- 1Pathogen Biology Laboratory, Department of Biotechnology and Bioinformatics, University of Hyderabad, Hyderabad, India
- 2Department of Microbiology, Dr. D. Y. Patil Medical College, Hospital and Research Centre (Dr. D. Y. Patil Vidyapeeth), Pune, India
- 3International Centre for Diarrhoeal Disease Research, Bangladesh (icddr,b), Dhaka, Bangladesh
- 4Robert Koch Institute, Berlin, Germany
Colibactin, a genotoxin, encoded by the pks pathogenicity island of Escherichia coli belonging to the B2 phylogroup has been reported as a determinant of bacterial pathogenicity. The present study was carried out to detect the pks pathogenicity island in extraintestinal pathogenic E. coli (ExPEC) isolated from a tertiary hospital in Pune, India. Of 462 isolates analyzed, the pks genomic island was detected in 35 (7.6%) isolates, which predominantly belonged to pathogenic phylogroup B2 (97%), and harbored virulence genes such as fimH, sfaD/E, and usp. Biofilm formation assay revealed 21 of the 35 pks-carrying isolates to be strong (SBF > 1.0), 10 isolates to be moderate (SBF = 0.5–1.0), and 4 as weak (SBF < 0.5) biofilm formers. All of the pks-carrying isolates proved resistant against bactericidal activity of human serum. Assays carried out to detect antimicrobial susceptibility revealed 11% of these isolates to be multidrug resistant, 37% producing ESBL and 25% were positive for blaCTX-M-15. The observed prevalence of multidrug resistance and colibactin producing characteristics among pathogenic E. coli belonging to phylogenetic group B2 advocate urgent need for broader surveillance in order to understand and prevent transmission of these ExPEC in community and hospital settings.
Introduction
Extraintestinal pathogenic Escherichia coli (ExPEC), apart from being a seasoned nosocomial pathogen is also an important cause of community-acquired (Johnson and Russo, 2002) and other infections such as urinary tract infection, sepsis, neonatal meningitis, and colibacillosis in humans and animals (Kaper et al., 2004; Smith et al., 2007). Genome analysis revealed that pathogenic E. coli have genome sizes in excess of about 1.0 Mb than the commensal strains mainly due to the presence of genes encoding multiple virulence factors, such as adhesins, toxins, invasins and siderophores that are absent or unlikely to be present in commensal strains (Croxen and Finlay, 2010). The virulence factors are mostly associated with phages and pathogenicity islands and undergo horizontal gene transfer which disseminates traits, thereby offering fitness advantages to recipient organisms (Ahmed et al., 2008).
Secreted toxins are the virulence factors that play an important role in long term colonization and pathogenesis of ExPEC. Some of these known secreted toxins comprise of important genotoxins, such as cytolethal distending toxins (CDT’s), cycle inhibiting factors (cif’s) and cytotoxic necrotizing factors (CNF’s) which can directly regulate the cell cycle of the host (Nougayrède et al., 2005). Colibactin is another important genotoxin produced by a 54-kb pathogenicity island known as pks island harbored by the members of Enterobacteriaceae (Nougayrède et al., 2006). This genomic island consists of a clbA-S gene cluster that encodes non-ribosomal peptides and polyketide synthases along with accessory and tailoring enzymes (Nougayrède et al., 2006). Colibactin acts as a cyclomodulin and blocks the eukaryotic cell cycle causing progressive enlargement of the nucleus as well as the cell body eventually leading to cell death (Cuevas-Ramos et al., 2010). The cytopathic effect of these genotoxins is mediated by live bacteria and requires a direct contact with the host cell (Nougayrède et al., 2006).
The pks island was first identified in sequenced genomes of ExPEC prototype strains and was detected predominantly in strains of phylogenetic group B2 (Johnson et al., 2008). The island was shown to display several signatures reminiscent of its horizontal acquisition (Putze et al., 2009; Messerer et al., 2017). The origin and prevalence of the colibactin island among enteric pathogens is largely unexplored. However, it was also found to be present in other members of Enterobacteriaceae such as Citrobacter koseri, Klebsiella pneumoniae, and Enterobacter aerogenes (Putze et al., 2009). Epidemiological studies demonstrated the prevalence of pks in ExPEC and their association with severe infections in different host populations of varied geographical locations (Johnson et al., 2008; Buc et al., 2013; Shimpoh et al., 2017).
Epidemiological studies on ExPEC in India have so far been focused on understanding antimicrobial resistance, evolution and presence of pandemic clones (Avasthi et al., 2011; Jadhav et al., 2011; Hussain et al., 2012, 2014; Shaik et al., 2017). However, a better understanding of ExPEC associated virulence factors may help in the development of therapeutic interventions, such as diagnostics and/or vaccines against ExPEC infections, and this would also facilitate risk assessment of ExPEC strains.
While colibactin is regarded as a virulence factor in ExPEC, not much is known in the context of its molecular epidemiology entailing Indian clinical isolates. Therefore, the present study was performed in order to investigate the prevalence and carriage of pks island, and to decipher the genotypic and functional characteristics of pks harboring clinical ExPEC isolates from India.
Materials and Methods
Bacterial Isolates
A total of 462 isolates of ExPEC were harnessed for this study. These isolates were originally collected by SJ and her colleagues from Dr D. Y. Patil University Hospital, Pune, India as a part of their routine diagnostic screening during the years 2009–2015 and were also described in our previous study (Ranjan et al., 2016). The bacterial collection of our study, essentially a sub-set of the collection studied by Ranjan et al. (2016), comprised 370 isolates cultured from urine, 63 from pus and 29 from other extra intestinal clinical samples. Standard microbiological laboratory methods were employed for the identification and preservation of these isolates (Jadhav et al., 2011). All isolates were collected, preserved and handled as per standard biosafety guidelines and according to the approvals of the Institutional Biosafety Committee (IBSC) of the University of Hyderabad (Ref. UH/IBSC/NA/12/7 dated 09/4/2012 and NA-N-32 dated 27/8/2015). The clinical information of the isolates is described in the Supplementary Table S1.
