
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 23 October 2018
Sec. Food Microbiology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.02540
 Karen G. Jarvis1*
Karen G. Jarvis1* Ninalynn Daquigan1
Ninalynn Daquigan1 James R. White2
James R. White2 Paul M. Morin3
Paul M. Morin3 Laura M. Howard3
Laura M. Howard3 Julia E. Manetas3
Julia E. Manetas3 Andrea Ottesen4
Andrea Ottesen4 Padmini Ramachandran4
Padmini Ramachandran4 Christopher J. Grim1
Christopher J. Grim1Food microbiome composition impacts food safety and quality. The resident microbiota of many food products is influenced throughout the farm to fork continuum by farming practices, environmental factors, and food manufacturing and processing procedures. Currently, most food microbiology studies rely on culture-dependent methods to identify bacteria. However, advances in high-throughput DNA sequencing technologies have enabled the use of targeted 16S rRNA gene sequencing to profile complex microbial communities including non-culturable members. In this study we used 16S rRNA gene sequencing to assess the microbiome profiles of plant and animal derived foods collected at two points in the manufacturing process; post-harvest/pre-retail (cilantro) and retail (cilantro, masala spice mixes, cucumbers, mung bean sprouts, and smoked salmon). Our findings revealed microbiome profiles, unique to each food, that were influenced by the moisture content (dry spices, fresh produce), packaging methods, such as modified atmospheric packaging (mung bean sprouts and smoked salmon), and manufacturing stage (cilantro prior to retail and at retail). The masala spice mixes and cucumbers were comprised mainly of Proteobacteria, Firmicutes, and Actinobacteria. Cilantro microbiome profiles consisted mainly of Proteobacteria, followed by Bacteroidetes, and low levels of Firmicutes and Actinobacteria. The two brands of mung bean sprouts and the three smoked salmon samples differed from one another in their microbiome composition, each predominated by either by Firmicutes or Proteobacteria. These data demonstrate diverse and highly variable resident microbial communities across food products, which is informative in the context of food safety, and spoilage where indigenous bacteria could hamper pathogen detection, and limit shelf life.
You are what you eat. This idea, that the food that you consume controls your health, is not novel, and may be attributed to an early monograph on gout by Brillat-Savarin (2010). In recent years, efforts led by the Human Microbiome Project and the MetaHIT consortium have greatly expanded our knowledge of the microbiomes associated with the human host, especially the gastrointestinal (GI) tract (Qin et al., 2010; Human Microbiome Project Consortium, 2012a,b). That knowledge appears to have important consequences for individual human health. This paper explores food microbiomes to identify bacteria that are associated with foods and therefore have the potential to colonize our gastrointestinal tract. These resident cohorts of bacteria on our food may also be a factor in food safety, affecting our ability to detect and identify pathogens during foodborne outbreaks, and contribute to the general transmission of bacteria between the environment and the human body.
More specifically, surveys of microbial populations in the human gut led Arumugam and colleagues to describe three distinct “Enterotypes” or microbial communities that can differ between individual human hosts (Arumugam et al., 2011; Costea et al., 2018). Enterotypes seem to be controlled by fetal microbiome composition, blood type, and the metabolic potential of the associated microbiome. Once established, the most likely source of new microbes joining our GI microbiome is the food we eat: each food stuff and commodity we consume likely contains a microbiome that passes through our bodies while nutritional ingredients and components are digested. It is unclear how much of this inoculum successfully passes through our gastric barrier, reaches our intestine and colon, resulting in colonization. Foods such as fresh produce that are often consumed raw with no kill step also have the potential to alter the microbial composition of our GI tract and, if they harbor pathogens, can cause illness. Thus, characterizing food microbiomes may provide reference points for understanding which bacteria are beneficial, how frequently our GI tract is exposed to foodborne pathogens, and how these bacterial communities interact with one another. Current methods to detect foodborne pathogens such as Salmonella, Escherichia coli, and Listeria rely on culturing these organisms directly from food, which requires at least 1 week for the isolation of a presumptive positive colony, followed by 2–4 days for culture confirmation. These methods are complex and challenging due to the wide variety of food matrices implicated in outbreaks and the presence of many different bacterial species residing in foods that grow along with adulterating organisms and may compromise pathogen detection.
Recent studies employing microbiome profiling have revealed that the microflora indigenous to tomatoes, cilantro, spinach, mung bean sprouts, and ice cream, can affect our ability to detect Salmonella, Listeria, and E. coli in these foods primarily because the incursion of a foodborne pathogen typically occurs at a very low abundance compared to the resident microflora (Ottesen et al., 2013; Jarvis et al., 2015; Leonard et al., 2015, 2016; Margot et al., 2016; Ottesen et al., 2016). For example, detection of Shiga Toxigenic Escherichia coli (STEC) in spinach is hampered by non-pathogenic E. coli and detection of Salmonella is greatly reduced in foods, such as cilantro, that also harbor closely related species such as Enterobacter and Citrobacter (Leonard et al., 2015; Daquigan et al., 2016; Grim et al., 2017). These studies suggest that both bacterial load and the composition of the indigenous microbiota can influence our ability to detect foodborne pathogens. Identifying the intrinsic bacterial biomass in foods and how those intrinsic bacterial communities are different from commodity to commodity is a fundamental first step for assessing the sensitivity and specificity of microbiome profiling for detecting pathogens in foods.
Analysis of food microbiomes can also provide insights into food quality by revealing the presence of spoilage associated microorganisms. Microbial spoilage is the most common cause of food spoilage which is considered as any change in food that renders it unacceptable to a consumer including sensory characteristics such as changes in color, texture, and development of off odors and colors (Gram et al., 2002). Some examples of bacteria commonly associated with food spoilage include Pseudomonas species in plant and animal derived foods, Xanthomonas in fresh produce, and Shewanella in chilled fish and meat (Blackburn, 2006). Enterobacteriaceae are natural inhabitants of many food plants and animals providing numerous opportunities for introduction into the food supply. For example, the production of pectic enzymes by Erwinia species result in soft rot of fresh vegetables, and the production of volatile compounds by Serratia and Proteus species results in the spoilage of dry cured meats (Blackburn, 2006). Food spoilage caused by Lactic Acid Bacteria (LAB) is influenced by intrinsic and extrinsic factors such as low pH and temperature or types of food packaging such as modified atmospheric packaging (MAP) that lower oxygen levels. Lactobacillus and Leuconostoc species have been associated with spoilage of vacuum packaged meat and fish, low pH foods such as, ketchup and salad dressings, and cheeses (Blackburn, 2006). Additional spoilage- associated bacteria include spore-formers such as Bacillus, and other genera such as Acinetobacter, Flavobacterium, Psychrobacter, and Brochothrix that are responsible for spoilage of particular food groups. For example, Acinetobacter have been isolated from soil, water, sewage, milk, vegetables, and chicken. Flavobacterium species, also commonly found in the environment in soil and water, prefer chilled foods such as milk, raw meats, and wild and farmed fresh-water fish (Blackburn, 2006).
The purpose of this study was to define the baseline microbiomes, employing 16S rRNA gene sequencing, of the plant-derived commodities cilantro, mung bean sprouts, cucumbers, and masala spice mixes, sourced from two points in the food supply chain. In order to have a non-plant-derived commodity for comparison, we analyzed three smoked salmon samples to demonstrate the value of microbiome profiling for assessing the microbiota associated with the smoking process. These commodities represent the global nature of our food supply and have been linked to foodborne outbreaks in the past. For example, a 2007 – 2009 survey of imported spices determined that the mean prevalence of Salmonella was 0.066 (95% CI 0.057–0.076) and included a variety of serotypes some with antimicrobial resistance profiles (Van Doren et al., 2013). Herbs and spices contaminated with Salmonella have resulted in numerous outbreaks including one in Germany due to contaminated paprika and another in the United States caused by contaminated cilantro (Lehmacher et al., 1995; Campbell et al., 2001). Since 2013, four Salmonella outbreaks have been attributed to domestic and imported cucumbers and epidemiological evidence identified four serotypes (Poona, Newport, Saint Paul, and Oslo) responsible for these outbreaks (Angelo et al., 2015; Bottichio et al., 2016). Seed and bean sprouts were linked to 33 outbreaks from 1998 through 2010 (Dechet et al., 2014); despite rigorous guidelines to reduce contamination these continue to be implicated in outbreaks of Salmonella and Escherichia coli (Mohle-Boetani et al., 2009; CDC, 2013; Bayer et al., 2014). Smoked salmon products have historically been associated with Salmonella and Listeria outbreaks (Rorvik, 2000; Friesema et al., 2014).
The Department of Agriculture and Rural Development in Lansing Michigan (MDARD) collected seven cilantro samples prior to retail distribution, in January, February, and March of 2015 (Supplementary Table 1). Six replicates of six MDARD cilantro samples (CIL27A, CIL27B, CIl27C, CIL28A, CIL28C, and CIL28D) and three replicates of one MDARD cilantro sample (CIL4Z) were sequenced. An additional set of cilantro samples and two brands of mung bean sprouts were purchased from a local retail store in April of 2015. The retail cilantro samples were combined into a homogenous composite and eighteen replicates (CILSB1 thru CILSB18) were sequenced. Three replicates of mung bean sprout brand A (SPA), and nine replicates of mung bean sprout brand B (SPB) were sequenced (Supplementary Table 1). Seven cucumber samples (CUC2, CUC3, CUC4, CUC5, CUC7, CUC8, and CUC9) were collected from a farm in New Jersey in November of 2014 and sequenced individually. We also sequenced Chicken Masala (MA), Fish Masala (MB), Meat Masala (MC), Egg Masala (MD), and Garam Masala (ME) spice mixtures. These masala spices were mixtures of 8–17 individual spices that were imported from India in November of 2014; two replicates of each spice mixture were sequenced (Supplementary Table 2). Three smoked salmon samples, SmkSalD, SmkSalE, and SmkSalF, were obtained from the California Department of Public Health, Richmond, CA, United States in January 2016, and two replicates of each were sequenced.
Culture enrichment media was used as “wash” media to remove the microbiome from each food as follows: twenty-five grams of each food sample was aseptically transferred to separate, sterile Whirlpak bags (Nasco, Fort Atkinson, WI, United States) and mixed with a sterile culture medium at a 1:10 (w/v). Cilantro samples were rinsed with modified Buffered Peptone Water (mBPW). Mung bean sprouts and smoked salmon samples were rinsed with Buffered Listeria Enrichment Broth. Cucumbers, and masala spice mixes were rinsed with Lactose Broth, and Trypticase Soy Broth, respectively. All samples were hand massaged for 2 min at room temperature in their respective broth to dislodge resident bacteria. Aliquots (15 ml) were removed and centrifuged at 7,100 rcf for 30 min and bacterial pellets were stored at -20°C.
