- 1College of Food Science and Technology, Nanjing Agricultural University, Nanjing, China
- 2Department of Food Science and Nutrition, University of Maryland, College Park, MD, United States
- 3Department of Plant Science and Landscape Architecture, University of Maryland, College Park, MD, United States
Pullulanase plays an important role as a starch hydrolysis enzyme in the production of bio-fuels and animal feed, and in the food industry. Compared to the methods currently used for pullulanase production, synthesis by Bacillus subtilis would be safer and easier. However, the current yield of pullulanase from B. subtilis is low to meet industrial requirements. Therefore, it is necessary to improve the yield of pullulanase by B. subtilis. In this study, we mined 10 highly active promoters from B. subtilis based on transcriptome and bioinformatic data. Individual promoters and combinations of promoters were used to improve the yield of pullulanase in B. subtilis BS001. Four recombinant strains with new promoters (Phag, PtufA, PsodA, and PfusA) had higher enzyme activity than the control (PamyE). The strain containing PsodA+fusA (163 U/mL) and the strain containing PsodA+fusA+amyE (336 U/mL) had the highest activity among the analyzed dual- and triple-promoter construct stains in shake flask, which were 2.29 and 4.73 times higher than that of the strain with PamyE, respectively. Moreover, the activity of the strain containing PsodA+fusA+amyE showed a maximum activity of 1,555 U/mL, which was 21.9 times higher than that of the flask-grown PamyE strain in a 50-liter fermenter. Our work showed that these four strong promoters mined from transcriptome data and their combinations could reliably increase the yield of pullulanase in quantities suitable for industrial applications.
Introduction
Industrial starch fermentation for the production of alcohols, amino acids, nucleotides, antibiotics, and high-glucose and high-maltose syrups (Ram and Venkatasubramanian, 1982; Malviya et al., 2010) relies on pullulanase to degrade α-1,6-glycosidic bonds to improve the efficiency of starch hydrolysis (Reddy et al., 2015). Pullulanase is also used to produce high-amylose starch, resistant starch, slow-digestion starch, maltooligosaccharides, and branched cyclodextrins (Shikaishi et al., 2014; Li et al., 2017).
Because pullulanase is used in these applications, a safe, low-cost, high-yield production method is needed. Although heterologous expression of pullulanase in Escherichia coli under various conditions has yielded as much as 580 U/mL (Nie et al., 2013) and 2523.5 U/mL (Zou et al., 2014), there are many restrictions for its use in foods, feeds, and pharmaceuticals because of the endotoxins and exotoxins produced by E. coli. Safer alternative species have been used for pullulanase production, including Bacillus subtilis (24.5 U/mL) (Song et al., 2016) and Pichia pastoris (350 U/mL) (Xu et al., 2006); however, the yields from these strains are relatively low. To solve these problems, researchers have isolated new types of pullulanase enzymes from various microorganisms, such as Bacillus sp. AV-7 (Kunamneni and Singh, 2006), Thermus thermophiles (Wu et al., 2014), Bacillus deramificans (Duan et al., 2013), Anoxybacillus sp. SK3-4 (Kahar et al., 2016), and Bacillus naganoensis. In addition, the yield of pullulanase has been increased through mutation breeding (2.82 U/ml) (Wang et al., 2014), protein engineering (46.9 U/ml) (Chen et al., 2016; Nisha and Satyanarayana, 2016), and the manipulation of culture condition (543 U/ml) (Zou et al., 2016). Despite these efforts, the yield of pullulanase from these strains is still too low to meet industrial demand.
B. subtilis is a viable species for improving pullulanase yield because it is a generally recognized as safe (GRAS) microbial-derived product (Ming et al., 2010). Therefore, we chose to use B. subtilis for our pullulanase production study. Protein yield is known to be closely related to the strength of the promoter; thus, a strong promoter is a necessary requirement for high protein yield (Blazeck et al., 2012). The most well-known promoter in B. subtilis is the cytidine deaminase (ccd) promoter P43 (Wu et al., 1991), which has been used to express GFP (Kong et al., 2009), β-galactosidase, staphylokinase (Kim et al., 2008) and alkaline protease (Kim et al., 2008). Yang et al. isolated a strong B. subtilis promoter (Plaps) that is 13 times stronger than the P43 promoter by using a promoter trapping system. Inducible promoters have also been widely used in B. subtilis; including promoters that are regulated by xylose, sucrose (Biedendieck et al., 2007), maltose (Biedendieck et al., 2007; Yue et al., 2017), starch, phosphates (Abdel-Fattah et al., 2005; Makarewicz et al., 2006), citric acid (Yamamoto et al., 2000), tetracycline (Geissendörfer and Hillen, 1990), and glycine (Phan and Schumann, 2007).
Moreover, promoters an also be combined to form a multiple-promoter complexes to further enhance the expression (Zhang et al., 2017), and have been shown to increase enzyme production 1.6- (Yang et al., 2013), and 12-fold (Kang et al., 2010). Zhang et al. (2017) designed a dual-promoter expression system, PhpaII-PamyQ. Using this system, they increased enzyme activity to 571.2 U/mL in a 3 L fermenter, which was 18.7 times the activity obtained in shake flasks. Guan C. R. et al. (2016) showed that amino peptidase could be expressed in B. subtilis by the synthetic dual promoter PgsiB-PHpaII. Using this system, the obtained enzyme activity was 88.86 U/mL in shake flasks and 205 U/mL in a 5 L fermenter. In addition, the core elements of promoters, including the −35 and −10 regions (Jiao et al., 2017) and ribosome recognition site (Wang and Doi, 1984), have been optimized to enhance the promoter strength.
One bioinformatic method for selecting candidate strong promoters to improve production efficiency is analyzing the amount of mRNA expressed in a transcriptome, which should represent the strength of the promoter (McCleary, 2009). This method has been used for other applications to increase production efficiency, thus saving time and reducing costs. For example, Liu et al. (2017) analyzed the top 10 most highly expressed genes and operons among 3,959 genes and 1,249 operons in transcriptome data from Bacillus licheniformis ATCC14580. Using this method, a novel high-efficiency promoter (PBL9) was identified, which showed 23% higher expression than P43 in B. subtilis. Geng et al. (2014) cloned a root-specific promoter and developed a high-yield screening system in peanut by establishing a simple digital expression profile based on Illumina sequencing data from peanut. However, no study has utilized transcriptome data to select highly active promoters in B. subtilis based on gene expression levels.
