- 1Faculdade de Medicina da Universidade de São Paulo, São Paulo, Brazil
- 2Centro de Bacteriologia, Instituto Adolfo Lutz, São Paulo, Brazil
- 3Faculdade de Medicina de Botucatu, Botucatu, Brazil
Dissemination of carbapenem-resistant Acinetobacter baumannii is currently one of the priority themes discussed around the world, including in Brazil, where this pathogen is considered endemic. A total of 107 carbapenem-resistant A. baumannii (CRAB) isolates were collected from patients with bacteraemia attended at a teaching hospital in Brazil from 2008 to 2014. From these samples, 104 (97.2%) carrie3d blaOXA−23−like, all of them associated with ISAba1 The blaOXA−231 (1.9%) and blaOXA−72 (0.9%) genes were also detected in low frequencies. All isolates were susceptible to minocycline, and 38.3% of isolates presented intermediate susceptibility to tigecycline (MIC = 4 μg/ml). Molecular typing assessed by multi-locus sequence typing demonstrated that the strains were mainly associated with clonal complexes CC79 (47.4%), followed by CC1 (16.9%), and CC317 (18.6%), belonging to different pulsotypes and in different prevalences over the years. Changes in the clones' prevalence reinforce the need of identifying and controlling CRAB in hospital settings to preserve the already scarce therapeutic options available.
Introduction
Emergence and dissemination of carbapenem-resistant Acinetobacter baumannii (CRAB) is currently one of the priority themes discussed around the world (Higgins et al., 2010; US Centers for Disease Control Prevention. Antibiotic Resistance Threats in the United States., 2013; World Health Organization, 2017). In Brazil, a continental country in which healthcare associated infections rates are distinctly high (Fortaleza et al., 2017), CRAB is considered endemic (Rossi, 2011) and Acinetobacter infections present the highest mortality rates among ICU patients with bacterial bloodstream infections (Marra et al., 2011). Carbapenem resistance in Acinetobacter is usually mediated by carbapenem-hydrolysing class D β-lactamase (CHDL), mainly codified by the blaOXA−23-like, blaOXA−24-like, blaOXA−58-like and blaOXA−143-like genes (Zarrillii et al., 2013).
Molecular epidemiology of CRAB highlights the prevalence of International Clone 1 (Clonal Complex CC1) worldwide, while International Clones 2 (CC2) and 3 (CC3) are prevalent in Europe and in the United States (Zarrilli et al., 2009; Karah et al., 2012; Zarrillii et al., 2013). In Brazil and other Latin American countries (Clímaco et al., 2013; Medeiros and Lincopan, 2013; Stietz et al., 2013; Rodríguez et al., 2016; Escandón-Vargas et al., 2017), clonal complexes CC1, CC15 are predominant, along with CC79, which has also been identified in Spain and in the United States (Villalón et al., 2011; Mosqueda et al., 2014; Kanamori et al., 2016).
Circulation of a limited number of lineages of multidrug- (MDR) and extensively-drug resistance (XDR) A. baumannii underscores the need for surveillance and effective implementation of measures to contain their dissemination.
To determine the occurrence of high-risk clones CRAB circulating in a Brazilian hospital, we evaluated their antimicrobial susceptibility and clonality in isolates recovered from bloodstream infections in patients attended at a teaching hospital in inner Brazil.
Materials and Methods
Epidemiological Design
This was an observational retrospective study performed with 107 carbapenem-resistant A. baumannii isolates recovered from not-repeated patients with bacteraemia attending at Botucatu Medical School Hospital/UNESP (BMSH/UNESP), from 2008 to 2014. The study was approved as a retrospective study by the Local Research Ethics Committee (Process CAAE 49985115.5.0000.0059). We were granted an exemption from the requirement to obtain written informed-consent from the participants and/or their legal guardians because the isolates included in the study had already been stored, on an ongoing basis, in the Culture Collection of the Department of Microbiology and Immunology, UNESP, Botucatu, São Paulo, Brazil.
Settings and A. baumannii Isolates
BMSH/UNESP is a 415-bed (52 intensive care unit beds) tertiary regional reference hospital, located in the inner of the State of Sao Paulo, Brazil. For this study, frozen isolates stocked in deep-freezer were recovered in Brain-Heart Infusion (BHI) broth and streaked onto BHI Agar plates. Acinetobacter isolates were initially identified by morphological and biochemical characteristics (Gram stain, oxidase-negative, catalase-positive, glucose oxidation, ability to grow at 42° and 44°C) (Vaneechoutte et al., 2015). A. baumannii species was screened by PCR detection of blaOXA−51−like (Woodford et al., 2006) and gltA genes (Wong et al., 2014). A subset of randomly selected isolates was submitted to ITS and/or rpoB gene sequencing (Chang et al., 2005; La Scola et al., 2006). As MLST was carried out for each pulsotype (see below), A. baumannii identification was also confirmed by this method.
Detection of Oxacillinase Genes and ISAba1
PCR for CHDL oxacillinases-encoding genes (blaOXA−23−like, blaOXA−24/40−like, blaOXA−143−like and blaOXA−58−like) and other carbapenemases (blaKPC, blaNDM, blaSPM, blaIMP, blaVIM, blaOXA−48) was performed in all isolates as previously described (Woodford et al., 2006; Higgins et al., 2010; Poirel et al., 2011). Full sequencing of the blaOXA−24-like and the blaOXA−143−like was carried out for allele determination (Héritier et al., 2005; Higgins et al., 2009; Cayô et al., 2014). Presence of the ISAba1 upstream the blaOXA genes was also investigated by PCR mapping, employing the ISAba1 forward primer (Segal et al., 2005) and the blaOXA reverse primers (Woodford et al., 2006).
