- 1Department of Molecular Microbiology, Faculty of Life Sciences, Tokyo University of Agriculture, Tokyo, Japan
- 2NODAI Genome Research Center, Tokyo University of Agriculture, Tokyo, Japan
- 3Department of Northern Biosphere Agriculture, Faculty of Bioindustry, Tokyo University of Agriculture, Hokkaido, Japan
Root-associated microbial communities are very important in the adaptation of halophytes to coastal environments. However, little has been reported on microbial community structures related to halophytes, or on comparisons of their compositions among halophytic plant species. Here, we studied the diversity and community structure of both rhizosphere and root endosphere bacteria in two halophytic plants: Glaux maritima and Salicornia europaea. We sampled the rhizosphere, the root endosphere, and bulk control soil samples, and performed bacterial 16S rRNA sequencing using the Illumina MiSeq platform to characterize the bacterial community diversities in the rhizosphere and root endosphere of both halophytes. Among the G. maritima samples, the richness and diversity of bacteria in the rhizosphere were higher than those in the root endosphere but were lower than those of the bulk soil. In contrast for S. europaea, the bulk soil, the rhizosphere, and the root endosphere all had similar bacterial richness and diversity. The number of unique operational taxonomic units within the root endosphere, the rhizosphere, and the bulk soil were 181, 366, and 924 in G. maritima and 126, 416, and 596 in S. europaea, respectively, implying habitat-specific patterns for each halophyte. In total, 35 phyla and 566 genera were identified. The dominant phyla across all samples were Proteobacteria and Bacteroidetes. Actinobacteria was extremely abundant in the root endosphere from G. maritima. Beneficial bacterial genera were enriched in the root endosphere and rhizosphere in both halophytes. Rhizobium, Actinoplanes, and Marinomonas were highly abundant in G. maritima, whereas Sulfurimonas and Coleofasciculus were highly abundant in S. europaea. A principal coordinate analysis demonstrated significant differences in the microbiota composition associated with the plant species and type of sample. These results strongly indicate that there are clear differences in bacterial community structure and diversity between G. maritima and S. europaea. This is the first report to characterize the root microbiome of G. maritima, and to compare the diversity and community structure of rhizosphere and root endosphere bacteria between G. maritima and S. europaea.
Introduction
Salinity affects more than 800 million hectares of the total agricultural land, leading to a decrease of approximately 1–2% of the global arid and semi-arid zones every year (Kouam and Marie Solange Mandou, 2017; Etesami and Beattie, 2018). Many agricultural crops are susceptible to salt stress (Glenn et al., 1991). Future agricultural production in salt-damaged fields, therefore, requires the development of salt-tolerant crops (Etesami and Beattie, 2018). To generate crops that are able to grow on salt-damaged fields, a large amount of basic research has focused on funding and characterizing salt-resistant-related genes in model plants, and using these to improve plant salt tolerance through genetic modification and editing. However, despite numerous studies, only minor success has been achieved, as these approaches have often overlooked the important role of plant–microbe interactions in response to salt stress conditions (Coleman-Derr and Tringe, 2014; Yuan et al., 2016). It is well-known that the plant-associated microbial community plays an important role in adapting plants to extreme environments (Redman et al., 2002; Yuan et al., 2016).
A large number of reports have shown that halotolerant plant growth-promoting rhizobacteria (PGPRs) isolated from halophytes enhance salt tolerance in their host plants. For example, the salt tolerance of Arthrocnemum macrostachyum is improved by its endophytic bacteria (Navarro-Torre et al., 2017; Tian and Zhang, 2017). As another example, in Salicornia strobilacea, rhizospheric bacteria which can stably colonize the rhizoplane are capable of improving plant growth (Marasco et al., 2016). Furthermore, several halotolerant PGPRs have also been shown to improve the growth of various agricultural crops under salt stress conditions (Etesami and Beattie, 2018). Micrococcus yunnanensis, Planococcus rifietoensis, and Variovorax paradoxus, which are found in halophytes, have been shown to significantly improve salt-stress tolerance in sugar beet (Zhou et al., 2017; Etesami and Maheshwari, 2018). Pseudomonas spp. from Suaeda salsa have also been shown to be responsible for increasing salt stress tolerance and plant growth in cucumber and rice plants (Yuan et al., 2016). However, further research on the diversity of bacterial communities present in the rhizosphere and the root endosphere of various halophyte species is required before these PGPRs can be used in saline soil-based agriculture (Etesami and Beattie, 2018). Of particular importance is the fact that the diversity of halotolerant PGPRs present in soil with high salt content, as well as the diversity of halophyte-associated endophytic bacteria, depend on both the soils chemical and physical properties, as well as the plant species (Qin et al., 2016; Szymańska et al., 2016). Therefore, we have studied the diversity and community structure of bacteria present in the rhizosphere and the root endosphere of two halophytic plants: Glaux maritima (Primulaceae) and Salicornia europaea (Chenopodiaceae), which have different degrees of salt tolerance. G. maritima grows in coastal salt marshes and coastal meadows which are periodically flooded by the sea (Freipica and Ievinsh, 2010). The growth and development of G. maritima explants is stimulated by 100 mM NaCl and decreased when the NaCl concentration is raised above 200 mM. S. europaea is one of the highest salt accumulating halophytes and is found in both coastal and inland saline sites. The growth of S. europaea is significantly enhanced under 3% NaCl conditions and is suppressed in the presence of 5% (856 mM) NaCl, but the explants are able to survive (Yamamoto et al., 2009; Zhao et al., 2016).
In this report, we describe the first study to characterize the rhizosphere and root endosphere bacteria related to G. maritima and to compare its diversity and community structure with those of S. europaea. Our results bring new insight into the complex bacteria community in coastal halophytes.
Materials and Methods
Sample Collection
Naturally growing G. maritima were collected from Lake Notoro in the eastern part of Hokkaido, Japan, (44°2′50″N/144°11′25″E) and S. europaea was collected in a nearby location (44°2′51″N/144°11′20″E) in July 2017 (Supplementary Figure S1). Sampling was conducted under permission from the Hokkaido Prefecture. Sampling was performed according to the method described by Schlaeppi et al. (2014). We excavated whole plants including the surrounding soil in blocks (∼20 cm in length, ∼20 cm wide, and 10–20 cm in depth). The plants in their soil cores were brought to the laboratory, and the root systems were sampled within 12 h of removing the plants from their natural habitat. A total of 20 individual samples representing quadruplicate samples of five plant species were obtained in total, and from these, the rhizosphere and root compartments were separated and used for bacterial community profiling.
Sample Preparation
Fractionation of the rhizosphere and the root endosphere was performed according to the methods described previously (Schlaeppi et al., 2014; D’Amico et al., 2018). Roots were collected and cut into 3 cm long segments starting 0.5 cm below the root base. The collected roots were placed into 15 mL sterile tubes containing 10 mL PBS-S buffer (130 mM NaCl, 7 mM Na2HPO4, 3 mM NaH2PO4, pH 7.0, 0.02% Silwet L-77), and then washed with shaking at 180 rpm for 20 min. The roots were then transferred to a new 15 mL sterile tube, and the soil suspension was centrifuged for 20 min at 4,000 ×g. The pellet generated was defined as the rhizosphere (soil-root interface), and frozen in liquid nitrogen for storage at -80°C. After subsequent washing using the same procedure as described above, the roots were transferred to a new 15 mL sterile tube with 10 mL PBS-S buffer and sonicated for 10 min with a water bath sonicator at 40 kHz (Model 5510, Branson Ultrasonics Corporation, Danbury, CT, United States) to enrich for endophytic bacteria present in the roots. The roots were washed in a fresh volume of 10 mL PBS-S buffer using the same procedure described above and then dried on 50 mm diameter Whatman filter paper (GE Healthcare, Pittsburgh, PA, United States), transferred to a new 15 mL sterile tube, and then frozen in liquid nitrogen for storage at -80°C. This root sample, which is enriched in root endophytic bacteria, was defined as the root endosphere (Re) as described by D’Amico et al. (2018). After all plants were harvested from the soil block, the bulk soil samples were collected from 0.5 to 3.5 cm from the soil surface corresponding to a 3 cm root length, frozen in liquid nitrogen, and stored at -80°C. The collected rhizosphere (Rh), root endosphere (Re), and bulk control soil (Bl) samples were used for DNA extraction.
DNA Extraction, PCR Amplification, and Gene Clone Library Construction
Samples (0.5 g) of each of the Rh and Bl samples were used for DNA extraction. Re samples were frozen in liquid nitrogen and ground to a fine powder with a sterilized mortar and pestle. Following this, 0.5 g of the Re sample was used for DNA extraction. Total DNA was extracted using a NucleoSpin Soil Kit (Macherey-Nagel, Düren, Germany) containing buffer SL1 and enhancer SX, which have been previously used to extract both a high quality and quantity of DNA from paddy soil (Knauth et al., 2013).
