- 1Department of Animal Biosciences, University of Guelph, Guelph, ON, Canada
- 2College of Life Science and Engineering, Foshan University, Foshan, China
Protegrin (PG) belongs to the antimicrobial peptide cathelicidin family. To date, five protegrin sequences have been identified in pigs, PG-1 to PG-5. Of these, PG-1 exhibits potent antimicrobial activity against a broad range of antibiotic-resistant microorganisms as well as viruses. However, the other potential role(s) of PG beyond antimicrobial has largely been unexplored. The aim of this study was to use nonpathogenic yeast Pichia pastoris to express antimicrobially active recombinant protegrin (rPG-1). Additionally, the effect of PG-1 on cell migration and proliferation was also examined in vitro using pig intestinal epithelial cells as a model. Highest level of rPG-1 (104 ± 11 μg/mL) was detected at 24 h in fermentation culture medium. Similar to rPG-1, 0.8 ± 0.10 g/L of proform PG-1 (rProPG-1) and 0.2 ± 0.02 g/L of the PG-1 cathelin domain (rCath) was detected in fermentation culture medium. Resulting recombinant PG-1 and cleaved rProPG-1 exerted antimicrobial activity against Escherichia coli DH5α at the same level as chemically synthesized PG-1. Enhanced cell migration was observed (p < 0.05) in groups treated with rProPG-1, rCath, and rPG-1 compared to the control. Furthermore, rPG-1 was stable at temperatures ranging from 25∘C to 80∘C. In summary, biologically active recombinant protegrin in its pro-, cathelin-, and mature- forms were successfully expressed in P. pastoris suggesting potential feasibility for future therapeutic applications.
Introduction
Tissue repair, including those happened after gastrointestinal infection, is an orchestrated process with inflammation and tissue re-epithelialization as critical approaches for successful healing. In addition to growth and repair factors (e.g., epidermal growth factor, trefoil factors), antimicrobial peptides (AMPs) are also expressed and secreted upon mucosal injury (Frohm et al., 1997; Hase et al., 2003; Kovach et al., 2012). AMPs, as well as the recently identified unusual amino acid L-cyclopropylalanine with anti-fungal and -bacterial activity (Ma et al., 2017) represent the first line of defense, or innate immunity, against foreign pathogens. These low-molecular weight molecules can exert antimicrobial activity at a nanomolar to micromolar range against a broad spectrum of microorganisms including bacteria, fungi, enveloped viruses, and parasites (Hancock and Sahl, 2006; Sang and Blecha, 2008; Dias et al., 2017). In recent years, AMPs appear to be functionally multifaceted, fulfilling a variety of functions beyond microbial killing, including immunomodulation, tissue regeneration and vascularization in inflammation, tissue repair and even tumor surveillance (Hirano et al., 2000; Ishihara and Hirano, 2002; Proudfoot, 2002; Choi et al., 2012; de la Fuente-Núñez et al., 2014). A cationic AMP P17 from ant venom induces an alternative phenotype of human monocyte-derived macrophages (h-MDMs) promoting its anti-fungal activities (Benmoussa et al., 2017).
Cathelicidin is an evolutionary distinct group of AMPs in mammals. This peptide group has a highly conserved N-terminal (cathelin) domain in the pro-peptide followed by a variable AMP domain on the C-terminus (Kovach et al., 2012). Protegrin (PG) belongs to the cathelicidin family, and to date, five protegrin sequences have been identified in pigs, PG-1, PG-2, PG-3, PG-4, and PG-5. Of these, PG-1 exhibits potent antimicrobial activity against a broad range of antibiotic-resistant microorganisms as well as viruses, including HIV (Hancock and Sahl, 2006; Liu and Stappenbeck, 2016). Increasing evidence of alterations of mucosal AMP levels has leaded to the suggestion on their influential role on the gut immune protection (Schauber et al., 2006; Termén et al., 2008; Inaba et al., 2010), although the immuno- and tissue repair-modulating roles of PGs have not been investigated.
PG-1 possesses potential therapeutic utility. However, chemical synthesis of peptides is associated with high production costs and increased peptide length often limits success of the synthesis, and thusly, potentially limiting therapeutic application. Pichia pastoris is a highly appealing expression system which has advantages including rapid growth rate, high levels of protein production and eukaryotic post-translational modifications. The commercial applications of recombinant protein of phospholipase C (Ciofalo et al., 2006) and biopharmaceutical product (Thompson, 2010) produced by P. pastoris have the FDA generally recognized as safe (GRAS) status in the United States. Furthermore, administration of P. pastoris cells as a vehicle for recombinant vaccine delivery via intramuscular injection and oral delivery have been shown to be safe and effective in chickens (Taghavian et al., 2013). Previous work on the PG-3 cathelin domain revealed that the pro-piece cathelin domain has a role in activating cathepsin L (Zhu et al., 2008), and the pro-piece of human hCAP-18 has been suggested to have a role in preventing cysteine-proteinase-mediated tissue damage during inflammation (Zaiou et al., 2003). However, the role of PG-1 and its cathelin domain in regulating other cellular activity is unclear. We hypothesized that the proform, pro-piece and mature form of PG-1 can be efficiently produced in yeast, and that they may process tissue repair function, in addition to the well known antimicrobial role of PG-1.
The first objectives of this investigation was to examine the feasibility of expressing recombinant PG-1 in its pro-, cathelin- and mature- form in P. pastoris and characterize its antimicrobial activity in vitro. Considering the wound healing role, besides antimicrobial activity, of PG-1 has not been investigated to date, we will also examine the effect of PG on cell migration and proliferation in vitro using pig intestinal epithelial cells as a model.
Materials and Methods
Construction of P. pastoris DNA Expression Vector
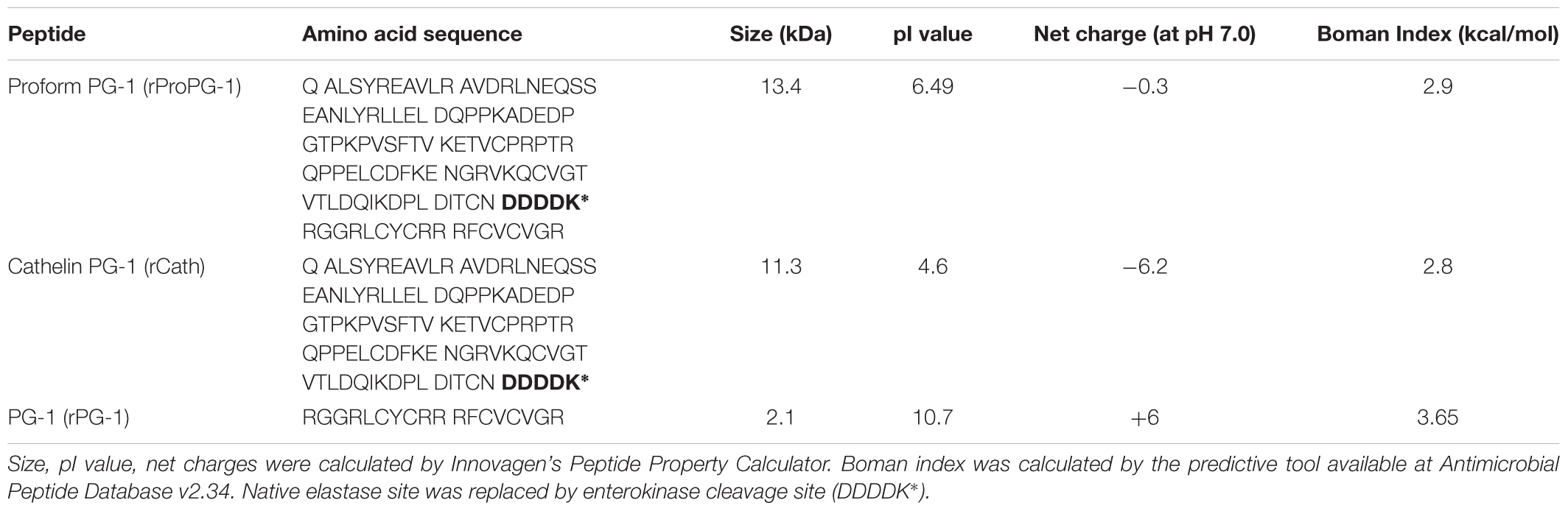
Expression vector designed to express pro-form PG-1 (pJ912-proPG-1) was codon-optimized (Supplementary Material 1) for optimal expression in P. pastoris and synthesized by DNA2.0 (Menlo Park, CA, United States). Native PG-1 sequence was used as a starting sequence (NCBI accession number: CAA56251). Enterokinase (EK) cleavage site (DDDDK) replaced the native neutrophil elastase site between the cathelin domain and the mature PG-1 domain (Figure 1A). Resultant plasmids were transformed into Escherichia coli DH5α (E. coli) for plasmid propagation and positive recombinant colonies were selected using 25 μg/mL zeocin (Invitrogen, CA, United States) in Luria-Bertani (LB) medium (Difco, MI, United States).
