
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 02 November 2018
Sec. Microbial Symbioses
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.02485
 Sanqiang Gong1
Sanqiang Gong1 GuangJun Chai1
GuangJun Chai1 Yilin Xiao1
Yilin Xiao1 Lijia Xu2
Lijia Xu2 Kefu Yu2
Kefu Yu2 Jinlong Li1
Jinlong Li1 Fang Liu1
Fang Liu1 Hao Cheng1
Hao Cheng1 Fengli Zhang1
Fengli Zhang1 Baolin Liao3
Baolin Liao3 Zhiyong Li1*
Zhiyong Li1*The coral symbiont Symbiodinium plays important roles in the adaptation of coral to environmental changes. However, coral-Symbiodinium symbiotic associations are not well-understood in the South China Sea (SCS) whilst considering environmental factors and host taxa. In this study, next-generation sequencing of the internal transcribed spacer region 2 (ITS2) marker gene was used to explore the symbiotic associations between Symbiodinium and five typical coral species across tropical and subtropical reef regions of the SCS. The results showed that Acropora sp., Galaxea fascicularis, Platygyra lamellina, and Sarcophyton glaucum exhibited distinct Symbiodinium compositions between tropical and subtropical reef regions, whereas Porites lutea had stable Symbiodinium compositions. More heterogeneous Symbiodinium compositions among different coral species were observed in the tropical region, but there were no statistically significant differences in Symbiodinium compositions among different coral species in subtropical reef regions. There was a correlation between the Symbiodinium compositions and environmental factors, except for the composition of P. lutea. Symbiodinium subclades D1, D2, C71, C71a, C21, C3b, and C161 were primarily explained by the seawater temperature, nitrate, ammonia, and phosphate. Several host-specific Symbiodinium subclades (e.g., C15, C15.6, and C91) were observed in P. lutea as well. The findings of this study demonstrate the relationship of Symbiodinium diversity with coral hosts and the environment are helpful for elucidating the adaptation of corals to global climate change and anthropogenic disturbance.
Coral symbiont Symbiodinium, a photosynthetic algae that is a dinoflagellate, plays a central role in the adaptation of corals and the maintenance of coral reefs (Kemp et al., 2014). Consequently, monitoring Symbiodinium compositions and their mutually symbiotic associations with different coral hosts under different environmental stresses is essential to further project the fate of corals and to develop coral reef conservation strategies.
At present, nine Symbiodinium clades (known as clades A–I) have been identified (Pochon et al., 2014). Clades A–D are the main Symbiodinium harbored by corals, and certain other clades, such as F/G, have also been found in corals (Baker, 2003). Within any given clade, different Symbiodinium subclades are mainly identified by the commonly used polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE) profiles of the less-conserved Symbiodinium internal transcribed spacer region 2 (ITS2) sequence (LaJeunesse et al., 2003). Previous studies have revealed that the environment could influence Symbiodinium compositions, as well as their mutually symbiotic associations with corals, and the breakdown of Symbiodinium-coral associations because of the effects of climate change and anthropogenic disturbance that would contribute to dramatic declines in coral reef cover worldwide (Hughes et al., 2003, 2017; Hoegh-Guldberg et al., 2007). In contrast, certain coral species still maintain highly stable coral-Symbiodinium symbiotic associations over broad geographical ranges and temperature gradients, as well as through bleaching events or after transplantation (LaJeunesse et al., 2004a,b; Stat et al., 2009; Thomas et al., 2014). Therefore, corals might adapt to environmental changes via different mechanisms (Warner et al., 2006). For example, most corals can adapt to environmental changes by changing their symbiont communities, but some corals (most notably the genus Porites) remain common on devastated reefs, even though they fail to change their symbiont communities.
The South China Sea (SCS) has extensive an coral reef development (at least 8,000 km2, including at least 571 coral species), and coral reefs mainly develop in the Nansha Islands, Zhongsha Islands, Xisha Islands, Hainan Island, Leizhou Peninsula, and Dongsha Islands (Yu, 2012; Huang et al., 2015). The Symbiodinium subclades associated with coral species in the Nansha Islands (Huang et al., 2006), Xisha Islands (Dong et al., 2009), Dongsha Islands (Dong et al., 2008a), Hainan Island (specifically the Luhuitou fringing reef, Dong et al., 2008b), and Taiwan Island (Chen et al., 2005a,b; Keshavmurthy et al., 2014, 2017) have mainly been investigated using DGGE-based analyses. However, the DGGE-based method is unable to detect Symbiodinium subclades with relative abundances of less than 10% and, thus, may lack sufficient sensitivity to detect the change in symbiosis between corals and Symbiodinium (Mieog et al., 2007). The next-generation sequencing of Symbiodinium ITS2 region offers a highly sensitive method that is able to detect low-abundance Symbiodinium subclades and is reliable enough for measurements of Symbiodinium subclade compositions in corals (Arif et al., 2014; Thomas et al., 2014; Boulotte et al., 2016; Cunning et al., 2017). Most recently, Tong et al. (2017) analyzed Symbiodinium subclade compositions in Galaxea fascicularis (scleractinian) and Montipora spp. (scleractinian) in tropical (Hainan Island and Xisha Island) and subtropical (Hong Kong) reef regions of the SCS by next-generation sequencing of Symbiodinium ITS2 region, suggesting that temperature is the main factor that drives coral-Symbiodinium symbiotic associations. A similar result was also reported by Zhou et al. (2017) in G. fascicularis in the SCS. However, knowledge gaps still remain. At present, little is known regarding the diversity of Symbiodinium and their symbiotic associations with abundant corals (such as Acropora and Porites) in the SCS whilst considering environmental factors and host taxa.
In this study, the dominant coral species of Acropora sp. (scleractinian) and Porites lutea (scleractinian) as well as the three common coral species of G. fascicularis, Platygyra lamellina (scleractinian), and Sarcophyton glaucum (alcyonacea) were sampled from different reef sites across tropical (Sanya Bay-SY, Hainan Island) and subtropical (Xuwen County-XW, Leizhou Peninsula, and/or Daya Bay-DY) reef regions of the SCS. The Symbiodinium compositions in these coral samples in the SY, XW, and DY reef regions were analyzed by next-generation sequencing of Symbiodinium ITS2 region, aiming to elucidate coral-Symbiodinium symbiotic associations across these regions. Furthermore, the relationship of Symbiodinium compositions with environmental factors and host taxa were analyzed.
Sampling regions of SY (E109.470°-109.489°, N18.200°-18.217°), XW (E109.867°-109.869°, N20.333°-20.332°), and DY (E114.621°-114.611°, N22.833°-22.762°) are located in the northern SCS (Figure 1). The SY reef region, Hainan Island, China has a typical tropical ocean climate, and the mean annual surface seawater temperature (SST) in 2015 was 26.59°C (Supplementary Table 1); this region has been listed as part of the National Coral Reef Natural Reserves since 1990 (Li et al., 2008). The XW reef region, Leizhou Peninsula, is located on the northern coast of the SCS with a subtropical climate (Yu, 2000), and the mean annual SST of XW reef region in 2015 was 23.71°C (Supplementary Table 1). The XW reef region has been listed as part of the National Coral Reef Natural Reserves since 2007. The DY reef region is located to the southeast of Shenzhen City of Guangdong Province, China and has a typical subtropical ocean climate (Chen et al., 2009), and the mean annual SST in this region in 2015 was 21.50°C (Supplementary Table 1).
