- 1Department of Biological Sciences and Technology, School of Environmental Studies, China University of Geosciences, Wuhan, China
- 2State Key Laboratory of Biogeology and Environmental Geology, China University of Geosciences, Wuhan, China
To exchange electrons with extracellular substrates, some microorganisms employ extracellular electron transfer (EET) pathways that physically connect extracellular redox reactions to intracellular metabolic activity. These pathways are made of redox and structural proteins that work cooperatively to transfer electrons between extracellular substrates and the cytoplasmic membrane. Crucial to the bacterial and archaeal EET pathways are the quinol oxidases and/or quinone reductases in the cytoplasmic membrane where they recycle the quinone/quinol pool in the cytoplasmic membrane during EET reaction. Up to date, three different families of quinol oxidases and/or quinone reductases involved in bacterial EET have been discovered. They are the CymA, CbcL/MtrH/MtoC, and ImcH families of quinol oxidases and/or quinone reductases that are all multiheme c-type cytochromes (c-Cyts). To investigate to what extent they are distributed among microorganisms, we search the bacterial as well as archaeal genomes for the homologs of these c-Cyts. Search results reveal that the homologs of these c-Cyts are only found in the Domain Bacteria. Moreover, the CymA homologs are only found in the phylum of Proteobacteria and most of them are in the Shewanella genus. In addition to Shewanella sp., CymA homologs are also found in other Fe(III)-reducing bacteria, such as of Vibrio parahaemolyticus. In contrast to CymA, CbcL/MtrH/MtoC, and ImcH homologs are much more widespread. CbcL/MtrH/MtoC homologs are found in 15 phyla, while ImcH homologs are found in 12 phyla. Furthermore, the heme-binding motifs of CbcL/MtrH/MtoC and ImcH homologs vary greatly, ranging from 3 to 23 and 6 to 10 heme-binding motifs for CbcL/MtrH/MtoC and ImcH homologs, respectively. Moreover, CymA and CbcL/MtrH/MtoC homologs are found in both Fe(III)-reducing and Fe(II)-oxidizing bacteria, suggesting that these families of c-Cyts catalyze both quinol-oxidizing and quinone-reducing reactions. ImcH homologs are only found in the Fe(III)-reducing bacteria, implying that they are only the quinol oxidases. Finally, some bacteria have the homologs of two different families of c-Cyts, which may improve the bacterial capability to exchange electrons with extracellular substrates.
Introduction
Many microorganisms can exchange electrons between the redox proteins in the cytoplasmic or inner membrane and extracellular substrates, such as metal ions associated with minerals, humic substances and electrodes, and microbial cells of the same or difference species. For example, the metal-reducing bacteria Geobacter spp. and Shewanella spp. use solid phase Fe(III)-containing minerals, humic substances and electrodes as terminal electron acceptors for anaerobic respiration (Lovley et al., 1987, 1996; Myers and Nealson, 1988; Bond and Lovley, 2003; Bretschger et al., 2007). The metal-oxidizing bacteria Mariprofundus ferrooxydans PV-1 and Rhodopseudomonas palustris TIE-1 use Fe(II) and electrodes as electron and/or energy source for growth (Jiao et al., 2005; Emerson et al., 2007; Summers et al., 2013; Bose et al., 2014). Moreover, Geobacter metallireducens transfers electrons directly to Geobacter sulfurreducens via conductive nanowires and multiheme c-type cytochromes (c-Cyts) (Summers et al., 2010). Shewanella oneidensis MR-1 transfers electrons from one cell to another via the multiheme c-Cyts on the outer membrane extension that connects the two cells (Gorby et al., 2006; Pirbadian et al., 2014; Subramanian et al., 2018). Electron exchange between the microbial cytoplasmic membrane and extracellular substrates or cells is often referred to as microbial extracellular electron transfer (EET) (Shi et al., 2016).
To exchange electrons extracellularly, some microorganisms form EET pathways that consist of structural and redox proteins. For example, S. oneidensis MR-1 possesses a metal-reducing (Mtr) EET pathway. The currently identified protein components of the Mtr pathway include six multiheme c-Cyts: the cytoplasmic membrane CymA, the periplasmic Fcc3 and STC and the outer membrane MtrA, MtrC and OmcA, and one outer membrane porin protein MtrB (Shi et al., 2016). The Mtr pathway starts at CymA that oxidizes the quinol in the cytoplasmic membrane (Marritt et al., 2012a,b; McMillan et al., 2012, 2013). The released electrons are transferred first through the periplasm by Fcc3 and STC (Firer-Sherwood et al., 2011; Fonseca et al., 2013; McMillan et al., 2013) and then across the outer membrane to the bacterial surface by MtrABC (Hartshorne et al., 2009; White et al., 2012, 2013). On the bacterial surface, OmcA interacts with MtrC to mediate the electron transfer to Fe(III)-containing minerals (Shi et al., 2006; Zhang et al., 2008, 2009). To date, the Mtr pathway of S. oneidensis MR-1 is the most rigorously characterized microbial EET pathway (Shi et al., 2016). Similar to the Mtr pathway, the porin-cytochrome (Pcc) EET pathways are proposed in G. sulfurreducens, which also includes multiheme c-Cyts and the outer membrane porin proteins (Shi et al., 2016). In the Pcc pathways, the porin-cytochrome protein complexes for transferring electrons across the outer membrane to the Fe(III)-containing minerals are functionally verified (Liu et al., 2014). Although they are biochemically and biophysically characterized (Lloyd et al., 2003; Morgado et al., 2010), the proposed EET role of the periplasmic multiheme c-Cyts PpcA and PpcD remain to be demonstrated. In addition, the proposed cytoplasmic membrane quinol oxidases CbcL and ImcH also remain to be characterized. The Mtr and Pcc pathways transfer electrons from the CymA, CbcL and ImcH in the cytoplasmic membrane to the extracellular Fe(III)-containing minerals, in which the CymA, CbcL, and ImcH sever or are proposed to sever as quinol oxidases. The metal-oxidizing (Mto) pathway of the Fe(II)-oxidizing bacterium Sideroxydans lithotrophicus ES-1 is, however, proposed to transfer electrons from extracellular electron donors to CymA in the cytoplasmic membrane where CymA is proposed to function as a quinone reductase (Liu et al., 2012; Shi et al., 2012). Thus, CymA, CbcL, and ImcH play or are proposed to play crucial roles in redox cycling of quinone/quinol pool in the cytoplasmic membrane during bacterial EET.
CymA, CbcL, and ImcH belong to different protein families and each exhibits its own characteristics. For example, as a member of the NapC/NirT family of quinol dehydrogenases, CymA of S. oneidensis MR-1 is a tetra-heme c-Cyt that possesses a short transmembrane domain at its N-terminus and a periplasmic domain at its C-terminal region (Figure 1A; Shi et al., 2007). Different from CymA, CbcL of G. sulfurreducens is predicted to have nine heme-binding motifs (CX2CH) of c-Cyts in its N-terminal half and six transmembrane helices in its C-terminal half (Figure 1B). The transmembrane region of CbcL also displays a high degree of similarity to the HydC/FdnI-like b-type cytochromes that contain two putative hemes (Zacharoff et al., 2015). Finally, ImcH of G. sulfurreducens is predicted to possess three transmembrane helices in its N-terminal region and seven CX2CH motifs in its C-terminal region (Figure 1C). It should be pointed out that part of ImcH, which includes the third transmembrane helix and the first three CX2CH motifs adjacent to the third helix, show a certain degree of similarity to the NapC/NirT family of quinol dehydrogenases (Levar et al., 2014).
