- 1Department of Soil, Plant and Food Sciences, University of Bari Aldo Moro, Bari, Italy
- 2Public Laboratory of Research SELGE Network No. 14, Bari, Italy
Trichoderma asperellum strain icc012 and Trichoderma gamsii strain icc080, the microbial active ingredients of RemedierTM (ISAGRO, Novara, Italy), are biocontrol agents (BCAs) employable for crop protection against a wide range of fungal pathogens, including soil-borne pathogens and fungi involved in grapevine trunk disease. In this study, single and duplex real-time quantitative PCR (qPCR) methods to detect and quantify T. asperellum and T. gamsii were developed. Primers/probe sets were designed on the T. asperellum and T. gamsii rpb2 genes and tested for specificity on a panel of microorganisms commonly associated with grape wood and soil. No differences were observed comparing single- and duplex-qPCR assays on different BCAs, 1 pg of target DNA was detected approximately at Cq = 34. R2-values and the efficiency were always equal to 0.99 and >80%, respectively. The detection limit of the duplex-qPCR assay on artificially inoculated samples was 2 × 103 and 4 × 104 conidia g-1 of grape wood tissue and soil, respectively. The methods will be useful to better schedule BCA application in the field and in grapevine nurseries, as well as for investigating the dynamic of BCA populations.
Introduction
The application of biocontrol agents (BCAs) in sustainable agriculture model represents an eco-friendly strategy compared with the use of synthetic plant protection products (PPPs) for managing weeds, insects and fungal pathogens including fungicide-resistant mutants (Jensen et al., 2016; Bruce et al., 2017; Rotolo et al., 2018).
The genus Trichoderma, a cosmopolitan inhabitant of soil and plant root ecological niches includes the most explored BCA species, representing over 60% of all the currently registered BCAs used for the management of plant pathogens (Benítez et al., 2004; Harman et al., 2004; Mukherjee et al., 2013; Hyder et al., 2017; Sharma et al., 2017). Their biological activity is closely related to the ability of: (i) producing a wide range of lysing enzymes; (ii) degrading substrates; (iii) possessing high resistance to microbial inhibitors; (iv) competing for nutrients and space, (v) acting directly through mycoparasitism, (vi) producing antifungal metabolites; (vii) inducing systemic resistance in plants (Strange, 1993; Harman et al., 2004; Shoresh et al., 2005; Reino et al., 2008; Lorito et al., 2010; Qualhato et al., 2013). Trichoderma spp. are fast-growing, strong spore producers and stimulate plant growth through the production of promoting molecules (e.g., Eziashi et al., 2007; Vinale et al., 2008; Hermosa et al., 2012; Singh et al., 2014).
Since combining two or more beneficial microbes in a biopesticide would be advantageous to BCA management (Raupach and Kloepper, 1998), the mixture of T. asperellum strain icc012 and T. gamsii strain icc080 is used in RemedierTM to increase the activity and widening the environmental adaptability (Liguori, 2016). This microbial pesticide is registered against soil-borne pathogens affecting horticultural crops and turfs, and it is the only BCA-based PPP allowed in Italy to control pathogens associated with grapevine trunk diseases (GTDs).
To date, 133 fungal species belonging to 34 genera have been associated with GTDs affecting, singularly or simultaneously, table and wine grapes as well as rootstocks. PPPs effective in controlling GTDs-associated fungi are still lacking, and the BCAs Trichoderma atroviride and Trichoderma harzianum were the ones most studied for their effectiveness (Gramaje et al., 2018). On the other hand, information on T. asperellum and T. gamsii refer mostly to their use for cutting wounds protection while the population dynamics has been scarcely studied and appropriate monitoring systems are lacking. Yet, the monitoring of these BCAs in natural environments is essential to evaluate their effectiveness and scheduling their applications (Torsvik and Øvreås, 2002).
Molecular detection and quantification of fungal species (Filion et al., 2003; Lievens et al., 2006; López-Mondéjar et al., 2010; Sharma and Salwan, 2017) are substituting for conventional techniques, such as those based on the assessment of colony forming units (CFU) and on chemical, biological and immunological assays (Thornton et al., 1994). In fact, the differentiation of Trichoderma using morphological characteristics is very difficult, due to the scarcity of specific traits (Błaszczyk et al., 2011; Devi et al., 2012). Different qPCR and qRT-PCR assays have therefore been proposed for the quantification of T. harzianum (Rubio et al., 2005; López-Mondéjar et al., 2010; Beaulieu et al., 2011), T. atroviride (Cordier et al., 2007; Savazzini et al., 2008) and Trichoderma spp. (Hagn et al., 2007; Kim and Knudsen, 2008).
This study aimed at developing a molecular qPCR tool for an easy detection and quantification of T. asperellum strain icc012 and T. gamsii strain icc080. Comparisons between single- and duplex-qPCR assays were performed, then the assays were validated on fungal DNA extracts from grapevine wood tissue and soil samples contaminated with different concentrations of BCAs conidia.
Materials and Methods
Strains and Media
Trichoderma asperellum icc012 and T. gamsii icc080 were kindly supplied by Isagro SpA (Novara, Italy). Non-target species of fungi, yeasts and bacteria used were from the microbial culture collection of our Department.
Fungi and yeasts were routinely grown on potato dextrose agar (PDA: infusion from 200 g peeled and sliced potatoes kept at 60 ± 1°C for 1 h, 20 g dextrose per liter of distilled water, pH adjusted to 6.5, and 20 g agar Oxoid No. 3) at 21 ± 1°C in the darkness. Alternatively, bacteria were routinely grown on Luria-Bertani medium (LB: 10 g tryptone-peptone, 5 g yeast extract, pH adjusted to 7.0, and 14 g agar per liter of distilled water) at 25 ± 1°C in the darkness.
