- 1State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan, China
- 2The Provincial Key Lab of Plant Pathology of Hubei Province, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, China
Coniothyrium minitans is a sclerotial parasite, which has been investigated for commercial control of crop diseases caused by Sclerotinia sclerotiorum. Previously, we obtained a T-DNA insertional mutant, ZS-1TN24363, which did not produce melanin during conidiation. To understand the function of melanin in C. minitans, we cloned the gene that was disrupted by the T-DNA insertion, and found that this gene, called CmMR1, encoded a putative protein of 1,011 amino acids, which is a homolog of the transcription factor MR. Full-length CmMR1 contains 3,167 bp, with three exons and two introns. To confirm that the disrupted gene is responsible for the melanin-deficiency of the mutant, CmMR1 was disrupted and three targeted knockout mutants were obtained. Biological assays showed that the phenotype of the targeted knockout mutants was similar to that of the T-DNA insertional mutant. Furthermore, gene complementation confirmed that CmMR1 is responsible for the mutant phenotype. CmMR1 disruption did not affect hyphal growth, conidiation, and parasitization of C. minitans, however, the ROS accumulation increased and tolerance to UV light decreased significantly in the mutants. Our result may enhance the understanding of melanin in the ecology of C. minitans on molecular level.
Introduction
Coniothyrium minitans is a mycoparasite of Sclerotinia spp. and certain species in other related genera (Campbell, 1947; Li et al., 2006), and it parasitizes both the hypha and sclerotia of Sclerotinia sclerotiorum. C. minitans has been proved to be able to control Sclerotinia rots of vegetable crops, stem rot of rapeseed (Brassica napus), and head rot of sunflower (Huang, 1981; Whipps and Gerlagh, 1992). C. minitans has been developed as a commercial biological agent and been widely used in the United States, European Union, and China. C. minitans is a coelomycete, which grows on sclerotia, and produces pycnidia with numerous conidia on or in the sclerotia (Whipps et al., 2008). It also grows well on media such as on potato dextrose agar (PDA) and wheat kernel and forms pycnidia. The pycnidia are initially white, and darken with time. Conidia are dispersed with water droplets from the pycnidia. C. minitans synthesizes melanin, which accumulates on the pycnidia, conidia, and the aging hyphae, lending a dark appearance to the entire mature colony.
Many fungi synthesize melanin (Bell and Wheeler, 1986). There are two melanin synthesis pathways; one uses 1,8-dihydroxynaphthalene (DHN) as the initial molecule whereas the other uses L-3,4-dihydroxyphenylalanine (DOPA) (Plonka and Grabacka, 2006). Several studies showed that the melanin synthesis pathways in fungi are regulated by the MR transcription factor. Two transcription factors involved in melanin synthesis were identified (CMR1 and PIG1) in Colletotrichum lagenarium and Magnaporthe grisea. Upon disruption of CMR1, the colony of C. lagenarium turned to reddish brown from dark brown. Furthermore, CMR1 up-regulates the expression of SCD1 and THR1, which encode scytalone dehydratase and T3HN reductase, respectively (Tsuji et al., 2000). Disruption of CMR1 in Cochliobolus heterostrophus leads to melanin shortage, change in colony color to orange or pink, and significant decrease in conidial production. The expression of SCD1, BRN1 (T3HN reductase), and BRN2 (T4HN reductase) could not be determined in the mutant, whereas the expression of PKS18 was moderately down-regulated (Eliahu et al., 2007). In Bipolaris oryzae, disruption of BMR1 caused albinism in the mutants, and over-expression of BMR1 in this fungus enhanced the expression of PKS, SCD1, and THR1. BMR1 levels in the overexpression transformants were ten times that of the wild-type strains (Kihara et al., 2008). Interestingly, disruption of Amr1 in Alternaria brassicicola resulted in impaired accumulation of melanin; however, the pathogenicity was significantly enhanced and many genes encoding hydrolases were up-regulated in the disruption mutants (Cho et al., 2012).
In previous research, we constructed a transfer DNA (T-DNA) insertional library of C. minitans strain ZS-1 (Li et al., 2005). A whitish mutant, ZS-1TN24363, was screened from this mutant library. A MR transcription factor gene was disrupted by the T-DNA insertion in ZS-1TN24363. We hypothesized that MR transcription factor regulated melanin in C. minitans. In this study, the relationship of the phenotype and gene disruption was confirmed and the potential ecological role of melanin in C. minitans was discussed.
Materials and Methods
Fungal Strains, Media, and Maintenance
Coniothyrium minitans strain ZS-1 was originally isolated from soil at Zhushan county, Hubei province, China; S. sclerotiorum strain 1980 is a genome-sequenced strain gifted by Dr. Weidong Chen of the Washington State University. Fungal strains were grown on PDA medium, maintained on PDA slants, and stored at 4°C. Bacteria used for molecular experiments were grown on Luria-Bertani (LB) medium. IM medium and Co-medium were used for fungal transformation mediated by Agrobacterium tumefaciens (Gong et al., 2007; Wei et al., 2016; Yang et al., 2016).
Gene Cloning, RT-PCR Amplification, and Phylogenetic Analysis
Mycelial mass of mutant ZS-1TN24363 was collected from the colonies cultured on PDA medium for DNA extraction using the cetyltrimethylammonium bromide (CTAB) method. HiTAIL-PCR was used to clone the flanking DNA segment of the T-DNA insertional site (Liu and Chen, 2007; Xiao et al., 2014). RNA samples were extracted using a RNA extraction kit of Invitrogen. Complementary DNA (cDNA) was synthesized using a cDNA synthesis kit provided by TransGen Biotech (Beijing, China); DNA sequencing analysis was performed by the TianYiHuiYuan (Guangzhou, China) and the DNA sequence was analyzed using the Basic Local Alignment Search Tool for Nucleotides (BLASTN) against the local database of C. minitans to retrieve sequence information of the putative disrupted gene; the expression of the putative disrupted gene was determined using RT-PCR with primer pairs CmMR1-F/CmMR1-R. Actin was checked as a control with Actin-F/Actin-R. The putative disrupted gene was further analyzed using the DNAMAN program to explore the encoded protein, and BLASTP analysis was performed using the National Center of Biotechnology Information’s (NCBI) (GenBank) database. Homologs of other fungi were collected and analyzed, and the phylogenetic tree was constructed using the neighbor-joining method of the MEGA 6.0 program. The expression pattern of melanin synthesis associated gene CmSCD, Cm3HNR and Cm4HNR in strain ZS-1 and the mutant ZS-1TN24363 was monitored by quantitative real time RT-PCR (qRT-PCR) amplification. Data was acquisited and analyzed by using the Bio-Rad CFX ManagerTM Software (version 2.0). The relative expression level of each target gene was quantified by the comparative CT method (2-ΔΔCt) of quantification. Primer pairs used in this study are listed in Table 1.
