- 1Department of Population Health and Pathobiology, College of Veterinary Medicine, NC State University, Raleigh, NC, United States
- 2Department of Clinical Sciences, College of Veterinary Medicine, NC State University, Raleigh, NC, United States
- 3Department of Molecular and Biomedical Sciences, College of Veterinary Medicine, NC State University, Raleigh, NC, United States
Objective: The intestinal concentrations of antimicrobial drugs that select for resistance in fecal bacteria of cattle are poorly understood. Our objective was to associate active drug concentrations in the intestine of steers with changes in the resistance profile and composition of the fecal microbiome.
Methods: Steers were administered either a single dose (12.5 mg/kg) or 3 multiple doses (5 mg/kg) of enrofloxacin subcutaneously every 24 h. Enrofloxacin and ciprofloxacin concentrations in intestinal fluid were measured over 96 h, and the abundance and MIC of E. coli in culture and the composition of the fecal microbiota by 16S rRNA gene sequencing were assessed over 192 h after initial treatment.
Results: Active drug concentrations in the ileum and colon exceeded plasma and interstitial fluid concentrations, but were largely eliminated by 48 h after the last dose. The concentration of E. coli in the feces significantly decreased during peak drug concentrations, but returned to baseline by 96 h in both groups. The median MIC of E. coli isolates increased for 24 h in the single dose group, and for 48 h in the multiple dose group. The median MIC was higher in the multiple dose group when compared to the single dose group starting 12 h after the initial dose. The diversity of the fecal microbiota did not change in either treatment group, and taxa-specific changes were primarily seen in phyla commonly associated with the rumen.
Conclusions: Both dosing regimens of enrofloxacin achieve high concentrations in the intestinal lumen, and the rapid elimination mitigates long-term impacts on fecal E. coli resistance and the microbiota.
Introduction
Some studies have demonstrated an association between antimicrobial administration and antimicrobial resistance (AMR) in fecal bacteria of cattle (Kanwar et al., 2013; Zaheer et al., 2013; Hog and Korsgaard, 2017). Yet others demonstrate little impact of antimicrobial therapy on AMR in fecal microbiota of cattle (Checkley et al., 2010; Morley et al., 2011; Schmidt et al., 2013). Methodological variation in identifying and characterizing antimicrobial resistance in the studies investigating AMR associated with treatment in feedlots prevents meaningful comparison of their results. Additionally, conclusions are often drawn from single bacterial isolates, selected resistance genes, or over varying sampling times, which further confounds interpretation and comparison between studies (Benedict et al., 2013, 2014).
Parenteral antibiotics administered to treat infections diffuse into the intestine and influence the population and susceptibility of the intestinal bacteria (Lindecrona et al., 2000; Ferran et al., 2013; Foster et al., 2016). However, there is little known about the duration of this effect, particularly on potential foodborne pathogens. Measurement of active, unbound drug concentrations in the intestinal lumen is necessary to correlate pharmacokinetic-pharmacodynamic (PK-PD) indices with microbial changes. Based on evidence that the exposure relationships are important for development of antimicrobial resistance (Martinez et al., 2012), it is possible that resistance can be affected by the dose and frequency of drug administration (Enne, 2010).
In the US, there are two approved dosing regimens for enrofloxacin for treating respiratory disease in cattle, but little is known about the relative impact of these dosing regimens on AMR. It is presumed that each dosing regimen has similar efficacy; therefore, impact of each regimen on AMR may provide a rational basis for choosing one regimen over the other. Further, understanding the relationships between dosing regimen of enrofloxacin and AMR is critical due to the importance of fluoroquinolones in human health. The objective of this study was to model the PK of the active concentrations of enrofloxacin and its metabolite, ciprofloxacin, in the intestine of steers, and correlate these PK indices with changes in the MIC of E. coli and fecal microbiome. Our hypothesis is that the single 12.5 mg/kg dose of enrofloxacin will achieve a greater maximum concentration of active drug within the intestine resulting in less selection for AMR bacteria and changes in the microbiota due to the brief drug exposure.
Materials and Methods
Animals and Treatments
This study was approved by the North Carolina State University Institutional Animal Care and Use Committee. Twelve healthy, 6-months-old Holstein steers (125 to 241 kg) were obtained as in previous studies (McKellar et al., 1999; TerHune et al., 2005; Davis et al., 2007; Foster et al., 2016). Sample size was determined based on previous PK-PD studies in cattle demonstrating differences in drug concentrations in different sampling locations (Davis et al., 2007; Foster et al., 2016). At 24–48 h post-probe placement (described below), steers received either a single subcutaneous injection of 12.5 mg/kg enrofloxacin (Baytril 100®; Bayer Animal Health, Shawnee Mission, KS) or three subcutaneous injections of 5.0 mg/kg enrofloxacin (Baytril 100®) administered once each day on 3 sequential days. Treatments were administered in a crossover design, with each calf receiving both treatments separated by a minimum washout period of 10 days, accounting for at least six drug half-lives from last dose. Half of the steers (n = 6) received the single dose regimen first, while the other half received the multiple dose regimen first.
Plasma Collection
Prior to drug administration, a jugular catheter (Intracath®, Becton Dickinson; Franklin Lakes, NJ) was inserted in the jugular vein. Blood samples were collected at time 0, 5, 10, 15, 30, 45, 60, 90 min and 2, 4, 6, 8, 24, 36, and 48 h after the single dose for optimum pharmacokinetic modeling and to encompass at least three drug half-lives, accounting for 90% of drug elimination from the plasma. In the multiple dose group, additional samples were collected at 2, 4, 6, 8, 12, and 24 h after the second dose, and 5, 10, 15, 30, 45, 60, 90 min and 2, 4, 6, 8, 24, 36, and 48 h after the third dose. The tubes were briefly stored on ice before centrifugation at 1,000 × g for 10 min to collect plasma and stored at −80°C until assayed.
Interstitial Fluid Collection
An in-vivo ultrafiltration probe (UF-3-12, BAS; Bioanalytical Systems, West Lafayette, IN, USA) was inserted in the subcutaneous space above the shoulders as described previously (Davis et al., 2007; Messenger et al., 2012). The interstitial fluid (ISF) was collected at time 0 and appropriate intervals for each drug, which will account for approximately three drug half-lives post-administration. The collected fluid was immediately frozen at −80°C until assayed.
Placement of Intestinal Ultrafiltration Probes
Surgical procedures took place 24–48 h prior to enrofloxacin administration over 4 days with three surgeries per day as previously described (Warren et al., 2014) with the only significant modification being the use of standing-restraint and local anesthesia instead of general anesthesia. Local anesthesia was provided by infusion 15 ml of 2% lidocaine above and below the transverse processes of lumbar vertebrae L1, L2, L3, and L4 in order to anesthetize the dorsal and ventral spinal nerves T13, L1 and L2. Adequacy of anesthesia was determined by pricking the skin of the flank prior to the incision. Steers received 2.2 mg/kg of flunixin meglumine intravenously immediately prior to surgery and 24 h later. The collecting loops of an ultrafiltration probe (UF-3-12, BAS; Bioanalytical Systems, West Lafayette, IN, USA) were inserted into the lumen of the ileum and spiral colon. The free ends of the probes were exteriorized through the abdominal wall. To collect the ultrafiltrate, a 3 mL evacuated tube with no additive (Vacutainer®, Becton-Dickinson) was inserted onto the needle of the vacuum vial needle holder at the end of the external tubing. Samples were collected from probes placed in the ileum and spiral colon at 0, 2, 4, 6, 8, 12, 24, 36, and 48 h post high-dose drug administration, and additionally 26, 28, 30, 32, 50, 52, 54, 56, 60, 72, 84, and 96 h after the first low-dose drug administration by changing the collection tubes at the predetermined time points. The collected fluid was aliquoted into cryogenic vials and immediately frozen at −80°C until assayed.