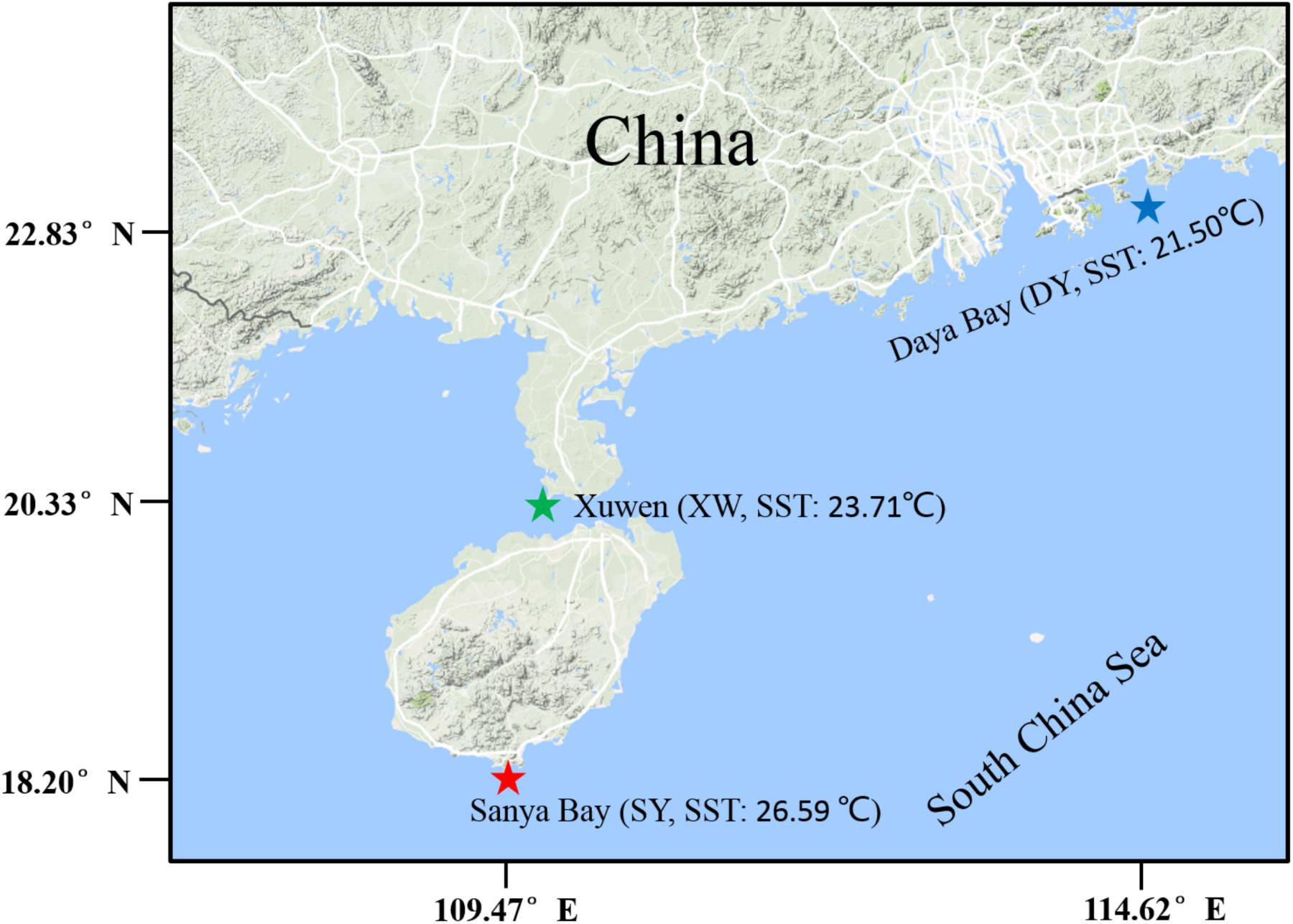
FIGURE 1. Sampling locations. Sampling regions are indicated by stars. The red star represents the tropical reef region of Sanya Bay (SY), the green star represents the subtropical reef region of Xuwen (XW) County, and the blue star represents the subtropical reef region of Daya Bay (DY).
Coral samples of Acropora sp., G. fascicularis, P. lamellina, P. lutea, and S. glaucum were obtained from the SY, XW, and DY reef regions in August and September 2015 (Table 1). These coral samples were identified based on the morphology and a molecular barcode analysis of the cytb gene. The cytb gene was amplified by PCR with previously reported primers for corals (Fukami et al., 2004), and it was cloned into the pEASY’T5 Zero Cloning Vector (Transgene Biotech, Beijing, China). Clone per coral sample was picked up for further Sanger sequencing. An alignment analysis of cytb gene was performed by BLASTN against the NCBI database1. Coral samples with cytb gene sequence similarity ≥98% and with similar morphology were identified as the same coral species. The representative sequences of cytb genes obtained in this study have been submitted to the NCBI database with accession numbers MH160422–MH160426 (Supplementary Data Sheet 1). At each reef region, coral colonies from different reef sites (at least three reef sites) separated by at least 100 m were collected. Fragments of approximately 1 cm2 were picked and washed with filtered seawater (0.22-μm polycarbonate membranes) at least three times.
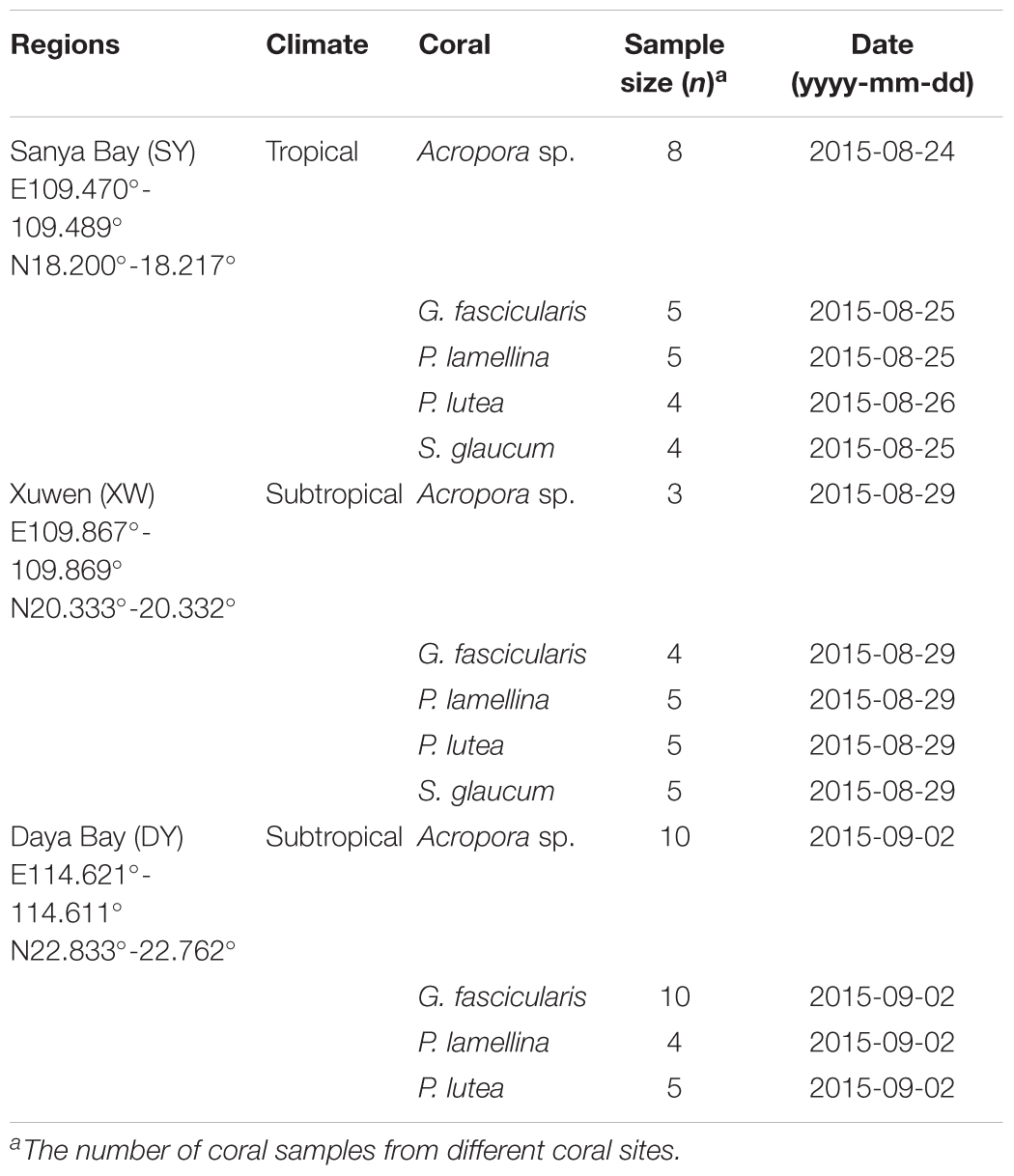
TABLE 1. Coral samples from SY, XW, and DY reef regions of the northern SCS, including sampling regions (latitudes and longitudes), climate, and coral samples information.
The seawater samples from at least five reef sites in each reef region were collected and filtered using 0.22-μm polycarbonate membranes (Whatman GF/F, 47 mm). Next, the filtered seawater from each of the different reef sites, that is, SY, XW, and DY reef regions, were mixed. All samples were preserved in RNAlater® at 4°C in the field and stored at -80°C in the lab until DNA extraction was conducted.
The mean annual SSTs of the SY, XW, and DY reef regions were calculated based on the SST of these reef regions in spring, summer, autumn, and winter of 2015 (Supplementary Table 1).
The SST, salinity, and depth of different reef regions (at least five reef sites in each reef region) were measured by OCEAN SEVEN 320 Plus CTD (Idronaut, Italy). Dissolved oxygen (DO) was detected using a YSI 6600V2-02 multi-parameter instrument (YSI, United States). Dissolved nutrients (nitrite-NO2-, nitrate-NO3-, ammonia-NH4+, and phosphate-PO43-) were analyzed using a Lachat QC8500 Flow Injection Autoanalyzer (Lachat Instruments, United States) (Supplementary Table 2). The differences in environmental factors among different coral reef regions were tested using a multiple comparison test (Tukey’s HSD) in the R software environment (R 3.1.2).
Total DNA of each coral sample was extracted using the Qiagen DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The integrity of the DNA was monitored by 0.8% (w/v) agarose gel electrophoresis stained with 1 × SYBR Safe (Invitrogen, Carlsbad, United States). The amount of DNA was determined by a Qubit® 3.0 Fluorometer (Life Technologies, United States). The purity of the DNA was determined by NanoDropTM 2000 Spectrophotometer (Thermo Scientific, United States). All DNA samples were stored at -20°C for further use.