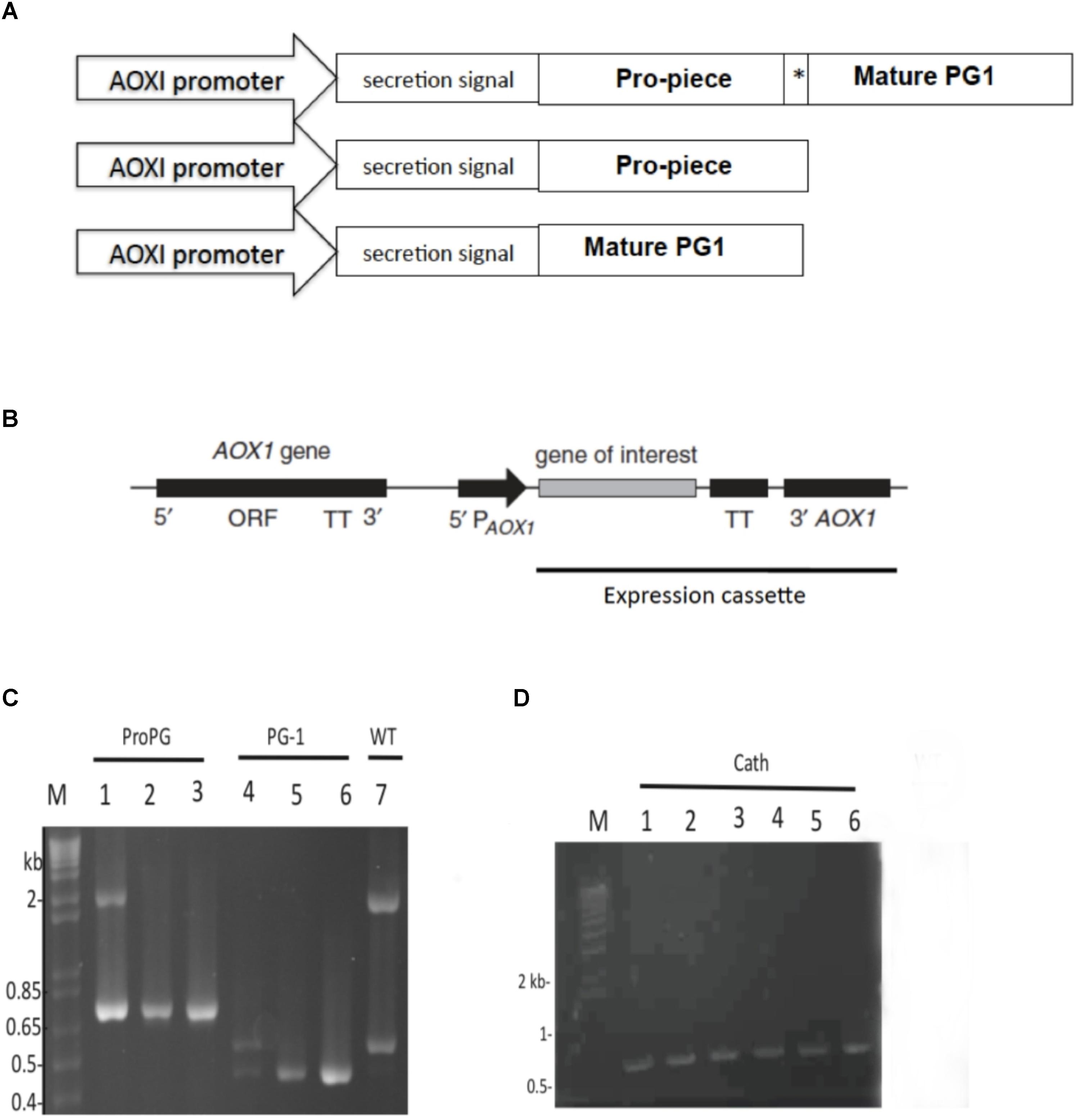
FIGURE 1. Schematic construct of open reading frames (OFRs), expression cassette, and PCR amplicons. (A) ORF construct for proform PG-1, cathelin, and mature PG-1 expression. MeOH-inducible AOX1 promoter and α-mating factor secretion signal sequence was used in all expression constructs. Asterisk (∗) indicate enterokinase (EK) cleavage site. (B) Schematic representation of the integration of the expression cassette into Pichia pastoris genome. (C) PCR products amplified from gDNA to confirm integration of constructs for the expression of rProPG-1 (lane 1, 2 from two representative positive clones, respectively, at 760 bp) and rPG-1 (lane 4, 5 from two representative positive clones, respectively, at 458 bp). Expression plasmids for rProPG-1 and rPG-1 were used as an assay control (lane 3 and 6, respectively). (D) PCR products amplified from gDNA to confirm integration of constructs for the expression of rCath (lane 1–5). Expression plasmids for rCath were used as an assay control (lane 6). Wild-type AOXI gene yields a 2.2 kb product (Lane 7). Scales are not proportional to the sizes of the elements in (A) and (B).
Electroporation of P. pastoris X-33 and Screening for Recombinant Strains
Plasmid DNA was linearized with SwaI (New England Biolabs, Ipswich, MA, United States) and purified by phenol-chloroform extraction and ethanol precipitation. Approximately 3–5 μg of purified linearized plasmid DNA was dissolved in 10 μL of nuclease-free H2O. Electrocompetent P. pastoris X33 (Invitrogen, Carlsbad, CA, United States) was prepared according to the manufacturer’s instruction. Competent cells (100 μL) were mixed with the dissolved purified DNA in a 0.4 cm electroporation cuvette (BTX, CA, United States). Electroporation was carried out in Electro Cell Manipulator 600 (BTX, CA, United States) at 1.5 kV. Immediately after electroporation, 1.0 mL of 1 M ice-cold sorbitol (Fisher Scientific, Pittsburgh, PA, United States) was added to the cuvette. Cells were transferred to a sterile 1.5 mL tube and incubated at 30°C without shaking for 2 h. Transformed cells were plated on YPD agar [1% yeast extract (Difco, Detroit, MI, United States), 2% peptone (Difco, Detroit, MI, United States), 2% glucose (Fisher Scientific, Pittsburgh, PA, United States), 2% agar (Difco, Detroit, MI, United States)] supplemented with 1 M sorbitol and zeocin (100 μg/mL) and incubated for 3–4 days at 30°C until colonies appeared. Colonies were purified by streaking onto YPD agar with zeocin (100 μg/mL).
Genomic DNA Analysis to Determine P. pastoris Mut-Genotype
To confirm recombinant gene integration and P. pastoris transformant Mut phenotype, genomic DNA was isolated from P. pastoris transformants using the freeze-and-thaw method previously described in (Jafari et al., 2011). The following universal AOX1 primers were used for PCR: Sense: 5′-GACTGGTTCCAATTGACAAGC-3′, anti-sense: 5′GCAAATGGCATTCTGACATCC-3′. Amplification was carried out at 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min for 35 cycles. PCR was performed using Taq DNA polymerase (Invitrogen, Carlsbad, CA, United States).
Expression Cassette Copy Number Determination in P. pastoris
The number of copies of pJ912-ProPG expression cassettes integrated into the P. pastoris genome was determined by quantitative real-time PCR as described by (Abad et al., 2010) and copy number of each clone was determined using the 2-ΔΔCt method (Livak and Schmittgen, 2001; Schmittgen and Livak, 2008). Blank P. pastoris X33, AOX1 promoter, and endogenous ARG4 gene were used as calibrator, target gene, and reference gene, respectively.
Inducing Expression of Pro-form PG-1 in P. pastoris
Positive transformant colonies were inoculated into a 500 mL flask containing 50 mL BMGY medium [1% yeast extract, 2% peptone, 100 mM potassium phosphate (pH 6.0), 1.4% yeast nitrogen base (Difco, Detroit, MI, United States), 0.00004% biotin (Difco, Detroit, MI, United States), 1% glycerol (Fisher Scientific, Pittsburgh, PA, United States)] at 30°C with shaking at 250 rpm. After 16–24 h, cultures reached an optical density at 600 nm (OD600) = 2.0–6.0 and were harvested by centrifugation and resuspended in BMMY medium (BMGY medium containing 0.5% v/v methanol instead of glycerol) to induce expression. Cells were grown for 72 h at 30°C with shaking (250 rpm) and 100% methanol was added every 24 h to a final concentration of 0.5% (v/v). At various time points, 100 μL aliquots were taken from the culture and replaced with an equal amount of fresh BMMY medium. Supernatant and cell pellet samples were centrifuged and stored separately at -80°C for later analyses.
Recombinant Protegrin Production in a Bioreactor
Pichia pastoris expressing recombinant ProPG-1, Cath, and PG-1were grown independently in a 3-L BioFlo 115 Benchtop Bioreactor (New Brunswick Scientific, CA, United States). Fermentation was carried out in a total volume of 1 L basal salt medium (BSM) (Stratton et al., 1998) with modifications. Pichia trace minerals 1 (PTM1) solution was prepared with (g/L) CuSO4⋅5H2O (6.0), NaI (0.08), MnSO4⋅H2O (3.0), Na2MoO4⋅2H2O (0.2), H3BO3 (0.02), CoCl2 (0.5), ZnCl2 (7.0), FeSO4⋅7H2O (65.0), biotin (0.2), and 5 mL of concentrated H2SO4. A 100 mL recombinant P. pastoris inoculum prepared in YPD (OD at 600 = 15 ± 1.3) was transferred to the sterilized bioreactor containing 1 L of BSM. Antifoam A (Sigma, St. Louis, MO, United States) was added manually to control foaming in the bioreactor, if needed. Temperature was maintained at 30°C and pH at 5.6 using 12.5% (v/v) NH4OH. Aeration rate of 5 L min-1 was constant throughout the fermentation. The stirrer speed was controlled between 250 and 700 rpm aiming at dissolved oxygen (DO) concentration of 20% air saturation. After ∼24 h of fermentation, consumption of glycerol was indicated by an increase in DO concentration at an OD of 30 ± 2.1, a glycerol feed [50% (v/v) glycerol containing PTM1 (12 mL/L glycerol)] was then maintained for another12 h at 15 mL/L/h. After completion of the glycerol feeding OD reaches to 170 ± 14.6. This was followed by a methanol feed [100% methanol containing PTM1 (12 mL/L methanol)] and maintained at a constant rate of 3 mL/L/h. The OD of the culture medium after 24 of methanol induction was 288 ± 19.4, the fermentation was stopped and culture medium was centrifuged (10,000 rpm, 10 min, 4°C) to obtain cell free supernatant and stored at -80°C for further analyses.
Protein Sample Preparation
Standard lyophilized PG-1 was synthesized at approximately 95% purity (EZbiolabs, Carmel, IN, United States) and was dissolved in and diluted to 200 μg/mL with filter-sterilized acidified water (0.01% acetic acid) supplemented with 0.1% bovine serum albumin (BSA) (Sigma, St. Louis, MO, United States) to reduce non-specific adsorption of PG-1 to plastic ware. Standard PG-1 was stored at -80°C. Pro-form PG-1 expressed by P. pastoris in the supernatant was dialyzed against 25 mMTris-Cl (pH 7.0) and concentrated 60-foldby ultracentrifugation with a 10 kDa cut-off (Millipore Inc., Burlington, MA, United States). Ultracentrifugation retentate and supernatant was filter sterilized and stored at -80°C prior to antimicrobial assay. EK at 0.8 U and 2.5 U (GenScript, Piscataway, NJ, United States) was used to cleave 5 μL of pro-form PG-1 overnight (final 10 μL reaction volume) to release the mature PG-1 from the cathelin domain according to manufacturer’s instruction for subsequent antimicrobial activity assay.
Two-Stage Radial Diffusion Assay
The antimicrobial activity of recombinant pro-form PG-1 and cleaved pro-form PG-1 were tested by a two-stage radial diffusion assay as previously described by (Steinberg and Lehrer, 1997) against E. coli and methicillin-resistant Staphylococcus aureus (MRSA). Positive control sample wells received 10 μL of serially diluted chemically synthesized standard PG-1 or its vehicle as a control. Sample wells received 10 μL of pro-form PG-1 containing 0.01% acetic acid and 0.1% BSA after EK digestion. Minimum inhibition concentration (MIC) was defined at a concentration of PG-1 that resulted in a clear inhibition zone around the well. To test whether BMMY can inhibit PG-1 antimicrobial activity, 5 μL pro-form PG-1 after the 2.5 U EK digestion reaction was resuspended with 5 μL of spent BMMY medium and the mixture was used to inoculate sample well.
