- 1Department of Agriculture, Iranian Research Organization for Science and Technology, Tehran, Iran
- 2Institute of Animal Nutrition and Functional Plant Compounds, Department for Farm Animals and Veterinary Public Health, University of Veterinary Medicine Vienna, Vienna, Austria
In the search for natural alternatives to antibiotic feed additives, we compared the efficacy of two doses of Scrophularia striata extract [S. striata-Low at 40 and S. striata-High at 80 mg g-1 dry matter (DM)] with monensin (monensin) and a negative control in the modulation of rumen fermentation, methane production and microbial abundance in vitro. Microbes were investigated using qPCR and 16S rRNA targeted sequencing. Data showed that the addition of S. striata increased production of total short chain fatty acids (SCFA) in comparison to both monensin and control (P = 0.04). The addition of S. striata increased acetate production, and increased propionate at the higher dosage (P < 0.001). Supplementation of S. striata lowered methane production (P < 0.001) compared to control but with no effect compared to monensin. Ammonia concentration decreased by 52% (P < 0.001) with S. striata-High supplementation (4.14 mmol L-1) compared to control, which was greater than that of monensin (36%). The diversity of rumen bacteria was reduced (P < 0.001) for monensin and S. striata for both the number of observed OTUs and the Chao1 index. Quantitative analysis of Protozoa showed a decrease in the monensin treatment (P = 0.05) compared to control. Archaea copy numbers decreased equally in both S. striata-High and monensin treatments compared to the control group. Supplementation with S. striata increased relative abundances of Fibrobacteres (P < 0.001) and Planctomycetes (P = 0.001) in comparison to both the control and monensin treatments. Significant negative correlations were observed between the abundances of Bacteroides, Fusobacterium, and Succinivibrio genera and methane (r > -0.71; P ≤ 0.001). The abundance of Fibrobacter genera and total SCFA (r = 0.86), acetate (r = 0.75), and valerate (r = -0.51; P < 0.001) correlated positively. These results suggest that S. striata supplementation at 80 mg g-1 DM inclusion, similar to monensin, supports rumen fermentation, lowers methane and ammonia production. However, S. striata supported rumen fermentation toward higher total SCFA and propionate production, while unlike monensin still supported a diverse rumen microbiome and an increase in cellulolytic bacteria such as Fibrobacter.
Introduction
Rumen microbiota are essential for the host, providing short-chain fatty acids (SCFA), which make up 75% of the total metabolizable energy for ruminants (Siciliano-Jones and Murphy, 1989; Bergman, 1990). In dairy cattle, of the major SCFA, propionate is critical for gluconeogenesis (den Besten et al., 2013) and therefore has particular importance for meeting the high glucose demands of lactation. In addition, the production of propionate is an electron-accepting pathway and serves as a H+ sink, lowering the substrate availability for methanogenesis (den Besten et al., 2013). The production of methane not only has high global warming potential, but also reduces feed energy efficiency in ruminants (Johnson and Johnson, 1995). Therefore, ruminant nutritionists have been working for decades to develop dietary strategies capable of modulating both the rumen fermentation profile to increase propionate production for high producing cattle and reducing methane production (McAllister and Newbold, 2008; Hristov et al., 2013).
In this regard, monensin, an ionophore widely used as a growth promoter (Shen et al., 2017), has been previously shown to inhibit ruminal methanogenesis and promote the production of propionate (Green et al., 1999; Duffield et al., 2008). However, because of the increasing concerns about antibiotic resistance and since the ban of antibiotics in the European Union (Broudiscou et al., 2000; Shen et al., 2017), considerable efforts have been devoted to investigating the potential use of plant phytochemicals such as essential oils, saponins and tannins as alternatives to antibiotics in ruminant nutrition (Śliwiński et al., 2002; Bodas et al., 2012). Scrophularia striata is a herbaceous flowering plant, commonly known as figwort, which is often used in traditional folk remedies against infections (Monsef-Esfahani et al., 2010; Mahboubi et al., 2013). Previous research has shown that the ethanolic extract of S. striata has antimicrobial activity against Staphylococcus aureus, Eschericia coli, Enterococcus faecalis, and Newcastle disease (Bahrami and Valadi, 2010; Bahrami, 2011). However, the effects of S. striata extract on the anaerobic microbiota of the rumen and the associated fermentation have not been previously reported.
Therefore, this study was designed to evaluate effects of S. striata on SCFA production, methane production as well as the diversity and abundance of the rumen microbiome compared to monensin in a semi continuous in vitro system. We hypothesized that feeding of S. striata, similar to monensin, could alter the rumen microbiome leading to beneficial modifications to rumen fermentation including increased propionate production and an inhibition of ruminal methane production.
Materials and Methods
Plant Material and Extraction
Scrophularia striata was purchased from a local herbalist’s shop in Ilam, a province in the west of Iran and was authenticated as S. striata Bioss family (Scrophulariaceae) with a voucher specimen being preserved in the pharmacy herbarium (TEH No: 6916) at Tehran University of Medical Science, Tehran, Iran. Whole plants containing stem and seed were ground in a centrifugal mill with 0.5 mm screen. Ground plant material was mixed with 600 ml L-1 aqueous ethanol solution at the ratio 1:25 (w:v) for 1 h in sonicator bath with cooling system (Sonorex RK 156H, Bandelin, Berlin, Germany). The mixture was filtered through filter paper and concentrated in a vacuum at 40°C by a rotary evaporator (Buchi, Flawil, consisting of water bath B-480, Rotovapor R-124, and Vacuum Controller B-72, Switzerland). After ethanol evaporation, the particulate was transferred to beaker flask, frozen at -20°C and then freeze-dried according to previously described methods (Goel et al., 2008).
Experimental Design, Rusitec Procedure, and Sample Collection
The experiment was designed as a completely randomized design with 2 experimental runs, 4 treatments, and 3 replicates per run, resulting in six independent measurements per treatment. The treatments consisted of either no additives (control), 0.83 mg g-1 DM of substrate monensin sodium salt (monensin; Sigma Chemical Co. Ltd.), 40 mg g-1 DM (S. striata-Low), or 80 mg g-1 DM (S. striata-High) of dried of S. striata extract. Monensin and S. striata were dissolved in 1 ml ethanol [90% (v/v), TechniSolv®, pure] and an equal volume of ethanol was added to the control treatment group (Cardozo et al., 2005).
All runs were comprised of 12 fermenters and lasted for 10 days, with sampling on the last 5 days of each run based on a 4 day adaptation period for the rumen microbes (Soliva and Hess, 2007). Initial inocula for each experimental run was collected from three non-lactating rumen-fistulated Holstein cows, housed at the Dairy Research Station of the University of Veterinary Medicine Vienna (Pottenstein, Austria). The use of donor cows was approved by the institutional ethics committee of the University of Veterinary Medicnie (Vetmeduni). Donor cattle were fed hay and grass silage ad libitum and free access to drinking water. Animals were cared for according to the Austrian guidelines for animal welfare (Federal Ministry of Health, 2004). Rumen contents of the three cows were mixed in equal parts, the liquor was first filtered through four layers of medical gauze (1-mm pore size) before adding to the fermenters and the solid digesta was filled (∼60 g) into nylon bags and placed into each fermenter on the first day of each experimental run according to the previously published methodology (Klevenhusen et al., 2015). In brief, each fermenter was filled with 100 ml artificial saliva and 600 ml rumen fluid on the first day of each experimental run (McDougall, 1948). New substrate was provided to each fermenter daily, based on a 50:50 forage-to-concentrate dairy ration (Supplementary Table S1) using nylon bags (140 mm × 70 mm, 150-μm pore size, Linker Industrie–Technik, Kassel, Germany). After 24 h of incubation, the bags containing solid digesta were replaced with fresh bags containing the experimental diet. Accordingly, every feed bag was incubated for 48 h, and then replaced. Artificial saliva buffer was continuously infused into each vessel (357 ± 19.2 ml/d) through a 12 channel peristaltic pump (Model ISM 932D, Ismatec, Index Health and science GmbH, Wertheim, Germany) in accordance with Soliva and Hess (2007). Feed additives were pipetted into the fermenter fluid every day at the time of feeding (0900 h) starting on day 0. After each feeding procedure the system was flushed with N2 gas for 3 min to maintain anaerobic condition inside the vessels. Daily effluent and fermentation gasses were collected in a 1 L volumetric flask located in an ice-filled box and reusable gas-tight aluminum bags (TECOBAG 8L, Tesseraux ContainerGmbH, Burstadt, Germany), respectively. On sampling days, directly before feed bag exchange, fermenter fluid samples were collected using a syringe equipped with a plastic tube. Part of the fluid samples were immediately analyzed for rumen fermentation parameters and aliquots were stored in separate tubes at -20°C for SCFA and ammonia measurements. Additional fluid samples were snap frozen in liquid nitrogen and stored at -80°C for DNA extraction. The feed residue bags were hand-washed with running water until the water was clear and kept at -20°C for chemical composition analysis.
