
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 24 September 2018
Sec. Microbial Physiology and Metabolism
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.02267
This article is part of the Research Topic Microbial Stress: From Sensing to Intracellular and Population Responses View all 25 articles
Small regulatory RNAs (sRNAs) are currently considered as major post-transcriptional regulators of gene expression in bacteria. The interplay between sRNAs and transcription factors leads to complex regulatory networks in which both transcription factors and sRNAs may appear as nodes. In cyanobacteria, the responses to nitrogen availability are controlled at the transcriptional level by NtcA, a CRP/FNR family regulator. In this study, we describe an NtcA-regulated sRNA in the cyanobacterium Nostoc sp. PCC 7120, that we have named NsrR1 (nitrogen stress repressed RNA1). We show sequence specific binding of NtcA to the promoter of NsrR1. Prediction of possible mRNA targets regulated by NsrR1 allowed the identification of nblA, encoding a protein adaptor for phycobilisome degradation under several stress conditions, including nitrogen deficiency. We demonstrate specific interaction between NsrR1 and the 5′-UTR of the nblA mRNA, that leads to decreased expression of nblA. Because both NsrR1 and NblA are under transcriptional control of NtcA, this regulatory circuit constitutes a coherent feed-forward loop, involving a transcription factor and an sRNA.
Small non-coding RNAs (sRNAs) are important players in bacterial regulatory networks. Most often, this type of molecules exert its function at the post-transcriptional level by fine-tuning responses to different environmental situations (Wagner and Romby, 2015). The interplay between transcription factors and small RNAs provides an additional level of control that, in many cases, results in feed-forward regulatory loops (coherent or incoherent) between a transcription factor, an sRNA and the regulated target (Mandin and Guillier, 2013; Nitzan et al., 2017).
Global nitrogen regulation in cyanobacteria is controlled by NtcA, a protein that belongs to the CRP/FNR family of transcriptional regulators (Herrero et al., 2001). Direct transcriptional regulation mediated by NtcA is operated by binding to conserved sequences (first defined as GTAN8TAC, later re-defined as GTN10AC) in the promoters of the regulated genes (Luque et al., 1994; Mitschke et al., 2011; Picossi et al., 2014; Giner-Lamia et al., 2017). NtcA has been described to act either as an activator or as a repressor of transcription, depending on the location of the binding site with respect to the regulated transcriptional start site (TSS) (Mitschke et al., 2011; Picossi et al., 2014). Most NtcA-activated promoters contain an NtcA binding site centered around position -41.5 with respect to the TSS (like Class II CRP-activated promoters). In contrast, NtcA-repressed genes bear NtcA binding sites that overlap critical elements of the promoter, including the -35 box, the -10 box or the TSS. Therefore, the mechanism of repression by NtcA involves interference with RNA polymerase binding. Similar to the case of other CRP-family regulators, NtcA has been shown to regulate expression of several sRNAs (Mitschke et al., 2011; Brenes-Álvarez et al., 2016) including NsiR4 (nitrogen-stress inducible RNA 4), which is involved in nitrogen assimilation control via regulation of the key enzyme glutamine synthetase (Klähn et al., 2015).
Non-diazotrophic cyanobacteria respond to nutrient deprivation by a process called chlorosis, involving degradation of photosynthetic pigments, including the phycobilisomes (PBS) (Grossman et al., 1993a,b). Degradation of PBS protects the photosynthetic apparatus under exposure to high light and also provides amino acids as a source of nitrogen under nitrogen deficiency. NblA (non-bleaching phenotype) is a protein adaptor involved in PBS degradation by targeting phycobiliproteins to a proteolytic complex (Karradt et al., 2008; Sendersky et al., 2014). The nblA gene is induced under stress situations that promote PBS degradation (Collier and Grossman, 1994). nblA has multiple promoters and is subject to complex regulation that includes transcription induction by NtcA under nitrogen deprivation (Luque et al., 2001; Mitschke et al., 2011) and regulation by FurA under iron deficiency (González et al., 2016). Although heterocystous cyanobacteria are ultimately able to overcome nitrogen deprivation by fixation of atmospheric nitrogen, their transient response to nitrogen deprivation also includes induction of the nblA gene and partial degradation of PBS (Baier et al., 2004; Karradt et al., 2008).
We have previously carried out a differential RNA-Seq-based genome-wide identification of TSSs in the heterocystous cyanobacteria Nostoc sp. PCC 7120 (also known as Anabaena sp. PCC 7120), which included a description of transcriptional responses corresponding to the NtcA regulon (Mitschke et al., 2011). Based on the RNA-Seq data, we have also conducted a global approach to the identification of conserved, potentially regulatory small non-coding RNAs (sRNAs) in Nostoc sp. PCC 7120. This work has lead to the prediction and verification of several phylogenetically conserved sRNAs, including nitrogen-regulated and heterocyst-specific sRNAs (Brenes-Álvarez et al., 2016).
In this work, we show transcription of a small, nitrogen-regulated RNA, which we have named NsrR1 (nitrogen stress-repressed RNA 1), in Nostoc sp. PCC 7120. The promoter of NsrR1 contains a putative NtcA binding site, whose position, overlapping the -10 box, is suggestive of NtcA-mediated repression. We demonstrate sequence specific binding of purified NtcA to the promoter of nsrR1. Phylogenetic analysis reveals that sequences encoding orthologs of NsrR1 and the nearby NtcA binding site overlapping the -10 box are conserved among heterocystous cyanobacteria. Computational target prediction for NsrR1 identified nblA mRNA as possibly interacting with NsrR1 and therefore being subjected to post-transcriptional regulation by this sRNA. Here, we verify that NsrR1 regulates expression of NblA by its interaction with the 5′-UTR of nblA mRNA, therefore participating in an NtcA-operated coherent feed-forward regulatory loop.
Cultures of wild-type and the different mutant strains of Nostoc sp. PCC 7120 (Supplementary Table S1) were bubbled with an air/CO2 mixture (1% v/v) and grown photoautotrophically at 30°C in BG11 medium (Rippka et al., 1979) containing ferric citrate instead of ammonium ferric citrate, lacking NaNO3 and containing 6 mM NH4Cl, 10 mM NaHCO3, and 12 mM N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid-NaOH buffer (pH 7.5). Nitrogen deficiency was induced by removal of combined nitrogen. Occasionally, 17.6 mM NaNO3 was used as nitrogen source. Solid media were solidified with 1% Difco Agar. Mutant strains were grown in the presence of appropriate antibiotics at the following concentrations: streptomycin (Sm) and spectinomycin (Sp), 2–3 μg/ml each (liquid medium) or 5 μg/ml each (solid medium), neomycin (Nm), 5 μg/ml (liquid medium) or 25 μg/ml (solid medium).
Escherichia coli strains (Supplementary Table S1) were grown in LB medium, supplemented with appropriate antibiotics (Sambrook and Russell, 2001).
To generate a strain lacking NsrR1 (ΔnsrR1), two overlapping fragments encompassing flanking sequences around NsrR1 were amplified by PCR using as template genomic DNA with oligonucleotides 170 and 171 and oligonucleotides 172 and 173, respectively (see Supplementary Table S2 for oligonucleotide sequences and description). The resulting products were then used as templates for a third PCR with oligonucleotides 170 and 173 resulting in the fusion of both fragments and the deletion of the sequences encoding NsrR1. The fragment was cloned into pSpark (Canvax Biotech), rendering pSAM319 and its sequence was verified by sequencing (Eurofins Genomics) (see Supplementary Table S3 for plasmids description). After digestion with BamHI at the sites provided by oligonucleotides 170 and 173, the fragment was cloned into BamHI-digested sacB-containing SmSpR vector pCSRO (Merino-Puerto et al., 2010), rendering pSAM325, which was transferred to Nostoc sp. strain PCC 7120 by conjugation (Elhai and Wolk, 1988) with selection for resistance to Sm and Sp. Cultures of the exconjugants obtained were used to select for clones resistant to 5% sucrose (Cai and Wolk, 1990), and individual sucrose resistant colonies were checked by PCR. Clones lacking the deleted region were named ΔnsrR1.
To establish controlled expression of NsrR1 in Nostoc, the nsrR1 gene was placed under control of the petE promoter, which mediates Cu2+-regulated transcription (Buikema and Haselkorn, 2001) and was cloned in a self-replicating plasmid. The petE promoter of Nostoc sp. PCC 7120 (genomic coordinates 278185 to 277848) was amplified with oligonucleotides 186 and 299 and fused to the nsrR1 fragment amplified with oligonucleotides 184 and 185 by a third PCR using the two fragments as templates and oligonucleotides 184 and 299. The product was cloned into pSpark, rendering pSAM329 and its sequence was verified. After digestion with ClaI and XhoI at the sites provided by oligonucleotides 299 and 184, the fragment was cloned into pSAM221, rendering pIAE17. pIAE17 was introduced into the ΔnsrR1 strain by conjugation as described above.
Wild type or ΔnsrR1 cells were grown under standard conditions in the presence of NH4+ or subjected to nitrogen deficiency for the number of hours indicated in each case. Spectra of the different cultures where taken between 500 and 750 nm on a JASCO V-650 spectrophotometer and normalized for chlorophyll content by the absorbance at 680 nm after subtraction of absorbance at 750 nm. Absorbance at 635 nm and 680 nm were used to estimate phycocyanin and chlorophyll content, respectively.
The predicted sequences of NsrR1 and its putative homologs were obtained from Brenes-Álvarez et al. (2016) and multiple sequence alignment was generated with T-Coffee (Notredame et al., 2000) at the EMBL-EBI web server (Li et al., 2015).
Target prediction of NsrR1 was performed using CopraRNA (Wright et al., 2013, 2014) with the NsrR1 homologs of Nostoc sp. PCC 7120, Anabaena cylindrica PCC 7122, Anabaena variabilis ATCC 29413 (recently renamed Trichormus variabilis NIES-23), Cylindrospermum stagnale PCC 7417, Nodularia spumigena CCY9414, ‘Nostoc azollae’ 0708, Nostoc punctiforme PCC 73102, Nostoc sp. PCC 7524, Rivularia sp. PCC 7116, Nostoc sp. PCC 7107, and Calothrix sp. PCC 7507.
The interaction between NsrR1 and the nblA mRNA of diverse cyanobacteria was analyzed with IntaRNA (Mann et al., 2017).