The hypervariable V3-V4 regions of the bacterial 16S rRNA gene were amplified using the following primer pairs: forward, 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACG GGNGGCWGCAG-3′ and reverse, 5′-GTCTCGTGGGCTCGG AGATGTGTATAAGAGACAG GGACTACHVGGGTWTCTAA T-3′, according to previous methods (Tian et al., 2015; Wei et al., 2017). The nucleotide sequences of the Illumina adapter overhang are shown as the underlined regions, whereas the gene-specific sequences targeting the V3-V4 regions of the prokaryotic 16S rRNA genes are not underlined (Chan et al., 2015). A PCR amplicon library was generated following the Illumina 16S sample preparation guide (16S Sample Preparation Guide, 15044223; Illumina. San Diego, CA, United States). The library quality was assessed on an Agilent 2200 Tapestation (Agilent Technologies, Inc.). The libraries were sequenced as paired-end, 300 bp reads on an Illumina MiSeq benchtop sequencer. All sequence data obtained in this study have been deposited in the DDBJ Sequence Read Archive (DRA) database under accession number: DRA006852.
Sequence Processing and Analysis
Sequence processing was performed according to the method described by Zabat et al. (2018). Raw paired-end FASTQ files were quality filtered, trimmed, de-noised, and merged using DADA2 (Callahan et al., 2016) in QIIME2 (ver. 2017.11)1. Chimeric sequences were identified and removed using the consensus method in DADA2. Using DADA2, sequences were clustered into operational taxonomic units (OTUs) at 100% identity. Taxonomic analysis of OTUs was performed using the QIIME2 q2-feature-classifier plugin with a pre-trained Naïve Bayes classifier on the SILVA 99% OTU database (version 128)2 trimmed to the V3-V4 region of the 16S rRNA gene (DeSantis et al., 2006). Multiple sequence alignment and phylogenetic reconstruction were carried out using MAFFT and FastTree, respectively (Price et al., 2009; Katoh and Standley, 2013).
Statistical Analysis
Subsequent analyses were performed in R ver. 3.4.3 using the phyloseq (McMurdie and Holmes, 2013), pheatmap (Kolde, 2015), and VennDiagram (Chen, 2012) packages. OTUs classified as chloroplast and mitochondria were filtered out. The alpha diversity was estimated using the Shannon index and the Chao1 index using an absolute abundance matrix. The relationship between bacterial community structures from the two halophytes were evaluated using a principal coordinate analysis (PCoA) and complete linkage clustering (CLC) based on weighted UniFrac distances from the relative abundance matrix, and were statistically confirmed using PERMANOVA.
Results
General Characteristics of the Amplicons and Sequencing Data
In this study, we obtained 14,666,766 raw reads from the Miseq sequencing analysis of 24 samples, ranging from 1,138,891 to 334,380 reads per sample. After read-quality filtering, a total of 6,792,261 quality-filtered reads were obtained, ranging from 535,377 to 142,027 reads per sample with an average length of 416–419 bp. A total of 64,665 OTUs were extracted, ranging from 1,074 to 5,040 reads per sample (Supplementary Table S1).
The rarefaction curves for the OTUs obtained from each sample are shown in Supplementary Figure S2. The highest richness was in the G. maritima Bl control soil sample, exhibiting significantly higher OTU (P = 0.0011), Chao1 (P = 0.0011), and Shannon (P = 0.0006) indices compared with the G. maritima Re samples. Furthermore, a comparison of alpha diversity metrics revealed a disparity in the OTU, Chao, and Shannon indices in the Rh samples from G. maritima compared with the Re samples (P = 0.0096, 0.0096, and 0.0081, respectively) (Figure 1 and Table 1). In contrast, the S. europaea Bl, Rh, and Re samples had similar numbers of OTUs, and both the Shannon and Chao1 indices exhibited a similar pattern to that of the OTUs (Figure 1 and Table 1).
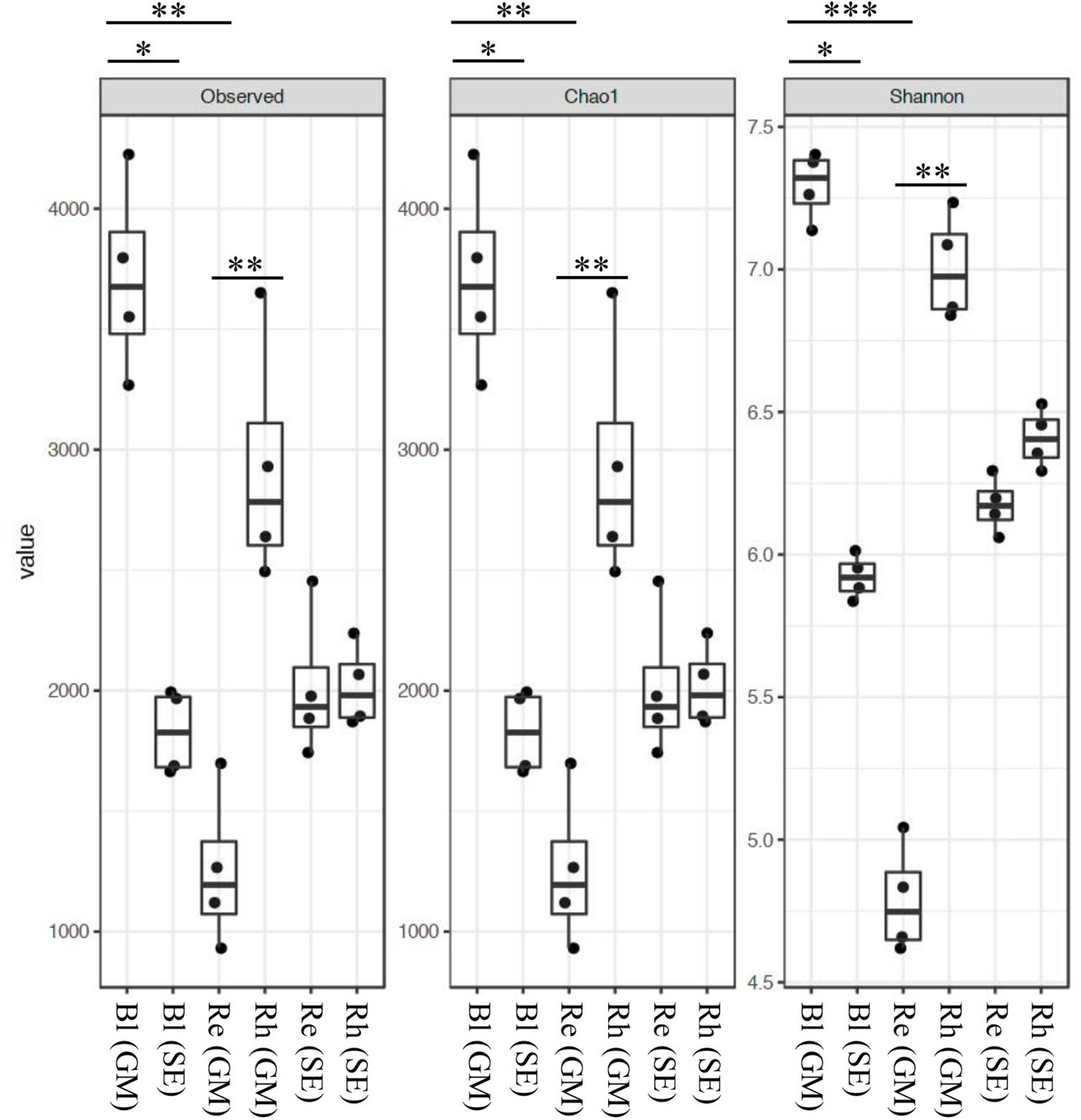
FIGURE 1. Alpha-diversity indices for the 16S rRNA gene sequences. Box plots of the observed OTUs, Chao1, and Shannon indices in bulk control soil (Bl), root endosphere (Re), and rhizosphere (Rh) samples from both G. maritima (GM) and S. europaea (SE). Whiskers represent the minimum and maximum values. All other points are contained within the box, and the bar represents the median. A Holm-adjusted P-value was calculated from Dunn’s test of multiple comparisons using rank sums. Asterisks indicate statistically significant differences between pairs of values (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001).
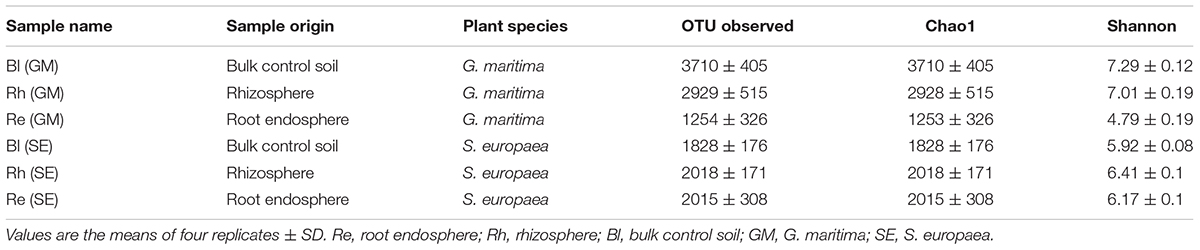
TABLE 1. Summary of the richness and diversity indices of the three samples from both G. maritima and S. europaea.
