- Department of Molecular Biology, Graduate School of Pharmaceutical Sciences, Kyushu University, Fukuoka, Japan
Chromosomal replication initiation requires dynamic mechanisms in higher-order nucleoprotein complexes that are constructed at the origin of replication. In Escherichia coli, DnaA molecules construct functional oligomers at the origin oriC, enabling localized unwinding of oriC and stable binding of DnaB helicases via multiple domain I molecules of oriC-bound DnaA. DnaA-bound DnaB helicases are then loaded onto the unwound region of oriC for construction of a pair of replisomes for bidirectional replication. However, mechanisms of DnaB loading to the unwound oriC remain largely elusive. In this study, we determined that His136 of DnaA domain III has an important role in loading of DnaB helicases onto the unwound oriC. DnaA H136A mutant protein was impaired in replication initiation in vivo, and in DnaB loading to the unwound oriC in vitro, whereas the protein fully sustained activities for oriC unwinding and DnaA domain I-dependent stable binding between DnaA and DnaB. Functional and structural analyses supported the idea that transient weak interactions between DnaB helicase and DnaA His136 within specific protomers of DnaA oligomers direct DnaB to a region in close proximity to single stranded DNA at unwound oriC bound to DnaA domain III of the DnaA oligomer. The aromatic moiety of His136 is basically conserved at corresponding residues of eubacterial DnaA orthologs, implying that the guidance function of DnaB is common to all eubacterial species.
Introduction
Chromosomal DNA replication is initiated by synergistic mechanisms involving multiple proteins with various functions. The initial steps of replication in Escherichia coli occur at the unique replication origin, oriC, which has a sophisticated structure that directs unwinding of duplex DNA and loading of replicative helicases (Kaguni, 2011; Leonard and Grimwade, 2015; Wolański et al., 2015; Katayama et al., 2017; Figure 1A). In these steps, the initiator DnaA molecules construct specific oligomers with the aid of DiaA (a DnaA-binding protein) and appropriately located DnaA-binding sequences (DnaA boxes) in the oriC DnaA-oligomerization region (DOR) (Leonard and Grimwade, 2015; Katayama et al., 2017). ATP-bound DnaA (ATP-DnaA), but not ADP-bound DnaA (ADP-DnaA), efficiently constructs homo-oligomers in a head-to-tail manner, with DnaA–DnaA interactions via the bound ATP and the Arg285 residue of flanking DnaA molecules (Kawakami et al., 2005; Erzberger et al., 2006; Ozaki et al., 2012a; Noguchi et al., 2015; Sakiyama et al., 2017; Figures 1B,C). The left-half DOR adjoins the oriC duplex unwinding element (DUE) and includes a specific binding site (IBS) for IHF (Integration host factor), a DNA-bending protein. The DnaA subcomplex including the left-half DOR and IHF causes localized unwinding of DNA in the DUE (Bramhill and Kornberg, 1988; Ozaki and Katayama, 2012; Figure 1B). The unwound single-stranded (ss) DUE is stabilized by binding to the DnaA oligomer, which is a prerequisite for DnaB loading (Ozaki et al., 2008; Ozaki and Katayama, 2012; Sakiyama et al., 2017).
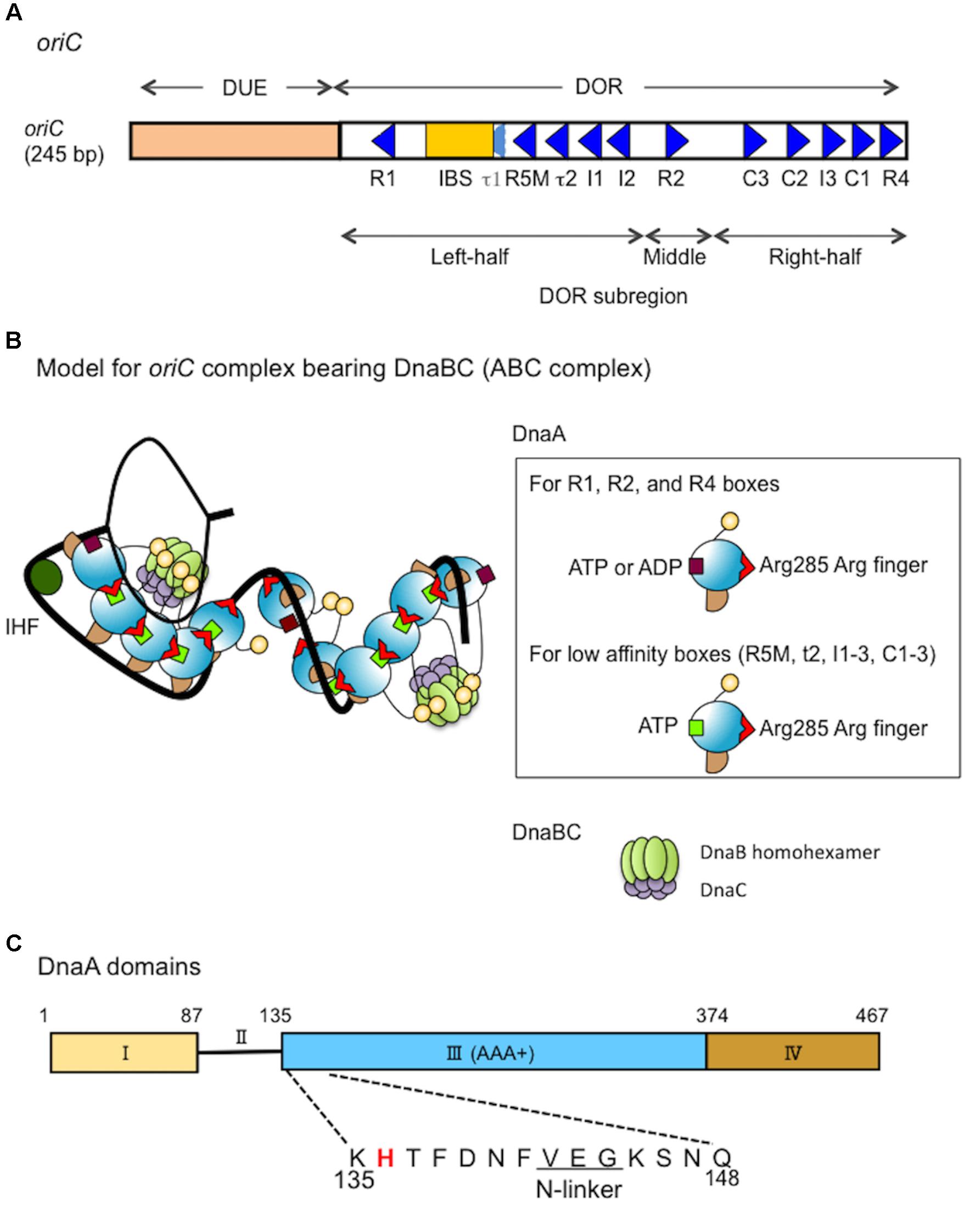
FIGURE 1. Structures of oriC, DnaA and the complex bearing DnaA and DnaBC. (A) Structure of Escherichia coli oriC. The duplex unwinding element (DUE) and DnaA-oligomerization region (DOR) are indicated. DnaA boxes (consensus sequence TTATNCACA) are shown by triangles, oriented to indicate sequence directionality (McGarry et al., 2004; Kawakami et al., 2005; Rozgaja et al., 2011; Shimizu et al., 2016). The R1 and R4 boxes have high affinity, the R2 box has moderate affinity and the others have low affinity for DnaA. A single integration host factor (IHF)-binding site (IBS) is present. When IHF binds to IBS, DnaA does not bind to the τ1 box (Sakiyama et al., 2017). (B) A possible structural model for oriC–DnaA complex carrying DnaBC complexes (ABC complex). DnaA domains are colored as in C. DnaA oligomers are constructed, and in one DnaA protomer, Arg285 “arginine finger” interacts with ATP bound to the flanking DnaA protomer (Kawakami et al., 2005; Erzberger et al., 2006). In the DnaA oligomers on oriC DOR (black line) (Rozgaja et al., 2011), Arg285 faces the middle of the DOR (Noguchi et al., 2015). DnaA domain IV can swivel using a short flexible loop present in its N-terminus, which supports DnaA oligomerization on DOR (Shimizu et al., 2016). In a crystal structure of the DNA-free oligomers of A. aeolicus DnaA domain III and domain IV, domain IV of one protomer interacts with domain III of the flanking protomer (Erzberger et al., 2002). By contrast, in molecular dynamics modeling of E. coli DnaA complexes at oriC, the structural changes induced by DNA binding and swiveling of domain IV prevents domain III-domain IV interaction (Shimizu et al., 2016). We propose that, near the time of initiation, when cellular ATP-DnaA levels peak (Kurokawa et al., 1999; Fujimitsu et al., 2009; Katayama et al., 2010), ATP-DnaA molecules bind to the low affinity DnaA box clusters R5M-τ2-I1-I2 and C3-C2-I3-C1 (Kawakami et al., 2005; Ozaki et al., 2008, 2012a; Keyamura et al., 2009; Sakiyama et al., 2017). We note that other studies (McGarry et al., 2004; Grimwade et al., 2007, 2018) using different methodologies to detect DnaA binding, report that R5M and C1 bind ATP-DnaA and ADP-DnaA with similar affinities. The illustration presents a model at the time of initiation. For stable DUE unwinding, ssDUE binds to DnaA domain III (Ozaki et al., 2008; Duderstadt et al., 2011; Sakiyama et al., 2017). Interactions between domain I of multiple DnaA protomers and each DnaB homohexamer promote stable binding (Abe et al., 2007; Keyamura et al., 2009). Deletion analysis of oriC supports the idea that each DnaA subcomplex (one on the left-half DOR and one on the right-half DOR) binds a single DnaB–DnaC complex (Ozaki and Katayama, 2012), although which DnaA protomer binds DnaB is unclear. In addition, the orientation of the DnaA subcomplex at the right-half DOR is important for efficient DnaB loading (Shimizu et al., 2016). When the DUE is unwound (thin black line), DnaB helicases are loaded onto the ssDNA regions and DnaC is released (Kaguni, 2011). For simplicity, DiaA is not shown in this model. (C) Domain structure of DnaA. His136 is highlighted in red and the N-linker motif is underlined. See text for details.
DnaB helicase-loading includes critical processes for transition from replication initiation to DNA elongation. DnaB–DnaC complexes bind to DnaA oligomers that are bound to oriC (Keyamura et al., 2009; Kaguni, 2011; Ozaki and Katayama, 2012; Soultanas, 2012; Zawilak-Pawlik et al., 2017; Figures 1B,C). DnaB is a replicative helicase with a homohexamer structure and ring (or spiral) configuration (Kaguni, 2011; Itsathitphaisarn et al., 2012; Strycharska et al., 2013). DnaC acts in the loading of DnaB by stably binding to DnaB and promoting conformational changes in DnaB hexamer that are required for its loading on the ssDNA (Davey and O’Donnell, 2003; Galletto et al., 2003; Biswas and Biswas-Fiss, 2006; Makowska-Grzyska and Kaguni, 2010; Kaguni, 2011). DnaB C-terminal domain (CTD) is suggested to bind DnaC (Galletto et al., 2003). A pair of DnaB–DnaC complexes is thought to bind to an oriC–DnaA complex, resulting in a higher-order complex (Keyamura et al., 2009; Ozaki and Katayama, 2012; Ozaki et al., 2012a; Shimizu et al., 2016; Figure 1B): a deletion analysis of oriC suggests that each DnaA subcomplex constructed on the left- and right-half subregions of DOR binds a single DnaB helicase (or DnaBC complex) (Ozaki and Katayama, 2012). Also, deletion and insertion analyses suggest that the orientation of DnaA subcomplex constructed on the right-half DOR is optimized for efficient DnaB loading (Shimizu et al., 2016). The two DnaB helicases in the complex are loaded onto the ssDNA region of oriC in opposite directions each other, enabling bidirectional migration, leading to loading of one replisome on each strand (Fang et al., 1999; Carr and Kaguni, 2001; Ozaki and Katayama, 2012; Soultanas, 2012; Bell and Kaguni, 2013).
Unlike hyperthermophile bacterium Aquifex aeolicus DnaC (Mott et al., 2008), E. coli DnaC does not stably bind to DnaA (Keyamura et al., 2009). In E. coli, DnaB in DnaBC complex binds to DnaA complexes (Abe et al., 2007; Keyamura et al., 2009) and this mechanism is suggested to be conserved even in A. aeolicus (Mott et al., 2008). In addition, we recently demonstrate that YfdR, a protein encoded by an E. coli cryptic prophage, binds to DnaA depending on domain I Phe46 (a primary DnaB-binding site; also see below) and competes with the DnaBC complex in DnaA binding (Noguchi and Katayama, 2016). These results also support the notion that, in E. coli, DnaC per se does not directly bind DnaA, but does so indirectly as part of the DnaB–DnaC complex, in which DnaB binds to DnaA. However, dynamic mechanisms involved in DnaA–DnaB interactions have remained to be further elucidated.
The initiator protein DnaA has four functional domains (Kaguni, 2011; Katayama et al., 2017; Figure 1C). Domain I contains a specific site for binding to DnaB and DiaA (Sutton et al., 1998; Seitz et al., 2000; Abe et al., 2007; Keyamura et al., 2009). Domain II is a flexible linker between domains I and III (Nozaki and Ogawa, 2008). Domain III is the AAA+ domain that participates in nucleotide binding, ssDNA recruitment, and DnaA–DnaA interactions (Felczak and Kaguni, 2004; Iyer et al., 2004; Kawakami et al., 2005; Ozaki and Katayama, 2009; Duderstadt et al., 2011; Ozaki et al., 2012a,b). In addition, DnaA domain III N-terminal Val142–Gly144 constitutes a specific N-linker motif, which is thought to structurally interact with the adenine moiety of ATP/ADP (Smith et al., 2004), and our previous results demonstrate that the well-conserved Glu143 residue is specifically important for stable ATP/ADP binding (Ozaki et al., 2012b). DnaA domains II–III are also thought to have a weak binding site for DnaB (Marszalek et al., 1996; Seitz et al., 2000), although the precise location of this site has not been determined. DnaA domain IV binds directly to DnaA boxes, which has the 9-mer consensus sequence of (5′)TTATNCACA(3′) (Sutton and Kaguni, 1997; Fujikawa et al., 2003; Kaguni, 2011). In the N-terminus of domain IV, a short flexible loop enables the swiveling of this domain (Erzberger et al., 2002; Shimizu et al., 2016).