Genomic DNA was prepared from the bacterial pellets of the microbiomes of masala spice mixes, cilantro, cucumber, mung bean sprouts, and smoked salmon using the DNAeasy Blood and Tissue (Qiagen, Germantown, MD, United States) protocol on the QIAcube (Qiagen, Germantown, MD, United States) automated sample preparation instrumentation.
16S rRNA gene library preparation, sequencing, data processing and analyses were performed as described by Daquigan et al. (2016). The PCR primers used in this study targeted the V1 – V3 hypervariable regions (508 bp) of the 16S rRNA gene. Amplicon libraries were sequenced in six separate MiSeq runs multiplexed at 96 samples per run except for the fifth and sixth MiSeq runs, which only contained 60 and 86 samples, respectively (Supplementary Table 3). To normalize across samples prior to downstream analyses, the 16S rRNA gene sequence profiles were subsampled to an even level of coverage resulting in 10,000 16S rRNA sequence reads for each cilantro, mung bean sprout, cucumber, and masala spice mix sample, and 30,000 16S rRNA sequence reads for each smoked salmon sample. High-quality 16S rRNA gene sequence reads were assigned to a taxonomic lineage using Resphera Insight (Resphera Biosciences, Baltimore, MD, United States; Daquigan et al., 2016; Abernethy et al., 2017; Drewes et al., 2017; Grim et al., 2017). Using these methodologies our sensitivity for cilantro, cucumbers, mung bean sprouts, cucumbers, and masala spice mixes with 10,000 sequence reads per sample we have a 95% chance of observing a pathogen with at least 0.03% abundance (Grim et al., 2017). In smoked salmon, with 30,000 sequence reads per sample, we have a 95% chance of observing a pathogen with at least 0.01% abundance (Grim et al., 2017).
16S rRNA gene sequence data from this study has been submitted to the FDA MetaGenomeTraker Project (NCBI Accession PRJNA390622).
The species richness in plant-derived foods was highest in the masala spice mixes with observed species ranging from 968 to 1097 (Supplementary Table 1). Mung bean sprouts, cilantro and cucumber observed species ranged from 226 to 340 (mung bean sprouts), 216 to 573 (cilantro), and 227 to 423 (cucumbers) (Supplementary Table 1). The Simpson reciprocal index measures of diversity corroborated that the species richness was highest in the masala spice mixes and lowest in the mung bean sprouts (Supplementary Table 1) (Simpson, 1949). The species richness of the smoked salmon samples was lower than the plant derived foods with observed species ranging from 89 to 181 and, as expected, the alpha diversity in these samples was low (Supplementary Table 1).
Unweighted UniFrac distance measures revealed striking variations among the microbiota of the plant-derived foods (Figure 1) (Lozupone and Knight, 2005). The first coordinate, accounting for 22% of the variation in beta-diversity among all food samples, clearly separates our samples into four groups corresponding to the type of food: cucumbers, cilantro, mung bean sprouts, and masala spice mixes (Figure 1). The second and third coordinates, representing 12 and 6.5% of the beta-diversity, further separate the cilantro, mung bean sprout, and masala spice microbiota indicating differences even within a single food type (Figure 1).
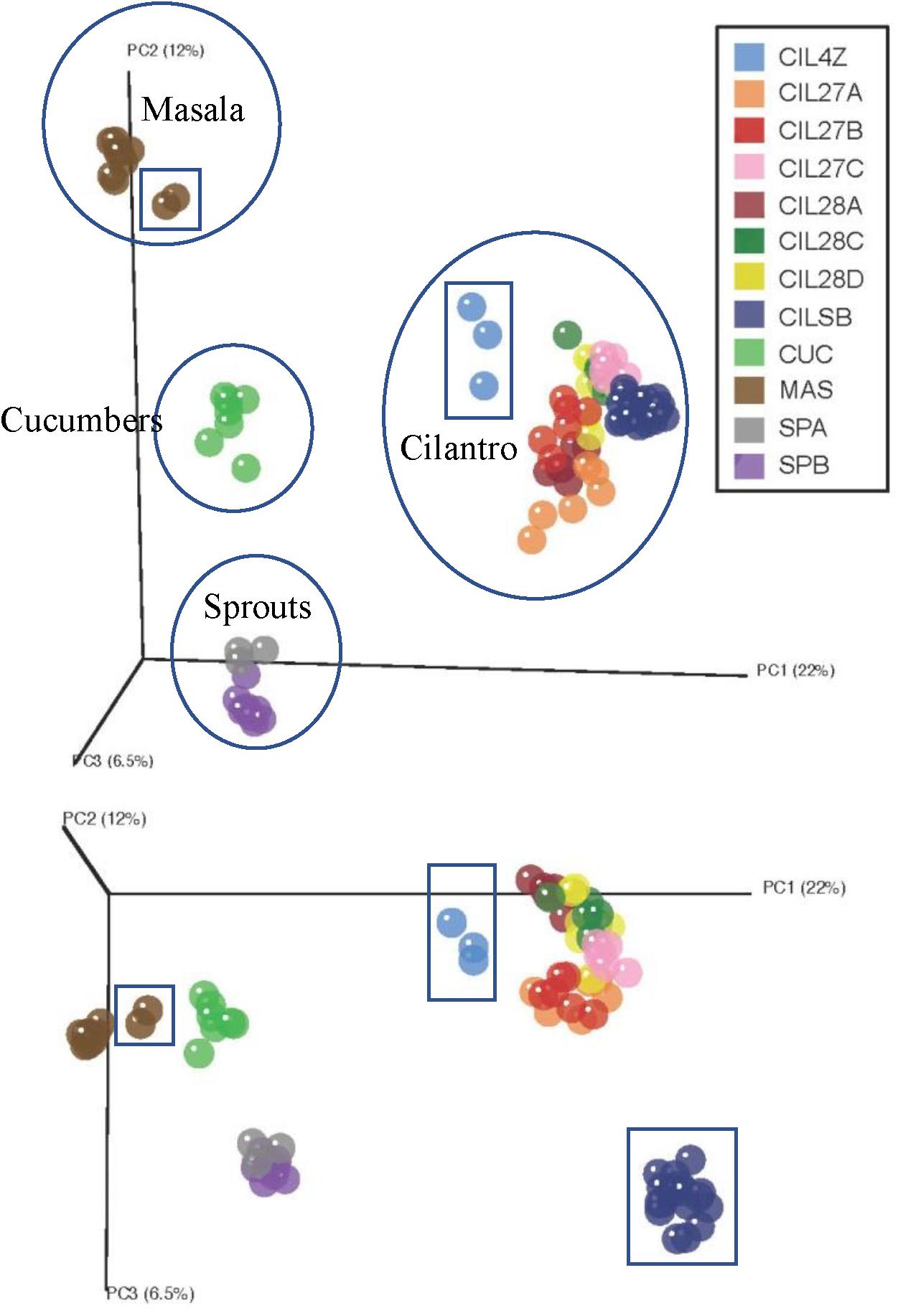
FIGURE 1. Principal coordinates analysis reveals clustering by commodity type. Each food is circled and squares indicate garam masala (brown), MDARD cilantro CIL4Z (light blue), and retail cilantro CILSB (dark blue).
Unsupervised hierarchical clustering of all foods revealed that Firmicutes were nearly absent in cilantro with proportional abundances of <1% in all but nine of the cilantro samples, and abundances in these nine samples only reached 2.3% (Figure 2). Furthermore, Pseudomonadaceae, Enterobacteriaceae, Flavobacteriaceae, and Oxalobacteraceae were the predominant taxa driving beta-diversity (Figure 2). For example, we observed that proportional abundances of Enterobacteriaceae vary greatly within each commodity; these are generally the lowest in cilantro (34 to 1%), and smoked salmon (7% to 0%) and the highest in SPB (69%) (Figure 2 and Supplementary Figure 1). Oxalobacteraceae and Flavobacteriaceae are nearly absent in all commodities, except cilantro, in which proportional abundances vary from 1% in CIL4Z to 29% in CIL28C for Oxalobacteraceae and 2% in CIL4Z to 34% in CILSB for Flavobacteriaceae (Figure 2 and Supplementary Figures 2, 3). Furthermore, cilantro had the highest proportional abundances of Pseudomonadaceae among all foods (44–79%) while the smoked salmon harbored the lowest abundances of these members (<1%) (Figure 2 and Supplementary Figure 4).
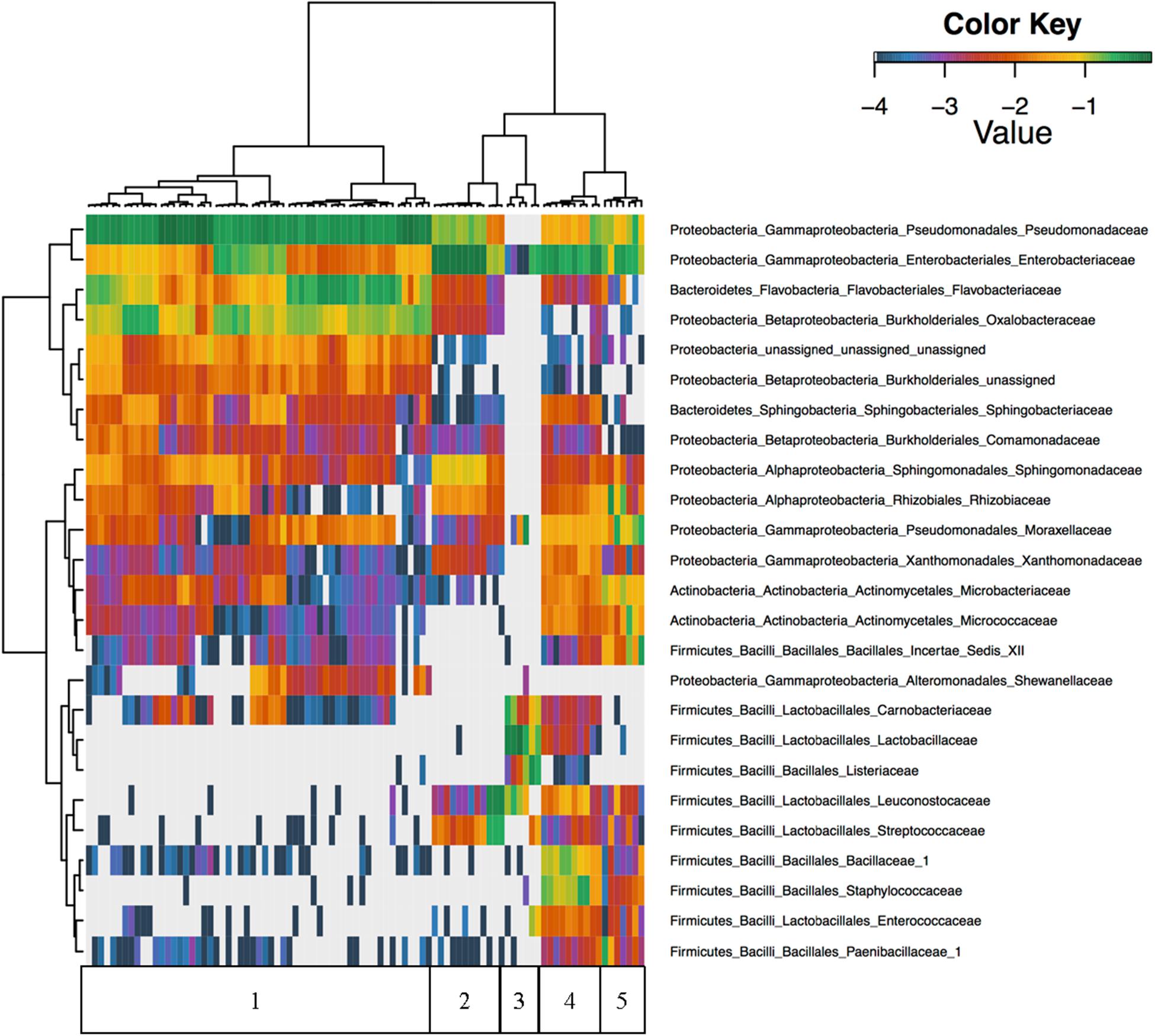
FIGURE 2. Unsupervised hierarchical clustering of the top 25 families across all samples. Values reflect proportional abundances. 1 = cilantro, 2 = Mung bean sprouts, 3 = Smoked Salmon, 4 = Masala spice mixes, 5 = Cucumbers.
