- 1Bioresource Collection and Research Center, Food Industry Research and Development Institute, Hsinchu, Taiwan
- 2Molecular Medicine Research Center, Chang Gung University, Taoyuan, Taiwan
- 3Department of Animal Science and Technology, College of Bioresources and Agriculture, National Taiwan University, Taipei, Taiwan
Lactobacillus casei, Lactobacillus paracasei, and Lactobacillus rhamnosus are phenotypically and genotypically closely related, and together comprise the L. casei group. Although the strains of this group are commercially valuable as probiotics, the taxonomic status and nomenclature of the L. casei group have long been contentious because of the difficulties in identifying these three species by using the most frequently used genotypic methodology of 16S rRNA gene sequencing. Long used as the gold standard for species classification, DNA–DNA hybridization is laborious, requires expert skills, and is difficult to use routinely in laboratories. Currently, genome-based comparisons, including average nucleotide identity (ANI) and digital DNA–DNA hybridization (dDDH), are commonly applied to bacterial taxonomy as alternatives to the gold standard method for the demarcating phylogenetic relationships. To establish quick and accurate methods for identifying strains in the L. casei group at the species and subspecies levels, we developed species- and subspecies-specific identification methods based on housekeeping gene sequences and whole-cell matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) spectral pattern analysis. By phylogenetic analysis based on concatenated housekeeping gene sequences (dnaJ, dnaK, mutL, pheS, and yycH), 53 strains were separated into four clusters corresponding to the four species: L. casei, L. paracasei and L. rhamnosus, and Lactobacillus chiayiensis sp. nov. A multiplex minisequencing assay using single nucleotide polymorphism (SNP)-specific primers based on the dnaK gene sequences and species-specific primers based on the mutL gene sequences provided high resolution that enabled the strains at the species level to be identified as L. casei, L. paracasei, and L. rhamnosus. By MALDI-TOF MS analysis coupled with an internal database and ClinProTools software, species- and subspecies-level L. casei group strains were identified based on reliable scores and species- and subspecies-specific MS peaks. The L. paracasei strains were distinguished clearly at the subspecies level based on subspecies-specific MS peaks. This article describes the rapid and accurate methods used for identification and classification of strains in the L. casei group based on housekeeping gene sequences and MALDI-TOF MS analysis as well as the novel speciation of this group including L. chiayiensis sp. nov. and ‘Lactobacillus zeae’ by genome-based methods.
Introduction
Lactobacillus is the largest genus of the family Lactobacillaceae. As of May 2018, it consists of 196 validly published species1, and these species are commonly isolated not only from environments associated with fermented food, such as fruits, meat, sourdough, vegetables, and wine, but also from the gastrointestinal and vaginal tracts of humans and animals (Watanabe et al., 2008; Hammes and Hertel, 2009; Pot et al., 2014; Tamang et al., 2015). Currently, lactobacilli are widely applied in fields related to food, feed, pharmaceuticals and biotechnology; for example, they are used as dairy starters, probiotics, vaccine carriers and silage inoculants (Seegers, 2002; Giraffa et al., 2010), which are among the most economically interesting applications of lactic acid bacteria (LAB). Lactobacilli are gram-positive, rod-shaped, facultatively anaerobic or microaerophilic, non-spore-forming, acid-tolerant, and catalase-negative bacteria with DNA G+C content that is usually less than 50 mol%. Lactobacillus casei, Lactobacillus paracasei, and Lactobacillus rhamnosus are phylogenetically and phenotypically closely related; together, they are regarded as the L. casei group. Members of this group are facultatively heterofermentative, have 45–47 mol% DNA G+C content, and have identical peptidoglycan types (L-Lys-D-Asp) (Salvetti et al., 2012). The widely known probiotic strains that are part of this group (such as L. casei strain Shirota and L. rhamnosus GG) are used worldwide in fermented dairy products or food supplements and as probiotics to enhance host health (Shida and Nomoto, 2013; Ashraf and Shah, 2014; Reid, 2015; Orlando et al., 2016). Although this group comprises many commercially valuable strains, its taxonomic status has long been contentiousas has its nomenclaturebecause methods with inadequate taxonomic resolution have been used, leading to species being mislabeled in products, publications and some publicly available DNA sequences.
The taxonomy of the L. casei group has been altered numerous times. In the Approved Lists of Bacterial Names (Skerman et al., 1980), L. casei described as a single species with five subspecies on the basis of phenotypic features: L. casei subsp. casei, L. casei subsp. alactosus, L. casei subsp. pseudoplantarum, L. casei subsp. tolerans, and L. casei subsp. rhamnosus. On the basis of DNA–DNA homology, this species was reclassified into three species and two subspecies (Collins et al., 1989): (i) L. casei (including strains previously belonging to L. casei subsp. casei); (ii) L. paracasei comprising two subspecies, L. paracasei subsp. paracasei (including the previous subspecies L. casei subsp. alactosus and L. casei subsp. pseudoplantarum) and L. paracasei subsp. tolerans (including the previous subspecies L. casei subsp. tolerans); and (iii) L. rhamnosus (including the previously classified subspecies L. casei subsp. rhamnosus).
The conventional taxonomy of the genus Lactobacillus has mostly depended on morphological, physiological, and biochemical traits, and scientists have principally depended on commercial identification kits, such as, the API 50 CHL system (Charteris et al., 2001; Boyd et al., 2005). Although phenotypic tests have been applied to determine the metabolic characteristics of each strain, the L. casei group members have numerous characteristics in common that result in remarkably similar phenotypes. This similarity in phenotypic traits may be influenced by the possible acquisition or depletion of plasmids encoding large number of carbohydrate fermentation traits or environmental conditions, and this may result in isolates with metabolic features that are atypical (Ahrné et al., 1989), rendering the established identification methods laborious and imprecise. Therefore, using molecular taxonomic methods to improve species identification of Lactobacillus strains has become common among researchers.
On the basis of analyses of cellular soluble protein patterns, randomly amplified polymorphic DNA (RAPD) fingerprinting, and DNA–DNA hybridization (DDH), Dellaglio et al. (1991) and Dicks et al. (1996) proposed that L. casei subsp. casei ATCC 393 and ‘Lactobacterium zeae’ ATCC 15820 should be reclassified as ‘Lactobacillus zeae’ nom. rev., designated the strain ATCC 334 as a neotype of L. casei subsp. casei, and forbade the name L. paracasei. In 2008, the Judicial Commission of the International Committee on Systematics of Prokaryotes (ICSP) stated that the current taxonomy of the L. casei group is comprised of three closely related species: L. casei (type strain: ATCC 393), L. paracasei subsp. paracasei (type strain: ATCC 25302) and L. paracasei subsp. tolerans (type strain: ATCC 25599), and L. rhamnosus (type strain: ATCC 7469) (Tindall, 2008). Huang et al. (2018) updated the taxonomy proposing a new species for the L. casei group based on whole-genome sequencing (WGS), multilocus sequence analysis (MLSA), matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), phenotypic characterization and species-specific PCR.
As far as the molecular taxonomic methods, such as RAPD-PCR (Huang and Lee, 2009) and temporal temperature gradient gel electrophoresis (TGGE) (Vásquez et al., 2001) have been applied to identify L. casei group strains. DDH has long been used as a method for delineation of bacterial species because of its status as a conventional gold standard genotypic assay (Wayne et al., 1987; Stackebrandt et al., 2002). However, DDH is laborious, requires expert skills, and is inconsistently reproducible. Furthermore, the data obtained are non-cumulative and difficult to apply to all microorganisms (Mehlen et al., 2004; Pontes et al., 2007). Recent progress in technology for DNA sequencing technology has enabled genome sequences to be applied relatively inexpensively, providing a high-quality and adequate bioinformatic tool for taxonomic studies of prokaryotes (Rosselló-Móra and Amann, 2015). Analyzing comparative sequencing of the 16S rRNA gene is currently a common route to identification and classification of bacteria. Since 1994, strains with similarity of more than 97% to the 16S rRNA gene sequence have been considered to belong to the same species (Stackebrandt and Goebel, 1994). The species-level cutoff percentage was evaluated to 98.7% and then 98.65% in 2006 and 2014, respectively (Stackebrandt and Ebers, 2006; Kim et al., 2014). However, in many cases, species that are closely related—such as those belonging to the Lactobacillus buchneri group (L. buchneri, L. kefiri, L. parabuchneri, and L. parakefiri), L. casei group (L. casei, L. paracasei, and L. rhamnosus), Lactobacillus plantarum group (L. fabifermentans, L. plantarum, L. paraplantarum, and L. pentosus) and Lactobacillus sakei group (L. curvatus, L. graminis, and L. sakei) are indistinguishable using 16S rRNA gene sequencing because of the high degree of similarity (as high as 99%) of the 16S rRNA gene sequences among the species (Torriani et al., 2001; Koort et al., 2004; Watanabe et al., 2009; Huang et al., 2010; Huang and Lee, 2011). Consequently, substantially conserved protein-encoding genes are useful as alternative molecular targets for distinguishing closely related species. Thus, a housekeeping sequence in addition to supplemental techniques including PCR-restriction fragment length polymorphism (RFLP), SNaPshot minisequencing, species- and subspecies-specific PCR, MLSA, and MALDI-TOF MS, has successfully been applied to achieve higher resolution within the L. casei group (Felis et al., 2001; Naser et al., 2007; Watanabe et al., 2008; Huang and Lee, 2009, 2011; Huang et al., 2011, 2014, 2015, 2018; Bottari et al., 2017).