Detection of pks-Genomic Island and Phylogroup Determination
The clinical isolates were screened for the presence of pks island by PCR using primers for the four representative genes of the genomic island so as to generate two flanking (clbB and clbQ) and two internal (clbA and clbN) amplicons in order to document presence of a complete island (Johnson et al., 2008). Heat killed bacterial lysates were used as DNA templates for PCR amplification, as described previously (Ranjan et al., 2017). PCR amplifications were carried out in 30 cycles at specific reaction conditions as described earlier (Johnson et al., 2008; Ranjan et al., 2017). The pks-positive isolates were assigned to one of the eight phylogroups based on multiplex-PCR amplification of four genes (chuA, yjaA, arpA, and TspE4.C2) as described elsewhere (Clermont et al., 2013).
Antibiotic Susceptibility and Extended Spectrum-β-Lactamase Production
Antibiotic susceptibility analysis was performed, as previously described, by Kirby-Bauer disk diffusion method, on Mueller Hinton agar plates (Qumar et al., 2017). Antimicrobial disks (Himedia, India) specific for fosfomycin (200 μg), chloramphenicol (30 μg), co-trimoxazole (20 μg), tetracycline (30 μg), gentamicin (10 μg), nalidixic acid (30 μg), doxycycline (30 μg), ciprofloxacin (5 μg), and colistin (10 μg) were used to determine the antibiotic susceptibility profile of the isolates. ESBL production was determined using disk synergy between clavulanic acid and indicator cephalosporins, CTX (cefotaxime) and CAZ (ceftazidime). Both the assays were performed in accordance with Clinical Laboratory Standards Institute (CLSI) guidelines (CLSI, 2013). Isolates exhibiting resistance to three or more antimicrobials were designated as multidrug resistant (MDR).
Virulence and Antimicrobial Resistance Genotyping
PCR based screening of virulence genes encoding bacterial adhesins (fimH, sfaD/E, afa), toxins (usp, cvaC, sat), iron acquisition system (iucD) and protectants (ibeA) were performed using primers and reaction conditions as previously described (Jadhav et al., 2011; Ranjan et al., 2017). ESBL gene blaCTX-M-15, (Monstein et al., 2007), genes conferring resistance to tetracycline (tetA), sulfonamides (sul1) and aminoglycoside acetyl transferases (aac(6′)-Ib) were also screened for using primers and PCR conditions as described in previous studies (Jadhav et al., 2011; Ranjan et al., 2016, 2017). The isolates were also screened by PCR using generic primers for TEM and amplified products were sequenced to identify the variants of the gene.
Determination of Siderophore Production, Biofilm Formation and Serum Resistance Assay
All the pks-positive isolates were screened for siderophore production using Chrome Azurol S Blue agar plates. A single colony of the bacterial isolate(s) was streaked on these plates and incubated overnight at 37°C. Colonies showing characteristic orange halos were identified as positive for siderophore production (Schwyn and Neilands, 1987).
All the 35 pks-positive isolates were analyzed for their biofilm forming capabilities as described previously (Nandanwar et al., 2014). Briefly, OD at 600 nm was taken for overnight grown bacterial cultures and all the isolates were diluted to an OD of 0.05 in fresh M63 minimal medium. An aliquot of 200 μL of the diluted culture was pipetted into flat-bottom 96 well sterile microtiter plates in triplicates. The plates were covered by a breathable sealing after obtaining OD at 600 nm [OD600(0 h)]. The plates were incubated for 48 h at stationary condition at 28°C. Following this, OD was obtained at 600 nm [OD600(48 h)]. Media was aspirated and wells washed thrice with 300 μL of deionized water. After air drying, bacteria were fixed using 250 μL of 99% methanol for 15 min and stained using 0.1% crystal violet solution for 30 min. Following staining, wells were washed thrice with deionized water and air dried. To solubilize the stained bacteria, 300 μL of Ethanol: Acetone (80:20) solution was added and incubated for 30 min at 100 rpm. OD at 570 nm was read in microtiter plate reader and specific biofilm formation was obtained using the following formula: SBF (specific biofilm formation) = (AB-CW)/G where AB = OD at 570 nm of attached and stained bacteria, CW = OD of control at 570 nm and G = OD600(48 h) - OD600(0 h), representing bacterial growth. The experiment was repeated twice in technical triplicates.
Serum resistance was also determined for all the 35 pks-positive E. coli isolates in vitro using 50% human serum as described earlier (Hussain et al., 2014). Briefly, 5 μL of overnight culture was added to 495 μl of LB broth and incubated in a shaking incubator at 37°C for 1 h at 200 rpm. The bacterial cultures were pelleted and resuspended in 1mL of 1X PBS; 30 μL of this inoculum was added to 270 μL of 50% human serum in triplicates in a 96 well microtiter plate. In each case, an initial sample was collected and plated after dilution on LB agar plates for enumerating the colony forming units (CFU) at 0 h. The inoculated plate was incubated for 3 h at 37°C at 100 rpm. After 3 h, samples from each well were serially diluted and plated on LB agar plates for 3 h count. Isolates which had equal or higher CFU counts at 3 h compared to 0 h were considered resistant to human serum. Growth was obtained by subtracting CFU counts of 0 h from that of 3 h. The experiment was repeated two times in technical triplicate(s).
Statistical Analysis
All statistical calculations were performed using GraphPad Prism (version 5.01). Non-parametric Mann–Whitney U test was performed for serum resistance assay. p-values ≤ 0.05 were considered to be significant and were denoted in the graph.
