- 1Department of Civil, Environmental, and Geodetic Engineering, The Ohio State University, Columbus, OH, United States
- 2Department of Civil and Environmental Engineering, Colorado State University, Fort Collins, CO, United States
- 3School of Earth Sciences, The Ohio State University, Columbus, OH, United States
- 4Department of Microbiology, The Ohio State University, Columbus, OH, United States
- 5Department of Civil and Environmental Engineering, University of New Hampshire, Durham, NH, United States
Hydraulic fracturing is the prevailing method for enhancing recovery of hydrocarbon resources from unconventional shale formations, yet little is understood regarding the microbial impact on biogeochemical cycling in natural-gas wells. Although the metabolisms of certain fermentative bacteria and methanogenic archaea that dominate in later produced fluids have been well studied, few details have been reported on microorganisms prevelant during the early flowback period, when oxygen and other surface-derived oxyanions and nutrients become depleted. Here, we report the isolation, genomic and phenotypic characterization of Marinobacter and Arcobacter bacterial species from natural-gas wells in the Utica-Point Pleasant and Marcellus Formations coupled to supporting geochemical and metagenomic analyses of produced fluid samples. These unconventional hydrocarbon system-derived Marinobacter sp. are capable of utilizing a diversity of organic carbon sources including aliphatic and aromatic hydrocarbons, amino acids, and carboxylic acids. Marinobacter and Arcobacter can metabolize organic nitrogen sources and have the capacity for denitrification and dissimilatory nitrate reduction to ammonia (DNRA) respectively; with DNRA and ammonification processes partially explaining high concentrations of ammonia measured in produced fluids. Arcobacter is capable of chemosynthetic sulfur oxidation, which could fuel metabolic processes for other heterotrophic, fermentative, or sulfate-reducing community members. Our analysis revealed mechanisms for growth of these taxa across a broad range of salinities (up to 15% salt), which explains their enrichment during early natural-gas production. These results demonstrate the prevalence of Marinobacter and Arcobacter during a key maturation phase of hydraulically fractured natural-gas wells, and highlight the significant role these genera play in biogeochemical cycling for this economically important energy system.
Introduction
Hydraulic fracturing has enhanced the extraction of oil and gas from previously inaccessible unconventional petroleum resources (e.g., shales, tight sands, coalbeds), with hydraulically fractured wells accounting for the production of 67% of the natural gas and 51% of the oil in the United States in 2015 (Kerr, 2010; Tour et al., 2010; EIA, 2016a,b). Hydraulic fracturing of unconventional systems is accomplished through the high-pressure injection of water (∼90%), proppant (∼9%), and chemical additives (∼1%) into subsurface formations (Arthur et al., 2009; Vidic et al., 2013). This process generates and sustains a network of fractures in the intrinsically low-permeability formation, liberating hydrocarbons which return through the wellbore to be collected at the surface (Arthur et al., 2009; Kerr, 2010; Tour et al., 2010). Injected fluids return to the surface over several months as they reach equilibrium with deep formation brines and undergo water-rock interactions (Vengosh et al., 2017). The fluids that return to the surface are referred to as flowback fluids (generally the first few weeks after hydraulic fracturing occurs) and produced fluids (several weeks to years after hydraulic fracturing occurs and throughout the production life cycle). Flowback fluids are characterized by high carbon concentrations (>100 mg/L), moderate to high salinities (1–8 NaCl%), and increasing concentrations of soluble nitrogen and sulfur species (Chapman et al., 2012; Barbot et al., 2013; Haluszczak et al., 2013; Warner et al., 2013; Cluff et al., 2014; Harkness et al., 2015; Lester et al., 2015). Produced fluids, in contrast, more closely resemble deep formation brines (>10% NaCl). Produced fluids are extremely high in total dissolved solids, have lower carbon content (50 mg/L and below), and contain elevated concentrations of reduced sulfur species (Chapman et al., 2012; Barbot et al., 2013; Haluszczak et al., 2013; Warner et al., 2013; Cluff et al., 2014; Harkness et al., 2015; Lester et al., 2015; Booker et al., 2017; Vengosh et al., 2017). Despite dramatic changes in solution chemistry during the gas extraction process, microbial life has been detected in flowback and produced fluids, and likely impacts fluid chemistry changes, infrastructure stability, and hydrocarbon recovery efficiency (Struchtemeyer and Elshahed, 2012; Cluff et al., 2014; Gaspar et al., 2014; Daly et al., 2016; Liang et al., 2016; Booker et al., 2017; Nixon et al., 2017; Lipus et al., 2018).
Microbial growth in oil and gas wells can contribute to reservoir souring, well corrosion, and clogged pore space, decreasing hydrocarbon productivity (Fichter et al., 2012; Gaspar et al., 2014). Although biocides are commonly added to control microbial growth, evidence of a dynamic bacterial and archaeal community has been observed in fluids collected during and after hydraulic fracturing from multiple unconventional systems in the United States. (Fichter et al., 2012; Struchtemeyer and Elshahed, 2012; Mohan et al., 2013; Cluff et al., 2014; Daly et al., 2016; Lipus et al., 2017, 2018; Borton et al., 2018). These studies suggest that microorganisms are injected from surface input fluids and persist despite extreme environmental conditions such as high salinities, limited access to terminal electron acceptors (e.g., oxygen, nitrate, sulfate), high pressures, and warmer temperatures than near-surface conditions (Mouser et al., 2016). Despite these ecosystem stressors, a low-diversity microbial community persists and plays a significant role in the biogeochemical cycles of unconventional systems, including low porosity shales and limestones (Fichter et al., 2012; Cluff et al., 2014; Daly et al., 2016; Liang et al., 2016; Booker et al., 2017; Nixon et al., 2017; Borton et al., 2018; Lipus et al., 2018).
Prior studies from our group revealed that Marcellus Shale-associated taxa Halanaerobium, Marinobacter, and Frackibacter may participate in carbon and nitrogen cycling through fermentation/transformation of both injected chemicals and microbially-produced osmoprotectants (Daly et al., 2016). Laboratory studies have suggested that Halanaerobium may also contribute to sulfide-induced well-souring and corrosion months to years after fracturing has occurred (Liang et al., 2016; Booker et al., 2017; Nixon et al., 2017; Lipus et al., 2018). The aforementioned studies have focused on taxa that dominate in later production phases characterized by elevated salinity and depleted organic carbon. Still, relatively little has been reported on the physiological and biogeochemical roles of dominant taxa within the first few months of production, a time period that may be critical for the establishment of later persisting taxa. During this natural-gas well maturation transition period, organic carbon is abundant (e.g., indigenous hydrocarbons and injected carbon), salinity is moderate in comparison to later time points (1–10% versus 10–30% in later produced fluids), and terminal electron acceptors (e.g., oxygen, nitrate, sulfate) remain available, allowing for a diversity of microbially-mediated redox reactions and enzymatic transformations not thermodynamically possible during later stages of production (Mouser et al., 2016).
Two genera, Marinobacter and Arcobacter, have been detected in produced fluids from numerous unconventional hydrocarbon-producing formations including the Barnett, Antrim, Haynesville, Utica, Point Pleasant, and Marcellus (Davis et al., 2012; Fichter et al., 2012; Struchtemeyer and Elshahed, 2012; Mohan et al., 2013; Wuchter et al., 2013; Cluff et al., 2014; Daly et al., 2016; Liang et al., 2016; Mouser et al., 2016). Although one study applied a metagenomics approach to infer Marinobacter sp. may be capable of utilizing organic nitrogen and hydrocarbon compounds (Daly et al., 2016), the metabolic capabilities of these taxa in unconventional systems are poorly defined. Here, we used laboratory experiments to investigate salinity tolerance and carbon substrate utilization of Marinobacter and Arcobacter strains cultivated from natural-gas well produced fluids. We complement these laboratory investigations with genomic, metagenomic, and geochemical data from hydraulically stimulated natural-gas wells, demonstrating the power of using both cultivation and cultivation-independent approaches for elucidating sources and use of carbon and energy for halotolerant microorganisms from a deep terrestrial ecosystem.
Materials and Methods
Methods Overview
Fluids were collected from six different hydraulically fractured natural-gas wells for geochemical and genomic analyses summarized in this manuscript: four natural-gas wells producing from the Utica-Pt. Pleasant Formation (Utica-3, Utica-6, Utica-7, and Utica-8), and two natural-gas wells producing from the Marcellus Shale (Marcellus-4 and Marcellus-5) (Table 1). Nitrogen and sulfur analyses were performed on all six wells while metagenomic analyses, near-full-length reconstruction of the 16S rRNA gene, NPOC, and CO2 analyses were performed on one well (Marcellus-4). Marinobacter sp. UTICA-S1B6 was isolated from Utica-3, whereas Arcobacter sp. UTICA-S4D1 was isolated from Utica-8, and Arcobacter sp. MARC-MIP3H16 was isolated from Marcellus-4. Additionally, seven 16S rRNA gene sequences reported in a previous study (Marcellus-1, Daly et al., 2016) were used in our phylogenetic analysis.
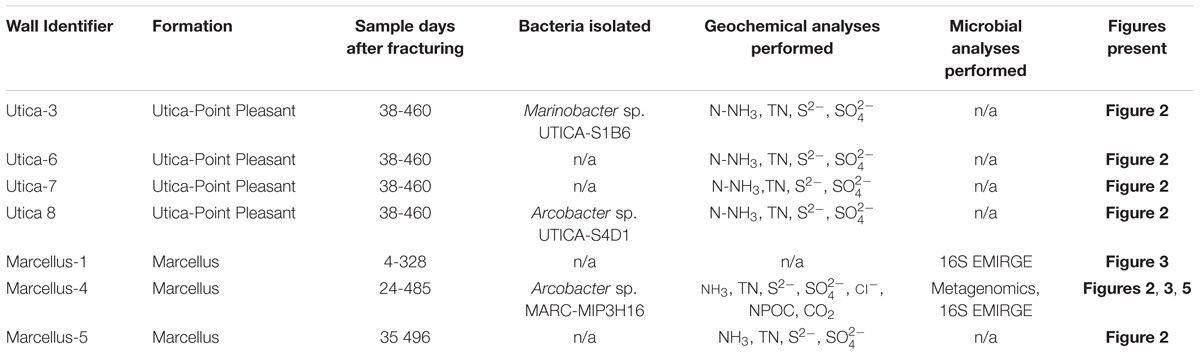
TABLE 1. Identifiers for hydraulically fractured natural-gas wells analyzed and/or summarized in this study.
Sample Collection
Produced fluid samples were collected from six hydraulically fractured natural-gas wells in the northern Appalachian Basin: four from the Utica-Point Pleasant Formation (Utica-3, Utica-6, Utica-7, and Utica-8) and two from the Marcellus Shale (Marcellus-4 and Marcellus-5). Liquid samples were taken from the gas-water separator. Samples were collected from the start of flowback at various intervals over a year into production (496 days) as previously reported (Luek et al., 2018). Samples for bacterial enumeration were collected in 50 mL polypropylene conical vials containing 5.0 mL of 25% paraformaldehyde and stored at 4°C until enumeration. Samples for isolation and culturing (25 mL) were collected in sterile serum bottles with no headspace and sealed with butyl rubber septa to maintain anaerobic conditions, then upon arrival at the laboratory purged with N2 gas and stored at ambient temperature in the dark until inoculation (Figure 1). Samples for geochemical analysis were collected in 1 L sterile containers (either HDPE or glass) with no headspace and stored at 4°C until analysis. Sulfide measurements were conducted on unfiltered samples (Hach method 8131, see following section). Samples for total dissolved nitrogen, ammonia, and sulfate were filtered using 0.45 μm PES filters (EMD Millipore, Burlington, MA, United States), with the exception of the Marcellus-4 and Marcellus-5 wells in which 0.22 μm PES filters were used, within 48 h of sampling and stored at 4°C until analysis.