The major objective of this study was to improve the yield of pullulanase production by B. subtilis using different promoters. To this end, we first chose three B. subtilis transcriptome data sets to screen for strong promoters. Next, pullulanase expression driven by the selected promoters was evaluated in B. subtilis BS001. Then, these promoters were combined to generate dual- or triple-promoter expression systems to improve yield.
Materials and Methods
Microbial Strains and Vectors
The bacterial strains used in this study are described in Table 1. E. coli was cultured in LB broth at 37°C. B. subtilis was cultured in CSA medium (maltose, 40 g/L; cotton seed powder, 10 g/L; soybean meal, 10 g/L; ammonium sulfate, 5 g/L; ammonium citrate, 10 g/L; dipotassium hydrogen phosphate, 9 g/L; magnesium sulfate, 0.2 g/L; manganese sulfate, 0.05 g/L; ferrous sulfate, 0.05 g/L; and calcium chloride, 1 g/L, which was adjusted to pH 6.0 before sterilization at 121°C for 20 min, pH 5.8 after sterilization) at 37°C.
The vectors used in this study are also listed in Table 1. The gene ID and position of all promoters are shown in Table 2. The sequence of pulA and promoters were shown in Supplementary Material. Super Pfu DNA polymerase, DNA markers, restriction endonucleases, reverse transcriptase, and TRIzol reagent (for RNA extraction) were purchased from TaKaRa Biotechnology (Dalian, China). The pullulanase gene (pulA) and primers (Table S1) were synthesized by Genscript (Nanjing, China).
Analysis of Transcriptome Data
Target gene yield is closely related to the strength of the promoter in the host strain. To select a strong promoter, three B. subtilis subsp. subtilis str. 168 transcriptome data sets were downloaded from the NCBI SRA database. The transcriptome accession numbers are ERR1223408 (https://trace.ncbi.nlm.nih.gov/Traces/sra/?run=ERR1223408), SRR3488633 (https://trace.ncbi.nlm.nih.gov/Traces/sra/?run=SRR3488633), and SRR3466199 (https://trace.ncbi.nlm.nih.gov/Traces/sra/?run=SRR3466199), which were used as controls in the respective studies (described in Table 3). The sequence data were processed by using the NGS QC Toolkit (2.3.3) to remove low-quality reads. The B. subtilis 168 genome (NC_000964.3) was used as a reference for transcript identification by Bowtie 2 (Version 2.2.9). Gene expression levels were analyzed by RPKM (Reads Per Kilo-bases per Million-reads), a standard method for the analysis of gene expression levels, in the HTSeq software package (Version 0.6.1). Functional annotation of the genes was based on databases, e.g., http://bacteria.ensembl.org/Bacillus_subtilis_subsp_subtilis_str_168/Info/Index, http://networks.systemsbiology.net/bsu, and http://genome2d.molgenrug.nl/. Then, the top 200 most highly expressed genes in each transcriptome were selected and analyzed. Genes present in all three sets were sorted by RPKM value. Finally, the promoters of these genes were predicted by Promoter Scan software (https://www.ncbi.nlm.nih.gov/Class/NAWBIS/Modules/DNA/dna21b.html), BPROM (http://www.softberry.com/berry.phtml?topic=bprom&group=programs&subgroup=gfindb), and BDGP (http://www.fruitfly.org/seq_tools/promoter.html) to identify the ribosome binding sites, transcription initiation sites, and TATA boxes.
Design of the Expression Box
The pullulanase expression unit in B subtilis can be divided into four parts, the promoter, the Shine-Dalgarno mRNA stabilizing sequence (STAB-SD), the signal peptide sequence, and the pullulanase gene (Figure 1). Several promoters, including Phag, PtufA, PcapD, PyqeY, PsodA, PfusA, PgapA, PahpF, PglnA, and Pmdh, were mined from the analyzed transcriptome data. The amyE promoter (PamyE) was used as a control promoter. The STAB-SD of cry3A from Bacillus thuringiensis was selected for use in the pullulanase expression system, as we hoped that this sequence could improve the stability of the mRNA and increase the yield of target gene (Park et al., 1999). The signal peptide was from B. subtilis 168 amyE. The reference pullulanase sequence was a type I pullulanase from Bacillus acidpullulyticus (Accession number: 2WAN_A, GI: 229597615). The codons of the gene were optimized for expression in B. subtilis based on the codon preference of B. subtilis 168 (GenBank accession number MH411123) by using codon optimization software (http://www.jcat.de/). Selected promoters were also combined into dual and/or triple promoter systems to increase expression.
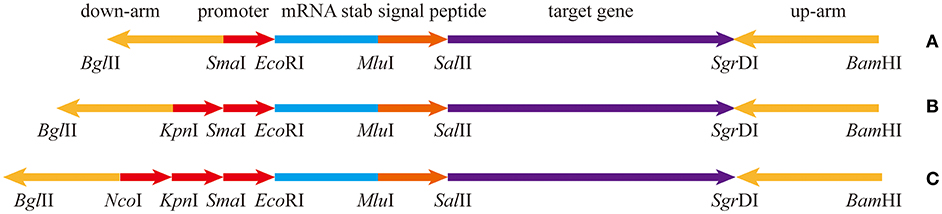
Figure 1. The expression box of pullulanase containing different number of promoters. (A) The expression box with single promoter. (B) The expression box with dual-promoter. (C) The expression box with triple-promoter. The upstream and downstream homology arms are upstream and downstream of amyE; mRNA stabilizing sequence is from Shine-Dalgarno mRNA stabilizing sequence of cryIIIA of B. thuringiensis; the signal peptide is from amyE of B. subtilis 168. The target gene is artificially synthesized codon-optimized pullulanase gene (pulA) based on the pullulanase gene of B. acidpullulyticus.
Pullulanase Expressed With Different Promoters and Combinations of Promoters
The homologous arms of the amylase gene (amyE) and the pulA expression box were ligated to the pCBS expression vector according to general methods (Sambrook and Russell, 2006). Then, the expression vectors were transformed into B. subtilis BS001 according to the method described by Dubnau (Gryczan et al., 1978). Recombinant strains were selected by resistance to erythromycin. Positive transformants were selected by blue-white screening after incubation at 45°C for 12 h. All engineered strains were cultured in CSA medium at 37°C and 180 rpm for 48 h. Extracellular enzyme activity was measured according to the method of Kahar et al. (2016), and the proteins in the supernatant were separated by SDS-PAGE according to “The Condensed Protocols from Molecular Cloning: a Laboratory Manual” (Sambrook and Russell, 2006). Protein content was determined by the Coomassie Brilliant Blue method. Specific enzyme activity (U/mg) was the enzyme activity (U/mL) divided by the protein content (mg/mL).