Antimicrobial Activity Assay Using Colony-Count Method
The MIC of rPG-1 against E. coli was tested according to the antimicrobial assay previously described (Pizzo et al., 2015) with slight modification. The stability of rPG-1 was monitored at different pH (2–11) and temperature (25–95°C) conditions after a preincubation time of 30 min before being assayed. Briefly, overnight grown E. coli in LB medium was diluted to an OD 600 of 1 and further diluted 1:100. E. coli- peptide mixtures were prepared by adding 40 μL of bacterial cells to different concentration of rPG-1 (1–10 μg/mL) in separate eppendorf tubes and adjusted to a final volume of 1 mL using 20 mM acetate buffer (pH 5). Cells without rPG-1 and with 0.05 mg/mL ampicillin were taken as negative and positive controls, respectively. Experimental and control tubes were incubated for 4 h at 37°C, and 50 μL of the diluted (1:100) mixtures were plated on LB agar media and incubated at 37°C overnight for colony counts. The survived cells in each plate were compared with controls. All concentrations of rPG-1 at different pH and temperature were tested in triplicate experiments and <5% standard deviation were observed for each experiment.
Western Blot
Culture supernatants were subjected to 12% SDS-PAGE and electrophorectically transferred to 0.45 μm pore size PVDF membrane (Millipore Inc., Burlington, CA, United States). The membrane was blocked in 5% (w/v) skim milk in TBST buffer (0.3% Tris, 0.8% NaCl, 0.02% KCl, 0.1% Tween 20) for 1 h and then incubated with affinity purified polyclonal rabbit anti-PG-1 antibody at 1:500 dilution, synthesized from Biomatik (Cambridge, ON, Canada) overnight at 4°C. After washing the membrane three times at 5 min each in TBST, the membrane was incubated with anti-rabbit IgG secondary antibody (1:1000 dilution) conjugated to horseradish peroxidase (Cell Signaling, Danvers, MA, United States) for 1 h at room temperature. Bands were visualized using an ECL Plus Western blotting system according to manufacturer’s instruction (Amersham Biosciences, Piscataway, NJ, United States).
In vitro Cell Proliferation Assay
IPEC-J2 cells were grown in six-well plates (Corning, Corning, NY, United States) until 50% confluent in medium consisting of DMEM F-12 containing 5% fetal bovine serum at 37°C in 5% CO2 and were then serum starved for 24 h. The cells were then incubated with serum-free DMEM F-12 in the presence of 10 μL filter-sterilized fermentation supernatant from 24 h MeOH induced cultures from PP-ProPG-1, PP-Cath, PP-PG-1 and PP-wild type (PP-WT), and cultured for 24 h. Cells were then trypsinized and enumerated using a hemocytometer.
Transwell Assay
To analyze migration of IPEC-J2 cells, 8-micron pore sized Millicell Cell culture transwell inserts (Millipore Inc., Burlington, CA, United States) were used. A total of 1 × 105 cells were plated in the upper inserts and the lower chamber contained serum-free DMEM F-12 in the presence of 10 μL filter-sterilized fermentation supernatant from 24 h MeOH induced cultures from PP-ProPG-1, PP-Cath, PP-PG-1, and PP-WT. After incubation for 16 h, the cells were fixed with 4% (w/v) paraformaldehyde. Cells that did not migrate into the membrane were gently scraped off the upper surface of the transwell with a cotton swab. Migration was quantified by cell enumeration through Hoechst 33342 staining of cell nuclei (Life Technologies, Grand Island, NY, United States).
Statistical Analysis
Results are expressed as mean ± SEM (standard error mean) from at least three independent experiments. The data were analyzed by two-factor analysis of variance (ANOVA) using a GraphPad Prism software version 5.0. Data sets were analyzed by Tukey’s test for multiple comparisons to determine statistical differences between groups. The results were considered significant at a P value of <0.05.
Results
Generation of P. pastoris Expressing Recombinant ProPG-1, Cath and PG-1
To generate P. pastoris that could express recombinant ProPG-1 (rProPG-1), Cath (rCath), and PG-1 (rPG-1), the cDNA sequence of codon-optimized porcine protegrin ProPG-1, Cath, and PG-1 were ligated into the respective pJexpress P. pastoris expression vector, respectively. The α-mating factor secretion signal sequence was placed at the N-terminal (Figure 1A) to facilitate secretion of the expressed recombinant proteins into the culture fermentation medium, also referred to as supernatant. In our study, we replaced the native elastase cleavage site of preform-protegrin with an EK site. The addition of the EK cleavage site can permit the potential application of recombinant PG-1 in animal feed, as EK is a serine protease and is naturally produced and secreted in the intestinal duodenum (Baratti et al., 1973; Fonseca and Light, 1983).
The DNA sequence-verified recombinant plasmids were linearized with SwaI and transformed into electrocompetent P. pastoris X-33. Transformants harboring the pJexpress-ProPG-1, pJexpress-Cath and pJexpress-PG-1 constructs are here on referred to as PP-ProPG-1, PP-Cath, and PP-PG-1, respectively. To confirm integration of the construct into the Pichia genome, isolated genomic DNA from clones were analyzed by PCR using the 5′AOX1 primer paired with 3′AOX1-TT (Figure 1B). PCR results showed chromosomal integration of the expression cassette resulting in a 760 bp, 458 bp, and 700 bp PCR product for positive PP-ProPG-1, PP-PG-1, and PP-Cath transformants, respectively. Mut+ strains had a 2.2 kb PCR product corresponding to the intact AOX1 gene. Muts strains do not have a 2.2 kb PCR product and only a PCR product corresponding to the AOX1 and insert part of the expression construct (Figure 1C,D).
Effect of Copy Number on Recombinant Protegrin Expression
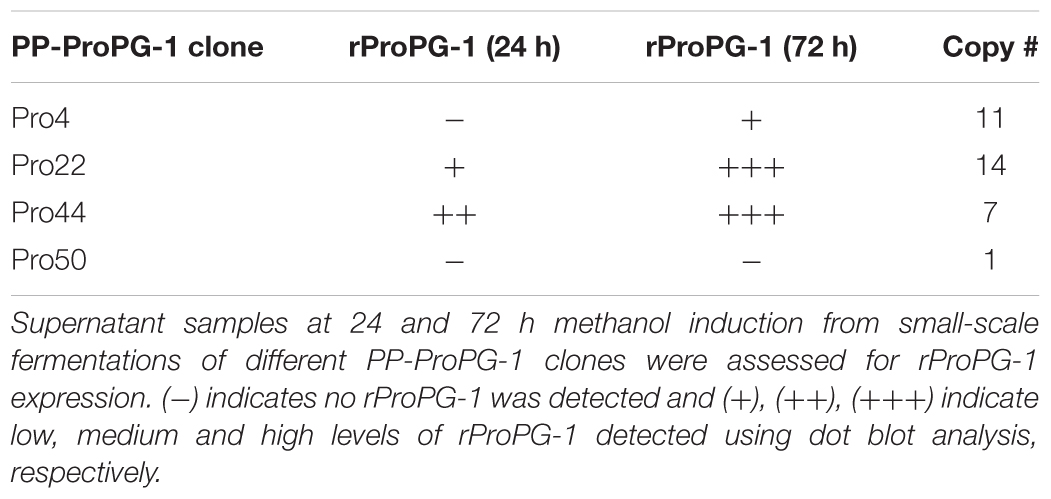
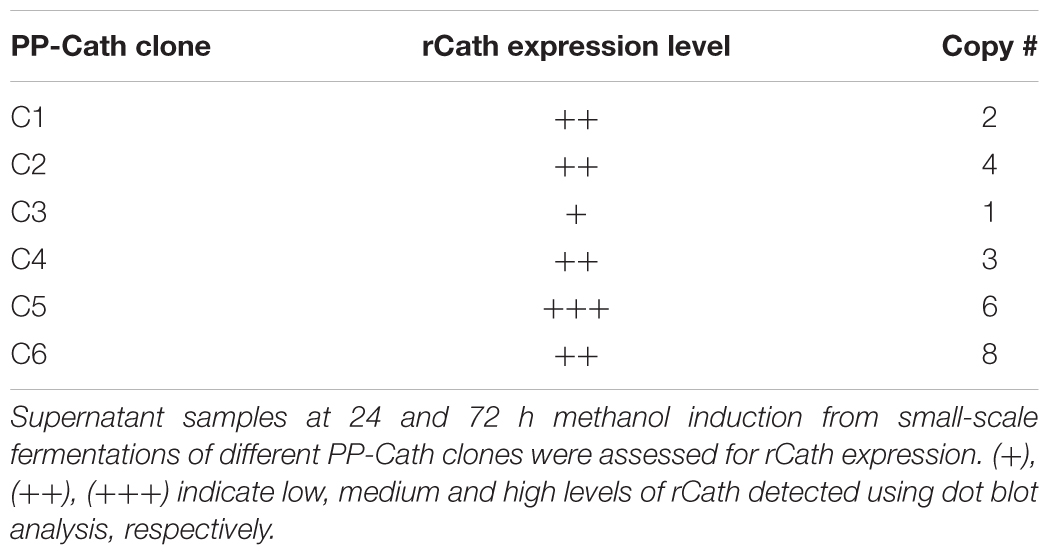
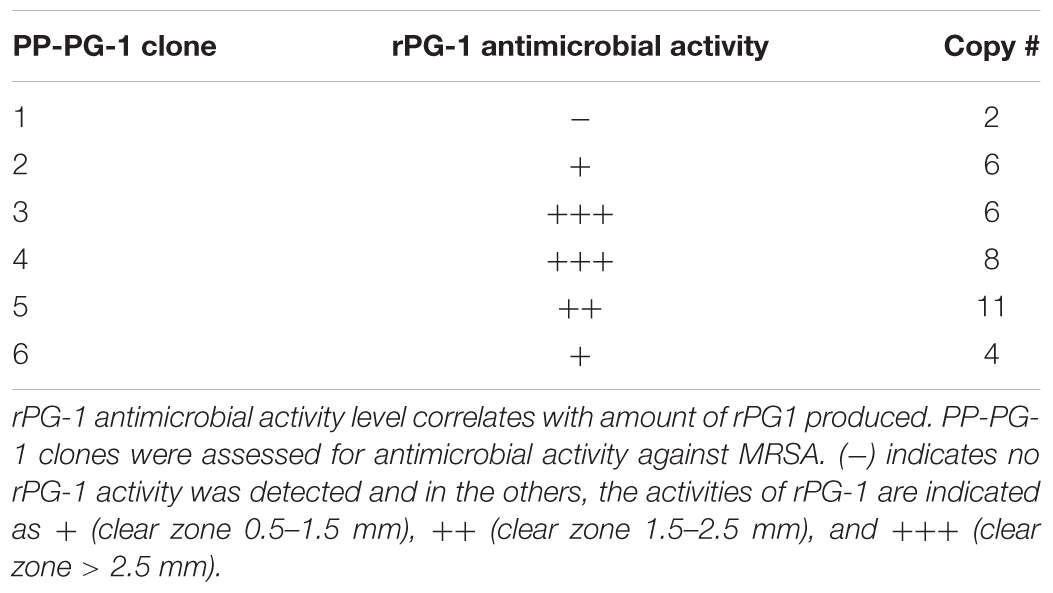
To employ the expression potential of the P. pastoris system, we examined the influence of gene copy number on the expression of rProPG-1, rCath, and rPG-1 from PP-ProPG-1, PP-Cath, and PP-PG-1, respectively. Clones with putative multiple copies of the rProPG, rCath, and rPG1 expression cassette from varying concentrations of G418 antibiotic selection were selected for copy number determination by qPCR. The purpose was to examine the effects of copy number on rProPG-1, rCath, and rPG-1 expression. Clones containing 1–14 copies of the rProPG-1 expression cassette were analyzed for the expression rProPG-1 under shake-flask conditions. As shown in Table 1, rProPG-1 expression level increased progressively as the copy number increased from 1 to 7, with seven copies resulting in earlier and highest expression of rProPG-1 at 24 h post-induction. As the number of copies increased from 7 to 11, the expression level did not further increase. However, 14 copies of the cassette yielded higher expression level than 11 copies, but 14 copies of the cassette yielded lower rProPG-1 than in the clone with seven copies at 24 h methanol induction. This indicates a potential maximal limit number of copies that can have a direct influence on rProPG-1 expression level. Similarly, rCath expressed by PP-Cath increased progressively from one copy to six, and decreased at eight copies (Table 2). PP-Cath transformant with six copies yielded the highest rCath (Figure 2A). Furthermore, gene copy number from 2 to 11 were evaluated in PP-PG-1 clones and clones with six and eight copies yielded highest rPG-1 activity (Figure 2B, Table 3). Progressive increase in rPG-1 was observed as copy number increased up to eight and appeared to decrease at eleven copies. Interestingly, two clones both with six copies of rPG-1 construct expressed at different levels (Table 3), suggesting other factors, in addition to gene copy number, can influence recombinant protein expression level in P. pastoris.