Primers/Probe Sets Design
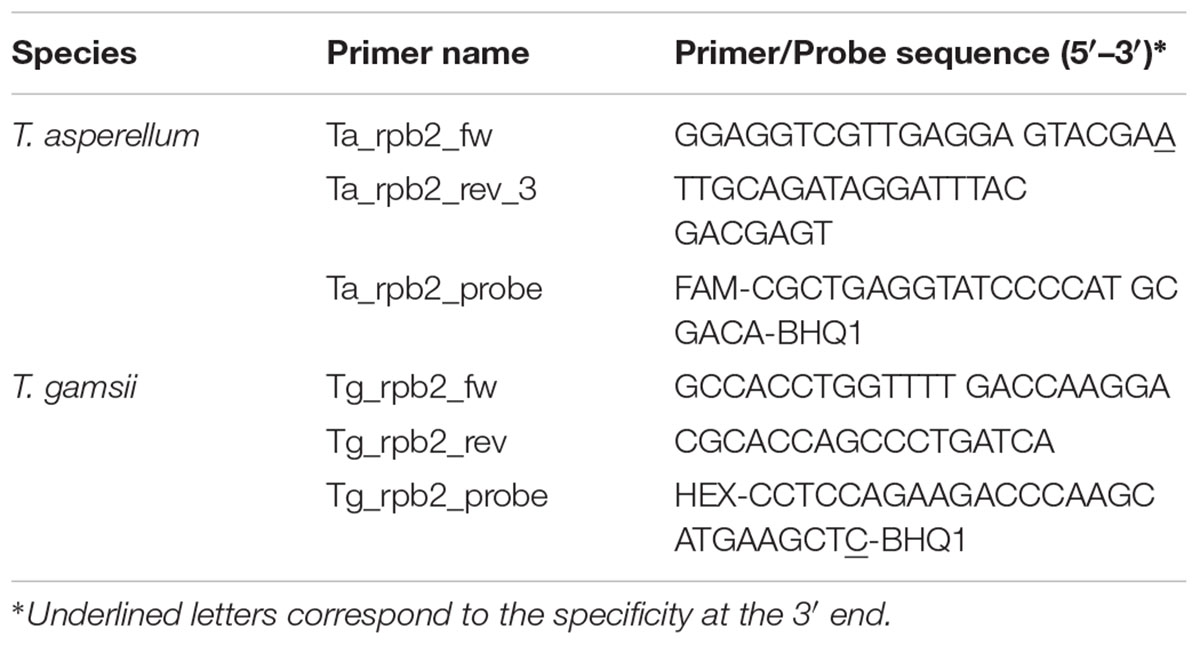
Sequences of the genes translation elongation factor 1-alpha (tef1), endochitinase 42 (ech42) and RNA polymerase B subunit II (rpb2) of target and non-target Trichoderma species were retrieved from GenBank1, and aligned using the SeqMan Pro software (DNASTAR, Lasergene, Madison, WI, United States). Based on the highest presence of species-specific single-nucleotide polymorphisms (SNPs), the rpb2 gene was selected and the sequences of different Trichoderma species (Supplementary Figure S1), including 64 sequences of T. asperellum and 10 sequences of T. gamsii, were aligned and examined in silico using the SeqMan Pro software (DNASTAR). The SNPs identified in T. asperellum and T. gamsii were used. The primers/probe sets (Table 1) were manually designed primarily in order to include the specific SNPs in the 3′ position of the primer forward (base 504 for T. asperellum reference sequence GenBank accession No. GU198278.1) and probe (base 806 for T. gamsii reference sequence GenBank accession No. KJ665270.1). Other SNPs in different positions of T. asperellum and T. gamsii primers/probe sets were also recorded. The absence of secondary structures and dimers and the feasibility of the use of Taq Man®-qPCR were verified using the Primer Express 3.0 software (Applied Biosystems, Foster City, CA, United States). Primers/probe sets were custom-synthesized (Macrogen, Seoul, South Korea) including FAM (6-carboxyfluorescein) and HEX (6-hexachlorofluorescein) fluorescent dyes to label the T. asperellum and T. gamsii probes, respectively.
DNA Extraction From Trichoderma and qPCR Conditions
Genomic DNA of both BCAs and non-target fungi and yeasts was extracted from 5-day-old colonies grown at 21 ± 1°C on cellophane disks overlaid on PDA, according to the protocol of De Miccolis Angelini et al. (2010). DNA from bacteria was extracted according to Rotolo et al. (2016). Quantity and quality of DNA was assessed using a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States).
Amplifications were performed in a CFX96TM Real-Time PCR Detection System Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, United States) whereas the CFX ManagerTM version 1.0 software (Bio-Rad Laboratories, Hercules, CA, United States) was used for experimental setup and data analysis.
PCR mixes consisted of 6.25 μL of Sso AdvancedTM Universal Probes Supermix (Bio-Rad Laboratories, Hercules, CA, United States), 250 nM of each primer, 150 nM of each probe, 1 (single-qPCR) or 2 (duplex-qPCR) μL of DNA template, and ultrapure water to 12.5 μL. Thermal cycling conditions were 95°C for 2 min, followed by 40 cycles of 95°C for 5 s and 64.5°C for 30 s. All qPCR assays were run with appropriate controls, including the non-template control (NTC). Two replicates of each sample were analyzed, and reactions were repeated at least twice.
The qPCR products were loaded on 1.5% (w/v) agarose gel (Bio-Rad Laboratories, Hercules, CA, United States), including GelRed (Società Italiana Chimici, Rome, Italy), electrophoresed for 110 min at 110 V, and visualized with a Bio-Rad Gel Doc XR 2.0 system (Bio-Rad Laboratories, Hercules, CA, United States).
Specificity and Sensitivity Assays
The specificity of the TaqMan-based duplex-qPCR was assessed on a panel of microorganisms commonly associated with grapevines (Table 2). Genomic grapevine and soil DNAs were also used as external negative control to exclude cross-reaction of the primers/probe sets, and all qPCR assays were run with appropriate controls, including the NTCs.