RNA samples were isolated from cells collected at different times after removing combined nitrogen from the media. Total RNA was isolated using hot phenol as described (Mohamed and Jansson, 1989) with modifications (Brenes-Álvarez et al., 2016). Northern blot hybridization was performed as previously described (Muro-Pastor et al., 1999; Steglich et al., 2008). Strand specific 32P-labeled probes for Northern blot were prepared with Taq DNA polymerase using a PCR fragment (oligonucleotides 158 and 159) as template in a reaction with α-32P-dCTP and one single oligonucleotide as primer (corresponding to the complementary strand of the sRNA or mRNA to be detected). Hybridization signals were quantified on a Cyclone Storage Phosphor System with Optiquant software.
To produce His-tagged NtcA protein, the ntcA gene was amplified using Nostoc DNA as template and primers 343 and 344, and the PCR product was cloned in vector pET-28a (+) (Novagen) using NcoI and XhoI, producing plasmid pSAM334 (see Supplementary Table S3 for plasmids description). Plasmid pSAM334, containing the ntcA gene fused to a sequence encoding a His6-tag under a T7 polymerase-dependent promoter, was transferred by electroporation to E. coli BL21-(DE3)-RIL, in which the gene encoding T7 RNA polymerase is under the control of an isopropyl-β-D-1-thiogalactopyranoside (IPTG)-regulated promoter. A 25-ml pre-inoculum of this strain was grown overnight in LB medium supplemented with chloramphenicol and kanamycin and used to inoculate 275 ml of the same medium. The culture was incubated at 37°C until OD600 = 0.6. Recombinant NtcA expression was induced by addition of 1 mM IPTG. After 4 h at 37°C, cells were collected and resuspended in 20 mM sodium phosphate buffer (pH 7.2) containing 500 mM NaCl, 20 mM imidazole and 1 mM phenylmethylsulfonyl fluoride (6 ml/g of cells). Cells were broken by sonication and after centrifugation at 15,000 × g (15 min, 4°C), the His6-NtcA protein was purified from the supernatant by chromatography through a 1-ml HisTrap HP column (GE Healthcare), using imidazole to elute the retained proteins. Samples obtained after purification were subjected to SDS-PAGE to assess purity and 10% glycerol was added to fractions before storage at -20°C.
A similar procedure was followed for expression and purification of NblA using oligonucleotides 358 and 359 for cloning of the nblA gene in pET-28a (+), rendering pIAE32. In this case, an additional purification step was carried out by size-exclusion chromatography in a HiLoad 16/60 Superdex 75 column (Pharmacia) using 20 mM sodium phosphate buffer (pH 7.2) containing 150 mM NaCl. A total amount of 3.5 mg of purified protein was used in seven subcutaneous injections of a rabbit to produce antibodies in the ‘Animal Production and Experimentation Center’, Universidad de Sevilla (Seville, Spain). Antiserum was recovered at several times up to 5 months after the first injection and stored at -80°C until used. Antibodies specific for NblA were purified from the serum by affinity chromatography on immobilized His-tagged NblA using the AminoLink Plus® Immobilization Kit (Thermo Fisher).
Electrophoresis mobility shift assays (EMSA) were carried out as described (Muro-Pastor et al., 1999) with a 295-bp PCR fragment amplified with oligonucleotides 153 and 183 and encompassing positions -239 to +50 with respect to the TSS of nsrR1. A mutated version of the same DNA fragment was obtained by overlap-extension PCR using mutagenic complementary oligonucleotides 418 and 419 and flanking oligonucleotides 153 and 183.
For Western blot analysis, E. coli cells were resuspended in SDS-PAGE loading buffer and the proteins fractionated on 15% SDS-PAGE. Antibodies against NblA (see above), GFP (Roche) and E. coli GroEL (Sigma-Aldrich) were used. The ECL Plus immunoblotting system (GE Healthcare) was used to detect the different antibodies using anti-rabbit (Sigma-Aldrich) or anti-mouse (Bio-Rad) horse-radish peroxidase conjugated secondary antibodies.
For the experimental target verification in E. coli, we used the reporter system described (Urban and Vogel, 2007) and the superfolder GFP (sfGFP) plasmid pXG10-SF (Corcoran et al., 2012). The 5′-UTR of nblA, from the TSS at position -106 with respect to the initiation codon (coordinates 5407397) to 60 nucleotides within the coding region, containing the predicted NsrR1 interaction sequence, was amplified from genomic DNA using primers 253 and 254. The information about the TSS was taken from Mitschke et al. (2011). The corresponding PCR product was cloned into the vector pXG-10-SF using NsiI and NheI, resulting in a translational fusion of a truncated NblA protein with sfGFP (plasmid pIAE11, Supplementary Table S4). For NsrR1 expression in E. coli, the nsrR1 gene was amplified from genomic DNA using primers 197 (5′-phosphorylated) and 198. The PCR product was digested with XbaI and fused to a plasmid backbone that was amplified from pZE12-luc with primers PLlacOB and PLlacOD and digested with XbaI, rendering pAVN1 (Supplementary Table S5).
For the mutagenesis of NsrR1 and the 5′-UTR of nblA, plasmids like those harboring the native versions were constructed using, in addition to the above-mentioned oligonucleotides, overlapping PCR with primers containing the desired changes (Supplementary Tables S2, S4, S5). Positions for mutations were selected on the basis of lowered hybridization energies predicted by IntaRNA (Mann et al., 2017). For testing various combinations of both plasmids, these were introduced into E. coli DH5α. Plasmid pJV300 was used as a control expressing an unrelated RNA. Fluorescence measurements were done by flow cytometry or with a microplate reader (Varioskan) using liquid cultures from eight individual colonies of each combination of plasmids, as described previously (Wright et al., 2013).
RNA transcripts were generated in vitro with MEGAscript High Yield Transcription Kit (AM1333, Ambion). The DNA templates for the transcription of NsrR1 (wild type or Mut-63 variant) and nblA 5′-UTR RNA (wild type and variant Comp-63) were generated by PCR with a forward primer that includes a T7 promoter sequence and three extra Gs upstream the 5′-end of the coded RNA, and a reverse primer matching the 3′ end of the RNA (Supplementary Tables S2, S6). The nblA 5′-UTR fragment extends from the TSS at position -106 to 60 nucleotides downstream the translational start. After transcription, RNAs were treated with DNase I and purified by phenol and chloroform extraction, ethanol-precipitated at -20°C, and washed with 70% ethanol.
For end labeling, 100 pmol of wild type or mutated variants of NsrR1 RNA was dephosphorylated with one unit of rAPid Alkaline Phosphatase (Roche) at 37°C for 1 h in a 20 μl reaction, followed by purification and precipitation as above. 20 pmol of dephosphorylated RNA was 5′-labeled in a 15 μl reaction with 2 μl of [γ-32P] ATP (10 mCi/ml, 3,000 Ci/mmol) and 15 units of polynucleotide kinase (Thermo) for 1 h at 37°C. Unincorporated nucleotides were removed with G-25 spin columns and the labeled RNAs were purified on a denaturing 8% polyacrylamide gel. The labeled RNA was visualized with a Cyclone Storage Phosphor System and the RNA bands excised and eluted overnight at 4°C in 300 μl of 20 mM Tris-HCl (pH 7.5), 0.25 M sodium acetate, 0.25% SDS, 1 mM EDTA. The eluate was ethanol-precipitated at -20°C, washed with 70% ethanol and resuspended in water.
0.1 pmol of labeled NsrR1 RNA was mixed in 7 μl with different amounts of unlabelled nblA 5′-UTR RNA, denatured for 1 min at 95°C and chilled on ice for 5 min, followed by addition of 1 μl of 1 mg/ml yeast RNA (Ambion AM7118) and 1 μl of 10x structure buffer (Ambion). The samples were incubated further for 15 min at 37°C.
For RNase treatment, 1 μl of 0.01 U/ml RNAse T1 (Ambion AM2283) or 1 μl of 0.01 U/ml RNAse A was added and the samples incubated for 15 min at room temperature. Reactions were stopped by addition of 20 μl of inactivation/precipitation buffer (Ambion) and incubation at -20°C for 15 min. The precipitate was washed with 70% ethanol and resuspended in 3–7 μl of denaturing formamide loading buffer.
Lead acetate treatment was performed by the addition of 1 μl of 25 mM lead(II) acetate (Sigma) and incubation for 1 min at 37°C. Reactions were stopped by addition of 1 μl 0.1 M EDTA and 22 μl of inactivation/precipitation buffer (Ambion) and incubation at -20°C for 15 min. The precipitate was washed with 70% ethanol and resuspended in 3–7 μl of denaturing formamide loading buffer.
Alkaline ladder was obtained by incubating 0.2 pmol of 5′-labeled RNA at 95°C for 3 min in 7.5 μl of alkaline hydrolysis buffer (Ambion) containing 1.5 μg of yeast RNA (Ambion AM7118). Reactions were stopped by the addition of 15 μl of denaturing formamide loading buffer.
RNase T1 G ladders were obtained by incubating 0.1 pmol of 5′-labeled RNA and 1 μl of 1 mg/ml yeast RNA (Ambion AM7118) in 9 μl sequencing buffer (Ambion) for 10 min at 50°C, followed by the addition of 1 μl of 0.1 U/ml RNAse T1 (Ambion AM2283) and incubation at room temperature for 15 min. Reactions were stopped by the addition of 20 μl of inactivation/precipitation buffer (Ambion) and incubation at -20°C for 15 min. The precipitate was washed with 70% ethanol and resuspended in 3–7 μl of denaturing formamide loading buffer.
All samples were run on 10% polyacrylamide, 7 M urea gels and bands visualized with a Cyclone Storage Phosphor System.
A nitrogen stress-responsive TSS was identified by differential RNA-Seq at position 2050703f of the Nostoc sp. PCC 7120 chromosome, in the 710-bp intergenic region between two annotated open reading frames (alr1709 and all1710). Transcription from this TSS is significantly repressed in the absence of combined nitrogen (log2 fold-change = -2.1) (Mitschke et al., 2011). Another RNA-Seq experiment available for Nostoc sp. PCC 7120 also showed nitrogen-regulated transcription in the intergenic region between alr1709 and all1710 (Flaherty et al., 2011).