Functional mechanisms of DnaA–DnaB interaction for loading of DnaB on the ssDUE are thought to include multiple steps (Sutton et al., 1998; Seitz et al., 2000; Abe et al., 2007; Keyamura et al., 2009). DnaA domain I is the primary source of weak affinity for the DnaB CTD (Sutton et al., 1998; Seitz et al., 2000). We previously determined that a patch of DnaA that includes the Glu21 and Phe46 residues is exposed on the surface of domain I, binds to DnaB, and supports stable DnaB binding when DnaA oligomers are constructed on oriC (Abe et al., 2007; Keyamura et al., 2009). As DnaB is a homohexamer, binding of a single DnaB hexamer to multiple domain I molecules of a DnaA oligomer would effectively increase its affinity for DnaA, stabilizing DnaA–DnaB binding (Abe et al., 2007; Keyamura et al., 2009; Ozaki and Katayama, 2009; Zawilak-Pawlik et al., 2017). Although DnaA domain I is suggested to interact with a site including the N-terminus and its flanking region of DnaB CTD (Seitz et al., 2000), specific amino acids in the region have not been determined.
In addition to domain I, DnaA domains II–III are thought to contain a second site for DnaB binding, which is important for DnaB loading on oriC DNA (Figure 1C). A previous study assessed specific inhibition with monoclonal anti-DnaA antibodies, and the results suggested that the DnaA Pro111–Gln148 region includes a site for DnaB interaction (Marszalek and Kaguni, 1994; Marszalek et al., 1996; Sutton et al., 1998). Another study assessed functional interactions of various truncated forms of DnaA, and the results suggested that the DnaA Ser130–Gln148 region has a specific interaction site for the DnaB N-terminal domain (NTD) (Seitz et al., 2000). Deletion analysis of domain II has demonstrated that the DnaA Ala99–Val134 region is largely dispensable for DnaA functions in replication initiation at oriC (Nozaki and Ogawa, 2008). Taken together, these results suggest that the DnaA domain III N-terminus spanning Lys135–Gln148 might contain the second essential site for DnaB interaction (Figure 1C).
In this study, to determine the role for the second DnaB-binding site of DnaA, we extended functional analysis of the DnaA domain III N-terminus to the region spanning Lys135–Gln148. Alanine-scanning experiments revealed that His136, Phe141, and Val142 are crucial for complementation of dnaA46 temperature-sensitive mutations. Our previous study indicated that Val142 is an essential constituent of the N-linker (Ozaki et al., 2012b), and Phe141 is its flanking bulky residue. Thus, in this study, we focused our attention on analyses on His136 and found that a substitution of this residue (H136A) impaired specifically DnaA-dependent loading of DnaB on ssDUE, without affecting its assembly at oriC, unwinding of DUE, or recruitment of ssDUE. A structural model for the oriC–DnaA complex is compatible with a predicted role for His136 in directing DnaB for loading on the ssDUE.
Materials and Methods
Nucleic Acids
Plasmid pKA234, a derivative of the pING vector which has an arabinose-inducible promoter, was used for overproduction of wild-type DnaA, and has been described previously (Ozaki et al., 2012b). Derivatives of pKA234 encoding DnaA variants with individual alanine substitutions for each residue from Lys135A to Gln148 were constructed with specific mutagenic primers and QuikChange site-directed mutagenesis protocol (Stratagene [Agilent], Agilent, La Jolla, CA, United States), as previously described (Ozaki et al., 2012b).
The DOR dsDNA fragment (ΔDUE) and 28-mer T-rich ssDUE strand have been described previously (Ozaki and Katayama, 2012). M13KEW101 and pBSoriC are oriC plasmids containing intact oriC, and pBSoriCΔR4-R2 is a derivative of pBSoriC with only the left half of oriC. M13KEW101, pBSoriC, and pBSoriCΔR4-R2 have been described previously (Kawakami et al., 2005; Ozaki and Katayama, 2012). pBSoriCΔDUE is a DUE-deleted derivative of pBSoriC, constructed by outward-directed PCR with pBSoriC as a template and primers ori-1 and MR28r1-r, as described previously (Ozaki et al., 2008; Ozaki and Katayama, 2012); the amplified DNA was digested with HincII and self-ligated, resulting in pBSoriCΔDUE. Biotinylated oriC DNA (bio-oriC) has been described previously (Keyamura et al., 2009). M13-A-site ssDNA is a derivative of M13 ssDNA with a hairpin structure that contains a DnaA-box sequence (Masai et al., 1990).
Buffers
Buffer G contained 20 mM HEPES-KOH (pH 7.6), 5 mM magnesium acetate, 1 mM EDTA, 10% glycerol, 0.1% Triton X-100, and 4 mM dithiothreitol (DTT). Buffer F contained 20 mM Tris–HCl (pH 7.5), 8 mM DTT, 10 mM magnesium acetate, 125 mM potassium glutamate, 3 mM ATP, and 0.5 mg/mL bovine serum albumin (BSA). Buffer ABC contained 20 mM Tris–HCl (pH 7.5), 0.1 mg/mL BSA, 8 mM DTT, 8 mM magnesium acetate, 0.01% Brij-58, and 125 mM potassium glutamate.
DnaA Proteins
Wild-type DnaA and DnaA H136A proteins were overproduced in E. coli strain KA450 [ΔoriC1071::Tn10 rnhA199(Am) dnaA17(Am)] from pKA234 or pH136A (a derivative of pKA234 encoding DnaA H136A), and purified as previously described (Ozaki et al., 2012b).
Flow-Cytometry Analysis
Flow cytometry was performed as described previously (Noguchi et al., 2015; Inoue et al., 2016; Sakiyama et al., 2017). Briefly, cells were grown in LB medium at 30°C to an absorbance at 660 nm of 0.2, then incubated at 42°C for 150 min. Before and after 42°C incubation, aliquots were withdrawn for analysis of cell mass (or cell volume) with a FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ, United States). The remaining aliquots of the cell cultures were further incubated in the presence of rifampicin and cephalexin for 4 h, followed by analysis of DNA content by flow cytometry.
oriC Plasmid Replication Assay
The assay was performed essentially as described previously (Keyamura et al., 2009). Briefly, a crude protein extract containing proteins required for oriC replication (except for DnaA) was prepared from E. coli strain WM433 (dnaA204) (Fuller et al., 1981). Reactions (25 μL) were performed with purified DnaA, M13KEW101 oriC plasmid (38 fmol as plasmid; 600 pmol nucleotides), and WM433 crude extract (200 μg), as described (Keyamura et al., 2009).
P1 Nuclease Assay for DUE Unwinding
The assay was performed essentially as described previously (Ozaki et al., 2012b). Briefly, ATP-DnaA or ADP-DnaA was incubated for 3 min at 38°C with M13KEW101 oriC plasmid (12 fmol as plasmid; 400 ng) and HU protein (16 ng), followed by brief incubation with P1 nuclease. DNA was purified and incubated with restriction enzyme BseRI, followed by analysis by agarose-gel electrophoresis, with ethidium bromide staining.
ssDUE-Recruitment Electrophoretic Mobility Shift Assay (EMSA)
The assay was performed as described previously (Ozaki and Katayama, 2012; Ozaki et al., 2012a; Sakiyama et al., 2017). Briefly, DnaA and the DUE-deleted oriC dsDNA fragment, DOR(ΔDUE) (5 nM) were incubated on ice for 5 min, followed by incubation for 5 min at 30°C with 32P-labeled 28-mer T-rich ssDUE strand (2.5 nM) in the presence of λphage DNA (25 ng). The resultant DNA complexes were analyzed by 4% polyacrylamide-gel electrophoresis at 4°C.
Form I∗ Assay
The assay was performed as previously described (Ozaki and Katayama, 2012; Noguchi et al., 2015; Shimizu et al., 2016). Briefly, ATP-DnaA and pBSoriC or pBSoriCΔR4-R2 (1.6 nM) were incubated for 15 min at 30°C in buffer (25 μL) containing 760 nM SSB, 76 nM GryA, 100 nM His-GyrB, 42 nM IHF, 100 nM His-DnaB, and 100 nM His-DnaC. Reactions were terminated by addition of 1% SDS, and the DNA samples were purified and analyzed by 0.65% agarose-gel electrophoresis and ethidium bromide staining.
Biotin-Tagged oriC Pull-Down Assay
The assay was performed as previously described (Keyamura et al., 2009; Ozaki and Katayama, 2012; Ozaki et al., 2012a; Noguchi and Katayama, 2016). Briefly, bio-oriC (419 bp, 100 fmol), including the entire oriC and flanking regions (Keyamura et al., 2007), was incubated on ice for 10 min in buffer G (10 μL) containing DnaA and 1 mM ATP. The bio-oriC and bound proteins were recovered by pull-down with streptavidin-coated beads (Promega, Madison, WI, United States), washed twice with buffer G (12.5 μL) containing 75 mM KCl, and dissolved in SDS sample buffer. Proteins were quantified using silver staining and quantitative protein standards, as we previously performed (Ozaki and Katayama, 2012; Noguchi and Katayama, 2016). The recovered amounts of bio-oriC were deduced by quantifying bio-oriC remaining in supernatants. For analysis of DnaB binding to oriC–DnaA complexes, bio-oriC was coincubated with DnaA and DnaB in the presence of absence of DnaC, followed by the wash step conducted only once with buffer G excluding KCl.
His-DnaB Pull-Down Assay
The assay was performed under similar conditions to the Form I∗ assay. ATP-DnaA (90 nM) and pBSoriC or pBSoriCΔDUE (1.6 nM) were incubated for 15 min at 30°C in buffer F (10 μL) containing 40 nM IHF, 200 nM His-DnaB K236A, and the native (non-tagged) DnaC (200 nM). After addition of Co2+-conjugated magnetic beads (1 μL bed volume: Dynabeads, Invitrogen, Carlsbad, CA, United States) and incubation for 15 min at 4°C, the beads and bound materials were collected by magnetic pull-down and washed in buffer F containing 100 mM NaCl and excluding BSA. His-DnaB-bound plasmid DNA was eluted in standard SDS sample buffer, and analyzed by 1% agarose-gel electrophoresis and ethidium bromide staining.
ABC Primosome Assay
The assay was performed as described previously (Abe et al., 2007; Keyamura et al., 2009). Briefly, the indicated amounts of DnaA were incubated at 30°C for 15 min in buffer ABC (25 μL) containing M13-A-site ssDNA (1.1 nM as ssDNA; 220 pmol nucleotides), 0.5 μg SSB, 65 ng DnaB, 65 ng DnaC, 72 ng DnaG, 108 ng DNA polymerase III∗, 26 ng β-clamp subunit, 1 mM ATP, 0.25 mM each of GTP, CTP, and UTP, and 0.1 mM each of dNTP and [α-32P]dATP. DNA polymerase III∗ is a subcomplex of DNA polymerase III holoenzyme lacking the β-clamp subunit. Reactions were stopped by addition of 1 mL 10% trichloroacetic acid, and the amounts of synthesized DNA were measured by liquid scintillation.
Results
DnaA His136 Is Essential for Initiation Activity in vivo
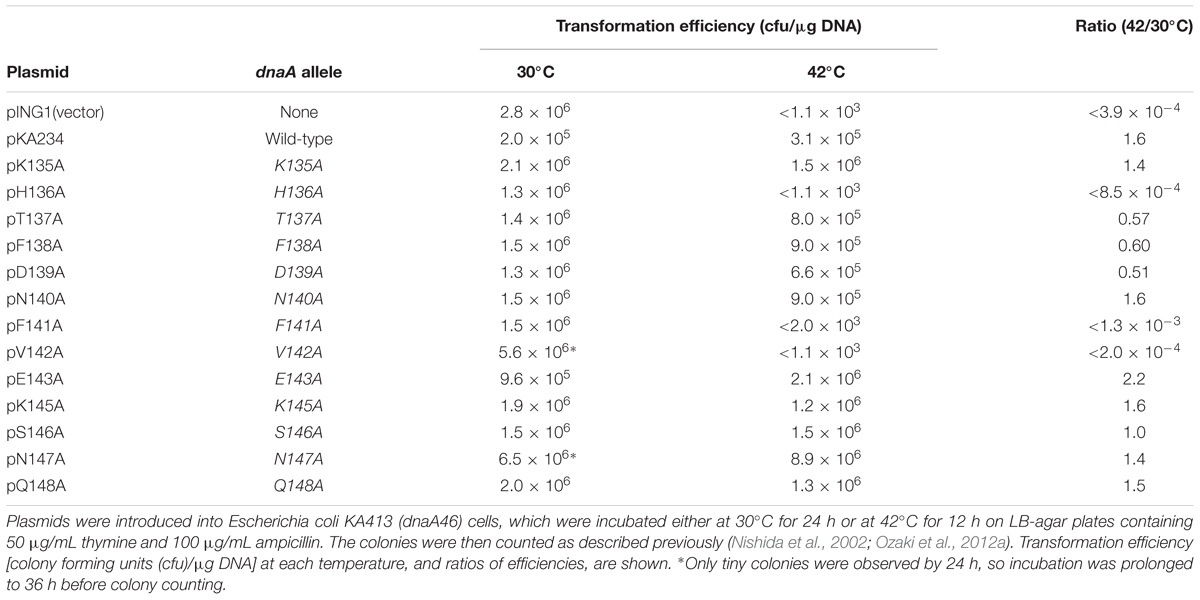
A putative DnaB interaction site has been speculated to reside in a DnaA domain III N-terminal region spanning Lys135–Gln148. We conducted an alanine-scanning analysis on all the amino acid residues in this region except for Gly144, which lacks a side chain. Plasmid pKA234 contains the wild-type dnaA-coding region downstream of the arabinose-inducible PBAD promoter (Kubota et al., 1997). Derivatives of pKA234 containing dnaA mutations encoding alanine substitutions were constructed and used for complementation tests with a temperature (42°C)-sensitive dnaA46 host strain (KA413). Even in the absence of the inducer arabinose, introduction of pKA234, but not the vector pING1, enabled the KA413 cells to grow at 42°C, presumably because of leaky expression of the pKA234 dnaA gene (Table 1). In similar experiments that we previously performed, DnaA amounts in cells bearing pKA234 was 1.3- to 2.8-fold higher than those in cells bearing pING1 (Kawakami et al., 2005; Ozaki et al., 2008). Unlike pKA234, plasmids containing dnaA alleles encoding H136A, F141A, or V142A substitutions did not complement the dnaA46 temperature (42°C) sensitive growth (Table 1).