Antimicrobial Susceptibility Testing
Minimum inhibitory concentration (MIC) values were determined for ampicillin-sulbactam, minocycline, and tetracycline (broth microdilution method), imipenem, meropenem, tigecycline, and polymyxin B (E-test, BioMeriéux, Marci l'Etoile, France). The susceptibility profile of the isolates was completed by disc-diffusion method to amikacin, cefepime, ceftazidime, cefotaxime, ciprofloxacin, gentamicin, levofloxacin, piperacillin-tazobactam, trimethoprim-sulfamethoxazole, ticarcillin-clavulanate, and tobramycin. Breakpoints employed to define susceptibility, intermediate or resistance followed CLSI recommendations (Clinical Laboratory Standards Institute, 2015), except for tigecycline, for which the US Food and Drug Administration breakpoints were applied (susceptible: ≤ 2 μg ml−1; resistant: ≥8 μg ml−1). Isolates were categorized as multidrug- (MDR, non-susceptibility to ≥1 agent in ≥3 antimicrobial categories among aminoglycosides, antipseudomonal carbapenems, antipseudomonal fluoroquinolones, antipseudomonal penicillins + β-lactamase inhibitors, extended-spectrum cephalosporins, folate pathway inhibitors, penicillins + β-lactamase inhibitors, polymyxins, tetracyclines) or extensively-drug-resistant (XDR, non-susceptibility to ≥1 agent in all but ≤ 2 categories, as described earlier) (Magiorakos et al., 2012).
Determination of the Electrophoretic Pattern by PFGE
Genetic diversity among all the 107 A. baumannii isolates were investigated by PFGE (Seifert et al., 2005). Macrorestriction was performed with ApaI (Promega) and DNA digested fragments were resolved using a CHEF-DR-III (Bio-Rad). Dendrogram was generated with BioNumerics v.7.6.2 (Applied Maths, Sint-Martens-Latem, Belgium) based on the Dice similarity using the UPGMA method, with tolerance and optimization parameters set at 1.5%. Clusters were defined as isolates with similarity ≥87% and named with capital letters (A to K) while pulsotypes were defined as each electrophoretic pattern with 100% similarity (named with capital letters and numbers, from A1 to K4).
MLST Analysis
MLST was performed in a representative isolate of each pulsotype as per the Institute Pasteur protocol (https://pubmlst.org/abaumannii/info/primers_Pasteur.shtml), in order to sequence the internal region of the genes gltA, fusA, recA, cpn60, pyrG, rplB and rpoB. PCR products were purified with enzyme ExoSAP-IT (Affymetrix) according to manufacturer's instructions. Nucleotide sequences were obtained using an Applied 3730 Automatic Sequencer (Applied Biosystems). The results were analyzed on BioNumerics v.7.6.2 (Applied Maths, Sint-Martens-Latem, Belgium), and compared with the Institute Pasteur database (https://pubmlst.org/abaumannii/); clonal complexes were determined by the eBurst algorithm (http://eburst.mlst.net/).
Results
A. baumannii Isolates and Detection of Oxacillinase Genes
The 107 isolates were recovered from non-repetitive patients with bacteremia attending a teaching hospital in the State of São Paulo, Brazil, between 2008 and 2014. These isolates were recovered from blood (96.3%) or vascular catheter (3.7%).
All isolates were confirmed as A. baumannii species by PCR detection of blaOXA−51−like and/or sequencing of the rpoB and ITS genes, as well as by the MLST analysis. Out of these isolates, 104 (97.2%) carried the blaOXA−23−like with ISAba1 upstream; remaining strains carried blaOXA−231 (blaOXA−143−like; n = 2; 1.9%) or blaOXA−72 (blaOXA−24−like; n = 1; 0.9%). The blaOXA−58−like gene was not detected, as well as the additional carbapenemases investigated.
Antimicrobial Susceptibility Testing
According to the susceptibility test, 39.3% of isolates were considered MDR, and 60.7% XDR. The entire population evaluated confirmed resistance to imipenem, meropenem, ciprofloxacin, piperacillin-tazobactam and levofloxacin, while susceptibility to minocycline was observed in all the isolates. Resistance rates to other antimicrobials tested by disc-diffusion method were: amikacin (89.7%), cefepime (98.1%), ceftazidime (96.2%), cefotaxime (99.0%), gentamicin (76.6%), trimethoprim-sulfamethoxazole (68.2%), ticarcillin-clavulanate (99.0%), and tobramycin (68.2%). The MIC50/MIC90 (μg/ml) values were calculated for each antimicrobial agent, as follows: imipenem (>32/>32), meropenem (>32/>32), tetracycline (8/16), ampicillin-sulbactam (16:8/32:16), minocycline (0.25/0.5) and tigecycline (2/3). Although resistance to tigecycline was not detected, 38.3% of isolates presented intermediate susceptibility to this drug (according to FDA breakpoints).
Distribution of susceptibility rates among the clones (Table 1) evidenced high-rates of non-susceptibility to cephalosporins, carbapenems, quinolones and ticarcillin-clavulanic acid. For the aminoglycosides, the highest susceptibility rate to amikacin was detected for isolates belonging to CC1 (54.5%), while gentamycin and tobramycin were more active against isolates belonging to CC317 (60.7% and 53.6% of susceptibility, respectively). Furthermore, isolates belonging to CC317 presented the highest susceptibility rates to sulfamethoxazole-trimethoprim (92.9%), tigecycline (75%), and ampicillin-sulbactam (53.6%).