Furthermore, a recent approach to the global identification of phylogenetically conserved small non-coding RNAs in Nostoc sp. PCC 7120 (Brenes-Álvarez et al., 2016) identified a putative conserved small RNA transcribed from the TSS at position 2050703f to a predicted Rho-independent terminator that ends at position 2050814f (Ionescu et al., 2010). We named the predicted sRNA NsrR1 (nitrogen stress-repressed RNA 1), not to be confused with the previously described NsiR1 (Ionescu et al., 2010). A schematic representation of the nsrR1 genomic region in Nostoc sp. PCC 7120 is shown in Figure 1A. Sequences putatively encoding homologs of NsrR1 were found in the genomes of heterocystous strains and two unicellular strains that are phylogenetically close to the heterocystous strains, but not in more distantly related cyanobacteria (Brenes-Álvarez et al., 2016). The alignment of the corresponding sequences allows the identification of putative Rho-independent transcriptional terminators in all cases, suggesting transcription of a phylogenetically conserved small RNA (Figure 1B).
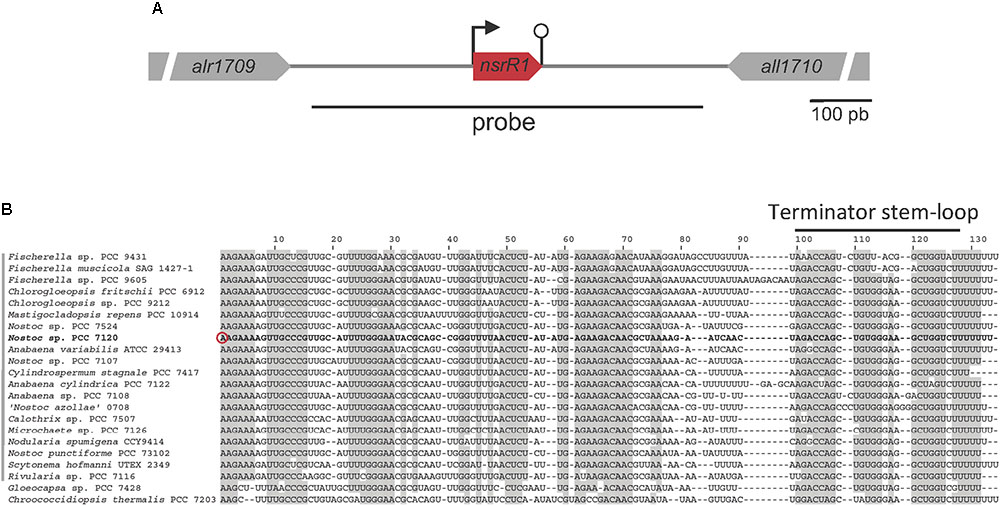
FIGURE 1. The nitrogen-stress repressed RNA 1 (NsrR1) in cyanobacteria. (A) Schematic representation of the region encoding NsrR1 in Nostoc sp. PCC 7120. The two flanking annotated open reading frames are indicated together with the region used as a probe in Northern blots (black line under the scheme). The bent arrow represents the transcriptional start at position 2050703f, and the stem-loop represents the transcriptional terminator. (B) Alignment of NsrR1 homologs identified in Brenes-Álvarez et al. (2016). The sequence from Nostoc sp. PCC 7120 is shown in bold. The verified TSS in Nostoc sp. PCC 7120 (Mitschke et al., 2011) is marked with a red circle and the terminator stem-loops with a black bar. Heterocystous strains are indicated with a gray bar on the left. Shadowed nucleotides indicate more than 90% conservation of the sequence.
To determine whether a small non-coding RNA was in fact transcribed in this region, Northern blot hybridization was performed using a strand specific DNA probe that covered the intergenic region between alr1709 and all1710 shown in Figure 1A.
Figure 2A shows expression of a small RNA of about 110 nt, consistent with RNA-Seq data and the prediction for transcriptional terminators. Upon removal of combined nitrogen, transcription was strongly repressed (90% after 9 h without nitrogen, Supplementary Figure S1). In the ntcA mutant strain repression was significantly lower at all time points (Figure 2A and Supplementary Figure S1), suggesting transcriptional repression operated by NtcA. The observation that repression, although to a lower extent, took place in the ntcA mutant strain suggests that NtcA is not the only component repressing nsrR1. Transcription of NsrR1 seemed de-repressed in the wild-type after 24 h of nitrogen stress, a situation in which heterocysts have already differentiated and atmospheric nitrogen is being fixed so that nitrogen stress is alleviated. We have also analyzed the time-course of NsrR1 transcription upon ammonium addition to cells that were stably growing with nitrate or dinitrogen as nitrogen sources. In both cases addition of ammonium led to de-repression of NsrR1 expression (Supplementary Figure S2). De-repression was faster upon ammonium addition to cells growing in the presence of nitrate (Supplementary Figure S2A) than upon ammonium addition to cells growing without a source of combined nitrogen (Supplementary Figure S2B).
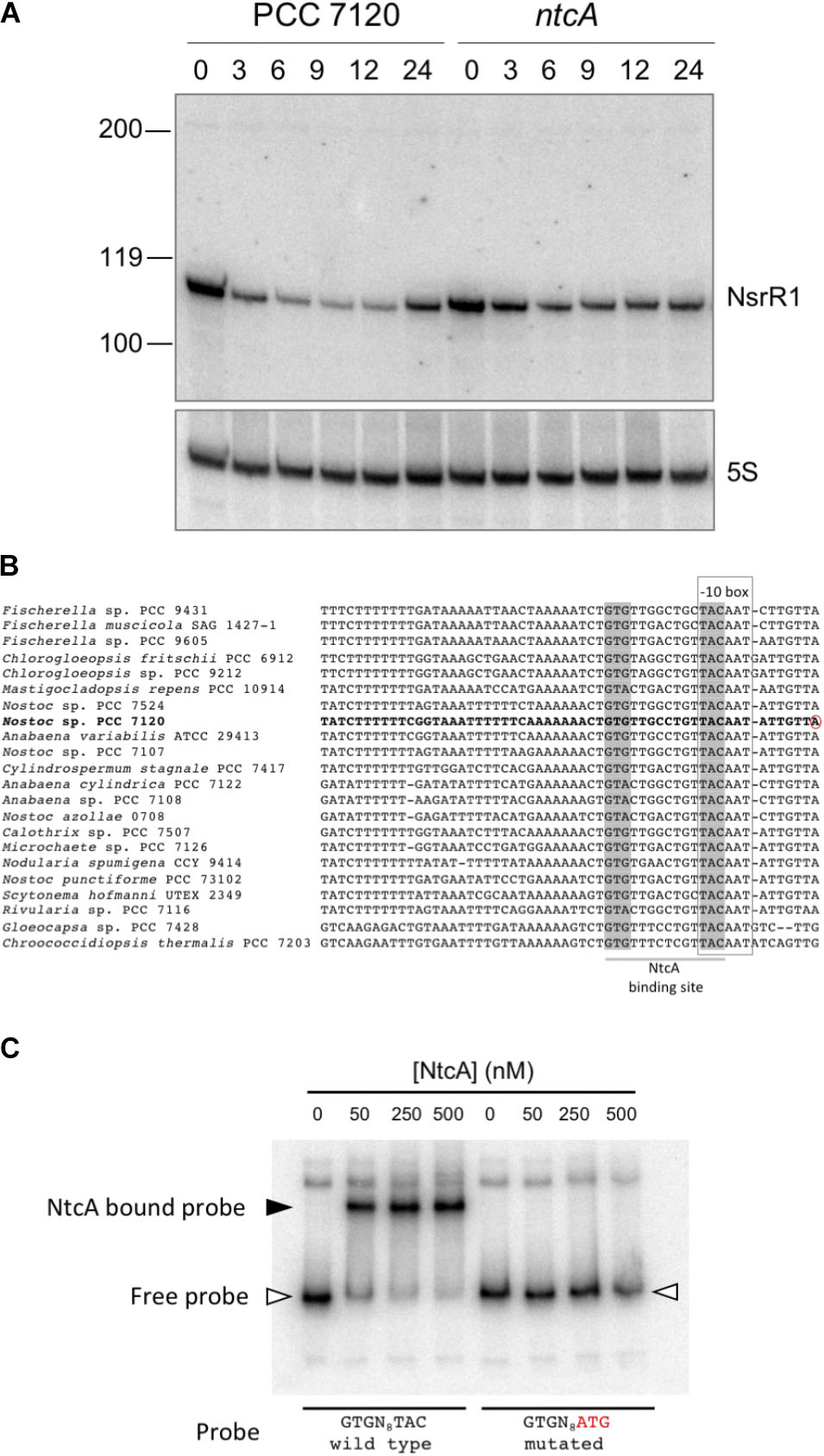
FIGURE 2. Transcription of NsrR1 is regulated by NtcA. (A) Nitrogen-responsive expression of NsrR1 in Nostoc sp. PCC 7120 and its ntcA mutant derivative CSE2. Expression was analyzed in cells grown in the presence of ammonium and transferred to medium containing no source of combined nitrogen (N2) for the number of hours indicated. The upper panel shows hybridization to the NsrR1 probe. The lower panel shows hybridization to a probe for 5S RNA used as loading and transfer control. Size markers are indicated on the left. (B) Comparison of promoter sequences upstream of nsrR1 homologs encoded in cyanobacterial genomes. The verified TSS in Nostoc sp. PCC 7120 (Mitschke et al., 2011) is marked with a red circle. Putative NtcA binding sites (highlighted in gray) and –10 boxes (framed) are indicated. (C) Electrophoretic mobility shift assays showing binding of purified His-tagged NtcA protein to a DNA fragment containing the wild type promoter of NsrR1 from Nostoc sp. PCC 7120 or a mutated version altered in the positions indicated in red. The position of retarded DNA fragments is indicated with a black triangle, whereas the free fragments are indicated with empty triangles.
Cyanobacterial nitrogen-responsive promoters are in many cases directly regulated by binding of the global nitrogen regulator NtcA. We have compared the promoter of NsrR1 from Nostoc sp. PCC 7120 with the sequences upstream from the homologs shown in Figure 1B. In all cases a putative NtcA binding motif (GTRN8TAC) partially overlaps the -10 box (Figure 2B), suggesting NtcA might directly repress transcription of NsrR1 by binding to these sequences. EMSA assays were performed with purified histidine-tagged NtcA and a DNA fragment extending from -239 to +50 with respect to the TSS of NsrR1 that included the putative NtcA binding sequence. A retarded band was observed that was not detected when the NtcA binding sequence was mutated (TAC changed to ATG) (Figure 2C). These results demonstrate sequence-specific binding of NtcA to sequences overlapping the -10 box of the promoter region of NsrR1. These observations are compatible with direct repression of transcription from the NsrR1 promoter by NtcA, although some other mechanism must be responsible for the additional repression that is still observed in the ntcA mutant upon nitrogen depletion (Figure 2A). Supplementary Figure S3 shows a comparison of the position of the NtcA binding site in the promoter of nsrR1 and in three previously described NtcA-repressed promoters.