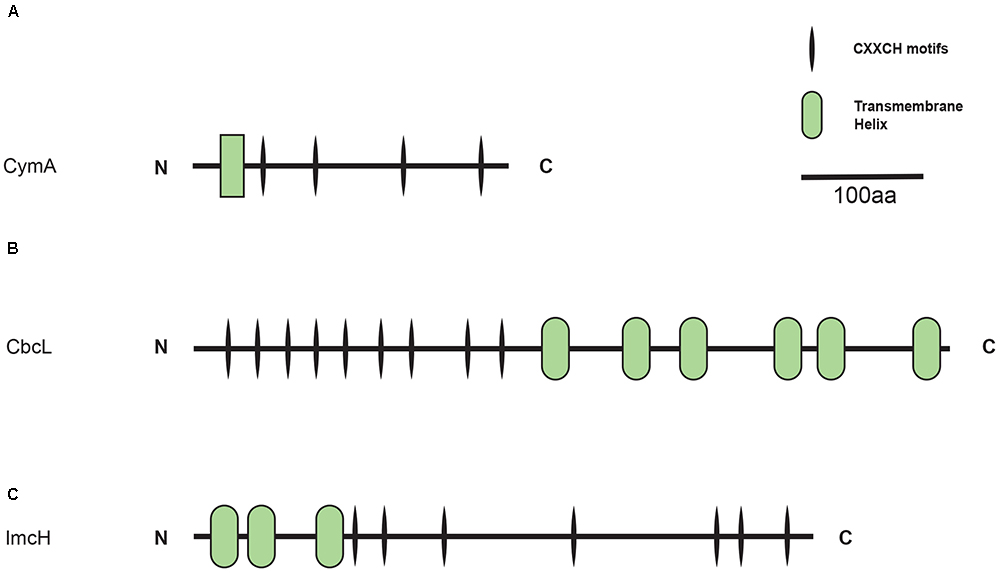
Figure 1. Distribution of CymA homologs. Inner layer, phylogenetic analysis of identified CymA homologs. A phylogenetic tree was constructed and graphically displayed as described in the text. Middle layer, schematic representation of length of and positions of heme-binding as well as transmembrane motifs in the polypeptides of the CymA homologs. Outer layer, bacterial genera from which CymA homologs were identified.
In addition to S. oneidensis MR-1 and S. lithotrophicus ES-1, CymA homologs also exist in other metal-reducing bacteria, such as Shewanella sp. strain ANA-3, Shewanella putrefaciens W3-18-1, Ferrimonas balearica and Rhodoferax ferrireducens (Murphy and Saltikov, 2007; Shi et al., 2012; Wei et al., 2016). Similarly, MtoC and MtrH, which are the homologs of CbcL, are also present in R. ferrireducens as well as the metal-oxidizing bacteria Dechloromonas aromatica RCB and Gallionella capsiferriformans ES-2 (Shi et al., 2012). Moreover, in R. ferrireducens, two cymA genes and a mtrH gene are found in the same gene cluster that also contains mtrABC genes and other c-Cyts-encoding genes (Shi et al., 2012). Given that they are found in both metal-reducing and metal-oxidizing bacteria, CbcL/MtoC/MtrH homologs are most likely involved in both quinol oxidation as well as quinone reduction. However, distribution of CymA and CbcL/MtoC/MtrH homologs in other bacteria have not been systemically investigated. Different from that from the homologs of CymA and CbcL/MtoC/MtrH, distribution of ImcH homologs in different bacteria has been searched. Search results showed that, in addition to Geobacter spp. ImcH homologs are found in a variety of other bacteria, including the Fe(III)-reducing bacteria Acidobacterium capsulatum, Anaeromxyobacter 2PC-C, Geoalkalibacter subterraneus, Geothrix fermantans, and Melioribacter roseus, where they are proposed to function as quinol oxidases (Levar et al., 2014; Shi et al., 2016). However, it remains unclear whether, similar to CymA and CbcL/MtoC/MtrH homologs, ImcH homologs also exist in the metal-oxidizing bacteria. Thus, in this study, we systematically searched the bacterial and archaeal genomes available in May 16, 2018 for the homologs of CymA, CbcL/MtoC/MtrH and ImcH to address the following questions: (1) To what extent CymA, CbcL/MtoC/MtrH and ImcH homologs are distributed among the sequenced microorganisms? (2) Is ImcH homolog also present in the metal-oxidizing bacteria?
Approach
Search for the CymA, MtrH/MtoC/CbcL and ImcH Homologs
The bacterial and archaeal genomes were searched for CymA, CbcL/MtoC/MtrH and ImcH homologs by the approach described before (Shi et al., 1998, 2012, 2014; Shi and Zhang, 2004). In the beginning, previously identified CymA homologs of F. balearica, R. ferrireducens, S. lithotrophicus ES-1 and S. oneidensis MR-1, CbcL/MtoC/MtrH homologs of D. aromatica RCB, G capsiferriformans ES-2, G. sulfurreducens and R. ferrireducens, as well as ImcH homologs of A. capsulatum, A. 2PC-C, G. subterraneus, G. sulfurreducens, G. fermantans, and M. roseus were used as templates to search for the open reading frames (ORFs) whose deduced polypeptide sequences exhibited similarity to the templates by BLAST (E < 0.01) (Altschul et al., 1990; Supplementary Tables S1–S3). The search was carried by using the blastp program of the National Center for Biotechnology Information (NCBI) and the BLAST program of the Universal Protein Resource (UniProt). The search parameters are: scoring matrix = BLOSUM62, gapopen = 0, and gapextend = 0. The databases searched were non-redundant protein sequences database (nr) and UniprotKB database. The tentatively identified homologs were subject to inspection with in-house Perl scripts for the CX2CH motifs as well as the analyses by a hidden Markov model-based TMHMM software for predicting the transmembrane helices (Krogh et al., 2001; Shi et al., 2012, 2014). The verified homologs were also used as templates for next round of genome search. Additionally, the ORFs adjacent to that of the verified homologs were also searched for those encoding putative c-Cyts (Shi et al., 2012, 2014).
Phylogenetic Reconstruction and Gene Cluster Identification
The polypeptide sequences were aligned by Clustal W (version 2.1) with the following settings: Gap Opening Penalty = 10; Gap Extension Penalty = 0.2; Protein matrix = BLOSUM series (Larkin et al., 2007). The aligned sequences of all verified CymA, CbcL/MtoC/MtrH or ImcH homologs were analyzed separately by MEGA7. Maximum Likelihood method was used to construct phylogenetic trees with a confidence level determined by 1000 bootstrap replications (Kumar et al., 2016). The reconstructions were performed by applying the best amino acid model of amino acid substitutions, which was determined by Close-Neighbor-Interchange model (Kumar et al., 2016). The Evolview v2 was used to graphically display the results of phylogenetic reconstruction (He et al., 2016). After the homologs were acquired, other genes, whose polypeptides had CX2CH motifs on upstream and downstream of the homologs, were also identified by the method described above. The figure was plotted by IBS (Liu et al., 2015).
Results and Discussion
Overview
As shown in Table 1, a total of 282 homologs were identified, which include 73 CymA homologs, 138 CbcL/MtoC/MtrH homologs, and 71 ImcH homologs. All of them are in the Domain Bacteria. Sixty-three percent of identified CymA homologs were found in the genus of Shewanella and the rest were found in 11 bacterial genera and 2 bacteria that were not classified at the genus level, which were all in the phylum of Proteobacteria (Table 1 and Supplementary Table S1). Compared to the CymA homologs that are found only one phylum, CbcL/MtoC/MtrH homologs were much more widespread and were identified in 15 bacterial phyla that included 33 genera and the bacteria whose classification could not be assigned to the genus level. Seventeen CbcL/MtoC/MtrH homologs were found in the genus of Geobacter (Table 1 and Supplementary Table S2). Similar to the CbcL/MtoC/MtrH homologs, ImcH homologs were also widespread and were found in 12 phyla, which included 19 genera and the bacteria not classified to the genus level. In the genus of Geobacter, 18 ImcH homologs were identified.(Table 1 and Supplementary Table S3).