Feces Collection
Feces was collected directly from the rectum of each steer 0, 12, 24, 36, 48, 72, 96, 120, 144, and 168 h post single-dose drug administration, and additionally 60, 84, and 192 h after the first multiple-dose drug administration. Samples were placed into bags (Whirlpak®, Nasco, Fort Atkinson, WI) and stored briefly on ice before microbiological analysis. An aliquot of each sample was stored in a 2 ml cryogenic vial and frozen at −80°C. To avoid the residual effects of enrofloxacin on fecal bacteria between crossover arms of the study, only fecal samples collected from the first arm of each crossover study were analyzed and reported here.
Determining Active Drug Concentration
For plasma samples, we used solid-phase extraction, identical to the method in our previous papers (Davis et al., 2007). Plasma and tissue fluid samples were analyzed by reverse-phase high-pressure liquid chromatography with fluorescence (enrofloxacin and its metabolite ciprofloxacin) detection to determine the active concentrations of each drug as previously described (Davis et al., 2007; Foster et al., 2016). All drug concentrations were determined from calibration curves made from fortified blank plasma, intestinal and interstitial fluid collected from the experimental calves prior to antibiotic administration. Calibration curves were prepared from fortifying the blank matrix with reference drug standards of enrofloxacin and ciprofloxacin (United States Pharmacopeia, Rockville, MD) to validate the HPLC analysis and perform quality control assessments during the assay.
Pharmacokinetic Analysis
The drug concentrations were analyzed using standard pharmacokinetic methods to determine the drug disposition for each drug in each calf. A computer program (Phoenix, V. 6.1; Pharsight Corporation, Certara, St. Louis MO) was used to determine pharmacokinetic parameters as well as deriving statistical values.
Plasma, ISF, and intestinal drug concentrations were plotted on linear and semi-logarithmic graphs for analysis and for visual assessment of the best model for pharmacokinetic analysis. Specific models (e.g., one, two, etc. compartments) were determined for best fit based on visual analysis for goodness of fit and by visual inspection of residual plots. The best model fit was based on the equation described in the following formula:
Where C is the plasma concentration, t is time, k01 is the non-IV absorption rate, assuming first-order absorption, k10 is the elimination rate constant, V is the apparent volume of distribution, and D is the non-IV dose. Secondary parameters calculated from the model included the peak concentration (CMAX), time to peak concentration (TMAX), area under the plasma-concentration vs. time profile (AUC), and the respective absorption and terminal half-lives (t½).
Data from some calves were analyzed using non-compartmental analysis (NCA) due to sparse sampling using the same pharmacokinetic program described above. For the NCA, the area under the plasma concentration vs. time curve (AUC) from time 0 to the last measured concentration (defined by the LOQ), was calculated using the log-linear trapezoidal method. The AUC from time 0 to infinity was calculated by adding the terminal portion of the curve, estimated from the relationship Cn/λZ, to the AUC0 Cn, where λZ is the terminal slope of the curve, and Cn is the last measured concentration point.
The relative drug transfer from the plasma compartment to the ISF and intestinal fluids was measured by calculation of a penetration factor. The penetration factor was determined by the ratio of AUC for the intestinal fluid to the AUC for plasma:
Quantification of E. coli From Feces
One gram of feces was inoculated into 9 ml EC broth (Oxoid Ltd., Basingstoke, Hampshire, England) and vortexed. One ml was removed and serially diluted 10-fold in sterile phosphate-buffered saline, and 100 μl was plated in triplicate onto HardyCHROM™ ECC Media (Hardy Diagnostics, Santa Maria, CA). Plates were incubated overnight at 37°C and pink-violet colonies were counted to determine the CFU/ml of E. coli. Dilutions that yielded between 30 and 300 colonies on each of the three plates were counted to quantify E. coli based on the dilution. These three replicates at the counted dilution were averaged to determine the quantity of E. coli at each time point. The remaining enrichment was incubated overnight at 37°C, and if no growth was observed on direct plates, the samples were streaked for isolation on ECC plates and incubated overnight at 37°C. From the quantified or enrichment plate, eight colonies were randomly picked and streaked onto Columbia agar with 5% sheep blood (Remel, Lenexa, KS) and incubated overnight at 37°C. Following incubation, each isolate was transferred to a 2 ml cryogenic vial containing LB Broth (Sigma-Aldrich, St. Louis, MO) supplemented with 25% glycerol, vortexed, and frozen at −80°C.
Determination of Minimum Inhibitory Concentration
The minimum inhibitory concentration (MIC) of each E. coli isolate to enrofloxacin was determined using broth microdilution according to Clinical and Laboratory Standards Institute guidelines (Clinical Laboratory Standards Institute, 2013). Each isolate was grown overnight on blood agar. A single colony was inocluated into Mueller Hinton broth and brought to a 0.5 McFarland Standard. Fifty μl of bacterial suspension was inoculated into 50 μl of 2-fold serial dilutions of enrofloxacin ranging in concentration from 0.03 to 32 mg/L (USP, Rockville, MD). Plates were incubated overnight (18 h) at 37°C. The first well with no visible growth within a given isolate was determined to be the MIC.
DNA Extraction and 16S rRNA Sequencing
Fresh feces collected at each time point were frozen in 1 gram aliquots for future analysis. From these samples, 50 mg of feces were extracted individually using a MoBio PowerMag Microbiome kit (Qiagen, Inc., Germanton, MD) optimized for the epMotion 5075 TMX (Eppendorf, Hauppauge, NY). The DNA libraries were prepared as described previously (Seekatz et al., 2015).
Illumina MiSeq Sequencing of Bacterial Communities
The V4 region of the 16S rRNA gene was amplified from each sample using the Dual indexing sequencing strategy (Kozich et al., 2013). Sequencing was done on the Illumina MiSeq platform, using a MiSeq Reagent Kit V2 500 cycles (2 × 250 bp; Illumina cat# MS102-2003), according to the manufacturer's instructions with modifications (Kozich et al., 2013). Accuprime High Fidelity Taq (Life Technologies cat # 12346094) was used. PCR was performed using the conditions (Standard or Touch Down) shown by Seekatz (Seekatz et al., 2015). If additional template was used, the water volume was changed accordingly. PCR products were visualized using an E-Gel 96 with SYBR Safe DNA Gel Stain, 2% (Life technologies cat# G7208-02). Libraries were normalized using SequalPrep Normalization Plate Kit (Life technologies cat #A10510-01) following the manufacturer's protocol for sequential elution. The concentration of the pooled samples was determined using Kapa Biosystems Library Quantification kit for Illumina platforms (KapaBiosystems KK4824). The sizes of the amplicons in the library were determined using the Agilent Bioanalyzer High Sensitivity DNA analysis kit (cat# 5067-4626). The final library consisted of equal molar amounts from each of the plates, normalized to the pooled plate at the lowest concentration. Sequencing libraries were prepared according to Illumina's protocol for Preparing Libraries for Sequencing on the MiSeq (part# 15039740 Rev. D) for 2 or 4 nM libraries. If the library concentration was below 1 nM, an alternative method was used for denaturation (Quail et al., 2008). PhiX and genomes were added in 16S amplicon sequencing to create diversity. Sequencing reagents were prepared according to the Schloss SOP, and custom read 1, read 2 and index primers were added to the reagent cartridge. FASTQ files were generated for paired end reads.
