- Department of Basic Sciences, College of Veterinary Medicine, Mississippi State University, Starkville, MS, United States
Edwardsiella ictaluri is an intracellular Gram-negative facultative pathogen causing enteric septicemia of catfish (ESC), a common disease resulting in substantial economic losses in the U.S. catfish industry. Previously, we demonstrated that several universal stress proteins (USPs) are highly expressed under in vitro and in vivo stress conditions, indicating their importance for E. ictaluri survival. However, the roles of these USPs in E. ictaluri virulence is not known yet. In this work, 10 usp genes of E. ictaluri were in-frame deleted and characterized in vitro and in vivo. Results show that all USP mutants were sensitive to acidic condition (pH 5.5), and EiΔusp05 and EiΔusp08 were very sensitive to oxidative stress (0.1% H2O2). Virulence studies indicated that EiΔusp05, EiΔusp07, EiΔusp08, EiΔusp09, EiΔusp10, and EiΔusp13 were attenuated significantly compared to E. ictaluri wild-type (EiWT; 20, 45, 20, 20, 55, and 10% vs. 74.1% mortality, respectively). Efficacy experiments showed that vaccination of catfish fingerlings with EiΔusp05, EiΔusp07, EiΔusp08, EiΔusp09, EiΔusp10, and EiΔusp13 provided complete protection against EiWT compared to sham-vaccinated fish (0% vs. 58.33% mortality). Our results support that USPs contribute E. ictaluri virulence in catfish.
Introduction
Enteric septicemia of channel catfish (ESC) is one of the most prevalent diseases of cultured catfish, causing significant losses (USDA, 2014). The most common practice in ESC treatment is use of feed medicated with oxytetracycline, sulfadimethoxine, or florfenicol. However, one of the earliest clinical signs of ESC is reduced appetite. Thus, these antimicrobials are only useful in limiting the spread of an outbreak and rather than treating the disease. Also, medicated feed may lead to the emergence of resistant Edwardsiella ictaluri strains (Tu et al., 2008).
The universal stress proteins (USP) have a conserved domain of 140–160 amino acids, and are present in archaea, bacteria, and plants (Nachin et al., 2005), but not in animals and human (Siegele, 2005). In Escherichia coli usp are involved in various functions from oxidative stress to adhesion and motility (Nachin et al., 2005). Under stress, USPs are overproduced and through a variety of mechanisms aid the survival of organism in stressful conditions (Heermann et al., 2009b). The uspA mutation caused decreased survival in E. coli (Tkaczuk et al., 2013). It is known that USPs are needed by pathogens (Hensel, 2009). USPs affect persistence and survival of Mycobacterium tuberculosis (Hingley-Wilson et al., 2010), and cause growth arrest and reduce the virulence in Salmonella typhimurium C5 (Liu et al., 2007) and Burkholderia pseudomallei (Al-Maleki et al., 2014). USPs are also necessary for the intracellular growth adaption of Listeria monocytogenes (Chatterjee et al., 2006). Similarly, Staphylococcus aureus virulence factors were downregulated in vivo while expression of uspA increased (Chaffin et al., 2012). Acinetobacter baumannii uspA is essential in pneumonia and pathogenesis (Elhosseiny et al., 2015).
Although increased expression of several usp genes in E. ictaluri under various stressors has been reported (Akgul et al., 2018), the role of USPs in E. ictaluri virulence is not known yet. Therefore, in this study, 10 E. ictaluri usp genes were studied by introducing in-frame deletions and determining their survival under acidic and oxidative stress conditions. Also, the virulence and protective properties of mutants against ESC infection were tested in catfish fingerlings.
Materials and Methods
Animals
All fish experiments were performed based on a protocol approved by the Mississippi State University Institutional Animal Care and Use Committee (protocol number 15-043). Channel catfish fingerlings were obtained from the fish hatchery at the College of Veterinary Medicine, Mississippi State University, and maintained at 25–28°C during experiments. Tricaine methanesulfonate (MS-222, Western, Chemical, Inc.) was used to sedate (100 mg/ml) or euthanize (400 mg/ml) the catfish.
Bacterial Strains, Plasmids, and Growth Conditions
Bacterial strains and plasmids used in this work are listed in Table 1. E. ictaluri 93–146 wild-type (WT) was grown at 30°C using Brain Heart Infusion (BHI) broth and agar (Difco, Sparks, MD, United States). E. coli strains were cultured at 37°C using Luria-Bertani (LB) broth and agar (Difco). E. coli CC118λpir was used for cloning and SM10λpir or BW19851 were used for transferring pMEG-375 or pAKgfplux1 into E. ictaluri. When required, the following antibiotics and reagents (Sigma-Aldrich, Saint Louis, MN, United States) were added to culture medium at the following concentrations: ampicillin (Amp: 100 μg/ml), colistin (Col: 12.5 μg/ml), sucrose (5%), and mannitol (0.35%).
Construction of In-Frame Deletion Mutants
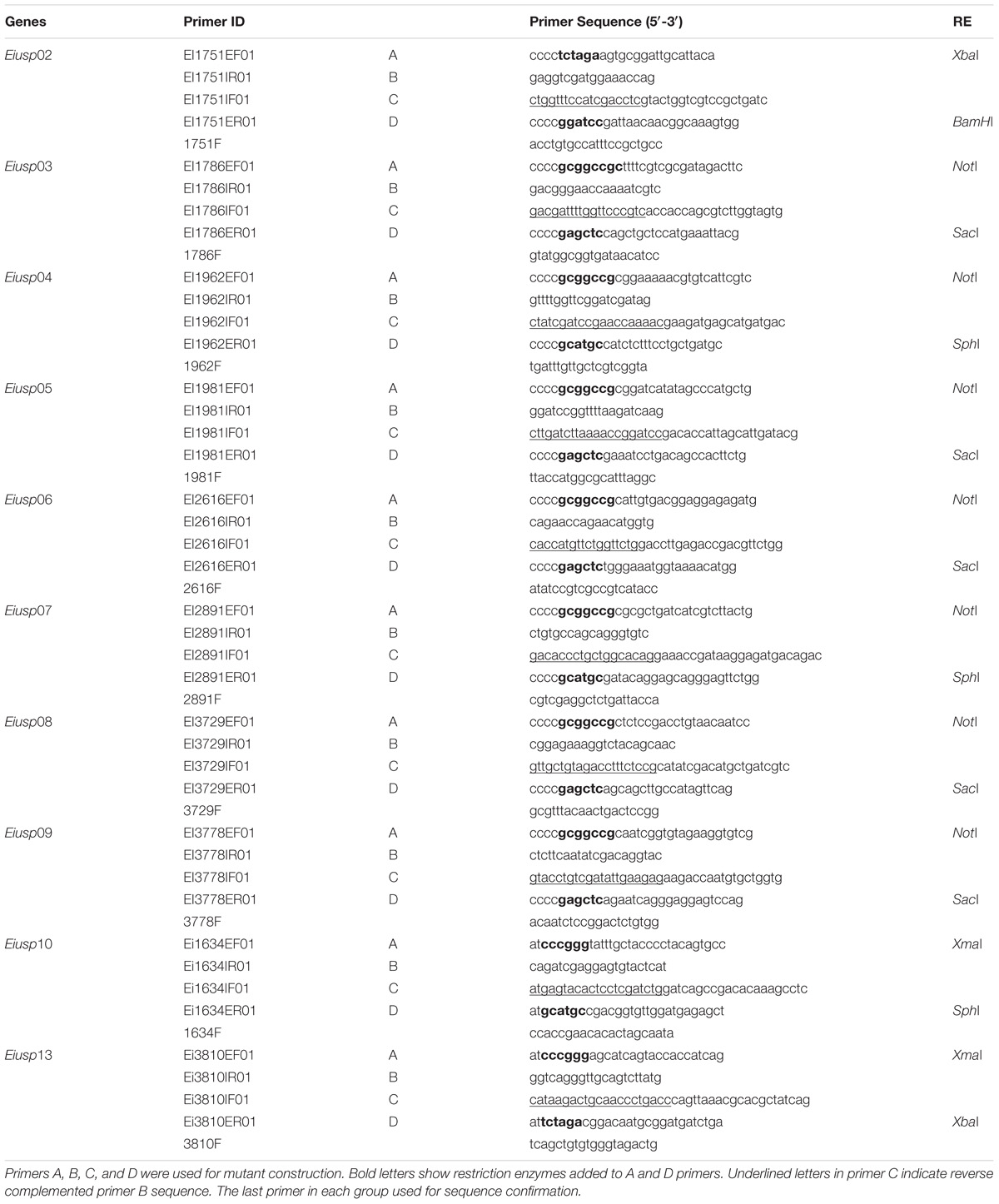
The nucleotide sequences of 10 E. ictaluri usp genes were obtained from the E. ictaluri 93–146 genome (GenBank accession: CP001600), and four primers were designed for each gene (Tables 2, 3). Restriction sites were included in forward and reverse primers. Overlap extension PCR was used to delete the functional usp genes from the E. ictaluri genome (Horton et al., 1990). Genomic DNA was isolated from E. ictaluri using a DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA, United States) and used as template in PCR. The upstream and downstream regions of each gene were amplified, and products were gel-extracted using a QIAquick Gel Extraction Kit (Qiagen). The amplified upstream and downstream fragments were mixed equally and used as a template in the subsequent overlap extension PCR to generate the in-frame deletion fragment for each gene. The in-frame deletion fragments were digested with appropriate restriction enzymes (NEB) (Table 2) and cleaned up. The suicide plasmid pMEG-375 was purified from an overnight E. coli culture by a QIAprep Spin Miniprep Kit (Qiagen) and digested with appropriate restriction enzymes respective to the inserts. The in-frame deletion fragments were ligated into the linearized pMEG-375 vector using T4 DNA Ligase (NEB) at 16°C overnight. E. coli CC118λpir was transformed by electroporation and plated on LB agar plus ampicillin. Resulting plasmids were isolated from the colonies and confirmed by size, restriction enzyme digestion, and finally by sequencing. The resulting plasmids named as pEiΔusp02-10 and pEiΔusp13 were transferred into E. coli SM10λpir or BW19851 by chemical transformation and mobilized into E. ictaluri WT by conjugation. First integration was selected by ampicillin, and ampicillin resistant colonies were propagated on BHI agar to allow for the second crossover allelic exchange. After this step, colonies were streaked on counter selective BHI plates with 5% sucrose, 0.35% mannitol, and colistin to allow loss of pMEG-375. Potential mutant colonies were tested for ampicillin sensitivity to ensure the loss of the plasmid, confirmed by PCR, and sequencing.