Results
Screening for pks Island and Phylogenetic Grouping
A total of 462 E. coli isolates from our collection were screened for the presence of pks island, of which 35 were found to be positive for all the four targeted genes (clbA, clbB, clbN and clbQ) amplified from flanking and internal regions. Of these, 30 were originally cultured from urine, four from pus and one from blood. The prevalence of pks-positive isolates was 7.6% of the total E. coli collection studied. Using multiplex PCR, identification of E. coli phylogenetic groups was performed and out of the 35 isolates, 34 belonged to phylogroup B2 while one was assigned to phylogroup D (Table 1).
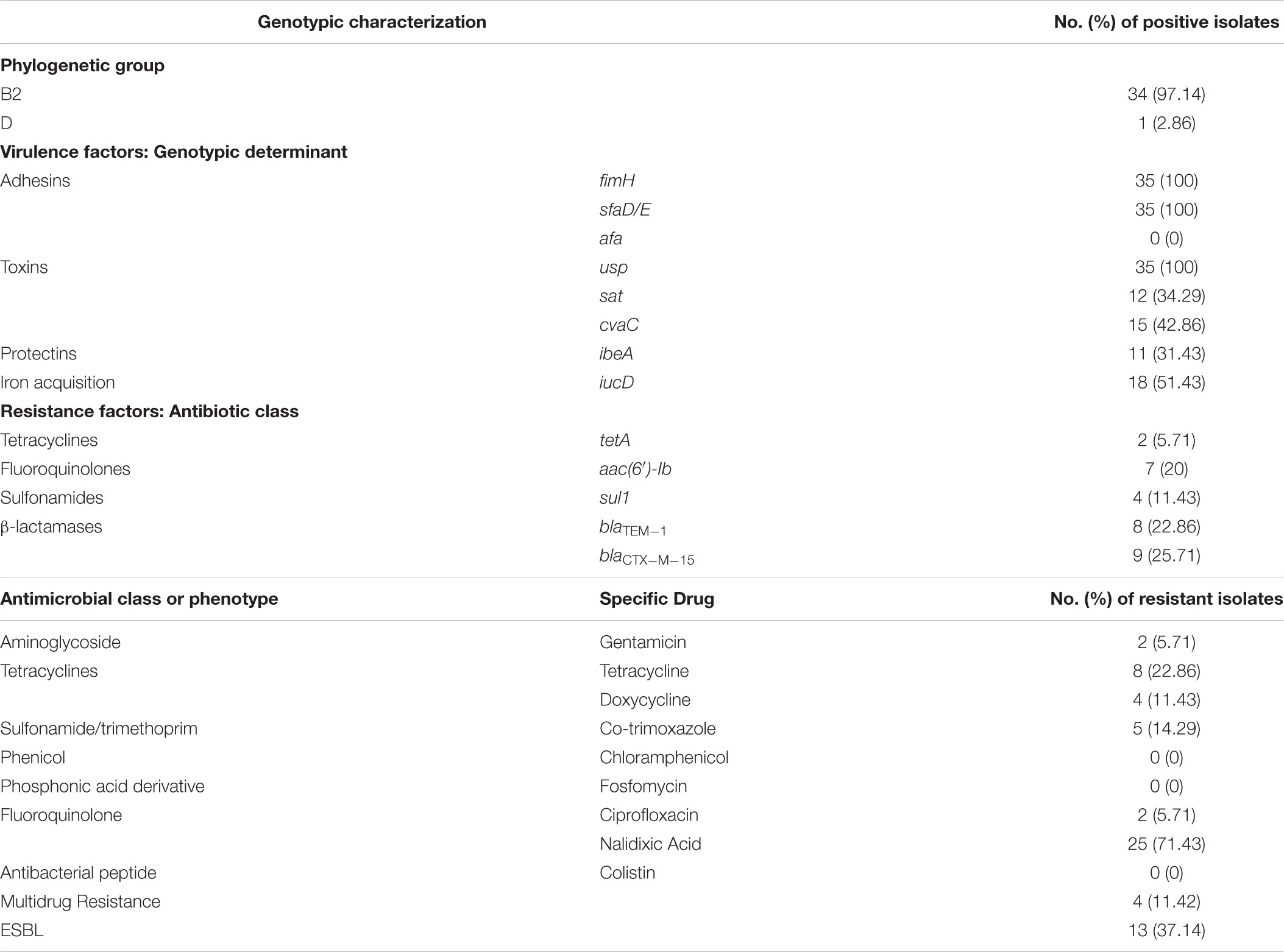
TABLE 1. Phylogroups, virulence and resistance genotypes, and antimicrobial resistance of pks-positive E. coli isolates.
Antimicrobial Susceptibility and ESBL Production
Antimicrobial susceptibility testing against nine different antimicrobial agents belonging to seven different antibiotic classes revealed that the isolates were only moderately resistant to antibiotics. Maximum resistance was observed against nalidixic acid (71.4%). The isolates were found to be less resistant to tetracycline (22.86%), co-trimoxazole (14.29%), doxycycline (11.43%), gentamicin (5.71%), and ciprofloxacin (5.71%) and were completely sensitive to fosfomycin, chloramphenicol, and colistin. Of the 35 E. coli isolates, 13 (37.14%) were found to be ESBL producers and 4 (11.42%) were multidrug resistant. The details of resistance profile(s) for each antibiotic tested are shown in Table 1.
Virulence and Resistance Genotyping
PCR based virulence and resistance genotyping of pks-positive isolates revealed their being relatively virulent as they possessed higher number of virulence genes compared to the resistance genes detected. Among bacterial adhesins tested, fimH and sfaD/E were 100% prevalent while afa was not detected in any of the isolates. Screening of toxin genes revealed presence of usp gene among all the isolates (100%), whereas cvaC and sat showed 42.86% and 34.29% prevalence, respectively. The genes iucD (iron acquisition system) and ibeA (protectant) were present in 51.43% and 31.43% of the isolates, respectively (Table 1).
