- Institute of Agricultural Resources and Regional Planning, Chinese Academy of Agricultural Sciences, Beijing, China
High temperature is a key limiting factor for mycelium growth and development in Pleurotus ostreatus. Thermotolerance includes the direct response to heat stress and the ability to recover from heat stress. To better understand the mechanism of thermotolerance in P. ostreatus, we used morphological and physiological analysis combined with an iTRAQ-based proteomics analysis of P. ostreatus subjected to 40°C for 48 h followed by recovery at 25°C for 3 days. High temperature increased the concentrations of thiobarbituric acid reactive substances (TBARS) indicating that the mycelium of P. ostreatus were damaged by heat stress. However, these physiological changes rapidly returned to control levels during the subsequent recovery phase from heat stress. In comparison to unstressed controls, a total of 204 proteins were changed during heat stress and/or the recovery phase. Wherein, there were 47 proteins that responded to both stress and recovery conditions, whereas 84 and 73 proteins were responsive to only heat stress or recovery conditions, respectively. Furthermore, quantitative real-time PCR (qRT-PCR) confirmed differential expression of nine candidate genes revealed that some of the proteins, such as a mitogen-activated protein kinase (MAPK), phenylalanine ammonia-lyase (PAL), and heat shock protein (HSP), were also regulated by heat stress at the level of transcription. These differentially expressed proteins (DEPs) in mycelium of P. ostreatus under heat stress were from 13 biological processes. Moreover, protein–protein interaction analysis revealed that proteins involved in carbohydrate and energy metabolism, signal transduction, and proteins metabolism could be assigned to three heat stress response networks. On the basis of these findings, we proposed that effective regulatory protein expression related to MAPK-pathway, antioxidant enzymes, HSPs, and other stress response proteins, and glycolysis play important roles in enhancing P. ostreatus adaptation to and recovery from heat stress. Of note, this study provides useful information for understanding the thermotolerance mechanism for basidiomycetes.
Introduction
Pleurotus ostreatus, also known as the oyster mushroom, is the third largest edible fungus produced in China. In 2015, the annual oyster mushroom production was estimated at 5.9 million tons, which represented 17% of the total edible fungi production for that year (data from China edible fungi association). P. ostreatus is highly valued for its superior texture, flavor, and nutritional quality as well as its demonstrated antioxidative, hypocholesterolemic, and antiatherogenic activities (Anandhi et al., 2013), antitumor properties (Jedinak and Sliva, 2008), and its ability to enhance the immune system (Jesenak et al., 2013). It is one of the most widely cultivated and consumed edible mushrooms in China due to its short growth time, high adaptability, and productivity.
High temperature stress or heat stress is defined as the temperature that when held beyond a critical threshold for a sufficient period of time will cause irreversible damage to growth and development. Heat stress for several days inhibits mycelium growth, impairs fruiting, and affects the quality of the mushroom (Chang and Miles, 2004). In China, P. ostreatus is usually cultivated within agricultural type facilities where it often encounters heat stress which reduces hyphae viability, delays fruiting, and leads to a decrease in production yield. Therefore, temperature is one of the crucial environmental factors that influence mushroom growth and development. Since tolerance to heat and other abiotic stressors is necessary for organisms to live in adverse environmental conditions and to function properly, the strategies of adaptation to high temperatures employed in P. ostreatus mycelium need further investigation. Previous studies exploring P. ostreatus response to high temperatures have only focused on physiological changes including cell programmed death (Song et al., 2014), cell membrane stability (Kong et al., 2012), mycelial micromorphology, and antioxidant systems (Meng et al., 2015), but few studies to date have investigated the changes in protein expression induced by heat stress during the thermotolerance response.
The present work aims to evaluate the quantitative changes in protein expression in the mycelium of P. ostreatus in response to heat stress using isobaric tags for relative and absolute quantitation (iTRAQ), an extremely powerful tool for identifying dynamic changes in proteomes on a global scale. Proteomic responses to abiotic stress have been widely studied in many plants and fungi including rice, wheat, barley, Populus euphratica, norway spruce, bitter gourd, grapevine (Liu et al., 2014), soybean (Das et al., 2016) Flammulina velutipes (Liu et al., 2017), Agaricus bisporus (Zhao-Ming et al., 2009), and Boletus edulis (Liang et al., 2007). iTRAQ has become a powerful method for investigating proteomic changes during various developmental stages (Hultinrosenberg et al., 2013). This technique has a high degree of sensitivity, and the lysine or N-terminal amine specific isobaric reagents of iTRAQ allow the identification and quantitation of multiple samples simultaneously.
In this study, iTRAQ labeling coupled with liquid chromatography-tandem mass spectrometry (LC-MS/MS) was used to identify differentially expressed proteins (DEPs) under heat stress and their subsequent recovery in order to better understand thermotolerance in mycelium of P. ostreatus. In addition, the morphological and physiological changes induced by heat stress were observed for each treatment. Moreover, we compared the changes at the proteomic and transcriptional levels under heat stress and their subsequent recovery conditions. These data might also provide new insights to the underlying molecular mechanisms of the proteins involved in thermotolerance in basidiomycetes.
Materials and Methods
Strain and Growth Conditions
Pleurotus ostreatus (CCMSSC 00389) was provided by the China Center for Mushroom Spawn Standards and Control. For all experiments, mycelia were grown in potato-dextrose agar (PDA) medium for 7 days at 28°C. Then 0.1 g of mycelia from solid medium were transferred to 100 mL of DifcoTM Potato Dextrose Broth medium in 250 mL erlenmeyer flasks. The mixture was dispersed using a liquid homogenizer, then returned to a culture flask, and incubated with shaking at 28°C and 160 rpm for 5 days.
Heat and Recovery Treatments
The experimental plates included four different treatments: control treatment 1 (CK1): cultures were incubated with shaking at 28°C and 160 rpm for 5 days then held stationary at 28°C for 48 h. Heat stress (HS): cultures were incubated with shaking at 28°C and 160 rpm for 5 days then held stationary and subjected to heat stress at 40°C for 48 h. Recovery (RC): following the heat stress, cultures were incubated with shaking at 28°C and 160 rpm for 3 days. Control treatment 2 (CK2): cultures were incubated with shaking at 28°C and 160 rpm for 5 days then held stationary at 28°C for 48 h followed by incubation with shaking at 28°C and 160 rpm for 3 days.
Measurement of Thiobarbituric Acid Reactive Substances (TBARS)
Thiobarbituric acid reactive substances (TBARS) were analyzed according to the method of Kong et al. (2012) with some modifications. The mycelia were ground into powder with liquid nitrogen, and then transferred into a 1.5 mL Eppendorf tube. Briefly, 0.5 mL of 5% TCA was added. Then the mixture was extracted for 10 min in ice water bath. The supernatants were collected by centrifuging at 10,000 × g for 10 min and mixed with 0.5 mL of 0.67% TBA in a new Eppendorf tube. The mixture was subsequently incubated at 95°C for 30 min, and then centrifuged at 10,000 × g for 10 min. The absorbance of the supernatant was measured at 532 and 600 nm wavelength using a UV-spectrophotometer (TU-1810, PERSEE, Beijing, China). All tests were performed in triplicate.
Protein Extraction and iTRAQ Labeling
Protein extraction was performed according to a modified version of the trichloroacetic acid (TCA) acetone precipitation method described by Pratt et al. (2006) with some modifications. Triplicates of the frozen mycelia were combined equally for iTRAQ analysis. Approximately 500 mg of each ground up mycelia sample was combined with 10 mL of 10% m/v TCA in acetone and the samples were incubated at -20°C for 12 h. The samples were then centrifuged at 10,000 g for 15 min at 4°C. The supernatant was discarded without disturbing the pellets. The washing step with pre-cooled acetone was repeated three times until the pellets were white. The dried pellets were lyzed with 1 mL protein extraction reagent (4% SDS, 100 mM DTT, and 150 mM Tris-HCl, pH8.0). The pellets were dissolved by ultrasound (pulse on 10 s, pulse off 15 s, power 50 W) using 10 repeats and incubated at 100°C for 5 min. The solution was centrifuged at 40,000 g for 30 min at 4°C to remove insoluble impurities. The concentration of the protein was determined by the Brandford method using bovine serum albumin as a standard (Bradford, 1976), and the protein samples were analyzed by SDS-PAGE. For each sample, 200 μg protein were dissolved in 5 μL of 1 M dithiothreitol solution and incubated for 1 h at 37°C. Then, 20 μL of 1 M iodoacetamide solution was added and the samples were incubated for 1 h in darkness at room temperature. All samples were added to the filters and centrifuged at 12,000 g for 10 min. The collected liquid was discarded after centrifugation. Then, the filters were washed twice with 100 μL of UA buffer (8 M urea, 100 mM Tris-HCl, pH 8.0) and then three times with 100 μL of dissolution buffer (0.5 M triethylammonium bicarbonate at pH 8.5). The protein suspensions were digested with 40 μL of trypsin buffer (2 μg trypsin in 40 μL dissolution buffer) and incubated at 37°C for 12–16 h. After digestion with trypsin, the obtained peptides were dried by vacuum centrifugation and 100 μg of them were reconstituted in the dissolution buffer (0.5 M triethylammonium bicarbonate at pH 8.5) and processed according to the manufacturer’s protocol for iTRAQ Reagent Multi-Plex Kit (Applied Biosystems). Peptides from the digestion of the treatment samples CK1, CK2, HS, and RC were separately labeled using iTRAQ reagents with molecular masses of 114, 115, 116, and 117 Da. The pooled mixtures of iTRAQ-labeled peptides for each of the treatment groups were fractionated by strong cation exchange (SCX) chromatography.
Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) and Data Analysis
Three replicates were run for the LC-MS/MS analysis. Digested peptide mixtures were pressure-loaded onto a fused silica capillary column packed with 3-μm dionex C18 material (RP; Phenomenex). The RP sections with 100 Å were 15 cm long and the column was washed with buffer A (water, 0.1% formic acid) and buffer B (acetonitrile, 0.1% formic acid). After desalting, a 5-mm, 300-μm C18 capture tip was placed in line with a quaternary HPLC (Agilent 1100) and analyzed using a 12-step separation.
The first step consisted of a 5-min gradient from 0 to 2% buffer B, followed by a 45-min gradient to 40% buffer B. Next, a 3-min gradient from 40 to 80% and 10-min 80% of buffer B was run followed by a 2-min buffer B gradient from 80 to 2%. Approximately 100 μg of tryptic peptide mixture was then loaded on to the columns and was separated at a flow rate of 0.5 μL/min using a linear gradient. As peptides were eluted from the micro-capillary column, they were electrosprayed directly into a micrOTOF-Q II mass spectrometer (BRUKER Scientific) with the application of a distal 180°C source temperature. The mass spectrometer was operated in the MS/MS (auto) mode. Survey MS scans were acquired in the TOF-Q II with the resolution set to a value of 20,000. Each survey scan (50–2,500) was followed by five data-dependent tandem mass (MS/MS) scans at 2HZ normalized scan speed.