Construction of Bioluminescent USP Mutants
The constructed USP mutants were made bioluminescence using pAKgfplux1 plasmid as described previously (Karsi and Lawrence, 2007). Briefly, the overnight culture of both recipient (USP mutants) and donor cells (E. coli SM10λpir carrying pAKgfplux1) were mixed at 1:2 ratio (donor : recipient) and centrifuged briefly. Pellet was transferred onto sterile 0.45 μM filter papers placed on a BHI agar and incubated at 30°C for 24 h. Bacteria on the filter paper were collected in BHI broth with ampicillin and colistin and then spread on BHI plates containing ampicillin and colistin. After incubation at 30°C for 24–48 h, ampicillin resistant bioluminescent E. ictaluri colonies carrying pAKgfplux1 appeared on plates.
Growth Kinetics of the E. ictaluri USP Mutants in BHI
Growth kinetics of the ten E. ictaluri USP mutants was compared to E. ictaluri WT in BHI medium as previously described (Abdelhamed et al., 2016). Each bacterial strain had four replicates. Overnight cultures were grown in a shaking incubator at 30°C for 18 h. The optical densities (OD600) were measured, and adjusted volumes were added to 15 ml fresh BHI (1:100 dilution). Cultures were grown for 24 h by sampling and measuring OD600 values at 2, 4, 8, 12, and 20 h.
Survival of E. ictaluri USP Mutants in Low pH Stress
Survival of bioluminescent USP mutants and EiWT was determined under acidic stress (pH 5.5) as previously described (Seifart Gomes et al., 2011). Bacteria were cultured overnight, and OD600 values were used to adjust culture volumes. The experiment was performed in 96 well black plates with four replicates at acidic and neutral pH. For each well, 5 μl of bacteria were inoculated into 195 μl of BHI broth plus ampicillin and colistin. The plates were incubated in Cytation 5 Cell Imaging Multi-Mode Reader (BioTek, Winooski, VT, United States), and the photon emissions were collected for 3 h at 30°C. Bioluminescence imaging (BLI) of the 96-well plate was taken using IVIS 100 Series (Caliper Corporation, Hopkinton, MA, United States). Three independent experiments were done and used for statistical analysis.
Survival of E. ictaluri USP Mutants in Oxidative Stress
The survival of the ten USP mutants in BHI supplemented with 0.1% of H2O2 were determined as previously described (Seifart Gomes et al., 2011). The experiment was performed in 96 well plates with four replicates under oxidative stress and normal conditions. The plates were incubated in Cytation 5 Cell Imaging Multi-Mode Reader, and the photon emissions were collected for 3 h at 30°C.
Virulence and Efficacy of E. ictaluri USP Mutants in Catfish Fingerlings
Virulence and vaccine efficacy trials were conducted as reported by our group (Karsi et al., 2009). Approximately 720 channel catfish fingerlings (average: 13.728 cm, 10.544 g) were stocked into 36 tanks at a rate of 20 fish/tank. Tanks were divided into twelve groups with three replicate tanks each group. The experiment included 10 E. ictaluri USP mutants, positive control (EiWT), and negative control (BHI exposed). After 1 week of acclimation, fish were challenged/vaccinated by immersion with 1.3 × 107 CFU/ml water for 1 h. Mortalities were recorded daily for 21 days, and the mean percent mortalities were calculated for each treatment group. Protective properties of USP mutants against EiWT infection was determined by challenging vaccinated catfish with EiWT (2.8 × 107 CFU/ml water). Fish mortalities were recorded daily, and the percent mortality was calculated for each group.
Statistical Analysis
For the growth kinetic experiment, significant differences between EiWT and USP mutants were determined by Student’s t-test. For acid and hydrogen peroxide assays, photon counts were log10 transformed t-tests were conducted. Percent reduction in bioluminescence was calculated by dividing mean photon emissions of USPs to mean photon emission of EiWT. For fish experiments, percent mortalities were arcsine transformed, and analysis of variance (ANOVA) was carried out using PROC GLM of SAS v9.4 (SAS Institute, Inc., Cary, NC, United States). In virulence/vaccination trial, the percent mortalities of USP mutants were compared to that of EiWT, while in efficacy trail, the comparisons were made to the sham-vaccinated group at the alpha level of 0.05.
Results
Construction of the E. ictaluri USP Mutants
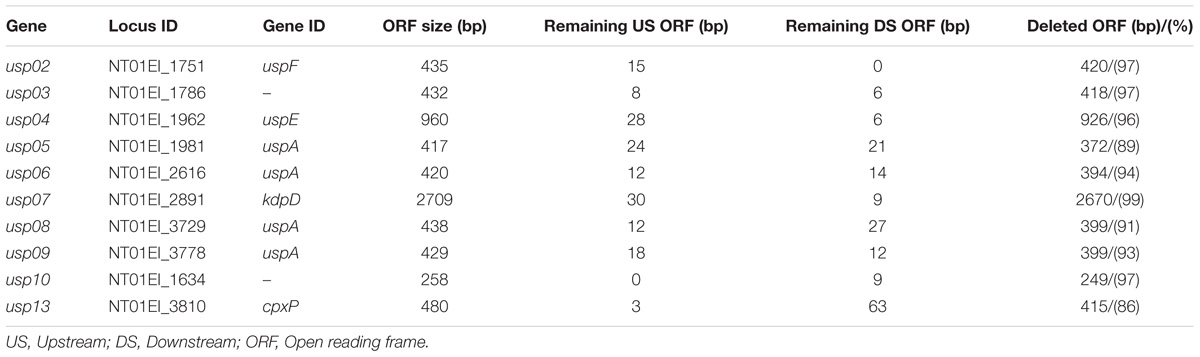
Thirteen universal stress proteins were identified in the E. ictaluri genome (Williams et al., 2012) by sequence similarity (Figure 1). They were scattered through the chromosome, and no operon structure was observed. We were able to delete 10 E. ictaluri usp genes in-frame, and mutants were verified by PCR (Figure 2) as well as sequencing. Properties of wild-type and mutated usp genes are shown in Table 3. In-frame deletion resulted in removal of a large portion (86–99%) of the wild-type usp genes (Table 3).