The ITS2 region of the ribosomal RNA gene of Symbiodinium was PCR amplified using a pair of barcoded primers: ITSintfor2 (5′-GAATTGCAGAACTCCGTG-3′) and ITS2-reverse (5′-GGGATCCATATGCTTAAGTTCAGCGGGT-3′) (LaJeunesse and Trench, 2000). The PCR was performed with 12.5 μl of a Qiagen Multiplex PCR reagent (Qiagen, Hilden, Germany), 0.1 μM primer, 50 ng of DNA, and DNase-free water to make a total volume of 25 μl. The following PCR conditions were used: initial denaturation for 3 min at 94°C, followed by 34 cycles of 98°C for 10 s, 51°C for 30 s, 68°C for 30 s, and a final extension step of 5 min at 68°C. The DNA libraries were validated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, United States) and were quantified using a Qubit® 3.0 Fluorometer (Life Technologies, United States). All qualified amplification products were mixed in equal amounts followed by sequencing on an Illumina MiSeq instrument (Illumina, San Diego, CA, United States) according to the manufacturer’s instructions using a 300 × 2 paired-end configuration. The raw data were submitted to the NCBI Sequence Read Archive (SUB2393447).
According to the methods and script supplied by Cunning et al. (2017), paired reads from each sample were merged using the illumina-utils software (Eren et al., 2013) with an enforced Q30-check. Only sequences with overlap ≥150 bp and mismatch ≤3 bp were retained. Chimeric sequences were removed using USEARCH 6.1 (Edgar, 2010) implemented in QIIME (Caporaso et al., 2010). Primers were trimmed using cutadapt (Martin, 2011), allowing three mismatches. Only sequences with both forward and reverse primer matches and length ≥250 and ≤380 bp after trimming were retained. Qualified ITS2 sequences were clustered into operational taxonomic units (OTUs) at 97% identity within samples (e.g., sequences from each sample clustered independently). The OTUs that represented ≤3 ITS2 sequences were removed. The most abundant sequence from each OTU was chosen as the representative sequence and classified as a Symbiodinium subclade against a custom reference database of Symbiodinium subclades (Tong et al., 2017) using the Needleman-Wunsch global alignment algorithm implemented in the Biostrings package (Pagès et al., 2016) in R (R Core Team, 2014). Each OTU was then assigned a Symbiodinium subclade name with the highest alignment score.
The resulting Symbiodinium subclades count tables of different samples were merged for downstream statistical analysis. The Shannon–Wiener (H′) diversity index was calculated in the R software environment (R 3.1.2) to assess the alpha-diversity of Symbiodinium subclades associated with coral samples. The differences in the Shannon–Wiener (H′) diversity index of Symbiodinium subclades among different samples was tested with the multiple comparison test (Tukey’s HSD) using R software (R 3.1.2). A rarefaction curve was plotted using “make_rarefaction_plots.py” script in the QIIME package based on the merged count table (Caporaso et al., 2010). To present the relationships of the Symbiodinium subclade compositions in different coral samples from different reef regions, a Bray–Curtis dissimilarity-based principal coordinates analysis (PCoA) of Symbiodinium subclade compositions was performed by the vegan package in R (Oksanen et al., 2016). To test the relationships among environmental factors, Symbiodinium subclade compositions, and reef regions, a canonical correspondence analysis (CCA) was conducted in R using the vegan package (Oksanen et al., 2016).
To identify the phylogenetic relationships of Symbiodinium subclades with coral hosts, phylogenetic trees were constructed using two different methods: the Bayesian inference (BI) and maximum likelihood (ML) methods. Analyses were performed with the Kimura 2-parameter model, and the evolutionary models that fit best for the datasets were calculated using the program JModelTest version 2.1.7 (Darriba et al., 2012). For BI, the chain was run for 10 million generations, saving every 1,000th generation in Mrbayes (Ronquist et al., 2012). For ML, we performed 1,000 replicates of the rapid bootstrapping algorithm in MEGA version 6.06 (Tamura et al., 2013). Nearly identical trees were obtained; thus, only the ML tree was presented in this study.
For 77 coral samples, a total of 4,458,888 Symbiodinium ITS2 sequences (average 57,907 Symbiodinium ITS2 sequences per coral sample) were retained after quality control. Thirty-two Symbiodinium subclades were classified (Supplementary Table 3 and Supplementary File 1), of which 13 Symbiodinium subclades (covering more than 95% of the total Symbiodinium ITS2 sequences) were selected for further analysis, while the rest (19 Symbiodinium subclades) were designated as others (Supplementary File 2).
For each coral sample, rarefaction curves (Supplementary Figure 1) showed that the number of observed Symbiodinium subclades reached an asymptote, indicating that the number of Symbiodinium ITS2 sequences from each coral sample was able to meet current needs for the diversity analysis of Symbiodinium subclades.
It was noted that Acropora sp., G. fascicularis, P. lamellina, and S. glaucum exhibited distinct Symbiodinium subclade compositions between tropical and subtropical reef regions, whereas P. lutea failed to exhibit differences in Symbiodinium subclade compositions between tropical and subtropical reef regions (Figure 2). Symbiodinium subclade compositions among different coral species were more heterogeneous in the SY reef region, but there were no notable differences in Symbiodinium subclade compositions among the explored coral species in the XW and DY reef regions. Specifically, the Symbiodinium subclade C3 was dominant in most explored corals, the subclade D1 was mainly detected in corals in the SY reef region, and the subclade C161 was mainly associated with corals in the XW and DY reef regions. In addition, nearly all Symbiodinium subclades observed in explored coral samples were also detected in seawater samples, implying a possible linkage between Symbiodinium subclades in coral hosts and those in seawater.
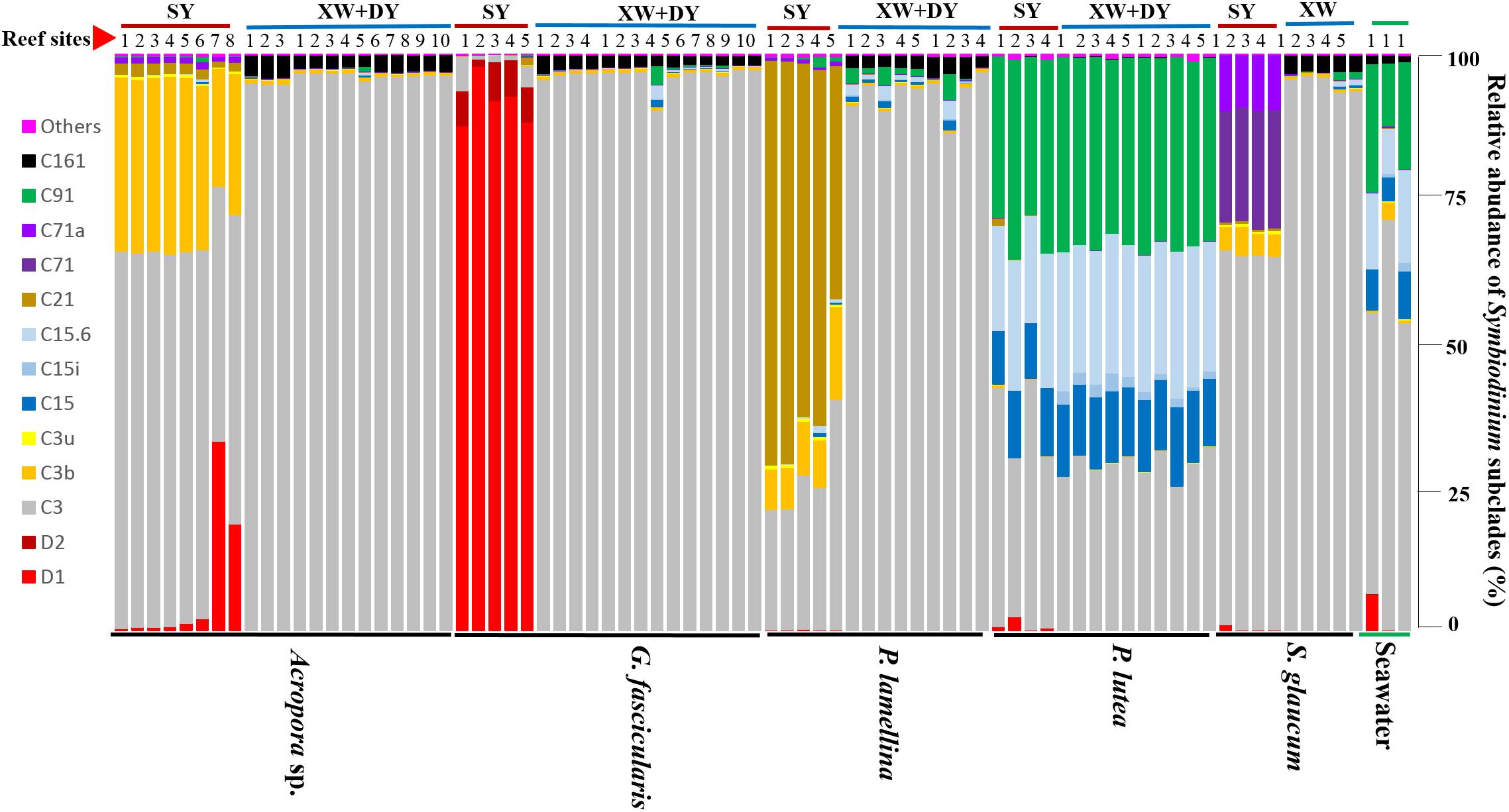
FIGURE 2. Bar plot of the relative abundance of different Symbiodinium subclades in coral samples and filters of seawaters from tropical (red color code) and subtropical (blue color code) reefs in the northern SCS. Each bar represents the relative abundance of different Symbiodinium subclades in one sample.