Data were processed by ProteinPilot v.4.5 software (AB Sciex) and compared with the UniProt database. A 1.5-fold change cut off was used to categorize proteins as significantly changed. Proteins with iTRAQ ratios > 1.5 were considered to be up-regulated, and proteins with iTRAQ ratios < 0.67 were considered to be down-regulated. Information from the Gene Ontology (GO) was applied to the functional analysis. GO categories with a P-value < 0.05 were considered to be significant.
Quantitative Real-Time PCR (qRT-PCR) Analysis
Total RNA was extracted from the mycelia using E.Z.N.A.TM Plant RNA Kit (Omega Bio-Tek) according to the manufacturer’s instructions. Briefly, 150 ng total cellular RNA was reverse transcribed using TIANScript RT Kit. The KAPA SYBR FAST qPCR Master Mix Kit (Kapa Biosystems, United States) and the ABI 7500 Real-Time PCR amplifier (Applied Biosystems, Foster City, CA, United States) were used for qPCR. All reactions were carried out in a total volume of 20 μL which contained 2 μL of diluted cDNA, 0.8 μL of primer mix (10 μM), 6.8 μL of nuclease-free water, 0.4 μL ROX Low, and 10 μL of SYBR Green mix. All reactions were performed in triplicate. The qPCR amplification procedures were as follows: 95°C for 3 min, 40 cycles of 95°C for 3 s, 60°C for 32 s, and a final extension at 72°C for 30 s. The GAPDH-encoding gene, gapdh, was used as the reference. Primers were designed using the DNAMAN software (Table 2) and were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China).
Bioinformatics Analyses
Functional classifications were performed using GO1, and pathway analysis was performed using KEGG2. The protein–protein interaction (PPI) network was analyzed using STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) software3. The relative expression of the genes was calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001).
Results
Effect of Heat Stress Treatment and Subsequent Recovery on Morphological and Physiological Changes
The four treatments were being incubated for 5 days at 28°C and then heat stress treatment for 48 h at 40°C (HS), 7 days at 28°C (CK1), 3 days at 28°C following the heat stress (RC), and 10 days at 28°C (CK2), respectively. The cultures for four treatments exhibited clearly different colony morphologies. Mycelia for CK1 produced vigorous aerial hyphae and the plate was almost fully colonized (Figure 1A), but the mycelium for HS treatment barely grew compared to the mycelium before heat stress (Figures 1B,C). Mycelia for CK2 grew thicker than that for CK1 and the plate was fully colonized (Figure 1D). Mycelia for RC treatment germinated vigorous aerial hyphae compared to that for following incubated at 40°C for 3 days again (Figures 1E,F). This result indicates that high temperature significantly inhibited the growth of mycelium.
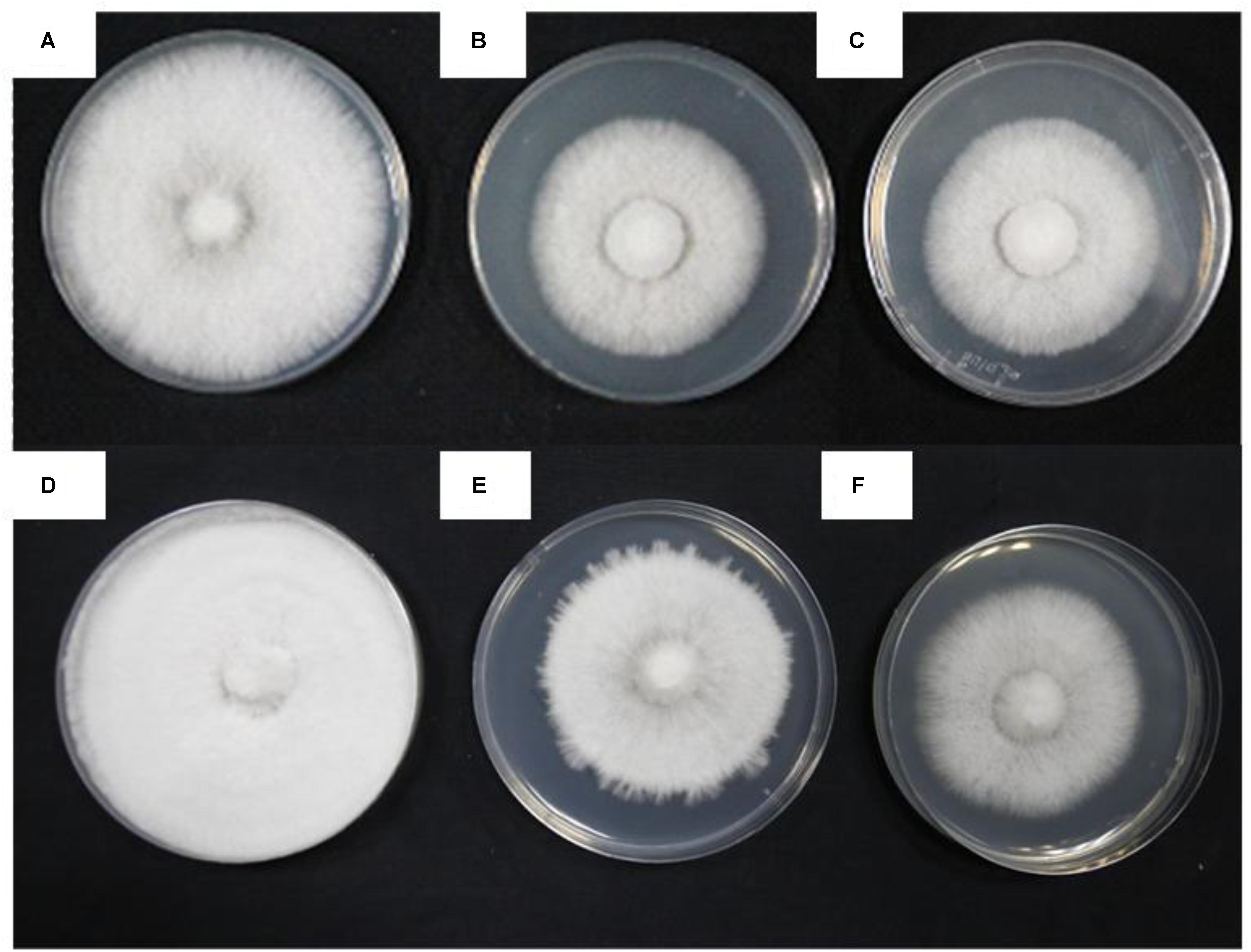
FIGURE 1. Different colony shapes and mycelial morphology of P. ostreatus in response to heat stress and recovery. (A) CK1; (B) incubated for 5 days at 28°C; (C) HS sample; (D) CK2; (E) RC sample; and (F) incubated for 3 days at 40°C following heat stress.
The present study investigated changes in cell membrane thermostability of P. ostreatus mycelium under heat stress and subsequent recovery. We used the TBARS concentration as an indicator of heat stress-induced peroxidation and destruction of lipid membranes (Kong et al., 2012). One-way ANOVA analysis showed that heat treatment (40°C for 48 h) significantly increased TBARS concentration in the mycelium compared with the CK 1 (Figure 2). TBARS content was as high as 3.586 nmol g-1 FW, 73.55% higher than that incubated at 28°C for 48 h (2.064 nmol g-1 FW). This result indicates that heat damages cell membranes by increasing the amount of reactive oxygen species (ROS) and that exposure to heat treatment for long periods of time may be lethal to the edible fungi mycelium. After subsequent recovery, there was no difference in TBARS concentration between RC (2.340 nmol g-1 FW) and control treatment (2.193 nmol g-1 FW; Figure 2), it is possible that the mycelia have a metabolic mechanism for repair of heat-induced cell membrane damage which allows a slow return to growth.
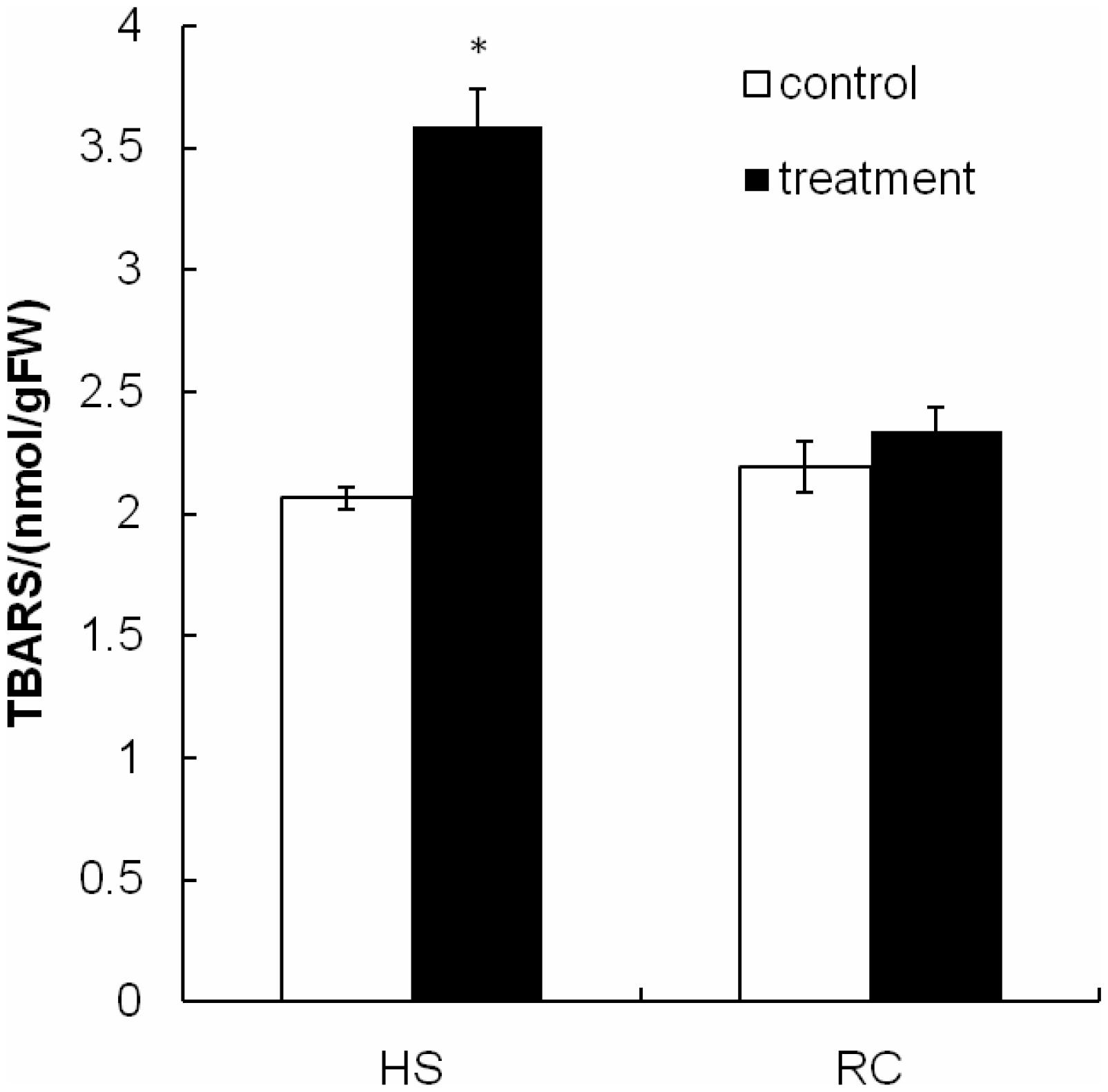
FIGURE 2. The TBARS concentration in mycelium after heat stress and recovery for P. ostreatus. HS group was cultivated for 5 days and subjected to heat stress for 2 days. RC group was allowed to recover for 3 days after exposure to heat stress. Data were analyzed by Duncan’s ANOVA test. Error bars represent the standard deviation of three replicates. The asterisks indicate the significance of differences between treatments and their corresponding controls (∗P < 0.05).