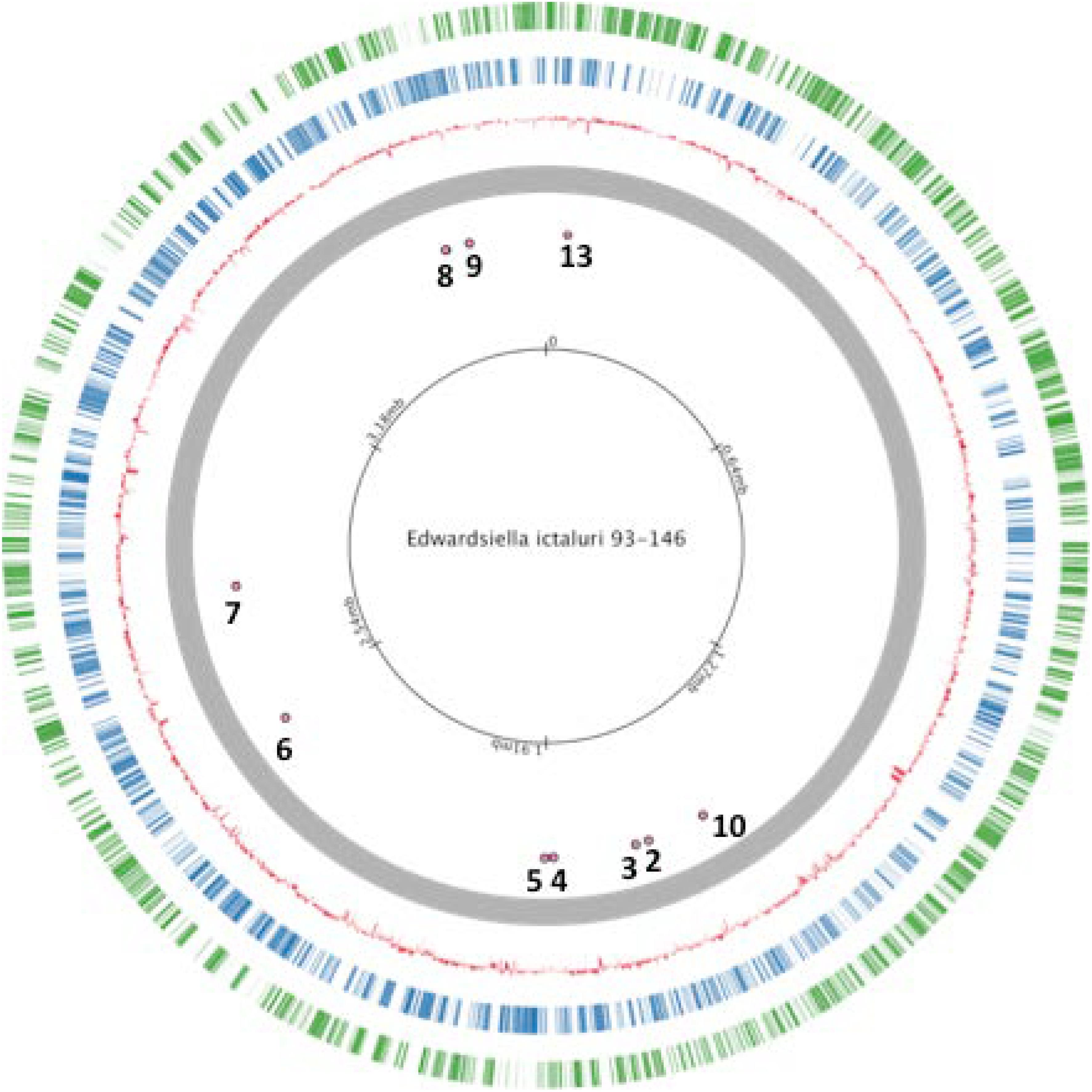
FIGURE 1. Locations of studied universal stress proteins in Edwardsiella ictaluri strain 93–146 genome.
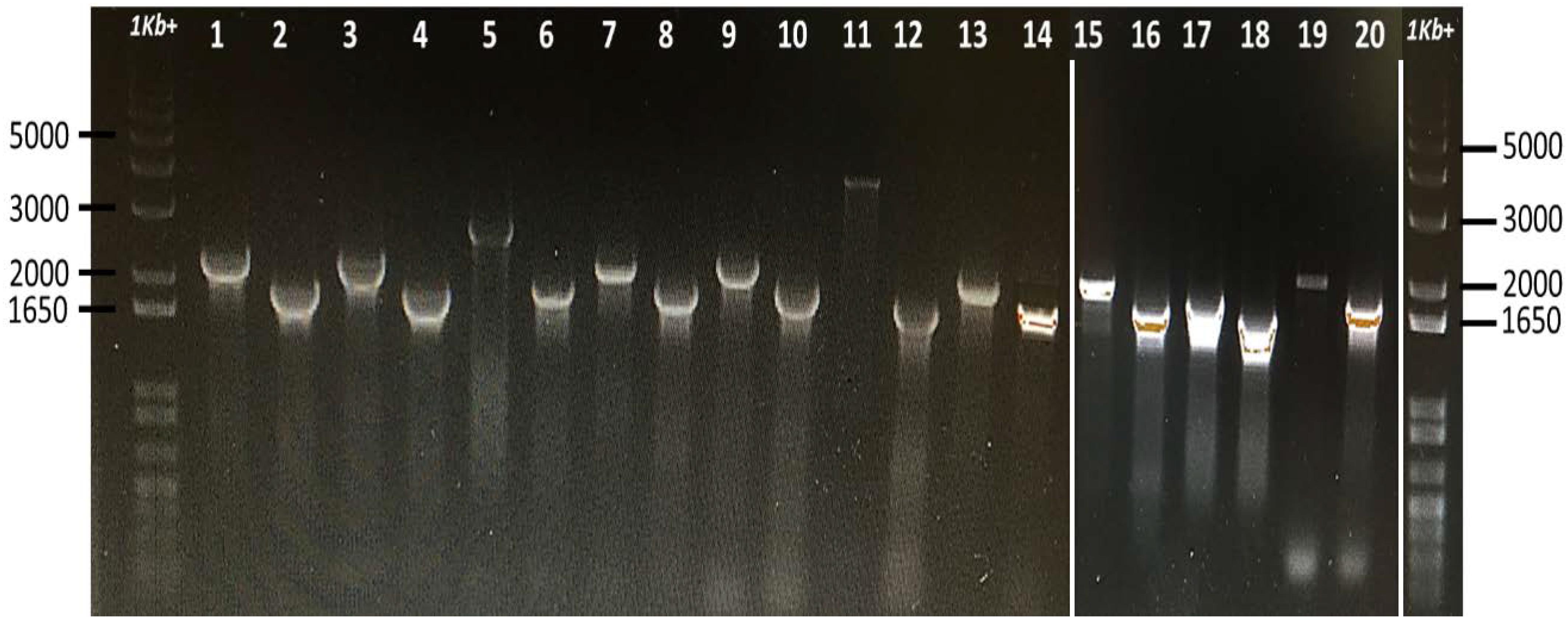
FIGURE 2. Confirmation of E. ictaluri USP mutants by using forward (A) and reverse (D) primers. Fragments were amplified from mutant and wild-type strains and separated on two different 1% agarose gels, which were then combined (white lines above indicate joints). A 1 Kb+ marker lane was also added to the end. Lane 1, EiWT (usp02) and lane 2, EiΔusp02; lane 3, EiWT (usp03) and lane 4, EiΔusp03; lane 5, EiWT (usp04) and lane 6, EiΔusp04; lane 7 is EiWT (usp05) and lane 8, EiΔusp05; lane 9 is EiWT (usp06) and lane 10 is EiΔusp06; lane 11 is EiWT (usp07) and lane 12 is EiΔusp07; lane 13 is EiWT (usp08) and lane 14 is EiΔusp08; lane 15 is EiWT (usp09) and lane 16 is EiΔusp09; lane 17 is EiWT (usp10) and lane 18 is EiΔusp10; lane 19 is EiWT (usp13) and lane 20 is EiΔusp13.
Growth Kinetics of the E. ictaluri USP Mutants in BHI
The growth of EiWT and USP mutants in BHI broth indicated that EiΔusp03 and EiΔusp04 have a significantly (p < 0.001) higher growth rate than EiWT. After 20 h incubation, the growth of EiWT was 23.6 and 17.42% lower than EiΔusp03 and EiΔusp04, respectively (Figure 3). Whereas, no significant differences were observed in the growth kinetics of EiWT and EiΔusp02, EiΔusp05, EiΔusp06, EiΔusp07, EiΔusp08, EiΔusp09, EiΔusp10, and EiΔusp13 strains at all tested time points.