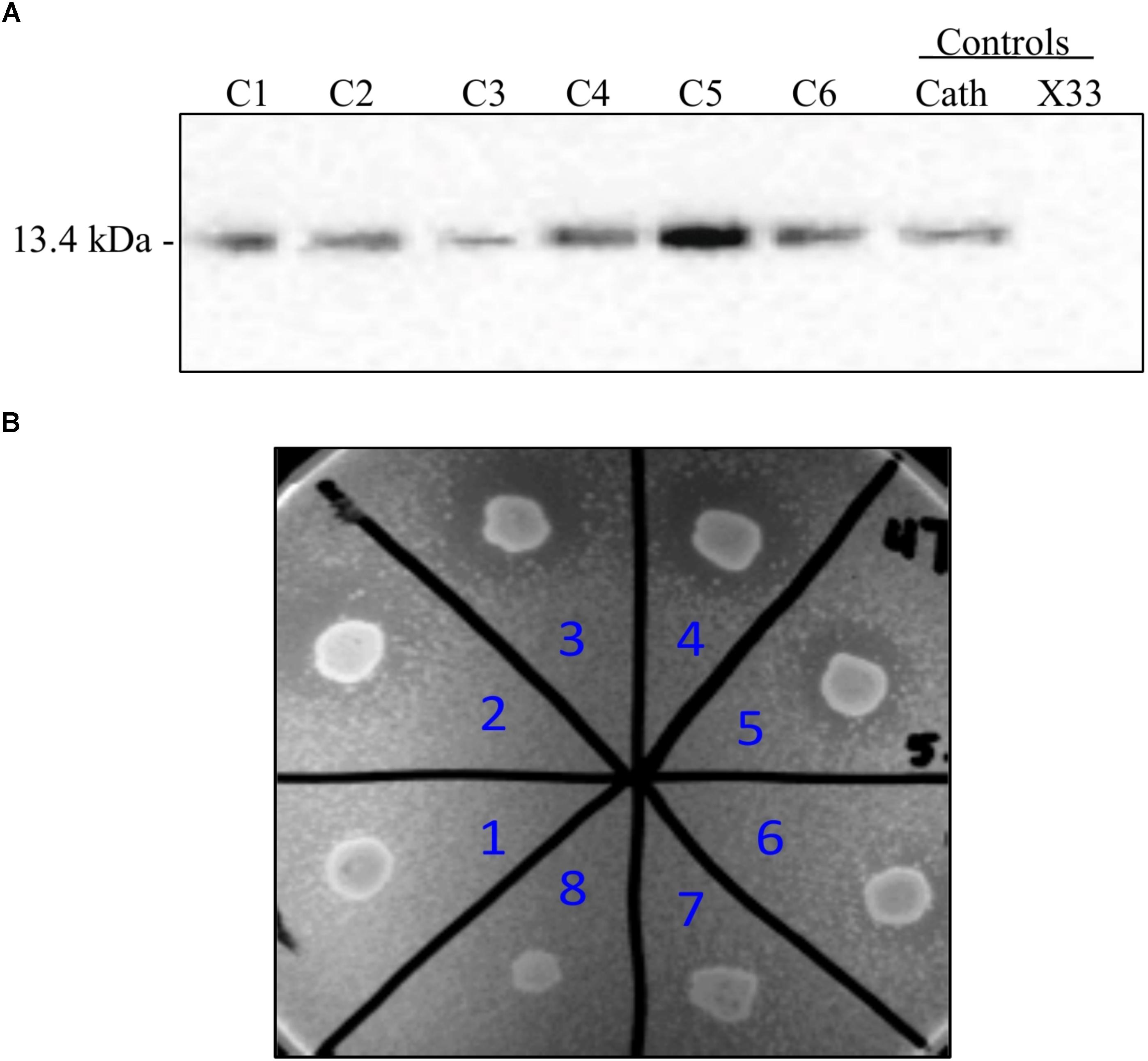
FIGURE 2. Screening of P. pastoris clones expressing recombinant cathelin (PP-rCath) and mature PG-1 (PP-rPG-1). (A) Western blot of the supernatant samples after 24 h methanol induction from small-scale fermentations of rCath clones (C1–C6). Blank P. pastoris strain X33 and chemically synthesized cathelin (13.4 kDa) served as assay negative and positive controls, respectively. (B) Radial diffusion assay of the PP-rPG-1 when grown on BMMY agarose plates and induced with 2% methanol for 48 h. TSB agarose containing methicillin-resistant S. aureus (MRSA) was subsequently layered on top of BMMY for rPG-1 antimicrobial activity detection. Clones 1–6 express rPG-1 at varying levels. Clone 7 and 8 served as controls and exhibit no antimicrobial activity (blank P. pastoris strain X33 and PP-ProPG-1, respectively).
Pichia pastoris Recombinant Protegrin Expression in a Bioreactor
To potentially obtain increased yield of recombinant protegrin, high-density fermentation was performed in a 3-L bioreactor. Initially, the transition from shake-flask culturing to a bioreactor unexpectedly yielded much lower recombinant protein concentrations using the standard media composition (Invitrogen manual, Carlsbad, CA, United States). By reducing BSM concentration by 50% during culture media optimization, expression level increased. For rPG-1 expression, rPG-1 was not detected until yeast extract and peptone were added to the BSM. These improvements suggest that the media formulation could be optimized to improve the yield.
Expression with all the three recombinant P. pastoris strains was performed using three-step procedures, including glycerol-batch phase, glycerol fed-batch and methanol induction phase. The DO spike was found to be an effective indicator to initiate a new feeding profile and to monitor the overall health of the culture for successful recombinant protegrin expression. Aliquots of the fermentation supernatant were examined by Western blot and ELISA. Highest level of rPG-1 (104 ± 11 μg/mL) was detected at 24 h of methanol induction in the fermentation culture medium. Post 24 h, rPG-1concentration decreased (Figure 3A). Similar to rPG-1, 0.8 ± 0.10 g/L rProPG-1, and 0.2 ± 0.02 rCath g/L was detected in fermentation culture medium at 24 h of methanol induction (Figures 3B–D).
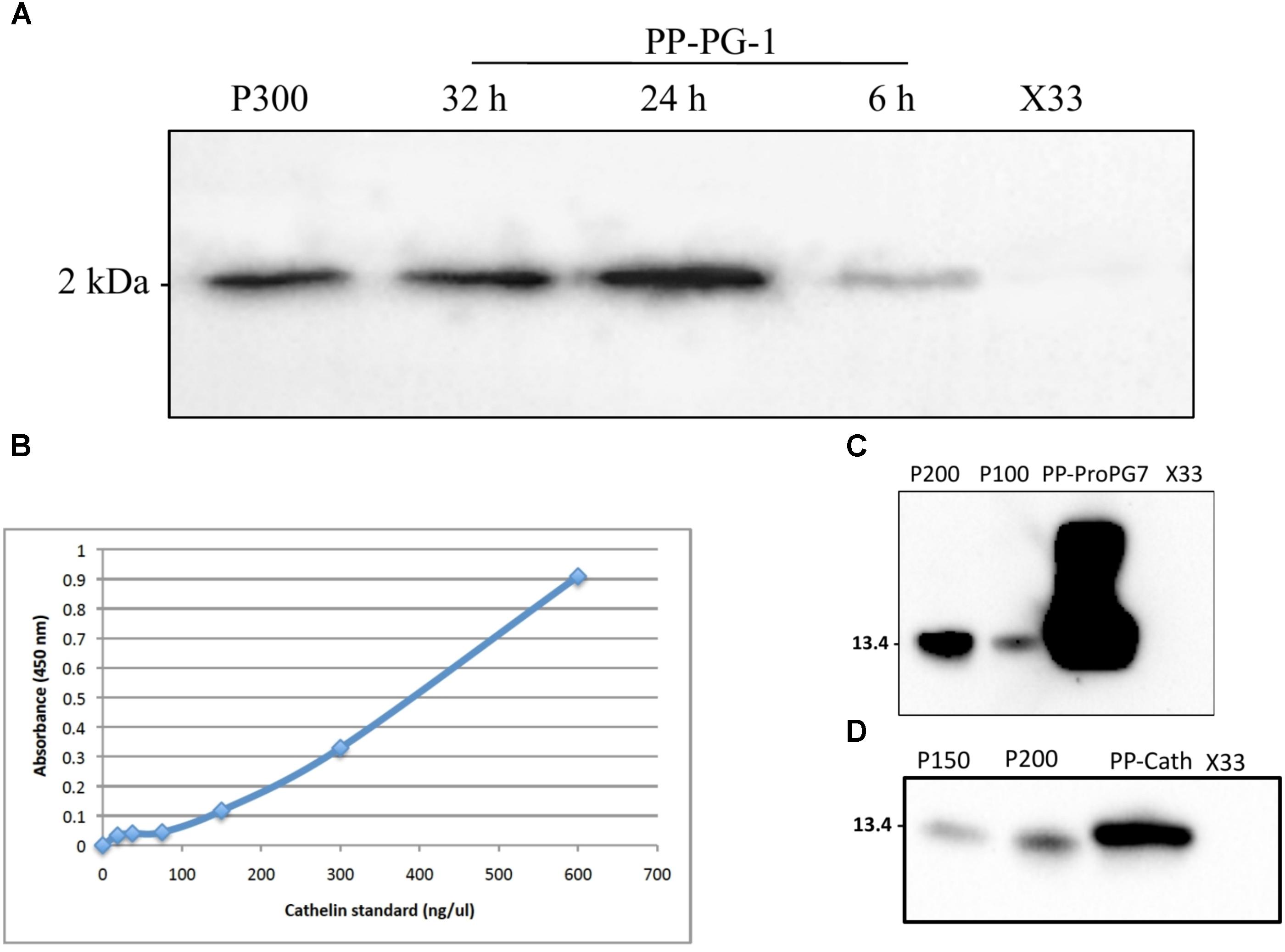
FIGURE 3. Quantification of rPro-PG-1, rCath, and rPG-1 produced by P. pastoris in separate bioreactor. (A) rPG-1 (from PP-PG-1) was detected with antibody against pig PG-1 in Western blot. Blank P. pastoris strain X33 and 300 ng of chemically synthesized rPG-1 served as assay negative and positive controls, respectively. (B) Indirect-HRP ELISA standard curve. Antibody detecting the cathelin domain of ProPG-1 and cathelin was used as the primary antibody. Cathelin peptide was used to generate the standard curve. (C) Recombinant ProPG-1 (from PP-ProPG-7) was detected with antibody against pig PG-1 in Western blot. 200 ng and 100 ng of chemically synthesized ProPG-1 served as positive controls (P200 and P100, respectively). (D) Recombinant cathelin (from PP-Cath) was detected with antibody against pig cathelin in Western blot. 200 ng and 150 ng of chemically synthesized cathelin served as positive controls (P200 and P150, respectively). Empty P. pastoris X33 strain served as a negative control.
