- 1Department of Life Science and Medical Bioscience, Waseda University, Tokyo, Japan
- 2Biomedical Research Institute, National Institute of Advanced Industrial Science and Technology (AIST), Ibaraki, Japan
Toxin-antitoxin (TA) systems are implicated in prokaryotic stress adaptation. Previously, bioinformatics analysis predicted that such systems are abundant in some slowly growing chemolithotrophs; e.g., Nitrosomonas europaea. Nevertheless, the molecular functions of these stress-response modules remain largely unclear, limiting insight regarding their physiological roles. Herein, we show that one of the putative MazF family members, encoded at the ALW85_RS04820 locus, constitutes a functional toxin that engenders a TA pair with its cognate MazE antitoxin. The coordinate application of a specialised RNA-Seq and a fluorescence quenching technique clarified that a unique triplet, UGG, serves as the determinant for MazF cleavage. Notably, statistical analysis predicted that two transcripts, which are unique in the autotroph, comprise the prime targets of the MazF endoribonuclease: hydroxylamine dehydrogenase (hao), which is essential for ammonia oxidation, and a large subunit of ribulose 1,5-bisphosphate carboxylase/oxygenase (rbcL), which plays an important role in carbon assimilation. Given that N. europaea obtains energy and reductants via ammonia oxidation and the carbon for its growth from carbon dioxide, the chemolithotroph might use the MazF endoribonuclease to modulate its translation profile and subsequent biochemical reactions.
Introduction
Toxin-antitoxin (TA) systems comprise small genetic modules that modulate microbial cell fates in stressful environments. During periods of low or no stress, antitoxins potently inactivate toxins, thus allowing microbes to grow normally. Under environmental stress, however, antitoxins are preferentially degraded, resulting in toxin activation and subsequent microbial growth arrest (Schuster and Bertram, 2013). Currently, these systems are grouped into six distinct classes depending on antitoxin features (Page and Peti, 2016; Hall et al., 2017). In type I and type III, the antitoxins comprise RNAs that mask toxin activities by repressing toxin translation or directly binding to corresponding toxins, respectively (Brantl, 2012; Goeders et al., 2016). In other TA systems, the antitoxins consist of proteins that counteract the toxins by forming a TA protein complex (type II) (Gerdes et al., 2005), functioning as an antagonist of the toxin (type IV) (Masuda et al., 2012a,b), inhibiting translation by destabilising the toxin mRNA (type V) (Wang et al., 2012), or serving as an adaptor molecule to introduce the toxin to cellular protease (type VI) (Aakre et al., 2013). Among these, type II systems have been rigorously investigated and are known to be ubiquitous and abundant in free-living microbes, especially in some chemolithotrophic prokaryotes (Pandey and Gerdes, 2005).
Nitrosomonas europaea, which is predicted to carry more than 50 type II TA pairs, is one such chemolithotroph (Shao et al., 2011; Xie et al., 2017). N. europaea obtains energy and reductants by oxidising ammonia to nitrite (Arp et al., 2002; Arp and Stein, 2003), inhabiting wastewater treatment and sediments where ammonia may be in abundant supply (Koops et al., 2006). As the oxidation process is important in terms of industrial, agricultural, and environmental nitrogen cycles (Bothe et al., 2000), the physiology, biochemistry, and molecular biology of this bacteria in varied environments has been investigated. It has been well documented that N. europaea is susceptible to numbers of environmental factors such as temperature, pH, nitrite and ammonia concentrations, heavy metals, and organic and inorganic compounds (Groeneweg et al., 1994; Stein and Arp, 1998; Koops et al., 2006; Wei et al., 2006a,b; Park and Ely, 2008a,b; Radniecki et al., 2009; Yu and Chandran, 2010; Pellitteri-Hahn et al., 2011). Considering that N. europaea harbours a very high number of putative TA pairs (Pandey and Gerdes, 2005), this bacterium might regulate cellular activities using type II toxins.
Among the putative N. europaea toxins, a great majority are predicted to catalyse cellular RNA decay (Pandey and Gerdes, 2005; Shao et al., 2011; Xie et al., 2017); Escherichia coli MazF, a sequence-specific toxin endoribonuclease (Zhang et al., 2003), provides a helpful basis to better understand these endoribonucleolytic toxins in N. europaea. In the absence of stresses, MazF-mediated RNA cleavage is neutralised by its cognate antitoxin MazE (Zhang et al., 2003). Environmental stresses such as starvation, high temperature, and oxidative stress (Hazan et al., 2004), however, trigger MazE degradation via the ClpAP protease (Aizenman et al., 1996), activating the MazF endoribonuclease. E. coli MazF then comprehensively alters the translation profile by cleaving transcripts at ACA sites (Amitai et al., 2009; Sauert et al., 2016). MazF homologues are widely distributed in the prokaryotic domain (Chopra et al., 2013), with a hallmark of these enzymes being their sequence-specificities (Masuda and Inouye, 2017; Schifano and Woychik, 2017). Typically, MazF toxins recognise three to seven specific-sequences (i.e., ACA, UACAU, and UUACUCA), suggesting that the biological roles of MazF homologues are diverse and differ across microbial species (Zhu et al., 2006, 2008, 2009; Park et al., 2011; Rothenbacher et al., 2012; Schifano et al., 2013, 2014; Schuster et al., 2013; Verma and Bhatnagar, 2014; Miyamoto et al., 2016b, 2017; Li et al., 2017).
Notably, five mazEF loci are predicted in the N. europaea genome, whereas only one is found in the majority of prokaryotes (Pandey and Gerdes, 2005; Chopra et al., 2013). The cleavage-specificities of these endoribonucleases are less well studied and largely unknown, except for one located at the ALW85_RS06130 locus (previously designated as NE1181) (Miyamoto et al., 2016b); therefore, detailed analysis is required to interpret their physiological functions. In the present work, we focused our study on the predicted MazF endoribonuclease located at ALW85_RS04820 (hereafter MazFne1; Pandey and Gerdes, 2005) and revealed that it codes for a functional toxin endoribonuclease. Our study suggests that MazFne1 may serve as a crucial regulator of translation and cellular activities.
Materials and Methods
Plasmids and Oligonucleotides
The pET24a expression vectors encoding mazEne1 and pET24a encoding mazFne1 were purchased from GenScript Japan (Tokyo, Japan). The sequences of both genes were optimised for recombinant protein expression in E. coli. PCR primers were purchased from Tsukuba Oligo Service (Ibaraki, Japan). All fluorescently labelled oligonucleotides were purchased from Japan Bio Services (Saitama, Japan).
Reverse Transcription (RT)-PCR
Nitrosomonas europaea ATCC 19718 was obtained from the Biological Resource Center (NBRC), National Institute of Technology and Evaluation (NITE), Japan. N. europaea was cultivated in the dark at 28°C. HEPES medium 829 was used as the growth medium, as described in the NBRC manual. Cells were harvested by centrifugation at 2900 g. The cell pellet was transferred to a 2 mL screw-cap tube containing glass beads (0.1 mm in diameter) and disrupted in TRI-Reagent (Zymo Research, Orange, CA, United States). Total RNA was extracted using the Direct-zolTM RNA MiniPrep Kit (Zymo Research) according to the manufacturer’s protocol. Extracted RNA was then incubated with 4U of TURBO DNase (Thermo Fisher Scientific, Waltham, MA, United States) in the TURBO DNase Buffer at 37°C for 1 h and purified with RNA Clean and ConcentratorTM-5 (Zymo Research). Resultant RNA was incubated at 65°C for 5 min and cDNA was synthesised using the SuperScript VILO cDNA Synthesis Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. PCR was carried out with Q5 High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA, United States) using the primers listed in Supplementary Table 1. PCR conditions were as follows: an initial denaturation at 98°C for 1 min, followed by 30 cycles of amplification (denaturation at 98°C for 10 s, annealing at 65°C for 15 s, and extension at 72°C for 20 s), and a final extension for 2 min at 72°C. The samples were separated on a 2% agarose gel and visualized by GelRedTM Nucleic Acid Gel Stain (Biotium, Inc., Fremont, CA, United States).