Identification of Differentially Expressed Proteins in Response to Heat Stress and/or Recovery in P. ostreatus Mycelium as Revealed by iTRAQ Analysis
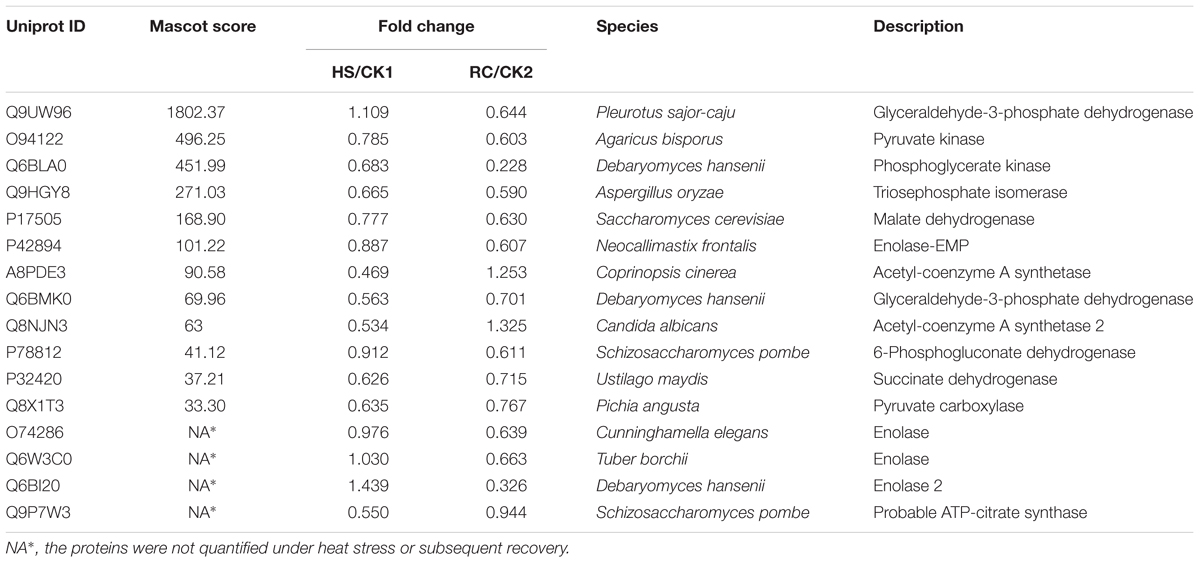
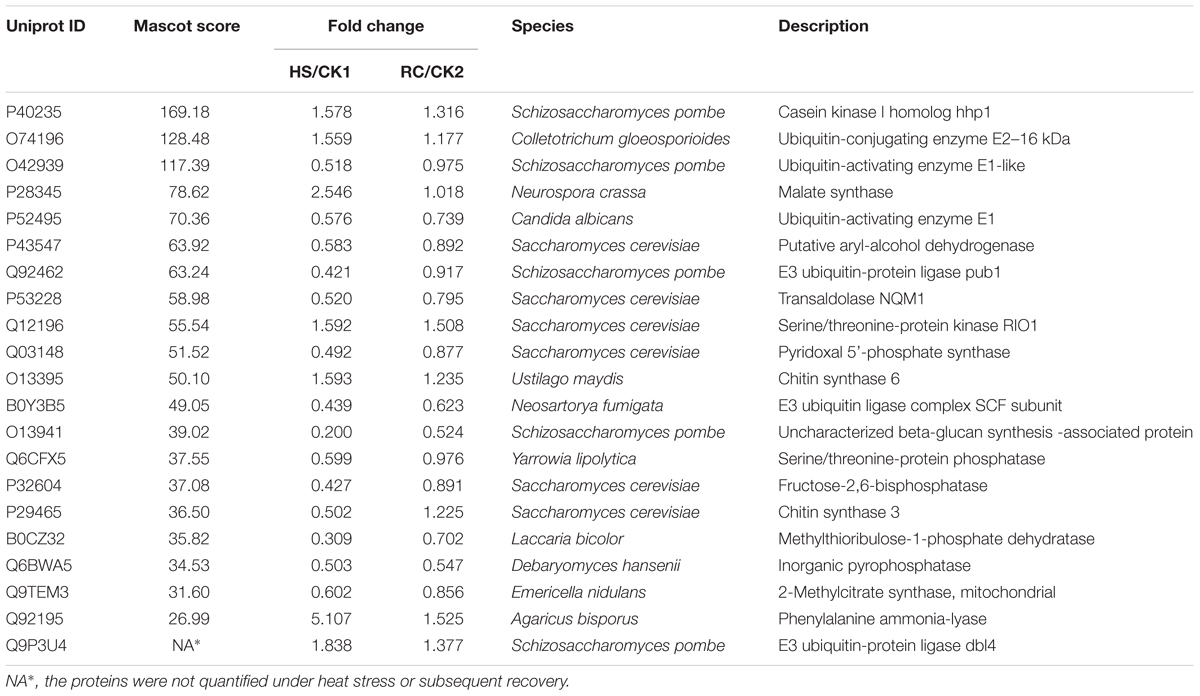
Total proteins from three biological replicates were extracted from each of the four treatment groups of P. ostreatus (CK1, HS, CK2, and RC) and subjected to iTRAQ labeling and 2D LC-MS/MS analysis. Six hundred and eighty-six proteins were quantified with at least one significant peptide sequence and 204 of these characterized proteins were differentially expressed. Heat stress and recovery affected protein expression levels in various ways. Compared to the corresponding control levels, heat stress was associated with 61 proteins that were up-regulated and 70 that were down-regulated. In contrast, 59 were up-regulated and 61 were down-regulated after recovery (Figure 3). There were 84 (35 up- and 49 down-regulated) proteins and 73 (34 up- and 39 down-regulated) proteins responding to only heat stress or recovery, respectively, whereas 47 proteins were differentially expressed in both heat stress and recovery. Among these 47 proteins, 23 proteins were up-regulated under both heat stress and recovery and 19 proteins were down-regulated under both conditions. Three proteins were up-regulated under heat stress and down-regulated during recovery, while two proteins were down-regulated under heat stress but up-regulated during recovery (Table 1).
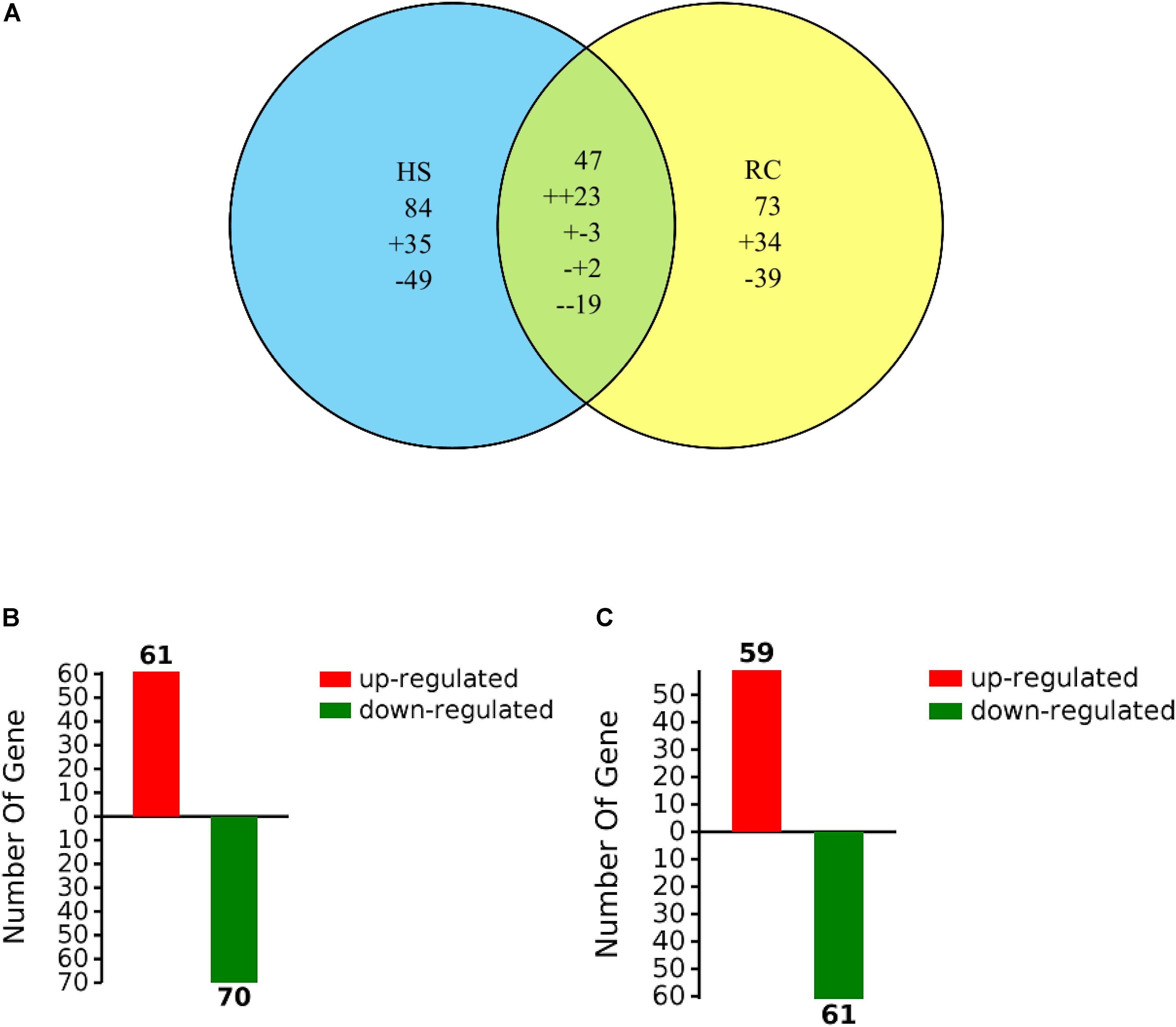
FIGURE 3. Venn diagram of differentially expressed proteins that were up- or down-regulated (A) by heat stress or recovery and total number (B,C) of identified DEPs from heat stress or recovery. The “+” and “-” indicate up- and down-regulated proteins, respectively. The “++” and “–” indicate up- and down-regulated under both heat stress and recovery, respectively. The “+-” indicates up-regulated under heat stress and down-regulated during recovery and the “-+” indicates down-regulated under heat stress but up-regulated during recovery.