Based on a Venn diagram analysis, 155 OTUs were found to be common to all G. maritima samples. There were 924, 366, and 181 OTUs that were exclusive to the Bl, Rh, and Re G. maritima samples, respectively (Figure 2A). Similar results were also obtained in S. europaea. All samples shared 209 OTUs with 596, 416, and 126 OTUs being unique in the Bl, Rh, and Re samples, respectively (Figure 2B).
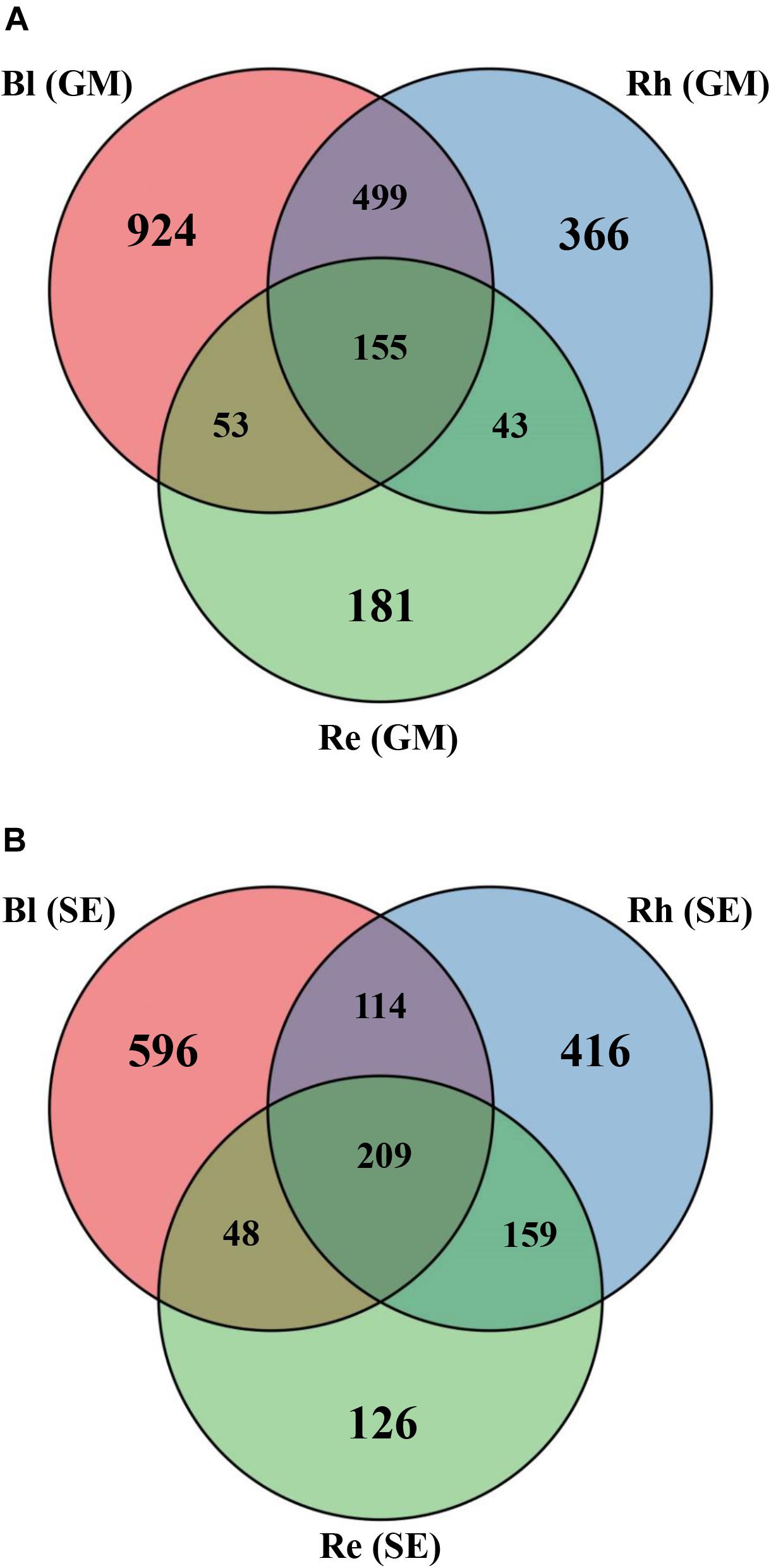
FIGURE 2. Venn diagrams showing the overlap of the OTU calculated separately for the (A) G. maritima and (B) S. europaea microbial communities. Re, root endosphere; Rh, rhizosphere; Bl, bulk control soil; GM, G. maritima; SE, S. europaea.
Microbial Taxonomic Analysis at the Phylum and Class Levels
Classification of the high-quality sequences also demonstrated differences in the bacterial communities among the different samples at the phylum level. A total of fifty-seven phyla were identified in all samples. The relative abundance of the top 15 phyla (>1% of relative abundance in at least one sample) are shown in Figure 3A and Supplementary Table S2. Proteobacteria and Bacteroidetes were the dominant phyla (>10% relative abundance) across all samples, accounting for 42.2–59.6% and 14.5–20.9% of the total high-quality sequences, respectively. Actinobacteria, Planctomycetes, and Chloroflexi were the sub-dominant phyla (>1% relative abundance) in all samples, accounting for 2.7–27.2, 3.7–8.9, and 1.2–6.0 of the total high-quality sequences, respectively. Interestingly, the abundance of Actinobacteria in the G. maritima Re sample was extremely high, compared with that in the other samples. Acidobacteria, Verrucomicrobia, Deferribacteres, Latescibacteria, Parcubacteria, Fibrobacteres, Ignavibacteriae, Cyanobacteria, Chlamydiae, and Spirochaetae were present at > 1% relative abundance in at least one sample. The other 42 phyla had much lower abundances (less than 1% of the high-quality sequences). These phyla, therefore, were defined as rare phyla, and are referred to as “others” in Figure 3A.
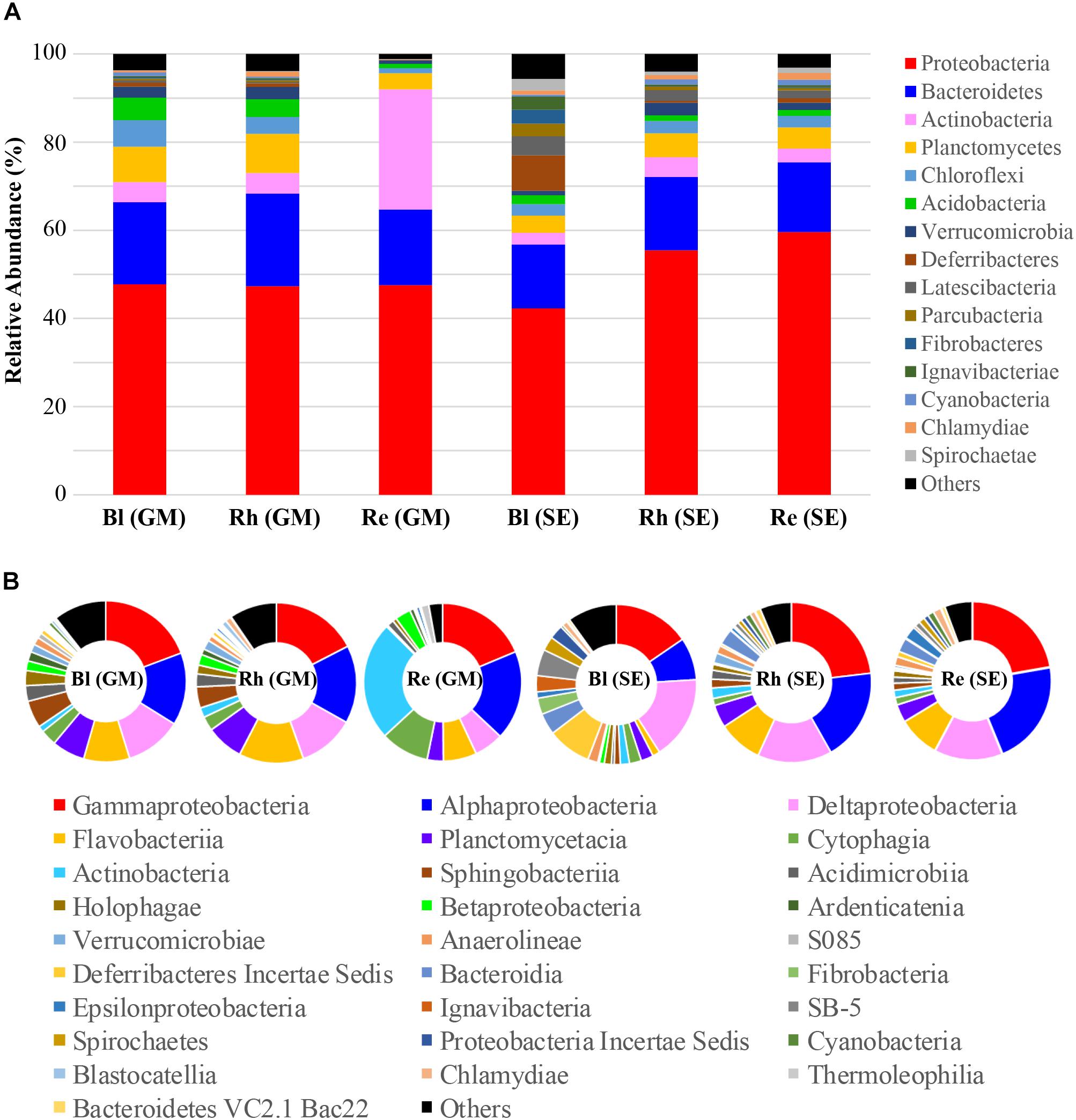
FIGURE 3. Average relative abundances of bacteria at (A) the phylum level and (B) the class level in the different samples. Re, root endosphere; Rh, rhizosphere; Bl, bulk control soil; GM, G. maritima; SE, S. europaea.