Microbiota Analysis
The median number of sequences per group was 24,866. Analysis of the V4 region of the 16S rRNA gene was done using mothur (version 1.37.4; Schloss et al., 2009). The standard operating procedure at http://www.mothur.org/wiki/MiSeq_SOP was followed to process the MiSeq data. The paired-end reads were assembled into contigs and then aligned to the SILVA 16S rRNA sequence database (Pruesse et al., 2007; Quast et al., 2013) and classified to the family taxonomic level using the Wang method and an 80% bootstrap minimum (Wang et al., 2007) and the mothur-adapted RDP training set v9. Chimeric sequences were removed using UCHIME (Edgar et al., 2011). Sequences were clustered into operational taxonomic units (OTU) using a 3% sequence similarity threshold. The percentage relative abundance of bacterial Phyla and Family members in each sample was calculated. The inverse Simpson index on OTUs was used as a measure of alpha diversity.
Additional high-resolution analysis was performed on exact amplicon sequence variants (ASVs) generated by the DADA2 method (Callahan et al., 2016, 2017). ASV generation followed the DADA2 tutorial (https://benjjneb.github.io/dada2/tutorial.html), but with samples pooled for the ASV inference step in order to increase sensitivity to rare sequence variants. E. coli ASVs were identified by exact matching to sequenced E. coli genomes.
Data Analysis
Differences in the E. coli concentration between groups were determined by a one way ANOVA. ANOVA on ranks was used to compare median MIC over time within treatment groups. A p-value < 0.05 was considered significant. Differential abundance testing was performed using the DESeq2 package (Love et al., 2014), and following the recommendations for its application to microbiome sequencing data in McMurdie and Holmes (McMurdie and Holmes, 2014). Briefly, DESeq2 controls for the over-dispersion typical in microbiome sequencing data by fitting the abundance-variance relationship over all ASVs, and using that shrinkage estimator of the variance to test for significance. Testing for the treatment effect was performed on paired samples from each steer, before and after treatment, and after controlling for the steer. False discovery rate (FDR) was controlled using the Benjamini-Hochberg procedure (Benjamini and Hochberg, 1995).
Deposition of Data
Sequence data will be deposited in the SRA database upon acceptance of the manuscript.
Results
Pharmacokinetic Modeling
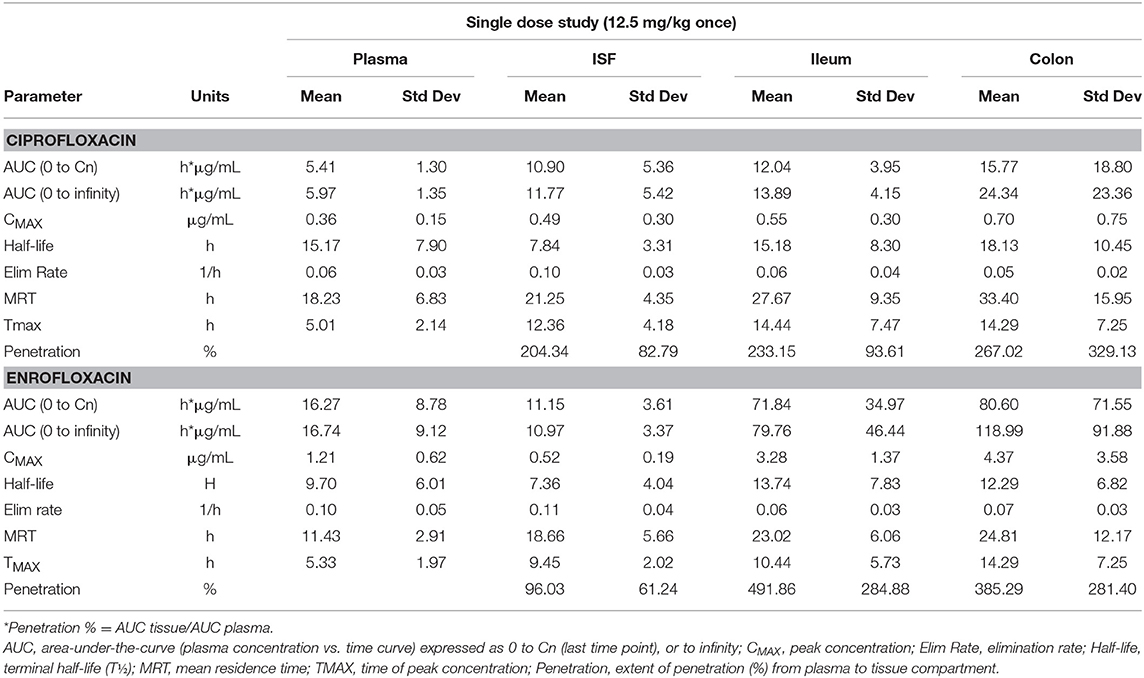
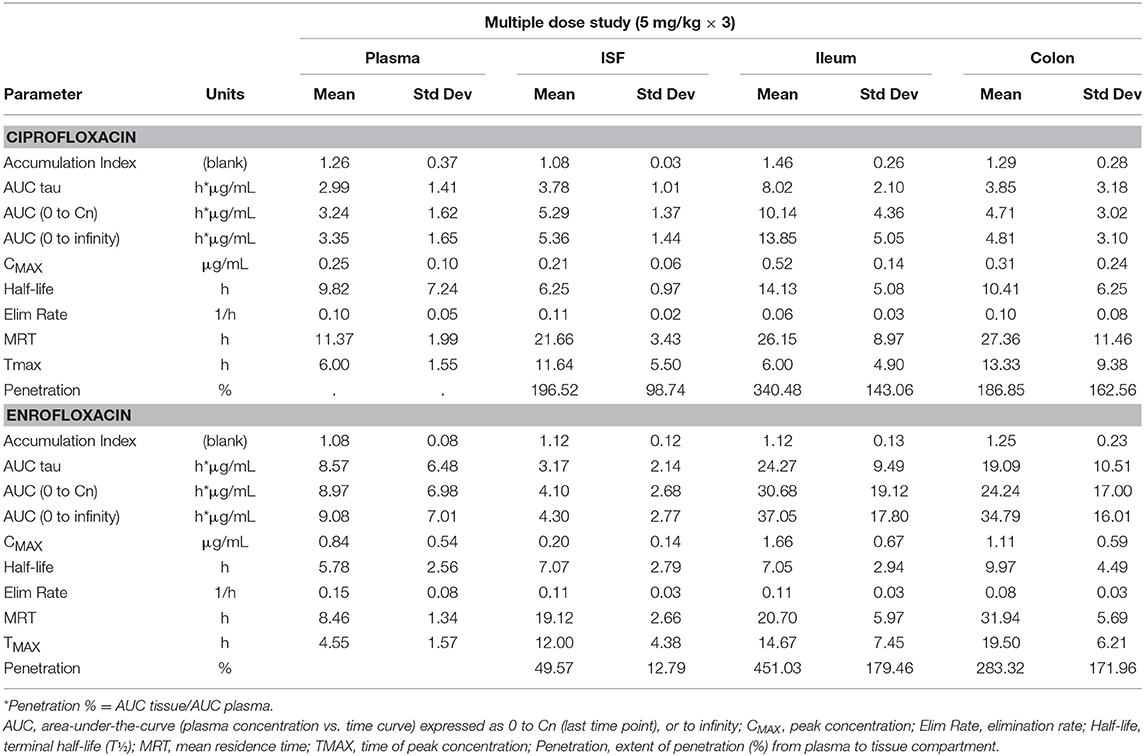
Tables 1, 2 depict the results of the PK analysis for enrofloxacin and ciprofloxacin for the single and multiple dose regimens. The ciprofloxacin concentration accounted for 25.8% of total fluoroquinolone concentrations in the single dose regimen and 26.7% of total fluoroquinolone concentrations in the multiple dose regimen. Ciprofloxacin penetration from the plasma into the ISF was two times greater than enrofloxacin in the single dose regimen and four times greater than enrofloxacin in the multiple dose regimen. Enrofloxacin penetration into the ileum from the plasma (single dose = 491.86%, multiple dose = 451.03%) and into the colon from the plasma (single dose = 385.29%, multiple dose = 293.32%) exceeded the penetration into the ISF from the plasma (single dose = 96.03%, multiple dose = 49.57%). Figure 1 depicts the combined concentrations of enrofloxacin and ciprofloxacin over time with the two dosing regimens in plasma, interstitial fluid, ileum and spiral colon.