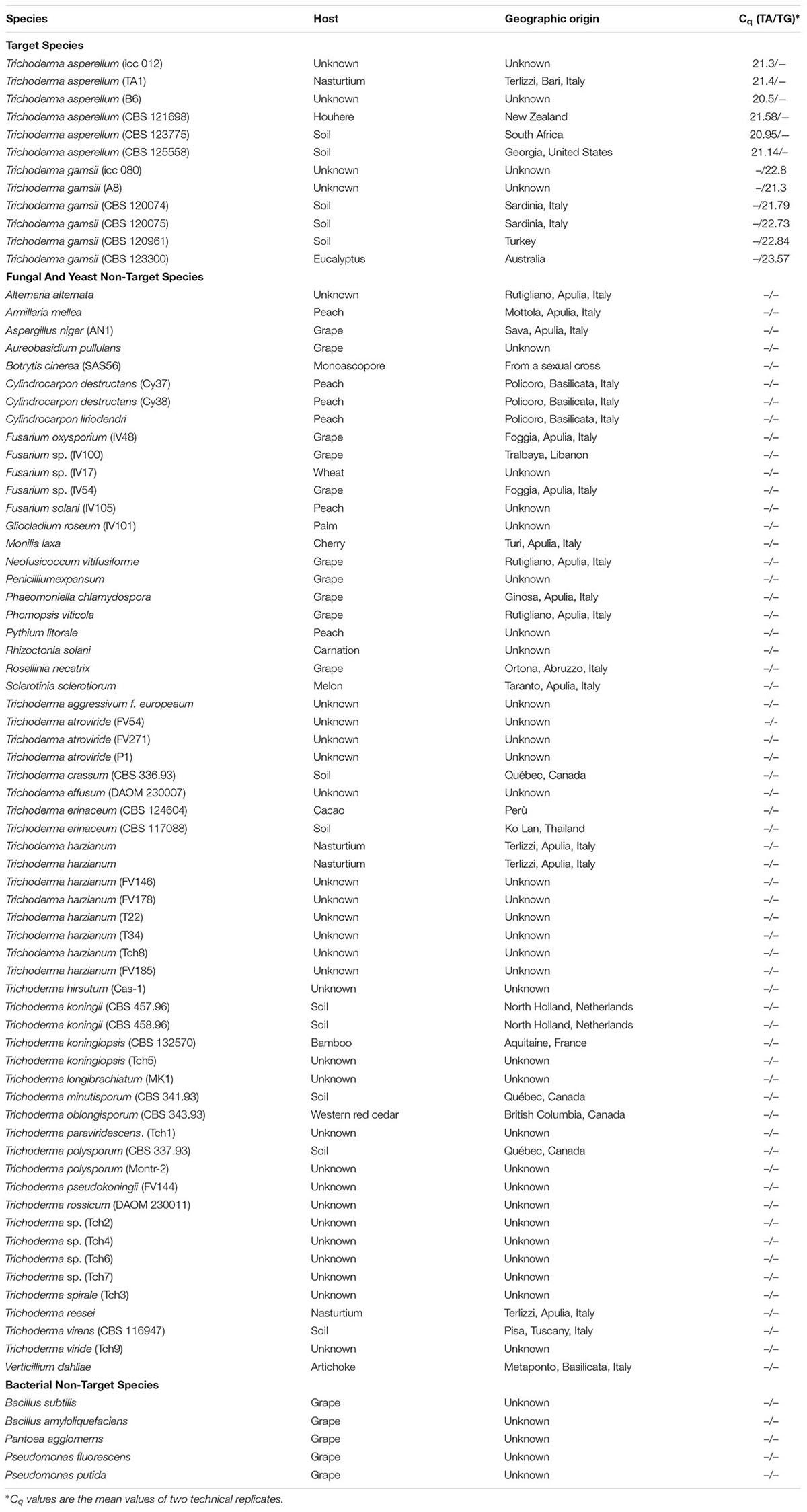
TABLE 2. Quantification cycle (Cq) values of T. asperellum (TA) and T. gamsii (TG) primers/probe sets tested in the specificity assay through duplex-qPCR.
Ten-fold serial dilutions (100 ng to 0.01 pg) of genomic DNA of T. asperellum icc012 and T. gamsii icc080 were used in sensitivity assays. The standard curves for each BCA were generated in both single- and duplex-qPCR by plotting the quantification cycle (Cq) values vs. the Log10 of 10-fold serial dilutions of DNA. Comparison between single- and duplex-qPCR was done for each species. Two replicates of each dilution were analyzed, and reactions were repeated at least twice. qPCR reactions were positive if Cq value was ≤35.
Preparation of Grape Wood and Soil Samples
Conidia of T. asperellum and T. gamsii were scraped from the surface of 7-days-old colonies grown on PDA at 25 ± 1°C in the dark and suspended in sterile distilled water containing 0.05% Tween 20. Mycelial fragments were removed through Miracloth (Calbiochem, Darmstadt, Germany). Aliquots (1 mL) of diluted conidial suspension (from 108 to 100 conidia mL-1) were used to infest 50 mg of grape wood chips (protocol 1) and 50 or 250 mg of clay soil (protocol 1 and protocol 2, respectively) previously sterilized, then ground and sieved at 2 mm to separate the gravel fraction. Samples were centrifuged (Eppendorf, Hamburg, Germany) at 14,000 rpm for 30 min and the pellet was subjected to DNA extraction. Five wood and soil samples artificially infested with T. asperellum or/and T. gamsii conidia were analyzed by qPCR and in the meantime, samples of T. asperellum- or T. gamsii-infested soil were placed on PDA and CFU were counted.
DNA Extraction and Purification, qPCR From Wood and Soil Samples
DNA extraction from wood chips and clay soil (protocol 1) was done using the CTAB method (Cullen et al., 2001), slightly modified as described below. Samples were homogenized in 600 μL extraction buffer (0.12 M Na2HPO4, 1.5 M NaCl, 2% CTAB) with 0.5 g acid-washed glass beads 425–600 μm (Sigma-Aldrich, St. Louis, MO, United States) and 2 steel spheres (5 mm diam.). The suspension was strongly shaken for 5 min at 1,500 oscillations min-1 using a Mixer Mill (MM301, Retsch GmbH, Haan, Germany). The supernatant, collected after centrifugation at 14,000 rpm for 15 min, was transferred in a new 2 mL micro-tube. Extraction was carried out in 750 μL of chloroform. Nucleic acids were collected by centrifugation for 15 min at 14,000 rpm, precipitated with 750 μL of isopropanol at -80 ± 3°C for 30 min, and recovered by centrifugation at 14,000 rpm for 15 min. The pellet, washed with 200 μL of ethanol (70%), was suspended in 50 μL of ultrapure water.
DNA extraction from soil was carried out according to Martin-Laurent et al. (2001, protocol 2).
Wood-DNA extract was purified using Sepharose 6B (Sigma-Aldrich, St. Louis, MO, United States)-columns, while the soil-DNA extract was purified on both Sepharose 6B- and polyvinylpolypyrrolidone (PVPP) (Sigma-Aldrich, St. Louis, MO, United States) columns, prepared as reported by Rotolo et al. (2016). DNA concentration and purity were estimated as described above and DNA was stored at -80°C until use. To verify the success of DNA extraction, the extract was amplified by using the universal primers ITS5/ITS4, and the PCR mixture and conditions previously described. DNA extracted was directly amplified in single- and duplex-qPCR. Curves were generated by plotting the Cq values vs. the Log10 of the number of conidia added to the samples and BCAs were finally quantified as conidia per g-1 of grapevine wood tissue or soil.
Results
Primers/Probe Sets Specificity
SNPs identified in intra- and external species alignments of the rpb2 gene sequences were used for species-specific primers/probe sets design. The 142 and 113 bp amplicons were confirmed for T. asperellum and T. gamsii, respectively (Figure 1).