We observed high abundances of Leuconostocaceae and Streptococcaceae in SPA, which include members associated with food spoilage, but not in SPB. Leuconostocaceae proportional abundances are also high in smoked salmon, which also harbored high abundances of spoilage associated Listeriaceae, Lactobacillaceae, and Carnobacteriaceae (Figure 2). Additionally, of the top 25 families identified across all foods, 19 were absent from the smoked salmon samples (Figure 2). Finally, the microbiota found on the masala spices included high proportional abundances of Bacillaceae_1 and Staphylococcaceae (Figure 2).
The five masala spice mixes, Chicken, Fish, Meat, Egg, and Garam, exhibited a potential correlation between the number of spices in each mix and levels of species diversity (P = 0.08, Pearson coefficient = 0.58). Specifically, the Meat masala, which contained 17 ingredients, had the highest diversity and the Fish masala, which contained the least number of ingredients had the lowest species diversity (Supplementary Table 2). Bacterial communities found in masala spice mixes consisted primarily of Proteobacteria (41–84%) and Firmicutes (9–54%), followed by Actinobacteria (5–12%) and Bacteroidetes (0.41–2%), although the Garam masala had higher abundances of Proteobacteria and lower abundances of Firmicutes than the rest of the spice mixtures (Supplementary Table 4). The difference in the Garam masala composition was due to higher proportional abundances of Pseudomonas (19%) and Pantoea (24%) members in conjunction with a lower proportional abundance of Staphylococcus; 3% in Garam as compared to 11–39% in the remaining four masala spice mixes (Figure 3 and Supplementary Figures 4, 5). The fennel seeds which are unique to the Garam mix, may account for the higher abundances of Pseudomonas and Pantoea in this spice mix (Supplementary Table 3). Unassigned Enterobacteriaceae Operational Taxonomic Units (OTUs) were observed in all of the spice mixes; the Egg masala had the highest proportional abundances (10%) which, likely represent additional Enterobacter species (Figure 3 and Supplementary Table 5). Bacillus species were also identified in all masala spices at proportional abundances as high as 13% in the Meat masala (Figure 3 and Supplementary Table 5).
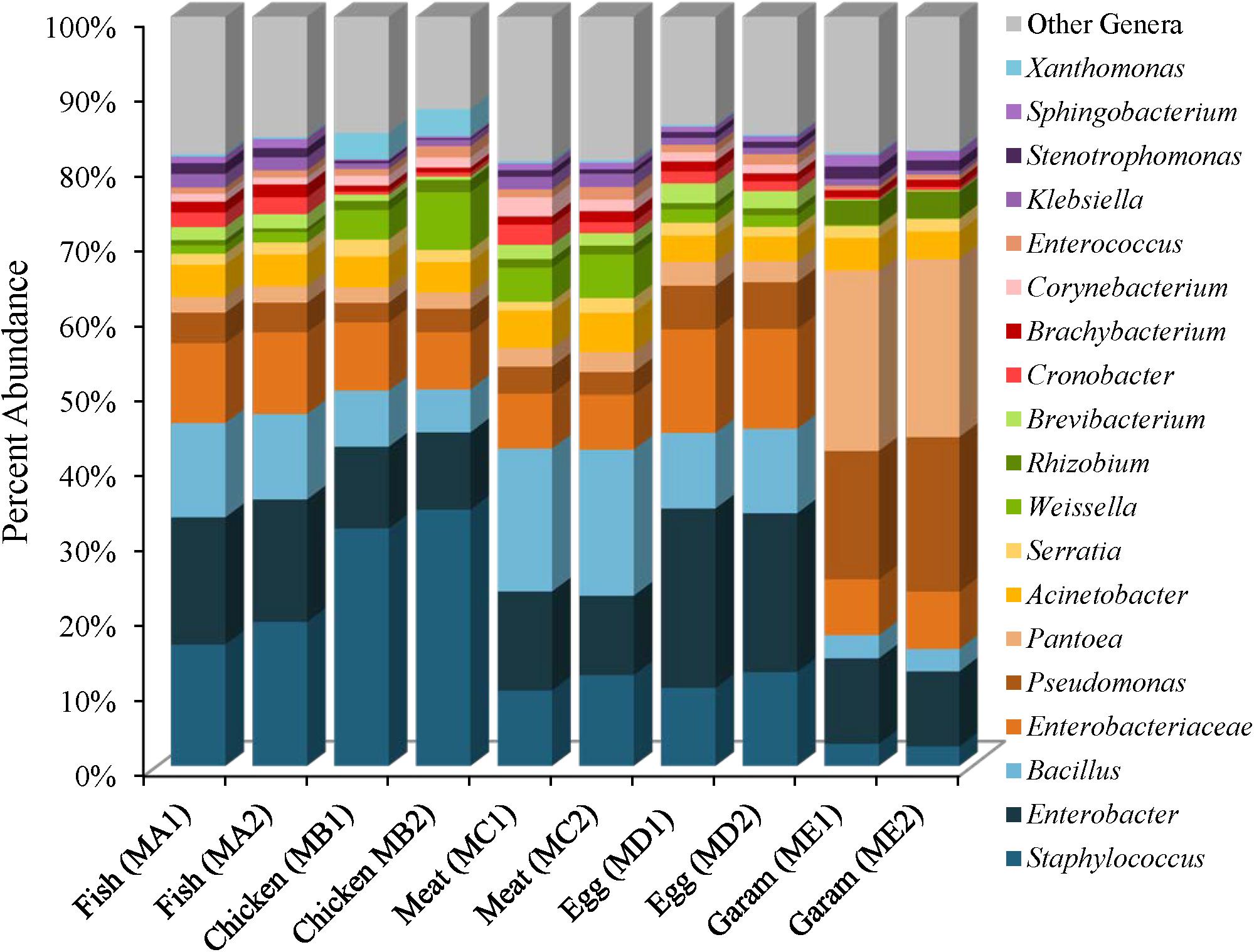
FIGURE 3. Genus level proportional abundances (≥1%) of masala spice mixes. Two replicates of each spice mixture were sequenced.
The cilantro microbiota consists primarily of Proteobacteria (54–99%), followed by Bacteroidetes (0.83–46%), Actinobacteria (0.03–5%) and Firmicutes (0–2%) (Supplementary Table 4). The Simpson reciprocal and Shannon diversity indexes for cilantro samples ranged from 3.97 to 33.89 and 3.81 to 6.34, respectively and the subsamples had similar levels of diversity (Supplementary Table 2). For example, the highest alpha diversity was observed in CIL27C, which also harbored the highest number of observed species.
Weighted UniFrac analysis resulted in three distinct cilantro clusters; (1) the retail samples, (2) 36 of the MDARD samples and (3) CIL4Z, also an MDARD sample (Figure 1). The presence of Flavobacterium at proportional abundances of 34% in the retail cilantro samples, CILSB and their absence in the MDARD samples led to distinct clustering of these groups (Figures 1, 4 and Supplementary Figure 3). In fact, CILSB, the retail cilantro, was the only food in this study that contained Flavobacterium (Figure 4 and Supplementary Figure 3). These retail cilantro samples harbored at least seven distinct Flavobacterium OTUs (Supplementary Table 5). The MDARD cilantro sample CIL4Z contained high proportional abundances of Pseudomonas (79%) that accounted for most of the Proteobacteria in this cilantro sample (Figure 4 and Supplementary Figure 4). Additionally, two members of the Oxalobacteraceae family, Janthinobacterium and Duganella, were absent in the microbiota of CIL4Z further distinguishing them from the rest of the cilantro samples (Figure 4, Supplementary Figure 2, and Supplementary Table 5).