Val142 is a component of the N-linker motif (Val142-Glu143-Gly144), a conserved sequence in AAA+ family proteins that is thought to interact with the adenine moiety of ATP (Smith et al., 2004). DnaA V142A has previously been shown to cause overinitiation of replication at 30°C in the absence of wild-type DnaA presumably because its binding to ADP is unstable, resulting in rapid exchange of bound ADP to ATP (Ozaki et al., 2012b), and here consistently, we observed that DnaA V142A resulted in slow colony formation at 30°C and severe inhibition of colony formation at 42°C (Table 1). DnaA F141A also resulted in growth inhibition at 42°C, suggesting that substitution of the hydrophobic aromatic side chain of phenylalanine resulted in DnaA structural changes that indirectly inhibited the function of the N-linker motif residues, such as Val142.
We previously indicated that Glu143 within the N-linker motif is important for stable binding of ATP and ADP, and DnaA E143A causes moderate overinitiation of replication at 30°C in the absence of wild-type DnaA, presumably because of rapid exchange of bound ADP to ATP (Ozaki et al., 2012b). When co-expressed with the wild-type DnaA, DnaA E143A was shown not to cause inhibition of cell growth (Ozaki et al., 2012b), which might be a consequence of formation of mixed complexes on oriC repressing overinitiation (Ozaki et al., 2012b). These previous observations are consistent with the current data of DnaA E143A at 30°C (Table 1). When cells were incubated at 42°C, a temperature at which replication initiation is stimulated even in wild-type cells, a severe inhibitory effect of DnaA E143A for cell growth by overinitiation might be suppressed. Based on these ideas, we excluded DnaA F141A and V142A for further analyses in this study.
To further elucidate initiation activity in vivo of DnaA H136A, we used flow cytometry to determine the chromosomal replication modes of dnaA46 cells containing derivatives of pKA234. Cells were cultured for exponential growth at 30°C and shifted to 42°C for inactivation of the intrinsic DnaA46. Further incubation with rifampicin and cefalexin enabled run-out replication of the chromosomes. At 30°C, distinct DNA peaks were seen for each strain (Figure 2). In dnaA46 cells with the pING vector, the two-chromosome peak predominated, with minor peaks for one, three, four, and five chromosomes, indicating asynchronous initiations (Skarstad et al., 1986, 1988). Introduction of wild-type dnaA (pKA234) into dnaA46 cells stimulated initiation and resulted in a predominant four-chromosome peak. DnaA F46A protein is impaired in stable DnaB binding and DnaB loading at oriC (Abe et al., 2007; Keyamura et al., 2009). Introduction of the dnaA F46A allele as a negative control (pF46A) gave a similar profile to that of dnaA46 cells with the pING vector, whereas moderate stimulation was detected with dnaA H136A (pH136A). This stimulation could be a consequence of mixture of DnaA46 and DnaA H136A proteins (see below). At 42°C, the profiles with pING and dnaA H136A were fundamentally similar with respect to inhibition of initiation, whereas moderate stimulation occurred with dnaA F46A, as previously observed (Abe et al., 2007; Keyamura et al., 2009). These results suggest that DnaA H136A is impaired in the initiation of chromosomal replication in vivo. In addition, the smeared peaks of dnaA H136A culture at 42°C suggest that progression of replication forks also are moderately inhibited by this mutation. Abnormal interaction between DnaA H136A and DnaB helicases might inhibit fork progression in addition to replication initiation in vivo.
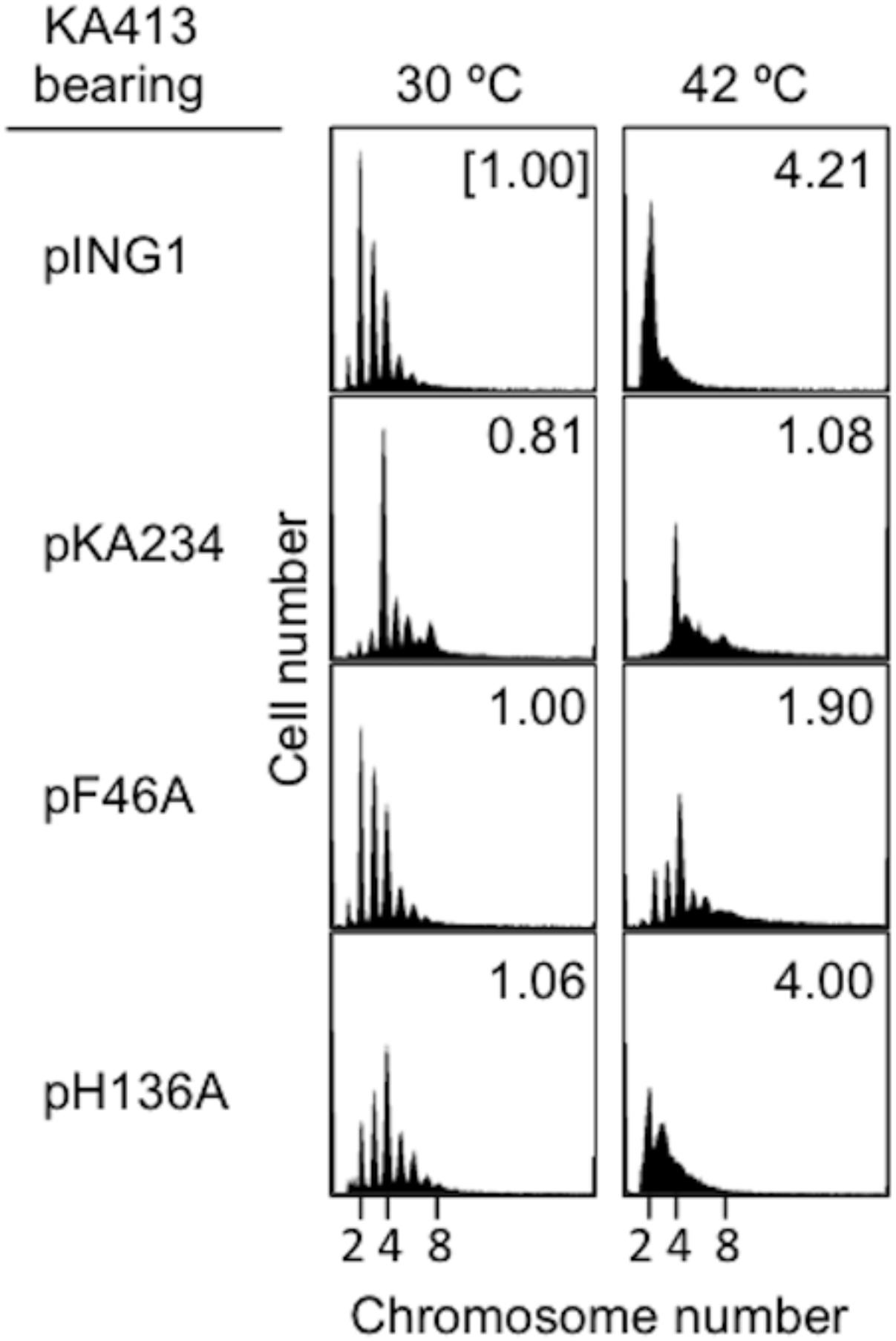
FIGURE 2. Flow-cytometry analysis. KA413 (dnaA46) cells containing the indicated plasmids were grown to exponential phase in LB medium containing 50 μg/mL thymine and 50 μg/mL ampicillin at 30°C, and further incubated at 42°C for 150 min. Before and after 42°C incubation, portions of the cultures were withdrawn for analyses of cell size (mass) and DNA content by flow cytometry. Chromosome numbers per cell corresponding to each peak are indicated. Mean cell mass relative to that of KA413 cells containing pING1 and grown at 30°C is indicated at the top right corner of each panel. pKA234 is a DnaA-expressing derivative of pING1. pF46A and pH136A are derivatives of pKA234 encoding variants of DnaA with single amino acid substitutions.
Purified DnaA H136A Protein Sustains ATP Binding, but Is Inactive in Initiation in vitro
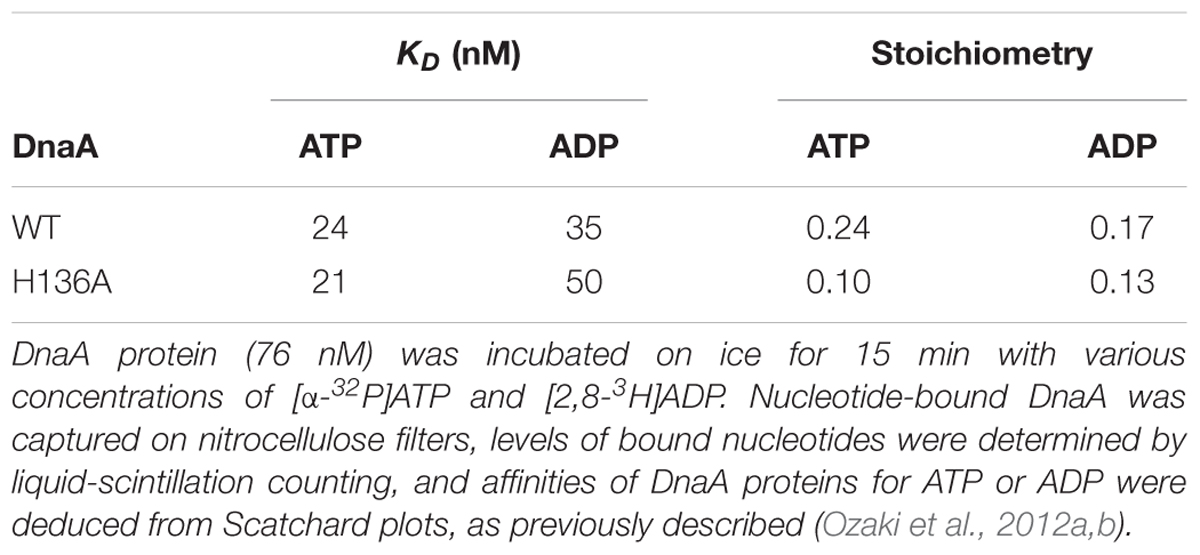
DnaA H136A protein was overproduced and purified, and shown to have high-affinity binding of ATP and ADP at levels similar to wild-type DnaA (Table 2), suggesting preservation of the overall protein structure of domain III. By contrast, when replication initiation was assessed with an oriC plasmid and a replicative-protein extract, even the ATP form of DnaA H136A was inactive, unlike that of wild-type DnaA (Figure 3A).