To identify possible mRNAs regulated by NsrR1, we have used the CopraRNA algorithm, that considers folding, energy of hybridization and phylogenetic conservation of the sRNA (Wright et al., 2013, 2014) (Table 1). Among the 20 best targets predicted by CopraRNA, nblA is the gene with a known function that has the highest score (p-value = 8.45 × 10-7). NsrR1 is predicted to interact with the 5′-UTR of nblA mRNA (Figure 3A). A potential interaction between NsrR1 and the 5′-UTR of nblA mRNA is conserved in many cyanobacteria that contain NsrR1 (Supplementary Figure S4). Although the exact position in the 5′-UTR predicted to interact with NsrR1 is not conserved, in most cases it is very close or overlapping the putative Shine-Dalgarno sequence.
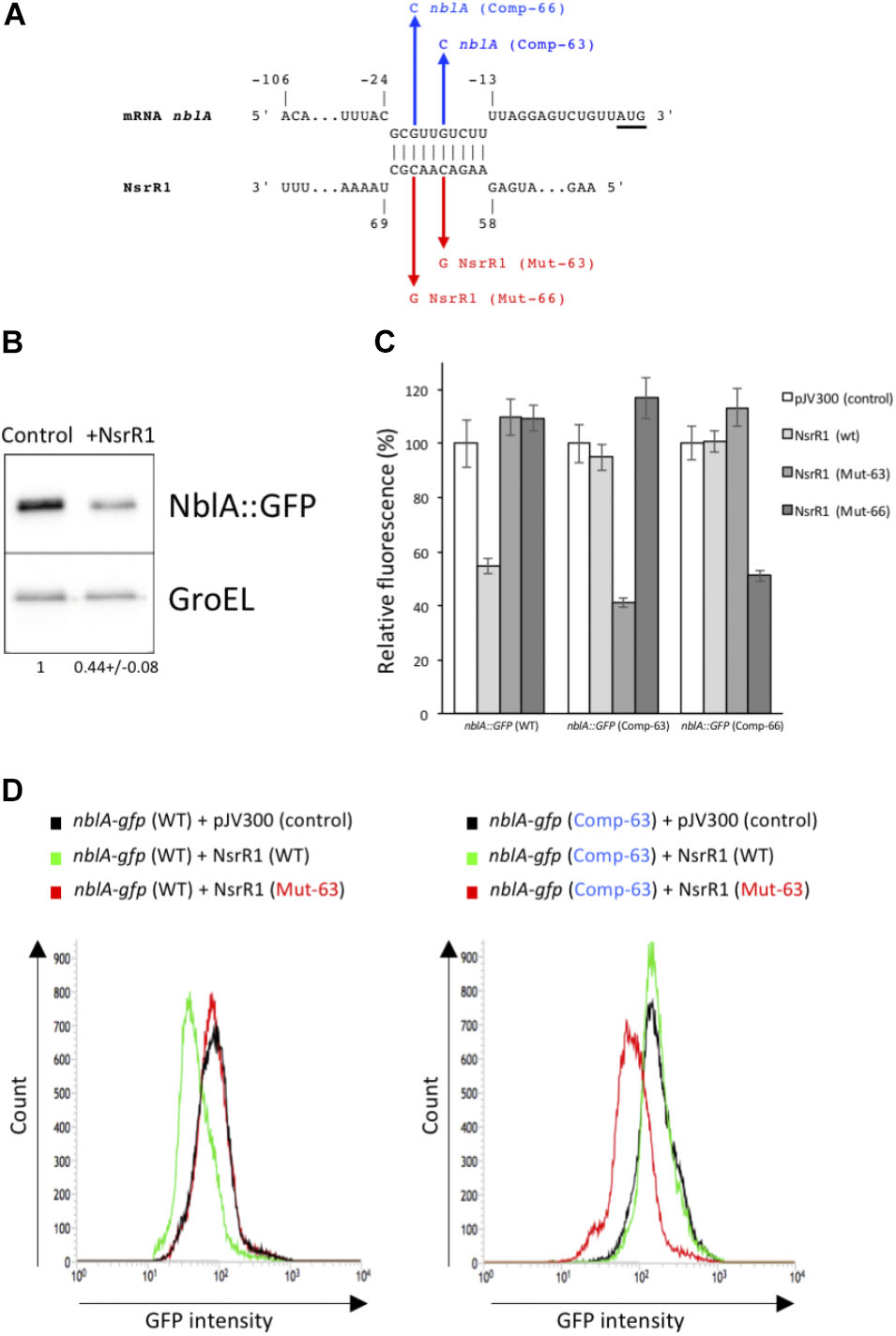
FIGURE 3. Verification of NsrR1 interaction with the 5′-UTR of nblA using an in vivo reporter system. (A) Predicted interaction between NsrR1 and the 5′-UTR of nblA according to CopraRNA (Wright et al., 2013, 2014). nblA nucleotides are numbered with respect to the start of the coding sequence (AUG codon is underlined). Mutations introduced in NsrR1 at position 63 (C to G, Mut-63) or 66 (C to G, Mut-66) and the corresponding compensatory mutations in nblA 5′-UTR position –18 (G to C, Comp-63) or –21 (G to C, Comp-66) are indicated in red and blue, respectively. (B) Accumulation of GFP protein in E. coli DH5α cultures bearing an nblA::sfgfp fusion combined with plasmid pJV300 (encoding a control RNA) or a plasmid encoding NsrR1. Western blots were carried out using antibodies against GFP or GroEL. Numbers at the bottom of the image indicate relative GFP levels with respect to control after normalization with GroEL (average of two experiments). (C) Fluorescence measurements of E. coli DH5α cultures with various combinations of plasmids expressing different versions of NsrR1 or nblA::sfgfp fusions. Plasmid pJV300 (encoding a control RNA) was used as control. The data are presented as the mean ± standard deviation of cultures from eight independent colonies after subtraction of fluorescence in cells bearing pXG-0. (D) FACS-based reporter assays. E. coli DH5α cells carrying combinations of plasmids bearing wild-type and mutated version Mut-63 of NsrR1 and wild type or mutated version Comp-63 of nblA::sfgfp were grown to stationary phase and subjected to flow cytometry analysis. Data were acquired on 50,000 cells per sample.
To verify the interaction between NsrR1 and the mRNA of nblA, we used a heterologous reporter assay (Corcoran et al., 2012), in which the shortest 5′-UTR of nblA, corresponding to transcription from position -106 (Mitschke et al., 2011) plus 60 nucleotides of the coding sequence of nblA were fused to the gene for sfGFP and co-expressed with NsrR1 or with a control RNA in E. coli. According to CopraRNA, the predicted interacting region (Figure 3A) was located very close to the translation initiation region of nblA and therefore was predicted to affect translation of the mRNA. Accumulation of GFP protein was measured in cells bearing different combinations of plasmids. Cells carrying the nblA::sfgfp translational fusion showed significant GFP fluorescence, as well as GFP protein determined by Western blot (Figure 3B), demonstrating that the translation initiation region of nblA was functional in E. coli. The GFP fluorescence of cells bearing the nblA::sfgfp fusion (and the amount of GFP protein) decreased to about 50% of the control when NsrR1 was co-expressed, indicating a direct interaction between NsrR1 and the 5′-UTR of nblA, which affects translation (Figures 3B,C). To verify the interaction at the predicted site, two different point mutations were introduced in NsrR1 in the middle of the predicted helix between NsrR1 and the 5′-UTR of nblA (nucleotide 63, C to G; nucleotide 66, C to G). Compensatory mutations were introduced in the corresponding positions of the 5′-UTR of nblA (nucleotide 89, G to C; nucleotide 86, G to C). Mutation of either one of those nucleotides in NsrR1 drastically reduced the interaction between NsrR1 and the 5′-UTR of nblA, as suggested by the absence of fluorescence reduction with respect to control (Figures 3C,D). When the mutated version of the sRNA was combined with the mutated version of the mRNA containing the compensatory change, base pairing was restored and again a significant fluorescence reduction was observed. These data support a direct interaction of NsrR1 with the 5′-UTR of the nblA mRNA and its effect on translation of NblA.
In addition, we have studied the interaction between NsrR1 and the nblA mRNA by in vitro footprinting analysis. 32P-labeled NsrR1 was incubated with unlabeled nblA mRNA (a fragment extending from positions -106 to +60 with respect to the start of the coding sequence) and probed with RNase T1 (Figure 4A) or lead(II) acetate (Figure 4B). When wild type NsrR1 RNA and nblA mRNA were used, a footprint was detected between positions 58 and 67 of NsrR1, in agreement with the bioinformatic prediction (Figure 3A). This footprint was not observed with the mutant version of nblA mRNA (Comp-63). However, when the mutant version of NsrR1 RNA (Mut-63) was used, only the mutant nblA mRNA (Comp-63) generated a footprint, but not the wild type nblA mRNA. These in vitro results are therefore in agreement with the in vivo results obtained in E. coli with the GFP fusion analysis.