Huys et al. (2006) reported that more than 28% of commercially available probiotic products were labeled incorrectly at the genus or species level because methods that limit taxonomic resolution were used. Correct identification of probiotic strains at the species level is essential for safety assessment as it allows a linkage to potentially relevant, species-related scientific, and technological information. Therefore, a method that quickly and accurately identifies starter cultures is required for the management and quality control of commercial probiotic products.
This article describes the rapid and accurate methods for identification and classification of strains in the L. casei group based on housekeeping gene sequences and MALDI-TOF MS analysis in addition to the current state in classification of the L. casei group, including a novel species identified using genome-based methods.
L. casei Group Housekeeping-Gene-Based Phylogenetic Analysis
Housekeeping genes exhibit high sequence variation and are feasible alternatives to the 16S rRNA gene in accurately classifying and identifying bacteria (Petti, 2007). For example, gene sequence analysis of the phenylalanyl t-RNA synthase alpha subunit (pheS) and RNA polymerase alpha subunit (rpoA) has been used to distinguish closely related LAB species of the genera Lactobacillus, Enterococcus, Leuconostoc, Pediococcus, and Weissella (Naser et al., 2005, 2007; De Bruyne et al., 2007, 2008, 2010; Chao et al., 2010, 2012, 2013; Oki et al., 2012; Nguyen et al., 2013; Nyanzi et al., 2013; Huang et al., 2018). To date, numerous protein-encoding genes for example, dnaJ, dnaK, hsp60, mutL, pheS, recA, rpoA, spxB, tuf, and yycH have been used as phylogenetic targets to discriminate among species of the L. casei group and have exhibited satisfactory resolution with a high level of discrimination (Felis et al., 2001; Naser et al., 2007; Huang and Lee, 2011; Huang et al., 2014, 2015; Yu et al., 2014; Sardaro et al., 2016; Bottari et al., 2017). However, horizontal gene transfer (HGT) or lateral gene transfer may cause problems in phylogenetic tree construction based on a single gene. Concatenation of several housekeeping genes may reduce the weight of HGT and recombination (Gogarten et al., 2002; Boucher et al., 2004; Macheras et al., 2011; Timilsina et al., 2015). Furthermore, it could accurately locate taxonomic positions for closely related species and strains (Glaeser and Kämpfer, 2015).
The ad hoc committee aimed at re-evaluation of the species definitions in bacteriology has determined that sequencing of several housekeeping genes (i.e., five or more) is a reliable method of examining deep-level phylogenies and complex species groups (Stackebrandt et al., 2002). MLSA based on housekeeping gene sequences provides high discriminative power and has the potential to replace DDH (Zeigler, 2003; Gevers et al., 2005; Vandamme and Peeters, 2014; Glaeser and Kämpfer, 2015). However, MLSA has several drawbacks; for example, no universal threshold for species definition, no common set of genes has yet been suggested, and few reference sequences have been released through public databases (Mulet et al., 2010; Lai et al., 2014; Li C. et al., 2014; Rosselló-Móra and Amann, 2015; Peeters et al., 2016; Liu et al., 2017). Therefore, MLSA must be improved to render it more feasible and applicable.
Multilocus sequence analysis has been increasingly used to identify and describe novel species (Killer et al., 2014, 2017; Chen et al., 2017; Doi et al., 2017; Liu et al., 2017; Ribeiro et al., 2017; Modesto et al., 2018). Mattarelli et al. (2014) stated that a minimum of two additional phylogenetic markers should be used to accurately identify isolates, especially for describing novel species in the genera Bifidobacterium and Lactobacillus.
Huang et al. (2018) used MLSA to characterize the L. casei group based on concatenated sequences of three housekeeping genes (dnaK, pheS, and yycH; 1627 bp), with the neighbor-joining, maximum-likelihood, and minimum-evolution methods used for phylogenetic analysis. The results showed that a novel species, L. chiayiensis sp. nov. (strains BCRC 81062T and BCRC 18859), comprised an independent cluster notably distinct from L. casei and ‘L. zeae.’ Furthermore, a split network tree based on the concatenated sequences of five housekeeping genes (dnaJ, dnaK, mutL, pheS, and yycH; 2567 bp) was performed, and the 53 strains of the L. casei group were divided into three principal clusters: Cluster A (comprising five strains of L. casei, ‘L. zeae,’ and L. chiayiensis sp. nov.); Cluster B (comprising 27 strains of L. paracasei); Cluster C (comprising 21 strains of L. rhamnosus). Split network analysis revealed that Cluster A comprised three subclusters: subclusters A-1 (L. casei), A-2 (‘L. zeae’), and A-3 (L. chiayiensis sp. nov.) (Figure 1). Thus, multilocus phylogenetic analysis of the sequence data of ‘L. zeae’ indicated that this microorganism could be reclassified as an independent species or a subspecies of L. casei.
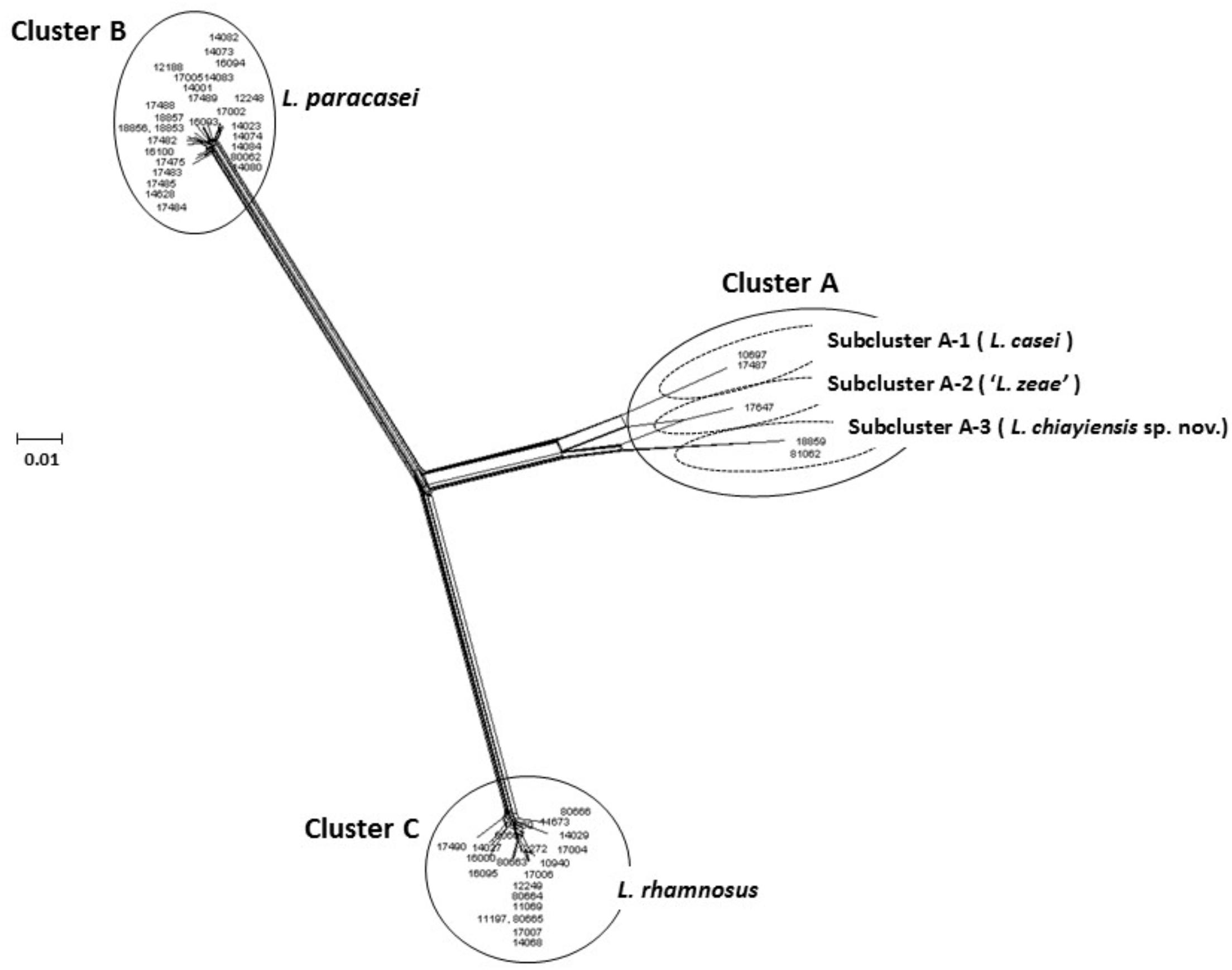
FIGURE 1. Concatenated split network tree based on five genes. The dnaJ, dnaK, mutL, pheS, and yycH gene sequences (2567 bp) from 53 L. casei group strains were concatenated and reconstructed using the SplitTree4 program. Sequence similarities were corrected using the Jukes-Cantor correction. Bar, 0.01 expected nucleotide substitutions per site.