The same coral species or genus in the SY reef region showed higher Shannon–Wiener (H′) index values of Symbiodinium subclades than corresponding corals in the XW or DY reef regions (P < 0.001) (Figure 3). One exception was P. lutea, as its Shannon–Wiener (H′) index values of Symbiodinium subclades were not statistically significantly different across the three reef regions (P > 0.05).
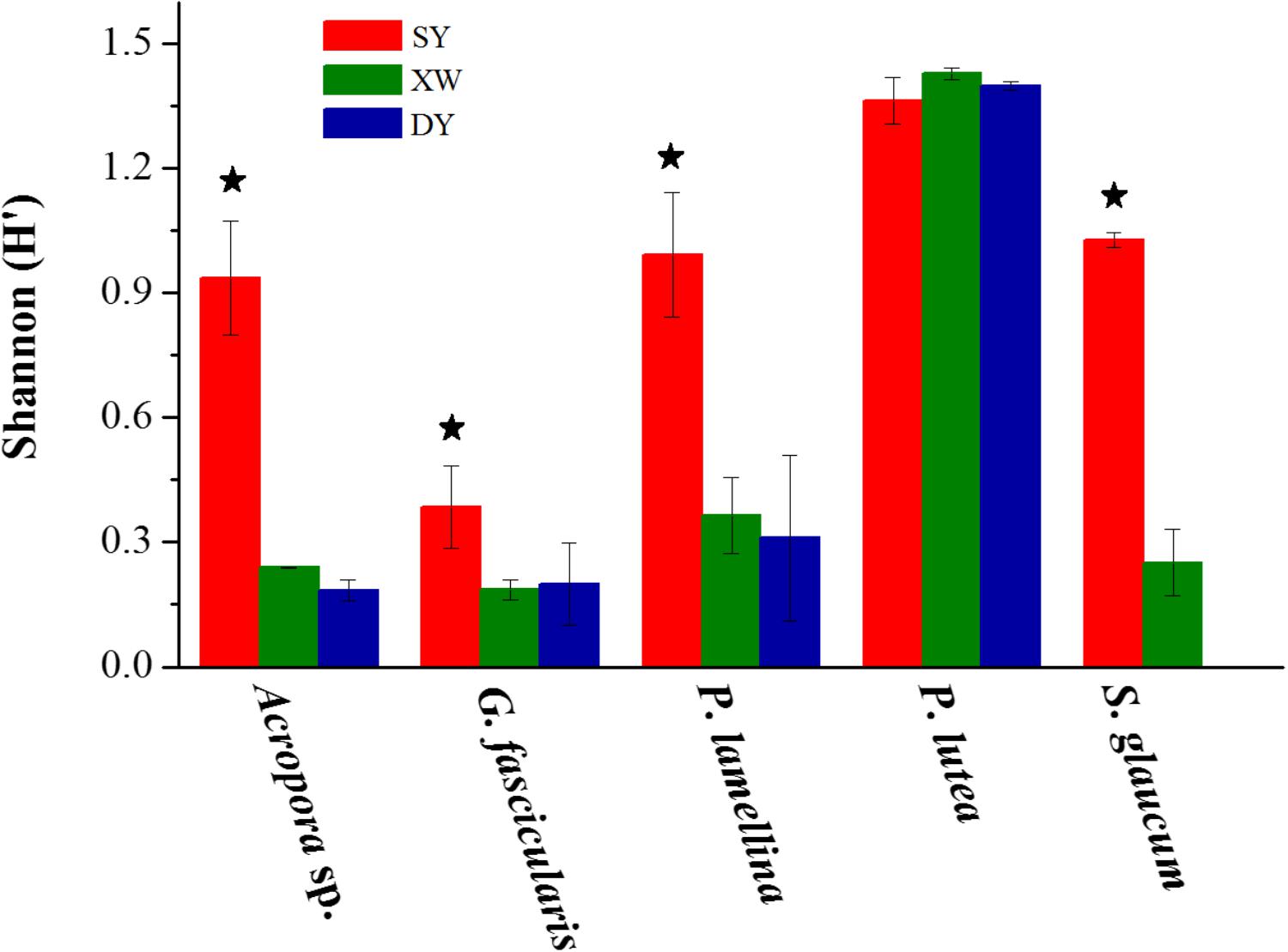
FIGURE 3. Bar plot of the Shannon (H′) diversity index values of Symbiodinium subclades in the same coral species or genus living in tropical SY and subtropical XW and DY reef regions in the northern SCS. The Shannon (H′) diversity index values are mean ± SE. Stars above the bar plots denote statistically significant differences between the same coral species or genus living in tropical SY and subtropical XW or DY (P < 0.001).
A PCoA based on Symbiodinium subclade counts of different coral samples in the SY, XW, and DY reef regions revealed a segregation of Symbiodinium subclade compositions (Figure 4). The Symbiodinium subclade compositions of Acropora sp., G. fascicularis, P. lamellina, and S. glaucum in the SY region were grouped together, whereas the Symbiodinium subclade compositions of Acropora sp., G. fascicularis, P. lamellina, and S. glaucum in the XW and DY reef regions were grouped together. However, the Symbiodinium subclade compositions of P. lutea in the SY, XW, and DY reef regions were grouped together. The Monte Carlo permutation test showed that the Symbiodinium subclade compositions among these three groups were significantly different (P = 0.002).
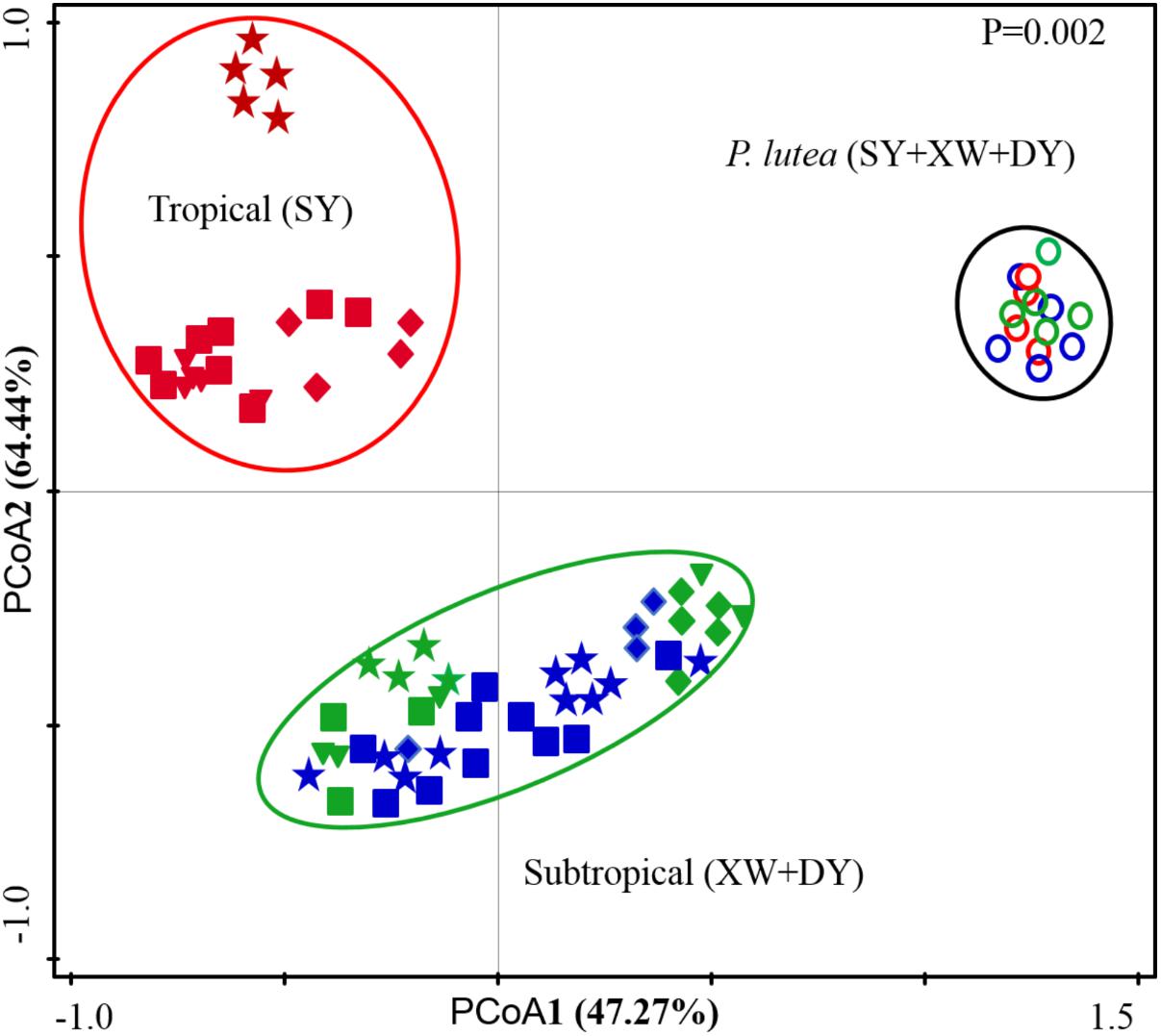
FIGURE 4. Principle coordinates analysis (PCoA) scores plot based on read counts of Symbiodinium subclades in coral samples from the tropical SY and subtropical XW and DY reefs in the northern SCS. The square (■) represents Acropora sp.; the star ( ) represents G. fascicularis; the diamond (♦) represents P. lamellina; the circle (
) represents G. fascicularis; the diamond (♦) represents P. lamellina; the circle ( ) represents P. lutea; and the triangle (
) represents P. lutea; and the triangle ( ) represents S. glaucum. For the same coral species or genus samples, different colors represent different reefs (red represents coral samples from tropical SY; green represents coral samples from subtropical XW; and blue represents coral samples from subtropical DY).