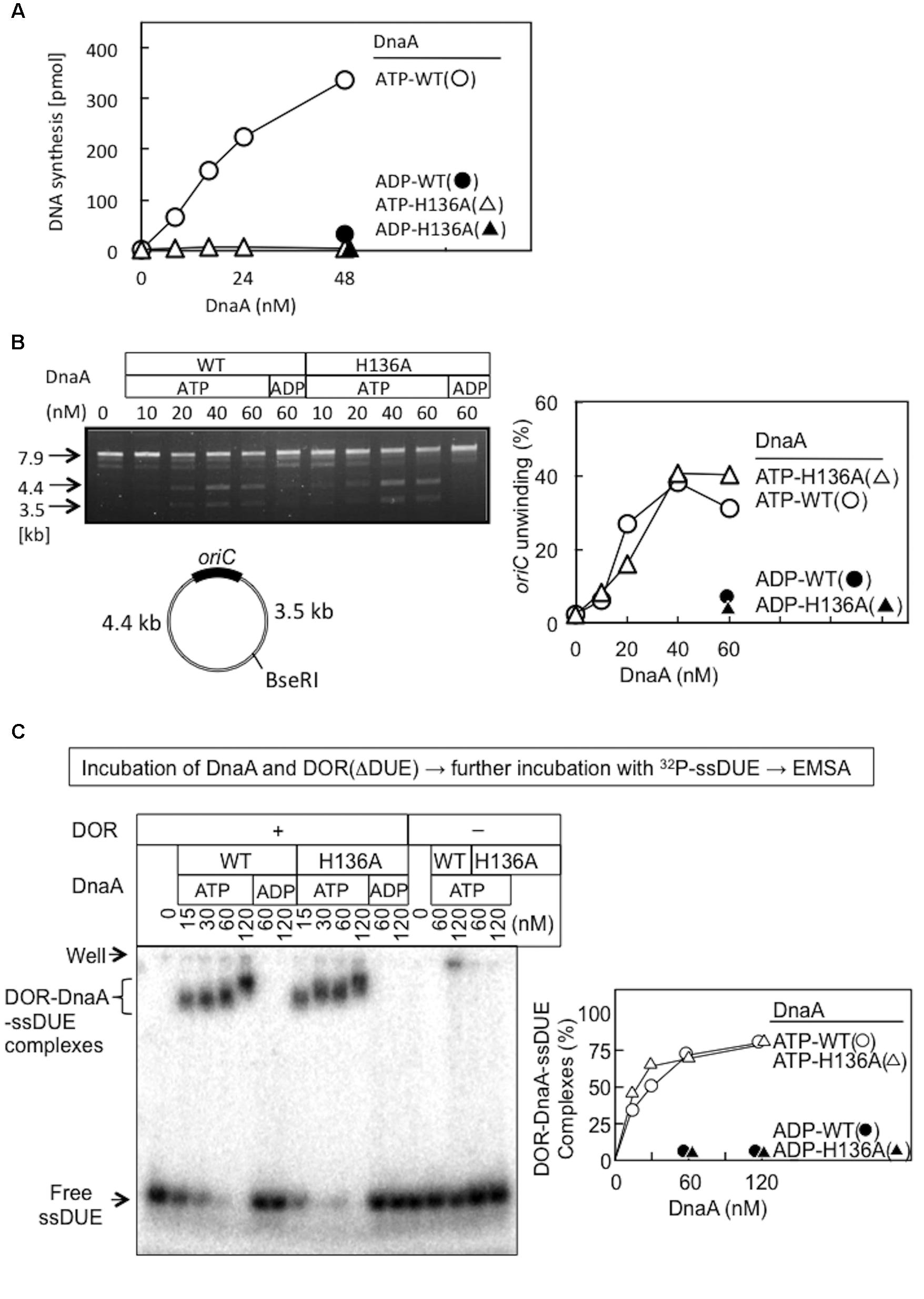
FIGURE 3. Activities of wild-type (WT) and H136A mutant DnaA in replication initiation and oriC unwinding. (A) In vitro oriC replication assay. Nucleotide-bound WT and H136A DnaA were incubated at 30°C for 20 min with a replicative WM433 (dnaA204) crude extract, M13KEW101 oriC plasmid (600 pmol nucleotides), and dNTPs containing [α -32P]dATP, as described previously (Nishida et al., 2002). Similar results were obtained in an independent experiment. (B) P1-nuclease assay. Nucleotide-bound forms of DnaA were incubated with M13KEW101 oriC plasmid and HU protein, followed by incubation with P1 nuclease and then with BseRI. The BseRI restriction site is located distally to oriC, so that two DNA fragments (3.5 and 4.4 kb) are produced if oriC is unwound and digested by P1 nuclease. Digested samples were analyzed by 1% agarose-gel electrophoresis. oriC unwinding (3.5 and 4.4 kb fragments) was quantified. (C) Nucleotide-bound forms of DnaA and an oriC DOR(ΔDUE) fragment (5 nM) were incubated on ice for 5 min, then for 5 min at 30°C with 32P-labeled 28-mer ssDUE (2.5 nM). Formation of protein–DNA complexes was analyzed by 4% polyacrylamide-gel electrophoresis, and quantified.
DnaA H136A Is Active in oriC Unwinding and ssDUE Binding
Functions of DnaA His136 were assessed with reconstituted systems for oriC unwinding and ssDUE binding. In the P1-nuclease assay, unwinding of oriC DUE by initiation complexes produces ssDNA that is sensitive to endonuclease P1. In this assay, the ATP forms of wild-type DnaA and DnaA H136A demonstrated similar activities in specific oriC unwinding (Figure 3B).
To determine the abilities of these proteins to stabilize the unwound DUE, we analyzed ssDUE-binding activity by electrophoretic mobility shift assay (EMSA) using ssDUE, oriC DOR, and DnaA. In this assay, ATP-DnaA molecules can construct homo-oligomers on the DOR and bind ssDUE with high affinity, producing DOR-DnaA-ssDUE ternary complexes (Ozaki et al., 2008; Ozaki and Katayama, 2012; Noguchi et al., 2015; Sakiyama et al., 2017). Construction of the ternary complexes has been shown to occur with ATP-DnaA (but not ADP-DnaA) and to require specific DnaA residues involved in construction of oriC open complexes (including AAA+ arginine-finger Arg285 and ssDUE-binding H/B motifs Val211 and Arg245) (Ozaki et al., 2008; Ozaki and Katayama, 2012). Here, the ATP form of DnaA H136A resulted in binding of the ssDUE to the DnaA–DOR complexes at a level similar to that seen with wild-type DnaA (Figure 3C). DnaA-ssDUE binding was dependent on DOR as we previously demonstrated. Faint signals in the gel wells were irregular aggregates of DnaA which involved 32P-ssDUE. Those were unstable and slowly degraded during electrophoresis, resulting in faint smeared bands (Ozaki et al., 2008; Ozaki and Katayama, 2012; Sakiyama et al., 2017). These results indicate that DnaA H136A is fully active in the primary reactions required for DUE unwinding and stable binding of ssDUE, which are prerequisites for DnaB loading on ssDUE.
DnaA H136A Is Impaired in DnaB Loading Onto Unwound oriC
We conducted the Form I∗ assay to determine DnaB loading onto the unwound strands of oriC (via indirect interactions with DnaC and DnaA) and its subsequent helicase action on DNA strands. Loading on ssDUE activates DnaB helicase, expanding the ssDNA region and introducing positive supercoils. Activation of gyrase then produces highly negatively supercoiled oriC plasmid (Form I∗), and this topoisomeric form is distinguished from Form I by gel electrophoresis (Baker et al., 1986).
Form I∗ production was severely impaired for DnaA H136A, even at high levels (Figure 4A). Compared with wild-type DnaA, DnaA H136A, and the negative control DnaA F46A (which is inactive in primary DnaB binding) only had low levels of Form I∗ production, even in the presence of excessive amounts of DnaBC proteins (Figure 4B). The left-half oriC is a minimal region for DUE unwinding and DnaB loading (Ozaki and Katayama, 2012; Sakiyama et al., 2017). Compared with wild-type DnaA, DnaA H136A, and DnaA F46A also produced only low levels of Form I∗ even with left-half oriC (Figure 4C), consistent with the idea that DnaB loading reactions per se are inhibited also with the full-length oriC, but not with the idea that DnaBC complexes binds simultaneously to left- and right-half DnaA–oriC subcomplexes, causing abortive interactions with each other, and inhibiting DnaB loading on ssDUE. Taken together, the results suggest that the DnaA His136 residue has a predominant role in the process of productive loading of DnaB to unwound oriC.
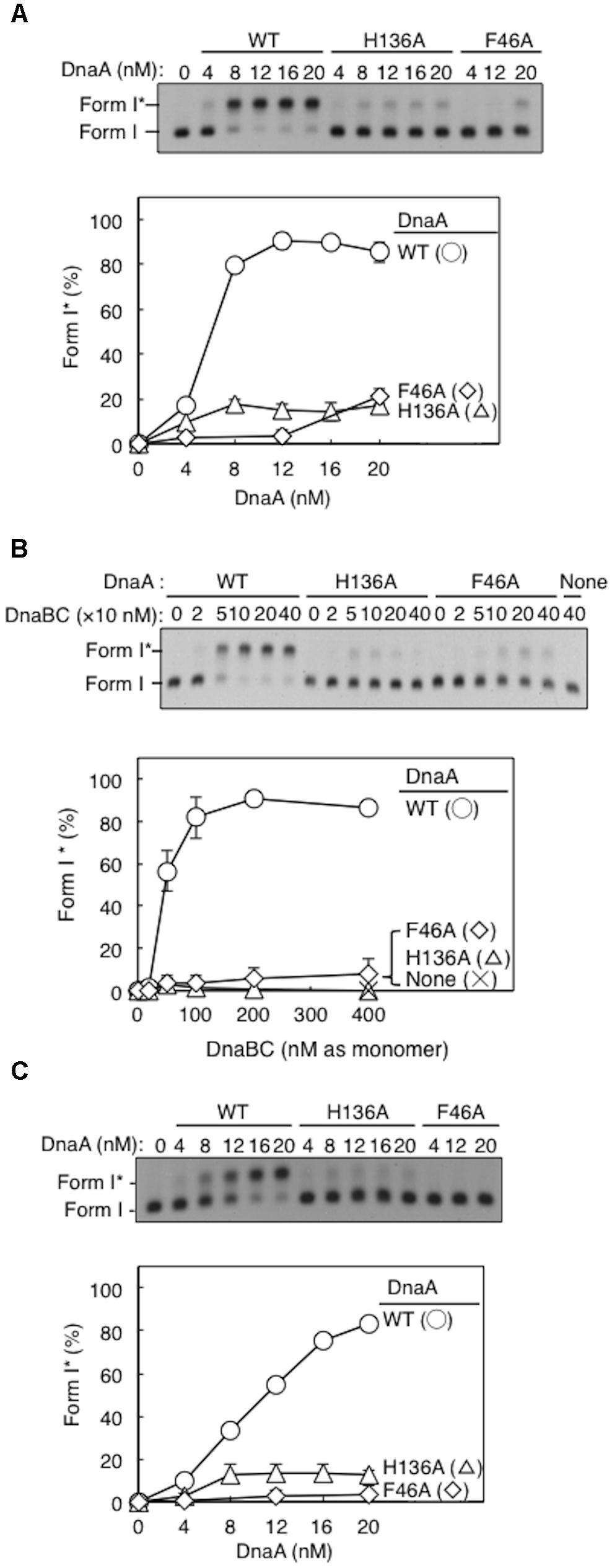
FIGURE 4. DnaA-directed loading of DnaB at oriC. Representative gel images are shown in black–white inverted mode, and migration positions of negatively supercoiled (Form I) and highly negatively supercoiled (Form I∗) plasmid DNA are indicated. Form I∗ is produced from Form I by DnaB helicase and DNA gyrase activities. Relative amounts of Form I∗ to input DNA were quantified as “Form I∗ (%),” and mean values with SD (n = 2) are shown in each graph. WT, wild-type DnaA; F46A, DnaA F46A; H136A, DnaA H136A; None, no DnaA. (A) Form I∗ assay with WT, F46A, and H136A DnaA. Indicated amounts of ATP-DnaA were incubated for 15 min at 30°C in buffer containing DnaB–DnaC (100 nM), SSB, gyrase, IHF, and pBSoriC oriC plasmid Form I. The resultant DNA forms were analyzed by agarose-gel electrophoresis. (B) Form I∗ assay with fixed concentration (20 nM) DnaA and various concentrations DnaB–DnaC complexes. Other conditions were as in (A). (C) Form I∗ assay, as in (A), but with pBSoriCΔR4-R2 plasmid.
A Subgroup of DnaA Molecules in an oriC Complex Requires His136 for DnaB Loading
Here, activity of mixtures of the wild-type DnaA and DnaA H136A or DnaA F46A proteins were analyzed using Form I∗ assay. In these experiments, complexes including both the wild-type DnaA and H136A (or F46A) proteins should be constructed on the same oriC molecule. Thus, if a subgroup of DnaA protomers is not used for DnaB loading, the partial inclusion of DnaA H136A (or F46A) which is fully active in DUE unwinding, might retain DUE unwinding and DnaB loading. DnaA F46A, although inactive for DnaB binding by itself, can help achieve optimal Form I∗ formation when wild-type DnaA is provided suboptimally (Figure 5A), as previously demonstrated (Keyamura et al., 2009), suggesting that DnaA F46A can contribute to DnaB loading when it is a part of a complex including the wild-type DnaA at oriC. This means that only a subgroup (but not all) of DnaA molecules assembled on oriC would require Phe46 for DnaB loading (Keyamura et al., 2009). Here, we obtained similar results with DnaA H136A in the presence of wild-type DnaA (Figure 5A).