Development of Species and Subspecies-Specific Primers
Detection Through Conventional PCR Assay
The bacterial 16S rRNA gene has nine hypervariable regions (V1–V9). These nine regions exhibit substantial sequence diversity among bacteria. The hypervariable regions contain species-specific sequences that are suitable for diagnostic assays (Van de Peer et al., 1996). Ward and Timmins (1999) have developed three PCR primer pairs that are specific for the L. casei group member (L. casei, L. paracasei, and L. rhamnosus) on the basis of variation in the V1 region of the 16S rRNA gene. These primer pairs were validated using 63 Lactobacillus isolates from cheeses. However, ambiguous results were obtained when these primers were used to analyze 76 wild isolates from dairy samples (milk and differently ripened cheeses) (Bottari et al., 2017). A species-specific primer for ‘L. zeae’ was designed by Desai et al. (2006) on the basis of the 16S rRNA gene. As well as the 16S rRNA gene, the 16S–23S rRNA intergenic spacer region was investigated to successfully identify L. paracasei and L. rhamnosus (Song et al., 2000). Protein-coding genes evolve rapidly to provide greater interspecies variation and can act as supplemental targets for designing species-specific primers. We previously developed several highly specific primer sets targeted the dnaK, mutL, and yycH genes for the species-level identification of L. casei, L. chiayiensis sp. nov., L. paracasei, and L. rhamnosus by using PCR-based methods (Huang et al., 2014, 2018; Huang and Huang, 2018). Studies have established a multiplex species-specific PCR assay on the basis of the 16S–23S rRNA spacer region and tuf, mutL, and pepR genes for identifying L. casei group members (Ventura et al., 2003; Kwon et al., 2004; Watanabe et al., 2008; Bottari et al., 2017).
Primer specificity for the selected species can be advanced only if known gene sequences exist that possess particular and satisfactory variations in nucleotides. By contrast, novel species-specific primers can be developed on the basis of sequence characterized amplified region (SCAR) markers through RAPD-PCR analysis. SCARs are detected by PCR using specific primers designed based on the sequence data of RAPD fragment. Multiplex PCR assay of SCAR makers is useful method for reducing the costs and time for routine identification. A combination of RAPD-PCR and the SCAR technique has been applied to develop the species- and subspecies-specific primers for L. paracasei subsp. tolerans, L. rhamnosus, and ‘L. zeae’ within the L. casei group (Huang and Lee, 2009).
Detection Using SNaPshot Minisequencing Assay
SNaPshot is a primer extension-based multiplex system for detecting single nucleotide polymorphisms (SNPs) by using fluorescently labeled dideoxynucleotides (ddNTPs). Each fluorescent ddNTP emits a unique wavelength, which is subsequently represented as a particular color in a base-dependent manner. The extension products with fluorescent labeling can be visualized using capillary electrophoresis with automated sequencing. Huang et al. (2011) designed a PCR primer pair specific to L. casei group by using rpoA gene sequence and performed a SNaPshot minisequencing assay to identify species belonging to the L. casei group. A SNaPshot minisequencing assay using dnaK as a target gene was also developed, and five species-specific SNP primers were subsequently designed by analyzing the conserved regions of the dnaK gene sequences. The specificity of this assay was evaluated using 63 L. casei group strains. SNaPshot minisequencing assay combined with group-specific PCR was confirmed to quickly, accurately, and inexpensively identify the L. casei group at the species level. By using species-specific PCR based on dnaJ gene sequences with SNaPshot minisequencing, strains of L. rhamnosus were identified with high accuracy at the species level; furthermore, the method determined the SNP haplotypes (Huang et al., 2015).
Multiplex minisequencing is a method for determining the exact nucleotide located at a particular site, particularly when numerous SNPs are screened simultaneously within a single reaction tube. Only 40 min is required for minisequencing products to undergo automated fluorescent capillary electrophoresis, making this method quicker than direct sequencing. A total of 2.5 h is required for sequencing products to undergo capillary electrophoresis.
Rapid Identification and Classification of L. casei Group Strains at the Species and Subspecies Levels Using MALDI-TOF MS
MALDI-TOF MS is a phenotype-based method that can be used for the identification, classification, and dereplication of large numbers of microorganisms on the basis of the specific proteomic profiles of these microorganisms (Ghyselinck et al., 2011). Applied in routine clinical diagnosis, and its use has been extended to environmental monitoring, food safety, biodiversity, and gut microbiology (Pavlovic et al., 2013; Samb-Ba et al., 2014; Santos et al., 2016). MALDI-TOF MS can typically discriminate between bacterial strains at both the genus and species levels using proteomics-based identification, and subspecies- and strain-level taxonomic resolution is possible when the method is combined with specific mass spectral (MS) peaks (Spinali et al., 2015). MALDI-TOF MS is straightforward, rapid, accurate, and inexpensive (although the initial outlay for the instrument is considerable), and it has high throughput (Neville et al., 2011; Sandrin et al., 2013; Tran et al., 2015). However, reference databases are mainly designed for routine clinical practice (Bizzini and Greub, 2010), and expansion of the database is necessary to increase identification rates through matching. In addition, and as a disadvantage associated with this method, procuring fresh, pure cultures and sufficient bacteria cells (105–107 cells) are required for protein extraction and analysis.
MALDI-TOF MS has been effectively used for identifying strains of lactobacilli isolated from dairy and meat products (Angelakis et al., 2011; Doan et al., 2012; Dušková et al., 2012; Nacef et al., 2017), fermented food (Kim et al., 2017), carious dentin from children (Callaway et al., 2013), human oral cavities and vaginas (Anderson et al., 2014), and poultry (Dec et al., 2014). A reliance on protein fingerprint profiling only could produce unclear findings for some closely related species or subspecies (e.g., Bacillus pumilus and Bacillus safensis, Lactobacillus johnsonii and Lactobacillus gasseri, and Lactobacillus plantarum subspecies plantarum, and Lactobacillus plantarum subspecies argentoratensis) (Šedo et al., 2013; Branquinho et al., 2014; Wieme et al., 2014). Thus, researchers recognize that particular MS peaks must be identified and developed for diagnosis. MS peaks from bacterial cell lysate consist mainly of ribosomal proteins and include several housekeeping proteins, such as the housekeeping genes used for MLSA to determine high-level taxonomy and phylogeny (Welker and Moore, 2011). A ribosomal protein typing-based method using MALDI-TOF MS has been proposed, and since then, it has been used to successfully differentiate Bacillus spp., Bifidobacterium animalis, Bifidobacterium longum, Lactococcus lactis, and the L. casei group (Tanigawa et al., 2010; Hotta et al., 2011; Sato et al., 2011, 2012; Ruiz-Moyano et al., 2012).