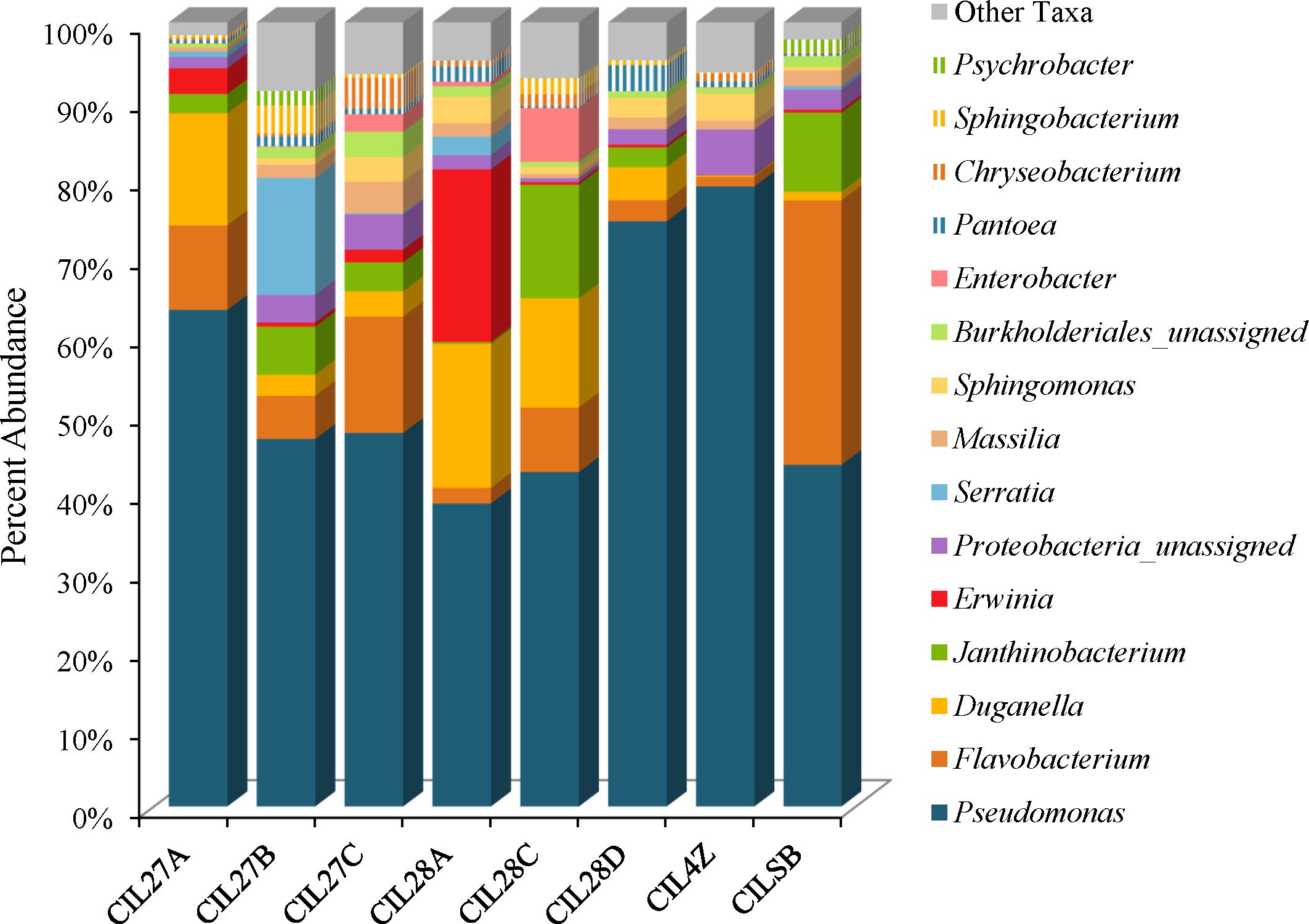
FIGURE 4. Genus level average proportional abundances (≥1%) of MDARD and retail cilantro samples. Proportional abundances were average for six replicates of MDARD samples CIL27A, CIL27B, CIl27C, CIL28A, CIL28C, and CIL28D and three replicates of MDARD sample CIL4Z. The proportional abundances for the retail cilantro samples CILSB1 thru CILSB18 were averaged.
Although cilantro harbored the lowest proportional abundances of Enterobacteriaceae in this study, MDARD samples CIL28A and CIL27B had notable abundances of Erwinia (14–29%) and Serratia (11–19%) (Figure 4, Supplementary Figure 1, and Supplementary Table 5).
The cucumber microbiota is comprised of Proteobacteria (45–85%), followed by Firmicutes (2–40%), Actinobacteria (8–31%), and Bacteroidetes (0–2%) (Supplementary Table 4). The Simpson reciprocal and Shannon alpha-diversity indices range from 7.9 (CUC4) to 34.1 (CUC9) for the Simpson reciprocal and 4.2 (CUC8) to 6.3 (CUC9) for the Shannon index (Supplementary Table 1). Although the weighted UniFrac clustering results indicate a distinct microbiota for cucumbers, we did observe variation in the abundances of predominant community members (Figures 1, 2, 5). For example, Acinetobacter were observed on all cucumbers, at proportional abundances ranging from 2 to 32% (Figure 5 and Supplementary Table 5). Rhizobium proportional abundances ranged from 50% in CUC4 to less than 0.5% in CUC7 and CUC9 (Figure 5 and Supplementary Table 5). Additionally, each cucumber harbored Pantoea at abundances of 4–25%, Microbacterium and Curtobacterium species (Figure 5, Supplementary Figure 5, and Supplementary Table 5). Like the masala spice mixes, unassigned Enterobacteriaceae OTUs were observed in four of the cucumber samples at proportional abundances ranging from 3% to 5%, most likely represent Enterobacter (Figure 5 and Supplementary Table 5). There were also a few distinguishing genera present at relatively high proportional abundances in some cucumbers. For example, Paenibacillus were observed at abundances of 31% in CUC5 and two Exiguobacterium OTUs were observed in cucumbers at proportional abundances of 14 and 20% in CUC8, and 5 and 9% in CUC7 (Figure 5 and Supplementary Table 5). In the Actinobacteria phyla, we observed Arthrobacter in CUC7 at proportional abundances of 25%, and 12% of these OTUs were classified as Arthrobacter defluvii (Figure 5 and Supplementary Table 5). Finally, some cucumbers harbored spoilage-associated bacteria such as Lactococcus, in CUC4 (2%) and CUC7 (1%), and Leuconostoc present in CUC3 (5%) (Figure 5 and Supplementary Table 5).
Species diversity in the mung bean sprouts were lower in SPA (brand A) than SPB (brand B). The Simpson reciprocal index ranges were 6.44–7.21 in SPA and 11.55–17.37 in SPB, and the Shannon indices ranged from 3.65 to 3.89 in SPA and 4.94 to 5.34 SPB (Supplementary Table 2). Firmicutes (81–85%) predominated in SPA followed by Proteobacteria (15–19%), Bacteroidetes (0.11–0.14%), and Actinobacteria (0–0.01%) while the SPB, harbored mostly Proteobacteria (96–99%) with proportionately less Firmicutes (0.42–2.72%); the Bacteroidetes (0.36–0.79%), and Actinobacteria (0–0.06%) abundances were similar to SPA (Supplementary Table 4). These differences were due to higher proportional abundances of spoilage-associated Leuconostoc (54%) and Lactococcus species (29%) in SPA that were present at very low levels in SPB, 0.04 and 1.3%, respectively (Figure 6 and Supplementary Table 5). Although the SPA microbiomes were less complex, we did identify 3% proportional abundances of Enterobacteriaceae incertae sedis OTUs, which likely represent Kosakonia cowanii, a recently classified species previously known as Enterobacter cowanii (Supplementary Table 5) (Brady et al., 2013).
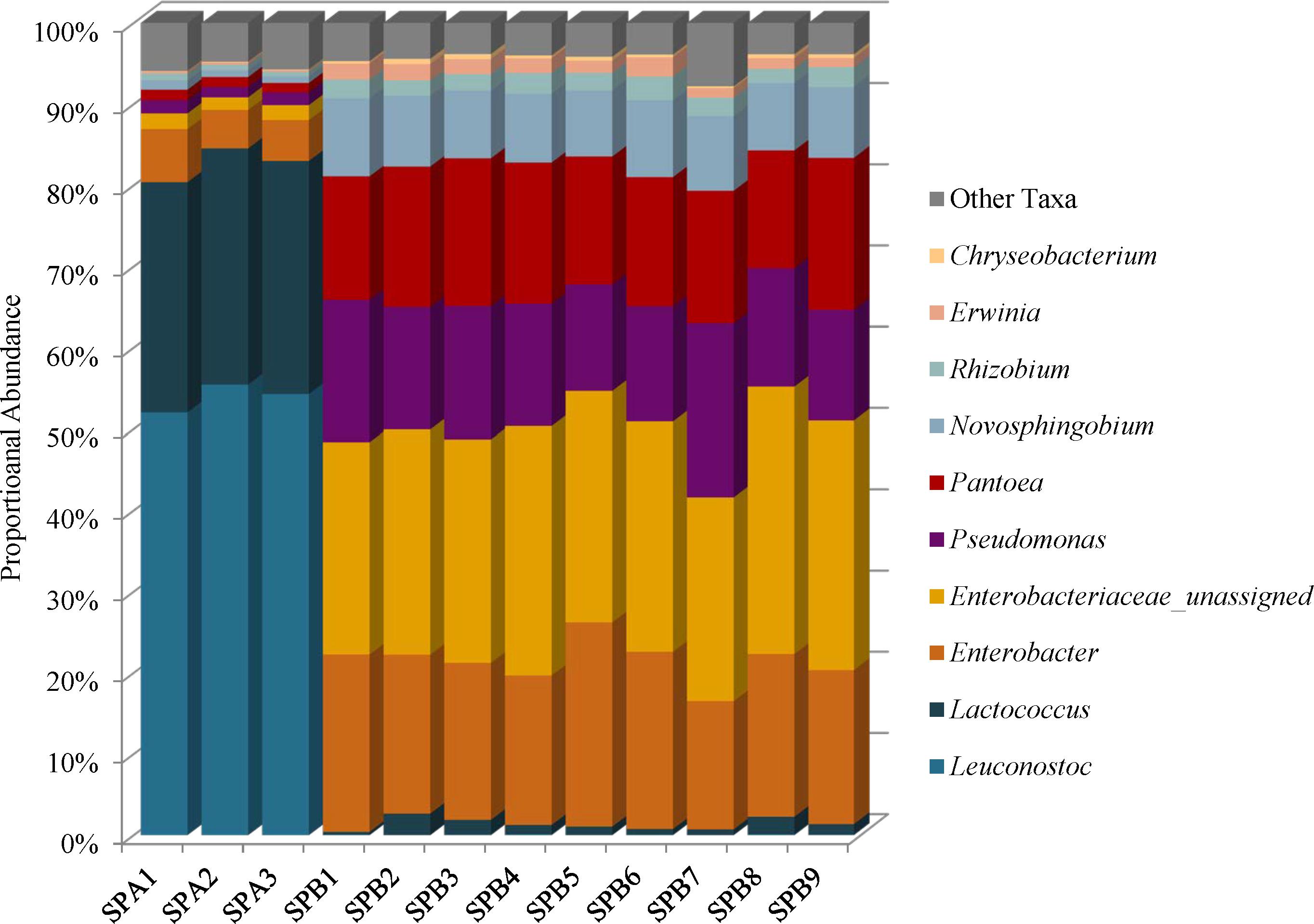
FIGURE 6. Genus level proportional abundances (≥1%) of three replicates of mung bean sprout brand A (SPA) and nine replicates of mung bean sprout B (SPB).
