- 1Laboratory for Molecular Microbiology, Institute of Molecular Genetics and Genetic Engineering, University of Belgrade, Belgrade, Serbia
- 2Faculty of Biology, University of Belgrade, Belgrade, Serbia
- 3Teagasc Food Research Centre, Moorepark, Fermoy, Ireland
- 4APC Microbiome Ireland, University College Cork, Cork, Ireland
The gene cluster responsible for the production of the aureocin A53-like bacteriocin, lactolisterin BU, is located on plasmid pBU6 in Lactococcus lactis subsp. lactis BGBU1-4. Heterologous expression of pBU6 confirmed that production and limited immunity to lactolisterin BU were provided by the plasmid. Comparative analysis of aureocin A53-like operons revealed that the structural genes shared a low level of identity, while other genes were without homology, indicating a different origin. Subcloning and expression of genes located downstream of the structural gene, lliBU, revealed that the lactolisterin BU cluster consists of four genes: the structural gene lliBU, the abcT gene encoding an ABC transporter, the accL gene encoding an accessory protein and the immL gene which provides limited immunity to lactolisterin BU. Reverse transcription analysis revealed that all genes were transcribed as one polycistronic mRNA. Attempts to split the lactolisterin BU operon, even when both parts were under control of the PlliBU promoter, were unsuccessful indicating that expression of lactolisterin BU is probably precisely regulated at the translational level by translational coupling and is possible only when all genes of the operon are in cis constellation. Two ρ-independent transcription terminators were detected in the lactolisterin BU operon: the first in the intergenic region of the lliBU and abcT genes and the second at the end of operon. Deletion of the second transcription terminator did not influence production of the bacteriocin in lactococci.
Introduction
Ribosomal synthesis of antimicrobial peptides or peptide complexes, known as bacteriocins, presents a successful strategy for the reduction of microbial competitors to obtain more nutrients and living space in the environment (Cotter et al., 2013). They can be active against closely related bacterial species or different bacterial genera, and the producer protects itself by synthesis of specific immunity proteins (Kjos et al., 2014). Bacteriocins produced by lactic acid bacteria (LAB) are generally regarded as natural and suitable for consumption as they have been present in many foods and beverages since ancient times (Cleveland et al., 2001). Numerous studies reported that bacteriocins may be applied in the food industry to extend food preservation time (Cotter et al., 2005; Yang et al., 2014; Ahmad et al., 2017). A number of studies consider potential clinical applications of bacteriocins for maintenance of human health as a part of anti-infection or even are able to cause apoptotic and necrotic death of cancer cells in vitro (Lagos et al., 2009; Yang et al., 2014; Ahmad et al., 2017; Baindara et al., 2017). In addition to being administered as food supplements, some bacteriocins may be produced at the site of infection by probiotic bacteria (Gotteland et al., 2008).
Bacteriocins differ in molecular mass, structure, biochemical and genetic characteristics, modes of action and target cell receptors. Based on these criteria, bacteriocins from LAB are generally classified into two main classes: Class I bacteriocins (lantibiotics) contain unusual amino acids such as lanthionine and dehydrated amino acids as a result of post-translational modifications and Class II bacteriocins consisting of unmodified or peptides with minor modifications. Furthermore, Class II bacteriocins are subdivided into four subclasses: pediocin-like bacteriocins (class IIa), two-peptide bacteriocins (class IIb), cyclic bacteriocins (IIc), and linear non-pediocin-like bacteriocins (class IId) (Cotter et al., 2013). Recently a new modified classification of LAB bacteriocins was proposed (Alvarez-Sieiro et al., 2016) with two main differences: (a) the introduction of class III which includes thermo-labile bacteriolysins and non-lytic bacteriocins with a molecular mass greater than 10 kDa and (b) leaderless bacteriocins are classified as subclass IIc. Class II bacteriocins, in general, are small (37–58 amino acids) cationic peptides with pIs varying from 8 to 11. They contain a relatively high number of small amino acids such as glycine and alanine, giving them a high degree of conformational freedom (Kaiser and Montville, 1996). Most bacteriocins are synthesized as a pro-peptide with an N-terminal leader sequence, which is cleaved by a dedicated ABC transporter or the Sec system during export from cells. However, there is a group of so-called leaderless bacteriocins, which are synthesized without an N-terminal leader sequence and the mechanism behind their export is still unknown. Leaderless bacteriocins are often relatively small (30–50 amino acids) and without modifications (Ovchinnikov et al., 2017).
Bacteriocin genetic systems can be located on chromosomes, plasmids or conjugative transposons (Engelke et al., 1992; Rauch and De Vos, 1992; De Vos et al., 1995; Kojic et al., 2005). Class II bacteriocins are organized in operons consisting of at least four genes: a structural gene, an immunity gene, a transporter gene and an accessory protein gene (Nes et al., 1996; Mesa-Pereira et al., 2017) or in gene clusters. The structural bacteriocin gene encodes a pre-form of the bacteriocin containing an N-terminal leader sequence (except leaderless bacteriocins). Each bacteriocin has its own protein conferring immunity encoded by an immunity gene located next to or close downstream of the bacteriocin structural gene. The ABC transporter protein is responsible for transport of the bacteriocin from the cytoplasm to the outside of the membrane and for extracellular activation by cleavage of the leader sequence. An additional gene encoding an accessory protein (approximately 470 amino acids) is usually located next to the ABC transporter and is apparently required for the ABC-transporter-dependent translocation process (Nes et al., 1996). Expression of bacteriocin genes is usually regulated by external induction factors, in most cases small peptides secreted by the producer strain itself or by its competitors for the ecological resources. The production of class II bacteriocins is transcriptionally regulated by a three component quorum sensing system, which consists of an induction factor, a membrane-associate protein histidine kinase, and response regulators (Diep et al., 2003). Recently, it has been described for the first time that the expression of LsbB, a leaderless bacteriocin, is regulated by a transcription terminator sequence located downstream of the structural gene (Uzelac et al., 2015). However, in some cases, bacteriocin production depends on environmental conditions such as temperature, pH and free peptide concentration.
In this study, we investigated the function of the hypothetical genes within the lactolisterin BU gene cluster of Lactococcus lactis subsp. lactis BGBU1-4. Previously, it was shown that strain BGBU1-4 produces a novel, leaderless bacteriocin, lactolisterin BU, with strong antimicrobial activity against many species of Gram-positive bacteria, including important food spoilage and food-borne pathogens such as Listeria monocytogenes, Staphylococcus aureus, Bacillus sp. and streptococci. The production of lactolisterin BU is related to the smallest plasmid pBU6 of 6.2 kb, which contains nine open reading frames (ORFs): repB, lliBU, abcT, hyp1, hyp2, hyp3, mobC, relM, and rnaY (Lozo et al., 2017). An identity (BLAST) search within the lactolisterin BU operon recognized the product of the abcT gene as a sugar transporter, while other genes were designated as hypothetical. The aim of this work was to determine the function of the hypothetical genes (hyp1, hyp2, and hyp3) and their involvement in bacteriocin activity.
Materials and Methods
Bacterial Strains and Growth Conditions
The strains, their derivatives and plasmids used in this study are listed in Table 1. Lactococcus lactis strains were grown in M17 medium (Merck GmbH, Darmstadt, Germany) supplemented with 0.5% glucose (GM17) at 30°C. Escherichia coli strains (DH5α and EC101) were grown in Luria Bertani broth (LB) at 37°C with aeration. To each medium, agar (1.5%; Torlak, Belgrade, Serbia) was added for use as a solid medium. The following antibiotic concentrations were used: ampicillin, 100 μg/ml (E. coli); erythromycin, 10 μg/ml (lactococci) and 300 μg/ml (E. coli); chloramphenicol, 10 μg/ml (lactococci) and 35 μg/ml (E. coli). The 5-bromo-4-chloro-3-indolyl-β-D-galacto-pyranoside (X-Gal) (Fermentas, Vilnius, Lithuania) was added to LB medium plates for blue/white color screening of colonies with cloned fragments at a final concentration of 80 μg/ml (lactococci) and 40 μg/ml (E. coli). Ortho-nitrophenyl-β-galactoside (ONPG) was used for the β-galactosidase assay (Sigma Chemical Co., St. Louis, MO, United States).
All strains carrying constructs were stored in growth medium containing 15% glycerol (Sigma Chemie GmbH, Deisenhofen, Germany) at −80°C.
DNA Manipulations
For plasmid isolation from E. coli a GeneJET Plasmid Miniprep kit was used according to the manufacturer’s recommendations (Thermo Fisher Scientific, Waltham, MA, United States). Plasmids from lactococci were isolated by a modification of the method described by O’Sullivan and Klaenhammer (1993). Digestion with restriction enzymes was conducted according to the supplier’s instructions (Thermo Fisher Scientific, Waltham, MA, United States). The DNA fragments from agarose gels were purified using QIAqick Gel extraction kit as described by the manufacturer (Qiagen, Hilden, Germany). DNA was ligated with T4 DNA ligase according to the manufacturer’s recommendations (Agilent Technologies, United States).
