- 1Department of Microbiology, University of Massachusetts, Amherst, MA, United States
- 2Department of Physical and Biological Sciences, Western New England University, Springfield, MA, United States
- 3Department of Biology, University of Southern Denmark, Odense, Denmark
- 4Assembly Biosciences, San Francisco, CA, United States
- 5Department of Biochemistry and Molecular Biology, Pennsylvania State University, University Park, PA, United States
Direct interspecies electron transfer (DIET) is important in diverse methanogenic environments, but how methanogens participate in DIET is poorly understood. Therefore, the transcriptome of Methanosarcina barkeri grown via DIET in co-culture with Geobacter metallireducens was compared with its transcriptome when grown via H2 interspecies transfer (HIT) with Pelobacter carbinolicus. Notably, transcripts for the F420H2 dehydrogenase, Fpo, and the heterodisulfide reductase, HdrABC, were more abundant during growth on DIET. A model for CO2 reduction was developed from these results in which electrons delivered to methanophenazine in the cell membrane are transferred to Fpo. The external proton gradient necessary to drive the otherwise thermodynamically unfavorable reverse electron transport for Fpo-catalyzed F420 reduction is derived from protons released from G. metallireducens metabolism. Reduced F420 is a direct electron donor in the carbon dioxide reduction pathway and also serves as the electron donor for the proposed HdrABC-catalyzed electron bifurcation reaction in which reduced ferredoxin (also required for carbon dioxide reduction) is generated with simultaneous reduction of CoM-S-S-CoB. Expression of genes for putative redox-active proteins predicted to be localized on the outer cell surface was higher during growth on DIET, but further analysis will be required to identify the electron transfer route to methanophenazine. The results indicate that the pathways for electron and proton flux for CO2 reduction during DIET are substantially different than for HIT and suggest that gene expression patterns may also be useful for determining whether Methanosarcina are directly accepting electrons from other extracellular electron donors, such as corroding metals or electrodes.
Introduction
The mechanisms by which methanogens conserve energy to support growth during direct interspecies electron transfer (DIET) are of interest because it is becoming increasingly apparent that DIET may be an important alternative to hydrogen interspecies transfer (HIT) for methane production in anaerobic digesters as well as methanogenic soils and sediments (Shrestha and Rotaru, 2014; Dubé and Guiot, 2015; Cheng and Call, 2016; Lovley, 2017c). A better understanding of DIET could help with the development of molecular approaches that can be used to detect DIET in methanogenic environments (Rotaru et al., 2014b; Holmes et al., 2017) and might lead to new approaches for promoting DIET to accelerate and stabilize anaerobic digestion (Cheng and Call, 2016; Barua and Dhar, 2017; Lovley, 2017b,c; Baek et al., 2018; Park et al., 2018).
Physiological studies of DIET require defined co-cultures. Geobacter metallireducens is an environmentally relevant pure culture model for electron-donating partners for DIET because Geobacter species function as the electron-donating partner in important methanogenic environments such as anaerobic digesters (Morita et al., 2011; Rotaru et al., 2014b) and terrestrial wetlands (Holmes et al., 2017). Studies with defined co-cultures in which G. metallireducens was the electron-donating partner for DIET (Shrestha et al., 2013; Rotaru et al., 2014a; Ueki et al., 2018) have suggested that c-type cytochromes and electrically conductive pili [e-pili] (Lovley, 2017a) facilitate electron transport from G. metallireducens to the electron accepting partner.
Outer-surface c-type cytochromes and e-pili are also involved in electron uptake by G. sulfurreducens when it is the electron-accepting partner in DIET-based co-cultures with G. metallireducens (Summers et al., 2010; Shrestha et al., 2013; Ueki et al., 2018). However, Methanosarcina barkeri and Methanothrix (formerly Methanosaeta) harundinacea, the only methanogens definitively shown to participate in DIET (Rotaru et al., 2014a,b), do not possess outer-surface c-type cytochromes or e-pili. Their outer surface electrical contacts for DIET are unknown.
The basic physiology and biochemistry of M. barkeri are much better understood than for Mt. harundinacea (Thauer et al., 2008; Gonnerman et al., 2013; Welte and Deppenmeier, 2014; Boone and Mah, 2015; Kulkarni et al., 2018; Mand et al., 2018). This makes M. barkeri the organism of choice for initial DIET mechanistic studies. Another advantage is that methods are available for genetic manipulation of M. barkeri (Kohler and Metcalf, 2012), but not Mt. harundinacea. However, one caveat for the study of DIET is that M. barkeri mutants have been previously constructed in a strain adapted to grow in high salt concentrations to prevent cell aggregation (Kohler and Metcalf, 2012). G. metallireducens, the only known electron-donating partner for M. barkeri, has yet to be adapted to grow at such high salt conditions.
Thus, at least at present, alternative approaches to evaluating the physiology of M. barkeri during DIET are required. Comparing the transcriptome of cells grown via DIET versus cells grown via HIT clearly reflected differences in electron uptake mechanisms in studies in which G. sulfurreducens functioned as the electron-accepting partner (Shrestha et al., 2013). G. sulfurreducens was grown by DIET with G. metallireducens as the electron-donating partner, or by HIT in co-culture with Pelobacter carbinolicus a microorganism closely related to G. metallireducens, but which is incapable of DIET (Shrestha et al., 2013). The G. sulfurreducens transcriptome demonstrated that cells were poised for growth on H2 when G. sulfurreducens was grown with P. carbinolicus and expressed genes for outer-surface proteins involved in direct uptake of electrons during DIET-based growth with G. metallireducens (Shrestha et al., 2013). M. barkeri can also be grown in co-culture with either G. metallireducens or P. carbinolicus (Rotaru et al., 2014a), providing an opportunity to compare M. barkeri gene expression patterns during growth via DIET and HIT.
Any model describing how the electron-accepting partner utilizes electrons derived from DIET must account for the uncoupling of the routes for interspecies electron and proton flux (Figure 1). e-Pili only transport electrons. Protons move between DIET partners by diffusion. This uncoupled transport of electrons and protons is in stark contrast to HIT in which H2 simultaneously transports both electrons and protons as the H2 diffuses between the two partners. When the H2 is oxidized in the cytoplasm with electron transfer to an electron acceptor, protons are also released and are immediately available to balance the negative charge transferred to the electron acceptor. This maintains charge balance within the cell (Figure 1). In contrast, in DIET, e-pili and associated electron transport proteins deliver electrons to cytoplasmic electron acceptors. Protons have to be translocated into the cytoplasm for charge balance (Figure 1). This proton consumption also prevents acidification of the extracellular matrix of the DIET aggregates. Thus, proposed mechanisms for electron uptake during DIET need to include an explanation for how protons are translocated into the cytoplasm of the electron-accepting partner.