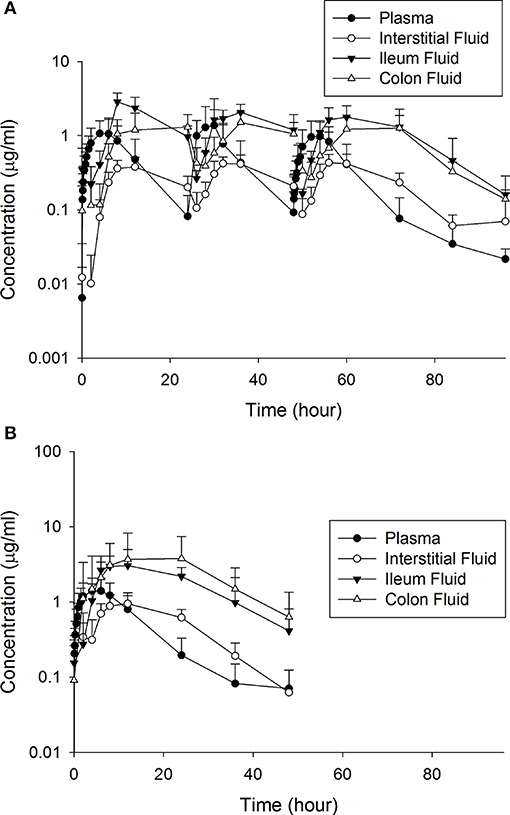
Figure 1. The combined concentrations of enrofloxacin and ciprofloxacin between the (A) multiple and (B) single dose treatment regimens.
Concentration of E. coli
The concentration of E. coli recovered at 12 (1.60 log10 CFU/g), 24 (0.75 log10 CFU/g) and 36 (1.11 log10 CFU/g) h was significantly different from 0 h (6.17 log10 CFU/g) in the single dose group (Figure 2A). By 72 h post-treatment, the E. coli concentration (4.72 log10 CFU/g) had returned to baseline. In animals administered the multiple dose regimen, the concentrations at 24, 36, 48, 60 and 72 h were significantly different from 0 h (Figure 2A), and then returned to baseline by 96 h post-treatment. The correlation between culturable and sequenced E. coli concentrations was 0.557 over all samples, increasing to 0.808 when considering only those samples with >500,000 CFUs/g, which roughly corresponds to the minimum detectable frequency of 0.0001 for the 16S sequencing in this study (~10 k reads per sample; Figure 2B). This correlation suggests that the culture results agree with the sequencing findings presented below.
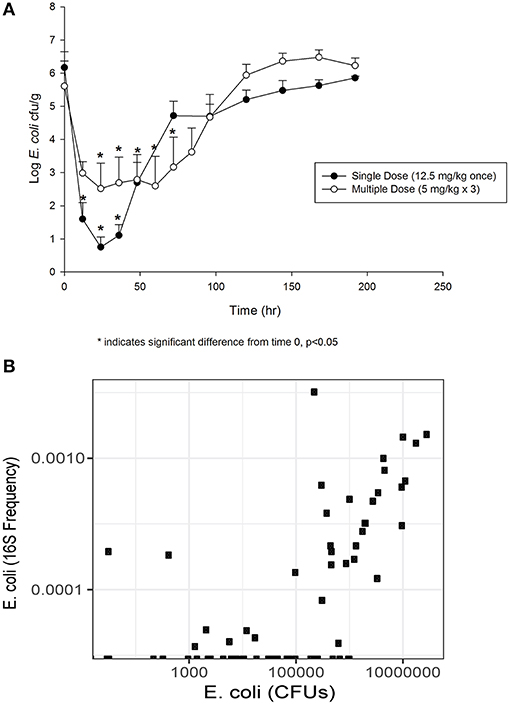
Figure 2. (A) Fecal E. coli concentration over time after treatment with either a single dose (12.5 mg/kg once) or multiple dose (5 mg/kg once a day for 3 days) of enrofloxacin. Mean ± SD. * indicates a significant difference from time 0, p < 0.05. (B) Comparison of culturable E. coli concentration and the frequency of the E. coli 16S sequence variant.
E. coli Minimum Inhibitory Concentration
Prior to treatment, 84% E. coli isolates had an MIC of 0.03 mg/L or less, which is below the epidemiological cutoff value of 0.12 mg/L (Figure 3). The median MIC (0.029 mg/L) was not different between the two groups at time 0. In the single dose group, the median MIC significantly increased at 24 h post-treatment (0.5 mg/L), but returned to baseline by 48 h (0.03 mg/L). The median MIC of E. coli isolates from the multiple dose group significantly increased by 12 h after treatment (0.75 mg/L), and remained increased through 48 h (1.0 mg/L; Figure 3). When comparing individual time points across treatment regimens, the median MIC of E. coli isolates from the multiple dose group was significantly higher than the single dose group starting at 12 h and remained higher through the end of the study (Figure 3).
