
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 01 October 2018
Sec. Fungi and Their Interactions
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.02335
 Hyunkyu Sang1
Hyunkyu Sang1 Alexander Witte1
Alexander Witte1 Janette L. Jacobs1
Janette L. Jacobs1 Hao-Xun Chang1
Hao-Xun Chang1 Jie Wang2
Jie Wang2 Mitchell G. Roth1,3
Mitchell G. Roth1,3 Martin I. Chilvers1*
Martin I. Chilvers1*The succinate dehydrogenase inhibitor (SDHI) fungicide, fluopyram, is used as a soybean seed treatment to manage Fusarium virguliforme, the casual agent of sudden death syndrome (SDS). More recently, other species within clade 2 of the Fusarium solani species, F. tucumaniae in South America and F. brasiliense in America and Africa, have been recognized as additional agents capable of causing SDS. To determine if fluopyram could be used for management of SDS caused by these species, in vitro sensitivity tests of the three Fusarium species to fluopyram were conducted. The mean EC50 values of F. brasiliense and F. virguliforme strains to fluopyram were 1.96 and 2.21 μg ml-1, respectively, but interestingly F. tucumaniae strains were highly sensitive (mean EC50 = 0.25 μg ml-1) to fluopyram compared to strains of the other two species. A sequence analysis of Sdh genes of Fusarium strains revealed that the F. tucumaniae strains contain an arginine at codon 277 in the SdhB gene instead of a glycine as in other Fusarium species. Replacement of glycine to arginine in SdhB-277 in a F. virguliforme wild-type strain Mont-1 through genetic transformation resulted in increased sensitivity to two SDHI fungicides, fluopyram and boscalid. Similar to a F. tucumaniae strain, the Mont-1 (SdhBG277R) mutant caused less SDS and root rot disease than Mont-1 on soybean seedlings with the fluopyram seed treatment. Our study suggests the amino acid difference in the SdhB in F. tucumaniae results in fluopyram being efficacious if used as a seed treatment for management of F. tucumaniae, which is the most abundant SDS causing species in South America. The establishment of baseline sensitivity of Fusarium species to fluopyram will contribute to effective strategies for managing Fusarium diseases in soybean and other pathosystems such as dry bean.
Soybean sudden death syndrome (SDS) caused by Fusarium virguliforme, is an economically devastating disease in North America. SDS ranked among the top forth yield-suppressing soybean diseases and 209.7 million bushels of soybean yield loss were attributed to SDS during 2010-2014 in the United States and Ontario, Canada (Allen et al., 2017). SDS outbreaks caused by F. virguliforme have been reported in nearly all major soybean producing States of the U.S. except North Dakota (Pennypacker, 1983; Rupe, 1989; Roy, 1997; Roy et al., 1997; Mulrooney et al., 2002; Aoki et al., 2003; Kurle et al., 2003; Ziems et al., 2006; Bernstein et al., 2007; Chilvers and Brown-Rytlewski, 2010). While F. virguliforme is the dominant SDS pathogen in North America, SDS outbreaks have predominantly been attributed to F. tucumaniae in South America and F. brasiliense especially in Brazil (O’Donnell et al., 2010). A recent survey of SDS symptomatic soybean fields in Michigan found that F. brasiliense was also present in the U.S. (Wang et al., unpublished). All of these SDS causing species belong to clade 2 of the Fusarium solani species complex (FSSC), which also contains species capable of causing root rot, but possibly little to no SDS (O’Donnell et al., 2010).
Due to the significant economic impact of SDS in soybean production, different management strategies such as utilizing partially resistant varieties, crop rotation, tillage and planting date have been investigated to manage SDS (Wrather et al., 1995; Leandro et al., 2018). Soybean seed treatments have been used routinely to protect from early infection by root rot pathogens but most seed treatment fungicides are not effective against SDS (Weems et al., 2015). In 2014, the succinate dehydrogenase inhibitor (SDHI), fluopyram, was registered as a seed treatment (ILeVO, Bayer Crop Science) and has been widely used in soybean fields because of its effective suppression of SDS (Kandel et al., 2016, 2018). Wang et al. (2017) established baseline sensitivity of F. virguliforme isolates from different states to fluopyram. The majority of F. virguliforme isolates characterized in the study displayed sensitivity to fluopyram (mean EC50 = 3.35 μg ml-1) (Wang et al., 2017). Although this information is useful in efforts to prolong the product life of fluopyram to F. virguliforme, the sensitivity of other important SDS pathogens like F. brasiliense and F. tucumaniae to fluopyram has not been determined.
The SDHIs are new generation fungicides and have been widely applied to manage many important plant pathogenic fungi (Sierotzki and Scalliet, 2013). Currently, 11 different chemical groups and 23 common name SDHIs are listed by the Fungicide Resistance Action Committee (FRAC, 2018a). These chemicals have a common site-specific mode of action within mitochondria, strongly binding to ubiquinone-binding sites (Qp) in the succinate dehydrogenase complex composed of four subunits (SdhA, SdhB, SdhC, and SdhD). The binding of SDHIs results in blockage of access to the substrate, which consequently prevents further catalyzation of the oxidation of succinate to fumarate and reduction of ubiquinone to quinone. This leads to reduced energy production and arrested fungal growth (Matsson and Hederstedt, 2001; Sierotzki and Scalliet, 2013). Resistance to SDHIs has been reported in 23 plant pathogenic fungi to date and is associated with mutation(s) in SdhB, SdhC, and/or SdhD genes (FRAC, 2015; FRAC, 2017; Popko et al., 2018). Because SDHIs have been widely applied as foliar fungicides, most field-developed SDHI-resistant pathogens (e.g., Alternaria alternata, Botrytis cinerea, Zymoseptoria tritici) are air-borne foliar pathogens (Sierotzki and Scalliet, 2013). SDHIs are also used as seed treatments, but knowledge in the exposure of soil-borne pathogens such as Fusarium species to SDHIs is limited and it is assumed that risk for development of SDHIs resistance for soil-borne pathogens is considerably less than for foliar pathogens. Despite the importance of soil-borne pathogens in agricultural production, Fusarium species such as the SDS casual pathogens have not been extensively studied in terms of SDHI resistance nor the function of their Sdh genes related to SDHIs.
In recent years, soybean SDS caused by F. virguliforme, F. brasiliense, and F. tucumaniae has become a significant problem not only in the United States but also in South America and South Africa (Aoki et al., 2003; Tewoldemedhin et al., 2017; Wang et al., unpublished). To protect soybean yields from losses caused by these SDS pathogens, an understanding of the pathogens’ sensitivity to fungicides and mechanism on target genes is crucial. Fluopyram as a seed treatment currently demonstrates the best field efficacy to manage F. virguliforme and the SDS that it causes. To address the concerns outlined above three objectives were established: (1) perform an in vitro sensitivity assay of F. virguliforme, F. brasiliense, and F. tucumaniae strains to fluopyram, (2) compare the sequences of their Sdh genes to investigate genetic factor(s) driving fungicide sensitivity, (3) validate the involvement of genetic factor(s) in fluopyram sensitivity using a reverse genetics approach and virulence on fluopyram treated soybean seed.
A total of 35 FSSC strains were used in this study. The species (n = number of strains used) are as follows: F. brasiliense (n = 17), F. tucumaniae (n = 4), F. virguliforme (n = 14). Thirteen Fusarium strains were obtained from the Agricultural Research Service Culture Collection (NRRL - Northern Regional Research Laboratory, United States). Additional strains were obtained from two different studies (Wang and Chilvers, 2016; Wang et al., unpublished). The origins and hosts are described in Table 1.
Thirty five strains of Fusarium species were grown on half-strength PDA (19.5 g of PDA and 7.5 g of agar in 1 L of distilled water) for 10 days. Half-strength PDA was used to increase the growth rate of Fusarium species. Agar plugs from actively growing colonies were inoculated on half-strength PDA without and with 0.5, 1, 3, 5, 7, 10, 50 μg ml-1 of fluopyram (Luna Privilege, Bayer CropScience, containing 43% of active ingredient). After 10 days, colonies were imaged using a scanner according to the method described in (Wang et al., 2017). The effective fungicide concentration to reduce mycelial growth rate by 50% (EC50) was calculated using R (R Core Team, 2015) package “drc” (Ritz and Streibig, 2005) with the four-parameter log-logistic model (LL.4). Two separate experiments and three replicates (petri plates) of each treatment per each experiment were conducted for each isolate.