Detection of Promoter Activity by qPCR
All engineered strains were cultured in flasks at 37°C for 24 and 48 h. Then, samples removed were centrifuged at 12,000 × g for 5 min, and the RNA was extracted by using TRIzol according to a previously described method (Sambrook and Russell, 2006). RNA was reverse transcribed into cDNA, and pullulanase expression was detected by qPCR and the ΔΔCT method. In this study, the reference gene was the 16S ribosome gene.
Pullulanase Yield From Engineered Strains in Shake Flasks and 50-L Fermenters
Engineered strains containing pullulanase under PamyE, PsodA+fusA, and PsodA+fusA+amyE were cultured in 250 mL flasks containing 50 mL of CSA medium (pH 5.8) at 37°C for 48 h. Extracellular enzyme activity was determined every 4 h. The strain containing PsodA+fusA+amyE was cultured in 1,000 mL flasks containing 200 mL of CSA medium at 37°C for ~12 h until the cell density (OD600) reached 20. Then, the cells were transferred to a 50 L fermenter (10% inoculum). The fermentation was conducted for 48 h under the following conditions: the total sugar content was maintained at 0.5–1.0% by adding 50% maltose syrup, dissolved oxygen was maintained at >20% by controlling stirring speed and ventilation, the pH was maintained at 5.8 or 6.5 by adding ammonia water, and the temperature was maintained at 37 or 33°C. Extracellular enzyme activity was determined every 4 h.
The pullulanase yield in 50-L fermenters was shown one experiment data in section The Pullulanase Yield From Engineered Strains in Flasks and 50-L Fermenters. The other experiments were repeated four times, and the data were analyzed by one-way analysis of variance (ANOVA) with Tukey's multiple comparison tests for post-hoc comparisons in SPSS (version 17.0). A p-value < 0.05 was considered statistically significant.
Results
Selection of Strong Promoters Based on Transcriptome Data
Three transcriptome data sets from B. subtilis, ERR1223408, SRR3488633, and SRR3466199 were analyzed by using bioinformatic methods. The genome of B. subtilis 168 was used to annotate the transcriptomes, and 4,217 genes were identified. The expression levels of most genes was low, and the RPKM values were <200 as shown in Figures 2A–C. The top 200 RPKM value of genes were identified in each of the three transcriptomes data sets, and within this group of genes, there were 105 that were present in all three transcriptomes. The RPKM value of the 10 most highly expressed genes that were represented in all three transcriptomes and amyE are shown in Figure 2D. The hag gene (flagellin) had the highest RPKM value (43159), which was 98 times that of amyE (439). The RPKM value of tufA gene (elongation factor Tu) and cspD gene (cold shock protein) were >30,000. The promoter regions of the selected genes was predicted by Promoter Scan, BPROM, and BDGP as described in Table 2, and the sequences were shown in Supplementary Material.
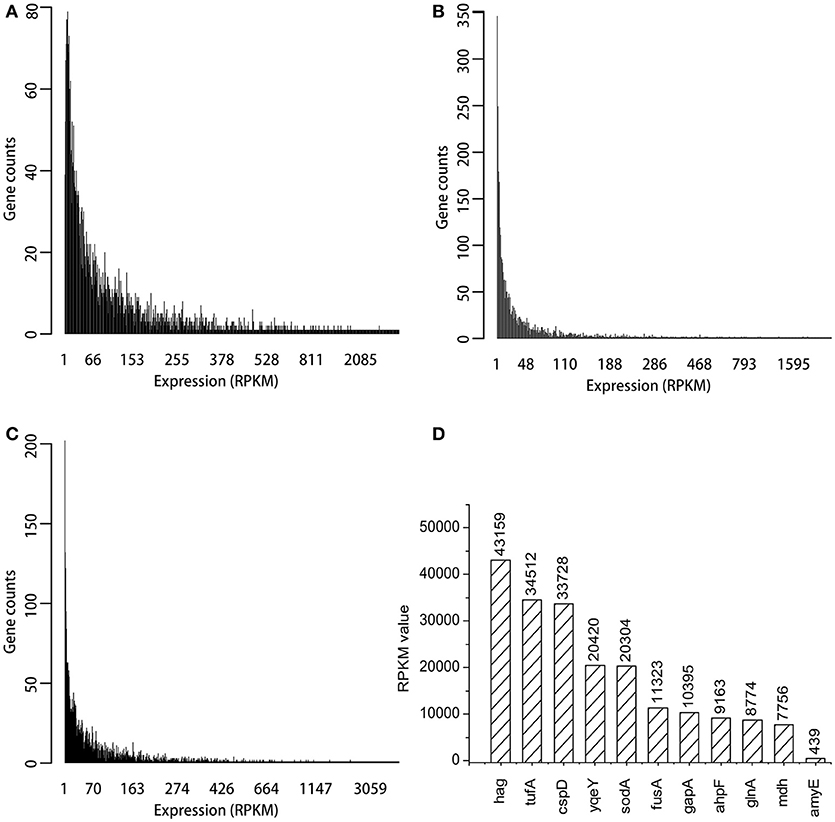
Figure 2. Expression distribution of all genes in three transcriptomes and RPKM values of top ten genes. (A) The expression distribution in ERR1223408. (B) The expression distribution in SRR3488633. (C) The expression distribution in SRR3466199. (D) The top 10 expressed genes and amyE based on RPKM values.
The Effect of Different Promoters and Their Combinations on the mRNA of Pullulanase
The amount of mRNA at 24 and 48 h of t strains were calculated by qPCR, and the result are shown in Figure 3. As the number of promoter's increases, the amount of mRNA increased continuously. The mRNA levels of the strains containing Phag, PtufA, PsodA, and PfusA were higher than the strain containing PamyE. Whereas the strains with dual promoters were higher the level in strains with single promoters, which were 15.5–19.2 times higher than that of the strain containing PamyE at 24 h and 11.6–17 times higher than that at 48 h, respectively (Figure 4). The strain containing PsodA+hag+tufA had the highest level of pullulanase mRNA, which was 53.5 and 37 times higher than that in the strain containing PamyE at 24 and 48 h, respectively. However, the expressed enzyme activity (285 U/ml) was lower than that in the strain containing PsodA+fusA+amyE (336 U/ml). It suggested that the enzyme activity would be not only related to the strength of the promoter, but also related to other factors.