Sample Analysis
Immediately after sampling pH and redox potential (seven MultiTM, Mettler-Toledo GmbH, Schwerzenbach, Switzerland) of the rumen fermenter liquid were measured. Fermentation gasses were also measured every 24 h during the sampling period. Gas volume was determined using the water displacement technique (Soliva and Hess, 2007), and the concentration of CH4 was measured using a portable infrared detector (ATEX biogas Monitor Check BM 2000, Ansyco, Karlsruhe, Germany). In brief, the biogas monitor is attached to the gas collection bag after a 24 h collection period, the sample is measured through pumping of gas into the monitor for 30 s to provide a consistent reading as per the manufacturer’s instructions.
Concentration and composition of SCFA (acetate, propionate, n-butyrate, isobutyrate, n-valerate, isovalerate, and caproate) were analyzed by GC, as described previously (Qumar et al., 2016). Briefly, incubation fluid samples were thawed and then centrifuged at 20,000 × g for 25 min at 4°C. The supernatant (0.6 mL) was transferred into a fresh tube and 0.2 mL of HCl (1.8 mol L-1) was added, followed by 0.2 mL of the internal standard (4-methylvaleric acid, Sigma-Aldrich Co. LLC., St. Louis, MI, United States). After centrifugation of the mixture at 20,000 × g for 25 min at 4°C, the clear supernatant was transferred into the GC vial. Analysis of SCFA concentrations was conducted via a gas chromatography apparatus (GC Model 8060 MS 172 DPFC, No.: 950713, Fisons, Rodano, Italy) which was equipped with a flame-ionization detector and a 30 m × 0.53 mm ID × 0.53 μm df capillary column (Trace TR Wax, Thermo Fisher Scientific, Waltham, MA, United States). Injector and detector were at temperatures of 170°C and 190°C, respectively. Helium was used as carrier gas with a flow rate of 6 mL min-1. Stratos Software (Stratos Version 4.5.0.0, Polymer Laboratories, Church Stretton, Shropshire, United Kingdom) was utilized for generation and evaluation of chromatograms.
The concentration of ammonia was determined using the indophenol reaction (Weatherburn, 1967). The fermenter fluid samples were thawed at room temperature and subsequently centrifuged at 15,115 × g for 10 min. The clear supernatant was diluted with distilled water to obtain a concentration within the standard calibration curve. Ammonia and phenol were oxidized by sodium hydroxide in the presence of sodium nitroprusside and dichloroisocyanuric acid. The absorbance was measured at 655 nm using U3000 Spectrophotometer (INULA GmbH, Vienna, Austria) after 90 min of reaction.
Analysis of the residual feeds after 48 h of incubation and the original feed samples were conducted in duplicate. Prior to analysis, samples were oven-dried at 65°C for 48 h and ground using through a 0.75 mm sieve (VDLUFA, 2007). The DM of feeds and feed residues was measured after oven drying at 104°C for 24 h and ash was analyzed by combustion of samples over night at 580°C. Crude protein was measured by Kjeldahl method (VDLUFA, 2007). Neutral detergent fiber (aNDFom; Udén et al., 2005) and acid detergent fiber (ADFom) contents were analyzed using Fibretherm FT12 (C. Gerhardt GMbH and Co. KG, Konigswinter, Germany) based on VDLUFA (2007). Heat stable α-amylase used in the NDF procedure and all fiber fractions were expressed exclusive residual ash. Disappearance of nutrients as a measure of degradability was calculated from the difference between content of nutrients in the diet before and after 48 h incubation in the Rusitec system.
DNA Extraction and qPCR
Frozen rumen fluid samples were thawed on ice, vortexed shortly to mix the sample, and then samples from d 5 to 10 were pooled per fermenter per run (n = 6 per treatment). From the pooled sample, 1 mL subsample was used for DNA isolation. DNA isolation was performed using the QIAamp Fast DNA Stool Mini Kit (QIAGEN, Hilden, Germany) and samples were mixed with 1 mL of InhibitEX buffer provided in the kit and heated at 95°C for 5 min to ensure proper lysis of bacteria. After heating, additional enzymatic (mutanolysin, 2.5 U μL-1; lysozym, 100 mg mL-1; proteinase K, 20 mg mL-1) and mechanical lysis (0.4 g sterile ceramic beads, diameter 1.4 mm) using a bead-beater (FastPrep-24 5G, MP Biomedicals, LLC, Santa Ana, CA, United States) was performed to disrupt bacterial cells according to previously published procedures (Kong et al., 2010). This was followed by chemical removal of cell debris and PCR inhibitors by centrifugation and supernatants were transferred to fresh tubes for column based isolation of total genomic DNA using the QIAcube robotic workstation. Total genomic DNA was eluted in 200 μL of low-salt buffer. The isolated DNA concentration was determined by a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, United States) using the Qubit dsDNA HS Assay Kit (Life Technologies) and stored at -20°C until further analysis.
Quantitative PCR Analysis
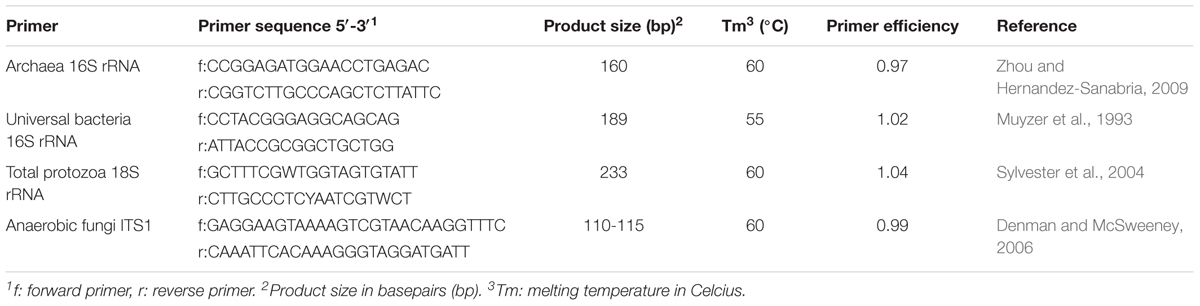
Using qPCR, the abundance of Protozoa, Bacteria, Archaea, and Fungi present in Rusitec samples were analyzed on a Stratagene Mx3000P real-time PCR System (Agilent Technologies, Santa Clara, CA, United States) using established primer pairs (Table 1). DNA samples and negative controls were assayed in duplicate in a 20 μL reaction mixture containing 10 μL the Fast-Plus EvaGreen Master Mix with Low ROX (Biotium, Hayward, CA, United States), 1 μL of each primer (Protozoa 400 nM; Bacteria 100 nM; Archaea and Fungi 200 nM), 7 μL of nuclease-free water and 1 μL DNA template (4 ng). The amplification program included an initial denaturation step at 95°C for 10 min, followed by 40 cycles of 95°C for 10 s, primer annealing at 60°C for 30 s, and elongation at 72°C for 30 s. Fluorescence was measured at the last step of each cycle. Melting curve analysis was performed to determine the specificity of the amplification. The dissociation of PCR products was monitored by slow heating with an increment of 0.1°C s-1 from 55 to 95°C, with fluorescence measurement at 0.1°C intervals. Standard curves for all primers were generated using 10-fold serial dilutions (108–103 molecules μL-1) of the purified and quantified PCR products generated by standard PCR using DNA from the pooled rumen fluid from both runs of the present experiment. The PCR efficiency was calculated: E = 10(-1/slope)-1 (Table 1). Results were analyzed using the associated software (Stratagene MxPro, QPCR Software, version 2.00).
Sequencing, Sequence Processing, and Bioinformatics Analysis
One 40 μL aliquot of each DNA sample was sent for amplicon sequencing using a MiSeq Illumina sequencing platform and paired-end technology (Microsynth AG, Balach, Switzerland). Sequencing targeted the V3–V5 hypervariable region of the 16S rRNA gene using the primer set 357F-HMP (5′-CCTACGGGAGGCAGCAG-3′) and 926R-HMP (5′-CCGTCAATTCMTTTRAGT-3′) to produce an amplicon size of ∼523 bp (Peterson et al., 2009). In brief, libraries were constructed by ligating sequencing adapters and indices onto purified PCR products (Nextera XT Sample Preparation Kit, Illumina, San Diego, CA, United States) according to the recommendations of the manufacturer. Equimolar amounts of each of the libraries were pooled together and sequenced on an Illumina MiSeq Personal Sequencer and the resulting paired ends were stitched together by Microsynth (Balach, Switzerland) resulting in a total of 3.8 million unfiltered reads with an average of 503 nt in length. Sequence quality control was performed using the QIIME pipeline (Caporaso et al., 2010). Sequences were first quality filtered following (Q = 20; 27) and then screened and filtered for chimeras using USEARCH and its database (USEARCH v8.1; Bokulich et al., 2013). Clustering and alignment were performed using UCLUST (Edgar, 2010) and OTUs were defined using PyNAST (Caporaso et al., 2010) and the SILVA database (v123; accessed July 19, 2017; Quast et al., 2012; Yilmaz et al., 2013). The degree of similarity between sequences was defined as 97% to obtain OTU. Any OTUs which clustered with less than 10 reads were manually removed. A total of 969,155 sequences (mean sequences per sample 38,504) clustered into 2,727 OTUs for further analysis. All OTUs with a relative abundance greater than 0.1% (135 OTUs) were taken in consideration for statistical analysis and statistically derived means for relative abundance were graphed in excel. For calculation of the non-parametric species richness estimators Chao1 and the observed OTUs per sample were used. Beta diversity analysis was performed using weighted Unifrac dissimilarity metrics and the principal coordinate analysis (PCoA) plotting in QIIME with rarefaction at 20,133 sequences, based on the lowest number of sequences in a sample. Sequencing data are available in BioProject SRA database under the accession number SRP140677.
