Corrigendum: Adenoviromics: Mining the Human Adenovirus Species D Genome
- 1Howe Laboratory, Massachusetts Eye and Ear, Harvard Medical School, Boston, MA, United States
- 2Molecular Virology Laboratory, Korea Zoonosis Research Institute, Jeonbuk National University, Jeonju, South Korea
- 3Department of Cell and Systems Biology, University of Toronto, Toronto, ON, Canada
- 4Department of Microbiology and Immunology, University of Oklahoma Health Sciences Center, Oklahoma City, OK, United States
- 5Bioinformatics and Computational Biology Program, School of Systems Biology, George Mason University, Manassas, VI, United States
Human adenovirus (HAdV) infections cause disease world-wide. Whole genome sequencing has now distinguished 90 distinct genotypes in 7 species (A-G). Over half of these 90 HAdVs fall within species D, with essentially all of the HAdV-D whole genome sequences generated in the last decade. Herein, we describe recent new findings made possible by mining of this expanded genome database, and propose future directions to elucidate new functional elements and new functions for previously known viral components.
Introduction
Human adenovirus (HAdV) infections represent a significant source of morbidity and mortality, world-wide and at all ages, through highly transmittable infections at mucosal sites, including the eye, and urinary, respiratory, and gastrointestinal tracts (Horwitz, 1996). HAdV causes fatal acute respiratory distress syndrome in healthy adults and is especially lethal in infants and the immune compromised (Bhanthumkosol, 1998; Ryu et al., 2003; Wallot et al., 2006; Engelmann et al., 2016; Tan et al., 2016; Zhang et al., 2016). No FDA-approved therapy for acute HAdV infection is available. At resolution of acute infection, persistence may develop within nasopharyngeal lymphoid tissue (Neumann et al., 1987; Garnett et al., 2002, 2009; Zhang et al., 2010; Assadian et al., 2016), as yet uncharacterized cells in the gastrointestinal tract (Roy et al., 2009), and possibly the ocular surface (Kaye et al., 2005), permitting evolution of new HAdVs through homologous recombination between two or more HAdVs infecting the same cell(s) (Lee et al., 2005, 2018; Echavarria et al., 2006; McCarthy et al., 2009; Seto et al., 2010).
HAdVs are divided phylogenetically into seven species (A-G), with a total of 90 recognized genotypes with whole genome sequences in GenBank, including the original 51 “serotypes”—determined by serum neutralization—which now all have been fully sequenced (Table 1) (Robinson et al., 2013a) Human adenovirus species D (HAdV-D) is the largest and most rapidly growing among all HAdV species, and contains viruses associated with epidemic keratoconjunctivitis (EKC), a severe, hyperacute ocular surface infection (Butt and Chodosh, 2006). A collaboration funded by the American Recovery and Reinvestment Act of 2009 came to fruition with the complete whole genome sequencing and analysis of all previously unsequenced HAdV-D serotypes (Robinson et al., 2013a), leading to a new understanding of adenovirus ontogeny (Jones et al., 2007; Robinson et al., 2008, 2009a,b; Robinson et al., 2011b,c; Robinson et al., 2013a,b; Walsh et al., 2009, 2010a,b; Arnold et al., 2010; Torres et al., 2010; Dehghan et al., 2011, 2013a,b; Walsh et al., 2011; Liu et al., 2011; Seto et al., 2011, 2013; Singh et al., 2012, 2013; Zhou et al., 2012)—including those HAdV-Ds associated with EKC (Robinson et al., 2008, 2009b, 2011b; Walsh et al., 2009; Zhou et al., 2012)—and ultimately to a new typing system for HAdV based on genomics (Seto et al., 2011).
Recent published work demonstrates how genome “mining,” in-depth analyses of the growing HAdV genome database, can bring about new realizations and add critical new information to prior ones. The trimeric fiber protein on adenoviruses mediates viral entry through interaction of the distal most “knob” structure on the fiber with host cell receptors. In a phylogenetic analysis of HAdV-D fiber genes, HAdV-D types associated with EKC were recently shown to form a unique clade (Ismail et al., 2016). By proteotyping, a new in silico methodology described in detail below, EKC virus-associated fiber knobs were uniquely shared, and signature amino acid positions distinguished EKC from non-EKC types. Remarkably, human corneal epithelial cell tropism could be predicted by the presence of a lysine or alanine at residue 240, and this amino acid residue in EKC viruses showed evidence for positive selection. These data added to the prior observation by Huang and coworkers that artificial mutation to a lysine at residue 240 in a non-EKC virus could confer infection of Chang cells, a conjunctiva derived continuous cell line (Huang et al., 1999). However, because Chang cells came later known to be contaminated by HeLa cells, the importance of residue 240 to ocular tropism was until this new observation, in some doubt.
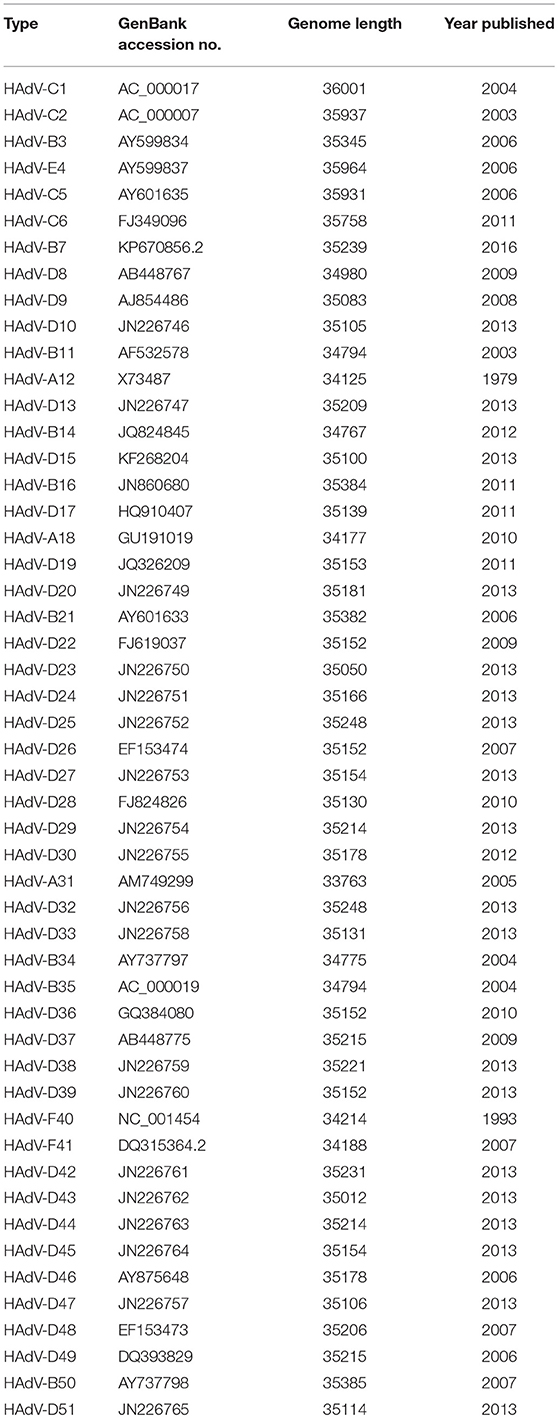
Another recently published effort provided further evidence of the importance and potential for HAdV genome mining. Late adenoviral gene expression is initiated by the adenovirus major late promoter (Ramke et al., 2017), followed by splicing of mRNAs to the viral tripartite leader for translation (Chow et al., 1977; Akusjärvi and Pettersson, 1978; Chow and Broker, 1978; Logan and Shenk, 1984). The HAdV tripartite leader is a 200-nucleotide 5' noncoding region that circumvents the requirement for eukaryotic initiation factor 4F or cap binding protein complex (Ziff and Evans, 1978; Akusjärvi and Pettersson, 1979; Dolph et al., 1988; Zhang et al., 1989), and permits translation of HAdV mRNAs at late times in infection when cap-dependent translation is blocked due to shut down of host cellular cap-dependent mRNA translation. HAdV 5′ untranslated regions (5′UTRs) are critical for cap-independent initiation, and impact mRNA localization and stability. The HAdV tripartite leader (TPL), composed of three introns (TPL 1-3), drives translation of HAdV late mRNA. The annotation of 72 HAdV genotypes for the HAdV TPL and another previously described leader, the i-leader, let to identification of newly identified polycistronic mRNAs for RID-α and RID-β within the E3 transcription unit, and a potential new open reading frame (ORF) within the i-leader sequence, with termination of this potential protein in TPL3 (Ramke et al., 2017). In addition, the authors also identified a potential new leader sequence embedded within the E3 region, tentatively named the j-leader (Figure 1).
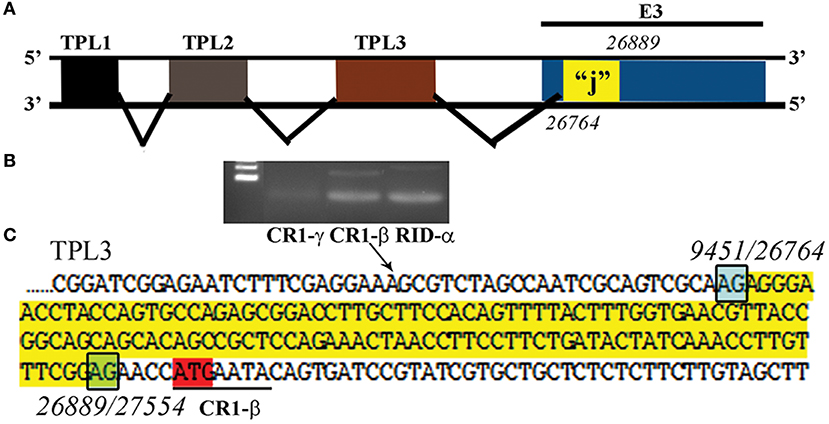
Figure 1. Putative “j”-leader located within the CR1-α E3 gene. (A) Schematic for the location of a newly detected leader (“j”-leader) embedded within the E3 CRI-α gene, experimentally determined to be spliced to some, but not all mRNAs of the E3 genes. (B) Gel photomicrograph of mRNA transcripts amplified with forward primer from TPL1 and reverse primers from CR1-γ, CR1-β, and RID-α. Primers were chosen to elicit similarly sized bands to facilitate subsequent sequencing. (C) Nucleotide sequence of the PCR product for CR1-β. The putative j-leader sequence and splice sites are shown in yellow and green, respectively. Note an additional 4 nucleotide 5′UTR (AACC) prior to the CR1-β start site (red). The 5′UTR in (C) prior to the splice site for the j-leader is from TPL3. Adapted from Ramke et al. (2017) with permission.