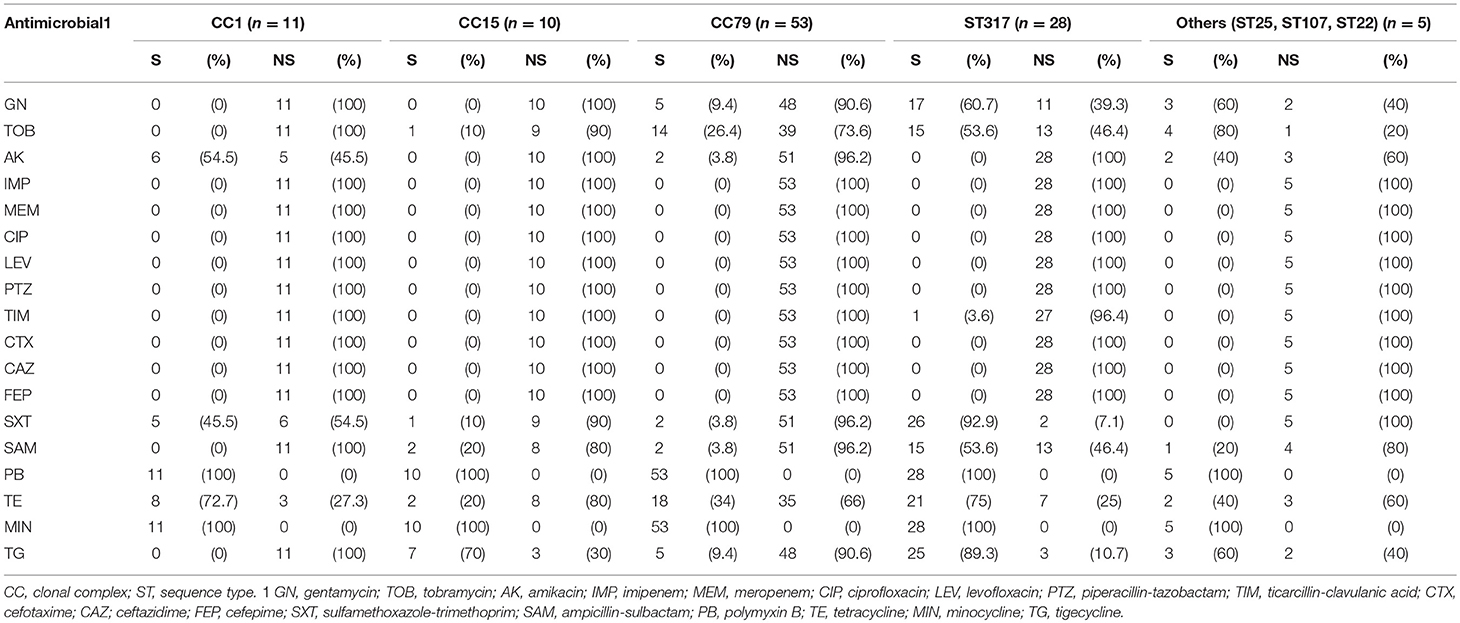
Table 1. Distribution (frequency and %) of antimicrobial susceptibility (S) and non-susceptibility (NS, intermediate + resistance) among the clones of A. baumannii.
Molecular Typing
The 107 CRAB isolates were distributed into 53 PFGE pulsotypes belonging to eleven clusters (A, B, C, D, E, F, G, H, I, J, K) and eight additional pulsotypes with a single isolate (Figure 1). By extrapolating the results of MLST to isolates presenting the same pulsotypes, we observed the frequencies of 49.5% of strains belonging to clonal complexes (and sequence types) CC79 (ST79, 8.4%; ST175, 0.9% and ST730, 40.2%); 26.2% for CC317 (ST317, 100%), 10.3% for CC1 (ST1, 9.3%; ST986, 0.9%); 9.3% for CC15 (ST15, 9.3%), 1.9% for CC25 (ST25, 0.9%; ST945, 0.9%), 1.9% for CC107 (ST107, 100%), and 0.9% for CC22 (ST22 0.9%). With the exception of the PFGE cluster I, which presented the blaOXA−231 allele, the blaOXA−23−like gene associated with upstream ISAba1 was present in all clusters (Figure 1).
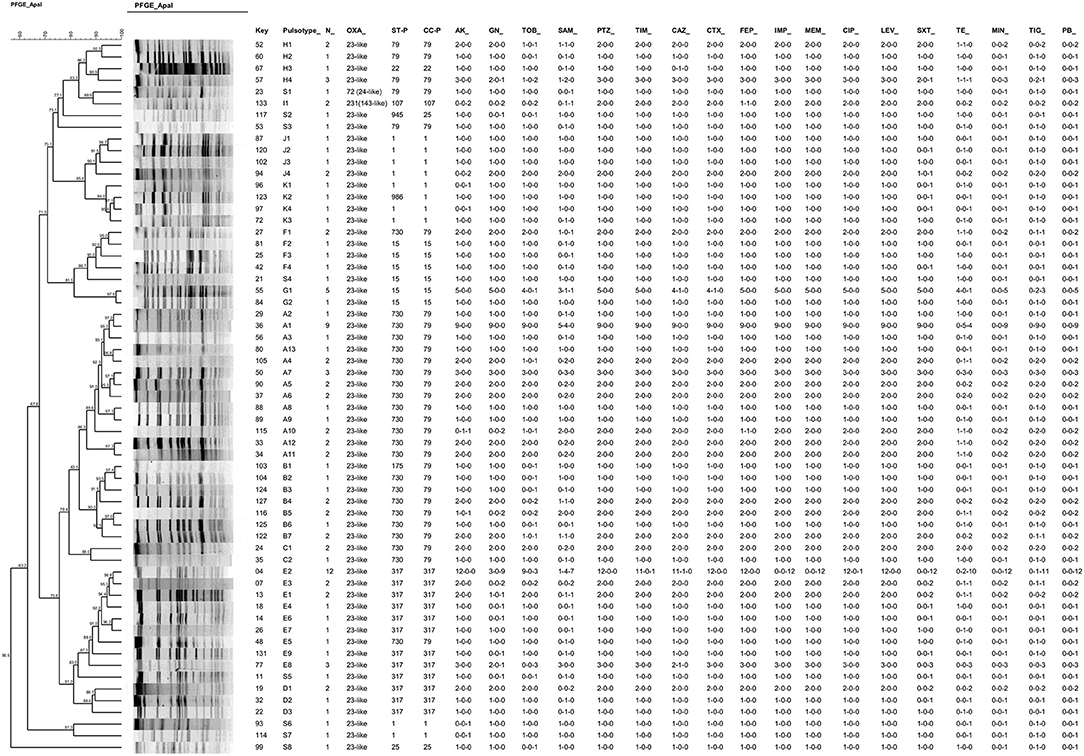
Figure 1. Dendrogram resulted from PFGE analysis of one representative isolate of each pulsotype (n = 61) combined with antimicrobial susceptibility results of each pulsotype. Information on additional OXA-enzymes, PFGE cluster, STs and CCs defined by MLST are shown. The susceptibility results are indicated in occurrences for each pulsotype in the following order: resistant-intermediate-susceptible number of strains (i.e., 1-1-0 for a given antimicrobial means 1 resistant strain; 1 intermediate and zero susceptible). AK, amikacin; GN, gentamycin; TOB, tobramycin; SAM, ampicillin-sulbactam; PTZ, piperacillin-tazobactam; TIM, ticarcillin-clavulanic acid; CAZ; ceftazidime; CTX, cefotaxime; FEP, cefepime; IMP, imipenem; MEM, meropenem; CIP, ciprofloxacin; LEV, levofloxacin; SXT, sulfamethoxazole-trimethoprim; TE, tetracycline; MIN, minocycline; TG, tigecycline; PB, polymyxin B. Key: univocal identification number; ST, sequence type; CC, Clonal Complex.
