyme system of the thermophilic bacterium Clostridium stercorarium: comparative characterisation and addition of new hemicellulolytic glycoside hydrolases. Biotechnol. Biofuels 11:229. doi: 10.1186/s13068-018-1228-3
Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V., and Henrissat, B. (2009). The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238. doi: 10.1093/nar/gkn663
Cavedon, K., Leschine, S. B., and Canale-Parola, E. (1990). Cellulase system of a free-living, mesophilic clostridium (strain C7). J. Bacteriol. 172, 4222–4230. doi: 10.1128/jb.172.8.4222-4230.1990
Chauve, M., Mathis, H., Huc, D., Casanave, D., Monot, F., and Lopes Ferreira, N. (2010). Comparative kinetic analysis of two fungal beta-glucosidases. Biotechnol. Biofuels 3:3. doi: 10.1186/1754-6834-3-3
Chen, S., Hong, Y., Shao, Z., and Liu, Z. (2010). A cold-active β-glucosidase (Bgl1C) from a sea bacteria Exiguobacterium oxidotolerans A011. World J. Microbiol. Biotechnol. 26, 1427–1435. doi: 10.1007/s11274-010-0317-7
Chen, Y., He, Y., Zhang, B., Yang, J., Li, W., Dong, Z., et al. (2011). Complete genome sequence of Alicyclobacillus acidocaldarius strain Tc-4-1. J. Bacteriol. 193, 5602–5603. doi: 10.1128/JB.05709-11
Cheng, G., Varanasi, P., Arora, R., Stavila, V., Simmons, B. A., Kent, M. S., et al. (2012). Impact of ionic liquid pretreatment conditions on cellulose crystalline structure using 1-ethyl-3-methylimidazolium acetate. J. Phys. Chem. B 116, 10049–10054. doi: 10.1021/jp304538v
Cox, J., and Mann, M. (2008). Maxquant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372. doi: 10.1038/nbt.1511
Da Costa, S. G., Pereira, O. L., Teixeira-Ferreira, A., Valente, R. H., da Rezende, S. T., Guimarães, V. M., et al. (2018). Penicillium citrinum UFV1 β-glucosidases: purification, characterization, and application for biomass saccharification. Biotechnol. Biofuels 11:226. doi: 10.1186/s13068-018-1226-5
Datta, S. (2016). Recent strategies to overexpress and engineer cellulases for biomass degradation. Curr. Metabolomics 4, 14–22. doi: 10.2174/2213235X03666150702155845
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, 29–37. doi: 10.1093/nar/gkr367
Florindo, R. N., Souza, V. P., Manzine, L. R., Camilo, C. M., Marana, S. R., Polikarpov, I., et al. (2018). Structural and biochemical characterization of a GH3 B-glucosidase from the probiotic bacteria, Bifidobacterium adolescentis. Biochimie 148, 107–115. doi: 10.1016/j.biochi.2018.03.007
Gu, S., Fang, L., and Xu, X. (2013). Using SOApaligner for short reads alignment. Curr. Protoc. Bioinformatics 44, 11.11.1–11.11.17. doi: 10.1002/0471250953.bi1111s44
Harnpicharnchai, P., Champreda, V., Sornlake, W., and Eurwilaichitr, L. (2009). A thermotolerant beta-glucosidase isolated from an endophytic fungi, Periconia sp., with a possible use for biomass conversion to sugars. Protein Expr. Purif. 67, 61–69. doi: 10.1016/j.pep.2008.05.022
Henrissat, B., Driguez, H., Viet, C., and Schülein, M. (1985). Synergism of cellulases from Trichoderma reesei in the degradation of cellulose. Nat. Biotechnol. 3, 722–726. doi: 10.1038/nbt0885-722
Hornburg, D., Drepper, C., Butter, F., Meissner, F., Sendtner, M., and Mann, M. (2014). Deep proteomic evaluation of primary and cell line motoneuron disease models delineates major differences in neuronal characteristics. Mol. Cell. Proteomics 13, 3410–3420. doi: 10.1074/mcp.M113.037291
Huang, J., Dong, C., Wei, Y., Wang, Q., Li, Z., Ying, C., et al. (2014). Direct ethanol production from lignocellulosic sugars and sugarcane bagasse by a recombinant Trichoderma reesei strain HJ48. Sci. World J. 2014:798683. doi: 10.1155/2014/798683
Ibrahim, M. F. (2011). Crude Cellulase Cocktail for Lignocellulosic Materials Degradation. Available at: https://www.researchgate.net/publication/2596779
Ja’afaru, M. I. (2013). Screening of fungi isolated from environmental samples for xylanase and cellulase production. ISRN Microbiol. 2013:283423. doi: 10.1155/2013/283423
Jayant, M., Rashmi, J., Shailendra, M., and Deepesh, Y. (2011). Production of cellulase by different co-culture of Aspergillus niger and Penicillium chrysogenum from waste paper, cotton waste and baggase. J. Yeast Fungal Res. 2, 24–27.