PlatinumTM Taq DNA Polymerase High Fidelity (Thermo Fisher Scientific, Waltham, MA, United States) was used to amplify DNA fragments using a GeneAmp PCR System 2700 thermal cycler (Applied Biosystems, Foster City, CA, United States). The PCR programs consisted of initial denaturation (5 min at 96°C), 30 cycles of denaturation (30 s at 96°C), annealing (30 s at the appropriate temperature (Table 2) and polymerization (2 min, 1 min or 30 s at 68°C; depending on the length of the amplified fragment), and an additional extension step of 5 min at 68°C. Sets of specific primers used in this study are listed in Table 2. PCR products were purified with a Thermo Scientific PCR Purification Kit according to the manufacturer’s protocol (Thermo Fisher Scientific, Waltham, MA, United States). For cloning PCR products a pBluescriptT/A (Uzelac et al., 2015) or pAZIL vector were used.
Construction of Derivatives
The SacI/PvuII fragment from pBU6, containing the lactolisterin BU operon (lliBU, abcT, hyp1 and hyp2), hyp3 and part of mobC was cloned into pAZIL vector predigested with SacI/SmaI restriction enzymes giving construct pAZIL-SP (Figure 1B and Table 1). The fragment SacI/BglII from pBU6, containing just the lactolisterin BU operon (lliBU, abcT, hyp1 and hyp2) was cloned into pAZIL vector predigested with SacI/BamHI restriction enzymes giving construct pAZIL-SB (Figure 1B and Table 1). The SacI/ScaI fragment from pBU6, containing lliBU and abcT was cloned into pAZIL vector predigested with SacI/SmaI restriction enzymes giving construct pAZIL-SS (Figure 1B and Table 1). In addition, (i) PvuII fragment from pBU6 (containing whole pBU6 plasmid; PvuII restriction site cut of the start of mobC) was cloned into pAZIL vector predigested with SmaI restriction enzyme. Obtained constructs were named pAZIL-pBU6P-1 and pAZIL-pBU6P-5 (both possible orientations) (Figure 1B and Table 1); (ii) BglII fragment from pBU6 (containing whole pBU6 plasmid; BglII restriction site is within the intergenic region of the hyp2 and hyp3 genes) was cloned into pAZIL vector predigested with BamHI restriction enzyme. Obtained constructs, were named pAZIL-pBU6B-2 and pAZIL-pBU6B-4 (both possible orientations) (Figure 1B and Table 1). MG7284 was transformed with each of the constructs and the obtained derivatives were used as bacteriocin producers in the agar well diffusion assay.
The lactolisterin BU promoter, PlliBU, was amplified by PCR using primers PBU1-4-Fw and PBU1-4-Rev where pBU6 was used as a template (Table 2). The obtained fragment was cloned into pBluescriptT/A vector giving a construct named pBluescriptT/A-PlliBU (Table 1). The hyp1 gene was amplified from pBU6 using the hyp1PstI-Fw and hyp1XbaI-Rev primers (Table 2) and cloned into pBluescriptT/A vector (giving the construct pBluescriptT/A-h1). The XbaI fragment was transferred, downstream of the PlliBU promoter, into the pBluescriptT/A-PlliBU predigested with XbaI restriction enzyme, giving the construct pBluescriptT/A-PlliBU-h1. The SacI/EcoRV fragment from pBluescriptT/A-PlliBU-h1 was transferred into pNZ8150 vector predigested with SacI/ScaI restriction enzymes giving the construct pNZ8150-h1 (Figure 1D and Table 1). The hyp2 gene was amplified from pBU6 plasmid using the hyp2PstI-Fw and hyp2XbaI-Rev primers (Table 2) and cloned into pBluescriptT/A vector giving the construct pBluescriptT/A-h2. The XbaI fragment was transferred, downstream of the PlliBU promoter, into the pBluescriptT/A-PlliBU predigested with XbaI restriction enzyme, giving the construct pBluescriptT/A-PlliBU-h2. The SacI/EcoRV fragment from pBluescriptT/A-PlliBU-h2 was transferred into pNZ8150 vector predigested with SacI/ScaI restriction enzymes giving the construct pNZ8150-h2 (Figure 1D and Table 1). A fragment containing the hyp1 and hyp2 genes was amplified from pBU6 plasmid using the hyp1PstI-Fw and hyp2XbaI-Rev primers (Table 2) and cloned into pBluescriptT/A vector giving the construct pBluescriptT/A-h1+h2. The XbaI fragment was transferred, downstream of the PlliBU promoter, into the pBluescriptT/A-PlliBU predigested with XbaI restriction enzyme, giving the construct pBluescriptT/A-PlliBU-h1+h2. The SacI/EcoRV fragment from pBluescriptT/A-PlliBU-h1+h2 was transferred into pNZ8150 vector predigested with SacI/ScaI restriction enzymes giving the construct pNZ8150-h1+h2 (Figure 1D and Table 1). The hyp3 gene (with its promoter – Phyp3) was amplified by PCR using primers hyp3BglII-Fw and hyp3-Rev where pBU6 was used as a template (Table 2) and cloned into pBluescriptT/A vector, giving the construct pBluescriptT/A-h3. The fragment from pBluescriptT/A-h3 was transferred into pNZ8150 as PstI/XbaI giving the construct pNZ8150-h3 (Table 1). Fragment containing the hyp3, mobC, relM and rnaY genes was amplified by PCR using primers hyp3BglII-Fw and rnaY-Rev where pBU6 was used as a template (Table 2) and cloned into pBluescriptT/A vector, giving the construct pBluescriptT/A-h3mobCrelMrnaY. The fragment from pBluescriptT/A-h3mobCrelMrnaY was transferred into pNZ8150 as PstI/XbaI fragment giving the construct pNZ8150-h3+mobC+relM+rnaY (Table 1). MG7284 and MG7284/pAZIL-SS were transformed with each of the constructs (pNZ8150-h1, pNZ8150-h2, pNZ8150-h1+h2, pNZ8150-h3 and pNZ8150-h3+mobC+relM+rnaY) and the obtained derivatives (listed in Table 1) were used as bacteriocin producers in the agar well diffusion assay. MG7284 and MG7284/pAZIL-SB were transformed with the following constructs (pNZ8150-h3 and pNZ8150-h3+mobC+relM+rnaY) and the obtained derivatives (listed in Table 1) were used as bacteriocin producers in the agar well diffusion assay. MG7284/pAZIL-SP was transformed with pNZ8150-h3 and the obtained derivative (Table 1) was used for further experiments.

FIGURE 1. Schematic representation of the construction of appropriate constructs. (A,F) Linear gene map of the pBU6 plasmid carrying the lactolisterin BU cluster; (B–E) the scheme of construction of clones used for analysis of influence of different genes/regions on the expression of the lliBU gene. Relevant restriction sites are indicated. The size and orientation of predicted ORFs are indicated by arrows. +/−, presence/absence of Trans (transformants); Bac, bacteriocin activity; Imm, expressed limited immunity.
The construct pAZIL-SB was partially digested with EcoRI to remove a fragment of 382 bp (without PlliBU and first part of the lliBU gene) giving the construct pAZIL-EEB. In order to obtain pNZ8150-EEB, XbaI fragment from pAZIL-EEB (carrying all lactolisterin BU operon with non-functional lliBU gene) was recloned into pNZ8150 vector. MG7284 and MG7284/pAZIL-SS were transformed with pNZ8150-EEB and the obtained derivatives (Table 1) were used for further experiments.
A section of the operon from the SacI restriction site (upstream of PlliBU) to the end of hyp1 gene was amplified by PCR using primers SacI_before_prom-Fw and hyp1-Rev where pBU6 was used as a template (Table 2) and cloned into pAZIL vector (predigested with SmaI restriction enzyme), giving the construct pAZIL-S-h1 (Figure 1C and Table 1). The constructs pAZIL-S-h1+h2-TT2, pAZIL-S-h1+h2+1/2TT2 and pAZIL-S-h1+h2+TT2 (Figure 1C and Table 1) were constructed as described above using the appropriate primers (SacI_before_prom-Fw/before_the_loop-hyp2-Rev, SacI_before_prom-Fw/middle_the_loop_hyp2-Rev and SacI_before_prom-Fw/after_the_loop-hyp2-Rev, respectively) (Table 2). MG7284 was transformed with each of the following constructs (pAZIL-S-h1, pAZIL-S-h1+h2-TT2, pAZIL-S-h1+h2+1/2TT2 and pAZIL-S-h1+h2+TT2) and the obtained derivatives (listed in Table 1) were used as bacteriocin producers in the agar well diffusion assay.
