- Biotechnology Research Institute for Drug Discovery, National Institute of Advanced Industrial Science and Technology (AIST), Tokyo, Japan
Sulfur is an essential element in all living organisms. In tRNA molecules, there are many sulfur-containing nucleosides, introduced post-transcriptionally, that function to ensure proper codon recognition or stabilization of tRNA structure, thereby enabling accurate and efficient translation. The biosynthesis of tRNA sulfur modifications involves unique sulfur trafficking systems that are closely related to cellular sulfur metabolism, and “modification enzymes” that incorporate sulfur atoms into tRNA. Herein, recent biochemical and structural characterization of the biosynthesis of sulfur modifications in tRNA is reviewed, with special emphasis on the reaction mechanisms of modification enzymes. It was recently revealed that TtuA/Ncs6-type 2-thiouridylases from thermophilic bacteria/archaea/eukaryotes are oxygen-sensitive iron-sulfur proteins that utilize a quite different mechanism from other 2-thiouridylase subtypes lacking iron-sulfur clusters such as bacterial MnmA. The various reaction mechanisms of RNA sulfurtransferases are also discussed, including tRNA methylthiotransferase MiaB (a radical S-adenosylmethionine-type iron-sulfur enzyme) and other sulfurtransferases involved in both primary and secondary sulfur-containing metabolites.
Introduction
Transfer RNA (tRNA) is an essential adaptor molecule that bridges genomic information from mRNAs to amino acid sequences in proteins. Precursor tRNA molecules undergo various maturation steps such as removal of leader, trailer, and intronic sequences, addition of 3′-CCA sequences, and chemical modification of nucleosides. More than 100 post-transcriptional modifications of tRNAs have been identified (Cantara et al., 2011; Väre et al., 2017; Boccaletto et al., 2018), among which sulfur modifications are especially important for tRNA functions. Four kinds of thionucleoside derivatives are found in tRNAs (Figures 1A,B): 4-thiouridine (s4U) at positions 8 and 9 (Lipsett, 1965; Singer and Smith, 1972; Griffey et al., 1986), 2-thiocytidine (s2C) at position 32 (Carbon et al., 1968; Murao et al., 1972), 2-thiouridine (s2U) at position 33 (Crain et al., 2002), 2-thiouridine derivatives (xm5s2U) at positions 34 (Carbon et al., 1968; Oashi et al., 1970) and 54 (Watanabe et al., 1974), and 2-methylthioadenosine derivatives (ms2x6A) at position 37 (Burrows et al., 1968; Ishikura et al., 1971) (where “x” represents several functional groups differing between species and organelles). At position 34, there is taurine (2-aminoethansulfonic acid)-containing modification at C5 carbon of U (Suzuki et al., 2002), 2-selenouridine derivatives (xm5se2U), and 2-geranyl-thiourideine derivatives (xm5ges2U) (Wittwer et al., 1984; Dumelin et al., 2012). The biosynthesis of tRNA sulfur modifications involves sulfur trafficking systems and “modification enzymes.” The sulfur trafficking systems used in RNA modification are closely related to and shared with cellular sulfur metabolism (Laxman et al., 2013), whereas modification enzymes recognize substrate tRNAs and incorporate sulfur atoms. Some sulfur-containing cofactors and secondary metabolites are depicted in Figure 1C.
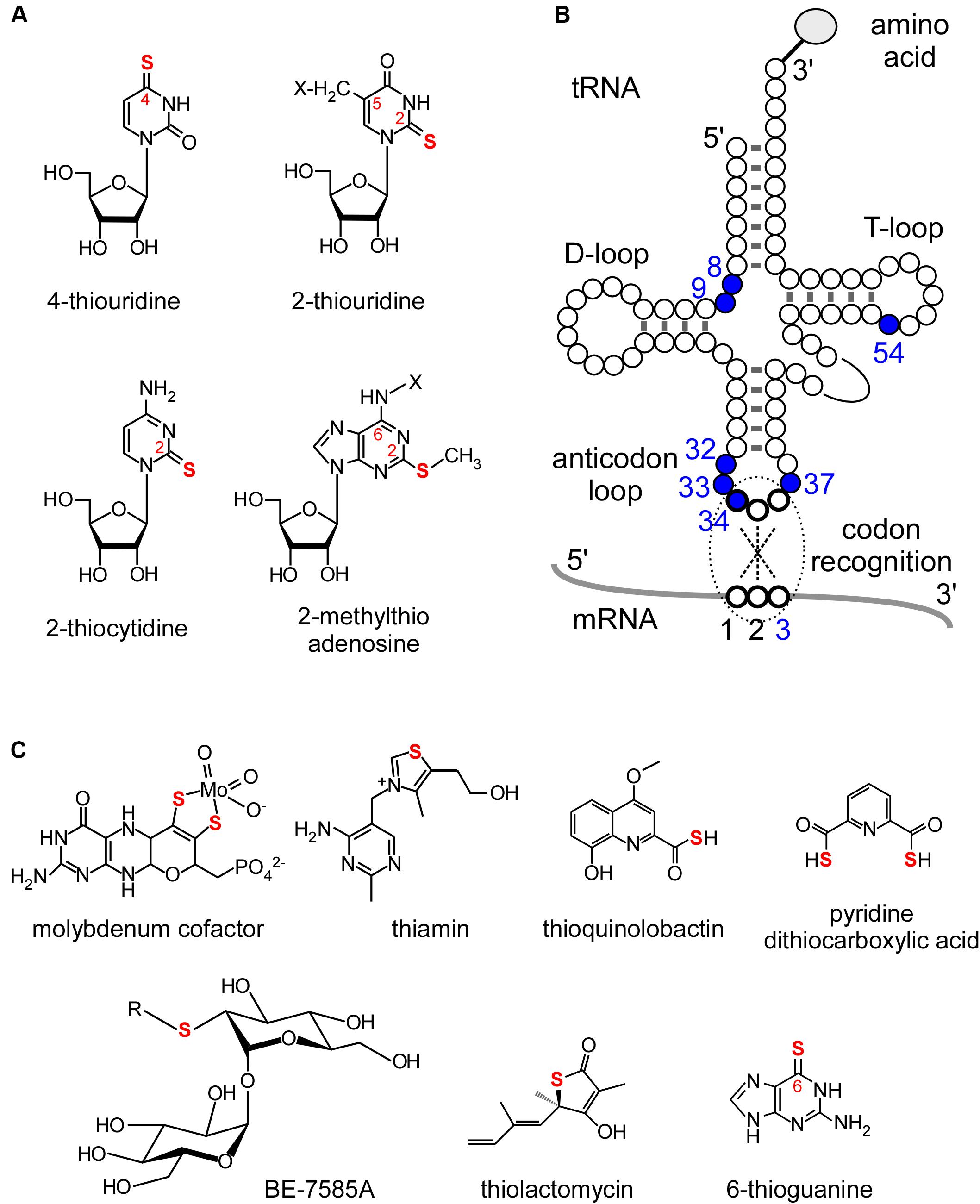
FIGURE 1. Sulfur modifications in tRNAs and other sulfur-containing metabolites. (A) Chemical structures of sulfur modifications in tRNAs. (B) Positions of thionucleosides in tRNAs. (C) Examples of sulfur-containing cofactors and secondary metabolites.
Functions of Sulfur Modifications in Trnas
The functions of sulfur modifications are briefly summarized in this section. For more detailed information, please refer to previous reviews (Shigi, 2014, 2016) and articles cited therein. Uridine at position 34 (the wobble base) of tRNAs for lysine, glutamic acid, and glutamine is almost universally modified to s2U derivatives, although the C5 carbon of uridine is also modified by functional groups that differ between species (Elseviers et al., 1984). Due to steric clashes between the bulky 2-thio group and the 2′-OH group of ribose, the ribose of s2U preferentially adopts the C3′-endo conformation (Yokoyama et al., 1985; Agris et al., 1992). Therefore, xm5s2U stabilizes base paring with NNA and NNG codons for lysine, glutamic acid, and glutamine (Agris et al., 1973; Murphy et al., 2004; Durant et al., 2005; Johansson et al., 2008). Absence of the 2-thio modification leads to ribosome stalling at AAA, CAA, and GAA codons in mRNAs. Interestingly, pausing of the ribosome causes protein misfolding and aggregation (Nedialkova and Leidel, 2015), suggesting that optimal codon translation by tRNA wobble modifications is very important for maintaining proteome integrity. tRNA modifications are proposed to control the translation efficiency of specific groups of genes with mRNA codon bias as a mechanism of adaptation to specific environments (Laxman et al., 2013; Tigano et al., 2015; Tyagi and Pedrioli, 2015; Chionh et al., 2016).