To compare the sequences of SdhB, SdhC, and SdhD genes of three Fusarium species, the genes were amplified and sequenced from DNA of seven F. brasiliense strains (NRRL 22678, MI-Mtc-A8, MI-Mtc-A17, MI-Mtc-B6, MI-Mtc-B9brn, MI-Mtc-B11, and MIMtc-C6), four F. tucumaniae strains (NRRL 31096, NRRL 31777, NRRL 31950, and NRRL 34549), and five F. virguliforme strains (Mont-1, NRRL 32392, NRRL 36898, NRRL 36900, and NRRL 54291). Sensitivities of F. virguliforme strains to fluopyram were similar, as such strains were randomly selected for sequencing. Strains of F. brasiliense demonstrated a range of sensitivity to fluopyram, as such strains were selected to represent this range for sequence analysis. The genome sequences of F. virguliforme (Mont1 (NRRL 22292), NRRL 34551, LL0009, and Clinton-1B), F. brasiliense (NRRL 31757), and F. tucumaniae (NRRL 31096, NRRL 31781, and NRRL 34546) (GenBank BioProject Accession: PRJNA63281 and PRJNA289542) (Srivastava et al., 2014; Huang et al., 2016) were obtained from NCBI. Sdh genes were mined from the genome sequences using BLAST+ (Camacho et al., 2009) and used for the reference sequences and primer design. DNA from the strains were extracted by a CTAB method (Cubero et al., 1999) and each gene was amplified using Phusion High-Fidelity DNA Polymerase mixtures (New England BioLabs) with primer pairs described in Supplementary Table S1. The final PCR amplicons were purified by ExoSAP-IT (Affymetrix) and sequenced by the Research Technology Support Facility at Michigan State University. MEGA v6.06 was used for sequencing analysis of the Sdh genes (Tamura et al., 2013).
The upstream and full length of SdhB (1,126 bp) was amplified from gDNA of F. virguliforme strain Mont-1 (NRRL 22292) using primers F_KpnI_FvSdhB and R_SpeI_FtSdhB. The amplified fragment contained 949A, instead of 949G, because the reverse primer was designed for this replacement. The downstream region of SdhB (1,000 bp) was amplified from gDNA of Mont-1 using primers F_NotI_downFvSdhB and R_NsiI_downFvSdhB. Amplicons of the SdhB gene and downstream region of SdhB were digested by restriction enzymes KpnI/SpeI and NotI/NsiI, respectively, and purified by Monarch DNA Gel Extraction Kit (New England BioLabs). Each purified fragment was inserted into Topo-hph plasmid (Sang et al., 2017) digested by KpnI/SpeI or NotI/NsiI to generate Topo-hph-FvSdhB(G277R) or Topo-hph-downFvSdhB. Using the two generated plasmids as a DNA template, two DNA constructs were amplified with primers F_FvSdhB/R_YG and F_HY/R_downFvSdhB, respectively, each containing 1,126 bp of the upstream and full length of SdhB and 731 bp of hph or 1 kb downstream of the SdhB and 1,126 bp of PtrpC-hph. The constructs were gel-purified using the Gel Extraction Kit, and the final two DNA constructs (each 2 μg) were transformed using a polyethylene glycol (PEG)-mediated transformation method in protoplasts from the F. virguliforme strain Mont-1. The protoplast generation and PEG-mediated transformation were conducted using a modified method from Mansouri et al. (2009) and Hallen-Adams et al. (2011). After the generation of hygromycin resistant F. virguliforme transformants, single spore isolation of each transformant was conducted and DNA of the transformants was extracted using a CTAB method (Cubero et al., 1999). The four primer sets (F_detFvSdhB/R_detFvSdhB; F_detFvSdhB/R_YG; F_HY/R_detFvSdhB; F_ptrpC/R_hgh) were used to confirm Mont-1(SdhBG277R) mutants and the fragment amplified by the primer pair (F_detFvSdhB/R_detFvSdhB) was sequenced.
To investigate the effect of a G277R substitution in the SdhB gene of the strain Mont-1, sensitivity of F. virguliforme mutants Mont-1(SdhBG277R)-1 and Mont1(SdhBG277R)-2 to two different SDHI fungicides fluopyram and boscalid was assayed. Boscalid was added in this assay to test whether the mutation in SdhB confers cross sensitivity to two different chemical groups of SDHI fungicides, carboxamide (boscalid) and pyridinylethyl-benzamide (fluopyram). Two F. virguliforme strains (Mont-1 and NRRL 54291) and two F. tucumaniae strains (NRRL 31096, and NRRL 34549) were included in this sensitivity assay as reference strains. The six strains were grown on PDA for 8 days and the agar plugs (3 mm in diameter) from the edge of a fungal colony were inoculated onto the half-strength PDA without or with fluopyram (1 μg ml-1) and boscalid (Endura, BASF, containing 70% of active ingredients; 50 μg ml-1). After 5 days, pictures of strains on the half-strength PDA and half-strength PDA amended with fluopyram or boscalid were taken and the diameters of each colony were measured. Two separate experiments and three replicates per each experiment were conducted.
The pathogenicity of F. virguliforme strains Mont-1 and Mont-1(SdhBG277R)-1 and F. tucumaniae NRRL 31096 were assayed on soybean seedlings from two different seed varieties (A and B) without treatment, with base seed treatment, and with base + fluopyram seed treatment. Base seed treatment (Bayer CropScience) contained prothioconazole + penflufen + Metalaxyl (EverGol Energy, 0.019 mg a.i./seed; Bayer CropScience), Metalaxyl (Allegiance, 0.02 mg a.i./seed; Bayer CropScience), and clothianodin + B. firmus (Poncho/VOTiVO, 0.13 mg a.i./seed; Bayer CropScience). The base + fluopyram included fluopyram at a rate of 0.15 mg a.i./seed. Pro-Ized red seed colorant (Gustafson LLC) and finisher (Peridiam Precise 1010; Bayer CropScience) were added at the rate of 32.6 and 65 ml/100kg of seed, respectively. Fungal inoculum was prepared by the method used in Wang et al. (2018). Briefly, each strain was grown on Nash-Snyder (NS) medium for 14 days at room temperature. A fungal colonized NS medium plate and a non-colonized NS medium plate, and 100 ml of autoclaved deionized water were mixed and homogenized in a sterile stainless-steel blender carafe for 30 s. The homogenized inoculum slurry was inoculated to autoclaved sorghum grain (1.8 kg) in spawn bags (Fungi perfecti) and allowed to colonize the grain for 4 weeks at room temperature. The grain inoculum in the bag were mixed by shaking every day. For seedling pathogenicity assay, twelve grams of infested grain inoculum or control grain were mixed with 250 ml of medium vermiculite and placed into a 354 ml paper cup (Solo) with three drainage holes on the bottom. Seventy ml of the medium vermiculite was poured over the inoculum-vermiculite mixture in the cup and six soybean seeds were sown in each cup. After that, seeds were covered with an additional 70 ml of the medium vermiculite. The cups were incubated in a growth chamber at 20°C with 14 h light period and watered as needed. After 28 days of incubation, SDS disease severity index was rated (scale 1–9): 1 = healthy plant, 2 = leaf showing slight yellowing and/or chlorotic flecks or blotches, 3 = leaf with interveinal chlorosis, 4 = leaf with necrosis along a portion (>2 cm) of its margin with interveinal chlorosis. 5 = necrosis along the entire margin of leaf with leaf or leaves cupped and/or irregular. 6 = interveinal necrosis and necrotic and/or leaf loss of most of the leaf area, 7 = most of the leaf area is necrotic and entire defoliated plants with new growth, 8 = entirely defoliated plants and most plants dead or dying, 9 = plant dead or dying. Roots were harvested and washed, and root rot disease severity was rated (scale:1–7) according to the modified method described previously by Schneider and Kelly (2000): 1 = healthy roots, 2 = small lesions on tap root or lateral root, 3 = increased size or coalescing of root lesion with 1–10% reduction in root mass, 4 = increased root lesion length with full encircling of root and 10–20% reduction in root mass, 5 = increase in root lesion elongation with tap root rotted and 20–50% reduction in root mass, 6 = intense root discoloration and root with 50–80% root mass reduction, 7 = pithy or hollow root with portion of root falling off with 80–100% root mass reduction and functionally dead. The washed roots were dried for 2 days at 55°C and dry root and shoot mass (g) were measured. The experiment with two different soybean varieties was performed with three replicate pots (six seeds in each pot).