Toxicity Assay
E. coli strain BL21 (DE3) cells (BioDynamics Laboratory Inc., Tokyo, Japan) were transformed with pET24a-mazFne1. These cells were pre-cultivated overnight in LB medium supplemented with 50 μg/mL kanamycin at 37°C. Cells were diluted 1:1000 in 10 mL of LB medium containing 50 μg/mL kanamycin. MazFne1 was induced by the addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) when the OD600 reached approximately 0.1. OD600 was measured at every 30 min.
Expression and Purification of MazEne1
Recombinant MazEne1 was obtained as outlined in our previous study (Miyamoto et al., 2016b) with slight modifications. E. coli strain BL21 (DE3) cells (BioDynamics Laboratory Inc.) were transformed with pET24a-mazEne1. These cells were pre-cultivated overnight in LB medium supplemented with 20 μg/mL kanamycin at 37°C. Subsequently, they were inoculated into 1 L of LB medium containing 20 μg/mL kanamycin. MazEne1 was induced by the addition of 1 mM IPTG when the OD600 reached approximately 0.6. After 3.5 h of incubation, the cells were harvested by centrifugation at 9200 g and stored at -80°C until further use. E. coli cells containing MazE were thawed on ice and resuspended in 18 mL of binding buffer (20 mM sodium phosphate buffer (pH 8.0), 300 mM NaCl, 40 mM imidazole, and 5 mM 2-mercaptoethanol). Subsequently, these cells were lysed by sonication and collected by centrifugation at 4400 g. The supernatant was filtered through a 0.45 μm membrane (Millex, Darmstadt, Germany), and applied to an equilibrated 1 mL His-Trap FF crude column (GE Healthcare, Little Chalfont, United Kingdom). The column was washed with 40 column volumes of binding buffer using AKTA pure 25 (GE Healthcare). Histidine-tagged MazE was selectively eluted with the elution buffer (20 mM sodium phosphate buffer (pH 8.0), 300 mM NaCl, 500 mM imidazole, and 5 mM 2-mercaptoethanol) using the following program: flow rate, 1 mL/min; linear elution gradient, 20 column volumes; fraction size, 0.5 mL. The 18th fraction from the beginning of the elution program was used for further experiments. The molecular weight and purity were confirmed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE). Protein concentration was determined using the Bio-Rad Protein Assay (Bio-Rad, Hercules, CA, United States).
Expression and Purification of MazFne1
Recombinant MazFne1 was expressed and purified as described previously with minor modifications (Miyamoto et al., 2016b). E. coli strain BL21 (DE3) cells (BioDynamics Laboratory Inc.) was transformed with pET24a-mazFne1. The transformant was cultivated overnight in LB medium supplemented with 20 μg/mL kanamycin at 37°C and inoculated into 1 L LB medium containing 20 μg/mL kanamycin. MazF was induced by the addition of 1 mM IPTG when the OD600 reached approximately 4.0. After 3.5 h of incubation, the cells were harvested by centrifugation at 9200 g and stored at -80°C. The cells containing MazFne1 were thawed on ice and resuspended in 18 mL of binding buffer (20 mM sodium phosphate buffer (pH 8.0), 300 mM NaCl, 40 mM imidazole, and 5 mM 2-mercaptoethanol). The cells were lysed by sonication and collected by centrifuging at 4400 g for 15 min. The supernatant was then filtered through a 0.45 μm membrane (Millex). After equilibrating a 1 mL His-Trap FF crude column (GE Healthcare), the supernatant was applied to the column and washed with 55 column volumes of binding buffer using AKTA pure 25 (GE Healthcare). Hexa-histidine tagged MazFne1 was selectively eluted using the following program: flow rate, 1 mL/min; linear elution gradient, 20 column volumes; fraction size, 0.5 mL. The elution buffer contained 20 mM sodium phosphate buffer (pH 8.0), 300 mM NaCl, 500 mM imidazole, and 5 mM 2-mercaptoethanol. The 17th and 18th fractions from the beginning of the elution program were used for further experiments. The molecular weight and purity were confirmed using SDS–PAGE. Protein concentration was determined using a Bio-Rad Protein Assay (Bio-Rad).
Enzymatic Activity of MazF and MazE
Synthetic RNA constructs were prepared as described in our previous study (Miyamoto et al., 2016a). RNA 500-2 (100 ng) was incubated with 1, 3, or 10 pmol of MazF at 37°C for 90 min in MazF reaction buffer [20 mM Tris–HCl (pH 8.0), 1 mM dithiothreitol, 0.01% Triton X-100, and 4 U of recombinant RNase inhibitor (TaKaRa, Shiga, Japan)] in a 50 μL reaction volume. As a control reaction, 10 pmol of MazF was pre-incubated with 50 pmol of MazE at 25°C for 10 min, and this mixture was incubated with 100 ng of RNA 500-2 at 37°C for 90 min in MazF reaction buffer. These RNAs were purified with RNA Clean and ConcentratorTM-5 (Zymo Research). Gel loading buffer II (Ambion, Austin, TX, United States) was added to each sample followed by incubation at 95°C for 5 min. Samples were separated on a 10% polyacrylamide gel containing 7 M urea. The RNA was stained using SYBR Gold (Life Technologies) and detected using a Typhoon 9210 imager (GE Healthcare).
Cleavage Sequence Identification
The cleavage sequence was identified using the protocols described in our previous study (Miyamoto et al., 2016a, 2017). Briefly, 0.625 pmol of eight RNA mixtures (500-2, 1000-1, 1000-2, 1000-3, 1000-4, 1000-5, 1500-1, and 2000-1) were incubated with 50 ng of MazFne1 at 37°C for 30 min in MazF reaction buffer. Phosphorylation, barcode ligation, and sequencing library construction were performed as described previously (Miyamoto et al., 2016a, 2017). Sequencing was performed using the MiSeq platform with the MiSeq 500 cycles reagent kit v2 (Illumina, San Diego, CA, United States) according to the manufacturer’s protocol. Sequence data were analysed using CLC Genomics 7.5.1 (Qiagen, Venlo, The Netherlands). The parameters described in our previous study (Miyamoto et al., 2016a, 2017) were used for the analysis. We added pseudo-counts to the nucleotide positions whose coverages were 0, and calculated the relative coverage increase, the value defined as the coverage at the (n + 1)th position divided by the coverage at the nth position. Nucleotide positions with coverage less than 100 were excluded from analysis. Of these nucleotide positions, those showing the overall top 50 relative coverage increases were selected from all references. The sequences five-base pairs upstream and downstream of these positions were extracted and aligned using WebLogo (Crooks et al., 2004). These sequence data were submitted to the DDBJ database under the accession number DRA006546.
Fluorometric Assay
The fluorometric assay was carried out as outlined in our previous study (Miyamoto et al., 2016a,b). Briefly, 0.7 pmol of MazFne1 or 100 ng of RNase A was incubated with 20 pmol of fluorescently labelled oligonucleotides in MazF reaction buffer in a total volume of 20 μL. As a control reaction, 0.7 pmol of MazFne1 was pre-incubated with 3.5 pmol of MazEne1 at 25°C for 10 min, and this mixture was incubated with 20 pmol of fluorescently labelled oligonucleotides in MazF reaction buffer in a total volume of 20 μL. All reactions were conducted at 37°C in triplicate and fluorescence intensity was recorded every 1 min using a Light Cycler 480 system (Roche, Basel, Switzerland) with 483 nm excitation and 533 nm detection filters.