A total of 151 bacterial classes were identified across all samples (Supplementary Table S3). There were 28 classes with a relative abundance of higher than 1% in at least one sample (Figure 3B and Supplementary Table S3). The other 123 classes with <1% abundance of the high-quality sequences, and are referred to as “others” in Figure 3B. Among these 28 classes, the more dominant classes (>5% relative abundance) in all samples were Gammaproteobacteria, Alphaproteobacteria, and Deltaproteobacteria, accounting for 14.0–21.9%, 7.8–20.7%, and 6.0–15.4% of the total high-quality sequences, respectively. The sub-dominant classes were Flavobacteria, Planctomycetacia, Cytophagia, and Actinobacteria, which had relative abundances of higher than 1% in all samples, especially the G. maritima Re sample, which possessed the highest abundance of Cytophagia and Actinobacteria, accounting for 9.9 and 24.3% of the total high-quality sequences, respectively. Blastocatellia, Chlamydiae, and Thermoleophilia had >1% abundance of high-quality sequences in the Rh and/or Re samples from G. maritima. Among the S. europaea Bl, Rh, and Re samples, the abundance of Verrucomicrobia, Cyanobacteria, Chlamydiae, and Bacteroidetes VC2.1 Bac22 were higher in the Rh and/or Re samples than in the Bl sample.
Comparison of Bacterial Community Structure at the Family and Genus Levels
Based on a heatmap analysis, the relative abundance of the top 50 classified families and genera clearly revealed that there were significant different bacterial community structures among the sample types. Figure 4 shows a clustering of the top 50 classified families. These classified families belonged to 11 phyla as shown in Supplementary Table S4. The distributions of the families differed greatly across the different samples. Hyphomonadaceae, Oceanospirillaceae, Methylophilaceae, Micromonosporaceae, Flammeovirgaceae, Alteromonadaceae, Rhizobiaceae, and Cellvibrionaceae were significantly abundant in the G. maritima Re samples, whereas only one family, Helicobacteraceae, was more abundant in the S. europaea Re samples. No family was significantly enriched in the G. maritima Rh samples. In contrast, Caldilineaceae, Rhodobacteraceae, and Chromatiaceae were more highly abundant in the Rh samples from S. europaea. CA002 was predominantly distributed in the G. maritima Bl samples. Thiotrichaceae, possible family 01, Ectothiorhodospiraceae, Desulfobacteraceae, and Spirochaetaceae were highly abundant in the S. europaea Bl samples.
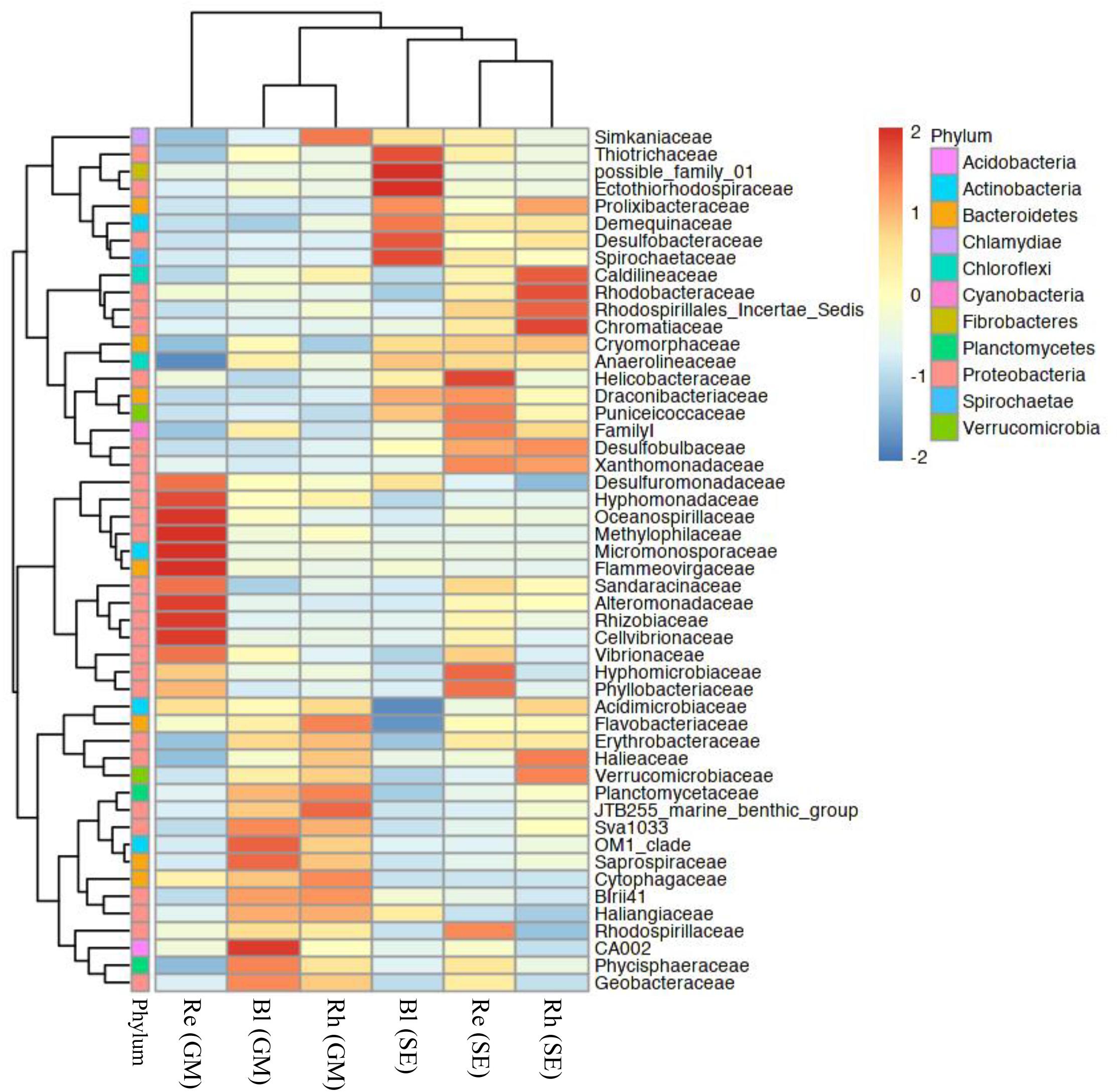
FIGURE 4. Heatmap of bacterial distribution of the top 50 abundant families in all sample types. The dendrogram shows complete-linkage agglomerative clustering based on a Euclidean distance. The heatmap color (blue to red) represents the row z-score of the mean relative abundance from low to high. Re, root endosphere; Rh, rhizosphere; Bl, bulk control soil; GM, G. maritima; SE, S. europaea.
At the genus level, the top 50 classified bacterial genera belonged to 10 phyla as shown in Supplementary Table S5. From the heatmap shown in Figure 5, nine genera (Labrenzia, Methylotenera, Rhizobium, Marinoscillum, Actinoplanes, Paraglaciecola, Simiduia, Marinomonas, and Pelobacter) had were highly abundant in the G. maritima Re samples, whereas Sulfurimonas, Coleofasciculus, and Aestuariispira were significantly more abundant in the S. europaea Re samples. Zeaxanthinibacter, Sneathiella, Blastocatella, and Halioglobus were more abundant in the G. maritima Rh samples. In contrast, Roseovarius and Halochromatium were highly abundant in the S. europaea Rh samples. Although there was no significantly enriched genus in the G. maritima Bl samples, the following seven genera were dominant the S. europaea Bl sample: Thiogranum, SEEP-SRB1, Caldithrix, Ignavibacterium, Sva008 sediment group, Candidatus Thiobios, and Spirochaeta 2.