All statistical analyses were conducted by the JMP software package, version 14.0 (SAS Institute Inc.).
In order to establish the sensitivity of three Fusarium species to fluopyram, EC50 values were measured using 35 strains from geographically different locations (Table 1). The mean EC50 values of F. brasiliense (n = 17), F. tucumaniae (n = 4), and F. virguliforme (n = 14) to fluopyram were 1.96 ± 0.2 μg ml-1, 0.25 ± 0.1 μg ml-1, and 2.21 ± 0.1 μg ml-1, respectively. The EC50 values among F. brasiliense, F. virguliforme, or F. tucumaniae strains were not statistically different (P = 0.091, P = 0.132, and P = 0.637, respectively). However, EC50 values among all Fusarium strains were statistically significant (P = 0.0013). The mean EC50 value of F. tucumaniae was significantly lower than the values of F. brasiliense (P = 0.0013) or F. virguliforme (P < 0.0001). There was no statistical difference between the mean EC50 values of F. brasiliense and F. virguliforme. The EC50 range of F. tucumaniae, F. brasiliense, and F. virgulforme strains were 0.06 to 0.4 μg ml-1, 0.8 to 4.24 μg ml-1, 1.53 to 2.99 μg ml-1 (Table 1).
To investigate the potential genetic factor(s) driving the sensitivity of F. brasiliense, F. tucumaniae, and F. virguliforme strains to fluopyram, the SDHI target genes (SdhB, SdhC, and SdhD) of 16 strains (F. brasiliense strains: NRRL 22678, MIMtc-A8, MIMtc-A17, MIMtc-B6, MIMtc-B9brn, MIMtc-B11, and MIMtc-C6; F. tucumaniae strains: NRRL 31096, NRRL 31777, NRRL 31950, and NRRL 34549; F. virguliforme strains: Mont-1, NRRL 32392, NRRL 36898, NRRL 36900, and NRRL 54291) were sequenced and compared with reference sequences from eight genome sequences of F. brasiliense, F. tucumaniae, and F. virguliforme strains. Amino acid sequences of SdhC and SdhD from F. brasiliense, F. tucumaniae, and F. virgulforme strains and the Fusarium genome sequences were identical, except SdhD sequence from one of genome sequences (NRRL 34546), which has a gap in the coding sequences of SdhD. However, two amino acid differences were detected in SdhB among three Fusarium species. Both F. tucumaniae and F. virgulforme strains have arginine (R) at codon 28 of SdhB but F. brasiliense strains (except for one strain, MI-Mtc-B6) contain cysteine (C) at the same position. F. brasiliense and F. virgulforme strains have glycine (G) at codon 277 of SdhB but F. tucumaniae strains contain arginine (R) at the same position (Figure 1). All amino acid sequences of SdhB, SdhC, and SdhD from F. brasiliense, F. tucumaniae, and F. virguliforme strains are presented in Supplementary Table S2.
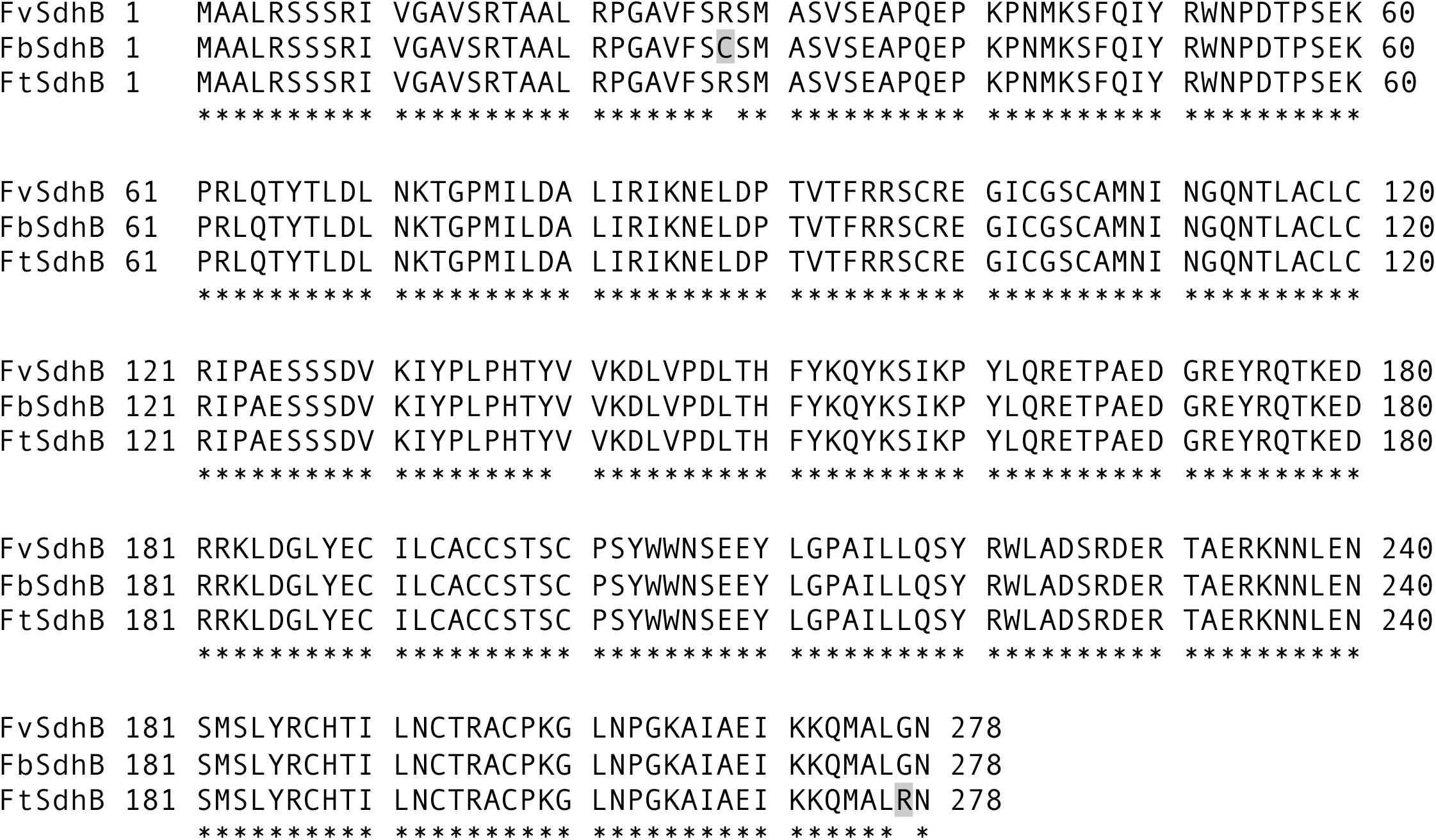
FIGURE 1. Alignment of the full length of SdhB amino acid sequences of F. brasiliense, F. virguliforme, and F. tucumaniae strains. FbSdhB, FvSdhB, and FtSdhB refer to SdhB amino acid sequence from F. brasiliense, F. virguliforme, and F. tucumaniae, respectively. The shaded letters indicate difference at amino acid position 28 in F. brasiliense from other species and 277 in F. tucumaniae from other species. Asterisks indicate identical amino acids among the strains compared.