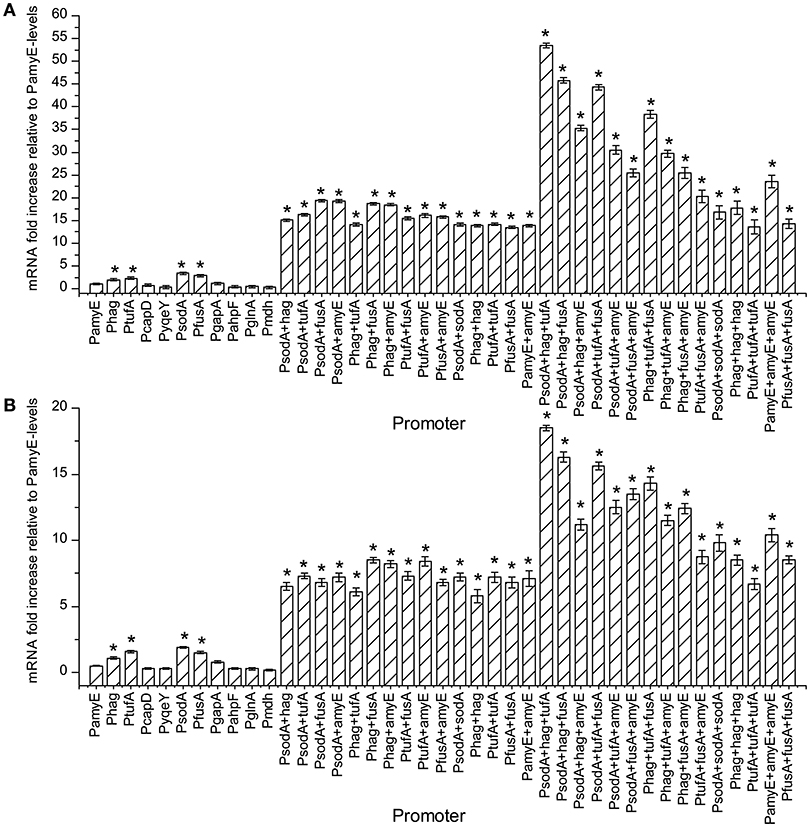
Figure 3. The effect of different promoters and their combinations on the amount of mRNA. Normalized gene expression (ΔΔCT) reported. The reference gene is the 16S ribosomal gene, PamyE is control, graphed relative to zero. Panel (A) is the sample cultured for 24 h. Panel (B) is the sample cultured for 48 h. All tests repeated three time. “*”means the amount of mRNA increased significantly (P < 0.05) compared to control promoter PamyE at 24 or 48 h, respectively.
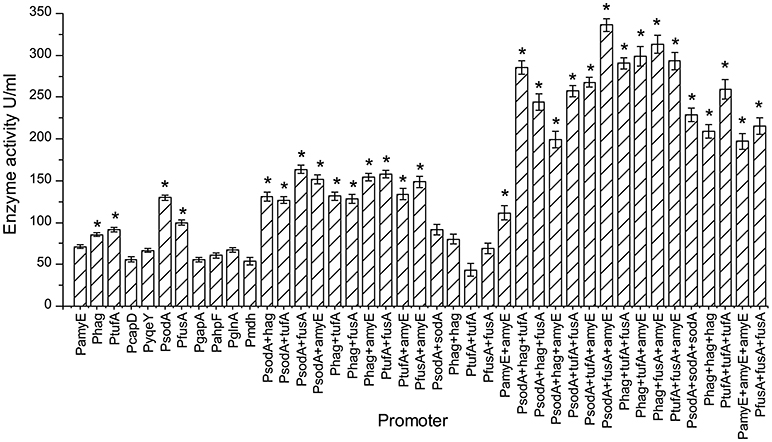
Figure 4. The pullulanase activity of all engineering strains. The effect of different promoters and their combinations on the activity of pullulanase. “*”means the activity of pullulanase increased significantly (P < 0.05) compared to control promoter PamyE.
The Effect of Different Promoters and Their Combinations on the Activity of Pullulanase
The expression of pullulanase from a single, dual, or triple promoter is shown in Figure 4. Recombinant strains were selected and cultured for 48 h in CSA medium at 37°C for 48 h. The pullulanase activity of strain containing triple promoter was higher than that in the strain containing single or dual promoter. Among the single promoter isolates, the enzyme activity obtained from the strain with PsodA was the highest (129.8 U/mL), which was 1.82 times higher than that of control strain with PamyE (71.1 U/ml). And the enzyme activity levels in the strains containing PsodA+fusA, PtufA+fusA, Phag+amyE, or PsodA+amyE were more than 150 U/mL, which was more than 2 times higher than that of the control strain with PamyE. Among the triple-promoter strains, the pullulanase activity in the strain containing PsodA+fusA+amyE was 336 U/mL, which is 4.72 times higher than that of the strain with PamyE (Figure 4).The results indicated that pullulanase activity was significantly improved by the multiple promoter combinations.
In addition, the pullulanase proteins expressed in the strain containing PsodA+fusA+amyE and the blank strain was confirmed by SDS-PAGE electrophoresis. It suggested that the pullulanase expressed successfully and a band was clearly shown in the engineered strains at 24 and 48 h, whereas the blank strain did not appeared the target band (Figure 5A). However, it is noteworthy that the pullulanase protein yield of the strain with PsodA+fusA+amyE reached a maximum, 14.3 g/L at 24 h, whereas its enzyme activity and specific enzyme activity (145.7 U/mL and 10.2 U/mg) was lower than those at 36 h (298.7 U/mL and 66.2 U/mg) and 48 h (336.4 U/mL and 76.5 U/mg), respectively (Figure 5B). This may be that a portion of pullulanase was misfolded or not modified at 24 h, and then the misfolded enzyme was likely degraded and was modified, thus, the specific enzyme activity was increased at 36 and 48 h.