) represents S. glaucum. For the same coral species or genus samples, different colors represent different reefs (red represents coral samples from tropical SY; green represents coral samples from subtropical XW; and blue represents coral samples from subtropical DY).
The distinct Symbiodinium subclade compositions in explored corals living between tropical (SY) and subtropical (XW and DY) reef regions, except those in P. lutea, were probably explained by the different environmental factors of the different reef regions (Figures 5, 6). According to the CCA (Figure 6), the SST and NO3-, NH4+, and PO43- served as the main factors affecting Symbiodinium subclade compositions in tropical (SY) and subtropical (XW and DY) reef regions. The Symbiodinium subclades D1, D2, C71, C71a, C21, and C3b mainly occurred in the SY reef region and were positively correlated with the SST and NO3-, NH4+, and PO43-. Conversely, the Symbiodinium subclade C161 occurred predominantly in the XW and DY reef regions and was negatively correlated with those environmental factors. However, there was no clear correlation between Symbiodinium subclades C15, C15.6, C91, and C15i (mainly associated with P. lutea) and all of the measured environmental factors.
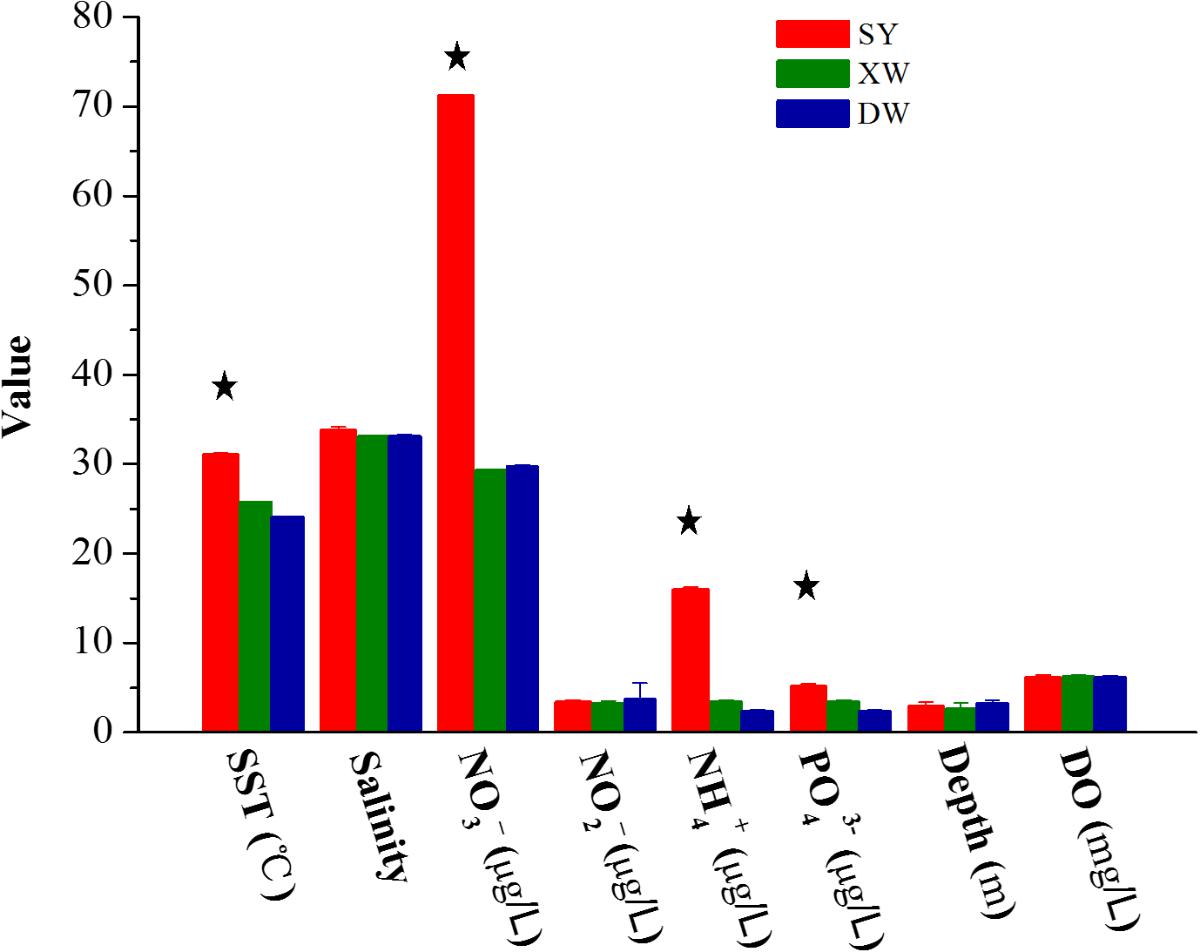
FIGURE 5. Bar plot of the value of environmental factors in the tropical SY and subtropical XW and DY reef regions in the northern SCS. The value of environmental factors is mean ± SE (SY, n = 8; XW, n = 5; DY, n = 10). Stars above bar plots denote statistically significant differences between tropical SY and subtropical XW or DY (P < 0.001).
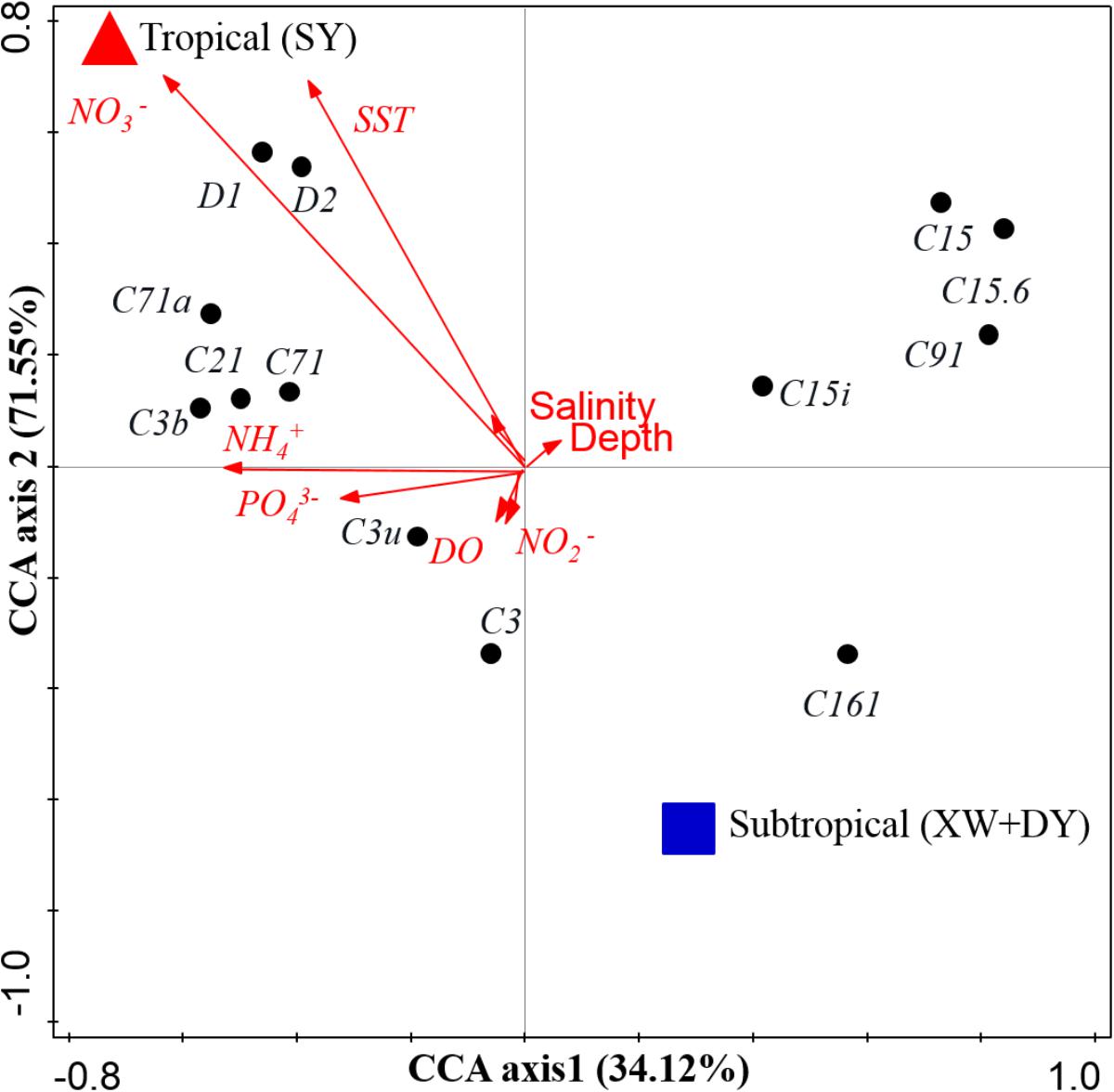
FIGURE 6. Biplot of the canonical correspondence analysis (CCA) of Symbiodinium subclades with environmental factors. Red triangles represent the tropical SY reef region, and blue squares represent the subtropical DY and XW reef regions.