Structure and Infection
The HAdV is non-enveloped, icosahedral in shape, and contains a double stranded DNA genome of ~36,000 base pairs (bp) with ~1 open reading frame (ORF) for every 1000 nucleotides. Viral DNA is associated with four (interior) core proteins including Mu, VII, V, and terminal protein. The histone-like protein (p) VII protects viral DNA from cellular DNA damage responses (Lischwe and Sung, 1977; Karen and Hearing, 2011; Avgousti et al., 2017). The outer protein coat (capsid) of the virus consists of 240 hexon capsomers and 12 penton capsomers, along with several minor capsid proteins. The latter include pVI, pIIIa, pVIII, and pIX and are important to capsid stability. Each penton capsomer contains a ring of five penton base proteins which bind and support the trimeric fiber protein with its distal fiber knob. During viral infection, the fiber knob binds to one of several host cell receptors (Nemerow, 2000; Goosney and Nemerow, 2003; Nemerow et al., 2009). The penton base protein contains two hypervariable loops. The interaction between fiber knob and a host cell receptor brings about secondary contact between the hypervariable loop 2 (HVL2) arginine-glycine-aspartic acid (RGD) motif in each penton base protein (five per penton base capsomer) with host cell integrins αvβ3, αvβ5, and αvβ1, that in turn induce endocytosis of the virus (Li et al., 1998a,b; Li et al., 2000). HAdV structural proteins can serve multiple functions. For example, the minor capsid structural protein VI (pVI) plays a critical role in at least three distinct aspects of the viral “life” cycle: endosomal escape during cell entry, nuclear assembly during viral replication, and stability of the intact, infectious virus outside the host (Wodrich et al., 2003; Wiethoff et al., 2005; Moyer et al., 2011, 2016). These findings suggest that, as with pVI, other HAdV structural proteins may have multiple functions yet to be elucidated.
Genomics and Evolution
The relatively large genome database for HAdV-D (over 50 unique viruses with available whole genome sequences) (Tables 1, 2) has permitted detailed analyses of genome relationships within this clinically important adenovirus species. HAdV-D genomes are highly conserved (>90%). However, whole genome analyses of HAdV-D have revealed specific loci of genetic hypervariability in the hexon, penton base, fiber, and E3 CR1α, β, and γ genes (Figure 2), dictating nonsynonymous amino acid changes in corresponding proteins (Figure 3). GC content confers genome stability and resistance to recombination (Gruss et al., 1991), and the genomes of HAdV-D have among the highest GC content among HAdV species (~56%). The hypervariable regions in HAdV-D were found to be sharply reduced in GC nucleotide content relative to the rest of the genome (Robinson et al., 2013a). Mutations in HAdV are relatively infrequent, with genome stability now documented in some types across decades (Hofmayer et al., 2009; Mahadevan et al., 2010; Seto et al., 2010; Dehghan et al., 2013b; Robinson et al., 2013a; Alkhalaf et al., 2015). However, those regions of the genome shown to be hypervariable and relatively low in GC content are the very same also shown to undergo homologous recombination (Robinson et al., 2009a, 2011b; Walsh et al., 2009; Zhou et al., 2012; Singh et al., 2013), driving the evolution of new genotypes.
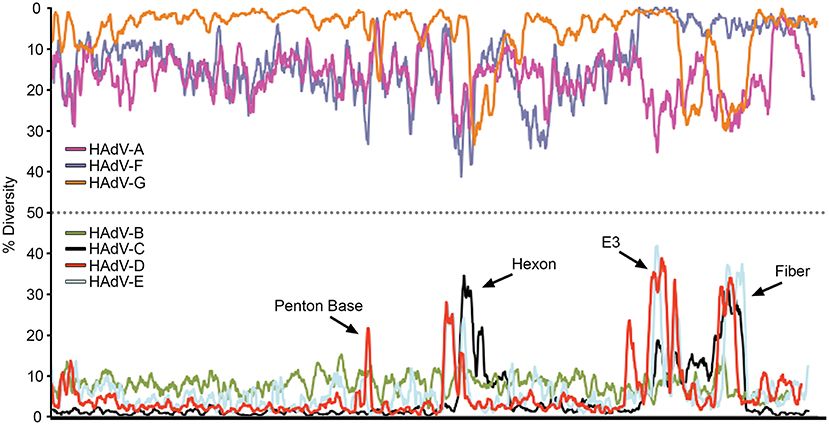
Figure 2. Nucleotide diversity plots, by HAdV species, generated with DnaSP, represent the average number of nucleotide differences per site between each type in every HAdV species. The % diversity is calculated on the y-axis; the x-axis illustrates the nucleotide position on the genome. HAdV-Ds (red line on bottom half of plot) show particular diversity in the penton base, hexon, E3, and fiber coding regions, with otherwise very high conservation. From Robinson et al. (2013a) with permission.
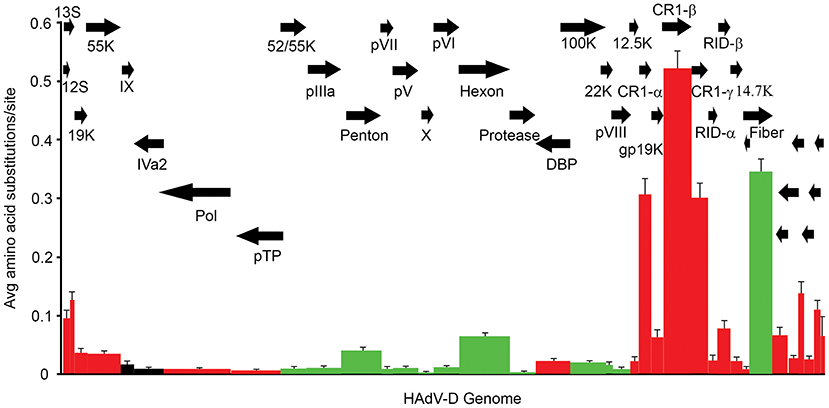
Figure 3. Amino acid diversity calculated in MEGA 4.02, measuring the average amino acid substitution for each HAdV-D protein. Each bar in the graph corresponds to a protein as represented by arrows. Red = early genes. Dark green = late genes. Black = intermediate genes. The hypervariable loops of penton base and hexon proteins were also analyzed separately (light green) and showed particularly high levels of amino acid substitutions. From Robinson et al. (2013a) with permission.
Adenoviruses recombine specifically during viral replication (Williams et al., 1975; Meinschad and Winnacker, 1980; Munz et al., 1983), and do so by both homologous and heterologous mechanisms (Young et al., 1984; Epstein and Young, 1991; Crawford-Miksza and Schnurr, 1996). However, the evidence for homologous recombination as the major mechanism driving HAdV-D evolution is unassailable (Robinson et al., 2013a; Singh et al., 2013). Specifically, recombination occurs in the two penton base hypervariable regions (these code for two hypervariable loops (HVLs) on the penton base protein, separated from one another by ~125 conserved amino acids), seven hexon hypervariable regions (these are closely adjacent in the hexon gene and determine two adjacent HVLs on the hexon protein), fiber (fiber gene and protein are entirely hypervariable), and E3 CR1α, β, and γ (each also entirely hypervariable). For homologous recombination between two HAdVs to occur, at least two virus types with high nucleotide sequence homology at corresponding locations in both genomes must co-infect the same cell, and viral DNA replication should be ongoing. Co-infection by two or more HAdVs has been well documented (Lee et al., 2005; Echavarria et al., 2006; Vora et al., 2006; McCarthy et al., 2009; Halstead et al., 2010; Seto et al., 2010), as has the presence of two HAdV types in archived clinical samples (Singh et al., 2012).
“Proteotyping” is a novel approach to the study of genome evolution (Obenauer et al., 2006), and has been applied to characterize recombination among HAdV-D (Robinson et al., 2013a; Singh et al., 2013). In this method, maximum likelihood trees are used to align amino acid sequences of hypervariable, frequently recombined proteins. Each amino acid is assigned a unique, arbitrary color. Consensus residues are colored white, and gaps in the alignment are colored black. A threshold of <10% sequence divergence is used to distinguish unique proteotypes. An example of proteotyping is shown (Figure 4), comparing an amino acid alignment from E3 14.7K, a highly conserved gene with one distinct proteotype, with the hypervariable E3 CR1α (Singh et al., 2013), with six distinct proteotypes observed among 38 HAdV-Ds. E3 14.7K is therefore not hypervariable and not recombinant. E3 CR1α is hypervariable and recombinant.
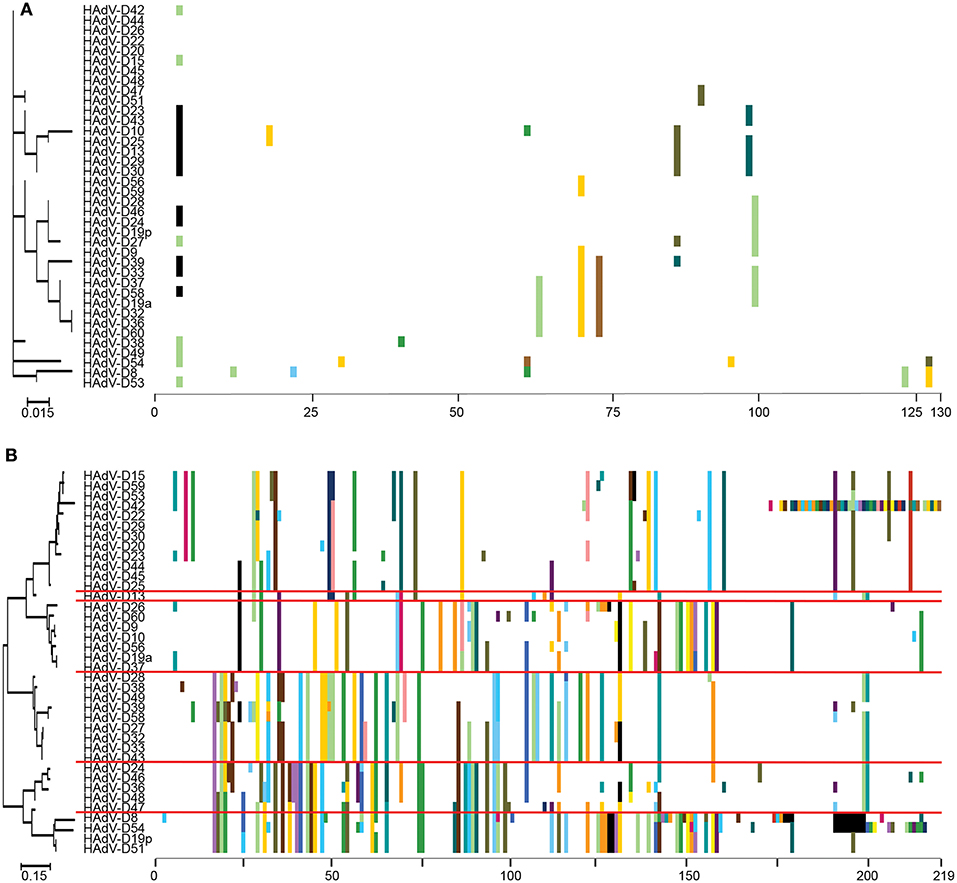
Figure 4. Proteotyping analysis comparing the HAdV-D E3 14.7K (A) and CR1α (B) proteins. The 14.7K protein was conserved, while CR1α demonstrated 6 unique proteotypes. Maximum likelihood phylogenetic trees are shown to the left for each putative protein, and amino acid signatures to the right. The scale bar at the bottom left of each sub-figure denotes the phylogenetic distance reflected in horizontal dimension of the respective tree. To construct the amino acid signatures shown, each amino acid was assigned a unique color (upper right corner), consensus amino acids at each position across all 38 viruses were assigned white, and gaps in the alignment were colored black. Horizontal red lines delineate distinct proteotypes. Adapted from. Singh et al. (2013) with permission.