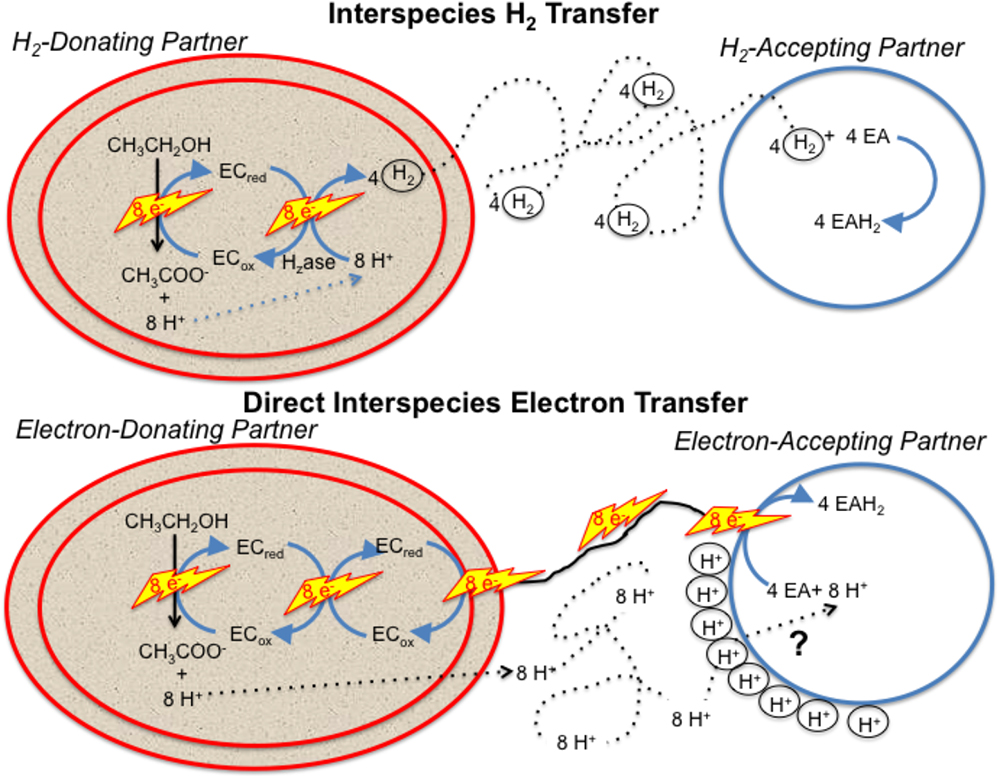
Figure 1. Generalized model for electron and proton flux during hydrogen interspecies electron transfer (HIT) and direct interspecies electron transfer (DIET) with growth on ethanol as an example. H2 diffusion shuttles both electrons and protons between cells and carries both electrons and protons into the cell when cytoplasmic electron acceptors are reduced. In contrast, electron and protons are transported by different mechanisms during DIET. Electron transfer is direct, through e-pili and other electrical contacts. Protons move by diffusion creating a positive proton pressure outside the cell. A mechanism for proton translocation into the cell is required for charge balance in the cytoplasm when cytoplasmic electron acceptors (EA) are reduced and to prevent acidification of the external space between cells. EC: electron carrier.
Here we report transcriptomic data from M. barkeri grown via DIET and HIT. The results suggest a mechanism for M. barkeri to utilize electrons and protons, derived from the electron-donating partner during DIET, to conserve energy to support growth from the reduction of carbon dioxide to methane.
Materials and Methods
Co-culture Incubation and mRNA Extraction
Triplicate replicates of co-cultures of G. metallireducens/M. barkeri and P. carbinolicus/M. barkeri were grown under strict anaerobic conditions as previously described (Rotaru et al., 2014a). Cultures were harvested during the exponential phase of growth and mRNA was isolated as previously described (Shrestha et al., 2013).
Illumina Sequencing and Assembly of Reads
Directional multiplex libraries were prepared with the ScriptSeqTM v2 RNA-Seq library preparation kit (Epicentre). Single end sequencing was performed with a Hi-Seq 2000 platform at the Deep Sequencing Core Facility at the University of Massachusetts Medical School in Worcester, MA, United States.
All raw data generated by Illumina sequencing were quality checked by visualization of base quality scores and nucleotide distributions with FASTQC1. Initial raw non-filtered forward and reverse sequencing libraries contained an average of 3892089 ± 134932 reads that were ∼100 basepairs long. Sequences from all of the libraries were trimmed and filtered with trimmomatic (Bolger et al., 2014) with the sliding window approach set to trim bases with quality scores lower than 3, strings of 3+N’s, and reads with a mean quality score lower than 20. Bases were also cut from the start and end of reads that fell below a threshold quality of 3, and any reads smaller than 50 bp were eliminated from the library. These parameters yielded an average of 2732020 ± 217212 quality reads per RNA-Seq library. SortMeRNA (Kopylova et al., 2012) was then used to separate all ribosomal RNA (rRNA) reads from the libraries. Databases used by SortMeRNA to identify all rRNA sequences included Rfam 5.8S Eukarya, Rfam 5S Archaea/Bacteria, SILVA 16S Archaea, SILVA 16S Bacteria, SILVA 23S Bacteria, SILVA 18S Eukarya, and SILVA 28S Eukarya (Burge et al., 2013; Quast et al., 2013).
Mapping of mRNA Reads
Trimmed and filtered mRNA reads from the triplicate samples for the two different co-culture conditions were mapped against the genome of M. barkeri strain MS DSM 800 downloaded from IMG/MER2. Mapped reads were normalized with the RPKM (reads assigned per kilobase of target per million mapped reads) method (Mortazavi et al., 2008; Klevebring et al., 2010) using ArrayStar software (DNAStar). Graphical analysis of reads from all three biological replicates for each condition demonstrated that the results were highly reproducible. Therefore, all reported values were obtained after merging and averaging replicates. Expression levels were considered significant only when the log2 RPKM value was higher than that of the median RPKM value.
Out of the 3,809 predicted protein-coding genes in the M. barkeri MS genome, 1,912 and 1,909 genes had expression levels that were higher than the median in DIET- and HIT-grown cells, respectively.
Genome Data Analysis
Sequence data for all of the bacterial genomes was acquired from the U.S. Department of Energy Joint Genome Institute3 or from GenBank at the National Center for Biotechnology Information (NCBI)4. Initial analyses were done with analysis tools available on the Integrated Microbial Genomes (IMG) website (see text footnote 2). Some protein domains were identified with NCBI conserved domain search (Marchler-Bauer et al., 2015) and Pfam search (Finn et al., 2016) functions. Transmembrane helices were predicted with TMpred (Hofmann and Stoffel, 1993), TMHMM (Krogh et al., 2001), and HMMTOP (Tusnady and Simon, 2001) and signal peptides were identified with PSORTb v. 3.0.2 (Yu et al., 2010) and Signal P v. 4.1 (Petersen et al., 2011).
Accession Number
Illumina sequence reads have been submitted to the NCBI database under project number PRJNA501858 and accession SAMN10346831-SAMN10346836.
Results and Discussion
As previously described (Rotaru et al., 2014a) co-cultures of G. metallireducens and M. barkeri that were well-adapted for growth via DIET required ca. 25 days to metabolize the 20 mM ethanol provided as substrate whereas P. carbinolicus/M. barkeri co-cultures required ca. 15 days. Both G. metallireducens and P. carbinolicus metabolized ethanol to acetate with either the production of H2 (P. carbinolicus) or extracellular electron transfer (G. metallireducens). M. barkeri metabolized acetate in both co-cultures, but in the initial growth phases of the cultures acetate production was faster than consumption, resulting in an accumulation of acetate (Rotaru et al., 2014a).