In addition to position 34, the distribution of sulfur modifications at other positions differs between species, and modifications at positions 32 and 37 in the anticodon loop are also important for precise codon recognition. The 2-methylthio modification at position 37 directly stabilizes mRNA-tRNA interactions with U in the anticodon third position and A in the codon first position, as revealed by structural analysis of mRNA-tRNA interactions in the ribosome (Jenner et al., 2010). The s4U modification at position 8 is responsible for near-ultraviolet light sensing in bacteria (Favre et al., 1969; Carre et al., 1974; Ryals et al., 1982). When the cell is irradiated with near-ultraviolet light, s4U crosslinks with cytidine 13, resulting in a disordered tRNA structure that leads to translational arrest. In some thermophilic microorganisms, such as Thermus thermophilus and Pyrococcus furiosus, 5-methyl-2-thiouridine (m5s2U) or 2-thioribothymidine (s2rT) is found at position 54 in almost all tRNA molecules (Watanabe et al., 1974; Kowalak et al., 1994), and the 2-thiolation content increases with increasing cultivation temperature (Watanabe et al., 1976; Kowalak et al., 1994). The m5s2U modification strengthens the duplex structure formed by the D-loop and T-loop, which stabilizes the overall tRNA structure (Horie et al., 1985). In T. thermophilus, m5s2U is indispensable for growth at high temperature (Shigi et al., 2006a). s2U at position 33 was also found in mitochondrial tRNATrp from Leishmania (Crain et al., 2002), the function of this modification has not been elucidated.
Biosynthetic Pathways for Sulfur Modifications in Trnas
In eukaryotes and bacteria, sulfur atoms in sulfur-containing molecules such as thionucleosides are derived from free L-cysteine in the cell. The sulfur atom of L-cysteine is activated by cysteine desulfurase, a pyridoxal-5′-phosphate (PLP)-dependent enzyme, via covalent attachment to its catalytic cysteine residue to generate the persulfide (R-SSH) form (Flint, 1996; Lauhon and Kambampati, 2000; Lauhon, 2002; Nilsson et al., 2002). Enzyme-linked persulfides are then transferred to downstream sulfur carrier proteins, and eventually transferred to the final sulfurtransferases in each pathway (Mueller, 2006; Shi et al., 2010). Thus, biosynthetic pathways for thionucleosides are part of a larger metabolic system involving other sulfur-containing molecules with iron-sulfur (Fe-S) clusters, thiamin, and the molybdenum cofactor (Moco) (Figure 1C; Schindelin et al., 2001; Settembre et al., 2003; Hidese et al., 2011). Moreover, each pathway is mutually influenced by others as part of a “sulfur trafficking” network (Maynard et al., 2012; Dahl et al., 2013). Other fascinating features include the involvement of numerous sulfur carrier proteins that deliver activated sulfur species such as R-SSH and thiocarboxylates (R-COSH), and the mechanisms by which they achieve the safe, directional flow of potentially harmful sulfur atoms (see below).
Thionucleoside synthesis can be classified into two types based on the involvement of Fe-S proteins, and hence the dependency on Fe-S cluster biosynthesis. The biosynthesis of s2C32, ms2A37, and m5s2U54 is dependent on Fe-S clusters (Lauhon et al., 2004; Leipuviene et al., 2004; Chen et al., 2017). Biosynthesis of s4U8 and s2U34 differs among species; the biosynthesis of s4U8 in bacteria, such as E. coli, Salmonella typhimurium, Bacillus subtilis (Lauhon et al., 2004; Leipuviene et al., 2004; Rajakovich et al., 2012), and some archaea, such as Thermoproteales, Thermoplasmatales, Halobacteriales, and Sulfolobales (Liu et al., 2012), is not dependent on Fe-S clusters, while in methanogenic archaea and some other archaea, such as Thermococcales, it is dependent on Fe-S clusters (Liu et al., 2012, 2016). The biosynthesis of s2U34 is not dependent on Fe-S clusters in bacteria (Lauhon et al., 2004; Leipuviene et al., 2004; Black and Dos Santos, 2015), but the opposite is true for archaeal and eukaryotic pathways (Nakai et al., 2007; Liu et al., 2016).
se2U34 is synthesized from s2U34 via geranylated intermediate (ges2U) by SelU (YbbB) (Chen et al., 2005; Dumelin et al., 2012; Sierant et al., 2018b). Desulfuration activity of 2-thiouracil by DUF523 domain protein in the cell has recently been reported (Aucynaite et al., 2018). Extensive in vitro analysis of desulfuration of s2U derivative as a form of nucleoside or within an RNA chain has been performed, hydrogen peroxide and cytochrome C or FeII -mediated reactions forms predominantly generate 4-pyrimidinone nucleoside (h2U), rather than U (Sochacka et al., 2013; Sierant et al., 2018a). These studies will lead to better understanding of the in vivo metabolism of thionucleosides.
The Role of Sulfurtransferase Mnma in 2-Thio U Synthesis
MnmA is a thiouridylase that catalyzes 2-thiolation of uridine at position 34 in bacteria (Kambampati and Lauhon, 2003). The homologous enzyme Mtu1 is involved in s2U formation in eukaryotic mitochondria (Umeda et al., 2005). In Escherichia coli, TusA, the TusBCD complex, and TusE are required for s2U formation (Ikeuchi et al., 2006). TusA interacts with cysteine desulfurase IscS, accepts the persulfide, and directs sulfur flow to this pathway. MnmA accepts the persulfide sulfur on its conserved Cys199 residue in the active site from TusA via TusD and TusE. However, in most species, there is no need for such intermediate persulfide carrier proteins (Black and Dos Santos, 2015). MnmA possesses a PP-loop motif and is a member of the ATP-pyrophosphatase family. This enzyme utilizes a two-step mechanism to form an adenylated intermediate (Figure 2A). Nucleophilic attack by the persulfide sulfur generates s2U and releases AMP. The modification enzyme ThiI involved in s4U synthesis also contains a PP-loop and utilizes a similar two-step mechanism (Mueller et al., 1998; Mueller and Palenchar, 1999; Neumann et al., 2014). A snapshot of s2U formation via the acyl-adenylated intermediate was clearly revealed in a structural analysis of the E. coli MnmA-tRNA complex (Figure 2B; Numata et al., 2006). In the catalytic pocket, which is separated from bulk solvent, the uridine reacts with ATP to form an acyl-adenylated intermediate that reacts with the terminal sulfur released from the persulfide on Cys199 with assistance from another conserved cysteine (Cys102).