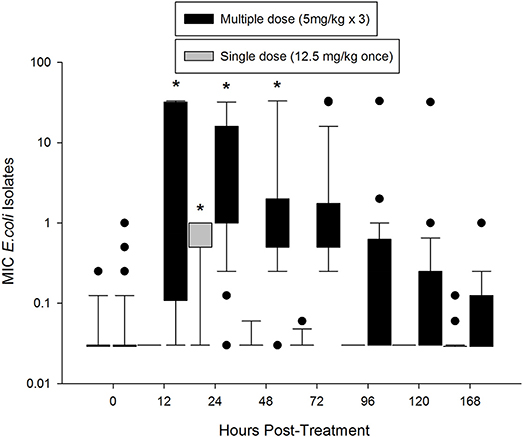
Figure 3. Median MIC of enrofloxacin in fecal E. coli isolates over time after treatment with either a single dose (12.5 mg/kg once) or multiple dose (5 mg/kg once a day for 3 days) of enrofloxacin. * indicates a significant difference from time 0, p < 0.05.
Alterations in the Fecal Microbiota
Alpha-diversity, a measure of the diversity of bacterial phyla within each sample, showed no significant change over time in either dosing regimen (Figures 4A,B). There were subtle changes to the fecal microbial community at the Phylum (Figures 4C,D), and Family (Figures 5A,B) level. One ASV from the Family Prevotellaceae significantly increased in frequency at 48 h, while several ASVs from the Rikenellaceae, Christensenellaceae, Erysipelotrichaceae, and Enterobacteriaceae Families significantly decreased in frequency at a FDR of 0.01 (Table 3). At 168 h, only one ASV from the Family Erysipelotrichaceae, Genus Faecalitalea that was differentially abundant even at a relaxed FDR threshold of 0.25 (data not shown).
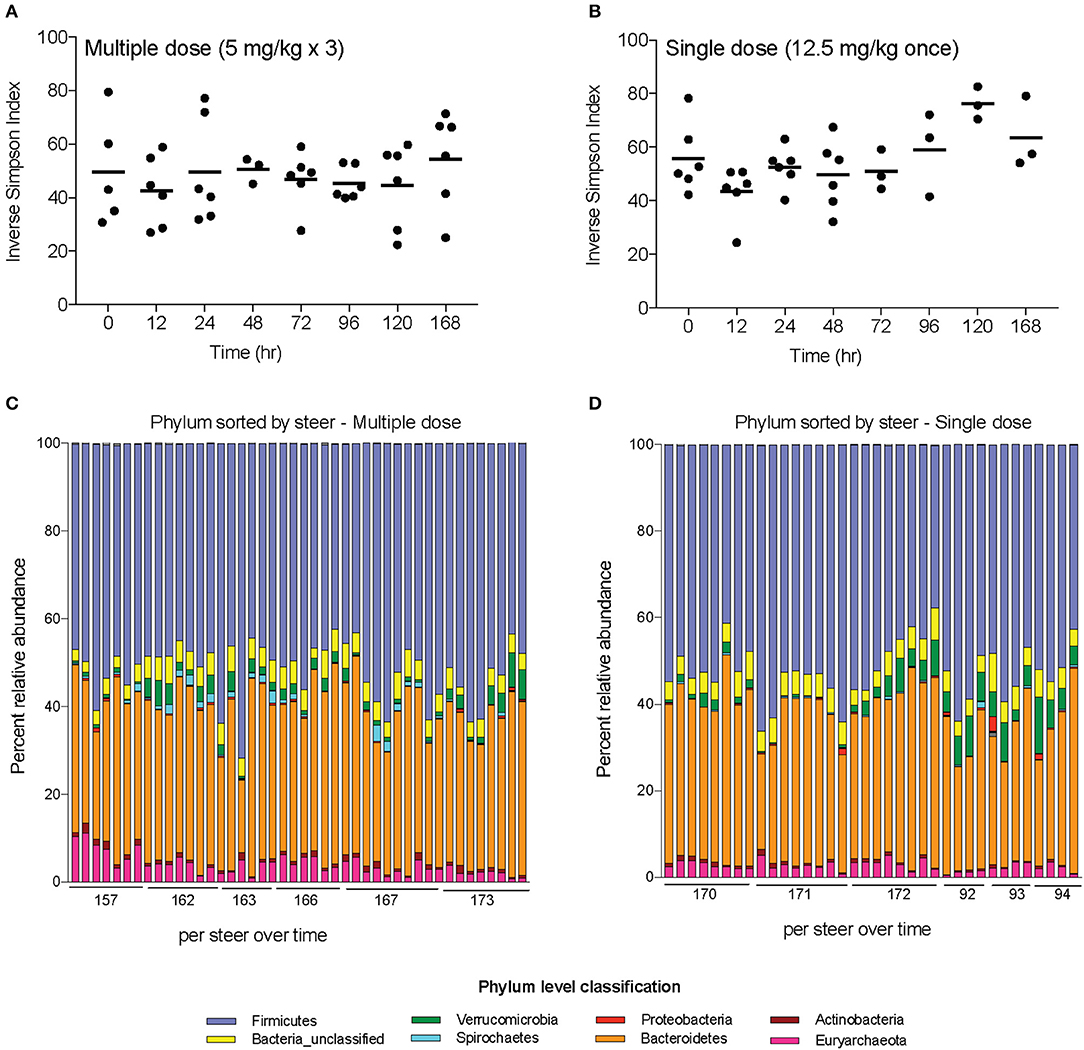
Figure 4. Changes in diversity and bacterial community membership in steers before, during, and after treatment with enrofloxacin. Alpha-diversity measurements in steers treated with (A) multiple and (B) single doses of antibiotics. Bar plots depict the mean percent abundances of the top bacterial Phyla in steers treated with (C) multiple and (D) single doses of antibiotics.