Antimicrobial resistance genotyping revealed that the ESBL gene blaCTX-M-15 was present in 25.71% (n = 9) of the isolates and all these isolates were phenotypically observed to be ESBL producers by double disk synergy test. Further, 22.86% (n = 8) of the isolates were positive for TEM, and sequencing of PCR products followed by BLAST analysis identified all the amplicons to be entailing blaTEM-1. The gene aac(6′)-Ib, which is involved in aminoglycoside resistance, was present in 20% of the pks-positive isolates. Occurrence of genes that confer sulfonamide (sul1) and tetracycline (tetA) resistance was observed to be at 11.43% and 5.71% of isolates, respectively (Table 1). Overall, we observed low prevalence of resistance genes in concordance with the phenotypic antibiotic resistances detected by disk diffusion assays.
Virulence Associated Phenotypes
The isolates were analyzed for siderophore production, biofilm formation, and serum resistance which are essential ExPEC virulence properties. All pks-positive isolates formed orange halos on Chrome Azurol S plates, confirming the production of siderophores. Biofilm formation assay was performed twice in triplicates for all the pks positive isolates in order to determine their biofilm forming capabilities and was documented by specific biofilm formation (SBF) values. Isolates showing SBF values greater than 1 were designated as strong biofilm formers, 0.5–1.0 as moderate biofilm formers and those showing less than 0.5 were considered as weak biofilm formers (Figure 1). Majority (21/35) of the isolates tested were strong biofilm formers while 10/35 isolates moderately formed the biofilm and 4/35 isolates were weak biofilm formers. Bacterial resistance to the bactericidal activity of human serum was also assessed and the number of CFU were found to be significantly higher for all the pks positive isolates after 3 h of incubation in the human serum as compared to the negative control, indicative of the fact that all the isolates were resistant to human serum. Serum resistance was performed twice in technical triplicate(s) (Figure 2) and p-values were obtained using Mann–Whitney U test.
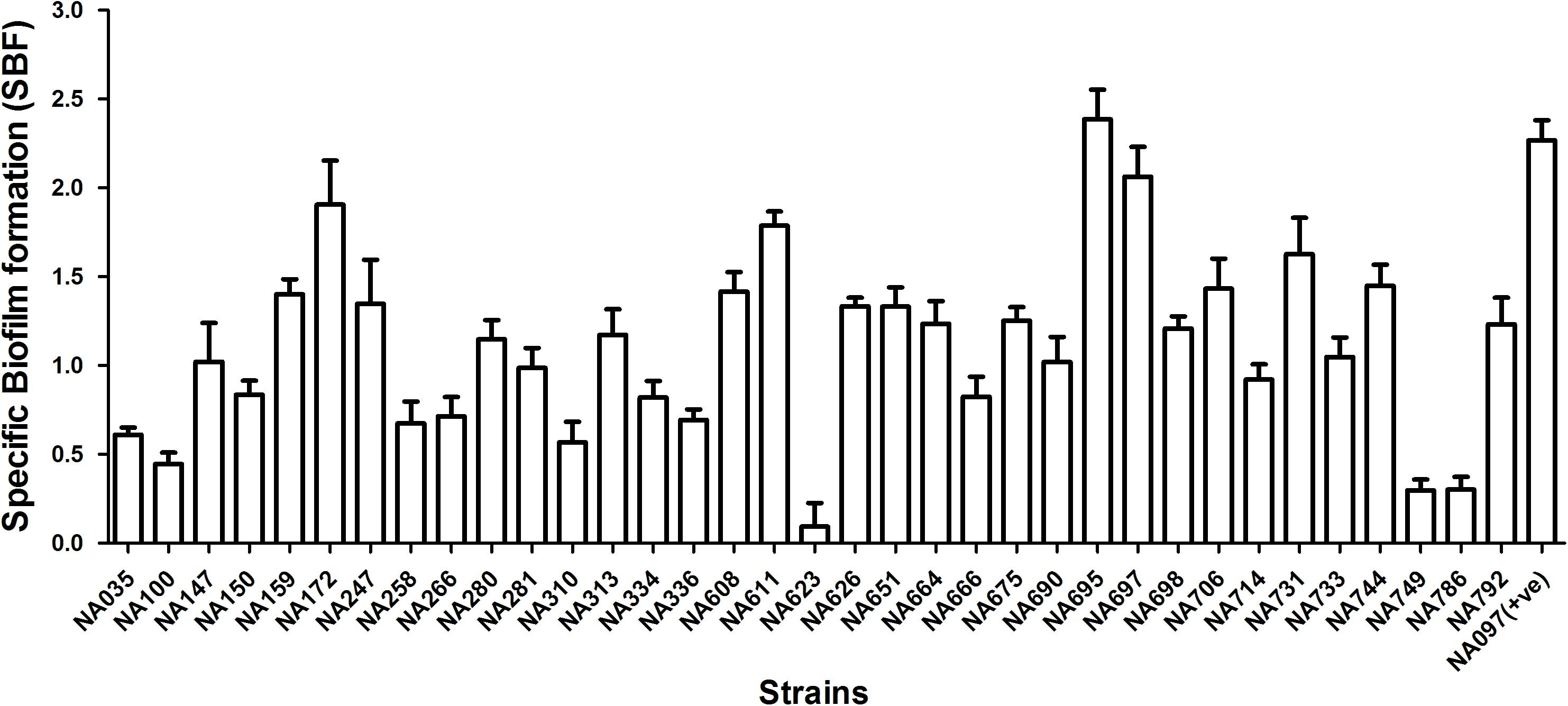
FIGURE 1. Biofilm formation in 35 pks positive E. coli isolates in M63 medium. Values are shown as mean of specific biofilm formation. Isolates demonstrating SBF values > 1.0 were considered as strong, 0.5–1.0 as moderate and <0.5 as weak biofilm formers. Majority of isolates (21/35) demonstrated strong biofilm formation; the remaining 10 and 4 isolates showed moderate and weak biofilm formation, respectively. NA097 was employed as a positive control in the biofilm formation assay.