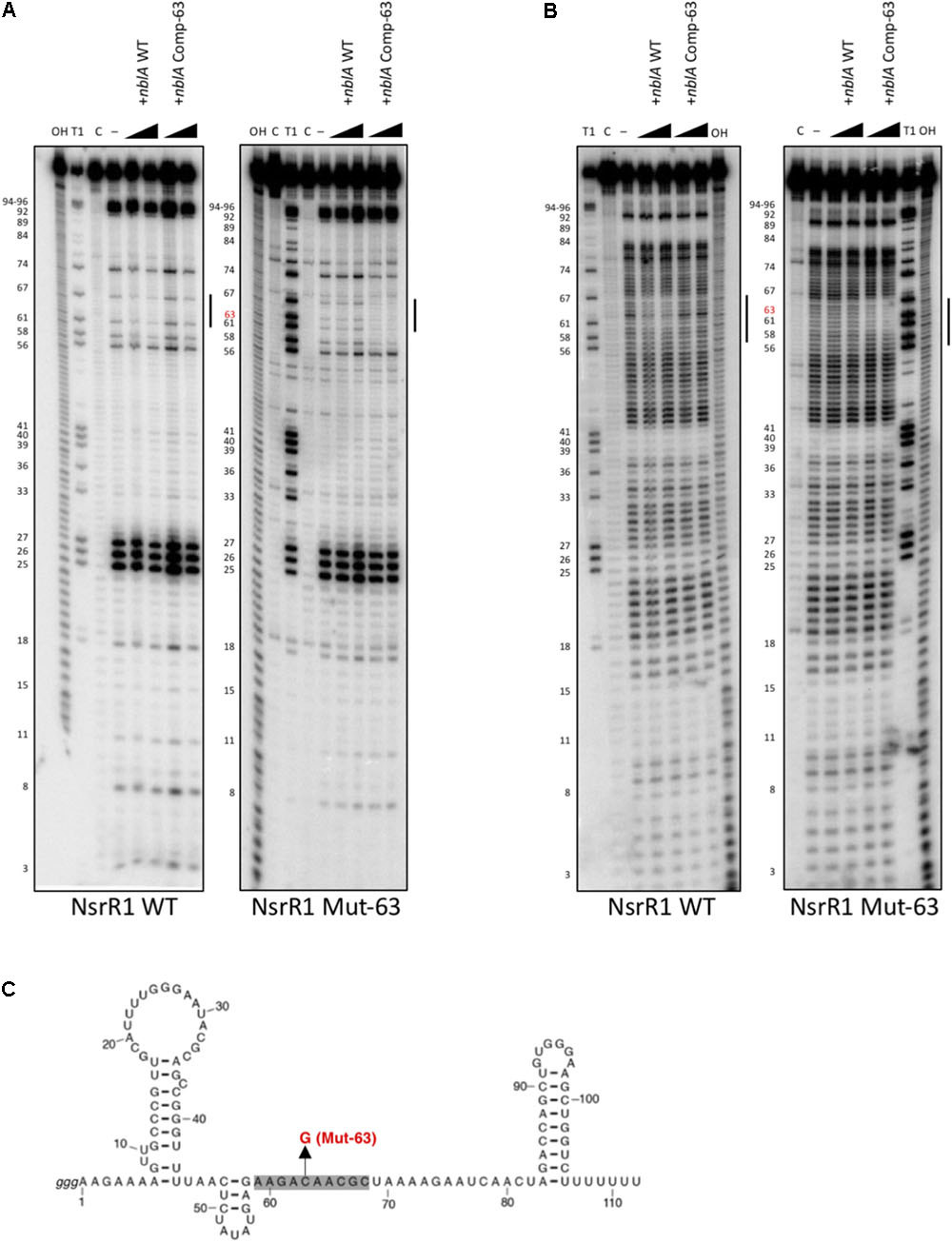
FIGURE 4. In vitro footprinting assay of the NsrR1/nblA 5′-UTR interaction. (A) RNase T1 footprinting of the interaction between wild type NsrR1 (left) or mutant C63G (Mut-63, right) with wild type and compensatory mutant (Comp-63) of the nblA 5′-UTR. (B) Pb(II) footprinting of the interaction between wild type NsrR1 (left) or mutant C63G (right) with wild type and compensatory mutant (Comp-63) of the nblA 5′-UTR. Two concentrations of nblA RNA were used (100 and 200 nM). The protected area is indicated by vertical bars in (A,B). NsrR1 was 5′ end-labeled. C, untreated control; OH, alkaline ladder; T1, RNase T1 ladder. Nucleotide positions of NsrR1 are shown on the left. (C) Secondary structure model of NsrR1 from Nostoc sp. PCC 7120 based on structure probing (Supplementary Figure S5). The area protected by nblA RNA is shaded. The nucleotide changed in Mut-63 is also indicated.
We have also performed in vitro structure probing analysis of NsrR1 (Supplementary Figure S5). The NsrR1 region involved in interaction with nblA mRNA is predicted to be single stranded, according to the pattern of sensitivity to RNase T1, RNase A, and lead(II) acetate (Figure 4 and Supplementary Figure S5).
Transcription of NsrR1 is only partially (and transiently) repressed upon nitrogen step down. Therefore, to study in vivo in Nostoc the possible effect of NsrR1 on nblA expression, we have generated a mutant strain lacking NsrR1 (ΔnsrR1) by deleting the DNA region containing nsrR1 through homologous recombination. Cells lacking NsrR1 have no apparent phenotype under normal growth conditions. Furthermore, there is no growth difference with respect to wild-type cells in medium containing nitrate, ammonia, or lacking a nitrogen source (Supplementary Figure S6). A complemented strain (ΔnsrR1 + PpetE::nsrR1) in which NsrR1 expression is under control of the Cu2+-inducible petE promoter (Buikema and Haselkorn, 2001) was generated, releasing NsrR1 expression from nitrogen control and allowing Cu2+-controlled expression instead.
nblA is transcribed from five TSS, three of them induced in response to nitrogen deprivation (Mitschke et al., 2011) (see a scheme in Figure 5A). Northern blot hybridization confirmed the NtcA dependent induction of transcription from three of the nblA promoters (Figure 5B). We have compared nblA expression between the ΔnsrR1 and the ΔnsrR1 + PpetE::nsrR1 strains during several hours after nitrogen step down (so that transcription of nblA is induced) and Cu2+ addition (to induce transcription of NsrR1 from the petE promoter in strain ΔnsrR1 + PpetE::nsrR1) (Figure 5C). Under these conditions transcription of all three N-regulated nblA transcripts was induced in the ΔnsrR1 strain in response to nitrogen deficiency. However, accumulation of the three transcripts was strongly reduced in the strain that produced NsrR1 from the petE promoter (Figure 5C). After 8 h in the absence on combined nitrogen the amount of nblA mRNA (adding all three transcripts together) in the ΔnsrR1 strain is about fourfold higher than in the ΔnsrR1 + PpetE::nsrR1 strain (Supplementary Figure S7), indicating that NsrR1 has a significant negative effect on the accumulation of nblA mRNA in Nostoc.
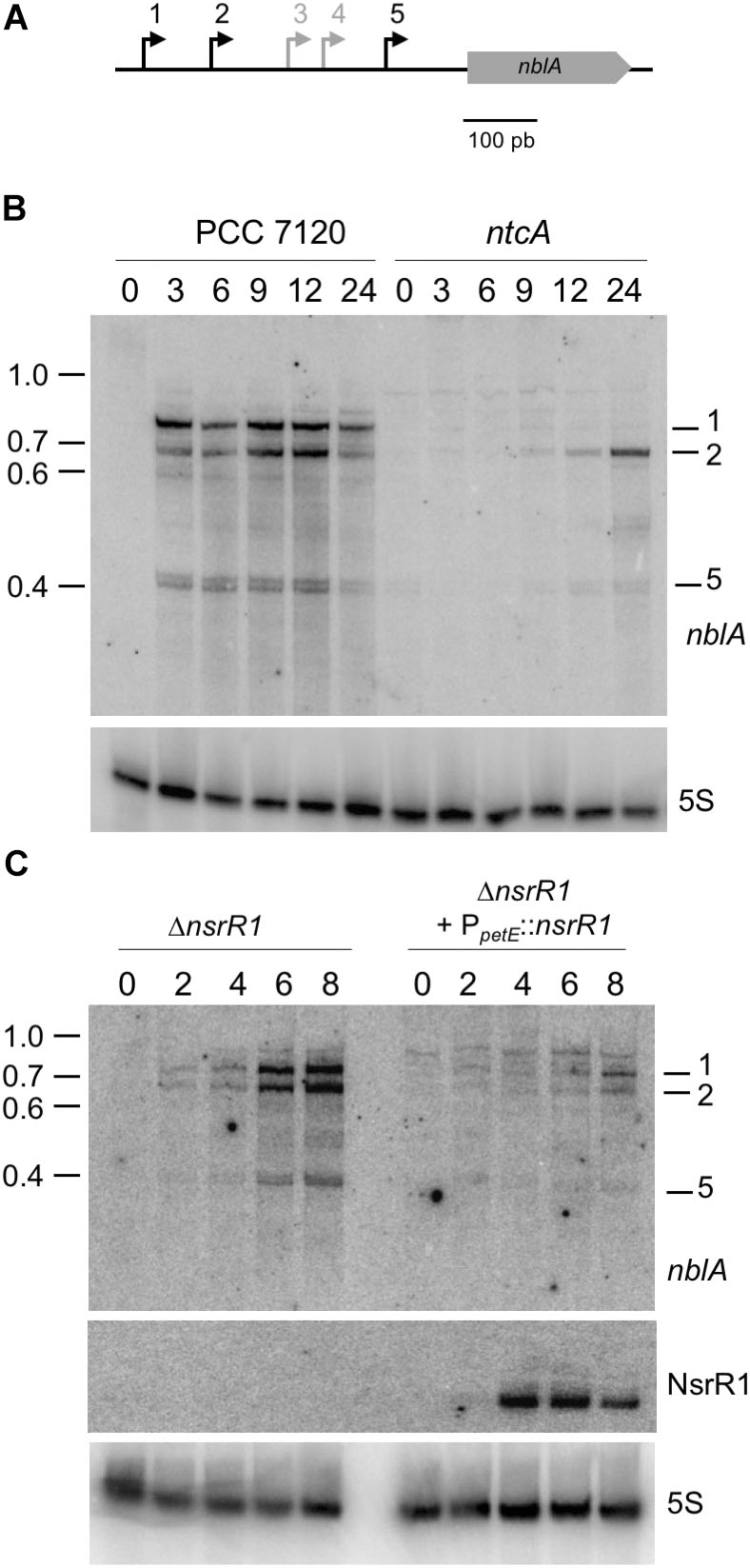
FIGURE 5. Nitrogen-regulated transcripts of nblA and their regulation by NsrR1 in Nostoc sp. PCC 7120. (A) Schematic representation of the five transcriptional start sites (bent arrows) previously defined for nblA (Mitschke et al., 2011), with the nitrogen-regulated transcriptional starts (1, 2, and 5) indicated in black. (B) Identification by Northern hybridization of the transcripts originated from nitrogen-regulated transcriptional starts in Nostoc sp. PCC 7120 and its ntcA mutant derivative CSE2. Expression was analyzed in cells grown in the presence of ammonium and transferred to medium containing no source of combined nitrogen for the number of hours indicated. The upper panel shows hybridization to the nblA probe. The lower panel shows hybridization to a probe for 5S RNA used as loading and transfer control. Size markers (kb) are indicated on the left. Transcripts corresponding to TSS 1, 2, and 5 are indicated. (C) Nitrogen-regulated expression of nblA in strains with altered levels of NsrR1. Expression was analyzed in a mutant strain lacking NsrR1 (ΔnsrR1) and in the same strain complemented with a plasmid bearing nsrR1 under control of the Cu2+-regulated promoter of the petE gene. Cells were grown in the presence of ammonium in medium lacking Cu2+ (to prevent expression of the petE promoter) and transferred to medium containing no source of combined nitrogen and 1.5 μM Cu2+ for the number of hours indicated. The upper panel shows hybridization to the nblA probe. The middle panel shows hybridization to the probe for NsrR1 indicated in Figure 1A. The lower panel shows hybridization to a probe for 5S RNA used as loading and transfer control.