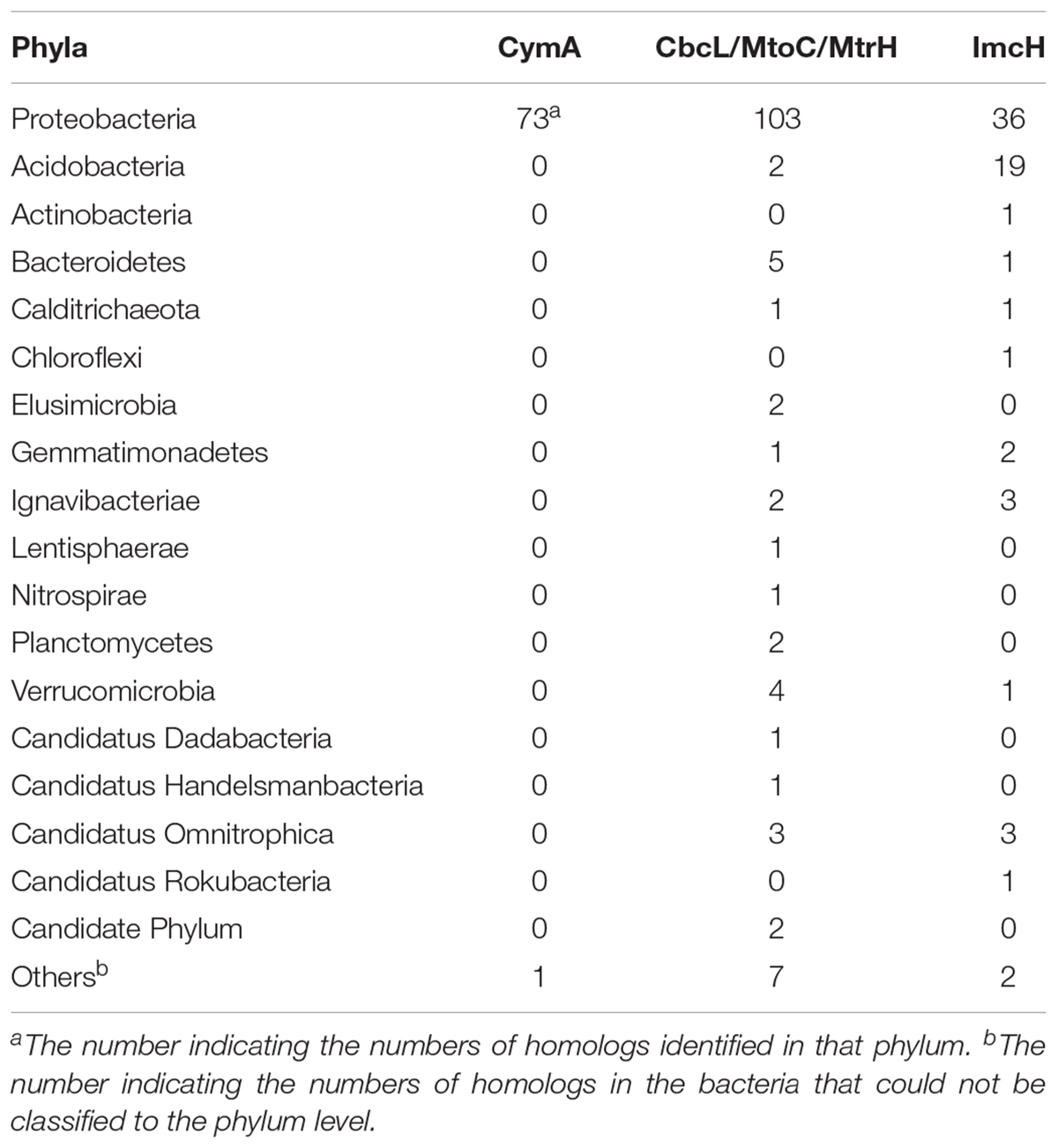
Table 1. Summary of identified CymA, CbcL/MtoC/MtrH, and ImcH homologs in different bacterial phyla.
The CymA Homologs
As shown in Figure 2, all CymA homologs of Shewanella sp. were clustered together and they were 58.8–100% identical. This observation is consistent with previous findings (Murphy and Saltikov, 2007) and suggests that the functional role of CymA homologs is well conserved in Shewanella sp. The identity between the CymA homologs of Shewanella sp. and those identified from other bacteria ranged from 22.5 to 96.7%. The polypeptides of nearly all CymA homologs from Shewanella sp. contained 187 amino acid residues, except those of Shewanella sp. GutDb and S. piezotoleransstrain WP3 whose polypeptides possessed186 and 188 amino acid residues, respectively (Figure 2 and Supplementary Table S1). The polypeptides of the CymA homologs from other bacteria were 187 to 364 amino acid residue long. Notably, the polypeptides of CymA homologs from 9 Vibrio sp., Aliivibrio sp. 1S175 and Trabulsiella guamensis had 363–364 amino acid residues, which were much longer than those of the rest of identified CymA homologs (Figure 2 and Supplementary Table S1). Moreover, compared to those with much shorter polypeptides, the CymA homologs with longer polypeptides possessed an extra CX2CH motif close to their C-termini (Figure 2 and Supplementary Table S1). Finally, a putative atypical heme-binding motif CX3CH and AX2CH was found in the polypeptide sequence close to the transmembrane domain for the two CymA homologs from R. ferrireducens and the one identified from the metagenome of a mine drainage, respectively (Shi et al., 2012). Thus, all identified CymA homologs contained 4 heme-binding motifs, except those from Vibrio sp., Aliivibrio sp. 1S175 and T. guamensis, which contained 5 heme-binding motifs (Figure 2 and Supplementary Table S1).
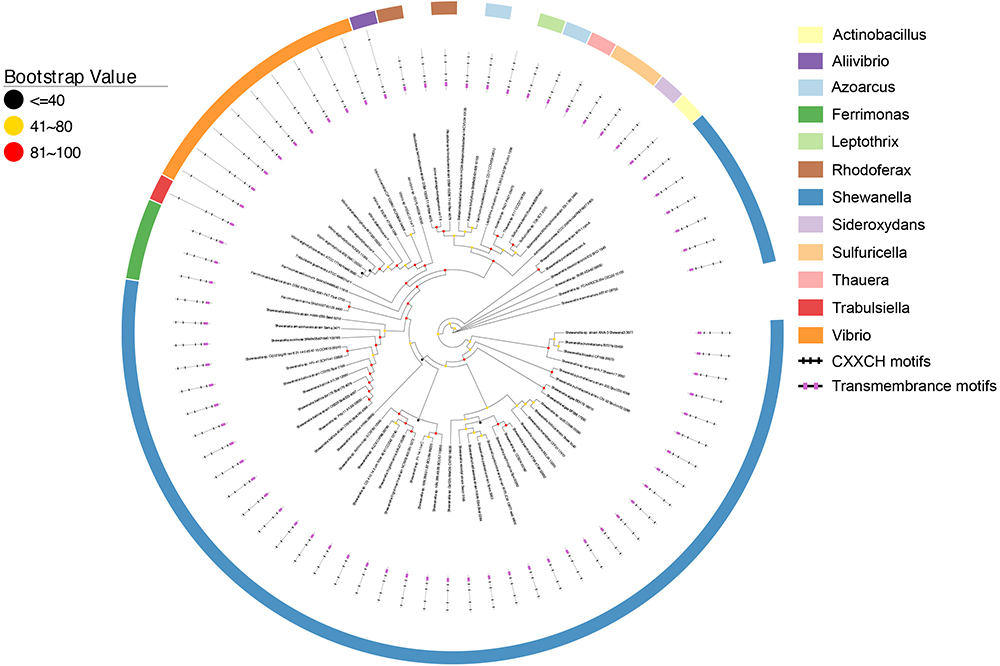
Figure 2. Distribution of CymA homologs. Inner layer, phylogenetic analysis of identified CymA homologs. A phylogenetic tree was constructed and graphically displayed as described in the text. Middle layer, schematic representation of length of and positions of heme-binding as well as transmembrane motifs in the polypeptides of the CymA homologs. Outer layer, bacterial genera from which CymA homologs were identified.
In addition to Shewanella sp., CymA homologs were identified from the bacteria that reduced solid phase Fe(III)-containing and/or Mn(IV)-containing minerals, such as F. balearica, R. ferrireducens, and Vibrio parahaemolyticus and the bacteria that oxidized Fe(II) or Mn(II), such as Sideroxydans lithotrophicus ES-1 and Leptothrix cholodnii (Emerson and Moyer, 1997; Finneran et al., 2003; Nolan et al., 2010; Takeda et al., 2012; Wee et al., 2014). Previous results showed that the cymA homologs of F. balearica, R. ferrireducens, and S. lithotrophicus ES-1 were part of gene clusters that also contained genes whose homologs were involved in extracellular electron transfer (Shi et al., 2012; Emerson et al., 2013; Figure 3). Further analysis of genes adjacent to the cymA genes revealed that, like the cymA found in F. balearica, R. ferrireducens, and S. lithotrophicus, a mtrB/mtoB-like gene and a mtrA/mtoA-like gene were also next to the cymA gene of Azoarcus tolulyticus (Figure 3). Thus, the CymA homologs found in these bacteria are most likely involved in EET. Furthermore, mtrABC homologs were also found in V. parahaemolyticus in which they participated in extracellular reduction of Fe(III)-containing and Mn(IV)-containing minerals (Shi et al., 2012; Wee et al., 2014). Although it is not part of the mtrABC gene cluster, which is similar to that in Shewanella sp., the cymA homolog of V. parahaemolyticusis also likely involved in extracellular reduction of Fe(III)-containing and Mn(IV)-containing minerals. Given that cymA and mtrABC homologs are found in several Vibrio sp. (Shi et al., 2012), EET may also be widespread in Vibrio sp. L. cholodnii oxidizes Mn(II) extracellularly (Takeda et al., 2012). However, whether the released electrons after Mn(II) oxidation are transferred inside bacterial cells remains unknown. Therefore, the role of CymA homolog of L. cholodnii in EET is also unclear.