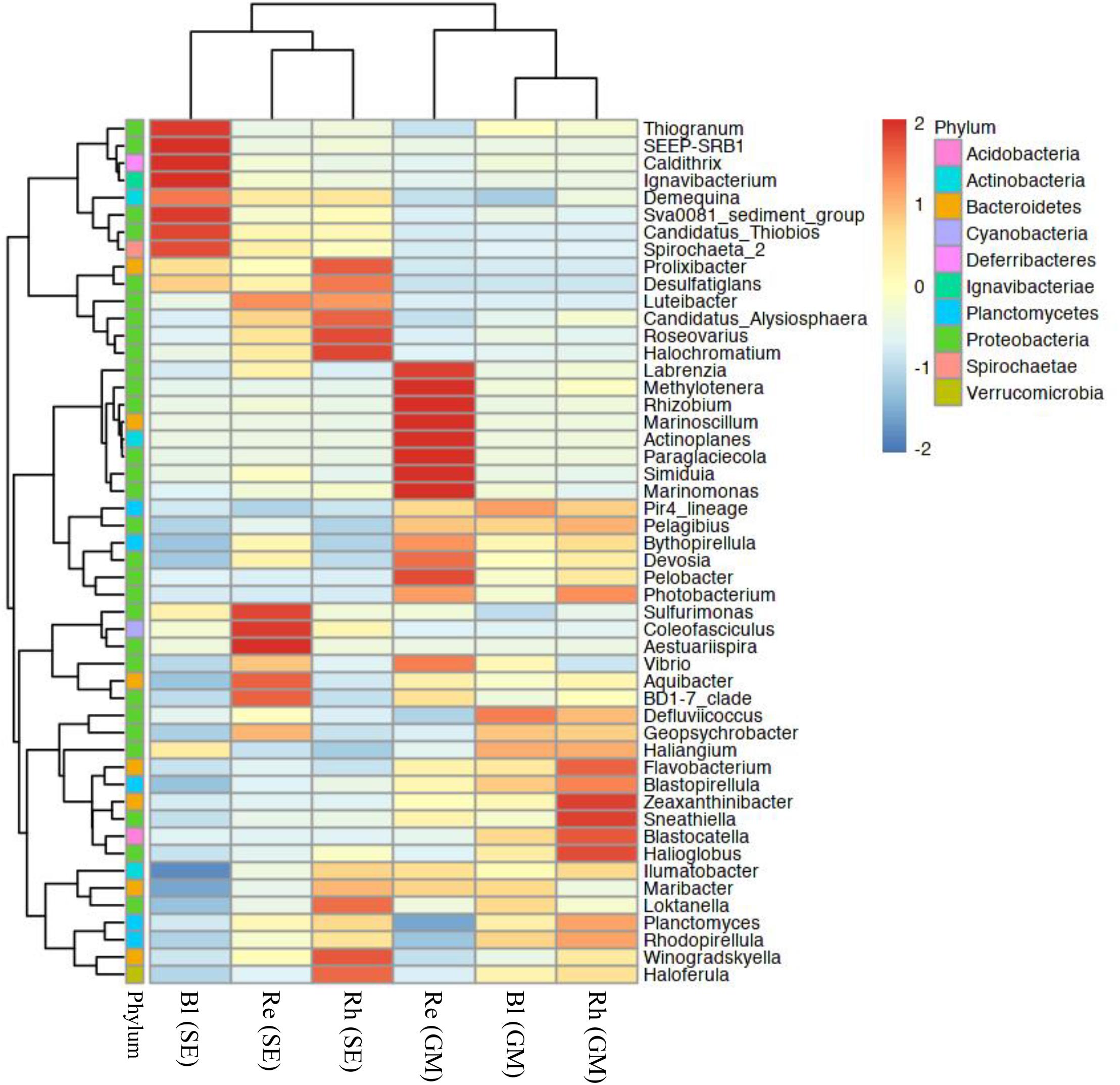
FIGURE 5. Heatmap of bacterial distribution of the top 50 abundant genera in all sample types. The dendrogram shows complete-linkage agglomerative clustering based on a Euclidean distance. The heatmap color (blue to red) represent the row z-score of the mean relative abundance from low to high. Re, root endosphere; Rh, rhizosphere; Bl, bulk control soil; GM, G. maritima; SE, S. europaea.
Comparative Analysis of Bacterial Communities in the Different Sample Groups
A beta-diversity analysis based on PCoA (Figure 6A) and CLC (Figure 6B) was performed to compare the bacterial compositions among the different samples. All the samples were clustered into two groups by PCoA: the first containing the G. maritima Bl control soil, Rh, and Re samples (group 1 in Figure 6A) and the second containing the same S. europaea samples (group 2 in Figure 6A). This result shows that there is a strong separation based on plant species, which explained 40.1% (axis 1) of the variation. The quadruplicate Bl control soil, Rh, and Re samples obtained for each halophyte also clustered together, with the exception of the Re sample from S. europaea, explaining 18.8% (axis 2) of the variation (Figure 6A). Similar results were also obtained for the CLC tree. As shown in Figure 6B, the Re, Rh, and Bl samples from G. maritima (group 1) and the same samples from S. europaea (group 2) were also separated into two different clusters in the CLC tree. These results indicate that the microbiota in the Re, Rh, and Bl samples from G. maritima are largely different from those from S. europaea.
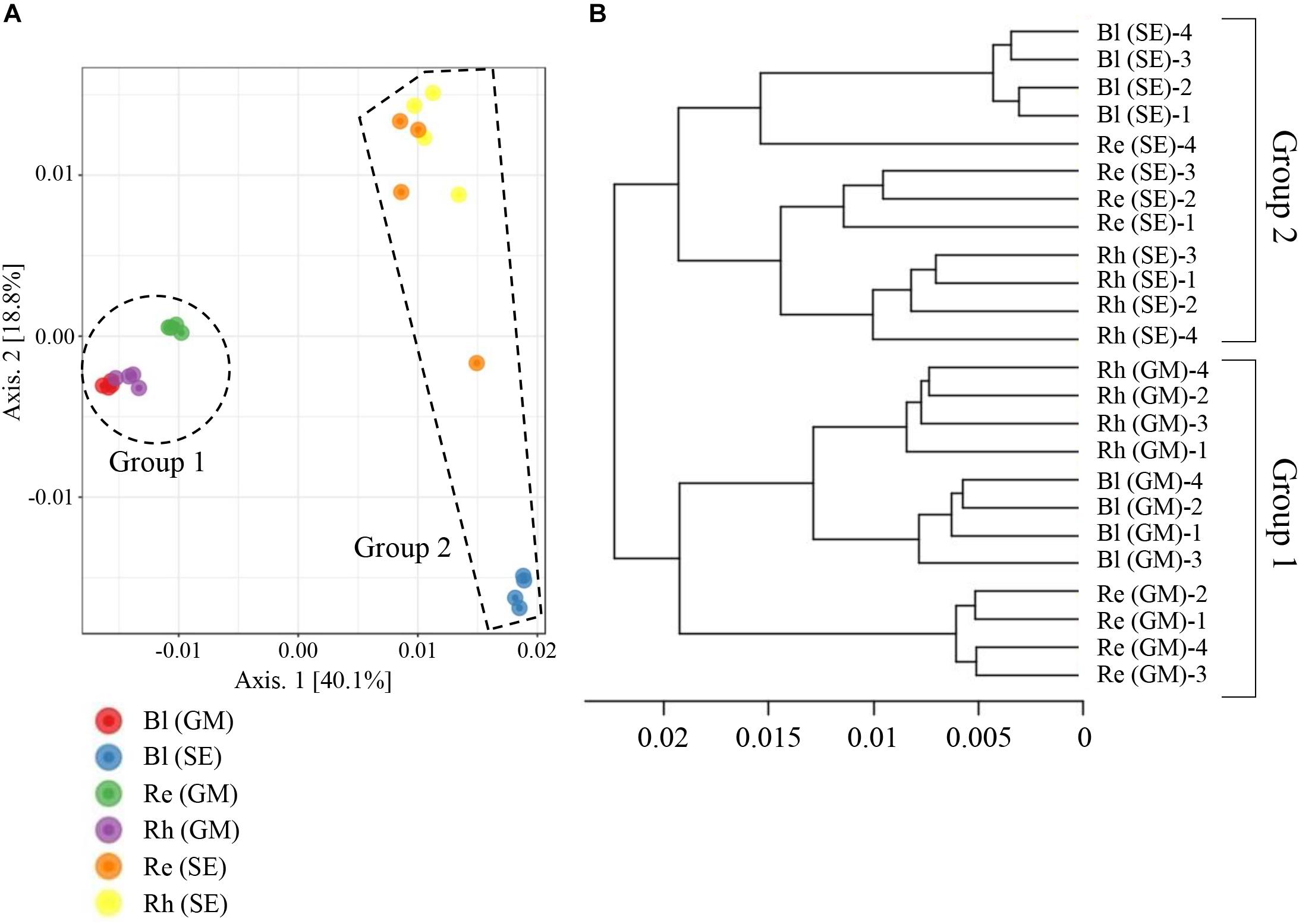
FIGURE 6. Principal coordinate analysis (PCoA) (A) and complete linkage clustering (CLC) (B) of the bacterial communities in different samples based on weighted UniFrac distances. Re, root endosphere; Rh, rhizosphere; Bl, bulk control soil; GM, G. maritima; SE, S. europaea.