Gene Knockout and Complementation
To investigate whether the disrupted gene was responsible for the change in phenotype of the T-DNA insertional mutant ZS-1TN24363, both gene knockout and gene complementation were performed. Split-marker recombination (Zhang et al., 2016) was used to knockout the potential gene. Two genomic DNA segments were amplified using the DNA sample from the wild-type strain ZS-1 as template; one fragment upstream of the target gene (fragment U) was amplified using primer pair U-F/R; the other fragment (fragment D) down-stream of the target gene was amplified using primer pair D-F/R. The hygromycin-resistance gene (HYG) was PCR amplified from the plasmid pKS1004 (Zhang et al., 2016) with primer pairs HYG-F/HYG-R. Fragments U and HYG were mixed as DNA template for PCR amplification with primer pairs U-F and HY-R, and the PCR product was named UHY; fragments D and HYG were mixed as DNA template for PCR amplification with primer pairs YG-F and D-R, and the PCR product was named YGD. The position of the primers was included in Supplementary Figure S3. The PCR products UHY and YGD were mixed and transformed in protoplasts of C. minitans wild-type strain ZS-1 using polyethylene glycol (PEG) mediated technique (Wei et al., 2013; Zhang et al., 2016). The transformed protoplasts were spread on regeneration medium containing 50 μg/μL hygromycin and placed in an incubator for 1 week; colonies growing on the medium were transferred to fresh hygromycin-amended PDA plates and further cultured as gene-knockout candidates. Four primers were designed to confirm these candidates; one pair (KOG-F/R) was designed on the target gene, another primer (KOU-F) on the up-stream region of the target gene, and the last primer (KOD-R) on the down-stream region of the target gene. DNA samples were extracted from candidate knockout mutants and amplified by PCR. The primer pairs KOG-F/R, KOU-F/HY-R, YG-F/KOD-R, and HYG-F/HY-R were used to confirm knockout of the target genes.
Based on the local genome database of C. minitans, the promoter of the target gene was predicted using the program on http://www.fruitfly.org/seq_tools/promoter.html. The promoter region is about 1,000 nucleotides up-stream of the ATG codon of the target gene. The primer pairs were designed based on the predicted results, and the genomic DNA of strain ZS-1 was used for PCR amplification with primer pairs CO-F/R. Then, the full-length gene with the promoter region was amplified by PCR and digested with KpnI. p3300 was also digested and dephosphorylated. Then PCR products were ligated on the dephosphorylated vector using the T4-DNA ligase to obtain a gene complementary vector. This vector was introduced into A. tumefaciens strain EHA105 for fungal transformation of the knockout mutants following the method described by Wei et al. (2016) and Yang et al. (2016). The candidate gene-complemented transformants were further confirmed using PCR amplification with primer pairs KOG-F/R, HYG-F/HY-R and NEO-F/R.
Biological Assay
Colony Morphology and Growth Rate Determination
Knockout mutants, complemented tranformants, ZS-1TN24363, and the wild-type strain ZS-1 were activated on PDA plates for 3–4 days. Next, hyphal agar disks were taken from the colony margin with a sterilized hole punch (diameter 5 mm), transferred to the center of fresh 20 mL PDA plates (diameter 90 mm), and incubated at 20°C. The colony diameter was measured every day from the 3rd day to the 10th day. Each strain had three replicates (three plates) and the experiment was repeated thrice.
Hyphal Tips Observation
To observe the hyphal tips, all strains mentioned above were activated and transferred on cellophane laid on PDA plates and incubated for 4 days. The colony on cellophane was cut into pieces, and hyphal tips were observed under a light microscope. Typical hyphal tips were imaged with a digital camera (Nikon). This experiment was repeated thrice.
Biomass Determination
To determine the biomass of the strains growing on PDA medium, all strains were activated on PDA plates, the hyphal agar disks were punched and placed on cellophane laid on PDA medium, and incubated at 20°C for 10 days. The whole mycelial mass was harvested by rolling the colonies. The weight of fresh mass was measured. Each strain had three replicates, and the experiment was repeated thrice.
Conidial Production
To determine the conidial production of the strains, the activated strains were grown on 20 mL PDA plates for 15 days. The whole colony and agar was placed in a mortar with 5 mL distilled water and finely ground, and the liquid was passed through three layers of lens-cleaning paper. The mass was washed again with 5 mL distilled water and the entire liquid was centrifuged at 5,000 rpm for 10 min. The pellet was re-suspended in 1 mL distilled water and the conidial concentration was calculated under the light microscope using a hemocytometer. The conidial production of the entire colony was then calculated. Three colonies of each strain were examined and the experiment was repeated thrice.
Parasitic Ability Assay
Two methods were used to determine the parasitic ability of the strains. One was dual culture with strain 1980 of S. sclerotiorum according to the method described by Zeng et al. (2014). Mutant or the wild-type strains were placed on one side of the PDA plate (diameter 90 mm) and strain 1980 on the opposite side of the same plate at a distance of 60 mm from the mutant or the wild-type strain. The dual culture was incubated for 20 days. The inhibition zone between the colony of C. minitans and that of S. sclerotiorum was determined, if any, and the morphologies of the two colonies were also observed. To quantify the parasitic ability of the mutants, S. sclerotiorum colony was divided into four iso-distance regions, and five disks from the same region were punched out and placed on fresh PDA plates. After 7 days of incubation, the emerging colonies were judged to be those of C. minitans or S. sclerotiorum.
Parasitism to sclerotia were further evaluated according to Jiang et al. (2000). S. sclerotiorum was inoculated on carrot rods in 250 mL flasks for 30 days to produce sclerotia. Sclerotia were harvested and dried at 37°C for 3 days, and sclerotia with similar sizes were selected to test the parasitic ability of the mutants. Conidia of C. minitans were collected from colonies growing on PDA plate for 25 days and diluted with sterilized double distilled water to obtain 106 spores/mL. Sclerotia were submerged in 50% bleach thrice for 5 min to remove the microorganisms on the surface in 70% ethanol for additional 5 min (which was repeated once more), and then washed with double distilled water twice. The surface-sterilized sclerotia were divided into 9 groups with 30 sclerotia per group, and then submerged into the conidial suspension of the wild-type strain ZS-1, mutant ZS-1TN24363, knockout mutants, and complemented transformants for 30 min. Next, the inoculated sclerotia were half-buried in sterilized wet-sand in plates. The plates were sealed and incubated for 30 days. The sclerotia were taken out from the sand and dissected using a surgical knife to rank the rot degree of sclerotia using a method described by Jiang et al. (2000). Each treatment was replicated thrice; desterilized double distilled water was used as the control. The experiments were repeated thrice.
NBT Staining Test
To determine the reactive oxygen species (ROS) content in the mutant colony, the mutants and the wild-type strain of C. minitans were activated and placed on fresh PDA plates for 3 days. Then, mycelia were incubated in 0.05 M phosphate buffer containing 0.05% NBT for 1 h and the reaction was stopped using water-free ethanol. The colonies were observed with naked eyes. Each treatment had three replicates and the experiment was repeated thrice.