Since the sensitivity of F. brasiliense and F. virgulforme strains to fluopyram was not statistically different, the amino acid difference at codon 28 of SdhB between F. brasiliense and F. virguliforme does not appear to affect their sensitivity. On the other hand, F. tucumaniae strains contained SdhB-277R and were significantly more sensitive to fluopyram than the strains of the other two Fusarium species. Therefore, the function of the amino acid difference in SdhB was characterized using a reverse genetic approach. The SdhB-277G of F. virguliforme strain Mont-1 was replaced with SdhB-277R using a split marker method and the target gene replacement in the two mutants was confirmed by PCR with four primer pairs (Figure 2A). A 2,058-bp fragment of the left flanking region and 2,171-bp fragment of right flanking region were amplified only in two SdhBG277R mutants by primer sets F_detSdhB/R_YG and F_HY/R_detSdhB, respectively (Figures 2B,C). The fragment containing upstream and full length SdhB, hygromycin resistance cassette, and downstream SdhB (3,763-bp) was amplified in two mutants by F_detSdhB/ R_detSdhB and the fragment containing only upstream and full length SdhB and downstream SdhB (2,372-bp) was amplified in the F. virguliforme strain Mont-1 (Figure 2D). The hygromycin resistance cassette (1,391-bp) was amplified by primer set F_ptrpC and R_hph and detected in only two mutants (Figure 2E). The sequence of the fragment amplified by F_detSdhB/ R_detSdhB in the mutants confirmed SdhB-277G was replaced by SdhB-277R (Figure 2F).
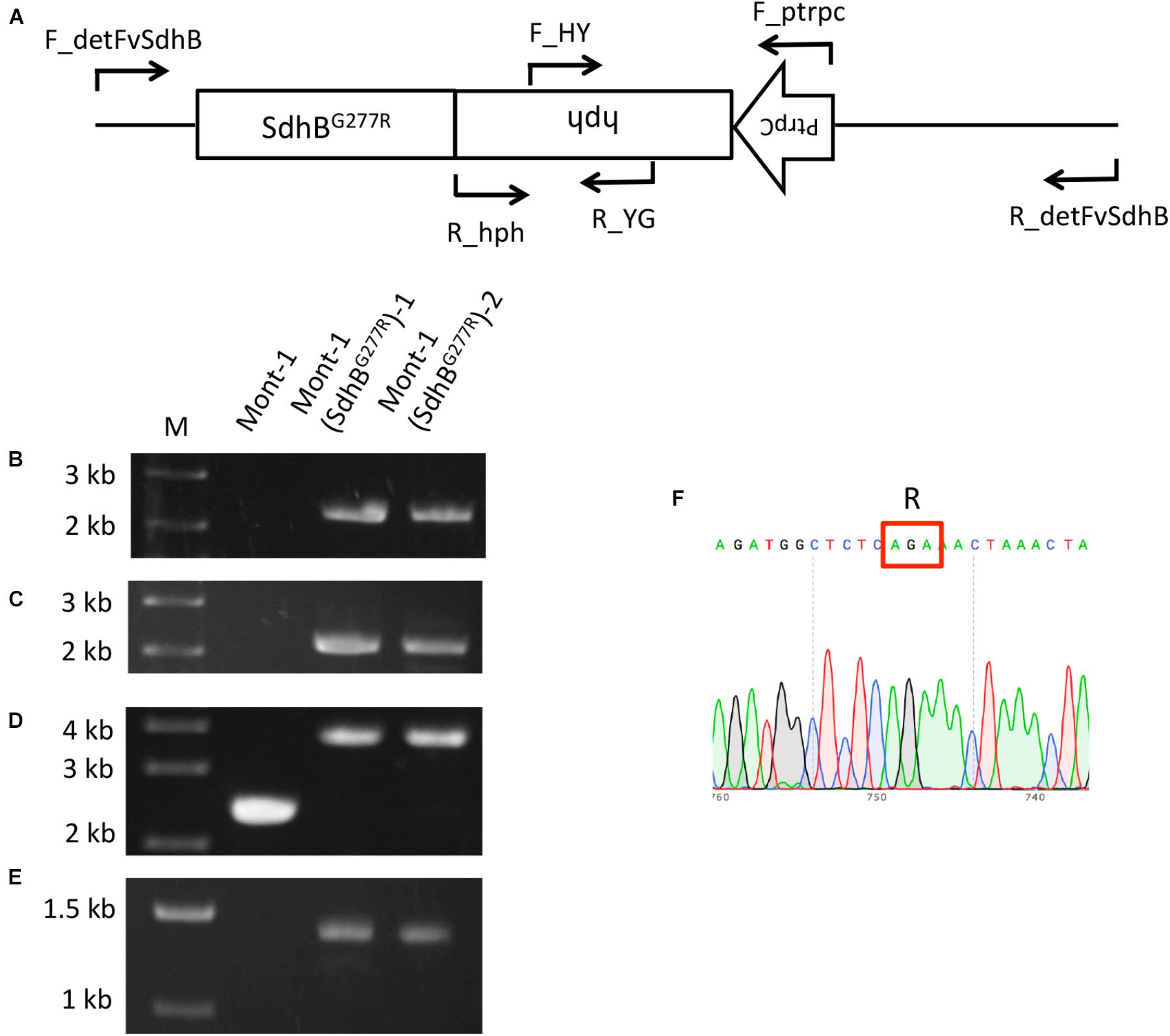
FIGURE 2. Validation of F. virguliforme mutants Mont-1(SdhBG277R)-1 and -2. (A) Schematic diagram of the SdhB gene and hygromycin resistance cassette (PtrpC and hph) and short arrows indicate primer binding sites. (B) Primer pair F_detFvSdhB/R_YG was used to amplify the 2,058-bp fragment of the left flanking region. (C) Primer pair F_HY /R_detFvSdhB was used to amplify the 2,171-bp fragment of right flanking region. (D) Primer pair F_detFvSdhB/R_detFvSdhB was used to amplify the fragment containing upstream and full length of SdhB, hygromycin resistance cassette, and downstream of SdhB (3,763-bp). (E) Primer pair F_ptrpC/R_hph was used to amplify the hygromycin resistance cassette (1,391-bp). (F) The sequence of the fragment from F_detSdhB/R_detSdhB in the strain Mont-1(SdhBG277R)-1 indicated the successful replacement from SdhB-277G to SdhB-277R.
On half-strength PDA, there was no statistical difference in growth rate of any strains used for in vitro sensitivity tests. Two mutants Mont-1(SdhBG277R)-1 and -2 and F. tucumaniae strains NRRL 31096 and NRRL 34549 were more sensitive to fluopyram (1 μg ml-1) and boscalid (50 μg ml-1) than the two wild type F. virguliforme strains Mont-1 and NRRL 54291. These results validate that the substitution SdhBG277R in F. virguliforme confers increased sensitivity to two different SDHI fungicides (Figure 3).