Huang and Huang (2018) used a MALDI-TOF MS instrument (Microflex LT, Bruker Daltonics) to rapidly identify L. casei group strains at the species and subspecies-level by using ethanol/formic acid/acetonitrile for protein extraction in a routine protocol (Chambers et al., 2015) with MS peaks analyzed using ClinProTools software (Bruker Daltonics). Type strains of the L. casei group were used to construct main spectral profiles (the highest quality spectra) for compiling an in-house database given the optimized culture conditions [cultivated anaerobically at 37°C on de Man, Rogosa and Sharpe (MRS) agar for 20 h]. The reference strains of L. casei group were used to validate the in-house database. Forty-eight (100%) strains had high scores (mean: 2.45 ± 0.1) and were accurately identified at the species level, which was consistent with the results obtained using genotypic methods (housekeeping gene sequencing and species-specific PCR).
During a study whose objective was to isolate lactobacilli from different environmental samples, strain BCRC 81062T was isolated from cow manure collected in Chiayi, Taiwan. Strain BCRC 18859 (=NRIC 1947) was isolated from coconut juice from the Philippines, which was distributed by the NODAI.
Research Institute Culture Collection (NRIC; Japan) as L. paracasei subsp. tolerans. These two strains could not be clearly identified as any recognized species of the genus Lactobacillus by 16S rRNA gene sequencing analysis.
Huang et al. (2018) performed the MALDI-TOF MS analysis for classification of these two novel strains. A dendrogram derived from UPGMA cluster analysis of MALDI-TOF MS spectra in the 2000–9500 m/z (224 peaks) region of strains in the L. casei group were divided into five distinct clusters: Cluster A comprised two novel L. chiayiensis strains (BCRC 81062T and BCRC 18859) and the ‘L. zeae’ strain; Cluster B comprised two strains of L. casei; Cluster C comprised 10 L. rhamnosus strains; Cluster D comprised 12 L. paracasei subsp. paracasei strains; Cluster E comprised two L. paracasei subsp. tolerans strains. Cluster A was divided into two subclusters: ‘L. zeae’ (BCRC 17942T) was the single strain located in independent subcluster A-1. Two strains of L. casei were located in subcluster A-2 (Figure 2). This result was in good concordance, with the exception of the subspeciation of L. paracasei in the MLSA analysis based on housekeeping gene sequences. In addition, species- and subspecies-specific MS peaks were observed in the MS profiles, and these peaks served as the specific markers for differentiating L. casei group strains at both species and subspecies levels (Table 1).
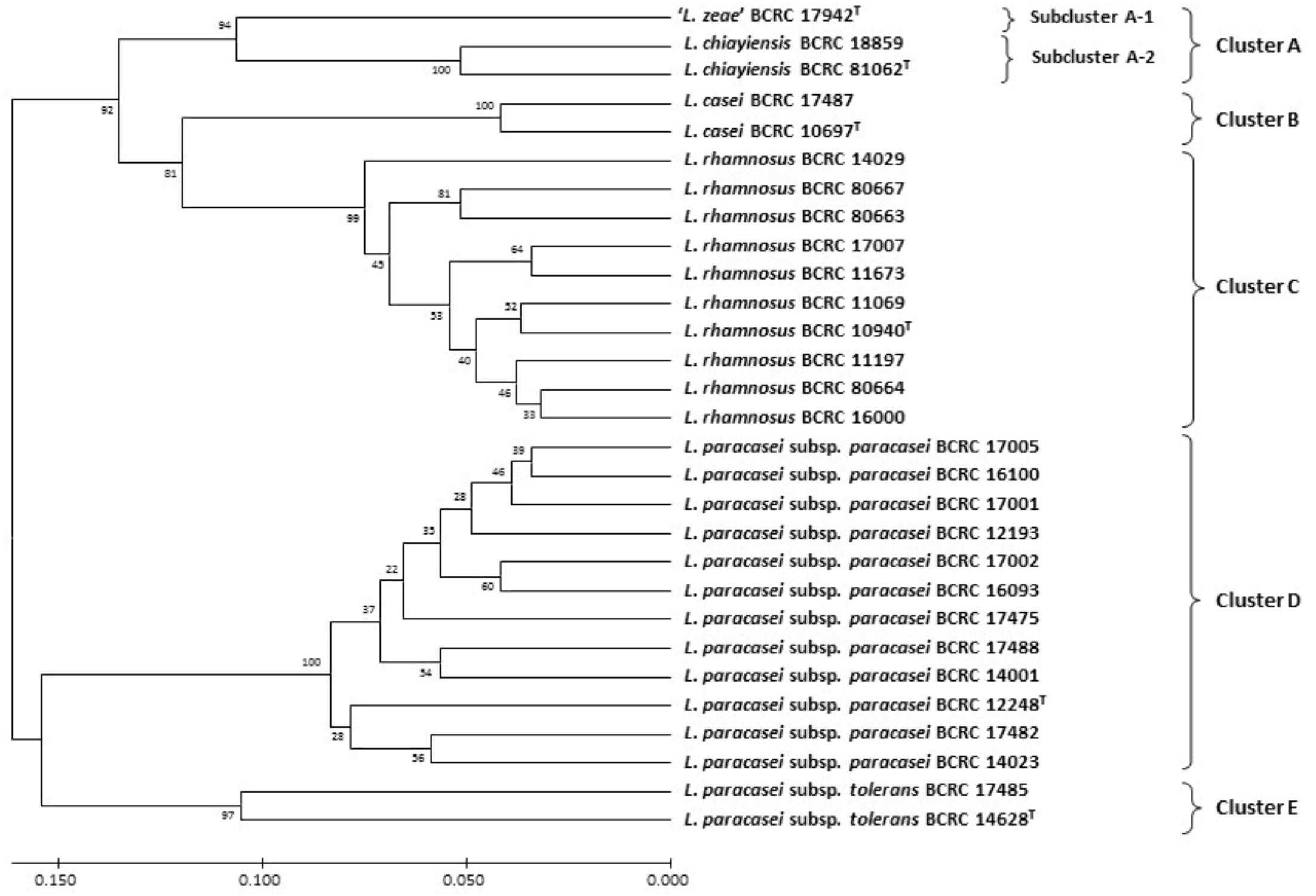
FIGURE 2. Dendrogram derived from UPGMA cluster analysis of MALDI-TOF MS spectra in the 2000–9500 m/z region (224 peaks) of strains in the L. casei group. Bootstrap values (%) based on 1000 replicates are given at the nodes. Bar, proportion of different peaks.
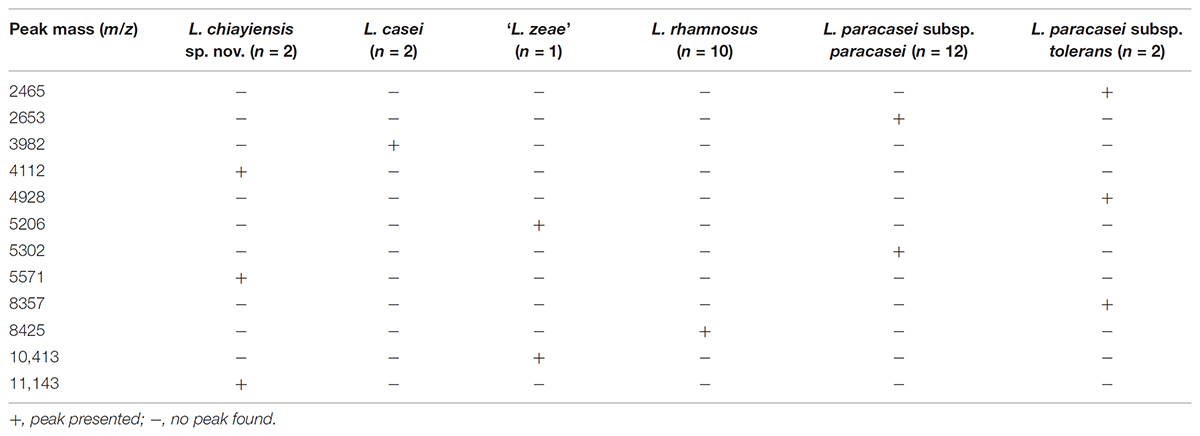
TABLE 1. Species- and subspecies-specific MALDI-TOF MS peaks used for differentiation of the Lactobacillus casei group.