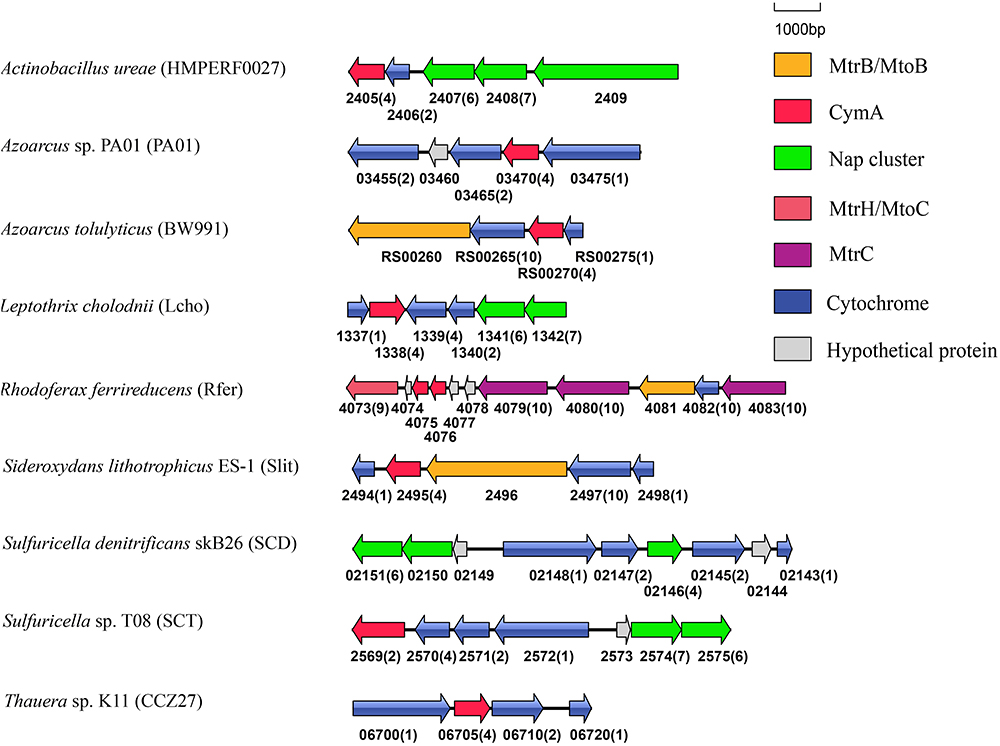
Figure 3. Genetic organization of cymA and its associated genes encoding c-Cyts and MtrB/MtoB. Shown are the relative positions of genes identified within the complete nucleotide sequences of these bacteria. The genes are labeled by arrows whose sizes and orientations indicate their relative lengths and directions in which they are presumed to be transcribed. The numbers displayed below the gene clusters are part of their locus tags whose letter parts are displayed in the parenthesis after species name. The numbers in parenthesis after the locus tag indicating the numbers of heme-binding motifs in the polypeptides deduced from c-Cyt-encoding genes.
CymA is a member of the NapC/NirT family of quinol dehydrogenases (Shi et al., 2007). In S. oneidensis MR-1, CymA is involved in extracellular reduction of Fe(III)- and Mn(IV)-containing minerals and DMSO as well as in reduction of fumarate, nitrate and nitrite in the periplasm (Myers and Myers, 1997, 2000; Schwalb et al., 2003; Lies et al., 2005; Gralnick et al., 2006). Furthermore, in Shewanella sp. strain ANA-3 and S. putrefaciens strain CN-32, CymA is involved in arsenate reduction (Murphy and Saltikov, 2007). Notably, cymA genes were often found next to the napG and napH genes that are involved in nitrate reduction (Figure 3; Richardson, 2000). The CymA homologs found in these bacteria, such as Actinobacillus ureae, Leptothrix cholodnii, Sulfuricella sp. T08 and Sulfuricella denitrificans skB26 are probably also involved in nitrate reduction. Finally, in Azoarcus sp. PA01 and Thauera sp. K11, cymA genes were also found next to the genes encoding putative multi-heme c-Cyts without defined functions (Figure 3).
Except the CymA homologs of S. lithotrophicus ES-1, no additional CymA homolog was found in the Fe(II)-oxidizing bacteria in this study. In S. lithotrophicus ES-1, the CymA homolog was proposed to function as a quinone reductase (Liu et al., 2012; Shi et al., 2012, 2016). Lack of additional CymA homologs from the metal-oxidizing bacteria renders it impossible to assess the difference between CymA of the metal-oxidizing bacteria with those of the metal-reducing bacteria. CymA of S. lithotrophicus ES-1 was 33.1% identical to CymA of S. oneidensis MR-1, which is a functional quinol oxidase (Marritt et al., 2012a,b; McMillan et al., 2012, 2013). The identities between CymA of S. lithotrophicus ES-1 and CymA homologs of other bacteria ranged from 29.4 to 66.4%. CymA of S. oneidensis MR-1 possesses intrinsic quinone reductase activty and its quinol oxidase activitiy is activated by its physical association with the periplasmic c-Cyt Fcc3 (McMillan et al., 2013). Thus, the catalytic activity and electron transfer direction of CymA of S. oneidensis MR-1 and probably the CymA homologs in other bacteria are regulated by their interaction with other proteins.
The CbcL/MtoC/MtrH Homologs
Figure 4 shows that CbcL homologs of Geobacter sp. are clustered together as well as with the homologs from other bacteria, such as Desulfuromonas sp. and Syntrophus sp. The identity among these homologs ranged from 57.6 to 100%, demonstrating the conserved functional roles of CbcL homologs in these bacteria. MtoC of the Fe(II)-oxidizing bacterium D. aromatica strain RCB was clustered with those identified from metagenomic sequencing the samples collected from underground water at the Horonobe Underground Research Laboratory (URL) (Shi et al., 2012). The identity among these homologs were 63.2–82.7%. As no apparent Fe(II)-oxidizing bacterium was found in the same site (Hernsdorf et al., 2017), the roles of MtoC homologs clustered with that of D. aromatica strain RCB remain uncertain. Additionally, MtoC of the Fe(II)-oxidizing bacterium G. capsiferriformans strain ES-2 was clustered with ones of a bacterium from Gallionellales, the sulfur-oxidizing bacterium Sulfuricaulis limicola and a bacterial community from a pilot plant for the treatment of acid mine drainage (AMD) from the lignite mining district in Lusatia, Germany. They were 58.2–91.9% identical. Notably, the MtoC homologs of D. aromatica strain RCB and G. capsiferriformans strain ES-2 were only 58.7% identical. Moreover, MtrH of the Fe(III)-reducing bacterium R. ferrireducens was clustered with the homologs of the bacteria with no apparent role in EET and was 55.4 and 55.2% identical to MtoCs of G. capsiferriformans strain ES-2 and D. aromatica, respectively. Finally, the identities between CbcL of G. sulfurreducens and MtrH of R. ferrireducens as well as MtoCs of D. aromatica RCB and G. capsiferriformans ES-2 were 35.7, 35.4, and 35.5%, respectively.