The similarities of ITS2 sequences of different Symbiodinium subclades used for phylogenetic tree construction were less than 97%, supporting the fact that the 13 Symbiodinium subclades from explored coral samples were distinct (Supplementary Data Sheet 2). The bootstrap values of correlated Symbiodinium subclades were less than 70% in the ML phylogenetic tree (Figure 7A), revealing low confidence to support branches among those Symbiodinium subclades. Based on the 13 Symbiodinium subclade compositions detected in different coral species in SY, XW, and DY reef regions, it was noted that different coral species could be associated with multiple Symbiodinium subclades and vice versa, suggesting flexible coral-Symbiodinium symbiotic associations. Interestingly, the Symbiodinium subclades C15, C15.6, and C91 seemed to be specific to P. lutea. The three subclades were the most dominant Symbiodinium in P. lutea, across tropical and subtropical reef regions, and they showed stable compositions (Figure 7).
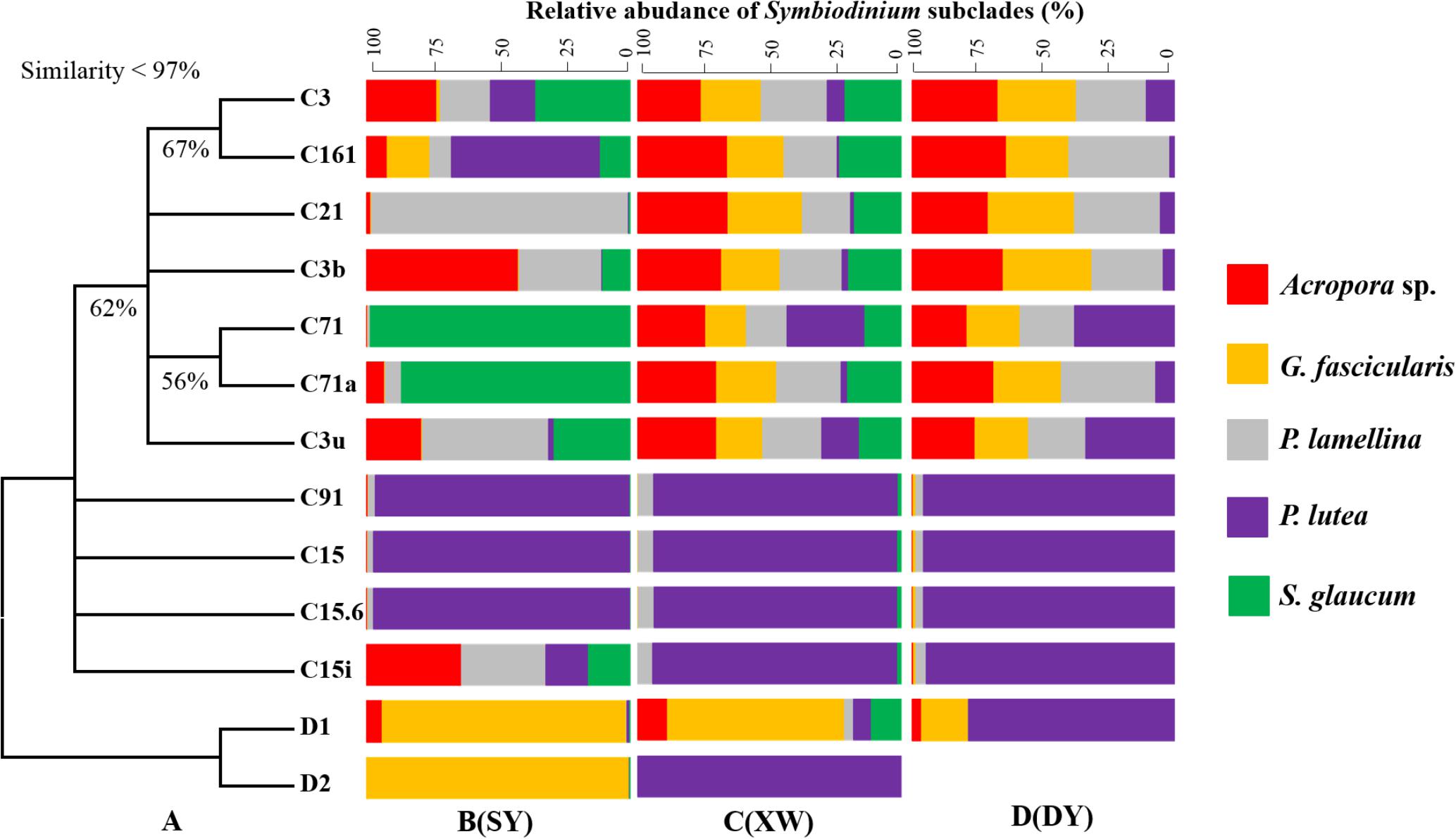
FIGURE 7. Combination chart of the phylogenetic tree (A) and bar plot of Symbiodinium subclades in explored corals living in the SY (B), XW (C), and DY (D) reef regions. The phylogenetic tree was drawn based on ITS2 sequences of different Symbiodinium subclades using the maximum likelihood and neighbor joining methods, and nearly identical trees were obtained. The presented tree is derived from the maximum likelihood method (bootstrap values below 50% are not shown). The bar plot was drawn based on the ITS2 sequences of Symbiodinium subclades in explored corals. Each bar represents the relative abundance (mean) of one Symbiodinium subclade in different coral species.
In this study, we investigated Symbiodinium subclade compositions in five typical coral species across the SY, XW, and DY reef regions in the northern SCS. Several studies have investigated Symbiodinium subclade compositions of certain corals in the SCS and suggested that temperature (SST) could shape coral-Symbiodinium symbiotic associations (Zhou and Huang, 2011; Tong et al., 2017; Zhou et al., 2017). According to this study, in addition to SST, other environmental factors, especially NO3-, NH4+, and PO43-, and host taxa affect coral-Symbiodinium symbiotic associations in the SCS as well.
It has been suggested that corals may adapt to environmental changes by regulating their hosted Symbiodinium (Cunning et al., 2015a,b). For example, Keshavmurthy et al. (2014) explored corals exposed to long-term elevated SST in the presence of a nuclear power plant outlet and found that corals changed their symbiotic Symbiodinium subclades from C1 and C3 (stress sensitive) to D1a (stress tolerant) or to a mixture of Symbiodinium subclades C1/C3/C21a/C15 and D1a. Corals from some extremely hot regions with high SST were found to host heat tolerant Symbiodinium, that is, subclades within clade D (Berkelmans and van Oppen, 2006; Lajeunesse et al., 2009; D’Angelo et al., 2015). Extremely high temperatures could lead to coral bleaching, but corals that hosted more heat tolerant Symbiodinium were less likely to bleach; several bleached corals gained more heat tolerant Symbiodinium after recovery (Kemp et al., 2014). The recent study by Tong et al. (2017) about G. fascicularis and Montipora spp. in the SCS showed that the putative heat tolerant Symbiodinium subclades within clade D were mainly detected in corals across tropical reef regions, and Symbiodinium subclade compositions in explored corals were distinct between tropical and subtropical reef regions, suggesting that the SST mainly shapes coral–algal symbiosis. Zhou et al. (2017) found that SST shaped G. fascicularis-algal symbiosis in the SCS. Based on this study, in addition to SST, other environmental factors, such as seawater NO3-, NH4+, and PO43-, and host taxa could also affect the coral-Symbiodinium symbiotic associations in the SCS. The compositions of Symbiodinium subclades in Acropora sp., G. fascicularis, P. lamellina, and S. glaucum from the tropical reef region (SY) with high SST, NO3-, NH4+, and PO43- were distinct from those from the subtropical reef regions (XW + DY), suggesting that nutrient inflow might also affect coral-Symbiodinium symbiotic associations in addition to SST. These results are consistent with Sawall et al. (2014), who suggested that nutrients have the potential to affect Symbiodinium community structures associated with Pocillopora verrucosa in the Red Sea.