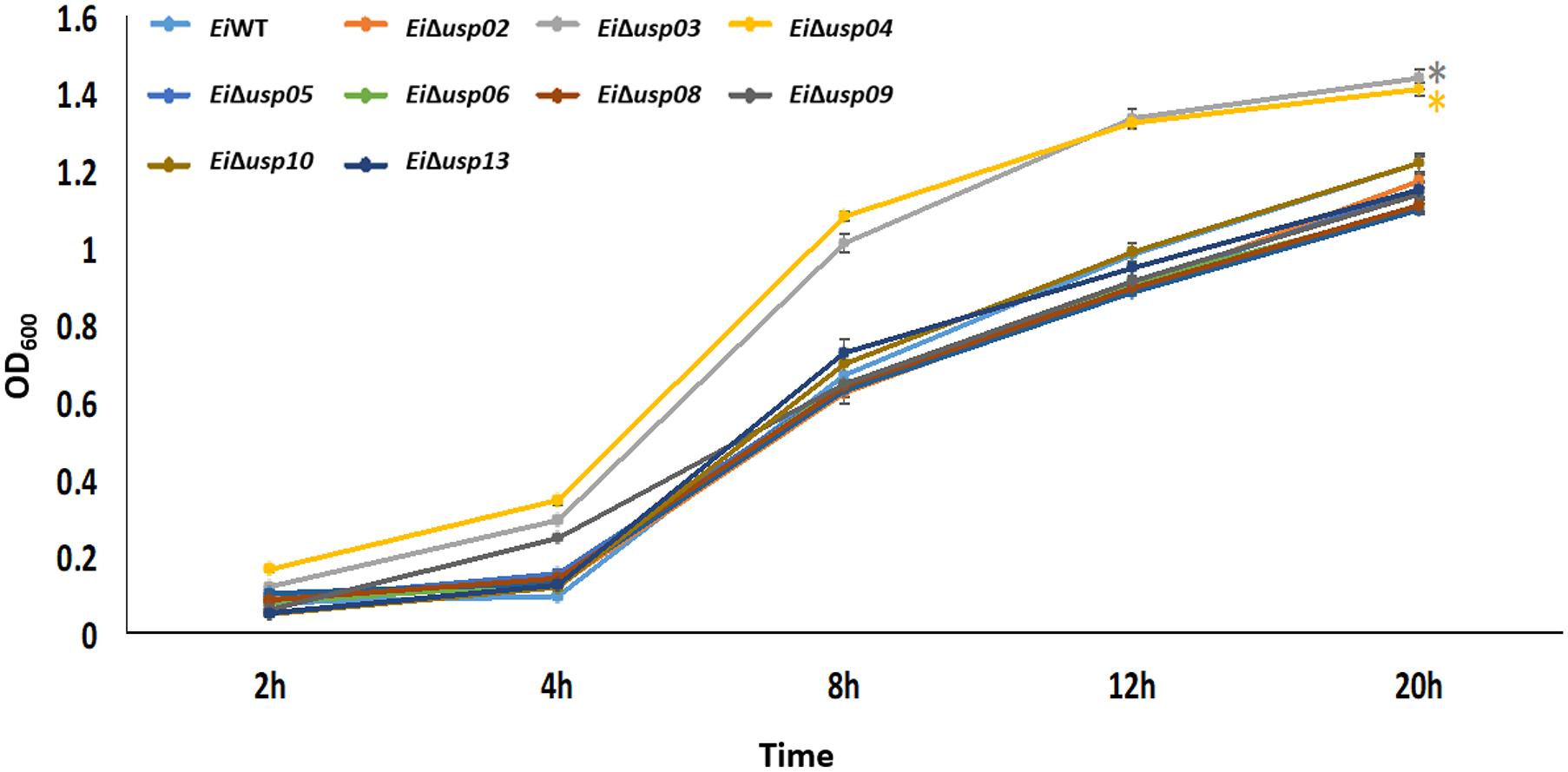
FIGURE 3. Growth of E. ictaluri USP mutants and WT in BHI broth. The data represent means of four replicates. EiΔusp03 and EiΔusp04 have a significantly (p < 0.001) higher growth rate than EiWT and other USP mutants, which indicated by a “∗.” No significant differences were observed in the growth kinetics of EiWT and EiΔusp02, EiΔusp05, EiΔusp06, EiΔusp07, EiΔusp08, EiΔusp09, EiΔusp10, and EiΔusp13 strains.
Survival of E. ictaluri USP Mutants in Low pH Stress
To evaluate the role of usp genes in survival and growth of E. ictaluri at low pH, mutants and EiWT were exposed to acidic pH (5.5) and neutral pH, and bacterial growth (quantified by bioluminescent signal) were calculated. The growth rate (photon numbers) of the all USP mutants in low pH was significantly lower than that of in neutral pH. In contrast, the growth of EiWT at low pH was lower but not significant (Figures 4A,B). The strongest effect of low pH was observed in EiΔusp03 growth (62% reduction) compared to EiWT. The order of susceptibility of USP mutants in low pH as follows: Δusp03 > Δusp07 > Δusp13 > Δusp09 > Δusp10 > Δusp08 > Δusp06 > Δusp04 > Δusp05 > Δusp02. The reduced growth of the USP mutants indicates that usp genes contribute E. ictaluri survival under acidic conditions.
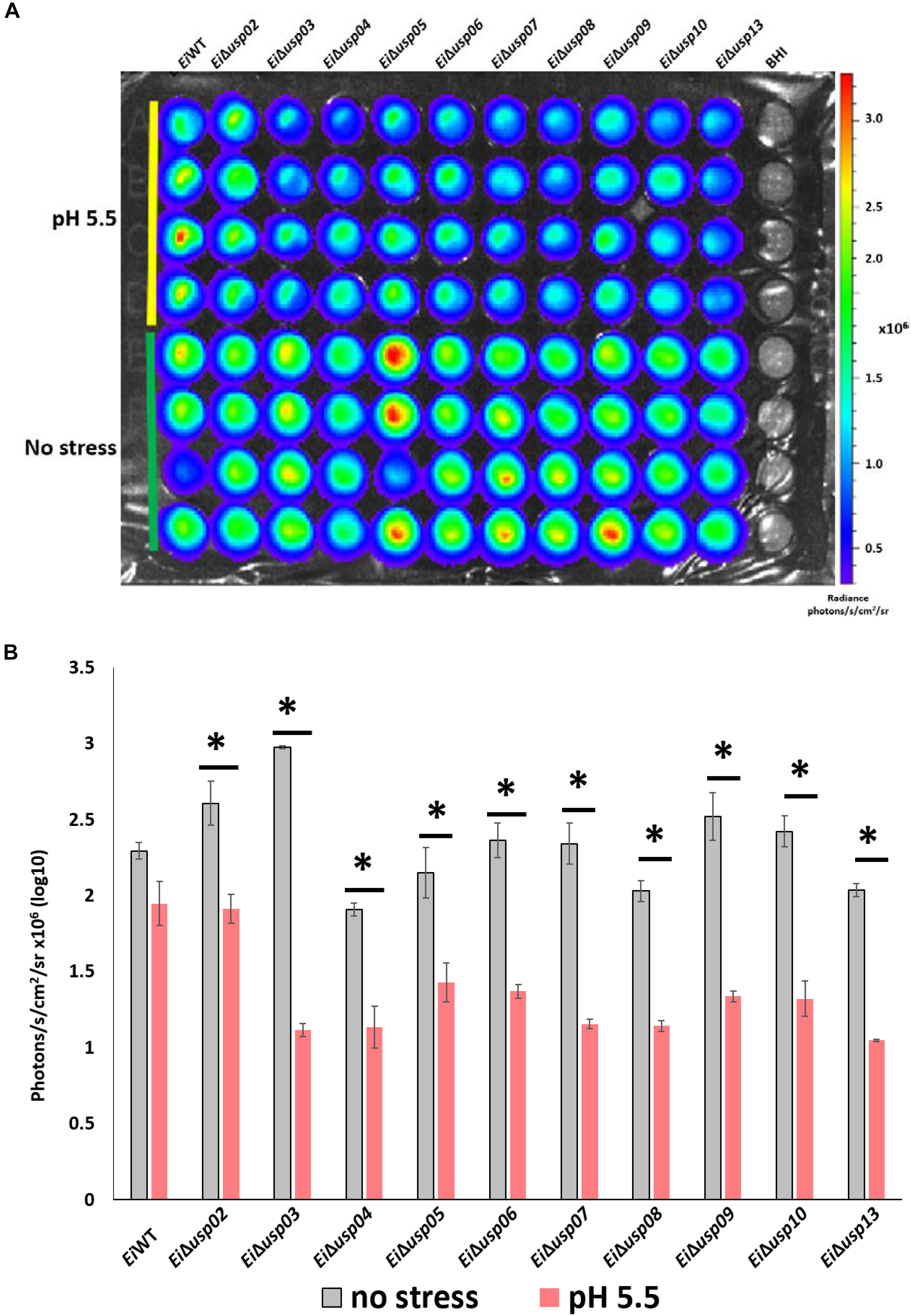
FIGURE 4. The survival assay of E. ictaluri WT and USP mutants in pH 5.5. (A) Each strain had four replicates (column A–D). Strains start with E. ictaluri WT, EiΔusp02-13 and BHI control. (B) The bars show the difference between bioluminescence of USP mutants and WT. ∗indicates a significant difference between stress and non-stress at P < 0.01.