Detection of Tolerance to Ultraviolet (UV) Light
To determine if the mutant conidia could tolerate UV light, conidia were harvested from 10-day-old colonies of mutants and the wild-type strain and diluted with sterilized double distilled water to obtain 105 spores/mL. Spore suspension (100 μL) was carefully spread on PDA plates and dried, and irradiated under UV light (20 J/cm2) for 0, 3, 6, and 9 min. The plates were incubated for 36 h, and the germinating conidia were counted under a light microscope. Each strain and mutant had three replicates and this experiment was repeated thrice.
Statistical Analysis
The one way analysis of variance (ANOVA) of the DPS software was used to analyze the significance of the differences between treatments. Means were separated using the Student Newman–Keuls multiple range test when ANOVA was significant at P < 0.01.
Results
Biological Characteristics of the T-DNA Insertional Mutant ZS-1TN24363 of C. minitans
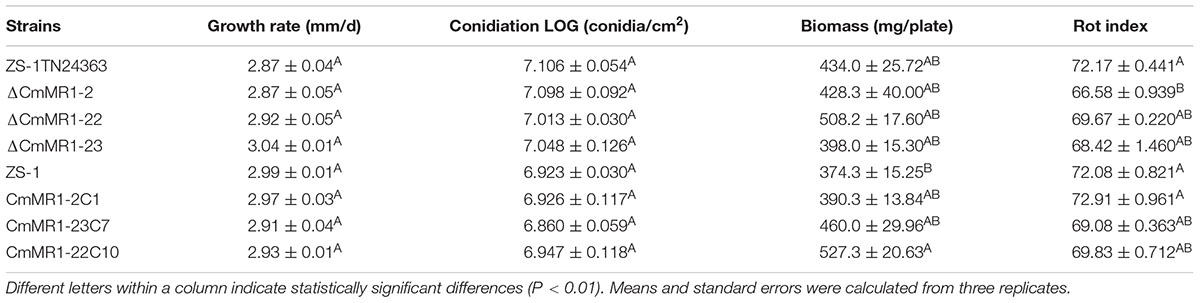
Similar to the wild-type strain, mutant ZS-1TN24363 could grow on PDA medium (Figure 1A). In addition, the hyphal tips of the mutant were similar to those of the wild-type strain (Figure 1B). The mutant could also form pycnidia and conidia like the wild-type. However, the pycnidia formed were whitish to orange in color, which was significantly different from that of the wild-type strain (dark pycnidia) (Figure 1A), suggesting that this mutant could not synthesize melanin. Further investigation revealed that this mutant could parasitize sclerotia and formed white pycnidia on sclerotia, suggesting that this mutant retained parasitic ability (Figure 1C).
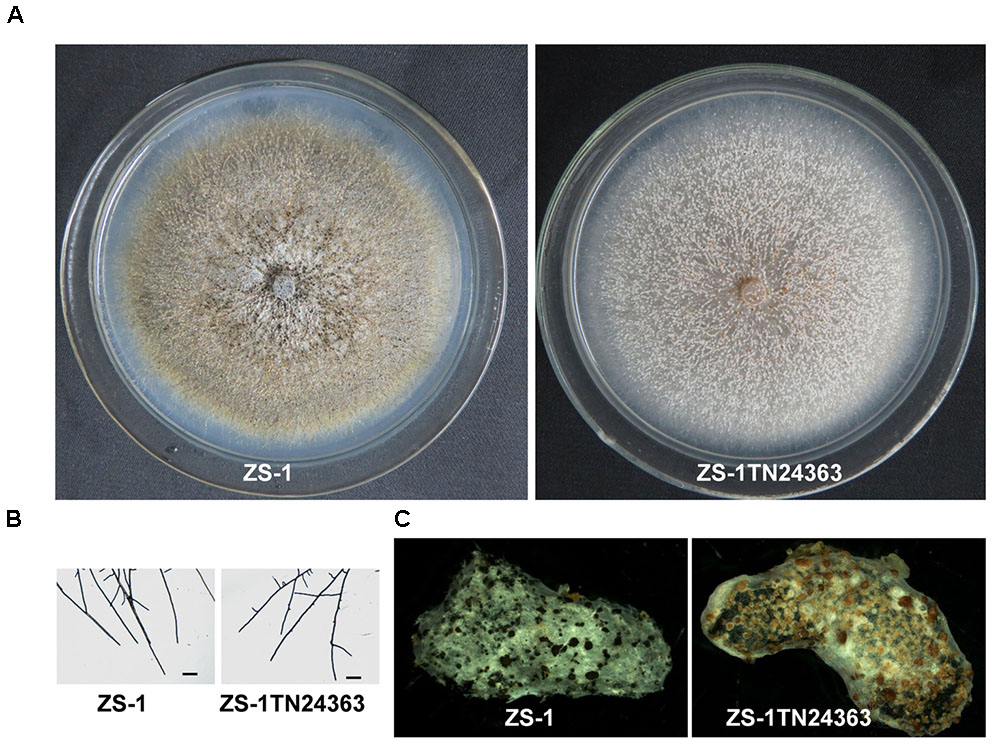
FIGURE 1. Morphology and parasitization of strain ZS-1 and mutant ZS-1TN24363. (A) Colony morphology (PDA, 20°C, 20 days). (B) Hyphal tips (PDA, 20°C, 4 days). (C) Pycnidia formed on sclerotia of S. sclerotiorum (20°C for 30 days). Scale bar means 50 μm.
The T-DNA Disrupted CmMR1 of C. minitans
The flanking 1,000 bp genomic DNA of the T-DNA insertion site was obtained by hiTAIL-PCR (Supplementary Figures S1A,B). This DNA fragment was sequenced and the T-DNA border was identified. The sequence of the flanking DNA was used for searching in a local genome database of C. minitans strain ZS-1. We found that the T-DNA insertion possibly disrupted a gene of 3,167 bp. This gene contained three exons and two introns, and encoded a 1,011 amino acid-long protein (Supplementary Figure S1C). BLASTP analysis on the NCBI database revealed that this protein contained three conserved domains, namely a Zn2Cys6 binuclear cluster domain, a Med3 domain that regulates the transcriptional activity of RNA polymerase II, and a fungal-TF-MHR domain that is generally present in a large family of fungal zinc cluster transcription factors (Supplementary Figure S1D). Thus, we presumed that this protein was a transcription factor that belonged to the MHR superfamily. The proteins of MHR superfamily are conserved in fungi. Phylogenetic analysis revealed that this protein was closely related to CMR1, BMR1, and AMR1 (Supplementary Figure S2), and we named this protein CmMR1 (GenBank Acc. No. MF166860).
To probe if CmMR1 was expressed in the wild-type and mutant strains, the total RNA was extracted from the mycelial mass collected from colonies growing for 48 to 96 h on PDA plates and RNA samples were examined with RT-PCR amplification. The results showed that CmMR1 expressed all the stage from the hyphal growth (48 h) to the pycnidial maturation (96 h) stage in the wild-type strain ZS-1. However, the expression could not be detected at all stage tested in ZS-1TN24363, suggesting that CmMR1 was disrupted by the T-DNA insertion in ZS-1TN24363 (Figure 2A).