In the Indo-Pacific Ocean, previous studies have shown that Porites-Symbiodinium symbiotic associations were stable over broad geographical scales and temperature ranges as well as through bleaching events or after transplantation (LaJeunesse et al., 2004a,b; Stat et al., 2009). Similar to these studies, our present results based on high-throughput sequencing show that the Symbiodinium subclade compositions of P. lutea (Porites) are not statistically and significantly different across subtropical and tropical reef regions in the SCS. In addition, there are no clear correlations of the Symbiodinium subclades C15, C15.6, and C91 mainly detected in P. lutea with all detected environmental factors. All of these results reveal that the host could also affect Symbiodinium subclade compositions, and the Symbiodinium subclades C15, C15.6, and C91 seem to be specific to P. lutea.
The adaptation of corals to future climate change and anthropogenic disturbances strongly relies on their hosted Symbiodinium. In the SCS, temperature is one of the measured environmental factors that could shape coral-Symbiodinium symbiotic associations. Therefore, the rise of SST is a potential threat to corals in the SCS. Previous studies have shown that heat-tolerant corals contain a greater number of clade D Symbiodinium. Similar to Tong et al. (2017), the present results also show that G. fascicularis in the SCS appears to host more heat-tolerant Symbiodinium subclades within clade D, which may lead to their dominance during future climate change. In addition, the present study shows that other environmental factors (especially NO3-, NH4+, and PO43-) can also serve as major factors affecting coral-Symbiodinium symbiotic associations. Increases in coastal nutrients are frequently linked to human activities (Doney, 2010), which suggests that human activities may also affect coral-Symbiodinium symbiotic associations. The present results suggest that seawater pollution may be another potential threat to the explored corals in the SCS. A “nugget of hope” for these threats is that most explored corals in this study have the potential to adapt to future climate change with the flexibility of their symbiosis with Symbiodinium. One exception is the coral P. lutea. The present results show that Symbiodinium compositions in P. lutea are probably affected by host itself, and the Symbiodinium subclades C15, C15.6, and C91 seem to be specific to P. lutea. According to Warner et al. (2006), this type of specificity favors the evolution of mutualism in symbiosis, thereby, increasing the persistence of intimate and long-term mutualisms. In fact, in recent years, certain dominant coral species have been declining, while certain subdominant coral species are becoming more dominant (Grottoli et al., 2014). For example, the coral community structure in SY (Luhuitou, China) changed dramatically into the current predominance of massive Porites (primarily P. lutea) (Zhao et al., 2016). Based on the above results, the specificity between Symbiodinium and P. lutea may be a reason for P. lutea being the dominant coral in the SCS.
This study demonstrates the multiple relationships of the Symbiodinium community with both the coral host and the environment. Corals might adapt to different environmental conditions by changing their Symbiodinium communities (e.g., taking up the more heat-tolerant clade D Symbiodinium). Conversely, certain corals, such as P. lutea, might be able to adapt to different environmental conditions (e.g., climate change and elevated nutrient inflow) via host-specific Symbiodinium subclades (e.g., C15, C15.6, and C91). Elucidating detailed mechanisms of corals’ adaptation to different environments warrants future investigation in both field and lab settings.
The raw sequence information in this paper has been deposited in the GenBank Sequence Read Archive with accession number SUB2393447.
SG and ZL conceived and designed the experiments. SG performed the experiments and analyzed the experimental data. SG, LX, JL, FL, HC, and BL contributed to reagents and materials. SG, ZL, GC, YX, FZ, and KY wrote the manuscript.
This work was supported by the National Key Basic Research Programme of China (2013CB956103).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer CA-G and handling Editor declared their shared affiliation at time of review.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02485/full#supplementary-material
Arif, C., Daniels, C., Bayer, T., Banguera-Hinestroza, E., Barbrook, A., Howe, C. J., et al. (2014). Assessing Symbiodinium diversity in scleractinian corals via next-generation sequencing-based genotyping of the ITS2 rDNA region. Mol. Ecol. 23, 4418–4433. doi: 10.1111/mec.12869
Baker, A. C. (2003). Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Annu. Rev. Ecol. Evol. Sci. 34, 661–689. doi: 10.1146/annurev.ecolsys.34.011802.132417
Berkelmans, R., and van Oppen, M. J. (2006). The role of zooxanthellae in the thermal tolerance of corals: a ’nugget of hope’ for coral reefs in an era of climate change. Proc. Biol. Sci. 273, 2305–2312. doi: 10.1098/rspb.2006.3567
Boulotte, N. M., Dalton, S. J., Carroll, A. G., Harrison, P. L., Putnam, H. M., Peplow, L. M., et al. (2016). Exploring the Symbiodinium rare biosphere provides evidence for symbiont switching in reef-building corals. ISME J. 10, 2693–2701. doi: 10.1038/ismej.2016.54
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., and Bushman, F. D. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chen, C. A., Wang, J. T., Fang, L. S., and Yang, Y. W. (2005a). Fluctuating algal symbiont communities in Acropora palifera (Cnidaria; Scleractinia) from Taiwan. Mar. Ecol. Prog. Ser. 295, 113–121. doi: 10.3354/meps295113
Chen, C. A., Yang, Y. W., Wei, N. V., Tsai, W. S., and Fang, L. S. (2005b). Symbiont diversity in scleractinian corals from tropical reefs and subtropical non-reef communities in Taiwan. Coral Reefs 24, 11–22. doi: 10.1007/s00338-004-0389-7
Chen, T., Yu, K., Shi, Q., Li, S., and Price, G. J. (2009). Twenty-five years of change in scleractinian coral communities of Daya Bay (northern South China Sea) and its response to the 2008 AD extreme cold climate event. Chin. Sci. Bull. 54, 2107–2117. doi: 10.1007/s11434-009-0007-8
Cunning, R., Gates, R. D., and Edmunds, P. J. (2017). Using high-throughput sequencing of ITS2 to describe Symbiodinium metacommunities in St. John, US Virgin Islands. PeerJ 5:e3472. doi: 10.7717/peerj.3472
Cunning, R., Silverstein, R. N., and Baker, A. C. (2015a). Investigating the causes and consequences of symbiont shuffling in a multi-partner reef coral symbiosis under environmental change. Proc. Biol. Sci. 282:20141725. doi: 10.1098/rspb.2014.1725
Cunning, R., Vaughan, N., Gillette, P., Capo, T. R., Matté, J. L., and Baker, A. C. (2015b). Dynamic regulation of partner abundance mediates response of reef coral symbioses to environmental change. Ecology 96, 1411–1420. doi: 10.1890/14-0449.1
D’Angelo, C., Hume, B. C. C., Burt, J., Smith, E. G., Achterberg, E. P., and Wiedenmann, J. (2015). Local adaptation constrains the distribution potential of heat-tolerant Symbiodinium from the Persian/Arabian Gulf. ISME J. 9, 2551–2560. doi: 10.1038/ismej.2015.80
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). Jmodeltest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
Doney, S. C. (2010). The growing human footprint on coastal and open-ocean biogeochemistry. Science 328, 1512–1516. doi: 10.1126/science.1185198
Dong, Z., Huang, H., Huang, L., and Li, Y. (2009). Diversity of symbiotic algae of the genus Symbiodinium in scleractinian corals of the Xisha Islands in the South China Sea. J. Syst. Evol. 47, 321–326. doi: 10.1111/j.1759-6831.2009.00034.x
Dong, Z. J., Huang, H., Huang, L. M., Li, Y. C., and Li, X. B. (2008a). PCR-RFLP analysis of large subunit rDNA of symbiotic dinoflagellates in scleractinian corals from Luhuitou fringing reef of Sanya, Hainan. Biodiver. Sci. 16, 498–502. doi: 10.3724/SP.J.1003.2008.08074
Dong, Z. J., Huang, H., Huang, L. M., Li, Y. C., and Zhou, G. W. (2008b). Molecular taxonomy and diversity of symbiotic dinoflagellates in scleractinian corals near the sea off the Dongshan Island in Fujian Province. J. Oceanogr. Taiwan Strait 27, 135–140.