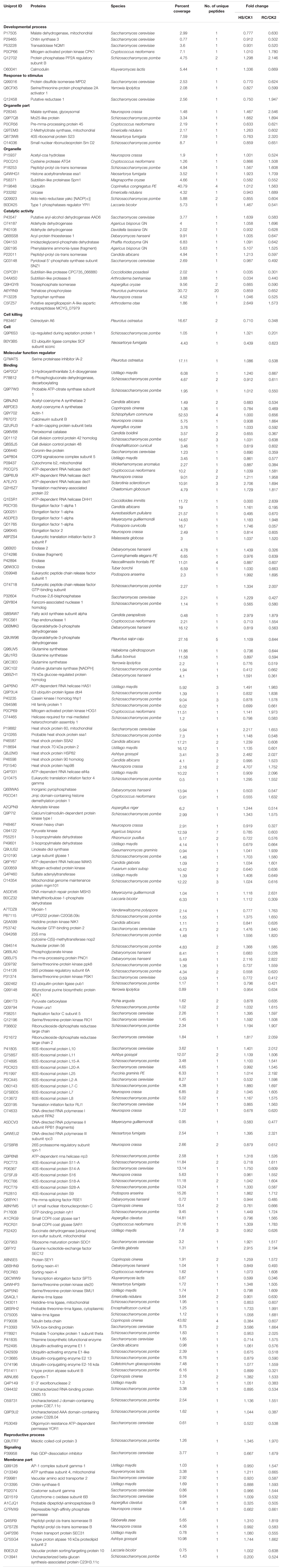
TABLE 1. Proteins with significant expression level changes in the mycelium under heat stress or subsequent recovery.
Functional Categorization Analysis
Among the 204 DEPs, eight were characterized as hypothetical or unknown proteins using P. ostreatus genomics information published in uniprot4. To gain functional information about these proteins, BLASTP5 was used to search for homologous proteins against the NCBI non-redundant protein database. GO annotations enrichment, which was classified into biological process, cell components, and molecular function. The results showed that the DEPs identified in the mycelium under heat stress and recovery were primarily involved in cellular, metabolic, multi-organism, reproductive, and developmental processes; biological regulation; localization; nitrogen utilization; cellular component organization or biogenesis; reproduction; response to stimulus; signaling biological processes, whereas growth biological processes detected in HS (Figure 4A), and cell killing and immune system process detected in RC (Figure 4B). With regard to the cellular components, most DEPs were associated with organelle, organelle part, protein-containing complex, supramolecular complex, cell, cell part, nucleiod, membrane-enclosed lumen, membrane part, membrane, extracellular region part, extracellular region, but the proportions of molecular function are different in each treatment (Figure 5). Under the category of molecular function, most DEPs in the mycelium under heat stress and recovery were correlated with catalytic activity; binding; molecular function regulator; signal transducer activity; structural molecule activity; transcription regulator activity; transporter activity; antioxidant activity, but the proportions of molecular function are different in each treatment (Figure 6).
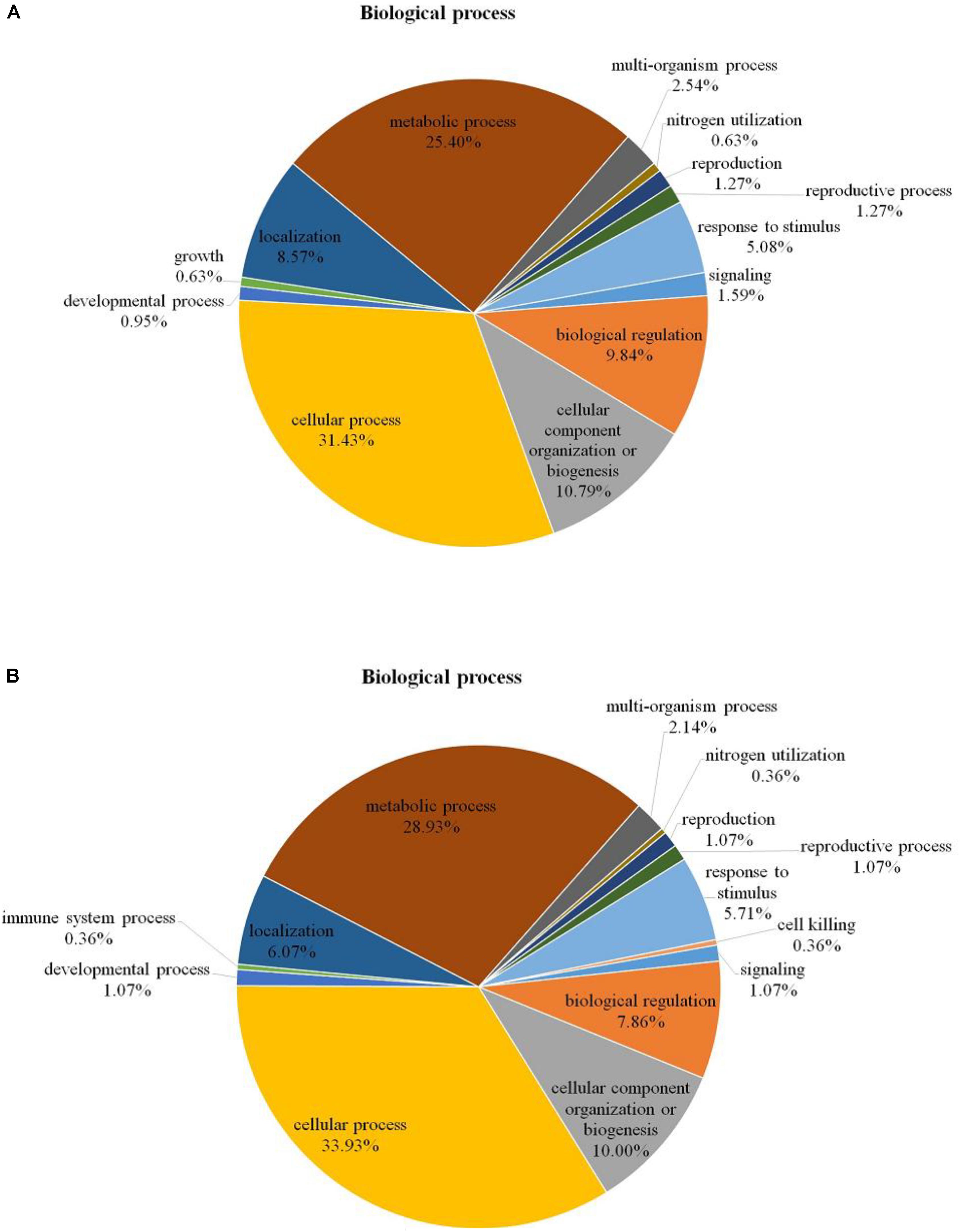
FIGURE 4. Bioinformatics analysis of DEPs responsive to heat stress (A) and subsequent recovery (B) in P. ostreatus mycelia compared to the control group through gene ontology (GO) in biological process (BP).
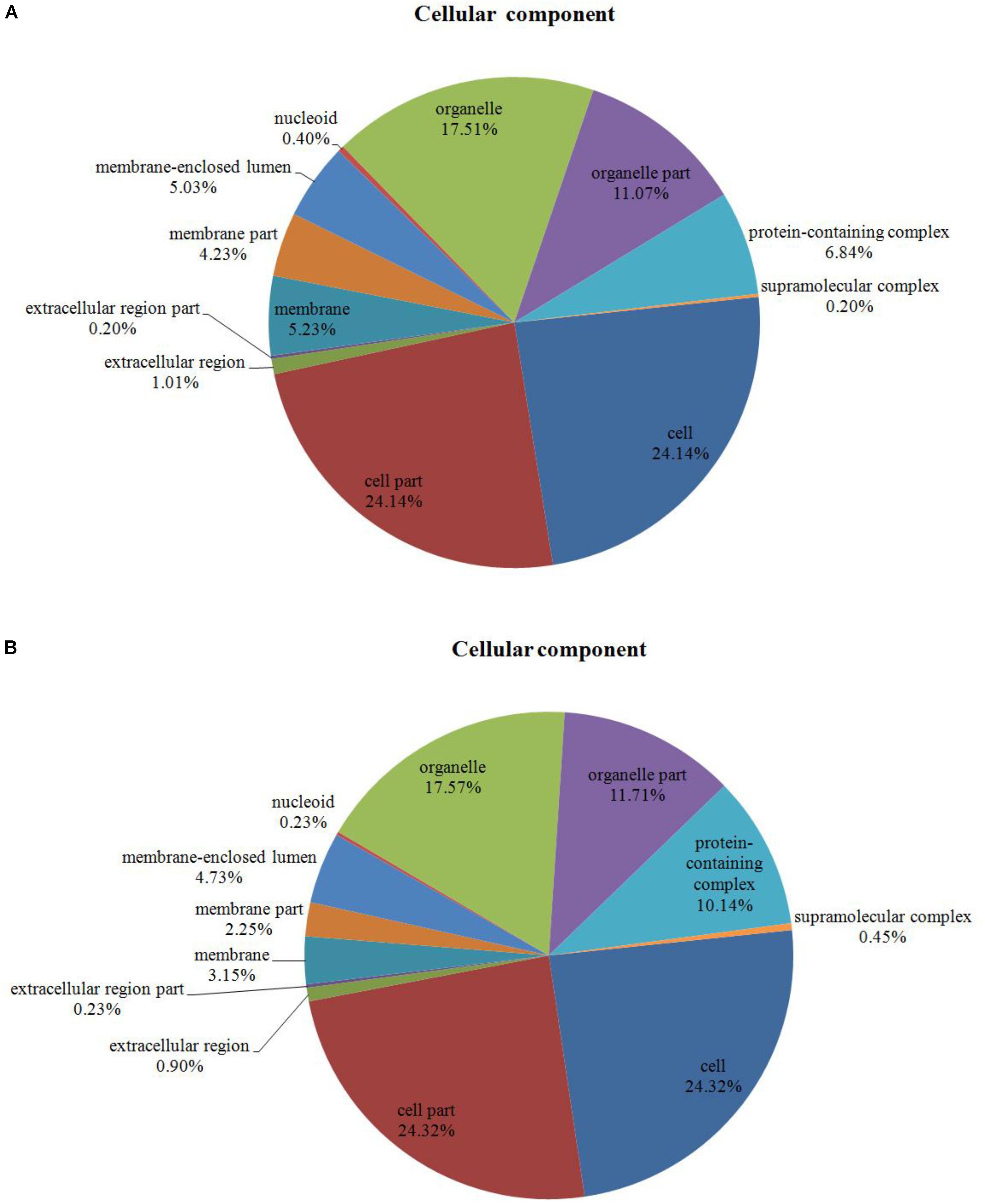
FIGURE 5. Bioinformatics analysis of DEPs responsive to heat stress (A) and subsequent recovery (B) in P. ostreatus mycelia compared to the control group through gene ontology (GO) in cell component (CC).