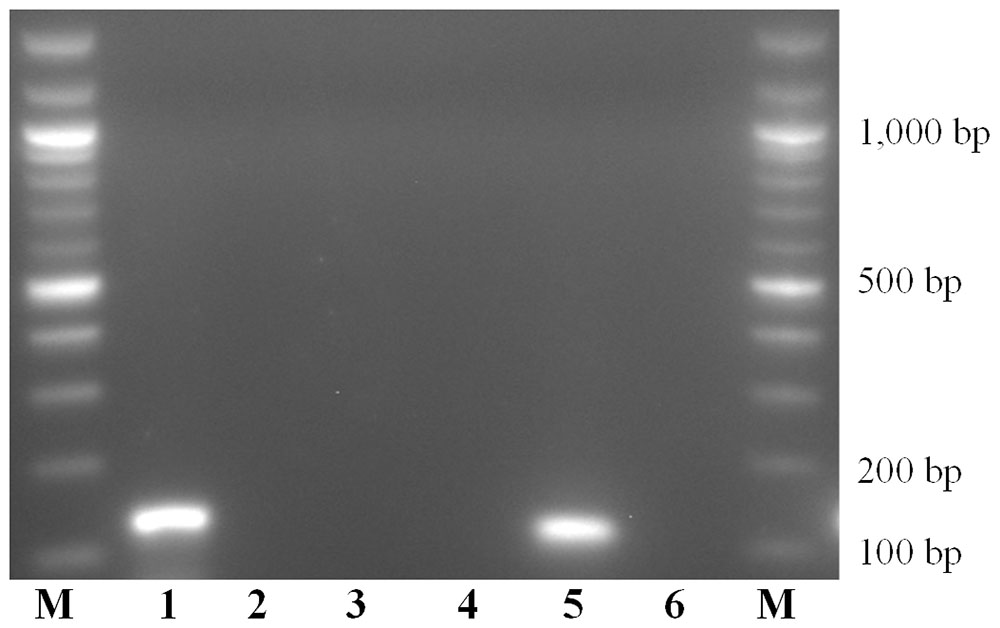
FIGURE 1. Amplicons obtained with Trichoderma asperellum and T. gamsii primers/probe sets. M: 100 bps marker; Amplification mixtures were: 1–3, T. asperellum primers/probe set; 4–6, T. gamsii primers/probe set. Samples analyzed was: 1 and 4, T. asperellum DNA; 2 and 5, T. gamsii DNA; 3 and 6, no template controls.
For duplex-qPCR assays, the best conditions to avoid unspecific amplification products were 64.5°C (annealing temperature), 250 nM/150 nM (primers/probes concentrations) and 35 cycles. The specificity of the assay was also tested against genomic DNA from 66 non-target organisms as well as from grapevines and soil. No amplicons were generated using non-target DNA from some Trichoderma spp. (T. atroviride, T. paraviridescens, and T. polysporium) not carrying the 3′ SNP in the primer forward (T. asperellum) or in the probe (T. gamsii) (Table 2 and Supplementary Figure S1).
Primers/Probe Sets Sensitivity
A linear response was observed from 100 ng to 1 pg of T. asperellum and T. gamsii DNA in single-qPCR (Figures 2A,B). R2 and efficiency of the standard curve were always equal to 0.99 and >80%, respectively, and the linear regression slopes were -3.10 and -3.11, respectively, for T. asperellum and T. gamsii. Both T. asperellum and T. gamsii primers/probe sets showed the same sensitivity when duplexed (Figure 2C). In the duplex-qPCR assay, 1 pg of target DNA for both species was also detected approximately at Cq 34. Unspecific amplification occurred beyond the 35th cycle. In duplex-qPCR, R2 and efficiency of the standard curves were also equal to 0.99 and >80% for both primer/probe sets, while the linear regression slopes were -3.21 and -3.13 for T. asperellum and T. gamsii, respectively.
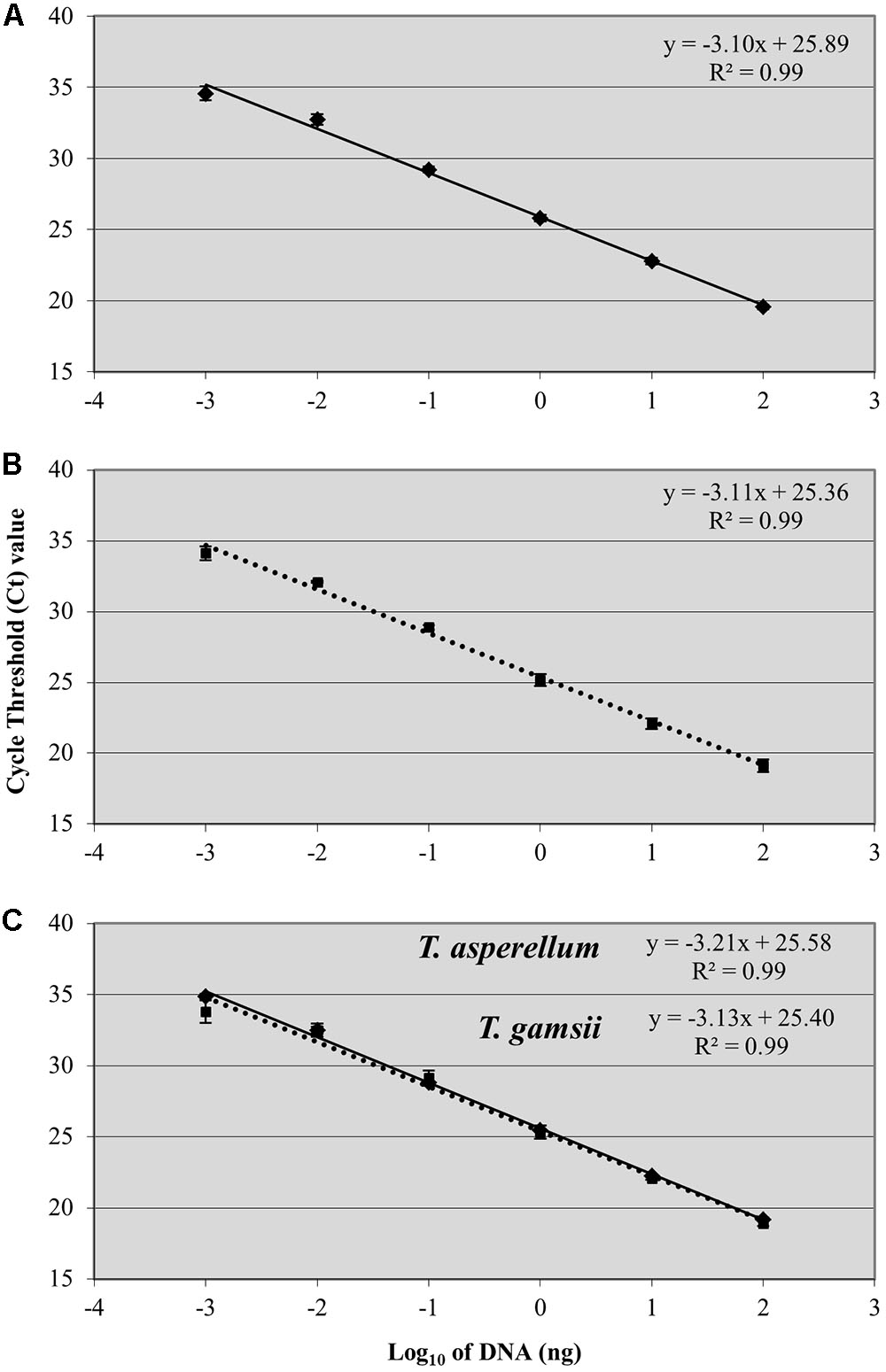
FIGURE 2. Standard curves generated by single-qPCR for Trichoderma asperellum (A) and T. gamsii (B) and duplex-qPCR (C). The Ct values were plotted against the DNA concentrations expressed on a logarithmic scale. Bars mean standard error of two technical replicates.