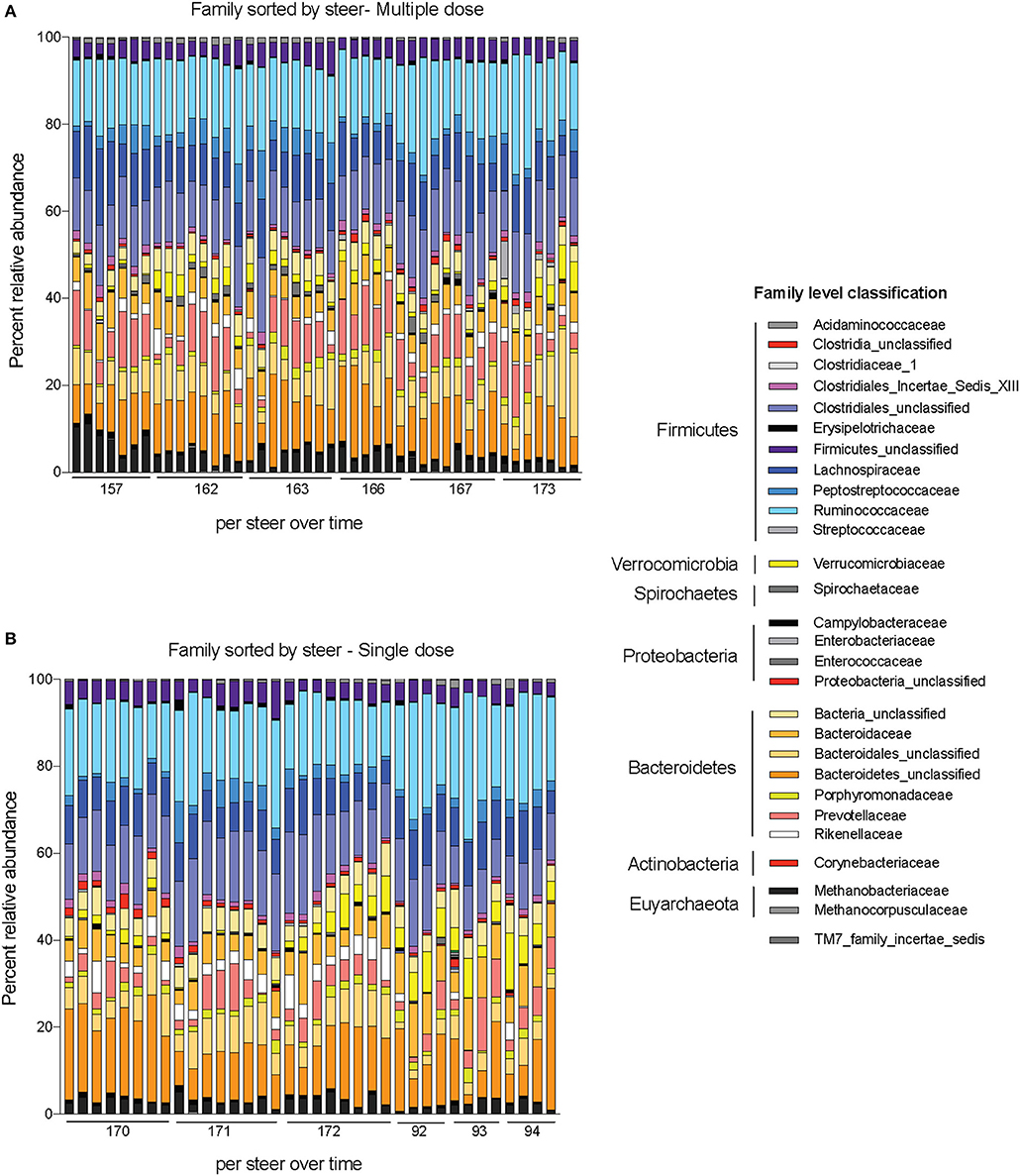
Figure 5. Changes to bacterial community membership in steers before, during, and after treatment with enrofloxacin. Bar plots depict the mean percent abundances of the top bacterial Families in steers treated with (A) multiple and (B) single doses of antibiotics.

Table 3. The relative abundances of each 16S rRNA gene sequence variant at 0 h (before treatment) and at 48 h (after treatment) were compiled for each steer.
Discussion
Enrofloxacin is commonly used for treatment of bovine respiratory disease (National Animal Health Monitoring System, 2011), but little is known about the relative impact of the two dosing regimens on AMR in fecal bacteria. In this study, subcutaneous administration of this fluoroquinolone at either dose resulted in high concentrations of active drug in the ileum and colon. Ciprofloxacin penetrates the ISF to a much larger extent than enrofloxacin. Our earlier studies showed that lower protein binding for ciprofloxacin (19 ± 5 vs. 46 ± 4%) might account for this difference in diffusion from plasma to interstitial fluid (Davis et al., 2007). However, this difference in protein binding does not appear to have the same effect on the penetration of enrofloxacin from the plasma to the intestine as enrofloxacin penetration was 450–490% into the ileum and 280–380% into the colon, similar to our previous study (Foster et al., 2016). This suggests that there may be a transporter protein or biliary excretion responsible for the high penetration of these fluoroquinolones into the intestine relative to both the plasma and ISF. Active secretion of fluoroquinolones into the intestine (Griffiths et al., 1993, 1994; Dautrey et al., 1999) and other tissues (Pulido et al., 2006; Alvarez et al., 2008; Mulgaonkar et al., 2013) has been demonstrated in other studies. Further, the difference in the AUC in the colon was greater than would be expected based on the different doses, indicating that this transport mechanism may be somewhat concentration dependent.
The concentration of E. coli shows an inverse relationship with the concentration of enrofloxacin, as the single dose quickly lowered the E. coli concentration by 5 log10 CFU/g of feces. By 96 h post-treatment, E. coli fecal concentrations returned back to baseline for both the single and multiple dose regimens. This precipitous decline and rapid re-establishment of E. coli concentrations with low MIC values toward enrofloxacin is consistent with the “inverted U” concept to describe the effects of fluoroquinolone concentration on bacterial resistance (Tam et al., 2007). With high fluoroquinolone exposure, as we observed in the intestine of these cattle, E. coli were significantly, but temporarily suppressed (Figure 2A). By 96 h after treatment, the median MIC of the isolates from the single dose regimen reverted to the MIC of a wild-type population as the E. coli concentration rebounded. The return to baseline concentration and MIC corresponded with elimination of the drug from the intestine. In the multiple dose group, the median MIC increased to a greater degree than in the single dose group. Again, the change we observed in MIC was temporary, suggesting a lack of fitness for bacteria with a high MIC in the intestine of these cattle. This time for return to a wild-type MIC range is well within the slaughter withdrawal time of 28 days (Weatherbee, 2012) established by the FDA to prevent drug residues. Our results indicates that when FDA-approved slaughter withdrawal times are observed for enrofloxacin, it may mitigate the risk of transfer of resistant E. coli isolates through the food supply at slaughter. As we only assessed the MIC of E. coli in this study, it is unclear if these findings can be extrapolated to other enteric bacteria.
While evaluating changes to the complete fecal microbiome, we demonstrated only minor changes in community structure after treatment, and there was no change in microbial diversity. The transient changes that were seen were primarily in bacteria predominantly associated with the rumen microbiota (Prevotellaceae, Rikenellaceae, Christensenellaceae, and Erysipelotrichaceae; Mao et al., 2015). Further, the relatively small, short-lived changes found in steers treated with enrofloxacin is in contrast to findings in chickens (Li et al., 2017), mice (Choo et al., 2017), and humans (Stewardson et al., 2015; Pop et al., 2016) administered a fluoroquinolone that demonstrate significant changes at the phylum and genus levels which persist for a week or more. Potentially, the large microbial population of the rumen provides a source of microbes to continually repopulate the intestinal environment, leading to minimal changes in the fecal microbiota in ruminants compared to simple-stomached animals. A similar approach to measure active drug concentrations in the rumen and assess the impact of parenteral drugs on the rumen microbiome would be valuable to address this question. Alternatively, differences in the dosing regimens used in these studies compared those used in cattle may also influence these findings as each of these studies administered the drugs for longer durations and by a different route.
The study was conducted in healthy Holstein steers, and the PK of enrofloxacin or associated microbiological changes may not represent that of sick calves. Nonetheless, our results are similar to findings in the National Antimicrobial Monitoring System that suggest resistance to fluoroquinolones in bovine E. coli isolates is rare in cattle at slaughter (Food and Drug Administration, 2018). The PK findings and short duration of changes to fecal bacteria found in this study may help explain the rare incidence of fluoroquinolone-resistant E. coli isolates in cattle at slaughter (1.2% of isolates in beef cattle in 2015). Resistance to fluoroquinolones in Salmonella isolates was similarly uncommon in beef cattle at slaughter, while rates of fluoroquinolone-resistance in Enterococcus spp. and Campylobacter spp. tended to be higher. As we only examined the impact on fecal E. coli, extrapolation of these findings to other fecal bacteria may not be appropriate, and warrants additional investigation.