Distribution of clones over the time evidenced the emergence of A. baumannii pulsotypes belonging to clonal complex 79 in 2010, which became endemic in the institution until 2014 (Figure 2). The CC79 strains (n = 53) were distributed into 29 pulsotypes (Figure 1), recovered from 2010 to 2014. Although the pulsotype A1 was the most numerous (9 isolates), it was distributed in 2011 (4 isolates), 2013 (4 isolates) and 2014 (1 isolate). The remaining 28 pulsotypes were represented by 1, 2, or 3 isolates, each, and were recovered from the period comprised between 2010 over 2014. The 2014 isolates (the year with the largest number of isolates (13) belonging to CC79) were represented by 9 different pulsotypes with only 1 or 2 isolates in each electrophoretic pattern.
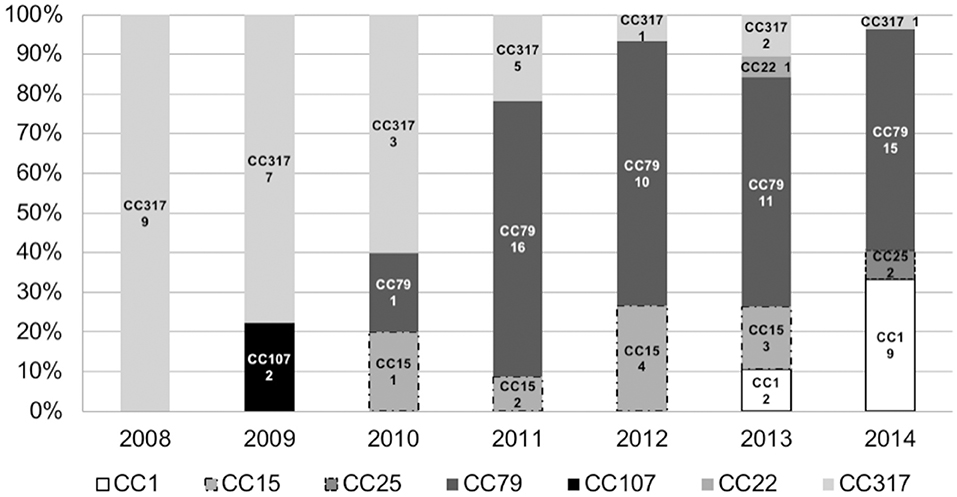
Figure 2. Distribution of Sequence Types (ST) or Clonal Complexes (CC) defined by MLST (Institute Pasteur protocol) over the years. Numbers below the CC or ST indicate number of occurrences (n = 107).
On the other hand, reduction in the frequency of CC317 was remarkable (Figure 2). The CC317 isolates (n = 28) were typed into 12 pulsotypes. A major clone (pulsotype E2) was identified comprising 12 isolates recovered from 2008 (6), 2009 (2), 2010 (2), 2011 (1), 2012 (1). Conversely, the remaining 11 pulsotypes were represented by 1, 2, or 3 isolates, each, recovered over the period of 2008–2013.
CC1 strains were detected in 2013 at frequency lower than 10% (2 isolates) but in 2014 this clonal complex represented more than 30% of the CRAB isolated from BSI (9 isolates belonging to 8 different pulsotypes), same year that CC25 strains were firstly detected.
Discussion
In this study, we verified the high prevalence of blaOXA−23−like in MDR and XDR strains isolated from patients with bacteraemia caused by carbapenem-resistant Acinetobacter baumannii in a teaching hospital in Brazil. In addition, changes in the clonal structure of circulating strains was verified, with predominance of Clonal Complexes CC1, CC15, CC79, and CC317.
The oxacillinase genes are differently distributed around the globe, with the blaOXA−23-like gene being predominant and widespread in several countries (Evans and Amyes, 2014). In this study, the blaOXA−23−like gene was identified in most of the isolates belonging to different PFGE restriction patterns and MLST clonal complexes, reinforcing the already known data from this country (Medeiros and Lincopan, 2013). In addition, we also detected the blaOXA−72 and blaOXA−231 genes, but in lower frequencies (n = 2, 1.9%; and n = 1; 0.9%, respectively), consistent with previous reports that detected both genes in this country over the last years (Gionco et al., 2012; Vasconcelos et al., 2015; Camargo et al., 2016; Pagano et al., 2017). Absence of another carbapenemases reinforces the role of oxacillinases among the isolates from Brazil, a country in which CRAB is considered highly prevalent (Rossi, 2011), but we cannot exclude the occurrence of other emergent carbapenemases (such as OXA-235-like or TMB-like), not sought in this study.