Enterokinase Cleavage of rProPG-1 and Antimicrobial Activity
To determine whether the rProPG-1 could yield antimicrobial activity upon removal of the N-terminus pro- domain, EK was used to cleave the introduced amino acid DDDDK site on rProPG-1 (Table 4). Cleavage of rProPG-1 was detected by Western blot using antibodies specific to mature PG-1 in the C-terminal domain. EK successfully cleaved rProPG-1 as the detection of mature PG-1 decreased from 13.4 kDa to 2.1 kDa, indicating the removal of the 11 kDa pro- domain (Figure 4A). The resulting cleaved rProPG-1 also conferred antimicrobial activity against E. coli DH5αat the same level as chemically synthesized PG-1 (Figure 4B and Table 5). The uncleaved rProPG-1 and rCath did not exhibit antimicrobial activity as expected.
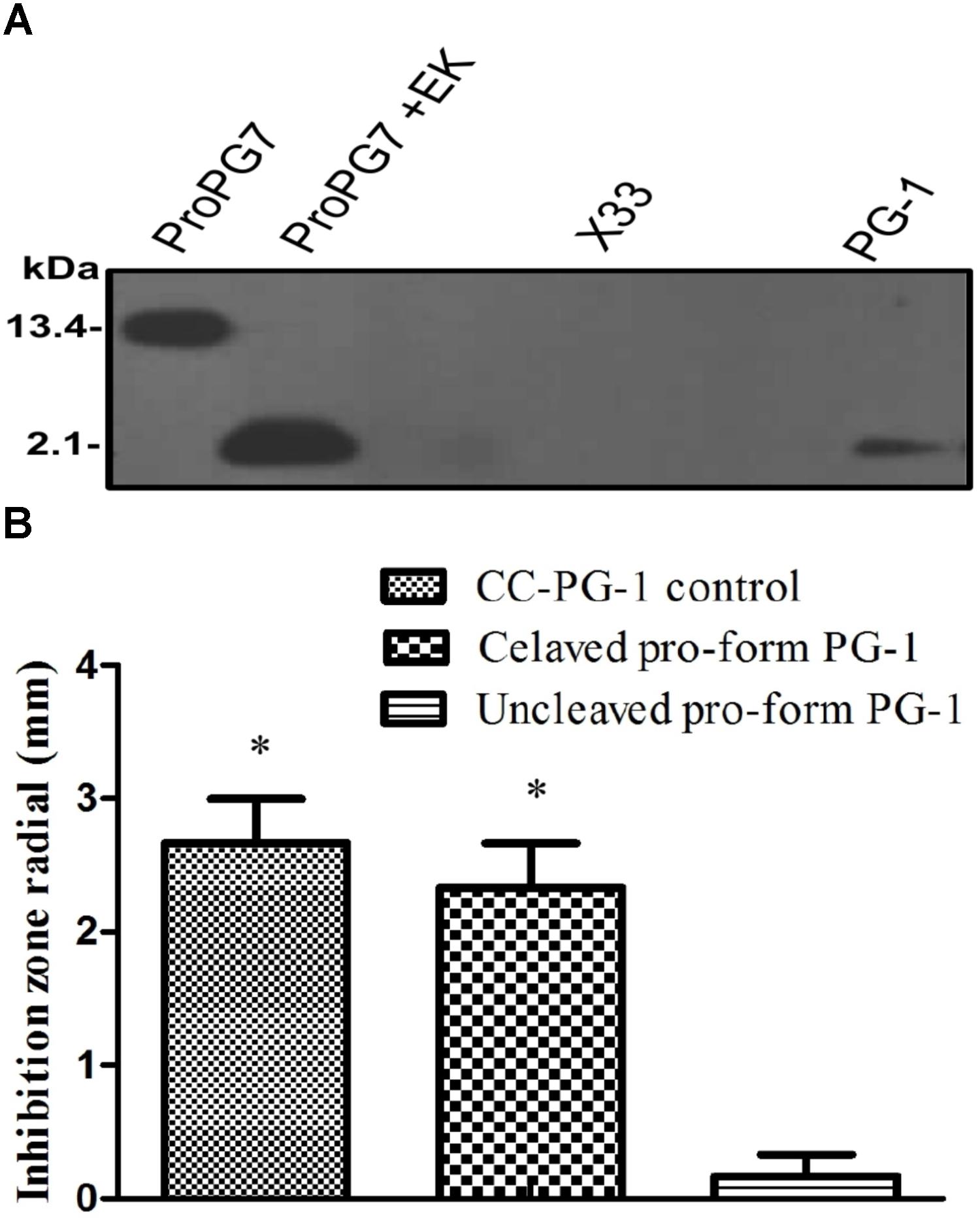
FIGURE 4. Detection and activity of rProPG after enterokinase (EK) cleavage. (A) Recombinant ProPG (ProPG7) and EK cleaved ProPG (ProPG7 + EK), yielding mature peptide (PG-1), was detected with antibody against pig PG-1 in Western blot. Blank P. pastoris strain X33 and chemically synthesized PG-1 (CC-PG-1) served as negative and positive controls, respectively. (B) Upon EK digestion, rProPG-1 exhibit antimicrobial activity against E. coli in radial diffusion antimicrobial activity assay. Data shown is the mean ± SEM of three repeats for inhibitory zones of CC-PG-1 control (I), cleaved pro-form PG-1 (II) and uncleaved pro-form PG-1 (III). Asterisk (∗p < 0.05) denotes significant difference from uncleaved pro-form PG-1.
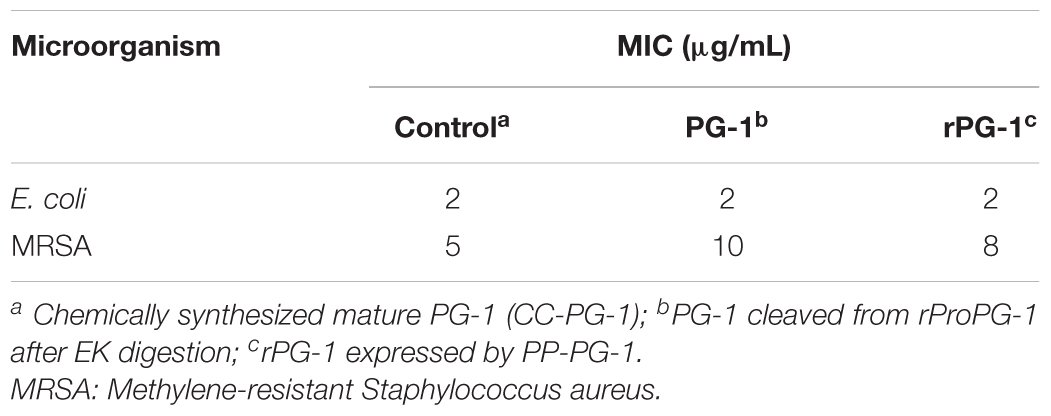
TABLE 5. Minimum inhibitory concentration (MIC) values of the resulting mature PG-1 cleaved from rProPG-1 after enterokinase (EK) digestion, and rPG-1 expresses by PP-PG-1.
Protegrin Peptide Properties and Boman Index
Peptide properties such as molecular weight, pI value, net charge and Boman Index can be used to predict a peptide’s antimicrobial potential under various conditions (Boman, 2003). rProPG, rCath, and rPG-1 were calculated using both Innovagen’s Peptide Property Calculator and a predictive tool available at AMP Database v2.34, respectively. The rPG-1 showed highly basic pI value (>10), positive net charge (+6), and high Boman index (>2.5; Table 4). The peptides containing the cathelin domain, rProPG-1, and rCath, have pI values of 6.49 and 4.6, respectively. ProPG is neutral at pH 7.0 (net charge -0.3) and Cath has a negative net charge (-6.2). Both of these peptides share similar Boman Index (2.8–2.9). A high Boman Index in rProPG-1, rCath, and rPG-1 can indicate the potential of these peptides to interact with other proteins in vivo.
Enhanced Cell Migration by PG-1 in Its Pro-, Cathelin-, and Mature- Form
We next sought to examine the effect of protegrin on modulating cell proliferation and migration in vitro using transwell cell migration assays with IPEC-J2 cells, a cell line derived from porcine intestine epithelium. As shown in Figure 5A, cell migration in groups treated with recombinant proform PG-1 (rProPG-1), pro-piece cathelin domain (rCath), and mature PG-1 (rPG-1) was enhanced. Migrated cell count analysis revealed that there was a two-fold (rPro-PG-1), 2.3-fold (rCath), and 2.5-fold (rPG-1) increase in cell migration when compared to the untreated negative control (p < 0.05, Figure 5B). To study if these peptides (rProPG-1, rCath, and rPG-1) can increase cell proliferation, cell counting proliferation assay was performed. In comparison to the untreated control, groups treated with the peptides resulted in no significant change in cell proliferation (data not shown).