For analysis of the PlliBU promoter activity, PlliBU was fused to lacZ1 gene in pNZ8150-lacZ1 vector. The PlliBU promoter transcriptional fusion was constructed as follows; PCR amplified fragments (primers are listed in Table 2) of promoters PlliBU and PlcnB (promoter of lcnB gene used as a control) were first cloned to pBluescriptT/A giving constructs (pBluescriptT/A-PlliBU and pBluescriptT/A-PlcnB) that were recloned as EcoRI-BamHI fragments to pNZ8150-lacZ1 (Table 1). The constructs, designated pNZ8150-PlliBU-lacZ1 and pNZ8150-PlcnB-lacZ1, were used for further experiments. MG7284 was transformed with constructs pNZ8150-PlliBU-lacZ1 and pNZ8150-PlcnB-lacZ1 separately, resulting in the strains MG7284/pNZ8150-PlliBU-lacZ1 and MG7284/pNZ8150-PlcnB-lacZ1. The resulting transformants were screened on solid GM17 medium supplemented with chloramphenicol and X-gal.
DNA Sequencing and Sequence Analysis
Cloned fragments of all relevant constructs were sequenced by Macrogen Sequencing Service (Macrogen Europe, Amsterdam, Netherlands). The DNA Strider 1.4f7 program was used for sequence analysis and ORF prediction.
Transformation of E. coli and L. lactis subsp. cremoris MG7284
Standard heat-shock transformation was used for plasmid transformation of E. coli competent cells (Hanahan, 1983). L. lactis subsp. cremoris MG7284 was transformed with plasmid constructs using a method described by Holo and Nes (1989) with modifications. To obtain competent cells, the overnight culture was diluted 100-fold in GM17 medium supplemented with 1% glycine. After growth at 30°C to an optical density at 600 nm OD600 = 0.6 to 1.0, the cells were harvested by centrifugation at room temperature at 5,000 × g. Following two washes in sterile MilliQ water, the cells were suspended in 1/100 culture volume of sterile MilliQ water. Aliquots (200 μl) of cells were mixed with 5–10 μl of DNA dissolved in 10 mM Tris-hydrochloride (pH 7.5), kept for 5 min at room temperature and then transferred to an electroporation cuvette (2-mm electrode gap) and exposed to a single electrical pulse. The pulse was delivered by a Gene Pulser Eporator (Eppendorf, Hamburg, Germany) set at 25 μF and normally at 2.0 kV. Immediately following the discharge 800 μl of GM17 medium was added to suspensions. After regeneration for 2 h at 30°C appropriate dilutions were spread on selective GM17 plates. Transformants were visible after 1 or 2 days of incubation at 30°C.
For clonal confirmation, pulse-field gel electrophoresis (PFGE) was performed, as described previously by Kojic et al. (2006).
Bacteriocin Activity Assay
For detection of bacteriocin activity, an agar-well diffusion assay was performed as described previously by Kojic et al. (1991). For testing the level of sensitivity to lactolisterin BU spot on the lawn assay was used; double serial dilutions of purified lactolisterin BU were spotted (5 μl) on the surface of the top agar inoculated with each indicator strain. After 24 h of incubation at 30°C plates were examined for the presence of inhibition zones. A clear zone of inhibition was taken as evidence of bacteriocin production. For monitoring of time dependent production of bacteriocin by different constructs all cultures were started with 1% inoculum from overnight culture and activity of cell free supernatant was tested after different time intervals.
Antimicrobial activity of cytoplasmic fraction was tested in the following way: cells from 50 ml of overnight cultures of bacteriocin producers (BGBU1-4 and MG7284/pAZIL-SB), MG7284/pAZIL-SS and MG7284/pAZIL as negative control were harvested by centrifugation (10 min 5,000 × g), washed with 10 ml of PP buffer (0.5 M sucrose, 40 mM NH4-acetate, 10 mM Mg-acetate, pH 7), treated with lysozyme (4 mg/ml in PP buffer) 30 min at 37°C to obtain protoplasts. Protoplasts were gently washed with PP buffer and diluted to obtain equal number of cells. 1 ml of each sample was sonicated on ice for different time period (20, 40, and 60 s) with intervals of 10 s per every 20 s of treatment, to disrupt membrane. Cell debris was removed by centrifugation (5 min 20,000 × g) and supernatant containing cytoplasmic proteins was used in bacteriocin activity assay. Also cell debris was resuspended in PP buffer and used as negative control in bacteriocin activity assay.
The bacteriocin activity assay was performed at least in two independent repetitions.
Enzymatic Activity: β-Galactosidase Assay
β-Galactosidase activity was determined essentially as described by Miller (1972) with the modification introduced by Uzelac et al. (2015). The resulting derivative MG7284/pNZ8150-PlliBU-lacZ1 and appropriate controls (MG7284/pNZ8150-PlcnB-lacZ1 and MG7284/pNZ8150-lacZ1) were analyzed with three separate bacterial cultures in two independent experiments.
RT-PCR
Total mRNA was isolated from Lactococcus lactis subsp. lactis BGBU1-4 at late logarithmic phase of growth using RNeasy Mini Kit as described by the manufacturer (Qiagen, Hilden, Germany). First strand cDNA synthesis with reverse transcriptase was carried out as previously published by Uzelac et al. (2015). The obtained cDNA was subsequently amplified by PCR using appropriate pairs of primers (Table 2). The size of the obtained PCR products was checked on a 1% agarose gel. Reproducibility of obtained PCR products was confirmed in two independent experiments.
Results and Discussion
Lactolisterin BU production is plasmid encoded. In a previous study, it was reported that Lactococcus lactis subsp. lactis BGBU1-4 contains a plasmid, pBU6, carrying the structural lactolisterin BU operon (Lozo et al., 2017). Based on comparisons with known bacteriocin clusters, it was hypothesized that the lactolisterin BU bacteriocin operon was organized into a four gene operon-like structure consisting of (lliBU) the structural gene; (abcT) an ABC transporter; and hyp1 and hyp2 genes to which the exact function has not been attributed.
Comparative Analysis of Aureocin A53-Like Clusters
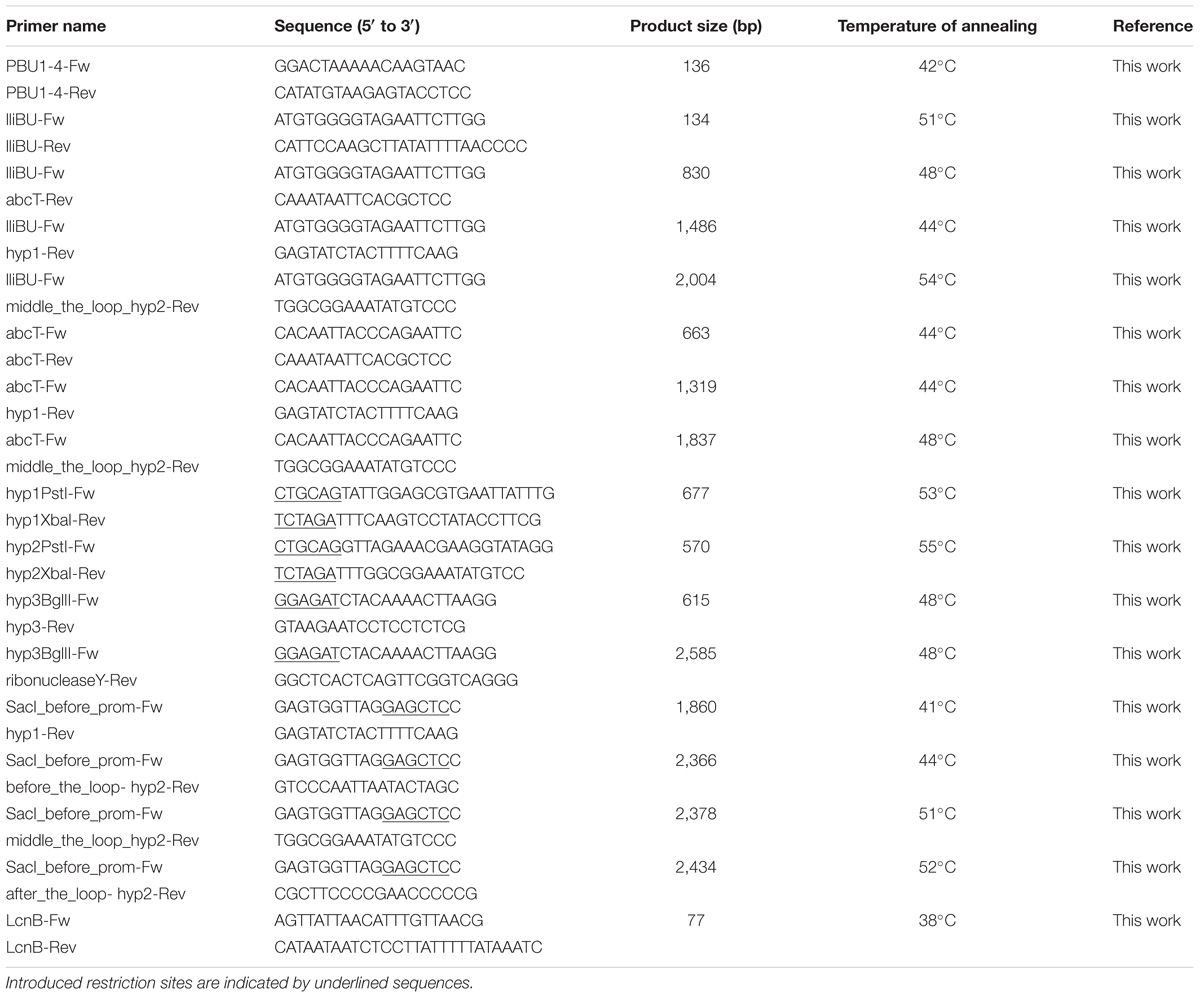
Since lactolisterin BU showed the highest identity with bacteriocins belonging to aureocin A53-like group (Lozo et al., 2017), the possibility of a common origin was considered. To determine the similarity of the organization of gene clusters among aureocin A53-bacteriocins, lactolisterin BU operon was compared with the three most similar aureocin A53-like bacteriocin clusters: BHT-B bacteriocin from Streptococcus ratti (GenBank: DQ145753.1), lacticin Z (GenBank: AB740019.1) and lacticin Q (GenBank: AB712393.1) from Lactococcus lactis QU 5. It was found that the lactolisterin BU operon has a unique organization that is distinct from the other aureocin A53-like clusters (Figure 2). Bearing in mind that the aureocin A53-like bacteriocin clusters do not show similarity, it can be assumed that the cluster does not have a common origin. Thus, comparative analysis with known aureocin A53-like clusters, did not help with defining/predicting the exact function of hypothetical genes of the lactolisterin BU cluster.