Statistical Analyses
Analysis of variance was conducted using the PROC MIXED procedure of SAS (Version 9.2, SAS Institute, Cary, NC, United States). Treatments were considered as fixed effects, while experimental run and fermenter within run were defined as random effects. Degrees of freedom estimation was done using Kenward-Rogers. All values were reported as least squares means. Comparison between the control and treatments was carried with Dunnett–Hsu. Differences were declared as significant at P ≤ 0.05. Correlation analysis using Pearsons correlation coefficient was performed using the PROC CORR procedure of SAS. Following Hinkle et al. (2003), the r was interpreted as follows: 0.00–0.30 as negligible; 0.30–0.50 as low; 0.50–0.70 as moderate; 0.70–0.90 as high, and 0.90–1.00 as substantial.
Results
Rumen Fermentation Profile, Methane Production, and Nutrient Degradability
The pH in the fermenters remained constant throughout both experimental runs (Supplementary Figure S1 and Table 2). However, redox potential in the fermenters was lower for all treatments when compared to the control (P = 0.007). Analysis showed that the ammonia concentration decreased (P < 0.001) with monensin and S. striata supplementation. The extent of ammonia reduction was 35, 26, and 52%, respectively, by monensin, S. striata-Low and S. striata-High treatments. Total SCFA production was decreased by monensin, while S. striata supplementation increased it compared with control (P = 0.001). All individual SCFA were affected by supplementation (Table 2). Monensin decreased acetate production by 9%, whereas S. striata had no effect in comparison with control. Propionate was increased by 186, 83, and 35% by feeding monensin, S. striata-High and S. striata-Low, respectively. As a result, monensin and S. striata-High had a lower acetate to propionate ratio (A:P) in comparison to control (P < 0.001). Supplementation decreased iso-butyrate concentrations in comparison to control (P < 0.001). Butyrate was decreased by monensin and S. striata-High (P < 0.001) in comparison to control. In contrast, the addition of S. striata-Low increased the concentration of iso-valerate above that of the control group (Table 2). Monensin decreased the proportion of iso-valerate to 8.1% of the control concentration and increased of n-valerate by 97% (P < 0.001). Supplementation with monensin, S. striata-Low and S. striata-High caused a drop in methane production by 67, 29, and 36%, respectively (P < 0.003). The DM, OM, and ADF degradation were not affected by supplementation (Table 2). However, S. striata-High decreased CP degradation (P = 0.03) and all additives decreased NDF degradability (P = 0.02) compared to the control group.
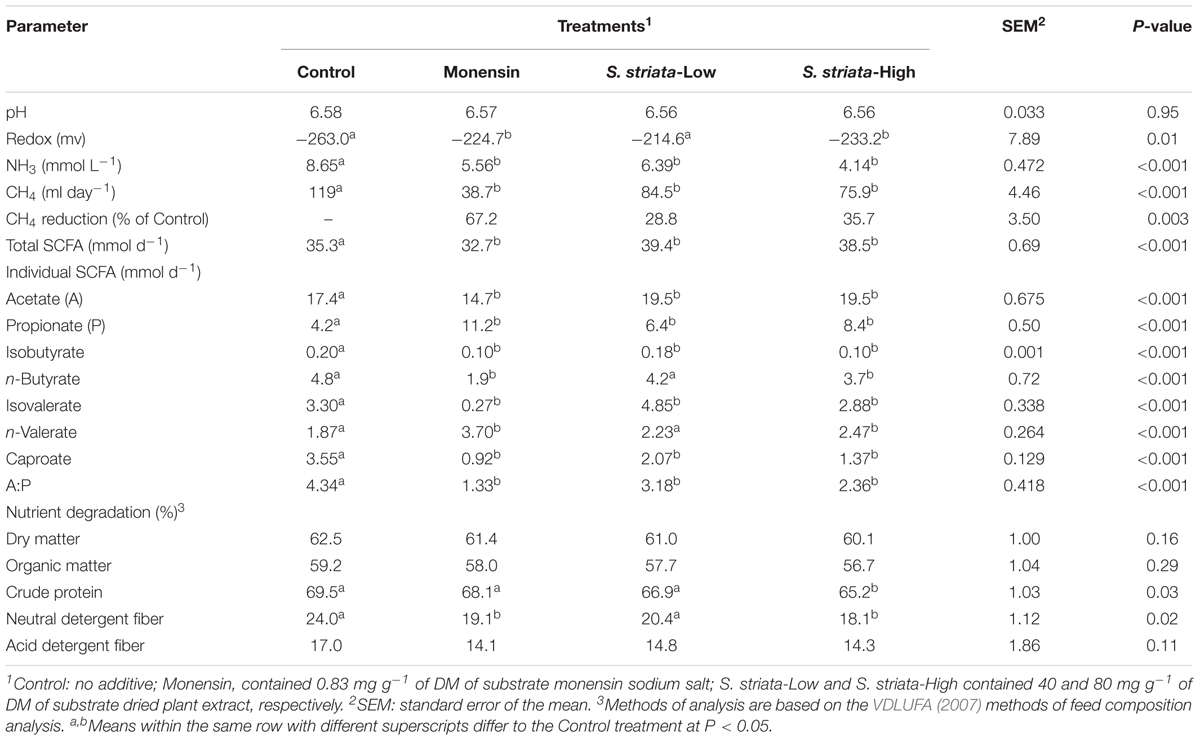
TABLE 2. Effect of supplementation of Scrophularia striata extract and monensin on rumen fermentation profile and nutrient degradation.
Quantitative Analysis at the Kingdom Level of Taxonomy
Results of qPCR analysis are presented in Figure 1. Supplementation did not affect the abundance of total Bacteria and Fungi populations compared to control. Monensin remarkably lowered the gene copy number of Protozoa (1.00 × 102), though it was significant only when compared to S. striata-High group (3.15 × 105; P = 0.05). Copy numbers of total Archaea were lower with S. striata-High and monensin compared to S. striata-Low (P = 0.001), and tended to be lower with monensin compared to control (P = 0.08).
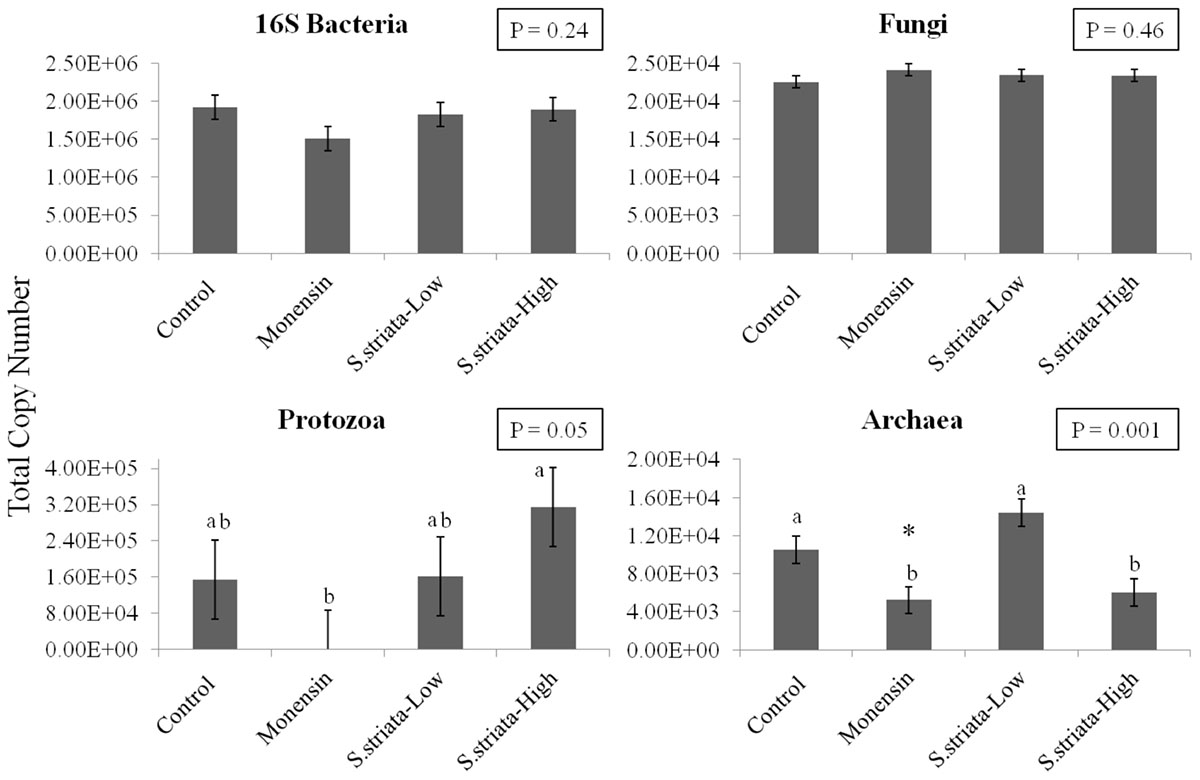
FIGURE 1. Effect of supplementation of Scrophularia striata extract, in two doses (S. striata-Low and S. striata-High), and monensin on copy number of Bacteria (16S rRNA), Fungi (ITS Gene 1), Protozoa (18S rRNA), and Archaea (16S rRNA) analyzed by qPCR. Data is presented in total copy numbers of the target gene. ∗In tendency lower than control (P = 0.08). a,bTreatments with different letters differ significantly to the Control P < 0.05.