Analysis of UGG Frequency in N. europaea Coding Sequences
Statistical analysis was performed as outlined in a previous study (Zhu et al., 2009). Protein-coding sequences of N. europaea were retrieved from the NCBI database. The CDS data as of February 10, 2018 were used for the analysis. The following parameters were taken into account: (i) p is the probability of TGG appearing in a N. europaea gene and represents the product of (percentage of U) multiplied by (percentage of G)2; (ii) L represents the length of each coding sequence (CDS); (iii) E is the expected number of TGG motifs in a CDS and is calculated as p(L – 2); (iv) K is the actual number of TGG sequence in a CDS; and (v) P represents the probability of each CDS containing TGG motifs at least K times. P is given by:
Accession Numbers
The GenBank accession numbers are as follows: artificially designed RNAs: 500-2 (AB610940), 1000-1 (AB610944), 1000-2 (AB610945), 1000-3 (AB610946), 1000-4 (AB610947), 1000-5 (AB610948), 1500-1 (AB610949), and 2000-1 (AB610950); N. europaea genome (NC_004757); MazEne1 (WP_041356592); MazFne1 (WP_011111532).
Results
MazFne1 Constitutes a Toxic Protein That Is Co-transcribed With Its Upstream Gene
Although the putative ORF existing immediately upstream of the mazF gene, encoded at ALW85_RS04820 (originally annotated as NE0921; mazFne1), was not initially annotated (Chain et al., 2003), bioinformatics analysis predicted that it codes for the MazE counterpart (MazEne1) (Supplementary Figure 1A) (Pandey and Gerdes, 2005), with MazEne1 and MazFne1 exhibiting 20.7 and 18.7% amino acid sequence identities to those of E. coli, respectively (Supplementary Figure 1B). Moreover, the mazEFne1 genes are organised in a manner fully consistent with the type II TA system (Sevin and Barloy-Hubler, 2007), with an 11 bp overlap between mazEne1 and mazFne1 (Supplementary Figure 2A).
A common feature of type II TA systems is their operon structure (Gerdes et al., 2005). To investigate whether the putative mazEF genes are co-transcribed in N. europaea, we carried out reverse transcription (RT)-PCR using three different primer sets (Supplementary Figure 2A, upper panel): one for the mazEne1 gene (primers 1 and 2), another for mazFne1 (primers 3 and 4), and the third for mazEFne1 (primers 1 and 4). In all cases, we observed amplification only in the presence of reverse transcriptase (Supplementary Figure 2A, lower panel), demonstrating that mazEne1 and mazFne1 are co-transcribed.
Another key feature of TA pairs is the effect of toxin expression on bacterial growth. To briefly investigate whether MazFne1 constitutes a toxin, we constructed an expression plasmid, in which the mazFne1 gene was under the control of an IPTG-inducible promoter, and examined whether MazFne1 overexpression retarded E. coli growth. As shown in Supplementary Figure 2B, MazFne1 induction led to stable growth arrest, indicating that mazFne1 indeed codes for a toxin. Taken together, these results suggest that MazEFne1 constitutes a TA system.
MazFne1 Forms a TA System With Its Cognate Antitoxin MazEne1
To evaluate whether MazEFne1 functions as a TA system at the molecular level, we purified these enzymes using affinity chromatography (Figure 1A). As shown in Figure 1B, dose-dependent RNA degradation was detected following treatment of the RNA substrate with MazFne1, suggesting that MazFne1 constitutes an endoribonuclease (Figure 1B, lanes 2–4). Notably, the substrate remained intact when MazEne1 was added prior to RNA exposure (Figure 1B, lane 5); thus, the observed RNA cleavage was catalysed by MazFne1 but not by contaminating RNases. From these results, we concluded that the mazEFne1 locus encodes a functional TA system.
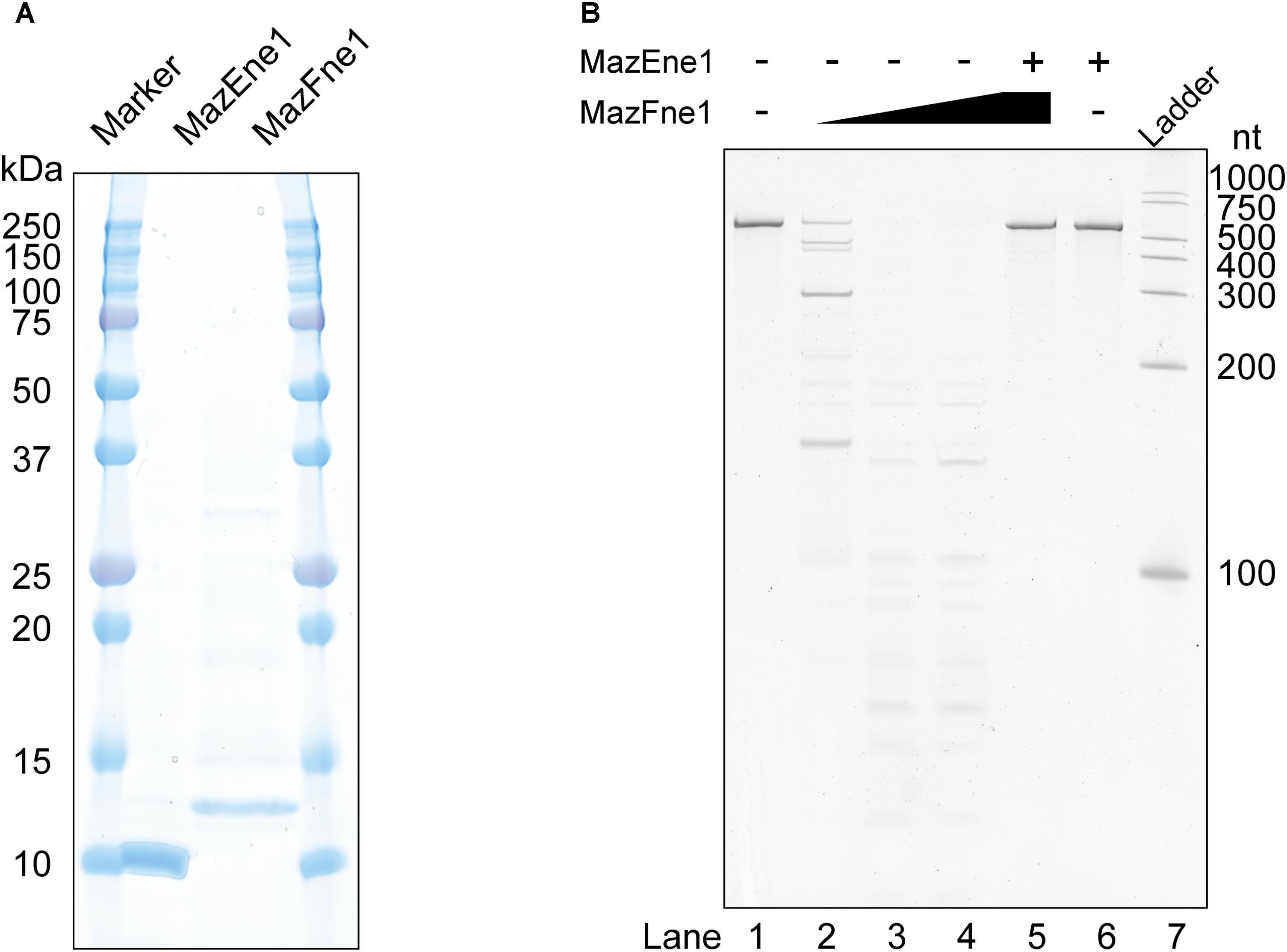
FIGURE 1. MazEF homologue isolated from N. europaea and their enzymatic activities. (A) Molecular weight and purity of MazEne1 and MazFne1. (B) A 533-nt artificially designed RNA (RNA 500-2) was incubated with MazEne1 and/or MazFne1: lane 1, a control reaction in the absence of enzymes; lanes 2–4, 1, 3, and 10 pmol of MazFne1 was added, respectively; lane 5, 10 pmol of MazFne1 was pre-incubated with 50 pmol of MazEne1 and RNA 500-2 was subsequently added; lane 6, 50 pmol of MazEne1 was added; lane 7, ladder.