Unassigned Enterobacteriaceae (27%), Enterobacter (20%), Pseudomonas (16%), and Pantoea (16%) predominate in SPB, and these samples harbor the highest proportional abundances of Enterobacteriaceae of all foods in this study (Figure 6 and Supplementary Figure 1). The unassigned Enterobacteriaceae in SPB are mostly likely members of the Enterobacter genera (Figure 6 and Supplementary Table 5).
To highlight potential differences between plant and animal derived commodities, we sequenced three smoked salmon samples. Smoked salmon species diversity was lower than plant derived food species diversity; Simpson reciprocal and Shannon diversity indices ranged from 2.47 to 7.90 and 1.34 to 2.65, respectively (Supplementary Table 2). The microbiomes were comprised almost entirely of Proteobacteria and Firmicutes at proportional abundances that varied between the three samples such that SmkSalD harbored extremely low levels of Proteobacteria (0.41 and 0.15%) as compared to SmkSalE (76 and 51%) and SmlSalF (50 and 25%) (Supplementary Table 6). Likewise, the Firmicutes abundances varied from 99% in SmkSalD, 24 to 49% in SmkSalE and 50 to 75% in SmkSalF (Supplementary Table 6).
The Firmicutes identified in smoked salmon: Lactobacillus, Brochothrix, Carnobacterium, Leuconostoc, Vagococcus, and Lactococcus, are commonly associated with spoilage of refrigerated high-protein foods, such as meat and fish (Figure 7 and Supplementary Table 6). Lactobacillus abundances were highest in the SmkSalD replicates (57 and 55%) and dropped as low as 0.48% in SmkSalF (Figure 7 and Supplementary Table 6). Brochothrix predominated in SmkSalF with proportional abundances of 39 and 44% in F1 and F2, respectively (Figure 7 and Supplementary Table 6). SmkSalE also harbored Brochothrix but at much lower proportional abundances, 5% in E1 and 13% in E2; SmkSalD was almost deplete of this taxa with abundances less than 1% (Figure 7 and Supplementary Table 6). Two OTUs of Carnobacterium were observed in smoked salmon at levels that suggest competition. One Carnobacterium OTU reached abundances of 5% in SmkSalD and SmkSalF but abundances in SmkSalE were <1%, and abundances of the second OTU were highest in SmkSalD reaching 8% in D1 and 6% in D2, while remaining very low (<1%) in SmkSalE and SmkSalF (Figure 7 and Supplementary Table 6). Leuconostoc abundances were highest in SmkSalD (26%), followed by SmkSalE, and notably absent in SmkSalF. Two Vagococcus OTUs were observed in SmkSalF at proportional abundances of 2 and 7% in F1 and F2, respectively (Figure 7 and Supplementary Table 6).
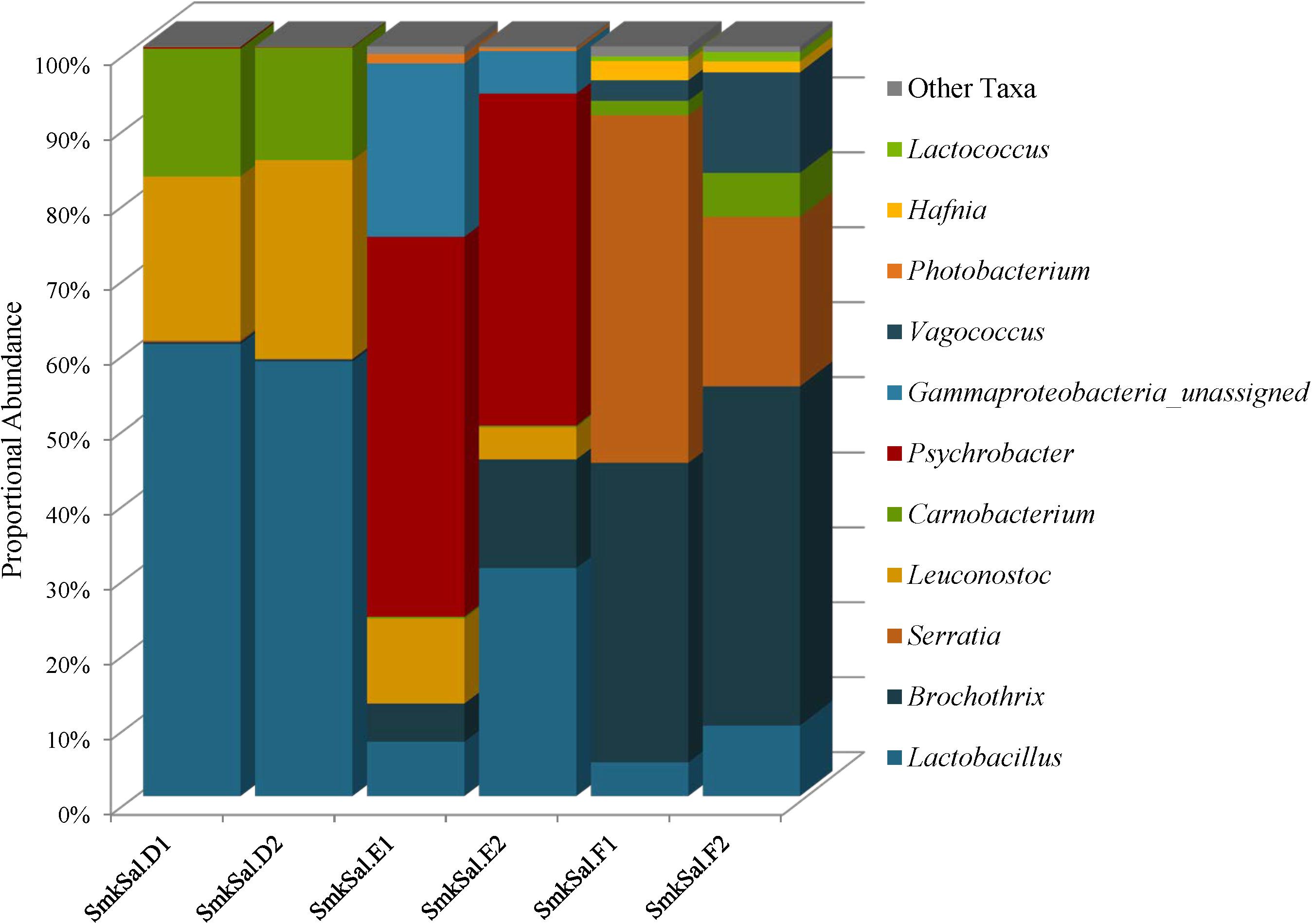
FIGURE 7. Genus level proportional abundances (≥1%) of three smoked salmon samples. Two replicates of each sample were sequenced.
Proteobacteria were only observed in SmkSalE and SmkSalF, and the members of this phyla were very different from those observed in plant derived foods. For example, Psychrobacter were observed in SmkSalE at proportional abundances of 22% (E1) and 23% (E2) (Figure 7 and Supplementary Table 6). SmkSalE also harbored unassigned Gammaproteobacteria OTUs at relatively high proportional abundances of 23% in E1 and 6% in E2, suggesting a novel community member or OTUs that could not be resolved beyond the class level with our analysis tool (Supplementary Table 6). The Proteobacteria in SmkSalF were distributed among three Serratia OTUs (Figure 7 and Supplementary Table 6). SmkSalF1 also harbored Hafnia OTUs at proportional abundances of 2% (Figure 7 and Supplementary Table 6).
Some genera were distinct to plant and animal derived foods. For example, Sphingomonas, Pseudomonas, Pantoea, Acinetobacter, and Erwinia were present in all the plant foods; the latter four cause food spoilage (Supplementary Table 7). Hafnia and Morganella, both commensals of the human gastrointestinal tract, Brochothrix, linked to meat spoilage, and Photobacterium, a bioluminescent marine bacteria of the Vibrionaceae family, were unique to smoked salmon. The masala spice mixes, which had the highest alpha diversity in our study, harbored 19 unique genera including food spoilage associated bacteria such as Bacillus, and Cronobacter which are known foodborne pathogens, and Pediococcus that is used in food fermentations (Supplementary Table 7). Mung bean sprout brand A harbored the spoilage associated bacteria, Leuconostoc and Klebsiella, as well as Comamonas genera that were not observed in brand B (Supplementary Table 7). Mung bean sprout brand B harbored six genera that we did not observe in brand A including Sphingomonas, Chryseobacterium, Lelliottia, Xanthomonas, Herbaspirillum, and Janthinobacterium (Supplementary Table 7). The cilantro samples harbored a unique cohort of Burkholderiales including Massilia, Duganella, Variovorax, and Acidovorax species (Supplementary Table 7). Other taxa unique to cilantro were Alkanindiges, Flavobacterium, Pedobacter, Shewanella, Paracoccus, Methylotenera, and Aquaspirillum (Supplementary Table 7). Carnobacterium and Psychrobacter were unique to smoked salmon and cilantro.
Identifying baseline microbiomes in foods is an important first step to observing bacteria that co-enrich with foodborne pathogens. Since the resident bacteria will resuscitate simultaneously with foodborne pathogens they may also compete for nutrients during the non-selective and selective culture enrichment steps and hamper pathogen detection. Thus, understanding these microbiome profiles may provide knowledge on improving culture enrichment methods. Subsequent to this study we incorporated heterotrophic plate counting into our workflow since it is possible and likely that some of the biomass we observed using 16S rRNA gene amplicon sequencing is from non-viable bacteria. Our results revealed bacterial loads of 4 log CFU/gm in smoked salmon, 5 log CFU/gm in garam masala spice mix, and 7–8 log CFU/gm in cilantro (unpublished data). Our findings support a previous microbiome study that demonstrated commodity driven bacterial diversity in conventionally and organically grown fresh fruits and vegetables (Leff and Fierer, 2013). For example, sprouts, spinach, lettuce, tomatoes, peppers, and strawberries, harbored high levels of Enterobacteriaceae and the microbiomes of apples, grapes, peaches, and mushrooms were predominated by Actinobacteria, Firmicutes, Bacteroidetes, and Proteobacteria (Leff and Fierer, 2013). The microbial community profiling in this study also expands our knowledge about bacteria that may hamper pathogen detection or impact shelf life (Daquigan et al., 2016; Ottesen et al., 2016; Grim et al., 2017).