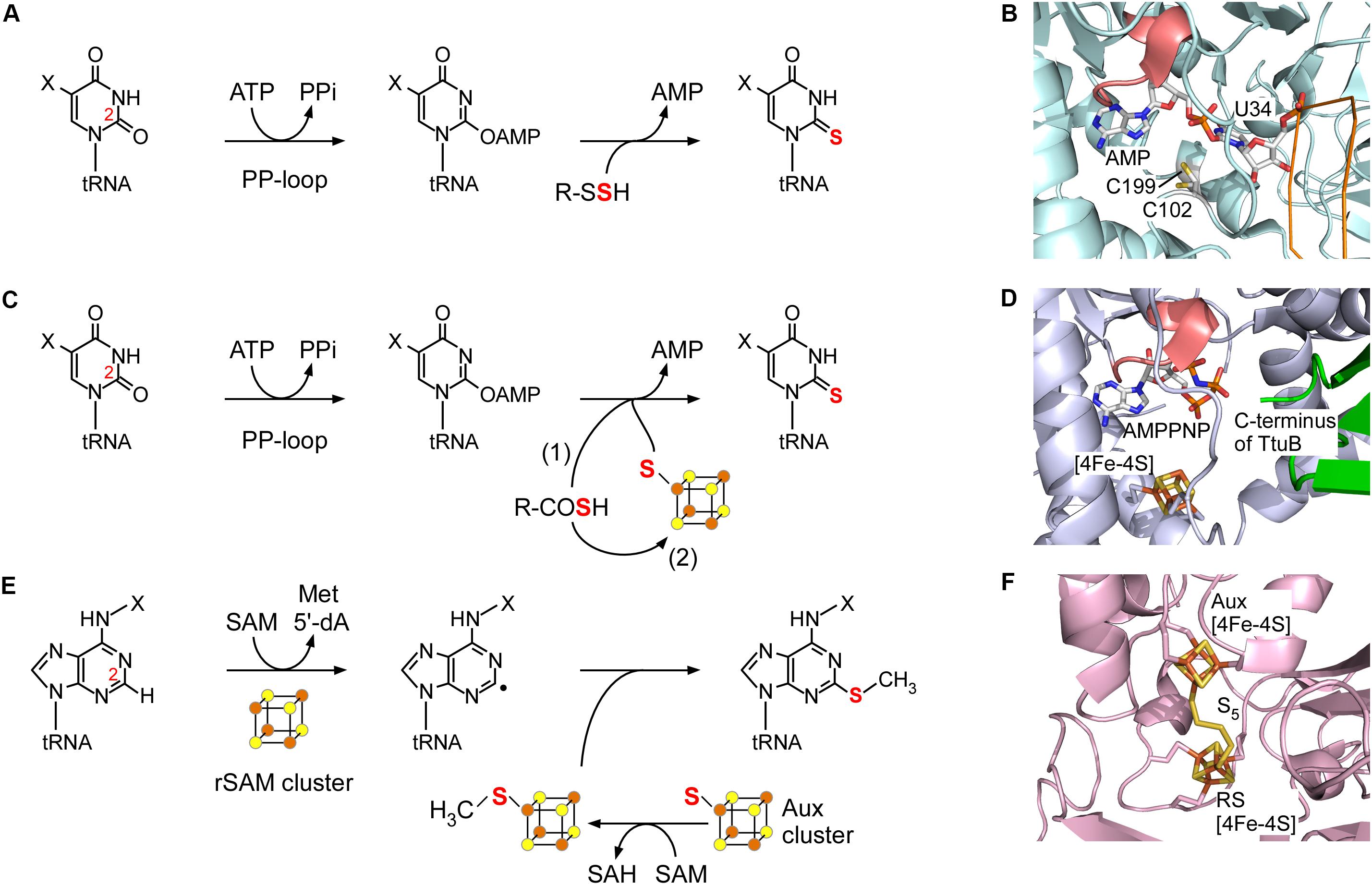
FIGURE 2. Two-step reaction mechanism of representative (methyl)sulfurtransferases. (A) The 2-thiouridylation reaction catalyzed by MnmA. A protein persulfide is utilized as the sulfur donor. (B) Crystal structure of Escherichia coli MnmA, showing the tRNA complex with an adenylated intermediate (PDB ID: 2deu). The backbone of the tRNA and the PP-loop of MnmA are colored orange and pink, respectively. The two catalytic cysteines (Cys102 and Cys199) are shown in stick representation. (C) The 2-thiouridylation reaction catalyzed by TtuA/Ncs6. A protein thiocarboxylate may be utilized directly as shown in scheme (1). Alternatively, a free sulfide from the thiocarboxylate may be utilized once trapped by the Fe-S cluster as shown in scheme (2). (D) Crystal structure of the Thermus thermophilus TtuA (blue)-TtuB (green) complex (modeled from PDB ID: 5b4e and 5gha). The ATP analog bound to the PP-loop (pink), and the C-terminus of TtuB and the Fe-S cluster encounter one another in the catalytic center of TtuA. Note that two residues of the C-terminus of TtuB are not visible. (E) The 2-methylthioadenylation reaction catalyzed by MiaB. A methylsufide formed on the auxiliary (Aux) cluster is utilized as a substrate. (F) Crystal structure of Thermotoga maritima RimO clearly showing the pentasulfide bridging the rSAM and Aux clusters (PDB ID: 4jc0).
The Role of Iron-Sulfur Protein Ncs6/Ttua in 2-Thio U Synthesis
In eukaryotes and archaea, Ncs6 and its archaeal homolog NcsA catalyze the 2-thiolation reaction of uridine at position 34 (Bjork et al., 2007; Chavarria et al., 2014). In eukaryotes, Ncs6 forms a heterocomplex with the Ncs2 protein that appears to have a role beyond catalysis (Esberg et al., 2006; Dewez et al., 2008). In some thermophilic bacteria and archaea, such as T. thermophilus, Thermotoga maritima, and Pyrococcus horikoshii, TtuA catalyzes the same 2-thiouridylation reaction at different positions (e.g., position 54) (Shigi et al., 2006a; Arragain et al., 2017). Although Ncs6/TtuA has a PP-loop motif and requires ATP for activity, the sulfur transfer mechanism (Figure 2C) is markedly different from that of MnmA in two aspects: Ncs6/TtuA utilizes an oxygen-sensitive Fe-S cluster and a unique thiocarboxylate (R-COSH) that is formed on the carboxy terminus of the sulfur carrier protein Urm1/TtuB, believed to be ancient ubiquitin-like post-translational modifiers (Shigi et al., 2008; Leidel et al., 2009; Shigi, 2012). The C-terminus of Urm1/TtuB is thiocarboxylated with a sulfur atom from free L-cysteine via an adenylated intermediate catalyzed by the E1-like enzyme Uba4/TtuC. Meanwhile, Tum1/TtuD enhances the activity of cysteine desulfurases and directs sulfur flow to s2U biosynthesis (Shigi et al., 2006b; Noma et al., 2009; Shigi et al., 2016).
In TtuA, a [4Fe-4S] cluster is ligated by three conserved cysteines, leaving one iron atom free for ligand binding, which may be important in the sulfur transfer reaction (Figure 2D; Nakagawa et al., 2013; Arragain et al., 2017; Chen et al., 2017). TtuA activates the C2 position of U54 by forming an acyl-adenylated intermediate. The thiocarboxylate of TtuB is subsequently attached near the adenylate by the iron-sulfur cluster, and the sulfur atom then attacks the C2 position of the uridine (Figure 2C (1)), forming the s2U product. In an alternative mechanism, a sulfide ion released from TtuB-COSH may bind to the free iron atom of the Fe-S cluster, and become incorporated into s2U (Figure 2C (2)). The latter pathway may be utilized by organisms lacking a TtuB homolog, and the sulfide could be derived from free sulfide ions in the cell. In support of this mechanism, sulfur atoms from free sulfides were incorporated in vitro, and a sulfide captured by the iron-sulfur cluster was observed in the crystal structure (Arragain et al., 2017), although direct proof in vivo has not yet been obtained. Changes in the electronic properties of the Fe-S cluster during the reaction should also be investigated to understand the role in sulfur transfer.
It has also been demonstrated that Saccharomyces cerevisiae Ncs6 and Methanococcus maripaludis NcsA can bind [3Fe-4S] clusters (Liu et al., 2016). TtcA, which has a 2-thiocytidylase activity at position 32 and belongs to a subgroup of the Ncs6/TtuA family, also requires the [4Fe-4S] cluster for catalysis (Jager et al., 2004; Bouvier et al., 2014). In addition, M. maripaludis ThiI has a [3Fe-4S] cluster that is essential for catalysis (Liu et al., 2016). Knowledge of the functional differences and distributions of [4Fe-4S], [3Fe-4S], and other cluster types in these enzymes may lead to a better understanding of the precise mechanisms of Fe-S cluster-dependent sulfurtransferases. Although it was revealed that TtuA recognizes a common T-loop sequence in tRNAs (Shigi et al., 2002), the structural basis of tRNA recognition by Ncs6/TtuA family enzymes remains to be elucidated.