Validation on Grapevine Wood and Soil
To validate the single- and duplex-qPCR assays, wood chips, and soil samples were artificially infested with a 10-fold dilution of conidial suspensions of T. asperellum and T. gamsii used singularly and in mixture. BCAs were only detected in all samples expected to be positive.
The detection limit in single- and duplex-qPCR was 2 × 103 conidia g-1 of grape wood chips for both BCAs. In single-qPCR, R2 and efficiency of the standard curves were 0.99 and 131.4% for T. asperellum and 0.99 and 137.3% for T. gamsii. In the same assays, slopes values were -2.74 and -2.66 for T. asperellum and T. gamsii, respectively (Table 3). In duplex-qPCR, R2 and efficiency of the standard curves were 0.97 and 127.3% for T. asperellum and 0.99 and 137.6% for T. gamsii, while slope values were -2.09 for T. asperellum and -2.66 for T. gamsii (Table 3).
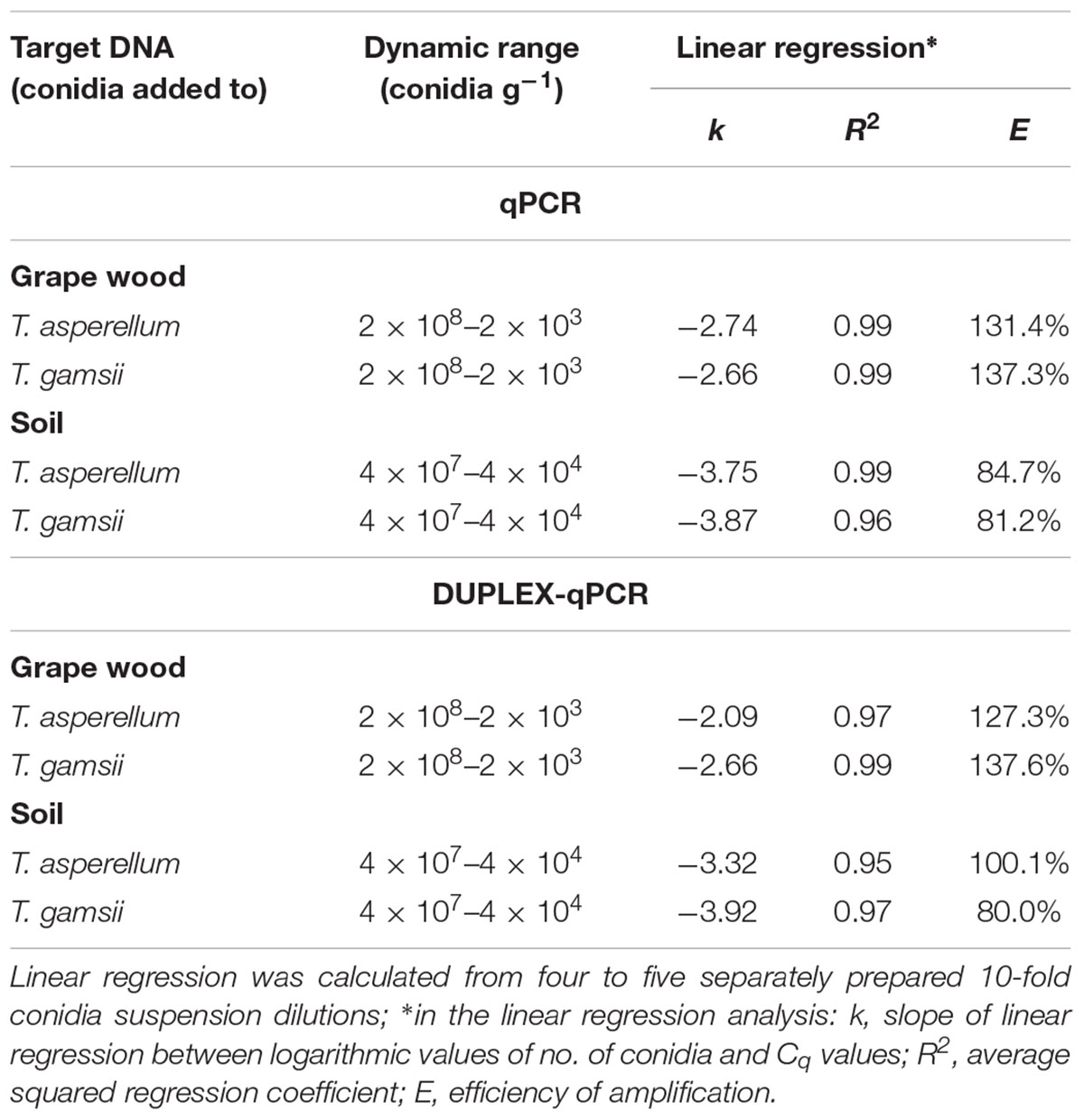
TABLE 3. Performance of the single- and duplex-qPCR assay for detection of T. asperellum and T. gamsii in grape wood and soil samples.
Two protocols for DNA extraction from soil were preliminarily compared using different concentrations of T. asperellum and T gamsii conidia and protocol 2 worked better than protocol 1 (Supplementary Table S1).
In both single- and duplex-qPCR, the detection limit for T. asperellum and T gamsii was 4 × 104 conidia g-1 of soil (Table 3). In single-qPCR, R2 and efficiency of the standard curves were 0.99 and 84.7% for T. asperellum and 0.96 and 81.2% for T. gamsii. In the same assays, slopes values were -3.75 and -3.87 for T. asperellum and T. gamsii, respectively (Table 3). On the other hand, in duplex-qPCR, R2 and efficiency of the standard curves were 0.95 and 100.1% for T. asperellum and 0.97 and 80.0% for T. gamsii. Slopes values were, respectively, -3.32 and -3.92 for T. asperellum and T. gamsii, respectively (Table 3).
BCAs quantification obtained by qPCR assays always agreed with the CFU formed on PDA medium.
Discussion
A fast reliable and sensitive species-specific method for detecting and quantifying the BCAs T. asperellum and T. gamsii, that are also the bioactive ingredients of the biofungicide RemedierTM (strains icc012 and icc080, respectively), used to control pathogens associated with GTDs as well as soil-borne and turf pathogens, was developed.
qPCR represents an alternative tool for an efficient quantification of individual fungal species through the estimation of DNA (Lievens and Thomma, 2005). In the current study, single- and duplex-qPCRs based on the uniqueness of SNPs identified on the single-copy gene encoding the rpb2 were set up and standardized.
rpb2 is a widely studied gene, whose many sequences are available in GenBank for homology comparisons. According to the in silico analysis, SNP in the base position 504 of T. asperellum and 806 of T. gamsii were recognized to specifically discriminate the target BCAs from the non-target fungi, including T. harzianum and T. viride, that are commonly detected in soil and currently used for crop protection (Druzhinina et al., 2011). Less specific SNPs were detected on the other two examined gene sequences corresponding to ech42 and tef1. However, the latter gene proved unsuitable for our purpose, although species-specific primers based on the tef1 gene had been proposed for the identification of T. asperellum, T. longibrachiatum and T. virens (Devi et al., 2012).