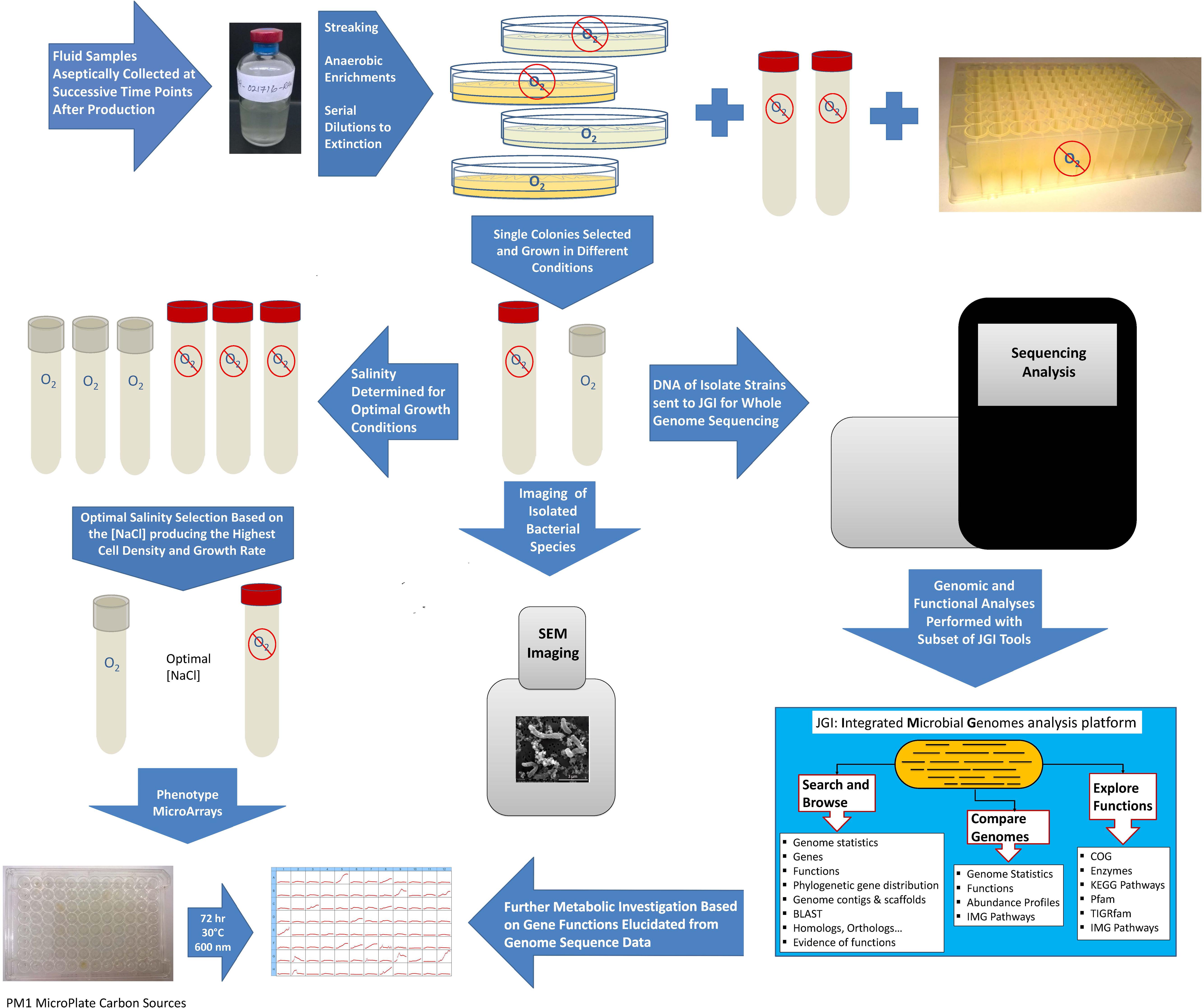
FIGURE 1. Flowchart summarizing methods used for isolating and characterizing Marinobacter and Arcobacter strains from produced fluid samples.
Non-purgeable Organic Carbon, Nitrogen, Sulfur, and Chloride Analysis
Non-purgeable organic carbon (NPOC) was measured in the Marcellus-4 samples by a TOC/TN analyzer equipped with autosampler (TOC-V CSN/TNM-1/ASI-V, Shimadzu, Kyoto, Japan). Total dissolved nitrogen (TDN) was analyzed in samples from all wells by a Shimadzu TOC/TN analyzer (see above), employing appropriate dilutions as needed for samples. Ammonia (NH3) in the produced fluid samples was analyzed colorimetrically using the modified Berthelot reaction method on a Skalar San++ continuous flow nutrient analyzer. Reagents were prepared following the method supplied by the manufacturer. Sulfide was measured using a Hach DR 900, method 8131. Sulfate and chloride were analyzed using a Thermo-Scientific Dionex ICS-2100 ion chromatograph (Waltham, MA, United States); samples were diluted by a factor of 100–1000 due to the high salinity.
Carbon Dioxide Analysis
Produced gas samples were collected from the Marcellus-4 natural-gas well over a period of 140 days. Produced gas samples were collected with negligible air contamination using thick-walled 0.95 cm (3/8 inch) outside diameter and 40.6 cm (16 inch) long refrigeration-grade copper tubes. The samples were collected by pumping produced fluid that had been collected using HDPE carboys from the producing natural-gas well through the copper tubes, with tubes tapped to remove air bubbles prior to collection. The copper tubes were then sealed using brass refrigeration clamps with a 0.762 mm (0.030 inch) gap (Kang et al., 2016; Harkness et al., 2017). Gas samples in the copper tubes were prepared for analysis by cold welding ∼2.5 cm (∼1 inch) splits of the copper tubing using stainless steel clamps. The copper tube was then attached to an ultra-high vacuum steel line (total pressure = 1–3 × 10−9 torr), which is monitored continuously using a 0–20 torr MKS capacitance monometer (accurate to the nearest thousandths) and an isolated ion gauge, using a 0.64 cm (1/4 inch) VCR connection and expanded to obtain aliquots for various gas geochemical analyses.
Carbon dioxide concentrations (CO2) were measured on a SRS Quadrupole MS and an SRI 8610C Multi-Gas 3+ gas chromatograph (GC) equipped with a flame ionization detector (FID) and thermal conductivity detector (TCD) at the Ohio State University Noble Gas Laboratory (Darrah et al., 2015; Heilweil et al., 2016; Moore et al., 2018). All reported CO2 concentrations were above the method detection limit. The average external precision was determined by measuring a series of synthetic natural gas standards obtained from Praxair and the DCG Partnership. Standard analytical errors for CO2 was less than ±1.06% based on daily replicate measurements during analyses.
Microbial Enumeration
Microbial (bacterial and archaeal) enumeration was performed using filtration and epifluorescent microscopy as described previously (Noble and Fuhrman, 1998; Weinbauer et al., 1998; Chen et al., 2001; Brussaard, 2004; Patel et al., 2007; Diemer et al., 2013). For each sample, the volume filtered was optimized such that at least two filters were prepared to yield approximately 10–200 cells per counting field. Between 1 and 15 mL of fixed sample was filtered through a 25 mm dia. 0.2 μm pore black PCTE filter (Sterlitech, Kent, WA, United States), stained with 2X SYBR Gold (Life Technologies, Carlsbad, CA, United States) in TE buffer, then mounted on a microscope slide with SlowFade reagent (Life Technologies, Carlsbad, CA, United States). The slide was viewed with a Labomed Lx500 epifluorescent microscope through a 40X air objective under 480 nm excitation. For each sample 20 randomly selected fields were counted per filter.
Bacterial Isolation
Marinobacter sp. UTICA-S1B6 was recovered from produced fluid collected from a Utica-Point Pleasant natural-gas well (Utica-3) on the first day of flowback using Difco Marine Broth 2216 (DM2216) medium supplemented with 40 mM nitrate at 30°C (Figure 1, Tummings et al., 2018). Arcobacter strains UTICA-S4D1 and MARC-MIP3H16 were isolated from produced fluids collected at 159 days (from the Utica-Point Pleasant formation, Utica-8) and 93 days (from Marcellus Shale, Marcellus-4) after hydraulic fracturing began, respectively (Figure 1, Panescu et al., 2018). Arcobacter strains were grown at 30°C in the dark on DM2216 at 5% NaCl purged with 80:20 N2/CO2. We tested growth under anaerobic and aerobic conditions; because cells grew better in the presence of oxygen, they were maintained under aerobic conditions for growth experiments (Figure 1). The purity of isolates was verified by 16S rRNA sequencing (see Isolation and Sequencing of DNA).
Salinity Growth Curves
Two of the three isolates, Marinobacter sp. UTICA-S1B6 and Arcobacter sp. MARC-MIP3H16, were tested for salinity tolerance (Figure 1 and Supplemental Table S2). Salinity experiments for both strains were conducted aerobically at 30°C in the dark with shaking. In addition, an anaerobic experiment was also carried out for Marinobacter at 37°C in the dark. For Arcobacter, a stock of Trypticase Soy Broth (Cat. No. 1.05459, EMD Millipore Corporation, Billerica, MA, United States) was amended with 1% DL Trace Mineral Solution and NaCl at the following concentrations: 0.5, 2, 4, 6, 8, 10, 12, and 14%. For Marinobacter, bacterial growth medium consisting of 25% DM2216 and 75% Lennox Broth (Bonis and Gralnick, 2015) with 1% NaCl, was prepared from commercially available powdered mixes. To these media, NaCl was added to obtain the following final concentrations: 0.9, 2.5, 5, 7.5, 10, 12.5, 15, and 20%. Singlet cultures were grown to mid-log phase, 2% was transferred into fresh media, then 2% was transferred again into triplicate cultures from which data were used for the salinity curves. The media was aliquoted into 9 mL increments in borosilicate glass tubes suitable for anaerobic culturing of size 18 × 150 mm purchased from Bellco Glass (Vineland, NJ, United States) and autoclaved at 121°C for 20 min; the tubes were used for both the aerobic and anaerobic cultures. Aerobic media tubes were covered with silicone foam stoppers while the anaerobic media tubes were closed with butyl rubber stoppers clamped with aluminum seals. Absorbance at a wavelength of 610 nm was measured with a Hach DR 900 Colorimeter (Loveland, CO, United States) fitted with a custom adapter.
A standard curve relating optical density to cell numbers was constructed. Cultures were grown to late log phase in the same test media at 2.5% NaCl, fixed (2.5% paraformaldehyde final concentration) and diluted or concentrated to obtain absorbance values of <0.1, ∼0.5, and ∼1.0 at 610 nm. Two aliquots from each concentration were diluted in 2.5% NaCl to yield 10–200 cells per counting field, and counted exactly as described above in the Section “Microbial Enumeration.” Growth rates were calculated using a first-order model based on cell numbers and time elapsed.
Utilization of Carbon Substrates
Culture plates containing DM2216 with 2% agar (Affymetrix USB, Tewksbury, MA, United States) were streaked with approximately 10 μL of cryogenic glycerol stock containing bacterial isolates and incubated at 30°C for 2–3 days. Individual 1–2 mm diameter colonies were picked, resuspended in liquid DM2216, re-streaked and re-grown as described above. Second generation colonies were resuspended and inoculated in Biolog PM1 (Hayward, CA, United States) inoculating fluid augmented with 5% NaCl solution, 20 μM pyruvate (carbon source) and 0.12X Marinobacter Iron Medium (MIM) (Bonis and Gralnick, 2015, AffymetrixUSB, Tewksbury, MA, United States) (Figure 1). The cultures were grown to late log phase (2–3 days) where 2% was transferred into fresh PM1-MIM-pyruvate medium. Once the second generation PM1-MIM-pyruvate culture reached the desired density, the cells were pelleted for 10 min at 5,000 × g, washed twice in 5% NaCl, and resuspended in 500 μL PM1-MIM with no carbon source. The washed culture was quantified on a NanoDrop 2000 spectrophotometer (Thermo Scientific) at 600 nm in an Eppendorf Uvette fitted with an adapter (50 μL of culture used), and diluted to 0.1 Absorbance units with PM1-MIM, of which 880 μL was combined with an additional 12 mL PM1-MIM inoculating fluid and distributed to a new PM1 substrate plate at 110 μL per well. All carbon substrate assessments were performed under aerobic conditions. The plate was sealed with a sterile, optically clear, non-cytotoxic, gas-permeable adhesive film (4titude, Wotton, England). Optical densities were measured using a plate reader (BioTek Synergy HTX Multi-Mode Reader, Winooski, VT, United States) at 600 nm every 2 h for 3 days while incubating at 30°C. Normalization took place by subtracting the optical densities of a control plate containing media with no cells.
Isolation and Sequencing of DNA
Marinobacter and Arcobacter genomic DNA was extracted using Qiagen DNA extraction kits (Qiagen DNA minikit and Qiagen DNeasy kit, respectively) (Hilden, Germany) as previously described (Panescu et al., 2018; Tummings et al., 2018). The 16S rRNA gene was amplified with universal primer sets for bacteria (27F, 1492R) and archaea (4Fa, 1492R). The 16S rRNA gene was sequenced using the Sanger method on a 3730 DNA Analyzer (Applied Biosystems) for quality control at the Comprehensive Cancer Center Genomics Shared Resource at the Ohio State University. After passing quality control, the isolates were subjected to full genome sequencing using an Illumina MiSeq at the Joint Genome Institute (JGI), Walnut Creek, CA, United States. Genome assemblies were constructed using Spades v. 3.6.2 with annotation performed by the JGI in the Integrated Microbial Genomes platform v. 4.12.1 (Figure 1, Huntemann et al., 2016; Panescu et al., 2018; Tummings et al., 2018).
Phylogenetic Analysis
Maximum-likelihood (ML) phylogenetic trees were constructed in RAxML using the full length 16S rRNA gene in Geneious v. 8 using a MUSCLE alignment with a maximum of 10,000 iterations (v.7.2.8, nucleotide model GTR gamma, rapid bootstrapping and search for best-scoring ML tree algorithm, with 1,000 bootstrap replicates). Nearly full-length 16S rRNA EMIRGE sequences used in the trees were reconstructed from related shale fluid metagenome samples as described previously (Miller et al., 2011; Daly et al., 2016).