To analyze the accumulation of NblA protein in the different strains, we have generated antibodies against recombinant purified Nostoc NblA protein. Unfortunately, although these antibodies could detect NblA in Western blots of extracts from E. coli cells overproducing NblA (Supplementary Figure S8), they were, however, unable to detect NblA in extracts of Nostoc under conditions of full induction of nblA expression. Similarly, a histidine-tagged version of NblA could not be detected in Nostoc using commercial anti-His antibodies. Lack of detection of NblA with either antibodies could likely be due to the presence of very low levels of NblA in cyanobacterial cells. In fact, it has been suggested that NblA is itself degraded by the cellular degradation machinery (Baier et al., 2014; Sendersky et al., 2014).
To assess possible effects of the ΔnsrR1 mutation on pigment content, whole cell spectra were taken of cells growing in the presence of NH4+ or 24 and 48 h after removal of combined nitrogen. ΔnsrR1 mutant cells have lower phycocyanin content in all cases, even in the presence of NH4+ (Figure 6). Phycocyanin content was further reduced in both strains by nitrogen starvation but while in the wild type strain it is similar at 24 and 48 h, in the ΔnsrR1 mutant strain the reduction is stronger at 48 h than at 24 h (Figure 6B).
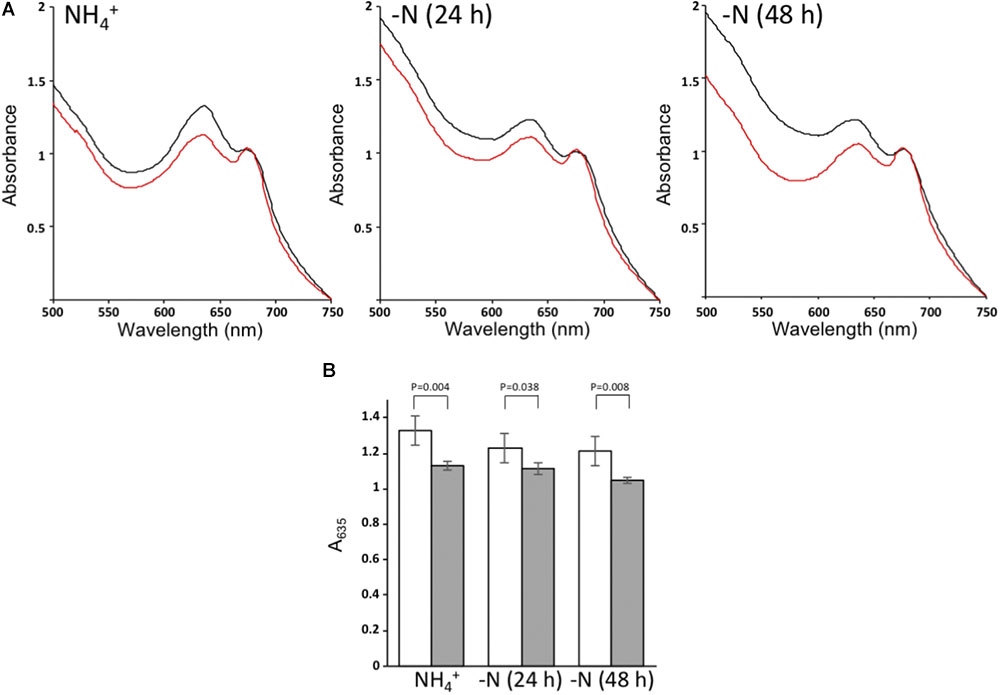
FIGURE 6. Whole cell absorption spectra. (A) Absorption spectra of wild type cells (black) or ΔnsrR1 cells (red) growing in the presence of NH4+ (left) or after 24 h (center) or 48 h (right) of nitrogen depletion, normalized by the chlorophyll content (A680). (B) Normalized A635 of cultures from wild type cells (white) or ΔnsrR1 cells (gray) growing in the presence of NH4+, or after 24 h or 48 h of nitrogen depletion. Error bars represent the standard error of four experiments. p-values (unpaired t-test) are indicated for the comparison of wild type and ΔnsrR1 cells at each time point.
Small regulatory RNAs are important players in the operation of regulatory effects exerted by classical transcription factors. In E. coli, every major transcription factor regulates transcription of one or several regulatory RNAs that, in turn, operate post-transcriptional regulation. FNR and CRP, the best studied members of the CRP/FNR family, are known to regulate the expression of several small regulatory RNAs, including CRP-induced CyaR (Papenfort et al., 2008; De Lay and Gottesman, 2009), CRP-repressed Spot 42 (Beisel and Storz, 2011) or FNR-induced FnrS (Durand and Storz, 2010), all of them contributing to the regulatory effects exerted by these two transcriptional regulators.
In response to nitrogen deficiency, expression of many cyanobacterial genes is regulated, either positively or negatively, by the global nitrogen regulator NtcA, a CRP-family regulator. In many cases the mechanism underlying such regulation seems to be direct activation or repression of gene expression, as deduced from the presence of consensus NtcA binding sites in the respective promoters. However, in cases where no obvious sequence elements can be identified as responsible for the observed regulation, the mechanism is assumed to be indirect. Transcriptomic analyses identifying NtcA-regulated non-coding RNAs suggest this type of molecules might also be involved in NtcA-mediated post-transcriptional regulation (Mitschke et al., 2011; Brenes-Álvarez et al., 2016). A feed-forward coherent regulatory loop involving NtcA and a nitrogen-regulated sRNA has been recently described in Synechocystis sp. PCC 6803. In this unicellular strain NtcA operates transcriptional regulation of the genes encoding glutamine synthetase and its inactivating factors IF7 and IF17, and also regulates transcription of NsiR4 (nitrogen stress inducible RNA 4), that modulates translation of IF7 (Klähn et al., 2015). A similar loop has been described for RpaB, that operates direct transcriptional regulation of several genes encoding proteins of the photosystem I and of PsrR1 (photosynthesis regulatory RNA 1), a high light-responsive small RNA that post-transcriptionally regulates translation of some of those photosystem I-related proteins (Georg et al., 2014). These regulatory loops, like the case of PmgR1 (photomixotrophic growth RNA 1), a sRNA that acts downstream of PmgA and is involved in regulation of glycogen accumulation (de Porcellinis et al., 2016), point to a relevant role of small regulatory RNAs in the regulation of cyanobacterial metabolism.
Here, we describe NsrR1, a small RNA whose transcription is regulated in response to nitrogen limitation in Nostoc sp. PCC 7120. Repression of NsrR1 transcription is at least partially operated by NtcA, as deduced from Northern analysis in the ntcA mutant and binding assays with purified NtcA protein (Figure 2). The observation that repression by NtcA is only partial suggests participation of some other regulator(s) integrating different signal(s). In this context, expression of NsrR1 might be additionally regulated by another repressor, perhaps in the CRP family. Conservation of a T immediately upstream of the GTN10AC conserved NtcA binding sequence suggests possible binding of an unidentified CRP-like regulator (Forcada-Nadal et al., 2014). In contrast to the wide distribution observed for some cyanobacterial sRNAs such as Yfr1 (Voss et al., 2007; Brenes-Álvarez et al., 2016), NsrR1 homologs are only found in the genomes from heterocystous cyanobacteria (and closely related unicellular strains), suggesting its possible function might be specifically related to some particular metabolic trait in this group of organisms.
Phycobilisomes account for a large amount of the total protein, and therefore nitrogen, in cyanobacterial cells. In unicellular cyanobacteria unable to fix N2, recycling of phycobiliproteins as a source of amino acids is an adaptive response to lack of available combined nitrogen, prior to cessation of growth. In heterocystous cyanobacteria, that are ultimately able to overcome nitrogen deficiency by nitrogen fixation, transcription of nblA is induced as a transient response to nitrogen deprivation but the extension of PBS degradation is much lower than in unicellular strains (Maldener et al., 1991; Baier et al., 2004). Using an in vivo reporter system established in E. coli we demonstrate a direct interaction between NsrR1 and the 5′-UTR of nblA, leading to reduced expression of an NblA::sfGFP fusion in the presence of NsrR1. Unfortunately, we have not succeeded in detecting the NblA protein in extracts from Nostoc sp. PCC 7120, and therefore cannot assess directly the effects of NsrR1 overexpression on NblA accumulation at the protein level. Antibodies generated and purified against NblA from Nostoc sp. PCC 7120 detected NblA when expressed in E. coli (Supplementary Figure S8) but not in cyanobacterial extracts, probably because of limited amount of NblA in these cells.
We have also demonstrated reduced accumulation of nblA transcripts in strains of Nostoc sp. PCC 7120 that over-express NsrR1 (Figure 5 and Supplementary Figure S7), which can be a consequence of the interaction of these two RNA molecules leading to reduced translation of NblA. Interaction of NsrR1 at the predicted region in the 5′-UTR of nblA has been verified by in vitro footprinting experiments (Figure 4). The predicted region targeted is located close to the translation initiation and is therefore common to all five possible nblA 5′-UTRs. Thus, although transcriptional regulation of nblA might be operated by different regulators integrating different signals, all possible transcripts would be susceptible to additional post-transcriptional regulation by NsrR1 and therefore the accumulation of NblA protein would be ultimately under control of nitrogen limitation. The reduced accumulation of nblA mRNAs in the presence of NsrR1 can be attributed to destabilization of the mRNA when translation is inhibited. A more direct mechanistic effect of NsrR1 on mRNA stability, recruitment of a ribonuclease for instance, cannot be excluded.