The specificity against fungal and bacterial species associated with different crops, with special reference to GTD pathogens, was tested in silico and by qPCR. The 142 and 113 bp amplicons identified for T. asperellum and T. gamsii, respectively, discriminated both the BCAs present in the biopesticide Remedier. The single-base mismatch in 3′ position of the primer forward or of the probe were sufficient for avoiding false negatives and for discrimination from the majority of other Trichoderma species and other fungi. Some Trichoderma species, which are occasionally found (Samuels et al., 2010; Braithwaite et al., 2017), did not carry the single-base mismatch in 3′ position (Supplementary Figure S1). In these species other SNPs present in different positions of the T. asperellum and T. gamsii primers/probe sets could allow the discrimination from the target species as observed for T. atroviride, T. paraviridescens, and T. polysporium analyzed in duplex-qPCR (Table 2).
Starting from DNA extracted from pure culture, the detection limit was 1 pg, in agreement with other studies using single-copy nuclear genes for qPCR (Ridgway and Stewart, 2000; Bluhm et al., 2002; Dodd et al., 2004; Fredlund et al., 2008; Scauflaire et al., 2012). According to provision for quantitative real-time PCR experiments, the linear dynamic range was extended to six different Log10 DNA amounts (Bustin et al., 2009). Slopes of the linear regression of qPCR assay ranged between -3.1 and -3.6, corresponding to a PCR efficiency of between 80 and 100%, and the R2-value was always ≥0.99.
When BCAs added to grapevines or soil samples were quantified in single- and duplex-qPCR, the R2-values were always ≥0.95, but the slope values (-2.0 to -2.7) indicated a slight decrease in the efficiency of the qPCR, caused by the presence of potential inhibitors (i.e., polyphenols, polysaccharides, humic acids, and metals) co-extracted with the DNA and inhibiting PCR reactions by decreasing the Taq polymerase activity or by limiting template’s availability (Kermekchiev et al., 2009; Opel et al., 2010). Assuming 100% DNA recovery, the protocol quantified up to 2 × 103 and 4 × 104 conidia g-1 in plants and soil, respectively. These results agree with those reported for fungi other than Trichoderma (Selma et al., 2008; Garrido et al., 2009) and for other Trichoderma species, such as T. virens (Dodd et al., 2004; Oskiera et al., 2017).
Conclusion
The duplex-qPCR assay represents a useful tool for the simultaneous detection and quantification of T. asperellum and T. gamsii and can assist in learning more about fungal activity, survival and spread in large-scale monitoring. Accordingly, BCA populations can be monitored on grapevines and different crops, both in the field and nursery.
Author Contributions
DG, SP, RDMA, and FF conceived and planned the experiments. DG and CR performed the experiments. DG and SP took the lead in writing the manuscript. SP, RDMA, and FF supervised the research. All authors contributed to the interpretation of the results, provided critical feedback and helped to shape the research, analysis, and manuscript.
Funding
This research was partially carried out in the framework of the Projects: (i) “Laboratory network for the selection, characterization, and conservation of germplasm and for preventing the spread of economically-relevant and quarantine pests (SELGE) No. 14,” funded by the Apulia Region, PO FESR 2007–2013 – Axis I, Line of intervention 1.2., Action 1.2.1 and (ii) Epidemiology, genetics of plant pathogens and development of molecular diagnostic methods granted by the University of Bari.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02073/full#supplementary-material
FIGURE S1 | SNPs identification in T. asperellum (A) and T. gamsii (B) rpb2 gene sequences for primers/probe sets design. ∗For each Trichoderma spp., GenBank accession number of representative rpb2 gene sequences is included between parenthesis. SNPs as compared to T. asperellum (A) and T. gamsii (B) are displayed with a different color.
TABLE S1 | Detection of T. asperellum DNA extracted with two extraction protocols by qPCR. ∗50 mg (protocol 1) and 250 mg (protocol 2). n.a., no amplification. Values are mean ± standard error of three biological replicates.
Footnotes
References
Beaulieu, R., López-Mondéjar, R., Tittarelli, F., Ros, M., and Pascual, J. A. (2011). qRT-PCR quantification of the biological control agent Trichoderma harzianum in peat and compost-based growing media. Bioresour. Technol. 102, 2793–2798. doi: 10.1016/j.biortech.2010.09.120
Benítez, T., Rincón, A. M., Limón, M. C., and Codón, A. C. (2004). Biocontrol mechanisms of Trichoderma strains. Int. Microbiol. 7, 249–260.
Błaszczyk, L., Popiel, D., Chełkowski, J., Koczyk, G., Samuels, G. J., Sobieralski, K., et al. (2011). Species diversity of Trichoderma in Poland. J. Appl. Genet. 52, 233–243. doi: 10.1007/s13353-011-0039-z
Bluhm, B. H., Flaherty, J. E., Cousin, M. A., and Woloshuk, C. P. (2002). Multiplex polymerase chain reaction assay for the differential detection of trichothecene and fumonisin-producing species of Fusarium in cornmeal. J. Food Prot. 65, 1955–1961. doi: 10.4315/0362-028X-65.12.1955
Braithwaite, M., Johnston, P. R., Ball, S. L., Nourozi, F., Hay, A. J., Shoukouhi, P., et al. (2017). Trichoderma down under: species diversity and occurrence of Trichoderma in New Zealand. Austr. Plant Pathol. 46, 11–30.