The Role of Radical S-Adenosylmethionine (Rsam) Enzyme Miab in 2-Methylthio a Synthesis
Methylthio-A37 methylthiotransferases such as MiaB in bacteria and its paralogs in eukaryotes (Esberg et al., 1999; Pierrel et al., 2002; Arragain et al., 2010) are a subgroup of rSAM enzymes that possesses two Fe-S clusters (Lanz and Booker, 2015). rSAM enzymes catalyze the reductive cleavage of SAM to methionine and the highly reactive 5′-deoxyadenosyl (5′-dA) radical using a [4Fe-4S] cluster, called the “rSAM cluster.” By abstracting a hydrogen atom from the substrate, the 5′-dA radical generates a substrate radical intermediate (Figure 2E). The rSAM cluster and an additional auxiliary [4Fe-4S] cluster (the “Aux cluster”) are ligated by two sets of three conserved cysteine residues, and located near each other; the distance between the two clusters is ∼8 Å in the structurally characterized related enzyme RimO (Forouhar et al., 2013; Figure 2F), which catalyzes the insertion of a methylthio group on the Asp89 residue of the bacterial ribosomal protein S12 (Anton et al., 2008). Interestingly, the two ligand-free iron atoms of the rSAM and Aux clusters are bridged by a pentasulfide chain in this structure. It was proposed that this bridging sulfur mimics the sulfur donor, and the sulfur does not appear to come from the iron-sulfur clusters themselves, but the exact nature of the sulfur donor remains to be determined. An explanation of the reaction mechanism has been proposed in which the transfer of a methyl group from another molecule of SAM to the sulfur atom of the tip of the polysulfide attached to the Aux cluster is followed by attack of a substrate radical on the methylated sulfur atom to generate ms2A (Landgraf et al., 2013; Figure 2E). Recently, a hypermodified nucleoside, 2-methylthiomethylenethio-A (msms2A), was identified in E. coli tRNAs, and MiaB is involved in msms2A synthesis (Dal Magro et al., 2018). MiaB may abstract a hydrogen radical from the methyl group of ms2A, which is introduced in the first step of the reaction, and a second methylthio transfer reaction could then follow.
Relationships With Other Sulfur-Containing Metabolites
The use of protein-thiocarboxylate intermediates (MoaD-COSH and ThiS-COSH) in the biosynthesis of sulfur-containing essential metabolites such as Moco and thiamin was revealed by pioneering research by the groups of Rajagopalan (Leimkuhler et al., 2011) and Begley (Settembre et al., 2003), respectively. As described above, tRNA sulfurtransferase Ncs6/TtuA also utilizes Urm1/TtuB-COSH as a sulfur donor. In some bacteria, thiocarboxylates are utilized, as demonstrated in the biosynthesis of L-cysteine in Mycobacterium tuberculosis (Burns et al., 2005) and L-methionine in Wolinella succinogenes (Krishnamoorthy and Begley, 2011).
In addition to these primary metabolites, there are numerous other C-S bond-containing natural products (Figure 1C), the biosynthesis of which has been comprehensively reviewed (Dunbar et al., 2017). Protein thiocarboxylates are utilized in the biosynthesis of siderophores in some Pseudomonas (Matthijs et al., 2004), such as pyridine-2,6-dithiocarboxylic acid and thioquinolobactin, and the antibiotic BE-7585A in Amycolatopsis orientalis, which contains a 2-thiosugar moiety (Sasaki et al., 2014). Although the biosynthetic gene clusters for these siderophores contain a pathway-specific ThiS paralog, the BE-7585A biosynthetic gene cluster does not. Instead, the 2-thiosugar is synthesized by borrowing sulfur carrier proteins from L-cysteine and Moco biosynthesis.
Interestingly, in the biosynthesis of the thiotetronate antibiotics such as thiolactomycin and Tü 3010 (Tao et al., 2016), the backbone polyketide is synthesized by a polyketide synthase (PKS) and a nonribosomal peptide synthase (NRPS) encoded in the biosynthetic operon. Remarkably, in vivo experiments showed that the sulfur atom in Tü 3010 may be incorporated by a cysteine desulfurase and MnmA, separately encoded from the biosynthetic operon, which are also probably involved in s2U biosynthesis in tRNAs. The involvement of these genes in s2U synthesis should be experimentally validated, it would therefore be interesting to decipher the mechanism by which MnmA specifically recognizes and incorporates the sulfur atom in the precursor of thiotetronate, in addition to its cognate tRNA substrates. Alternatively, an additional sulfur carrier protein(s) may mediate between the MnmA and thiotetronate biosynthesis machinery. The 6-thioguanosine (s6G) modification is a virulence factor in the plant pathogen Erwinia amylovorans, and two proteins are required for the formation of s6G both in vivo and in vitro (Litomska et al., 2018). The first, YcfC, is distantly related to PLP-dependent transferases such as cysteine desulfurases and carbon-sulfur lyases. The second, YcfA, is a PP-loop-containing ATPase distantly related to, and perhaps evolved from, tRNA modification enzymes such as MnmA, ThiI, Ncs6/TtuA, and TtcA. Furthermore, YcaO forms a thiolate phosphorylated intermediate similar to that formed by the PP-loop ATPase, representing another interesting example of the biosynthesis of thioamide-containing natural products (Mahanta et al., 2018).
Perspectives
This review summarizes recent advances in our understanding of the sulfur-related modification of RNA. Recent studies reveal the widespread involvement of modification enzymes with Fe-S clusters in all three domains of life. Because Fe-S clusters and sulfur modifications themselves (Sierant et al., 2018a) are sensitive to cellular oxidative stress, sulfur modifications may carefully be regulated by cellular oxidative status. Differences in the stability of protein persulfides and protein thiocarboxylates in cells may be important and require investigation in the future. Cellular sulfur donors mediating sulfur-related modification of tRNAs are believed to be derived from free L-cysteine (Lauhon and Kambampati, 2000; Lauhon, 2002; Nilsson et al., 2002; Shigi et al., 2006b), and numerous types of cellular-free persulfides such as L-Cys-SSH have been discovered (Ida et al., 2014; Akaike et al., 2017). The ms2A modification is regulated by L-Cys-SSH in mammalian cells (Takahashi et al., 2017), while free sulfide is proposed to serve as a sulfur donor in archaea (Liu et al., 2010). Because sulfur atoms can adopt diverse chemical forms and partake in a wide range of reactions, it is important to exercise great caution when attempting to identify the actual in vivo sulfur donors responsible for the biosynthesis of sulfur-containing biomolecules.
The roles of thiocarboxylate sulfur-carrier proteins have been characterized in the biosynthesis of primary metabolites, leading to the discovery of their roles in those of secondary metabolites. Regarding sulfur carriers and/or other components shared by several biosynthetic pathway, the regulation mechanism of sulfur-flow to each pathway may be interesting and worthy of exploration, especially between primary and secondary metabolites. Strategy utilizing carrier proteins are not limited to the biosynthesis pathway of sulfur-containing molecules, it is more general strategies in life. In L-lysine synthesis in Thermus thermophilus (Horie et al., 2009) and L-lysine/L-arginine synthesis in Sulfolobus acidocaldarius (Ouchi et al., 2013), “amino-group carrier proteins (AmCPs)” are utilized for carrying reaction intermediates, which prevents unwanted intramolecular reactions and enables successive reaction steps to proceed efficiently. Recently, AmCPs have also been identified as parts of the machinery producing the natural product diamino-dihydroxy-heptanoic acid in Streptomyces species (Matsuda et al., 2017).
The biosynthetic pathways underpinning sulfur modification of RNA in all domains of life share many aspects in common; hence research on bacteria can strengthen our understanding of this process in eukaryotes, including humans. Dysfunctional RNA modification, especially involving anticodons, can lead to diseases (Shigi, 2016). Abnormalities in RNA modification are caused by three main factors: (1) mutations in genes encoding modification enzymes, (2) mutations in substrate tRNAs, and (3) alterations in metabolites acting as substrates. The cytosolic ms2A modification is required for the production of proinsulin, which explains why single-nucleotide polymorphisms (SNPs) in the Cdkal1 gene (a MiaB homolog) are a risk factor for type II diabetes (Wei et al., 2011). Mutation of the Mtu1 gene causes abnormalities in s2U modifications, and leads to the mitochondrial disease reversible infantile liver failure (RILF) (Wu et al., 2016). Similarly, in the mitochondrial disease myoclonic epilepsy with red ragged fibers (MERRF), a point mutation in mt-tRNA-Lys leads to the abnormal modification of its anticodon, resulting in disease (Kirino and Suzuki, 2005).