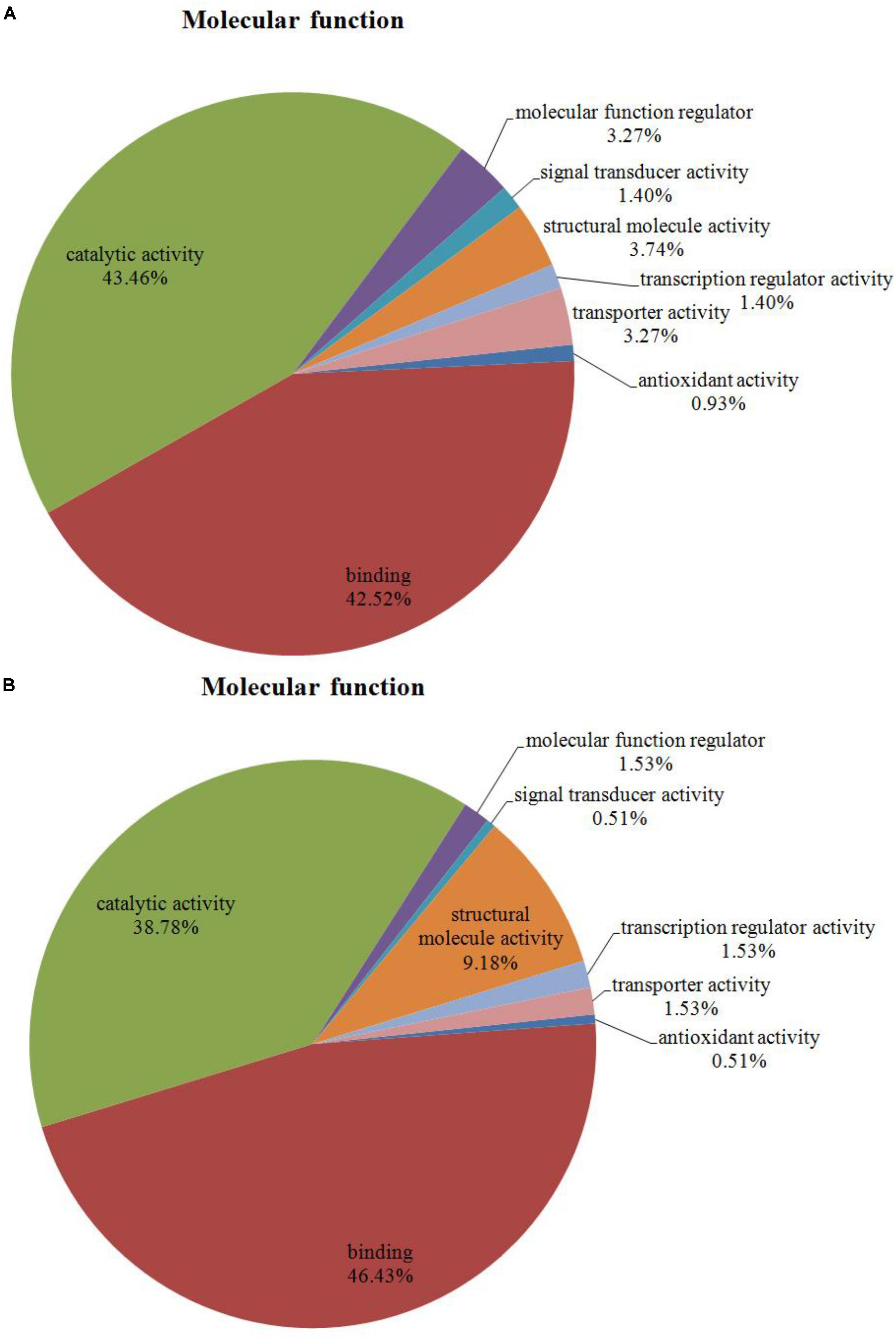
FIGURE 6. Bioinformatics analysis of DEPs responsive to heat stress (A) and subsequent recovery (B) in P. ostreatus mycelia compared to the control group through gene ontology (GO) in molecular function (MF).
The KEGG pathway and enrichment analysis indicated that the DEPs in the mycelium under heat stress were highly enriched in AGE-RAGE signaling pathway in diabetic complications; carbon metabolism; citrate cycle (TCA cycle); MAPK signaling pathway; glyoxylate and dicarboxylate metabolism; protein processing in endoplasmic reticulum; nitrogen metabolism; ubiquitin mediated proteolysis; biosynthesis of amino acids; fructose and mannose metabolism (Figure 7A). While the DEPs in the mycelium under recovery were highly enriched in pyrvate metabolism; ribosome; protein processing in endoplasmic reticulum; glycolysis/gluconeogenesis; tryptophan metabolism; purine metabolism; longevity regulating pathway; phagosome; and biosynthesis of amino acids (Figure 7B).
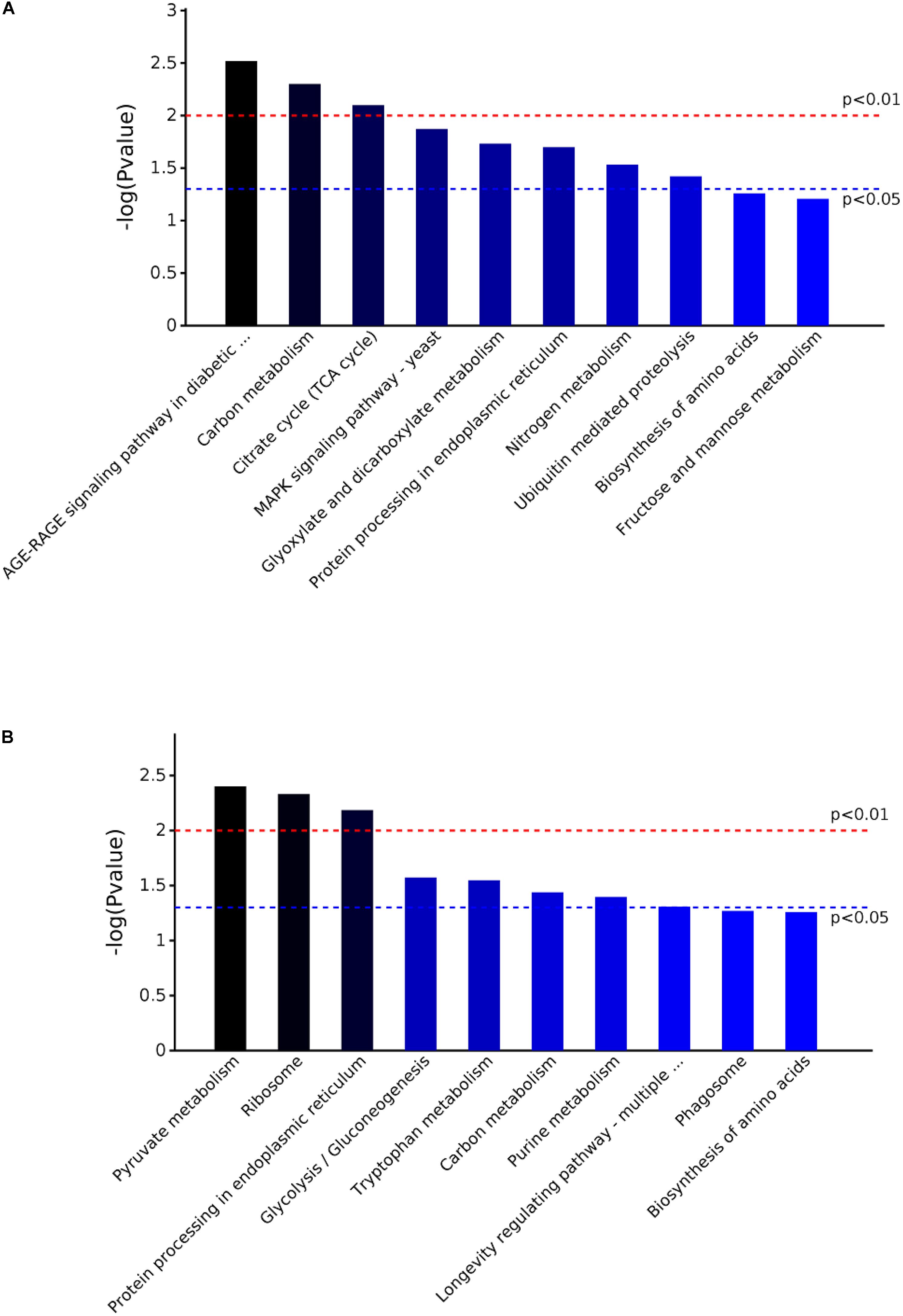
FIGURE 7. The KEGG pathway enrichment analysis of the DEPs in P. ostreatus mycelium under heat stress (A) and subsequent recovery (B). Top 10 enrichment in KEGG pathway maps of the DEPs. P-value was calculated using Fisher’s exact test.
String Analysis of Protein–Protein Interactions for DEPs
The PPIs whose combined score was >0.9 were used to build network using Cytoscape tool in each group. It was of note that the DEPs in the mycelium under heat stress of top 10 enrichment in KEGG pathway formed three subsets of protein interaction networks: carbohydrate and energy metabolism, signal transduction, and proteins metabolism (Figure 8A), while in the mycelium under recovery of top 10 enrichment in KEGG pathway formed differently compared to HS (Figure 8B). This indicated that proteins in this network played important functions in redox homeostasis, response to stress, signal transduction, and protein metabolism.
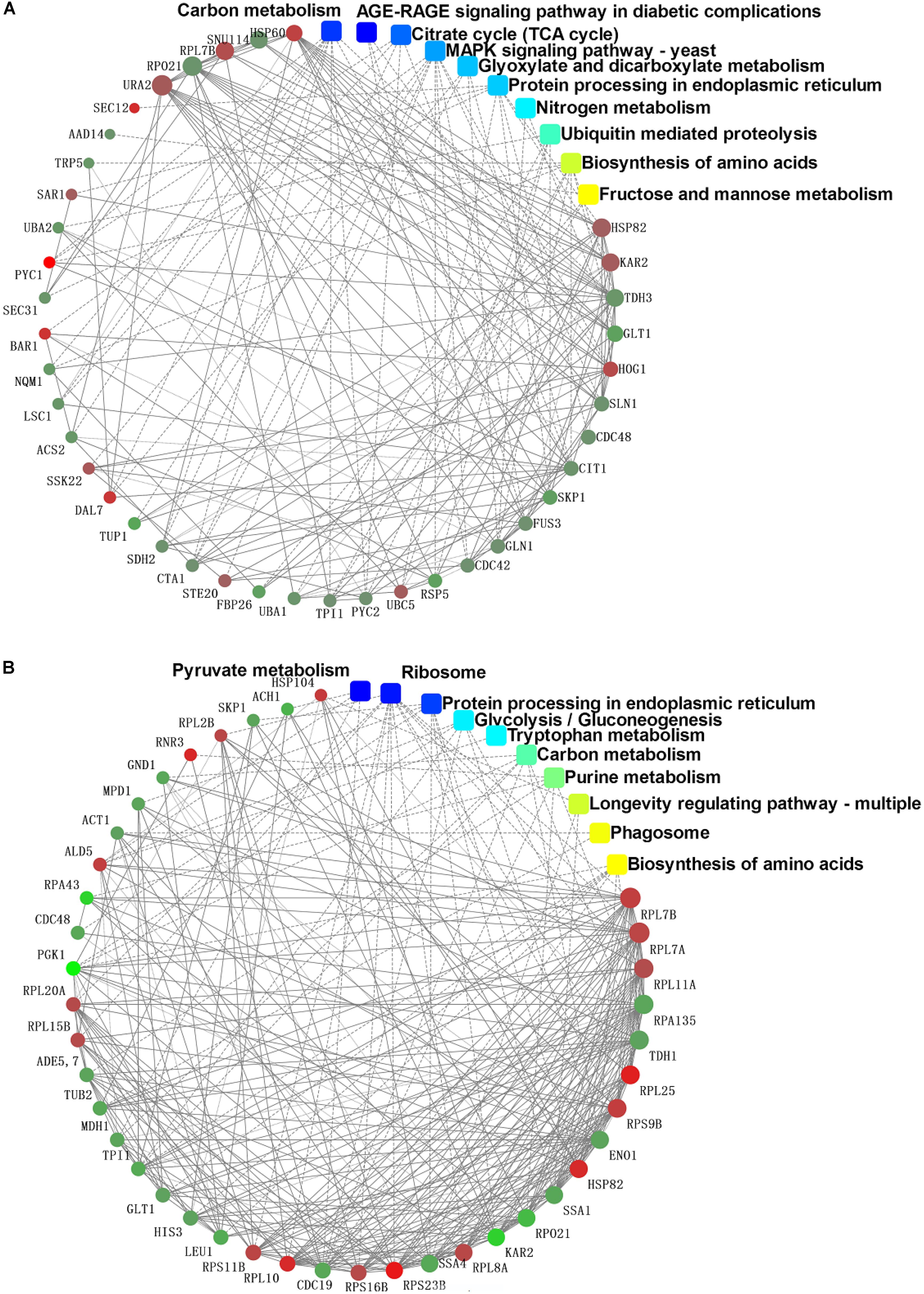
FIGURE 8. Protein–protein interaction network analysis among the significantly expressed proteins in P. ostreatus mycelium under heat stress (A) and subsequent recovery (B) using String software.