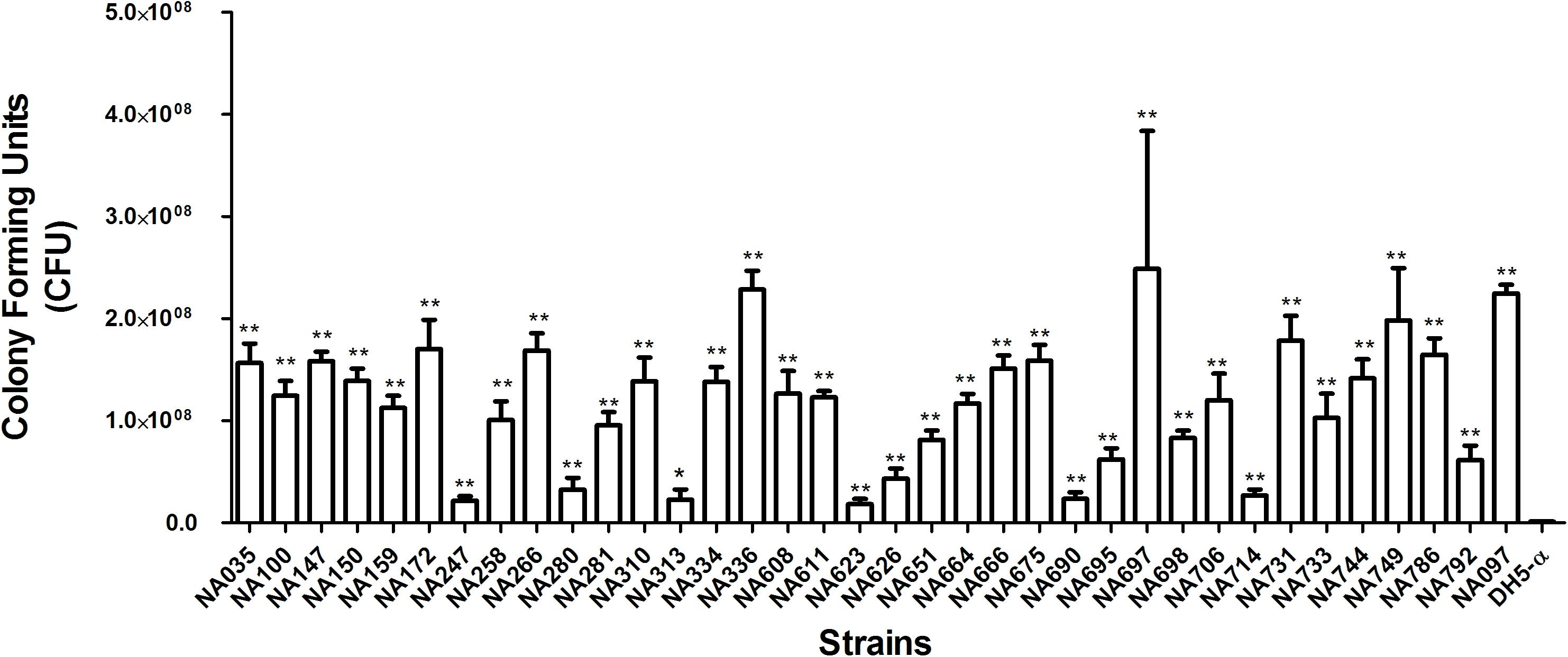
FIGURE 2. Serum resistance assay of pks-positive isolates in human serum. Mann–Whitney U test was carried out for calculating the significant differences. Significant differences were indicated by asterisks and p ≤ 0.05 was considered to be significant. ∗p-value ≤ 0.05, ∗∗p-value ≤ 0.01. NA097 was taken as the positive control, while DH5-α served as the negative control.
Discussion
The pks island encodes enzymes that are able to synthesize colibactin, a genotoxin that could induce host DNA damage and its presence may contribute to increased virulence and severe disease outcomes. Ever since the description of colibactin by Nougayrède et al. (2006), many studies were undertaken to develop a comprehensive understanding of this bacterial genotoxin (Bossuet-Greif et al., 2018; Faïs et al., 2018). However, epidemiological data on the prevalence of the same and factors associated with pks-positive E. coli isolates, particularly from the Southern World have not been documented. The present study showed that 35 out of the 462 clinical ExPEC isolates were positive for all the four genes, indicating the presence of complete pks island(s) which might be able to synthesize functional colibactin. Thus, the overall prevalence was found to be 7.6% and this constitutes first epidemiological data on pks island harboring E. coli from India. In contrast, reports from other countries, such as those by Johnson et al. (2008) and Shimpoh et al. (2017) demonstrated high pks prevalence among clinical E. coli.
Our previous studies have suggested that majority of the pks negative clinical E. coli from India were genetically diverse and had a high prevalence of antibiotic resistance; these studies also identified and characterized the clonally evolving pandemic sequence type 131 E. coli isolates in India (Jadhav et al., 2011; Ranjan et al., 2016; Hussain et al., 2017; Shaik et al., 2017). However, the role of colibactin in the emergence of lineage specific virulence in E. coli that were comparatively less resistant to antibiotics has been shown herein for the first time from India. Such lineages could become a matter of public health concern and analysis of the underlying strains would be of great importance given the high burden of infectious diseases in this region. Thus, the findings of this study have implications for better understanding of the epidemiological context of pathogenic E. coli in human diseases.
Escherichia coli harboring pks island was reported to be strongly associated with bacteremia and human colorectal tumors (Johnson et al., 2008; Buc et al., 2013). Recent reports suggest that mice infected with colibactin positive E. coli had significantly lower survival rates compared to those infected with isogenic colibactin-negative mutant(s) (Marcq et al., 2014). It has been further demonstrated that pks-positive E. coli infection induces cellular senescence and concurrently produces growth factors which promote tumor growth (Secher et al., 2013; Cougnoux et al., 2014; Dalmasso et al., 2015). The pks-positive E. coli isolates in the present study were detected in different specimen types including urine, blood and pus. Therefore, it can be surmised that the pks-positive E. coli might contribute in many invasive and non-invasive infections at different anatomic sites.