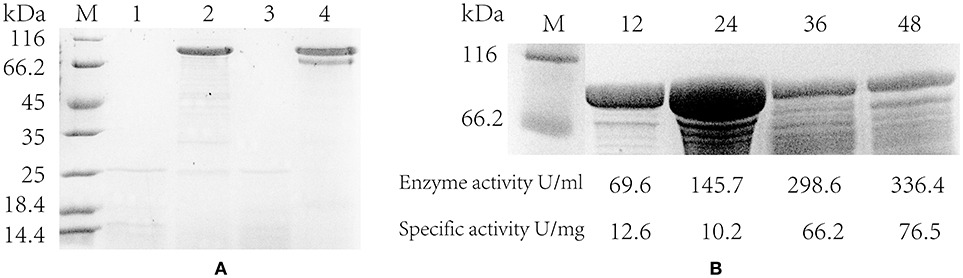
Figure 5. The extracellular protein and specific activity of engineered strain. (A) Detecting the expression of pullulanase in flasks by SDS-PAGE. lane 1 and 3 are the blank control (host strain without pulA gene); lane 2 and 4 are the engineering strain with PamyE; lane 1–2 are the extracellular protein in the supernatant at 24 h; lane 3–4 are the extracellular protein at 48 h. (B) The SDS-PAGE of engineering strains with PsodA+fusA+amyE in flasks, lane 1 is protein marker, lane 2–5 are the extracellular protein in the supernatant at 12, 24, 36, and 48 h. Below the figure is the data of enzyme activity and specific activity.
The Pullulanase Yield From Engineered Strains in Flasks and 50-L Fermenters
The activity of pullulanase in the strain with PsodA+fusA+amyE over 48 h in shake flasks was increased from 71 to 336.4 U/mL following optimization of the growth conditions (Figure 6A). The strains with PamyE, PsodA+fusA, and PsodA+fusA+amyE were cultured in 250 mL flasks containing 50 mL of medium. The results of the enzyme activity assay indicated that the enzyme was produced starting at ~12, and reached a maximum at 40–44. The strain with PsodA+fusA+amyE was subsequently cultured in a 50-L fermenter at either 37 or 33°C, and the pH was maintained at either 5.8 or 6.5. At pH 5.8, enzyme activity was higher at 33°C (1,555 U/mL) than at 37°C (1,005 U/mL). In addition, enzyme activity was higher at pH 5.8 (1,555 U/mL) than at pH 6.5 (1,122 U/mL) at 33°C (Figure 6B).
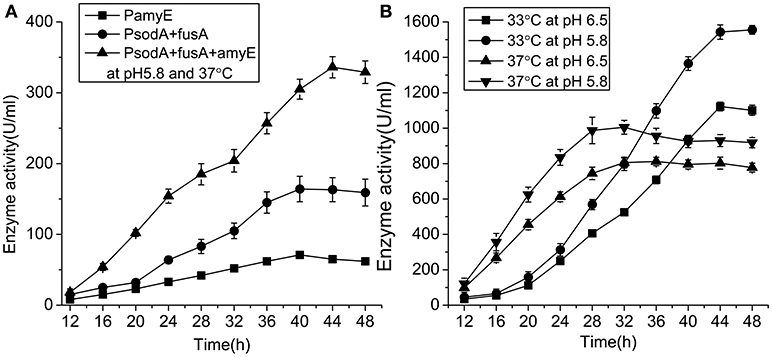
Figure 6. The enzyme activity of engineered strains. (A) The enzyme activity of engineered strains with different promoter in 250 ml flasks at pH 5.8 and 37°C. (B) The enzyme activity of engineered strain with the promoter PsodA+fusA+amyE in 50-Liter fermenter under different conditions.
Discussion
We analyzed three transcriptome data sets from B. subtilis to select the genes with the highest expression levels. The promoters of the top 10 genes were determined through predictive bioinformatic analyses and were used to express pullulanase in B. subtilis. Four promoters (PsodA, Phag, PtufA, and PfusA) were stronger than PamyE. Transcriptome mining, with the goal of engineering promoter-induced modifications to increase gene expression, has been previously reported. For example, Liu et al. (2017) selected a promoter, PBL9, from 3,595 genes and 1,249 operons in a B. licheniformis transcriptome, that was 23% stronger than P43. Liao et al. (2015) screened the candidate promoter Pr2 (the promoter of sigW) from 288 genes with higher expression levels (RPKM values) than the control gene P43, and observed the strongest β-galactosidase activity in post-log phase. Park et al. (2007) identified a cadmium-inducible promoter via transcriptome analysis of Hansenula polymorpha SEO1 that had broad specificity for heavy metals and was also responsive to arsenic and mercury. This study of pullulanase expression in B. subtilis reinforces the idea that selecting promoters from transcriptome data is a good approach for identifying strong promoters and can be used to optimize the expression of industrially important microbial products, saving time, reducing costs, and improving safety. This study is also the first to select promoters based on B. subtilis transcriptome data, showing that it is a viable option to modify expression of pullulanase.
The yield of pullulanase from a single promoter was unsatisfactory because of insufficient strength. Some researchers proved that optimizing the −35 and −10 regions of promoter could enhance promoter strength. Jiao et al. (2017) constructed a super-strong promoter, Pg3, by −35 and −10 regions mutations, which was 1.63 times higher than that before mutation in B. subtilis. In addition, Feng et al. (2017) generated P43 promoter variants, which was 1.77 times higher than P43 promoter. Research has also shown that artificial dual-promoters are typically stronger than single promoters. For example, the dual promoter PgsiB-PhpaII was shown to be stronger than PhpaII, PyxiE, P43, PgsiB, Pluxs, or PaprE alone (Guan C. et al., 2016). In addition, the strength of the dual promoters PhpaII-PamyR and PhpaII-Pblma was 11- to 12-fold higher than the single promoter PhpaII in B. subtilis (Kang et al., 2010). Sinah et al. (2012) also constructed a set of two promoters for high protein expression in both E. coli and S. cerevisiae. Therefore, we combined strong single promoters to generate artificial multiple-promoter systems to increase the yield of the target protein. Pullulanase mRNA transcript levels and enzyme activity were significantly increased with the number of promoters (Figure 5).
PsodA+fusA+amyE was a semi-constitutive promoter constructed from PsodA and PfusA, which are constitutive promoters, and PamyE, which is a starch- and maltose-inducible promoter. Therefore, this triple promoter system could be induced in CSA medium and did not require an inducer. Constitutive promoters are advantageous in large-scale industrial production because they do not require an inducer. This simplifies the composition of the medium and the fermentation conditions, thus reducing production costs. Although the activity of pullulanase heterologously expressed under various conditions was as high as 580 U/mL (Nie et al., 2013) and 2523.5 U/mL (Zou et al., 2014) in E. coli, it was comparatively low in B. subtilis, at 5.7 U/mL (Chen et al., 2001), 2.82 U/mL (Wang et al., 2014), and 24.5 U/mL (Song et al., 2016). In this study, the yield of pullulanase was as high as 1,555 U/mL, which is the highest yield reported to date. In addition, we recently improved pullulanase activity to 2,180 U/mL by optimizing the medium composition and controlling the fermentation conditions.