Scanning Electron Microscopy (SEM) Sample Preparation and Imaging
Cells from cultures grown to log phase were fixed by combining 178 μL culture with 22 μL 25% PFA and allowed to equilibrate for 1–2 h in a microcentrifuge tube. Cells were next pelleted at 5,000 × g for 10 min, supernatant was removed, and cell pellet was washed with 1.0 mL PBS and resuspended in 200 μL PBS. For each isolate, ∼100 μL of the resuspended culture was placed on a 0.2 μm Nucleopore polycarbonate filter (Whatman, GE Healthcare, Chicago, IL, United States) mounted on a 25 mm diameter Swinnex syringe filter holder (EMD Millipore, Darmstadt, Germany) connected to flexible tubing. The culture was embedded into the filter by applying gentle vacuum (manually with a disposable syringe or by using an electric vacuum pump). The filter was next rinsed with 300–500 μL of PBS at 2% NaCl followed by additional vacuum application. After the culture was embedded onto the filter and all excess liquid was removed, the filter was allowed to dry. Dried samples were subjected to successive 5 min rinses of increasing concentrations of 35–100% ethanol. After evaporation, the filter was covered in hexamethyldisilazane, covered with a petri dish lid and allowed to dry overnight. The filters were subsequently mounted with carbon tape on aluminum stubs and imaged with a FEI Quanta FEG 250 scanning electron microscope at the Subsurface Energy Materials Characterization and Analysis Laboratory (SEMCAL), School of Earth Sciences, The Ohio State University (Figure 1).
Metagenomic Sequencing, Assembly, and Annotation
Metagenomic sequencing, assembly, and annotation was performed as previously described (Borton et al., 2018). In short, produced fluid samples collected at 7 time points (drilling mud sample prior to fracturing, days after fracturing 25, 34, 35, 79, 93, 142) from Marcellus Shale natural-gas well Marcellus-4 (300–1000 ml) were concentrated on a 0.22-μm PES filter (Nalgene; Fisher Scientific). Total nucleic acids were extracted using a modified phenol-chloroform extraction method. Briefly, libraries were created and quantified using an Illumina Library creation kit (KAPA Biosystems) with solid-phase reversible 402 immobilization size selection. Libraries were then sequenced on the Illumina HiSeq 2500 sequencing platform utilizing a TruSeq Rapid paired-end 404 cluster kit. Fastq files were generated with CASSAVA 1.8.2. Illumina sequences from each sample were first trimmed from both the 5′ and 3′ ends with Sickle and then each sample was assembled individually with IDBA-UD (Wrighton et al., 2012; Brown et al., 2015) using default parameters. Scaffolds were annotated as described previously by predicting open reading frames with MetaProdigal (Hyatt et al., 2012). Sequences were compared with USEARCH (Edgar, 2010) to KEGG, UniRef90, and InterProScan (Quevillon et al., 2005) with single and reverse best hit (RBH) matches of >60 bases reported.
Genomic and Metagenomic Analyses
Inquiry into genes contained in isolate genomes was performed using IMG/MER from JGI (Figure 1, Chen et al., 2017). Genes in isolate genomes were pulled from IMG/MER using EC numbers and KEGG Orthology numbers after genomic annotation following the JGI genome annotation pipeline (Huntemann et al., 2016). Homologous genes in assembled metagenomes were mined by KEGG orthology number, EC number, and name with a cutoff of >200 bit score and >45.9% identity similarity to genus Marinobacter/Arcobacter at day 34 (after fracturing) for Marinobacter and Arcobacter and days 34 and 93 for Arcobacter (Booker et al., 2017). Arcobacter and Marinobacter normalized relative abundances through time (days 25, 34, 35, 79, 93, 142) were tracked using the 16S rRNA gene reconstructed from the metagenomic data using EMIRGE (Miller et al., 2011; Daly et al., 2016).
Data Accession
Whole genome sequences for Arcobacter sp. MARC-MIP3H16 and UTICA-S4D1 were deposited in NCBI under accession numbers PTIW01000000 and FUYO01000000 and at the JGI IMG/M database under genome IDs 2700989666 and 2703719342, respectively (Pansecu et al., 2018). Whole genome sequences for Marinobacter sp. UTICA-S1B3, UTICA-S1B6, UTICA-S1B9 are available in NCBI under accession numbers PTIV00000000, PTIT00000000, and PTIU00000000, respectively, and at JGI IMG/M database under genome IDs 2700989663, 2700989662, and 2700989665 (Tummings et al., 2018). All metagenomic sequencing data has been deposited under Bioproject PRJNA308326.
Results
Marinobacter and Arcobacter Persist in Unconventional Source Ecosystem for Weeks to Months After Hydraulic Fracturing
Using near-full-length 16S rRNA gene sequences reconstructed from 7 sample metagenomes using EMIRGE, we tracked the relative abundance of two key taxa, Marinobacter and Arcobacter, during the first 140 days after hydraulic fracturing in a Marcellus Shale natural-gas well (Marcellus-4). Marinobacter were detected in drill muds (6.3%) used during natural-gas well development, and produced fluid samples collected in the first 4 weeks after hydraulic fracturing, reaching up to 20% relative abundance on day 34, but decreased thereafter to levels below 0.5% (Figure 2A). We compared a representative 16S rRNA gene sequence reconstructed from metagenomes in the Marcellus natural-gas well (Marcellus-4) using EMIRGE (Day 38 EMIRGE Marinobacter sp, Figure 3A) to (i) near-full-length 16S rRNA gene sequences reconstructed from metagenomes in another previously studied Marcellus Shale natural-gas well (Marcellus-1) (Figure 3A, Daly et al., 2016), (ii) 16S rRNA gene sequences from genomes available for Marinobacter strains isolated from the Utica-Point Pleasant Formation (Utica-3) (Day 0 after flowback began), and (iii) 16S rRNA gene sequences acquired from NCBI for Marinobacter strains isolated from other marine environments. Representative Marinobacter sampled from Marcellus-4 were highly similar to the three isolates recovered from the Utica-Pt. Pleasant Formation (97–99% shared nucleotide identity) (UTICA-S1B3; UTICA-S1B6; UTICA-S1B9) and cluster closely to 16S rRNA gene sequences collected from another Marcellus Shale natural-gas well (Marcellus-1, Figure 3A).
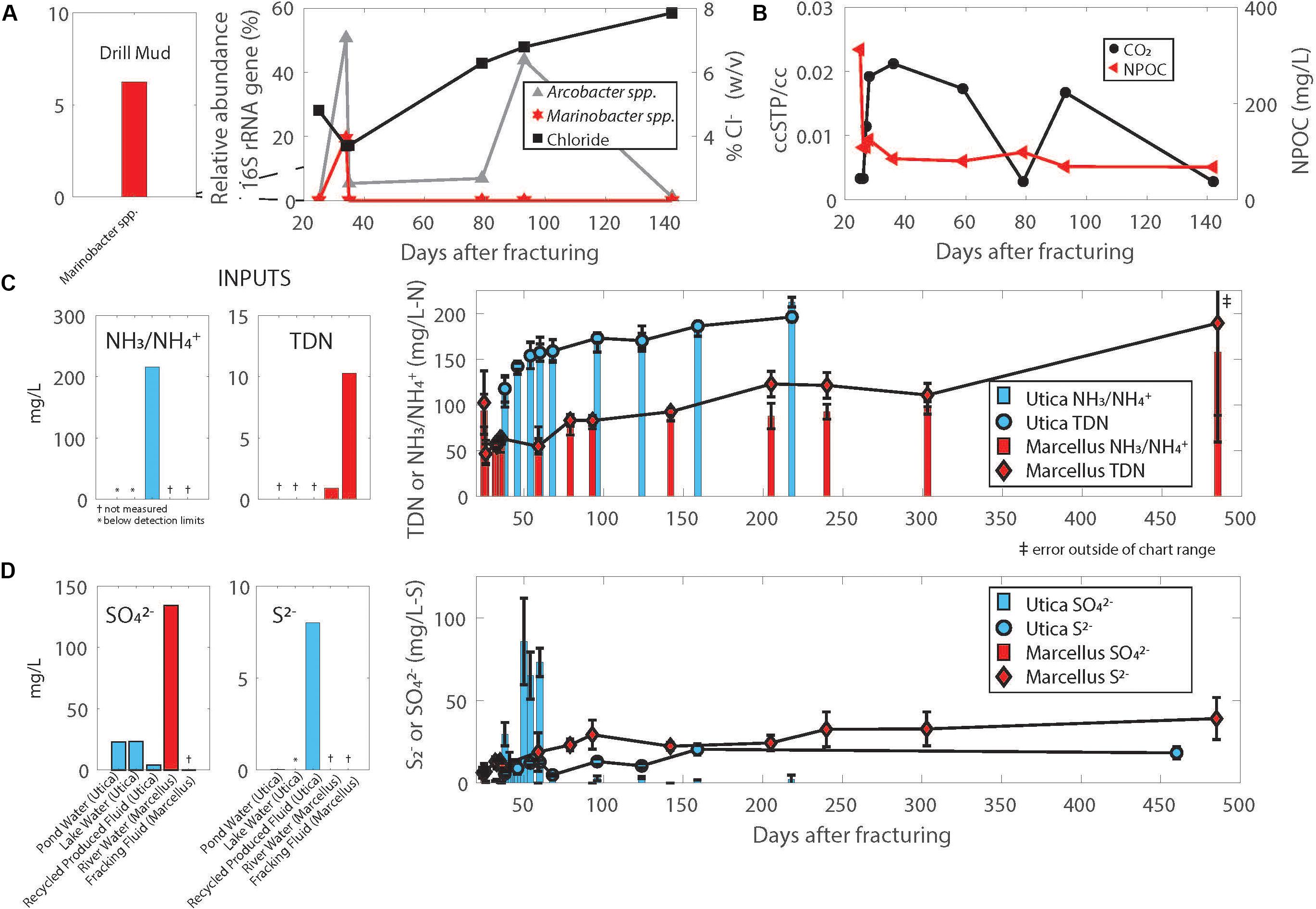
FIGURE 2. (A) Relative abundance of the 16S rRNA gene corresponding to Arcobacter and Marinobacter reconstructed from metagenomes using EMIRGE combined with chloride trends in the same Marcellus Shale natural-gas well (Marcellus-4). (B) CO2 and NPOC (non-purgeable organic carbon) trends in the Marcellus Shale natural-gas well (Marcellus-4). Temporal trends in nitrogen (C) and sulfur (D) in produced fluids from Utica-Point Pleasant formation and Marcellus Shale natural-gas wells. Plotted values are averages of 4 Utica-Point Pleasant natural-gas wells (Utica-3, Utica-6, Utica-7, Utica-8) and 2 Marcellus natural-gas wells (Marcellus-4 and Marcellus-5); error bars are standard deviations between measurements for Utica wells, range for Marcellus wells. Averages were calculated using the same day after flowback began and converted to days after fracturing. Offset graphs to the left of main graphs indicate measurements in drill muds, injected fluid, or source waters. NH3/ indicates total ammonia/ammonium which are both measured in this method.

FIGURE 3. SEM images of (A) Marinobacter sp. UTICA-S1B6 and (B) Arcobacter sp. MARC-MIP3H16 and phylogenetic placement of (A) Marinobacter sp. UTICA-S1B6 and (B) Arcobacter spp. MARC-MIP3H16 and UTICA-S4D1. Orange text denotes near-full-length 16S rRNA gene sequences reconstructed from Marcellus Shale natural-gas well Marcellus-4 metagenomes using EMIRGE; blue text denotes near-full-length 16S rRNA gene sequences reconstructed from metagenomes using EMIRGE in a previous study, Marcellus-1; red text denotes full-length 16S rRNA gene sequences from Marcellus or Utica Pt. Pleasant natural-gas well isolates. Branches marked with blue dots indicate bootstrap support greater than or equal to 80%.
Based on visual analysis of SEM micrographs, Marinobacter sp. UTICA-S1B6 is rod-shaped and approximately 2–4 μm long by 0.5 μm thick (Figure 3A). The closest related strain to M. UTICA-S1B6, Marinobacter persicus, is a strict aerobe isolated from a hypersaline lake and capable of tolerating salinities up to 20% NaCl (Bagheri et al., 2013). Other closely related strains M. aquaeolei and M. hydrocarbonoclasticus were discovered in environments with high concentrations of hydrocarbons (oil well, marine petroleum spill, respectively) (Gauthier et al., 1992; Huu et al., 1999).