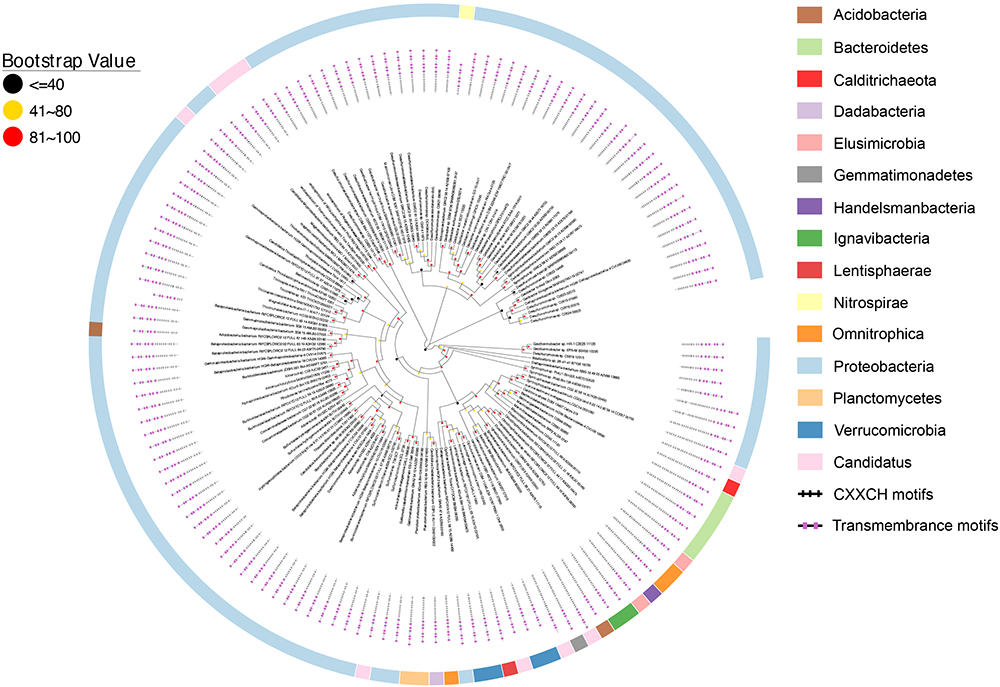
Figure 4. Distribution of CbcL/MtrH/MtoA homologs. Inner layer, phylogenetic analysis of identified CbcL/MtrH/MtoA homologs. A phylogenetic tree was constructed and graphically displayed as described in the text. Middle layer, schematic representation of length of and positions of heme-binding as well as transmembrane motifs in the polypeptides of the CbcL/MtrH/MtoA homologs. Outer layer, bacterial phyla from which CbcL/MtrH/MtoA homologs were identified.
The overall identities among identified CbcL/MtrH/MtoC homologs ranged from 28.6 to 100%. The polypeptide length of identified CbcL/MtrH/MtoC homologs ranged from 416 to 1034 amino acid residues (Figure 4 and Supplementary Table S2). In addition, their N-terminal portions also varied substantially, of which the CX2CH motifs ranged from 3 to 23 (Figure 4 and Supplementary Table S2). Moreover, most identified CbcL/MtrH/MtoC homologs are predicted to possess six transmembrane motifs. However, four homologs whose accession numbers are PKN12990.1, PLY03765.1, OGT98815.1, and OQX95005.1, respectively, have five transmembrane motifs and one homolog with an accession number of OGU12690.1 contains only four transmembrane motifs (Figure 4 and Supplementary Table S2). Finally, the three histidine residues that might be the ligands for b-type hemes were conserved in the transmembrane regions.
Similar to cymA, mtrH and mtoC genes are often associated with other c-Cyt genes. For instance, in the genome of R. ferrireducens, mtrH is part of a gene cluster that also contains cymA, mtrA, mtrB, mtrC, and mtrH (Figure 5). In the genomes of D. aromatica RCB and G. capsiferriformans, mtoC is in a gene cluster that contains mtoA, mtoB, and mtoD (Figure 5; Shi et al., 2012). Our results from this study showed that mtrH/mtoC genes of 7 bacterial species were also part of a gene cluster containing mtrAB/mtoAB genes (Figure 5). Given that mtrAB/mtoAB genes function to transfer electrons across the outer membrane (Hartshorne et al., 2009; Richardson et al., 2012; Shi et al., 2012), the mtrH/mtoC genes found with mtrAB/mtoAB genes are most likely involved in EET. Moreover, the cbcL/mtrH/mtoC genes of other 11 bacterial species were also adjacent to c-Cyt-encoding genes (Figure 5). Further comparisons between CbcL of G. sulfurreducens/MtrH of R. ferrireducens with MtoC of D. aromatica RCB and G. capsiferriformans ES-2, however, did not reveal any unique feature for those catalyzing quinol oxidation or quinone reduction.

Figure 5. Genetic organization of cbcL, mtrH, mtoC and their associated genes encoding c-Cyts and MtrB/MtoB. Shown are the relative positions of genes identified within the complete nucleotide sequences of these bacteria. The genes are labeled by arrows whose sizes and orientations indicate their relative lengths and directions in which they are presumed to be transcribed. The numbers displayed below the gene clusters are part of their locus tags whose letter parts are displayed in the parenthesis after species name. The numbers in parenthesis after the locus tag indicating the numbers of heme-binding motifs in the polypeptides deduced from c-Cyt-encoding genes.
The ImcH Homologs
Similar to the CbcL homologs of Geobacter sp., the ImcH homologs of Geobacter sp. were also clustered together as well as with those from other bacteria, such as Desulfuromonas sp. and Syntrophus sp. (Figure 6). The identities among these homologs were 53.6–100%, also suggesting a conserved functional role among these homologs. The identities between this group of ImcH homologs and those from other bacteria were 31.5–49%. The lengths of polypeptides of ImcH homologs identified from Geobacter sp. and their associated homologs ranged from 495 to 519 amino acid residues. They all contained 3 transmembrane motifs at their N-terminal part, an atypical CX3CH motif next to the transmembrane domain and six typical CX2CH motifs (Figure 6 and Supplementary Table S3). Similar to the ImcH homologs of Geobacter sp., the ImcH homologs identified from other bacteria all had 3 transmembrane motifs and an atypical CX3CH motif, except for the ones of Intrasporangium calvum and Anaerolinea thermophila which contained only one transmembrane motif (Figure 6 and Supplementary Table S3). Moreover, the CX2CH motifs of identified ImcH homologs varied, ranging from 5 to 9 (Figure 6 and Supplementary Table S3). Similar to cymA and cbcL/mtrH/mtoC, imcH genes were also often associated with the genes encoding c-Cyts. Different from cymA and cbcL/mtrH/mtoC, however, no mtrAB/mtoAB were found to be adjacent to imcH (Figure 7).
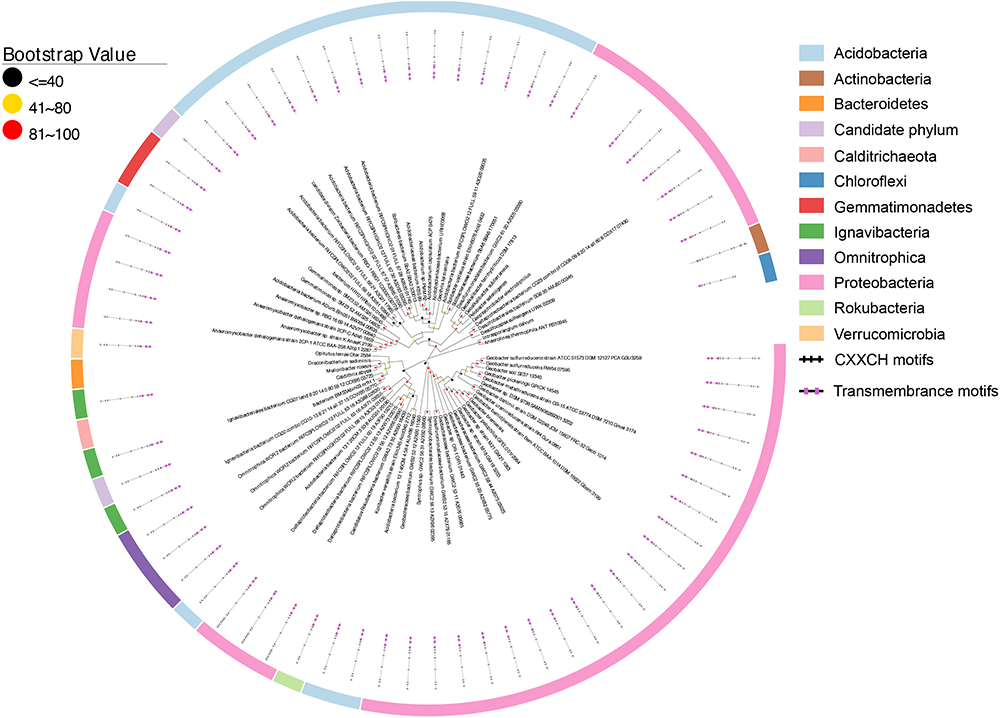
Figure 6. Distribution of ImcH homologs. Inner layer, phylogenetic analysis of identified ImcH homologs. A phylogenetic tree was constructed and graphically displayed as described in the text. Middle layer, schematic representation of length of and positions of heme-binding as well as transmembrane motifs in the polypeptides of the ImcH homologs. Outer layer, bacteria phyla from which ImcH homologs were identified.