Detecting and describing taxonomic novelty and reclassifying microbes has also been accomplished using MALDI-TOF MS (Kudo et al., 2012; Wieme et al., 2012; Li L. et al., 2014; Praet et al., 2015; Yanokura et al., 2015).
Phylogenetic Analysis of the L. casei Group Based on Whole-Genome Sequences
Because of the recent technological advancement in the field of WGS, dry-lab experiments using in silico methods based on genome-to-genome comparison can be used instead of the usual wet-lab DDH and conventional method for determining the DNA G+C content and sequencing of 16S rRNA and housekeeping genes of type strains (Richter and Rosselló-Móra, 2009; Chun and Rainey, 2014; Rosselló-Móra and Amann, 2015). WGS is now officially recognized as a source of taxonomic information (Whitman, 2016). Average nucleotide identity (ANI), digital DNA–DNA hybridization (dDDH) and tetranucleotide usage patterns (TETRA) are the most commonly used in the silico methods for species demarcation of phylogenetic relationships (Teeling et al., 2004; Goris et al., 2007; Meier-Kolthoff et al., 2013). ANI and dDDH values of 95–96% and 70%, respectively, are equivalent to DDH value of 70% and may be applied as boundaries in species delineation. In addition, dDDH values of 79–80% have been suggested for delineating subspecies within the domains Bacteria and Archaea on the basis of an investigation of genome-sequenced strains from more than 100 genera (Auch et al., 2010; Kim et al., 2014; Meier-Kolthoff et al., 2014; Richter et al., 2016). TETRA was used as an alignment-free parameter correlated with ANI because oligonucleotide frequencies are species-specific (Richter and Rosselló-Móra, 2009). Next-generation sequencing is responsible for an exponential rise in the quantity of available genome sequences of bacterial strains. However, the dubious quality of several genome sequences available in public databases (Ricker et al., 2012) is a substantial drawback. Chun et al. (2018) recommended minimal standards for utilizing genome data for taxonomic analysis of prokaryote and recommended quality requirements for genome sequences to address the aforementioned drawback.
Wuyts et al. (2017) identified three taxonomic clades (clade A: L. paracasei, clade B: L. casei, and clade C: L. rhamnosus) in the L. casei group based on a core genome phylogenetic tree, DNA G+C content analysis, and pairwise genome distances (ANIb and TETRA). These researchers also suggested that the heme-dependent catalase gene and the SOD-encoding gene play a role for molecular markers for the accurate identification of L. casei and L. paracasei.
Huang et al. (2018) analyzed the draft whole genome sequences of the novel two strains (BCRC 81062T and BCRC 18859). The ANI values were estimated using orthologous average nucleotide identity (OrthoANI) (Lee et al., 2016), and dDDH was performed using the genome to genome distance calculator (GGDC) with recommended Formula 2 (Meier-Kolthoff et al., 2013). The distance matrix generated using Gegenees software v.2.2.1 (Ågren et al., 2012) was plotted as a heat map, and the interspecies similarity between the novel strains and closely related type strains ranged from 7 to 44% (Figure 3A). A split network tree based on the fragmented all-against-all comparison of the whole genome sequences using Gegenees software to separate the novel strains and four type strains in the L. casei group could be separated into four clusters: Cluster A consisting of L. casei ATCC 393T (Accession No. GCA_000829055.1) and ‘L. zeae’ DSM 20178T (GCA_001433745.1); Cluster B consisting of two novel L. chiayiensis strains (BCRC 81062T: MSSM00000000; BCRC 18859: NOXN00000000); Cluster C consisting of L. rhamnosus JCM 1136T (GCA_000615245.1); Cluster D consisting of L. paracasei subsp. paracasei JCM 8130T (GCA_000829035.1) and L. paracasei subsp. tolerans DSM 20258T (NZ_AYYJ00000000) (Figure 3B). The ANI and dDDH values between L. chiayiensis sp. nov. BCRC 81062T and type strains in the L. casei group ranged from 77.4 to 88.5, and 22.3 to 36.1%, respectively, which were markedly lower than the commonly used cut-off threshold (i.e., 95–96% for ANI, and 70% for dDDH) for delineation of prokaryotic species. Phenotypic and chemotaxonomic characterization as well as MLSA based on three housekeeping genes (dnaK, pheS, and yycH), species-specific PCR and whole-cell MALDI-TOF MS spectral pattern analyses revealed that the novel strains (BCRC 81062T and BCRC 18859) represented a single, novel species within the L. casei group, for which the name L. chiayiensis sp. nov. is proposed.
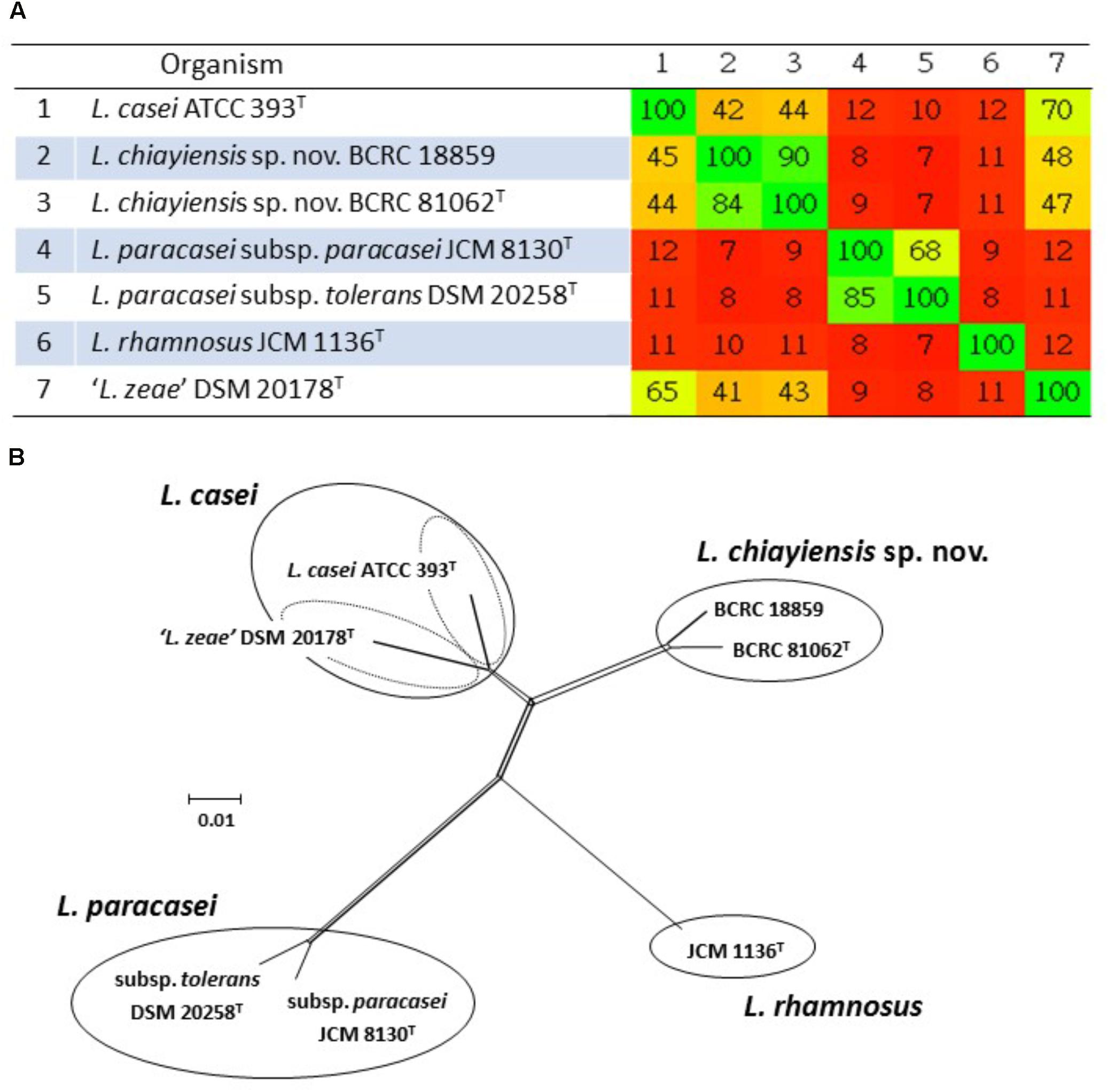
FIGURE 3. Phylogenomic analysis of the L. casei group. The values generated in the Gegenees software shown in the heat map indicate the percentage of similarity between the analyzed genomes; the colors varies from red (low similarity) to green (high similarity) (A). The split network tree was constructed using the SplitTree4 program based on the fragmented all-against-all comparison with the Gegenees software (B).