Examining food microbiomes in a culture independent manner provides a comprehensive characterization of resident microbial communities and their dynamics that can be utilized in many ways in food-related pursuits. For example, microbiome characterization is extensively used in the study of fermented foods, to help bring about enhancements in the finished product. Detection of adulteration or contamination, whether it be economically based or accidental exposure to a microbial pathogen, and the study of spoilage events are other areas where food microbiome research is growing. Historically, the assessment of quality and safety of food commodities, such as spices, was achieved using standard microbiological methods such as aerobic plate counts, and biochemical phenotypic typing assays to quantify and identify target bacteria (Pafumi, 1986; Satchell et al., 1989; Garcia et al., 2001; Sagoo et al., 2009; Sospedra et al., 2010). In this study, we examined the bacterial microbiomes associated with several food commodities, to establish baseline data, which will be available publicly at the FDA MetagenomeTrakr bioproject at NCBI.
In our masala spice mix samples, we observed a high prevalence of Enterobacter and Bacillus species with 16S rRNA gene sequencing, in agreement with previous spice investigations that measured total bacterial loads ranging from 102 to 107 CFU/gm, with a predominance of Enterobacter and Bacillus (Pafumi, 1986; Satchell et al., 1989; Garcia et al., 2001; Sagoo et al., 2009; Sospedra et al., 2010). A survey of black peppercorns, white peppercorns, coriander, and fennel seeds identified Enterobacter cloacae as the most common Enterobacteriaceae member, which is consistent with our findings where proportional abundances of Enterobacter species ranged from 11 to 23% with additional abundances (7–13%) of unidentified Enterobacteriaceae that are likely Enterobacter species (Satchell et al., 1989). The high bacterial diversity and number of unique species (19) we observed in masala spice was expected since the spice mixtures tested were comprised of as many as 17 different ingredients and most published studies were of individual spices. Factors noted as likely contributors to variations in bacterial loads in spices included pre- and post-harvest practices and packaging methods. For example, a review of spices from Mexican markets that were non-packaged or packaged in polyethylene bags or glass containers, revealed relatively low levels of total and fecal coliform counts in the glass and non-packaged spices while 70% of the spices packaged in polyethylene bags contained 103 CFU/gm of these bacteria (Garcia et al., 2001). Spice harvest practices such as laying spices on the ground to dry and the necessity of human handling are likely responsible for the Staphylococcal abundances observed in this study as was previously shown in a survey of herbs and spices collected in Spain (Sospedra et al., 2010). Although in our study we observed Staphylococcus sciuri which is a commensal animal-associated bacteria suggesting animals as a route of adulteration (Nemeghaire et al., 2014). An advantage of 16S rRNA gene sequencing over microbiological testing is the identification of the entirety of a much more diverse population that includes organisms such as Pseudomonas, Pantoea, Acinetobacter, and Serratia that may be overlooked using standard microbiological culture methods since these methods tend to select for particular bacterial populations.
We extended upon our previous work with cilantro in this study in two ways: first, we employed paired end sequencing using the Illumina MiSeq which is much more robust than the 454-pyrosequencing used in our original study and second, we sequenced samples from two points in the supply chain, post-harvest (MDARD) and retail (Jarvis et al., 2015). We confirmed our original microbiome composition of mostly Proteobacteria with low levels of Firmicutes and Actinobacteria. We also observed differences between MDARD and retail cilantro with the retail samples containing higher proportional abundances of Flavobacterium (34%), commonly recovered from terrestrial (soil, rhizosphere, and phyllosphere) and aquatic environments (Kolton et al., 2013). A study of niche adaptation among Flavobacterium found evidence that terrestrial species have adapted to rhizosphere and phyllosphere niches where they can preferentially metabolize plant carbohydrates while aquatic species had larger genomes with higher ratios of peptide and protein utilization genes (Kolton et al., 2013). Most of the species we observed in the retail cilantro were from aquatic environments. Further research is required to identify handling practices that may have contributed to the higher abundances of Flavobacterium in CILSB retail samples, which could also be indicative of a more advanced state of spoilage in these samples since they were collected at a later stage in the farm to fork continuum than the MDARD cilantro (Blackburn, 2006; Wareing et al., 2010).
The cucumbers each harbored similar bacterial community members at different proportional abundances. Since cucumbers grow on the ground, it is likely that the local soil and rhizosphere bacteria contribute heavily to the cucumber microbiota. The predominance of Proteobacteria on all cucumbers tested in our study is corroborated by a study specific to cucumber rhizospheres, and other studies focused on the bacterial diversity of soil and rhizosphere biology (Spain et al., 2009; Mendes et al., 2013; Tian and Gao, 2014). Our identification of Acinetobacter on cucumbers and in the masala spice mixes, at much lower abundances, is notable since some species are known to cause nosocomial infections. Species of Acinetobacter are also known to harbor genes conferring antimicrobial resistance and, in a recent study focused on screening ready-to-eat produce (lettuce, apples, pears, bananas, and strawberries), Acinetobacter was isolated from 77.9% of the samples screened (Carvalheira et al., 2017). Furthermore, antimicrobial susceptibility screening of the Acinetobacter isolated from lettuce and fruits revealed resistance to 14 antimicrobial agents, including colistin, in 13.3% of the strains (Carvalheira et al., 2017). The notion of vegetables as transmission vehicles of Acinetobacter is not new as a vegetable study in 1999 identified antimicrobial resistant species of the A. calcoaceticus–A. baumannii complex in 17% of the vegetables tested (Berlau et al., 1999). The presence of Acinetobacter in cucumber microbiomes is also an indicator of decomposition since they are known to cause food spoilage. The cucumbers in our study also harbored the highest proportional abundances of gram-positive soil associated Actinobacteria (8–31%) such as Curtobacterium, Microbacterium, and Arthrobacter among the foods sequenced. Finally, the presence of Paenibacillus at an abundance of 31% is CUC5 is notable from a food safety perspective as members of this genera have been linked to inhibiting Salmonella growth (Lorentz et al., 2006; Luo et al., 2013; Allard et al., 2014; Knolhoff et al., 2015; Sprando et al., 2017). The FDA relies on culture methods to recover Salmonella from food and it is possible that Paenibacillus could hamper Salmonella recovery from contaminated commodities during outbreak investigations.
Our findings with the SPA mung bean sprout sample that contained high proportional abundances of Leuconostoc and Lactococcus, suggests that these sprouts were in an advanced stage of spoilage, compared to SPB. This is noteworthy, since we did not observe visible compositional changes to either mung bean sprout sample. Our analysis identified a mixture of Leuconostoc OTUs at proportional abundances reaching 50% which likely represent one of three L. gelidum subspecies; L. gelidum subsp. gelidum, L. gelidum subsp. gasicomitatum, and L. gelidum subsp. aenigmaticum that are associated with spoilage in modified-atmosphere-packaged (MAP) foods (Rahkila et al., 2015). These psychrotrophic lactic acid bacteria have been associated with spoilage in packaged meat and vegetable products (Rahkila et al., 2015). L. citreum, another lactic acid bacteria, was also presumptively identified in SPA at proportional abundances of 14%. Interestingly, both L. gelidum and L. citreum are favorable members of food microbial communities since they produce bacteriocins active against Listeria monocytogenes (Hastings et al., 1991; Van Belkum and Stiles, 1995; Pujato et al., 2014). The SPA samples also harbored high proportional abundances of Lactococcus species (29%), presumptively identified as L. raffinolactis (13%) and L. piscium (13%). L. piscium has been isolated from vacuum packaged refrigerated beef (Sakala et al., 2002) and it is the only Lactococcus species associated with food spoilage in MAP packaged beef and vegetables (Rahkila et al., 2012; Pothakos et al., 2014). The second set of mung bean sprout samples, SPB, had a similar microbiota composition to the one defined by Margot et al. (2016) and Leff and Fierer (2013) comprised mostly Proteobacteria. However, SPB contained predominantly Enterobacter species while the microbiota in Margot et al. (2016) had a predominance of Janthinobacterium and the bean sprouts sequence by Leff and Fierer (2013) harbored mostly Pantoea.
Our microbiome observations in smoked salmon revealed a predominance of Lactobacillus, Brochothrix, Leuconostoc, and Carnobacterium species, indicative of spoilage which was not surprising since these samples had been opened for at least 1 week by the time our laboratory received them and spoilage of smoked salmon happens rather rapidly even at refrigerated conditions (Chaillou et al., 2015). These species parallel those found in a microbiological study where the smoking process and type of packaging were implicated in shaping the spoilage-associated microbiota in smoked salmon (Leroi et al., 1998; Emborg et al., 2002; Olofsson et al., 2007; Mace et al., 2013). A recent 16S rRNA gene sequencing comparison of fresh and spoiled smoked salmon corroborate our findings of low species richness (Chaillou et al., 2015). Chaillou et al. (2015) reported fluctuations in alpha diversity from 189 ± 58 operational taxonomic units (OTUs) in fresh seafood to 27 ± 12 OTUs in spoiled products. The alpha diversity of the observed species in our smoked salmon samples were 180 (E1) and 181(E2), 141 (F1) and 132 (F2), and 89 in both SmkSalD replicates, indicating that they may have been at different stages of spoilage even though all three samples harbored high abundances of spoilage associated bacteria. However, the types of spoilage associated bacteria in our study varied between samples such that SmkSalD contained the highest levels of Lactobacillus, SmkSalE harbored high levels of Psychrobacter, and Brochothrix and SmkSalF harbored a predominance of Serratia. Chaillou et al. (2015) identified Brochothrix thermosphacta as the most ubiquitous and abundant species among all of the meat and seafood they tested.
All of the foods in this study harbored microbiome profiles that would eventually lead to bacterial spoilage. For example, the presence of Flavobacterium, Erwinia, Pantoea, Acinetobacter, and Pseudomonas species in all plant food microbiomes supports the notion that spoilage can be predicted by assessing food microbiome profiles since they are all known to contribute to food spoilage (Blackburn, 2006; Wareing et al., 2010; Parlapani et al., 2013; Gu et al., 2018). Additionally, psychrotolerant species such as Acinetobacter, Pseudomonas, Psychrobacter and others, that are capable of surviving and growing at cold temperatures will increase in numbers after harvest in cucumbers, cilantro, and mung bean sprouts that have cold storage requirements. The high abundances of spoilage associated Enterobacteriaceae (Erwinia and Pantoea), Pseudomonas, and Acinetobacter in mung bean sprout brand B and Leuconostoc and Lactococcus in brand A demonstrate different stages or types of bacterial spoilage in MAP foods. We also observed lactic acid bacteria with known biopreservative functions such as Lactobacillus and Carnobacterium. These genera are known to produce antimicrobial substances (organic acids, peroxides, and antimicrobial peptides) and are purposely added to some foods to preserve and protect against spoilage and pathogenic bacteria (Calix-Lara et al., 2014; Spanu et al., 2017). Not surprisingly the masala spices which had the highest alpha-diversity among the foods we sequenced also harbored the most diverse population of spoilage associated bacteria and they were the only food that contained Bacillus species.