Figure 7. Genetic organization of imcH and its associated genes encoding c-Cyts. Shown are the relative positions of genes identified within the complete nucleotide sequence of these bacteria. The genes are labeled by arrows whose sizes and orientations indicate their relative lengths and directions in which they are presumed to be transcribed. The numbers displayed below the gene clusters are part of their locus tags whose letter parts are displayed in the parenthesis after species name. The numbers in parenthesis after the locus tag indicating the numbers of heme-binding motifs in the polypeptides deduced from c-Cyt-encoding genes.
Previous results showed that ImcH was found in other bacteria with EET capability, including several Acidobacteria sp. and Geobacter sp. as well as Anaeromyxobacter dehalogenans 2CP-1, G. ferrihydriticus, G. subterraneus, Geopsychrobacter electrodiphilus, Geothrix fermentans, M. roseus, and Pelobacterselenii genes, suggesting a broad involvement of ImcH in EET (Levar et al., 2014). However, all of these bacteria transfer electrons from cytoplasmic membrane to extracellular electron acceptors. If they are involved in EET, the ImcHs of these bacteria would function as quinol oxidases. Search results from this study did not identify additional ImcH from the bacteria with EET capability. Thus, different from CymA and CbcL/MtrH/MtoC that are implicated in both quinol oxidation and quinone reduction, ImcH has only been implicated in quinol oxidation so far.
Previous results also showed that the homologs of CymA and CbcL/MtrH/MtoC families or the homologs of CbcL/MtrH/MtoC and ImcH families could be found in the same bacterium (Shi et al., 2012; Levar et al., 2014; Zacharoff et al., 2015). In G. sulfurreducens, CbcL and ImcH participate in EET to the terminal electron acceptors with different redox potential (Levar et al., 2014, 2017; Zacharoff et al., 2015). Results from this study demonstrate that most Geobacter sp. investigated as well as other metal-reducing bacteria also have both CbcL and ImcH, where they may participate in EET to the terminal electron acceptors with different redox potentials. Moreover, in the metal-reducing bacterium R. ferrireducens, 2 cymAs and a mtrH are in the same gene cluster that also has mtrABC and the genes encoding for other c-Cyts. The two CymAs of R. ferrireducens are only 55.5% identical, which suggests that these two CymAs may oxidize different quinols. Thus, it is possible the CymAs and MtrH of R. ferrireducens are also involved in regulating EET to different terminal electron acceptors. Possessions of different quinol oxidases for EET would render the microorganisms better adapted to the changing environment. Finally, it should also be noted that homologs of CymA, CbcL/MtrH/MtoC, and ImcH families are not found in the same microorganism in this study.
Conclusion
Crucial to bacterial and probably archaeal EET are the quinol oxidases and quinone reductases in the cytoplasmic membrane. These groups of enzymes function either as the first step during electron transfer from the cytoplasmic membrane to extracellular electron acceptors and/or the last step during electron transfer from extracellular electron donors to the cytoplasmic membrane. CymA, CbcL/MtrH/MtoC and ImcH were the only three protein families that were known to be involved in these crucial EET steps during this search. Survey of bacterial genomes from this study reveals that CymA family is only restricted to the phylum of Proteobacteria and most of the homologs are found in the Shewanella genus, while CbcL/MtrH/MtoC and ImcH protein family are much more widespread among different bacterial phyla, demonstrating much broader distributions of latter two protein families in bacterial EET.
A unique feature for bacterial EET is its bidirectional nature (Shi et al., 2012). This is reflected by the involvements of Mtr homologs in both Fe(II) oxidation and Fe(III) reduction (Beliaev and Saffarini, 1998; Jiao and Newman, 2007), quinol oxidation and quinone reduction capability of CymA (Marritt et al., 2012a; McMillan et al., 2013) and reversible conductivity with nearly equal efficiency of MtrF and its homologs, such as MtrC (Breuer et al., 2012, 2014). Indeed, S. oneidensis MR-1 employs the Mtr pathway for transferring electron from the cytoplasmic membrane to electrode surfaces as well as from electrode surfaces to the cytoplasmic membrane under anoxic condition (Ross et al., 2011). Identification of CbcL/MtrH/MtoC homologs in both Fe(II)-oxidizing and Fe(III)-reducing bacteria are consistent with the bidirectional nature of bacterial EET. However, ImcH homologs have been identified only in the Fe(III)-reducing bacteria to date. The reason for this observation is currently unknown. It may be attributed to limited homologs identified, lack of insightful understanding of bacterial Fe(II) oxidation process or inability to reduce quinone by ImcH.
Lack of apparently distinctive feature between CbcL/MtrH homologs and MtoC homologs also indicate that the direction of catalysis and electron transfer of CbcL/MtrH/MtoC may be regulated by protein–protein interactions. Indeed, CbcL, MtoC and probably MtrH need to interact with other periplasmic redox proteins, such as c-Cyts, in order to exchange electrons with extracellular substrates (Shi et al., 2012, 2016). Previous results also demonstrated the importance of biochemical characterization of CymA from S. oneidensis MR-1 in understanding the underlying mechanisms of this type of redox enzymes for regulating their catalytic activity and electron transfer reactions. Thus, biochemical characterization of CbcL and ImcH homologs will be one of the future directions for investigating the catalytic properties of these new families of redox enzymes crucial for bacterial EET.
Author Contributions
LS designed the experiments. YZ performed the experiments. YZ and LS analyzed the data and prepared the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank National Natural Science Foundation of China (NSFC91851211 and 41772363), One Hundred Talents Program of Hubei Province and China University of Geosciences-Wuhan for their supports.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.03029/full#supplementary-material
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Beliaev, A. S., and Saffarini, D. A. (1998). Shewanella putrefaciens mtrB encodes an outer membrane protein required for Fe(III) and Mn(IV) reduction. J. Bacteriol. 180, 6292–6297.
Bond, D. R., and Lovley, D. R. (2003). Electricity production by Geobacter sulfurreducens attached to electrodes. Appl. Environ. Microbiol. 69, 1548–1555. doi: 10.1128/AEM.69.3.1548-1555.2003
Bose, A., Gardel, E. J., Vidoudez, C., Parra, E. A., and Girguis, P. R. (2014). Electron uptake by iron-oxidizing phototrophic bacteria. Nat. Commun. 5:3391. doi: 10.1038/ncomms4391
Bretschger, O., Obraztsova, A., Sturm, C. A., Chang, I. S., Gorby, Y. A., Reed, S. B., et al. (2007). Current production and metal oxide reduction by Shewanella oneidensis MR-1 wild type and mutants. Appl. Environ. Microbiol. 73, 7003–7012. doi: 10.1128/AEM.01087-07
Breuer, M., Rosso, K. M., and Blumberger, J. (2014). Electron flow in multiheme bacterial cytochromes is a balancing act between heme electronic interaction and redox potentials. Proc. Natl. Acad. Sci. U.S.A. 111, 611–616. doi: 10.1073/pnas.1316156111
Breuer, M., Zarzycki, P., Blumberger, J., and Rosso, K. M. (2012). Thermodynamics of electron flow in the bacterial deca-heme cytochrome MtrF. J. Am. Chem. Soc. 134, 9868–9871. doi: 10.1021/ja3027696
Emerson, D., Field, E. K., Chertkov, O., Davenport, K. W., Goodwin, L., Munk, C., et al. (2013). Comparative genomics of freshwater Fe-oxidizing bacteria: implications for physiology, ecology, and systematics. Front. Microbiol. 4:254. doi: 10.3389/fmicb.2013.00254
Emerson, D., and Moyer, C. (1997). Isolation and characterization of novel iron-oxidizing bacteria that grow at circumneutral pH. Appl. Environ. Microbiol. 63, 4784–4792.