Modified RNA-Seq Indicates That UGG Is Necessary for MazFne1 Cleavage
Having confirmed the endoribonuclease activity of MazFne1, we proceeded to define its cleavage-specificity. Toward this end, we utilised a specialised RNA-Seq that we have recently developed (Miyamoto et al., 2016a). In this approach, we digested eight artificially designed RNAs containing diverse 500–2000 nt sequences (500-2, 1000-1, 1000-2, 1000-3, 1000-4, 1000-5, 1500-1, and 2000-1) with MazFne1, and analysed the 5′-end sequences of the fragmented RNAs. As MazF digests RNA at specific sequence motifs, reads carrying particular sequences at their 5′ boundaries, which match the MazF-cleaved sites, are enriched. Accordingly, the coverage tends to increase at corresponding nucleotide positions (Figure 2A; Miyamoto et al., 2016a).
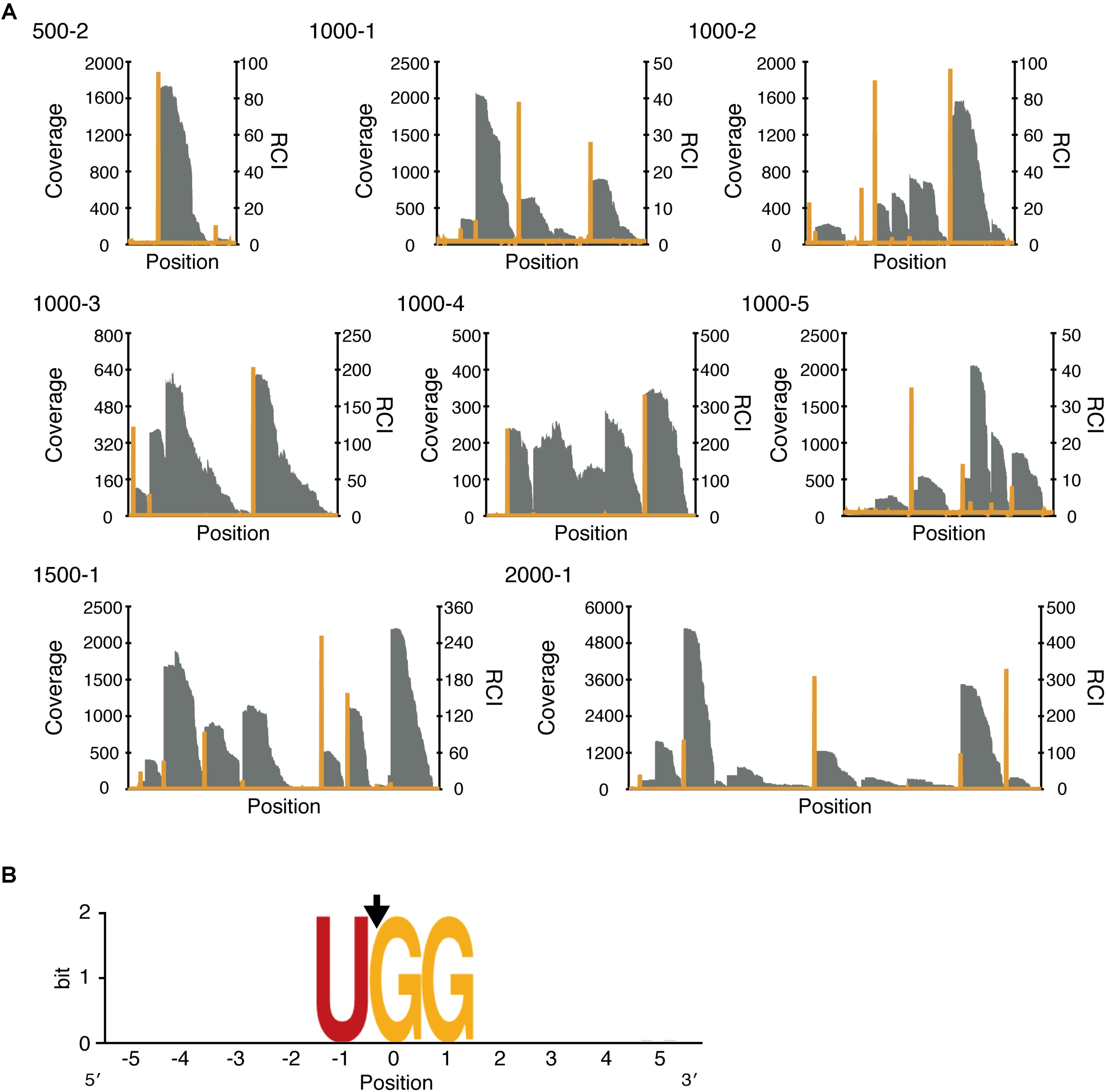
FIGURE 2. Analysis of the cleavage sequence of MazFne1. (A) Coverage (grey bar) and relative coverage increase (RCI; orange line), which is the value defined as the ratio of coverage at the (n + 1)th position to coverage at the nth position. (B) Nucleotide appearance frequency was visualised using WebLogo (Crooks et al., 2004). The black arrow indicates the position of the cleavage site.
To estimate the sequence determinants for MazFne1 cleavage, we extracted the sequences five bases up- and downstream of the identified nucleotide positions. After aligning the overall top 50 sequences, we found a strong consensus motif, UGG, from -1 to 1 positions (where the nucleotide position with coverage increase was numbered as zero; Figure 2B, Supplementary Table 2), suggesting that MazFne1 requires UGG triplets for its endoribonuclease activity and cut RNAs between U and the first G.
UGG Is the Target of MazFne1
To further determine the sequence requirements of MazFne1, we adopted a fluorescence quenching technique and prepared oligonucleotides (listed in Table 1) that were modified with 6-carboxyfluorescein (6-FAM) on the 5′-end and black hole quencher-1 (BHQ-1) on the 3′-end. Because these 2 dyes are tethered by nucleotides and are in close vicinity, the fluorescence of 6-FAM is dampened. As these oligonucleotides are degraded, however, unquenched 6-FAM molecules accumulate, resulting in an increase in fluorescence intensity (Wang and Hergenrother, 2007).
We sought to determine the MazFne1 target by monitoring the signal from 6-FAM. To corroborate the RNA-Seq result, we firstly utilised DR-13-UGG, which is a DNA-RNA chimeric oligonucleotide containing a UGG triplet in the middle of adenine repeats. As we anticipated, the oligonucleotide was sensitive to MazFne1 (Figure 3A, green plot). The cleavage was specifically caused by MazFne1 and was considered to occur at RNA nucleotide sites, because (i) cleavage activity was completely blocked upon MazEne1 addition (Figure 3A, yellow plot), and (ii) no detectable change in fluorescence intensity was observed when D-13-AAA, a DNA oligonucleotide, was used instead as the substrate for MazFne1 (Figure 3B).