Arcobacter spp. generally persist in the first weeks to months after hydraulic fracturing and remain abundant for a longer time period than Marinobacter before replacement by other abundant taxa commonly identified in produced fluids including Halanaerobium and Methanohalophilus as natural gas and produced fluid production continues and the natural-gas well matures (Davis et al., 2012; Mohan et al., 2013; Wuchter et al., 2013; Cluff et al., 2014; Liang et al., 2016). Although we did not detect Arcobacter in drill muds, it comprised over 50% of the microbial community in produced fluids from the Marcellus-4 natural-gas well collected on day 34, and subsequently ranged in relative abundance from 1 to 44% until day 142 (Figure 2A). In a similar fashion to Marinobacter discussed above, we compared these Arcobacter 16S rRNA sequences reconstructed from the Marcellus-4 natural-gas well metagenomes using EMIRGE (Days 94, 98 EMIRGE Arcobacter sp.) to (i) near full-length 16S rRNA sequences reconstructed from metagenomes in another previously studied Marcellus Shale natural-gas well (Marcellus-1) (Days 0, 7, Figure 3B, Daly et al., 2016), (ii) 16S rRNA gene sequences for two Arcobacter strains isolated from the Utica-Point Pleasant Formation (Utica-8) (Day 159 after hydraulic fracturing occurred) and Marcellus Shale natural-gas well Marcellus-4 (Day 93 after hydraulic fracturing), and (iii) 16S rRNA gene sequences acquired from NCBI for Arcobacter strains isolated from other marine environments. Representative Arcobacter sequences recovered from both Marcellus-4 (Day 94 Marcellus-4 and Day 98 Marcellus-4) and Marcellus-1 (T0_EMIRGE36 and T7_EMIRGE21) were very similar to Arcobacter spp. MARC-MIP3H16 and UTICA-S4D1 isolates. In particular, sequence from an initial flowback sample (Day 0 Marcellus-1) and an uncultured clone were most closely related (99.6%) to laboratory cultured strains (Figure 3B).
Arcobacter sp. MARC-MIP3H16 is a curved rod approximately 1–3 μm long by 0.3–0.5 μm thick with a polar flagellum (not shown) (Figure 3B). The closest related strains to A. MARC-MIP3H16, Arcobacter sp. solar lake and Arcobacter marinus, were both isolated from saline aquatic environments (Donachie et al., 2005; Kim et al., 2010). Arcobacter marinus can grow in 3–5% NaCl under both aerobic and microaerophilic conditions. Another closely related strain, Arcobacter halophilus, isolated from a hypersaline lagoon, is capable of growth across aerobic to anaerobic conditions, provided media contains at least 2% NaCl (Donachie et al., 2005). The close clustering of isolate and sample DNA despite lithogical and geographical differences demonstrates the prevalence of both Marinobacter and Arcobacter in early produced fluid from hydraulically fractured natural-gas wells.
Shale Marinobacter and Arcobacter Are Slight to Moderate Halophiles
The highest relative abundance of 16S rRNA gene copy numbers detected for Marinobacter and Arcobacter was associated with lower salt concentrations in the Marcellus-4 natural-gas well (approximately 40 g/L Cl− or 4% Cl−, Figure 2A). Therefore, to better understand the adaptability of Marinobacter and Arcobacter to increasing salinity levels, we tested the tolerance of both isolates across a broad range of salinities under aerobic (Arcobacter, Marinobacter) and anaerobic (Marinobacter) conditions (Figure 4A). Marinobacter sp. UTICA-S1B6 grew in concentrations between 0.9 and 15% Cl−, with optimal growth at 10% Cl− when oxygen was present (Figure 4A). In the absence of oxygen, Marinobacter growth rate improved at a lower salinity (5% Cl−). Arcobacter sp. MARC-MIP3H16 grew in salt concentrations between 2 and 12%, with an optimal growth rate at 4% Cl− (0.6 hr−1), considerably higher than growth at all other salinities (<0.25 hr−1) (Figure 4A). Our laboratory findings were therefore consistent with field salinity levels when Marinobacter and Arcobacter were highest in relative abundance during the first 140 days of production in the Marcellus-4 natural-gas well (4–8%). However, Arcobacter growth rate was significantly higher than Marinobacter in this range, which may partially explain its dominance and persistence during this transitional period in Marcellus-4 natural gas and produced fluid well maturation.
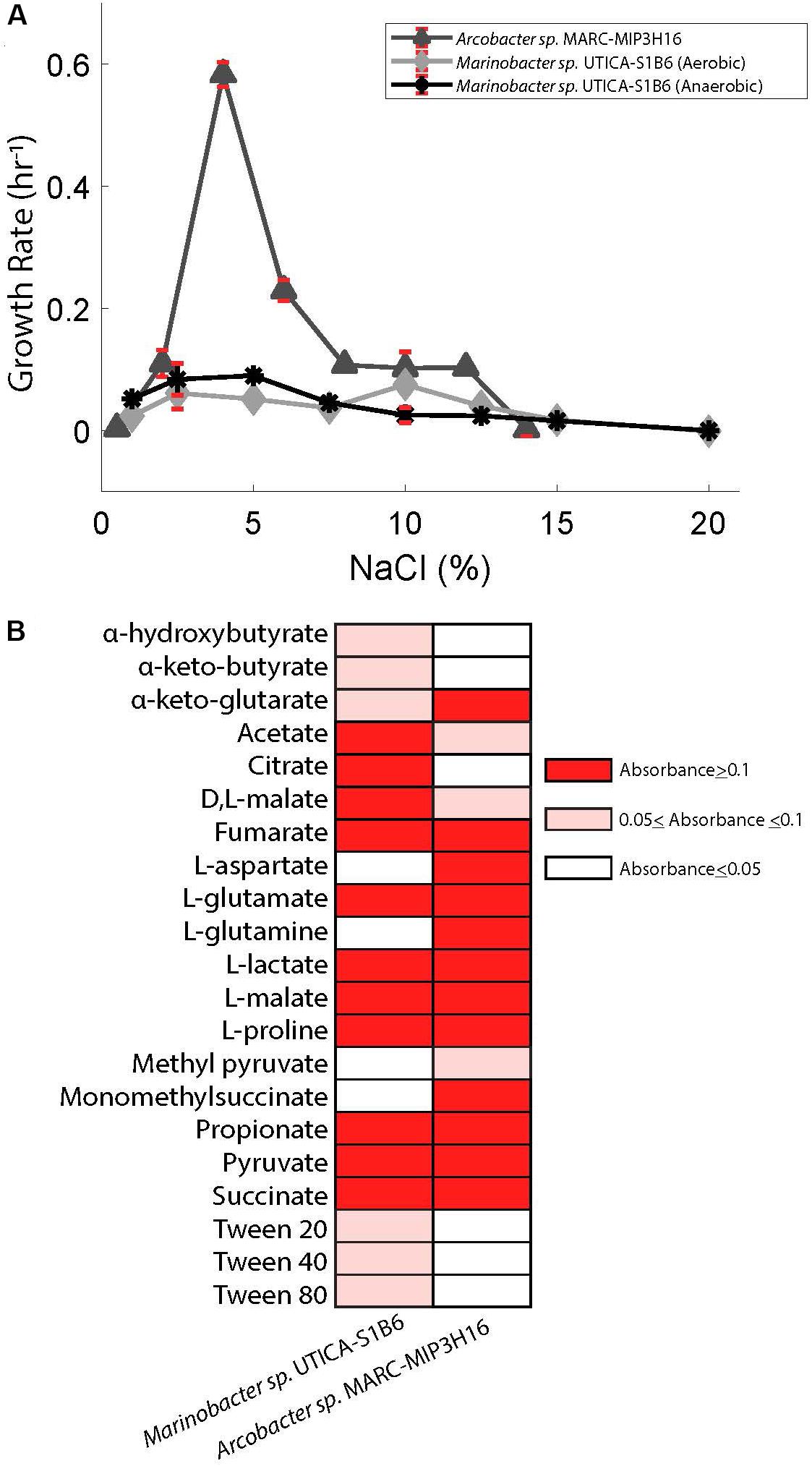
FIGURE 4. (A) Superimposed salinity growth rate curves for Arcobacter sp. MARC-MIP3H16 and Marinobacter sp. UTICA-S1B6. Error bars denote standard deviation from triplicate measurements. (B) Carbon sources utilized by Marinobacter sp. UTICA-S1B6 and Arcobacter sp. MARC-MIP3H16. Negative controls registered absorbance values of 0.023 and 0.014, respectively. Any absorbance readings under 0.05 were considered negative and assigned a white color in this figure.
To assess possible osmoadaptation mechanisms, we searched the isolate genomes for relevant genes associated with osmoprotectant synthesis and transport, and the movement of ions across the cell membrane (Daly et al., 2016). Genomic evidence of the isolated culture indicated that Marinobacter. sp. UTICA-S1B6 has a glycine betaine transporter for osmolyte uptake as well as the dehydrogenases responsible for conversion of choline into glycine betaine under aerobic conditions for de novo osmoprotectant synthesis. We also discovered several genes responsible for the synthesis of ectoine, L-ectoine synthase (ectC) and ectoine hydroxlase (ectD), and anti-porter genes (H+, Na+, K+, Cl−) responsible for the osmotic regulation within the cell (Figure 5, Oren, 2008). Arcobacter spp. UTICA-S4D1 and MARC-MIP3H16 both have the putative ability to synthesize ectoine via ectD, but not ectC. These Arcobacter strains do not have the functional potential for synthesis of osmolytes other than ectoine, but instead contain anti-porters (H+, Na+, K+, Cl−) that may be used for adaptation to changing concentrations of NaCl via the “salt-in” strategy, as well as transporters for glycine betaine and polar amino acids for uptake of extracellular osmolytes (Figure 5, Oren, 2008; Daly et al., 2016). Neither Marinobacter sp. UTICA-S1B6 nor Arcobacter spp. UTICA-S4D1 and MARC-MIP3H16 have genes responsible for uptake or synthesis of osmolytes sorbitol/mannitol, or trehalose.
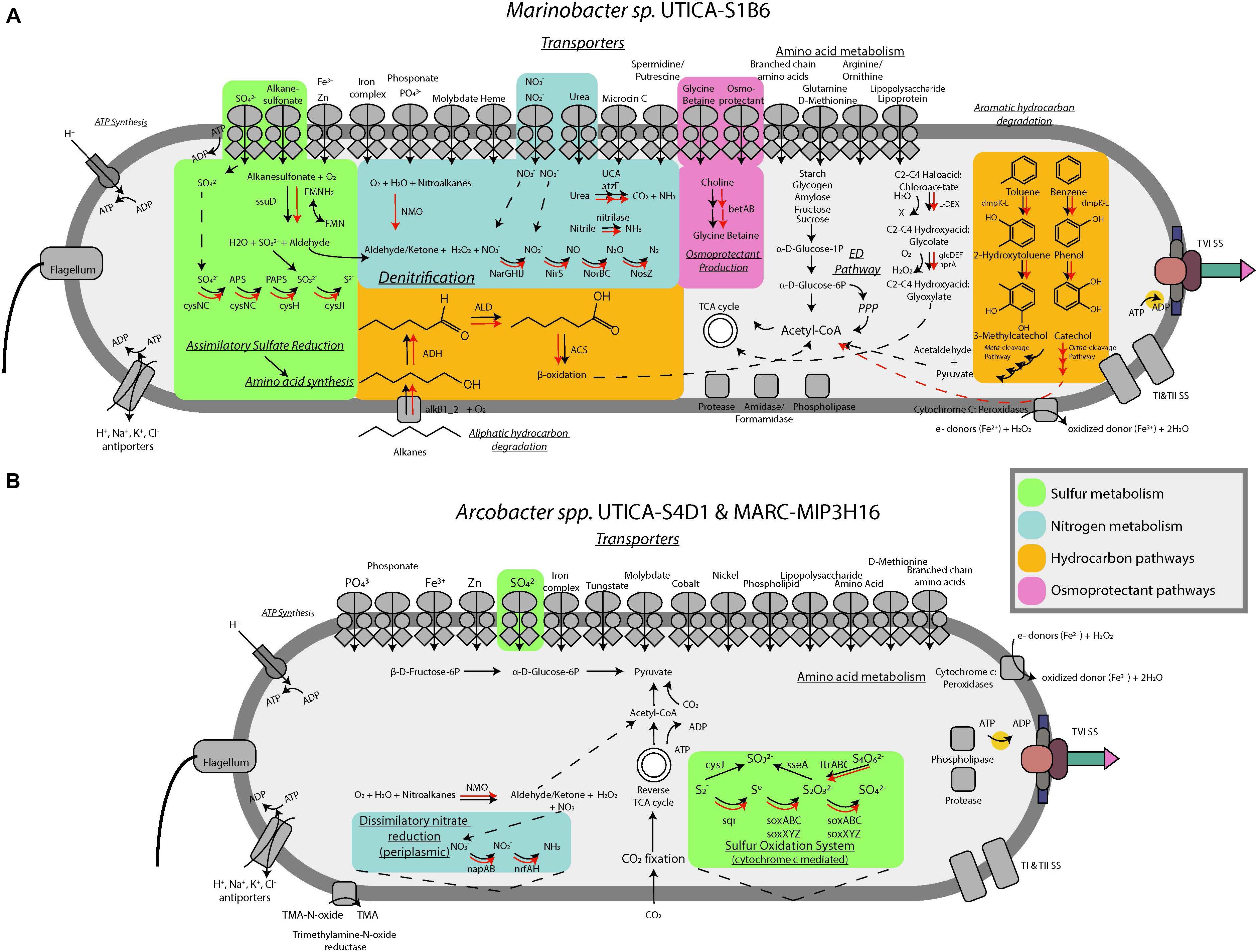
FIGURE 5. Conceptual metabolic models for (A) Marinobacter and (B) Arcobacter isolates incorporating isolate genomic (black arrows) and metagenome data (red arrows).