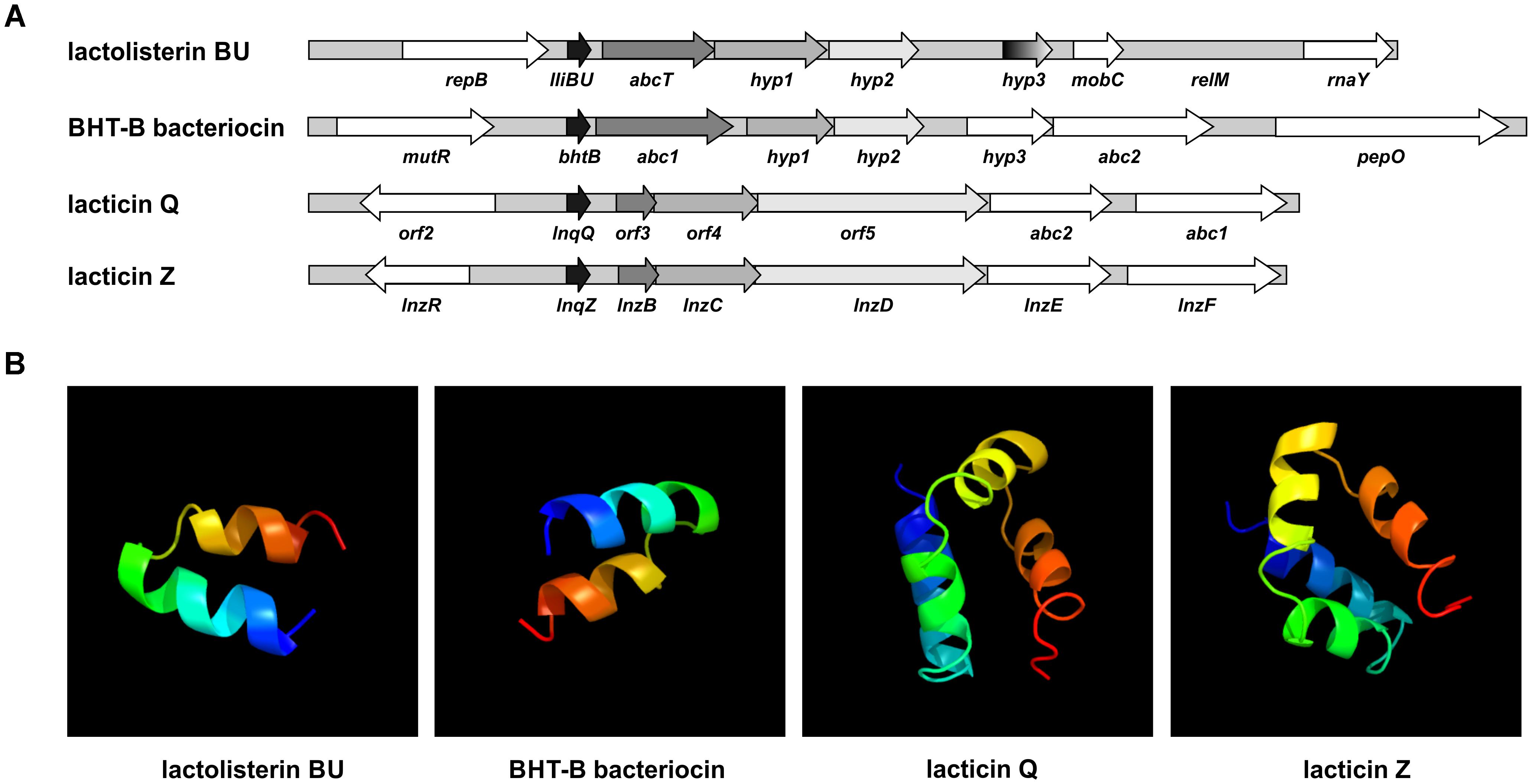
FIGURE 2. Comparative analysis of aureocin A53-like clusters. (A) Schematic presentation of different aureocin A53-like bacteriocins gene clusters. (B) Predicted 3D peptide structures for different aureocin A53-like bacteriocins using Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2).
In silico Analysis of Genes in Lactolisterin BU Cluster
Downstream of the lactolisterin BU (lliBU) structural gene are located an abcT gene encoding an ABC transporter ATP-binding protein (domain architecture ID 11438412) and three undefined hyp1, hyp2, and hyp3 genes, upstream of the mobC gene, predicted to be involved in plasmid mobilization. In silico analyses of these three hypothetical genes revealed that hyp1 is predicted to encode a putative sugar ABC transporter permease of 212 amino acid residues, but that hyp2 and hyp3 encode proteins of 169 and 92 amino acids that share low level similarity with uncharacterized proteins from LAB.
TMpred program prediction1 analysis of those three proteins revealed the presence of transmembrane helices (Hyp1 = 6; Hyp2 = 5 and Hyp3 = 2), indicating a cytoplasmic membrane location for these proteins. In addition, it was found that all three proteins have high pI values (Hyp1 = 10.52; Hyp2 = 9.77; and Hyp3 = 11.05) and are rich in the amino acid leucine (20.5; 11.8; and 15.5%, respectively).
In the lactolisterin BU cluster, two genes were predicted to encode possible transporter/enzymes involved in sugar catabolism, indicating that the lactolisterin BU peptide determinant was somehow introduced upstream of a sugar-related cluster and uses these proteins for its own transport.
The absence or existence of small intergenic regions between the genes of lactolisterin BU operon indicated the synthesis of a single polycistronic RNA (the intergenic region between the lliBU and abcT genes is 64 nucleotides; the abcT and hyp1 genes are overlapped within 8 nucleotides; the intergenic region between the hyp1 and hyp2 genes is only 3 nucleotides).
In silico analysis of the upstream region of lactolisterin BU operon using the program Bacterial Promoter Prediction2 revealed the presence of five putative promoter structures of which (TGAATATTTGCAATTTTTATATTAAATT) had the highest score (1) and, thus the greatest likelihood of being active.
Analysis to detect the presence of transcriptional terminators (TT) using3 revealed the presence of two apparent transcriptional terminators in the lactolisterin BU operon: TT1 within the intergenic region of the lliBU and abcT genes (GGTTAAAATATAATCTCGGAATGCATTAATATGCATCCCGAGATTATATTTTAACC) and second a TT2 after the hyp2 gene at the end of operon (TTTCCGCCAAAAAAAGTAAGCTATTCGCTTACTTTTTTTGGCGGAAA). The presence of two transcription terminators in the lactolisterin BU operon also pointed to the possibility that the first TT1 was introduced together with the bacteriocin structural gene during insertion into the sugar operon. We hypothesize that it was later modified in order to provide a precise regulation of expression by transcriptional attenuation.
Upstream of the lliBU, abcT, hyp1 and hyp2 at a variable distance from the ATG translation start codon non-perfect ribosomal binding sequences (RBSs) were detected (Table 3). These RBS could be involved in translational regulation of expression of each gene in the cluster.
Transcriptional Analysis of the Lactolisterin BU Operon
In order to test if all genes (lliBU, abcT, hyp1, and hyp2) for lactolisterin BU activity form an operon (transcribed as one polycistronic mRNA), their expression was investigated by RT-PCR using one forward primer from the lliBU gene and different reverse primers positioned in the lliBU, abcT, hyp1 or hyp2 genes. Due to the presence of a strong transcriptional terminator immediately after the lliBU gene, an additional forward primer from the abcT gene was designed and used for transcriptional analysis. The results of RT-PCR analysis unambiguously showed that all genes of the lactolisterin BU cluster were transcribed as a single polycistronic mRNA (Figure 3). This was in line with the previously characterized bacteriocin cluster for enterocin AS-48 that is expressed by two polycistronic mRNA in coordinate expression (Diaz et al., 2003).