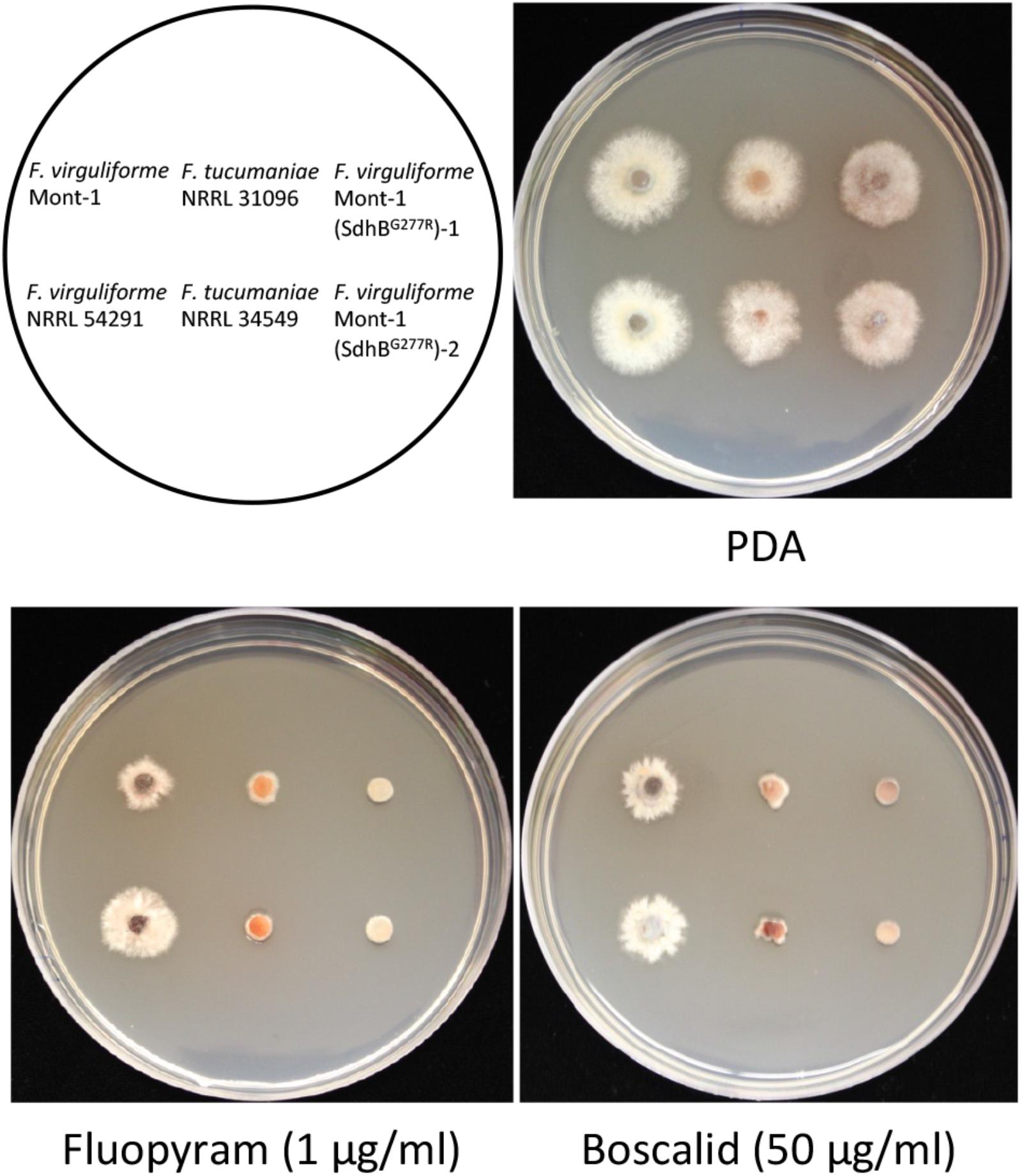
FIGURE 3. Sensitivity of two F. virguliforme strains (Mont-1 and NRRL 54291) and two mutants (Mont-1(SdhBG277R)-1 and -2) and two F. tucumaniae strains (NRRL 31096 and NRRL 34549) to two SDHI fungicides, fluopyram and boscalid. The pictures were taken after 5 days of growth on half strength PDA without or with fluopyram (1 μg ml-1) or boscalid (50 μg ml-1).
To assay whether the SdhBG277R substitution in F. virguliforme affects soybean health in seed treated with fluopyram, a soybean seedling pathogenicity assay was conducted using two soybean varieties and three different treatments: no seed treatment, a base treatment, and base + fluopyram treatment. On plants with no seed treatment, F. virguliforme strain Mont-1, F. tucumaniae NRRL 31096, and F. virguliforme mutant strain Mont-1(SdhBG277R)-1 caused interveinal chlorosis and necrosis typical symptoms of SDS (Figures 4A,B). Regardless of soybean variety, without seed treatment or with only a base treatment the F. virguliforme strains Mont-1 and Mont-1(SdhBG277R)-1 and F. tucumaniae NRRL 31096 caused significantly higher SDS disease severity index than the sorghum control. However, with the base + fluopyram treatment, F. virguliforme Mont-1(SdhBG277R)-1 and F. tucumaniae NRRL 31096 caused significantly less SDS symptoms on the two different varieties of soybean than the F. virguliforme wild type strain Mont-1 (Figures 4C,D).

FIGURE 4. Soybean sudden death syndrome (SDS) caused by F. virguliforme strains Mont-1 and Mont-1(SdhBG277R)-1 and F. tucumaniae strains NRRL 31096 without and with fluopyram. (A) The picture of soybean SDS from variety A with no treatment and with base + fluopyram treatment infected by strains Mont-1, Mont-1(SdhBG277R)-1, NRRL 31096, and sorghum control 28 days after inoculation. (B) The picture of soybean SDS from variety B with no treatment and with base + fluopyram treatment infected by strains Mont-1, Mont-1(SdhBG277R)-1, NRRL 31096, and sorghum control 28 days after inoculation. (C) SDS disease severity index (DSI) on soybean variety A with no treatment, base treatment, and base + fluopyram treatment infected by three Fusarium strains. Means followed by the same letter are not significantly different (P < 0.05) using Fisher’s Protected Least Significant Difference test. (D) SDS disease severity index (DSI) on soybean variety B with no treatment, base treatment, and base + fluopyram treatment infected by three Fusarium strains.
Fusarium virguliforme strains Mont-1 and Mont-1(SdhBG277R)-1 and F. tucumaniae NRRL 31096 caused significant root rot disease on both varieties of soybean seedlings without a seed treatment or with only a base treatment than the sorghum control (Figure 5). In the base + fluopyram treatment, both varieties of soybean seedlings infected by strains F. virguliforme Mont-1(SdhBG277R)-1 and F. tucumaniae NRRL 31096 displayed significantly lower root rot disease than the ones infected by wild type strain F. virguliforme Mont-1 (Figures 5C,D).
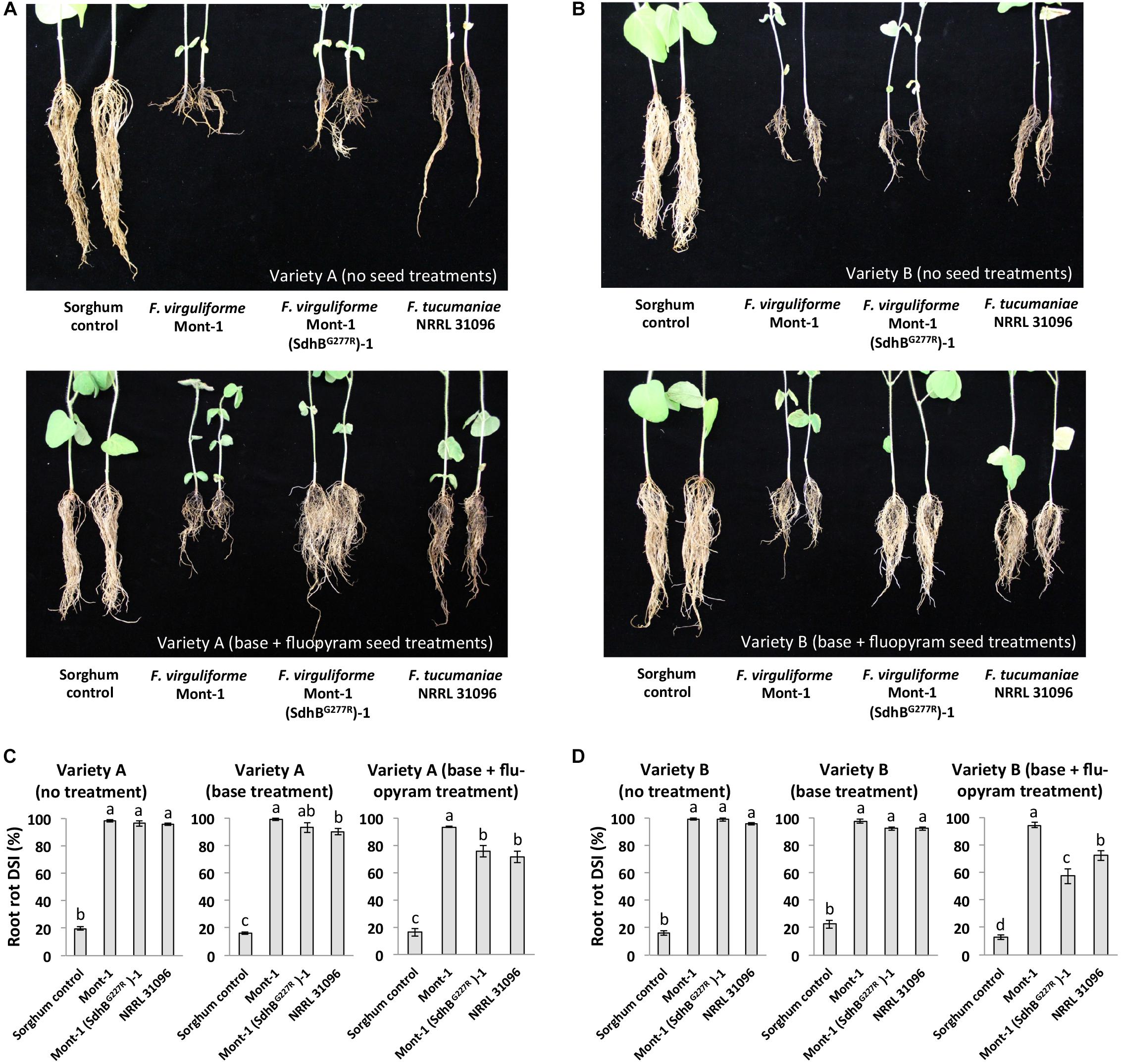
FIGURE 5. Root rot caused by F. virguliforme strains Mont-1 and Mont-1(SdhBG277R)-1 and F. tucumaniae strains NRRL 31096 without and with fluopyram. (A) The picture of soybean root rot from variety A with no treatment and with base + fluopyram treatment infected by strains Mont-1, Mont-1(SdhBG277R)-1, NRRL 31096, and sorghum control 28 days after inoculation. (B) The picture of soybean root rot from variety A with no treatment and with base + fluopyram treatment infected by strains Mont-1, Mont-1(SdhBG277R)-1, NRRL 31096, and sorghum control 28 days after inoculation. (C) Root rot disease severity index (DSI) on soybean variety A with no treatment, base treatment, and base + fluopyram treatment infected by three Fusarium strains. Means followed by the same letter are not significantly different (P < 0.05) using Fisher’s Protected Least Significant Difference test. (D) Root rot disease severity index (DSI) on soybean variety B with no treatment, base treatment, and base + fluopyram treatment infected by three Fusarium strains. Means followed by the same letter are not significantly different (P < 0.05) using Fisher’s Protected Least Significant Difference test.