Marinobacter and Arcobacter Utilize a Wide Variety of Carbon Sources
Marinobacter sp. UTICA-S1B6 can utilize a variety of organic compounds for carbon and energy sources, as evidenced by isolate genomic potential and laboratory physiological assessments. We detected genes for the Entner-Doudoroff pathway for conversion of glucose to pyruvate in M. UTICA-S1B6, however, this pathway was not complete as it lacked all genes responsible for phosphorylation of glucose (EC 3.1.3.9, 2.7.1.1, 2.7.2.2, and 2.7.1.199). Consistent with our genomic inferences, we confirmed the isolate’s inability to grow on glucose under oxic laboratory conditions (Supplementary Figure S1). We detected multiple amino acid ABC transporters and di/tricarboxylate transporters in the Marinobacter isolate genome, providing evidence that amino acid uptake is important for this strain. In complementary laboratory experiments, Marinobacter sp. UTICA-S1B6 grew on two amino acids (L-glutamate, L-proline) and several carboxylic acids (OD>0.1) as the sole carbon source, with citrate, L-malate, and acetate as three of the more optimally utilized compounds (Figure 4B). In addition, genes responsible for breakdown and utilization of starch, glycogen, amylose, fructose, and sucrose are present in the isolate genome (Figure 5A); however, M. UTICA-S1B6 did not grow on sucrose or fructose as a sole carbon source in the laboratory (Supplementary Figure S1).
Our genomic investigation also showed that Marinobacter sp. UTICA-S1B6 has the potential to degrade aliphatic and aromatic hydrocarbons. The strain contains the alkane-1-monooxygenase gene (alkB) responsible for activating alkanes to a primary alcohol using oxygen, as well as the alcohol and aldehyde dehydrogenases that would be necessary for substrate transformation before β-oxidation and processing through the TCA cycle (Figure 5A). Although we did not test the strain’s growth on aliphatic hydrocarbons, M. UTICA-S1B6 grew at mid-range optical densities (0.05–0.1) on polysorbate non-ionic surfactants Tween 20, 40, and 80, which contain long-chain hydrocarbons (Figure 4B). Genes known to catalyze the anaerobic breakdown of hydrocarbons (benzylsuccinate synthase, alkylsuccinate synthase) through fumarate addition (Agrawal and Gieg, 2013) were not identified in the genome, making it unlikely that Marinobacter sp. UTICA-S1B6 degrades hydrocarbons in the absence of oxygen.
Besides aliphatic hydrocarbons, Marinobacter sp. UTICA-S1B6 has the putative ability to degrade benzene and toluene, both of which have been found in produced fluids from the Marcellus Shale at concentrations in the low part per million range (Hayes et al., 2012). The pathway involving benzene and toluene degradation employs the dmpK-L gene family, found in the M. UTICA-S1B6 genome, which converts benzene to phenol and later to catechol, or toluene to 2-hydroxytoluene and later 3-methylcatechol (Kanehisa and Goto, 2000). Catechol and 3-methylcatechol are further metabolized by the xylE-J gene family to acetaldehyde and pyruvate, both of which can be converted to acetyl-CoA and into central metabolism (e.g., the TCA cycle) (Kanehisa and Goto, 2000). This strain does not have the capacity to utilize other common aromatic compounds such as xylene, ethylbenzene, or PAHs (e.g., naphthalene). Additionally, we found a haloacid dehalogenase gene (L-DEX) in the M. UTICA-S1B6 genome, encoding for the conversion of (S)- 2-haloacids (e.g., haloacetates, halopropionates, halobutyrates containing fluoro, chloro, bromo, or iodo substiutions) to (R)-2-hydroxyacids (Goldman et al., 1968). Production of 2-hydroxyacids such as glycolate could be further broken down into glyoxylate, which can be utilized in the TCA cycle (Figure 5A). The genes responsible for this pathway, glcDEF and hprA, were detected in the M. UTICA-S1B6 genome as well as the Marcellus-4 metagenomes (Figure 5A).
Our genomic investigation revealed Arcobacter spp. MARC-MIP3H16 and UTICA-S4D1 may fix CO2 via the reverse TCA cycle, in a similar fashion to other reported Arcobacter strains (Figure 5A) (Wirsen et al., 2002; Hügler et al., 2005; Klatt and Polerecky, 2015; Roalkvam et al., 2015). These two Arcobacter strains contained all nine enzymes involved in the reductive TCA cycle (Hügler et al., 2005). We followed trends in both organic [non-purgeable organic carbon (NPOC)] and inorganic carbon in the form of CO2 gas from the Marcellus-4 natural-gas well. Both CO2 and NPOC fluctuate over time, but there is a strong association between higher CO2 concentrations and higher relative abundance of Arcobacter based on the 16S rRNA gene around day 35 and 80 (Figures 2A,B). Although growth on inorganic carbon was not tested with these Arcobacter isolates, and CO2 trends in natural-gas wells are likely influenced by a variety of biogeochemical factors, this association suggests higher concentrations of CO2 may enable growth of Arcobacter through an autotrophic metabolism or chemosynthesis of reduced sulfur species during certain times of production.
In addition to the genomic potential to fix inorganic carbon, Arcobacter sp. MARC-MIP3H16 utilized 25 different amino acids and carboxylic acids (Figure 4B). In the genome, we found amino acid ABC transporters, monosaccharide transporters, dicarboxylate transporters, and C4-transporters, confirming the ability to uptake these compounds. Amino acids L-glutamine, L-glutamate, L-aspartate, and L-proline were optimally utilized as sole carbon sources, (OD>0.1). The carboxylic acids L-malate, propionate, pyruvate, L-lactate, and succinate were optimally converted for growth as sole carbon sources while several other carboxylic acids were minimally utilized (optical density changes between 0.05 and 0.1), such as acetate and methyl pyruvate.
Marinobacter and Arcobacter Influence Nitrogen, Sulfur, and Iron Cycles in Unconventional Systems
We tracked total dissolved nitrogen (TDN) and ammonium/ammonia in six northern Appalachian Basin natural-gas wells and observed a consistent trend after hydraulic fracturing. TDN levels increased from under 5 mg/L in initial flowback samples to above 100 mg/L within 6 months of production in natural-gas wells from both the Utica-Point Pleasant Formation and Marcellus Shale (Figure 2C). The majority of TDN (59% or greater at all time points) was comprised of fixed sources of nitrogen (ammonia/ammonium). Nitrate was analyzed but was below detection (<1 ppm N) at all time points in flowback and produced fluid samples. Other possible sources of nitrogen in the system include organic nitrogen compounds, which were not measured.
Although nitrate was not detected in ppm concentrations for these natural-gas wells, nitrate reduction is a possible mode of energy generation for both Marinobacter and Arcobacter, especially at levels below our analytical detection limits. Our genomic analysis showed that Marinobacter sp. UTICA-S1B6 is capable of denitrification, possessing the proper transporters and catalytic genes for complete conversion of nitrate to N2 (Figure 5A). In addition to utilizing inorganic nitrogen as an electron acceptor, M. UTICA-S1B6 can convert urea to ammonia with urease, which is especially interesting given urea is a component in the hydraulic fracturing fluid formulation for one of the Marcellus natural-gas wells in this study (Supplementary Figure S2). Arcobacter spp. MARC-MIP3H16 and UTICA-S4D1, on the other hand, are capable of dissimilatory nitrate reduction to ammonia. Like Marinobacter sp. UTICA-S1B6, Arcobacter spp. MARC-MIP3H16 and UTICA-S4D1 can also oxidize nitroalkanes via the NMO gene, which may serve as an alternative source of nitrate within the cell.
The genome of Marinobacter sp. UTICA-S1B6 contains genes responsible for importing and assimilating sulfur from both inorganic (e.g., sulfate) and organic sulfur forms. Specifically, M. UTICA-S1B6 can putatively utilize alkanesulfonates via the ssuD gene, producing sulfite and an aldehyde group (Figure 5A) (Eichhorn et al., 1999). The ability to convert alkane-derived nutrients to sources of carbon and macronutrients (e.g., sulfate, nitrate) is consistent in the Marcellus-4 metagenomes that cluster closely to our Marinobacter isolate (Figure 5A and Supplementary Table S1).
One of the more unique aspects of Arcobacter sp. MARC-MIP3H16 relative to other taxa derived from these hydrocarbon bearing systems is its ability to completely oxidize reduced sulfur compounds (inc. sulfide, elemental sulfur, and thiosulfate) to sulfate using the sox pathway (Figure 5B). Like other Arcobacter, A. MARC-MIP3H16 may be coupling this electron transfer process to chemosynthesis (Friedrich et al., 2005; Klatt and Polerecky, 2015; Roalkvam et al., 2015). Our geochemical analysis of sulfate and sulfide in produced fluids from natural-gas wells in both the Utica-Point Pleasant Formation and Marcellus Shale supports the potential for these metabolisms to occur in these systems. Sulfate was present at part per million concentrations (20–140 mg/L) in source waters used for hydraulic fracturing of these natural-gas wells (Figure 2D). In the Marcellus natural-gas wells, within 10 days after hydraulic fracturing, sulfate dropped below 1 mg/L and remained below detection thereafter (Figure 2D). In the Utica-Point Pleasant natural-gas wells, sulfate concentrations varied between 11 and 60 mg/L during the first 59 days post-fracturing. After this time, sulfate concentrations decreased to below 6 mg/L while sulfide increased to between 10–22 mg/L until about day 150 after hydraulic fracturing. Sulfide levels plateaued at levels of 20–35 mg/L as the natural-gas well matured (150 to >450 days, Figure 2D). This period of intermediate sulfate and sulfide concentrations was associated with a higher abundance of 16S rRNA gene sequences for Arcobacter (Figure 2A) and could be related to the reoxidation of reduced sulfur species during this time. Also present in the Arcobacter genomes was the sqr gene responsible for oxidation of sulfide to polysulfides, the pshABC gene which reduces sulfur and thiosulfate to sulfide, and the cysIJ gene which oxidizes sulfide to sulfite (Kanehisa and Goto, 2000). Both isolates are also capable of reducing tetrathionate to thiosulfate (ttr gene) (Kanehisa and Goto, 2000). One isolate, Arcobacter sp. MARC-MIP3H16, contains the gene responsible for oxidation of thiosulfate to sulfite (sseA) (Kanehisa and Goto, 2000).
We detected 36 cytochromes in the genome of Marinobacter sp. UTICA-S1B6, some of which may be involved in extracellular electron transfer to redox active species (e.g., iron and oxygen). One of these cytochromes was a (per)oxidase also detected in Arcobacter sp. MARC-MIP3H16. Additionally, Marinobacter contained a thiol (per)oxidase and a catalase (per)oxidase. Closely related Marinobacter subterrani is known to oxidize Fe(II), and Marinobacter aquaeolei oxidizes Fe(II) from metal structures during growth in biofilms (Singer et al., 2011; Bonis and Gralnick, 2015). The (per)oxidase cytochrome is one of many cytochromes thought to be responsible for iron oxidation, while a type II secretion system is associated with Marinobacter aquaeolei’s ability to form a biofilm that interacts with metal surfaces (Singer et al., 2011). Indeed, the (per)oxidase encoding gene (pfam 00141) in the genome of Marinobacter sp. UTICA-S1B6 is homologous to the peroxidase encoding gene of Marinobacter subterrani JG233 (82% identity, 1234 bit score) and Marinobacter aquaeolei VT8 (88% identity, bit score 1321). Interestingly, Marinobacter sp. UTICA-S1B6 also contains the type II secretion system, suggesting a capacity to attach to reduced iron surfaces (e.g., minerals such as clays, silicates, and carbonates common in shale, carbon steel casings) and oxidize reduced forms of iron abundant within the organic-rich subsurface systems responsible for the generation of petroleum (i.e., source rocks such as shale, mudstones, organic-rich limestones, etc.) (Singer et al., 2011).
Marinobacter and Arcobacter Possess Unique Virulence Strategies in Produced Fluid
During our assessment of these Marinobacter and Arcobacter genomes, we identified genes comprising a type VI secretion system (Figures 5A,B). This system is best described as a phage-like apparatus which can be used to lyse other cells by injecting enzymes designed to degrade cellular components. Although energetically costly (Alteri and Mobley, 2016), this virulence strategy may provide a competitive advantage to these taxa under nutrient limited conditions or environmental stress (Chakraborty et al., 2011; Alteri and Mobley, 2016). Said strategy could also contribute to horizontal gene transfer within this system (Borgeaud et al., 2015). To date, no literature has been published identifying this virulence survival mechanism in the environment that occurs following hydraulic fracturing, although there is strong genomic evidence for persistent viral attacks to these taxa (Daly et al., 2016). Indeed, the genomes of M. UTICA-S1B6, A. UTICA-S4D1, and A. MARC-MIP3H16 all contain genes indicative of a type I CRISPR-Cas system which serve as acquired immunity to predatory bacteriophage (Makarova et al., 2015).