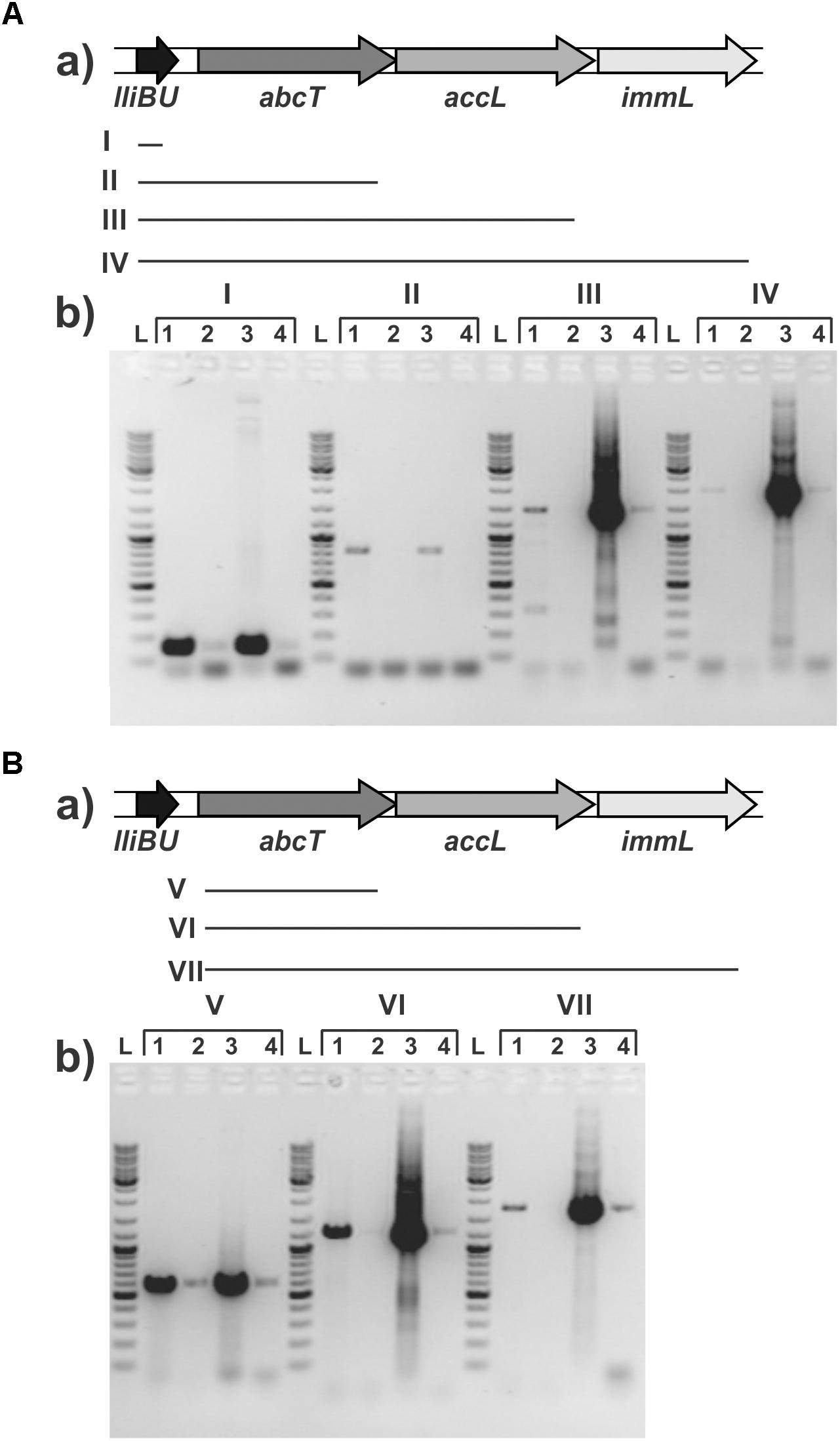
FIGURE 3. Transcriptional analysis of the lactolisterin BU operon. Amplicons obtained after RT-PCR using (A) lliBU-Fw and (I) lliBU-Rev; (II) abcT-Rev; (III) accBU-Rev; (IV) immBU-Rev; (B) abcT-Fw and (V) abcT-Rev; (VI) accBU-Rev; (VII) immBU-Rev primers; (a) schematic presentation of primer positions and length of PCR products in lactolisterin BU operon; (b) electrophoresis of obtained PCR products on 1% agarose gel; L – GeneRuler DNA Ladder Mix 0.1–10 kb (Thermo Fisher Scientific); 1 – cDNA of BGBU1-4; 2 – RT-PCR negative control (without reverse transcriptase); 3 – total DNA of BGBU1-4; 4 – PCR negative control (without DNA of BGBU1-4).
Considering that production of lactolisterin BU in the strain L. lactis subsp. lactis BGBU1-4 is balanced at low level since higher production of bacteriocin cause the death of the producer (Lozo et al., 2017), we wanted to determine the level of expression at the transcriptional level through the construction of the promoter PlliBU-lacZ fusion and the analysis of the activity. The promoter PlcnB of the lactococcin B gene was used as a positive control. Results of β-galactosidase activity of transcriptional fusions revealed that promoter PlliBU had activity comparable to promoter PlcnB (the difference in activity between the MG7284/pNZ8150-PlcnB-lacZ1 and MG7284/pNZ8150-PlliBU-lacZ1 did not have statistical significance; 183 ± 21: 169 ± 26 MU, respectively). This result indicates that regulation of expression was more likely to be at the translational level.
Cloning of Lactolisterin BU Operon in the pAZIL Vector
Lactolisterin BU is encoded by a gene cluster located on the smallest plasmid in strain BGBU1-4, pBU6 of 6.2 kb (Figure 1A). To characterize a minimal bacteriocin cluster, different fragments of pBU6 plasmid were cloned in a pAZIL shuttle vector (Figure 1B and Table 1). The smallest fragment providing bacteriocin activity in heterologous strain MG7284 was the 2,552 bp SacI-BglII fragment containing the lliBU, abcT, hyp1 and hyp2 genes (Figures 1B, 4). It was interesting that the construct pAZIL-SB (pAZIL vector containing SacI-BglII fragment) exhibited stronger antimicrobial activity than the wild type strain and transformants of MG7284 with pBU6 plasmid. Construct pAZIL-SP (pAZIL vector containing the SacI-PvuII fragment) showed even better antimicrobial activity (higher zone of inhibition; Figure 4) than a transformant with pAZIL-SB construct indicating the influence of other genes/structures on bacteriocin activity. Plasmid construct pAZIL-SS (pAZIL containing SacI–ScaI fragment of 1,284 bp) carrying only the lliBU and abcT genes did not provide antimicrobial activity, indicating that additional gene(s) were needed for bacteriocin production. In order to prove this conclusion we performed bacteriocin activity assay with cytoplasmic fraction of different strains: bacteriocin producers (BGBU1-4 and MG7284/pAZIL-SB), MG7284/pAZIL-SS and MG7284/pAZIL as negative control. Very low bacteriocin activity (1 mm of inhibition zone) was obtained only with strains that already produce bacteriocin in supernatant (BGBU1-4 and MG7284/pAZIL-SB), which was originated from residual bacteriocin molecules that were not exported (no bacteriocin activity was obtained with cell debris) strongly indicated that no bacteriocin synthesis occurs inside of the cells carrying pAZIL-SS construct, although the structural bacteriocin gene is present and an active promoter. According to both in silico analysis (all four genes are transcribed as polycistronic mRNA) and subcloning results, it can be concluded that the operon is located on the SacI-BglII fragment and consists of four structural genes: lliBU, abcT, hyp1 and hyp2. In addition, the time-dependent production of bacteriocin, that was noted in the parental BGBU1-4 strain (Lozo et al., 2017), was also tested in derivatives MG7284/pAZIL-SP and MG7284/pAZIL-SB. It was observed that derivative MG7284/pAZIL-SB retained time-dependent production of bacteriocin, like the parental strain (although bacteriocin production was higher), while MG7284/pAZIL-SP lost time-dependent production of the bacteriocin (Figure 4A). In addition to the role of specific genes, the influence of the orientation of the fragment in the vector on bacteriocin expression was observed (Figure 4B) indicating that some (promoter or other) sequences from the vector could boost the expression. Thus, it was noticed that depending on the orientation of the same fragment (BglII linearized plasmid pBU6), expression of bacteriocin could drastically differ.
FIGURE 4. Antimicrobial activity of constructed derivatives on Lactococcus lactis subsp. cremoris MG7284. (A) Time dependent antimicrobial activity of MG7284/pAZIL 1 – SP, 2 –SB, 3 – BGBU1-4, 4 – MG7284. (B) Dependence of cluster orientation cloned into pAZIL vector on antimicrobial activity: MG7284/pAZIL 1 – pBU6P-1, 2 – pBU6P-5, 3 – pBU6B-2, 4 – pBU6B-4, 5 – SP, 6 – SB. (C) Bacteriocin activity of lactolisterin BU cluster expressed in trans (a) MG7284/pAZIL 1 – SP, 2 – SB, MG7284/3 – pNZ8150-h1, 4 – pAZIL-SS+pNZ8150-h1, 5 – pNZ8150-h2, 6 – pAZIL-SS+pNZ8150-h2, 7 – pNZ8150-h1+h2, 8 – pAZIL-SS+pNZ8150-h1+h2, 9 – pNZ8150-EEB, 10 – pAZIL-SS+pNZ8150-EEB, 11 – pNZ8150-h3, 12 – pAZIL-SS+pNZ8150-h3, 13 – pAZIL-SS, 14 – MG7284; (b) MG7284/pAZIL 1 – SP, 2 – SP+pNZ8150-h3, 3 – SB, 4 – SB+pNZ8150-h3.