Emerson, D., Rentz, J. A., Lilburn, T. G., Davis, R. E., Aldrich, H., Chan, C., et al. (2007). A novel lineage of proteobacteria involved in formation of marine Fe-oxidizing microbial mat communities. PLoS One 2:e667. doi: 10.1371/journal.pone.0000667
Finneran, K. T., Johnsen, C. V., and Lovley, D. R. (2003). Rhodoferax ferrireducens sp. nov., a psychrotolerant, facultatively anaerobic bacterium that oxidizes acetate with the reduction of Fe(III). Int. J. Syst. Evol. Microbiol. 53(Pt 3), 669–673.
Firer-Sherwood, M. A., Bewley, K. D., Mock, J. Y., and Elliott, S. J. (2011). Tools for resolving complexity in the electron transfer networks of multiheme cytochromes c. Metallomics 3, 344–348. doi: 10.1039/c0mt00097c
Fonseca, B. M., Paquete, C. M., Neto, S. E., Pacheco, I., Soares, C. M., and Louro, R. O. (2013). Mind the gap: cytochrome interactions reveal electron pathways across the periplasm of Shewanella oneidensis MR-1. Biochem. J. 449, 101–108. doi: 10.1042/BJ20121467
Gorby, Y. A., Yanina, S., McLean, J. S., Rosso, K. M., Moyles, D., Dohnalkova, A., et al. (2006). Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc. Natl. Acad. Sci. U.S.A. 103, 11358–11363. doi: 10.1073/pnas.0604517103
Gralnick, J. A., Vali, H., Lies, D. P., and Newman, D. K. (2006). Extracellular respiration of dimethyl sulfoxide by Shewanella oneidensis strain MR-1. Proc. Natl. Acad. Sci. U.S.A. 103, 4669–4674. doi: 10.1073/pnas.0505959103
Hartshorne, R. S., Reardon, C. L., Ross, D., Nuester, J., Clarke, T. A., Gates, A. J., et al. (2009). Characterization of an electron conduit between bacteria and the extracellular environment. Proc. Natl. Acad. Sci. U.S.A. 106, 22169–22174. doi: 10.1073/pnas.0900086106
He, Z., Zhang, H., Gao, S., Lercher, M. J., Chen, W. H., and Hu, S. (2016). Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 44, W236–W241. doi: 10.1093/nar/gkw370
Hernsdorf, A. W., Amano, Y., Miyakawa, K., Ise, K., Suzuki, Y., Anantharaman, K., et al. (2017). Potential for microbial H2 and metal transformations associated with novel bacteria and archaea in deep terrestrial subsurface sediments. ISME J. 11, 1915–1929. doi: 10.1038/ismej.2017.39
Jiao, Y., Kappler, A., Croal, L. R., and Newman, D. K. (2005). Isolation and characterization of a genetically tractable photoautotrophic Fe(II)-oxidizing bacterium, Rhodopseudomonas palustris strain TIE-1. Appl. Environ. Microbiol. 71, 4487–4496. doi: 10.1128/AEM.71.8.4487-4496.2005
Jiao, Y., and Newman, D. K. (2007). The pio operon is essential for phototrophic Fe(II) oxidation in Rhodopseudomonas palustris TIE-1. J. Bacteriol. 189, 1765–1773. doi: 10.1128/JB.00776-06
Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Levar, C. E., Chan, C. H., Mehta-Kolte, M. G., and Bond, D. R. (2014). An inner membrane cytochrome required only for reduction of high redox potential extracellular electron acceptors. mBio 5:e02034-14. doi: 10.1128/mBio.02034-14
Levar, C. E., Hoffman, C. L., Dunshee, A. J., Toner, B. M., and Bond, D. R. (2017). Redox potential as a master variable controlling pathways of metal reduction by Geobacter sulfurreducens. ISME J. 11, 741–752. doi: 10.1038/ismej.2016.146
Lies, D. P., Hernandez, M. E., Kappler, A., Mielke, R. E., Gralnick, J. A., and Newman, D. K. (2005). Shewanella oneidensis MR-1 uses overlapping pathways for iron reduction at a distance and by direct contact under conditions relevant for biofilms. Appl. Environ. Microbiol. 71, 4414–4426. doi: 10.1128/AEM.71.8.4414-4426.2005
Liu, J., Wang, Z., Belchik, S. M., Edwards, M. J., Liu, C., Kennedy, D. W., et al. (2012). Identification and characterization of MtoA: a decaheme c-type cytochrome of the neutrophilic Fe(II)-oxidizing bacterium Sideroxydans lithotrophicus ES-1. Front. Microbiol. 3:37. doi: 10.3389/fmicb.2012.00037
Liu, W., Xie, Y., Ma, J., Luo, X., Nie, P., Zuo, Z., et al. (2015). IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics 31, 3359–3361. doi: 10.1093/bioinformatics/btv362
Liu, Y., Wang, Z., Liu, J., Levar, C., Edwards, M. J., Babauta, J. T., et al. (2014). A trans-outer membrane porin-cytochrome protein complex for extracellular electron transfer by Geobacter sulfurreducens PCA. Environ. Microbiol. Rep. 6, 776–785. doi: 10.1111/1758-2229.12204
Lloyd, J. R., Leang, C., Hodges Myerson, A. L., Coppi, M. V., Cuifo, S., Methe, B., et al. (2003). Biochemical and genetic characterization of PpcA, a periplasmic c-type cytochrome in Geobacter sulfurreducens. Biochem. J. 369(Pt 1), 153–161. doi: 10.1042/BJ20020597
Lovley, D. R., Coates, J. D., Blunt-Harris, E. L., Phillips, E. J. P., and Woodward, J. C. (1996). Humic substances as electron acceptors for microbial respiration. Nature 382, 445–448. doi: 10.1038/382445a0
Lovley, D. R., Stolz, J. F., Nord, G. L., and Phillips, E. J. P. (1987). Anaerobic production of magnetite by a dissimilatory iron-reducing microorganism. Nature 330, 252–254. doi: 10.1038/330252a0
Marritt, S. J., Lowe, T. G., Bye, J., McMillan, D. G., Shi, L., Fredrickson, J., et al. (2012a). A functional description of CymA, an electron-transfer hub supporting anaerobic respiratory flexibility in Shewanella. Biochem. J. 444, 465–474. doi: 10.1042/BJ20120197
Marritt, S. J., McMillan, D. G., Shi, L., Fredrickson, J. K., Zachara, J. M., Richardson, D. J., et al. (2012b). The roles of CymA in support of the respiratory flexibility of Shewanella oneidensis MR-1. Biochem. Soc. Trans. 40, 1217–1221. doi: 10.1042/BST20120150
McMillan, D. G., Marritt, S. J., Butt, J. N., and Jeuken, L. J. (2012). Menaquinone-7 is specific cofactor in tetraheme quinol dehydrogenase CymA. J. Biol. Chem. 287, 14215–14225. doi: 10.1074/jbc.M112.348813
McMillan, D. G., Marritt, S. J., Firer-Sherwood, M. A., Shi, L., Richardson, D. J., Evans, S. D., et al. (2013). Protein-protein interaction regulates the direction of catalysis and electron transfer in a redox enzyme complex. J. Am. Chem. Soc. 135, 10550–10556. doi: 10.1021/ja405072z
Morgado, L., Bruix, M., Pessanha, M., Londer, Y. Y., and Salgueiro, C. A. (2010). Thermodynamic characterization of a triheme cytochrome family from Geobacter sulfurreducens reveals mechanistic and functional diversity. Biophys. J. 99, 293–301. doi: 10.1016/j.bpj.2010.04.017
Murphy, J. N., and Saltikov, C. W. (2007). The cymA gene, encoding a tetraheme c-type cytochrome, is required for arsenate respiration in Shewanella species. J. Bacteriol. 189, 2283–2290. doi: 10.1128/JB.01698-06
Myers, C. R., and Myers, J. M. (1997). Cloning and sequence of cymA, a gene encoding a tetraheme cytochrome c required for reduction of iron(III), fumarate, and nitrate by Shewanella putrefaciens MR-1. J. Bacteriol. 179, 1143–1152. doi: 10.1128/jb.179.4.1143-1152.1997
Myers, C. R., and Nealson, K. H. (1988). Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science 240, 1319–1321. doi: 10.1126/science.240.4857.1319