Venn diagram analysis based on genome sequences of six type strains in the L. casei group showed that 148 (4.9%) and 109 (3.6%) of the ORFs in ‘L. zeae’ DSM 20178T were orthologous to those of L. casei ATCC 393T and L. chiayiensis sp. nov. BCRC 81062T, respectively. This result indicated that ‘L. zeae’ DSM 20178T is more closely related to these two strains than other strains (Figure 4). However, the results of polyphasic characterization implied that the current L. casei group has at least four distinct species; L. casei, L. chiayiensis sp. nov., L. paracasei, and L. rhamnosus. L. paracasei comprises two subspecies: L. paracasei subsp. paracasei and L. paracasei subsp. tolerans (Figure 5). The species ‘L. zeae’ was described by Dicks et al. (1996) and was thereafter rejected by the Judicial Commission of ICSP (Tindall, 2008). However, the ANI and dDDH values between L. casei ATCC 393T and ‘L. zeae’ DSM 20178T, were 94.6 and 57.3%, respectively, which were lower than the species delineation threshold. This has a possibility that ‘L. zeae’ DSM 20178T could be reclassified as the independent species or subspecies.
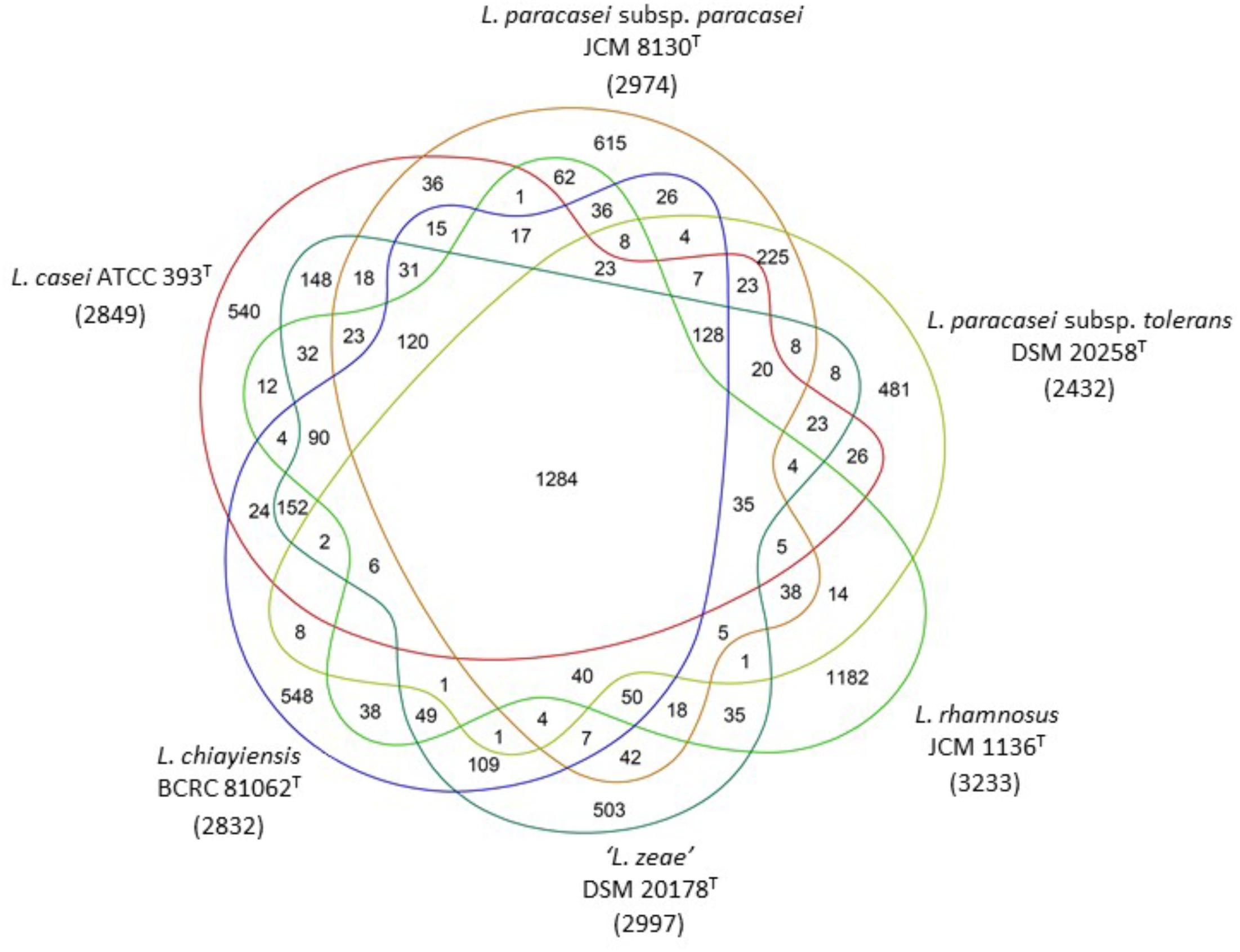
FIGURE 4. Ortholog analysis among six type strains in the L. casei group. Gene prediction and annotation were performed by RAST automated web server. Venn diagram showing the number of core, shared and unique genes for each genome. Numerals in parentheses under strain name are the numbers of the predicted ORFs.
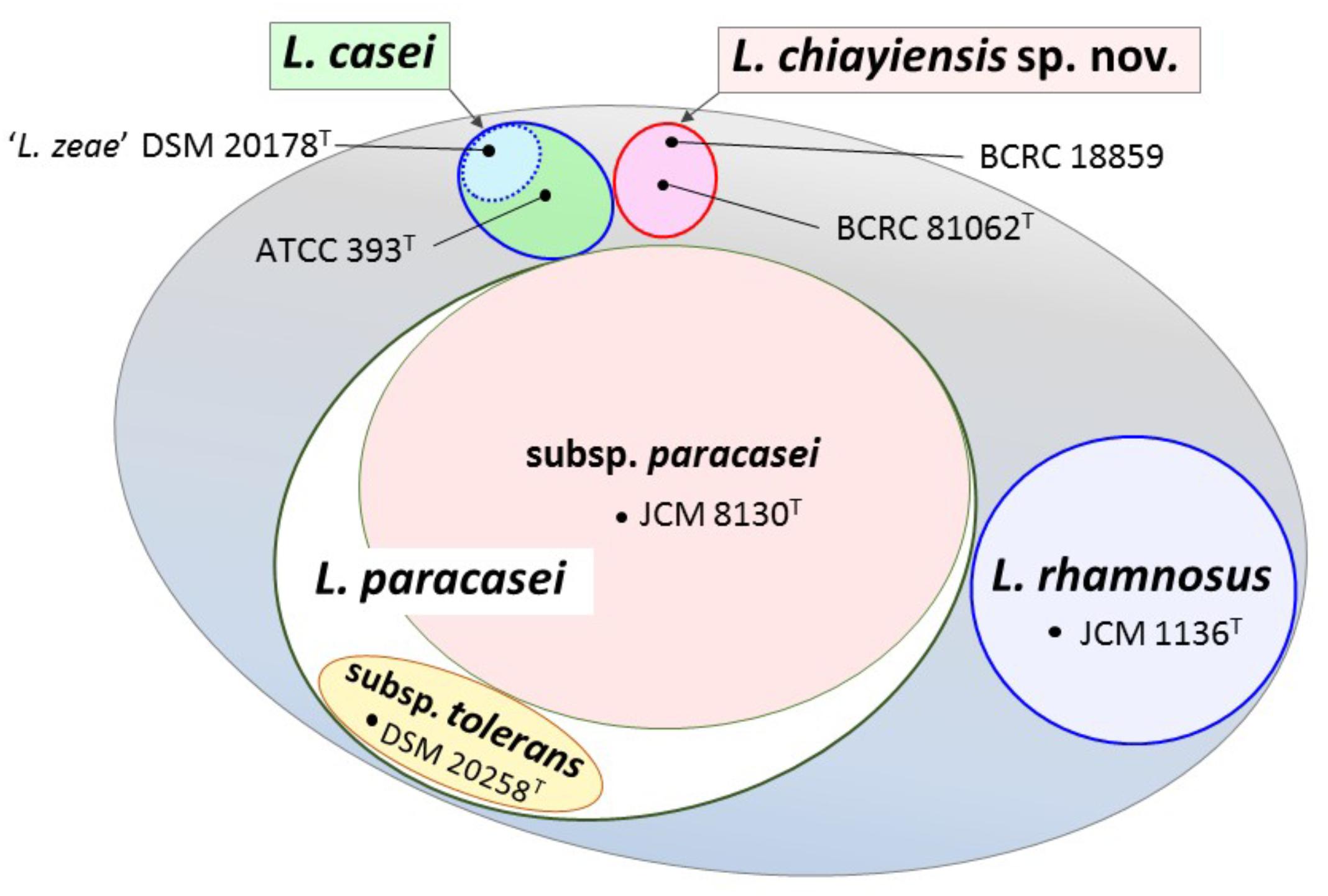
FIGURE 5. View showing the current state in classification of the L. casei group. The L. casei group has at least four distinct species; L. casei, L. chiayiensis sp. nov., L. paracasei, and L. rhamnosus. L. paracasei is comprised of two subspecies; L. paracasei subsp. paracasei and L. paracasei subsp. tolerans. ‘L. zeae’ has a possibility to be reclassified as the independent new taxon.