Alpha and Beta Microbial Diversity
Statistical analysis of both the intra- (alpha diversity; Table 3) and inter- (beta diversity; Figure 2) sample diversity was performed. Despite no differences in the total numbers of sequences found within a sample (P = 0.91), there were differences between treatments in the number of OTUs (P < 0.001) and the Chao1 index (P < 0.001). For both indices, the diversity within a sample was highest in the control, followed by S. striata-Low, S. striata-High, and monensin showed the least microbial diversity. PCoA showed a clear clustering of microbial communities for each treatment group. Principle component 1 described 40.8% of the diversity between samples between original rumen fluid and experimental treatments, with the monensin treatment clustering independently (Figure 2). Principle component 2 represented 30.2% of the diversity with a separation of the S. striata samples from the control and monensin groups. Principle component 3, representing 10.3% of the variation between samples, was related to the original pooled rumen fluid sample.

TABLE 3. Effect of supplementation of Scrophularia striata extract and monensin on number of sequences, observed operational taxonomic units (OTU) and the Chao1 index.
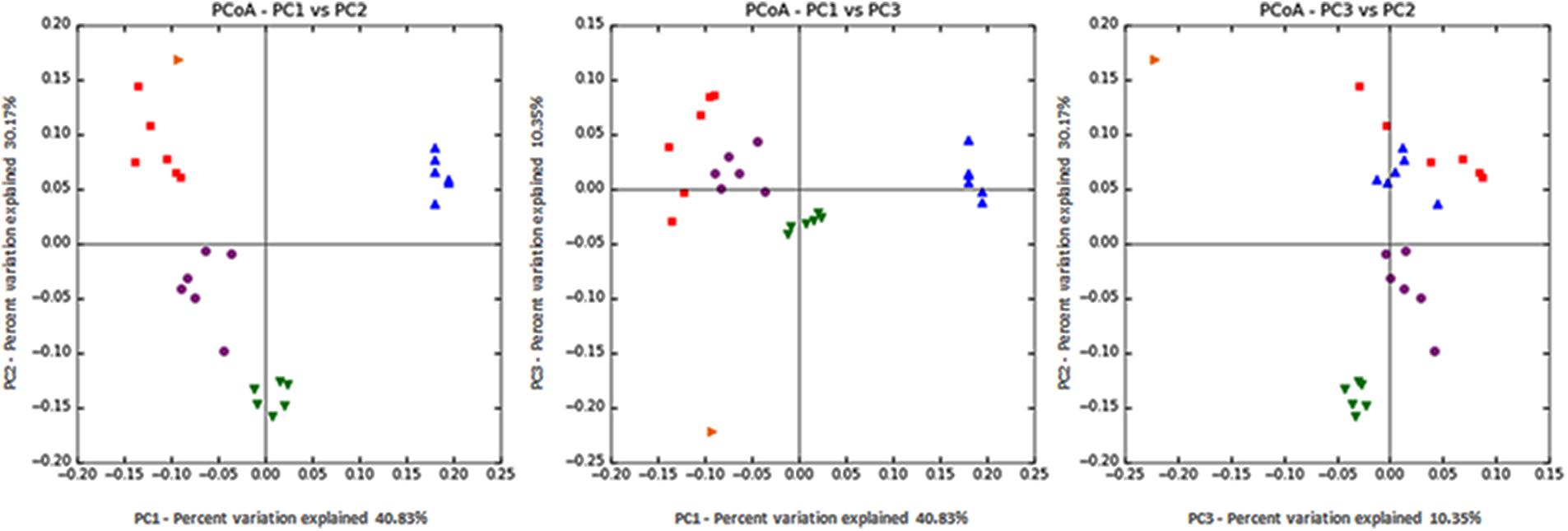
FIGURE 2. Principal coordinate analysis (PCoA) of 16S rRNA using the weighted Unifrac distance metric (PC1 = 40.8%, PC2 = 30.2%, and PC3 = 10.3%). Orange: original rumen fluid sample taken from cows, Red: control, Blue: monensin, Purple: S. striata-Low, and Green: S. striata-High. S. striata: Scrophularia striata extract.
16S rRNA Taxonomy
In total, 15 phyla, predominated by Bacteroidetes, Firmicutes, and Proteobacteria, could be identified into 42 genera. All phyla and all genera were significantly affected by treatments (Figure 3). Supplementation with S. striata-High increased the total Bacteroidetes and reduced the Firmicutes taxa, whereas monensin increased Fusobacteria, Synergistetes, and Proteobacteria phyla (Figure 3A). The addition of all additives decreased the relative abundance of Elusimicrobia and TM7 in comparison to control. Fusobacterium and Firmicutes were, respectively, 4 and 13% more abundant in the monensin in comparison to the S. striata-High supplementation (Figure 3). At the genera level, Prevotella was the most abundant genus ranging from 38.5% in control to 46.1% in S. striata-High and was significantly increased with S. striata supplementation. Bacteroidetes-associated genera represented the largest proportion of the in vitro microbiota despite the fact that samples were taken from the rumen liquid (Figure 3B). Firmicutes had the largest diversity in identified genera with a significant effect of supplementation (Figure 3C). A drastic increase in Fusobacterium spp. was found in monensin supplementation compared to the other treatments (Figure 3D). Direct comparison of the monensin and S. striata-High treatments at the phyla level for significant differences in the least square means showed of the 15 phyla, eight were significantly different between the two treatment groups (Figure 4). Of the 30 most abundant OTUs, 29 were significantly affected by the supplementation treatment. Bacteroidetes and Firmicutes accounted for 16 and 8 of the OTUs, respectively (Figure 5). Three OTUs were identified to the species level, Prevotella copri-like OTU2, Bacteroides uniformis-like OTU10 and Fibrobacter succinogenes-like OTU24. In comparison to the control, OTU2 was 4.2-fold and four-fold increased in monensin and S. striata-High, respectively (P < 0.001). OTU10 accounted for 5.13% of the relative microbial abundance in the monensin supplemented group but was less than 0.6% of the abundance in the other treatments. F. succinogenes-like OTU24 was increased in both the S. striata-Low (1.05%) and S. striata-High (0.87%) supplemented groups compared to both control and monensin.
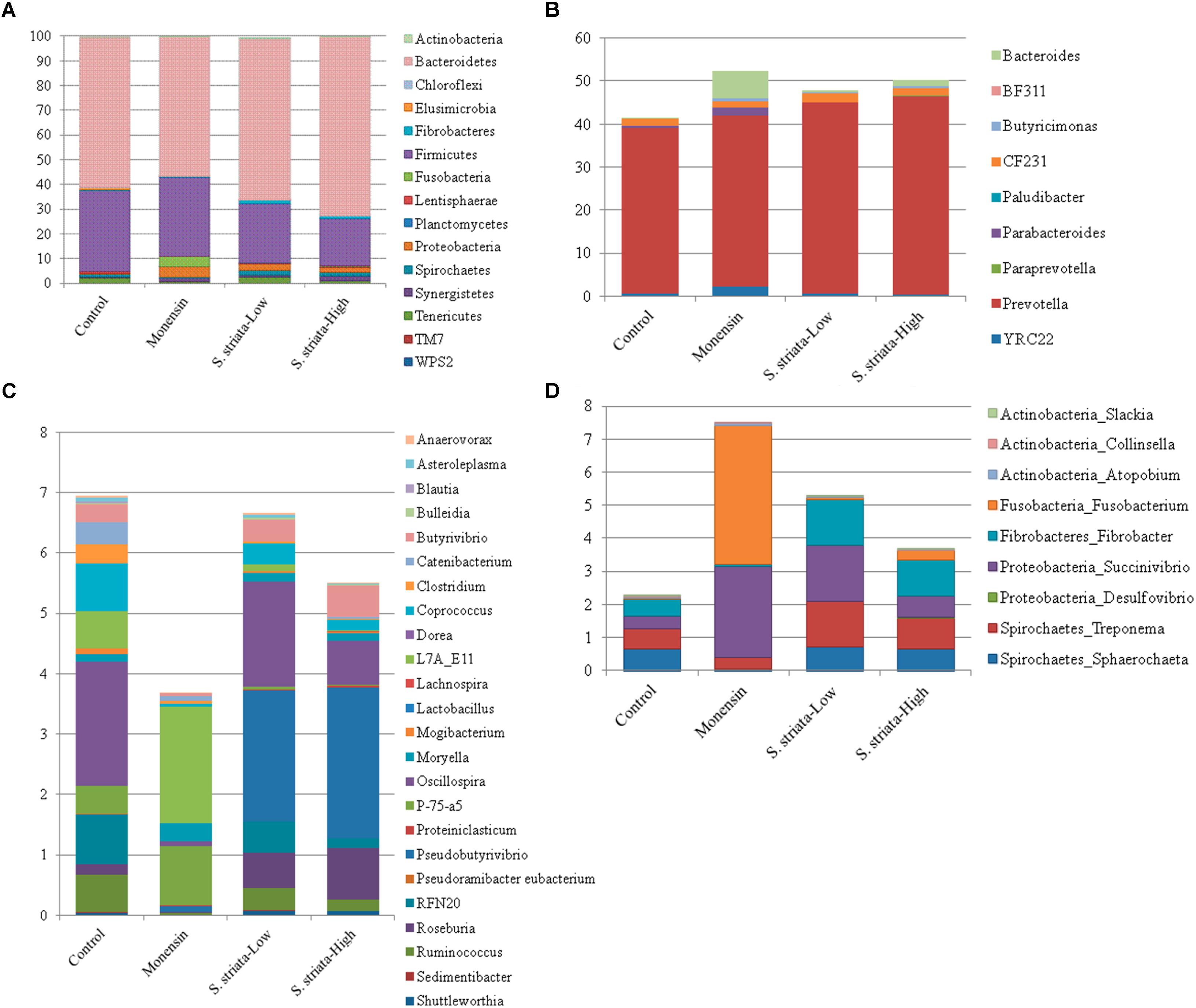
FIGURE 3. Mean relative abundance of 16S rRNA sequences identified to 97% with the SILVA128 database at the phyla level (A) and genera level for Bacteroidetes (B), Firmicutes (C), and other groups (D). All phyla and genera have a significant effect of supplementation (P ≤ 0.05).