Previous studies reported that the pks island was majorly detected in E. coli phylogenetic group B2 strains, which are mainly documented as extraintestinal pathogens (Taieb et al., 2016; Sarshar et al., 2017). Our results were in line with these observations as the pks-positive isolates in our study also belonged predominantly to phylogroup B2 (97%), except for one isolate which belonged to phylogroup D (3%) (Table 1). The pathogenic strains of E. coli mainly belong to group B2 and, to a lesser extent, group D and frequently harbor higher number of virulence-factors than group A and group B1 strains (Picard et al., 1999; Johnson et al., 2008).
Several studies have demonstrated a strong correlation between the presence of virulence genes and the pathogenic spectrum of E. coli strains (Bien et al., 2012). These include multiple ExPEC-associated virulence genes such as adhesins, invasins, secretory toxins, and iron scavenging systems (Johnson, 1991). Accordingly, the virulent nature of pks-positive E. coli isolates was supported by our findings as 100% of them were positive for fimH (D mannose specific adhesin of minor fimbrial component), sfaD/E (s-fimbrial adhesin) and usp (uropathogenic specific protein) while ≥ 30% of them were positive for iucD (enzyme for siderophore aerobactin synthesis), sat (secreted autotransporter vacuolating cytotoxin), cvaC (colicin-V precursor) and ibeA (invasion protein). The afimbrial adhesin gene, afa was found to be completely absent (Table 1). These findings could be attributed to the pathogenic potential of the pks-positive strains along with the presence of many other virulence determinants. Furthermore, in the present study, siderophore production was detected in all pks-positive isolates; this observation was consistent with previous reports on the positive correlation between the presence of pks island and iron scavenging systems among the B2 E. coli strains (Martin et al., 2017). The localization of pks island within the High-Pathogenicity Island (HPI) and its physical association with siderophore biosynthesis gene cluster has also been described in other members of Enterobacteriaceae (Putze et al., 2009). A majority of pks-positive E. coli isolates demonstrated high biofilm forming capabilities in M63 medium (Figure 1) and all the pks-positive isolates tested were found to be resistant to serum bactericidal activity (Figure 2). We speculate that these genotypic and phenotypic virulence traits expressed by B2 pks-positive E. coli could act as fitness factors in order to colonize and initiate/establish infection in intestinal and extra-intestinal sites.
Antibiotic susceptibility of the pks-positive isolates was performed and the isolates were observed to be uniquely associated with low antimicrobial resistance, this finding is consistent with the previous reports which have suggested that pks island harboring E. coli exhibit reduced antibiotic resistance (Chen et al., 2017; Sarshar et al., 2017). This observation could likely be due to opportunist E. coli infections arising from intestinal microbiota. Additionally, detailed characterization of such isolates from fecal and clinical samples is needed to certainly understand the reason behind such an observation. Multidrug resistance and ESBL production was observed in only 11.42 and 37% isolates, respectively (Table 1). Previous studies from our group have shown MDR rates of 95% for clinical ST131 strains, and 91% for clinical and stool non-ST131 strains (Hussain et al., 2014). We have also previously reported MDR rates of 100% for metallo-(β-lactamase (MBL) producing E. coli isolates (Ranjan et al., 2016) and 67% for isolates from skin and soft tissue infections (Ranjan et al., 2017). The pks positive isolates in contrast demonstrated lesser rates of antimicrobial resistance compared to the pks negative isolates characterized from the similar settings in our previous studies. All nine blaCTXM-15 positive E. coli isolates were observed to be ESBL producers (Table 1), although further screening for other groups of CTX-M genes, and other ESBL classes are warranted. In concordance with the antimicrobial susceptibility results, molecular detection of antimicrobial resistance genes showed a low prevalence of sul1 (sulphonamide resistance), tetA (tetracycline resistance), aac(6′)-Ib (aminoglycoside resistance), blaCTX-M-15 (extended spectrum-β-lactamases) and blaTEM-1 (broad spectrum-β-lactamases) genes (Table 1).
Conclusion
The prevalence of colibactin producing E. coli was found to be moderate among clinical E. coli isolates in our collection. These isolates harbored multiple virulence genes/traits and demonstrated relatively low antimicrobial resistance. These findings comprise essential baseline data required to understand aspects of functional molecular infection epidemiology of possibly genotoxic phenotypes of E. coli and their clinical significance. We hope to extend these studies at genomic and landscape scales to gain further insights into evolution, adaptation and dissemination of such isolates at clinical, community and ecosystem levels.