The presence of NsrR1 in ammonium growing cells would suppress transcriptional noise due to leaky transcription initiation of nblA, reducing background production of NblA that would have undesirable consequences on phycobilisome stability. In fact, RNA-Seq and primer extension assays (Mitschke et al., 2011) indicate above-background transcription of nblA from promoters 2, 4, and 5 in ammonium growing cells, that is not readily detectable in the less sensitive Northern blot experiments (Figure 5). Another protein directly involved in PBS degradation, NblB, has been shown to be present under nitrogen sufficient conditions in Synechococcus elongatus (Levi et al., 2018). The RNA-Seq data available for Nostoc sp. PCC 7120 (Flaherty et al., 2011; Mitschke et al., 2011) suggest that in this organism nblB is also expressed constitutively. Therefore, it would be of utmost importance to avoid any background accumulation of NblA under nitrogen sufficient conditions that, together with NblB, would compromise PBS stability. In addition, the presence of NsrR1 would lead to a delay in NblA production after NtcA induction of nblA transcription due to a threshold effect (Levine and Hwa, 2008), creating a delay in the onset of phycobilisome degradation and providing stability by avoiding an inappropriate response under fluctuating environmental conditions. Controlled degradation of PBS, although not essential for adaptation to transient nitrogen deficiency in N2-fixing strains, might be an advantage in natural environments subjected to changes in different factors such as light intensity or availability of other nutrients.
The changes detected in pigment content between wild type and ΔnsrR1 mutant cells agree with the proposed repression of NblA accumulation by NsrR1, and with our model of regulation (Figure 7). We speculate that the slightly, but significant, lower phycobilin/chlorophyll ratio of the ΔnsrR1 mutant compared with wild type (Figure 6B) when growing in the presence of NH4+ is due to background expression of nblA in the absence of NsrR1, resulting in reduced phycobilin stability. After 48 h of nitrogen depletion heterocysts are fully functional and nitrogen status should be more similar to NH4+ growing cells. In fact, in wild type cells the phycocyanin/chlorophyll ratio is stabilized while in the ΔnsrR1 it drops further, suggesting extended presence of NblA in the absence of NsrR1.
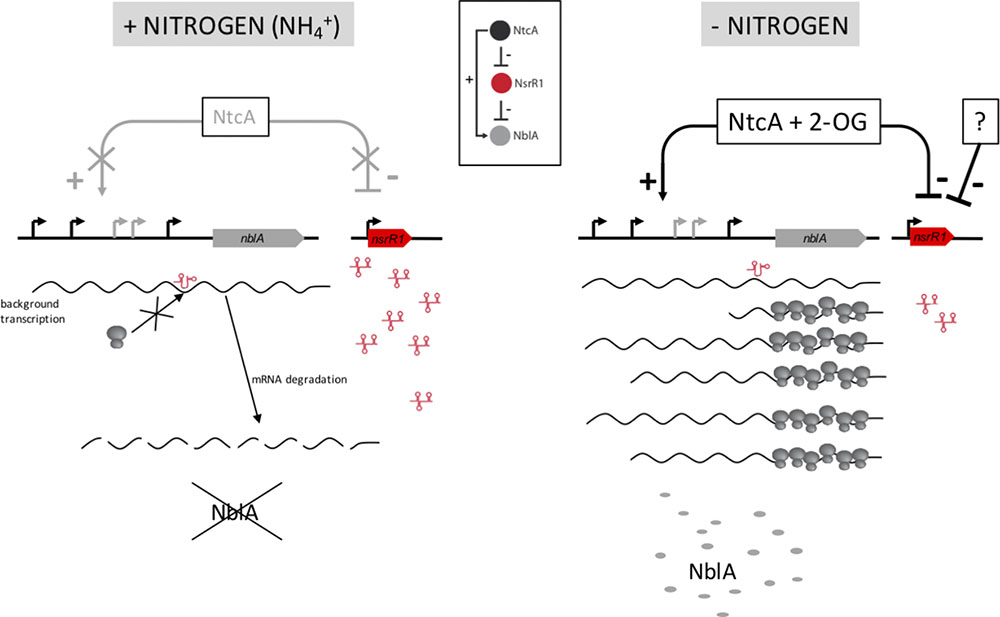
FIGURE 7. Model for the function of NsrR1 in nblA regulation. Under nitrogen sufficient conditions (Left) intracellular 2-oxoglutarate (2-OG) concentration is low and NtcA cannot activate transcription of nblA, while NsrR1 is expressed at high levels. Therefore, under these conditions NsrR1 is in excess of any small amount of nblA mRNA that could be present due to background transcription. Binding of NsrR1 to the mRNA blocks translation, resulting in degradation of the mRNA and preventing any production of NblA protein under nitrogen sufficient conditions. When the nitrogen source is removed (Right) 2-OG concentration increases and NtcA can activate transcription of nblA, while nsrR1 is partially repressed. As a result, there is not enough NsrR1 RNA to repress translation of an excess of nblA transcripts and NblA protein can accumulate. Inset: Scheme of a type 4 feed-forward loop (Nitzan et al., 2017) as applied to the NtcA-NsrR1-NblA regulatory circuit.
The regulatory circuit we propose (Figure 7) represents a type 4 coherent feed-forward loop (Alon, 2007; Nitzan et al., 2017). This type of loop, in which a transcription factor is the top regulator, provides a delayed increase in the level of the target protein (NblA in this case) when the transcription factor (NtcA in this case) is activated, and an accelerated decrease in target protein level when the transcription factor is deactivated (Beisel and Storz, 2011). Dual regulation at the transcriptional and post-transcriptional levels should provide strict control of the levels of the NblA proteolytic adaptor. In addition, because the top regulator (NtcA) is itself subjected to differential accumulation in heterocysts (Olmedo-Verd et al., 2006; Sandh et al., 2014), the feed-forward loop is likely reinforced in heterocysts vs. vegetative cells. Higher amounts of NtcA in heterocysts could promote stronger transcription of nblA. The combination of increased transcription of nblA with low levels of NsrR1 (affecting its translation) would lead to increased amounts of NblA protein producing faster PBS degradation specifically in developing heterocysts.
IÁ-E contributed to the acquisition, analysis, or interpretation of the data. AM-P and AV contributed to the acquisition, analysis, interpretation of the data, and writing of the manuscript.
This work was supported by grants BFU2013-48282-C2-1-P from Ministerio de Economía y Competitividad, and BFU2016-74943-C2-1-P from Agencia Estatal de Investigación (AEI), Ministerio de Economía, Industria y Competitividad, both co-financed by FEDER. IÁ-E is the recipient of a predoctoral contract from Ministerio de Economía y Competitividad, Spain (BES-2014-068488).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank M. Isabel Muro-Pastor (IBVF-CSIC) for help with protein purification, Jörg Vogel (University of Würzburg) for providing plasmids for the analysis of interactions in E. coli, and Alicia Orea (IBVF-CSIC) for technical assistance with flow cytometry.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02267/full#supplementary-material
Alon, U. (2007). Network motifs: theory and experimental approaches. Nat. Rev. Genet. 8, 450–461. doi: 10.1038/nrg2102
Baier, A., Winkler, W., Korte, T., Lockau, W., and Karradt, A. (2014). Degradation of phycobilisomes in Synechocystis sp. PCC6803: evidence for essential formation of an NblA1/NblA2 heterodimer and its codegradation by A Clp protease complex. J. Biol. Chem. 289, 11755–11766. doi: 10.1074/jbc.M113.520601
Baier, K., Lehmann, H., Stephan, D. P., and Lockau, W. (2004). NblA is essential for phycobilisome degradation in Anabaena sp. strain PCC 7120 but not for development of functional heterocysts. Microbiology 150, 2739–2749. doi: 10.1099/mic.0.27153-0
Beisel, C. L., and Storz, G. (2011). The base-pairing RNA spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol. Cell 41, 286–297. doi: 10.1016/j.molcel.2010.12.027
Brenes-Álvarez, M., Olmedo-Verd, E., Vioque, A., and Muro-Pastor, A. M. (2016). Identification of conserved and potentially regulatory small RNAs in heterocystous cyanobacteria. Front. Microbiol. 7:48. doi: 10.3389/fmicb.2016.00048
Buikema, W. J., and Haselkorn, R. (2001). Expression of the Anabaena hetR gene from a copper-regulated promoter leads to heterocyst differentiation under repressing conditions. Proc. Natl. Acad. Sci. U.S.A. 98, 2729–2734. doi: 10.1073/pnas.051624898
Cai, Y. P., and Wolk, C. P. (1990). Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 172, 3138–3145. doi: 10.1128/jb.172.6.3138-3145.1990
Collier, J. L., and Grossman, A. R. (1994). A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. EMBO J. 13, 1039–1047. doi: 10.1002/j.1460-2075.1994.tb06352.x
Corcoran, C. P., Podkaminski, D., Papenfort, K., Urban, J. H., Hinton, J. C., and Vogel, J. (2012). Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol. Microbiol. 84, 428–445. doi: 10.1111/j.1365-2958.2012.08031.x
De Lay, N., and Gottesman, S. (2009). The crp-activated small noncoding regulatory RNA CyaR (RyeE) links nutritional status to group behavior. J. Bacteriol. 191, 461–476. doi: 10.1128/JB.01157-08
de Porcellinis, A. J., Klahn, S., Rosgaard, L., Kirsch, R., Gutekunst, K., Georg, J., et al. (2016). The non-coding RNA Ncr0700/PmgR1 is required for photomixotrophic growth and the regulation of glycogen accumulation in the cyanobacterium Synechocystis sp. PCC 6803. Plant. Cell Physiol. 57, 2091–2103. doi: 10.1093/pcp/pcw128
Durand, S., and Storz, G. (2010). Reprogramming of anaerobic metabolism by the FnrS small RNA. Mol. Microbiol. 75, 1215–1231. doi: 10.1111/j.1365-2958.2010.07044.x
Elhai, J., and Wolk, C. P. (1988). Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 167, 747–754. doi: 10.1016/0076-6879(88)67086-8
Flaherty, B. L., Van Nieuwerburgh, F., Head, S. R., and Golden, J. W. (2011). Directional RNA deep sequencing sheds new light on the transcriptional response of Anabaena sp. strain PCC 7120 to combined-nitrogen deprivation. BMC Genomics 12:332. doi: 10.1186/1471-2164-12-332
Forcada-Nadal, A., Forchhammer, K., and Rubio, V. (2014). SPR analysis of promoter binding of Synechocystis PCC6803 transcription factors NtcA and CRP suggests cross-talk and sheds light on regulation by effector molecules. FEBS Lett. 588, 2270–2276. doi: 10.1016/j.febslet.2014.05.010
Georg, J., Dienst, D., Schurgers, N., Wallner, T., Kopp, D., Stazic, D., et al. (2014). The small regulatory RNA SyR1/PsrR1 controls photosynthetic functions in cyanobacteria. Plant Cell 26, 3661–3679. doi: 10.1105/tpc.114.129767
Giner-Lamia, J., Robles-Rengel, R., Hernández-Prieto, M. A., Muro-Pastor, M. I., Florencio, F. J., and Futschik, M. E. (2017). Identification of the direct regulon of NtcA during early acclimation to nitrogen starvation in the cyanobacterium Synechocystis sp. PCC 6803. Nucleic Acids Res. 45, 11800–11820. doi: 10.1093/nar/gkx860
González, A., Bes, M. T., Peleato, M. L., and Fillat, M. F. (2016). Expanding the role of FurA as essential global regulator in cyanobacteria. PLoS One 11:e0151384. doi: 10.1371/journal.pone.0151384
Grossman, A. R., Schaefer, M. R., Chiang, G. G., and Collier, J. L. (1993a). Environmental effects on the light-harvesting complex of cyanobacteria. J. Bacteriol. 175, 575–582. doi: 10.1128/jb.175.3.575-582.1993
Grossman, A. R., Schaefer, M. R., Chiang, G. G., and Collier, J. L. (1993b). The phycobilisome, a light-harvesting complex responsive to environmental conditions. Microbiol. Rev. 57, 725–749.