Author Contributions
NS designed the study and wrote the manuscript.
Funding
This study was supported in part by Grants-in-Aid for Scientific Research (16K07311) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, the Precise Measurement Technology Promotion Foundation, and the Takeda Science Foundation.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I would like to thank all collaborators, including Dr. Tsutomu Suzuki and Dr. Kimitsuna Watanabe (University of Tokyo), Dr. Shigeyuki Yokoyama (RIKEN), and Dr. Yoshikazu Tanaka (Tohoku University), members of their laboratories, and members of my research group. I would also like to thank Dr. Kenjyo Miyauchi (University of Tokyo) for comments on the manuscript.
References
Agris, P. F., Sierzputowska-Gracz, H., Smith, W., Malkiewicz, A., Sochacka, E., and Nawrot, B. (1992). Thiolation of uridine carbon-2 restricts the motional dynamics of the transfer RNA wobble position nucleoside. J. Am. Chem. Soc. 114, 2652–2656. doi: 10.1021/ja00033a044
Agris, P. F., Soll, D., and Seno, T. (1973). Biological function of 2-thiouridine in Escherichia coli glutamic acid transfer ribonucleic acid. Biochemistry 12, 4331–4337. doi: 10.1021/bi00746a005
Akaike, T., Ida, T., Wei, F. Y., Nishida, M., Kumagai, Y., Alam, M. M., et al. (2017). Cysteinyl-tRNA synthetase governs cysteine polysulfidation and mitochondrial bioenergetics. Nat. Commun. 8:1177. doi: 10.1038/s41467-017-01311-y
Anton, B. P., Saleh, L., Benner, J. S., Raleigh, E. A., Kasif, S., and Roberts, R. J. (2008). RimO, a MiaB-like enzyme, methylthiolates the universally conserved Asp88 residue of ribosomal protein S12 in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 105, 1826–1831. doi: 10.1073/pnas.0708608105
Arragain, S., Bimai, O., Legrand, P., Caillat, S., Ravanat, J. L., Touati, N., et al. (2017). Nonredox thiolation in tRNA occurring via sulfur activation by a [4Fe-4S] cluster. Proc. Natl. Acad. Sci. U.S.A. 114, 7355–7360. doi: 10.1073/pnas.1700902114
Arragain, S., Handelman, S. K., Forouhar, F., Wei, F. Y., Tomizawa, K., Hunt, J. F., et al. (2010). Identification of eukaryotic and prokaryotic methylthiotransferase for biosynthesis of 2-methylthio-N6-threonylcarbamoyladenosine in tRNA. J. Biol. Chem. 285, 28425–28433. doi: 10.1074/jbc.M110.106831
Aucynaite, A., Rutkiene, R., Gasparaviciute, R., Meskys, R., and Urbonavicius, J. (2018). A gene encoding a DUF523 domain protein is involved in the conversion of 2-thiouracil into uracil. Environ. Microbiol. Rep. 10, 49–56. doi: 10.1111/1758-2229.12605
Bjork, G. R., Huang, B., Persson, O. P., and Bystrom, A. S. (2007). A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA 13, 1245–1255. doi: 10.1261/rna.558707
Black, K. A., and Dos Santos, P. C. (2015). Abbreviated pathway for biosynthesis of 2-thiouridine in Bacillus subtilis. J. Bacteriol. 197, 1952–1962. doi: 10.1128/JB.02625-14
Boccaletto, P., Machnicka, M. A., Purta, E., Piatkowski, P., Baginski, B., Wirecki, T. K., et al. (2018). MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307. doi: 10.1093/nar/gkx1030
Bouvier, D., Labessan, N., Clemancey, M., Latour, J. M., Ravanat, J. L., Fontecave, M., et al. (2014). TtcA a new tRNA-thioltransferase with an Fe-S cluster. Nucleic Acids Res. 42, 7960–7970. doi: 10.1093/nar/gku508
Burns, K. E., Baumgart, S., Dorrestein, P. C., Zhai, H., McLafferty, F. W., and Begley, T. P. (2005). Reconstitution of a new cysteine biosynthetic pathway in Mycobacterium tuberculosis. J. Am. Chem. Soc. 127, 11602–11603. doi: 10.1021/ja053476x
Burrows, W. J., Armstrong, D. J., Skoog, F., Hecht, S. M., Boyle, J. T., Leonard, N. J., et al. (1968). Cytokinin from soluble RNA of Escherichia coli: 6-(3-methyl-2-butenylamino)-2-methylthio-9-beta-D-ribofuranosylpurine. Science 161, 691–693. doi: 10.1126/science.161.3842.691
Cantara, W. A., Crain, P. F., Rozenski, J., McCloskey, J. A., Harris, K. A., Zhang, X., et al. (2011). The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res. 39, D195–D201. doi: 10.1093/nar/gkq1028
Carbon, J., David, H., and Studier, M. H. (1968). Thiobases in Escherchia coli Transfer RNA: 2-Thiocytosine and 5-Methylaminomethyl-2-thiouracil. Science 161, 1146–1147. doi: 10.1126/science.161.3846.1146
Carre, D. S., Thomas, G., and Favre, A. (1974). Conformation and functioning of tRNAs: cross-linked tRNAs as substrate for tRNA nucleotidyl-transferase and aminoacyl synthetases. Biochimie 56, 1089–1101. doi: 10.1016/S0300-9084(74)80097-0
Chavarria, N. E., Hwang, S., Cao, S., Fu, X., Holman, M., Elbanna, D., et al. (2014). Archaeal Tuc1/Ncs6 homolog required for wobble uridine tRNA thiolation is associated with ubiquitin-proteasome, translation, and RNA processing system homologs. PLoS One 9:e99104. doi: 10.1371/journal.pone.0099104
Chen, M., Asai, S. I., Narai, S., Nambu, S., Omura, N., Sakaguchi, Y., et al. (2017). Biochemical and structural characterization of oxygen-sensitive 2-thiouridine synthesis catalyzed by an iron-sulfur protein TtuA. Proc. Natl. Acad. Sci. U.S.A. 114, 4954–4959. doi: 10.1073/pnas.1615585114
Chen, P., Crain, P. F., Nasvall, S. J., Pomerantz, S. C., and Bjork, G. R. (2005). A “gain of function” mutation in a protein mediates production of novel modified nucleosides. EMBO J. 24, 1842–1851. doi: 10.1038/sj.emboj.7600666
Chionh, Y. H., McBee, M., Babu, I. R., Hia, F., Lin, W., Zhao, W., et al. (2016). tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence. Nat. Commun. 7:13302. doi: 10.1038/ncomms13302
Crain, P. F., Alfonzo, J. D., Rozenski, J., Kapushoc, S. T., McCloskey, J. A., and Simpson, L. (2002). Modification of the universally unmodified uridine-33 in a mitochondria-imported edited tRNA and the role of the anticodon arm structure on editing efficiency. RNA 8, 752–761. doi: 10.1017/S1355838202022045
Dahl, J. U., Radon, C., Buhning, M., Nimtz, M., Leichert, L. I., Denis, Y., et al. (2013). The sulfur carrier protein TusA has a pleiotropic role in Escherichia coli that also affects molybdenum cofactor biosynthesis. J. Biol. Chem. 288, 5426–5442. doi: 10.1074/jbc.M112.431569
Dal Magro, C., Keller, P., Kotter, A., Werner, S., Duarte, V., Marchand, V., et al. (2018). A vastly increased chemical variety of RNA modifications containing a thioacetal structure. Angew. Chem. Int. Ed. Engl. 57, 7893–7897. doi: 10.1002/anie.201713188
Dewez, M., Bauer, F., Dieu, M., Raes, M., Vandenhaute, J., and Hermand, D. (2008). The conserved Wobble uridine tRNA thiolase Ctu1-Ctu2 is required to maintain genome integrity. Proc. Natl. Acad. Sci. U.S.A. 105, 5459–5464. doi: 10.1073/pnas.0709404105
Dumelin, C. E., Chen, Y., Leconte, A. M., Chen, Y. G., and Liu, D. R. (2012). Discovery and biological characterization of geranylated RNA in bacteria. Nat. Chem. Biol. 8, 913–919. doi: 10.1038/nchembio.1070
Dunbar, K. L., Scharf, D. H., Litomska, A., and Hertweck, C. (2017). Enzymatic carbon-sulfur bond formation in natural product biosynthesis. Chem. Rev. 117, 5521–5577. doi: 10.1021/acs.chemrev.6b00697
Durant, P. C., Bajji, A. C., Sundaram, M., Kumar, R. K., and Davis, D. R. (2005). Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: the effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry 44, 8078–8089. doi: 10.1021/bi050343f
Elseviers, D., Petrullo, L. A., and Gallagher, P. J. (1984). Novel E. coli mutants deficient in biosynthesis of 5-methylaminomethyl-2-thiouridine. Nucleic Acids Res. 12, 3521–3534. doi: 10.1093/nar/12.8.3521
Esberg, A., Huang, B., Johansson, M. J., and Bystrom, A. S. (2006). Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell. 24, 139–148. doi: 10.1016/j.molcel.2006.07.031
Esberg, B., Leung, H. C., Tsui, H. C., Bjork, G. R., and Winkler, M. E. (1999). Identification of the miaB gene, involved in methylthiolation of isopentenylated A37 derivatives in the tRNA of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 181, 7256–7265.