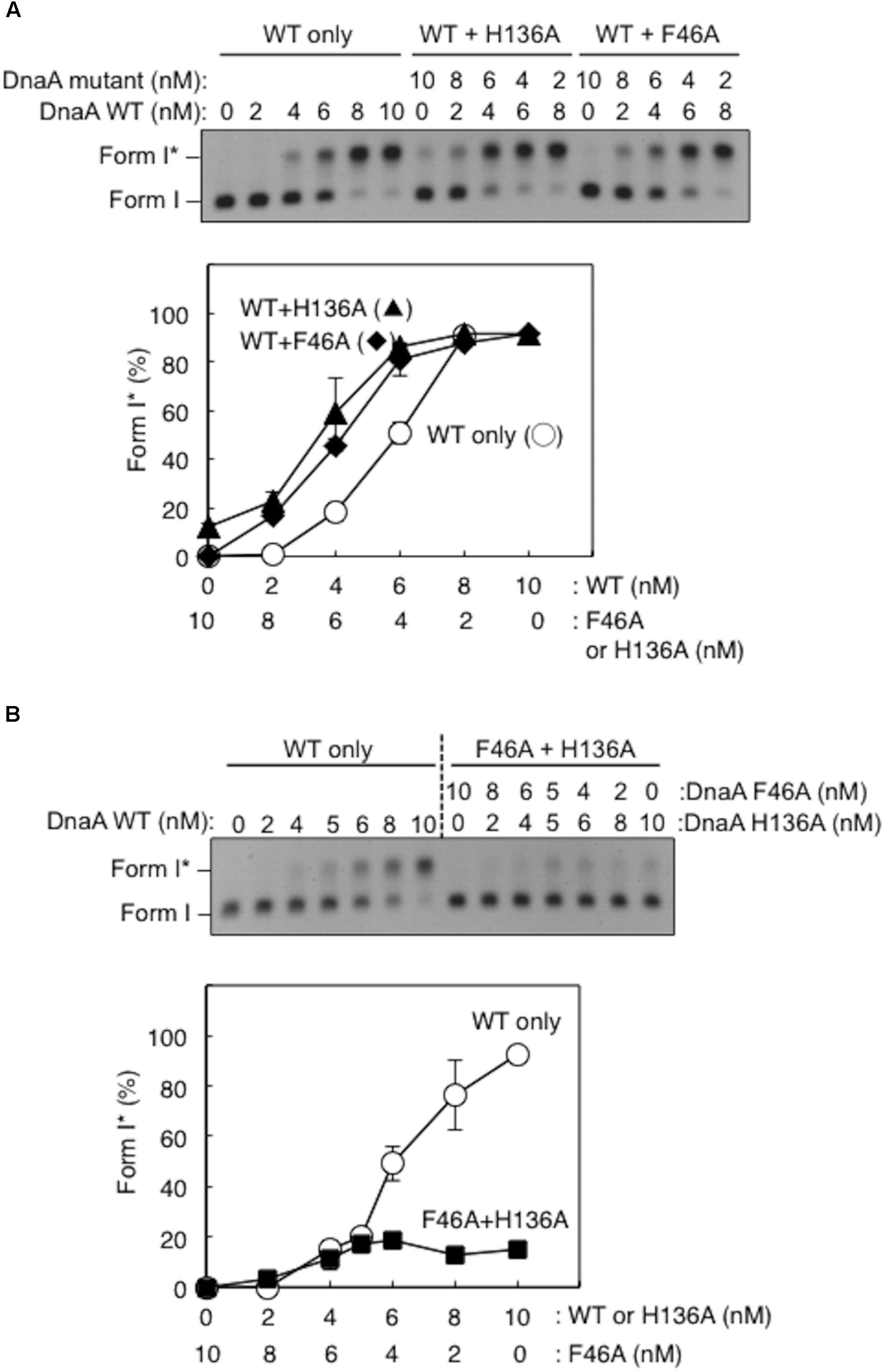
FIGURE 5. Activities of DnaB loading by mixtures of wild-type (WT) and mutant DnaA. Form I∗ assays were performed with different combinations of WT, F46A, and H136A DnaA, with pBSoriC oriC plasmid. Form I∗ is highly negatively supercoiled, and is produced from Form I by DnaB helicase and DNA gyrase activities. Reactions were incubated for 15 min at 30°C. Representative gel images are shown in black–white inverted mode. Amounts of Form I∗ relative to the input DNA were quantified as “Form I∗ (%),” and mean values with SD (n = 2) are shown. (A) Mixtures of WT DnaA and DnaA F46A or H136A were used. (B) Mixtures of DnaA F46A and DnaA H136A were used.
DnaA H136A and DnaA F46A proteins were mixed at various ratios and assessed by Form I∗ assay, but did not produce substantial amounts of Form I∗ (Figure 5B). Only weak Form I∗ formation due to the residual activity of DnaA H136A was observed. These results suggest that, at least in a subgroup of DnaA molecules assembled on oriC, both the His136 and Phe46 residues must be present in the same DnaA protomer for DnaB loading.
DnaA H136A Forms Stable oriC–DnaA Complexes, but Results in Impaired Loading of DnaB on Unwound oriC
DnaA R281A is impaired in stable DnaA assembly on oriC, resulting in largely reduced binding of DnaB compared with wild-type DnaA, although DUE unwinding activity is sustained (Felczak and Kaguni, 2004). Arg281 is a constituent of the AAA+ Box VII motif, which is thought to reside at the interface of DnaA oligomers, supporting stable DnaA–DnaA interactions. To assess the activities of DnaA H136A in stable construction of DnaA assembly on oriC, we performed a pull-down assay using biotin-tagged oriC fragments. DnaA H136A was recovered by oriC fragment pull-down at a similar level to wild-type DnaA (Figure 6A), indicating that DnaA H136A is competent for DnaA assembly, which is consistent with its activities in DUE unwinding and ssDUE binding. In the presence of DnaA H136A and DnaB, oriC pull-down of DnaB was similar to that in the presence of wild-type DnaA and DnaB, indicating stable binding of DnaB helicase by DnaA H136A (Figure 6B). Also, even when DnaC was coincubated, DnaA H136A stably bound DnaB at a level similar to wild-type DnaA (Figure 6C). As previously shown (Keyamura et al., 2007), recovery of DnaB was increased by the co-incubation of DnaC, suggesting a conformational change of DnaB by binding of DnaC. Recovery of DnaC was moderately less than that of DnaB, which might be caused by moderate dissociation of DnaC during the wash step in this pull-down experiment.
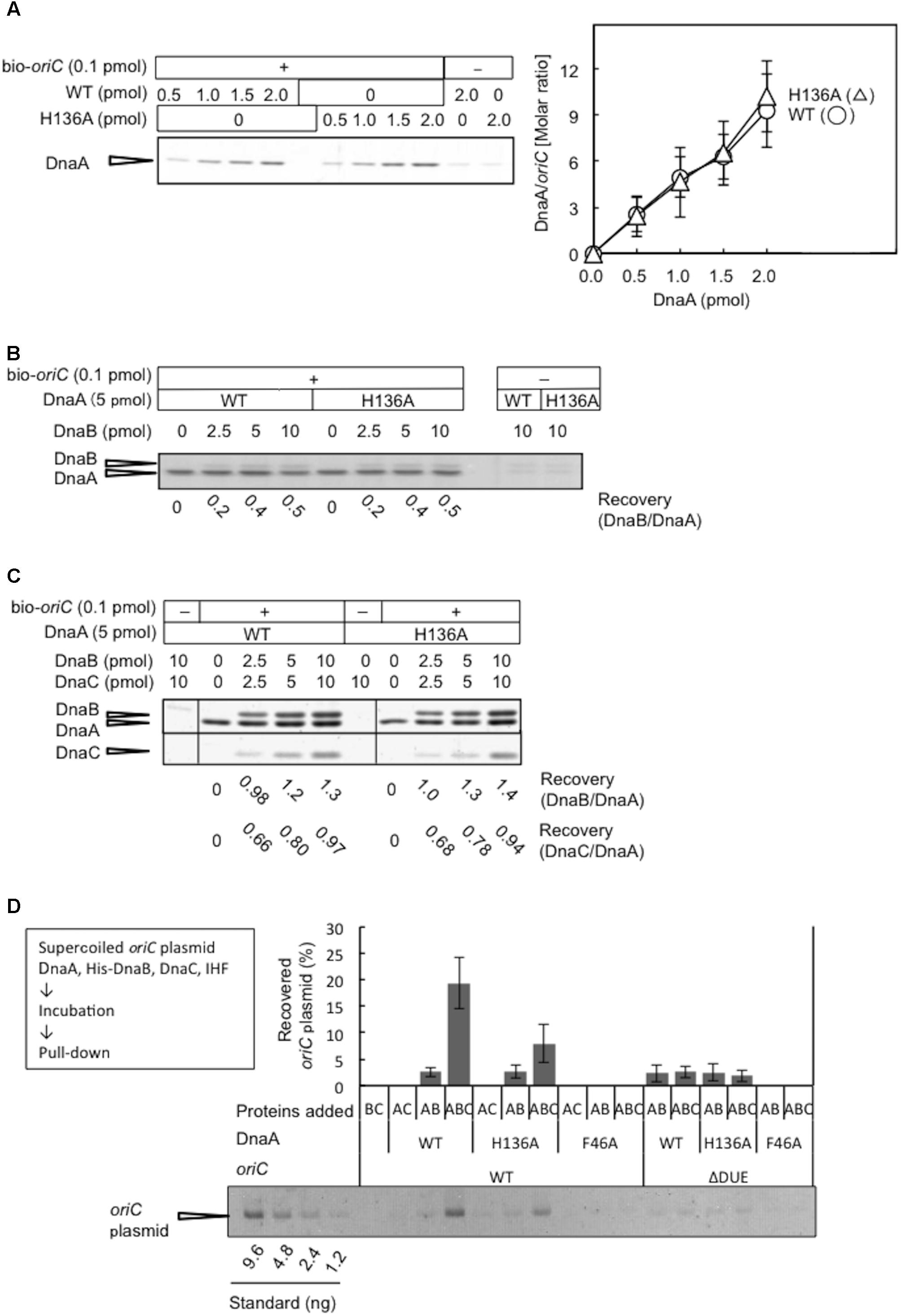
FIGURE 6. DnaA assembly on oriC and DnaB loading on unwound oriC. (A) Biotinylated oriC DNA (bio-oriC) pull-down assay with wild-type (WT) and H136A mutant DnaA. The bio-oriC (0.1 pmol) was incubated on ice for 10 min with indicated amounts of ATP-DnaA, and bound materials were recovered with streptavidin-coated beads. Recovered DnaA was analyzed by SDS-11% polyacrylamide gel electrophoresis, with silver staining (left panel) and the number of DnaA molecules bound to oriC was deduced using quantitative standards (right panel). Experiments were performed in triplicate, and a representative gel image and the means with SD are shown. (B) bio-oriC pull-down of DnaA and DnaB proteins. Similar to the above, bio-oriC was incubated with 5 pmol ATP-DnaA (WT or H136A) and different amounts of DnaB, and bound materials were recovered with streptavidin-coated beads. Ratios of bound DnaB (as monomers) and DnaA were calculated using quantitative standards. (C) bio-oriC pull-down of DnaA, DnaB, and DnaC proteins. Similar experiments were performed in the presence of DnaC. Ratios of bound DnaC (as monomers) and DnaA as well as bound DnaB (as monomers) and DnaA were calculated, using quantitative standards. (D) His-DnaB pull-down assay. ATP-DnaA (90 nM, WT, H136A, or F46A) was incubated for 15 min at 30°C in Form I∗ buffer containing pBSoriC or pBSoriCΔDUE (1.6 nM), IHF (40 nM), and His-DnaB (200 nM), with or without non-tagged DnaC (200 nM). His-DnaB-bound oriC plasmids were collected with magnetic beads, and analyzed by 1% agarose-gel electrophoresis. The percentages of recovered DNA relative to the input DNA were quantified and indicated as “Recovered oriC plasmid (%),” as means with SD (n = 2).
Loading of DnaB was further examined by a novel pull-down assay using His-tagged DnaB, DnaC, DnaA, IHF, and oriC plasmid. In this assay, oriC is unwound by DnaA complexes, DnaB undergoes DnaC-dependent loading on the ssDUE region, and the resultant DnaB-bound oriC plasmids are recovered with Co2+-conjugated beads. The use of wild-type DnaA and oriC in this assay resulted in a high level of recovery of oriC plasmid, which was dependent on the inclusion of DnaB and DnaC (Figure 6D), suggesting that stable complexes with DnaB hexameric rings (or spirals) encircling the ssDNA of oriC are necessary for oriC recovery. A low level of recovery of oriC plasmid in the absence of DnaC presumably represented direct binding of DnaB to DnaA oligomers on oriC involving DnaA Phe46. These complexes were likely to have been largely dissociated by the use of wash buffer. In agreement with this idea, the use of wild-type DnaA and mutant oriC with DUE deletion (ΔDUE), which is inactive for unwinding, only resulted in a low level of recovery of oriC plasmid. Moreover, the use of DnaA F46A resulted in no observable recovery of oriC plasmid (Figure 6D). These results were consistent with DnaA Phe46-dependent binding between DnaB and DnaA oligomers constructed on oriC being responsible for the basal recovery level, with considerable enhancement of recovery resulting from DnaB loading on ssDNA.
The inclusion of DnaA H136A and wild-type oriC in this assay in the presence of DnaB and DnaC resulted in moderate inhibition in oriC recovery, compared to the level seen with wild-type DnaA (Figure 6D). A low level of recovery (similar to that with wild-type DnaA) was seen with DnaA H136A in the absence of DnaC or in the presence of ΔDUE. These results further support the idea that DnaA H136 residue is specifically important for functional DnaB loading to the unwound site of oriC (see also section “Discussion”).
DnaA H136A Is Active in DnaB Loading in a Simplified System
DnaA–DnaB interactions were also studied using the simplified reconstituted ABC primosome system, which uses M13-phage-derived ssDNA with a hairpin structure containing the DnaA box R1 sequence (A-site ssDNA) (Masai et al., 1990; Masai and Arai, 1995; Carr and Kaguni, 2002; Figure 7). DnaA binding to the hairpin structure enables recruitment of DnaBC complex and DnaB loading to the SSB-coated ssDNA, followed by primer and DNA synthesis by DnaG primase and DNA polymerase III holoenzyme. Whereas DnaA F46A was essentially inactive in this system, DnaA H136A was fully active relative to wild-type DnaA (Figure 7), indicating that DnaA H136A promoted DnaB loading to ssDNA in this simple system. Notably, unlike the more complex oriC, only 2–4 DnaA molecules bind to this M13 hairpin region, with its single DnaA box (Carr and Kaguni, 2002), and except for the hairpin region, the whole template is single-stranded. These specific features may cause the different requirement for His136 between oriC and A-site ssDNA (see section “Discussion”).