Conclusion and Future Perspectives
Identifying closely related taxonomic groups (e.g., the L. casei group species) involves two steps. First, 16S rRNA gene sequencing is used to ascertain the species-group level of an unknown strain. This is subsequently the foundation used to define the housekeeping genes (dnaJ, dnaK, mutL, pheS, and yycH) used to ascertain the species level of the strain by MLSA. Group- and species-specific primers in combination with conventional PCR and SNaPshot minisequencing analyses are used; these techniques are useful for direct identification and differentiation of strains in the L. casei group, in particular, the multiplex primer set based on housekeeping gene is the simplest and quickest method. Nevertheless, proteomic identification of species within the L. casei group, based on the in-house database and applying ClinProTools to MALDI-TOF MS data, exhibits superior discriminatory power (at the subspecies level) and requires shorter analysis duration than DNA sequencing and PCR-based methods. This method also facilitates efficient quality control of probiotic products. Due to technological advancement, DNA sequencing can be performed inexpensively, and high-quality, useful bioinformatic tools are available for classifying and identifying prokaryotes. Whole-genome-based comparisons, such as ANI and dDDH, have become the gold standards for demarcation of phylogenetic relationships as well as for delineating new species; these methods can replace traditional DDH, conventional determination of DNA G+C content, and 16S rRNA and housekeeping genes sequencing.
Author Contributions
C-HH analyzed the data and wrote the manuscript. S-WL analyzed the genome data. LH reviewed and revised the manuscript. KW designed this review, wrote the manuscript, and is a corresponding author. All authors read and approved the manuscript.
Funding
This study was supported by the Ministry of Science and Technology, Taiwan (Project No. MOST 106-2320-B-080-001) and Ministry of Economic Affairs, Taiwan (Project No. 107-EC-17-A-22-0525).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Chii-Cherng Liao and Dr. Gwo-Fang Yuan (Food Industry Research and Development Institute, Hsinchu, Taiwan) for their support during the course of this research activity.
Footnotes
References
Ågren, J., Sundström, A., Håfström, T., and Segerman, B. (2012). Gegenees: fragmented alignment of multiple genomes for determining phylogenomic distances and genetic signatures unique for specified target groups. PLoS One 7:e39107. doi: 10.1371/journal.pone.0039107
Ahrné, S., Molin, G., and StÅhl, S. (1989). Plasmids in Lactobacillus strains isolated from meat and meat products. Syst. Appl. Microbiol. 11, 320–325. doi: 10.1016/S0723-2020(89)80031-1
Anderson, A. C., Sanunu, M., Schneider, C., Clad, A., Karygianni, L., Hellwig, E., et al. (2014). Rapid species-level identification of vaginal and oral lactobacilli using MALDI-TOF MS analysis and 16S rDNA sequencing. BMC Microbiol. 14:312. doi: 10.1186/s12866-014-0312-5
Angelakis, E., Million, M., Henry, M., and Raoult, D. (2011). Rapid and accurate bacterial identification in probiotics and yoghurts by MALDI-TOF mass spectrometry. J. Food Sci. 76, M568–M572. doi: 10.1111/j.1750-3841.2011.02369.x
Ashraf, R., and Shah, N. P. (2014). Immune system stimulation by probiotic microorganisms. Crit. Rev. Food Sci. Nutr. 54, 938–956. doi: 10.1080/10408398.2011.619671
Auch, A. F., von Jan, M., Klenk, H.-P., and Göker, M. (2010). Digital DNA–DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genomic Sci. 2, 117–134. doi: 10.4056/sigs.531120
Bizzini, A., and Greub, G. (2010). Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin. Microbiol. Infect. 16, 1614–1619. doi: 10.1111/j.1469-0691.2010.03311.x
Bottari, B., Felis, G. E., Salvetti, E., Castioni, A., Campedelli, I., Torriani, S., et al. (2017). Effective identification of Lactobacillus casei group species: genome-based selection of the gene mutL as the target of a novel multiplex PCR assay. Microbiology 163, 950–960. doi: 10.1099/mic.0.000497
Boucher, Y., Douady, C. J., Sharma, A. K., Kamekura, M., and Doolittle, W. F. (2004). Intragenomic heterogeneity and intergenomic recombination among haloarchaeal rRNA genes. J. Bacteriol. 186, 3980–3990. doi: 10.1128/JB.186.12.3980-3990.2004
Boyd, M. A., Antonio, M. A. D., and Hillier, S. L. (2005). Comparison of API 50 CH strips to whole-chromosomal DNA probes for identification of Lactobacillus species. J. Clin. Microbiol. 43, 5309–5311. doi: 10.1128/JCM.43.10.5309-5311.2005
Branquinho, R., Sousa, C., Lopes, J., Pintado, M. E., Peixe, L. V., and Osório, H. (2014). Differentiation of Bacillus pumilus and Bacillus safensis using MALDI-TOF-MS. PLoS One 9:e110127. doi: 10.1371/journal.pone.0110127
Callaway, A., Kostrzewa, M., Willershausen, B., Schmidt, F., Thiede, B., Küpper, H., et al. (2013). Identification of lactobacilli from deep carious lesions by means of species-specific PCR and MALDI-TOF mass spectrometry. Clin. Lab. 59, 1372–1379. doi: 10.7754/Clin.Lab.2013.121225
Chambers, T., Culak, R., Gharbia, S. E., and Shah, H. (2015). Minor differences in the proteome of Bacillus subtilis and Bacillus mojavensis based upon high abundance/conserved protein mass spectra; implications for rapid, improved identification of two pathogen genetically closely related. J. Proteomics Enzymol. 4:1. doi: 10.4172/2470-1289.1000119
Chao, S.-H., Huang, H.-Y., Kang, Y.-H., Watanabe, K., and Tsai, Y.-C. (2013). The diversity of lactic acid bacteria in a traditional Taiwanese millet alcoholic beverage during fermentation. LWT Food Sci. Technol. 51, 135–142. doi: 10.1016/j.lwt.2012.09.015
Chao, S.-H., Kudo, Y., Tsai, Y.-C., and Watanabe, K. (2012). Lactobacillus futsaii sp. nov., isolated from traditional fermented mustard products of Taiwan, fu-tsai and suan-tsai. Int. J. Syst. Evol. Microbiol. 62, 489–494. doi: 10.1099/ijs.0.030619-0
Chao, S.-H., Sasamoto, M., Kudo, Y., Fujimoto, J., Tsai, Y.-C., and Watanabe, K. (2010). Lactobacillus odoratitofui sp. nov., isolated from stinky tofu brine. Int. J. Syst. Evol. Microbiol. 60, 2903–2907. doi: 10.1099/ijs.0.019307-0
Charteris, W. P., Kelly, P. M., Morelli, L., and Collins, J. K. (2001). Quality control Lactobacillus strains for use with the API 50CH and API ZYM systems at 37°C. J. Basic Microbiol. 41, 241–251. doi: 10.1002/1521-4028(200110)41:5<241::AID-JOBM241>3.0.CO;2-2