Minocycline was active against all the isolates, even if they presented MDR or XDR phenotypes. Although the study performed by Wang and colleagues had demonstrated a higher susceptibility rate to minocycline by microdilution method in CRAB compared to epsilometric method, MIC values identified in the isolates of this study were far below the intermediate value for this drug, confirming their susceptibilities (Wang et al., 2016). This antimicrobial is commonly effective against carbapenem-resistant Acinetobacter baumannii (Lashinsky et al., 2017; Poirel et al., 2017), although resistance is already being observed (Cheah et al., 2016; Pournaras et al., 2017; Vasconcellos et al., 2017a). On the other hand, resistance rates to other antimicrobial classes were remarkable. Aminoglycosides, for instance, presented susceptibilities rates ranging from 31.8% to tobramycin to only 9.3% to amikacin. Despite the controversies, tigecycline can be considered one of the few therapeutic options for treatment of MDR infection in skin and soft tissue infections and meningitis (Montravers et al., 2013; Kooli et al., 2016; Lauretti et al., 2017). Still tigecycline is not recommended for the treatment of ventilator-associated pneumonia and BSI (http://www.fda.gov/drugs/drugsafety/ucm224370.htm), we evaluate the tigecycline activity in our BSI isolates with surveillance purposes and no resistance was observed, even this phenotype becomes progressively more common in A. baumannii in several countries (Navon-Venezia et al., 2007; Al-Sweih et al., 2011; Montravers et al., 2013; Sun et al., 2013; Provasi Cardoso et al., 2016; Vasconcellos et al., 2017a; Royer et al., 2018).
Among the CRAB isolates from a single hospital, we identified the predominance of CC79 (49.5%), CC1 (10.3%), and CC15 (9.3%), corresponding to 69.2% of all the isolates evaluated over the entire period. This finding is well documented in Brazil as well as the occurrence of isolates belonging to the CC25, which seems to configure an emerging clone in our country (Chagas et al., 2014; Campos et al., 2015; Camargo et al., 2016; Provasi Cardoso et al., 2016; Vasconcellos et al., 2017a,b; Royer et al., 2018), although ST25 has already been detected worldwide (Sahl et al., 2015). ST107 was also identified, corresponding to the two isolates carrying OXA-231, which reinforces the association between ST107 and OXA-231 in Brazil (Camargo et al., 2016; Rodrigues-Costa et al., 2018). Remarkably, we detected a ST barely reported in studies around the world, the ST317, belonging to the still small CC317. CC317 was identified in 26.2% of all isolates (belonging to 11 different pulsotypes) in this study, representing the most frequent ST in BSI over the years 2008 to 2010. According to MLST database, the first ST317 isolate was detected in 2009 from an unknown sample in Rio de Janeiro, Brazil (https://pubmlst.org/abaumannii/). The only other strain deposited at the MLST database was detected in the State of São Paulo, isolated from upper respiratory tract secretion in 2011 (Camargo et al., 2016), likely indicating a limited spread of this clone, only in Brazil. Although very prevalent among the isolates studied herein, CC317 seemed to be replaced by other clones (ST79 and ST730), which belong to CC79. Conversely, analysis of our data indicate that CC317 presented less pronounced antimicrobial resistance to sulfamethoxazole-trimethoprim, tigecycline, and ampicillin-sulbactam, indicating that, at least in parts, antimicrobial resistance can drive changes in prevalence of clones in specific settings under selective pressure.
In the latest years of the study, however, the emergence of strains belonging to CC1 likely indicate that another epidemiologic shift has occurred. Changes in clonal structure of Acinetobacter strains were reported in other hospitals (Park et al., 2012; Villalón et al., 2015), but the reasons to explain this shift remain to be totally understood. In our study, a remarkable diversity of pulsotypes identified into each of these clonal complexes suggests that selective pressure, instead of a strict clone spread, plays a more decisive role in the emergence of CRAB.
Conclusion
In summary, we observed a change in the prevalence of CRAB clones in a single hospital, despite the persistence of OXA-23-producing isolates with MDR or XDR phenotypes, possibly driven by antimicrobial resistance and selective pressure. Longitudinal studies, as the present, provide this type of temporal observation, making possible to track the dispersion dynamics of successful clones associated with the persistence of well-stablished resistance phenotypes in Acinetobacter baumannii and to propose measures to contain its dissemination.
Ethics Statement
This study was submitted and approved by the Adolfo Lutz Institute Ethics Committee under the register number CAAE 49985115.5.0000.0059.
Author Contributions
LT and CC conceived and designed the study, analyzed the data, and wrote the paper. LT, FV, and WS performed the experiments. MT-C, CC, TR, ALM, AF, ACM, and TS contributed reagents, materials, analysis tools.
Funding
This study was supported by grants n°. 2015/13179-4 and 2018/23620-8 São Paulo Research Foundation (FAPESP), São Paulo, Brazil.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Al-Sweih, N. A., Al-Hubail, M. A., and Rotimi, V. O. (2011). Emergence of tigecycline and colistin resistance in Acinetobacter species isolated from patients in Kuwait hospitals. J. Chemother. 23, 13–16. doi: 10.1179/joc.2011.23.1.13
Camargo, C. H., Tiba, M. R., Saes, M. R., Vasconcellos, F. M., Santos, L. F., Romero, E. C., et al. (2016). Population structure analysis of carbapenem-resistant Acinetobacter baumannii clinical isolates from Brazil reveals predominance of clonal complexes 1, 15, and 79. Antimicrob. Agents Chemother. 60, 2545–2547. doi: 10.1128/AAC.02186-15
Campos, J., Pereira, M., Yub, E. A., and Sampaio, J. (2015). “First report of NDM-1 producing Acinetobacter radioresistens and Acinetobacter ursingii from Brazil,” in 25th European Congress of Clinical Microbiology and Infectious Disease (ECCMID) (Copenhagen).