Author Contributions
AS designed and performed all experiments with assistance from AR, SJ, AH, and SS. AS was responsible for all of the data and served as the guarantor for the manuscript. RB, MA, and LW participated in detailed discussions, advised on the interpretation of some of the results and, contributed to the writing and editing of the manuscript. NA conceived the study and provided overarching supervision, laboratory facilities and resources, interpreted and discussed results, and wrote/edited the draft and final versions of the manuscript. All authors contributed to the development of the manuscript and its display items.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
AS acknowledges the junior research fellowship (JRF) from CSIR, India and would like to thank icddr,b for a short research stay support. We would like to thank all the members of Pathogen Biology Lab for their constructive comments on the manuscript and suggestions, especially to Nishant Nandanwar. icddr,b would like to thank its core donors: Sweden (SIDA), Bangladesh, Canada (CIDA and GAC), the United Kingdom (DFID).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02631/full#supplementary-material
References
Ahmed, N., Dobrindt, U., Hacker, J., and Hasnain, S. E. (2008). Genomic fluidity and pathogenic bacteria: applications in diagnostics, epidemiology and intervention. Nat. Rev. Microbiol. 6, 387–394. doi: 10.1038/nrmicro1889
Avasthi, T. S., Kumar, N., Baddam, R., Hussain, A., Nandanwar, N., Jadhav, S., et al. (2011). Genome of multidrug-resistant uropathogenic Escherichia coli strain NA114 from India. J. Bacteriol. 193, 4272–4273. doi: 10.1128/JB.05413-11
Bien, J., Sokolova, O., and Bozko, P. (2012). Role of uropathogenic escherichia coli virulence factors in development of urinary tract infection and kidney damage. Int. J. Nephrol. 2012:681473. doi: 10.1155/2012/681473
Bossuet-Greif, N., Vignard, J., Taieb, F., Mirey, G., Dubois, D., Petit, C., et al. (2018). The colibactin genotoxin generates DNA interstrand cross-links in infected cells. mBio 9:e02393-17. doi: 10.1128/mBio.02393-17
Buc, E., Dubois, D., Sauvanet, P., Raisch, J., Delmas, J., Darfeuille-Michaud, A., et al. (2013). High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS One 8:e56964. doi: 10.1371/journal.pone.0056964
Chen, Y.-T., Lai, Y.-C., Tan, M.-C., Hsieh, L.-Y., Wang, J.-T., Shiau, Y.-R., et al. (2017). Prevalence and characteristics of pks genotoxin gene cluster-positive clinical Klebsiella pneumoniae isolates in Taiwan. Sci. Rep. 7:43120. doi: 10.1038/srep43120
Clermont, O., Christenson, J. K., Denamur, E., and Gordon, D. M. (2013). The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5, 58–65. doi: 10.1111/1758-2229.12019
CLSI (2013). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals; Approved Standard. VET01-A4, 4 Edn. Wayne, PA: Clin. Lab. Stand. Inst.
Cougnoux, A., Dalmasso, G., Martinez, R., Buc, E., Delmas, J., Gibold, L., et al. (2014). Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut 63, 1932–1942. doi: 10.1136/gutjnl-2013-305257
Croxen, M. A., and Finlay, B. B. (2010). Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 8, 26–38. doi: 10.1038/nrmicro2265
Cuevas-Ramos, G., Petit, C. R., Marcq, I., Boury, M., Oswald, E., and Nougayrède, J.-P. (2010). Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 107, 11537–11542. doi: 10.1073/pnas.1001261107
Dalmasso, G., Cougnoux, A., Delmas, J., Darfeuille-Michaud, A., and Bonnet, R. (2015). The bacterial genotoxin colibactin promotes colon tumor growth by modifying the tumor microenvironment. Gut Microbes 5, 675–680. doi: 10.4161/19490976.2014.969989
Faïs, T., Delmas, J., Barnich, N., Bonnet, R., and Dalmasso, G. (2018). Colibactin: more than a new bacterial toxin. Toxins 10:151. doi: 10.3390/toxins10040151
Hussain, A., Ewers, C., Nandanwar, N., Guenther, S., Jadhav, S., Wieler, L. H., et al. (2012). Multi-resistant uropathogenic Escherichia coli from an endemic zone of urinary tract infections in India: genotypic and phenotypic characteristics of ST131 isolates of the CTX-M-15 extended-spectrum-beta-lactamase producing lineage. Antimicrob. Agents Chemother. 56, 6358–6365. doi: 10.1128/AAC.01099-12
Hussain, A., Ranjan, A., Nandanwar, N., Babbar, A., Jadhav, S., and Ahmed, N. (2014). Genotypic and phenotypic profiles of Escherichia coli isolates belonging to clinical sequence type 131 (ST131), clinical non-ST131, and fecal non-ST131 lineages from India. Antimicrob. Agents Chemother. 58, 7240–7249. doi: 10.1128/AAC.03320-14
Hussain, A., Shaik, S., Ranjan, A., Nandanwar, N., Tiwari, S. K., Majid, M., et al. (2017). Risk of transmission of antimicrobial resistant Escherichia coli from commercial broiler and free-range retail chicken in India. Front. Microbiol. 8:2120. doi: 10.3389/fmicb.2017.02120
Jadhav, S., Hussain, A., Devi, S., Kumar, A., Parveen, S., Gandham, N., et al. (2011). Virulence characteristics and genetic affinities of multiple drug resistant uropathogenic Escherichia coli from a semi urban locality in India. PLoS One 6:e18063. doi: 10.1371/journal.pone.0018063
Johnson, J. R. (1991). Virulence factors in Escherichia coli urinary tract infection. Clin. Microbiol. Rev. 4, 80–128. doi: 10.1128/CMR.4.1.80.Updated
Johnson, J. R., Johnston, B., Kuskowski, M. A., Nougayrede, J.-P., and Oswald, E. (2008). Molecular epidemiology and phylogenetic distribution of the Escherichia coli pks genomic island. J. Clin. Microbiol. 46, 3906–3911. doi: 10.1128/JCM.00949-08
Johnson, J. R., and Russo, T. A. (2002). Extraintestinal pathogenic Escherichia coli: “The other bad E coli”. J. Lab. Clin. Med. 139, 155–162. doi: 10.1067/mlc.2002.121550
Kaper, J. B., Nataro, J. P., and Mobley, H. L. (2004). Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2, 123–140. doi: 10.1038/nrmicro818
Marcq, I., Martin, P., Payros, D., Cuevas-Ramos, G., Boury, M., Watrin, C., et al. (2014). The genotoxin colibactin exacerbates lymphopenia and decreases survival rate in mice infected with septicemic Escherichia coli. J. Infect. Dis. 210, 285–294. doi: 10.1093/infdis/jiu071
Martin, P., Tronnet, S., Garcie, C., and Oswald, E. (2017). Interplay between siderophores and colibactin genotoxin in Escherichia coli. IUBMB Life 69, 435–441. doi: 10.1002/iub.1612
Messerer, M., Fischer, W., and Schubert, S. (2017). Investigation of horizontal gene transfer of pathogenicity islands in Escherichia coli using next-generation sequencing. PLoS One 12:e0179880. doi: 10.1371/journal.pone.0179880
Monstein, H. J., Östholm-Balkhed,Å, Nilsson, M. V., Nilsson, M., Dornbusch, K., and Nilsson, L. E. (2007). Multiplex PCR amplification assay for the detection of blaSHV, blaTEM and blaCTX-M genes in Enterobacteriaceae. APMIS 115, 1400–1408. doi: 10.1111/j.1600-0463.2007.00722.x
Nandanwar, N., Janssen, T., Kühl, M., Ahmed, N., Ewers, C., and Wieler, L. H. (2014). Extraintestinal pathogenic Escherichia coli (ExPEC) of human and avian origin belonging to sequence type complex 95 (STC95) portray indistinguishable virulence features. Int. J. Med. Microbiol. 304, 835–842. doi: 10.1016/j.ijmm.2014.06.009
Nougayrède, J.-P., Homburg, S., Taieb, F., Boury, M., Brzuszkiewicz, E., Gottschalk, G., et al. (2006). Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313, 848–851. doi: 10.1126/science.1127059
Nougayrède, J.-P., Taieb, F., De Rycke, J., and Oswald, E. (2005). Cyclomodulins: bacterial effectors that modulate the eukaryotic cell cycle. Trends Microbiol. 13, 103–110. doi: 10.1016/j.tim.2005.01.002
Picard, B., Garcia, J. S., Gouriou, S., Duriez, P., Brahimi, N., Bingen, E., et al. (1999). The link between phylogeny and virulence in Escherichia coli extraintestinal infection? Infect. Immun. 67, 546–553.