Soybean seedling dry root and shoot mass (g) from both varieties of seedlings infected by F. virguliforme Mont-1 and Mont-1(SdhBG277R)-1 and F. tucumaniae NRRL 31096 were statistically less than seedlings grown with the sorghum control, except for seedlings from seed variety B with base + fluopyram treatment (Tables 2, 3). The seedlings from seed variety A with base + fluopyram treatment challenged with strains F. virguliforme Mont-1(SdhBG277R)-1 and F. tucumaniae NRRL 31096 had greater dry root and shoot mass than seedlings challenged with the wild type F. virguliforme strain Mont-1. Although the dry root and shoot mass from seed variety B with base + fluopyram was not statistically different regardless of strains used, the strains F. virguliforme Mont-1(SdhBG277R)-1 and F. tucumaniae NRRL 31096 resulted in a numerically greater dry root and shoot mass than the F. virguliforme wild type strain Mont-1 (Tables 2, 3).
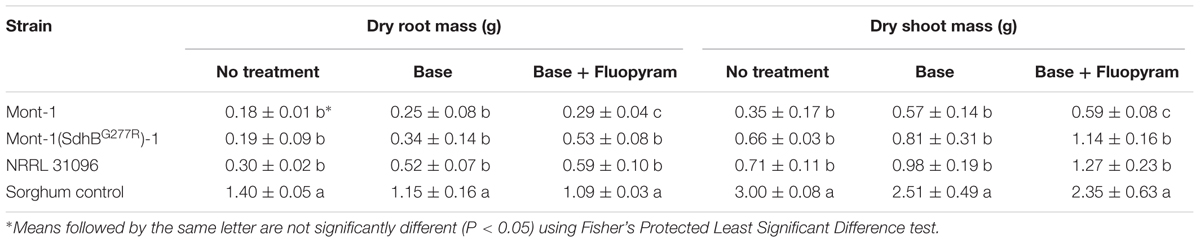
TABLE 2. Dry root and shoot mass (g) of soybean variety A caused by Fusarium virguliforme strains Mont-1 and Mont-1(SdhBG277R)-1 and F. tucumaniae strain NRRL 31096.
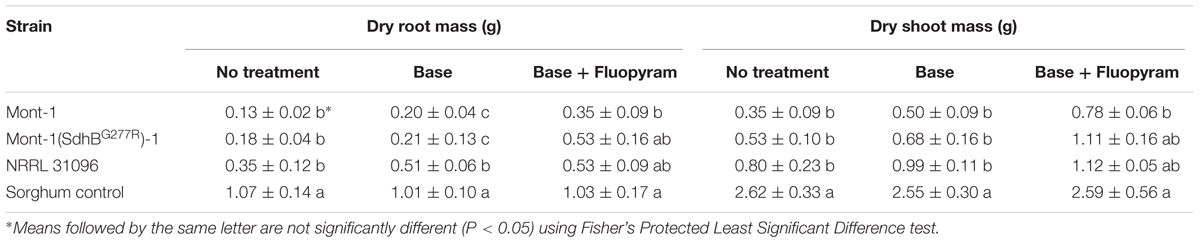
TABLE 3. Dry root and shoot mass (g) of soybean variety B caused by Fusarium virguliforme strains Mont-1 and Mont-1(SdhBG277R)-1 and F. tucumaniae strain NRRL 31096.
Fluopyram as a soybean seed treatment is increasingly being implemented throughout North America to manage SDS. Our findings indicate that F. tucumaniae is the most sensitive species to fluopyram among the three main causal species of Fusarium responsible for soybean SDS. The greater sensitivity in F. tucumaniae was attributed to the amino acid (277R) in SdhB, which was confirmed by F. virguliforme SdhBG277R mutants displaying increased sensitivity to fluopyram and boscalid. Significantly, the F. virguliforme SdhBG277R mutant reduced SDS foliar symptoms and root rot severity and increased shoot and root mass in the soybean seedlings from seeds treated with fluopyram compared to a wild type F. virguliforme strain. These findings suggest that a single amino acid difference in the SDHI target complex between two major SDS pathogens significantly affects their sensitivity to the fungicide. This discovery is valuable to both the soybean industry and farmers in the use of fluopyram as a seed treatment, especially in South America, where F. tucumaniae is more prevalent than F. virguliforme.
Fusarium virguliforme and F. brasiliense strains displayed similar sensitivity to fluopyram. The mean EC50 values of F. virguliforme and F. brasiliense to fluopyram were 1.96 μg ml-1 and 2.21 μg ml-1, respectively, which are close to the baseline sensitivity of 113 F. virguliforme isolates conducted by Wang et al. (2017). The EC50 values of these 113 out of 130 isolates were lower than 5 μg ml-1 of fluopyram, with the remaining 17 isolates being insensitive (>5 μg ml-1) to fluopyram (Wang et al., 2017). The strains used in the current study were sensitive to fluopyram (lower than 5 μg ml-1), which were collected before registration of fluopyram or from a field location with no history of fluopyram applications. Therefore, the sensitivity of the present panel of strains could be used as a baseline sensitivity of F. virguliforme and F. brasiliense for future fungicide resistance monitoring to fluopyram in soybean fields, and also other important legume crops such as dry bean, where production is constrained by F. brasiliense (Jacobs et al., 2018).