Favre, A., Yaniv, M., and Michelson, A. M. (1969). The photochemistry of 4-thiouridine in Escherichia coli t-RNA Val1. Biochem. Biophys. Res. Commun. 37, 266–271. doi: 10.1016/0006-291X(69)90729-3
Flint, D. H. (1996). Escherichia coli contains a protein that is homologous in function and N-terminal sequence to the protein encoded by the nifS gene of Azotobacter vinelandii and that can participate in the synthesis of the Fe-S cluster of dihydroxy-acid dehydratase. J. Biol. Chem. 271, 16068–16074.
Forouhar, F., Arragain, S., Atta, M., Gambarelli, S., Mouesca, J. M., Hussain, M., et al. (2013). Two Fe-S clusters catalyze sulfur insertion by radical-SAM methylthiotransferases. Nat. Chem. Biol. 9, 333–338. doi: 10.1038/nchembio.1229
Griffey, R. H., Davis, D. R., Yamaizumi, Z., Nishimura, S., Hawkins, B. L., and Poulter, C. D. (1986). 15N-labeled tRNA. Identification of 4-thiouridine in Escherichia coli tRNASer1 and tRNATyr2 by 1H-15N two-dimensional NMR spectroscopy. J. Biol. Chem. 261, 12074–12078.
Hidese, R., Mihara, H., and Esaki, N. (2011). Bacterial cysteine desulfurases: versatile key players in biosynthetic pathways of sulfur-containing biofactors. Appl. Microbiol. Biotechnol. 91, 47–61. doi: 10.1007/s00253-011-3336-x
Horie, A., Tomita, T., Saiki, A., Kono, H., Taka, H., Mineki, R., et al. (2009). Discovery of proteinaceous N-modification in lysine biosynthesis of Thermus thermophilus. Nat. Chem. Biol. 5, 673–679. doi: 10.1038/nchembio.198
Horie, N., Hara-Yokoyama, M., Yokoyama, S., Watanabe, K., Kuchino, Y., Nishimura, S., et al. (1985). Two tRNAIle1 species from an extreme thermophile, Thermus thermophilus HB8: effect of 2-thiolation of ribothymidine on the thermostability of tRNA. Biochemistry 24, 5711–5715. doi: 10.1021/bi00342a004
Ida, T., Sawa, T., Ihara, H., Tsuchiya, Y., Watanabe, Y., Kumagai, Y., et al. (2014). Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc. Natl. Acad. Sci. U.S.A. 111, 7606–7611. doi: 10.1073/pnas.1321232111
Ikeuchi, Y., Shigi, N., Kato, J., Nishimura, A., and Suzuki, T. (2006). Mechanistic insights into sulfur relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol. Cell. 21, 97–108. doi: 10.1016/j.molcel.2005.11.001
Ishikura, H., Yamada, Y., and Nishimura, S. (1971). The nucleotide sequence of a serine tRNA from Escherichia coli. FEBS Lett. 16, 68–70. doi: 10.1016/0014-5793(71)80688-9
Jager, G., Leipuviene, R., Pollard, M. G., Qian, Q., and Bjork, G. R. (2004). The conserved Cys-X1-X2-Cys motif present in the TtcA protein is required for the thiolation of cytidine in position 32 of tRNA from Salmonella enterica Serovar typhimurium. J. Bacteriol. 186, 750–757. doi: 10.1128/JB.186.3.750-757.2004
Jenner, L. B., Demeshkina, N., Yusupova, G., and Yusupov, M. (2010). Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol. 17, 555–560. doi: 10.1038/nsmb.1790
Johansson, M. J., Esberg, A., Huang, B., Bjork, G. R., and Bystrom, A. S. (2008). Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol. Cell. Biol. 28, 3301–3312. doi: 10.1128/MCB.01542-07
Kambampati, R., and Lauhon, C. T. (2003). MnmA and IscS are required for in vitro 2-thiouridine biosynthesis in Escherichia coli. Biochemistry 42, 1109–1117. doi: 10.1021/bi026536+
Kirino, Y., and Suzuki, T. (2005). Human mitochondrial diseases associated with tRNA wobble modification deficiency. RNA Biol. 2, 41–44. doi: 10.4161/rna.2.2.1610
Kowalak, J. A., Dalluge, J. J., McCloskey, J. A., and Stetter, K. O. (1994). The role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry 33, 7869–7876. doi: 10.1021/bi00191a014
Krishnamoorthy, K., and Begley, T. P. (2011). Protein thiocarboxylate-dependent methionine biosynthesis in Wolinella succinogenes. J. Am. Chem. Soc. 133, 379–386. doi: 10.1021/ja107424t
Landgraf, B. J., Arcinas, A. J., Lee, K. H., and Booker, S. J. (2013). Identification of an intermediate methyl carrier in the radical S-adenosylmethionine methylthiotransferases RimO and MiaB. J. Am. Chem. Soc. 135, 15404–15416. doi: 10.1021/ja4048448
Lanz, N. D., and Booker, S. J. (2015). Auxiliary iron-sulfur cofactors in radical SAM enzymes. Biochim. Biophys. Acta 1853, 1316–1334. doi: 10.1016/j.bbamcr.2015.01.002
Lauhon, C. T. (2002). Requirement for IscS in biosynthesis of all thionucleosides in Escherichia coli. J. Bacteriol. 184, 6820–6829. doi: 10.1128/JB.184.24.6820-6829.2002
Lauhon, C. T., and Kambampati, R. (2000). The iscS gene in Escherichia coli is required for the biosynthesis of 4-thiouridine, thiamin, and NAD. J. Biol. Chem. 275, 20096–20103. doi: 10.1074/jbc.M002680200
Lauhon, C. T., Skovran, E., Urbina, H. D., Downs, D. M., and Vickery, L. E. (2004). Substitutions in an active site loop of Escherichia coli IscS result in specific defects in Fe-S cluster and thionucleoside biosynthesis in vivo. J. Biol. Chem. 279, 19551–19558. doi: 10.1074/jbc.M401261200
Laxman, S., Sutter, B. M., Wu, X., Kumar, S., Guo, X., Trudgian, D. C., et al. (2013). Sulfur amino acids regulate translational capacity and metabolic homeostasis through modulation of tRNA thiolation. Cell 154, 416–429. doi: 10.1016/j.cell.2013.06.043
Leidel, S., Pedrioli, P. G., Bucher, T., Brost, R., Costanzo, M., Schmidt, A., et al. (2009). Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature 458, 228–232. doi: 10.1038/nature07643
Leimkuhler, S., Wuebbens, M. M., and Rajagopalan, K. V. (2011). The history of the discovery of the molybdenum cofactor and novel aspects of its biosynthesis in bacteria. Coord. Chem. Rev. 255, 1129–1144. doi: 10.1016/j.ccr.2010.12.003
Leipuviene, R., Qian, Q., and Bjork, G. R. (2004). Formation of thiolated nucleosides present in tRNA from Salmonella enterica Serovar typhimurium occurs in two principally distinct pathways. J. Bacteriol. 186, 758–766. doi: 10.1128/JB.186.3.758-766.2004
Lipsett, M. N. (1965). The isolation of 4-thiouridylic acid from the soluble ribonucleic acid of Escherichia coli. J. Biol. Chem. 240, 3975–3978.