Bruce, T. J., Smart, L. E., Birch, A. N. E., Blok, V. C., MacKenzie, K., Guerrieri, E., et al. (2017). Prospects for plant defence activators and biocontrol in IPM–Concepts and lessons learnt so far. Crop Prot. 97, 128–134. doi: 10.1016/j.cropro.2016.10.003
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Cordier, C., Edel-Hermann, V., Martin-Laurent, F., Blal, B., Steinberg, C., and Alabouvette, C. (2007). SCAR-based real-time PCR to identity a biocontrol strain (T1) of Trichoderma atroviride and study its population dynamics in soils. J. Microbiol. Method 68, 60–68. doi: 10.1016/j.mimet.2006.06.006
Cullen, D. W., Lees, A. K., Toth, I. K., and Duncan, J. M. (2001). Conventional PCR and real-time quantitative PCR detection Helminthosporium solani in soil and on potato tubers. Eur. J. Plant Pathol. 107, 387–398. doi: 10.1023/A:1011247826
De Miccolis Angelini, R. M., Habib, W., Rotolo, C., Pollastro, S., and Faretra, F. (2010). Selection, characterization and genetic analysis of laboratory mutants of Botryotinia fuckeliana (Botrytis cinerea) resistant to the fungicide boscalid. Eur. J. Plant Pathol. 128, 185–199. doi: 10.1007/s10658-010-9643-8
Devi, P., Prabhakaran, N., Kamil, D., Pandey, P., and Borah, J. L. (2012). Characterization of Indian native isolates of Trichoderma spp. and assessment of their bio-control efficiency against plant pathogens. Afr. J. Biotechnol. 11, 15150–15160. doi: 10.5897/AJB12.2007
Dodd, L. S., Hill, R. A., and Stewart, A. (2004). A duplex-PCR bioassay to detect a Trichoderma virens biocontrol isolate in non-sterile soil. Soil Biol. Biochem. 36, 1955–1965. doi: 10.1016/j.soilbio.2004.03.012
Druzhinina, I. S., Seidl-Seiboth, V., Herrera-Estrella, A., Horwitz, B. A., Kenerley, C. M., Monte, E., et al. (2011). Trichoderma: the genomics of opportunistic success. Nat. Rev. Microbiol. 16, 749–759. doi: 10.1038/nrmicro2637
Eziashi, E. I., Omamor, I. B., and Odigie, E. E. (2007). Antagonism of Trichoderma viride and effects of extracted water-soluble compounds from Trichoderma species and benlate solution on Ceratocystis paradoxa. Afr. J. Biotechnol. 6, 388–392. doi: 10.5897/AJB2007.000-2019
Filion, M., St-Arnaud, M., and Jabaji-Hare, S. H. (2003). Direct quantification of fungal DNA from soil substrate using real-time PCR. J. Microbiol. Method 53, 67–76. doi: 10.1016/S0167-7012(02)00225-7
Fredlund, E., Gidlund, A., Olsen, M., Börjesson, T., Spliid, N. H. H., and Simonsson, M. (2008). Method evaluation of Fusarium DNA extraction from mycelia and wheat for down-stream real-time PCR quantification and correlation to mycotoxin levels. J. Microbiol. Method 73, 33–40. doi: 10.1016/j.mimet.2008.01.007
Garrido, C., Carbú, M., Fernández-Acero, F. J., Boonham, N., Colyer, A., Cantoral, J. M., et al. (2009). Development of protocols for detection of Colletotrichum acutatum and monitoring of strawberry anthracnose using real time PCR. Plant Pathol. 58, 43–51. doi: 10.1111/j.1365-3059.2008.01933.x
Gramaje, D., Úrbez-Torres, J. R., and Sosnowski, M. R. (2018). Managing grapevine trunk disease with respect to etiology and epidemiology: current strategies and future prospects. Plant Dis. 102, 12–39. doi: 10.1094/PDIS-04-17-0512-FE
Hagn, A., Wallisch, S., Radl, V., Munch, J. C., and Schloter, M. (2007). A new cultivation independent approach to detect and monitor common Trichoderma species in soils. J. Microbiol. Method 69, 86–92. doi: 10.1016/j.mimet.2006.12.004
Harman, G. E., Howell, C. R., Viterbo, A., Chet, I., and Lorito, M. (2004). Trichoderma species—opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2, 43–56. doi: 10.1038/nrmicro797
Hermosa, R., Viterbo, A., Chet, I., and Monte, E. (2012). Plant-beneficial effects of Trichoderma and of its genes. Microbiology 158, 17–25. doi: 10.1099/mic.0.052274-0
Hyder, S., Inam-ul-Haq, M., Bibi, S., Humayun, A., Malik, A. H., Ghuffar, S., et al. (2017). Novel potential of Trichoderma spp. as biocontrol agent. J. Entomol. Zool. Stud. 5, 214–222.
Jensen, D. F., Karlsson, M., Sarrocco, S., and Vannacci, G. (2016). “Biological control using microorganisms as an alternative to disease resistance,” in Biotechnology for Plant Disease Control, ed. D. B. Collinge (New York, NY: Wiley), 341–363.
Kim, T. G., and Knudsen, G. R. (2008). Quantitative real-time PCR effectively detects and quantifies colonization of sclerotia of Sclerotinia sclerotiorum by Trichoderma spp. Appl. Soil Ecol. 40, 100–108. doi: 10.1016/j.apsoil.2008.03.013
Kermekchiev, M. B., Kirilova, L. I., Vail, E. E., and Barnes, W. M. (2009). Mutants of Taq DNA polymerase resistant to PCR inhibitors allow DNA amplification from whole blood and crude soil samples. Nucleic Acids Res. 37:e40. doi: 10.1093/nar/gkn1055
Lievens, B., and Thomma, B. P. (2005). Recent developments in pathogen detection arrays: implications for fungal plant pathogens and use in practice. Phytopathology 95, 1374–1380. doi: 10.1094/PHYTO-95-1374
Lievens, B., Brouwer, M., Vanachter, A. C. R. C., Cammue, B. P. A., and Thomma, B. P. H. J. (2006). Real-time PCR for detection and quantification of fungal and oomycete tomato pathogens in plant and soil samples. Plant Sci. 171, 155–165. doi: 10.1016/j.plantsci.2006.03.009
Liguori, R. (2016). The role of research and innovation in crop protection: the contribution of Isagro. J. Plant Pathol. 98, S75.
López-Mondéjar, R., Antón, A., Raidl, S., Ros, M., and Pascual, J. A. (2010). Quantification of the biocontrol agent Trichoderma harzianum with real-time TaqMan PCR and its potential extrapolation to the hyphal biomass. Bioresour. Technol. 101, 2888–2891. doi: 10.1016/j.biortech.2009.10.019
Lorito, M., Woo, S. L., Harman, G. E., and Monte, E. (2010). Translational research on Trichoderma: from Omics to the field. Annu. Rev. Phytopathol. 48, 395–417. doi: 10.1146/annurev-phyto-073009-114314
Martin-Laurent, F., Philippot, L., Hallet, S., Chaussod, R., Germon, J. C., Soulas, G., et al. (2001). DNA extraction from soils: old bias for new microbial diversity analysis methods. Appl. Environ. Microb. 67, 2354–2359. doi: 10.1128/AEM.67.5.2354-2359.2001
Mukherjee, P. K., Horwitz, B. A., Herrera-Estrella, A., Schmoll, M., and Kenerley, C. M. (2013). Trichoderma research in the genome era. Annu. Rev. Phytopathol. 51, 105–129. doi: 10.1146/annurev-phyto-082712-102353
Opel, K. L., Chung, D., and McCord, B. R. (2010). A study of PCR inhibition mechanisms using real time PCR. J. Forensic Sci. 55, 25–33. doi: 10.1111/j.1556-4029.2009.01245.x
Oskiera, M., Szczech, M., Stêpowska, A., Smolińska, U., and Bartoszewski, G. (2017). Monitoring of Trichoderma species in agricultural soil in response to application of biopreparations. Biol. Control 113, 65–72. doi: 10.1016/j.biocontrol.2017.07.005
Qualhato, T. F., Lopes, F. A. C., Steindorff, A. S., Brandão, R. S., Jesuino, R. S. A., and Ulhoa, C. J. (2013). Mycoparasitism studies of Trichoderma species against three phytopathogenic fungi: evaluation of antagonism and hydrolytic enzyme production. Biotechnol. Lett. 35, 1461–1468. doi: 10.1007/s10529-013-1225-3
Raupach, G. S., and Kloepper, J. W. (1998). Mixtures of plant growth-promoting rhizobacteria enhance biological control of multiple cucumber pathogens. Phytopathology 88, 1158–1164. doi: 10.1094/PHYTO.1998.88.11.1158
Ridgway, H. J., and Stewart, A. (2000). Molecular marker assisted detection of the mycoparasite Coniothyrium minitans A69 in soil. N. Z. Plant Protect. 53, 114–117.