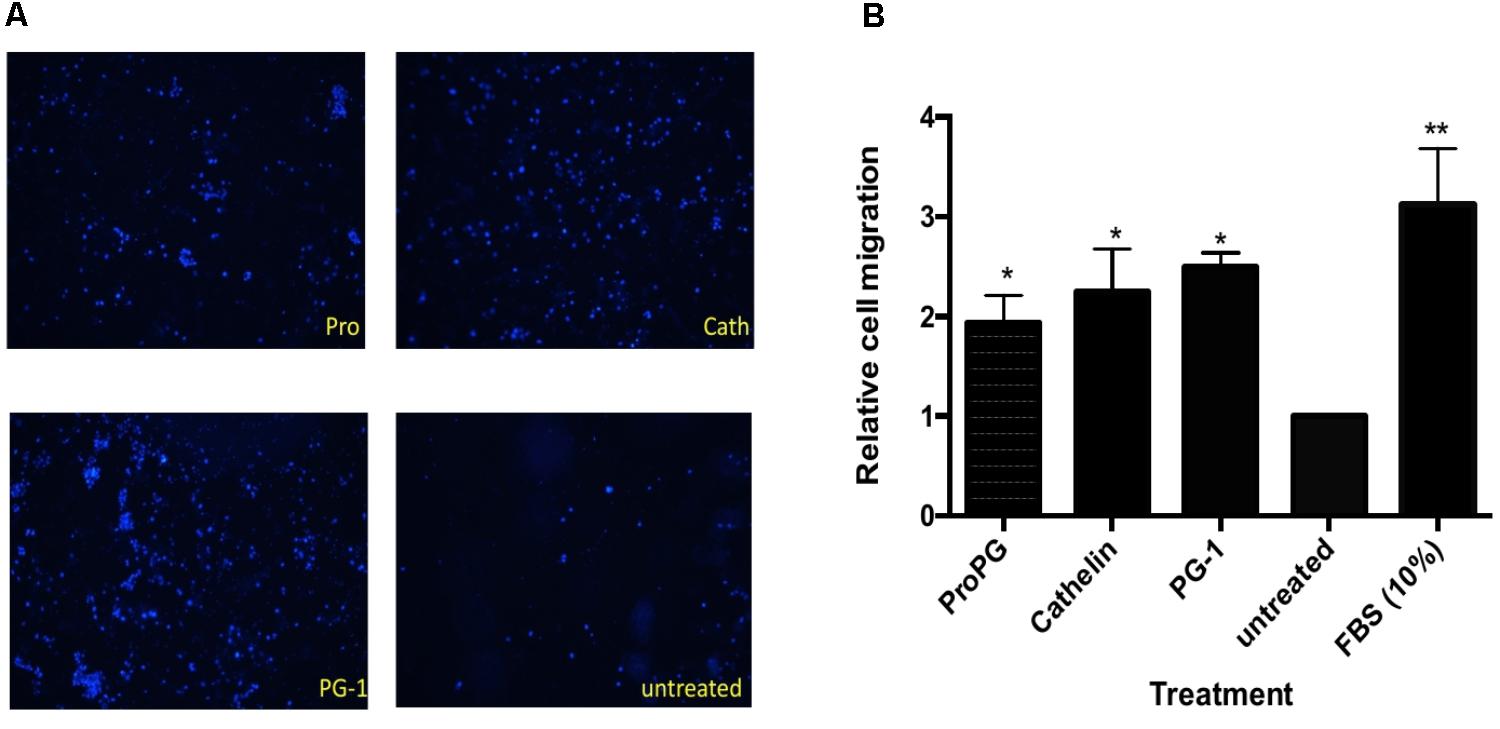
FIGURE 5. Effect of proform PG-1, cathelin domain and mature PG-1 on cell migration. (A) Representative images of transwell migrated IPEC-J2 cells stained with Hoechst 33342. (B) Quantification of cell migration from transwell migration assay. Bars represent the mean ± SEM of three experiments. Asterisk ∗ (p < 0.05) and ∗∗ (p < 0.01) denote significant difference from untreated control group.
Antimicrobial Activity of rPG-1 and Its Stability at Various pH and Temperature
The minimum inhibitory concentration (MIC) values of the resulting mature peptide (rPG-1) expressed by PP-PG-1 was determined by a two-stage radial diffusion assay. The lowest concentration of the rPG-1 that inhibited the visible growth of E. coli and MRSA after overnight incubation is the MIC (Table 6). rPG-1 conferred similar antimicrobial activity against E. coli at the same level as chemically synthesized PG-1 (CS-PG-1). The MIC value for rPG-1 expressed by PP-rPG-1 against MRSA is 37.5% lower than the CS-PG-1 at 8 μg/mL versus 5 μg/mL, respectively (Table 5). Differences in MIC may be attributed to differences in quantification methods of the chemically synthesized and recombinant peptides (e.g., empirical weighing versus western blot densitometry). Various pH and temperature conditions were used to investigate the stability of rPG-1 after an incubation time of 30 min. As shown in Table 6, rPG1 retained potent activity in both acidic and alkaline (pH 2.0, 4.0, 6.0, and 11.0), and temperature conditions (25–80°C). Antimicrobial activity was quite stable over a pH range of 2.0–11.0, although the MIC at a higher temperature (90°C) increased by 25% compared to the MIC observed at 25°C.
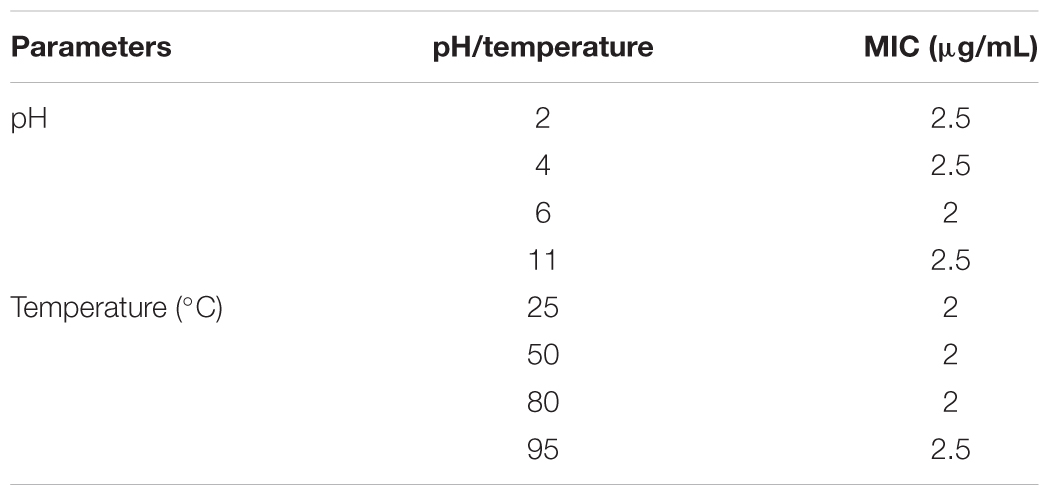
TABLE 6. Stability of the rPG-1 at different pH and temperature in terms of MIC against E. coli after a pre-incubation time of 30 min, antimicrobial activity assay using colony count method was performed to determine the antimicrobial activity.
Discussion
The use of P. pastoris as an expression system has numerous advantages such as rapid and high-density growth in defined and relatively inexpensive media, high recombinant protein yield, and post-translational modification abilities similar to that of mammalian cells. P. pastoris also secrete a low number of endogenous proteins, making downstream protein recovery and purification more feasible. AMPs that have been expressed by P. pastoris such as hPAB-β, Ch-penaeidin, MP1102 (Li et al., 2005; Chen et al., 2011; Zhang et al., 2015) are driven by the AOX promoter system. These studies indicate methylotrophic yeast-inducible system is suitable for high-level expression of active AMPs. Pro-healing and immune-modulating aspect of cathelicidin AMPs are primarily based on the human and mouse cathelicidin AMP. In our work, we investigated the feasibility of expressing recombinant PG-1 in its pro-, cathelin-, and mature- form in P. pastoris and characterized their antimicrobial activity in vitro. We then examined the effect of recombinant PG-1 in its various forms on cell migration and proliferation in vitro. To our knowledge, our finding is the first report on the role of PG-1 on cell migration and thus tissue repair potential. Interestingly, a recent report showed an increased expression of porcine β-Defensin2 (PBD2), pBD3, pBD114, pBD129, and protegrins (PG) 1-5 in IPEC-J2 cells when exposed to a host defense peptide (HDP)- synthesizing Lactobacillus reuteri I5007 (Liu et al., 2017). Similarly, butyrate and its analogs showed induction of porcine HDP gene expression with approximately 20-, 45-, 60-, and 80-fold induction was observed for pBD2, PG1-5, pEP2C, and pBD3, respectively, using IPEC-J2 intestinal epithelial cells (Zeng et al., 2013). These findings together with our findings on the PG-1 tissue damage healing potential suggest that PG-1 may be one of the components of innate defense during infection.
To investigate the feasibility of producing rProPG-1, rCath, and rPG-1 for downstream studies and application, we constructed plasmids for the expression and secretion of these three peptides in P. pastoris. Stable clones were generated via integration of the constructs into the P. pastoris genome. This will also eliminate the need for constant selective pressure during fermentation. With integration, it is possible to generate strains harboring more than one copy of the introduced expression cassette via repeated single crossover DNA recombinant events (Clare et al., 1991a; Nordén et al., 2011). Insertion of multiple recombinant gene copies is often desired to enhance recombinant protein yield, as low transcript levels can be a limiting factor in protein production (Nordén et al., 2011). To increase the number of integrated expression cassettes, we initially attempted to construct a known number of concatemers of the expression cassette prior to transformation into P. pastoris, similar to Mansur et al. (2005). We found the stable generation of long concatemers of the expression construct difficult to achieve. Instead, we then enriched for strains with an increased number of vector copies by plating transformants on medium containing increasing levels of the selection drug (e.g., G418), where higher drug resistant clones can correlate with higher gene dosage (Nordén et al., 2011). Clones with putative multiple copies of the rProPG-1, rCath, and rPG-1 expression cassette from varying concentrations of G418 antibiotic selection were then selected to evaluate recombinant protein expression levels. Although it was evident that there is a positive correlation between gene dosage level and level of expression (Tables 1–3), an upper limit of gene dosage was observed. This bell-shaped correlation curve between gene copy number and protein yield has been reported in P. pastoris, where production and secretion level gradually decline with increased gene dosage (Zhu et al., 2009). This upper limit of gene dosage has been suggested to be related to endoplasmic reticulum (ER) stress with the accumulation of recombinant protein within the ERs, also further indicating there may be other bottleneck factors limiting recombinant protein production in addition to gene dosage.