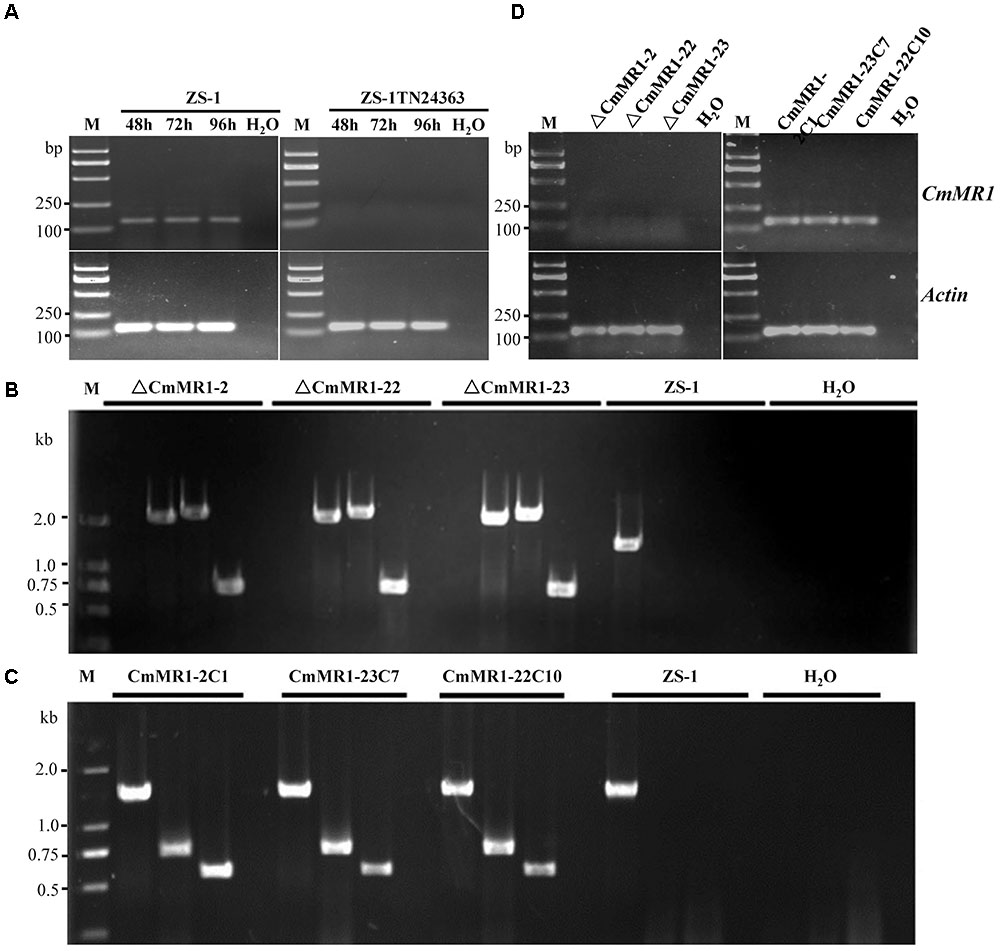
FIGURE 2. Targeted knockout and complementary of CmMR1 in C. minitans. (A) RT-PCR analysis, showing the expression pattern of CmMR1 from 48 hour post inoculation (hpi) to 96 hpi in strain ZS-1 and in the mutant ZS-1TN24363 with primer pairs CmMR1-F/CmMR1-R. Actin was checked as a control with Actin-F/Actin-R. M means DL2000 Marker. (B) Identification of the CmMR1 knockout transformants via PCR amplification; For each strain, the knockout event was confirmed by PCR amplification using primer pairs KOG-F/R (for gene CmMR1, 1,381 bp), HYG-F/HY-R (for HYG, 740 bp), KOU-F/HY-R (for upstream, 2,236 bp), and YG-F/KOD-R (for downstream, 2,499 bp), individually. (C) Identification of the complemented transformants via PCR by primer pairs KOG-F/R, HYG-F/HY-R and NEO-F/R (neomycin-resistance gene fragment, 582 bp). (D) Expression of CmMR1 at 72 hpi in the knockout and complemented transformants via RT-PCR amplification. Actin was checked as a control with Actin-F/Actin-R. M means DL2000 Marker.
Target Knockout and Gene Complementation
To confirm that CmMR1 is responsible for the change in colony morphology in ZS-1TN24363, both targeted gene knockout and gene complementation were conducted. The strategy for targeted knockout of CmMR1 is shown in Supplementary Figure S3. Three knockout candidates were obtained and named ΔCmMR1-2, ΔCmMR1-22, and ΔCmMR1-23, respectively. PCR amplification with three primer pairs confirmed the targeted knockout event. In all candidates, a 1,381 bp fragment could not be amplified with the primer pair KOG-F/R, but could be amplified from the genomic DNA of the wild-type strain ZS-1; furthermore, the HYG fragment (740 bp), and fusion DNA fragments (2,236 bp for upstream and 2,499 bp for downstream) could be amplified from the genomic DNA of the candidates, but not from the wild-type strain ZS-1 (Figure 2B). The expression of CmMR1 could not be detected at 72 hpi in the candidates (Figure 2D).
The full length gene was amplified from the genomic DNA of the wild-type strain ZS-1 and transformed into the targeted knockout mutants via Agrobacterium-mediated transformation. Strains growing on G418 and hygromycin-containing medium were considered as transformants. Most transformants could produce dark pycnidia, and three of them, CmMR1-2C1, CmMR1-23C7, and CmMR1-22C10, were randomly selected for PCR amplification. The results showed that a 1,381 bp DNA fragment could be successfully amplified from the transformants and strain ZS-1. Furthermore, genes for both hygromycin-resistance fragment (740 bp) and neomycin-resistance gene fragment (582 bp) could be amplified (Figure 2C) and the expression of CmMR1 could be detected at 72 hpi in the transformants (Figure 2D).
Colony Morphology of the Target Knockout Mutant of CmMR1
Compared to the wild-type strain, the targeted knockout mutants did not accumulate melanin and their colonies appeared whitish to orange in color. The mutants produced pycnidia and conidia similar to the wild-type strain; however, their pycnidia were not dark but white to orange in color (Figure 3A). The hyphal tips of the mutants were similar to those of the wild-type strain (Figure 3B). These characteristics were similar to that of the T-DNA insertional mutant (Figure 1B). The gene complemented transformants showed similar phenotypes as the wild-type strain ZS-1 (Figures 3A, 1B) and they could form dark pycnidia on medium. Thus, disruption of CmMR1 was responsible for the phenotype of mutant ZS-1TN24363.