Litomska, A., Ishida, K., Dunbar, K. L., Boettger, M., Coyne, S., and Hertweck, C. (2018). Enzymatic thioamide formation in bacterial antimetabolite pathway. Angew Chem. Int. Ed. Engl. 57, 36. doi: 10.1002/anie.201804158
Liu, Y., Sieprawska-Lupa, M., Whitman, W. B., and White, R. H. (2010). Cysteine is not the sulfur source for iron-sulfur cluster and methionine biosynthesis in the methanogenic archaeon Methanococcus maripaludis. J. Biol. Chem. 285, 31923–31929. doi: 10.1074/jbc.M110.152447
Liu, Y., Vinyard, D. J., Reesbeck, M. E., Suzuki, T., Manakongtreecheep, K., Holland, P. L., et al. (2016). A [3Fe-4S] cluster is required for tRNA thiolation in archaea and eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 113, 12703–12708. doi: 10.1073/pnas.1615732113
Liu, Y., Zhu, X., Nakamura, A., Orlando, R., Soll, D., and Whitman, W. B. (2012). Biosynthesis of 4-thiouridine in tRNA in the methanogenic archaeon Methanococcus maripaludis. J. Biol. Chem. 287, 36683–36692. doi: 10.1074/jbc.M112.405688
Mahanta, N., Liu, A., Dong, S., Nair, S. K., and Mitchell, D. A. (2018). Enzymatic reconstitution of ribosomal peptide backbone thioamidation. Proc. Natl. Acad. Sci. U.S.A. 115, 3030–3035. doi: 10.1073/pnas.1722324115
Matsuda, K., Hasebe, F., Shiwa, Y., Kanesaki, Y., Tomita, T., Yoshikawa, H., et al. (2017). Genome mining of amino group carrier protein-mediated machinery: discovery and biosynthetic characterization of a natural product with unique hydrazone unit. ACS Chem. Biol. 12, 124–131. doi: 10.1021/acschembio.6b00818
Matthijs, S., Baysse, C., Koedam, N., Tehrani, K. A., Verheyden, L., Budzikiewicz, H., et al. (2004). The Pseudomonas siderophore quinolobactin is synthesized from xanthurenic acid, an intermediate of the kynurenine pathway. Mol. Microbiol. 52, 371–384. doi: 10.1111/j.1365-2958.2004.03999.x
Maynard, N. D., Macklin, D. N., Kirkegaard, K., and Covert, M. W. (2012). Competing pathways control host resistance to virus via tRNA modification and programmed ribosomal frameshifting. Mol. Syst. Biol. 8:567. doi: 10.1038/msb.2011.101
Mueller, E. G. (2006). Trafficking in persulfides: delivering sulfur in biosynthetic pathways. Nat. Chem. Biol. 2, 185–194. doi: 10.1038/nchembio779
Mueller, E. G., Buck, C. J., Palenchar, P. M., Barnhart, L. E., and Paulson, J. L. (1998). Identification of a gene involved in the generation of 4-thiouridine in tRNA. Nucleic Acids Res. 26, 2606–2610. doi: 10.1093/nar/26.11.2606
Mueller, E. G., and Palenchar, P. M. (1999). Using genomic information to investigate the function of ThiI, an enzyme shared between thiamin and 4-thiouridine biosynthesis. Protein Sci. 8, 2424–2427. doi: 10.1110/ps.8.11.2424
Murao, K., Tanabe, T., Ishii, F., Namiki, M., and Nishimura, S. (1972). Primary sequence of arginine transfer RNA from Escherichia coli. Biochem. Biophys. Res. Commun. 47, 1332–1337. doi: 10.1016/0006-291X(72)90218-5
Murphy, F. V., Ramakrishnan, V., Malkiewicz, A., and Agris, P. F. (2004). The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat. Struct. Mol. Biol. 11, 1186–1191. doi: 10.1038/nsmb861
Nakagawa, H., Kuratani, M., Goto-Ito, S., Ito, T., Katsura, K., Terada, T., et al. (2013). Crystallographic and mutational studies on the tRNA thiouridine synthetase TtuA. Proteins 81, 1232–1244. doi: 10.1002/prot.24273
Nakai, Y., Nakai, M., Lill, R., Suzuki, T., and Hayashi, H. (2007). Thio modification of yeast cytosolic tRNA is an iron-sulfur protein-dependent pathway. Mol. Cell. Biol. 27, 2841–2847. doi: 10.1128/MCB.01321-06
Nedialkova, D. D., and Leidel, S. A. (2015). Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell 161, 1606–1618. doi: 10.1016/j.cell.2015.05.022
Neumann, P., Lakomek, K., Naumann, P. T., Erwin, W. M., Lauhon, C. T., and Ficner, R. (2014). Crystal structure of a 4-thiouridine synthetase-RNA complex reveals specificity of tRNA U8 modification. Nucleic Acids Res. 42, 6673–6685. doi: 10.1093/nar/gku249
Nilsson, K., Lundgren, H. K., Hagervall, T. G., and Bjork, G. R. (2002). The cysteine desulfurase IscS is required for synthesis of all five thiolated nucleosides present in tRNA from Salmonella enterica Serovar typhimurium. J. Bacteriol. 184, 6830–6835. doi: 10.1128/JB.184.24.6830-6835.2002
Noma, A., Sakaguchi, Y., and Suzuki, T. (2009). Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 37, 1335–1352. doi: 10.1093/nar/gkn1023
Numata, T., Ikeuchi, Y., Fukai, S., Suzuki, T., and Nureki, O. (2006). Snapshots of tRNA sulphuration via an adenylated intermediate. Nature 442, 419–424. doi: 10.1038/nature04896
Oashi, Z., Saneyoshi, M., Harada, F., Hara, H., and Nishimura, S. (1970). Presumed anticodon structure of glutamic acid tRNA from E. coli: a possible location of a 2-thiouridine derivative in the first position of the anticodon. Biochem. Biophys. Res. Commun. 40, 866–872. doi: 10.1016/0006-291X(70)90983-6
Ouchi, T., Tomita, T., Horie, A., Yoshida, A., Takahashi, K., Nishida, H., et al. (2013). Lysine and arginine biosyntheses mediated by a common carrier protein in Sulfolobus. Nat. Chem. Biol. 9, 277–283. doi: 10.1038/nchembio.1200
Pierrel, F., Bjork, G. R., Fontecave, M., and Atta, M. (2002). Enzymatic modification of tRNAs: MiaB is an iron-sulfur protein. J. Biol. Chem. 277, 13367–13370. doi: 10.1074/jbc.C100609200
Rajakovich, L. J., Tomlinson, J., and Dos Santos, P. C. (2012). Functional analysis of Bacillus subtilis genes involved in the biosynthesis of 4-Thiouridine in tRNA. J. Bacteriol. 194, 4933–4940. doi: 10.1128/JB.00842-12
Ryals, J., Hsu, R. Y., Lipsett, M. N., and Bremer, H. (1982). Isolation of single-site Escherichia coli mutants deficient in thiamine and 4-thiouridine syntheses: identification of a nuvC mutant. J. Bacteriol. 151, 899–904.