Cayô, R., Merino, M., Ruiz Del Castillo, B., Cano, M. E., Calvo, J., and Bou, G. (2014). OXA-207, a novel OXA-24 variant with reduced catalytic efficiency against carbapenems in Acinetobacter pittii from Spain. Antimicrob. Agents Chemother. 58, 4944–4948. doi: 10.1128/AAC.02633-13
Chagas, T. P., Carvalho, K. R., de Oliveira Santos, I. C., Carvalho-Assef, A. P., and Asensi, M. D. (2014). Characterization of carbapenem-resistant Acinetobacter baumannii in Brazil (2008).-2011): countrywide spread of OXA-23-producing clones (CC15 and CC79). Diagn. Microbiol. Infect. Dis. 79, 68–72. doi: 10.1016/j.diagmicrobio.2014.03.006
Chang, H. C., Wei, Y. F., Dijkshoorn, L., Vaneechoutte, M., Tang, C. T., and Chang, T. C. (2005). Species-level identification of isolates of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex by sequence analysis of the 16S-23S rRNA gene spacer region. J. Clin. Microbiol. 43, 1632–1639. doi: 10.1128/JCM.43.4.1632-1639.2005
Cheah, S. E., Li, J., Tsuji, B. T., Forrest, A., Bulitta, J. B., and Nation, R. L. (2016). Colistin and polymyxin B dosage regimens against Acinetobacter baumannii: differences in activity and the emergence of resistance. Antimicrob. Agents Chemother. 60, 3921–3933. doi: 10.1128/AAC.02927-15
Clímaco, E. C., Oliveira, M. L., Pitondo-Silva, A., Oliveira, M. G., Medeiros, M., Lincopan, N., et al. (2013). Clonal complexes 104, 109 and 113 playing a major role in the dissemination of OXA-carbapenemase-producing Acinetobacter baumannii in Southeast Brazil. Infect. Genet. Evol. 19, 127–133. doi: 10.1016/j.meegid.2013.06.024
Clinical and Laboratory Standards Institute (2015). Performance Standards for Antimicrobial Susceptibility Testing. Vol. M100-S25. Wayne: Clinical and Laboratory Standards Institute.
Escandón-Vargas, K., Reyes, S., Gutiérrez, S., and Villegas, M. V. (2017). The epidemiology of carbapenemases in Latin America and the Caribbean. Expert Rev. Anti Infect. Ther. 15, 277–297. doi: 10.1080/14787210.2017.1268918
Evans, B. A., and Amyes, S. G. (2014). OXA β-lactamases. Clin. Microbiol. Rev. 27, 241–263. doi: 10.1128/CMR.00117-13
Fortaleza, C. M. C. B., Padoveze, M. C., Kiffer, C. R. V., Barth, A. L., Carneiro, I. C. D. R. S., Giamberardino, H. I. G., et al. (2017). Multi-state survey of healthcare-associated infections in acute care hospitals in Brazil. J. Hosp. Infect. 96, 139–144. doi: 10.1016/j.jhin.2017.03.024
Gionco, B., Pelayo, J. S., Venancio, E. J., Cay,ô, R., Gales, A. C., and Carrara-Marroni, F. E. (2012). Detection of OXA-231, a new variant of blaOXA−143, in Acinetobacter baumannii from Brazil: a case report. J. Antimicrob. Chemother. 67, 2531–2532. doi: 10.1093/jac/dks223
Héritier, C., Poirel, L., Fournier, P. E., Claverie, J. M., Raoult, D., and Nordmann, P. (2005). Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob. Agents Chemother. 49, 4174–4179. doi: 10.1128/AAC.49.10.4174-4179.2005
Higgins, P. G., Lehmann, M., and Seifert, H. (2010). Inclusion of OXA-143 primers in a multiplex polymerase chain reaction (PCR) for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 35:305. doi: 10.1016/j.ijantimicag.2009.10.014
Higgins, P. G., Poirel, L., Lehmann, M., Nordmann, P., and Seifert, H. (2009). OXA-143, a novel carbapenem-hydrolyzing class D beta-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 53, 5035–5038. doi: 10.1128/AAC.00856-09
Kanamori, H., Parobek, C. M., Weber, D. J., van Duin, D., Rutala, W. A., Cairns, B. A., Juliano, J. J., et al. (2016). Next-generation sequencing and comparative analysis of sequential outbreaks caused by multidrug-resistant Acinetobacter baumannii at a large academic burn center. Antimicrob. Agents Chemother. 60, 1249–1257. doi: 10.1128/AAC.02014-15
Karah, N., Sundsfjord, A., Towner, K., and Samuelsen, Ø. (2012). Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii. Drug Resist. Updat. 15, 237–247. doi: 10.1016/j.drup.2012.06.001
Kooli, I., Brahim, H. B., Kilani, M., Gannouni, C., Aouam, A., Toumi, A., et al. (2016). Successful treatment of postoperative multidrug-resistant Acinetobacter baumannii meningitis by tigecycline. J. Glob. Antimicrob. Resist. 5:62–63. doi: 10.1016/j.jgar.2015.12.003
La Scola, B., Gundi, V. A., Khamis, A., and Raoult, D. (2006). Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J. Clin. Microbiol. 44, 827–832. doi: 10.1128/JCM.44.3.827-832.2006
Lashinsky, J. N., Henig, O., Pogue, J. M., and Kaye, K. S. (2017). Minocycline for the treatment of multidrug and extensively drug-resistant, A. baumannii: a review. Infect. Dis. Ther. 6, 199–211. doi: 10.1007/s40121-017-0153-2
Lauretti, L., D'Alessandris, Q. G., Fantoni, M., D'Inzeo, T., Fernandez, E., and Pallini, R. (2017). First reported case of intraventricular tigecycline for meningitis from extremely drug-resistant Acinetobacter baumannii. J. Neurosurg. 127, 370–373. doi: 10.3171/2016.6
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., and Giske, C. G. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Marra, A. R., Camargo, L. F., Pignatari, A. C., Sukiennik, T., Behar, P. R., Medeiros, E. A., et al. (2011). Nosocomial bloodstream infections in Brazilian hospitals: analysis of 2,563 cases from a prospective nationwide surveillance study. J. Clin. Microbiol. 49, 1866–1871. doi: 10.1128/JCM.00376-11