Survival of E. ictaluri USP Mutants in Oxidative Stress
Exposure to hydrogen peroxide (0.1% H2O2) significantly reduced growth of EiΔusp05 and EiΔusp08 compared to no stress group (91 and 35% reduction, respectively), while growth of EiΔusp02 and EiΔusp03 increased under oxidative stress (Figures 5A,B). No differences for EiΔusp04, EiΔusp06, EiΔusp07, EiΔusp09, EiΔusp10, and EiΔusp13 strains were observed.
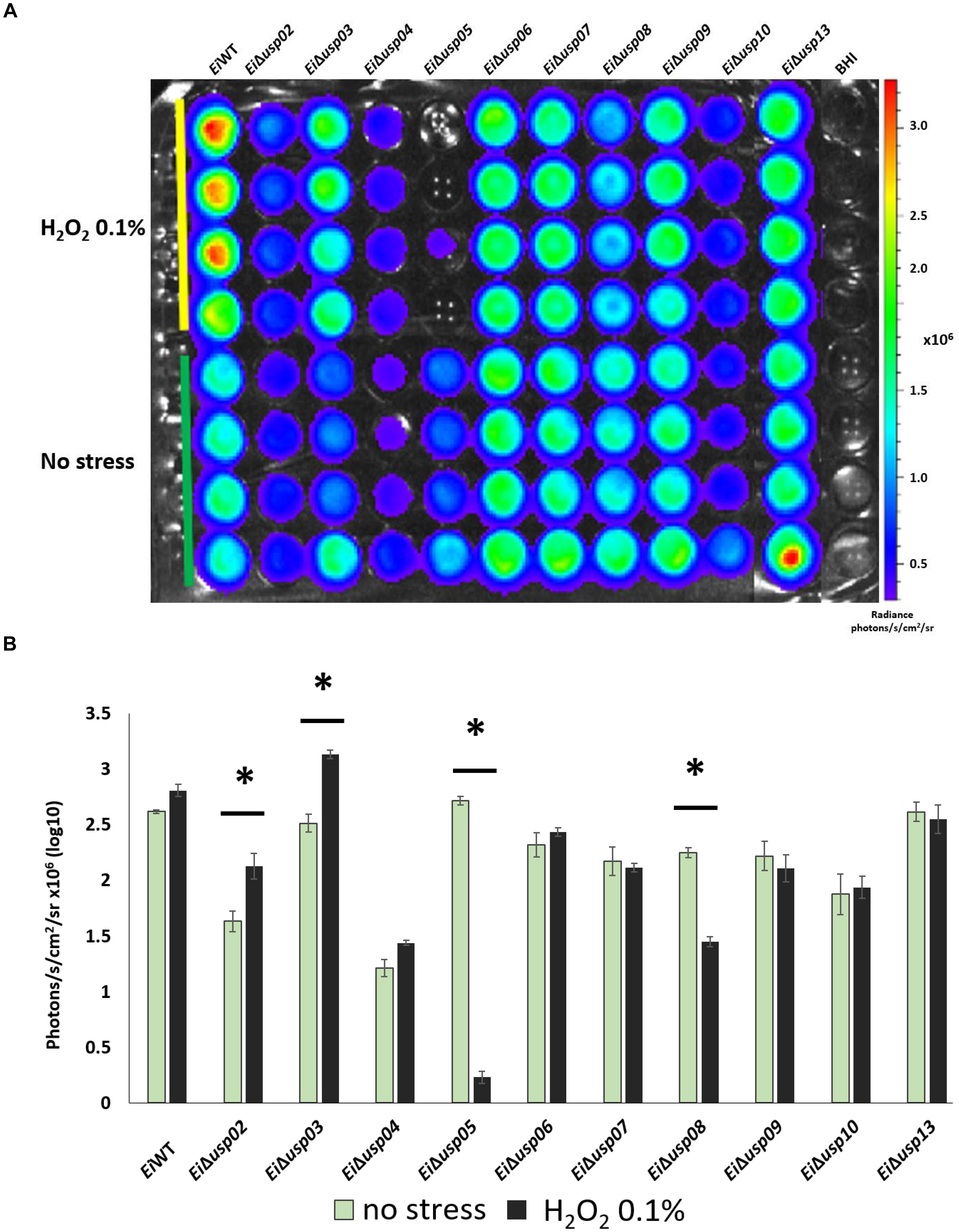
FIGURE 5. The survival assay of E. ictaluri WT and USP mutants exposed to 0.1% H2O2. (A) Each strain had four replicates (column A–D). Strains start with E. ictaluri WT, EiΔusp02-13 and BHI control. (B) The bars show the difference between bioluminescence of USP mutants and WT. ∗indicates a significant difference between stress and non-stress at P < 0.01.
Virulence and Efficacy of E. ictaluri USP Mutants in Catfish Fingerlings
The percent mortalities in catfish challenged with EiΔusp05, EiΔusp07, EiΔusp08, EiΔusp09, EiΔusp10, and EiΔusp13 were significantly lower than that of EiWT (20, 44.8, 20, 20, 55, and 10% vs. 74.1% mortality, respectively) (Figure 6A). In contrast, no significant differences between EiΔusp02, EiΔusp03, EiΔusp04, and EiΔusp06 and EiWT (79.8, 84.4, 74.6, and 79.82% vs. 74.1% mortality, respectively) were observed (Figure 6A). The order of attenuation in the 10 USP mutants are as following: EiΔusp13 > EiΔusp05 > EiΔusp08 > EiΔusp09 > EiΔusp07 > EiΔusp10 > EiΔusp04 > EiΔusp06 > EiΔusp02 > EiΔusp03.