Transcriptome Reflects Faster Growth During HIT and Possible Greater Importance of Membrane and Outer-Surface Proteins During DIET
Transcript abundances for M. barkeri genes involved in amino acid biosynthesis, protein synthesis, and enzymes in the methane production pathways from both carbon dioxide and acetate were generally higher in the P. carbinolicus/M. barkeri co-cultures than in the G. metallireducens/M. barkeri co-cultures (Figure 2 and Supplementary Tables S1, S2). This is consistent with the faster growth of the P. carbinolicus/M. barkeri co-cultures. The highest proportion of genes that were more highly expressed during DIET-based growth were genes for proteins predicted to be associated with the membrane or cell surface (Figure 2).
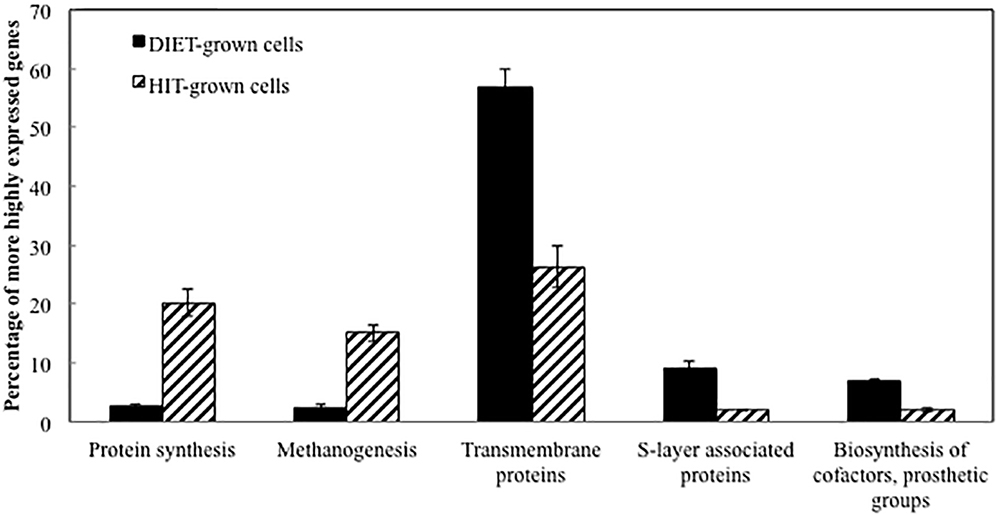
Figure 2. Comparison of relative expression of genes for different major classes of proteins during growth via DIET and HIT.
Genes for all three functional M. barkeri hydrogenases [Ech, Frh, and Vht (Mand et al., 2018)] were more abundant during HIT-based growth (Table 1). However, the results suggest that it will not be possible to use hydrogenase gene transcript levels to diagnose whether M. barkeri is participating in HIT or DIET in microbial communities. The increase in hydrogenase gene expression in HIT-grown cells was comparable to the general increase in expression of genes for other methanogenesis enzymes, such as Mcr (Table 1 and Supplementary Table S2), suggesting that there was not a specific upregulation of hydrogenase genes in response to growth via HIT.
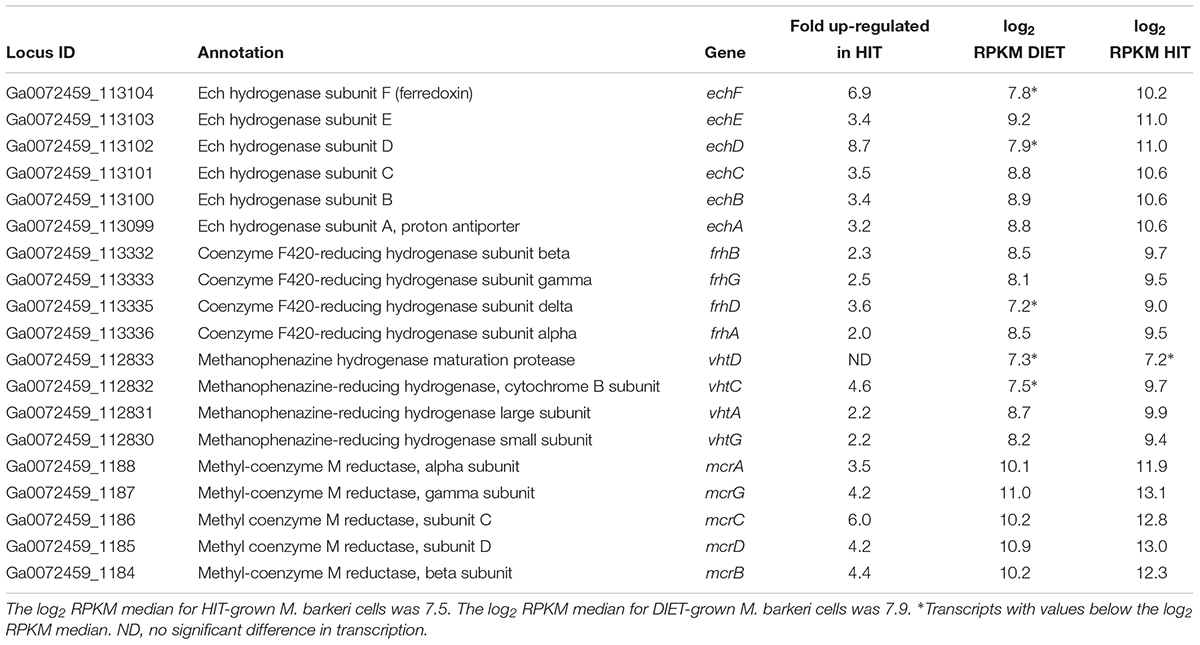
Table 1. Comparison of transcripts from genes coding for hydrogenase protein complexes (Ech, Frh, and Vht) and genes from the methyl coenzyme M reductase protein complex (Mcr) in M. barkeri cells growing via HIT in co-culture with P. carbinolicus or via DIET in co-culture with G. metallireducens.
Considering that gene expression for many metabolic genes was generally lower in DIET-grown cells, any genes for which transcript abundance was higher during DIET, or even comparable to HIT-grown cells, are of considerable interest. In the following sections, genes with higher expression during growth on DIET are examined further. The results are placed in the context of a working model (Figure 3) for generating the reduced co-factors required for carbon dioxide reduction to methane (F420H2, reduced ferredoxin) while also providing a mechanism for CoM-S-S-CoB reduction and a chemiosmotic potential to provide energy to support DIET-based growth.