Medeiros, M., and Lincopan, N. (2013). Oxacillinase (OXA)-producing Acinetobacter baumannii in Brazil: clinical and environmental impact and therapeutic options. J. Bras. Patol. Med. Lab. 49, 391–405. doi: 10.1590/S1676-24442013000600003
Montravers, P., Bassetti, M., Dupont, H., Eckmann, C., Heizmann, W. R., Guirao, X., et al. (2013). Efficacy of tigecycline for the treatment of complicated skin and soft-tissue infections in real-life clinical practice from five European observational studies. J. Antimicrob. Chemother. 68(Suppl. 2):ii15–24. doi: 10.1093/jac/dkt141
Mosqueda, N., Gato, E., Roca, I., López, M., de Alegría, C. R., Fernández Cuenca, F., et al. (2014). Characterization of plasmids carrying the blaOXA−24/40 carbapenemase gene and the genes encoding the AbkA/AbkB proteins of a toxin/antitoxin system. J. Antimicrob. Chemother. 69, 2629–2633. doi: 10.1093/jac/dku179
Navon-Venezia, S., Leavitt, A., and Carmeli, Y. (2007). High tigecycline resistance in multidrug-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 59, 772–774. doi: 10.1093/jac/dkm018
Pagano, M., Rocha, L., Sampaio, J. L., Martins, A. F., and Barth, A. L. (2017). Emergence of OXA-72-producing Acinetobacter baumannii Belonging to High-Risk Clones (CC15 and CC79) in Different Brazilian States. Infect. Control Hosp. Epidemiol. 38, 252–254. doi: 10.1017/ice.2016.287
Park, Y. K., Jung, S. I., Park, K. H., Kim, D. H., Choi, J. Y., and Kim, S. H. (2012). Changes in antimicrobial susceptibility and major clones of Acinetobacter calcoaceticus-baumannii complex isolates from a single hospital in Korea over 7 years. J. Med. Microbiol. 61, 71–79. doi: 10.1099/jmm.0.033852-0
Poirel, L., Jayol, A., and Nordmann, P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30, 557–596. doi: 10.1128/CMR.00064-16
Poirel, L., Walsh, T. R., Cuvillier, V., and Nordmann, P. (2011). Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70, 119–123. doi: 10.1016/j.diagmicrobio.2010.12.002
Pournaras, S., Dafopoulou, K., Del Franco, M., Zarkotou, O., Dimitroulia, E., Protonotariou, E., et al. (2017). Predominance of international clone 2 OXA-23-producing-Acinetobacter baumannii clinical isolates in Greece, 2015: results of a nationwide study. Int. J. Antimicrob. Agents 49, 749–753. doi: 10.1016/j.ijantimicag.2017.01.028
Provasi Cardoso, J., Cay,ô, R., Girardello, R., and Gales, A. C. (2016). Diversity of mechanisms conferring resistance to β-lactams among OXA-23-producing Acinetobacter baumannii clones. Diagn. Microbiol. Infect. Dis. 85, 90–97. doi: 10.1016/j.diagmicrobio.2016.01.018
Rodrigues-Costa, F., Cayô, R., Matos, A. P., Girardello, R., Martins, W. M. B. S., Carrara-Marroni, F. E., et al. (2018). Temporal evolution of Acinetobacter baumannii ST107 clone: conversion of blaOXA−143 into blaOXA−231 coupled with mobilization of ISAba1 upstream occAB1. Res. Microbiol. doi: 10.1016/j.resmic.2018.07.001. [Epub ahead of print].
Rodríguez, C. H., Balderrama Yarhui, N., Nastro, M., Nuñez Quezada, T., Castro Cañarte, G., Magne Ventura, R., et al. (2016). Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii in South America. J. Med. Microbiol. 65, 1088–1091. doi: 10.1099/jmm.0.000328
Rossi, F. (2011). The challenges of antimicrobial resistance in Brazil. Clin. Infect. Dis. 9, 1138–1143. doi: 10.1093/cid/cir120
Royer, S., de Campos, P. A., Araújo, B. F., Ferreira, M. L., Gonçalves, I. R., and Batistão, D. W. D. F. (2018). Molecular characterization and clonal dynamics of nosocomial blaOXA−23 producing XDR Acinetobacter baumannii. PLoS ONE 13:e0198643. doi: 10.1371/journal.pone.0198643
Sahl, J. W., Del Franco, M., Pournaras, S., Colman, R. E., Karah, N., Dijkshoorn, L., et al. (2015). Phylogenetic and genomic diversity in isolates from the globally distributed Acinetobacter baumannii ST25 lineage. Sci. Rep. 5:15188. doi: 10.1038/srep15188
Segal, H., Garny, S., and Elisha, B. G. (2005). Is IS(ABA-1) customized for Acinetobacter? FEMS Microbiol. Lett. 243, 425–429. doi: 10.1016/j.femsle.2005.01.005
Seifert, H., Dolzani, L., Bressan, R., van der Reijden, T., van Strijen, B., and Stefanik, D. (2005). Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J. Clin. Microbiol. 43, 4328–4335. doi: 10.1128/JCM.43.9.4328-4335.2005
Stietz, M. S., Ramírez, M. S., Vilacoba, E., Merkier, A. K., Limansky, A. S., and Centrón, D. (2013). Acinetobacter baumannii extensively drug resistant lineages in Buenos Aires hospitals differ from the international clones I-III. Infect. Genet. Evol. 14, 294–301. doi: 10.1016/j.meegid.2012.12.020
Sun, Y., Cai, Y., Liu, X., Bai, N., Liang, B., and Wang, R. (2013). The emergence of clinical resistance to tigecycline. Int. J. Antimicrob. Agents 41, 110–116. doi: 10.1016/j.ijantimicag.2012.09.005
US Centers for Disease Control Prevention. Antibiotic Resistance Threats in the United States. (2013). Available online at: http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf (accessed November 20, 2017).