Transcriptional Expression Analysis of Selected Proteins as Revealed by qRT-PCR
The data used in this study were subjected to rigorous statistical and bioinformatics analysis to eliminate possible errors as by Liu et al. (2017). To provide further information of the correspondence between proteins and their mRNA expression patterns, quantitative real-time PCR (qRT-PCR) was performed to investigate the dynamic transcriptional expression patterns of nine representative DEPs. The summarized primer data of nine representative DEPs are shown in Table 2. After heat treatment and recovery, the changes of the mRNA levels in eight genes correlated with changes at the protein levels as indicated by iTRAQ analysis, this included a mapkHOG1, β-gs, pal, m-1-pd, hsp60, grp78, hsp90, and hsp104 (Figures 9A–C, E–I). The expression of the genes agreed with proteomics results (Table 1). The mRNA of ms showed a up-regulated trend in the mycelium under recovery (Figure 9D); however, ms had a lower protein expression level (Table 1). The expression of ms genes was not in accordance with proteomics due to translational or post-translational regulation. The result is generally consistent with those of a previous report (Vedeler et al., 2012; Liu et al., 2017).
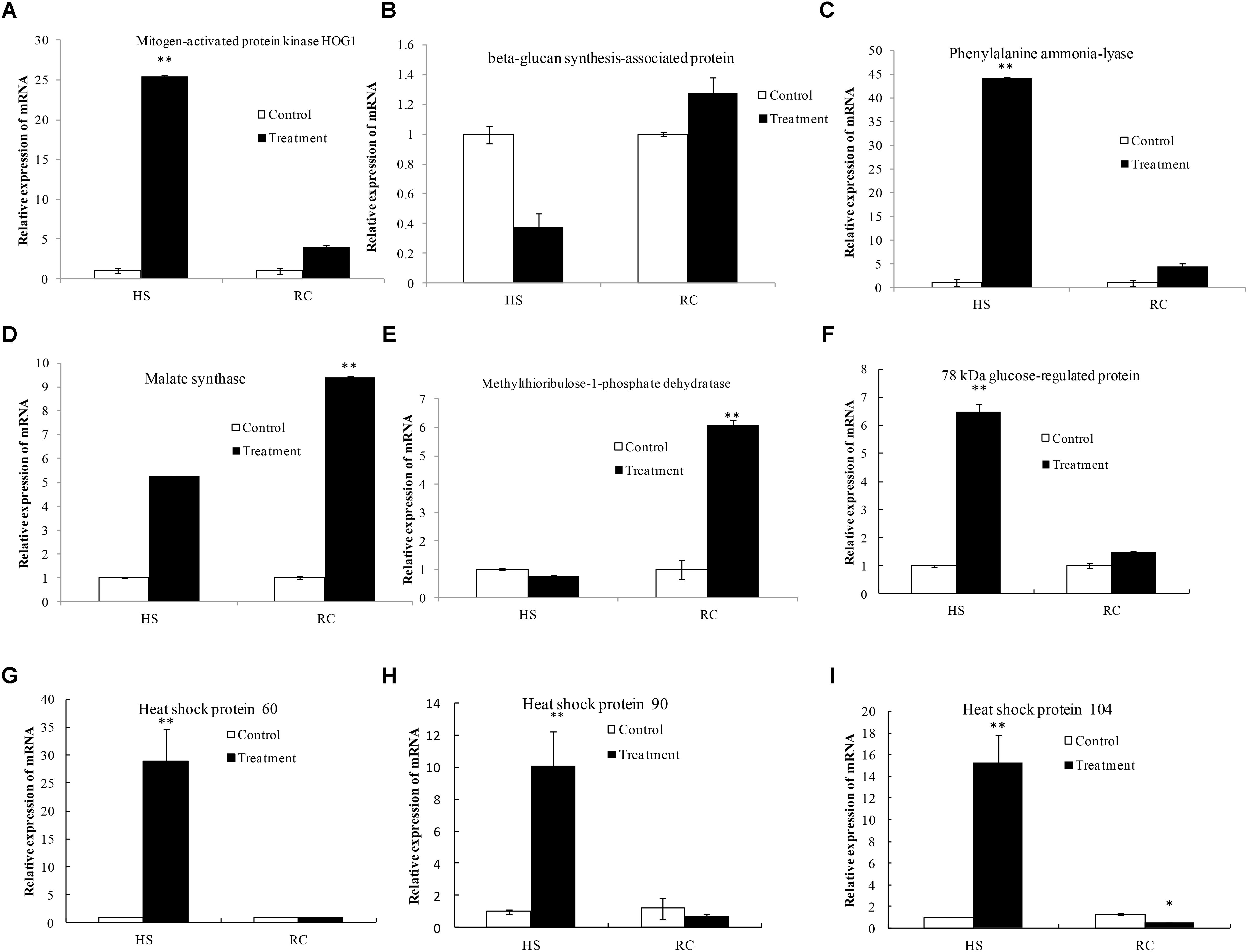
FIGURE 9. Transcriptional expression analysis of representative proteins as revealed by qRT-PCR. The relative mRNA expression levels of matched differentially abundant proteins including mitogen-activated protein kinase HOG1 (A), beta-glucan synthesis-associated protein (B), phenylalanine ammonia-lyase (C), malate synthase (D), methylthioribulose-1-phosphate dehydratase (E), 78 kDa glucose-regulated protein homolog (F), heat shock protein 60 (G), heat shock protein 90 (H), and heat shock protein 104 (I). Gapdh was used as the reference gene. Mean values and standard deviations of three biological replicates are shown. The asterisks indicate the significance of differences between treatments and their corresponding controls (∗∗P < 0.01, ∗P < 0.05).
Discussion
One of the many locations for heat stress injury in cell is the membrane. TBARS is the product of lipid peroxidation in fungi. With the increase of temperature, the levels of membrane lipid peroxidation will be increased (Kong et al., 2012). In this study, we investigated the morphological and TBARS content of the mycelium in P. ostreatus under heat stress and subsequent recovery (Figures 1, 2). These results showed that the mycelium of P. ostreatus were damaged under heat stress at 40°C for 48 h, but they subsequently recovered at 25°C for 3 days. These results indicated that P. ostreatus mycelia suffered greater damage on membrane lipid after high temperature (40°C) and P. ostreatus mycelia treated with 40°C for 48 h was a suitable treatment for studying changes in extracellular metabolites.
In this study, taking advantage of iTRAQ-based quantitative proteomics technology, we investigated the response of P. ostreatus to heat stress and recovery on a proteome-scale. More than 204 proteins, which were almost 29.73% of all detected 686 proteins, were up- or down-regulated in heat-treated and recovery in P. ostreatus, indicating that heat strongly influences fungi physiology. The biological relevance of these DEPs in the P. ostreatus under heat stress and subsequent recovery are discussed below.
Carbohydrate and Energy Metabolism
Heat stress alters the abundance of many proteins involved in carbohydrate and energy metabolism, which was mainly included the citrate cycle (TCA cycle), glycolysis, glyoxylate and dicarboxylate metabolism, and nitrogen metabolism in P. ostreatus mycelia. The TCA cycle is an important aerobic pathway involved in the conversion of carbohydrates, fats, and proteins to form energy (Cetica et al., 2003), which starts with acetyl-CoA, the activated form of acetate, derived from glycolysis and pyruvate oxidation for carbohydrates and from beta oxidation of fatty acids, and it is noteworthy that four proteins involved in the TCA process, including 2-methylcitrate synthase, succinate dehydrogenase, ATP-citrate synthase, and pyruvate dehydrogenase had lower expression levels in mycelia after heat stress but recovered to control levels after subsequent recovery. Pyruvate dehydrogenase is an enzyme component of the multienzyme pyruvate dehydrogenase complex and is involved in the formation of cellular energy during the TCA cycle. 2-Methylcitrate synthase catalyzes the synthesis of (2S,3S)-2-methylcitrate from propionyl-CoA and oxaloacetate and also from acetyl-CoA. In this study, the abundance of Pyruvate dehydrogenase and 2-methylcitrate synthase decreased under heat stress. This suggests that the TCA cycle was inhibited in P. ostreatus after 48 h of heat stress treatment (Rice and Bayles, 2008). As shown in Table 3, there are complex protein abundance change patterns in acute normal culture to heat stress transfer in mycelia of P. ostreatus at the molecular level. There were six kinds of enzymes involved in glycolysis that showed no significant change in expression under heat stress, but were down-regulated after subsequent recovery; these included glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase, pyruvate kinase, and enolase. Overall, the results indicate that the glycolytic pathway was not affected by heat stress and that the TCA process was suppressed by the heat stress despite the return to control levels during recovery. These results suggest that the glycolytic pathway is more heat-resistant than the TCA cycle in the respiration of mycelium of P. ostreatus during heat stress.