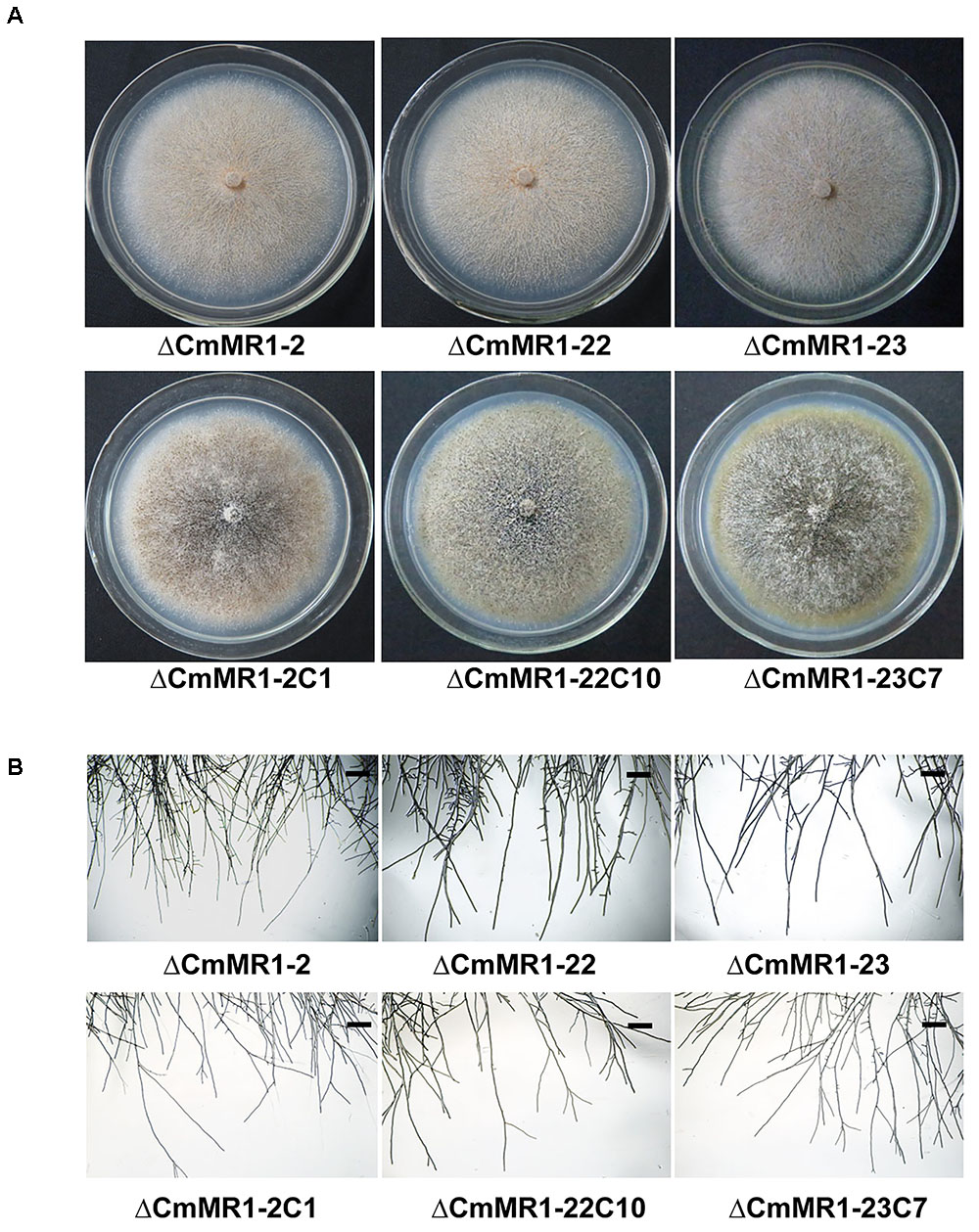
FIGURE 3. Comparison of colony morphology and hyphal tips of CmMR1 targeted knockout mutants and complemented transformants. (A) Colony morphology. Images were taken 20 days after incubation on PDA at 20°C. (B) Hyphal tips of on PDA at 20°C for 4 days. Scale bar means 50 μm.
Disruption of CmMR1 Did Not Affect Hyphal Growth, Conidiation, and Parasitic Ability
Compared to the wild-type strain ZS-1, the T-DNA insertional mutants, the targeted knockout mutants, and the gene complemented transformants differed negligibly regarding hyphal growth, mycelial mass, and conidiation (Table 2). When dual cultured with S. sclerotiorum on PDA plates, all the strains tested could parasitize the hyphae of S. sclerotiorum, with a slight stronger parasitic ability in the T-DNA insertional mutant (Figure 4). However, when inoculated on sclerotia of S. sclerotiorum, the parasitic ability did not differ significantly among all the mutants and the wild-type strain (Table 2). These results suggested that CmMR1 had negligible effects on hyphal growth, conidiation, and parasitic ability.
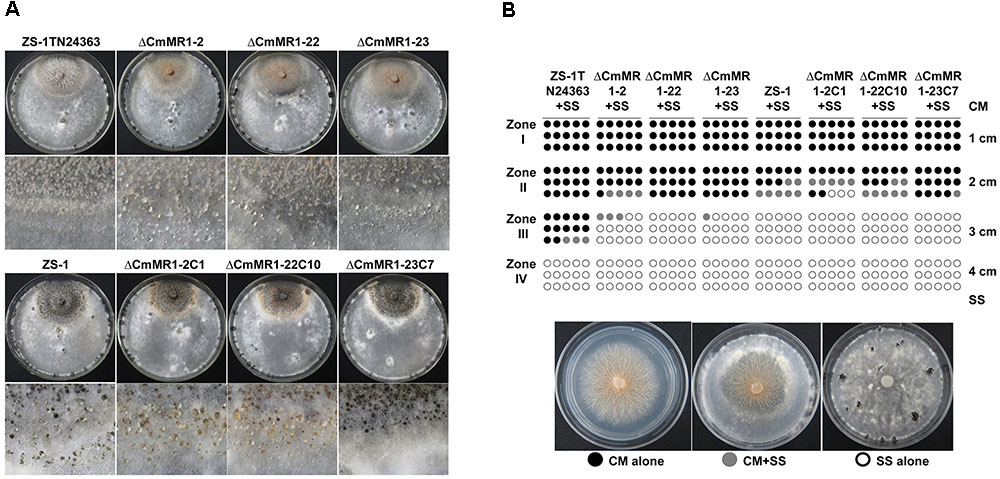
FIGURE 4. Parasitic activity assays of C. minitans to sclerotia of S. sclerotiorum. (A) A dual culture of C. minitans and S. sclerotiorum (PDA, 20°C, 20 days). (B) A schematic diagram showing the number of agar disks that gave rise to either C. minitans, S. sclerotiorum or both. Each circle represents a colony developed from a mycelial agar disk sampled from zones I, II, III or IV between the inoculation sites of C. minitans and S. sclerotiorum in a dual culture. Appearance of S. sclerotiorum colonies indicated that C. minitans could not parasitize S. sclerotiorum in this region. Similarly, the presence of C. minitans colonies suggested that S. sclerotiorum was eliminated by C. minitans. Occurrence of colonies of both C. minitans and S. sclerotiorum indicated that C. minitans was present at this region but could not destroy the hypha of S. sclerotiorum.