Vaneechoutte, M., Nemec, A., KÄmpfer, P., Cools, P., and Wauters, G. (2015).). “Acinetobacter, Chryseobacterium, Moraxella, and Other Nonfermentative Gram-Negative Rods*,” in Manual of Clinical Microbiology, 11th Edn., eds J. Jorgensen, M. Pfaller, K. Carroll, G. Funke, M. Landry, S. Richter, and D. Warnock (Washington, DC: ASM Press), 813–837. doi: 10.1128/9781555817381.ch44
Vasconcellos, F. M., Casas, M. R., Tavares, L. C., Garcia, D. O., and Camargo, C. H. (2017a). In vitro activity of antimicrobial agents against multidrug- and extensively drug-resistant Acinetobacter baumannii. J. Med. Microbiol. 66, 98–102. doi: 10.1099/jmm.0.000422
Vasconcellos, F. M., Tiba-Casas, M. R., Tavares, L. C. B., Souza, W. V., Garcia, D. O., and Camargo, C. H. (2017b). Evaluation of a new trilocus sequence-based multiplex-PCR to detect major Acinetobacter baumannii clonal complexes circulating in Brazil. Infect. Genet. Evol.. 54, 4–6. doi: 10.1016/j.meegid.2017.06.009
Vasconcelos, A. T., Barth, A. L., Zavascki, A. P., Gales, A. C., Levin, A. S., and Lucarevschi, B. R. (2015). The changing epidemiology of Acinetobacter spp. producing OXA carbapenemases causing bloodstream infections in Brazil: a BrasNet report. Diagn Microbiol. Infect. Dis. 83, 382–385. doi: 10.1016/j.diagmicrobio.2015.08.006
Villalón, P., Valdezate, S., Cabezas, T., Ortega, M., Garrido, N., and Vindel, A. (2015). Endemic and epidemic Acinetobacter baumannii clones: a twelve-year study in a tertiary care hospital. BMC Microbiol. 15:47. doi: 10.1186/s12866-015-0383-y
Villalón, P., Valdezate, S., Medina-Pascual, M. J., Rubio, V., Vindel, A., and Saez-Nieto, J. A. (2011). Clonal diversity of nosocomial epidemic Acinetobacter baumannii strains isolated in Spain. J. Clin. Microbiol. 49, 875–882. doi: 10.1128/JCM.01026-10
Wang, P., Bowler, S. L., Kantz, S. F., Mettus, R. T., Guo, Y., McElheny, C. L., et al. (2016). Comparison of minocycline susceptibility testing methods for carbapenem-resistant Acinetobacter baumannii. J. Clin. Microbiol. 54, 2937–2941. doi: 10.1128/JCM.01810-16
Wong, Y. P., Chua, K. H., and Thong, K. L. (2014). One-step species-specific high resolution melting analysis for nosocomial bacteria detection. J. Microbiol. Methods 107, 133–137. doi: 10.1016/j.mimet.2014.10.001
Woodford, N., Ellington, M. J., Coelho, J. M., Turton, J. F., Ward, M. E., Brown, S., et al. (2006). Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27, 351–353. doi: 10.1016/j.ijantimicag.2006.01.004
World Health Organization (2017). Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Geneva: World Health Organization.
Zarrilli, R., Giannouli, M., Tomasone, F., Triassi, M., and Tsakris, A. (2009). Carbapenem resistance in Acinetobacter baumannii: the molecular epidemic features of an emerging problem in health care facilities. J. Infect. Dev. Ctries. 3, 335–341. doi: 10.3855/jidc.240
Keywords: Acinetobacter baumannii, oxacillinases, resistance epidemiology, healthcare associated infections, clonal complexes, MLST
Citation: Tavares LCB, Vasconcellos FM, Sousa WV, Rocchetti TT, Mondelli AL, Ferreira AM, Montelli AC, Sadatsune T, Tiba-Casas MR and Camargo CH (2019) Emergence and Persistence of High-Risk Clones Among MDR and XDR A. baumannii at a Brazilian Teaching Hospital. Front. Microbiol. 9:2898. doi: 10.3389/fmicb.2018.02898
Received: 28 July 2018; Accepted: 12 November 2018;
Published: 04 January 2019.
Edited by:
Maria Alejandra Mussi, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaReviewed by:
Benjamin Andrew Evans, University of East Anglia, United KingdomGuillermo Daniel Repizo, CONICET Instituto de Biología Molecular y Celular de Rosario (IBR), Argentina
Copyright © 2019 Tavares, Vasconcellos, Sousa, Rocchetti, Mondelli, Ferreira, Montelli, Sadatsune, Tiba-Casas and Camargo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carlos Henrique Camargo, Y2FybG9zLmNhbWFyZ29AaWFsLnNwLmdvdi5icg==
 Laís Calissi Brisolla Tavares1,2
Laís Calissi Brisolla Tavares1,2 Francielli Mahnic de Vasconcellos
Francielli Mahnic de Vasconcellos Monique Ribeiro Tiba-Casas
Monique Ribeiro Tiba-Casas Carlos Henrique Camargo
Carlos Henrique Camargo