FIGURE 4. Comparison of the mean relative abundance of phyla between treatment groups supplemented with Scrophularia striata extract at 80 mg g-1 DM (S. striata-High) and those given monensin at 0.83 mg g-1 DM. Differences in the P-value for least square means between monensin and S. striata-High are Bacteroidetes (P < 0.001), Firmicutes (P < 0.001), Fusobacteria (P < 0.001), Proteobacteria (P = 0.001), Fibrobacteres (P = 0.001), Spirochaetes (P = 0.001). Other includes Lentisphaerae (P = 0.04), Planctomycetes (P = 0.001), Actinobacteria (P = 0.96), Chloroflexi (P = 0.65), Elusimicrobia (P = 0.99), Synergistetes (P = 0.69), Tenericutes (P = 0.37), TM7 (P = 0.99), WPS-2 (P = 0.28). ∗Individual phyla within the “Other” group do not have means which are significantly (P < 0.05).
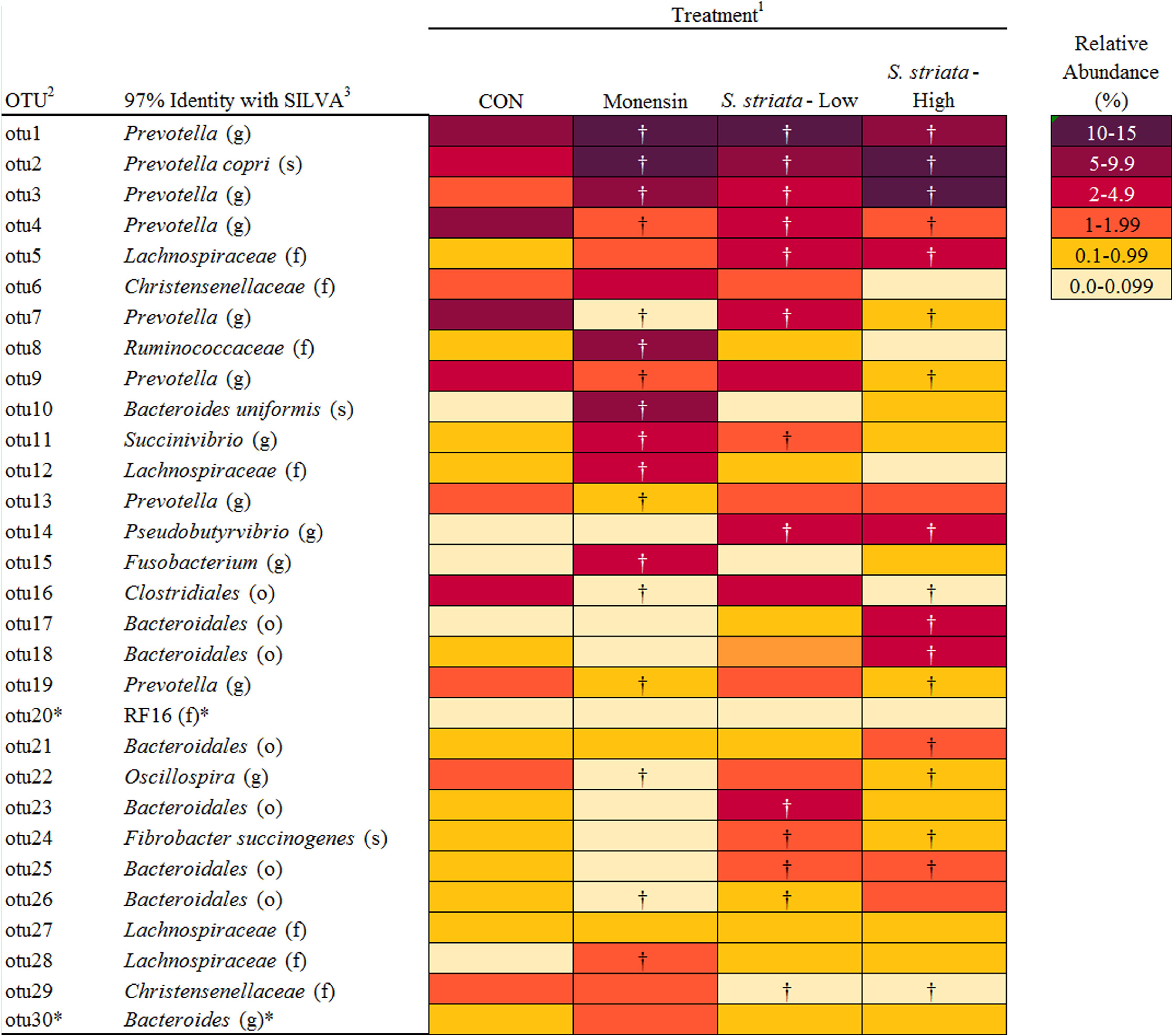
FIGURE 5. Effect of supplementation of Scrophularia striata extract and monensin on percent relative abundance and taxonomic classification of 30 most abundant operational taxonomic units (OTUs). 1Control: no additive; monensin: monensin, contained 0.83 mg g-1 of DM of substrate monensin sodium salt; S. striata-Low and S. striata-High contained 40 and 80 mg g-1 of DM of substrate dried plant extract, respectively. 2OTUs listed in order of relative abundance. 3Name of the closest relative based on 97% similarity in the sequence database at the class (c), order (o), family (f), genus (g), or species (s) level. †Adjusted mean separation based on Dunnett-Hsu shows a significant variation from the Control. ∗Means are not significantly different (P < 0.05).
Correlations Between Genera and Rumen Fermentation
Several correlations between rumen fermentation parameters and microbial communities significantly altered by the supplementations were found (Table 4). Fusobacterium spp. showed the greatest number of highly (0.70 < r < 0.90) negative correlations, including methane, acetate, butyrate, isovalerate and total SCFA production (P < 0.001) and positive correlations to propionate (P < 0.001) and valerate (P < 0.001). Similarly, Bacteroides (phyla Bacteroidetes) and Succinivibro (phyla Proteobacteria) genera also showed an inverse relationship to methane production (r = -0.76 and r = -0.71, respectively) and a positive correlation to valerate concentrations in vitro (P < 0.001). Furthermore, Bacteroides spp. was also highly correlated with decreases in butyrate and isovalerate, similar to Fusobacterium spp. Succinivibrio showed similar correlations as Bacteroides (Table 4). The strongest correlation was that of Fibrobacter spp. with total SCFA concentrations (r = 0.86; P < 0.001), which was supported by additional positive correlations with acetate and isovalerate concentrations. Lachnospira spp. showed a highly negative correlation to ammonia concentrations (P < 0.001).
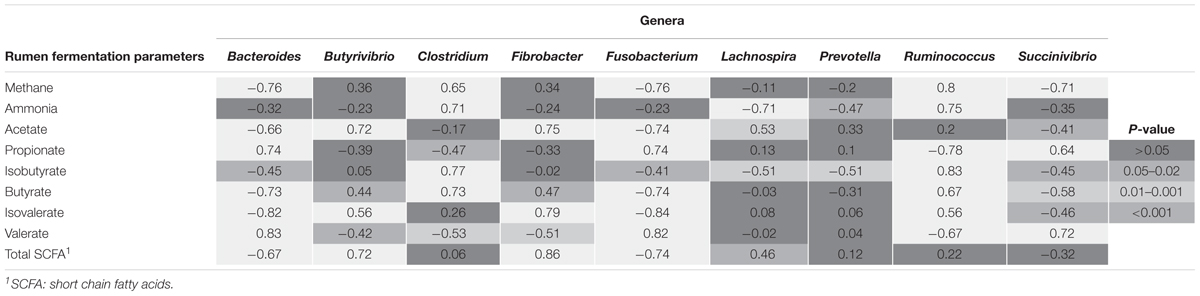
TABLE 4. Pearson correlation coefficients (r) between rumen fermentation parameters and genera significantly impacted by supplementation.