Another way to interpret the analyses for those proteins like E3 CR1α, with more than one proteotype is that those proteotypes containing more than one HAdV type have previously recombined in nature, while those proteotypes with only one HAdV type are those that have not (yet) been shown to recombine in nature. HAdV-D37 and 29 fall within different hexon proteotypes (Figure 5). HAdV-D37 shares a hexon proteotype with HAdV-D13 and 30 (Robinson et al., 2013a), while HAdV-D29 shares a hexon proteotype with HAdV-D15, 56, and 69 (Singh et al., 2015). These two hexon proteotypes therefore have undergone prior homologous recombination. In contrast, the hexon proteins of HAdV-D10 and 28 are each in a proteotype with only one member; hexon recombination for these two viruses has therefore not yet been documented in nature. In sum, these data show by independent means that homologous recombination within HAdV-D is common, and confirm previously recognized patterns of homologous recombination among HAdV-D (Robinson et al., 2009a, 2011b, 2013a,b; Walsh et al., 2009, 2010a; Singh et al., 2012, 2013; Zhou et al., 2012; Gonzalez et al., 2014).
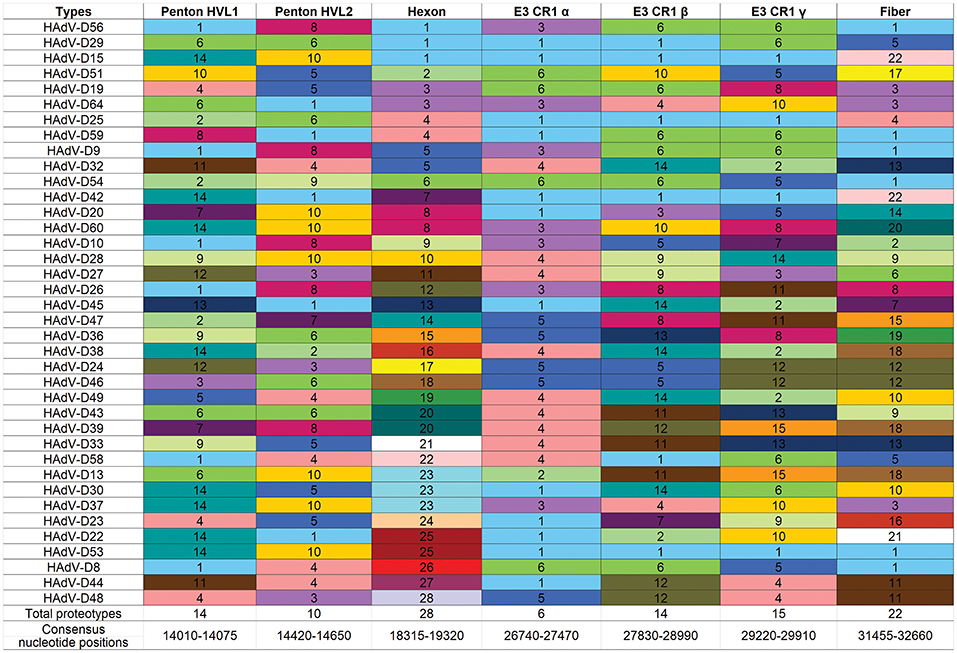
Figure 5. Proteotyping for 38 HAdV-Ds, sorted for the hexon proteotype column. Numbers and colors are arbitrary, and distinguish distinct proteotypes. Recombinants can be identified by rows. For example, HadV-D56, -D29, and -D15 fall within the same proteotype and are predicted to share highly similar nucleotide sequences for their respective hexon hypervariable regions (as confirmed by Singh et al., 2015). For HAdV-D29 and –D15, the recombination event extended through the E3 CR1β ORF gene and then ended.
The local sequence and/or structure of DNA in regions flanking recombinogenic sites is significant for directing cellular recombination machinery to those regions. In bacteria, a signal for recombination between homologous DNA is the crossover hotspot instigator, or Chi nucleotide sequence. This was first discovered in bacteriophage lambda, then in bacterial DNA, and later shown to mediate recombination between them (Stahl, 1998). The Chi sequence in E. coli (ChiEC) is 5′-GCTGGTGG-3′ (Smith et al., 1981; Bianco and Kowalczykowski, 1997), and its presence induces the conversion of the RecBCD enzyme from a helicase to an exonuclease, producing ssDNA that can invade homologous dsDNA during recombination (Taylor et al., 1985). The RecA enzyme of E. coli is loaded onto unwound ssDNA by RecBCD and promotes ssDNA exchange/recombination with homologous dsDNA (Cox, 1999; Smith, 2012). RecA has significant homology to eukaryotic Rad51 and its paralogs (Suwaki et al., 2011), enzymes that repair dsDNA breaks in human cells, facilitate homologous recombination, and during adenovirus infection, bind to the E2 DNA binding protein (Tookman et al., 2016). In our study of the region just 5′ to HVL2 on the penton base gene, a recombination hot-spot for HAdV-D (Robinson et al., 2009a), we found Chi-like sequences (ChiAD), e.g., 5′-ACTTCTGA-3′ in the proteotype containing HAdV-D64, and 5′-TCTCCTGA-3′ in the proteotype including HAdV-D37 (Lee et al., 2018). The putative ChiAD sequences we identified in HAdV-D were found within the GC-rich component of GC/AT transition zones that precede and include HVL2, and were conserved within each proteotype. In vitro, E. coli lysates containing RecA protein increased recombination of two HAdV-D genotypes with the same penton base HVL2 proteotype. RecA was shown by ChIP to bind specifically to ChiAD nucleotide sequence in the same regions, and also colocalize with adenovirus DNA within infected cell nuclei. These data suggest that Chi-like nucleotide sequences adjacent to the junction of conserved and hypervariable gene segments in HAdV-D may be an important signal for homologous recombination, and provide evidence in support of the idea that local bacterial flora might enhance natural recombination through Chi-like nucleotide sequences at HAdV-D recombination hotspots.
Another explanation for homologous recombination between HAdV, not exclusive of a role for ChiAD, is the potential for GC-low (AT-rich) single stranded DNA (ssDNA) to form hairpin loops (Nagy and Bujarski, 1997; Ohshima et al., 2007), a physical nonlinearity that would facilitate binding of ssDNA of one HAdV-D type to a homologous segment of ss or dsDNA from a physically adjacent but different HAdV-D type during co-infection of the same cell. Hairpin loops and other alterations in the physical configuration of ssDNA during DNA replication might also contribute to polymerase jumping (Jennings et al., 1983; Spaan et al., 1983; Pääbo et al., 1990; Viswanathan et al., 1999), in which physical constraints to polymerization lead to translocation of the DNA polymerase to an adjacent DNA from a different virus, resulting in a recombined DNA. Polymerase jumping has been shown to occur during HAdV DNA replication (King et al., 1997; de Jong et al., 2003), although it has not been suggested previously as a mechanism for HAdV-D evolution. Analysis of 38 HAdV-D whole genome sequences identified instances of 15 nucleotide-long GC-rich sequence adjacent to 15 nucleotide-long AT-rich sequence (sometimes with a 15 or 30 nucleotide-long GC-moderate sequence intervening), located just 5′ and 3′ to frequently recombined gene segments, and which were shown by in silico analysis of their corresponding ssDNA to form hairpin loops (Robinson et al., 2013a). Taken together, these data suggest covariant effects of nucleotide sequence and ssDNA secondary structures on homologous recombination between two HAdV-Ds.
Transcriptome
Regions of the HAdV-D genome currently thought to be “noncoding,” may contain functional elements. Because viruses exist on the nano-scale, viral genomes are by necessity constrained by size, and “junk” nucleotide sequences represent an extravagance. The National Human Genome Research Institute project to identify functional elements in the human genome (Encyclopedia of DNA Elements, or ENCODE) identified functionality in much of the human genome previously without known utility (Consortium et al., 2007; Qu and Fang, 2013; Kellis et al., 2014) The double-stranded DNA genomes of HAdV also contain regions with no known function. Transcriptional profiling of host gene expression has been studied after HAdV infection (Dorer et al., 2011) However, although viral transcriptomes have been reported for several viruses, most notably dengue, varicella zoster, and Epstein-Barr viruses (Ortmann et al., 2008; Ertl et al., 2011; Nagel et al., 2011, 2013; Arvey et al., 2013; Sujayanont et al., 2014), a de novo HAdV transcriptome has not been reported. Wu and coworkers used deep RNA sequencing to confirm known bat AdV transcripts (Wu et al., 2013), but did not investigate “noncoding” regions. In silico ORF prediction in HAdV can be difficult due to splice variants and inconsistencies in banked gene annotations (Davison et al., 2003), but in a prior annotation of HAdV-D37, ~60 new additional ORFs were predicted using in combination, the NCBI ORF finder, TIGR annotation engine, and GeneMark Heuristic model (Robinson et al., 2008) Putative genes were found within the large regions of noncoding DNA on the complementary strand opposite to established HAdV genes (Figure 6), in smaller regions on the coding strand within established transcription units but between confirmed genes, and overlapping or completely within established genes. Work is in progress in our laboratories to identify putative new genomic elements in HAdV by high-throughput sequencing of the viral transcriptome of HAdV-D37.
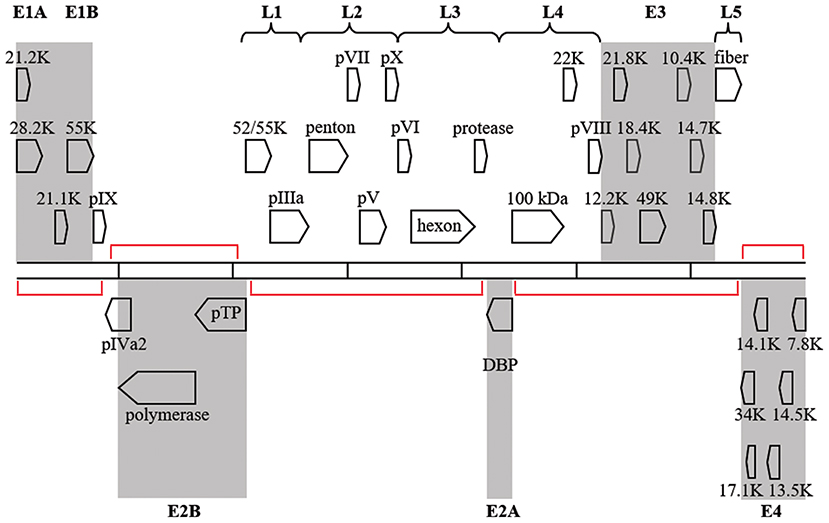
Figure 6. Transcription map for HAdV-D37. Genes are divided by early (shaded) or late expression. Red brackets denote large areas of “noncoding” DNA, but many additional, smaller, potential coding regions exist between and within known genes. Adapted from Robinson et al. (2008) with permission.