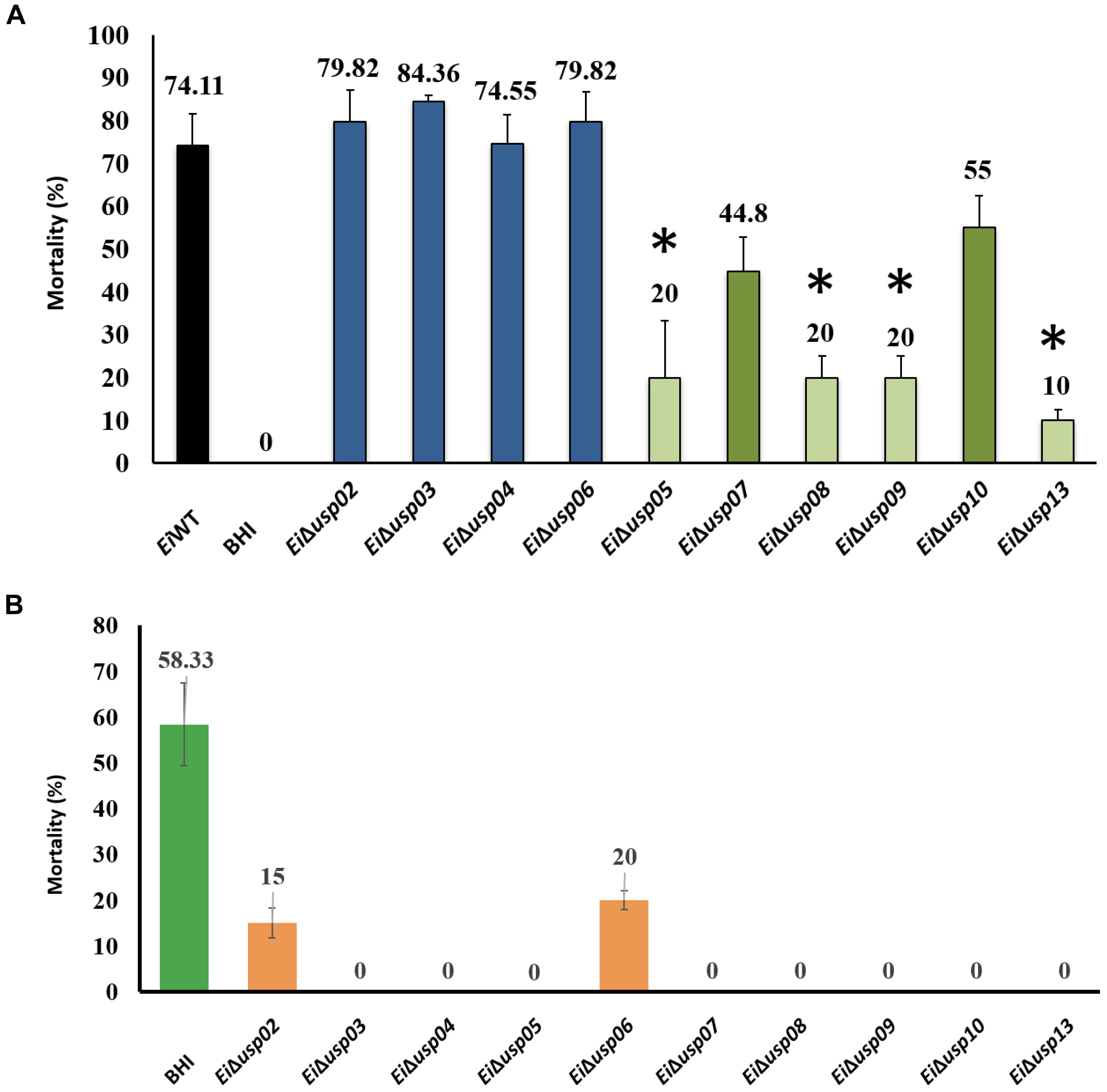
FIGURE 6. Vaccination tests of USP mutants in channel catfish fingerlings. (A) Percent mortalities seen after vaccination. (B) Percent mortalities of channel catfish fingerlings immunized with USP mutants and re-challenged with E. ictaluri wild-type 3 weeks post immunization. ∗indicates significant differences between mutant and WT at P < 0.01.
At 3 weeks post-immunization, EiΔusp05, EiΔusp07, EiΔusp08, EiΔusp09, EiΔusp10, and EiΔusp13 provided significant protection against EiWT challenges (no mortalities; p < 0.01) compared to sham-vaccinated fish (58.33% mortality) (Figure 6B). Although immunization with EiΔusp03 and EiΔusp04 protected catfish significantly, they were not safe. EiΔusp05, EiΔusp08, EiΔusp09, and EiΔusp13 were both safe and protective among all USP mutants.
Figure 7 provides overall summary of the results.
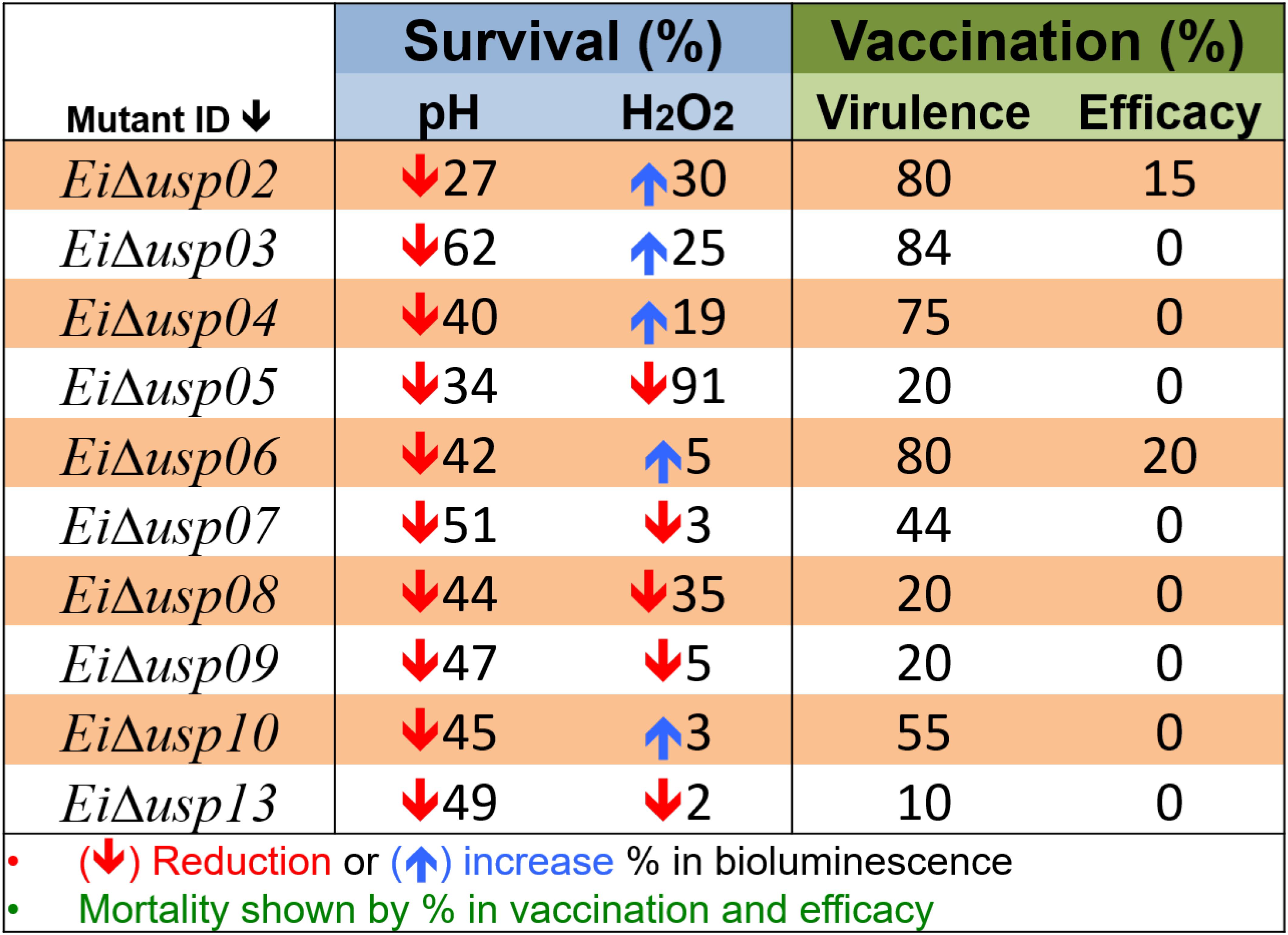
FIGURE 7. Overall summary of results. Survival percent under acidic (pH) and oxidative stress (H2O2) conditions was calculated based on changes in bioluminescence signal. The downward direction arrow indicates reduction in survival percent between mutant strain compared with wild type. The upward direction indicates increase in survival percent. Virulence percent is based on catfish mortality after immersion challenge with USP mutant strains. Efficacy perecent is based of mortality after re-challenge the immunized fish with E. ictaluri WT at 21 day post-immunization.
Discussion
Several previous studies reported that universal stress proteins (USPs) play a role in different bacteria to respond to different stress conditions, such as heat, substrate starvation, exposure to antimicrobial agents, acidic stress, and oxidative stress (Seifart Gomes et al., 2011). The objective of this study was to determine the role of E. ictaluri usp genes in acidic and oxidative stresses as well as in virulence. Also, mutants’ vaccine potentials were determined.
The uspA gene among usp genes has been studied in different bacterial strains. Deletion of the uspA genes resulted in decreased virulence in Salmonella typhimurium C5, Listeria monocytogenes, and Acinetobacter baumannii (Liu et al., 2007; Seifart Gomes et al., 2011; Elhosseiny et al., 2015). Also, uspA affected the host invasion and survival in Salmonella enterica and Mycobacterium tuberculosis (Hensel, 2009; Hingley-Wilson et al., 2010). In the present study, there were four usp genes (usp05, usp06, usp08, and usp09) with high similarity to uspA. The growth rate of EiΔusp05, EiΔusp06, EiΔusp08, and EiΔusp09 were similar to E. ictaluri WT. However, EiΔusp05 and EiΔusp08 showed reduced growth in oxidative and acidic stresses compared to EiWT. Virulence data showed that EiΔusp05, EiΔusp08, and EiΔusp09 were significantly attenuated compared to E. ictaluri WT. However, EiΔusp06 was not attenuated. These results are consistent with a previous study in L. monocytogenes where not all uspA are involved in reduced virulence (Seifart Gomes et al., 2011). Previously, our group reported that transposon insertion mutants in usp05 reduced E. ictaluri virulence in catfish and provided better protection against ESC (Kalindamar, 2013). Additionally, expressions of usp05 were very high in response to host stress or high level of H2O2 in E. ictaluri (Akgul et al., 2018). The usp05 gene (uspA) is an important regulator of survival and virulence in many pathogens (Tkaczuk et al., 2013). In E. coli, uspA mutant caused a survival defect under a variety of growth-arrested conditions, whereas overexpression induced growth in the growth-arrested state. Our data suggest that usp05, usp08, and usp09 are important virulence genes in E. ictaluri.