Signal Transduction
Reactive oxygen species are found in normal living organisms where they are constantly being produced under the oxidative stress caused by toxic heavy metals, heat shock, inflammation, ionizing irradiation, immune responses, and environmental stimuli (Zhai et al., 2018). Studies have shown that antioxidant enzymes can remove and reduce ROS produced by metabolic stress conditions in an attempt to maintain homeostatic equilibrium. As shown in Table 4, 18 dysregulated proteins involved in the heat stress response were detected. Four of the key proteins involved in the redox reactions, i.e., peroxisomal catalase, thiamine biosynthetic bifunctional enzyme, linoleate diol synthase (LDS), and uricase which play a role in protecting against oxidative stress resulted up-regulated during heat stress. For example, expression of LDS is increased by 1.94-fold under heat stress, which converted oleic acid, linoleic acid, and α-linolenic acid to 7,8-dihydroxy fatty acids, but this enzyme showed no activity when γ-linolenic acid, eicosatrienoic acid, arachidonic acid, and eicosapentaenoic acid were used as substrates (Brodowskys et al., 1992). Catalase, universal in many fungi, rapidly catalyzes the decomposition of hydrogen peroxide into less-reactive gaseous oxygen and water molecules protecting the cell from oxidative damage due to accumulation of ROS (Isobe et al., 2006). In our study, the expression of CAT was not significantly changed under heat stress; however, the expression was significantly lower after recovery. Similar results were observed for Po-cat2 activity under heat stress which may be caused by the inhibition of the overall protein synthesis under stressful conditions or by alternative H2O2 detoxification pathways function (Wang et al., 2017). CAT and ascorbate peroxidase (APX), another key detoxifying enzyme, act together to alleviate the aggregation of H2O2 and other ROS resulting from uric acid oxidation catalyzed by uricase. Uricase is increased 1.7-fold under heat stress. In addition, another redox enzyme, thiamine biosynthetic bifunctional enzyme, is increased 1.6-fold under heat stress (Table 3). It is clear that these key enzymes participate in the removal of ROS and protecting the cells from oxidation damage (Sun et al., 2013).
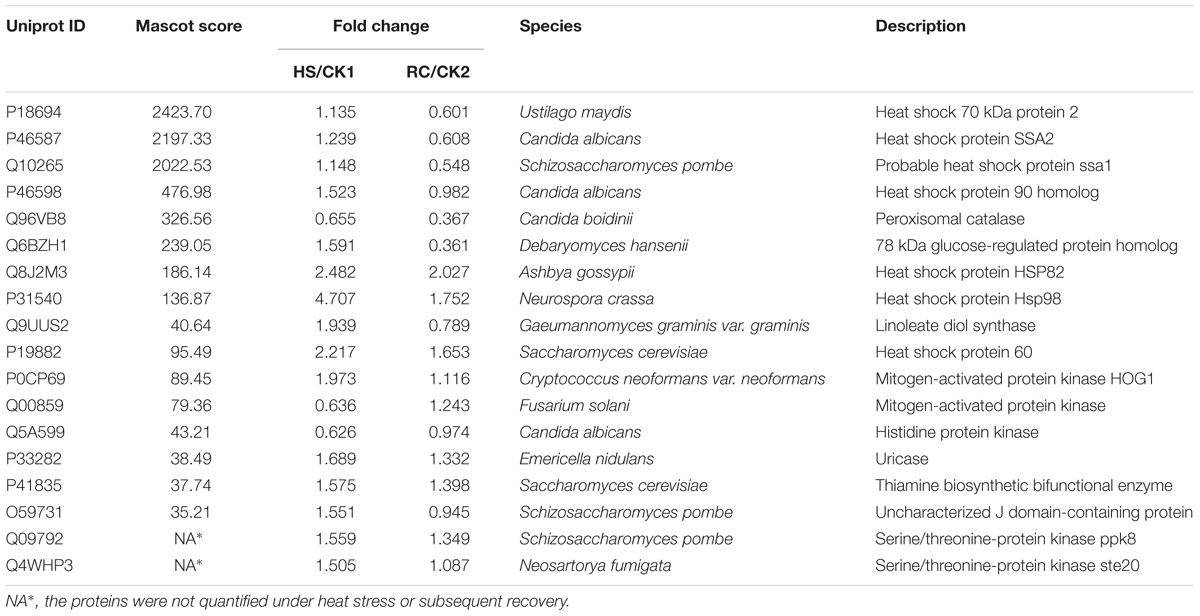
TABLE 4. Variation of proteins involved in abiotic stress and redox under heat stress and/or subsequent recovery.
Most of the proteins involved in oxidative stress are heat shock proteins (HSPs) with chaperone activity that belong to five conserved classes, HSP60, HSP70, HSP90, HSP100, and the small heat shock proteins (sSHPs). In fungi as well as most eukaryotic cells, HSPs are involved in various routine biological processes such as transcription, translation and post-translational modifications, protein folding, and aggregation and disaggregation of proteins (Tiwari et al., 2015). In our experiments, the expression of Hsp60 increased 2.2-fold under heat stress. This result agrees with results from Paracoccidioides brasiliensis which showed that Hsp60 is also up-regulated in response to thermal stress (Felipe et al., 2005). This might suggest that Hsp60 may have important functions in alleviating heat stress in P. ostreatus mycelium. The P. brasiliensis study also identified additional heat shock proteins which are essential for cell viability: Hsp70-2, 70-kDa HSPs of the SSA subfamily, Hsp70/SSA1 and Hsp70/SSA2, as well as glucose-regulated protein 78 kDa (GRP78). The Hsp70 protein family both under normal or environmental conditions of stress prevent protein aggregation and promote protein folding (Frydman, 2001). In addition, they participate in protein input and transfer processes and promote the degradation of unstable proteins. Moreover, Hsp70 has been reported to accumulate during the heat stress response in several organisms (Sørensen et al., 2003; Lee et al., 2007), and the expression of GRP78, a member of the Hsp70 family, increased by 1.6-fold under heat stress and then decreased to 0.36-fold after recovery. Interestingly, it has been shown that GRP78 promotes endoplasmic reticulum protein complex assembly6. Two Hsp90 family proteins, Hsp82 and Hsp90 homolog, were also evaluated during heat stress and recovery. Hsp82 expression increased 2.5-fold under heat stress and then decreased to twofold after recovery. In contrast, the Hsp90 homolog was not affected by thermal stress. Members of the Hsp90 family are molecular chaperones that mediate the folding of a defined set of signaling proteins involved in repair, signal transduction, cell-cycle regulation, protein degradation, and transport (Richter and Buchner, 2001; Pratt et al., 2006). Studies have shown that when P. euphratica was subjected to high temperature stress, Hsp90 was significantly increased and then returned to normal levels (Ferreira et al., 2007). In addition, our study has identified one Hsp104 protein belonging to the Hsp100 family which has been shown to be a molecular chaperone in plants (Gurley, 2000), yeast (Glover and Lindquist, 1998), and bacteria (Queitsch et al., 2000). In fact, it has been reported that Hsp104 is the most crucial thermotolerance-related protein of Saccharomyces cerevisiae, enhancing survival after exposure to extreme heat or high concentrations of ethanol (Glover and Lindquist, 1998). In our study, similar results were observed. Hsp104 was increased by fourfold under heat stress and then decreased to 1.8-fold after recovery. In mycelium of P. ostreatus, Hsp104 is highly expressed and is one of the most important factors for heat resistance. Moreover, Hsp104 provides mycelia with a strong resistance to stress by alleviating the pressure of protein aggregation and promoting degradation of denatured peptide polymers (Bösl et al., 2006). Our study also shows that certain thermo-induced transcription factors show no change in expression under heat stress, but decline in expression levels when returned to normal temperatures. This finding may indicate that these thermo-induced transcription factors may not play a direct role in response to heat stress. In summary, our study suggests that HSPs are key players in P. ostreatus heat resistance, and that these components deserve further in-depth study.
The mitogen-activated protein kinases (MAPK) signal pathway is an important signaling system to mediate cell responses (Zhao et al., 2017). The DPs identified in the mycelium under heat stress were found annotating pathway related to MAPK signal pathway, including the cell division control protein 42 homolog, E3 ubiquitin-protein ligase pub1, serine/threonine protein kinase ste20, peroxisomal catalase, MAPK, and MAPK HOG1 involved in maintaining cellular homeostasis. As a signal/pheromone stress regulator protein, MAPK was increased by 2.0-fold under heat stress and then decreased to 1.1-fold after recovery, the expression of this proteins returned to normal level, indicating that MAPK is an important resistant substance in high temperature stress. Moreover, the expression of histidine protein kinase which plays an important role in the hyphal formation and virulence effect decreased to 0.62-fold under heat stress.
Proteins Metabolism
In our study, it can be seen that many of the proteins involved in metabolism are down-regulated under heat stress suggesting that high temperature affects mycelial metabolism (Table 5). However, the expression of phenylalanine ammonia-lyase (PAL) is increased by 5.1-fold under heat stress, and declined 1.5-fold after following recovery, compared to controls. This indicates that PAL may also play a role in the mycelium of P. ostreatus under heat stress. PAL catalyzes the first step in the general pathway of biosynthesis of polyphenolic compounds including lignin, cinnamate esters, and flavonoids, and is one of the key enzymes in the metabolism of these compounds. The activity of PAL increases dramatically in response to various stimuli (Jones, 1984). A previous study in Pea Leaf showed that PAL activity has no significant change within 12–14 h, and the activity maximum was at 36–48 h after wounding or jasmonic acid (JA) application. PAL activation induced by wounding or JA lagged far behind the H2O2 burst. Moreover, the data imply that plasma membrane NADPH oxidase-originated H2O2 burst is essential for wounding or JA-induced PAL activation (Liu et al., 2008). In our study, similar results were obtained, which might indicate that the accumulation of H2O2, O2-, OH- induced by heat stress prompts a significant increase in the expression and synthesis of PAL.
Two additional enzymes involved in cell wall metabolism are chitin synthase (CHS) 3 which is responsible for chitin synthesis and CHS 6 which is involved in its degradation. Chitin production involves a dynamic balance between CHS and the chitin degradation enzyme, chitinase (Rogg et al., 2012). Interestingly, the expression of these two proteins are opposite in response to thermal stress. CHS 3 is reduced by 0.5-fold under heat stress, and CHS 6 is increased 1.6-fold, indicating that CHS 6 plays a dominant role in cell wall integrity and stress. Another protein involved in cell wall synthesis and degradation is the uncharacterized beta-glucan synthesis-associated protein. Its expression declined fivefold under heat stress, indicating that the cell wall of hyphae may have suffered serious damage under heat stress. In addition, triose phosphate isomerase, glutamate synthetase, and affinity phosphate permease, and inorganic pyrophosphatase are down-regulated. Again, this supports the hypothesis that high temperature stress affects hyphal biosynthesis and metabolism.
Conclusion
An iTRAQ-based proteomic technique was employed to compare the abundance of proteins in heat stress and/or subsequent recovery of P. ostreatus mycelium culture for 48 h. Two hundred and four DEPs were identified. These DEPs are mainly involved in the biological processes of cellular, metabolic, multi-organism, reproductive, and developmental processes; biological regulation; localization; nitrogen utilization; cellular component organization or biogenesis; reproduction; response to stimulus; signaling and growth biological processes. The diverse array of proteins affected by heat stress conditions and subsequent recovery indicates that there is a remarkable flexibility in mycelium metabolism, which may contribute to its survival in heat stress. The morphological combined with physiological analysis that the iTRAQ-based proteomic technique is sufficiently reliable for the identification and quantification of a large number of mycelium. qRT-PCR results suggest that the expression of some proteins (e.g., malate synthase) can be regulated by post-transcriptional modifications. With iTRAQ-based proteomic technique, many new heat-responsive proteins, such as PAL, LDS, and MAPK, were identified from P. ostreatus mycelium. These novel proteins provide a good starting point for further research into their functions using genetic or other approaches. These findings significantly improve the understanding of the molecular mechanisms involved in the tolerance of fungi to heat stress.