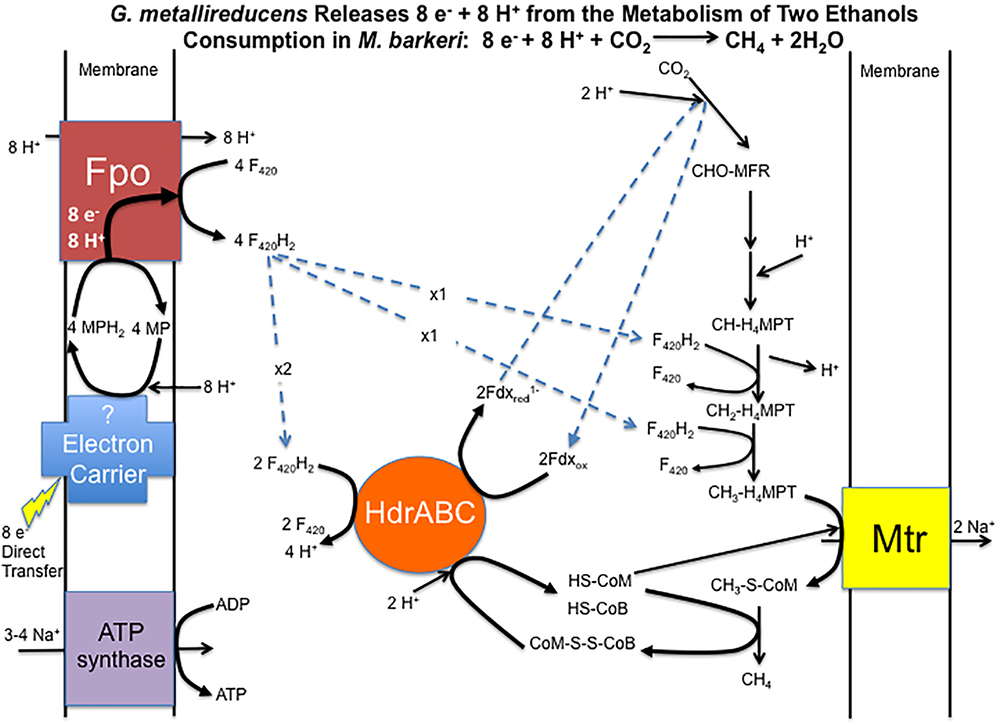
Figure 3. Model for electron and proton flux for carbon dioxide reduction to methane in Methanosarcina barkeri during DIET-based growth. Each two moles of ethanol oxidized to acetate by G. metallireducens releases eight electrons and eight protons. Electrons delivered to methanophenazine in the cell membrane are transferred to Fpo. Proton translocation drives Fpo-catalyzed reduction of F420 to F420H2. Half of the F420H2 produced serves as a reductant in the carbon dioxide reduction pathway. The remaining F420H2 is the electron donor for HdrABC, which reduces ferredoxin and CoM-S-S-CoB in an electron bifurcation reaction. The steps in carbon dioxide reduction, including the role of reduced ferredoxin, CoM-SH, and CoB-SH, as well as sodium pumping with Mtr, are as previously described (Thauer et al., 2008) for M. barkeri.
Proton-Driven Reverse Electron Transport to Reduce F420 With Fpo
Transcripts for genes for most of the subunits for the membrane-bound F420H2 dehydrogenase, Fpo, were higher in DIET-grown cells (Table 2). Considering that transcripts for most genes for methanogenesis were more abundant in HIT-grown cells, these results suggest that Fpo plays a key role in electron transport for carbon dioxide reduction to methane during DIET. During methylotrophic methanogenesis Fpo oxidizes F420H2 with the reduction of methanophenazine in the membrane, coupled with vectorial proton translocation to the outside of the membrane (Welte and Deppenmeier, 2014; Kulkarni et al., 2018; Mand et al., 2018). However, under some conditions Fpo may catalyze the reverse reaction in which reduced methanophenazine serves as the electron donor for the reduction of F420 (Mand et al., 2018). In this direction, proton translocation through Fpo into the cytoplasm is required in order to make the reaction thermodynamically favorable.
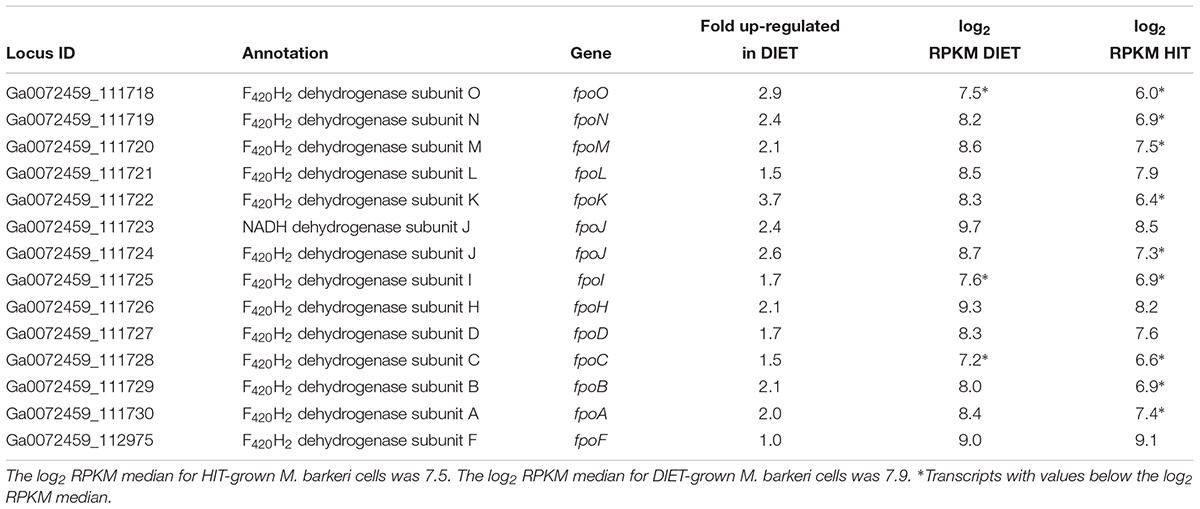
Table 2. Comparison of transcripts from genes coding for subunits of Fpo dehydrogenase in M. barkeri cells growing via HIT in co-culture with P. carbinolicus or via DIET in co-culture with G. metallireducens.
Therefore, it is proposed that electrons derived from DIET reduce methanophenazine in the oxidized state (MP) to MPH2 and that MPH2 is the electron donor for Fpo to reduce F420 in the cytoplasm (Figure 3). A proton gradient to drive the reaction is available from the protons released into the extracellular matrix from G. metallireducens metabolism in direct proportion to electrons transported from G. metallireducens through e-pili. The proton flux through Fpo does not acidify the cytoplasm because an equivalent number of protons are consumed from the cytoplasm when MP is reduced to MPH2 (Figure 3). The protons required to produce MPH2 are transferred to F420 during the Fpo-catalyzed reaction MPH2 + F420 → MP + F420H2. In this way electron transfer through methanophenazine to F420 is achieved with charge balance.
Possible Increased Methanophenazine Production to Support DIET
The proposed generation of F420H2 by Fpo with electrons derived from DIET requires an abundance of reduced methanophenazine (Figure 3). The pathway involved in biosynthesis of methanophenazine has not been identified, however, it is likely to resemble those of respiratory quinones because both have a polyprenyl side-chain connected to a redox-active moiety. In fact, studies have shown that a farnesylgeranyl pyrophosphate synthetase from the terpenoid backbone biosynthesis pathway is required for methanophenazine biosynthesis in M. mazei (Ogawa et al., 2010). Nine genes predicted to code for proteins involved in ubiquinone/menaquinone biosynthesis; six UbiE methyltransferase proteins, UbiA prenyltransferase, phenylacrylic acid decarboxylase (UbiD), and a ubiquinone biosynthesis protein (UbiB) were >2 fold more highly expressed in DIET-grown cells and another 11 putative ubiquinone biosynthesis genes were >1.5 fold up in DIET grown cells (Table 3). Given that M. barkeri does not contain ubiquinone or menaquinone, it seems likely that these genes code for enzymes involved in methanophenazine synthesis.