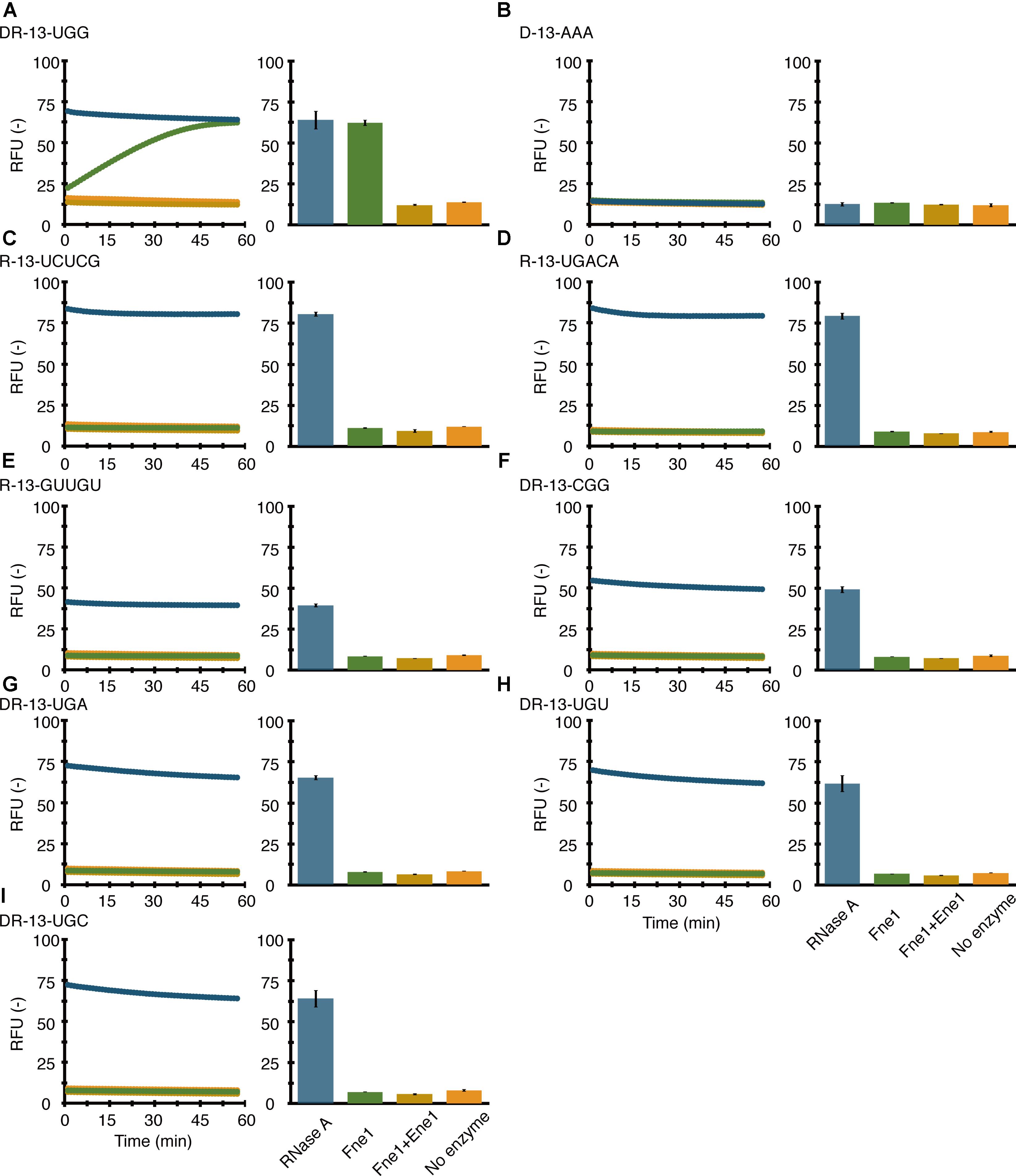
FIGURE 3. Sequence-specific RNA cleavage with MazFne1; 0.7 pmol of MazFne1 (green) or MazFne1 pre-incubated with 3.5 pmol of MazEne1 (yellow) was incubated with 20 pmol of fluorogenic oligonucleotides: (A) DR-13-UGG, (B) D-13-AAA, (C) R-13-UCUCG, (D) R-13-UGACA, (E) R-13-GUUGU, (F) DR-13-CGG, (G) DR-13-UGA, (H) DR-13-UGU, or (I) DR-13-UGC. In the control reactions, fluorescence intensities in the presence of 100 ng of RNase A (blue) and absence of enzymes (orange) at each time point (left) and the end point (right) were measured.
We next prepared three additional RNA oligonucleotides (R-13-UCUCG, R-13-UGACA, and R-13-GUUGU), whose sequences are identical to that of RNA 1000-4 (one of the substrates used in the modified RNA-Seq) but lack the UGG motif. As expected, these RNAs were also tolerant to MazFne1 (Figures 3C–E). The results were concordant with the result from the modified RNA-Seq, suggesting that UGG is the target of MazFne1.
Lastly, we synthesised four DNA-RNA chimeric oligonucleotides (DR-13-CGG, DR-13-UGA, DR-13-UGU, and DR-13-UGC) to investigate whether MazFne1 cleaves 3-base motifs similar to UGG. As shown in Figures 3F–I, none of these oligonucleotides were degraded. Taken together, our findings demonstrated that MazFne1 constitutes an endoribonuclease that specifically cleaves the UGG triplet.
The UGG Motif Is Abundant in Genes That Are Unique in the Ammonia-Oxidising Bacteria
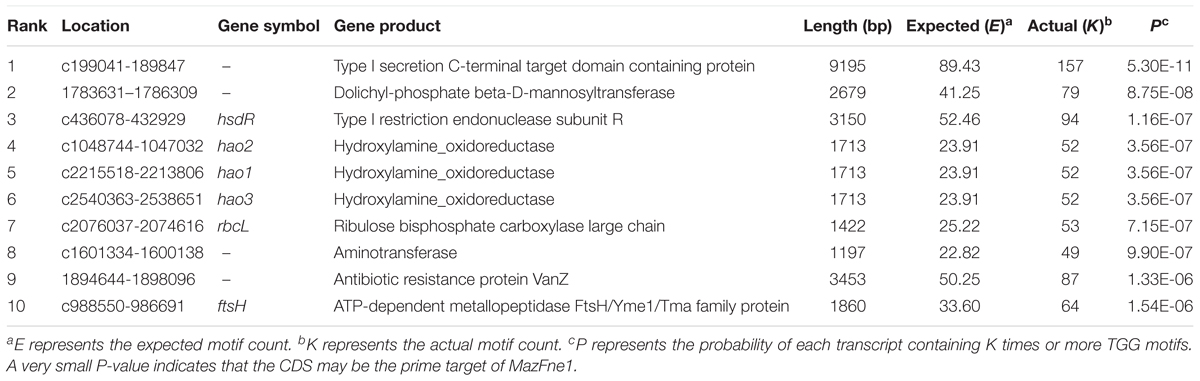
The number of cleavage sequences in a transcript has been shown to correlate with mRNA stabilities (Zhu et al., 2009; Schifano et al., 2014). To evaluate the potential effect of MazFne1 on N. europaea cells, we performed statistical analysis and searched for genes containing multiple TGG sequences (Zhu et al., 2009) among the 2,646 coding sequences (CDS) identified in this organism. Specifically, we counted the actual number of TGG triplets in each CDS (K) and calculated the probabilities of each transcript containing K times or more TGG motifs (P), on the basis of nucleotide contents and the length of each CDS (Figure 4). Notably, when we extracted the top 10 CDS with the smallest P-values, the hao and rbcL genes, which are indispensable for ammonia oxidation and CO2 fixation, respectively, were detected (Table 2). As the CDS showing small P-values are considered to be preferentially digested, MazFne1 may serve to efficiently repress N. europaea cellular activities (see Discussion).