We demonstrated that EiΔusp03 and EiΔusp04 have a faster growth rate than EiWT and other USP mutants. However, lack of usp genes did not cause growth differences in Listeria monocytogenes (Seifart Gomes et al., 2011), E. coli (Nystrom and Neidhardt, 1993) or other bacteria when cultured in conventional media (Liu et al., 2007; Hingley-Wilson et al., 2010). Indeed, EiΔusp03 and EiΔusp04 did not show any virulence attenuation in E. ictaluri, which was similar to USP mutant Rv2623 in Mycobacterium tuberculosis (Hingley-Wilson et al., 2010). This study suggested that usp genes might play a role in latency and persistence of chronic TB infection. We think that usp03 and usp04 are not involved in virulence but may play other roles in stress responses in E. ictaluri.
Edwardsiella ictaluri can survive and continue growth in up to 3 mM of H2O2 and low acidic pH 5.5. When the USP mutants and EiWT exposed to low pH, growth rates did not change significantly. As shown previously, L. monocytogenes ATP Binding USPs exhibited role in the response to acid stress during exponential growth phase (Tremonte et al., 2016).
Our results indicated that E. ictaluri usp07 contributes to virulence of E. ictaluri. Mortality was significantly decreased in the EiΔusp07 mutant compared to EiWT strain. The usp07 is a KdpD protein, and it contains a uspA domain (Heermann et al., 2009a). We included whole KdpD as usp07 because USP domain is located between the N-terminal sensor domain and C-terminal catalytic domain of this Osmo-sensitive K+ channel histidine kinase. Mutant KdpD in Salmonella typhimurium is attenuated in animal infection model and macrophage survival experiments. It also promotes resistance to osmotic, oxidative and antimicrobial stresses (Alegado et al., 2011). KdpD is also involved in oxidative-osmotic stress, response to host, and virulence (Freeman et al., 2013). In our gene expression study after host stress, usp07 showed a very high expression level (Akgul et al., 2018). It is important to note that usp07 involved in E. ictaluri virulence and acid stress response.
The usp13 was described as a universal stress protein and extra cytoplasmic adaptor protein (CpxP) like protein (Williams et al., 2012). The usp13 (CpxP) is placed in the inner membrane with histidine kinase CpxA and CpxR, a response regulator (Vogt and Raivio, 2012; Debnath et al., 2013). CpxP is the most highly inducible member of the Cpx regulon, and it has elevated expression in response to both envelope stress and entry into stationary phase growth (Motohashi et al., 1999; DiGiuseppe and Silhavy, 2003). The CPX system is important and required for virulence in both Gram-negative and -positive bacteria (Raju et al., 2012). Previously, we determined that E. ictaluri, usp13 is highly expressed when exposed low acidic pH (5.5) and the catfish invasion (Akgul et al., 2018). The usp13 (cpxP) is an essential regulator of cell membrane stress in bacteria during host infection. Therefore, it is involved in the virulence of E. ictaluri with a very high reduction in virulence (Figure 6).
The expression of E. coli usp genes is controlled by some effector proteins and signaling molecules, such as SOS repose proteins (Gustavsson et al., 2002; Kvint et al., 2003; Persson et al., 2007). However, mechanisms of USPs in other bacterial species are not known entirely. Overall our results are in line with studies from various species that USPs were crucial for protecting the cells from the damaging effects of reactive oxygen species (ROS) (Nachin et al., 2005; Liu et al., 2007; Seifart Gomes et al., 2011; Elhosseiny et al., 2015; Figure 7).
Conclusion
Our lab aims to develop live attenuated vaccines to protect catfish against E. ictaluri infections. Live attenuated bacterial should be both safe and confer full protection against wild-type infections. This study identified that EiΔusp05, EiΔusp08, EiΔusp09, and EiΔusp13 strains have vaccine potential and further efforts, such as constructing double mutants to improve their safety, could be pursued. The data presented in this study display that USPs are essential for both stress physiology and pathogenesis in E. ictaluri.
Author Contributions
AK and ML conceived the project and designed the experiments. AA, SN, SK, HT, and HA conducted the experiments. AA wrote the manuscript. SN, SK, HT, HA, ML, and AK reviewed the manuscript.
Funding
This project was supported by Agriculture and Food Research Initiative Competitive grant no. 2016-67015-24909 from the USDA National Institute of Food and Agriculture.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Laboratory Animal Resources and Care at the College of Veterinary Medicine for providing the SPF channel catfish. AA was supported by a fellowship from the Republic of Turkey.
References
Abdelhamed, H., Lu, J., Lawrence, M. L., and Karsi, A. (2016). Involvement of tolQ and tolR genes in Edwardsiella ictaluri virulence. Microb. Pathog. 100, 90–94. doi: 10.1016/j.micpath.2016.09.011
Akgul, A., Akgul, A., Lawrence, M. L., and Karsi, A. (2018). Stress-related genes promote Edwardsiella ictaluri pathogenesis. PLoS One 13:e0194669. doi: 10.1371/journal.pone.0194669
Alegado, R. A., Chin, C. Y., Monack, D. M., and Tan, M. W. (2011). The two-component sensor kinase KdpD is required for Salmonella typhimurium colonization of Caenorhabditis elegans and survival in macrophages. Cell. Microbiol. 13, 1618–1637. doi: 10.1111/j.1462-5822.2011.01645.x
Al-Maleki, A. R., Mariappan, V., Vellasamy, K. M., Shankar, E. M., Tay, S. T., and Vadivelu, J. (2014). Enhanced intracellular survival and epithelial cell adherence abilities of Burkholderia pseudomallei morphotypes are dependent on differential expression of virulence-associated proteins during mid-logarithmic growth phase. J. Proteomics 106, 205–220. doi: 10.1016/j.jprot.2014.04.005
Chaffin, D. O., Taylor, D., Skerrett, S. J., and Rubens, C. E. (2012). Changes in the Staphylococcus aureus transcriptome during early adaptation to the lung. PLoS One 7:e41329. doi: 10.1371/journal.pone.0041329
Chatterjee, S. S., Hossain, H., Otten, S., Kuenne, C., Kuchmina, K., Machata, S., et al. (2006). Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 74, 1323–1338. doi: 10.1128/IAI.74.2.1323-1338.2006
Debnath, I., Norton, J. P., Barber, A. E., Ott, E. M., Dhakal, B. K., Kulesus, R. R., et al. (2013). The Cpx stress response system potentiates the fitness and virulence of uropathogenic Escherichia coli. Infect. Immun. 81, 1450–1459. doi: 10.1128/IAI.01213-12
DiGiuseppe, P. A., and Silhavy, T. J. (2003). Signal detection and target gene induction by the CpxRA two-component system. J. Bacteriol. 185, 2432–2440. doi: 10.1128/JB.185.8.2432-2440.2003
Dozois, C. M., Daigle, F., and Curtiss, R. III (2003). Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc. Natl. Acad. Sci. U.S.A. 100, 247–252. doi: 10.1073/pnas.232686799
Elhosseiny, N. M., Amin, M. A., Yassin, A. S., and Attia, A. S. (2015). Acinetobacter baumannii universal stress protein A plays a pivotal role in stress response and is essential for pneumonia and sepsis pathogenesis. Int. J. Med. Microbiol. 305, 114–123. doi: 10.1016/j.ijmm.2014.11.008
Freeman, Z. N., Dorus, S., and Waterfield, N. R. (2013). The KdpD/KdpE two-component system: integrating K+ Homeostasis and virulence. PLoS Pathog. 9:e1003201. doi: 10.1371/journal.ppat.1003201
Gustavsson, N., Diez, A., and Nystrom, T. (2002). The universal stress protein paralogues of Escherichia coli are coordinately regulated and co-operate in the defence against DNA damage. Mol. Microbiol. 43, 107–117. doi: 10.1046/j.1365-2958.2002.02720.x
Heermann, R., Lippert, M.-L., and Jung, K. (2009a). Domain swapping reveals that the N-terminal domain of the sensor kinase KdpD in Escherichia coli is important for signaling. BMC Microbiol. 9:133. doi: 10.1186/1471-2180-9-133
Heermann, R., Weber, A., Mayer, B., Ott, M., Hauser, E., Gabriel, G., et al. (2009b). The universal stress protein UspC scaffolds the KdpD/KdpE signaling cascade of Escherichia coli under salt stress. J. Mol. Biol. 386, 134–148. doi: 10.1016/j.jmb.2008.12.007
Hensel, M. (2009). “Secreted proteins and virulence in Salmonella enterica,” in Bacterial Secreted Proteins: Secretory Mechanisms and Role in Pathogenesis, ed. K. Wooldridge (Nottingham: Caister Academic Press).