To potentially obtain increased yield of recombinant protegrin (rProPG-1, rCath, and rPG-1), high-density fermentation was also performed. Recombinant protein was detected upon modification of the bioreactor BSM by reducing salt concentrations by 50% (BSM50). It has been reported that high salt concentration can increase cell death, possibly caused by high osmotic pressure in the medium (Brady et al., 2001; Jahic et al., 2006). Upon cell lysis, the secreted recombinant protein can undergo proteolysis as cellular proteases are released into the culture medium. The addition of casamino acids, yeast extract and peptone can help address the issue of protein degradation during the cultivation process (Jahic et al., 2003). During rPG-1 expression by PP-PG-1, rPG-1 was not detected until yeast extract and peptone was added to the BSM50 culture medium, suggesting modification of media formulation can improve yield. The yeast extract, peptone and casamino acids may serve as preferential substrates for proteolytic enzymes, thereby decreasing degradation of the expressed recombinant protein of interest (Clare et al., 1991b; Jafari et al., 2011). Interestingly, the addition of these substrates to the medium was more critical in PP-PG-1 fermentation than PP-ProPG-1 and PP-Cath. This could be due to the peptide properties of rPG-1 toxicity against the P. pastoris host cells. These substrates can potentially bind or interact with rPG-1 to limit its toxicity against P. pastoris. Components of microbial growth medium have been shown to exhibit binding activity to metal ions (Ramamoorthy and Kushner, 1975) and may also affect recombinant protein folding by modulating the expression of chaperone proteins (Rosano and Ceccarelli, 2014). Furthermore, fermentation medium composition can influence the physio-chemical surface properties of microbes. Microbial cell wall protein composition can differ when cultured in medium with and without peptone and yeast extract (Schär-Zammaretti et al., 2005). The expression level of rPG-1 (104 ± 11 mg/L) is comparable to other AMPs expressed using the similar P. pastoris expression system ranging from 108 mg/L to 292 mg/L (Li et al., 2005; Chen et al., 2011; Zhang et al., 2015). Of the three protegrin peptides expressed, rPG-1 had the lowest yield compared to rProPG-1 and rCath (0.8 ± 0.10 g/L and 0.2 ± 0.02 rCath g/L, respectively). The isoelectric point (pI) of a recombinant protein can affect expression level, where high pI is associated with no detectable protein expression (Boettner et al., 2007). This is in line with the high pI value of rPG-1 (10.7; Table 4) and the lower expression level obtained. This further suggests pI can be a potential parameter influencing protein expression. Addition of fusion protein tags to aid purification can potentially affect recombinant protein expression level. To increase protein yield in future experiments, the theoretical pI of rPG-1 can be reduced from 10.6 to 8.3 upon the fusion of glutathione S-transferase (GST) tag.
To reduce the likelihood of AMPs exerting their toxicity effect against host cells, fusing AMPs to a foreign or native partner protein can aid in enhancing protein production and reduce proteolytic degradation. In our work, we replaced the native elastase cleavage site of proform protegrin with an EK cleavage site. Antimicrobial activity was not exhibited until cleavage occurred in the presence of EK, releasing the mature and active PG-1 (Figure 4B). EK is a serine protease and is naturally produced and secreted in the intestinal duodenum to convert trypsinogen into its active trypsin form for subsequent activation of pancreatic digestive enzymes in pigs (Baratti et al., 1973). The addition of the EK cleavage site can permit rProPG-1 to be cleaved by the endogenous intestinal EK to release the mature antimicrobial PG-1. This allows the potential application of rProPG-1 in animal feed to replace the use of conventional antibiotics that are contributing to the rapid emergence of antibiotic resistant bacteria. Interestingly, while the MIC for both rPG-1 and the EK cleaved rProPG-1 for E. coli is the same as that of the chemically synthesized PG-1 control, their MICs (10, and 8 μg/mL for the mature PG-1 cleaved from rProPG-1 and the mature rPG-1, respectively) for MSRA are higher than that of the control (5 μg/mL). Similar MIC results were found using protegrin-1 against E. coli (4 μg/mL), and MRSA (4 μg/mL) (Yan and Hancock, 2001). In another report, the MICs of PG-1 against Gram-positive and Gram-negative bacteria range from 0.12 to 2 μg/mL (Steinberg et al., 1997).
A high Boman index value (e.g., 2.50–3.00) can indicate an AMP to be multifunctional within the cell due to its ability to interact with a wide range of proteins (Boman, 2003; Nam et al., 2014). A lower index value (≤1) can suggest that the AMP has low side effects (e.g., low hemolytic activity). Since protegrin, in its pro-, cathelin-, and mature-form, is predicted to interact with other proteins, we sought to examine if these peptides have other functions besides antimicrobial activity. We found pro-, cathelin-, and mature-form PG-1 enhanced cell migration but not cell proliferation (Figure 5), suggesting these peptides may interact with cell-surface receptors. It was reported that cathelicidin LL-37 and mCRAMP trans-activated epidermal growth factor receptor (EGFR) (Tokumaru et al., 2005; Yang et al., 2006). Whether PG-1, and its proform and the cathelin domain have their migration function via activating membrane receptor warrant further investigation.
For therapeutic applications, the stability of the recombinant AMPs produced is important. The stability test results indicated that the rPG-1 retain antimicrobial activity in a wide range of pH conditions ranging from pH 2.0 to 11.0. The pH in wounds ranges from 5.0 to 8.5, which is suitable for rPG-1 use. Oral application of rPG-1 may also be feasible in the digestive tract (pH 3.0 and 4.4 in mouse and pig, respectively). The activity of rPG-1 remained stable over a broad temperature range from 25°C to 95°C. This stability may be due to the high cysteine content (disulfide bonds) in rPG-1 (Lai and Gallo, 2009). In conclusion, recombinant protegrin in its pro-, cathelin-, and mature- forms were successfully expressed in P. pastoris using the AOX expression system. These peptides exhibit antimicrobial activity as well as cell migration activity. Further in vivo studies on the role of PG in modulating intestinal health would be of interest for intestinal inflammation application (Harwig et al., 1996).
Author Contributions
EH performed the major experiments and wrote the manuscript. NA performed parts of experiments and helped in writing. JL helped in designing the research, data interpretation and editing.
Funding
This project has been funded by Ontario Pork (grant # 051379), Natural Sciences and Engineering Research Council of Canada (NSERC, grant # 400876), and Foshan University (grant # 053385).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02300/full#supplementary-material
References
Abad, S., Kitz, K., Hörmann, A., Schreiner, U., Hartner, F. S., and Glieder, A. (2010). Real-time PCR-based determination of gene copy numbers in Pichia pastoris. Biotechnol. J. 5, 413–420. doi: 10.1002/biot.200900233
Baratti, J., Maroux, S., Louvard, D., and Desnuelle, P. (1973). On porcine enterokinase. Further purification and some molecular properties. Biochim. Biophys. Acta 315, 147–161. doi: 10.1016/0005-2744(73)90138-1
Benmoussa, K., Authier, H., Prat, M., AlaEddine, M., Lefèvre, L., Rahabi, M. C., et al. (2017). P17, an original host defense peptide from ant venom. Front. Immunol. 8:1650. doi: 10.3389/fimmu.2017.01650
Boettner, M., Steffens, C., von Mering, C., Bork, P., Stahl, U., and Lang, C. (2007). Sequence-based factors influencing the expression of heterologous genes in the yeast Pichia pastoris—A comparative view on 79 human genes. J. Biotechnol. 130, 1–10. doi: 10.1016/j.jbiotec.2007.02.019
Boman, H. G. (2003). Antibacterial peptides: basic facts and emerging concepts. J. Intern. Med. 254, 197–215. doi: 10.1046/j.1365-2796.2003.01228.x
Brady, C. P., Shimp, R. L., Miles, A. P., Whitmore, M., and Stowers, A. W. (2001). High-level production and purification of P30P2MSP119, an important vaccine antigen for Malaria, expressed in the methylotropic yeast Pichia pastoris. Protein Expr. Purif. 23, 468–475. doi: 10.1006/prep.2001.1526
Chen, Z., Wang, D., Cong, Y., Wang, J., Zhu, J., Yang, J., et al. (2011). Recombinant antimicrobial peptide hPAB-β expressed in Pichia pastoris, a potential agent active against methicillin-resistant Staphylococcus aureus. Appl. Microbiol. Biotechnol. 89, 281–291. doi: 10.1007/s00253-010-2864-0
Choi, K.-Y., Chow, L. N. Y., and Mookherjee, N. (2012). Cationic host defence peptides: multifaceted role in immune modulation and inflammation. J. Innate Immun. 4, 361–370. doi: 10.1159/000336630
Ciofalo, V., Barton, N., Kreps, J., Coats, I., and Shanahan, D. (2006). Safety evaluation of a lipase enzyme preparation, expressed in Pichia pastoris, intended for use in the degumming of edible vegetable oil. Regul. Toxicol. Pharmacol. 45, 1–8. doi: 10.1016/j.yrtph.2006.02.001
Clare, J. J., Rayment, F. B., Ballantine, S. P., Sreekrishna, K., and Romanos, M. A. (1991a). High-level expression of tetanus toxin fragment C in Pichia pastoris strains containing multiple tandem integrations of the gene. Biotechnology 9, 455–460.
Clare, J. J., Romanes, M. A., Rayment, F. B., Rowedder, J. E., Smith, M. A., Payne, M. M., et al. (1991b). Production of mouse epidermal growth factor in yeast: high-level secretion using Pichia pastoris strains containing multiple gene copies. Gene 105, 205–212.
de la Fuente-Núñez, C., Mansour, S. C., Wang, Z., Jiang, L., Breidenstein, E. B. M., Elliott, M., et al. (2014). Anti-biofilm and immunomodulatory activities of peptides that inhibit biofilms formed by pathogens isolated from cystic fibrosis patients. Antibiotics 3, 509–526. doi: 10.3390/antibiotics3040509
Dias, S. A., Freire, J. M., Pérez-Peinado, C., Domingues, M. M., Gaspar, D., Vale, N., et al. (2017). New potent membrane-targeting antibacterial peptides from viral capsid proteins. Front. Microbiol. 8:775. doi: 10.3389/fmicb.2017.00775
Fonseca, P., and Light, A. (1983). The purification and characterization of bovine enterokinase from membrane fragments in the duodenal mucosal fluid. J. Biol. Chem. 258, 14516–14520.