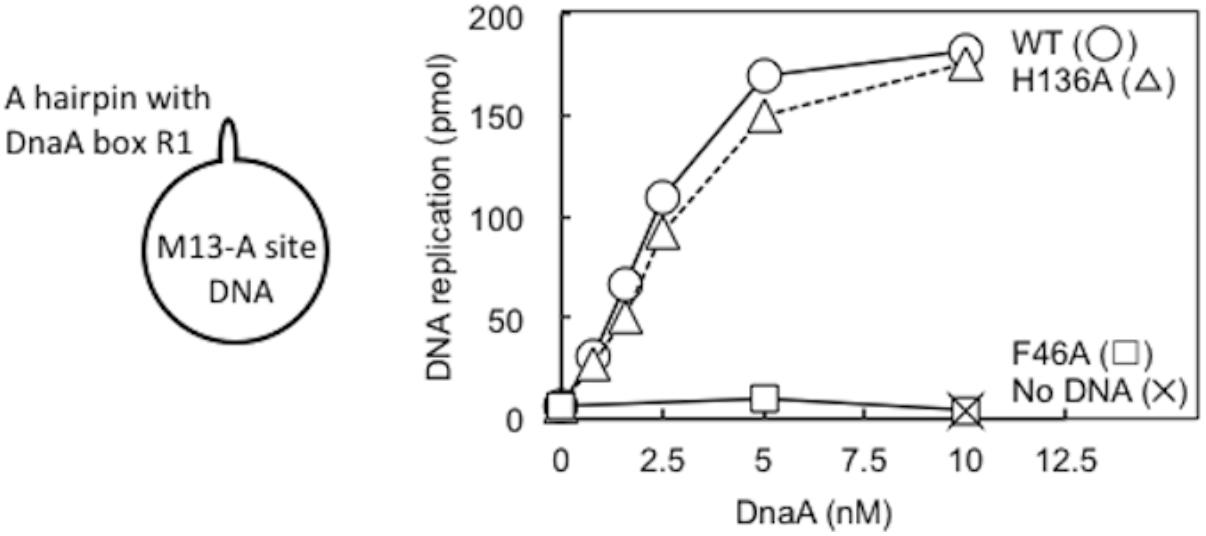
FIGURE 7. ABC primosome assay. The indicated concentrations of wild-type (WT) or F46A or H136A mutant DnaA were incubated for 15 min at 30°C in buffer containing DnaB, DnaC, DnaG, SSB, NTPs, dNTPs, and DNA polymerase III holoenzyme, with or without M13 A-site ssDNA (220 pmol nucleotides). DNA replication was quantified by measurement of incorporation of radiolabeled dATP. Similar results were obtained when reactions were incubated at 37 or 42°C.
Evolutional Conservation of DnaA His136
Among bacterial DnaA orthologs, the position corresponding to E. coli DnaA His136 is generally occupied by an aromatic residue, such as tyrosine, phenylalanine, or histidine (Figure 8). Even in the hyperthermophile A. aeolicus DnaA ortholog, the corresponding residue is tyrosine (Erzberger et al., 2002). In γ proteobacteria including E. coli, the histidine residue predominates (Figure 8). Thus, the aromatic moiety at the position of E. coli DnaA His136 appears to have an evolutionarily conserved role in the loading of DnaB helicases at oriC.
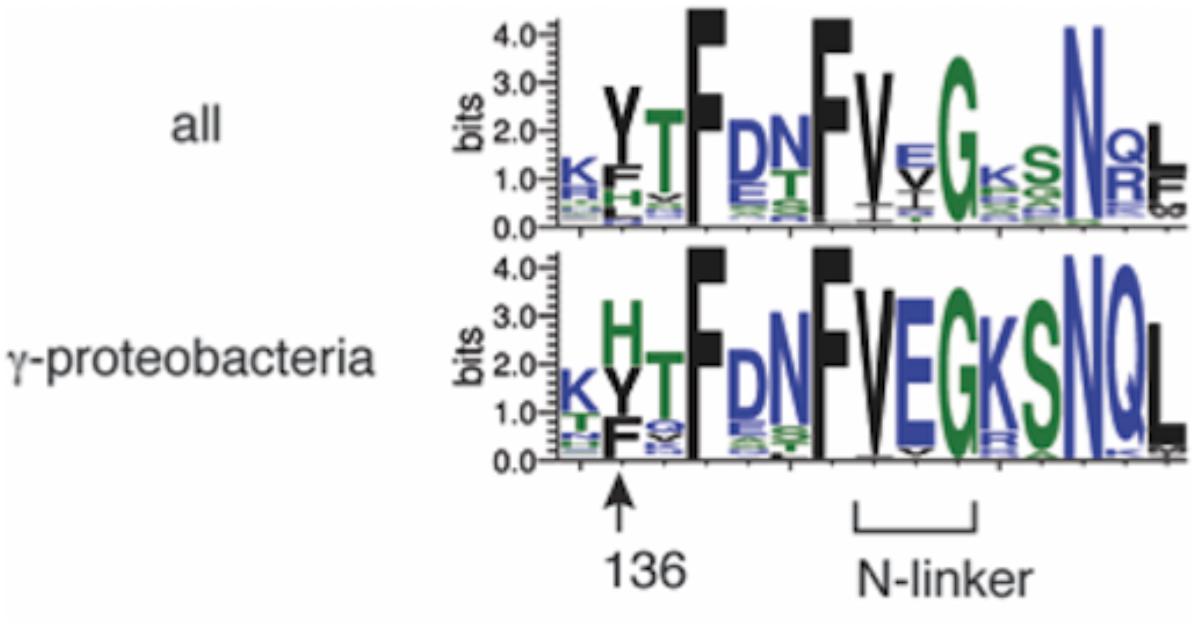
FIGURE 8. Bacterial DnaA orthologs possess a conserved aromatic residue at the position corresponding to DnaA His136. Sequence logos of DnaA His136 and its surrounding residues are shown based on the sequence alignment of 581 DnaA homologs from all bacterial phyla (top) and 162 DnaA homologs from γ proteobacteria (bottom). The protein sequences were obtained from the NCBI database and the logos were created using WebLogo (http://weblogo.threeplusone.com).
Discussion
Loading of replicative helicases on unwound origin DNA region is a crucial step in replication initiation of chromosomes. In E. coli, this step depends on dynamic interactions between the initiator DnaA and DnaB replicative helicase. Stable binding between the two depends on a site including DnaA Glu21 and Phe46 of domain I. This site is suggested to interact with a specific site of DnaB, which resides in a region including the N-terminus and its franking region of DnaB CTD. In addition, the DnaA region Lys135–Gln148 within domain III has been implicated as having weak physical contact with DnaB NTD. However, the biological importance of this interaction has not previously been determined. Here, we used alanine scanning of these DnaA residues to highlight the role of His136 in replication initiation in vivo. The plasmid complementation test and flow cytometry analysis demonstrated that the dnaA H136A allele has only low in vivo initiation activity. In-depth biochemical characterization of DnaA H136A revealed that His136 is indispensable for DnaB loading at oriC, but that DUE unwinding, ssDUE binding, and assembly on oriC do not depend on this residue. Intriguingly, DnaA H136A was fully active in an oriC-independent DnaB loading system for ssDNA replication (i.e., ABC primosome system). These observations suggest that DnaA His136 residue has a distinct role in DnaB loading at oriC. Because DnaA H136A-oriC complexes maintain the primary contact between DnaA Phe46 and DnaB, we conclude that a secondary, weak contact via His136 might allow DnaA-directed DnaB loading on the ssDUE region. The conservation of His136 suggests that its role is conserved in eubacterial DnaA orthologs.
Loading of DnaB at oriC relies on stable formation of DnaA oligomers on oriC. DnaA Arg281 residue indirectly facilitates DnaB binding and loading by contributing to stable binding of DnaA molecules to oriC (Felczak and Kaguni, 2004). By contrast, the results of the pull-down assay using an oriC fragment indicated that the numbers of DnaA and DnaB molecules bound to oriC were similar irrespective of whether wild-type DnaA or DnaA H136A was used. Thus, unlike DnaA K281A, DnaA H136A constructs stable complexes at oriC which are fully competent in stable DnaB binding. In our highly detailed structural model of DnaA (Shimizu et al., 2016), the location of the His136 residue suggests that it is unlikely to be involved in interactions between DnaA protomers, unlike the Arg281 residue, which resides at the inter-protomer interface (Figure 9).
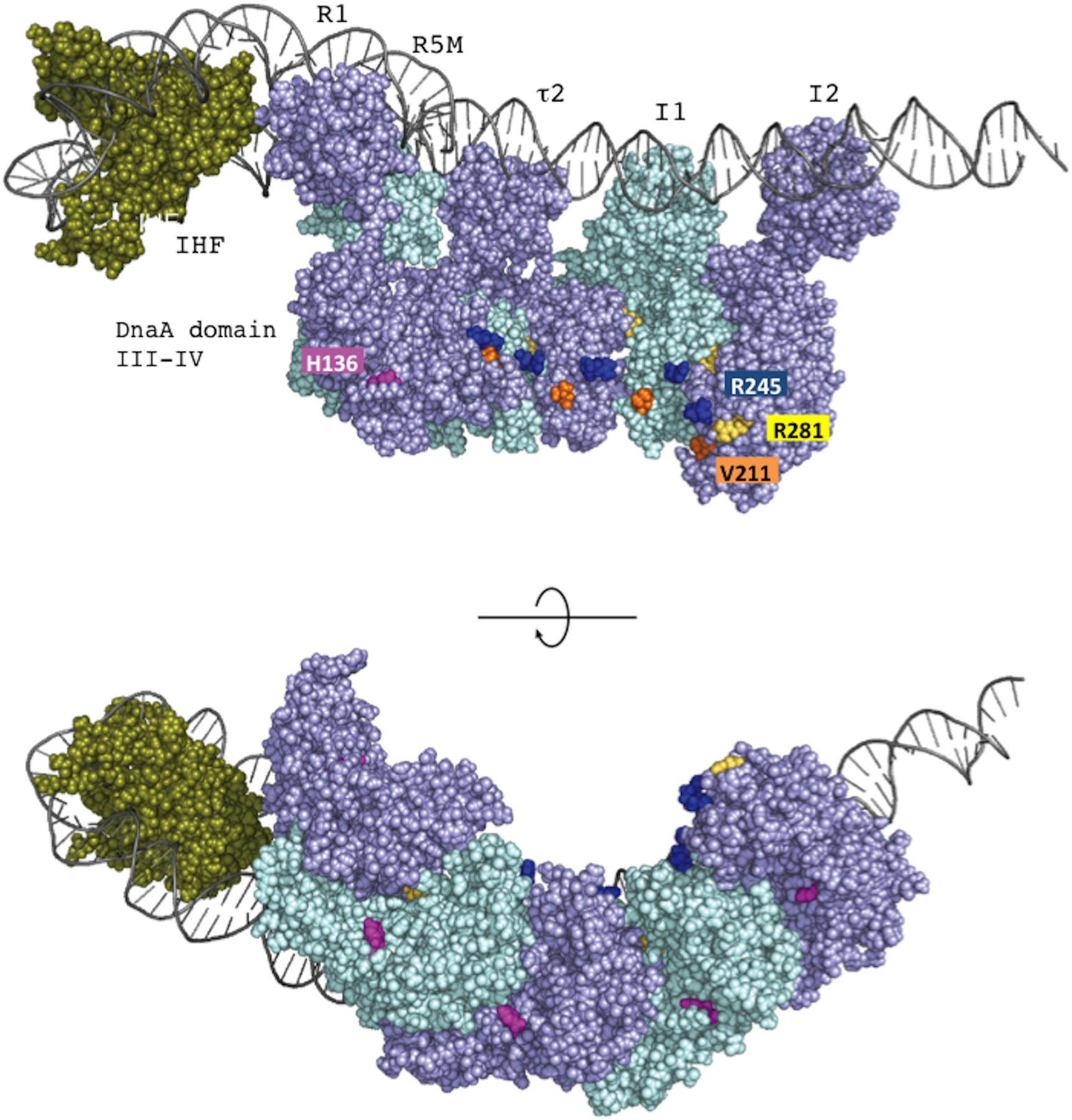
FIGURE 9. The position of DnaA H136 residue in the left-half DnaA subcomplex. On the basis of a molecular-dynamics-simulation near-atomic model of the Escherichia coli initiation complex (Shimizu et al., 2016), the left-half oriC complex and important functional residues are shown. In this model, ATP-DnaA domain III–IV molecules are shown complexed with five DnaA boxes (R1, R5M, τ2, I1, and I2) in the left-half DnaA-oligomerization region (DOR). Also, a DNA-bending protein integration host factor (IHF) binds to a unique IHF-binding site. The left-half oriC DNA is shown as silver lines. DnaA domain III and IV molecules are shown in violet and light blue. His136 residues are colored pink, and Arg281 residues are colored yellow. ssDUE-binding H/B-motif residues Val211 and Arg245 are shown in orange and dark blue, respectively. IHF is shown in dark green. Two sides of the complexes are shown (upper and lower panels).
Our previous in vitro and in vivo studies as well as the present data are in support of the ssDUE-recruitment mechanism in which DnaA oligomers constructed on DOR stably bind ssDUE using DnaA domain III H/B-motifs (Figure 1B; Ozaki and Katayama, 2012; Sakiyama et al., 2017). Recent structural analysis also supports this mechanism (Shimizu et al., 2016). In the paper of Duderstadt et al. (2010), DnaA oligomer formation was moderately stimulated by 25-mer ssDNA and largely decreased by the addition of 13-mer dsDNA bearing a single R1 box, resulting in an increase of DnaA monomers. It should be noted in these experiments that glutaraldehyde cross-linking was used because of the instability of ssDNA binding to DnaA and that this cross-linking produced considerable amounts of DnaA oligomers (dimers to tetramers) even in the absence of ssDNA, causing high background levels. Those results mean that addition of 13-mer dsDNA bearing the R1 box inhibits oligomerization of DnaA and the resultant R1-bound DnaA monomers (but not oligomers) do not bind stably to the ssDNA (and do not mean that dsDNA binding and ssDNA binding of DnaA mutually exclusive). This is consistent with our previous data. We demonstrated that unlike DnaA oligomers constructed on DOR fragments bearing multiple DnaA-binding sites, DnaA bound to a short DNA bearing only a single R1 box is inactive for stable ssDUE binding (Ozaki and Katayama, 2012). In addition, we demonstrated that two T-rich regions in the ssDUE are essential for the binding of ssDUE to DnaA oligomers constructed on the DOR (Ozaki et al., 2008). Consistently, we recently showed that two DnaA molecules bound to the R1 and R5M boxes are crucial for ssDUE binding (Sakiyama et al., 2017). As previously explained (Katayama et al., 2017; Sakiyama et al., 2017), these results concordantly support our idea that when DnaA molecules form an oligomer on the left-half DOR, stable ssDUE binding occur as a result of the linkage effect that enhances cooperative binding (Stauffer and Chazin, 2004; Figure 1B).