The hyp1 Gene Encodes an Accessory Protein
In order to determine the role of the hyp1 gene, we made additional constructs (Figures 1B–E and Table 1). The most important construct for determining the function of the hyp1 gene was pAZIL-S-h1 (containing a cloned PCR fragment carrying PlliBU, lliBU, abcT, and hyp1 genes). Transformants were successfully obtained in E. coli, but no transformants were obtained in MG7284 after three independent attempts. In control transformations with the same competent cells, but with other constructs (pAZIL-SS, pAZIL-SB, and pAZIL-SP), we obtained between 6 × 104 and 2 × 105/μg of plasmid DNA. By combining the obtained results with respect to the activity of subclones, it can be concluded that the hyp1 gene encoded for the accessory protein and was designed as the accL gene (Figure 1F). The combined evidence is as follows (i) the pAZIL-SS construct did not provide antimicrobial activity because it lacked a key protein for delivery of bacteriocin, an accessory protein, (ii) construct pAZIL-S-h1 contained all genes necessary for synthesis and transport of bacteriocin and provided transformants to produce active bacteriocin that killed themselves due to the absence of the immunity protein. These results explain why in the first case transformants that lacked bacteriocin production capacity were generated and in the second, we were not able to get them at all. The location of an accessory protein downstream from the gene for ABC transporter was previously confirmed for the bacteriocin BacSJ (Kojic et al., 2010) and many other class II bacteriocins (Nes et al., 1996) and that it is essential for export of bacteriocins in addition to the ABC transporter. In lactolisterin BU and BHT-B operons accessory protein resembles permease enzymes which can contribute to streamlining its role in bacteriocin transport and interaction with ABC transporter.
The hyp2 Gene Is Responsible for Immunity to Lactolisterin BU
A construct containing the hyp2 gene was the only one that had the ability to synthesize and increase resistance to lactolisterin BU. In addition, for construct pAZIL-S-h1, which contains the operon without the hyp2 gene, it was not possible to obtain any transformants due to the apparent absence of immunity.
To determine if the hyp2 gene product (alone) can provide immunity to lactolisterin BU, this gene was amplified and cloned under the control of the PlliBU promoter, transferred to pNZ8150 vector and expressed in MG7284. The transformants carrying this construct were exposed to different concentrations (500, 250, 125, 62.5, 31.2, 15.6, 7.8, and 3.9 μg/ml) of purified lactolisterin BU and it was found that all of them were as sensitive as the sensitive strain MG7284 (Table 4). To further investigate the possible role of the hyp2 gene product in providing immunity to lactolisterin BU all constructs were exposed to different concentrations (500, 250, 125, 62.5, 31.2, 15.6, 7.8, and 3.9 μg/ml) of purified lactolisterin BU in a spot on the lawn bacteriocin assay. It was observed that constructs containing a functional hyp2 gene showed approximately four times higher resistance to bacteriocin then “sensitive strains”- i.e., the same or even higher than the parental strain-producer of lactolisterin BU (Figure 5 and Table 4). Since the function of the hyp2 gene was determined to be to provide immunity to lactolisterin BU, the gene was renamed immL (Figure 1F). Notably, a limited level of protection against its own synthesized antimicrobial molecule was noted for aureocin A53-like bacteriocins previously (Iwatani et al., 2007). Most likely it is the limiting factor for bacteriocin production, which must be well balanced in order not to kill itself. It could be assumed that low level of immunity to lactolisterin BU is result of balanced translational regulation or still ongoing transition of sugar to bacteriocin operon.
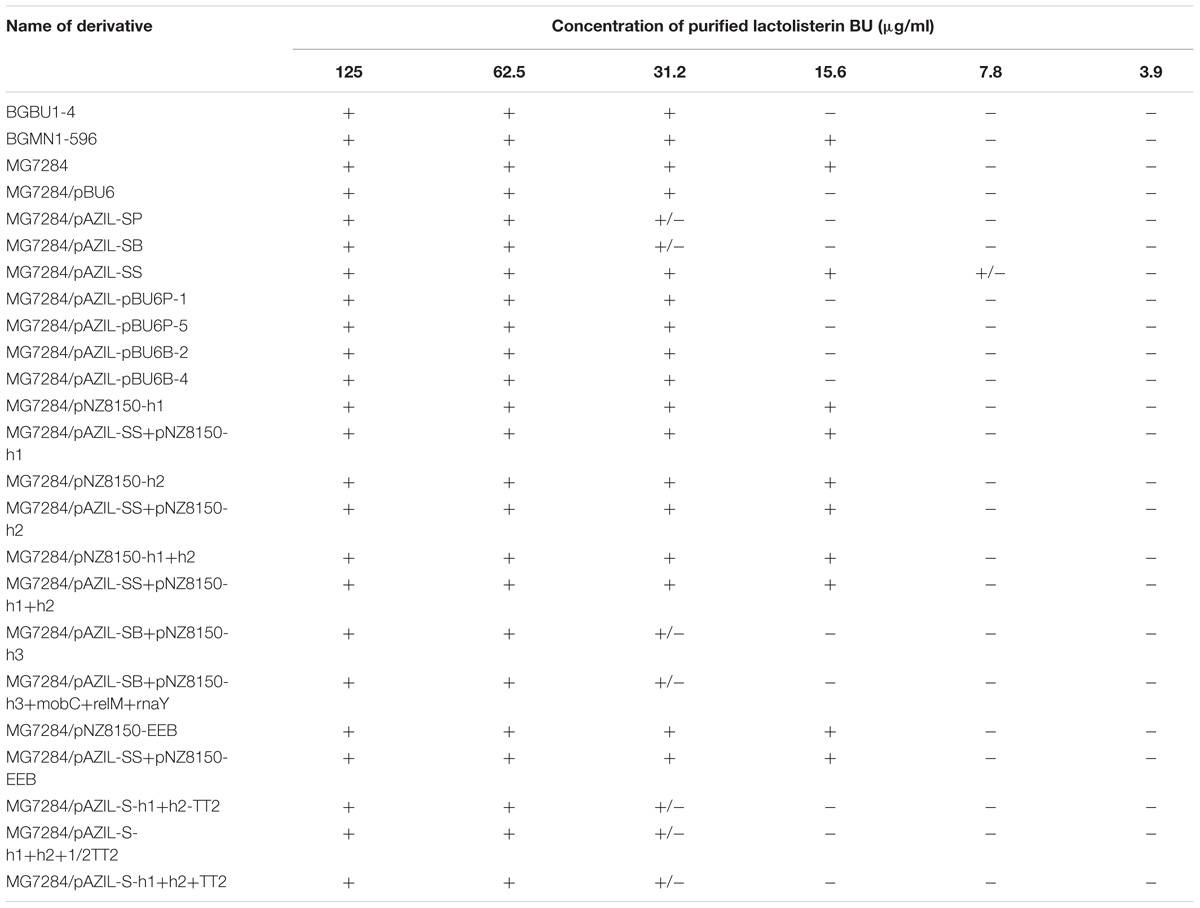
TABLE 4. Sensitivity of all analyzed derivatives to different concentrations of purified lactolisterin BU in spot on the lawn bacteriocin assay.