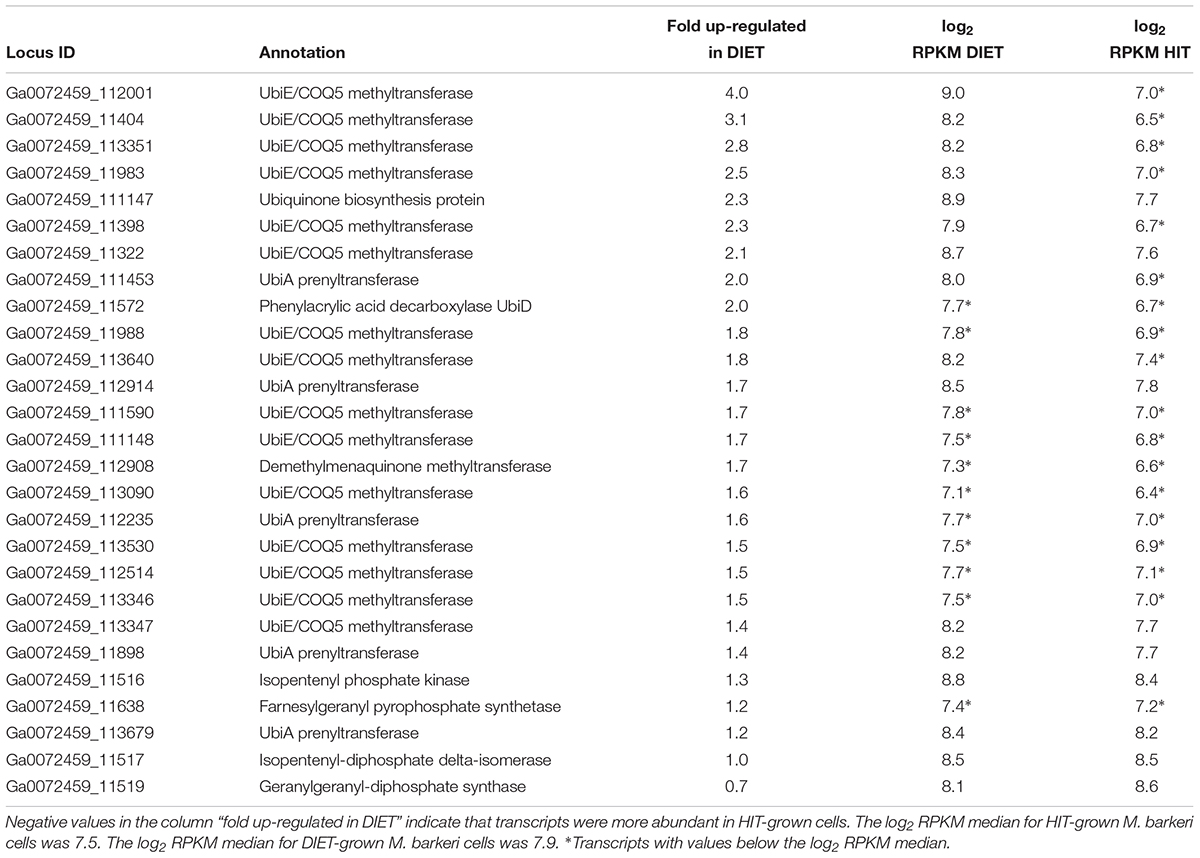
Table 3. Comparison of transcripts from genes coding for enzymes from the terpenoid backbone or terpenoid/quinone biosynthesis pathways in M. barkeri cells growing via DIET in co-culture with G. metallireducens or via HIT in co-culture with P. carbinolicus.
It has been calculated that the concentration of methanophenazine in membranes of M. acetivorans grown on methanol is sufficient to convert the membrane into an “electrically quantitized” conductive material (Duszenko and Buan, 2017). Methanophenazine concentrations in membranes of methanol-grown M. barkeri were too low for this effect (Duszenko and Buan, 2017). However, increased methanophenazine synthesis during growth via DIET might also yield an electrically quantitized membrane in M. barkeri, alleviating the need for redox-active proteins to aid in electron transport through the membrane during DIET.
Generating Reduced Ferredoxin and Reducing CoM-S-S-CoB With HdrABC
In addition to F420H2, M. barkeri needs to generate reduced ferredoxin during DIET. It is required for the first step in carbon dioxide reduction to methane (Thauer et al., 2008). One of the few soluble protein complexes with higher gene transcript abundance during DIET is the heterodisulfide reductase HdrA1B1C1 (Table 4), suggesting that it is important for DIET. Transcript abundance for the genes for subunits of the homologous HdrA2B2C2 was more comparable to that during growth on HIT, with the transcripts for the genes of the A2 and B2 slightly higher during DIET and lower transcripts for the C2 subunit. When the general pattern of higher gene transcript abundance for soluble proteins in HIT-grown cells is considered, these results suggest that HdrA2B2C2 might also be important in DIET.
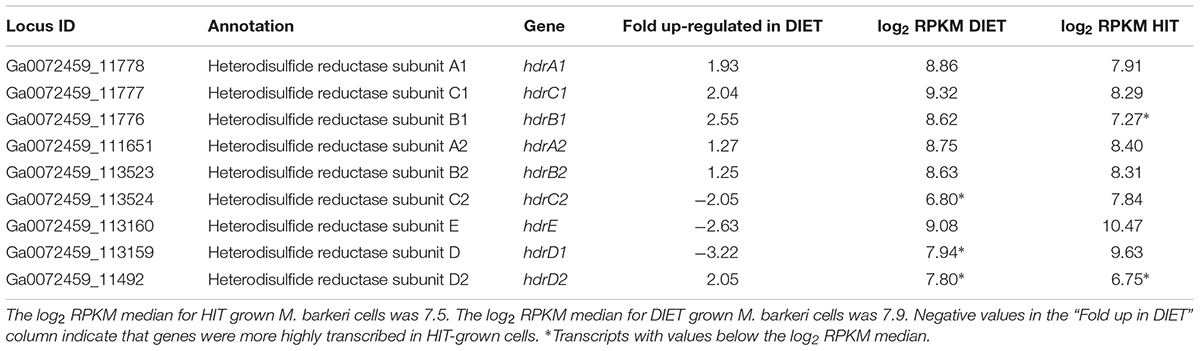
Table 4. Comparison of transcripts from genes coding for heterodisulfide reductase complexes HdrA1B1C1 and HdrA2B2C2 in M. barkeri cells growing via HIT in co-culture with P. carbinolicus or via DIET in co-culture with G. metallireducens.
In vitro purified HdrA2B2C2 from M. acetivorans oxidizes F420H2 with the reduction of ferredoxin and CoB-S-S-CoM through flavin-based electron bifurcation (Yan et al., 2017). The phenotypes for various Methanosarcina mutants have suggested that HdrA1B1C1 can couple the oxidation of reduced ferredoxin with the reduction of both F420 and CoB-S-S-CoM (Buan and Metcalf, 2010; Gonnerman et al., 2013). However, this reaction has not been verified biochemically (Yan and Ferry, 2018) and the direction of electron flow for the HdrA1B1C1 complex could be similar to that demonstrated for the HdrA2B2C2 complex, especially under conditions in which there is substantial production of reduced F420 and limited routes for generating reduced ferredoxin. An electron bifurcation reaction in this direction would also be consistent with the electron bifurcation from flavin with the reduction of CoB-S-S-CoM and ferredoxin associated with the MvhADG/HdrABC complexes found in methanogens that specialize in growth with H2/CO2 (Kaster et al., 2011).