Jiao, J. Y., Liu, L., Zhou, E. M., Wei, D. Q., Ming, H., Xian, W. D., et al. (2015). Actinomadura amylolytica sp. nov. and Actinomadura cellulosilytica sp. nov., isolated from geothermally heated soil. Antonie van Leeuwenhoek 108, 75–83. doi: 10.1007/s10482-015-0465-8
Joo, H., and Chang, C. (2010). Oxidant and SDS-stable alkaline protease from a halo-tolerant Bacillus clausii I-52: enhanced production and simple purification. J. Appl. Microbiol. 98, 491–497. doi: 10.1111/j.1365-2672.2004.02464.x
Kemner, J. M., Liang, X., and Nester, E. W. (1997). The Agrobacterium tumefaciens virulence gene chvE is part of a putative ABC-type sugar transport operon. J. Bacteriol. 179, 2452–2458. doi: 10.1128/jb.179.7.2452-2458.1997
Khandeparker, R., and Numan, M. T. (2008). Bifunctional xylanases and their potential use in biotechnology. J. Ind. Microbiol. Biotechnol. 35, 635–644. doi: 10.1007/s10295-008-0342-9
Liu, S. Y., Shibu, M. A., Jhan, H. J., Lo, C. T., and Peng, K. C. (2010). Purification and characterization of novel glucanases from Trichoderma harzianum ETS 323. J. Agric. Food Chem. 58, 10309–10314. doi: 10.1021/jf1029338
Madden, T. (2002). “The BLAST sequence analysis tool,” in The NCBI Handbook Internet, ed. J. McEntyre (Bethesda, MD: National Library of Medicine).
Mallerman, J., Papinutti, L., and Levin, L. (2014). Characterization of β-glucosidase produced by the white rot fungus flammulina velutipes. J. Microbiol. Biotechnol. 25, 57–65. doi: 10.4014/jmb.1401.01045
Miller, G. L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Biochem. 31, 426–428. doi: 10.1021/ac60147a030
Nieves, R. A., Ehrman, C. I., Adney, W. S., Elander, R. T., and Himmel, M. E. (1998). Technical communication: survey and analysis of commercial cellulase preparation suitable for biomass conversion to ethanol. World J. Microbiol. Biotechnol. 14, 301–304. doi: 10.1023/A:1008871205580
Ouyang, J., Yan, M., Kong, D., and Xu, L. (2006). A complete protein pattern of cellulase and hemicellulase genes in the filamentous fungus Trichoderma reesei. Biotechnol. J. 1, 1266–1274. doi: 10.1002/biot.200600103
Peterson, R., and Nevalainen, H. (2012). Trichoderma reesei RUT-C30–thirty years of strain improvement. Microbiology 158, 58–68. doi: 10.1099/mic.0.054031-0
Petit, E., Coppi, M. V., Hayes, J. C., Tolonen, A. C., Warnick, T., Latouf, W. G., et al. (2015). Genome and transcriptome of clostridium phytofermentans, catalyst for the direct conversion of plant feedstocks to fuels. PLoS One 10:e0118285. doi: 10.1371/journal.pone.0118285
Rahman, Z., Shida, Y., Furukawa, T., Suzuki, Y., Okada, H., Ogasawara, W., et al. (2009). Application of Trichoderma reesei cellulase and xylanase promoters through homologous recombination for enhanced production of extracellular beta-glucosidase I. Biosci. Biotechnol. Biochem. 73, 1083–1089. doi: 10.1271/bbb.80852
Robison, P. D. (1984). Cellulase and xylanase production by Trichoderma reesei rut C-30. Biotechnol. Lett. 6, 119–122. doi: 10.1007/BF00127301
Saier, M. H. Jr., Tran, C. V., and Barabote, R. D. (2006). TCDB: the transporter classification database for membrane transport protein analyses and information. Nucleic Acids Res. 34, D181–D186. doi: 10.1093/nar/gkj001
Sali, A., and Blundell, T. L. (1993). Comparative protein modelling by satisfaction of spatial restraints. Mol. Med. Today 1, 270–277.