Discussion
The halophytic plant-associated microbial community, and halotolerant PGPRs, play important roles in allowing hosts to adapt to a costal environment (Redman et al., 2002; Yuan et al., 2016; Etesami and Beattie, 2018). However, the diversity of halotolerant PGPRs present in saline containing soils and in halophyte-associated endophytic bacteria depends on the soil’s chemical and physical properties, as well as the plant species (Qin et al., 2016; Szymańska et al., 2016). To extend our knowledge about bacterial diversity related to halophytes, here we investigated the diversity and community structure of bacteria present in the rhizospheres and root endospheres of G. maritima and S. europaea. This study showed that the diversity of bacterial communities, including possible halotolerant PGPRs candidates in the rhizosphere and root endosphere, depends on the halophytic plant species and the sampling site.
The diversity and richness of bacteria in the rhizosphere from G. maritima were higher than those in the root endosphere. Previous reports have suggested that microbial density is generally higher in the rhizosphere than in the root, and that bacterial diversity and richness gradually decreases from the soil to the root-endosphere (Bulgarelli et al., 2012, 2015; Lundberg et al., 2012; Edwards et al., 2015; Hacquard et al., 2015). On the other hand, we found there were no significant differences in bacterial diversity and richness between the bulk control soil, rhizosphere, and the root endosphere for S. europaea. A similar result has also been reported for Arabidopsis thaliana (Lundberg et al., 2012) and Bistorta vivipara (Vik et al., 2013). Vik et al. (2013) concluded that there is a similarity in the micro-niches available for bacteria in plant roots and soil. However, a previous report has demonstrated that the bacterial diversity in the endosphere of S. europaea was lower than that in the rhizosphere of S. europaea (Shi et al., 2015). This difference indicates that the local environment has a complex effect on the bacterial community.
In all samples, the dominant bacterial phyla were Proteobacteria, Bacteroidetes, Actinobacteria Planctomycetes, and Chloroflexi (>1% of high-quality sequences). The bacterial species belonging to Proteobacteria, Bacteroidetes, and Actinobacteria could play important roles in the ecology of G. maritima and S. europaea, as these phyla are also dominant in other halophytes (Shi et al., 2015; Mukhtar et al., 2017; Tian and Zhang, 2017). Mukhtar et al. (2017) reported that Planctomycetes was also a dominant phylum in Salsola stocksii, similar to our results. Interestingly, the root endosphere of G. maritima has been shown to possess high levels of bacterial species from Actinobacteria. Actinobacteria have also been shown to be enriched in metabolically active cells in the Arabidopsis root (Bulgarelli et al., 2012; Reinhold-Hurek et al., 2015). Bulgarelli et al. (2013) suggested that the wide range of antimicrobial compounds secreted by members of Actinobacteria could play an important role, in that such compounds could indirectly protect sugar beet against soil-borne fungal pathogens. Therefore, it is possible that some members of Actinobacteria present in the root endosphere of G. maritima could also secrete antimicrobial compounds that could protect the host plant from fungal pathogens. Similar to our results, Chloroflexi was found in high abundance in mangrove sediments at a depth of 10 cm (Mendes and Tsai, 2014). Bacterial species in the phylum Chloroflexi, therefore, could play important roles in the decomposition of organic compounds in the coastal environment, as has been reported previously (Yamada et al., 2005; Mendes and Tsai, 2014). The dominant bacterial classes were Gammaproteobacteria, Alphaproteobacteria, Deltaproteobacteria, Flavobacteria, Planctomycetacia, Cytophagia, and Actinobacteria (>1% of high-quality sequences) in all samples. Unlike our study, Yuan et al. (2016) reported that Deltaproteobacteria, Alphaproteobacteria, Bacteroidetes, and Verrucomicrobia were the dominant classes in the halophyte S. salsa using two primer sets that amplified the V3-V4 and V5-V9 regions. The choice of primers might have a significant effect on the observed differences in bacterial communities, as has been described previously (Klindworth et al., 2013; Thijs et al., 2017), as we only selected primers spanning the V3-V4 region of the bacterial 16S rRNA gene in this study. In addition, this difference might be a reflection of differences in plant species, sampling methods, and/or sampling area. In this regard, in a pristine mangrove, the bacterial composition has been shown to change dramatically according to the depth of the sediment samples (Mendes and Tsai, 2014).
The beta-diversity analyses showed that bacterial communities varied across the different plant species. These results were also supported by heatmap analyses at the family and genus levels (Figures 4, 5). As demonstrated by a PCoA, plant species explained 40.1% of the variation, whereas sample type explained 18.8% of the variation (Figure 6A). Similar to this, Dong et al. (2018) reported that the community structures of the root microbiome were significantly different among four different sugarcane species. Therefore, the data from this study and previous investigations indicate that plant species have a significant influence on bacterial communities.
For the top 50 genera, we have summarized the possible functions of the abundant bacterial genera in the rhizosphere and root endosphere from the two halophytes, as shown in Tables 2A,B. In the root endosphere and rhizosphere of G. maritima, many species of Actinoplanes, Marinomonas, and Rhizobium have been reported to have beneficial effects for host plants, such as the production of indole-3-acetic acid (IAA), indole-3-pyruvic acid (IPYA), and gibberellic acid (GA3), antifungal activity, N2-fixation, phosphate solubilization, siderophore production, and 1-aminocyclopropane-1-carboxylate (ACC) deaminase activity (Table 2A). Furthermore, several species of Rhizobium have been isolated as halotolerant PGPRs from three halophytes, Psoralea corylifolia, Salicornia brachiate, and Salicornia bigelovii (Etesami and Beattie, 2018). These three genera, therefore, could be possible candidates as halotolerant PGPRs for G. maritima. In addition, Pelobacter was also a dominant genus in the root endosphere from G. maritima. Pelobacter propionicus can use C2 compounds such as lactate, pyruvate, 2,3-butanediol, acetoin, and ethanol for growth under strictly anaerobic conditions, and can thereby induce propionate formation (Li et al., 2010). Methyl 3-(4-hydroxyphenyl) propionate functions as a modulator of the architecture of the root system (Liu et al., 2016). The propionate induced by Pelobacter, therefore, might regulate root formation in G. maritima. On the other hand, the Coleofasciculus (Microcoleus sp.) genus, which is related to N2-fixation, suggests a possible function in the root endosphere of S. europaea (Table 2B). Toledo et al. (1995) reported that Coleofasciculus can form a biofilm on the root of the black mangrove, in which the cyanobacterial filaments are embedded and may be capable of N2-fixation, although 15N assimilation tests have not yet been performed to confirm this. In addition, the sulfur-oxidizing genera, Sulfurimonas and Halochromatium, were significantly abundant in the root endosphere and rhizosphere, respectively, in S. europaea (Table 2B). According to a previous report, Sulfurimonas might be related to host detoxification by oxidizing sulfide and producing sulfate as an end product, suggesting that the accumulation of these bacteria around the rhizosphere might be critical for the host tolerance of coastal environments (Fahimipour et al., 2017). Hasler-Sheetal and Holmer (2015) also suggested that the oxidation of sulfide and the production of non-toxic S0 in the aerenchymous tissue of seagrass is involved in the detoxification mechanisms of the host plants. Thus, Coleofasciculus and Sulfurimonas could play important roles as halotolerant PGPRs in S. europaea. The possible function of any of the other bacteria in their host plants, unfortunately, is still unknown (Tables 2A,B).

TABLE 2A. Possible function of the abundant bacterial genera in the root endosphere and rhizosphere in G. maritima.

TABLE 2B. Possible function of the abundant bacterial genera in the root endosphere and rhizosphere in S. europaea.
Taken together, our results suggest that there are apparent differences in the bacterial communities, and in the different varieties of beneficial bacteria, found between G. maritima and S. europaea. Further research is required to clarify whether there are specific interactions between the bacteria enriched in the two halophytes and their host.
Conclusion
This report is the first to clarify the bacterial diversity and community structure of a halophyte, G. maritima, using next-generation sequencing technology and to compare the diversity and composition with those of S. europaea. The bacterial community structures varied among the three sample types, namely the root endosphere, the rhizosphere, and the bulk control soil in each halophyte. The more dominant bacteria phyla associated with both G. maritima and S. europaea were Proteobacteria and Bacteroidetes. Beneficial bacterial genera were enriched in the root endosphere and the rhizosphere in both halophytes. Actinoplanes, Marinomonas, and Rhizobium were found to be significantly abundant in G. maritima, whereas Sulfurimonas and Coleofasciculus were more highly abundant in S. europaea. Our results indicate that there are clear differences in bacterial diversity and community structure between G. maritima and S. europaea. These results provide a new insight into the complex bacterial community structures of halophytes. In the future, we plan to investigate the functional roles of these potential beneficial bacteria in the interaction between plants and microbes in coastal areas.