Eren, A. M., Vineis, J. H., Morrison, H. G., and Sogin, M. L. (2013). A filtering method to generate high quality short reads using Illumina paired-end technology. PLoS One 8:e66643. doi: 10.1371/journal.pone.0066643
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Fukami, H., Budd, A. F., Paulay, G., Solécava, A., and Allen, C. C. (2004). Conventional taxonomy obscures deep divergence between pacific and atlantic corals. Nature 427, 832–835. doi: 10.1038/nature02339
Grottoli, A. G., Warner, M. E., Levas, S. J., Aschaffenburg, M. D., and Schoepf, V. (2014). The cumulative impact of annual coral bleaching can turn some coral species winners into losers. Glob. Chang. Biol. 20, 3823–3833. doi: 10.1111/gcb.12658
Hoegh-Guldberg, O., Mumby, P. J., Hooten, A. J., Steneck, R. S., and Greenfield, P. (2007). Coral reefs under rapid climate change and ocean acidification. Science 318, 1737–1742. doi: 10.1126/science.1152509
Huang, D., Licuanan, W. Y., Hoeksema, B. W., Chen, C. A., and Ang, P. O. (2015). Extraordinary diversity of reef corals in the South China Sea. Mar. Biodiver. 45, 157–168. doi: 10.1007/s12526-014-0236-1
Huang, H., Dong, Z. J., Huang, L. M., and Zhang, J. B. (2006). Restriction fragment length polymorphism analysis of large subunit rDNA of symbiotic dinoflagellates from scleractinian corals in the Zhubi Coral Reef of the Nansha Islands. J. Integr. Plant. Biol. 48, 148–152. doi: 10.1111/j.1744-7909.2006.00148.x
Hughes, T. P., Baird, A. H., Bellwood, D. R., Card, M., and Connolly, S. R. (2003). Climate change, human impacts, and the resilience of coral reefs. Science 301, 929–933. doi: 10.1126/science.1085046
Hughes, T. P., Kerry, J. T., Alvarez-Noriega, M., Alvarez-Romero, J. G., and Anderson, K. D. (2017). Global warming and recurrent mass bleaching of corals. Nature 543, 373–377. doi: 10.1038/nature21707
Kemp, D. W., Hernandez-Pech, X., Iglesias-Prieto, R., Fitt, W. K., and Schmidt, G. W. (2014). Community dynamics and physiology of Symbiodinium spp. before, during, and after a coral bleaching event. Limnol. Oceanogr. 59, 788–797. doi: 10.4319/lo.2014.59.3.0788
Keshavmurthy, S., Meng, P. J., Wang, J. T., Kuo, C. Y., and Yang, S. Y. (2014). Can resistant coral-Symbiodinium associations enable coral communities to survive climate change? A study of a site exposed to long-term hot water input. PeerJ 2:e327. doi: 10.7717/peerj.327
Keshavmurthy, S., Tang, K. H., Hsu, C. M., Gan, C. H., and Kuo, C. Y. (2017). Symbiodinium spp. associated with scleractinian corals from Dongsha Atoll (Pratas), Taiwan, in the South China Sea. PeerJ 5:e2871. doi: 10.7717/peerj.2871
LaJeunesse, T. C., Bhagooli, R., Hidaka, M., deVantier, L., Done, T., Schmidt, G. W., et al. (2004a). Closely related Symbiodinium spp. differs in relative dominance in coral reef host communities across environmental, latitudinal andbiogeographic gradients. Mar. Ecol. Prog. Ser. 284, 147–161. doi: 10.3354/meps284147
LaJeunesse, T. C., Thornhill, D. J., Cox, E. F., Stanton, F. G., Fitt, W. K., and Schmidt, G. W. (2004b). High diversity and host specificity observed among symbiotic dinoflagellates in reef coral communities from Hawaii. Coral Reefs 23, 596–603. doi: 10.1007/s00338-004-0428-4
LaJeunesse, T. C., Loh, W., van Woesik, R., Hoegh-Guldberg, O., Schmidt, G. W., and Fitt, W. K. (2003). Low symbiont diversity in southern Great Barrier Reef corals, relative to those of the Caribbean. Limnol. Oceanogr. 48, 2046–2054. doi: 10.4319/lo.2003.48.5.2046
LaJeunesse, T. C., Smith, R. T., Finney, J., and Oxenford, H. (2009). Outbreak and persistence of opportunistic symbiotic dinoflagellates during the 2005 caribbean mass coral ‘bleaching’ event. Proc. Biol. Sci. 276, 4139–4148. doi: 10.1098/rspb.2009.1405
LaJeunesse, T. C., and Trench, R. K. (2000). Biogeography of two species of Symbiodinium (Freudenthal) inhabiting the intertidal sea anemone Anthopleura elegantissima (Brandt). Biol. Bull. 199, 126–134. doi: 10.2307/1542872
Li, S., Yu, K., Shi, Q., Chen, T., Zhao, M., and Zhao, J. (2008). Interspecies and spatial diversity in the symbiotic zooxanthellae density in corals from northern South China Sea and its relationship to coral reef bleaching. Chin. Sci. Bull. 53, 295–303. doi: 10.1007/s11434-007-0514-4
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Mieog, J. C., van Oppen, M. J. H., Cantin, N. E., Stam, W. T., and Olsen, J. L. (2007). Real-time PCR reveals a high incidence of Symbiodinium clade D at low levels in four scleractinian corals across the Great Barrier Reef: implications for symbiont shuffling. Coral Reefs 26, 449–457. doi: 10.1007/s00338-007-0244-8
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchinet, P. R., et al. (2016). Vegan: Community Ecology Package. R Package Version 2.3– 5. Available at: http://CRAN.R-project.org/package=vegan
Pagès, H., Aboyoun, P., Gentleman, R., and DebRoy, S. (2016). Biostrings: String Objects Representing Biological Sequences, and Matching Algorithms. R Package Version 2.40.2. Available at: http://bioconductor.org/packages/Biostrings/
Pochon, X., Putnam, H. M., and Gates, R. D. (2014). Multi-gene analysis of Symbiodinium dinoflagellates: a perspective on rarity, symbiosis, and evolution. PeerJ 2:e394. doi: 10.7717/peerj.394
R Core Team (2014). R: A Language and Environment for Statistical Computing. Available at: http://www.R-project.org/
Ronquist, F., Teslenko, M., van der Mark, P., Darling, A., and Höhna, S. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Sawall, Y., Al-Sofyani, A., Banguera-Hinestroza, E., and Voolstra, C. R. (2014). Spatio-temporal analyses of Symbiodinium physiology of the coral Pocillopora verrucosa along large-scale nutrient and temperature gradients in the Red Sea. PLoS One 9:e103179. doi: 10.1371/journal.pone.0103179
Stat, M., Pochon, X., Cowie, R. O. M., and Gates, R. D. (2009). Specificity in communities of Symbiodinium in corals from Johnston Atoll. Mar. Ecol. Prog. Ser. 386, 83–96. doi: 10.3354/meps08080
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Thomas, L., Kendrick, G. A., Kennington, W. J., Richards, Z. T., and Stat, M. (2014). Exploring Symbiodinium diversity and host specificity in Acropora corals from geographical extremes of Western Australia with 454 amplicon pyrosequencing. Mol. Ecol. 23, 3113–3126. doi: 10.1111/mec.12801
Tong, H., Cai, L., Zhou, G., Yuan, T., and Zhang, W. (2017). Temperature shapes coral-algal symbiosis in the South China Sea. Sci. Rep. 7:40118. doi: 10.1038/srep40118
Warner, M. E., LaJeunesse, T. C., Robison, J. D., and Thur, R. M. (2006). The ecological distribution and comparative photobiology of symbiotic dinoflagellates from reef corals in Belize: potential implications for coral bleaching. Limnol. Oceanogr. 51, 1887–1897. doi: 10.4319/lo.2006.51.4.1887
Yu, K. F. (2000). The sea surface temperature changing trend of the last 40 years in the Lei-Qiong sea area. Trop. Geogr. 2, 111–115.
Yu, K. F. (2012). Coral reefs in the South China Sea: their response to and records on past environmental changes. Sci. China Earth Sci. 55, 1217–1229. doi: 10.1007/s11430-012-4449-5
Zhao, M., Bernhard, R., Yu, K., Shi, Q., and Zhang, Q. (2016). Model suggests potential forporitescoral population recovery after removal of anthropogenic disturbance (luhuitou, hainan, south china sea). Sci. Rep. 6:33324. doi: 10.1038/srep33324
Zhou, G., Cai, L., Li, Y., Tong, H., and Jiang, L. (2017). Temperature-driven local acclimatization of Symbiodinium hosted by the coral Galaxea fascicularis at Hainan Island, China. Front. Microbiol. 8:2487. doi: 10.3389/fmicb.2017.02487
Keywords: corals, Symbiodinium, symbiosis, flexible symbiotic association, tropical, subtropical
Citation: Gong S, Chai G, Xiao Y, Xu L, Yu K, Li J, Liu F, Cheng H, Zhang F, Liao B and Li Z (2018) Flexible Symbiotic Associations of Symbiodinium With Five Typical Coral Species in Tropical and Subtropical Reef Regions of the Northern South China Sea. Front. Microbiol. 9:2485. doi: 10.3389/fmicb.2018.02485
Received: 06 June 2018; Accepted: 28 September 2018;
Published: 02 November 2018.
Edited by:
David William Waite, The University of Auckland, New ZealandReviewed by:
Beate Slaby, Helmholtz-Gemeinschaft Deutscher Forschungszentren (HZ), GermanyCopyright © 2018 Gong, Chai, Xiao, Xu, Yu, Li, Liu, Cheng, Zhang, Liao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiyong Li, enlsaUBzanR1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.