The Completed Pathway for Energy Conservation During DIET
Therefore, it is proposed that half of the F420H2 generated with Fpo is the electron donor for HdrABC (one or both homologs) to produce reduced ferredoxin with the simultaneous reduction of CoM-S-S-CoB (Figure 3). In this way the coupled activity of Fpo- and HdrABC-catalyzed reactions deliver the eight moles of electrons derived from the oxidation of two moles of ethanol to each required step in the carbon dioxide reduction pathway (Figure 3).
As noted above, the Fpo-catalyzed reduction of F420 is proton balanced. Protons are released from F420H2 oxidation by HdrABC, but an equivalent number of protons are consumed in other reactions in the carbon dioxide reduction pathway (Figure 3). Thus, the model also balances proton flux.
The proposed model generates a chemiosmotic gradient to produce ATP through the activity of the Mtr complex that is known to pump sodium across the membrane during methyl transfer in the carbon dioxide reduction pathway (Thauer et al., 2008). There are two possibilities for ATP generation from the sodium gradient. Genes for both the A1A0 ATP synthase and the F1F0 ATP synthase were expressed during DIET (Supplementary Table S3). The available evidence suggests that both can translocate sodium (Schlegel and Muller, 2013). Genes for several components of the F1F0 ATP synthase were more highly expressed during growth on DIET and others were expressed at levels comparable to HIT-grown cells (Supplementary Table S3). This suggests that the F1F0 ATP synthase may play a more important role during growth on DIET, but at present there is not enough information on F1F0 ATP function in M. barkeri to speculate why.
Transcriptomics Suggests Potential Outer Surface Electrical Contacts
A number of genes predicted to encode redox active proteins expected to be associated with the M. barkeri membrane and/or cell surface were more highly expressed in cells grown via DIET (Table 5). However, it is premature to speculate on their possible role in mediating electron transfer into the cell in the absence of further biochemical characterization to determine whether important characteristics, such as redox potential and cellular localization, are appropriate for proposed roles.
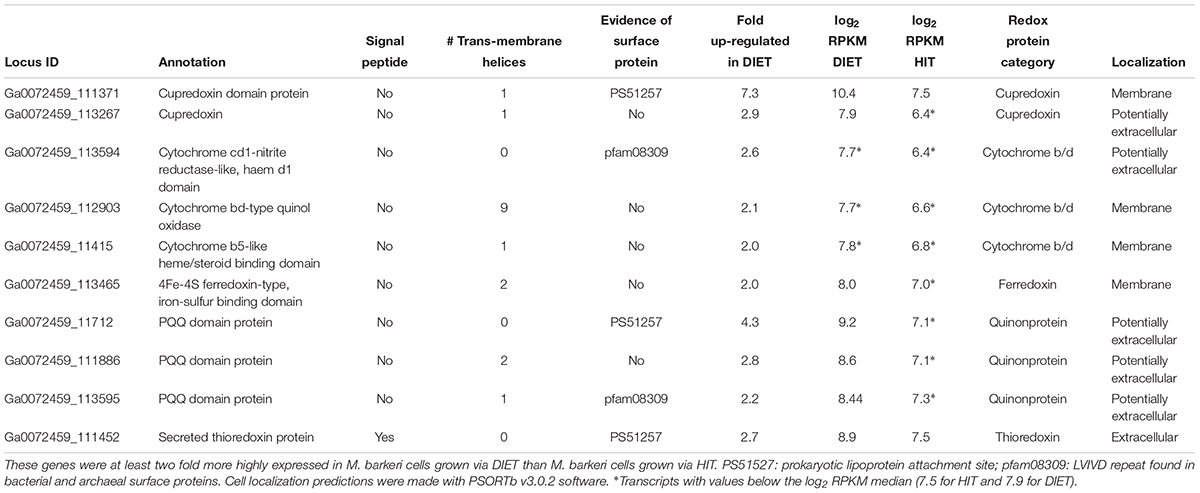
Table 5. Genes coding for putative transmembrane and/or surface associated electron transport proteins potentially involved in electron up-take during DIET.
For example, a gene putatively encoding a membrane-bound protein with a cupredoxin domain (Ga0072459_111371) was highly expressed specifically during DIET (Table 5). The cupredoxins rusticyanin and sulfocyanin play important roles in electron transfer into cells of Acidithiobacillus and Sulfolobus species during Fe(II) and S0 oxidation (Komorowski and Schafer, 2001; Dennison, 2005) and thus might play a similar role in electron transport into M. barkeri. The mid-point potentials of known cupredoxins (150 to 680 mV) are more positive than that expected for an electron carrier involved in electron transport to methanophenazine [mid-point potential of -165 mV (Tietze et al., 2003)]. However, modifications in cupredoxin structure and environment may greatly influence their mid-point potential (Marshall et al., 2009) and thus a role in electron transport into the cell is conceivable.
In a similar manner, genes encoding proteins that putatively incorporate pyrroloquinoline quinone (PQQ) as a co-factor were highly expressed during growth via DIET (Table 5). Like rusticyanin and sulfocyanin, proteins with PQQ-binding domains are involved in electron transport into the cell during oxidation of Fe(II) or Mn(II) (Croal et al., 2007; Johnson and Tebo, 2008). The mid-point potential of proteins with PQQ domains (∼90–100 mV) is too positive to play a role in electron transfer to methanophenazine. However, genes for PQQ biosynthesis were not found in the M. barkeri genome. Thus, it is possible that these proteins with predicted PQQ domains may incorporate another co-factor. Methanophenazine is one possibility. Further analysis of these proteins and others with higher gene expression during DIET (Table 5) is warranted. The expression of genes for a number of soluble electron carriers/co-factors were higher in DIET-grown cells, further suggesting differences in electron flux during DIET (Supplementary Table S4), but more analysis will be required to evaluate their role/significance.
Implications
The results suggest a pathway for electron and proton flux in M. barkeri during DIET that is significantly different than during HIT-based growth. The increased expression of genes for key components, including Fpo and HdrABC, and considerations of electron and proton transport during DIET, suggest an electron- and proton-balanced model in which the required electron donors are generated for each of the reductive steps of carbon dioxide reduction to methane while conserving energy to support growth (Figure 3). This model provides hypotheses that can be further evaluated experimentally with the appropriate M. barkeri mutants. However, as noted in the Introduction, this will require the discovery or development of an electron-donating strain capable of growing in the high salt medium that is used to generate M. barkeri mutants (Kohler and Metcalf, 2012). An alternative approach might be to adapt M. barkeri mutants to lower salt conditions, but this would require a time-consuming, labor-intensive adaption of each M. barkeri mutant strain with the risk of additional mutations arising during the adaption process.
The DIET transcriptome did not conclusively identify electrical contacts for DIET beyond the cell membrane. One potential reason for this is that M. barkeri might constitutively express these contacts. It is difficult to envision how Geobacter or other electron-donating partners could make electrical contacts with the outer surface of M. barkeri unless those contacts were expressed in advance of the initial Geobacter-M. barkeri electrical interaction. M. barkeri’s low affinity for H2 makes it a poor competitor for H2 in many environments (Thauer et al., 2008). Constitutive production of outer surface electrical contacts could poise M. barkeri for DIET and provide a competitive advantage in utilizing this alternative source of electrons for carbon dioxide reduction.