Transcription Factor Binding Sites
HAdV uses host TFs nuclear factor I and III (NF-I and NF-III) as part of the viral DNA replication complex (Pruijn et al., 1988; Mul et al., 1990; Hatfield and Hearing, 1991, 1993) Simian AdVs typically lack the NF-I binding site, while human viruses express it. It was previously reported that HAdV-E4, originally isolated in 1952, is a product of recombination between HAdV-B16 and the simian AdV, SAdV-E26. Clinical strains of HAdV-E4 isolated recently contain a NF-I binding site in the inverted terminal repeat (Houng et al., 2006; Dehghan et al., 2013a,b) that is absent in the original isolate (Purkayastha et al., 2005), suggesting that NF-I binding may be important to viral fitness in humans. To further elucidate mechanisms of viral gene expression, we are exploring novel TF binding sites on HAdV-D DNA, using ENCODE validated methodologies (Gerstein et al., 2012; Landt et al., 2012).
Protein Interactomes
The major HAdV capsid structural proteins—hexon, penton base, and fiber—interact directly with extracellular mediators of host immunity (Gahéry-Ségard et al., 1998; Molinier-Frenkel et al., 2002; Schoggins and Falck-Pedersen, 2006; Tamanini et al., 2006; Kalyuzhniy et al., 2008; Chintakuntlawar et al., 2010; Bradshaw et al., 2012; Flatt et al., 2013). The hexon, penton base, and fiber proteins also exhibit distinct amino acid signatures, characterizing discrete proteotypes (Robinson et al., 2013a). Gene products from the E3 transcription unit of HAdV function in viral immune evasion (Horwitz, 2004; Lichtenstein et al., 2004b; Windheim et al., 2004). In HAdV-D, the open reading frames for three of eight E3 genes—CR1α, CR1β, and CR1γ–are uniquely hypervariable compared to the other ORFs within the E3 transcription unit, and also segregate into discrete proteotypes (Singh et al., 2013). Highly conserved genes, such as DNA binding protein, DNA polymerase, and E3 14.7K, show no such variability (Robinson et al., 2013a; Singh et al., 2013).
While it may be assumed that hypervariablity in major capsid and E3 proteins is driven through evolutionary selection by the extracellular interactome, amino acid differences in a hypervariable protein can also lead to differences in that protein's intracellular interactome, the set of intrinsic host cell proteins which network with the viral protein, as was recently confirmed for E3 CR1 genes across HAdV species (Martinez-Martin et al., 2016). Viral capsid structural proteins are critical to virion stability. For the nonenveloped HAdV, fiber and penton base proteins on the external surface of the capsid serve as ligands for attachment to the host cell (Huang et al., 1999) and initiate viral entry (Wickham et al., 1993), respectively. HAdVs are typically internalized via endosomes. Endosomal acidification leads to structural instability of the capsid and endosomal release into the cytosol. HAdV capsid is then transported by microtubules to the nuclear membrane. Viral DNA then enters the nucleus through nuclear pores, leaving almost all the viral structural proteins in the cytosol (Henaff et al., 2011). Viral capsid proteins within the cell are eventually targeted for ubiquitination (Ko et al., 2010; Marvin and Wiethoff, 2012; Horan et al., 2013; Li et al., 2013) and degraded (Greber et al., 1993), but there are many opportunities for interaction with intracellular host cellular proteins during entry, trafficking, translation, assembly, and egress. Penton base HVL2, with its RGD motif, is critical to viral internalization through the interaction with host cell integrins,; (Wickham et al., 1993) but function of penton base HVL1 is unknown, and might be revealed though knowledge of its protein interactome. The closely adjacent hexon HVL1 and 2 form the epsilon epitope that determines serum neutralization, and interactions between the hexon protein and serum coagulation factor X confers liver tropism to HAdV-C5 (Sumarheni et al., 2014). However, nothing is known about potential hexon interactions with intracellular proteins during infection.
The E3 transcription unit of HAdV codes for proteins that mediate immune evasion by the virus (Horwitz, 2004). Although E3 is labeled as an early transcription region, its transcripts are expressed both early and late during viral infection (Chow et al., 1977; Chow and Broker, 1978; Bhat and Wold, 1986), and there is evidence for at least one E3 protein that late transcripts are translated (Robinson et al., 2011a). E3 gene products are not required for viral replication in cultured cells (Morin et al., 1987), but inhibit cellular and cytokine mediated host immune responses to infection (Horwitz, 2004; Lichtenstein et al., 2004b; Windheim et al., 2004). Almost all of what is known about the function of specific E3 proteins derives from studies on HAdV-C. For example, HAdV-C2 E3 CR1α directs another E3 protein (19K) to the endoplasmic reticulum of cytotoxic T cells (Wilson-Rawls et al., 1994), where 19K binds to and retains MHC class I proteins (Jefferies and Burgert, 1990), preventing presentation of viral peptides within MHC class I at the cell surface (Burgert and Kvist, 1985, 1987; Andersson et al., 1987; Burgert et al., 1987; Cox et al., 1991). CR1α, RIDα, and RIDβ proteins cooperate to evade TNFα-related apoptosis through TRAIL (Elsing and Burgert, 1998; Tollefson et al., 1998; Benedict et al., 2001; Lichtenstein et al., 2004a). CR1β (Wold et al., 1984), also called the adenovirus death protein (Tollefson et al., 1992), is required for cell lysis (Tollefson et al., 1996) and viral spread (Doronin et al., 2003). The ORF size of each E3 gene varies across HAdV species (Figure 7) (Robinson et al., 2011c). Similarly, immune evasion functions of E3 gene products may not be the same across HAdV species, or function similarly in all cell types (Routes and Cook, 1990). Windheim and coworkers recently showed that the CR1β protein of the eye pathogen HAdV-D64 suppresses natural killer cell function (Windheim et al., 2013). The E3 CR1 genes are uniquely hypervariable within HAdV-D, and as predicted, overlapping but distinguishable intracellular interactomes across proteotypes were recently reported by Martinez-Martin and colleagues, who used protein microarrays to identify novel CR1β binding partners (Martinez-Martin et al., 2016).
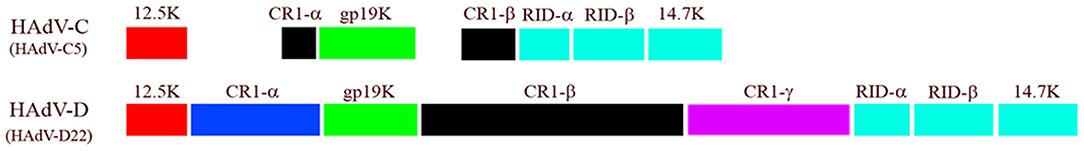
Figure 7. Comparison of E3 transcription unit from HAdV-C and -D. Note in particular the difference in ORF size between CR1β of the two HAdV species. Adapted from Robinson et al. (2011c) with permission.
Conclusions
HAdV was critical to the dual discoveries of viral oncogenesis and RNA splicing (Berget et al., 1977; Chow et al., 1977; Whyte et al., 1988). HAdV is also a significant agent of disease for which there is no approved treatment. Recent mining of HAdV genomes has been highly productive, and there is ample evidence to suggest that further whole genome analyses will elucidate new and fundamental mechanisms in HAdV biology. In the last decade, of 27 newly identified HAdVs, 19 were HAdV-Ds, suggesting the continuing evolution of new pathogens from species D. Analyses of 38 fully sequenced HAdV-D whole genomes identified homologous recombination of specific regions within the hexon, penton base, fiber, and E3 CR1 genes as the major mechanism behind HAdV-D evolution, a new finding (Robinson et al., 2013a; Singh et al., 2013). Stereotypical reductions in GC content at the junction of conserved and hypervariable regions, along with Chi-like sequence motifs (also a new finding), appear likely to augment the intrinsic tendency of HAdV to undergo homologous recombination in vivo (Lee et al., 2018).
Recently, the whole genome sequences of 85 HAdVs from archives and current collections were determined, including both historical and circulating strains, respectively (Ismail et al., 2018). Of these, 3 novel recombinants within HAdV-B and 15 within HAdV-D were identified. Only two of the 15 HAdV-Ds were found to contain novel genes (penton base and fiber); these were subsequently typed as HAdV-D71 and 72. Isolates of HAdV-D53 and HAdV-D58, two novel genotypes recently recognized, were also identified, adding confidence in their clinical importance. Fully genotyped HAdVs now number 90, with more awaiting type numbers, and the scientific community has a 10-fold larger database of unique HAdV genomes than available only 15 years ago. Published and validated ENCODE methodologies can now be applied, and comparisons made across disparate HAdV genomes. We suggest that the HAdV genome contains previously uncharacterized functional elements, and that every HAdV protein has pleiotropic interactions. Current technologies should afford a wave of new and important discoveries that may lead to needed therapies against adenoviral diseases.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was funded by National Institutes of Health grants EY013124, EY021558, and EY014104, a Senior Scientific Investigator Award grant (to JC) from Research to Prevent Blindness, Inc., New York, NY, The Falk Foundation, and the Massachusetts Lions Eye Research Fund.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Akusjärvi, G., and Pettersson, U. (1978). Nucleotide sequence at the junction between the coding region of the adenovirus 2 hexon messenger RNA and its leader sequence. Proc. Natl. Acad. Sci. U.S.A. 75, 5822–5826. doi: 10.1073/pnas.75.12.5822
Akusjärvi, G., and Pettersson, U. (1979). Sequence analysis of adenovirus DNA: complete nucleotide sequence of the spliced 5' noncoding region of adenovirus 2 hexon messenger RNA. Cell 16, 841–850. doi: 10.1016/0092-8674(79)90099-0
Alkhalaf, M. A., Guiver, M., and Cooper, R. J. (2015). Genome stability of adenovirus types 3 and 7 during a simultaneous outbreak in Greater Manchester, UK. J. Med. Virol. 87, 117–124. doi: 10.1002/jmv.23969
Andersson, M., McMichael, A., and Peterson, P. A. (1987). Reduced allorecognition of adenovirus-2 infected cells. J. Immunol. 138, 3960–3966.