Sequencing food microbiomes reveals key features about food safety and quality. For example, in foods with highly diverse and complex microbiomes such as cilantro, studies have shown that pathogen detection is hampered when closely related species are present (Grim et al., 2017). A spinach microbiome study corroborated this for Escherichia coli detection (Leonard et al., 2016). Our findings of various food spoilage bacteria in the mung bean sprouts and smoked salmon demonstrate the ability to assess food quality. The identification of clinically significant bacteria such as Staphylococcus and Acinetobacter suggest that food could play a role in transmission of organisms from the environment to humans. Finally, there is the notion that the microbiomes of foods that are consumed raw could have favorable contributions to our gut microbiomes. Supporting these hypotheses by defining core microbiomes of foods derived from different ecological niches will provide information that can be used to assess food health in a similar fashion to the application of human microbiome findings to human health. In this regard, we can also begin to understand the impact that food microbiomes have on gut health.
KJ, CG, AO, PR, and PM designed the project. KJ, ND, CG, PM, LH, and JM conducted the experiments. JW performed the bioinformatic analyses. All authors read and revised the manuscript.
This work was funded by the United States Food and Drug Administration.
JW is founder of Resphera Biosciences and has an equity position in the company.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank the Department of Agriculture and Rural Development in Lansing Michigan, the California Department of Public Health, and the FDA Northeast Food and Feed Laboratory for providing the food samples used in this study. We would like to thank Lili Fox Velez for reviewing and revising this manuscript and Marianna Sala-Rhatigan for technical assistance and support with genome sequencing. ND is an Oak Ridge Institute for Science and Education fellow, and we also thank the Department of Energy for their support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02540/full#supplementary-material
Abernethy, M. G., Rosenfeld, A., White, J. R., Mueller, M. G., Lewicky-Gaupp, C., and Kenton, K. (2017). Urinary microbiome and cytokine levels in women with interstitial cystitis. Obstet. Gynecol. 129, 500–506. doi: 10.1097/AOG.0000000000001892
Allard, S., Enurah, A., Strain, E., Millner, P., Rideout, S. L., Brown, E. W., et al. (2014). In situ evaluation of Paenibacillus alvei in reducing carriage of Salmonella enterica serovar newport on whole tomato plants. Appl. Environ. Microbiol. 80, 3842–3849. doi: 10.1128/AEM.00835-14
Angelo, K. M., Chu, A., Anand, M., Nguyen, T. A., Bottichio, L., Wise, M., et al. (2015). Outbreak of Salmonella newport infections linked to cucumbers–United States, 2014. Morb. Mortal. Wkly. Rep. 64, 144–147.
Arumugam, M., Raes, J., Pelletier, E., Le Paslier, D., Yamada, T., Mende, D. R., et al. (2011). Enterotypes of the human gut microbiome. Nature 473, 174–180. doi: 10.1038/nature09944
Bayer, C., Bernard, H., Prager, R., Rabsch, W., Hiller, P., and Malorny, B. (2014). An outbreak of Salmonella newport associated with mung bean sprouts in Germany and the Netherlands, October to November 2011. Euro Surveill. 19:20665. doi: 10.2807/1560-7917.ES2014.19.1.20665
Berlau, J., Aucken, H. M., Houang, E., and Pitt, T. L. (1999). Isolation of Acinetobacter spp including A. baumannii from vegetables: implications for hospital-acquired infections. J. Hosp. Infect. 42, 201–204. doi: 10.1053/jhin.1999.0602
Blackburn, C. (2006). Food Spoilage Microorganisms. Cambridge: Woodhead Publishing Limited. doi: 10.1201/9781439824573
Bottichio, L., Medus, C., Sorenson, A., Donovan, D., Sharma, R., Dowell, N., et al. (2016). Outbreak of Salmonella oslo infections linked to persian cucumbers - United States, 2016. Morb. Mortal. Wkly. Rep. 65, 1430–1433. doi: 10.15585/mmwr.mm655051a3
Brady, C., Cleenwerck, I., Venter, S., Coutinho, T., and De Vos, P. (2013). Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov. as Lelliottia nimipressuralis comb. nov. and Lelliottia amnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov. as Pluralibacter gergoviae comb. nov. and Pluralibacter pyrinus comb. nov., respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov. as Kosakonia cowanii comb. nov., Kosakonia radicincitans comb. nov., Kosakonia oryzae comb. nov. and Kosakonia arachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis nom. nov., Cronobacter helveticus comb. nov. and Cronobacter pulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacter. Syst. Appl. Microbiol. 36, 309–319. doi: 10.1016/j.syapm.2013.03.005
Brillat-Savarin, J. A. (2010). A Handbook of Gastronomy. Morrisville, NC: Lulu Enterprises Incorporated.
Calix-Lara, T. F., Rajendran, M., Talcott, S. T., Smith, S. B., Miller, R. K., Castillo, A., et al. (2014). Inhibition of Escherichia coli O157:H7 and Salmonella enterica on spinach and identification of antimicrobial substances produced by a commercial lactic acid bacteria food safety intervention. Food Microbiol. 38, 192–200. doi: 10.1016/j.fm.2013.09.006
Campbell, J. V., Mohle-Boetani, J., Reporter, R., Abbott, S., Farrar, J., Brandl, M., et al. (2001). An outbreak of Salmonella serotype Thompson associated with fresh cilantro. J. Infect. Dis. 183, 984–987. doi: 10.1086/319254
Carvalheira, A., Silva, J., and Teixeira, P. (2017). Lettuce and fruits as a source of multidrug resistant Acinetobacter spp. Food Microbiol. 64, 119–125. doi: 10.1016/j.fm.2016.12.005
CDC (2013). Outbreak of Escherichia coli O104:H4 infections associated with sprout consumption - Europe and North America. May-July 2011. Morb. Mortal. Wkly. Rep. 62, 1029–1031.
Chaillou, S., Chaulot-Talmon, A., Caekebeke, H., Cardinal, M., Christieans, S., Denis, C., et al. (2015). Origin and ecological selection of core and food-specific bacterial communities associated with meat and seafood spoilage. ISME J. 9, 1105–1118. doi: 10.1038/ismej.2014.202
Costea, P. I., Hildebrand, F., Arumugam, M., Backhed, F., Blaser, M. J., Bushman, F. D., et al. (2018). Enterotypes in the landscape of gut microbial community composition. Nat. Microbiol. 3, 8–16. doi: 10.1038/s41564-017-0072-8
Daquigan, N., Grim, C. J., White, J. R., Hanes, D. E., and Jarvis, K. G. (2016). Early recovery of Salmonella from food using a 6-Hour non-selective pre-enrichment and reformulation of tetrathionate broth. Front. Microbiol. 7:2103. doi: 10.3389/fmicb.2016.02103
Dechet, A. M., Herman, K. M., Chen Parker, C., Taormina, P., Johanson, J., Tauxe, R. V., et al. (2014). Outbreaks caused by sprouts, United States, 1998-2010: lessons learned and solutions needed. Foodborne Pathog. Dis. 11, 635–644. doi: 10.1089/fpd.2013.1705
Drewes, J. L., White, J. R., Dejea, C. M., Fathi, P., Iyadorai, T., Vadivelu, J., et al. (2017). High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes 3:34. doi: 10.1038/s41522-017-0040-3
Emborg, J., Laursen, B. G., Rathjen, T., and Dalgaard, P. (2002). Microbial spoilage and formation of biogenic amines in fresh and thawed modified atmosphere-packed salmon (Salmo salar) at 2 degrees C. J. Appl. Microbiol. 92, 790–799. doi: 10.1046/j.1365-2672.2002.01588.x
Friesema, I., De Jong, A., Hofhuis, A., Heck, M., Van Den Kerkhof, H., De Jonge, R., et al. (2014). Large outbreak of Salmonella Thompson related to smoked salmon in the Netherlands, August to December 2012. Euro Surveill. 19:20918. doi: 10.2807/1560-7917.ES2014.19.39.20918
Garcia, S., Iracheta, F., Galvan, F., and Heredia, N. (2001). Microbiological survey of retail herbs and spices from Mexican markets. J. Food Prot. 64, 99–103. doi: 10.4315/0362-028X-64.1.99
Gram, L., Ravn, L., Rasch, M., Bruhn, J. B., Christensen, A. B., and Givskov, M. (2002). Food spoilage—interactions between food spoilage bacteria. Int. J. Food Microbiol. 78, 79–97. doi: 10.1016/S0168-1605(02)00233-7
Grim, C. J., Daquigan, N., Lusk Pfefer, T. S., Ottesen, A. R., White, J. R., and Jarvis, K. G. (2017). High-resolution microbiome profiling for detection and tracking of Salmonella enterica. Front. Microbiol. 8:1587. doi: 10.3389/fmicb.2017.01587
Gu, G., Ottesen, A., Bolten, S., Ramachandran, P., Reed, E., Rideout, S., et al. (2018). Shifts in spinach microbial communities after chlorine washing and storage at compliant and abusive temperatures. Food Microbiol. 73, 73–84. doi: 10.1016/j.fm.2018.01.002
Hastings, J. W., Sailer, M., Johnson, K., Roy, K. L., Vederas, J. C., and Stiles, M. E. (1991). Characterization of leucocin A-UAL 187 and cloning of the bacteriocin gene from Leuconostoc gelidum. J. Bacteriol. 173, 7491–7500. doi: 10.1128/jb.173.23.7491-7500.1991
Human Microbiome Project Consortium (2012a). A framework for human microbiome research. Nature 486, 215–221. doi: 10.1038/nature11209
Human Microbiome Project Consortium (2012b). function and diversity of the healthy human microbiome. Nature 486, 207–214. doi: 10.1038/nature11234
Jarvis, K. G., White, J. R., Grim, C. J., Ewing, L., Ottesen, A. R., Beaubrun, J. J., et al. (2015). Cilantro microbiome before and after nonselective pre-enrichment for Salmonella using 16S rRNA and metagenomic sequencing. BMC Microbiol. 15:160. doi: 10.1186/s12866-015-0497-2