Author Contributions
KY contributed to the conception of the study, performed the experiments and data analyses, and wrote the manuscript. YS performed data analyses. TI performed the experiments and data analyses. HkS collected the samples. KT, MU, NT, SO, HmS, and ST contributed to a fruitful discussion. All authors contributed to manuscript revision and have read and approved the submitted version.
Funding
This research was supported by “Grant-in-Aid for Research Activity start-up” (17H07127) promoted by the Japan Society for the Promotion of Science and by MEXT-Supported Program for the Strategic Research Foundation at Private Universities (S1311017).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Editage (www.editage.jp) for English language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02878/full#supplementary-material
Footnotes
References
Asker, D., Beppu, T., and Ueda, K. (2007). Zeaxanthinibacter enoshimensis gen. nov., sp. nov., a novel zeaxanthin-producing marine bacterium of the family Flavobacteriaceae, isolated from seawater off enoshima island, japan. Int. J. Syst. Evol. Microbiol. 57, 837–843. doi: 10.1099/ijs.0.64682-0
Bibi, F., Jeong, J. H., Chung, E. J., Jeon, C. O., and Chung, Y. R. (2014). Labrenzia suaedae sp. nov., a marine bacterium isolated from a halophyte, and emended description of the genus labrenzia. Int. J. Syst. Evol. Microbiol. 64, 1116–1122. doi: 10.1099/ijs.0.052860-0
Bulgarelli, D., Garrido-Oter, R., Münch, P. C., Weiman, A., Dröge, J., Pan, Y., et al. (2015). Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 17, 392–403. doi: 10.1016/j.chom.2015.01.011
Bulgarelli, D., Rott, M., Schlaeppi, K., Ver Loren van Themaat, E., Ahmadinejad, N., Assenza, F., et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95. doi: 10.1038/nature11336
Bulgarelli, D., Schlaeppi, K., Spaepen, S., Ver Loren van Themaat, E., and Schulze-Lefert, P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838. doi: 10.1146/annurev-arplant-050312-120106
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Chan, C. S., Chan, K. G., Tay, Y. L., Chua, Y. H., and Goh, K. M. (2015). Diversity of thermophiles in a malaysian hot spring determined using 16S rRNA and shotgun metagenome sequencing. Front. Microbiol. 6:177. doi: 10.3389/fmicb.2015.00177
Chen, H. B. (2012). Venndiagram: generate high-resolution venn and euler plots. R Package Version 113.
Coleman-Derr, D., and Tringe, S. G. (2014). Building the crops of tomorrow: advantages of symbiont-based approaches to improving abiotic stress tolerance. Front. Microbiol. 5:283. doi: 10.3389/fmicb.2014
D’Amico, F., Candela, M., Turroni, S., Biagi, E., Brigidi, P., Bega, A., et al. (2018). The rootstock regulates microbiome diversity in root and rhizosphere compartments of Vitis vinifera Cultivar Lambrusco. Front. Microbiol. 9:2240. doi: 10.3389/fmicb.2018.02240
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/AEM.03006-05
Dong, M., Yang, Z., Cheng, G., Peng, L., Xu, Q., and Xu, J. (2018). Diversity of the bacterial microbiome in the roots of four Saccharum species: S. spontaneum, S. robustum, S. barberi, and S. officinarum. Front. Microbiol. 9:267. doi: 10.3389/fmicb.2018.00267
Edwards, J., Johnson, C., Santos-Medellín, C., Lurie, E., Podishetty, N. K., Bhatnagar, S., et al. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. U.S.A. 112, E911–E920. doi: 10.1073/pnas.1414592112
El-Tarabily, K. A., Nassar, A. H., Hardy, G. E., and Sivasithamparam, K. (2009). Plant growth promotion and biological control of Pythium aphanidermatum, a pathogen of cucumber, by endophytic actinomycetes. J. Appl. Microbiol. 106, 13–26. doi: 10.1111/j.1365-2672.2008.03926.x
Etesami, H., and Beattie, G. A. (2018). Mining halophytes for plant growth-promoting halotolerant bacteria to enhance the salinity tolerance of non-halophytic crops. Front. Microbiol. 9:148. doi: 10.3389/fmicb.2018.00148
Etesami, H., and Maheshwari, D. K. (2018). Use of plant growth promoting rhizobacteria (PGPRs) with multiple plant growth promoting traits in stress agriculture: action mechanisms and future prospects. Ecotoxicol. Environ. Saf. 156, 225–246. doi: 10.1016/j.ecoenv.2018.03.013
Fahimipour, A. K., Kardish, M. R., Lang, J. M., Green, J. L., Eisen, J. A., and Stachowicz, J. J. (2017). Global-scale structure of the eelgrass microbiome. Appl. Environ. Microbiol. 83, e03391–16. doi: 10.1128/AEM.03391-16
Fidalgo, C., Henriques, I., Rocha, J., Tacão, M., and Alves, A. (2016). Culturable endophytic bacteria from the salt marsh plant Halimione portulacoides: phylogenetic diversity, functional characterization, and influence of metal(loid) contamination. Environ. Sci. Pollut. Res. Int. 23, 10200–10214. doi: 10.1007/s11356-016-6208-1
Freipica, I., and Ievinsh, G. (2010). Relative NaCl tolerance of rare and endangered coastal plant species in conditions of tissue culture. Environ. Exp. Biol. 8, 35–42.
Glenn, E. P., O’Leary, J. W., Watson, M. C., Thompson, T. L., and Kuehl, R. O. (1991). Salicornia bigelovii Torr.: an oilseed halophyte for seawater irrigation. Science 251, 1065–1067. doi: 10.1126/science.251.4997.1065
Hacquard, S., Garrido-Oter, R., González, A., Spaepen, S., Ackermann, G., Lebeis, S., et al. (2015). Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe. 17, 603–616. doi: 10.1016/j.chom.2015.04.009
Hasler-Sheetal, H., and Holmer, M. (2015). Sulfide intrusion and detoxification in the seagrass Zostera marina. PLoS One 10:e0129136. doi: 10.1371/journal.pone.0129136
Jha, B., Gontia, I., and Hartmann, A. (2012). The roots of the halophyte Salicornia brachiata are a source of new halotolerant diazotrophic bacteria with plant growth-promoting potential. Plant Soil 356, 265–277. doi: 10.1007/s11104-011-0877-9
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41:e1. doi: 10.1093/nar/gks808
Knauth, S., Schmidt, H., and Tippkötter, R. (2013). Comparison of commercial kits for the extraction of DNA from paddy soils. Lett. Appl. Microbiol. 56, 222–228. doi: 10.1111/lam.12038
Kolde, R. (2015). Pheatmap: Pretty Heatmaps. R package version 1.0.8. Available at: https://CRAN.R-project.org/package=pheatmap.