Arnold, J., Jánoska, M., Kajon, A. E., Metzgar, D., Hudson, N. R., Torres, S., et al. (2010). Genomic characterization of human adenovirus 36, a putative obesity agent. Virus. Res. 149, 152–161. doi: 10.1016/j.virusres.2010.01.011
Arvey, A., Tempera, I., and Lieberman, P. M. (2013). Interpreting the Epstein-Barr Virus (EBV) epigenome using high-throughput data. Viruses 5, 1042–1054. doi: 10.3390/v5041042
Assadian, F., Sandström, K., Bondeson, K., Laurell, G., Lidian, A., Svensson, C., et al. (2016). Distribution and molecular characterization of human adenovirus and epstein-barr virus infections in tonsillar lymphocytes isolated from patients diagnosed with tonsillar diseases. PLoS ONE 11:e0154814. doi: 10.1371/journal.pone.0154814
Avgousti, D. C., Della Fera, A. N., Otter, C. J., Herrmann, C., Pancholi, N. J., and Weitzman, M. D. (2017). Adenovirus core protein VII down-regulates the DNA damage response on the host genome. J Virol. 91:e01089–17. doi: 10.1128/JVI.01089-17
Benedict, C. A., Norris, P. S., Prigozy, T. I., Bodmer, J. L., Mahr, J. A., Garnett, C. T., et al. (2001). Three adenovirus E3 proteins cooperate to evade apoptosis by tumor necrosis factor-related apoptosis-inducing ligand receptor-1 and−2. J. Biol. Chem. 276, 3270–3278. doi: 10.1074/jbc.M008218200
Berget, S. M., Moore, C., and Sharp, P. A. (1977). Spliced segments at the 5' terminus of adenovirus 2 late mRNA. Proc. Natl. Acad. Sci. U. S. A. 74, 3171–3175. doi: 10.1073/pnas.74.8.3171
Bhanthumkosol, D. (1998). Fatal adenovirus infections in infants probably infected with HIV. J. Med. Assoc. Thai. 81, 214–222.
Bhat, B. M., and Wold, W. S. (1986). Genetic analysis of mRNA synthesis in adenovirus region E3 at different stages of productive infection by RNA-processing mutants. J. Virol. 60, 54–63.
Bianco, P. R., and Kowalczykowski, S. C. (1997). The recombination hotspot Chi is recognized by the translocating RecBCD enzyme as the single strand of DNA containing the sequence 5'-GCTGGTGG-3'. Proc. Natl. Acad. Sci. U. S. A. 94, 6706–6711. doi: 10.1073/pnas.94.13.6706
Bradshaw, A. C., Coughlan, L., Miller, A. M., Alba, R., van Rooijen, N., Nicklin, S. A., et al. (2012). Biodistribution and inflammatory profiles of novel penton and hexon double-mutant serotype 5 adenoviruses. J Control Release 164, 394–402. doi: 10.1016/j.jconrel.2012.05.025
Burgert, H. G., and Kvist, S. (1985). An adenovirus type 2 glycoprotein blocks cell surface expression of human histocompatibility class I antigens. Cell 41, 987–997. doi: 10.1016/S0092-8674(85)80079-9
Burgert, H. G., and Kvist, S. (1987). The E3/19K protein of adenovirus type 2 binds to the domains of histocompatibility antigens required for CTL recognition. EMBO J. 6, 2019–2026.
Burgert, H. G., Maryanski, J. L., and Kvist, S. (1987). “E3/19K” protein of adenovirus type 2 inhibits lysis of cytolytic T lymphocytes by blocking cell-surface expression of histocompatibility class I antigens. Proc. Natl. Acad. Sci. U.S.A. 84, 1356–1360. doi: 10.1073/pnas.84.5.1356
Butt, A. L., and Chodosh, J. (2006). Adenoviral keratoconjunctivitis in a tertiary care eye clinic. Cornea 25, 199–202. doi: 10.1097/01.ico.0000170693.13326.fb
Chintakuntlawar, A. V., Zhou, X., Rajaiya, J., and Chodosh, J. (2010). Viral capsid is a pathogen-associated molecular pattern in adenovirus keratitis. PLoS Pathog. 6:e1000841. doi: 10.1371/journal.ppat.1000841
Chow, L. T., and Broker, T. R. (1978). The spliced structures of adenovirus 2 fiber message and the other late mRNAs. Cell 15, 497–510. doi: 10.1016/0092-8674(78)90019-3
Chow, L. T., Gelinas, R. E., Broker, T. R., and Roberts, R. J. (1977). An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA. Cell 12, 1–8. doi: 10.1016/0092-8674(77)90180-5
Consortium, E. P., Birney, E., Stamatoyannopoulos, J. A., Dutta, A., Guigó, R., Gingeras, T. R., et al. (2007). Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816. doi: 10.1038/nature05874
Cox, J. H., Bennink, J. R., and Yewdell, J. W. (1991). Retention of adenovirus E19 glycoprotein in the endoplasmic reticulum is essential to its ability to block antigen presentation. J. Exp. Med. 174, 1629–1637. doi: 10.1084/jem.174.6.1629
Cox, M. M. (1999). Recombinational DNA repair in bacteria and the RecA protein. Prog. Nucleic Acid Res. Mol. Biol. 63, 311–366. doi: 10.1016/S0079-6603(08)60726-6
Crawford-Miksza, L. K., and Schnurr, D. P. (1996). Adenovirus serotype evolution is driven by illegitimate recombination in the hypervariable regions of the hexon protein. Virology 224, 357–367. doi: 10.1006/viro.1996.0543
Davison, A. J., Benko, M., and Harrach, B. (2003). Genetic content and evolution of adenoviruses. J. Gen. Virol. 84, 2895–2908. doi: 10.1099/vir.0.19497-0
de Jong, R. N., van der Vliet, P. C., and Brenkman, A. B. (2003). Adenovirus DNA replication: protein priming, jumping back and the role of the DNA binding protein DBP. Curr. Top. Microbiol. Immunol. 272, 187–211. doi: 10.1007/978-3-662-05597-7_7
Dehghan, S., Seto, J., Hudson, N. R., Robinson, C. M., Jones, M. S., Dyer, D. W., et al. (2011). Complete genome sequence of human adenovirus prototype 17. J. Virol. 85, 11540–11541. doi: 10.1128/JVI.06051-11
Dehghan, S., Seto, J., Jones, M. S., Dyer, D. W., Chodosh, J., and Seto, D. (2013a). Simian adenovirus type 35 has a recombinant genome comprising human and simian adenovirus sequences, which predicts its potential emergence as a human respiratory pathogen. Virology 447, 265–273. doi: 10.1016/j.virol.2013.09.009
Dehghan, S., Seto, J., Liu, E. B., Walsh, M. P., Dyer, D. W., Chodosh, J., et al. (2013b). Computational analysis of four human adenovirus type 4 genomes reveals molecular evolution through two interspecies recombination events. Virology 443, 197–207. doi: 10.1016/j.virol.2013.05.014
Dolph, P. J., Racaniello, V., Villamarin, A., Palladino, F., and Schneider, R. J. (1988). The adenovirus tripartite leader may eliminate the requirement for cap-binding protein complex during translation initiation. J. Virol. 62, 2059–2066.
Dorer, D. E., Holtrup, F., Fellenberg, K., Kaufmann, J. K., Engelhardt, S., and Hoheiselet, J. D. al. (2011). Replication and virus-induced transcriptome of HAdV-5 in normal host cells versus cancer cells–differences of relevance for adenoviral oncolysis. PLoS ONE 6, e27934. doi: 10.1371/journal.pone.0027934
Doronin, K., Toth, K., Kuppuswamy, M., Krajcsi, P., Tollefson, A. E., and Wold, W. S. (2003). Overexpression of the ADP (E3-11.6K) protein increases cell lysis and spread of adenovirus. Virology 305, 378–387. doi: 10.1006/viro.2002.1772
Echavarria, M., Maldonado, D., Elbert, G., Videla, C., Rappaport, R., and Carballal, G. (2006). Use of PCR to demonstrate presence of adenovirus species B, C, or F as well as coinfection with two adenovirus species in children with flu-like symptoms. J. Clin. Microbiol. 44, 625–627. doi: 10.1128/JCM.44.2.625-627.2006
Elsing, A., and Burgert, H. G. (1998). The adenovirus E3/10.4K-14.5K proteins down-modulate the apoptosis receptor Fas/Apo-1 by inducing its internalization. Proc. Natl. Acad. Sci. U.S.A. 95, 10072–10077. doi: 10.1073/pnas.95.17.10072
Engelmann, I., Coiteux, V., Heim, A., Magro, L., Dewilde, A., Dulery, R., et al. (2016). Severe adenovirus pneumonia followed by bacterial septicaemia: relevance of co-infections in allogeneic hematopoietic stem cell transplantation. Infect Disord Drug Targets 16, 69–76. doi: 10.2174/1871526516666160407114623
Epstein, L. H., and Young, C. S. (1991). Adenovirus homologous recombination does not require expression of the immediate-early E1a gene. J. Virol. 65, 4475–4479.
Ertl, R., Birzele, F., Hildebrandt, T., and Klein, D. (2011). Viral transcriptome analysis of feline immunodeficiency virus infected cells using second generation sequencing technology. Vet. Immunol. Immunopathol. 143, 314–324. doi: 10.1016/j.vetimm.2011.06.010
Flatt, J. W., Kim, R., Smith, J. G., Nemerow, G. R., and Stewart, P. L. (2013). An intrinsically disordered region of the adenovirus capsid is implicated in neutralization by human alpha defensin 5. PLoS ONE 8:e61571. doi: 10.1371/journal.pone.0061571
Gahéry-Ségard, H., Farace, F., Godfrin, D., Gaston, J., Lengagne, R., Tursz, T., et al. (1998). Immune response to recombinant capsid proteins of adenovirus in humans: antifiber and anti-penton base antibodies have a synergistic effect on neutralizing activity. J. Virol. 72, 2388–2397.
Garnett, C. T., Erdman, D, Xu, W., and Gooding, L. R. (2002). Prevalence and quantitation of species C adenovirus DNA in human mucosal lymphocytes. J. Virol. 76, 10608–16. doi: 10.1128/JVI.76.21.10608-10616.2002
Garnett, C. T., Talekar, G., Mahr, J. A., Huang, W., Zhang, Y., Ornelles, D. A., et al. (2009). Latent species C adenoviruses in human tonsil tissues. J. Virol. 83, 2417–2428. doi: 10.1128/JVI.02392-08
Gerstein, M. B., Kundaje, A., Hariharan, M., Landt, S. G., Yan, K. K., Cheng, C., et al. (2012). Architecture of the human regulatory network derived from ENCODE data. Nature 489, 91–100. doi: 10.1038/nature11245
Gonzalez, G., Koyanagi, K. O., Aoki, K., Kitaichi, N., Ohno, S., Kaneko, H., et al. (2014). Intertypic modular exchanges of genomic segments by homologous recombination at universally conserved segments in human adenovirus species D. Gene 547, 10–17. doi: 10.1016/j.gene.2014.04.018
Goosney, D. L., and Nemerow, G. R. (2003). Adenovirus infection: taking the back roads to viral entry. Curr. Biol. 13, R99–R100. doi: 10.1016/S0960-9822(03)00037-X
Greber, U. F., Willetts, M., Webster, P., and Helenius, A. (1993). Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75, 477–486. doi: 10.1016/0092-8674(93)90382-Z
Gruss, A., Moretto, V., Ehrlich, S. D., Duwat, P., and Dabert, P. (1991). GC-rich DNA sequences block homologous recombination in vitro. J. Biol. Chem. 266, 6667–6669.