Reino, J. L., Guerrero, R. F., Hernández-Galán, R., and Collado, I. G. (2008). Secondary metabolites from species of the biocontrol agent Trichoderma. Phytochem. Rev. 7, 89–123. doi: 10.1007/s11101-006-9032-2
Rotolo, C., De Miccolis Angelini, R. M., Pollastro, S., and Faretra, F. (2016). A TaqMan-based qPCR assay for quantitative detection of the biocontrol agents Bacillus subtilis strain QST713 and Bacillus amyloliquefaciens subsp. plantarum strain D747. BioControl 61, 91–101. doi: 10.1007/s10526-015-9701-4
Rotolo, C., De Miccolis Angelini, R. M., Dongiovanni, C., Pollastro, S., Fumarola, G., Di Carolo, M., et al. (2018). Use of biocontrol agents and botanicals in integrated management of Botrytis cinerea in table grape vineyards. Pest Manage. Sci. 74, 715–725. doi: 10.1002/ps.4767
Rubio, M. B., Hermosa, M. R., Keck, E., and Monte, E. (2005). Specific PCR assays for the detection and quantification of DNA from the biocontrol strain Trichoderma harzianum 2413 in soil. Microb. Ecol. 49, 25–33. doi: 10.1007/s00248-003-0171-3
Samuels, G. J., Ismaiel, A., Bon, M. C., De Respinis, S., and Petrini, O. (2010). Trichoderma asperellum sensu lato consists of two cryptic species. Mycologia 102, 944–966. doi: 10.3852/09-243
Savazzini, F., Longa, C. M. O., Pertot, I., and Gessler, C. (2008). Real-time PCR for detection and quantification of the biocontrol agent Trichoderma atroviride strain SC1 in soil. J. Microbiol. Method 73, 185–194. doi: 10.1016/j.mimet.2008.02.004
Scauflaire, J., Godet, M., Gourgue, M., Liénard, C., and Munaut, F. (2012). A multiplex real-time PCR method using hybridization probes for the detection and the quantification of Fusarium proliferatum, F. subglutinans, F. temperatum, and F. verticillioides. Fungal Biol.-UK 116, 1073–1080. doi: 10.1016/j.funbio.2012.07.011
Selma, M. V., Martínez-Culebras, P. V., and Aznar, R. (2008). Real-time PCR based procedures for detection and quantification of Aspergillus carbonarius in wine grapes. Int. J. Food Microbiol. 122, 126–134. doi: 10.1016/j.ijfoodmicro.2007.11.049
Sharma, V., and Salwan, R. (2017). “Molecular markers and their use in taxonomic characterization of Trichoderma spp. Molecular Markers in Mycology,” in Fungal Biology, eds B. Singh and V. Gupta (Cham: Springer), 37–52. doi: 10.1007/978-3-319-34106-4_2
Sharma, V., Salwan, R., Sharma, P. N., and Gulati, A. (2017). Integrated translatome and proteome: approach for accurate portraying of widespread multifunctional aspects of Trichoderma. Front. Microbiol. 8:1602. doi: 10.3389/fmicb.2017.01602
Shoresh, M., Yedidia, I., and Chet, I. (2005). Involvement of jasmonic acid/ethylene signaling pathway in the systemic resistance induced in cucumber by Trichoderma asperellum T203. Phytopathology 95, 76–84. doi: 10.1094/PHYTO-95-0076
Singh, A., Sarma, B., and Sing, H. (2014). “Trichoderma: a silent worker of plant rhizosphere,” in Biotechnology and Biology of Trichoderma, eds V. G. Gupta, M. Schmoll, A. Herrera-Estrella, R. S. Upadhyay, I. Druzhinina, and M. Tuohy (Oxford: Newnes), 533–542. doi: 10.1016/B978-0-444-59576-8.00040-0
Strange, R. (1993). Plant Disease Control Towards Environmentally Acceptable Methods. London: Chapman & Hall. doi: 10.1007/978-1-4899-4632-4
Thornton, C. R., Dewey, F. M., and Gilugan, C. A. (1994). Development of a monoclonal antibody-based enzyme-linked immunosorbent assay for the detection of live propagules of Trichoderma harzianum in a peat-bran medium. Soil Biol. Biochem. 26, 909–920. doi: 10.1016/0038-0717(94)90307-7
Torsvik, V., and Øvreås, L. (2002). Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol. 5, 240–245. doi: 10.1016/S1369-5274(02)00324-7
Keywords: biocontrol agents, esca, grapevine, soil, probe, real time PCR
Citation: Gerin D, Pollastro S, Raguseo C, De Miccolis Angelini RM and Faretra F (2018) A Ready-to-Use Single- and Duplex-TaqMan-qPCR Assay to Detect and Quantify the Biocontrol Agents Trichoderma asperellum and Trichoderma gamsii. Front. Microbiol. 9:2073. doi: 10.3389/fmicb.2018.02073
Received: 13 April 2018; Accepted: 14 August 2018;
Published: 31 August 2018.
Edited by:
Raffaella Balestrini, Consiglio Nazionale delle Ricerche (CNR), ItalyReviewed by:
László Kredics, University of Szeged, HungaryFabiano Sillo, Università degli Studi di Torino, Italy
Copyright © 2018 Gerin, Pollastro, Raguseo, De Miccolis Angelini and Faretra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefania Pollastro, stefania.pollastro@uniba.it
 Donato Gerin
Donato Gerin Stefania Pollastro
Stefania Pollastro Celeste Raguseo
Celeste Raguseo Rita M. De Miccolis Angelini1,2
Rita M. De Miccolis Angelini1,2 Francesco Faretra
Francesco Faretra