Saloheimo, M., Kujapanula, J., Ylösmäki, E., Ward, M., and Penttilä, M. (2002). Enzymatic Properties and intracellular localization of the novel Trichoderma reesei β-glucosidase BGLII (Cel1A). Appl. Environ. Microbiol. 68, 4546–4553. doi: 10.1128/AEM.68.9.4546-4553.2002
Santos, C. A., Zanphorlin, L. M., Crucello, A., Tonoli, C. C. C., Ruller, R., Horta, M. A. C., et al. (2016). Crystal structure and biochemical characterization of the recombinant ThBgl, a GH1 β-glucosidase overexpressed in Trichoderma harzianum under biomass degradation conditions. Biotechnol. Biofuels 9:71. doi: 10.1186/s13068-016-0487-0
Schwanhäusser, B., Busse, D., Li, N., Dittmar, G., Schuchhardt, J., Wolf, J., et al. (2013). Global quantification of mammalian gene expression control. Nature 495, 126–127. doi: 10.1038/nature11848
Shinoda, S., Kurosaki, M., Kokuzawa, T., Hirano, K., Takano, H., Ueda, K., et al. (2018). Comparative biochemical analysis of cellulosomes isolated from Clostridium clariflavum DSM 19732 and Clostridium thermocellum ATCC 27405 grown on plant biomass. Appl. Biochem. Biotechnol. doi: 10.1007/s12010-018-2864-6 [Epub ahead of print].
Shiratori, H., Sasaya, K., Ohiwa, H., Ikeno, H., Ayame, S., Kataoka, N., et al. (2009). Clostridium clariflavum sp. nov. and Clostridium caenicola sp. nov., moderately thermophilic, cellulose-/cellobiose-digesting bacteria isolated from methanogenic sludge. Int. J. Syst. Evol. Microbiol. 59, 1764–1770. doi: 10.1099/ijs.0.003483-0
Sun, S. N., Cao, X. F., Xu, F., Sun, R. C., Jones, G. L., and Baird, M. (2014). Structure and thermal property of alkaline hemicelluloses from steam exploded Phyllostachys pubescens. Carbohydr. Polym. 101, 1191–1197. doi: 10.1016/j.carbpol.2013.09.109
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tatusov, R. L., Fedorova, N. D., Jackson, J. D., Jacobs, A. R., Kiryutin, B., Koonin, E. V., et al. (2003). The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. doi: 10.1186/1471-2105-4-41
Thomas, L., Ram, H., and Singh, V. P. (2017). Evolutionary relationships and taxa-specific conserved signature indels among cellulases of archaea, bacteria, and eukarya. J. Comput. Biol. 24, 1029–1042. doi: 10.1089/cmb.2016.0161
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Thongpoo, P., McKee, L. S., Araujo, A. C., Kongsaeree, P. T., and Brumer, H. (2013). Identification of the acid/base catalyst of a glycoside hydrolase family 3 (GH3) beta-glucosidase from Aspergillus niger ASKU28. Biochim. Biophys. Acta 1830, 2739–2749. doi: 10.1016/j.bbagen.2012.11.014
Van Dyk, J. S., and Pletschke, B. I. (2012). A review of lignocellulose bioconversion using enzymatic hydrolysis and synergistic cooperation between enzymes–factors affecting enzymes, conversion and synergy. Biotechnol. Adv. 30, 1458–1480. doi: 10.1016/j.biotechadv.2012.03.002
Vanee, N., Brooks, J. P., and Fong, S. S. (2017). Metabolic profile of the cellulolytic industrial actinomycete Thermobifida fusca. Metabolites 7:57. doi: 10.3390/metabo7040057
Ventorino, V., Ionata, E., Birolo, L., Montella, S., Marcolongo, L., De Chiaro, A., et al. (2016). Lignocellulose-adapted endo-cellulase producing streptomyces strains for bioconversion of cellulose-based materials. Front. Microbiol. 7:2061. doi: 10.3389/fmicb.2016.02061