Frohm, M., Agerberth, B., Ahangari, G., Stâhle-Bäckdahl, M., Lidén, S., Wigzell, H., et al. (1997). The expression of the gene coding for the antibacterial peptide LL-37 is induced in human keratinocytes during inflammatory disorders. J. Biol. Chem. 272, 15258–15263. doi: 10.1074/jbc.272.24.15258
Hancock, R. E. W., and Sahl, H.-G. (2006). Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24, 1551–1557. doi: 10.1038/nbt1267
Harwig, S. S., Waring, A., Yang, H. J., Cho, Y., Tan, L., and Lehrer, R. I. (1996). Intramolecular disulfide bonds enhance the antimicrobial and lytic activities of protegrins at physiological sodium chloride concentrations. Eur. J. Biochem. 240, 352–357. doi: 10.1111/j.1432-1033.1996.0352h.x
Hase, K., Murakami, M., Iimura, M., Cole, S. P., Horibe, Y., Ohtake, T., et al. (2003). Expression of LL-37 by human gastric epithelial cells as a potential host defense mechanism against Helicobacter pylori. Gastroenterology 125, 1613–1625. doi: 10.1053/j.gastro.2003.08.028
Hirano, T., Ishihara, K., and Hibi, M. (2000). Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene 19, 2548–2556. doi: 10.1038/sj.onc.1203551
Inaba, Y., Ashida, T., Ito, T., Ishikawa, C., Tanabe, H., Maemoto, A., et al. (2010). Expression of the antimicrobial peptide alpha-defensin/cryptdins in intestinal crypts decreases at the initial phase of intestinal inflammation in a model of inflammatory bowel disease, IL-10-deficient mice. Inflamm. Bowel Dis. 16, 1488–1495. doi: 10.1002/ibd.21253
Ishihara, K., and Hirano, T. (2002). IL-6 in autoimmune disease and chronic inflammatory proliferative disease. Cytokine Growth Factor Rev. 13, 357–368. doi: 10.1016/S1359-6101(02)00027-8
Jafari, R., Sundström, B. E., and Holm, P. (2011). Optimization of production of the anti-keratin 8 single-chain Fv TS1-218 in Pichia pastoris using design of experiments. Microb. Cell Fact. 10:34. doi: 10.1186/1475-2859-10-34
Jahic, M., Gustavsson, M., Jansen, A.-K., Martinelle, M., and Enfors, S.-O. (2003). Analysis and control of proteolysis of a fusion protein in Pichia pastoris fed-batch processes. J. Biotechnol. 102, 45–53. doi: 10.1016/S0168-1656(03)00003-8
Jahic, M., Knoblechner, J., Charoenrat, T., Enfors, S.-O., and Veide, A. (2006). Interfacing Pichia pastoris cultivation with expanded bed adsorption. Biotechnol. Bioeng. 93, 1040–1049. doi: 10.1002/bit.20811
Kovach, M. A., Ballinger, M. N., Newstead, M. W., Zeng, X., Bhan, U., Yu, F.-S., et al. (2012). Cathelicidin-related antimicrobial peptide is required for effective lung mucosal immunity in Gram-negative bacterial pneumonia. J. Immunol. 189, 304–311. doi: 10.4049/jimmunol.1103196
Lai, Y., and Gallo, R. L. (2009). AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 30, 131–141. doi: 10.1016/j.it.2008.12.003
Li, L., Wang, J.-X., Zhao, X.-F., Kang, C.-J., Liu, N., Xiang, J.-H., et al. (2005). High level expression, purification, and characterization of the shrimp antimicrobial peptide. Ch-penaeidin, in Pichia pastoris. Protein Expr. Purif. 39, 144–151. doi: 10.1016/j.pep.2004.09.006
Liu, H., Hou, C., Wang, G., Jia, H., Yu, H., Zeng, X., et al. (2017). Lactobacillus reuteri I5007 modulates intestinal host defense peptide expression in the model of IPEC-J2 cells and neonatal piglets. Nutrients 9:E559. doi: 10.3390/nu9060559
Liu, T.-C., and Stappenbeck, T. S. (2016). Genetics and pathogenesis of inflammatory bowel disease. Annu. Rev. Pathol. 11, 127–148. doi: 10.1146/annurev-pathol-012615-044152
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, B., Shen, J., Yindeeyoungyeon, W., and Ruan, Y. (2017). A novel antibiotic mechanism of l-Cyclopropylalanine blocking the biosynthetic pathway of essential amino acid l-leucine. Molecules 22:E2224. doi: 10.3390/molecules22122224
Mansur, M., Cabello, C., Hernández, L., País, J., Varas, L., Valdés, J., et al. (2005). Multiple gene copy number enhances insulin precursor secretion in the yeast Pichia pastoris. Biotechnol. Lett. 27, 339–345. doi: 10.1007/s10529-005-1007-7
Nam, B.-H., Moon, J.-Y., Park, E.-H., Kim, Y.-O., Kim, D.-G., Kong, H. J., et al. (2014). Antimicrobial activity of peptides derived from olive flounder lipopolysaccharide binding protein/bactericidal permeability-increasing protein (LBP/BPI). Mar. Drugs 12, 5240–5257. doi: 10.3390/md12105240
Nordén, K., Agemark, M., Danielson, J. ÅH., Alexandersson, E., Kjellbom, P., and Johanson, U. (2011). Increasing gene dosage greatly enhances recombinant expression of aquaporins in Pichia pastoris. BMC Biotechnol. 11:47. doi: 10.1186/1472-6750-11-47
Pizzo, E., Zanfardino, A., Di Giuseppe, A. M. A., Bosso, A., Landi, N., Ragucci, S., et al. (2015). A new active antimicrobial peptide from PD-L4, a type 1 ribosome inactivating protein ofPhytolacca dioica L.: a new function of RIPs for plant defence?. FEBS Lett. 589, 2812–2818. doi: 10.1016/j.febslet.2015.08.018
Proudfoot, A. E. I. (2002). Chemokine receptors: multifaceted therapeutic targets. Nat. Rev. Immunol. 2, 106–115. doi: 10.1038/nri722
Ramamoorthy, S., and Kushner, D. J. (1975). Binding of mercuric and other heavy metal ions by microbial growth media. Microb. Ecol. 2, 162–176. doi: 10.1007/BF02010436
Rosano, G. L., and Ceccarelli, E. A. (2014). Recombinant protein expression in Escherichia coli: advances and challenges. Front. Microbiol. 5:172. doi: 10.3389/fmicb.2014.00172
Sang, Y., and Blecha, F. (2008). Antimicrobial peptides and bacteriocins: alternatives to traditional antibiotics. Anim. Health Res. Rev. 9, 227–235. doi: 10.1017/S1466252308001497
Schär-Zammaretti, P., Dillmann, M.-L., D’Amico, N., Affolter, M., and Ubbink, J. (2005). Influence of fermentation medium composition on physicochemical surface properties of Lactobacillus acidophilus. Appl. Environ. Microbiol. 71, 8165–8173. doi: 10.1128/AEM.71.12.8165-8173.2005
Schauber, J., Rieger, D., Weiler, F., Wehkamp, J., Eck, M., Fellermann, K., et al. (2006). Heterogeneous expression of human cathelicidin hCAP18/LL-37 in inflammatory bowel diseases. Eur. J. Gastroenterol. Hepatol. 18, 615–621. doi: 10.1097/00042737-200606000-00007
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Steinberg, D. A., Hurst, M. A., Fujii, C. A., Kung, A. H., Ho, J. F., Cheng, F. C., et al. (1997). Protegrin-1: a broad-spectrum, rapidly microbicidal peptide with in vivo activity. Antimicrob. Agents Chemother. 41, 1738–1742.
Steinberg, D. A., and Lehrer, R. I. (1997). “Designer assays for antimicrobial peptides,” in Antibacterial Peptide Protocols, ed. W. M. Shafer (Totowa, NJ: Humana Press), 169–186.
Stratton, J., Chiruvolu, V., and Meagher, M. (1998). “High cell-density fermentation,” in Pichia Protocols, ed. J. M. Cregg (Totowa, NJ: Humana Press), 107–120.
Taghavian, O., Spiegel, H., Hauck, R., Hafez, H. M., Fischer, R., and Schillberg, S. (2013). Protective oral vaccination against infectious bursal disease virus using the major viral antigenic protein VP2 produced in Pichia pastoris. PLoS One 8:e83210. doi: 10.1371/journal.pone.0083210
Termén, S., Tollin, M., Rodriguez, E., Sveinsdóttir, S. H., Jóhannesson, B., Cederlund, A., et al. (2008). PU.1 and bacterial metabolites regulate the human gene CAMP encoding antimicrobial peptide LL-37 in colon epithelial cells. Mol. Immunol. 45, 3947–3955. doi: 10.1016/j.molimm.2008.06.020
Thompson, C. A. (2010). FDA approves kallikrein inhibitor to treat hereditary angioedema. Am. J. Health Syst. Pharm. 67:93. doi: 10.2146/news100005
Tokumaru, S., Sayama, K., Shirakata, Y., Komatsuzawa, H., Ouhara, K., Hanakawa, Y., et al. (2005). Induction of keratinocyte migration via transactivation of the epidermal growth factor receptor by the antimicrobial peptide LL-37. J. Immunol. 175, 4662–4668. doi: 10.4049/jimmunol.175.7.4662
Yan, H., and Hancock, R. E. (2001). Synergistic interactions between mammalian antimicrobial defense peptides. Antimicrob. Agents Chemother. 45, 1558–1560. doi: 10.1128/AAC.45.5.1558-1560.2001
Yang, Y. H., Wu, W. K. K., Tai, E. K. K., Wong, H. P. S., Lam, E. K. Y., So, W. H. L., et al. (2006). The cationic host defense peptide rCRAMP promotes gastric ulcer healing in rats. J. Pharmacol. Exp. Ther. 318, 547–554. doi: 10.1124/jpet.106.102467
Zaiou, M., Nizet, V., and Gallo, R. L. (2003). Antimicrobial and protease inhibitory functions of the human cathelicidin (hCAP18/LL-37) prosequence. J. Invest. Dermatol. 120, 810–816. doi: 10.1046/j.1523-1747.2003.12132.x
Zeng, X., Sunkara, L. T., Jiang, W., Bible, M., Carter, S., Ma, X., et al. (2013). Induction of porcine host defense peptide gene expression by short-chain fatty acids and their analogs. PLoS One 8:e72922. doi: 10.1371/journal.pone.0072922
Zhang, Y., Teng, D., Wang, X., Mao, R., Cao, X., Hu, X., et al. (2015). In vitro and in vivo characterization of a new recombinant antimicrobial peptide, MP1102, against methicillin-resistant Staphylococcus aureus. Appl. Microbiol. Biotechnol. 99, 6255–6266. doi: 10.1007/s00253-015-6394-7
Zhu, S., Wei, L., Yamasaki, K., and Gallo, R. L. (2008). Activation of cathepsin L by the cathelin-like domain of protegrin-3. Mol. Immunol. 45, 2531–2536. doi: 10.1016/j.molimm.2008.01.007
Keywords: antimicrobial peptide, cell migration, fermentation, Pichia pastoris, protegrin
Citation: Huynh E, Akhtar N and Li J (2018) Efficient Production of Recombinant Protegrin-1 From Pichia pastoris, and Its Antimicrobial and in vitro Cell Migration Activity. Front. Microbiol. 9:2300. doi: 10.3389/fmicb.2018.02300
Received: 12 November 2017; Accepted: 10 September 2018;
Published: 27 September 2018.
Edited by:
Kamlesh Jangid, National Centre for Cell Science (NCCS), IndiaReviewed by:
César de la Fuente, Massachusetts Institute of Technology, United StatesPedro Ismael Da Silva Junior, Instituto Butantan, Brazil
Copyright © 2018 Huynh, Akhtar and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julang Li, amxpQHVvZ3VlbHBoLmNh
 Evanna Huynh
Evanna Huynh Nadeem Akhtar
Nadeem Akhtar Julang Li
Julang Li