Isolate Genomes Largely Agree With Metagenomic Data
The key pathways summarized above for the isolate genome of Marinobacter sp. UTICA-S1B6 were also discovered with high homology in the Marinobacter metagenome sampled from fluids produced on day 34 from Marcellus Shale natural-gas well Marcellus-4 (bit score >200, identity >45.9%). One notable exception was the difference in the pathway for aromatic hydrocarbon degradation. The metagenome contained genes associated with catechol degradation via ortho-cleavage as opposed to the isolate genome which contained genes encoding for the meta-cleavage pathway (Kanehisa and Goto, 2000; Caspi et al., 2014). Several studies report that the meta-pathway is used for both non-substituted aromatics in addition to alkyl and halo- substituted aromatics such as toluene and chlorobenzene, whereas the ortho-pathway is generally observed exclusively during growth on non-substituted benzene or phenol (Mars et al., 1997; Veenagayathri and Vasudevan, 2011). The exclusive possession of the ortho-pathway by Marinobacter in the produced fluid metagenome may indicate sole preference for non-substituted or hydroxylated aromatic hydrocarbons in the environment. In addition, the gene for utilizing nitroalkanes through conversion to an aldehyde or ketone plus nitrate was discovered in the metagenomes with high homology to Marinobacter, but was absent in the genomes.
Largely, genes belonging to Arcobacter mined from the metagenome of the Marcellus Shale natural-gas well Marcellus-4 at days 34 and 93 meeting homology cutoffs agreed with the isolate genomes. However, a few exceptions are noted (Supplementary Table S1) including the absence of two sulfur genes in the metagenomes, cysJ (sulfide to sulfite) and sseA (thiosulfate to sulfite), as well as TMO (transforms trimethylamine n-oxide to trimethylamine), which were all present in the Arcobacter isolate genome.
Discussion
Marinobacter and Arcobacter Dominate Early Produced Fluid Communities Through Unique Osmoadaptations and Defense Mechanisms
Marinobacter are one of a dozen microbial taxa commonly found in producing natural-gas wells from hydraulically stimulated formations across the United States. (Mouser et al., 2016). The presence of this taxa during early production in Marcellus-4 is consistent with trends reported for other geographically and lithologically distinct unconventional hydrocarbon-producing systems (e.g., shales, mudstones, organic-rich limestones), which show Marinobacter present in early flowback fluid samples (<49 days after flowback began) but largely absent after that time (>49 days) (Davis et al., 2012; Fichter et al., 2012; Struchtemeyer and Elshahed, 2012; Mohan et al., 2013; Cluff et al., 2014). Similarly, the temporal trends reported here for Arcobacter abundance (Figure 2A) are consistent with those reported by Wuchter et al. (2013), who detected Arcobacter marinus in produced fluids from the Antrim Shale ranging from 35 to 85% of the total ε-proteobacteria abundance during the 5 months after hydraulic fracturing. Previous studies on fluids produced from the Marcellus Shale report Arcobacter abundance peaking (upwards of 66% of total reported microbial community based on the 16S rRNA gene) within the first 2 weeks after production begins and decreasing thereafter (Mohan et al., 2013; Cluff et al., 2014). This consistent detection of Marinobacter during the first few weeks after fracturing and Arcobacter during the first few months after fracturing suggests these taxa play an important, albeit fleeting, role in the subsurface following natural-gas well completion and hydraulic fracturing.
Based on genomic and metagenomic inference, there are several survival strategies these organisms possess that could enable their persistence in fluids derived from tight shales and other unconventional hydrocarbon-producing systems during this transitional biogeochemical phase. Arguably, the largest bottleneck for the survival of microorganisms in an ecosystem such as those generated following fracturing of shales or other unconventional systems is the high salinity observed in produced fluids from these formations, which is known to increase as hydrocarbon production proceeds. Both Arcobacter and Marinobacter possess strategies for adapting to high salt, namely via anti-porters using a “salt-in” strategy, where cells import ions into the cytoplasm to avoid osmotic stress under increasing concentrations in the environment. Both taxa can also synthesize the osmolyte ectoine and import extracellular osmolytes. Additionally, Marinobacter can produce osmoprotectant glycine betaine in response to high salinity, which can subsequently be fermented by other microbial community members to fuel methanogenesis (Daly et al., 2016).
The ability to prey upon other bacterial taxa is a survival strategy that could allow predatorial taxa to have a significant advantage in subsurface systems with strong competition for nutrient or electron acceptor resources. The type VI secretion system detected in both Marinobacter and Arcobacter genomes may allow these taxa to attack neighboring cells with a phage-like appendage, insert effector proteins, and capture cellular materials from the victim including lipids, polysaccharides, proteins, amino acids, and nucleic acids. Besides antagonism, the type VI secretion system may be used for cell signaling, biofilm remodeling, or lysing phage-infected bacteria (Russell et al., 2014). Interestingly, Marinobacter hydrocarbonoclasticus grown on hexadecane overexpressed type VI secretion system proteins relative to controls. The authors concluded a role for the type VI secretion system in alkane assimilation (Vaysse et al., 2009). As both Arcobacter and Marinobacter possess this system, its presence could confer their advantage in fluids derived from tight shales and other unconventional hydrocarbon-producing systems through biofilm formation, virulence, or hydrocarbon assimilation. Furthermore, the presence of CRISPR-associated encoding genes in the genomes of all three isolates may confer additional immunity in this system in the presence of lysogenic phage (Daly et al., 2016).
Marinobacter and Arcobacter Are Carbon Opportunists
Our investigation into the metabolic potential of Marinobacter sp. UTICA-S1B6 revealed the capacity for complex carbon oxidation, including aliphatic compounds, aromatic compounds, and haloacids. Our phylogenetic analysis based on the 16S rRNA gene showed M. UTICA-S1B6 is closely related to Marinobacter hydrocarbonoclasticus (96.6%), a bacterium isolated from seawater collected near an oil refinery in the Mediterranean Sea (Mounier et al., 2014). M. hydrocarbonoclasticus is able to degrade a wide variety of aliphatic and aromatic hydrocarbons through oleolytic biofilm formation. Marinobacter sp. UTICA-S1B6 could access and transform hydrocarbons in a similar manner based on our genomic evaluation by coupling these reactions to oxygen and/or nitrate reduction, enabling its persistence during the first few weeks of flowback before oxygenated resources are depleted. Furthermore, Marinobacter may persist as labile carbon substrates are depleted due to its ability to utilize a wide range of organic carbon sources much like other opportunistic Marinobacter species (Singer et al., 2011).
An autotrophic metabolism potentially confers advantage for Arcobacter species among a primarily heterotrophic and fermentative microbial community. As the natural-gas well matures and the sources of injected labile carbon dwindle, Arcobacter may alternate between heterotrophic and autotrophic metabolisms. Further experimentation is required to confirm its ability to fix CO2, but geochemical and genomic evidence suggest this enzymatic pathway may be important for the persistence of Arcobacter during the first few months following hydraulic fracturing and hydrocarbon production within a given natural-gas well. Much like opportunistic Marinobacter, Arcobacter species in unconventional systems may also utilize diverse carbon substrates (organic and inorganic), electron acceptors (oxygen, nitrate), and electron donors (reduced sulfur and iron species) that are generally present during early flowback.
Importance of Marinobacter and Arcobacter on Biogeochemical Cycling in Flowback and Produced Fluids
Nitrate is added in certain oil and gas systems to thermodynamically control growth of sulfate reducers and fermenters, thereby reducing biocorrosion damage to infrastructure through sulfide and/or acid generation (Voordouw, 2008; Arensdorf et al., 2009; Voordouw et al., 2009; An et al., 2017). As a result, there is need to understand the utilization of both organic and inorganic nitrogen species by microorganisms in subsurface environments such as shale or other unconventional hydrocarbon-bearing systems. In contrast to other subsurface terrestrial systems like deep groundwater environments that have limited nitrogen sources (Kutvonen et al., 2015; Bomberg and Ahonen, 2017), excess sources of fixed nitrogen are available in fluids produced from shale or other unconventional hydrocarbon resources.
Our results show that both TDN and ammonia increase significantly as production proceeds in six hydraulically fractured natural-gas wells in the northern Appalachian Basin. The individual constituents of TDN may originate from a variety of anthropogenic and indigenous sources. Since nitrate was below detection in produced fluids, it is not a considerable source of TDN here. In the case of the (Utica-3, Utica-6, Utica-7, and Utica-8) natural-gas wells, produced fluids from other hydraulically fractured natural-gas wells were used as a source of slickwater for hydraulic fracturing (i.e., recycled produced fluid), which may partially explain high initial TDN values. In terms of other nitrogen sources, organic nitrogen is frequently added to fracture fluid formulations for several different purposes, including clay stabilization, scale inhibition, friction reduction, gel formation, and as surfactants and solvents (Elsner and Hoelzer, 2016). For example, urea was one chemical component of the hydraulic fracturing fluid in the Marcellus Shale natural-gas well Marcellus-4, and its enzymatic conversion to ammonia and carbon dioxide may contribute to TDN and ammonia trends in these wells. Ammonia is also a potential byproduct of (poly)acrylamide and related polymer degradation (biotic or abiotic). Polyacrylamides are contained in many slickwater fracturing fluid recipes (Elsner and Hoelzer, 2016), and are listed on the FracFocus report for the three Marcellus and Utica-Point Pleasant natural-gas wells where these three strains were isolated (Marinobacter sp. UTICA-S1B6, Arcobacter sp. S4D1, and Arcobacter sp. MARC-MIP3H16) (Supplementary Figures S2–S4). Interestingly, Marinobacter contains amidase-encoding genes, suggesting that this taxa could degrade amide-based polymers in this system, such as polyacrylamides. Furthermore, both Marinobacter and Arcobacter can produce nitrate from nitroalkanes, a pre-cursor biochemical process that could enable denitrification or dissimilatory nitrate reduction despite low (<1 ppm) nitrate concentrations in produced fluids.
Nitrogen fixation is energetically costly, and the presence of bioavailable nitrogen sources (e.g., nitrate, urea, amines, ammonia) makes it unlikely nitrogen fixation occurs in these unconventional hydrocarbon-producing systems (Houlton et al., 2008). Fixed sources of nitrogen must therefore be biologically produced or sourced within the rock (e.g., shale). Arcobacter may produce ammonia during early production through dissimilatory nitrate reduction. A methanogen that thrives during later production, Methanohalophilus, may also contribute to ammonia production during methanogenesis fueled by methylamines (Borton et al., 2018). Altogether, it is plausible that Marinobacter and Arcobacter in early production of hydraulically fractured natural-gas wells contribute to marked increases in total nitrogen concentrations and ammonia in produced fluids by reducing nitrate and/or degrading xenobiotic organic nitrogen sources injected by well operators.
The primary cause of natural-gas well infrastructure fouling is attributed to the acidic and reactive properties of sulfides, making the sulfur cycle a vitally important process to understand in this system (Pirzadeh et al., 2014; Booker et al., 2017). Previous studies indicate thiosulfate reduction by Halanaerobium spps. as the primary mechanism for sulfide formation in later produced fluids (Liang et al., 2016; Booker et al., 2017). Contributing to this biogeochemical cycle, Arcobacter spp. have the capacity to refuel sulfide production, at least temporarily, through oxidation of reduced sulfur species (e.g., sulfides, sulfur) to thiosulfate and sulfate. This process would initially reduce sulfide levels until oxygen and/or nitrate are depleted from the system and provide electron acceptors (e.g., thiosulfate, sulfate) for other dominant taxa. Although Arcobacter spp. studied here have the ability to oxidize sulfide to sulfate, we propose thiosulfate to be the primary endpoint of Arcobacter sulfur metabolism due to (1) the lack of sulfate present in later produced fluids, and (2) the presence of known thiosulfate reducers in these wells (e.g. Halanaerobium).
Conclusion
Bacterial taxa common to early production stages of hydraulically fractured natural-gas wells play significant roles in carbon, nitrogen, and sulfur cycling in flowback and produced fluids. Marinobacter can oxidize hydrocarbons as well as capture carbon, nitrate, and sulfur species from alkane-derived hydrocarbons, which are most likely present from fracture fluid additives and/or brines produced from unconventional hydrocarbon systems during the early stages of natural-gas production. Arcobacter uniquely participates in carbon and sulfur cycling through coupling chemosynthesis to oxidation of reduced sulfur species, which may fuel heterotrophic, fermentative, and/or thiosulfate-reducing microorganisms. These taxa may further contribute to high ammonia concentrations in these natural-gas wells through urea conversion or nitrate reduction. Our genomic and experimental investigations of Marinobacter and Arcobacter physiology reveals the impact these slight to moderate halophilic taxa have on biogeochemical cycles during early production stages in hydraulic fractured natural-gas wells as oxygen and carbon resources are diminishing and salinities increase.
Significance
This work contributes to our understanding of carbon, nitrogen, and sulfur microbial metabolisms in saline produced fluids from fractured natural-gas wells.