Kouam, E., and Marie Solange Mandou, E. L. T. (2017). Effects of salinity stress (NaCl) on growth attributes and some nutrient accumulation in cowpea (Vigna unguiculata). Curr. Bot. 8, 164–170. doi: 10.19071/cb.2017.v8.3282
Li, Y. H., Zhu, J. N., Zhai, Z. H., and Zhang, Q. (2010). Endophytic bacterial diversity in roots of Phragmites australis in constructed beijing cuihu wetland (China). FEMS. Microbiol. Lett. 309, 84–93. doi: 10.1111/j.1574-6968.2010.02015.x
Lin, S. Y., Shieh, W. Y., Chen, J. S., and Tang, S. L. (2013). Complete genome sequence of Simiduia agarivorans SA1(T), a marine bacterium able to degrade a variety of polysaccharides. Genome Announc. 1:e00039–12. doi: 10.1128/genomeA.00039-12
Liu, Y., Wang, R., Zhang, P., Chen, Q., Luo, Q., Zhu, Y., et al. (2016). The nitrification inhibitor methyl 3-(4-hydroxyphenyl)propionate modulates root development by interfering with auxin signaling via the NO/ROS pathway. Plant Physiol. 71, 1686–1703. doi: 10.1104/pp.16.00670
Lundberg, D. S., Lebeis, S. L., Paredes, S. H., Yourstone, S., Gehring, J., Malfatti, S., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90. doi: 10.1038/nature11237
Marasco, R., Mapelli, F., Rolli, E., Mosqueira, M. J., Fusi, M., Bariselli, P., et al. (2016). Salicornia strobilacea (Synonym of Halocnemum strobilaceum) grown under different tidal regimes selects rhizosphere bacteria capable of promoting plant growth. Front. Microbiol. 7:1286. doi: 10.3389/fmicb.2016.01286
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Mendes, L. W., and Tsai, S. M. (2014). Variations of bacterial community structure and composition in mangrove sediment at different depths in southeastern brazil. Diversity 6, 827–843. doi: 10.3390/d6040827
Mesa, J., Mateos-Naranjo, E., Caviedes, M. A., Redondo-Gómez, S., Pajuelo, E., and Rodríguez-Llorente, I. D. (2015). Endophytic cultivable bacteria of the metal bioaccumulator Spartina maritima improve plant growth but not metal uptake in polluted marshes soils. Front. Microbiol. 6:1450. doi: 10.3389/fmicb.2015.01450
Montoya, L., Lozada-Chávez, I., Amils, R., Rodriguez, N., and Marín, I. (2011). The sulfate-rich and extreme saline sediment of the ephemeral tirez lagoon: a biotope for acetoclastic sulfate-reducing bacteria and hydrogenotrophic methanogenic archaea. Int. J. Microbiol. 2011:753758. doi: 10.1155/2011/753758
Mukhtar, S., Ishaq, A., Hassan, S., Mehnaz, S., Mirza, M. S., and Malik, K. A. (2017). Comparison of microbial communities associated with halophyte (Salsola stocksii) and non-halophyte (Triticum aestivum) using culture-independent approaches. Pol. J. Microbiol. 66, 353–364. doi: 10.5604/01.3001.0010.4866
Navarro-Torre, S., Barcia-Piedras, J. M., Mateos-Naranjo, E., Redondo-Gómez, S., Camacho, M., Caviedes, M. A., et al. (2017). Assessing the role of endophytic bacteria in the halophyte Arthrocnemum macrostachyum salt tolerance. Plant Biol. 19, 249–256. doi: 10.1111/plb.12521
Osman, J. R., Fernandes, G., Michael, S., and DuBow, M. S. (2017). Bacterial diversity of the rhizosphere and nearby surface soil of rice (Oryza sativa) growing in the camargue (France). Rhizosphere 3, 112–122. doi: 10.1016/j.rhisph.2017.03.002
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). Fast tree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Qin, Y., Druzhinina, I. S., Pan, X., and Yuan, Z. (2016). Microbially mediated plant salt tolerance and microbiome-based solutions for saline agriculture. Biotechnol. Adv. 34, 1245–1259. doi: 10.1016/j.biotechadv.2016.08.005
Redman, R. S., Sheehan, K. B., Stout, R. G., Rodriguez, R. J., and Henson, J. M. (2002). Thermotolerance generated by plant/fungal symbiosis. Science 298:1581. doi: 10.1126/science.1072191
Reinhold-Hurek, B., Bünger, W., Burbano, C. S., Sabale, M., and Hurek, T. (2015). Roots shaping their microbiome: global hotspots for microbial activity. Annu. Rev. Phytopathol. 53, 403–424. doi: 10.1146/annurev-phyto-082712-102342
Schlaeppi, K., Dombrowski, N., Oter, R. G., Ver Loren van Themaat, E., and Schulze-Lefert, P. (2014). Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc. Natl. Acad. Sci. U.S.A. 111, 585–592. doi: 10.1073/pnas.1321597111
Shi, Y. W., Lou, K., Li, C., Wang, L., Zhao, Z. Y., Zhao, S., et al. (2015). Illumina-based analysis of bacterial diversity related to halophytes Salicornia europaea and Sueada aralocaspica. J. Microbiol. 53, 678–685. doi: 10.1007/s12275-015-5080-x
Sorty, A. M., Meena, K. K., Choudhary, K., Bitla, U. M., Minhas, P. S., and Krishnani, K. K. (2016). Effect of plant growth promoting bacteria associated with halophytic weed (Psoralea corylifolia L) on germination and seedling growth of wheat under saline conditions. Appl. Biochem. Biotechnol. 180, 872–882. doi: 10.1007/s12010-016-2139-z
Szymańska, S., Płociniczak, T., Piotrowska-Seget, Z., Złoch, M., Ruppel, S., and Hrynkiewicz, K. (2016). Metabolic potential and community structure of endophytic and rhizosphere bacteria associated with the roots of the halophyte Aster tripolium L. Microbiol. Res. 182, 68–79. doi: 10.1016/j.micres.2015.09.007
Thijs, S., Op De Beeck, M., Beckers, B., Truyens, S., Stevens, V., Van Hamme, J. D., et al. (2017). Comparative evaluation of four bacteria-specific primer pairs for 16S rRNA gene surveys. Front. Microbiol. 8:494. doi: 10.3389/fmicb.2017.00494
Tian, B. Y., Cao, Y., and Zhang, K. Q. (2015). Metagenomic insights into communities, functions of endophytes, and their associates with infection by root-knot nematode, Meloidogyne incognita, in tomato roots. Sci. Rep. 5:17087. doi: 10.1038/srep17087
Tian, X. Y., and Zhang, C. S. (2017). Illumina-based analysis of endophytic and rhizosphere bacterial diversity of the coastal halophyte Messerschmidia sibirica. Front. Microbiol. 8:2288. doi: 10.3389/fmicb.2017.02288
Toledo, G., Bashan, Y., and Soeldner, A. (1995). In vitro colonization and increase in nitrogen fixation of seedling roots of black mangrove inoculated by a filamentous cyanobacteria. Can. J. Microbiol. 41, 1012–1020. doi: 10.1139/m95-140
Vik, U., Logares, R., Blaalid, R., Halvorsen, R., Carlsen, T., Bakke, I., et al. (2013). Different bacterial communities in ectomycorrhizae and surrounding soil. Sci. Rep. 3:3471. doi: 10.1038/srep03471
Wei, Z., Hu, X., Li, X., Zhang, Y., Jiang, L., Li, J., et al. (2017). The rhizospheric microbial community structure and diversity of deciduous and evergreen forests in taihu lake area, China. PLoS One 12:e0174411. doi: 10.1371/journal.pone.0174411
Yamada, T., Sekiguchi, Y., Imachi, H., Kamagata, Y., Ohashi, A., and Harada, H. (2005). Diversity localization, and physiological properties of filamentous microbes belonging to Chloroflexi sub phylum in mesophilic and thermophilic methanogenic sludge granules. Appl. Environ. Microbiol. 71, 7493–7503. doi: 10.1128/AEM.71.11.7493-7503.2005
Yamamoto, K., Oguri, S., Chiba, S., and Momonoki, Y. S. (2009). Molecular cloning of acetylcholinesterase gene from Salicornia europaea L. Plant Signal. Behav. 4, 361–366. doi: 10.4161/psb.4.5.8360
Yuan, Z., Druzhinina, I. S., Labbé, J., Redman, R., Qin, Y., Rodriguez, R., et al. (2016). Specialized microbiome of a halophyte and its role in helping non-host plants to withstand salinity. Sci. Rep. 6:32467. doi: 10.1038/srep32467
Zabat, M. A., Sano, W. H., Cabral, D. J., Wurster, J. I., and Belenky, P. (2018). The impact of vegan production on the kimchi microbiome. Food Microbiol. 74, 171–178. doi: 10.1016/j.fm.2018.04.001
Zhao, S., Zhou, N., Zhao, Z. Y., Zhang, K., Wu, G. H., and Tian, C. Y. (2016). Isolation of endophytic plant growth-promoting bacteria associated with the halophyte Salicornia europaea and evaluation of their promoting activity under salt stress. Curr. Microbiol. 73, 574–581. doi: 10.1007/s00284-016-1096-7
Keywords: bacterial diversity, Glaux maritima, Salicornia europaea, halophyte, endophytic bacteria, rhizosphere bacteria, 16S rRNA, metagenome
Citation: Yamamoto K, Shiwa Y, Ishige T, Sakamoto H, Tanaka K, Uchino M, Tanaka N, Oguri S, Saitoh H and Tsushima S (2018) Bacterial Diversity Associated With the Rhizosphere and Endosphere of Two Halophytes: Glaux maritima and Salicornia europaea. Front. Microbiol. 9:2878. doi: 10.3389/fmicb.2018.02878
Received: 21 May 2018; Accepted: 09 November 2018;
Published: 28 November 2018.
Edited by:
Suhelen Egan, University of New South Wales, AustraliaReviewed by:
Migun Shakya, Dartmouth College, United StatesDilfuza Egamberdieva, Leibniz-Zentrum für Agrarlandschaftsforschung (ZALF), Germany
Copyright © 2018 Yamamoto, Shiwa, Ishige, Sakamoto, Tanaka, Uchino, Tanaka, Oguri, Saitoh and Tsushima. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kosuke Yamamoto, a3kyMDA2ODNAbm9kYWkuYWMuanA=
 Kosuke Yamamoto
Kosuke Yamamoto Yuh Shiwa1,2
Yuh Shiwa1,2 Taichiro Ishige
Taichiro Ishige Hikaru Sakamoto
Hikaru Sakamoto Keisuke Tanaka
Keisuke Tanaka