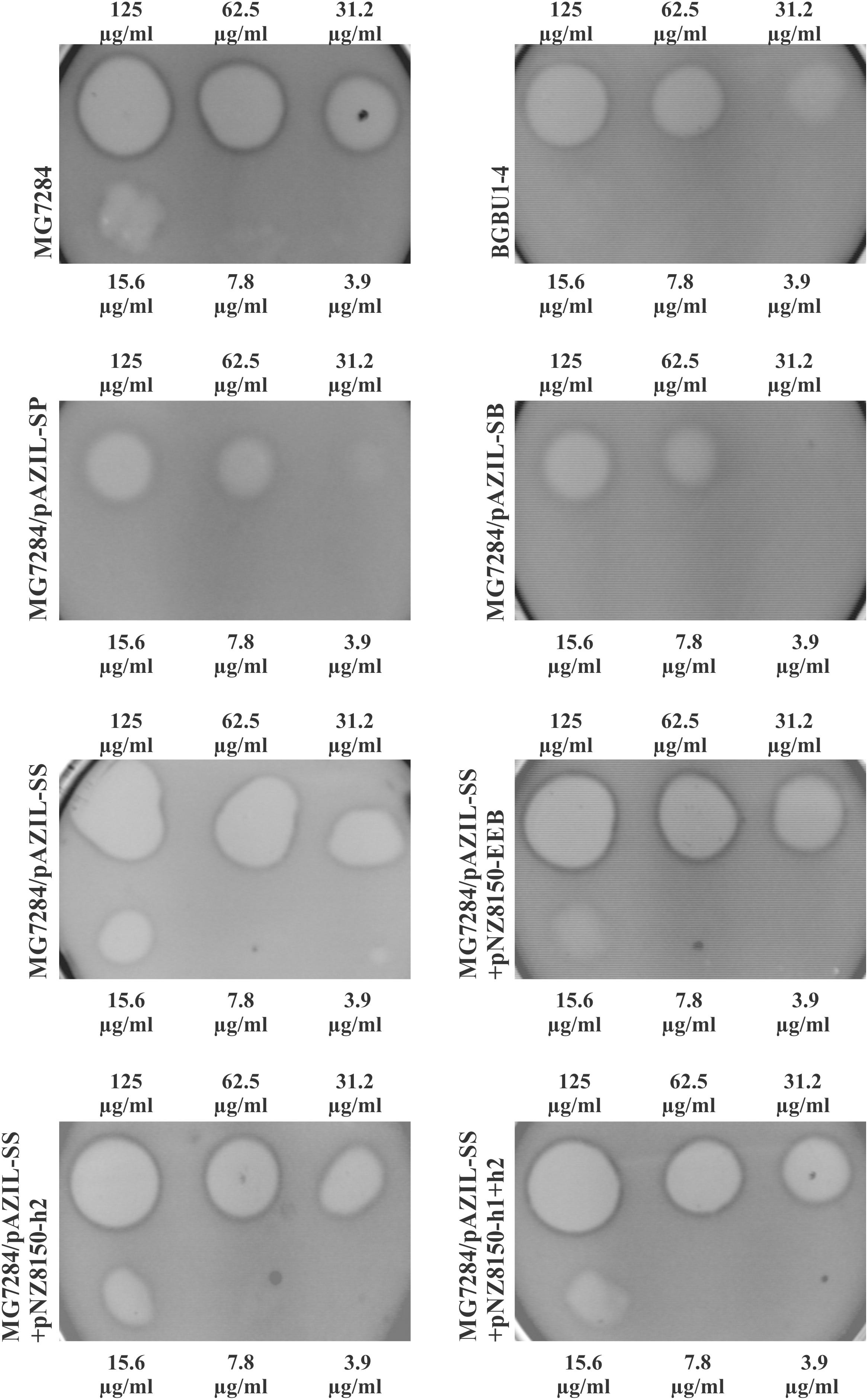
FIGURE 5. Sensitivity of representative strains and derivatives to different concentrations of purified lactolisterin BU in spot on the lawn bacteriocin assay.
The Transcription Terminator TT2 Did Not Influence the Expression of Lactolisterin BU in Lactococci
It has been documented that transcription terminators can be involved in regulation of gene expression by transcriptional attenuation or impacting on the stability of mRNA originating from bacteriocin operons (Uzelac et al., 2015). Different constructs were made to determine the possible involvement of TT2 in the expression of lactolisterin BU (Figure 1C). It was found that constructs without TT2 expressed the same level of bacteriocin (the same size of the zone of inhibition), indicating that TT2 was not involved in additional regulation of lactolisterin BU expression.
Product of the hyp3 Gene Modulates Expression of Lactolisterin BU
The possible involvement of the hyp3 gene in expression of lactolisterin BU was also analyzed (Figure 1E). During subcloning of the pBU6 plasmid, it was noticed that construct pAZIL-SP provided a larger zone of inhibition than pAZIL-SB (that contains the perceived complete lactolisterin gene cluster), despite the growth of bacteriocin producer MG7284/pAZIL-SP being lower. In addition, when the hyp3 gene was subcloned into the pNZ8150 vector and co-expressed with pAZIL-SB in MG7284, it provided the same level of lactolisterin BU production as MG7284/pAZIL-SP (Figure 4Cb). The hyp3 gene did not belong to lactolisterin BU operon, but may somehow modulate expression or transport of lactolisterin BU, possibly due its membrane location.
The Lactolisterin BU Operon Expressed in trans Does Not Provide Bacteriocin Activity
Subcloning experiments with the pBU6 plasmid showed that the lactolisterin BU gene cluster consists of four genes (lliBU, abcT, accL and immL) clustered as one transcriptional unit. To test the possibility of in trans expression by different combinations of lactolisterin BU genes, the following experiments were completed: the lactolisterin BU operon was split and cloned into two compatible vectors with different selection markers (pAZIL and pNZ8150) carrying different parts (genes) of the cluster (lliBU + abcT, hyp1 and hyp2; lliBU and abcT + hyp1 and hyp2; lliBU and abcT + hyp1; lliBU and abcT + hyp2) (Figures 1E, 4Ca). Transcription of the first part of the operon was driven by the natural promoter PlliBU, while the second part was also constructed to be transcribed under the same promoter. Results of bacteriocin testing showed that combinations of in trans co-expressed lactolisterin BU genes were not able to restore bacteriocin activity, indicating an obligate in cis gene constellation. This indicates that bacteriocin expression is controlled by a translational coupling mechanism, providing an optimal ratio of proteins within the operon to obtain enough bacteriocin molecules for combating competitors, but not so much as to kill itself due to limited immunity.
The results of this study highlight that sophisticated mechanisms of regulation were involved in expression of genes within the lactolisterin BU cluster, which require further study. Elucidation of regulatory mechanisms involved in the expression of lactolisterin BU could help in making constructs for the overexpression of bacteriocins and/or other proteins of interest.
Conclusion
The lactolisterin BU cluster consists of four genes (lliBU, abcT, accL, and immL) clustered as one transcriptional unit under the PlliBU promoter. Gene product functions were determined to be: lliBU – the bacteriocin structural gene, abcT – the ABC transporter, accL – the transporter accessory protein and immL – an immunity protein which provides limited protection. PlliBU promoter activity was determined to be as strong as that of PlcnB, indicating that expression is predominantly regulated at the translational level. The absence of activity when the gene cluster was divided and expressed in trans indicates that bacteriocin expression is controlled to provide an optimum ratio of protein expression levels within the operon. The operon functions in cis constellation of genes ensuring the optimal amount of each product for the cell most probably by a translational coupling mechanism. In addition, it was found that the presence of the hypothetical gene/protein 3, located downstream of the lactolisterin BU operon (possesses its own promoter) has a positive effect on the bacteriocin expression.
Author Contributions
MaM and MK conceived, designed, and coordinated this study, interpreted all of results and contributed to the preparation of the figures, and wrote this paper. JL, NM, MiM, and BJ provided experimental assistance and contributed to the preparation of the figures and manuscript. PO performed experiments of bacteriocin purification. PO and PC provided technical assistance and contributed to the preparation of this paper. All authors reviewed the results and approved the final version of the manuscript.
Funding
This work was funded by The Ministry of Education, Science and Technological Development of the Republic of Serbia, Republic of Serbia (Grant No. 173019).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
- ^ https://embnet.vital-it.ch/software/TMPRED_form.html
- ^ http://www.bacpp.bioinfoucs.com/ferramenta
- ^ http://rna.igmors.u-psud.fr/toolbox/arnold/
References
Ahmad, V., Khan, M. S., Jamal, Q. M. S., Alzohairy, M. A., Al Karaawi, M. A., and Siddiqui, M. U. (2017). Antimicrobial potential of bacteriocins: in therapy, agriculture and food preservation. Int. J. Antimicrob. Agents 49, 1–11. doi: 10.1016/j.ijantimicag.2016.08.016
Alvarez-Sieiro, P., Montalbán-López, M., Mu, D., and Kuipers, O. P. (2016). Bacteriocins of lactic acid bacteria: extending the family. Appl. Microbiol. Biotechnol. 100, 2939–2951. doi: 10.1007/s00253-016-7343-9
Baindara, P., Gautam, A., Raghava, G. P. S., and Kapole, S. (2017). Anticancer properties of a defensin like class IId bacteriocin Laterosporulin10. Sci. Rep. 7:46541. doi: 10.1038/srep46541
Cleveland, J., Montville, T. J., Nes, I. F., and Chikindas, M. L. (2001). Bacteriocins: safe, natural antimicrobials for food preservation. Int. J. Food Microbiol. 71, 1–20. doi: 10.1016/S0168-1605(01)00560-8
Cotter, P. D., Hill, C., and Ross, P. R. (2005). Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3, 777–788. doi: 10.1038/nrmicro1273
Cotter, P. D., Ross, R. P., and Hill, C. (2013). Bacteriocins – a viable alternative to antibiotics? Nat. Rev. Microbiol. 11, 95–105. doi: 10.1038/nrmicro2937
De Vos, W. M., Beerthuyzen, M. M., Luesink, E. L., and Kuipers, O. P. (1995). Genetics of the nisin operon and the sucrose-nisin conjugative transposon Tn5276. Dev. Biol. Stand. 85, 617–625.