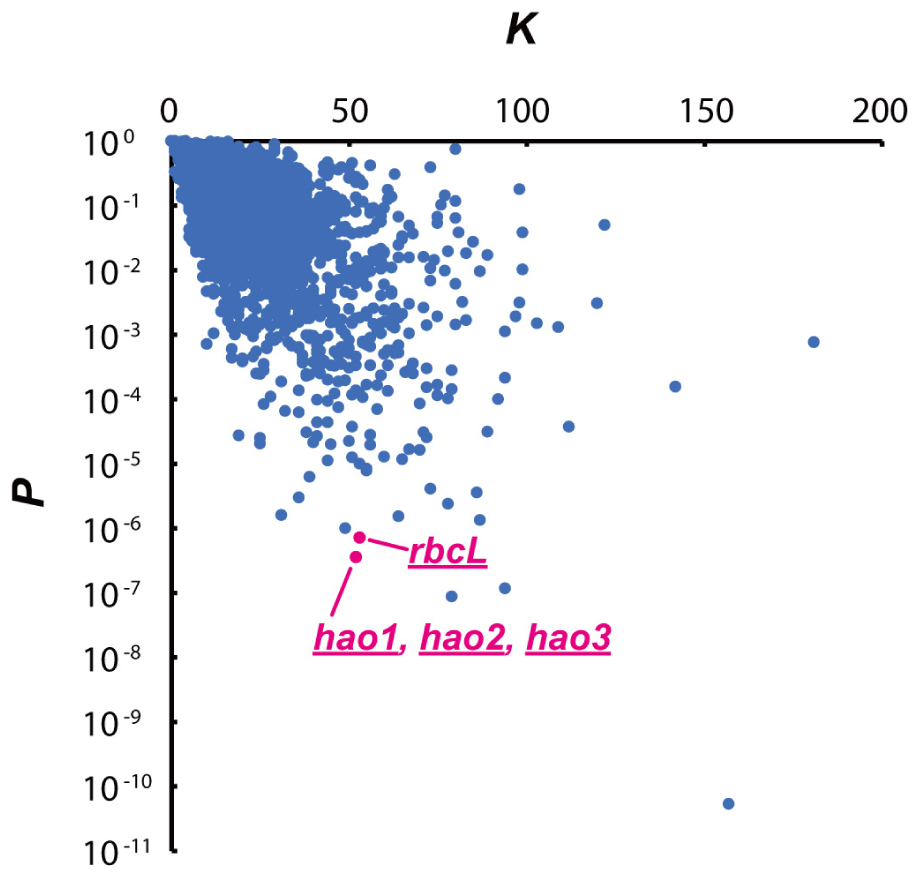
FIGURE 4. Relationship between the actual number of TGG triplets (K) and P-values in each CDS. A very small P-value indicates that the CDS is preferentially degraded by MazFne. Dots correspond to hao, and rbcL transcripts are highlighted.
Discussion
Microbes have to cope with sudden environmental alterations. A propitious strategy to survive in fluctuating surroundings is to regulate growth, with prokaryotic TA systems being a well-known example of such a strategy (Hall et al., 2017). In the present study, we characterised one of these representative TA systems, the MazEF family, encoded on the N. europaea chromosome. We found that MazEFne1 shares typical features of type II TA pairs: first, mazEne1 and mazFne1 are co-transcribed in N. europaea (Supplementary Figure 2A); second, ectopic expression of MazFne1 is toxic to bacterial cells (Supplementary Figure 2B); third, MazFne1 possesses endoribonuclease activity and MazEne1 counteracts its enzymatic activity (Figure 1); and fourth, MazFne1 specifically cleaves RNA at the UGG motif (Figures 2, 3).
Five MazF family members are encoded in the N. europaea chromosome. Although the sequence-specificities of these enzymes are not completely elucidated, they are, similar to other MazF homologues, considered to have different targets, triggers, and roles (Sat et al., 2003; Hazan et al., 2004; Tiwari et al., 2015; Gupta et al., 2017; Schifano and Woychik, 2017), because of their low sequence similarities and distinct loci. Indeed, we have previously shown that the N. europaea MazF encoded at ALW85_RS06130 (MazFne3; Supplementary Figure 1A) is an AAU-specific cutter. To the best of our knowledge, no stresses that activate MazFne1 have been identified. Further studies are needed to understand how these enzymes are beneficial to environmental fluctuations.
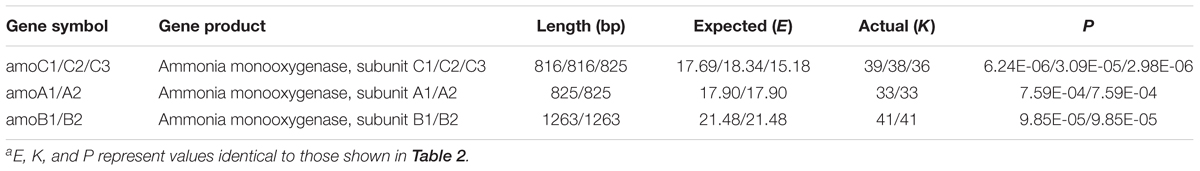
N. europaea production of reductant and energy via ammonia oxidation has been well elucidated. The process comprises a sequential oxidation reaction of ammonia to hydroxylamine (NH3 to NH2OH) and hydroxylamine to nitrite (NH2OH to NO2-); the former is catalysed by ammonia monooxygenase (AMO) and the latter by hydroxylamine oxidoreductase (recently designated as hydroxylamine dehydrogenase) (HAO) and putatively by nitric oxide oxidase (Caranto and Lancaster, 2017). Four electrons are released through hydroxylamine oxidation, half of which are used for ammonia oxidation and the other half for proton gradient formation, followed by ATP and NADH production (Arp et al., 2002; Arp and Stein, 2003). In the present study, we found that TGG triplets are strikingly abundant in the three copies of the hao genes (Table 2). Notably, these triplets are also relatively rich in amo genes (Table 3). Conceivably, the AMO and HAO transcripts are destabilised when MazFne1 is activated in N. europaea cells, thereby inhibiting cellular activities requiring energy and reductant.
Moreover, in addition to ammonia oxidation, carbon fixation would also likely be impaired. N. europaea utilises the Calvon-Benson-Bassham (CBB) cycle to fix carbon dioxide (Chain et al., 2003), with this step being initiated by a type I ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCo), which is composed of a large subunit and small subunits (Wei et al., 2004). In our analysis, the rbcL gene, encoding the large subunit of RubisCo, is predicted to be sensitive to MazFne1 (Table 2). Taken together with the fact that in N. europaea, the CBB cycle is powered by ATP and NADH generated from ammonia oxidation (Chain et al., 2003), MazFne1 would supposedly severely compromise the biosynthesis processes of this bacterium.
It has been previously observed that type II TA systems are abundant in some slowly growing bacteria, including N. europaea (Pandey and Gerdes, 2005). Although it remains unclear why these seemingly redundant TA pairs exist in this bacterium, the number of TA pairs might be associated with the growth rate. Indeed, 99.9% of N. europaea coding sequences contain at least one TGG motif. Presumably, MazFne1 arrests N. europaea growth by directly shutting down almost all protein synthesis, in addition to the indirect downregulation of many biochemical reactions through the inhibition of ammonia oxidation and carbon fixation. Given that a low metabolic state represents a common bacterial strategy to improve stress resistance and that TA systems are implicated in the formation of such states, referred to as “dormant and/or viable but non-culturable” (Helaine et al., 2014; Li et al., 2014; Ayrapetyan et al., 2015; Tiwari et al., 2015), N. europaea might withstand environmental stresses with the aid of MazFne1.
In summary, we found that MazFne1 codes for a functional endonucleolytic toxin and constitutes a TA pair with its cognate antitoxin, MazEne1. Additionally, MazFne1 digests RNAs at UGG sites in a ribosome-independent manner. Our finding indicates that N. europaea might benefit from the growth modulation mediated by MazFne1 under stressful conditions.
Author Contributions
TM, AY, YO, MT, RA, ST, and NN conceived and designed the experiments. TM, AY, YO, and MT performed the experiments. TM, AY, YO, MT, and RA analyzed the data. TM wrote the paper.