The amino acid sequences of SdhB, SdhC, and SdhD from strains of three Fusarium species were highly conserved because the three species are evolutionarily closely related, belonging to FSSC clade 2. Although most F. brasiliense strains harbor the different amino acid (SdhB-28C) from strains of F. virguliforme and F. tucumaniae, the sensitivity of F. brasiliense and F. virguliforme strains to fluopyram were not statistically different and the position of SdhB-28C is not close to the quinone binding sites (Fraaije et al., 2012). Interestingly, the SdhB-277G is a conserved amino acid in this position among Fusarium species (Supplementary Figure S1) but F. tucumaniae is the only species containing SdhB-277R. This position is adjacent to putative SDHI fungicides’ binding sites in SdhB (Fraaije et al., 2012; Scalliet et al., 2012), thus SdhB-277R and the substitution SdhBG277R might affect the binding of fluopyram to SdhB in F. tucumaniae and F. virguliforme mutants developed in this study, respectively. Some mutations (e.g., SdhB-H277Y in Alternaria alternate and SdhB-H267Y in Sclerotinia homoeocarpa) confer resistance to carboximide fungicides such as boscalid but increased sensitivity to fluopyram (Avenot and Michailides, 2010; Ishii et al., 2011; Popko et al., 2018), because fluopyram binds to a different cavity than other SDHIs (Fraaije et al., 2012). However, field and lab strains containing mutations in SdhB (e.g., P225F and H272L in Botrytis cinerea, H267L and N271K in Mycosphaerella graminicola) displayed reduced sensitivity to both carboximide and fluopyram (Scalliet et al., 2012; Veloukas et al., 2013). F. virguliforme SdhBG277R mutants in this study exhibited increased sensitivity to both fluopyram and boscalid, suggesting that the SdhB(277R) is associated with cross sensitivity to different SDHIs. Indeed, F. tucumaniae strains were also more sensitive to both fluopyram and boscalid compared to F. virguliforme strains. To our knowledge, this is the first report for SdhB-277R as a causal genetic variant and functional verification for its novel mechanistic function involving SDHI sensitivity in fungi.
The single amino acid substitution (G277R) in SdhB of F. virguliforme and 277R of SdhB in F. tucumaniae positively affects soybean health in the presence of fluopyram. Reduced severity of SDS and root rot by fluopyram increased the amount of shoots and roots, enhancing uptake of nutrients and water in the soil. In addition, the in vitro sensitivity of F. tucumaniae and F. brasiliense to fluopyram indicates that this seed treatment will likely have efficacy in South America where all three species can cause SDS. In North America fluopyram has been demonstrated to reduce SDS severity and protect yields from SDS caused by F. virguliforme (Kandel et al., 2016, 2018). Field studies of soybean with the fluopyram seed treatment against these other Fusarium species (F. tucumaniae and F. brasiliense) should be conducted to evaluate efficacy of fluopyram for management of SDS under field conditions.
Fungal pathogens have quickly evolved fungicide resistance to single site mode of action fungicides when continuously exposed to selection pressure (Brent and Hollomon, 2007; Jo et al., 2008). Plant pathogenic fungi have already developed resistance to many different classes of fungicides such as methyl benzimidazole carbamates, demethylation inhibitors, quinone outside inhibitors, and SDHIs (FRAC, 2018b). Although fluopyram has been used as a soybean seed treatment and applied once per year, F. virguliforme isolates with insensitivity to fluopyram are already present in soybean fields and it may imply potential risk for accumulating insensitive isolates by selection pressure (Wang et al., 2017). Therefore, a robust fungicide monitoring system with large number of isolates and SDS casual Fusarium species from different locations is needed to prevent possible accumulation of insensitivity isolates, which might lead to disease management failure. In addition, different combinations of seed treatments should be tested in the field for a long-term management of these important fungal pathogens.
HS, AW, JJ, H-XC, and MC designed the research. HS and AW performed the experiments and analyzed the data. JJ, JW, H-XC, and MR provided input on the experiments. HS and MC wrote the manuscript. AW, JJ, JW, H-XC, and MR edited the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The study was partially funded by Michigan Soybean Promotion Committee, the North Central Soybean Research Program, Bayer Crop Sciences. We thank Dr. Jennifer Riggs of Bayer Crop Sciences for discussion and in-kind support of product.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02335/full#supplementary-material
Allen, T. W., Bradley, C. A., Sisson, A. J., Byamukama, E., Chilvers, M. I., Coker, C. M., et al. (2017). Soybean yield loss estimates due to diseases in the United States and Ontario, Canada from 2010 to 2014. Plant Health Prog. 18, 19–27. doi: 10.1094/PHP-RS-16-0066
Aoki, T., O’donnell, K., Homma, Y., and Lattanzi, A. R. (2003). Sudden-death syndrome of soybean is caused by two morphologically and phylogenetically distinct species within the Fusarium solani species complex-F. virguliforme in North America and F. tucumaniae in South America. Mycologia 95, 660–684.
Avenot, H. F., and Michailides, T. J. (2010). Progress in understanding molecular mechanisms and evolution of resistance to succinate dehydrogenase inhibiting (SDHI) fungicides in phytopathogenic fungi. Crop Protection 29, 643–651. doi: 10.1016/j.cropro.2010.02.019
Bernstein, E. R., Atallah, Z. K., Koval, N. C., Hudelson, B. D., and Grau, C. R. (2007). First report of sudden death syndrome of soybean in Wisconsin. Plant Dis. 91:1201. doi: 10.1094/PDIS-91-9-1201B
Brent, K. J., and Hollomon, D. W. (2007). Fungicide Resistance in Crop Pathogens: How Can it Be Managed? FRAC Monograph No. 1, 2nd Edn. Brussels: CropLife International.
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST + : architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Chilvers, M. I., and Brown-Rytlewski, D. E. (2010). First report and confirmed distribution of soybean sudden death syndrome caused by Fusarium virguliforme in southern Michigan. Plant Dis. 94:1164. doi: 10.1094/PDIS-94-9-1164B
Cubero, O. F., Crespo, A. N. A., Fatehi, J., and Bridge, P. D. (1999). DNA extraction and PCR amplification method suitable for fresh, herbarium-stored, lichenized, and other fungi. Plant Syst. Evol. 216, 243–249. doi: 10.1007/BF01084401
Fraaije, B. A., Bayon, C., Atkins, S., Cools, H. J., Lucas, J. A., and Fraaije, M. W. (2012). Risk assessment studies on succinate dehydrogenase inhibitors, the new weapons in the battle to control Septoria leaf blotch in wheat. Mol. Plant Pathol. 13, 263–275. doi: 10.1111/j.1364-3703.2011.00746.x
FRAC (2015). List of Species Resistant to SDHIs April 2015. Available at: http://www.frac.info/docs/default-source/sdhi-wg/sdhi-references/list-of-species-resistant-to-sdhis-april-2015.pdf?sfvrsn=2d144a9a_2
FRAC (2017). Lists of Resistant Plant Pathogenic Organisms Dec 2017. Available at: http://www.frac.info/publications/downloads
FRAC (2018a). FRAC Code List, 2018: Fungicides Sorted by Mode of Action (including FRAC Code Numbering). Available at: http://www.frac.info/publications/downloads
FRAC (2018b). List of Plant Pathogenic Organisms Resistant to Disease Control Agents. Available at: http://www.frac.info/publications/downloads
Hallen-Adams, H. E., Cavinder, B. L., and Trail, F. (2011). “Fusarium graminearum from expression analysis to functional assays,” in Fungal Genomics: Methods and Protocol, eds J. Xu and B. H. Bluhm (New York, NY: Springer), 79–102.