Diaz, M., Valdivia, E., Martínez-Bueno, M., Fernández, M., Soler-González, A. S., Ramírez-Rodrigo, H., et al. (2003). Characterization of a new operon, as-48EFGH, from the as-48 gene cluster involved in immunity to enterocin AS-48. Appl. Environ. Microbiol. 69, 1229–1236. doi: 10.1128/AEM.69.2.1229-1236.2003
Diep, D. B., Myhre, R., Johnsborg, O., Aakra, A., and Nes, I. F. (2003). Inducible bacteriocin production in Lactobacillus is regulated by differential expression of the pln operons and by two antagonizing response regulators, the activity of which is enhanced upon phosphorylation. Mol. Microbiol. 47, 483–494. doi: 10.1046/j.1365-2958.2003.03310.x
Engelke, G., Gutowski-Eckel, Z., Hammelmann, M., and Entian, K. D. (1992). Biosynthesis of the lantibiotic nisin: genomic organization and localization of the NisB protein. Appl. Environ. Microbiol. 58, 3730–3743.
Gajic, O., Kojic, M., Banina, A., and Topisirovic, L. (1999). Characterization of natural isolate Lactococcus lactis subsp. lactis BGMN1-5, a strain producing two bacteriocins, cell wall-associated proteinase and showing clumping phenotype. Arch. Biol. Sci. 51, 69–78.
Gasson, M. J. (1983). Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154, 1–9.
Golić, N., Čadež, N., Terzić-Vidojević, A., Šuranská, H., Beganović, J., Lozo, J., et al. (2013). Evaluation of lactic acid bacteria and yeast diversity in traditional white pickled and fresh soft cheeses from the mountain regions of Serbia and lowland regions of Croatia. Int. J. Food Microbiol. 166, 294–300. doi: 10.1016/j.ijfoodmicro.2013.05.032
Gotteland, M., Andrews, M., Toledo, M., Muñoz, L., Caceres, P., Anziani, A., et al. (2008). Modulation of Helicobacter pylori colonization with cranberry juice and Lactobacillus johnsonii La1 in children. Nutrition 24, 421–426. doi: 10.1016/j.nut.2008.01.007
Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580.
Holo, H., and Nes, I. F. (1989). High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55, 3119–3123.
Iwatani, S., Zendo, T., Yoneyama, F., Nakayama, J., and Sonomoto, K. (2007). Characterization and structure analysis of a novel bacteriocin, lacticin Z, produced by Lactococcus lactis QU 14. Biosci. Biotechnol. Biochem. 71, 1984–1992. doi: 10.1271/bbb.70169
Kaiser, A. L., and Montville, T. J. (1996). Purification of the bacteriocin bavaricin MN and characterization of its mode of action against Listeria monocytogenes Scott A cells and lipid vesicles. Appl. Environ. Microbiol. 62, 4529–4535.
Kjos, M., Oppegård, C., Diep, D. B., Nes, I. F., Veening, J. W., Nissen-Meyer, J., et al. (2014). Sensitivity to the two-peptide bacteriocin lactococcin G is dependent on UppP, an enzyme involved in cell-wall synthesis. Mol. Microbiol. 92, 1177–1187. doi: 10.1111/mmi.12632
Kojic, M., Jovcic, B., Strahinic, I., Begovic, J., Lozo, J., Veljovic, K., et al. (2011). Cloning and expression of novel lactococcal aggregation factor from Lactococcus lactis subsp. lactis BGKP1. BMC Microbiol. 11:265. doi: 10.1186/1471-2180-11-265
Kojic, M., Lozo, J., Jovcic, B., Strahinic, I., and Topisirovic, L. (2010). Construction of a new shuttle vector and its use for cloning and expression of two plasmid-encoded bacteriocins from Lactobacillus paracasei subsp. paracasei BGSJ2-8. Int. J. Food Microbiol. 140, 117–124. doi: 10.1016/j.ijfoodmicro.2010.04.010
Kojic, M., Strahinic, I., Fira, D., Jovcic, B., and Topisirovic, L. (2006). Plasmid content and bacteriocin production by five strains of Lactococcus lactis isolated from semi-hard homemade cheese. Can. J. Microbiol. 52, 1110–1120. doi: 10.1139/w06-072
Kojic, M., Strahinic, I., and Topisirovic, L. (2005). Proteinase PI and lactococcin A genes are located on the largest plasmid in Lactococcus lactis subsp. lactis bv. diacetylactis S50. Can. J. Microbiol. 51, 305–314. doi: 10.1139/w05-009
Kojic, M., Svircevic, J., Banina, A., and Topisirovic, L. (1991). Bacteriocin-producing strain of Lactococcus lactis subsp. diacitilactis S50. Appl. Environ. Microbiol. 57, 1835–1837.
Lagos, R., Tello, M., Mercado, G., Garcia, V., and Monasterio, O. (2009). Antibacterial and antitumorigenic properties of microcin E492, a pore-forming bacteriocin. Curr. Pharm. Biotechnol. 10, 74–85. doi: 10.2174/138920109787048643
Law, J., Buist, G., Haandrikman, A., Kok, J., Venema, G., and Leenhouts, K. (1995). A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177, 7011–7018.
Lozo, J., Mirkovic, N., O’Connor, P. M., Malesevic, M., Miljkovic, M., Polovic, N., et al. (2017). Lactolisterin BU, a novel class II broad spectrum bacteriocin from Lactococcus lactis subsp. lactis bv. diacetylactis BGBU1-4. Appl. Environ. Microbiol. 83:e01519-17. doi: 10.1128/AEM.01519-17
Mesa-Pereira, B., O’Connor, P. M., Rea, M. C., Cotter, P. D., Hill, C., and Ross, R. P. (2017). Controlled functional expression of the bacteriocins pediocin PA-1 and bactofencin A in Escherichia coli. Sci. Rep. 7:3069. doi: 10.1038/s41598-017-02868-w
Mierau, I., and Kleerebezem, M. (2005). 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol. 68, 705–717. doi: 10.1007/s00253-005-0107-6
Miller, J. M. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
Nes, I. F., Diep, D. B., Håvarstein, L. S., Brurberg, M. B., Eijsink, V., and Holo, H. (1996). Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Van Leeuwenhoek 70, 113–128.
O’Sullivan, D. J., and Klaenhammer, T. R. (1993). Rapid mini-prep isolation of high-quality plasmid DNA from Lactococcus and Lactobacillus spp. Appl. Environ. Microbiol. 59, 2730–2733.
Ovchinnikov, K. V., Kristiansen, P. E., Straume, D., Jensen, M. S., Aleksandrzak-Piekarczyk, T., Nes, I. F., et al. (2017). The leaderless bacteriocin enterocin K1 is highly potent against Enterococcus faecium: a study on structure, target spectrum and receptor. Front. Microbiol. 8:774. doi: 10.3389/fmicb.2017.00774
Rauch, P. J., and De Vos, W. M. (1992). Characterization of the novel nisin-sucrose conjugative transposon Tn5276 and its insertion in Lactococcus lactis. J. Bacteriol. 174, 1280–1287.
Uzelac, G., Miljkovic, M., Lozo, J., Radulovic, Z., Tosic, N., and Kojic, M. (2015). Expression of bacteriocin LsbB is dependent on a transcription terminator. Microbiol. Res. 17, 45–53. doi: 10.1016/j.micres.2015.06.011
Vukotic, G., Mirkovic, N., Jovcic, B., Miljkovic, M., Strahinic, I., Fira, D., et al. (2015). Proteinase PrtP impairs lactococcin LcnB activity in Lactococcus lactis BGMN1-501: new insights into bacteriocin regulation. Front. Microbiol. 6:92. doi: 10.3389/fmicb.2015.00092
Keywords: bacteriocin, lactolisterin BU operon, ABC transporter, limited immunity, translational coupling
Citation: Miljkovic M, Lozo J, Mirkovic N, O’Connor PM, Malesevic M, Jovcic B, Cotter PD and Kojic M (2018) Functional Characterization of the Lactolisterin BU Gene Cluster of Lactococcus lactis subsp. lactis BGBU1-4. Front. Microbiol. 9:2774. doi: 10.3389/fmicb.2018.02774
Received: 22 August 2018; Accepted: 29 October 2018;
Published: 15 November 2018.
Edited by:
Aldo Corsetti, Università degli Studi di Teramo, ItalyReviewed by:
Bettina Buttaro, Temple University, United StatesLorena Ruiz, Instituto de Productos Lácteos de Asturias (IPLA), Spain
Manuel A. Ortega, Massachusetts Institute of Technology, United States
Copyright © 2018 Miljkovic, Lozo, Mirkovic, O’Connor, Malesevic, Jovcic, Cotter and Kojic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marija Miljkovic, bWFyaWphbWlsamtvdmljQGltZ2dlLmJnLmFjLnJz
 Marija Miljkovic
Marija Miljkovic Jelena Lozo
Jelena Lozo Nemanja Mirkovic
Nemanja Mirkovic Paula M. O’Connor
Paula M. O’Connor Milka Malesevic1
Milka Malesevic1 Branko Jovcic
Branko Jovcic Paul D. Cotter
Paul D. Cotter Milan Kojic
Milan Kojic