Interestingly, the strain containing PsodA+hag+tufA had the highest mRNA expression, but not the highest enzyme activity (Figures 3, 5). In fact, the enzyme activity in the strain containing PsodA+hag+tufA was only 84.8% of that in the strain containing PdosA+fusA+amyE (Figure 3). This suggests that post-transcriptional modifications may modulate enzyme levels or activity. Alternatively, the overexpressed mRNA might not be used as a template for translation due to limited amounts of tRNA or ribosomes (Yuan and Wong, 1995; Wu et al., 1998). Further, the concentration of pullulanase protein was 14.3 g/L at 24 h, while its specific activity was only 10.2 U/mg. This indicates that some of the pullulanase might be misfolded and have no enzymatic activity (Li et al., 2004; Yan and Wu, 2017). In the future studies, we would like to determine the structure of pullulanase by NMR or X-ray crystallography to confirm protein misfolding and explore how changes in the culture conditions or chaperones can be employed to improve the folding rate.
In this study, four strong promoters from B. subtilis were identified by analyzing transcriptome data in GenBank, and these promoters were used to express pullulanase in B. subtilis BS001. Both gene expression and protein production increased significantly with increasing tandem combinations of promoters. The enzyme activity of the strain with the triple promoter complex PsodA+fusA+amyE reached 336.4 U/mL in a shake flask and 1,555 U/mL in a 50-L fermenter, which was 4.73 times higher than that of the strain with PamyE. The strain with PsodA+hag+tufA showed the highest mRNA levels, which were 53.5 and 37 times higher than that of the strain with PamyE at 24 and 48 h, respectively. Taken together, these results demonstrate that bioinformatic analysis in combination with genetic recombination technology can be used to develop microbial bioproduct advancements that can quickly and safely benefit industrial production in a cost-effective way. We will furtherly upgrade the expression strength by promoter mutation and optimize fermentation conditions in order to furtherly enhance the pullulanase production.
Availability of Data and Materials
The datasets used and/or analyzed during the current study are available from the corresponding authors on reasonable request.
Author Contributions
FM designed and performed the experiments, analyzed the data, and wrote the manuscript. XZ and TN constructed plasmids and transformed into host strain B.subtilis B001. FL and XB conceived the project, designed the experiments. YL and FT wrote a part of the Discussion section and helped with language editing. ZL designed the research content and analyzed the data. All authors read and approved the final manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (No. 31571887& 31771948).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mr. Y. Wu for measuring the activity of the enzyme and Mr. Y. Wang for the control of the fermentation parameters at 50-liter fermenter. We also thank all of our colleagues for their support and suggestions. We would like to thank Editage (https://www.editage.com/) for English language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02635/full#supplementary-material
Abbreviations
P, promoter; PamyE, promoter amyE; RPKM, Reads Per Kilo-bases per Million-reads; STAB-SD, the Shine-Dalgarno mRNA stabilizing sequence.
References
Abdel-Fattah, W. R., Chen, Y., Eldakak, A., and Hulett, F. M. (2005). Bacillus subtilis phosphorylated PhoP: direct activation of the E sigma(A)- and repression of the E sigma(E)-responsive phoB-PS+V promoters during pho response. J. Bacteriol. 187, 5166–5178. doi: 10.1128/Jb.187.15.5166-5178.2005
Biedendieck, R., Gamer, M., Jaensch, L., Meyer, S., Rohde, M., Deckwer, W. D., et al. (2007). A sucrose-inducible promoter system for the intra- and extracellular protein production in Bacillus megaterium. J. Biotechnol. 132, 426–430. doi: 10.1016/j.jbiotec.2007.07.494
Blazeck, J., Garg, R., Reed, B., and Alper, H. S. (2012). Controlling promoter strength and regulation in Saccharomyces cerevisiae using synthetic hybrid promoters. Biotechnol. Bioeng. 109, 2884–2895. doi: 10.1002/bit.24552
Chen, A., Sun, Y., Zhang, W., Peng, F., Zhan, C., Liu, M., et al. (2016). Downsizing a pullulanase to a small molecule with improved soluble expression and secretion efficiency in Escherichia coli. Microb. Cell Fact. 15:9. doi: 10.1186/s12934-015-0403-5
Chen, J. T., Chen, M. C., Chen, L. L., and Chu, W. S. (2001). Structure and expression of an amylopullulanase gene from Bacillus stearothermophilus TS-23. Biotechnol. Appl. Biochem. 33, 189–199. doi: 10.1042/Ba20010003
Duan, X., Chen, J., and Wu, J. (2013). Improving the thermostability and catalytic efficiency of Bacillus deramificans pullulanase by site-directed mutagenesis. Appl. Environ. Microbiol. 79, 4072–4077. doi: 10.1128/AEM.00457-13
Feng, Y., Liu, S., Jiao, Y., Gao, H., Wang, M., Du, G., et al. (2017). Enhanced extracellular production of L-asparaginase from Bacillus subtilis 168 by B. subtilis WB600 through a combined strategy. Appl. Microbiol. Biotechnol. 101, 1509–1520. doi: 10.1007/s00253-016-7816-x
Forrest, D., James, K., Yuzenkova, Y., and Zenkin, N. (2017). Single-peptide DNA-dependent RNA polymerase homologous to multi-subunit RNA polymerase. Nat. Commun. 8:15774. doi: 10.1038/ncomms15774.
Geissendörfer, M., and Hillen, W. (1990). Regulated expression of heterologous genes in Bacillus-subtilis using the Tn10 encoded tet regulatory elements. Appl. Microbiol. Biotechnol. 33, 657–663.
Geng, L., Duan, X., Liang, C., Shu, C., Song, F., and Zhang, J. (2014). Mining tissue-specific contigs from peanut (Arachis hypogaea L.) for promoter cloning by deep transcriptome sequencing. Plant Cell Physiol. 55, 1793–1801. doi: 10.1093/pcp/pcu111
Gryczan, T. J., Contente, S., and Dubnau, D. (1978). Characterization of Staphylococcus aureus plasmids introduced by transformation into Bacillus subtilis. J. Bacteriol. 134, 318–329.