Discussion
We hypothesized that the supplementation of S. striata, a plant with known antimicrobial properties (Monsef-Esfahani et al., 2010; Mahboubi et al., 2013), would elicit similar propionigenic and methano-static effects to the ionophore monensin as a result of changes in the rumen microbiome. The most important findings of this study were a decrease in methane production, an enhanced propionate proportion and a higher total concentration of SCFA as a result of S. striata supplementation. The increasing effect of S. striata on propionate concentration, while decreasing effect on methane and ammonia production, resembles the effects of monensin. However, changes to the rumen microbiome as a result of S. striata supplementation were less dramatic than those seen under monensin supplementation.
In the current study, the degradation of aNDFom and CP decreased with S. striata-High supplementation in vitro. This agrees with previously reported data showing lower degradation of DM by addition of some flavonoid-rich plant extracts and pure flavonid compounds (Oskoueian et al., 2013; Kim et al., 2015). Previous studies have also reported reductions in DM and NDF digestibility with monensin (Poos et al., 1979; Anassori et al., 2012). However, in this study, S. striata-High supplementation had a similar effect on degradation as monensin with the exception of CP. This discrepancy among the different studies may be related to several factors such as diet composition, the period of adaptation to the product, the time when the samples were collected, the method of analysis used and the type and concentrations of the feed additives (Bodas et al., 2012). Ionophores are also generally microbial inhibitors and reduce both cellulose digestion and microbial yield (Demeyer and Van Nevel, 1979). Therefore, the effect of supplementation was assumed to be associated with a decrease in bacterial abundance. However, bacterial abundance as measured by copy number and total number of sequences was not affected by supplementation with either monensin or S. striata. There is evidence showing that improved feed efficiency is correlated with lower richness in the rumen microbiota (Shabat et al., 2016). Therefore, a possible reason for improved feed efficiency in previous in vivo studies with monensin might have been due to lower diversity of the microbiome. As a similar reduction in microbial diversity was observed with S. striata supplementation, it can be inferred that S. striata would also improve feed efficiency. In line with this, we found that S. striata improved efficiency of fermentation (mM SCFA per unit DM degraded).
In the present study, both monensin and S. striata additions caused an increase in propionate concentration, resulting in a decrease in the ratio of acetate to propionate. Effect of S. striata on decreasing acetate to propionate ratio confirmed results of a previous study reporting that feeding flavonoids compounds increased propionate and decreased the acetate to propionate ratio in dairy cows fed a high concentrate diet (Balcells et al., 2012). These results are also consistent with those of previous in vitro studies (Callaway et al., 1997; Shen et al., 2016, 2017). Ionophores work by inhibiting hydrogen producers such as Ruminococcus and Butyrivibrio and thereby favor propionate and succinate producers (Schären et al., 2017; Shen et al., 2017). The suppression of hydrogen transport diverts carbon away from acetate production toward propionate and increases irreversible acetate loss in the synthesis of butyrate. This same mode of action is hypothesized for S. striata based on its antimicrobial properties. In this study, monensin decreased both Butyrivibrio and Ruminococcus species (butyrate and acetate producers, respectively) and increased the total populations of Bacteroides, Fusobacterium, and Succinivibrio, which are recognized as major propionate producers (Russell, 2002; Puniya et al., 2015). However, supplementation with S. striata, despite decreasing methane production did not show the same increases in Bacteroides, Fusobacterium, and Succinivibrio and only minor decreases in Ruminococcus. In contrast, S. striata supplementation increased the relative abundance of Fibrobacter spp., of which the largest family member is F. succinogenes (Jeyanathan et al., 2014). F. succinogenes similar to Succinivibrio (Pope et al., 2011) produces succinate as its principal fermentation end product, while other rumen microbes (e.g., Selenomonas ruminantium; Scheifinger and Wolin, 1973) produce propionate via the succinate pathway (Shen et al., 2017). While in this study Fibrobacter spp. was significantly increased in the S. striata treatments, lactate-producing microbes such as Selenomonas were not found to be significantly affected by treatments, indicating that other microbes are potentially responsible for the production of propionate in the S. striata supplemented groups. One possibility for the differences between groups is the changes in the Bacteroides group, which was significantly increased in the S. striata supplementation and is predominated by the genus Prevotella spp. Prevotella spp. is a well known group of bacteria with extremely diverse metabolic capabilities (Bekele et al., 2010). It is possible that a member of this group is able to convert succinate produced by the Fibrobacter spp. to propionate, explaining the variation between the monensin and S. striata groups. Interestingly, the total number of Butyrivibrio also increased with increasing dosages of S. striata, which is in direct contrast to the effects of monensin supplemented groups found in this experiment. Similar to a previous study (Shen et al., 2017), we observed that the relative abundance of Butyrivibrio and Pseudobutyrivibrio in the S. striata treatments was significantly higher than in the monensin treatment. B. fibrisolvens (Stewart et al., 1997) and Pseudobutyrivibrio spp. (Kopečný et al., 2003) are important butyrate-producing species in the rumen and their presence may explain the higher butyrate levels in the S. striata groups compared to monensin group. There is also some evidence of growth promoting effects of phenolic compounds on several pure cultures of cellulolytic and non-cellulolytic bacteria resulting in enhanced acetate production in vitro (Borneman et al., 1986). In particular, the cellulolytic bacteria Treponema and Fibrobacter increased by S. striata in the current study, which could explain comparable acetate levels to the control. Accordingly, the abundance of Fibrobacter positively correlated with acetate concentration. The positive influence of S. striata on total SCFA production is potentially of great benefit to performance animals in order to meet the increased energy requirements of growth and production.
The potential of plant secondary metabolites to affect methane production has varied extensively ranging from -30 to +30% in a screening study with 450 plant species (Bodas et al., 2008). In the present study, the extent of methane reduction was, respectively, 29–36% in S. striata-Low and S. striata-High which, based on a study that reviewed enteric methane mitigation options, falls within the range of medium to high decrement (Hristov et al., 2013). Scrophularia extract supplemented treatments showed a decreased overall abundance of Firmicutes, which are important ruminal H2-producing microorganisms (Shen et al., 2017), whereas monensin supplementation showed no effect. These results contradict previous research which demonstrated that monensin can directly inhibit H2-producing bacteria, thereby indirectly decreasing methane production (Russell and Houlihan, 2003). Despite differences in microbial abundance between monensin and S. striata supplemented groups, both groups had decreases in CH4 and increases in propionate productions, with a dose dependant response being seen in the S. striata supplementation. However, the mechanism of action on methane production is probably different between monensin and S. striata. In contrast to S. striata, supplementation with monensin decreased the copy numbers of both Archaea and Protozoa populations. This supports previous research which noted a dose dependant response in Protozoa to monensin (Dennis et al., 1986). Therefore, the antiprotozoal effect of monensin seen in this experiment may have resulted indirectly in the reduction of the methanogenic Archaea due to their attachment to and dependency on Protozoa for hydrogen (Morgavi et al., 2010). In S. striata-High supplementation groups, Protozoa copy numbers were significantly increased and yet Archaea numbers were decreased. Since differences were seen in the copy numbers in the S. striata-Low supplemented groups for both Protozoa and Archaea, this indicated a dose dependant response in these groups to S. striata supplementation. These results again contradict previous research which demonstrated that monensin does not directly inhibit methanogens (Russell and Houlihan, 2003).
Since defaunation decreases rumen ammonia concentration resulting in lower deamination, an increased uptake of ammonia by the bacterial population and a smaller recycling of microbial proteins, low Protozoa counts could explain the decreased ammonia concentrations with monensin. However, in the S. striata-High treatment, Protozoa populations were highest and ammonia concentrations lowest. Two major groups of bacteria are responsible for ammonia production in the rumen; bacteria with low activity and high number and those with low numbers and high specific activity (Russell et al., 1991). Clostridium aminophilum, Clostridium sticklandii (Russell et al., 1988), and Peptostreptococcus anaerobius (McIntosh et al., 2003) have been recognized as important hyper-ammonia producing bacteria. The sensitivity of C. sticklandii and P. anaerobius to essential oils has been previously reported (McIntosh et al., 2003). Therefore, the lower ammonia production and CP degradation in S. striata treatments could be attributed to lower abundance of genus Clostridium spp. Ruminococcus also showed positive significant correlations with ammonia concentration and their abundances decreased with supplementation of monensin and S. striata. Lower ammonia concentrations in S. striata-High compared to monensin suggests that a higher dose of S. striata had greater impact on ammonia producing species. Inhibition of ruminal protein degradation by S. striata plant extract has important implications on improving post ruminal delivery of true protein (Szumacher-Strabel and Cieślak, 2010).
Conclusion
Addition of hydro-alcoholic extract of S. striata to rumen fluid in vitro elicited similar propionigenic, and methanostatic effects to monensin and decreased the acetate:propionate ratio. Microbial analysis, however, showed that S. striata and monensin have distinctly different impacts on relative abundances of the total populations of Protozoa and Archaea, as well as effects on the diversity and taxonomy of the bacterial population. The potential of high level of S. striata to decrease ammonia concentration and protein degradability at a greater extent than monensin is promising as it could be translated to more efficacious utilization of nitrogen by the host animal. However, these promising results necessitate the investigation of using S. striata in in vivo feeding trials.