Halstead, D. C., Gray, G. C., Meyer, K. S., Stanciu, S. R., and Gorospe, W. C. (2010). Recombinant adenovirus (AdV) Type 3 and Type 14 isolated from a fatal case of pneumonia. Rev. Med. Microbiol. 21, 28–30. doi: 10.1097/MRM.0b013e3283393523
Hatfield, L., and Hearing, P. (1991). Redundant elements in the adenovirus type 5 inverted terminal repeat promote bidirectional transcription in vitro and are important for virus growth in vivo. Virology 184, 265–276. doi: 10.1016/0042-6822(91)90843-Z
Hatfield, L., and Hearing, P. (1993). The NFIII/OCT-1 binding site stimulates adenovirus DNA replication in vivo and is functionally redundant with adjacent sequences. J. Virol. 67, 3931–3939.
Henaff, D., Salinas, S., and Kremer, E. J. (2011). An adenovirus traffic update: from receptor engagement to the nuclear pore. Future Microbiol. 6, 179–192. doi: 10.2217/fmb.10.162
Hofmayer, S., Madisch, I., Darr, S., Rehren, F., and Heim, A. (2009). Unique sequence features of the human adenovirus 31 complete genomic sequence are conserved in clinical isolates. BMC Genom. 10:557. doi: 10.1186/1471-2164-10-557
Horan, K. A., Hansen, K., Jakobsen, M. R., Holm, C. K., Søby, S., Unterholzner, L., et al. (2013). Proteasomal degradation of herpes simplex virus capsids in macrophages releases DNA to the cytosol for recognition by DNA sensors. J. Immunol. 190, 2311–2319. doi: 10.4049/jimmunol.1202749
Horwitz, M. S. (1996). “Adenoviruses,” in Fields Virology, eds B. N. Fields, D. M. Knipe, P. M. Howley (Philadelphia, PA: Lippincott-Raven), 2149–2171
Horwitz, M. S. (2004). Function of adenovirus E3 proteins and their interactions with immunoregulatory cell proteins. J. Gene. Med. 6 (Suppl 1), S172–S183. doi: 10.1002/jgm.495
Houng, H. S., Clavio, S., Graham, K., Kuschner, R., Sun, W., Russell, K. L., et al. (2006). Emergence of a new human adenovirus type 4 (Ad4) genotype: identification of a novel inverted terminal repeated (ITR) sequence from majority of Ad4 isolates from US military recruits. J. Clin. Virol. 35, 381–387. doi: 10.1016/j.jcv.2005.11.008
Huang, S., Reddy, V., Dasgupta, N., and Nemerow, G. R. (1999). A single amino acid in the adenovirus type 37 fiber confers binding to human conjunctival cells. J. Virol. 73, 2798–2802.
Ismail, A. M., Cui, T., Dommaraju, K., Singh, G., Dehghan, S., Seto, J., et al. (2018). Genomic analysis of a large set of currently-and historically-important human adenovirus pathogens. Emerg. Microbes Infect. 7:10. doi: 10.1038/s41426-017-0004-y
Ismail, A. M., Lee, J. S., Dyer, D. W., Seto, D., Rajaiya, J., and Chodosh, J. (2016). Selection pressure in the human adenovirus fiber knob drives cell specificity in epidemic keratoconjunctivitis. J. Virol. 90, 9598–9607. doi: 10.1128/JVI.01010-16
Jefferies, W. A., and Burgert, H. G. (1990). E3/19K from adenovirus 2 is an immunosubversive protein that binds to a structural motif regulating the intracellular transport of major histocompatibility complex class I proteins. J. Exp. Med. 172, 1653–1664. doi: 10.1084/jem.172.6.1653
Jennings, P. A., Finch, J. T., Winter, G., and Robertson, J. S. (1983). Does the higher order structure of the influenza virus ribonucleoprotein guide sequence rearrangements in influenza viral RNA? Cell 34, 619–627. doi: 10.1016/0092-8674(83)90394-X
Jones, M. S., Harrach, B., Ganac, R. D., Gozum, M. M., Dela Cruz, W. P., Riedel, B., et al. (2007). New adenovirus species found in a patient presenting with gastroenteritis. J. Virol. 81, 5978–5984. doi: 10.1128/JVI.02650-06
Kalyuzhniy, O., Di Paolo, N. C., Silvestry, M., Hofherr, S. E., Barry, M. A., Stewart, P. L., et al. (2008). Adenovirus serotype 5 hexon is critical for virus infection of hepatocytes in vivo. Proc. Natl. Acad. Sci. U.S.A. 105, 5483–5488. doi: 10.1073/pnas.0711757105
Karen, K. A., and Hearing, P. (2011). Adenovirus core protein VII protects the viral genome from a DNA damage response at early times after infection. J. Virol. 85, 4135–4142. doi: 10.1128/JVI.02540-10
Kaye, S. B., Lloyd, M., Williams, H., Yuen, C., Scott, J. A., O'Donnell, N., et al. (2005). Evidence for persistence of adenovirus in the tear film a decade following conjunctivitis. J. Med. Virol. 77, 227–231. doi: 10.1002/jmv.20440
Kellis, M., Wold, B., Snyder, M. P., Bernstein, B. E., Kundaje, A., Marinov, G. K., et al. (2014). Defining functional DNA elements in the human genome. Proc. Natl. Acad. Sci. U.S.A. 111, 6131–6138. doi: 10.1073/pnas.1318948111
King, A. J., Teertstra, W. R., and van der Vliet, P. C. (1997). Dissociation of the protein primer and DNA polymerase after initiation of adenovirus DNA replication. J. Biol. Chem. 272, 24617–24623. doi: 10.1074/jbc.272.39.24617
Ko, A., Lee, E. W., Yeh, J. Y., Yang, M. R., Oh, W., Moon, J. S., et al. (2010). MKRN1 induces degradation of West Nile virus capsid protein by functioning as an E3 ligase. J. Virol. 84, 426–436. doi: 10.1128/JVI.00725-09
Landt, S. G., Marinov, G. K., Kundaje, A., Kheradpour, P., Pauli, F., Batzoglou, S., et al. (2012). ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22, 1813–1831. doi: 10.1101/gr.136184.111
Lee, J. A., Kim, N. H., Kim, S. J., Choi, E. H., and Lee, H. J. (2005). Rapid identification of human adenovirus types 3 and 7 from respiratory specimens via multiplex type-specific PCR. J. Clin. Microbiol. 43, 5509–5514. doi: 10.1128/JCM.43.11.5509-5514.2005
Lee, J. Y., Lee, J. S., Materne, E. C., Rajala, R., Ismail, A. M., Seto, D, et al. (2018). Bacterial RecA protein promotes adenoviral recombination during in vitro infection. mSphere 3:e00105–18. doi: 10.1128/mSphere.00105-18
Li, C., He, Y., Nicolson, S., Hirsch, M., Weinberg, M. S., Zhang, P., et al. (2013). Adeno-associated virus capsid antigen presentation is dependent on endosomal escape. J. Clin. Invest. 123, 1390–1401. doi: 10.1172/JCI66611
Li, E., Stupack, D., Bokoch, G. M., and Nemerow, G. R. (1998a). Adenovirus endocytosis requires actin cytoskeleton reorganization mediated by Rho family GTPases. J. Virol. 72, 8806–8812.
Li, E., Stupack, D., Klemke, R., Cheresh, D. A., and Nemerow, G. R. (1998b). Adenovirus endocytosis via alpha(v) integrins requires phosphoinositide-3-OH kinase. J. Virol. 72, 2055–2061.
Li, E., Stupack, D. G., Brown, S. L., Klemke, R., Schlaepfer, D. D., and Nemerow, G. R. (2000). Association of p130CAS with phosphatidylinositol-3-OH kinase mediates adenovirus cell entry. J. Biol. Chem. 275, 14729–14735. doi: 10.1074/jbc.275.19.14729
Lichtenstein, D. L., Doronin, K., Toth, K., Kuppuswamy, M., Wold, W. S., and Tollefson, A. E. (2004a). Adenovirus E3-6.7K protein is required in conjunction with the E3-RID protein complex for the internalization and degradation of TRAIL receptor 2. J. Virol. 78, 12297–12307. doi: 10.1128/JVI.78.22.12297-12307.2004
Lichtenstein, D. L., Toth, K., Doronin, K., Tollefson, A. E., and Wold, W. S. (2004b). Functions and mechanisms of action of the adenovirus E3 proteins. Int. Rev. Immunol. 23, 75–111. doi: 10.1080/08830180490265556
Lischwe, M. A., and Sung, M. T. (1977). A histone-like protein from adenovirus chromatin. Nature 267, 552–554. doi: 10.1038/267552a0
Liu, E. B., Ferreyra, L., Fischer, S. L., Pavan, J. V., Nates, S. V., Hudson, N. R., et al. (2011). Genetic analysis of a novel human adenovirus with a serologically unique hexon and a recombinant fiber gene. PLoS ONE 6:e24491. doi: 10.1371/journal.pone.0024491
Logan, J., and Shenk, T. (1984). Adenovirus tripartite leader sequence enhances translation of mRNAs late after infection. Proc. Natl. Acad. Sci. U.S.A. 81, 3655–3659. doi: 10.1073/pnas.81.12.3655
Mahadevan, P., Seto, J., Tibbetts, C., and Seto, D. (2010). Natural variants of human adenovirus type 3 provide evidence for relative genome stability across time and geographic space. Virology 397, 113–118. doi: 10.1016/j.virol.2009.10.052
Martinez-Martin, N., Ramani, S. R., Hackney, J. A., Tom, I., Wranik, B. J., Chan, M., et al. (2016). The extracellular interactome of the human adenovirus family reveals diverse strategies for immunomodulation. Nat. Commun. 7:11473. doi: 10.1038/ncomms11473
Marvin, S. A., and Wiethoff, C. M. (2012). Emerging roles for ubiquitin in adenovirus cell entry. Biol. Cell 104, 188–198. doi: 10.1111/boc.201100096
McCarthy, T., Lebeck, M. G., Capuano, A. W., Schnurr, D. P., and Gray, G. C. (2009). Molecular typing of clinical adenovirus specimens by an algorithm which permits detection of adenovirus coinfections and intermediate adenovirus strains. J. Clin. Virol. 46, 80–84. doi: 10.1016/j.jcv.2009.06.008
Meinschad, C., and Winnacker, E. L. (1980). Recombination in adenovirus. I. Analysis of recombinant viruses under non-selective conditions. J. Gen. Virol. 48, 219–224. doi: 10.1099/0022-1317-48-1-219
Molinier-Frenkel, V., Lengagne, R., Gaden, F., Hong, S. S., Choppin, J., Gahery-Ségard, H., et al. (2002). Adenovirus hexon protein is a potent adjuvant for activation of a cellular immune response. J. Virol. 76, 127–135. doi: 10.1128/JVI.76.1.127-135.2002
Morin, J. E., Lubeck, M. D., Barton, J. E., Conley, A. J., Davis, A. R., and Hung, P. P. (1987). Recombinant adenovirus induces antibody response to hepatitis B virus surface antigen in hamsters. Proc. Natl. Acad. Sci. U.S.A. 84, 4626–4630. doi: 10.1073/pnas.84.13.4626
Moyer, C. L., Besser, E. S., and Nemerow, G. R. (2016). A single maturation cleavage site in adenovirus impacts cell entry and capsid assembly. J. Virol. 90, 521–532. doi: 10.1128/JVI.02014-15
Moyer, C. L., Wiethoff, C. M., Maier, O., Smith, J. G., and Nemerow, G. R. (2011). Functional genetic and biophysical analyses of membrane disruption by human adenovirus. J. Virol. 85, 2631–2641. doi: 10.1128/JVI.02321-10
Mul, Y. M., Verrijzer, C. P., and van der Vliet, P. C. (1990). Transcription factors NFI and NFIII/oct-1 function independently, employing different mechanisms to enhance adenovirus DNA replication. J. Virol. 64, 5510–5518.