Author Contributions
PM and JP conceived and designed the experiments with assistance from MW and KW. ME wrote the manuscript. JP performed the laboratory-based experiments. JP, SW, and JS acquired the SEM images. NN compiled geochemical data and contributed a figure. JP and ME performed the genomic analyses. DC, MW, KW, and PM designed field experiments while AH, RD, SW, and JS carried out field sampling and performed the geochemical measurements. KW and RD extracted DNA and processed metagenomes. ME performed the metagenomic analyses and assisted with geochemical measurements. TD performed the CO2 analysis.
Funding
This research was supported by funding from the National Science Foundation Dimensions of Biodiversity (Award No. 1342701) and DOE National Energy Technology Laboratory through the Marcellus Shale Energy and Environmental Laboratory (project #DE-FE0024297) under a subcontract from West Virginia University. We are especially grateful for support from Northeast Natural Energy for site access and sample support. Sequencing was performed under an award to KW, MW, and PM (No. 1931), and conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, supported by the Office of Science of the U.S. Department of Energy under contract no. DE-AC02-05CH11231.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02646/full#supplementary-material
References
Agrawal, A., and Gieg, L. M. (2013). In situ detection of anaerobic alkane metabolites in subsurface environments. Front. Microbiol. 4:140. doi: 10.3389/fmicb.2013.00140
Alteri, C. J., and Mobley, H. L. T. (2016). The versatile type VI secretion system. Microbiol. Spectr. 4, 337–356. doi: 10.1128/microbiolspec.VMBF-0026-2015
An, B. A., Shen, Y., and Voordouw, G. (2017). Control of sulfide production in high salinity Bakken shale oil reservoirs by halophilic bacteria reducing nitrate to nitrite. Front. Microbiol. 8:1164. doi: 10.3389/fmicb.2017.01164
Arensdorf, J. J., Miner, K., Ertmoed, R., Clay, W. K., Stadnicki, P., and Voordouw, G. (2009). “Mitigation of reservoir souring by nitrate in a produced-water reinjection system in Alberta,” in Proceedings of the SPE International Symposium on Oilfield Chemistry, Richardson, TX.
Arthur, J. D., Bohm, B., and Layne, M. (2009). Hydraulic Fracturing Considerations for Natural Gas wells of the Marcellus Shale. Available at: http://archives.datapages.com/data/gcags_pdf/2009/arthetal.htm
Bagheri, M., Amoozegar, M. A., Didari, M., Makhdoumi-Kakhki, A., Schumann, P., Spröer, C., et al. (2013). Marinobacter persicus sp. nov., a moderately halophilic bacterium from a saline lake in Iran. Antonie Van Leeuwenhoek 104, 47–54. doi: 10.1007/s10482-013-9923-3
Barbot, E., Vidic, N. S., Gregory, K. B., and Vidic, R. D. (2013). Spatial and temporal correlation of water quality parameters of produced waters from Devonian-age shale following hydraulic fracturing. Environ. Sci. Technol. 47, 2562–2569. doi: 10.1021/es304638h
Bomberg, M., and Ahonen, L. (2017). Editorial: geomicrobes: life in terrestrial deep subsurface. Front. Microbiol. 8:103. doi: 10.3389/fmicb.2017.00103
Bonis, B. M., and Gralnick, J. A. (2015). Marinobacter subterrani, a genetically tractable neutrophilic Fe(II)-oxidizing strain isolated from the Soudan Iron Mine. Front. Microbiol. 6:719. doi: 10.3389/fmicb.2015.00719
Booker, A. E., Borton, M. A., Daly, R. A., Welch, S. A., Nicora, C. D., Hoyt, D. W., et al. (2017). Sulfide generation by dominant Halanaerobium microorganisms in hydraulically fractured shales. mSphere 2:e00257-17. doi: 10.1128/mSphereDirect.00257-17
Borgeaud, S., Metzger, L. C., Scrignari, T., and Blokesch, M. (2015). The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science 347, 63–67. doi: 10.1126/science.1260064
Borton, M. A., Hoyt, D. W., Roux, S., Daly, R. A., Welch, S. A., Nicora, C. D., et al. (2018). Coupled laboratory and field investigations resolve microbial interactions that underpin persistence in hydraulically fractured shales. Proc. Natl. Acad. Sci. U.S.A. 115, E6585–E6594. doi: 10.1073/pnas.1800155115
Brown, C. T., Hug, L. A., Thomas, B. C., Sharon, I., Castelle, C. J., Singh, A., et al. (2015). Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523, 208–211. doi: 10.1038/nature14486
Brussaard, C. P. D. (2004). Optimization of procedures for counting viruses by flow cytometry. Appl. Environ. Microbiol. 70, 1506–1513. doi: 10.1128/AEM.70.3.1506-1513.2004
Caspi, R., Altman, T., Billington, R., Dreher, K., Foerster, H., Fulcher, C. A., et al. (2014). The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 42, D459–D471. doi: 10.1093/nar/gkt1103
Chakraborty, S., Sivaraman, J., Leung, K. Y., and Mok, Y.-K. (2011). Two-component PhoB-PhoR regulatory system and ferric uptake regulator sense phosphate and iron to control virulence genes in Type III and VI secretion systems of Edwardsiella tarda. J. Biol. Chem. 286, 39417–39430. doi: 10.1074/jbc.M111.295188
Chapman, E. C., Capo, R. C., Stewart, B. W., Kirby, C. S., Hammack, R. W., Schroeder, K. T., et al. (2012). Geochemical and strontium isotope characterization of produced waters from Marcellus Shale natural gas extraction. Environ. Sci. Technol. 46, 3545–3553. doi: 10.1021/es204005g
Chen, F., Lu, J., Binder, B. J., Liu, Y., and Hodson, R. E. (2001). Application of digital image analysis and flow cytometry to enumerate marine viruses stained with SYBR Gold. Appl. Environ. Microbiol. 67, 539–545. doi: 10.1128/AEM.67.2.539-545.2001
Chen, I.-M. A., Markowitz, V. M., Chu, K., Palaniappan, K., Szeto, E., Pillay, M., et al. (2017). IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 45, D507–D516. doi: 10.1093/nar/gkw929
Cluff, M. A., Hartsock, A., MacRae, J. D., Carter, K., and Mouser, P. J. (2014). Temporal changes in microbial ecology and geochemistry in produced water from hydraulically fractured Marcellus shale gas wells. Environ. Sci. Technol. 48, 6508–6517. doi: 10.1021/es501173p
Daly, R. A., Borton, M. A., Wilkins, M. J., Hoyt, D. W., Kountz, D. J., Wolfe, R. A., et al. (2016). Microbial metabolisms in a 2.5-km-deep ecosystem created by hydraulic fracturing in shales. Nat. Microbiol. 1:16146. doi: 10.1038/nmicrobiol.2016.146
Darrah, T. H., Jackson, R. B., Vengosh, A., Warner, N. R., Whyte, C. J., Walsh, T. B., et al. (2015). The evolution of Devonian hydrocarbon gases in shallow aquifers of the northern Appalachian Basin: insights from integrating noble gas and hydrocarbon geochemistry. Geochim. Cosmochim. Acta 170, 321–355. doi: 10.1016/j.gca.2015.09.006
Davis, J. P., Struchtemeyer, C. G., and Elshahed, M. S. (2012). Bacterial communities associated with production facilities of two newly drilled thermogenic natural gas wells in the Barnett shale (Texas, USA). Microb. Ecol. 64, 942–954. doi: 10.1007/s00248-012-0073-3
Diemer, G., Kyle, J. E., and Stedman, K. (2013). Counting Viruses Using Polycarbonate Track EtchTM Membrane Filters as an Alternative to AnodiscTM Membrane Filters. Available at: http://web.pdx.edu/kstedman/PCTE_virus_counting_protocol.pdf
Donachie, S. P., Bowman, J. P., On, S. L., and Alam, M. (2005). Arcobacter halophilus sp. nov., the first obligate halophile in the genus Arcobacter. Int. J. Syst. Evol. Microbiol. 55, 1271–1277. doi: 10.1099/ijs.0.63581-0
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
EIA (2016a). Hydraulic Fracturing Accounts for About Half of Current U.S. Crude Oil Production - Today in Energy. Washington, DC: U.S. Energy Information Administration.
EIA (2016b). Hydraulically Fractured Wells Provide Two-Thirds of U.S. Natural Gas Production - Today in Energy. Washington, DC: U.S. Energy Information Administration.
Eichhorn, E., van der Ploeg, J. R., and Leisinger, T. (1999). Characterization of a two-component alkanesulfonate monooxygenase from Escherichia coli. J. Biol. Chem. 274, 26639–26646. doi: 10.1074/jbc.274.38.26639
Elsner, M., and Hoelzer, K. (2016). Quantitative survey and structural classification of hydraulic fracturing chemicals reported in unconventional gas production. Environ. Sci. Technol. 50, 3290–3314. doi: 10.1021/acs.est.5b02818
Fichter, J., Wunch, K., Moore, R., Summer, E., Braman, S., and Holmes, P. (2012). How Hot is Too Hot for Bacteria? A Technical Study Assessing Bacterial Establishment in Downhole Drilling, Fracturing and Fracturing Operations. Houston, TX: NACE International.
Friedrich, C. G., Bardischewsky, F., Rother, D., Quentmeier, A., and Fischer, J. (2005). Prokaryotic sulfur oxidation. Curr. Opin. Microbiol. 8, 253–259. doi: 10.1016/j.mib.2005.04.005
Gaspar, J., Mathieu, J., Yang, Y., Tomson, R., Leyris, J. D., and Gregory, K. B. (2014). Microbial dynamics and control in shale gas production. Environ. Sci. Technol. Lett. 1, 465–473. doi: 10.1021/ez5003242
Gauthier, M. J., Lafay, B., Christen, R., Fernandez, L., Acquaviva, M., Bonin, P., et al. (1992). Marinobacter hydrocarbonoclasticus gen. nov., sp. nov., a new, extremely halotolerant, hydrocarbon-degrading marine bacterium. Int. J. Syst. Bacteriol. 42, 568–576. doi: 10.1099/00207713-42-4-568
Goldman, P., Milne, G. W. A., and Keister, D. B. (1968). Carbon-halogen bond cleavage III. Studies on bacterial halidohydrolases. J. Biol. Chem. 243,428–434.