Author Contributions
MBV, FK, and QZ designed the experiments. MBV and FK performed the experiments. RP analyzed and interpreted the data. RP and MBV drafted the paper. All authors read and approved the final version of the manuscript.
Funding
This work has been supported by the Center for International Scientific Studies and Collaboration (CISSC), Ministry of Science, Research and Technology of Iran, and Institute of Animal Nutrition and Functional Plant Compounds, Department for Farm Animals and Veterinary Public Health, University of Veterinary Medicine, Vienna, Austria. The authors acknowledge the funding support of the Vetmeduni Post Doc Program.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Prof. R. Chizzola (Institute of Animal Nutrition and Functional Plant Compounds), S. Bauer (Institute of Meat Hygiene, Meat Technology and Food Science), and Dr. H. J. Busse (Institute of Bacteriology, Mycology and Hygiene) for helping with extraction. The skillful assistance of P. Pourazad, M. Qumar, A. Dockner, S. Sharma, M. Wild, S. Leiner, and A. Sener in this experiment is greatly appreciated.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02164/full#supplementary-material
FIGURE S1 | Mean fermenter pH for each treatment group, on each day of the experimental period, averaged from all runs.
TABLE S1 | Ingredients and chemical composition of the concentrate and basal diet used as substrate in the RUSITEC.
References
Anassori, E., Dalir-Naghadeh, B., Pirmohammadi, R., Taghizadeh, A., Asri-Rezaei, S., Farahmand-Azar, S., et al. (2012). In vitro assessment of the digestibility of forage based sheep diet, supplemented with raw garlic, garlic oil and monensin. Vet. Res. Forum 3, 5–11.
Bahrami, A. (2011). The effectiveness of Scrophularia Striata on Newcastle disease. Aust. J. Basic Appl. Sci. 5, 2883–2888.
Bahrami, A., and Valadi, A. (2010). Effect of Scrophularia ethanolic leaves extracts on Staphylococcus aureus. Int. J. Pharm. 6, 393–396. doi: 10.3923/ijp.2010.431.434
Balcells, J., Aris, A., Serrano, A., Seradj, A. R., Crespo, J., and Devant, M. (2012). Effects of an extract of plant flavonoids (Bioflavex) on rumen fermentation and performance in heifers fed high-concentrate diets. J. Anim. Sci. 90, 4975–4984. doi: 10.2527/jas2011-4955
Bekele, A. Z., Koike, S., and Kobayashi, Y. (2010). Genetic diversity and diet specificity of ruminal prevotella revealed by 16S rRNA gene-based analysis. FEMS Microbiol. Lett. 305, 49–57. doi: 10.1111/j.1574-6968.2010.01911.x
Bergman, E. N. (1990). Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 70, 567–590. doi: 10.1152/physrev.1990.70.2.567
Bodas, R., López, S., Fernandez, M., García-González, R., Rodríguez, A. B., Wallace, R. J., et al. (2008). In vitro screening of the potential of numerous plant species as antimethanogenic feed additives for ruminants. Anim. Feed Sci. Technol. 145, 245–258. doi: 10.1016/j.anifeedsci.2007.04.015
Bodas, R., Prieto, N., García-González, R., Andrés, S., Giráldez, F. J., and López, S. (2012). Manipulation of rumen fermentation and methane production with plant secondary metabolites. Anim. Feed Sci. Technol. 176, 78–93. doi: 10.1016/j.anifeedsci.2012.07.010
Bokulich, N. A., Subramanian, S., Faith, J. J., Gevers, D., Gordon, J. I., Knight, R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–66. doi: 10.1038/nmeth.2276
Borneman, W. S., Akin, D. E., and VanEseltine, W. P. (1986). Effect of phenolic monomers on ruminal bacteria. Appl. Environ. Microbiol. 52, 1331–1339.
Broudiscou, L. P., Papon, Y., and Broudiscou, A. F. (2000). Effects of dry plant extracts on fermentation and methanogenesis in continuous culture of rumen microbes. Anim. Feed Sci. Technol. 87, 263–277. doi: 10.1016/S0377-8401(00)00193-0
Callaway, T. R., De Melo, A. M., and Russell, J. B. (1997). The effect of nisin and monensin on ruminal fermentations in vitro. Curr. Microbiol. 35, 90–96. doi: 10.1007/s002849900218
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Cardozo, P. W., Calsamiglia, S., Ferret, A., and Kamel, C. (2005). Screening for the effects of natural plant extracts at different pH on in vitro rumen microbial fermentation of a high-concentrate diet for beef cattle 1. J. Anim. Sci. 83, 2572–2579. doi: 10.2527/2005.83112572x
Demeyer, D. I., and Van Nevel, C. J. (1979). Effect of defaunation on the metabolism of rumen micro-organisms. Br. J. Nutr. 42, 515–524. doi: 10.1079/BJN19790143
den Besten, G., van Eunen, K., Groen, A. K., Venema, K., Reijngoud, D. J., and Bakker, B. M. (2013). The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid. Res. 54, 2325–2340. doi: 10.1194/jlr.R036012
Denman, S. E., and McSweeney, C. S. (2006). Development of a real-time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. FEMS Microbiol. Ecol. 58, 572–582.
Dennis, S. M., Nagaraja, T. G., and Dayton, A. D. (1986). Effect of lasalocid, monensin and thiopeptin on rumen protozoa. Res. Vet. Sci. 41, 251–256. doi: 10.1016/S0034-5288(18)30608-8
Duffield, T. F., Rabiee, A. R., and Lean, I. J. (2008). A meta-analysis of the impact of Monensin in lactating dairy cattle. Part 2. Production effects. J. Dairy Sci. 91, 1347–1360. doi: 10.3168/jds.2007-0608
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Federal Ministry of Health (2004). Verordnung der Bundesministerin für Gesundheit und Frauen über die Mindestanforderungen für die Haltung von Pferden und Pferdeartigen, Schweinen, Rindern, Schafen, Ziegen, Schalenwild, Lamas, Kaninchen, Hausgeflügel, Straußen und Nutzfischen (1. Tierhaltungsverordnung) StF: BGBl. II Nr. 485/2004. Vienna: Bundeskanzleramt Österreich.
Goel, G., Makkar, H. P. S., and Becker, K. (2008). Changes in microbial community structure, methanogenesis and rumen fermentation in response to saponin-rich fractions from different plant materials. J. Appl. Microbiol. 105, 770–777. doi: 10.1111/j.1365-2672.2008.03818.x
Green, B. L., McBride, B. W., Sandals, D., Leslie, K. E., Bagg, R., and Dick, P. (1999). The impact of a Monensin controlled-release capsule on subclinical ketosis in the transition dairy cow. J. Dairy Sci. 82, 333–342. doi: 10.3168/jds.S0022-0302(99)75240-9
Hinkle, D. E., Wiersma, W., and Jurs, S. G. (2003). Applied Statistics for the Behavioral Sciences. Boston, MA: Houghton Mifflin College Division.
Hristov, A. N., Oh, J., Firkins, J. L., Dijkstra, J., Kebreab, E., Waghorn, G., et al. (2013). Special topics—mitigation of methane and nitrous oxide emissions from animal operations: I. A review of enteric methane mitigation options. J. Anim. Sci. 91, 5045–5069. doi: 10.2527/jas.2013-6583
Jeyanathan, J., Martin, C., and Morgavi, D. P. (2014). The use of direct-fed microbials for mitigation of ruminant methane emissions: a review. Animal. 8, 250–261. doi: 10.1017/S1751731113002085
Johnson, K. A., and Johnson, D. E. (1995). Methane emissions from cattle. J. Anim. Sci. 73, 2483–2492. doi: 10.2527/1995.7382483x
Kim, E. T., Guan, L. L., Lee, S. J., Lee, S. M., Lee, S. S., Lee, I. D., et al. (2015). Effects of flavonoid-rich plant extracts on in vitro ruminal methanogenesis, microbial populations and fermentation characteristics. Asian Aust. J. Anim. Sci. 28, 530–537. doi: 10.5713/ajas.14.0692
Klevenhusen, F., Deckardt, K., Sizmaz,Ö., Wimmer, S., Muro-Reyes, A., Khiaosa-Ard, R., et al. (2015). Effects of black seed oil and Ferula elaeochytris supplementation on ruminal fermentation as tested in vitro with the rumen simulation technique (Rusitec). Anim. Prod. Sci. 55, 736–744. doi: 10.1071/AN13332
Kong, Y., Teather, R., and Forster, R. (2010). Composition, spatial distribution, and diversity of the bacterial communities in the rumen of cows fed different forages. FEMS Microbiol. Ecol. 74, 612–622. doi: 10.1111/j.1574-6941.2010.00977.x
Kopečný, J., Zorec, M., Mrazek, J., Kobayashi, Y., and Marinšek-Logar, R. (2003). Butyrivibrio hungatei sp. nov. and Pseudobutyrivibrio xylanivorans sp. nov., butyrate-producing bacteria from the rumen. Int. J. Syst. Evol. Microbiol. 53(Pt 1), 201–209.