Disruption of CmMR1 Led to High Accumulation of ROS and Decreased Tolerance to UV Light
The ROS content in CmMR1 mutants was determined using NBT staining, and the results showed that T-DNA insertional mutant ZS-1TN24363 and the gene targeted knockout mutants had deeper color, whereas the wild-type strain ZS-1 and the gene complemented transformants were brown in color (Figure 5). This result suggested that the CmMR1 disrupt mutants might accumulate higher levels of ROS. Without radiation treatment, the conidial germination of all mutants tested and the wild-type strain was similar at 36 h, and more than 90% conidia germinated. When treated with UV light for 3 min, 50% conidia of the wild-type strain and the gene complemented transformants did not germinate, while only 10% conidia of ZS-1TN24363 and ΔCmMR1-23 germinated. After treatment for 6 min, ZS-1TN24363 and ΔCmMR1-23 conidia could not germinate; however, 10%–25% of the strain ZS-1 and ΔCmMR1-23C7 conidia still germinated. A few of conidia of strain ZS-1 and ΔCmMR1-23C7 still germinated when treated for 9 min. Thus, results of ROS determination and UV treatment suggested that CmMR1 might affect the UV and ROS tolerance of C. minitans.
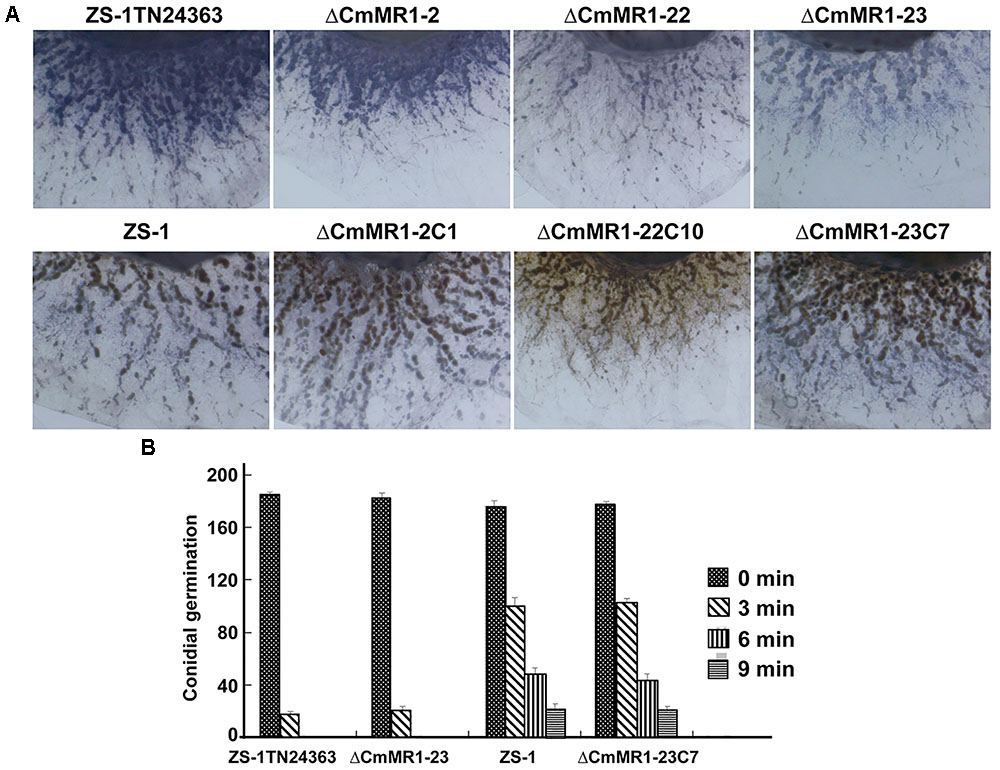
FIGURE 5. ROS production and tolerance to UV light of CmMR1 disrupted mutants of C. minitans. (A) ROS production in strains was monitored by NBT staining (colonies were developed on PDA for 3 days under 20°C). (B) Conidial germination of all strains irradiated with UV light (20 J/cm2) for 0, 3, 6, and 9 min (20°C, 36 h).
Expression of Melanin Synthesis Associated Gene CmSCD, Cm3HNR, and Cm4HNR Was Significantly Down-Regulated
The expression of melanin synthesis associated genes was monitored by qRT-PCR. CmSCD, Cm3HNR and Cm4HNR were significantly down-regulated at the conidial production stage (96 hpi) in the mutant ZS-1TN24363 compared to those in strain ZS-1 (Figure 6), while expression of CmLAC1 could not be detected either in strain ZS-1 or in mutant ZS-1TN24363.
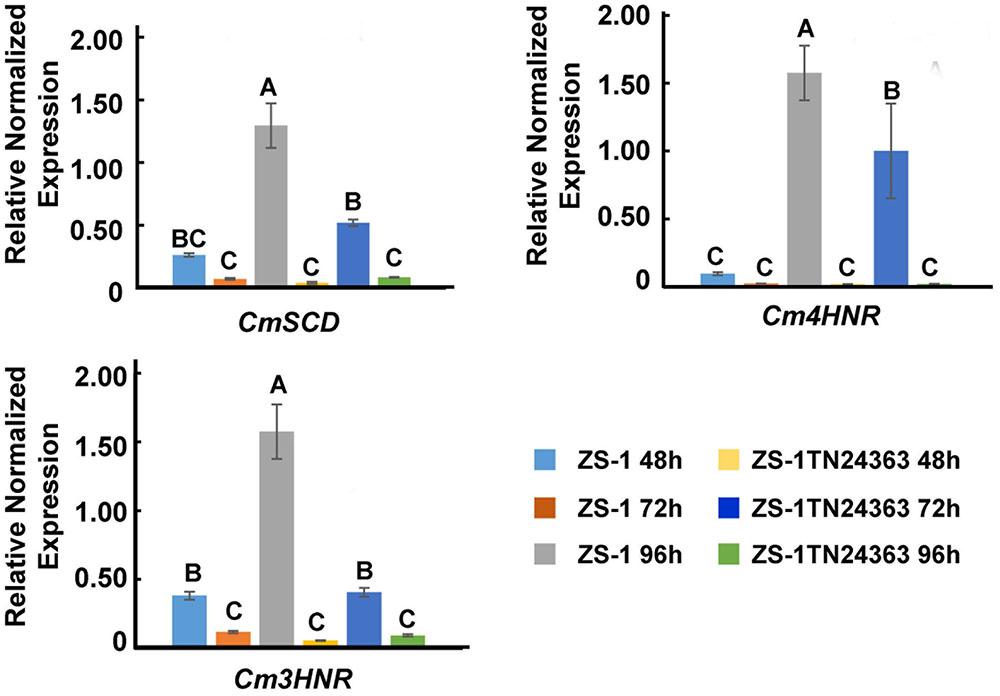
FIGURE 6. The expression of CmSCD, Cm3HNR and Cm4HNR. qRT-PCR amplification was used to check the expression of the genes from 48 to 96 hpi in strain ZS-1 and in mutant ZS-1TN24363. The Actin was used as an internal control to normalize the expression data with Actin-F/Actin-R. Significant tests were conducted with ANOVA and multiple comparing (significance level alpha = 0.01).