Guan, C., Cui, W., Cheng, J., Liu, R., Liu, Z., Zhou, L., et al. (2016). Construction of a highly active secretory expression system via an engineered dual promoter and a highly efficient signal peptide in Bacillus subtilis. N. Biotechnol. 33, 372–379. doi: 10.1016/j.nbt.2016.01.005
Guan, C., Cui, W., Cheng, J., Zhou, L., Liu, Z., and Zhou, Z. (2016). Development of an efficient autoinducible expression system by promoter engineering in Bacillus subtilis. Microbial. Cell Fact. 15:66. doi: 10.1186/s12934-016-0464-0
Jiao, S., Li, X., Yu, H., Yang, H., Li, X., and Shen, Z. (2017). In situ enhancement of surfactin biosynthesis in Bacillus subtilis using novel artificial inducible promoters. Biotechnol. Bioeng. 114, 832–842. doi: 10.1002/bit.26197
Kahar, U. M., Ng, C. L., Chan, K. G., and Goh, K. M. (2016). Characterization of a type I pullulanase from Anoxybacillus sp. SK3-4 reveals an unusual substrate hydrolysis. Appl. Microbiol. Biotechnol. 100, 6291–6307. doi: 10.1007/s00253-016-7451-6
Kang, H. K., Jang, J. H., Shim, J. H., Park, J. T., Kim, Y. W., and Park, K. H. (2010). Efficient constitutive expression of thermostable 4-alpha-glucanotransferase in Bacillus subtilis using dual promoters. World J. Microbiol. Biotechnol. 26, 1915–1918. doi: 10.1007/s11274-010-0351-5
Kim, J. H., Hwang, B. Y., Roh, J., Lee, J. K., Kim, K., Wong, S. L., et al. (2008). Camparison of P-aprE, P-amyE, and P-P43 promoter strength for beta-galactosidase and staphylokinase expression in Bacillus subtilis. Biotechnol. Bioprocess Eng. 13, 313–318. doi: 10.1007/s12257-007-0102-0
Kong, H. G., Choi, K. H., Heo, K. R., Lee, K. Y., Lee, H. J., Moon, B. J., et al. (2009). Generation of a constitutive green fluorescent protein expression construct to mark biocontrol bacteria using p43 promoter from Bacillus subtilis. Plant Pathol. J. 25, 136–141. doi: 10.5423/Ppj.2009.25.2.136
Kunamneni, A., and Singh, S. (2006). Improved high thermal stability of pullulanase from a newly isolated thermophilic Bacillus sp AN-7. Enzyme Microbial. Technol. 39, 1399–1404. doi: 10.1016/j.enzmictec.2006.03.023
Li, W., Zhou, X., and Lu, P. (2004). Bottlenecks in the expression and secretion of heterologous proteins in Bacillus subtilis. Res. Microbiol. 155, 605–610. doi: 10.1016/j.resmic.2004.05.002
Li, X., Fu, J., Wang, Y., Ma, F., and Li, D. (2017). Preparation of low digestible and viscoelastic tigernut (Cyperus esculentus) starch by Bacillus acidopullulyticus pullulanase. Int. J. Biol. Macromol. 102, 651–657. doi: 10.1016/j.ijbiomac.2017.04.068.
Liao, Y., Huang, L., Wang, B., Zhou, F., and Pan, L. (2015). The global transcriptional landscape of Bacillus amyloliquefaciens XH7 and high-throughput screening of strong promoters based on RNA-seq data. Gene 571, 252–262. doi: 10.1016/j.gene.2015.06.066
Liu, X., Yang, H., Zheng, J., Ye, Y., and Pan, L. (2017). Identification of strong promoters based on the transcriptome of Bacillus licheniformis. Biotechnol. Lett. 39, 873–881. doi: 10.1007/s10529-017-2304-7
Makarewicz, I., Dubrac, S., Msadek, T., and Borriss, R. (2006). Dual role of the PhoP similar to P response regulator: Bacillus amyloliquefaciens FZB45 phytase gene transcription is directed by positive and negative interactions with the phyC promoter. J. Bacteriol. 188, 6953–6965. doi: 10.1128/Jb.00681-06
Malviya, S. N., Malakar, R., and Tiwari, A. (2010). Pullulanase: a potential enzyme for industrial application. Int. J. Biomed. Res. 1, 10–20. doi: 10.7439/ijbr.v1i2.53
McCleary, W. R. (2009). Application of promoter swapping techniques to control expression of chromosomal genes. Appl. Microbiol. Biotechnol. 84, 641–648. doi: 10.1007/s00253-009-2137-y
Ming, Y. M., Wei, Z. W., Lin, C. Y., and Sheng, G. Y. (2010). Development of a Bacillus subtilis expression system using the improved Pglv promoter. Microb. Cell Fact. 9:55. doi: 10.1186/1475-2859-9-55
Mojardín, L., and Salas, M. (2016). Global Transcriptional Analysis of virus-host interactions between Phage varphi29 and Bacillus subtilis. J. Virol. 90, 9293–9304. doi: 10.1128/JVI.01245-16
Nagler, K., Krawczyk, A. O., De Jong, A., Madela, K., Hoffmann, T., Laue, M., et al. (2016). Identification of differentially expressed genes during Bacillus subtilis spore outgrowth in high-salinity environments using RNA sequencing. Front. Microbiol. 7:1564. doi: 10.3389/fmicb.2016.01564
Nie, Y., Yan, W., Xu, Y., Chen, W. B., Mu, X. Q., Wang, X., et al. (2013). High-level expression of Bacillus naganoensis pullulanase from recombinant Escherichia coli with auto-induction: effect of lac operator. PLoS ONE 8:e78416. doi: 10.1371/journal.pone.0078416
Nisha, M., and Satyanarayana, T. (2016). Characteristics, protein engineering and applications of microbial thermostable pullulanases and pullulan hydrolases. Appl. Microbiol. Biotechnol. 100, 5661–5679. doi: 10.1007/s00253-016-7572-y
Park, H., Bideshi, D. K., Johnson, J. J., and Federici, B. A. (1999). Differential enhancement of Cry2A versus Cry11A yields in Bacillus thuringiensis by use of the cry3A STAB mRNA sequence. Fems Microbiol. Lett. 181, 319–327. doi: 10.1016/S0378-1097(99)00555-8