Elucidating the role of Methanosarcina species in DIET in complex natural environments is complicated by the possibility that H2 must also be considered as a potential electron donor for carbon dioxide reduction (Holmes et al., 2017). The differences in gene expression patterns between DIET- and HIT-grown cells suggest that metatranscriptional analysis is a route to better characterize the extent to which Methanosarcina are involved in DIET. It has been suggested that M. barkeri as well as other methanogens, can directly accept electrons from other extracellular sources such as electrodes, conductive carbon materials, and metals, but it has been difficult to rule out the possibility that H2 might be an intermediary electron carrier (Cheng and Call, 2016; Blasco-Gomez et al., 2017; Lovley, 2017b,c). The finding that transcriptome patterns in cells directly accepting electrons from an external source differ substantially from cells utilizing H2 as an electron donor suggests that the transcriptomic analysis approach described here could also help resolve this question.
Author Contributions
DH, A-ER, PS, and DL conceived the study. A-ER grew the co-cultures. PS extracted and processed the nucleic acids for sequences. DH re-annotated the genome as necessary and analyzed the transcriptome data. DH and DL wrote the initial version of the manuscript. All authors made important modifications and additions.
Funding
This research was supported by the Army Research Office and was accomplished under Grant Number W911NF-17-1-0345. The views and conclusions contained in this document are those of the authors and should not be interpreted as representing the official policies, either expressed or implied, of the Army Research Office or the United States Government. During the writing of this manuscript A-ER was supported by two Danish Research Council Grants (Sapere Aude no. 418100203 and Innovationsfonden no. 410600017) and the Novo Nordisk Foundation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.03109/full#supplementary-material
Footnotes
- ^http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- ^http://img.jgi.doe.gov
- ^http://www.jgi.doe.gov
- ^http://www.ncbi.nlm.nih.gov
References
Baek, G., Kim, J., Kim, J., and Lee, C. (2018). Role and potential of direct interspecies electron transfer in anaerobic digestion. Energies 11:107. doi: 10.3390/en11010107
Barua, S., and Dhar, B. R. (2017). Advances towards understanding and engineering direct interspecies electron transfer in anaerobic digestion. Bioresour. Technol. 244, 698–707. doi: 10.1016/j.biortech.2017.08.023
Blasco-Gomez, R., Batlle-Vilanova, P., Villano, M., Balaguer, M. D., Colprim, J., and Puig, S. (2017). On the edge of research and technological application: a critical review of electromethanogenesis. Int. J. Mol. Sci. 18:874. doi: 10.3390/ijms18040874
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boone, D. R., and Mah, R. A. (2015). “Methanosarcina,” in Bergey’s Manual of Systematics of Archaea and Bacteria, ed. W. B. Whitman (Hoboken, NJ: John Wiley & Sons Ltd), 1–15.
Buan, N., and Metcalf, W. (2010). Methanogenesis by Methanosarcina acetivorans involves two structurally and functionally distinct classes of heterodisulfide reductase. Mol. Microbiol. 75, 843–853. doi: 10.1111/j.1365-2958.2009.06990.x
Burge, S. W., Daub, J., Eberhardt, R., Tate, J., Barquist, L., Nawrocki, E. P., et al. (2013). Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 41, D226–D232. doi: 10.1093/nar/gks1005
Cheng, Q., and Call, D. (2016). Hardwiring microbes via direct interspecies electron transfer: mechanisms and applications. Environ. Sci. Process. Impacts 18, 968–980. doi: 10.1039/c6em00219f
Croal, L. R., Jiao, Y. Q., and Newman, D. K. (2007). The fox operon from Rhodobacter strain SW2 promotes phototrophic fe(II) oxidation in Rhodobacter capsulatus SB1003. J. Bacteriol. 189, 1774–1782. doi: 10.1128/JB.01395-06
Dennison, C. (2005). Investigating the structure and function of cupredoxins. Coord. Chem. Rev. 249, 3025–3054. doi: 10.1002/pro.3310
Dubé, C.-D., and Guiot, S. R. (2015). Direct interspecies electron transfer in anaerobic digestion: a review. Adv. Biochem. Eng. 151, 101–115. doi: 10.1007/978-3-319-21993-6_4
Duszenko, N., and Buan, N. R. (2017). Physiological evidence for isopotential tunneling in the electron transport chain of methane-producing archaea. Appl. Environ. Microbiol. 83:e00950-17. doi: 10.1128/AEM.00950-17
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. doi: 10.1093/nar/gkv1344
Gonnerman, M. C., Benedict, M. N., Feist, A. M., Metcalf, W. W., and Price, N. D. (2013). Genomically and biochemically accurate metabolic reconstruction of Methanosarcina barkeri Fusaro, iMG746. Biotechnol. J. 8, 1070–1079. doi: 10.1002/biot.201200266
Hofmann, K., and Stoffel, W. (1993). TMbase - a database of membrane spanning proteins segments. Biol. Chem. Hoppe Seyler 374:166.
Holmes, D. E., Shrestha, P. M., Walker, D. J. F., Dang, Y., Nevin, K. P., Woodard, T. L., et al. (2017). Metatranscriptomic evidence for direct interspecies electron transfer between Geobacter and Methanothrix species in rice paddy soils. Appl. Environ. Microbiol. 83:e00223-17. doi: 10.1128/AEM.00223-17
Johnson, H. A., and Tebo, B. M. (2008). In vitro studies indicate a quinone is involved in bacterial Mn(II) oxidation. Arch. Microbiol. 189, 59–69. doi: 10.1007/s00203-007-0293-y
Kaster, A. K., Moll, J., Parey, K., and Thauer, R. K. (2011). Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea. Proc. Natl. Acad. Sci. U.S.A. 108, 2981–2986. doi: 10.1073/pnas.1016761108
Klevebring, D., Bjursell, M., Emanuelsson, O., and Lundeberg, J. (2010). In-depth transcriptome analysis reveals novel TARs and prevalent antisense transcription in human cell lines. Plos One 5:e9762. doi: 10.1371/journal.pone.0009762
Kohler, P. R. A., and Metcalf, M. W. (2012). Genetic manipulation of Methanosarcina spp. Front. Microbiol. 3:259. doi: 10.3389/fmicb.2012.00259
Komorowski, L., and Schafer, G. (2001). Sulfocyanin and subunit II, two copper proteins with novel features, provide new insight into the archaeal SoxM oxidase supercomplex. FEBS Lett. 487, 351–355. doi: 10.1016/S0014-5793(00)02343-7
Kopylova, E., Noe, L., and Touzet, H. (2012). SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28, 3211–3217. doi: 10.1093/bioinformatics/bts611