Munz, P. L., Young, C., and Young, C. S. (1983). The genetic analysis of adenovirus recombination in triparental and superinfection crosses. Virology 126, 576–586. doi: 10.1016/S0042-6822(83)80014-2
Nagel, M. A., Choe, A., Gilden, D., Traina-Dorge, V., Cohrs, R. J., and Mahalingam, R. (2013). GeXPS multiplex PCR analysis of the simian varicella virus transcriptome in productively infected cells in culture and acutely infected ganglia. J. Virol. Methods 193, 151–158. doi: 10.1016/j.jviromet.2013.05.020
Nagel, M. A., Choe, A., Traktinskiy, I., Cordery-Cotter, R., Gilden, D., and Cohrs, R. J. (2011). Varicella-zoster virus transcriptome in latently infected human ganglia. J. Virol. 85, 2276–2287. doi: 10.1128/JVI.01862-10
Nagy, P. D., and Bujarski, J. J. (1997). Engineering of homologous recombination hotspots with AU-rich sequences in brome mosaic virus. J. Virol. 71, 3799–3810.
Nemerow, G. R. (2000). Cell receptors involved in adenovirus entry. Virology 274, 1–4. doi: 10.1006/viro.2000.0468
Nemerow, G. R., Pache, L., Reddy, V., and Stewart, P. L. (2009). Insights into adenovirus host cell interactions from structural studies. Virology 384, 380–388. doi: 10.1016/j.virol.2008.10.016
Neumann, R., Genersch, E., and Eggers, H. J. (1987). Detection of adenovirus nucleic acid sequences in human tonsils in the absence of infectious virus. Virus Res. 7, 93–97. doi: 10.1016/0168-1702(87)90060-8
Obenauer, J. C., Denson, J., Mehta, P. K., Su, X., Mukatira, S., Finkelstein, D. B., et al. (2006). Large-scale sequence analysis of avian influenza isolates. Science 311, 1576–1580. doi: 10.1126/science.1121586
Ohshima, K., Tomitaka, Y., Wood, J. T., Minematsu, Y., Kajiyama, H., Tomimura, K., et al. (2007). Patterns of recombination in turnip mosaic virus genomic sequences indicate hotspots of recombination. J. Gen. Virol. 88, 298–315. doi: 10.1099/vir.0.82335-0
Ortmann, A. C., Brumfield, S. K., Walther, J., McInnerney, K., Brouns, S. J., van de Werken, H. J., et al. (2008). Transcriptome analysis of infection of the archaeon Sulfolobus solfataricus with Sulfolobus turreted icosahedral virus. J. Virol. 82, 4874–4883. doi: 10.1128/JVI.02583-07
Pääbo, S., Irwin, D. M., and Wilson, A. C. (1990). DNA damage promotes jumping between templates during enzymatic amplification. J. Biol. Chem. 265, 4718–4721.
Pruijn, G. J., van Miltenburg, R. T., Claessens, J. A., and van der Vliet, P. C. (1988). Interaction between the octamer-binding protein nuclear factor III and the adenovirus origin of DNA replication. J. Virol. 62, 3092–3102.
Purkayastha, A., Su, J., McGraw, J., Ditty, S. E., Hadfield, T. L., Seto, J., et al. (2005). Genomic and bioinformatics analyses of HAdV-4vac and HAdV-7vac, two human adenovirus (HAdV) strains that constituted original prophylaxis against HAdV-related acute respiratory disease, a reemerging epidemic disease. J. Clin. Microbiol. 43, 3083–3094. doi: 10.1128/JCM.43.7.3083-3094.2005
Qu, H., and Fang, X. (2013). A brief review on the human encyclopedia of DNA elements (ENCODE) project. Genomics Proteomics Bioinformatics 11, 135–141. doi: 10.1016/j.gpb.2013.05.001
Ramke, M., Lee, J. Y., Dyer, D. W., Seto, D., Rajaiya, J., and Chodosh, J. (2017). The 5'UTR in human adenoviruses: leader diversity in late gene expression. Sci. Rep. 7:618. doi: 10.1038/s41598-017-00747-y
Robinson, C. M., Rajaiya, J., Walsh, M. P., Seto, D., Dyer, D. W., Jones, M. S., et al. (2009a). Computational analysis of human adenovirus type 22 provides evidence for recombination among species D human adenoviruses in the penton base gene. J. Virol. 83, 8980–8985. doi: 10.1128/JVI.00786-09
Robinson, C. M., Rajaiya, J., Zhou, X., Singh, G., Dyer, D. W., and Chodosh, J. (2011a). The E3 CR1-gamma gene in human adenoviruses associated with epidemic keratoconjunctivitis. Virus Res. 160, 120–127. doi: 10.1016/j.virusres.2011.05.022
Robinson, C. M., Seto, D., Jones, M. S., Dyer, D. W., and Chodosh, J. (2011c). Molecular evolution of human species D adenoviruses. Infect. Genet. Evol. 11, 1208–1217. doi: 10.1016/j.meegid.2011.04.031
Robinson, C. M., Shariati, F., Gillaspy, A. F., Dyer, D. W., and Chodosh, J. (2008). Genomic and bioinformatics analysis of human adenovirus type 37: new insights into corneal tropism. BMC Genom. 9:213. doi: 10.1186/1471-2164-9-213
Robinson, C. M., Shariati, F., Zaitshik, J., Gillaspy, A. F., Dyer, D. W., and Chodosh, J. (2009b). Human adenovirus type 19: genomic and bioinformatics analysis of a keratoconjunctivitis isolate. Virus Res. 139, 122–126. doi: 10.1016/j.virusres.2008.10.001
Robinson, C. M., Singh, G., Henquell, C., Walsh, M. P., Peigue-Lafeuille, H., Seto, D., et al. (2011b). Computational analysis and identification of an emergent human adenovirus pathogen implicated in a respiratory fatality. Virology 409, 141–147. doi: 10.1016/j.virol.2010.10.020
Robinson, C. M., Singh, G., Lee, J. Y., Dehghan, S., Rajaiya, J., Liu, E. B., et al. (2013a). Molecular evolution of human adenoviruses. Sci. Rep. 3:1812. doi: 10.1038/srep01812
Robinson, C. M., Zhou, X., Rajaiya, J., Yousuf, M. A., Singh, G., DeSerres, J. J., et al. (2013b). Predicting the next eye pathogen: analysis of a novel adenovirus. MBio 4, e00595–e00512. doi: 10.1128/mBio.00595-12
Routes, J. M., and Cook, J. L. (1990). Resistance of human cells to the adenovirus E3 effect on class I MHC antigen expression. Implications for antiviral immunity. J. Immunol. 144, 2763–2770.
Roy, S., Vandenberghe, L. H., Kryazhimskiy, S., Grant, R., Calcedo, R., Yuan, X., et al. (2009). Isolation and characterization of adenoviruses persistently shed from the gastrointestinal tract of non-human primates. PLoS Pathog. 5:e1000503. doi: 10.1371/journal.ppat.1000503
Ryu, J. S., Cho, J. H., Han, H. S., Jung, M. H., Yoon, Y. H., Song, E. S., et al. (2003). Acute respiratory distress syndrome induced by adenovirus in an otherwise healthy woman. Yonsei Med. J. 44, 732–735. doi: 10.3349/ymj.2003.44.4.732
Schoggins, J. W., and Falck-Pedersen, E. (2006). Fiber and penton base capsid modifications yield diminished adenovirus type 5 transduction and proinflammatory gene expression with retention of antigen-specific humoral immunity. J. Virol. 80, 10634–10644. doi: 10.1128/JVI.01359-06
Seto, D., Chodosh, J., Brister, J. R., Jones, M. S., and Members of the Adenovirus Research, C. (2011). Using the whole-genome sequence to characterize and name human adenoviruses. J. Virol. 85, 5701–5702. doi: 10.1128/JVI.00354-11
Seto, D., Jones, M. S., Dyer, D. W., and Chodosh, J. (2013). Characterizing, typing, and naming human adenovirus type 55 in the era of whole genome data. J. Clin. Virol. 58, 741–742. doi: 10.1016/j.jcv.2013.09.025
Seto, J., Walsh, M. P., Metzgar, D., and Seto, D. (2010). Computational analysis of adenovirus serotype 5 (HAdV-C5) from an HAdV coinfection shows genome stability after 45 years of circulation. Virology 404, 180–186. doi: 10.1016/j.virol.2010.05.010
Singh, G., Robinson, C. M., Dehghan, S., Jones, M. S., Dyer, D. W., Seto, D., et al. (2013). Homologous recombination in E3 genes of human adenovirus species D. J. Virol. 87, 12481–12488. doi: 10.1128/JVI.01927-13
Singh, G., Robinson, C. M., Dehghan, S., Schmidt, T., Seto, D., Jones, M. S., et al. (2012). Overreliance on the hexon gene, leading to misclassification of human adenoviruses. J. Virol. 86, 4693–4695. doi: 10.1128/JVI.06969-11
Singh, G., Zhou, X., Lee, J. Y., Yousuf, M. A., Ramke, M., et al. (2015). Recombination of the epsilon determinant and corneal tropism: human adenovirus species D types 15, 29, 56, and 69. Virology 485, 452–459. doi: 10.1016/j.virol.2015.08.018
Smith, G. R. (2012). How RecBCD enzyme and Chi promote DNA break repair and recombination: a molecular biologist's view. Microbiol. Mol. Biol. Rev. 76, 217–228. doi: 10.1128/MMBR.05026-11
Smith, G. R., Kunes, S. M., Schultz, D. W., Taylor, A., and Triman, K. L. (1981). Structure of chi hotspots of generalized recombination. Cell 24, 429–436. doi: 10.1016/0092-8674(81)90333-0
Spaan, W., Delius, H., Skinner, M., Armstrong, J., Rottier, P., Smeekens, S., et al. (1983). Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 2, 1839–1844.