Discussion
Coniothyrium minitans is a well-known biological control agent. Previous studies have mainly focused on its growth, conidiation, and parasitical mechanism; however, studies regarding the survival property of this fungus are limited. C. minitans produces abundant melanin, which accumulates on the pycnidia and conidia; however, the melanin synthesis pathway and its functions are not known. In this study, we screened a T-DNA insertional melanin-deficient mutant ZS-1TN24363 and identified the transcription factor-encoding gene CmMR1. MR1 is a conserved fungal transcription factor, and in other fungi, this protein was also found to be associated with melanin synthesis. Here, we observed that the expression of CmSCD, Cm3HNR and Cm4HNR were significantly down-regulated at the conidial production stage (96 hpi) in mutant ZS-1T24363, suggesting that these genes were regulated by CmMR1. Previously, we demonstrated that disruption of a gene encoding PKS of C. minitans exhibited similar phenotypes as the CmMR1 target knockout mutants (Xiang et al., unpublished), suggesting that the PKS pathway is involved in melanin synthesis.
Melanin is involved in the pathogenicity of pathogenic fungi (Henson et al., 1999). Melanin accumulates in the appressorium of M. griesa, and a melanin layer was present around the plasma membrane of the appressorium, which was essential for its pathogenicity (Chumley and Valent, 1990; Soanes et al., 2012). The melanin layers were also found in the appressorium of C. lagenarium. Melanin is also essential for the pathogenicity of human pathogenic fungi as melanin deficiency leads to reduction or complete loss in pathogenicity (Langfelder et al., 2003). Moreover, in A. brassicicola, the transcription factor AMR1 induces melanin biosynthesis and increases fungal virulence, and was speculated to be important for both competitive saprophytic and plant parasitic activities (Cho et al., 2012). In our study, CmMR1 did not significantly affect the ability of C. minitans to parasitize S. sclerotiorum. This is possibly because C. minitans does not form an appressorium for penetrating host hypha (Huang and Kokko, 1988). The functions of the fungal transcription factor CMR1 may be slightly different; for example, in addition to the control of melanization, the A. alternata CmrA was reported to control spore development. The hyphal diameter and spore morphology changed and the production of conidia was significantly reduced when CmrA was knocked out (Fetzner et al., 2014). Fungi require melanin to tolerate environmental stress, such as UV irradiation and presence of oxidants. UV light induces the expression of CMR1, PIG1, and BMR1 in C. lagenarium, M. grisea, and B. oryzae, and their hypha were melanized (Tsuji et al., 2000; Kihara et al., 2008). Recently, the transcription factor homolog of BMR1 was identified in the basidiomycetous fungus Grifola frondosa, and it was induced by near-ultraviolet light and blue light (Kurahashi et al., 2015). In our study, we found that the conidia of CmMR1 mutants were highly sensitive to UV light irradiation. C. minitans parasitized the sclerotia of S. sclerotiorum under the soil. Since C. minitans grows under similar temperature conditions to S. sclerotiorum, it could also grow on the lesions induced by S. sclerotiorum on the aerial parts of plants where it may be irradiated by UV light. Further, application of conidia on the aerial parts of crops is an important way of successfully controlling Sclerotinia diseases in the field, and enhancing conidial melaninization is a potential way of improving the on-field survival ability of crops.
Conclusion
In this research, CmMR1, encoding a transcription factor MR, was cloned and the functions were studied in C. minitans, a mycoparasite of S. sclerotiorum. Knockout of CmMR1 led to the shortage of melanin during conidiation and decrease of tolerance to UV light. Our result may enhance the understanding of melanin in the ecology of C. minitans on molecular level.
Author Contributions
CL, DJ, and YF designed the research and wrote the paper. CL, XY, CQ, HZ, and JC executed the experiments. JC, TC, JX, and YF performed the data and bioinformatics analyses. All authors read and approved the final manuscript.
Funding
The research was financially supported by the National Key R&D Program of China (2017YFD0200401), the National Natural Science Foundation of China (Grant 31572048) and the earmarked fund for China Agriculture Research System (CAS-13).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the reviewers for their kind suggestions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02658/full#supplementary-material
FIGURE S1 |CmMR1 disrupted in the T-DNA insertional mutant ZS-1TN24363 of C. minitans. (A) The result of hiTAIL-PCR. Arrow shows the target product of the secondary PCR. (B) The sketch of the flanking genomic DNA segment and the T-DNA border. (C) T-DNA insertional event in mutant ZS-1TN24363. (D) Analysis of putative conserved domains coded by CmMR1.
FIGURE S2 | Phylogenetic analysis of CmMR1 and proteins of other fungi. The alignment was performed with Neighbor-joining method of MEGA 6.0 program. Bootstrap values (%) obtained with 1000 replicates are indicated on branches and branch lengths correspond to genetic distance; A scale bar at the lower left corresponds to a genetic distance of 0.1.
FIGURE S3 | The strategy of knockout with split-marker recombination.
References
Bell, A. A., and Wheeler, M. H. (1986). Biosynthesis and functions of fungal melanins. Annu. Rev. Phytopathol. 24, 411–451. doi: 10.1146/annurev.py.24.090186.002211
Campbell, W. A. (1947). A new species of Coniothyrium parasitic on sclerotia. Mycologia 39, 190–195. doi: 10.1080/00275514.1947.12017603
Cho, Y., Srivastava, A., Ohm, R. A., Lawrence, C. B., Wang, K. H., Grigoriev, I. V., et al. (2012). Transcription factor Amr1 induces melanin biosynthesis and suppresses virulence in Alternaria brassicicola. PLoS Pathog. 8:e1002974. doi: 10.1371/journal.ppat.1002974
Chumley, F. G., and Valent, B. (1990). Genetic analysis of melanin-deficient, nonpathogenic mutants of Magnaporthe grisea. Mol. Plant Microbe Interact. 3:135. doi: 10.1094/MPMI-3-135
Eliahu, N., Igbaria, A., Rose, M. S., Horwitz, B. A., and Lev, S. (2007). Melanin biosynthesis in the maize pathogen Cochliobolus heterostrophus depends on two mitogen-activated protein kinases, Chk1and Mps1, and the transcription factor Cmr1. Eukaryo. Cell 6, 421–429. doi: 10.1128/EC.00264-06
Fetzner, R., Seither, K., Wenderoth, M., Herr, A., and Fischer, R. (2014). Alternaria alternata transcription factor CmrA controls melanization and spore development. Microbiology 160, 1845–1854. doi: 10.1099/mic.0.079046-0
Gong, X. Y., Fu, Y. P., Jiang, D. H., Li, G. Q., Yi, X. H., and Peng, Y. L. (2007). L-Arginine is essential for conidiation in the filamentous fungus Coniothyrium minitans. Fungal Genet. Biol. 44, 1368–1379.