Krogh, A., Larsson, B., Von Heijne, G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Kulkarni, G., Mand, T. D., and Metcalf, W. W. (2018). Energy conservation via hydrogen cycling in the methanogenic archaeon Methanosarcina barkeri. mBio 9:e01256-18. doi: 10.1128/mBio.01256-18
Lovley, D. R. (2017a). Electrically conductive pili: biological function and potential applications in electronics. Curr. Opin. Electrochem. 4, 190–198. doi: 10.1016/j.coelec.2017.08.015
Lovley, D. R. (2017b). Happy together: microbial communities that hook up to swap electrons. ISME J. 11, 327–336. doi: 10.1038/ismej.2016.136
Lovley, D. R. (2017c). Syntrophy goes electric: direct interspecies electron transfer. Ann. Rev. Microbiol. 71, 643–664. doi: 10.1146/annurev-micro-030117-020420
Mand, T. D., Kulkarni, G., and Metcalf, W. W. (2018). Genetic, biochemical, and molecular characterization of Methanosarcina barkeri mutants lacking three distinct classes of hydrogenase. J. Bacteriol. 200:e0342-18. doi: 10.1128/JB.00342-18
Marchler-Bauer, A., Derbyshire, M. K., Gonzales, N. R., Lu, S., Chitsaz, F., Geer, L. Y., et al. (2015). CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43, D222–D226. doi: 10.1093/nar/gku1221
Marshall, N. M., Garner, D. K., Wilson, T. D., Gao, Y. G., Robinson, H., Nilges, M. J., et al. (2009). Rationally tuning the reduction potential of a single cupredoxin beyond the natural range. Nature 462, 113–127. doi: 10.1038/nature08551
Morita, M., Malvankar, N. S., Franks, A. E., Summers, Z. M., Giloteaux, L., Rotaru, A. E., et al. (2011). Potential for direct interspecies electron transfer in methanogenic wastewater digester aggregates. mBio 2:e00159-11. doi: 10.1128/mBio.00159-11
Mortazavi, A., Williams, B. A., Mccue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Ogawa, T., Yoshimura, T., and Hemmi, H. (2010). Geranylfarnesyl diphosphate synthase from Methanosarcina mazei: different role, different evolution. Biochem. Biophys. Res. Commun. 393, 16–20. doi: 10.1016/j.bbrc.2010.01.063
Park, J. H., Kang, H. J., Park, K. H., and Park, H. D. (2018). Direct interspecies electron transfer via conductive materials: a perspective for anaerobic digestion applications. Bioresour. Technol. 254, 300–311. doi: 10.1016/j.biortech.2018.01.095
Petersen, T. N., Brunak, S., Von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rotaru, A.-E., Shrestha, P. M., Liu, F., Nevin, K. P., and Lovley, D. R. (2014a). Direct interspecies electron transfer during syntrophic growth of Geobacter metallireducens and Methanosarcina barkeri on ethanol. Appl. Environ. Microbiol. 80, 4599–4605. doi: 10.3389/fmicb.2016.00236
Rotaru, A.-E., Shrestha, P. M., Liu, F., Shrestha, M., Shrestha, D., Embree, M., et al. (2014b). A new model for electron flow during anaerobic digestion: direct interspecies electron transfer to Methanosaeta for the reduction of carbon dioxide to methane. Energy Environ. Sci. 7, 408–415. doi: 10.1039/C3EE42189A
Schlegel, K., and Muller, V. (2013). Evolution of Na+ and H+ bioenergetics in methanogenic archaea. Biochem. Soc. Trans. 41, 421–426. doi: 10.1042/BST20120294
Shrestha, P. M., and Rotaru, A.-E. (2014). Plugging in or going wireless: strategies for interspecies electron transfer. Front. Microbiol. 5:237. doi: 10.3389/fmicb.2014.00237
Shrestha, P. M., Rotaru, A.-E., Summers, Z. M., Shrestha, M., Liu, F., and Lovley, D. R. (2013). Transcriptomic and genetic analysis of direct interspecies electron transfer. Appl. Environ. Microbiol. 79, 2397–2404. doi: 10.1128/AEM.03837-12
Summers, Z. M., Fogarty, H., Leang, C., Franks, A. E., Malvankar, N. S., and Lovley, D. R. (2010). Direct exchange of electrons within aggregates of an evolved syntrophic co-culture of anaerobic bacteria. Science 330, 1413–1415. doi: 10.1126/science.1196526
Thauer, R. K., Kaster, A.-K., Seedork, H., Buckel, W., and Hedderich, R. (2008). Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 8, 579–591. doi: 10.1038/nrmicro1931
Tietze, M., Beuchle, A., Lamla, I., Orth, N., Dehler, M., Greiner, G., et al. (2003). Redox potentials of methanophenazine and CoB-S-S-CoM, factors involved in electron transport in methanogenic archaea. Chembiochem 4, 333–335. doi: 10.1002/cbic.200390053
Tusnady, G. E., and Simon, I. (2001). The HMMTOP transmembrane topology prediction server. Bioinformatics 17, 849–850. doi: 10.1093/bioinformatics/17.9.849
Ueki, T., Nevin, K. P., Rotaru, A.-E., Wang, L.-Y., Ward, J. E., Woodard, T. L., et al. (2018). Geobacter strains expressing poorly conductive pili reveal constraints on direct interspecies electron transfer mechanisms. mBio 9:e01273-18. doi: 10.1128/mBio.01273-18
Welte, C., and Deppenmeier, U. (2014). Bioenergetics and anaerobic respiratory chains of aceticlastic methanogens. Biochim. Biophys. Acta 1837, 1130–1147. doi: 10.1016/j.bbabio.2013.12.002
Yan, Z., and Ferry, J. G. (2018). Electron bifurcation and confurcation in methanogenesis and reverse methanogenesis. Front. Microbiol. 9:1322. doi: 10.3389/fmicb.2018.01322
Yan, Z., Wang, M. Y., and Ferry, J. G. (2017). A ferredoxin-and F420H2-dependent, electron-bifurcating, heterodisulfide reductase with homologs in the domains bacteria and archaea. mBio 8:e02285-16. doi: 10.1128/mBio.02285-16
Yu, N. Y., Wagner, J. R., Laird, M. R., Melli, G., Rey, S., Lo, R., et al. (2010). PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26, 1608–1615. doi: 10.1093/bioinformatics/btq249
Keywords: syntrophy, methanogenesis, F420 dehydrogenase, heterodisulfide reductase, transcriptomics
Citation: Holmes DE, Rotaru A-E, Ueki T, Shrestha PM, Ferry JG and Lovley DR (2018) Electron and Proton Flux for Carbon Dioxide Reduction in Methanosarcina barkeri During Direct Interspecies Electron Transfer. Front. Microbiol. 9:3109. doi: 10.3389/fmicb.2018.03109
Received: 30 July 2018; Accepted: 30 November 2018;
Published: 13 December 2018.
Edited by:
Obulisamy Parthiba Karthikeyan, University of Michigan, United StatesReviewed by:
William Lanzilotta, University of Georgia, United StatesEric D. van Hullebusch, UMR7154 Institut de Physique du Globe de Paris (IPGP), France
Copyright © 2018 Holmes, Rotaru, Ueki, Shrestha, Ferry and Lovley. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dawn E. Holmes, ZGhvbG1lc0BtaWNyb2Jpby51bWFzcy5lZHU= Derek R. Lovley, ZGxvdmxleUBtaWNyb2Jpby51bWFzcy5lZHU=
 Dawn E. Holmes
Dawn E. Holmes Amelia-Elena Rotaru
Amelia-Elena Rotaru Toshiyuki Ueki
Toshiyuki Ueki Pravin M. Shrestha1,4
Pravin M. Shrestha1,4 James G. Ferry
James G. Ferry Derek R. Lovley
Derek R. Lovley