Mahboubi, M., Kazempour, N., and Nazar, A. R. B. (2013). Total phenolic, total flavonoids, antioxidant and antimicrobial activities of Scrophularia striata Boiss extracts. Jundishapur J. Nat. Pharm. Prod. 8, 15–19. doi: 10.17795/jjnpp-7621
McAllister, T. A., and Newbold, C. J. (2008). Redirecting rumen fermentation to reduce methanogenesis. Aust. J. Exp. Agric. 48, 7–13. doi: 10.1071/EA07218
McDougall, E. I. (1948). Studies on ruminant saliva. 1. The composition and output of sheep’s saliva. Biochem. J. 43, 99–109. doi: 10.1042/bj0430099
McIntosh, F. M., Williams, P., Losa, R., Wallace, R. J., Beever, D. A., and Newbold, C. J. (2003). Effects of essential oils on ruminal microorganisms and their protein metabolism. Appl. Environ. Microbiol. 69, 5011–5014. doi: 10.1128/AEM.69.8.5011-5014.2003
Monsef-Esfahani, H. R., Hajiaghaee, R., Shahverdi, A. R., Khorramizadeh, M. R., and Amini, M. (2010). Flavonoids, cinnamic acid and phenyl propanoid from aerial parts of Scrophularia striata. Pharm. Biol. 48, 333–336. doi: 10.3109/13880200903133829
Morgavi, D. P., Forano, E., Martin, C., and Newbold, C. J. (2010). Microbial ecosystem and methanogenesis in ruminants. Animal 4, 1024–1036. doi: 10.1017/S1751731110000546
Muyzer, G., De Waal, E. C., and Uitterlinden, A. G. (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59, 695–700.
Oskoueian, E., Abdullah, N., and Oskoueian, A. (2013). Effects of flavonoids on rumen fermentation activity, methane production, and microbial population. Biomed. Res. Int. 2013, 1–8. doi: 10.1155/2013/349129
Peterson, J., Garges, S., Giovanni, M., McInnes, P., Wang, L., Schloss, J. A., et al. (2009). The NIH human microbiome project. Genome Res. 19, 2317–2323. doi: 10.1101/gr.096651.109
Poos, M. I., Hanson, T. L., and Klopfenstein, T. J. (1979). Monensin effects on diet digestibility, ruminal protein bypass and microbial protein synthesis 1. J. Anim. Sci. 48, 1516–1524. doi: 10.2527/jas1979.4861516x
Pope, P. B., Smith, W., Denman, S., Tringe, S. G., Barry, K., Hugenholtz, P., et al. (2011). Isolation of Succinivibrionaceae implicated in low methane emissions from Tammar wallabies. Science 333, 646–648. doi: 10.1126/science.1205760
Puniya, A. K., Salem, A. Z., Kumar, S., Dagar, S. S., Griffith, G. W., Puniya, A., et al. (2015). Role of live microbial feed supplements with reference to anaerobic fungi in ruminant productivity: a review. J. Integr. Agric. 14, 550–560. doi: 10.1016/S2095-3119(14)60837-6
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Qumar, M., Khiaosa-ard, R., Pourazad, P., Wetzels, S. U., Klevenhusen, F., Kandler, W., et al. (2016). Evidence of in vivo absorption of lactate and modulation of short chain fatty acid absorption from the reticulorumen of non-lactating cattle fed high concentrate diets. PLoS One 11:e0164192. doi: 10.1371/journal.pone.0164192
Russell, J. B. (2002). Rumen Microbiology, and Its Role in Ruminant Nutrition. Ithaca, NY: Cornell University.
Russell, J. B., and Houlihan, A. J. (2003). Ionophore resistance of ruminal bacteria and its potential impact on human health. FEMS Microbiol. Rev. 27, 65–74. doi: 10.1016/S0168-6445(03)00019-6
Russell, J. B., Onodera, R., and Hino, T. (1991). “Ruminal protein fermentation: new perspectives on previous contradictions,” in Physiological Aspects of Digestion and Metabolism in Ruminants, eds T. Tsuda, Y. Sasaki, and R. Kawashima (San Diego, CA: Academic Press), 681–697. doi: 10.1016/B978-0-12-702290-1.50034-5
Russell, J. B., Strobel, H. J., and Chen, G. J. (1988). Enrichment and isolation of a ruminal bacterium with a very high specific activity of ammonia production. Appl. Environ. Microbiol. 54, 872–877.
Schären, M., Drong, C., Kiri, K., Riede, S., Gardener, M., Meyer, U., et al. (2017). Differential effects of Monensin and a blend of essential oils on rumen microbiota composition of transition dairy cows. J. Dairy Sci. 100, 2765–2783. doi: 10.3168/jds.2016-11994
Scheifinger, C. C., and Wolin, M. J. (1973). Propionate formation from cellulose and soluble sugars by combined cultures of Bacteroides succinogenes and Selenomonas ruminantium. Appl. Microbiol. 26, 789–795.
Shabat, S. K. B., Sasson, G., Doron-Faigenboim, A., Durman, T., Yaacoby, S., Miller, M. E. B., et al. (2016). Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 10, 2958–2972. doi: 10.1038/ismej.2016.62
Shen, J., Liu, Z., Yu, Z., and Zhu, W. (2017). Monensin and nisin affect rumen fermentation and microbiota differently in vitro. Front. Microbial. 8:1111. doi: 10.3389/fmicb.2017.01111
Shen, J. S., Liu, Z., Chen, Y. Y., Lv, P. A., and Zhu, W. Y. (2016). Effects of nisin on in vitro fermentation, methanogenesis and functional microbial populations of the rumen. Acta Microbiol. Sin. 56, 1348–1357.
Siciliano-Jones, J., and Murphy, M. R. (1989). Production of volatile fatty acids in the rumen and cecum–colon of steers as affected by forage:concentrate and forage physical form. J. Dairy Sci. 72, 485–492. doi: 10.3168/jds.S0022-0302(89)79130-X
Śliwiński, B. J., Soliva, C. R., Machmüller, A., and Kreuzer, M. (2002). Efficacy of plant extracts rich in secondary constituents to modify rumen fermentation. Anim. Feed Sci. Technol. 101, 101–114. doi: 10.1016/S0377-8401(02)00139-6
Soliva, C., and Hess, D. (2007). “Measuring methane emission of ruminants by in vitro and in vivo techniques,” in Measuring Methane Production from Ruminants, eds H. P. S. Makkar and P. E. Vercoe (Dordrecht: Springer), 15–135. doi: 10.1007/978-1-4020-6133-2_2
Stewart, C. S., Flint, H. J., and Bryant, M. P. (1997). The Rumen bacteria. In the Rumen Microbial Ecosystem. Dordrecht: Springer, 10–72. doi: 10.1007/978-94-009-1453-7_2
Sylvester, J. T., Karnati, S. K., Yu, Z., Morrison, M., and Firkins, J. L. (2004). Development of an assay to quantify rumen ciliate protozoal biomass in cows using real-time PCR. J. Nutr. 134, 3378–3384. doi: 10.1093/jn/134.12.3378
Szumacher-Strabel, M., and Cieślak, A. (2010). Potential of phytofactors to mitigate rumen ammonia and methane production. J. Anim. Feed Sci. 19, 319–337. doi: 10.22358/jafs/66296/2010
Udén, P., Robinson, P. H., and Wiseman, J. (2005). Use of detergent system terminology and criteria for submission of manuscripts on new, or revised, analytical methods as well as descriptive information on feed analysis and/or variability. Anim. Feed Sci. Technol. 118, 181–186. doi: 10.1016/j.anifeedsci.2004.11.011
VDLUFA (2007). Handbuch der Landwirtschaftlichen Versuchs-und Untersuchungsmethodik (VDLUFA-Methodenbuch), Bd. III Die Chemische Untersuchung von Futtermitteln. Darmstadt: VDLUFA-Verlag.
Weatherburn, M. W. (1967). Phenol-hypochlorite reaction for determination of ammonia. Anal. Chem. 39, 971–974. doi: 10.1021/ac60252a045
Yilmaz, P., Parfrey, L. W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., et al. (2013). The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Res. 28, D643–D648. doi: 10.1093/nar/gkt1209
Keywords: greenhouse gasses, fermentation profile, phytogenic compounds, ionophores, rumen microbiome, Rusitec
Citation: Bagheri Varzaneh M, Klevenhusen F, Zebeli Q and Petri R (2018) Scrophularia striata Extract Supports Rumen Fermentation and Improves Microbial Diversity in vitro Compared to Monensin. Front. Microbiol. 9:2164. doi: 10.3389/fmicb.2018.02164
Received: 17 April 2018; Accepted: 23 August 2018;
Published: 19 September 2018.
Edited by:
Sharon Ann Huws, Aberystwyth University, United KingdomReviewed by:
Alex V. Chaves, The University of Sydney, AustraliaLeticia Abecia, Center for Cooperative Research in Biosciences, Spain
Copyright © 2018 Bagheri Varzaneh, Klevenhusen, Zebeli and Petri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Renee Petri, cmVuZWUucGV0cmlAdmV0bWVkdW5pLmFjLmF0
 Maryam Bagheri Varzaneh
Maryam Bagheri Varzaneh Fenja Klevenhusen
Fenja Klevenhusen Qendrim Zebeli
Qendrim Zebeli Renee Petri
Renee Petri