Henson, J. M., Butler, M. J., and Day, A. W. (1999). The drak side of the mycelium: melanins of phytopathogenic fungi. Annu. Rev. Phytopathol. 37, 447–471. doi: 10.1146/annurev.phyto.37.1.447
Huang, H. C. (1981). Distribution of Coniothyrium minitans in Manitoba sunflower fields. Can. J. Plant Pathol. 3, 219–222. doi: 10.1080/07060668109501352
Huang, H. C., and Kokko, E. G. (1988). Penetration of hyphae of Sclerotinia sclerotiorum by Coniothyrium minitans without the formation of appressoria. J. Phytopathol. 123, 133–139. doi: 10.1111/j.1439-0434.1988.tb04460.x
Jiang, D., Li, G., Fu, Y., Yi, X., and Wang, D. (2000). Biocontrol of re-infection of oilseed rape stem rot caused by Sclerotinia sclerotiorum by Coniothyrium minitans and its survival on leaf of oilseed rape (Brassica napus). Acta Phytopathol. Sin. 30, 60–65.
Kihara, J., Moriwaki, A., Tanaka, N., Tanaka, C., Ueno, M., and Arase, S. (2008). Characterization of the BMR1 gene encoding a transcription factor for melanin biosynthesis genes in the phytopathogenic fungus Bipolaris oryzae. FEMS Microbiol. Lett. 281:221. doi: 10.1111/j.1574-6968.2008.01101.x
Kurahashi, A., Shimoda, T., Sato, M., Fujimori, F., Hiramad, J., and Nishibori, K. (2015). A putative transcription factor Gf.BMR1 in Grifola frondosa, the homolog of BMR1 in Bipolaris oryzae, was strongly induced by near-ultraviolet light and blue light. Mycoscience 56, 177–182. doi: 10.1016/j.myc.2014.05.005
Langfelder, K., Streibel, M., Jahn, B., Haase, G., and Brakhage, A. A. (2003). Biosynthesis of fungal melanins and their importance for human pathogenic fungi. Fungal Genet. Biol. 38, 143–158. doi: 10.1016/S1087-1845(02)00526-1
Li, G., Huang, H., Miao, H., Erickson, R. S., Jiang, D., and Xiao, Y. (2006). Biological control of sclerotinia diseases of rapeseed by aerial applications of the mycoparasite Coniothyrium minitans. Eur. J. Plant Pathol. 114, 345–355. doi: 10.1007/s10658-005-2232-6
Li, M., Gong, X., Zheng, J., Jiang, D., Fu, Y., and Hou, M. (2005). Transformation of Coniothyrium minitans, a parasite of Sclerotinia sclerotiorum, with Agrobacterium tumefaciens. FEMS Microbiol. Lett. 243, 323–329. doi: 10.1016/j.femsle.2004.12.033
Liu, Y. G., and Chen, Y. L. (2007). High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Bio. Tech. 43, 649–656. doi: 10.2144/000112601
Plonka, P. M., and Grabacka, M. (2006). Melanin synthesis in microorganisms- biotechnological and medical aspects. Acta Biochim. Pol. 53, 429–443.
Soanes, D. M., Chakrabarti, A., Paszkiewicz, K. H., Dawe, A. L., and Talbot, N. J. (2012). Genome-wide transcriptional profiling of appressorium development by the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 8:e1002514. doi: 10.1371/journal.ppat.1002514
Tsuji, G., Kenmochi, Y., Takano, Y., Sweigard, J., Farrall, L., and Furusawa, I. (2000). Novel fungal transcriptional activators, Cmr1p of Colletotrichum lagenarium and Pig1p of Magnaporthe grisea, contain Cys2His2 zinc finger and Zn(II)2Cys6 binuclear cluster DNA-binding motifs and regulate transcription of melanin biosynthesis genes in a developmentally specific manner. Mol. Microbiol. 38, 940–954. doi: 10.1046/j.1365-2958.2000.02181.x
Wei, W., Zhu, W., Cheng, J., Xie, J., Li, B., Jiang, D., et al. (2013). CmPEX6, a gene involved in peroxisome biogenesis, is essential for parasitism and conidiation by the sclerotial parasite Coniothyrium minitans. Appl. Environ. Microbl. 79, 3658–3666. doi: 10.1128/AEM.00375-13
Wei, W., Zhu, W., Cheng, J., Xie, J., Li, B., Jiang, D., et al. (2016). Nox complex signal and MAPK cascade pathway are cross-linked and essential for pathogenicity and conidiation of mycoparasite Coniothyrium minitans. Sci. Rep. 6:24325. doi: 10.1038/srep24325
Whipps, J. M., and Gerlagh, M. (1992). Biology of Coniothyrium minitans and its potential for use in disease biocontrol. Mycol. Res. 96, 897–907. doi: 10.1016/S0953-7562(09)80588-1
Whipps, J. M., Sreenivasaprasad, S., Muthumeenakshi, S., Rogers, C. W., and Challen, M. P. (2008). Use of Coniothyrium minitans as a biocontrol agent and some molecular aspects of sclerotial mycoparasitism. Eur. J. Plant Pathol. 121, 323–330. doi: 10.1007/s10658-007-9238-1
Xiao, X., Xie, J., Cheng, J., Yi, X., Li, B., Jiang, D., et al. (2014). Novel secretory protein Ss-Caf1 of the plant-pathogenic fungus Sclerotinia sclerotiorum is required for host penetration and normal sclerotial development. Mol. Plant Microbe Interact. 27, 40–55. doi: 10.1094/MPMI-05-13-0145-R
Yang, X., Cui, H., Cheng, J., Xie, J., Jiang, D., Hsiang, T., et al. (2016). A HOPS protein, CmVps39, is required for vacuolar morphology, autophagy, growth, conidiogenesis and mycoparasitistic functions of Coniothyrium minitans. Environ. Microbiol. 18, 3785–3797. doi: 10.1111/1462-2920.13334
Zeng, L., Zhang, J., Han, Y., Yang, L., Wu, M., Jiang, D., et al. (2014). Degradation of oxalic acid by the mycoparasite Coniothyrium minitans plays an important role in interacting with Sclerotinia sclerotiorum. Environ. Microbiol. 16, 2591–2610. doi: 10.1111/1462-2920.12409
Keywords: Coniothyrium minitans, melanin, transcription factor MR, ROS, UV-tolerance
Citation: Luo C, Zhao H, Yang X, Qiang C, Cheng J, Xie J, Chen T, Jiang D and Fu Y (2018) Functional Analysis of the Melanin-Associated Gene CmMR1 in Coniothyrium minitans. Front. Microbiol. 9:2658. doi: 10.3389/fmicb.2018.02658
Received: 22 July 2018; Accepted: 18 October 2018;
Published: 08 November 2018.
Edited by:
Hector Mora Montes, Universidad de Guanajuato, MexicoReviewed by:
Luis Castillo, University of La Serena, ChileChing-Hsuan Lin, National Taiwan University, Taiwan
Copyright © 2018 Luo, Zhao, Yang, Qiang, Cheng, Xie, Chen, Jiang and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanping Fu, eWFucGluZ2Z1QG1haWwuaHphdS5lZHUuY24=
 Chenwei Luo
Chenwei Luo Huizhang Zhao
Huizhang Zhao Xiaoxiang Yang
Xiaoxiang Yang Cuicui Qiang
Cuicui Qiang Jiasen Cheng
Jiasen Cheng Jiatao Xie
Jiatao Xie Tao Chen
Tao Chen Daohong Jiang
Daohong Jiang Yanping Fu
Yanping Fu