Myers, J. M., and Myers, C. R. (2000). Role of the tetraheme cytochrome CymA in anaerobic electron transport in cells of Shewanella putrefaciens MR-1 with normal levels of menaquinone. J. Bacteriol. 182, 67–75. doi: 10.1128/JB.182.1.67-75.2000
Nolan, M., Sikorski, J., Davenport, K., Lucas, S., Del Rio, T. G., Tice, H., et al. (2010). Complete genome sequence of Ferrimonas balearica type strain (PAT). Stand. Genomic Sci. 3, 174–182. doi: 10.4056/sigs.1161239
Pirbadian, S., Barchinger, S. E., Leung, K. M., Byun, H. S., Jangir, Y., Bouhenni, R. A., et al. (2014). Shewanella oneidensis MR-1 nanowires are outer membrane and periplasmic extensions of the extracellular electron transport components. Proc. Natl. Acad. Sci. U.S.A. 111, 12883–12888. doi: 10.1073/pnas.1410551111
Richardson, D. J. (2000). Bacterial respiration: a flexible process for a changing environment. Microbiology 146(Pt 3), 551–571. doi: 10.1099/00221287-146-3-551
Richardson, D. J., Butt, J. N., Fredrickson, J. K., Zachara, J. M., Shi, L., Edwards, M. J., et al. (2012). The ’porin-cytochrome’ model for microbe-to-mineral electron transfer. Mol. Microbiol. 85, 201–212. doi: 10.1111/j.1365-2958.2012.08088.x
Ross, D. E., Flynn, J. M., Baron, D. B., Gralnick, J. A., and Bond, D. R. (2011). Towards electrosynthesis in Shewanella: energetics of reversing the mtr pathway for reductive metabolism. PLoS One 6:e16649. doi: 10.1371/journal.pone.0016649
Schwalb, C., Chapman, S. K., and Reid, G. A. (2003). The tetraheme cytochrome CymA is required for anaerobic respiration with dimethyl sulfoxide and nitrite in Shewanella oneidensis. Biochemistry 42, 9491–9497. doi: 10.1021/bi034456f
Shi, L., Chen, B., Wang, Z., Elias, D. A., Mayer, M. U., Gorby, Y. A., et al. (2006). Isolation of a high-affinity functional protein complex between OmcA and MtrC: two outer membrane decaheme c-type cytochromes of Shewanella oneidensis MR-1. J. Bacteriol. 188, 4705–4714. doi: 10.1128/JB.01966-05
Shi, L., Dong, H., Reguera, G., Beyenal, H., Lu, A., Liu, J., et al. (2016). Extracellular electron transfer mechanisms between microorganisms and minerals. Nat. Rev. Microbiol. 14, 651–662. doi: 10.1038/nrmicro.2016.93
Shi, L., Fredrickson, J., and Zachara, J. (2014). Genomic analyses of bacterial porin-cytochrome gene clusters. Front. Microbiol. 5:657. doi: 10.3389/fmicb.2014.00657
Shi, L., Potts, M., and Kennelly, P. J. (1998). The serine, threonine, and/or tyrosine-specific protein kinases and protein phosphatases of prokaryotic organisms: a family portrait. FEMS Microbiol. Rev. 22, 229–253. doi: 10.1111/j.1574-6976.1998.tb00369.x
Shi, L., Rosso, K. M., Zachara, J. M., and Fredrickson, J. K. (2012). Mtr extracellular electron-transfer pathways in Fe(III)-reducing or Fe(II)-oxidizing bacteria: a genomic perspective. Biochem. Soc. Trans. 40, 1261–1267. doi: 10.1042/BST20120098
Shi, L., Squier, T. C., Zachara, J. M., and Fredrickson, J. K. (2007). Respiration of metal (hydr)oxides by Shewanella and Geobacter: a key role for multihaem c-type cytochromes. Mol. Microbiol. 65, 12–20. doi: 10.1111/j.1365-2958.2007.05783.x
Shi, L., and Zhang, W. (2004). Comparative analysis of eukaryotic-type protein phosphatases in two streptomycete genomes. Microbiology 150(Pt 7), 2247–2256. doi: 10.1099/mic.0.27057-0150/7/2247
Subramanian, P., Pirbadian, S., El-Naggar, M. Y., and Jensen, G. J. (2018). Ultrastructure of Shewanella oneidensis MR-1 nanowires revealed by electron cryotomography. Proc. Natl. Acad. Sci. U.S.A. 115, E3246–E3255. doi: 10.1073/pnas.1718810115
Summers, Z. M., Fogarty, H. E., Leang, C., Franks, A. E., Malvankar, N. S., and Lovley, D. R. (2010). Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria. Science 330, 1413–1415. doi: 10.1126/science.1196526
Summers, Z. M., Gralnick, J. A., and Bond, D. R. (2013). Cultivation of an obligate Fe(II)-oxidizing lithoautotrophic bacterium using electrodes. mBio 4:e00420-12. doi: 10.1128/mBio.00420-12
Takeda, M., Kawasaki, Y., Umezu, T., Shimura, S., Hasegawa, M., and Koizumi, J. (2012). Patterns of sheath elongation, cell proliferation, and manganese(II) oxidation in Leptothrix cholodnii. Arch. Microbiol. 194, 667–673. doi: 10.1007/s00203-012-0801-6
Wee, S. K., Burns, J. L., and DiChristina, T. J. (2014). Identification of a molecular signature unique to metal-reducing Gammaproteobacteria. FEMS Microbiol. Lett. 350, 90–99. doi: 10.1111/1574-6968.12304
Wei, H., Dai, J., Xia, M., Romine, M. F., Shi, L., Beliav, A., et al. (2016). Functional roles of CymA and NapC in reduction of nitrate and nitrite by Shewanella putrefaciens W3-18-1. Microbiology 162, 930–941. doi: 10.1099/mic.0.000285
White, G. F., Shi, Z., Shi, L., Dohnalkova, A. C., Fredrickson, J. K., Zachara, J. M., et al. (2012). Development of a proteoliposome model to probe transmembrane electron-transfer reactions. Biochem. Soc. Trans. 40, 1257–1260. doi: 10.1042/BST20120116
White, G. F., Shi, Z., Shi, L., Wang, Z., Dohnalkova, A. C., Marshall, M. J., et al. (2013). Rapid electron exchange between surface-exposed bacterial cytochromes and Fe(III) minerals. Proc. Natl. Acad. Sci. U.S.A. 110, 6346–6351. doi: 10.1073/pnas.1220074110
Zacharoff, L., Chan, C. H., and Bond, D. R. (2015). Reduction of low potential electron acceptors requires the CbcL inner membrane cytochrome of Geobacter sulfurreducens. Bioelectrochemistry 107, 7–13. doi: 10.1016/j.bioelechem.2015.08.003
Zhang, H., Tang, X., Munske, G. R., Tolic, N., Anderson, G. A., and Bruce, J. E. (2009). Identification of protein-protein interactions and topologies in living cells with chemical cross-linking and mass spectrometry. Mol. Cell Proteomics 8, 409–420. doi: 10.1074/mcp.M800232-MCP200
Zhang, H., Tang, X., Munske, G. R., Zakharova, N., Yang, L., Zheng, C., et al. (2008). In vivo identification of the outer membrane protein OmcA-MtrC interaction network in Shewanella oneidensis MR-1 cells using novel hydrophobic chemical cross-linkers. J. Proteome Res. 7, 1712–1720. doi: 10.1021/pr7007658
Keywords: quinol oxidase, quinone reductase, multiheme c-type cytochromes, extracellular electron transfer pathways, the cytoplasmic membrane
Citation: Zhong Y and Shi L (2018) Genomic Analyses of the Quinol Oxidases and/or Quinone Reductases Involved in Bacterial Extracellular Electron Transfer. Front. Microbiol. 9:3029. doi: 10.3389/fmicb.2018.03029
Received: 03 October 2018; Accepted: 23 November 2018;
Published: 10 December 2018.
Edited by:
Fanghua Liu, Yantai Institute of Coastal Zone Research (CAS), ChinaReviewed by:
Tobias Goris, Friedrich-Schiller-Universität Jena, GermanySantosh Kr Karn, Sardar Bhagwan Singh Post Graduate Institute of Biomedical Science & Research, India
Copyright © 2018 Zhong and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liang Shi, bGlhbmcuc2hpQGN1Zy5lZHUuY24=
 Yuhong Zhong
Yuhong Zhong Liang Shi
Liang Shi