Wang, L., Ridgway, D., Gu, T., and Moo-Young, M. (2005). Bioprocessing strategies to improve heterologous protein production in filamentous fungal fermentations. Biotechnol. Adv. 23, 115–129. doi: 10.1016/j.biotechadv.2004.11.001
Wilson, D. B. (2009). The first evidence that a single cellulase can be essential for cellulose degradation in a cellulolytic microorganism. Mol. Microbiol. 74, 1287–1288. doi: 10.1111/j.1365-2958.2009.06889.x
Wilson, D. B. (2010). Studies of Thermobifida fusca plant cell wall degrading enzymes. Chem. Rec. 4, 72–82. doi: 10.1002/tcr.20002
Wu, D., Hugenholtz, P., Mavromatis, K., Pukall, R., Dalin, E., Ivanova, N. N., et al. (2009). A phylogeny-driven genomic encyclopaedia of bacteria and archaea. Nature 462, 1056–1060. doi: 10.1038/nature08656
Wu, J., Geng, A., Xie, R., Wang, H., and Sun, J. (2018). Characterization of cold adapted and ethanol tolerant β-glucosidase from Bacillus cellulosilyticus and its application for directed hydrolysis of cellobiose to ethanol. Int. J. Biol. Macromol. 109, 872–879. doi: 10.1016/j.ijbiomac.2017.11.072
Yang, F., Yang, X., Li, Z., Du, C., Wang, J., and Li, S. (2015). Overexpression and characterization of a glucose-tolerant beta-glucosidase from T. aotearoense with high specific activity for cellobiose. Appl. Microbiol. Biotechnol. 99, 8903–8915. doi: 10.1007/s00253-015-6619-9
Ye, Y., Li, X., and Zhao, J. (2017). Production and characteristics of a novel xylose- and alkali-tolerant GH 43 beta-xylosidase from Penicillium oxalicum for promoting hemicellulose degradation. Sci. Rep. 7:11600. doi: 10.1038/s41598-017-11573-7
Yin, Y. R., Meng, Z. H., Hu, Q. W., Jiang, Z., Xian, W. D., Li, L. H., et al. (2017). The hybrid strategy of Thermoactinospora rubra YIM 77501(T) for utilizing cellulose as a carbon source at different temperatures. Front. Microbiol. 8:942. doi: 10.3389/fmicb.2017.00942
Yin, Y. R., Zhang, F., Hu, Q. W., Xian, W. D., Hozzein, W. N., Zhou, E. M., et al. (2015). Heterologous expression and characterization of a novel halotolerant, thermostable, and alkali-stable GH6 endoglucanase from Thermobifida halotolerans. Biotechnol. Lett. 37, 857–862. doi: 10.1007/s10529-014-1742-8
Keywords: Actinomadura amylolytica, glucose tolerance, β-glucosidase, GH1, cellulose degradation
Citation: Yin Y-R, Sang P, Xian W-D, Li X, Jiao J-Y, Liu L, Hozzein WN, Xiao M and Li W-J (2018) Expression and Characteristics of Two Glucose-Tolerant GH1 β-glucosidases From Actinomadura amylolytica YIM 77502T for Promoting Cellulose Degradation. Front. Microbiol. 9:3149. doi: 10.3389/fmicb.2018.03149
Received: 18 October 2018; Accepted: 04 December 2018;
Published: 18 December 2018.
Edited by:
Dirk Tischler, Ruhr-Universität Bochum, GermanyReviewed by:
Jack Christopher Leo, University of Oslo, NorwayCopyright © 2018 Yin, Sang, Xian, Li, Jiao, Liu, Hozzein, Xiao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Min Xiao, eGlhb21pbjhAbWFpbC5zeXN1LmVkdS5jbg== Wen-Jun Li, bGl3ZW5qdW4zQG1haWwuc3lzdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.