In conclusion, enrofloxacin penetrates the ileum and colon of cattle at high concentrations with either the single or multiple dosing regimens causing a significant reduction in fecal E. coli concentration that resolved within 4 days of treatment. With the multiple dose regimen, there was a small increase in median MIC. Yet, the median MIC returned to baseline by 2 weeks post-treatment. Assessment of the microbiome after treatment did not show clinically significant changes with either dosing regimen. From this study, it appears that dosing regimens in which antimicrobial concentrations rapidly achieve pharmacodynamic targets and are then quickly eliminated from the GI tract will minimize the effect on the fecal microbiota of cattle.
Author Contributions
MJ, CT, MP, and DF contributed to study design, data collection and analysis, and manuscript preparation. KF, BC, and TP contributed to data collection and analysis, and manuscript preparation.
Conflict of Interest Statement
DF, MJ, and MP have received research funding from and consulted for Bayer Animal Health.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by work performed by The University of Michigan Microbial Systems Molecular Biology Laboratory. This research was supported in part by United States Department of Agriculture National Institute of Food and Agriculture, Animal Health (project 1007631).
References
Alvarez, A. I., Pérez, M., Prieto, J. G., Molina, A. J., Real, R., and Merino, G. (2008). Fluoroquinolone efflux mediated by ABC transporters. J. Pharm. Sci. 97, 3483–3493. doi: 10.1002/jps.21233
Benedict, K. M., Gow, S. P., Checkley, S., Booker, C. W., McAllister, T. A., and Morley, P. S. (2013). Methodological comparisons for antimicrobial resistance surveillance in feedlot cattle. BMC Vet. Res. 9:216. doi: 10.1186/1746-6148-9-216
Benedict, K. M., Gow, S. P., Reid-Smith, R. J., Booker, C. W., McAllister, T. A., and Morley, P. S. (2014). Latent class comparison of test accuracy when evaluating antimicrobial susceptibility using disk diffusion and broth microdilution to test Escherichia coli and Mannheimia haemolytica isolates recovered from beef feedlot cattle. Epidemiol. Infect. 142, 2314–2325. doi: 10.1017/S0950268813003300
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300.
Callahan, B. J., McMurdie, P. J., and Holmes, S. P. (2017). Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 11, 2639–2643. doi: 10.1038/ismej.2017.119
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Checkley, S. L., Campbell, J. R., Chirino-Trejo, M., Janzen, E. D., and Waldner, C. L. (2010). Associations between antimicrobial use and the prevalence of antimicrobial resistance in fecal Escherichia coli from feedlot cattle in western canada. Can. Vet. J. 51, 853–861.
Choo, J. M., Kanno, T., Zain, N. M., Leong, L. E., Abell, G. C., Keeble, J. E., et al. (2017). Divergent relationships between fecal microbiota and metabolome following distinct antibiotic-induced disruptions. mSphere 2:e00005–17. doi: 10.1128/mSphere.00005-17
Clinical Laboratory Standards Institute, CLSI. (2013). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals; approved Standard, 4th Edn. Wayne, PA: Clinical Laboratory Standards Institute, CLSI.
Dautrey, S., Felice, K., Petiet, A., Lacour, B., Carbon, C., and Farinotti, R. (1999). Active intestinal elimination of ciprofloxacin in rats: modulation by different substrates. Br. J. Pharmacol. 127, 1728–1734. doi: 10.1038/sj.bjp.0702703
Davis, J. L., Foster, D. M., and Papich, M. G. (2007). Pharmacokinetics and tissue distribution of enrofloxacin and its active metabolite ciprofloxacin in calves. J. Vet. Pharmacol. Ther. 30, 564–571. doi: 10.1111/j.1365-2885.2007.00914.x
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Enne, V. I. (2010). Reducing antimicrobial resistance in the community by restricting prescribing: can it be done? J. Antimicrob. Chemother. 65, 179–182. doi: 10.1093/jac/dkp443
Ferran, A. A., Bibbal, D., Pellet, T., Laurentie, M., Gicquel-Bruneau, M., Sanders, P., et al. (2013). Pharmacokinetic/pharmacodynamic assessment of the effects of parenteral administration of a fluoroquinolone on the intestinal microbiota: comparison of bactericidal activity at the gut versus the systemic level in a pig model. Int. J. Antimicrob. Agents 42, 429–435. doi: 10.1016/j.ijantimicag.2013.07.008
Food Drug Administration (2018). NARMS Now. Rockville, MD: US Department of Health and Human Services. Available online at: https://www.fda.gov/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/ucm416741.htm (Accessed May, 2018).
Foster, D. M., Jacob, M. E., Warren, C. D., and Papich, M. G. (2016). Pharmacokinetics of enrofloxacin and ceftiofur in plasma, interstitial fluid, and gastrointestinal tract of calves after subcutaneous injection, and bactericidal impacts on representative enteric bacteria. J. Vet. Pharmacol. Ther. 39, 62–71. doi: 10.1111/jvp.12236
Griffiths, N. M., Hirst, B. H., and Simmons, N. L. (1993). Active secretion of the fluoroquinolone ciprofloxacin by human intestinal epithelial caco-2 cell layers. Br. J. Pharmacol. 108, 575–576. doi: 10.1111/j.1476-5381.1993.tb12844.x
Griffiths, N. M., Hirst, B. H., and Simmons, N. L. (1994). Active intestinal secretion of the fluoroquinolone antibacterials ciprofloxacin, norfloxacin and pefloxacin; a common secretory pathway? J. Pharmacol. Exp. Ther. 269, 496–502.
Hog, B. B., and Korsgaard, H. (2017). DANMAP 2016; Use of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria From Food Animals, Food and Humans in Denmark.
Kanwar, N., Scott, H. M., Norby, B., Loneragan, G. H., Vinasco, J., McGowan, M., et al. (2013). Effects of ceftiofur and chlortetracycline treatment strategies on antimicrobial susceptibility and on tet(A), tet(B), and bla CMY-2 resistance genes among E. coli isolated from the feces of feedlot cattle. PLoS ONE 8:e80575. doi: 10.1371/journal.pone.0080575
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13
Li, J., Hao, H., Cheng, G., Liu, C., Ahmed, S., Shabbir, M. A. B., et al. (2017). Microbial shifts in the intestinal microbiota of salmonella infected chickens in response to enrofloxacin. Front. Microbiol. 8:1711. doi: 10.3389/fmicb.2017.01711
Lindecrona, R. H., Friis, C., and Nielsen, J. P. (2000). Pharmacokinetics and penetration of danofloxacin into the gastrointestinal tract in healthy and in Salmonella typhimurium infected pigs. Res. Vet. Sci. 68, 211–216. doi: 10.1053/rvsc.1999.0361
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Mao, S., Zhang, M., Liu, J., and Zhu, W. (2015). Characterising the bacterial microbiota across the gastrointestinal tracts of dairy cattle: membership and potential function. Sci. Rep. 5:16116. doi: 10.1038/srep16116
Martinez, M. N., Papich, M. G., and Drusano, G. L. (2012). Dosing regimen matters: the importance of early intervention and rapid attainment of the pharmacokinetic/pharmacodynamic target. Antimicrob. Agents Chemother. 56, 2795–2805. doi: 10.1128/AAC.05360-11
McKellar, Q., Gibson, I., Monteiro, A., and Bregante, M. (1999). Pharmacokinetics of enrofloxacin and danofloxacin in plasma, inflammatory exudate, and bronchial secretions of calves following subcutaneous administration. Antimicrob. Agents Chemother. 43, 1988–1992.