Stahl, F. W. (1998). Recombination in phage lambda: one geneticist's historical perspective. Gene 223, 95–102. doi: 10.1016/S0378-1119(98)00246-7
Sujayanont, P., Chininmanu, K., Tassaneetrithep, B., Tangthawornchaikul, N., Malasit, P., and Suriyaphol, P. (2014). Comparison of phi29-based whole genome amplification and whole transcriptome amplification in dengue virus. J. Virol. Methods 195, 141–147. doi: 10.1016/j.jviromet.2013.10.005
Sumarheni, S., Hong, S. S., Josserand, V., Coll, J. L., Boulanger, P., Schoehn, G., et al. (2014). Human full-length coagulation factor X and a GLA domain-derived 40-mer polypeptide bind to different regions of the adenovirus serotype 5 hexon capsomer. Hum. Gene. Ther. 25, 339–349. doi: 10.1089/hum.2013.222
Suwaki, N., Klare, K., and Tarsounas, M. (2011). RAD51 paralogs: roles in DNA damage signalling, recombinational repair and tumorigenesis. Semin. Cell Dev. Biol. 22, 898–905. doi: 10.1016/j.semcdb.2011.07.019
Tamanini, A., Nicolis, E., Bonizzato, A., Bezzerri, V., Melotti, P., Assael, B. M., et al. (2006). Interaction of adenovirus type 5 fiber with the coxsackievirus and adenovirus receptor activates inflammatory response in human respiratory cells. J. Virol. 80, 11241–11254. doi: 10.1128/JVI.00721-06
Tan, D., Zhu, H., Fu, Y., Tong, F., Yao, D., Walline, J., et al. (2016). Severe community-acquired pneumonia caused by human adenovirus in immunocompetent adults: a multicenter case series. PLoS ONE 11:e0151199. doi: 10.1371/journal.pone.0151199
Taylor, A. F., Schultz, D. W., Ponticelli, A. S., and Smith, G. R. (1985). RecBC enzyme nicking at Chi sites during DNA unwinding: location and orientation-dependence of the cutting. Cell 41, 153–163. doi: 10.1016/0092-8674(85)90070-4
Tollefson, A. E., Hermiston, T. W., Lichtenstein, D. L., Colle, C. F., Tripp, R. A., Dimitrov, T., et al. (1998). Forced degradation of Fas inhibits apoptosis in adenovirus-infected cells. Nature 392, 726–730. doi: 10.1038/33712
Tollefson, A. E., Scaria, A., Hermiston, T. W., Ryerse, J. S., Wold, L. J., and Wold, W. S. (1996). The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J. Virol. 70, 2296–2306.
Tollefson, A. E., Scaria, A., Saha, S. K., and Wold, W. S. (1992). The 11,600-MW protein encoded by region E3 of adenovirus is expressed early but is greatly amplified at late stages of infection. J. Virol. 66, 3633–3642.
Tookman, L. A., Browne, A. K., Connell, C. M., Bridge, G., Ingemarsdotter, C. K., Dowson, S., et al. (2016). RAD51 and BRCA2 enhance oncolytic adenovirus Type 5 activity in ovarian cancer. Mol. Cancer Res. 14, 44–55. doi: 10.1158/1541-7786.MCR-15-0188-T
Torres, S., Chodosh, J., Seto, D., and Jones, M. S. (2010). The revolution in viral genomics as exemplified by the bioinformatic analysis of human adenoviruses. Viruses 2, 1367–1381. doi: 10.3390/v2071367
Viswanathan, V. K., Krcmarik, K., and Cianciotto, N. P. (1999). Template secondary structure promotes polymerase jumping during PCR amplification. Biotechniques 27, 508–511. doi: 10.2144/99273st04
Vora, G. J., Lin, B., Gratwick, K., Meador, C., Hansen, C., Tibbetts, C., et al. (2006). Co-infections of adenovirus species in previously vaccinated patients. Emerg. Infect. Dis. 12, 921–930. doi: 10.3201/eid1206.050245
Wallot, M. A., Dohna-Schwake, C., Auth, M., Nadalin, S., Fiedler, M., Malag,ó, M., et al. (2006). Disseminated adenovirus infection with respiratory failure in pediatric liver transplant recipients: impact of intravenous cidofovir and inhaled nitric oxide. Pediatr. Transplant. 10, 121–127. doi: 10.1111/j.1399-3046.2005.00411.x
Walsh, M. P., Chintakuntlawar, A., Robinson, C. M., Madisch, I., Harrach, B., Hudson, N. R., et al. (2009). Evidence of molecular evolution driven by recombination events influencing tropism in a novel human adenovirus that causes epidemic keratoconjunctivitis. PLoS ONE 4:e5635. doi: 10.1371/journal.pone.0005635
Walsh, M. P., Seto, J., Jones, M. S., Chodosh, J., Xu, W., and Seto, D. (2010a). Computational analysis identifies human adenovirus type 55 as a re-emergent acute respiratory disease pathogen. J. Clin. Microbiol. 48, 991–993. doi: 10.1128/JCM.01694-09
Walsh, M. P., Seto, J., Liu, E. B., Dehghan, S., Hudson, N. R., Lukashev, A. N., et al. (2011). Computational analysis of two species C human adenoviruses provides evidence of a novel virus. J. Clin. Microbiol. 49, 3482–3490. doi: 10.1128/JCM.00156-11
Walsh, M. P., Seto, J., Tirado, D., Chodosh, J., Schnurr, D., Seto, D., et al. (2010b). Computational analysis of human adenovirus serotype 18. Virology 404, 284–292. doi: 10.1016/j.virol.2010.05.013
Whyte, P., Buchkovich, K. J., Horowitz, J. M., Friend, S. H., Raybuck, M., Weinberg, R. A., et al. (1988). Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature 334, 124–129. doi: 10.1038/334124a0
Wickham, T. J., Mathias, P., Cheresh, D. A., and Nemerow, G. R. (1993). Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell 73, 309–319. doi: 10.1016/0092-8674(93)90231-E
Wiethoff, C. M., Wodrich, H., Gerace, L., and Nemerow, G. R. (2005). Adenovirus protein VI mediates membrane disruption following capsid disassembly. J. Virol. 79, 1992–2000. doi: 10.1128/JVI.79.4.1992-2000.2005
Williams, J., Grodzicker, T., Sharp, P., and Sambrook, J. (1975). Adenovirus recombination: physical mapping of crossover events. Cell 4, 113–119. doi: 10.1016/0092-8674(75)90117-8
Wilson-Rawls, J., Deutscher, S. L., and Wold, W. S. (1994). The signal-anchor domain of adenovirus E3-6.7K, a type III integral membrane protein, can direct adenovirus E3-gp19K, a type I integral membrane protein, into the membrane of the endoplasmic reticulum. Virology 201, 66–76. doi: 10.1006/viro.1994.1266
Windheim, M., Hilgendorf, A., and Burgert, H. G. (2004). Immune evasion by adenovirus E3 proteins: exploitation of intracellular trafficking pathways. Curr. Top. Microbiol. Immunol. 273, 29–85. doi: 10.1007/978-3-662-05599-1_2
Windheim, M., Southcombe, J. H., Kremmer, E., Chaplin, L., Urlaub, D., Falk, C. S., et al. (2013). A unique secreted adenovirus E3 protein binds to the leukocyte common antigen CD45 and modulates leukocyte functions. Proc. Natl. Acad. Sci. U.S.A. 110, E4884–E4893. doi: 10.1073/pnas.1312420110
Wodrich, H., Guan, T., Cingolani, G., Von Seggern, D., Nemerow, G., and Gerace, L. (2003). Switch from capsid protein import to adenovirus assembly by cleavage of nuclear transport signals. EMBO J. 22, 6245–6255. doi: 10.1093/emboj/cdg614
Wold, W. S., Cladaras, C., Magie, S. C., and Yacoub, N. (1984). Mapping a new gene that encodes an 11,600-molecular-weight protein in the E3 transcription unit of adenovirus 2. J. Virol. 52:307–313.
Wu, L., Zhou, P., Ge, X., Wang, L. F., Baker, M. L., and Shi, Z. (2013). Deep RNA sequencing reveals complex transcriptional landscape of a bat adenovirus. J. Virol. 87, 503–511. doi: 10.1128/JVI.02332-12
Young, C. S., Cachianes, G., Munz, P., and Silverstein, S. (1984). Replication and recombination in adenovirus-infected cells are temporally and functionally related. J. Virol. 51, 571–577.
Zhang, S. Y., Luo, Y. P., Huang, D. D., Fan, H., Lu, Q. B., Wo, Y., et al. (2016). Fatal pneumonia cases caused by human adenovirus 55 in immunocompetent adults. Infect Dis. (Lond). 48, 40–47. doi: 10.3109/23744235.2015.1055585
Zhang, Y., Dolph, P. J., and Schneider, R. J. (1989). Secondary structure analysis of adenovirus tripartite leader. J. Biol. Chem. 264, 10679–10684.
Zhang, Y., Huang, W., Ornelles, D. A., and Gooding, L. R. (2010). Modeling adenovirus latency in human lymphocyte cell lines. J. Virol. 84, 8799–8810. doi: 10.1128/JVI.00562-10
Zhou, X., Robinson, C. M., Rajaiya, J., Dehghan, S., Seto, D., Jones, M. S., et al. (2012). Analysis of human adenovirus type 19 associated with epidemic keratoconjunctivitis and its reclassification as adenovirus type 64. Invest. Ophthalmol. Vis. Sci. 53, 2804–2811. doi: 10.1167/iovs.12-9656
Keywords: adenovirus, genome, evolution, transcription factor, interactome
Citation: Ismail AM, Lee JS, Lee JY, Singh G, Dyer DW, Seto D, Chodosh J and Rajaiya J (2018) Adenoviromics: Mining the Human Adenovirus Species D Genome. Front. Microbiol. 9:2178. doi: 10.3389/fmicb.2018.02178
Received: 20 July 2018; Accepted: 24 August 2018;
Published: 11 September 2018.
Edited by:
Qiwei Zhang, Southern Medical University, ChinaReviewed by:
Jun Hang, Walter Reed Army Institute of Research, United StatesGabriel Gonzalez, Hokkaido University, Japan
Copyright © 2018 Ismail, Lee, Lee, Singh, Dyer, Seto, Chodosh and Rajaiya. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jaya Rajaiya, amF5YV9yYWphaXlhQG1lZWkuaGFydmFyZC5lZHU=
 Ashrafali M. Ismail
Ashrafali M. Ismail Ji Sun Lee1
Ji Sun Lee1 Donald Seto
Donald Seto James Chodosh
James Chodosh