Herrero, A., Muro-Pastor, A. M., and Flores, E. (2001). Nitrogen control in cyanobacteria. J. Bacteriol. 183, 411–425. doi: 10.1128/JB.183.2.411-425.2001
Ionescu, D., Voss, B., Oren, A., Hess, W. R., and Muro-Pastor, A. M. (2010). Heterocyst-specific transcription of NsiR1, a non-coding RNA encoded in a tandem array of direct repeats in cyanobacteria. J. Mol. Biol. 398, 177–188. doi: 10.1016/j.jmb.2010.03.010
Karradt, A., Sobanski, J., Mattow, J., Lockau, W., and Baier, K. (2008). NblA, a key protein of phycobilisome degradation, interacts with ClpC, a HSP100 chaperone partner of a cyanobacterial Clp protease. J. Biol. Chem. 283, 32394–32403. doi: 10.1074/jbc.M805823200
Klähn, S., Schaal, C., Georg, J., Baumgartner, D., Knippen, G., Hagemann, M., et al. (2015). The sRNA NsiR4 is involved in nitrogen assimilation control in cyanobacteria by targeting glutamine synthetase inactivating factor IF7. Proc. Natl. Acad. Sci. U.S.A. 112, E6243–E6252. doi: 10.1073/pnas.1508412112
Levi, M., Sendersky, E., and Schwarz, R. (2018). Decomposition of cyanobacterial light harvesting complexes: NblA-dependent role of the bilin-lyase homolog NblB. Plant J. 94, 813–821. doi: 10.1111/tpj.13896
Levine, E., and Hwa, T. (2008). Small RNAs establish gene expression thresholds. Curr. Opin. Microbiol. 11, 574–579. doi: 10.1016/j.mib.2008.09.016
Li, W., Cowley, A., Uludag, M., Gur, T., McWilliam, H., Squizzato, S., et al. (2015). The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 43, W580–W584. doi: 10.1093/nar/gkv279
Luque, I., Flores, E., and Herrero, A. (1994). Molecular mechanism for the operation of nitrogen control in cyanobacteria. EMBO J. 13, 2862–2869. doi: 10.1002/j.1460-2075.1994.tb06580.x
Luque, I., Zabulon, G., Contreras, A., and Houmard, J. (2001). Convergence of two global transcriptional regulators on nitrogen induction of the stress-acclimation gene nblA in the cyanobacterium Synechococcus sp. PCC 7942. Mol. Microbiol. 41, 937–947.
Maldener, I., Lockau, W., Cai, Y., and Wolk, C. P. (1991). Calcium-dependent protease of the cyanobacterium Anabaena: molecular cloning and expression of the gene in Escherichia coli, sequencing and site-directed mutagenesis. Mol. Gen. Genet. 225, 113–120. doi: 10.1007/BF00282649
Mandin, P., and Guillier, M. (2013). Expanding control in bacteria: interplay between small RNAs and transcriptional regulators to control gene expression. Curr. Opin. Microbiol. 16, 125–132. doi: 10.1016/j.mib.2012.12.005
Mann, M., Wright, P. R., and Backofen, R. (2017). IntaRNA 2.0: enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 45, W435–W439. doi: 10.1093/nar/gkx279
Merino-Puerto, V., Mariscal, V., Mullineaux, C. W., Herrero, A., and Flores, E. (2010). Fra proteins influencing filament integrity, diazotrophy and localization of septal protein SepJ in the heterocyst-forming cyanobacterium Anabaena sp. Mol. Microbiol. 75, 1159–1170. doi: 10.1111/j.1365-2958.2009.07031.x
Mitschke, J., Vioque, A., Haas, F., Hess, W. R., and Muro-Pastor, A. M. (2011). Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC7120. Proc. Natl. Acad. Sci. U.S.A. 108, 20130–20135. doi: 10.1073/pnas.1112724108
Mohamed, A., and Jansson, C. (1989). Influence of light on accumulation of photosynthesis-specific transcripts in the cyanobacterium Synechocystis 6803. Plant. Mol. Biol. 13, 693–700. doi: 10.1007/BF00016024
Muro-Pastor, A. M., Valladares, A., Flores, E., and Herrero, A. (1999). The hetC gene is a direct target of the NtcA transcriptional regulator in cyanobacterial heterocyst development. J. Bacteriol. 181, 6664–6669.
Nitzan, M., Rehani, R., and Margalit, H. (2017). Integration of bacterial small RNAs in regulatory networks. Annu. Rev. Biophys. 46, 131–148. doi: 10.1146/annurev-biophys-070816-034058
Notredame, C., Higgins, D. G., and Heringa, J. (2000). T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217. doi: 10.1006/jmbi.2000.4042
Olmedo-Verd, E., Muro-Pastor, A. M., Flores, E., and Herrero, A. (2006). Localized induction of the ntcA regulatory gene in developing heterocysts of Anabaena sp. strain PCC 7120. J. Bacteriol. 188, 6694–6699. doi: 10.1128/JB.00509-06
Papenfort, K., Pfeiffer, V., Lucchini, S., Sonawane, A., Hinton, J. C., and Vogel, J. (2008). Systematic deletion of Salmonella small RNA genes identifies CyaR, a conserved CRP-dependent riboregulator of OmpX synthesis. Mol. Microbiol. 68, 890–906. doi: 10.1111/j.1365-2958.2008.06189.x
Picossi, S., Flores, E., and Herrero, A. (2014). ChIP analysis unravels an exceptionally wide distribution of DNA binding sites for the NtcA transcription factor in a heterocyst-forming cyanobacterium. BMC Genomics 15:22. doi: 10.1186/1471-2164-15-22
Rippka, R., Deruelles, J., Waterbury, J. B., Herdman, M., and Stanier, R. Y. (1979). Generic assignments, strain stories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111, 1–61.
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory Press.
Sandh, G., Ramstrom, M., and Stensjo, K. (2014). Analysis of the early heterocyst Cys-proteome in the multicellular cyanobacterium Nostoc punctiforme reveals novel insights into the division of labor within diazotrophic filaments. BMC Genomics 15:1064. doi: 10.1186/1471-2164-15-1064
Sendersky, E., Kozer, N., Levi, M., Garini, Y., Shav-Tal, Y., and Schwarz, R. (2014). The proteolysis adaptor, NblA, initiates protein pigment degradation by interacting with the cyanobacterial light-harvesting complexes. Plant J. 79, 118–126. doi: 10.1111/tpj.12543
Steglich, C., Futschik, M. E., Lindell, D., Voss, B., Chisholm, S. W., and Hess, W. R. (2008). The challenge of regulation in a minimal photoautotroph: non-coding RNAs in Prochlorococcus. PLoS Genet. 4:e1000173. doi: 10.1371/journal.pgen.1000173
Urban, J. H., and Vogel, J. (2007). Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 35, 1018–1037. doi: 10.1093/nar/gkl1040
Voss, B., Gierga, G., Axmann, I. M., and Hess, W. R. (2007). A motif-based search in bacterial genomes identifies the ortholog of the small RNA Yfr1 in all lineages of cyanobacteria. BMC Genomics 8:375. doi: 10.1186/1471-2164-8-375
Wagner, E. G., and Romby, P. (2015). Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv. Genet. 90, 133–208. doi: 10.1016/bs.adgen.2015.05.001
Wright, P. R., Georg, J., Mann, M., Sorescu, D. A., Richter, A. S., Lott, S., et al. (2014). CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains. Nucleic Acids Res. 42, W119–W123. doi: 10.1093/nar/gku359
Keywords: regulatory RNA, cyanobacteria, NtcA, feed-forward loop, post-transcriptional regulation
Citation: Álvarez-Escribano I, Vioque A and Muro-Pastor AM (2018) NsrR1, a Nitrogen Stress-Repressed sRNA, Contributes to the Regulation of nblA in Nostoc sp. PCC 7120. Front. Microbiol. 9:2267. doi: 10.3389/fmicb.2018.02267
Received: 25 May 2018; Accepted: 05 September 2018;
Published: 24 September 2018.
Edited by:
Daniela De Biase, Università degli Studi di Roma La Sapienza, ItalyReviewed by:
Rakefet Schwarz, Bar-Ilan University, IsraelCopyright © 2018 Álvarez-Escribano, Vioque and Muro-Pastor. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Agustín Vioque, dmlvcXVlQHVzLmVz
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.