Author Contributions
YZ, MZ, and JQ conceived, designed, and performed the experiments, analyzed the data, and wrote and revised the manuscript. JZ conceived and designed the experiments.
Funding
This work was supported by the National Basic Research Program of China (Grant No. 2014CB138303) and the National Natural Science Foundation of China (31601805).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
- ^https://david.ncifcrf.gov/
- ^http://www.genome.jp/kegg/mapper.html
- ^https://string-db.org/
- ^https://www.uniprot.org/
- ^http://www.ncbi.nlm.nih.gov/BLAST/
- ^http://www.uniprot.org/uniprot/Q6BZH1-Function
References
Anandhi, R., Annadurai, T., Anitha, T. S., Muralidharan, A. R., Najmunnisha, K., Nachiappan, V., et al. (2013). Antihypercholesterolemic and antioxidative effects of an extract of the oyster mushroom, Pleurotus ostreatus, and its major constituent, chrysin, in Triton WR-1339-induced hypercholesterolemic rats. J. Physiol. Biochem. 69, 313–323. doi: 10.1007/s13105-012-0215-6
Bösl, B., Grimminger, V., and Walter, S. (2006). The molecular chaperone Hsp104—A molecular machine for protein disaggregation. J. Struct. Biol. 156, 139–148. doi: 10.1016/j.jsb.2006.02.004
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Brodowskys, I. D., Hambergi, M., and Oliw, E. H. (1992). A linoleic acid (8R) -dioxygenase and hydroperoxide isomerase of the Fungus Gaeumannom yces graminis. Ann. N. Y. Acad. Sci. 744:314. doi: 10.1111/j.1749-6632.1994.tb52750.x
Cetica, P., Pintos, L., Dalvit, G., and Beconi, M. (2003). Involvement of enzymes of amino acid metabolism and tricarboxylic acid cycle in bovine oocyte maturation in vitro. Reproduction 126, 753–763. doi: 10.1530/rep.0.1260753
Chang, S. T., and Miles, P. G. (2004). Mushrooms: cultivation, nutritional value, medicinal effect and environmental impact. Am. J. Orthopsychiatry 38, 688–692.
Das, A., Eldakak, M., Paudel, B., Kim, D. W., Hemmati, H., Basu, C., et al. (2016). Leaf proteome analysis reveals prospective drought and heat stress response mechanisms in soybean. BioMed. Res. Int. 2016:6021047. doi: 10.1155/2016/6021047
Felipe, M. S., Andrade, R. V., Arraes, F. B., Nicola, A. M., Maranhão, A. Q., Torres, F. A., et al. (2005). Transcriptional profiles of the human pathogenic fungus Paracoccidioides brasiliensis in mycelium and yeast cells. J. Biol. Chem. 280, 24706–24714. doi: 10.1074/jbc.M500625200
Ferreira, A. S., Tótola, M. R., and Borges, A. C. (2007). Physiological implications of trehalose in the ectomycorrhizal fungus Pisolithus sp. under thermal stress. J. Therm. Biol. 32, 34–41. doi: 10.1016/j.jtherbio.2006.08.009
Frydman, J. (2001). Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 70, 603–647. doi: 10.1146/annurev.biochem.70.1.603
Glover, J. R., and Lindquist, S. (1998). Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94, 73–82. doi: 10.1016/S0092-8674(00)81223-4
Gurley, W. B. (2000). HSP101: a key component for the acquisition of thermotolerance in plants. Plant Cell 12, 457–460. doi: 10.1105/tpc.12.4.457
Hultinrosenberg, L., Forshed, J., Branca, R. M., Lehtiö, J., and Johansson, H. J. (2013). Defining, comparing, and improving iTRAQ quantification in mass spectrometry proteomics data. Mol. Cell. Proteomics 12, 2021–2031. doi: 10.1074/mcp.M112.021592
Isobe, K., Inoue, N., Takamatsu, Y., Kamada, K., and Wakao, N. (2006). Production of catalase by fungi growing at low pH and high temperature. J. Biosci. Bioeng. 101, 73–76. doi: 10.1263/jbb.101.73
Jedinak, A., and Sliva, D. (2008). Pleurotus ostreatus inhibits proliferation of human breast and colon cancer cells through p53-dependent as well as p53-independent pathway. Int. J. Oncol. 33, 1307–1313.
Jesenak, M., Majtan, J., Rennerova, Z., Kyselovic, J., Banovcin, P., and Hrubisko, M. (2013). Immunomodulatory effect of pleuran (β-glucan from Pleurotus ostreatus) in children with recurrent respiratory tract infections. Int. Immunopharmacol. 15, 395–399. doi: 10.1016/j.intimp.2012.11.020
Jones, D. H. (1984). Phenylalanine ammonia-lyase: regulation of its induction, and its role in plant development. Phytochemistry 23, 1349–1359. doi: 10.1016/S0031-9422(00)80465-3
Kong, W., Huang, C., Chen, Q., Zou, Y., and Zhang, J. (2012). Nitric oxide alleviates heat stress-induced oxidative damage in Pleurotus eryngii var. tuoliensis. Fungal Genet. Biol. 49, 15–20. doi: 10.1016/j.fgb.2011.12.003
Lee, D. G., Ahsan, N., Lee, S. H., Kang, K. Y., Bahk, J. D., Lee, I. J., et al. (2007). A proteomic approach in analyzing heat-responsive proteins in rice leaves. Proteomics 7, 3369–3383. doi: 10.1002/pmic.200700266
Liang, Y., Chen, H., Tang, M., and Shen, S. (2007). Proteome analysis of an ectomycorrhizal fungus Boletus edulis under salt shock. Mycol. Res. 111, 939–946. doi: 10.1016/j.mycres.2007.06.005
Liu, G. T., Ma, L., Duan, W., Wang, B. C., Li, J. H., Xu, H. G., et al. (2014). Differential proteomic analysis of grapevine leaves by iTRAQ reveals responses to heat stress and subsequent recovery. BMC Plant Biol. 14:110. doi: 10.1186/1471-2229-14-110
Liu, J. Y., Men, J. L., Chang, M. C., Feng, C. P., and Yuan, L. G. (2017). iTRAQ-based quantitative proteome revealed metabolic changes of Flammulina velutipes mycelia in response to cold stress. J. Proteomics 156, 75–84. doi: 10.1016/j.jprot.2017.01.009
Liu, Y., Pan, Q. H., Yang, H. R., Liu, Y. Y., and Huang, W. D. (2008). Relationship between H 2 O 2 and jasmonic acid in pea leaf wounding response. Russ. J. Plant Physiol. 55:765. doi: 10.1134/S1021443708060058
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT methods. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Meng, L.-J., Kong, W.-W., Wu, X.-L., Liu, X.-M., Huang, C.-Y., and Zhang, J.-X. (2015). Biochemical pathway analysis of exogenous NO improving heat-tolerance of Pleurotus eryngii var. tuoliensis. Mycosystema 34, 632–639. doi: 10.13346/j.mycosystema.150038
Pratt, J. M., Simpson, D. M., Doherty, M. K., Rivers, J., Gaskell, S. J., and Beynon, R. J. (2006). Multiplexed absolute quantification for proteomics using concatenated signature peptides encoded by QconCAT genes. Nat. Protoc. 1, 1029–1043. doi: 10.1038/nprot.2006.129
Queitsch, C., Hong, S. W., Vierling, E., Lindquist, S., and Hong, S. W. (2000). Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12, 479–492. doi: 10.1105/tpc.12.4.479
Rice, K. C., and Bayles, K. W. (2008). Molecular control of bacterial death and lysis. Microbiol. Mol. Biol. Rev. Mmbr. 72, 85–109 doi: 10.1128/MMBR.00030-07
Richter, K., and Buchner, J. (2001). Hsp90: chaperoning signal transduction. J. Cell. Physiol. 188, 281–290. doi: 10.1002/jcp.1131
Rogg, L. E., Fortwendel, J. R., Juvvadi, P. R., and Steinbach, W. J. (2012). Regulation of expression, activity and localization of fungal chitin synthases. Med. Mycol. 50, 2–17 doi: 10.3109/13693786.2011.577104
Song, C., Chen, Q., Wu, X., Zhang, J., and Huang, C. (2014). Heat Stress Induces Apoptotic-Like Cell Death in Two Pleurotus Species. Curr. Microbiol. 69, 611–616. doi: 10.1007/s00284-014-0634-4
Sørensen, J. G., Kristensen, T. N., and Loeschcke, V. (2003). The evolutionary and ecological role of heat shock proteins. Ecol. Lett. 6, 1025–1037. doi: 10.1046/j.1461-0248.2003.00528.x
Sun, Y., Wang, Q., Li, Z., Hou, L., Dai, S., and Liu, W. (2013). Comparative proteomics of peanut gynophore development under dark and mechanical stimulation. J. Proteome Res. 12, 5502–5511. doi: 10.1021/pr4003504
Tiwari, S., Thakur, R., and Shankar, J. (2015). Role of Heat-Shock proteins in cellular function and in the biology of fungi. Biotechnol. Res. Int. 2015, 1–11. doi: 10.1155/2015/132635
Vedeler, A., Hollås, H., Grindheim, A. K., and Raddum, A. M. (2012). Multiple roles of annexin A2 in post-transcriptional regulation of gene expression. Curr. Protein Pept. Sci. 13, 401–412. doi: 10.2174/138920312801619402
Wang, L., Wu, X., Gao, W., Zhao, M., Zhang, J., and Huang, C. (2017). Differential expression patterns of Pleurotus ostreatus catalase genes during developmental stages and under heat stress. Genes 8, 335–346. doi: 10.3390/genes8110335
Zhai, Q., Fu, Z., Hong, Y., Yu, X., Han, Q., Lu, K., et al. (2018). iTRAQ-Based comparative proteomic analysis of adultschistosoma japonicumfrom water buffalo and yellow cattle. Front. Microbiol. 9:99. doi: 10.3389/fmicb.2018.00099
Zhao, B., Zhao, Z., Sun, X., Zhang, Y., Guo, Y., Tian, P., et al. (2017). Effect of micro strain stress on proliferation of endothelial progenitor cells in vitro by the MAPK-ERK1/2 signaling pathway. Biochem. Biophys Res. Commun. 492, 206–211. doi: 10.1016/j.bbrc.2017.08.050
Keywords: Pleurotus ostreatus, heat stress, TBARS, proteomics, recovery
Citation: Zou Y, Zhang M, Qu J and Zhang J (2018) iTRAQ-Based Quantitative Proteomic Analysis Reveals Proteomic Changes in Mycelium of Pleurotus ostreatus in Response to Heat Stress and Subsequent Recovery. Front. Microbiol. 9:2368. doi: 10.3389/fmicb.2018.02368
Received: 22 February 2018; Accepted: 14 September 2018;
Published: 09 October 2018.
Edited by:
Raffaella Balestrini, Consiglio Nazionale delle Ricerche (CNR), ItalyReviewed by:
Massimo Reverberi, Università degli Studi di Roma La Sapienza, ItalyElisa Bona, Università degli Studi del Piemonte Orientale, Italy
Copyright © 2018 Zou, Zhang, Qu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinxia Zhang, emhhbmdqaW54aWFAY2Fhcy5jbg==
 Yajie Zou
Yajie Zou Jinxia Zhang
Jinxia Zhang