Knolhoff, A. M., Zheng, J., Mcfarland, M. A., Luo, Y., Callahan, J. H., Brown, E. W., et al. (2015). Identification and structural characterization of naturally-occurring broad-spectrum cyclic antibiotics isolated from Paenibacillus. J. Am. Soc. Mass Spectrom. 26, 1768–1779. doi: 10.1007/s13361-015-1190-2
Kolton, M., Sela, N., Elad, Y., and Cytryn, E. (2013). Comparative genomic analysis indicates that niche adaptation of terrestrial Flavobacteria is strongly linked to plant glycan metabolism. PLoS One 8:e76704. doi: 10.1371/journal.pone.0076704
Leff, J. W., and Fierer, N. (2013). Bacterial communities associated with the surfaces of fresh fruits and vegetables. PLoS One 8:e59310. doi: 10.1371/journal.pone.0059310
Lehmacher, A., Bockemuhl, J., and Aleksic, S. (1995). Nationwide outbreak of human salmonellosis in Germany due to contaminated paprika and paprika-powdered potato chips. Epidemiol. Infect. 115, 501–511. doi: 10.1017/S0950268800058660
Leonard, S. R., Mammel, M. K., Lacher, D. W., and Elkins, C. A. (2015). Application of metagenomic sequencing to food safety: detection of Shiga Toxin-producing Escherichia coli on fresh bagged spinach. Appl. Environ. Microbiol. 81, 8183–8191. doi: 10.1128/AEM.02601-15
Leonard, S. R., Mammel, M. K., Lacher, D. W., and Elkins, C. A. (2016). Strain-level discrimination of shiga toxin-producing Escherichia coli in spinach using metagenomic sequencing. PLoS One 11:e0167870. doi: 10.1371/journal.pone.0167870
Leroi, F., Joffraud, J. J., Chevalier, F., and Cardinal, M. (1998). Study of the microbial ecology of cold-smoked salmon during storage at 8 degrees C. Int. J. Food Microbiol. 39, 111–121. doi: 10.1016/S0168-1605(97)00126-8
Lorentz, R. H., Artico, S., Silveira, A. B., Einsfeld, A., and Corcao, G. (2006). Evaluation of antimicrobial activity in Paenibacillus spp. strains isolated from natural environment. Lett. Appl. Microbiol. 43, 541–547. doi: 10.1111/j.1472-765X.2006.01995.x
Lozupone, C., and Knight, R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005
Luo, Y., Wang, C., Allard, S., Strain, E., Allard, M. W., Brown, E. W., et al. (2013). Draft genome sequences of Paenibacillus alvei A6-6i and TS-15. Genome Announc. 1:e00673-13. doi: 10.1128/genomeA.00673-13
Mace, S., Joffraud, J. J., Cardinal, M., Malcheva, M., Cornet, J., Lalanne, V., et al. (2013). Evaluation of the spoilage potential of bacteria isolated from spoiled raw salmon (Salmo salar) fillets stored under modified atmosphere packaging. Int. J. Food Microbiol. 160, 227–238. doi: 10.1016/j.ijfoodmicro.2012.10.013
Margot, H., Stephan, R., and Tasara, T. (2016). Mungo bean sprout microbiome and changes associated with culture based enrichment protocols used in detection of Gram-negative foodborne pathogens. Microbiome 4, 1–13. doi: 10.1186/s40168-016-0193-y
Mendes, R., Garbeva, P., and Raaijmakers, J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. doi: 10.1111/1574-6976.12028
Mohle-Boetani, J. C., Farrar, J., Bradley, P., Barak, J. D., Miller, M., Mandrell, R., et al. (2009). Salmonella infections associated with mung bean sprouts: epidemiological and environmental investigations. Epidemiol. Infect. 137, 357–366. doi: 10.1017/S0950268808000411
Nemeghaire, S., Argudín, M. A., Feßler, A. T., Hauschild, T., Schwarz, S., and Butaye, P. (2014). The ecological importance of the Staphylococcus sciuri species group as a reservoir for resistance and virulence genes. Vet. Microbiol. 171, 342–356. doi: 10.1016/j.vetmic.2014.02.005
Olofsson, T. C., Ahrne, S., and Molin, G. (2007). The bacterial flora of vacuum-packed cold-smoked salmon stored at 7 degrees C, identified by direct 16S rRNA gene analysis and pure culture technique. J. Appl. Microbiol. 103, 109–119. doi: 10.1111/j.1365-2672.2006.03216.x
Ottesen, A., Ramachandran, P., Reed, E., White, J. R., Hasan, N., Subramanian, P., et al. (2016). Enrichment dynamics of Listeria monocytogenes and the associated microbiome from naturally contaminated ice cream linked to a listeriosis outbreak. BMC Microbiol. 16:275. doi: 10.1186/s12866-016-0894-1
Ottesen, A. R., Gonzalez, A., Bell, R., Arce, C., Rideout, S., Allard, M., et al. (2013). Co-enriching microflora associated with culture based methods to detect Salmonella from tomato phyllosphere. PLoS One 8:e73079. doi: 10.1371/journal.pone.0073079
Pafumi, J. (1986). Assessment of the microbiological quality of spices and herbs. J. Food Prot. 49, 958–963. doi: 10.4315/0362-028X-49.12.958
Parlapani, F. F., Meziti, A., Kormas, K. A., and Boziaris, I. S. (2013). Indigenous and spoilage microbiota of farmed sea bream stored in ice identified by phenotypic and 16S rRNA gene analysis. Food Microbiol. 33, 85–89. doi: 10.1016/j.fm.2012.09.001
Pothakos, V., Snauwaert, C., De Vos, P., Huys, G., and Devlieghere, F. (2014). Psychrotrophic members of Leuconostoc gasicomitatum, Leuconostoc gelidum and Lactococcus piscium dominate at the end of shelf-life in packaged and chilled-stored food products in Belgium. Food Microbiol. 39, 61–67. doi: 10.1016/j.fm.2013.11.005
Pujato, S. A., Del, L. Q. A., Candioti, M. C., Reinheimer, J. A., and Guglielmotti, D. M. (2014). Leuconostoc citreum MB1 as biocontrol agent of Listeria monocytogenes in milk. J. Dairy Res. 81, 137–145. doi: 10.1017/S002202991300068X
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. doi: 10.1038/nature08821
Rahkila, R., Johansson, P., Sade, E., Paulin, L., Auvinen, P., and Bjorkroth, J. (2015). Multilocus sequence typing of Leuconostoc gelidum subsp. gasicomitatum, a Psychrotrophic lactic acid bacterium causing spoilage of packaged perishable foods. Appl. Environ. Microbiol. 81, 2474–2480. doi: 10.1128/AEM.04013-14
Rahkila, R., Nieminen, T., Johansson, P., Säde, E., and Björkroth, J. (2012). Characterization and evaluation of the spoilage potential of Lactococcus piscium isolates from modified atmosphere packaged meat. Int. J. Food Microbiol. 156, 50–59. doi: 10.1016/j.ijfoodmicro.2012.02.022
Rorvik, L. M. (2000). Listeria monocytogenes in the smoked salmon industry. Int. J. Food Microbiol. 62, 183–190. doi: 10.1016/S0168-1605(00)00334-2
Sagoo, S. K., Little, C. L., Greenwood, M., Mithani, V., Grant, K. A., Mclauchlin, J., et al. (2009). Assessment of the microbiological safety of dried spices and herbs from production and retail premises in the United Kingdom. Food Microbiol. 26, 39–43. doi: 10.1016/j.fm.2008.07.005
Sakala, R. M., Hayashidani, H., Kato, Y., Kaneuchi, C., and Ogawa, M. (2002). Isolation and characterization of Lactococcus piscium strains from vacuum-packaged refrigerated beef. J. Appl. Microbiol. 92, 173–179. doi: 10.1046/j.1365-2672.2002.01513.x
Satchell, F. B., Bruce, V. R., Allen, G., Andrews, W. H., and Gerber, H. R. (1989). Microbiological survey of selected imported spices and associated fecal pellet specimens. J. Assoc. Off. Anal. Chem. 72, 632–637.
Sospedra, I., Soriano, J. M., and Manes, J. (2010). Assessment of the microbiological safety of dried spices and herbs commercialized in Spain. Plant Foods Hum. Nutr. 65, 364–368. doi: 10.1007/s11130-010-0186-0
Spain, A. M., Krumholz, L. R., and Elshahed, M. S. (2009). Abundance, composition, diversity and novelty of soil Proteobacteria. ISME J. 3, 992–1000. doi: 10.1038/ismej.2009.43
Spanu, C., Scarano, C., Piras, F., Spanu, V., Pala, C., Casti, D., et al. (2017). Testing commercial biopreservative against spoilage microorganisms in MAP packed Ricotta fresca cheese. Food Microbiol. 66, 72–76. doi: 10.1016/j.fm.2017.04.008
Sprando, R. L., Black, T., Olejnik, N., Keltner, Z., Topping, V., Ferguson, M., et al. (2017). Assessing the effect of oral exposure to Paenibacillus alvei, a potential biocontrol agent, in male, non-pregnant, pregnant animals and the developing rat fetus. Food Chem. Toxicol. 103, 203–213. doi: 10.1016/j.fct.2017.03.009
Tian, Y., and Gao, L. (2014). Bacterial diversity in the rhizosphere of cucumbers grown in soils covering a wide range of cucumber cropping histories and environmental conditions. Microb. Ecol. 68, 794–806. doi: 10.1007/s00248-014-0461-y
Van Belkum, M. J., and Stiles, M. E. (1995). Molecular characterization of genes involved in the production of the bacteriocin leucocin A from Leuconostoc gelidum. Appl. Environ. Microbiol. 61, 3573–3579.
Van Doren, J. M., Neil, K. P., Parish, M., Gieraltowski, L., Gould, L. H., and Gombas, K. L. (2013). Foodborne illness outbreaks from microbial contaminants in spices, 1973-2010. Food Microbiol. 36, 456–464. doi: 10.1016/j.fm.2013.04.014
Keywords: microbiome, 16S rRna, food, metagenomics, produce, spices, seafood
Citation: Jarvis KG, Daquigan N, White JR, Morin PM, Howard LM, Manetas JE, Ottesen A, Ramachandran P and Grim CJ (2018) Microbiomes Associated With Foods From Plant and Animal Sources. Front. Microbiol. 9:2540. doi: 10.3389/fmicb.2018.02540
Received: 18 May 2018; Accepted: 04 October 2018;
Published: 23 October 2018.
Edited by:
David Rodriguez-Lazaro, University of Burgos, SpainReviewed by:
Julien Tremblay, National Research Council Canada (NRC-CNRC), CanadaCopyright © 2018 Jarvis, Daquigan, White, Morin, Howard, Manetas, Ottesen, Ramachandran and Grim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Karen G. Jarvis, a2FyZW4uamFydmlzQGZkYS5oaHMuZ292
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.