Funding
This work was supported by Japan Society for the Promotion of Science KAKENHI Grant Nos. 15K14241 and 16J09473.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02386/full#supplementary-material
References
Aakre, C. D., Phung, T. N., Huang, D., and Laub, M. T. (2013). A bacterial toxin inhibits DNA replication elongation through a direct interaction with the beta sliding clamp. Mol. Cell. 52, 617–628. doi: 10.1016/j.molcel.2013.10.014
Aizenman, E., Engelberg-Kulka, H., and Glaser, G. (1996). An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3’,5’-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. U.S.A. 93, 6059–6063. doi: 10.1073/pnas.93.12.6059
Amitai, S., Kolodkin-Gal, I., Hananya-Meltabashi, M., Sacher, A., and Engelberg-Kulka, H. (2009). Escherichia coli MazF leads to the simultaneous selective synthesis of both “death proteins” and “survival proteins”. PLoS Genet. 5:e1000390. doi: 10.1371/journal.pgen.1000390
Arp, D. J., Sayavedra-Soto, L. A., and Hommes, N. G. (2002). Molecular biology and biochemistry of ammonia oxidation by Nitrosomonas europaea. Arch. Microbiol. 178, 250–255. doi: 10.1007/s00203-002-0452-0
Arp, D. J., and Stein, L. Y. (2003). Metabolism of inorganic N compounds by ammonia-oxidizing bacteria. Crit. Rev. Biochem. Mol. Biol. 38, 471–495. doi: 10.1080/10409230390267446
Ayrapetyan, M., Williams, T. C., Baxter, R., and Oliver, J. D. (2015). Viable but nonculturable and persister cells coexist stochastically and are induced by human serum. Infect. Immun. 83, 4194–4203. doi: 10.1128/IAI.00404-15
Bothe, H., Jost, G., Schloter, M., Ward, B. B., and Witzel, K.-P. (2000). Molecular analysis of ammonia oxidation and denitrification in natural environments. FEMS Microbiol. Rev. 24, 673–690. doi: 10.1111/j.1574-6976.2000.tb00566.x
Brantl, S. (2012). Bacterial type I toxin-antitoxin systems. RNA Biol. 9, 1488–1490. doi: 10.4161/rna.23045
Caranto, J. D., and Lancaster, K. M. (2017). Nitric oxide is an obligate bacterial nitrification intermediate produced by hydroxylamine oxidoreductase. Proc. Natl. Acad. Sci. U.S.A. 114, 8217–8222. doi: 10.1073/pnas.1704504114
Chain, P., Lamerdin, J., Larimer, F., Regala, W., Lao, V., Land, M., et al. (2003). Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J. Bacteriol. 185, 2759–2773. doi: 10.1128/jb.185.9.2759-2773.2003
Chopra, N., Saumitra, Pathak, A., Bhatnagar, R., and Bhatnagar, S. (2013). Linkage, mobility, and selfishness in the MazF family of bacterial toxins: a snapshot of bacterial evolution. Genome Biol. Evol. 5, 2268–2284. doi: 10.1093/gbe/evt175
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Gerdes, K., Christensen, S. K., and Løbner-Olesen, A. (2005). Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3, 371–382. doi: 10.1038/nrmicro1147
Goeders, N., Chai, R., Chen, B., Day, A., and Salmond, G. P. (2016). Structure, evolution, and functions of bacterial type III toxin-antitoxin systems. Toxins 8:E282. doi: 10.3390/toxins8100282
Groeneweg, J., Sellner, B., and Tappe, W. (1994). Ammonia oxidation in Nitrosomonas at NH3 concentrations near km: effects of pH and temperature. Water Res. 28, 2561–2566. doi: 10.1016/0043-1354(94)90074-4
Gupta, A., Venkataraman, B., Vasudevan, M., and Gopinath, B. K. (2017). Co-expression network analysis of toxin-antitoxin loci in Mycobacterium tuberculosis reveals key modulators of cellular stress. Sci. Rep. 7:5868–5881. doi: 10.1038/s41598-017-06003-7
Hall, A. M., Gollan, B., and Helaine, S. (2017). Toxin-antitoxin systems: reversible toxicity. Curr. Opin. Microbiol. 36, 102–110. doi: 10.1016/j.mib.2017.02.003
Hazan, R., Sat, B., and Engelberg-Kulka, H. (2004). Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 186, 3663–3669. doi: 10.1128/JB.186.11.3663-3669.2004
Helaine, S., Cheverton, A. M., Watson, K. G., Faure, L. M., Matthews, S. A., and Holden, D. W. (2014). Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343, 204–208. doi: 10.1126/science.1244705
Koops, H. P., Purkhold, U., Pommerening-Röser, A., Timmermann, G., and Wagner, M. (2006). “The lithoautotrophic ammonia-oxidizing bacteria,” in The Prokaryotes, Vol. 5, ed. E. Stackebrandt (New York, NY: Springer-Verlag), 778–811.
Li, L., Mendis, N., Trigui, H., Oliver, J. D., and Faucher, S. P. (2014). The importance of the viable but non-culturable state in human bacterial pathogens. Front. Microbiol. 5:258. doi: 10.3389/fmicb.2014.00258
Li, T., Weng, Y., Ma, X., Tian, B., Dai, S., Jin, Y., et al. (2017). Deinococcus radiodurans toxin-antitoxin MazEF-dr mediates cell death in response to DNA damage stress. Front. Microbiol. 8:1427. doi: 10.3389/fmicb.2017.01427
Masuda, H., and Inouye, M. (2017). Toxins of prokaryotic toxin-antitoxin systems with sequence-specific endoribonuclease activity. Toxins 9:E140. doi: 10.3390/toxins9040140
Masuda, H., Tan, Q., Awano, N., Wu, K. P., and Inouye, M. (2012a). YeeU enhances the bundling of cytoskeletal polymers of MreB and FtsZ, antagonizing the CbtA (YeeV) toxicity in Escherichia coli. Mol. Microbiol. 84, 979–989. doi: 10.1111/j.1365-2958.2012.08068.x
Masuda, H., Tan, Q., Awano, N., Yamaguchi, Y., and Inouye, M. (2012b). A novel membrane-bound toxin for cell division, CptA (YgfX), inhibits polymerization of cytoskeleton proteins, FtsZ and MreB, in Escherichia coli. FEMS Microbiol. Lett. 328, 174–181. doi: 10.1111/j.1574-6968.2012.02496.x
Miyamoto, T., Kato, Y., Sekiguchi, Y., Tsuneda, S., and Noda, N. (2016a). Characterization of MazF-mediated sequence-specific RNA cleavage in Pseudomonas putida using massive parallel sequencing. PLoS One 11:e0149494. doi: 10.1371/journal.pone.0149494
Miyamoto, T., Yokota, A., Tsuneda, S., and Noda, N. (2016b). AAU-specific RNA cleavage mediated by MazF toxin endoribonuclease conserved in Nitrosomonas europaea. Toxins 8:E174. doi: 10.3390/toxins8060174
Miyamoto, T., Ota, Y., Yokota, A., Suyama, T., Tsuneda, S., and Noda, N. (2017). Characterization of a Deinococcus radiodurans MazF: a UACA-specific RNA endoribonuclease. Microbiologyopen 6:e00501. doi: 10.1002/mbo3.501
Page, R., and Peti, W. (2016). Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 12, 208–214. doi: 10.1038/nchembio.2044
Pandey, D. P., and Gerdes, K. (2005). Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33, 966–976. doi: 10.1093/nar/gki201
Park, J. H., Yamaguchi, Y., and Inouye, M. (2011). Bacillus subtilis MazF-bs (EndoA) is a UACAU-specific mRNA interferase. FEBS Lett. 585, 2526–2532. doi: 10.1016/j.febslet.2011.07.008
Park, S., and Ely, R. L. (2008a). Candidate stress genes of Nitrosomonas europaea for monitoring inhibition of nitrification by heavy metals. Appl. Environ. Microbiol. 74, 5475–5482. doi: 10.1128/AEM.00500-08