Herrero, M., de Lorenzo, V., and Timmis, K. N. (1990). Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in Gram-negative bacteria. J. Bacteriol. 172, 6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990
Hingley-Wilson, S. M., Lougheed, K. E., Ferguson, K., Leiva, S., and Williams, H. D. (2010). Individual Mycobacterium tuberculosis universal stress protein homologues are dispensable in vitro. Tuberculosis 90, 236–244. doi: 10.1016/j.tube.2010.03.013
Horton, R. M., Cai, Z. L., Ho, S. N., and Pease, L. R. (1990). Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8, 528–535.
Kalindamar, S. (2013). Identification of the Edwardsiella Ictaluri Genes Causing Impaired Growth in Complex Medium. Master thesis, Mississippi State University, Starkville, MS.
Karsi, A., Gulsoy, N., Corb, E., Dumpala, P. R., and Lawrence, M. L. (2009). High-throughput bioluminescence-based mutant screening strategy for identification of bacterial virulence genes. Appl. Environ. Microbiol. 75, 2166–2175. doi: 10.1128/AEM.02449-08
Karsi, A., and Lawrence, M. L. (2007). Broad host range fluorescence and bioluminescence expression vectors for gram-negative bacteria. Plasmid 57, 286–295. doi: 10.1016/j.plasmid.2006.11.002
Karsi, A., Menanteau-Ledouble, S., and Lawrence, M. L. (2006). Development of bioluminescent Edwardsiella ictaluri for noninvasive disease monitoring. FEMS Microbiol. Lett. 260, 216–223. doi: 10.1111/j.1574-6968.2006.00310.x
Kvint, K., Nachin, L., Diez, A., and Nyström, T. (2003). The bacterial universal stress protein: function and regulation. Curr. Opin. Microbiol. 6, 140–145. doi: 10.1016/S1369-5274(03)00025-0
Lawrence, M. L., Cooper, R. K., and Thune, R. L. (1997). Attenuation, persistence, and vaccine potential of an Edwardsiella ictaluri purA mutant. Infect. Immun. 65, 4642–4651.
Liu, W.-T., Karavolos, M. H., Bulmer, D. M., Allaoui, A., Hormaeche, R. D., Lee, J. J., et al. (2007). Role of the universal stress protein UspA of Salmonella in growth arrest, stress and virulence. Microb. Pathog. 42, 2–10. doi: 10.1016/j.micpath.2006.09.002
Metcalf, W. W., Jiang, W., and Wanner, B. L. (1994). Use of the rep technique for allele replacement to construct new Eschericia coli hosts for maintenance of R6K gamma origin plasmids at different copy numbers. Gene 138, 1–7. doi: 10.1016/0378-1119(94)90776-5
Miller, V. L., and Mekalanos, J. J. (1988). A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170, 2575–2583.
Motohashi, K., Watanabe, Y., Yohda, M., and Yoshida, M. (1999). Heat-inactivated proteins are rescued by the DnaK.J-GrpE set and ClpB chaperones. Proc. Natl. Acad. Sci. U.S.A. 96, 7184–7189. doi: 10.1073/pnas.96.13.7184
Nachin, L., Nannmark, U., and Nystrom, T. (2005). Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J. Bacteriol. 187, 6265–6272. doi: 10.1128/JB.187.18.6265-6272.2005
Nystrom, T., and Neidhardt, F. C. (1993). Isolation and properties of a mutant of Escherichia coli with an insertional inactivation of the uspA gene, which encodes a universal stress protein. J. Bacteriol. 175, 3949–3956. doi: 10.1128/jb.175.13.3949-3956.1993
Persson, O., Valadi, A., Nystrom, T., and Farewell, A. (2007). Metabolic control of the Escherichia coli universal stress protein response through fructose-6-phosphate. Mol. Microbiol. 65, 968–978. doi: 10.1111/j.1365-2958.2007.05838.x
Raju, R. M., Goldberg, A. L., and Rubin, E. J. (2012). Bacterial proteolytic complexes as therapeutic targets. Nat. Rev. Drug Discov. 11, 777–789. doi: 10.1038/nrd3846
Seifart Gomes, C., Izar, B., Pazan, F., Mohamed, W., Mraheil, M. A., Mukherjee, K., et al. (2011). Universal stress proteins are important for oxidative and acid stress resistance and growth of Listeria monocytogenes EGD-e In vitro and In vivo. PLoS One 6:e24965. doi: 10.1371/journal.pone.0024965
Siegele, D. A. (2005). Universal stress proteins in Escherichia coli. J. Bacteriol. 187, 6253–6254. doi: 10.1128/JB.187.18.6253-6254.2005
Tkaczuk, K. L., Shumilin, A., Chruszcz, M., Evdokimova, E., Savchenko, A., and Minor, W. (2013). Structural and functional insight into the universal stress protein family. Evol. Appl. 6, 434–449. doi: 10.1111/eva.12057
Tremonte, P., Succi, M., Coppola, R., Sorrentino, E., Tipaldi, L., Picariello, G., et al. (2016). Homology-based modeling of universal stress protein from Listeria innocua Up-regulated under acid stress conditions. Front. Microbiol. 7:1998. doi: 10.3389/fmicb.2016.01998
Tu, T. D., Haesebrouck, F., Nguyen, A. T., Sorgeloos, P., Baele, M., and Decostere, A. (2008). Antimicrobial susceptibility pattern of Edwardsiella ictaluri isolates from natural outbreaks of bacillary necrosis of Pangasianodon hypophthalmus in Vietnam. Microb. Drug Resist. 14, 311–316. doi: 10.1089/mdr.2008.0848
USDA (2014). Development of Approaches to Prevent and Ameliorate Diseases of Catfish (Agricultural Research Service Annual Report). Washington, DC: United States Department of Agriculture.
Vogt, S. L., and Raivio, T. L. (2012). Just scratching the surface: an expanding view of the Cpx envelope stress response. FEMS Microbiol. Lett. 326, 2–11. doi: 10.1111/j.1574-6968.2011.02406.x
Williams, M. L., Gillaspy, A. F., Dyer, D. W., Thune, R. L., Waldbieser, G. C., Schuster, S. C., et al. (2012). Genome sequence of Edwardsiella ictaluri 93-146, a strain associated with a natural channel catfish outbreak of enteric septicemia of catfish. J. Bacteriol. 194, 740–741. doi: 10.1128/JB.06522-11
Keywords: stress, ESC, USP, mutation, vaccine
Citation: Akgul A, Nho SW, Kalindamar S, Tekedar HC, Abdalhamed H, Lawrence ML and Karsi A (2018) Universal Stress Proteins Contribute Edwardsiella ictaluri Virulence in Catfish. Front. Microbiol. 9:2931. doi: 10.3389/fmicb.2018.02931
Received: 11 May 2018; Accepted: 14 November 2018;
Published: 28 November 2018.
Edited by:
Martin Stephen LLewellyn, University of Glasgow, United KingdomReviewed by:
Ulisses Padua Pereira, Universidade Estadual de Londrina, BrazilHetron Mweemba Munang’andu, Norwegian University of Life Sciences, Norway
Copyright © 2018 Akgul, Nho, Kalindamar, Tekedar, Abdalhamed, Lawrence and Karsi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Attila Karsi, a2Fyc2lAY3ZtLm1zc3RhdGUuZWR1
 Ali Akgul
Ali Akgul Seong Won Nho
Seong Won Nho Safak Kalindamar
Safak Kalindamar Hasan C. Tekedar
Hasan C. Tekedar Hossam Abdalhamed
Hossam Abdalhamed Mark L. Lawrence
Mark L. Lawrence Attila Karsi
Attila Karsi