McMurdie, P. J., and Holmes, S. (2014). Waste not, want not: Why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 10:e1003531. doi: 10.1371/journal.pcbi.1003531
Messenger, K. M., Papich, M. G., and Blikslager, A. T. (2012). Distribution of enrofloxacin and its active metabolite, using an in vivo ultrafiltration sampling technique after the injection of enrofloxacin to pigs. J. Vet. Pharmacol. Ther. 35, 452–459. doi: 10.1111/j.1365-2885.2011.01338.x
Morley, P. S., Dargatz, D. A., Hyatt, D. R., Dewell, G. A., Patterson, J. G., Burgess, B. A., et al. (2011). Effects of restricted antimicrobial exposure on antimicrobial resistance in fecal Escherichia coli from feedlot cattle. Foodborne Pathog. Dis. 8, 87–98. doi: 10.1089/fpd.2010.0632
Mulgaonkar, A., Venitz, J., Gründemann, D., and Sweet, D. H. (2013). Human organic cation transporters 1 (SLC22A1), 2 (SLC22A2), and 3 (SLC22A3) as disposition pathways for fluoroquinolone antimicrobials. Antimicrob. Agents Chemother. 57, 2705–2711. doi: 10.1128/AAC.02289-12
National Animal Health Monitoring System (2011). Beef Feedlot. Fort Collins, CO: USDA-APHIS-VS-CEAH-NAHMS.
Pop, M., Paulson, J. N., Chakraborty, S., Astrovskaya, I., Lindsay, B. R., Li, S., et al. (2016). Individual-specific changes in the human gut microbiota after challenge with enterotoxigenic Escherichia coli and subsequent ciprofloxacin treatment. BMC Genomics 17:440. doi: 10.1186/s12864-016-2777-0
Pruesse, E., Quast, C., Knittel, K., Fuchs, B. M., Ludwig, W., Peplies, J., et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196. doi: 10.1093/nar/gkm864
Pulido, M. M., Molina, A. J., Merino, G., Mendoza, G., Prieto, J. G., and Alvarez, A. I. (2006). Interaction of enrofloxacin with breast cancer resistance protein (BCRP/ABCG2): influence of flavonoids and role in milk secretion in sheep. J. Vet. Pharmacol. Ther. 29, 279–287. doi: 10.1111/j.1365-2885.2006.00744.x
Quail, M. A., Kozarewa, I., Smith, F., Scally, A., Stephens, P. J., Durbin, R., et al. (2008). A large genome center's improvements to the illumina sequencing system. Nat. Methods 5, 1005–1010. doi: 10.1038/nmeth.1270
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Schmidt, J. W., Griffin, D., Kuehn, L. A., and Brichta-Harhay, D. M. (2013). Influence of therapeutic ceftiofur treatments of feedlot cattle on fecal and hide prevalences of commensal Escherichia coli resistant to expanded-spectrum cephalosporins, and molecular characterization of resistant isolates. Appl. Environ. Microbiol. 79, 2273–2283. doi: 10.1128/AEM.03592-12
Seekatz, A. M., Theriot, C. M., Molloy, C. T., Wozniak, K. L., Bergin, I. L., and Young, V. B. (2015). Fecal microbiota transplantation eliminates clostridium difficile in a murine model of relapsing disease. Infect. Immun. 83, 3838–3846. doi: 10.1128/IAI.00459-15
Stewardson, A. J., Gaïa, N., François, P., Malhotra-Kumar, S., Delémont, C., Martinez de Tejada, B., et al. (2015). Collateral damage from oral ciprofloxacin versus nitrofurantoin in outpatients with urinary tract infections: a culture-free analysis of gut microbiota. Clin. Microbiol. Infect. 21, 344.e1–344.e11. doi: 10.1016/j.cmi.2014.11.016
Tam, V. H., Louie, A., Deziel, M. R., Liu, W., and Drusano, G. L. (2007). The relationship between quinolone exposures and resistance amplification is characterized by an inverted U: a new paradigm for optimizing pharmacodynamics to counterselect resistance. Antimicrob. Agents Chemother. 51, 744–747. doi: 10.1128/AAC.00334-06
TerHune, T. N., Skogerboe, T. L., Shostrom, V. K., and Weigel, D. J. (2005). Comparison of pharmacokinetics of danofloxacin and enrofloxacin in calves challenged with Mannheimia haemolytica. Am. J. Vet. Res. 66, 342–349. doi: 10.2460/ajvr.2005.66.342
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Warren, C. D., Prange, T., Campbell, N. B., Gerard, M. P., Martin, L. G., Jacob, M. E., et al. (2014). Implantation of an ultrafiltration device in the ileum and spiral colon of steers to continuously collect intestinal fluid. Res. Vet. Sci. 97, 611–615. doi: 10.1016/j.rvsc.2014.10.012
Weatherbee, T. G. (2012). Baytril 100 (Enrofloxacin) 100 mg/ml Antimicrobial Injectable Solution. Shawnee Mission, KS: Bayer Animal Health.
Zaheer, R., Cook, S. R., Klima, C. L., Stanford, K., Alexander, T., Topp, E., et al. (2013). Effect of subtherapeutic vs. therapeutic administration of macrolides on antimicrobial resistance in Mannheimia haemolytica and enterococci isolated from beef cattle. Front. Microbiol. 4:133. doi: 10.3389/fmicb.2013.00133
Keywords: antimicrobial resistance, fluoroquinolone, cattle, microbiome, pharmacokinetics
Citation: Ferguson KM, Jacob ME, Theriot CM, Callahan BJ, Prange T, Papich MG and Foster DM (2018) Dosing Regimen of Enrofloxacin Impacts Intestinal Pharmacokinetics and the Fecal Microbiota in Steers. Front. Microbiol. 9:2190. doi: 10.3389/fmicb.2018.02190
Received: 24 May 2018; Accepted: 27 August 2018;
Published: 19 September 2018.
Edited by:
Etienne Giraud, Institut National de la Recherche Agronomique de Toulouse, FranceReviewed by:
Peter Damborg, University of Copenhagen, DenmarkLudovic Pelligand, Royal Veterinary College, United Kingdom
Copyright © 2018 Ferguson, Jacob, Theriot, Callahan, Prange, Papich and Foster. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Derek M. Foster, ZGVyZWtfZm9zdGVyQG5jc3UuZWR1
 Kaitlyn M. Ferguson1
Kaitlyn M. Ferguson1 Benjamin J. Callahan
Benjamin J. Callahan Derek M. Foster
Derek M. Foster