Sasaki, E., Zhang, X., Sun, H. G., Lu, M. Y., Liu, T. L., Ou, A., et al. (2014). Co-opting sulphur-carrier proteins from primary metabolic pathways for 2-thiosugar biosynthesis. Nature 510, 427–431. doi: 10.1038/nature13256
Schindelin, H., Kisker, C., and Rajagopalan, K. V. (2001). Molybdopterin from molybdenum and tungsten enzymes. Adv. Protein Chem. 58, 47–94. doi: 10.1016/S0065-3233(01)58002-X
Settembre, E., Begley, T. P., and Ealick, S. E. (2003). Structural biology of enzymes of the thiamin biosynthesis pathway. Curr. Opin. Struct. Biol. 13, 739–747. doi: 10.1016/j.sbi.2003.10.006
Shi, R., Proteau, A., Villarroya, M., Moukadiri, I., Zhang, L., Trempe, J. F., et al. (2010). Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions. PLoS Biol. 8:e1000354. doi: 10.1371/journal.pbio.1000354
Shigi, N. (2012). Posttranslational modification of cellular proteins by a ubiquitin-like protein in bacteria. J. Biol. Chem. 287, 17568–17577. doi: 10.1074/jbc.M112.359844
Shigi, N. (2014). Biosynthesis and functions of sulfur modifications in tRNA. Front Genet 5:67. doi: 10.3389/fgene.2014.00067
Shigi, N. (2016). “Sulfur modifications in tRNA: function and implications for human disease,” in Modified Nucleic Acids in Biology and Medicine, eds S. Jurga, V. A. Erdmann, and J. Barciszewski (Berlin: Springer), 55–71. doi: 10.1007/978-3-319-34175-0_3
Shigi, N., Asai, S. I., and Watanabe, K. (2016). Identification of a rhodanese-like protein involved in thiouridine biosynthesis in Thermus thermophilus tRNA. FEBS Lett. 590, 4628–4637. doi: 10.1002/1873-3468.12499
Shigi, N., Sakaguchi, Y., Asai, S., Suzuki, T., and Watanabe, K. (2008). Common thiolation mechanism in the biosynthesis of tRNA thiouridine and sulphur-containing cofactors. EMBO J. 27, 3267–3278. doi: 10.1038/emboj.2008.246
Shigi, N., Sakaguchi, Y., Suzuki, T., and Watanabe, K. (2006a). Identification of two tRNA thiolation genes required for cell growth at extremely high temperatures. J. Biol. Chem. 281, 14296–14306. doi: 10.1074/jbc.M511675200
Shigi, N., Suzuki, T., Tamakoshi, M., Oshima, T., and Watanabe, K. (2002). Conserved bases in the TPsi C loop of tRNA are determinants for thermophile-specific 2-thiouridylation at position 54. J. Biol. Chem. 277, 39128–39135. doi: 10.1074/jbc.M207323200
Shigi, N., Suzuki, T., Terada, T., Shirouzu, M., Yokoyama, S., and Watanabe, K. (2006b). Temperature-dependent biosynthesis of 2-thioribothymidine of Thermus thermophilus tRNA. J. Biol. Chem. 281, 2104–2113. doi: 10.1074/jbc.M510771200
Sierant, M., Kulik, K., Sochacka, E., Szewczyk, R., Sobczak, M., and Nawrot, B. (2018a). Cytochrome c catalyzes the hydrogen peroxide-assisted oxidative desulfuration of 2-Thiouridines in Transfer RNAs. Chembiochem 19, 687–695. doi: 10.1002/cbic.201700692
Sierant, M., Leszczynska, G., Sadowska, K., Komar, P., Radzikowska-Cieciura, E., Sochacka, E., et al. (2018b). Escherichia coli tRNA 2-selenouridine synthase (SelU) converts S2U-RNA to Se2U-RNA via S-geranylated-intermediate. FEBS Lett. 592, 2248–2258. doi: 10.1002/1873-3468.13124
Singer, C. E., and Smith, G. R. (1972). Histidine regulation in Salmonella typhimurium. 13. Nucleotide sequence of histidine transfer ribonucleic acid. J. Biol. Chem. 247, 2989–3000.
Sochacka, E., Bartos, P., Kraszewska, K., and Nawrot, B. (2013). Desulfuration of 2-thiouridine with hydrogen peroxide in the physiological pH range 6.6-7.6 is pH-dependent and results in two distinct products. Bioorg. Med. Chem. Lett. 23, 5803–5805. doi: 10.1016/j.bmcl.2013.08.114
Suzuki, T., Suzuki, T., Wada, T., Saigo, K., and Watanabe, K. (2002). Taurine as a constituent of mitochondrial tRNAs: new insights into the functions of taurine and human mitochondrial diseases. EMBO J. 21, 6581–6589. doi: 10.1093/emboj/cdf656
Takahashi, N., Wei, F. Y., Watanabe, S., Hirayama, M., Ohuchi, Y., Fujimura, A., et al. (2017). Reactive sulfur species regulate tRNA methylthiolation and contribute to insulin secretion. Nucleic Acids Res. 45, 435–445. doi: 10.1093/nar/gkw745
Tao, W., Yurkovich, M. E., Wen, S., Lebe, K. E., Samborskyy, M., Liu, Y., et al. (2016). A genomics-led approach to deciphering the mechanism of thiotetronate antibiotic biosynthesis. Chem. Sci. 7, 376–385. doi: 10.1039/c5sc03059e
Tigano, M., Ruotolo, R., Dallabona, C., Fontanesi, F., Barrientos, A., Donnini, C., et al. (2015). Elongator-dependent modification of cytoplasmic tRNALysUUU is required for mitochondrial function under stress conditions. Nucleic Acids Res. 43, 8368–8380. doi: 10.1093/nar/gkv765
Tyagi, K., and Pedrioli, P. G. (2015). Protein degradation and dynamic tRNA thiolation fine-tune translation at elevated temperatures. Nucleic Acids Res. 43, 4701–4712. doi: 10.1093/nar/gkv322
Umeda, N., Suzuki, T., Yukawa, M., Ohya, Y., Shindo, H., Watanabe, K., et al. (2005). Mitochondria-specific RNA-modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs. Implications for the molecular pathogenesis of human mitochondrial diseases. J. Biol. Chem. 280, 1613–1624. doi: 10.1074/jbc.M409306200
Väre, V. Y., Eruysal, E. R., Narendran, A., Sarachan, K. L., and Agris, P. F. (2017). Chemical and conformational diversity of modified nucleosides affects tRNA structure and function. Biomolecules 7:E29. doi: 10.3390/biom7010029
Watanabe, K., Oshima, T., Saneyoshi, M., and Nishimura, S. (1974). Replacement of ribothymidine by 5-methyl-2-thiouridine in sequence GT psi C in tRNA of an extreme thermophile. FEBS Lett. 43, 59–63. doi: 10.1016/0014-5793(74)81105-1
Watanabe, K., Shinma, M., Oshima, T., and Nishimura, S. (1976). Heat-induced stability of tRNA from an extreme thermophile, Thermus thermophilus. Biochem. Biophys. Res. Commun. 72, 1137–1144. doi: 10.1016/S0006-291X(76)80250-1
Wei, F. Y., Suzuki, T., Watanabe, S., Kimura, S., Kaitsuka, T., Fujimura, A., et al. (2011). Deficit of tRNA(Lys) modification by Cdkal1 causes the development of type 2 diabetes in mice. J. Clin. Invest. 121, 3598–3608. doi: 10.1172/JCI58056
Wittwer, A. J., Tsai, L., Ching, W. M., and Stadtman, T. C. (1984). Identification and synthesis of a naturally occurring selenonucleoside in bacterial tRNAs: 5-[(methylamino)methyl]-2-selenouridine. Biochemistry 23, 4650–4655. doi: 10.1021/bi00315a021
Wu, Y., Wei, F. Y., Kawarada, L., Suzuki, T., Araki, K., Komohara, Y., et al. (2016). Mtu1-Mediated thiouridine formation of mitochondrial tRNAs is required for mitochondrial translation and is involved in reversible infantile liver injury. PLoS Genet. 12:e1006355. doi: 10.1371/journal.pgen.1006355
Keywords: biosynthesis, iron-sulfur cluster, post-transcriptional modification, radical SAM enzyme, sulfurtransferase, sulfur modification, tRNA
Citation: Shigi N (2018) Recent Advances in Our Understanding of the Biosynthesis of Sulfur Modifications in tRNAs. Front. Microbiol. 9:2679. doi: 10.3389/fmicb.2018.02679
Received: 04 August 2018; Accepted: 19 October 2018;
Published: 01 November 2018.
Edited by:
Alexander Loy, Universität Wien, AustriaReviewed by:
Patricia Coutinho Dos Santos, Wake Forest University, United StatesJaunius Urbonavičius, Vilnius Gediminas Technical University, Lithuania
Copyright © 2018 Shigi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Naoki Shigi, bmFva2ktc2hpZ2lAYWlzdC5nby5qcA==
 Naoki Shigi
Naoki Shigi