Huang, X., Das, A., Sahu, B. B., Srivastava, S. K., Leandro, L. F., O’donnell, K., et al. (2016). Identification of highly variable supernumerary chromosome segments in an asexual pathogen. PLoS One 11:e0158183. doi: 10.1371/journal.pone.0158183
Ishii, H., Miyamoto, T., Ushio, S., and Kakishima, M. (2011). Lack of cross resistance to a novel succinate dehydrogenase inhibitor, fluopyram, in highly boscalid resistant isolates of Corynespora cassiicola and Podosphaera xanthii. Pest Manag. Sci. 67, 474–482. doi: 10.1002/ps.2092
Jacobs, J. L., Oudman, K., Sang, H., and Chilvers, M. I. (2018). First report of Fusarium brasiliense causing root rot of dry bean in the United States.Plant Dis. doi: 10.1094/PDIS-02-18-0332-PDN
Jo, Y. K., Chang, W. S., Boehm, M., and Jung, G. (2008). Rapid development of fungicide resistance by Sclerotinia homoeocarpa on Turfgrass. Phytopathology 98, 1297–1304. doi: 10.1094/PHYTO-98-12-1297
Kandel, Y. R., McCarville, M. T., Adee, E. A., Bond, J. P., Chilvers, M. I., Conley, S. P., et al. (2018). Benefits and profitability of fluopyram-amended seed treatments for suppressing sudden death syndrome and protecting soybean yield: a meta-analysis. Plant Dis. 102, 1093–1110. doi: 10.1094/PDIS-10-17-1540-RE
Kandel, Y. R., Wise, K. A., Bradley, C. A., Chilvers, M. I., Tenuta, A. U., and Mueller, D. S. (2016). Fungicide and cultivar effects on sudden death syndrome and yield of soybean. Plant Dis. 100, 1339–1350. doi: 10.1094/PDIS-11-15-1263-RE
Kurle, J. E., Gould, S. L., Lewandowski, S. M., Li, S., and Yang, X. B. (2003). First report of sudden death syndrome (Fusarium solani f. sp. glycines) of soybean in Minnesota. Plant Dis. 87:449. doi: 10.1094/PDIS.2003.87.4.449B
Leandro, L. F. S., Eggeberger, S., Chen, C., Williams, J., Beattie, G. A., and Liebman, M. (2018). Cropping system diversification reduces severity and incidence of soybean sudden death syndrome caused by Fusarium virguliforme. Plant Dis. 102, 1748–1758. doi: 10.1094/PDIS-11-16-1660-RE
Mansouri, S., Wijk, R., Rep, M., and Fakhoury, A. M. (2009). Transformation of Fusarium virguliforme, the causal agent of sudden death syndrome of soybean. J. Phytopathol. 157, 319–321. doi: 10.1094/MPMI-11-10-0271
Matsson, M., and Hederstedt, L. (2001). The carboxin-binding site on Paracoccus denitrificans succinate: quinone reductase identified by mutations. J. Bioenerg. Biomembr. 33, 99–105. doi: 10.1023/A:1010744330092
Mulrooney, R. P., Gregory, N. F., Walker, S. D., and Webster, A.-M. (2002). First report of sudden death syndrome of soybean in Delaware and eastern shore of Maryland. Plant Dis. 86:696. doi: 10.1094/PDIS.2002.86.6.696C
O’Donnell, K., Sink, S., Scandiani, M., Luque, A., Colletto, A., Biasoli, M., et al. (2010). Soybean sudden death syndrome species diversity within North and South America revealed by multilocus genotyping. Phytopathology 100, 58–71. doi: 10.1094/PHYTO-100-1-0058
Pennypacker, B. W. (1983). First report of sudden death syndrome caused by Fusarium solani f. sp. glycines on soybean in Pennsylvania. Plant Dis. 83:879. doi: 10.1094/PDIS.1999.83.9.879B
Popko, J. T., Sang, H., Lee, J., Yamada, T., Hoshino, Y., and Jung, G. (2018). Resistance of Sclerotinia homoeocarpa field isolates to succinate dehydrogenase inhibitor fungicides. Plant Dis. doi: 10.1111/mpp.12174
R Core Team (2015). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Ritz, C., and Streibig, J. C. (2005). Bioassay analysis using R. J. Stat. Softw. 12, 1–22. doi: 10.18637/jss.v012.i05
Roy, K. W. (1997). Fusarium solani on soybean roots: nomenclature of the causal agent of sudden death syndrome and identity and relevance of F. solani form B. Plant Dis. 81, 259–266. doi: 10.1094/PDIS.1997.81.3.259
Roy, K. W., Rupe, J. C., Hershman, D. E., and Abney, T. S. (1997). Sudden death syndrome of soybean. Plant Dis. 81, 1100–1111. doi: 10.1094/PDIS.1997.81.10.1100
Rupe, J. C. (1989). Frequency and pathogenicity of Fusarium solani recovered from soybeans with sudden death syndrome. Plant Dis. 73, 581–584. doi: 10.1094/PD-73-0581
Sang, H., Popko, J. T., Chang, T., and Jung, G. (2017). Molecular mechanisms involved in qualitative and quantitative resistance to the dicarboximide fungicide iprodione in Sclerotinia homoeocarpa field isolates. Phytopathology 107, 198–207. doi: 10.1094/PHYTO-05-16-0211-R
Scalliet, G., Bowler, J., Luksch, T., Kirchhofer-Allan, L., Steinhauer, D., Ward, K., et al. (2012). Mutagenesis and functional studies with succinate dehydrogenase inhibitors in the wheat pathogen Mycosphaerella graminicola. PLoS One 7:e35429. doi: 10.1371/journal.pone.0035429
Schneider, K. A., and Kelly, J. D. (2000). A Greenhouse screening protocol for Fusarium root rot in bean. Hortscience 35, 1095–1098.
Sierotzki, H., and Scalliet, G. (2013). A review of current knowledge of resistance aspects for the next-generation succinate dehydrogenase inhibitor fungicides. Phytopathology 103, 880–887. doi: 10.1094/PHYTO-01-13-0009-RVW
Srivastava, S. K., Huang, X., Brar, H. K., Fakhoury, A. M., Bluhm, B. H., and Bhattacharyya, M. K. (2014). The genome sequence of the fungal pathogen Fusarium virguliforme that causes sudden death syndrome in soybean. PLoS One 9:e81832. doi: 10.1371/journal.pone.0081832
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tewoldemedhin, Y. T., Lamprecht, S. C., Vaughan, M. M., Doehring, G., and O’donnell, K. (2017). Soybean SDS in South Africa is caused by Fusarium brasiliense and a novel undescribed Fusarium sp. Plant Dis. 101, 150–157. doi: 10.1094/PDIS-05-16-0729-RE
Veloukas, T., Markoglou, A. N., and Karaoglanidis, G. S. (2013). Differential effect of SdhB gene mutations on the sensitivity to SDHI fungicides in Botrytis cinerea. Plant Dis. 97, 118–122. doi: 10.1094/PDIS-03-12-0322-RE
Wang, J., Bradley, C. A., Stenzel, O., Pedersen, D. K., Reuter-Carlson, U., and Chilvers, M. I. (2017). Baseline sensitivity of Fusarium virguliforme to fluopyram fungicide. Plant Dis. 101, 576–582. doi: 10.1094/PDIS-09-16-1250-RE
Wang, J., and Chilvers, M. I. (2016). Development and characterization of microsatellite markers for Fusarium virguliforme and their utility within clade 2 of the Fusarium solani species complex. Fungal Ecol. 20, 7–14. doi: 10.1016/j.funeco.2015.09.013
Wang, J., Jacobs, J. J., Roth, M. G., and Chilvers, M. I. (2018). Temporal dynamics of Fusarium virguliforme colonization of soybean roots. Plant Dis. doi: 10.1094/PDIS-03-18-0384-RE
Weems, J. D., Haudenshield, J. S., Bond, J. P., Hartman, G. L., Ames, K. A., and Bradley, C. A. (2015). Effect of fungicide seed treatments on Fusarium virguliforme infection of soybean and development of sudden death syndrome. Can. J. Plant Pathol. 37, 435–447. doi: 10.1080/07060661.2015.1115781
Wrather, J. A., Kendig, S. R., Anand, S. C., Niblack, T. L., and Smith, G. S. (1995). Effects of tillage, cultivar, and planting date on percentage of soybean leaves with symptoms of sudden death syndrome. Plant Dis. 79, 560–562. doi: 10.1094/PD-79-0560
Keywords: fluopyram, Fusarium brasiliense, Fusarium tucumaniae, Fusarium virguliforme, root rot, SdhB, soybean sudden death syndrome
Citation: Sang H, Witte A, Jacobs JL, Chang H-X, Wang J, Roth MG and Chilvers MI (2018) Fluopyram Sensitivity and Functional Characterization of SdhB in the Fusarium solani Species Complex Causing Soybean Sudden Death Syndrome. Front. Microbiol. 9:2335. doi: 10.3389/fmicb.2018.02335
Received: 27 June 2018; Accepted: 12 September 2018;
Published: 01 October 2018.
Edited by:
Angel Medina, Cranfield University, United KingdomReviewed by:
Carlo Vito Bruschi, International Centre for Genetic Engineering and Biotechnology, ItalyCopyright © 2018 Sang, Witte, Jacobs, Chang, Wang, Roth and Chilvers. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martin I. Chilvers, Y2hpbHZlcnNAbXN1LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.