Haluszczak, L. O., Rose, A. W., and Kump, L. R. (2013). Geochemical evaluation of flowback brine from Marcellus gas wells in Pennsylvania, USA. Appl. Geochem. 28, 55–61. doi: 10.1016/j.apgeochem.2012.10.002
Harkness, J. S., Darrah, T. H., Warner, N. R., Whyte, C. J., Moore, M. T., Millot, R., et al. (2017). The geochemistry of naturally occurring methane and saline groundwater in an area of unconventional shale gas development. Geochim. Cosmochim. Acta 208, 302–334. doi: 10.1016/j.gca.2017.03.039
Harkness, J. S., Dwyer, G. S., Warner, N. R., Parker, K. M., Mitch, W. A., and Vengosh, A. (2015). Iodide, bromide, and ammonium in hydraulic fracturing and oil and gas wastewaters: environmental implications. Environ. Sci. Technol. 49, 1955–1963. doi: 10.1021/es504654n
Hayes, T., Severin, B. F., Engineer, P. S. P., and Okemos, M. I. (2012). Barnett and Appalachian Shale Water Management and Reuse Technologies. Research Partnership to Secure Energy for America, Report no. 8122-05. Available at: https://www.netl.doe.gov/File%20Library/Research/Oil-Gas/Natural%20Gas/shale%20gas/08122-05-final-report.pdf
Heilweil, V. M., Solomon, D. K., Darrah, T. H., Gilmore, T. E., and Genereux, D. P. (2016). Gas-tracer experiment for evaluating the fate of methane in a coastal plain stream: degassing versus in-stream oxidation. Environ. Sci. Technol. 50, 10504–10511. doi: 10.1021/acs.est.6b02224
Houlton, B. Z., Wang, Y.-P., Vitousek, P. M., and Field, C. B. (2008). A unifying framework for dinitrogen fixation in the terrestrial biosphere. Nature 454, 327–330. doi: 10.1038/nature07028
Hügler, M., Wirsen, C. O., Fuchs, G., Taylor, C. D., and Sievert, S. M. (2005). Evidence for autotrophic CO2 fixation via the reductive tricarboxylic acid cycle by members of the ε subdivision of Proteobacteria. J. Bacteriol. 187, 3020–3027. doi: 10.1128/JB.187.9.3020-3027.2005
Huntemann, M., Ivanova, N. N., Mavromatis, K., Tripp, H. J., Paez-Espino, D., Tennessen, K., et al. (2016). The standard operating procedure of the DOE-JGI Metagenome Annotation Pipeline (MAP v.4). Stand. Genomic Sci. 11:17. doi: 10.1186/s40793-016-0138-x
Huu, N. B., Denner, E. B. M., Ha, D. T. C., Wanner, G., and Stan-Lotter, H. (1999). Marinobacter aquaeolei sp. nov., a halophilic bacterium isolated from a Vietnamese oil-producing well. Int. J. Syst. Evol. Microbiol. 49, 367–375. doi: 10.1099/00207713-49-2-367
Hyatt, D., LoCascio, P. F., Hauser, L. J., and Uberbacher, E. C. (2012). Gene and translation initiation site prediction in metagenomic sequences. Bioinformatics 28, 2223–2230. doi: 10.1093/bioinformatics/bts429
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kang, M., Christian, S., Celia, M. A., Mauzerall, D. L., Bill, M., Miller, A. R., et al. (2016). Identification and characterization of high methane-emitting abandoned oil and gas wells. Proc. Natl. Acad. Sci. U.S.A. 113, 13636–13641. doi: 10.1073/pnas.1605913113
Kerr, R. A. (2010). Energy. Natural gas from shale bursts onto the scene. Science 328, 1624–1626. doi: 10.1126/science.328.5986.1624
Kim, H. M., Hwang, C. Y., and Cho, B. C. (2010). Arcobacter marinus sp. nov. Int. J. Syst. Evol. Microbiol. 60, 531–536. doi: 10.1099/ijs.0.007740-0
Klatt, J. M., and Polerecky, L. (2015). Assessment of the stoichiometry and efficiency of CO2 fixation coupled to reduced sulfur oxidation. Front. Microbiol. 6:484. doi: 10.3389/fmicb.2015.00484
Kutvonen, H., Rajala, P., Carpén, L., and Bomberg, M. (2015). Nitrate and ammonia as nitrogen sources for deep subsurface microorganisms. Front. Microbiol. 6:1079. doi: 10.3389/fmicb.2015.01079
Lester, Y., Ferrer, I., Thurman, E. M., Sitterley, K. A., Korak, J. A., Aiken, G., et al. (2015). Characterization of hydraulic fracturing flowback water in Colorado: implications for water treatment. Sci. Total Environ. 512–513, 637–644. doi: 10.1016/j.scitotenv.2015.01.043
Liang, R., Davidova, I. A., Marks, C. R., Stamps, B. W., Harriman, B. H., Stevenson, B. S., et al. (2016). Metabolic capability of a predominant Halanaerobium sp. in hydraulically fractured gas wells and its implication in pipeline corrosion. Front. Microbiol. 7:988. doi: 10.3389/fmicb.2016.00988
Lipus, D., Roy, D., Khan, E., Ross, D., Vikram, A., Gulliver, D., et al. (2018). Microbial communities in Bakken region produced water. FEMS Microbiol. Lett. 365:fny107. doi: 10.1093/femsle/fny107
Lipus, D., Vikram, A., Ross, D., Bain, D., Gulliver, D., Hammack, R., et al. (2017). Predominance and metabolic potential of Halanaerobium in produced water from hydraulically fractured Marcellus shale wells. Appl. Environ. Microbiol. 83:e02659-16. doi: 10.1128/AEM.02659-16
Luek, J. L., Harir, M., Schmitt-Kopplin, P., Mouser, P. J., and Gonsior, M. (2018). Temporal dynamics of halogenated organic compounds in Marcellus Shale flowback. Water Res. 136, 200–206. doi: 10.1016/j.watres.2018.02.055
Makarova, K. S., Wolf, Y. I., Alkhnbashi, O. S., Costa, F., Shah, S. A., Saunders, S. J., et al. (2015). An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 13, 722–736. doi: 10.1038/nrmicro3569
Mars, A. E., Kasberg, T., Kaschabek, S. R., van Agteren, M. H., Janssen, D. B., and Reineke, W. (1997). Microbial degradation of chloroaromatics: use of the meta-cleavage pathway for mineralization of chlorobenzene. J. Bacteriol. 179, 4530–4537. doi: 10.1128/jb.179.14.4530-4537.1997
Miller, C. S., Baker, B. J., Thomas, B. C., Singer, S. W., and Banfield, J. F. (2011). EMIRGE: reconstruction of full-length ribosomal genes from microbial community short read sequencing data. Genome Biol. 12:R44. doi: 10.1186/gb-2011-12-5-r44
Mohan, A. M., Hartsock, A., Bibby, K. J., Hammack, R. W., Vidic, R. D., and Gregory, K. B. (2013). Microbial community changes in hydraulic fracturing fluids and produced water from shale gas extraction. Environ. Sci. Technol. 47, 13141–13150. doi: 10.1021/es402928b
Moore, M. T., Vinson, D. S., Whyte, C. J., Eymold, W. K., Walsh, T. B., and Darrah, T. H. (2018). Differentiating between biogenic and thermogenic sources of natural gas in coalbed methane reservoirs from the Illinois Basin using noble gas and hydrocarbon geochemistry. Geol. Soc. Lond. Spec. Publ. 468, 151–188. doi: 10.1144/SP468.8
Mounier, J., Camus, A., Mitteau, I., Vaysse, P.-J., Goulas, P., Grimaud, R., et al. (2014). The marine bacterium Marinobacter hydrocarbonoclasticus SP17 degrades a wide range of lipids and hydrocarbons through the formation of oleolytic biofilms with distinct gene expression profiles. FEMS Microbiol. Ecol. 90, 816–831. doi: 10.1111/1574-6941.12439
Mouser, P. J., Borton, M., Darrah, T. H., Hartsock, A., and Wrighton, K. C. (2016). Hydraulic fracturing offers view of microbial life in the deep terrestrial subsurface. FEMS Microbiol. Ecol. 92:fiw166. doi: 10.1093/femsec/fiw166
Nixon, S. L., Walker, L., Streets, M. D. T., Eden, B., Boothman, C., Taylor, K. G., et al. (2017). Guar gum stimulates biogenic sulfide production at elevated pressures: implications for shale gas extraction. Front. Microbiol. 8:679. doi: 10.3389/fmicb.2017.00679
Noble, R., and Fuhrman, J. (1998). Use of SYBR green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14, 113–118. doi: 10.3354/ame014113
Oren, A. (2008). Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Systems 4:2. doi: 10.1186/1746-1448-4-2
Panescu, J., Daly, R. A., Wrighton, K. C., and Mouser, P. J. (2018). Draft genome sequences of two chemosynthetic Arcobacter strains isolated from hydraulically fractured wells in marcellus and Utica shales. Genome Announc. 6:e00159-18. doi: 10.1128/genomeA.00159-18
Patel, A., Noble, R. T., Steele, J. A., Schwalbach, M. S., Hewson, I., and Fuhrman, J. A. (2007). Virus and prokaryote enumeration from planktonic aquatic environments by epifluorescence microscopy with SYBR Green I. Nat. Protoc. 2, 269–276. doi: 10.1038/nprot.2007.6
Pirzadeh, P., Lesage, K. L., and Marriott, R. A. (2014). Hydraulic fracturing additives and the delayed onset of hydrogen sulfide in shale gas. Energy Fuels 28, 4993–5001. doi: 10.1021/ef501059k
Quevillon, E., Silventoinen, V., Pillai, S., Harte, N., Mulder, N., Apweiler, R., et al. (2005). InterProScan: protein domains identifier. Nucleic Acids Res. 33, W116–W120. doi: 10.1093/nar/gki442
Roalkvam, I., Drønen, K., Stokke, R., Daae, F. L., Dahle, H., and Steen, I. H. (2015). Physiological and genomic characterization of Arcobacter anaerophilus IR-1 reveals new metabolic features in Epsilonproteobacteria. Front. Microbiol. 6:987. doi: 10.3389/fmicb.2015.00987
Russell, A. B., Peterson, S. B., and Mougous, J. D. (2014). Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Microbiol. 12, 137–148. doi: 10.1038/nrmicro3185
Singer, E., Webb, E. A., Nelson, W. C., Heidelberg, J. F., Ivanova, N., Pati, A., et al. (2011). Genomic potential of Marinobacter aquaeolei, a biogeochemical “opportunitroph.” Appl. Environ. Microbiol. 77, 2763–2771. doi: 10.1128/AEM.01866-10
Struchtemeyer, C. G., and Elshahed, M. S. (2012). Bacterial communities associated with hydraulic fracturing fluids in thermogenic natural gas wells in North Central Texas, USA. FEMS Microbiol. Ecol. 81, 13–25. doi: 10.1111/j.1574-6941.2011.01196.x
Tour, J. M., Kittrell, C., and Colvin, V. L. (2010). Green carbon as a bridge to renewable energy. Nat. Mater. 9, 871–874. doi: 10.1038/nmat2887
Tummings, S., Panescu, J., Daly, R. A., Wrighton, K. C., and Mouser, P. J. (2018). Draft genome sequences of Marinobacter strains recovered from Utica shale-produced fluids. Genome Announc. 6:e00155-18. doi: 10.1128/genomeA.00155-18
Vaysse, P.-J., Prat, L., Mangenot, S., Cruveiller, S., Goulas, P., and Grimaud, R. (2009). Proteomic analysis of Marinobacter hydrocarbonoclasticus SP17 biofilm formation at the alkane-water interface reveals novel proteins and cellular processes involved in hexadecane assimilation. Res. Microbiol. 160, 829–837. doi: 10.1016/j.resmic.2009.09.010
Veenagayathri, K., and Vasudevan, N. (2011). Ortho and meta cleavage dioxygenases detected during the degradation of phenolic compounds by a moderately halophilic bacterial consortium. Int. Res. J. Microbiol. 2, 406–414.
Vengosh, A., Kondash, A., Harkness, J., Lauer, N., Warner, N., and Darrah, T. H. (2017). The geochemistry of hydraulic fracturing fluids. Proc. Earth Planet. Sci. 17, 21–24. doi: 10.1016/j.proeps.2016.12.011
Vidic, R. D., Brantley, S. L., Vandenbossche, J. M., Yoxtheimer, D., and Abad, J. D. (2013). Impact of shale gas development on regional water quality. Science 340:1235009. doi: 10.1126/science.1235009
Voordouw, G. (2008). “Impact of nitrate on the sulfur cycle in oil fields,” in Microbial Sulfur Metabolism, eds C. Dahl and C. G. Friedrich (Berlin: Springer), 296–302.
Voordouw, G., Grigoryan, A. A., Lambo, A., Lin, S., Park, H. S., Jack, T. R., et al. (2009). Sulfide remediation by pulsed injection of nitrate into a low temperature Canadian heavy oil reservoir. Environ. Sci. Technol. 43, 9512–9518. doi: 10.1021/es902211j
Warner, N. R., Christie, C. A., Jackson, R. B., and Vengosh, A. (2013). Impacts of shale gas wastewater disposal on water quality in western Pennsylvania. Environ. Sci. Technol. 47, 11849–11857. doi: 10.1021/es402165b
Weinbauer, M. G., Beckmann, C., and Höfle, M. G. (1998). Utility of green fluorescent nucleic acid dyes and aluminum oxide membrane filters for rapid epifluorescence enumeration of soil and sediment bacteria. Appl. Environ. Microbiol. 64, 5000–5003.
Wirsen, C. O., Sievert, S. M., Cavanaugh, C. M., Molyneaux, S. J., Ahmad, A., Taylor, L. T., et al. (2002). Characterization of an autotrophic sulfide-oxidizing marine Arcobacter sp. that produces filamentous sulfur. Appl. Environ. Microbiol. 68, 316–325. doi: 10.1128/AEM.68.1.316-325.2002
Wrighton, K. C., Thomas, B. C., Sharon, I., Miller, C. S., Castelle, C. J., VerBerkmoes, N. C., et al. (2012). Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337, 1661–1665. doi: 10.1126/science.1224041
Keywords: natural gas, hydraulic fracturing, dark biosphere, deep subsurface, cultivation, characterization, genome, shale
Citation: Evans MV, Panescu J, Hanson AJ, Welch SA, Sheets JM, Nastasi N, Daly RA, Cole DR, Darrah TH, Wilkins MJ, Wrighton KC and Mouser PJ (2018) Members of Marinobacter and Arcobacter Influence System Biogeochemistry During Early Production of Hydraulically Fractured Natural Gas Wells in the Appalachian Basin. Front. Microbiol. 9:2646. doi: 10.3389/fmicb.2018.02646
Received: 18 July 2018; Accepted: 17 October 2018;
Published: 15 November 2018.
Edited by:
D’Arcy Renee Meyer-Dombard, University of Illinois at Chicago, United StatesReviewed by:
Trinity L. Hamilton, University of Minnesota Twin Cities, United StatesKim Marie Handley, The University of Auckland, New Zealand
Melanie R. Mormile, Missouri University of Science and Technology, United States
Copyright © 2018 Evans, Panescu, Hanson, Welch, Sheets, Nastasi, Daly, Cole, Darrah, Wilkins, Wrighton and Mouser. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paula J. Mouser, UGF1bGEuTW91c2VyQHVuaC5lZHU=
†Present address: Rebecca A. Daly, Michael J. Wilkins, and Kelly C. Wrighton, Department of Soil and Crop Sciences, Colorado State University, Fort Collins, CO, United States
 Morgan V. Evans
Morgan V. Evans Jenny Panescu
Jenny Panescu Andrea J. Hanson2
Andrea J. Hanson2 Julia M. Sheets
Julia M. Sheets Nicholas Nastasi
Nicholas Nastasi Rebecca A. Daly
Rebecca A. Daly David R. Cole
David R. Cole Michael J. Wilkins
Michael J. Wilkins Kelly C. Wrighton
Kelly C. Wrighton Paula J. Mouser
Paula J. Mouser