Our present results highlight a functional relationship between DnaA His136 and Phe46 (Figure 5). We previously reported that the DnaA domain I Phe46 exhibits high-affinity binding to DnaB when DnaA oligomers are constructed on oriC (Keyamura et al., 2009). In addition, although DnaA F46A alone has little or no activity for DnaB loading, DnaA F46A contributes to DnaB loading in the presence of wild-type DnaA. Similarly, we have now found that DnaA H136A alone has little or no activity for DnaB loading, but it participates in helicase loading at oriC when mixed with wild-type DnaA. This result suggests that His136 and Phe46 are only required in a subset of protomers in DnaA oligomers constructed on oriC. Further studies are required to determine which protomers in the DnaA subcomplexes might functionally interact with DnaB during the loading processes.
In the oriC plasmid-pull down assay using His-DnaB (Figure 6D), inhibition by DnaA H136A was moderate compared to the severe inhibition of Form I∗ production (Figure 4A). Given that DnaA Phe46-dependent DnaB interaction is intact even in DnaA H136A, a possible explanation to this is that interaction of DnaB with DnaA H136A at oriC results in abortive loading complexes of DnaB that are not competent for helicase activity. Alternatively, loading orientation of DnaB helicases on ssDUE or DUE strand-specific loading of DnaB might be compromised by the DnaA H136A mutation, resulting in abortive complexes.
Two distinct DnaB-binding modes of DnaA seem to be required for the loading of DnaB onto ssDUE. Notably, heterologous complexes formed at oriC by a combination of DnaA domain I F46A and domain III H136A are substantially inactive in DnaB loading (Figure 5B), suggesting that both Phe46 and His136 must be present in identical DnaA protomers for helicase loading at oriC. Although it has previously been suggested that a weak contact between DnaB and a DnaA region consisting of residues 111–148 (which includes His136) may precede stable binding involving DnaA domain I (Sutton et al., 1998), our results demonstrated that DnaA H136A fully sustained DnaA domain I-dependent DnaB binding. In a previous paper (Sutton et al., 1998) in which Surface Plasmon Resonance analysis was employed, the immobilization of DnaB on the sensor tip may have spatially occluded the DnaA domain I-binding site of DnaB, preventing DnaB interaction with DnaA. Taken together, a likely process is that DnaB binds to DnaA–oriC complexes via a primary contact mediated by DnaA domain I, followed by a secondary, weak contact mediated by DnaA His136. Binding of DnaB to DnaA domain I might bring DnaB into proximity with DnaA domain III, enabling the secondary weak contact through DnaA His136 of the same protomer (Figure 1B).
We now suggest a model in which the secondary, weak contact via His136 facilitates accessibility of DnaB to the ssDUE (Figure 10). Because the T-rich strand of the ssDUE directly binds to the H/B motifs of DnaA domain III (Ozaki et al., 2008; Duderstadt et al., 2011), DnaB bound to DnaA domain I can be brought in close proximity to the ssDUE through the physical contact with DnaA His136 and structural change of the flexible linker domain II (Figure 10). Notably, in our high-definition structural model of DnaA–oriC complexes, DnaA His136 residues are exposed on the surface of the DnaA protomers, suggesting that they are accessible to DnaB without physically obstructing ssDUE binding to the H/B-motifs (Shimizu et al., 2016; Figure 9).
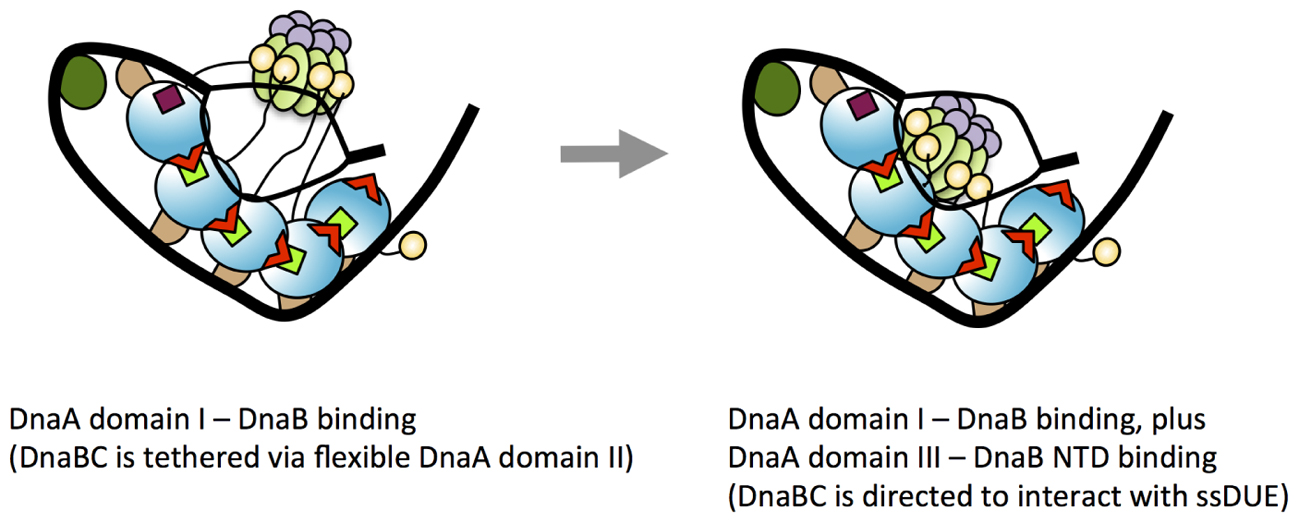
FIGURE 10. A model for dynamic DnaA–DnaB interactions toward DnaB loading to ssDUE. Possible processes of DnaB loading to the unwound DUE are shown. In the unwound complex, T-rich stand of ssDUE is bound to DnaA subcomplex constructed on the right-half DOR. At the first stage of DnaB loading, DnaB helicase stably binds to multiple molecules of DnaA bound to the DOR as shown in the left panel of this figure (for simplicity, the left-half oriC–DnaA subcomplex is shown here). At this point, DnaB in DnaBC complex binds to DnaA domain I and DnaBC is tethered via the flexible DnaA domain II loop, allowing for dynamic motion of DnaBC. In the next step (the right panel), DnaB specifically interacts with domain III His136 of the DOR-bound DnaA, which regulates the position and orientation of DnaBC, facilitating interaction with ssDUE (A-rich strand) and loading of DnaB. For DnaBC tethered to the right-half DOR–DnaA subcomplex, similar changes depending on DnaA His136 occur for DnaB loading to the expanded ssDUE (for details, see Ozaki and Katayama, 2012; Shimizu et al., 2016). Interaction between DnaA domain III and DnaB NTD as well as that between DnaA domain I and the DnaB CTD N-terminus-flanking region is suggested (Seitz et al., 2000). DnaC is suggested to bind to DnaB CDT (Galletto et al., 2003).
A role for DnaA His136 in regulation of loading of DnaB helicases to the ssDUE might explain why this residue is dispensable for DnaB loading in the ABC primosome system. The ssDNA template in the ABC primosome system most likely minimizes spatial constraints for DnaB loading, thereby bypassing the strict requirement for H136-mediated positioning of the helicase. In other words, ssDNA replication by the ABC primosome system does not require unwinding of dsDNA, regulated DnaA assembly like that on the left-half DOR–IHF complex or loading of two DnaB helicases in opposite directions. By contrast, at oriC, DnaB loading must occur on a short singled-stranded region of the DUE. At the M13-A site, the ssDNA region is much larger and thus DnaB loading is less spatially restricted; i.e., DnaB-loading may not be strictly regulated in the ABC promosome system; this may explain the differences in the roles of His136 at oriC and in the ABC promosome. Alternatively, SSB that coats the template ssDNA might have an auxiliary role in directing DnaB helicase for ssDNA loading in the ABC primosome system. Functional interaction between SSB and DnaB is reported to stimulate DnaB helicase activity (Biswas et al., 2002).
The linkage effect means that the presence of multiple contact points can result in a drastic increase in binding avidity (total affinity) even if each individual contact only has weak affinity (Stauffer and Chazin, 2004). A similar mechanism might underlie the DnaB interaction that depends on DnaA H136 residues in the oriC system. DnaA H136 residues are predicted to be regularly arrayed on the oriC-bound DnaA oligomers (Figure 9), which might enable formation of multiple contact points with a single DnaB homohexamer. By contrast, in the ABC primosome system, only a few DnaA protomers are involved in a region bearing only a single DnaA box (Masai et al., 1990; Carr and Kaguni, 2002), suggesting that the specific DnaA oligomer structure causing the His136-dependent linkage effect would not be constructed.
Whereas DnaA His136 was dispensable in the ABC primosome system, DnaA domain III is important. Specifically, two residues (Lys234 and Arg285) exposed on the protein surface that are important for DnaA–DnaA interaction stimulate ssDNA replication in the ABC primosome system (Kawakami et al., 2005; Ozaki et al., 2008). Binding of a few DnaA molecules to the hairpin structure of the template ssDNA (Carr and Kaguni, 2002) might be stimulated by domain III-dependent DnaA–DnaA interactions.
Dynamic protein complexes are constructed and change structurally at oriC for duplex unwinding and helicase loading. This study reveals the essential function for DnaA domain III His136 in the loading of DnaB replicative helicases on the ssDUE. Further analyses are required to dissect the DnaA protomers responsible for the DnaB interaction during DnaB loading on ssDUE and the DnaA-interacting sites on DnaB as well as to uncover the dynamic mechanisms of DnaA–DnaB complexes underlying the loading orientation of DnaB on ssDUE and DUE strand-specific loading of DnaB.
Author Contributions
YS, MN, SO, and TK conceived the experiments and wrote the paper. YS, MN, YA, and CH performed the experiments. All authors analyzed the data.
Funding
This work was supported by JSPS KAKENHI Grant Numbers JP26291004, JP17H03656, and JP16H00775. YS was supported by JSPS fellowship JP16J02075.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Research Support Center, Graduate School of Medical Sciences, Kyushu University for DNA sequencing.