Park, J. N., Sohn, M. J., Oh, D. B., Kwon, O., Rhee, S. K., Hur, C. G., et al. (2007). Identification of the cadmium-inducible Hansenula polymorpha SEO1 gene promoter by transcriptome analysis and its application to whole-cell heavy-metal detection systems. Appl. Environ. Microbiol. 73, 5990–6000. doi: 10.1128/AEM.00863-07
Phan, T. T., and Schumann, W. (2007). Development of a glycine-inducible expression system for Bacillus subtilis. J. Biotechnol. 128, 486–499. doi: 10.1016/j.jbiotec.2006.12.007
Ram, K. A., and Venkatasubramanian, K. (1982). Enhancement of starch conversion efficiency with free and immobilized pullulanase and alpha-1,4-glucosidase. Biotechnol. Bioeng. 24, 355–369. doi: 10.1002/bit.260240209
Reddy, C. K., Pramila, S., and Haripriya, S. (2015). Pasting, textural and thermal properties of resistant starch prepared from potato (Solanum tuberosum) starch using pullulanase enzyme. J. Food Sci. Technol. 52, 1594–1601. doi: 10.1007/s13197-013-1151-3
Sambrook, J., and Russell, D. W. (2006). The Condensed Protocols from Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Shikaishi, T., Fujimoto, D., and Sakano, Y. (2014). Synthesis of branched α-cyclodextrin carrying a side chain longer than maltose using Bacillus acidopullulyticus pullulanase. Agri. Biol. Chem. 53, 3093–3095. doi: 10.1080/00021369.1989.10869780
Sinah, N., Williams, C. A., Piper, R. C., and Shields, S. B. (2012). A set of dual promoter vectors for high throughput cloning, screening, and protein expression in eukaryotic and prokaryotic systems from a single plasmid. BMC Biotechnol. 12:54. doi: 10.1186/1472-6750-12-54
Song, W., Nie, Y., Mu, X. Q., and Xu, Y. (2016). Enhancement of extracellular expression of Bacillus naganoensis pullulanase from recombinant Bacillus subtilis: effects of promoter and host. Protein Expr. Purif. 124, 23–31. doi: 10.1016/j.pep.2016.04.008.
Wang, P. Z., and Doi, R. H. (1984). Overlapping promoters transcribed by bacillus subtilis sigma 55 and sigma 37 RNA polymerase holoenzymes during growth and stationary phases. J. Biol. Chem. 259, 8619–8625.
Wang, Y., Liu, Y., Wang, Z., and Lu, F. (2014). Influence of promoter and signal peptide on the expression of pullulanase in Bacillus subtilis. Biotechnol. Lett. 36, 1783–1789. doi: 10.1007/s10529-014-1538-x
Wu, H., Yu, X., Chen, L., and Wu, G. (2014). Cloning, overexpression and characterization of a thermostable pullulanase from Thermus thermophilus HB27. Protein Expr. Purif. 95, 22–27. doi: 10.1016/j.pep.2013.11.010
Wu, S. C., Ye, R., Wu, X. C., Ng, S. C., and Wong, S. L. (1998). Enhanced secretory production of a single-chain antibody fragment from Bacillus subtilis by coproduction of molecular chaperones. J. Bacteriol. 180, 2830–2835.
Wu, X. C., Lee, W., Tran, L., and Wong, S. L. (1991). Engineering a Bacillus subtilis expression-secretion system with a strain deficient in 6 extracellular proteases. J. Bacteriol. 173, 4952–4958.
Xu, B., Yang, Y. J., and Huang, Z. X. (2006). Cloning and overexpression of gene encoding the pullulanase from Bacillus naganoensis in Pichia pastoris. J. Microbiol. Biotechnol. 16, 1185–1191.
Yamamoto, H., Murata, M., and Sekiguchi, J. (2000). The CitST two-component system regulates the expression of the Mg-citrate transporter in Bacillus subtilis. Mol. Microbiol. 37, 898–912. doi: 10.1046/j.1365-2958.2000.02055.x
Yan, S., and Wu, G. (2017). Bottleneck in secretion of alpha-amylase in Bacillus subtilis. Microb. Cell Fact. 16:124. doi: 10.1186/s12934-017-0738-1
Yang, M., Zhang, W., Ji, S., Cao, P., Chen, Y., and Zhao, X. (2013). Generation of an artificial double promoter for protein expression in Bacillus subtilis through a promoter trap system. PLoS ONE 8:e56321. doi: 10.1371/journal.pone.0056321
Yuan, G., and Wong, S. L. (1995). Isolation and characterization of Bacillus subtilis groE regulatory mutants: evidence for orf39 in the dnaK operon as a repressor gene in regulating the expression of both groE and dnaK. J. Bacteriol. 177, 6462–6468.
Yue, J., Fu, G., Zhang, D. W., and Wen, J. P. (2017). A new maltose-inducible high-performance heterologous expression system in Bacillus subtilis. Biotechnol. Lett. 39, 1237–1244. doi: 10.1007/s10529-017-2357-7
Zhang, K., Su, L., Duan, X., Liu, L., and Wu, J. (2017). High-level extracellular protein production in Bacillus subtilis using an optimized dual-promoter expression system. Microb. Cell Fact. 16:32. doi: 10.1186/s12934-017-0649-1
Zou, C., Duan, X., and Wu, J. (2014). Enhanced extracellular production of recombinant Bacillus deramificans pullulanase in Escherichia coli through induction mode optimization and a glycine feeding strategy. Bioresour. Technol. 172, 174–179. doi: 10.1016/j.biortech.2014.09.035
Keywords: Bacillus, transcriptome, pullulanase, multi-promoter, fermentation
Citation: Meng F, Zhu X, Nie T, Lu F, Bie X, Lu Y, Trouth F and Lu Z (2018) Enhanced Expression of Pullulanase in Bacillus subtilis by New Strong Promoters Mined From Transcriptome Data, Both Alone and in Combination. Front. Microbiol. 9:2635. doi: 10.3389/fmicb.2018.02635
Received: 09 August 2018; Accepted: 16 October 2018;
Published: 02 November 2018.
Edited by:
C. Perry Chou, University of Waterloo, CanadaCopyright © 2018 Meng, Zhu, Nie, Lu, Bie, Lu, Trouth and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaoxin Lu, Zm1iQG5qYXUuZWR1LmNu
 Fanqiang Meng1
Fanqiang Meng1 Xiaomei Bie
Xiaomei Bie Zhaoxin Lu
Zhaoxin Lu