Putze, J., Hennequin, C., Nougayrède, J. P., Zhang, W., Homburg, S., Karch, H., et al. (2009). Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae. Infect. Immun. 77, 4696–4703. doi: 10.1128/IAI.00522-09
Qumar, S., Majid, M., Kumar, N., Tiwari, S. K., Semmler, T., Devi, S., et al. (2017). Genome dynamics and molecular infection epidemiology of multidrug-resistant Helicobacter pullorum isolates obtained from broiler and free-range chickens in India. Appl. Environ. Microbiol. 83:e02305-16. doi: 10.1128/AEM.02305-16
Ranjan, A., Shaik, S., Mondal, A., Nandanwar, N., Hussain, A., Semmler, T., et al. (2016). Molecular epidemiology and genome dynamics of New Delhi Metallo-β-Lactamase-producing extraintestinal pathogenic Escherichia coli strains from India. Antimicrob. Agents Chemother. 60, 6795–6805. doi: 10.1128/AAC.01345-16
Ranjan, A., Shaik, S., Nandanwar, N., Hussain, A., Tiwari, S. K., Semmler, T., et al. (2017). Comparative genomics of Escherichia coli isolated from skin and soft tissue and other extraintestinal infections. mBio 8:e01070-17. doi: 10.1128/mBio.01070-17
Sarshar, M., Scribano, D., Marazzato, M., Ambrosi, C., Aprea, M. R., Aleandri, M., et al. (2017). Genetic diversity, phylogroup distribution and virulence gene profile of pks positive Escherichia coli colonizing human intestinal polyps. Microb. Pathog. 112, 274–278. doi: 10.1016/j.micpath.2017.10.009
Schwyn, B., and Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Secher, T., Samba-Louaka, A., Oswald, E., and Nougayrède, J. P. (2013). Escherichia coli producing colibactin triggers premature and transmissible senescence in mammalian cells. PLoS One 8:e77157. doi: 10.1371/journal.pone.0077157
Shaik, S., Ranjan, A., Tiwari, S. K., Hussain, A., Nandanwar, N., Kumar, N., et al. (2017). Comparative genomic analysis of globally dominant ST131 clone with other epidemiologically successful extraintestinal pathogenic Escherichia coli (ExPEC) lineages. mBio 8:e01596-17. doi: 10.1128/mBio.01596-17
Shimpoh, T., Hirata, Y., Ihara, S., Suzuki, N., Kinoshita, H., Hayakawa, Y., et al. (2017). Prevalence of pks-positive Escherichia coli in Japanese patients with or without colorectal cancer. Gut Pathog. 9:35. doi: 10.1186/s13099-017-0185-x
Smith, J. L., Fratamico, P. M., and Gunther, N. W. (2007). Extraintestinal pathogenic Escherichia coli. Foodborne Pathog. Dis. 4, 134–163. doi: 10.1089/fpd.2007.0087
Keywords: genotoxins, pks island, colibactin, extraintestinal pathogenic E. coli (ExPEC), virulence
Citation: Suresh A, Ranjan A, Jadhav S, Hussain A, Shaik S, Alam M, Baddam R, Wieler LH and Ahmed N (2018) Molecular Genetic and Functional Analysis of pks-Harboring, Extra-Intestinal Pathogenic Escherichia coli From India. Front. Microbiol. 9:2631. doi: 10.3389/fmicb.2018.02631
Received: 05 August 2018; Accepted: 16 October 2018;
Published: 15 November 2018.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Yasufumi Matsumura, Kyoto University, JapanMaojun Zhang, National Institute for Communicable Disease Control and Prevention (China CDC), China
Shaofu Qiu, Institute of Disease Control and Prevention, Academy of Military Medical Sciences, China
Copyright © 2018 Suresh, Ranjan, Jadhav, Hussain, Shaik, Alam, Baddam, Wieler and Ahmed. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Niyaz Ahmed, bml5YXouYWhtZWRAdW9oeWQuYWMuaW4=; bml5YXouYWhtZWRAaWNkZHJiLm9yZw==
 Arya Suresh
Arya Suresh Amit Ranjan
Amit Ranjan Savita Jadhav2
Savita Jadhav2 Arif Hussain
Arif Hussain Sabiha Shaik
Sabiha Shaik Munirul Alam
Munirul Alam Ramani Baddam
Ramani Baddam Lothar H. Wieler
Lothar H. Wieler Niyaz Ahmed
Niyaz Ahmed