References
Abe, Y., Jo, T., Matsuda, Y., Matsunaga, C., Katayama, T., and Ueda, T. (2007). Structure and function of DnaA N-terminal domains: specific sites and mechanisms in inter-DnaA interaction and in DnaB helicase loading on oriC. J. Biol. Chem. 282, 17816–17827. doi: 10.1074/jbc.M701841200
Baker, T. A., Sekimizu, K., Funnell, B. E., and Kornberg, A. (1986). Extensive unwinding of the plasmid template during staged enzymatic initiation of DNA replication from the origin of the Escherichia coli chromosome. Cell 45, 53–64. doi: 10.1016/0092-8674(86)90537-4
Bell, S. P., and Kaguni, J. M. (2013). Helicase loading at chromosomal origins of replication. Cold Spring Harb. Perspect. Biol. 5:a010124. doi: 10.1101/cshperspect.a010124
Biswas, E. E., Chen, P. H., and Biswas, S. B. (2002). Modulation of enzymatic activities of Escherichia coli DnaB helicase by single-stranded DNA-binding proteins. Nucleic Acids. Res. 30, 2809–2816. doi: 10.1093/nar/gkf384
Biswas, S. B., and Biswas-Fiss, E. E. (2006). Quantitative analysis of binding of single-stranded DNA by Escherichia coli DnaB helicase and the DnaB-DnaC complex. Biochemistry 45, 11505–11513. doi: 10.1021/bi060118d
Bramhill, D., and Kornberg, A. (1988). A model for initiation at origins of DNA replication. Cell 54, 915–918. doi: 10.1016/0092-8674(88)90102-X
Carr, K. M., and Kaguni, J. M. (2001). Stoichiometry of DnaA and DnaB protein in initiation at the Escherichia coli chromosomal origin. J. Biol. Chem. 276, 44919–44925. doi: 10.1074/jbc.M107463200
Carr, K. M., and Kaguni, J. M. (2002). Escherichia coli DnaA protein loads a single DnaB helicase at a DnaA box hairpin. J. Biol. Chem. 277, 39815–39822. doi: 10.1074/jbc.M205031200
Davey, M. J., and O’Donnell, M. (2003). Replicative helicase loaders: ring dispatch breakers and ring makers. Curr. Biol. 13, R594–R596. doi: 10.1016/S0960-9822(03)00523-2
Duderstadt, K. E., Chuang, K., and Berger, J. M. (2011). DNA stretching by bacterial initiators promotes replication origin opening. Nature 478, 209–213. doi: 10.1038/nature10455
Duderstadt, K. E., Mott, M. L., Crisona, N. J., Chuang, K., Yang, H., and Berger, J. M. (2010). Origin remodeling and opening in bacteria rely on distinct assembly states of the DnaA initiator. J. Biol. Chem. 285, 28229–28239. doi: 10.1074/jbc.M110.147975
Erzberger, J. P., Mott, M. L., and Berger, J. M. (2006). Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 13, 676–683. doi: 10.1038/nsmb1115
Erzberger, J. P., Pirruccello, M. M., and Berger, J. M. (2002). The structure of bacterial DnaA: implications for general mechanisms underlying DNA replication initiation. EMBO J. 21, 4763–4773. doi: 10.1093/emboj/cdf496
Fang, L., Davey, M. J., and O’Donnell, M. (1999). Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol. Cell 4, 541–553. doi: 10.1016/S1097-2765(00)80205-1
Felczak, M. M., and Kaguni, J. M. (2004). The box VII motif of Escherichia coli DnaA protein is required for DnaA oligomerization at the E. coli replication origin. J. Biol. Chem. 279, 51156–51162. doi: 10.1074/jbc.M409695200
Fujikawa, N., Kurumizaka, H., Nureki, O., Terada, T., Shirouzu, M., Katayama, T., et al. (2003). Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 31, 2077–2086. doi: 10.1093/nar/gkg309
Fujimitsu, K., Senriuchi, T., and Katayama, T. (2009). Specific genomic sequences of E. coli promote replicational initiation by directly reactivating ADP-DnaA. Genes Dev. 23, 1221–1233. doi: 10.1101/gad.1775809
Fuller, R. S., Kaguni, J. M., and Kornberg, A. (1981). Enzymatic replication of the origin of the Escherichia coli chromosome. Proc. Natl. Acad. Sci. U.S.A. 78, 7370–7374. doi: 10.1073/pnas.78.12.7370
Galletto, R., Jezewska, M. J., and Bujalowski, W. (2003). Interactions of the Escherichia coli DnaB helicase hexamer with the replication factor the DnaC protein: effect of nucleotide cofactors and the ssDNA on protein–protein interactions and the topology of the complex. J. Mol. Biol. 329, 441–465. doi: 10.1016/S0022-2836(03)00435-2
Grimwade, J. E., Rozgaja, T. A., Gupta, R., Dyson, K., Rao, P., and Leonard, A. C. (2018). Origin recognition is the predominant role for DnaA-ATP in initiation of chromosome replication. Nucleic Acids Res. 46, 6140–6151. doi: 10.1093/nar/gky457
Grimwade, J. E., Torgue, J. J.-C., McGarry, K. C., Rozgaja, T., Enloe, S. T., and Leonard, A. C. (2007). Mutational analysis reveals Escherichia coli oriC interacts with both DnaA-ATP and DnaA-ADP during pre-RC assembly. Mol. Microbiol. 66, 428–439. doi: 10.1111/j.1365-2958.2007.05930.x
Inoue, Y., Tanaka, H., Kasho, K., Fujimitsu, K., Oshima, T., and Katayama, T. (2016). Chromosomal location of the DnaA-reactivating sequence DARS2 is important to regulate timely initiation of DNA replication in Escherichia coli. Genes Cells 21, 1015–1023. doi: 10.1111/gtc.12395
Itsathitphaisarn, O., Wing, R. A., Eliason, W. K., Wang, J., and Steitz, T. A. (2012). The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell 151, 267–277. doi: 10.1016/j.cell.2012.09.014
Iyer, L. M., Leipe, D. D., Koonin, E. V., and Aravind, L. (2004). Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 146, 11–31. doi: 10.1016/j.jsb.2003.10.010
Kaguni, J. M. (2011). Replication initiation at the Escherichia coli chromosomal origin. Curr. Opin. Chem. Biol. 15, 606–613. doi: 10.1016/j.cbpa.2011.07.016
Katayama, T., Kasho, K., and Kawakami, H. (2017). The DnaA cycle in Escherichia coli: activation, function and inactivation of the initiator protein. Front. Microbiol. 8:2496. doi: 10.3389/fmicb.2017.02496
Katayama, T., Ozaki, S., Keyamura, K., and Fujimitsu, K. (2010). Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 8, 163–170. doi: 10.1038/nrmicro2314
Kawakami, H., Keyamura, K., and Katayama, T. (2005). Formation of an ATP-DnaA-specific initiation complex requires DnaA Arginine 285, a conserved motif in the AAA+ protein family. J. Biol. Chem. 280, 27420–27430. doi: 10.1074/jbc.M502764200
Keyamura, K., Abe, Y., Higashi, M., Ueda, T., and Katayama, T. (2009). DiaA dynamics are coupled with changes in initial origin complexes leading to helicase loading. J. Biol. Chem. 284, 25038–25050. doi: 10.1074/jbc.M109.002717
Keyamura, K., Fujikawa, N., Ishida, T., Ozaki, S., Su’etsugu, M., Fujimitsu, K., et al. (2007). The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP DnaA-specific initiation complexes. Genes Dev. 21, 2083–2099. doi: 10.1101/gad.1561207
Kubota, T., Katayama, T., Ito, Y., Mizushima, T., and Sekimizu, K. (1997). Conformational transition of DnaA protein by ATP: structural analysis of DnaA protein, the initiator of Escherichia coli chromosome replication. Biochem. Biophys. Res. Commun. 232, 130–135. doi: 10.1006/bbrc.1997.6244
Kurokawa, K., Nishida, S., Emoto, A., Sekimizu, K., and Katayama, T. (1999). Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J. 18, 6642–6652. doi: 10.1093/emboj/18.23.6642
Leonard, A. C., and Grimwade, J. E. (2015). The orisome: structure and function. Front. Microbiol. 6:545. doi: 10.3389/fmicb.2015.00545
Makowska-Grzyska, M., and Kaguni, J. M. (2010). Primase directs the release of DnaC from DnaB. Mol. Cell 37, 90–101. doi: 10.1016/j.molcel.2009.12.031
Marszalek, J., and Kaguni, J. M. (1994). DnaA protein directs the binding of DnaB protein in initiation of DNA replication in Escherichia coli. J. Biol. Chem. 269, 4883–4890.
Marszalek, J., Zhang, W., Hupp, T. R., Margulies, C., Carr, K. M., Cherry, S., et al. (1996). Domains of DnaA protein involved in interaction with DnaB protein, and in unwinding the Escherichia coli chromosomal origin. J. Biol. Chem. 271, 18535–18542. doi: 10.1074/jbc.271.31.18535
Masai, H., and Arai, K. (1995). DnaA-dependent assembly of the ABC primosome at the A site, a single-stranded DNA hairpin containing a dnaA box. Eur. J. Biochem. 230, 384–395. doi: 10.1111/j.1432-1033.1995.tb20573.x
Masai, H., Nomura, N., and Arai, K. (1990). The ABC-primosome. A novel priming system employing dnaA, dnaB, dnaC, and primase on a hairpin containing a dnaA box sequence. J. Biol. Chem. 265, 15134–15144.
McGarry, K. C., Ryan, V. T., Grimwade, J. E., and Leonard, A. C. (2004). Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc. Natl. Acad. Sci. U.S.A. 101, 2811–2816. doi: 10.1073/pnas.0400340101
Mott, M. L., Erzberger, J. P., Coons, M. M., and Berger, J. M. (2008). Structural synergy and molecular crosstalk between bacterial helicase loaders and replication initiators. Cell 135, 623–634. doi: 10.1016/j.cell.2008.09.058
Nishida, S., Fujimitsu, K., Sekimizu, K., Ohmura, T., Ueda, T., and Katayama, T. (2002). A nucleotide switch in the Escherichia coli DnaA protein initiates chromosomal replication: evidence from a mutant DnaA protein defective in regulatory ATP hydrolysis in vitro and in vivo. J. Biol. Chem. 277, 14986–14995. doi: 10.1074/jbc.M108303200
Noguchi, Y., and Katayama, T. (2016). The Escherichia coli cryptic prophage protein YfdR binds to DnaA and initiation of chromosomal replication is inhibited by overexpression of the gene cluster yfdQ-yfdR-yfdS-yfdT. Front. Microbiol. 7:239. doi: 10.3389/fmicb.2016.00239
Noguchi, Y., Sakiyama, Y., Kawakami, H., and Katayama, T. (2015). The Arg fingers of key DnaA protomers are oriented inward within the replication origin oriC and stimulate DnaA subcomplexes in the initiation complex. J. Biol. Chem. 290, 20295–20312. doi: 10.1074/jbc.M115.662601
Nozaki, S., and Ogawa, T. (2008). Determination of the minimum domain II size of Escherichia coli DnaA protein essential for cell viability. Microbiology 154, 3379–3384. doi: 10.1099/mic.0.2008/019745-0
Ozaki, S., and Katayama, T. (2009). DnaA structure, function, and dynamics in the initiation at the chromosomal origin. Plasmid 62, 71–82. doi: 10.1016/j.plasmid.2009.06.003
Ozaki, S., and Katayama, T. (2012). Highly organized DnaA-oriC complexes recruit the single-stranded DNA for replication initiation. Nucleic Acids Res. 40, 1648–1665. doi: 10.1093/nar/gkr832
Ozaki, S., Kawakami, H., Nakamura, K., Fujikawa, N., Kagawa, W., Park, S.-Y., et al. (2008). A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J. Biol. Chem. 283, 8351–8362. doi: 10.1074/jbc.M708684200
Ozaki, S., Noguchi, Y., Hayashi, Y., Miyazaki, E., and Katayama, T. (2012a). Differentiation of the DnaA-oriC subcomplex for DNA unwinding in a replication initiation complex. J. Biol. Chem. 287, 37458–37471. doi: 10.1074/jbc.M112.372052
Ozaki, S., Noguchi, Y., Nishimura, M., and Katayama, T. (2012b). Stable nucleotide binding to DnaA requires a specific glutamic acid residue within the AAA+ box II motif. J. Struct. Biol. 179, 242–250. doi: 10.1016/j.jsb.2012.05.001
Rozgaja, T. A., Grimwade, J. E., Iqbal, M., Czerwonka, C., Vora, M., and Leonard, A. C. (2011). Two oppositely oriented arrays of low-affinity recognition sites in oriC guide progressive binding of DnaA during Escherichia coli pre-RC assembly. Mol. Microbiol. 82, 475–488. doi: 10.1111/j.1365-2958.2011.07827.x
Sakiyama, Y., Kasho, K., Noguchi, Y., Kawakami, H., and Katayama, T. (2017). Regulatory dynamics in the ternary DnaA complex for initiation of chromosomal replication in Escherichia coli. Nucleic Acids Res. 45, 12354–12373. doi: 10.1093/nar/gkx914
Seitz, H., Weigel, C., and Messer, W. (2000). The interaction domains of the DnaA and DnaB replication proteins of Escherichia coli. Mol. Microbiol. 37, 1270–1279. doi: 10.1046/j.1365-2958.2000.02096.x
Shimizu, M., Noguchi, Y., Sakiyama, Y., Kawakami, H., Katayama, T., and Takada, S. (2016). Near-atomic structural model for bacterial DNA replication initiation complex and its functional insights. Proc. Natl. Acad. Sci. U.S.A. 113, E8021–E8030. doi: 10.1073/pnas.1609649113
Skarstad, K., Boye, E., and Steen, H. B. (1986). Timing of initiation of chromosome replication in individual Escherichia coli cells. EMBO J. 5, 1711–1717.
Skarstad, K., von Meyenburg, K., Hansen, F. G., and Boye, E. (1988). Coordination of chromosome replication initiation in Escherichia coli: effects of different dnaA alleles. J. Bacteriol. 170, 852–858. doi: 10.1128/jb.170.2.852-858.1988
Smith, G. R., Contreras-Moreira, B., Zhang, X., and Bates, P. A. (2004). A link between sequence conservation and domain motion within the AAA+ family. J. Struct. Biol. 146, 189–204. doi: 10.1016/j.jsb.2003.11.022
Soultanas, P. (2012). Loading mechanisms of ring helicases at replication origins. Mol. Microbiol. 84, 6–16. doi: 10.1111/j.1365-2958.2012.08012.x
Stauffer, M. E., and Chazin, W. J. (2004). Structural mechanisms of DNA replication, repair, and recombination. J. Biol. Chem. 279, 30915–30918. doi: 10.1074/jbc.R400015200
Strycharska, M. S., Arias-Palomo, E., Lyubimov, A. Y., Erzberger, J. P., O’Shea, V. L., Bustamante, C. J., et al. (2013). Nucleotide and partner-protein control of bacterial replicative helicase structure and function. Mol. Cell 52, 844–854. doi: 10.1016/j.molcel.2013.11.016
Sutton, M. D., Carr, K. M., Vicente, M., and Kaguni, J. M. (1998). Escherichia coli DnaA protein. The N-terminal domain and loading of DnaB helicase at the E. coli chromosomal origin. J. Biol. Chem. 273, 34255–34262. doi: 10.1074/jbc.273.51.34255
Sutton, M. D., and Kaguni, J. M. (1997). Threonine 435 of Escherichia coli DnaA protein confers sequence-specific DNA binding activity. J. Biol. Chem. 272, 23017–23024. doi: 10.1074/jbc.272.37.23017
Wolański, M., Donczew, R., Zawilak-Pawlik, A., and Zakrzewska-Czerwińska, J. (2015). oriC-encoded instructions for the initiation of bacterial chromosome replication. Front. Microbiol. 5:735. doi: 10.3389/fmicb.2014.00735
Keywords: E. coli, oriC, DnaA, helicase, AAA+ family, protein–protein interaction
Citation: Sakiyama Y, Nishimura M, Hayashi C, Akama Y, Ozaki S and Katayama T (2018) The DnaA AAA+ Domain His136 Residue Directs DnaB Replicative Helicase to the Unwound Region of the Replication Origin, oriC. Front. Microbiol. 9:2017. doi: 10.3389/fmicb.2018.02017
Received: 29 May 2018; Accepted: 09 August 2018;
Published: 31 August 2018.
Edited by:
Feng Gao, Tianjin University, ChinaReviewed by:
Alan Leonard, Florida Institute of Technology, United StatesGregory Marczynski, McGill University, Canada
Dhruba Chattoraj, National Institutes of Health (NIH), United States
Copyright © 2018 Sakiyama, Nishimura, Hayashi, Akama, Ozaki and Katayama. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tsutomu Katayama, katayama@phar.kyushu-u.ac.jp
†These authors have contributed equally to this work
‡Present address: Yukari Sakiyama, Daiichi Sankyo Co., Tokyo, Japan Masahiro Nishimura, Fukuoka Prefectural Office, Fukuoka, Japan
 Yukari Sakiyama†‡
Yukari Sakiyama†‡ Masahiro Nishimura
Masahiro Nishimura Shogo Ozaki
Shogo Ozaki Tsutomu Katayama
Tsutomu Katayama