Park, S., and Ely, R. L. (2008b). Genome-wide transcriptional responses of Nitrosomonas europaea to zinc. Arch. Microbiol. 189, 541–548. doi: 10.1007/s00203-007-0341-7
Pellitteri-Hahn, M. C., Halligan, B. D., Scalf, M., Smith, L., and Hickey, W. J. (2011). Quantitative proteomic analysis of the chemolithoautotrophic bacterium Nitrosomonas europaea: comparison of growing- and energy-starved cells. J. Proteomics 74, 411–419. doi: 10.1016/j.jprot.2010.12.003
Radniecki, T. S., Semprini, L., and Dolan, M. E. (2009). Expression of merA, amoA and hao in continuously cultured Nitrosomonas europaea cells exposed to zinc chloride additions. Biotechnol. Bioeng. 102, 546–553. doi: 10.1002/bit.22069
Rothenbacher, F. P., Suzuki, M., Hurley, J. M., Montville, T. J., Kirn, T. J., Ouyang, M., et al. (2012). Clostridium difficile MazF toxin exhibits selective, not global, mRNA cleavage. J. Bacteriol. 194, 3464–3474. doi: 10.1128/JB.00217-12
Sat, B., Reches, M., and Engelberg-Kulka, H. (2003). The Escherichia coli mazEF suicide module mediates thymineless death. J. Bacteriol. 185, 1803–1807. doi: 10.1128/JB.185.6.1803-1807.2003
Sauert, M., Wolfinger, M. T., Vesper, O., Müller, C., Byrgazov, K., and Moll, I. (2016). The MazF-regulon: a toolbox for the post-transcriptional stress response in Escherichia coli. Nucleic Acids Res. 44, 6660–6675. doi: 10.1093/nar/gkw115
Schifano, J. M., Edifor, R., Sharp, J. D., Ouyang, M., Konkimalla, A., Husson, R. N., et al. (2013). Mycobacterial toxin MazF-mt6 inhibits translation through cleavage of 23S rRNA at the ribosomal A site. Proc. Natl. Acad. Sci. U.S.A. 110, 8501–8506. doi: 10.1073/pnas.1222031110
Schifano, J. M., Vvedenskaya, I. O., Knoblauch, J. G., Ouyang, M., Nickels, B. E., and Woychik, N. A. (2014). An RNA-seq method for defining endoribonuclease cleavage specificity identifies dual rRNA substrates for toxin MazF-mt3. Nat. Commun. 5:3538. doi: 10.1038/ncomms4538
Schifano, J. M., and Woychik, N. A. (2017). Cloaked dagger: tRNA slicing by an unlikely culprit. RNA Biol. 14, 15–19. doi: 10.1080/15476286.2016.1255396
Schuster, C. F., and Bertram, R. (2013). Toxin-antitoxin systems are ubiquitous and versatile modulators of prokaryotic cell fate. FEMS Microbiol. Lett. 340, 73–85. doi: 10.1111/1574-6968.12074
Schuster, C. F., Park, J. H., Prax, M., Herbig, A., Nieselt, K., Rosenstein, R., et al. (2013). Characterization of a mazEF toxin-antitoxin homologue from Staphylococcus equorum. J. Bacteriol. 195, 115–125. doi: 10.1128/JB.00400-12
Sevin, E. W., and Barloy-Hubler, F. (2007). RASTA-Bacteria: a web-based tool for identifying toxin-antitoxin loci in prokaryotes. Genome Biol. 8:R155. doi: 10.1186/gb-2007-8-8-r155
Shao, Y., Harrison, E. M., Bi, D., Tai, C., He, X., Ou, H. Y., et al. (2011). TADB: a web-based resource for Type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res. 39, D606–D611. doi: 10.1093/nar/gkq908
Stein, L. Y., and Arp, D. J. (1998). Loss of ammonia monooxygenase activity in Nitrosomonas europaea upon exposure to nitrite. Appl. Environ. Microbiol. 64, 4098–4102.
Tiwari, P., Arora, G., Singh, M., Kidwai, S., Narayan, O. P., and Singh, R. (2015). MazF ribonucleases promote Mycobacterium tuberculosis drug tolerance and virulence in guinea pigs. Nat. Commun. 6:6059. doi: 10.1038/ncomms7059
Verma, S., and Bhatnagar, R. (2014). MoxT toxin of Bacillus anthracis exhibits sequence specific ribonuclease activity. Biochem. Biophys. Res. Commun. 450, 998–1004. doi: 10.1016/j.bbrc.2014.06.092
Wang, N. R., and Hergenrother, P. J. (2007). A continuous fluorometric assay for the assessment of MazF ribonuclease activity. Anal. Biochem. 371, 173–183. doi: 10.1016/j.ab.2007.07.017
Wang, X., Lord, D. M., Cheng, H. Y., Osbourne, D. O., Hong, S. H., Sanchez-Torres, V., et al. (2012). A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat. Chem. Biol. 8, 855–861. doi: 10.1038/nchembio.1062
Wei, X., Sayavedra-Soto, L. A., and Arp, D. J. (2004). The transcription of the cbb operon in Nitrosomonas europaea. Microbiology 150, 1869–1879. doi: 10.1099/mic.0.26785-0
Wei, X., Vajrala, N., Hauser, L., Sayavedra-Soto, L. A., and Arp, D. J. (2006a). Iron nutrition and physiological responses to iron stress in Nitrosomonas europaea. Arch. Microbiol. 186, 107–118. doi: 10.1007/s00203-006-0126-4
Wei, X., Yan, T., Hommes, N. G., Liu, X., Wu, L., McAlvin, C., et al. (2006b). Transcript profiles of Nitrosomonas europaea during growth and upon deprivation of ammonia and carbonate. FEMS Microbiol. Lett. 257, 76–83. doi: 10.1111/j.1574-6968.2006.00152.x
Xie, Y., Wei, Y., Shen, Y., Li, X., Zhou, H., Tai, C., et al. (2017). TADB 2.0: an updated database of bacterial type II toxin-antitoxin loci. Nucleic Acids Res. 46, D749–D753. doi: 10.1093/nar/gkx1033
Yu, R., and Chandran, K. (2010). Strategies of Nitrosomonas europaea 19718 to counter low dissolved oxygen and high nitrite concentrations. BMC Microbiol. 10:70. doi: 10.1186/1471-2180-10-70
Zhang, Y. L., Zhang, J. J., Hoeflich, K. P., Ikura, M., Qing, G. L., and Inouye, M. (2003). MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12, 913–923. doi: 10.1016/S1097-2765(03)00402-7
Zhu, L., Inoue, K., Yoshizumi, S., Kobayashi, H., Zhang, Y., Ouyang, M., et al. (2009). Staphylococcus aureus MazF specifically cleaves a pentad sequence, UACAU, which is unusually abundant in the mRNA for pathogenic adhesive factor SraP. J. Bacteriol. 191, 3248–3255. doi: 10.1128/JB.01815-08
Zhu, L., Phadtare, S., Nariya, H., Ouyang, M., Husson, R. N., and Inouye, M. (2008). The mRNA interferases, MazF-mt3 and MazF-mt7 from Mycobacterium tuberculosis target unique pentad sequences in single-stranded RNA. Mol. Microbiol. 69, 559–569. doi: 10.1111/j.1365-2958.2008.06284.x
Keywords: toxin-antitoxin system, MazEF, sequence-specificity, RNase, Nitrosomonas europaea, ammonia oxidation, carbon fixation
Citation: Miyamoto T, Yokota A, Ota Y, Tsuruga M, Aoi R, Tsuneda S and Noda N (2018) Nitrosomonas europaea MazF Specifically Recognises the UGG Motif and Promotes Selective RNA Degradation. Front. Microbiol. 9:2386. doi: 10.3389/fmicb.2018.02386
Received: 01 August 2018; Accepted: 18 September 2018;
Published: 08 October 2018.
Edited by:
Martin G. Klotz, Washington State University, United StatesReviewed by:
Jeanette M. Norton, Utah State University, United StatesLisa Y. Stein, University of Alberta, Canada
Copyright © 2018 Miyamoto, Yokota, Ota, Tsuruga, Aoi, Tsuneda and Noda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Satoshi Tsuneda, c3RzdW5lZGFAd2FzZWRhLmpw Naohiro Noda, bm9kYS1uYW9oaXJvQGFpc3QuZ28uanA=
†These authors have contributed equally to this work
 Tatsuki Miyamoto
Tatsuki Miyamoto Akiko Yokota
Akiko Yokota Yuri Ota1,2
Yuri Ota1,2 Satoshi Tsuneda
Satoshi Tsuneda