Chen, Y.-S., Wang, L.-T., Liao, Y.-J., Lan, Y.-S., Chang, C.-H., Chang, Y.-C., et al. (2017). Lactobacillus musae sp. nov., a novel lactic acid bacterium isolated from banana fruits. Int. J. Syst. Evol. Microbiol. 67, 5144–5149. doi: 10.1099/ijsem.0.002429
Chun, J., Oren, A., Ventosa, A., Christensen, H., Arahal, D. R., da Costa, M. S., et al. (2018). Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 68, 461–466. doi: 10.1099/ijsem.0.002516
Chun, J., and Rainey, F. A. (2014). Integrating genomics into the taxonomy and systematics of the Bacteria and Archaea. Int. J. Syst. Evol. Microbiol. 64, 316–324. doi: 10.1099/ijs.0.054171-0
Collins, M. D., Phillips, B. A., and Zanoni, P. (1989). Deoxyribonucleic acid homology studies of Lactobacillus casei, Lactobacillus paracasei sp. nov., subsp. paracasei and subsp. tolerans, and Lactobacillus rhamnosus sp. nov., comb. nov. Int. J. Syst. Bacteriol. 39, 105–108. doi: 10.1099/00207713-39-2-105
De Bruyne, K., Camu, N., De Vuyst, L., and Vandamme, P. (2010). Weissella fabaria sp. nov., from a Ghanaian cocoa fermentation. Int. J. Syst. Evol. Microbiol. 60, 1999–2005. doi: 10.1099/ijs.0.019323-0
De Bruyne, K., Franz, C. M., Vancanneyt, M., Schillinger, U., Mozzi, F., de Valdez, G. F., et al. (2008). Pediococcus argentinicus sp. nov. from Argentinean fermented wheat flour and identification of Pediococcus species by pheS, rpoA and atpA sequence analysis. Int. J. Syst. Evol. Microbiol. 58, 2909–2916. doi: 10.1099/ijs.0.65833-0
De Bruyne, K., Schillinger, U., Caroline, L., Boehringer, B., Cleenwerck, I., Vancanneyt, M., et al. (2007). Leuconostoc holzapfelii sp. nov., isolated from Ethiopian coffee fermentation and assessment of sequence analysis of housekeeping genes for delineation of Leuconostoc species. Int. J. Syst. Evol. Microbiol. 57, 2952–2959. doi: 10.1099/ijs.0.65292-0
Dec, M., Urban-Chmiel, R., Gnat, S., Puchalski, A., and Wernicki, A. (2014). Identification of Lactobacillus strains of goose origin using MALDI-TOF mass spectrometry and 16S–23S rDNA intergenic spacer PCR analysis. Res. Microbiol. 165, 190–201. doi: 10.1016/j.resmic.2014.02.003
Dellaglio, F., Dicks, L., Du Toit, M., and Torriani, S. (1991). Designation of ATCC 334 in place of ATCC 393 (NCDO 161) as the neotype strain of Lactobacillus casei subsp. casei and rejection of the name Lactobacillus paracasei (Collins et al., 1989). Int. J. Syst. Evol. Microbiol. 41, 340–342. doi: 10.1099/00207713-41-2-340
Desai, A. R., Shah, N. P., and Powell, I. B. (2006). Discrimination of dairy industry isolates of the Lactobacillus casei group. J. Dairy Sci. 89, 3345–3351. doi: 10.3168/jds.S0022-0302(06)72371-2
Dicks, L. M. T., Du Plessis, E. M., Dellaglio, F., and Lauer, E. (1996). Reclassification of Lactobacillus casei subsp. casei ATCC 393 and Lactobacillus rhamnosus ATCC 15820 as Lactobacillus zeae nom. rev., designation of ATCC 334 as the neotype of L. casei subsp. casei, and rejection of the name Lactobacillus paracasei. Int. J. Syst. Bacteriol. 46, 337–340. doi: 10.1099/00207713-46-1-337
Doan, N. T. L., Van Hoorde, K., Cnockaert, M., De Brandt, E., Aerts, M., Le Thanh, B., et al. (2012). Validation of MALDI-TOF MS for rapid classification and identification of lactic acid bacteria, with a focus on isolates from traditional fermented foods in Northern Vietnam. Lett. Appl. Microbiol. 55, 265–273. doi: 10.1111/j.1472-765X.2012.03287.x
Doi, H., Osawa, I., Adachi, H., and Kawada, M. (2017). Vibrio japonicus sp. nov., a novel member of the Nereis clade in the genus Vibrio isolated from the coast of Japan. PLoS One 12:e0172164. doi: 10.1371/journal.pone.0172164
Dušková, M., Šedo, O., Kšicová, K., Zdráhal, Z., and Karpíšková, R. (2012). Identification of lactobacilli isolated from food by genotypic methods and MALDI-TOF MS. Int. J. Food Microbiol. 159, 107–114. doi: 10.1016/j.ijfoodmicro.2012.07.029
Felis, G. E., Dellaglio, F., Mizzi, L., and Torriani, S. (2001). Comparative sequence analysis of a recA gene fragment brings new evidence for a change in the taxonomy of the Lactobacillus casei group. Int. J. Syst. Evol. Microbiol. 51, 2113–2117. doi: 10.1099/00207713-51-6-2113
Gevers, D., Cohan, F. M., Lawrence, J. G., Spratt, B. G., Coenye, T., Feil, E. J., et al. (2005). Opinion: re-evaluating prokaryotic species. Nat. Rev. Microbiol. 3, 733–739. doi: 10.1038/nrmicro1236
Ghyselinck, J., Van Hoorde, K., Hoste, B., Heylen, K., and De Vos, P. (2011). Evaluation of MALDI-TOF MS as a tool for high-throughput dereplication. J. Microbiol. Methods 86, 327–336. doi: 10.1016/j.mimet.2011.06.004
Giraffa, G., Chanishvili, N., and Widyastuti, Y. (2010). Importance of lactobacilli in food and feed biotechnology. Res. Microbiol. 161, 480–487. doi: 10.1016/j.resmic.2010.03.001
Glaeser, S. P., and Kämpfer, P. (2015). Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Syst. Appl. Microbiol. 38, 237–245. doi: 10.1016/j.syapm.2015.03.007
Gogarten, J. P., Doolittle, W. F., and Lawrence, J. G. (2002). Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 19, 2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046
Goris, J., Klappenbach, J. A., Vandamme, P., Coenye, T., Konstantinidis, K. T., and Tiedje, J. M. (2007). DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Hammes, W. P., and Hertel, C. (2009). “Genus I. Lactobacillus Beijerink, 1901,” in Bergey’s Manual of Systematic Bacteriology, eds P. De Vos, G. M. Garrity, D. Jones, N. R. Krieg, W. Ludwig, F. A. Rainey, et al. (Berlin: Springer), 465–510.
Hotta, Y., Sato, J., Sato, H., Hosoda, A., and Tamura, H. (2011). Classification of the genus Bacillus based on MALDI-TOF MS analysis of ribosomal proteins coded in S10 and spc operons. J. Agric. Food Chem. 59, 5222–5230. doi: 10.1021/jf2004095
Huang, C.-H., Chang, M.-T., and Huang, L. (2014). Use of highly variable gene (yycH) as DNA marker to resolve interspecific relationships within the Lactobacillus casei group and a target for developing novel species-specific PCR primers. Eur. Food Res. Technol. 239, 719–724. doi: 10.1007/s00217-014-2278-9
Huang, C.-H., Chang, M.-T., Huang, L., and Chu, W.-S. (2015). Simultaneous discrimination of species and strains in Lactobacillus rhamnosus using species-specific PCR combined with multiplex mini-sequencing technology. Mol. Cell. Probes 29, 531–533. doi: 10.1016/j.mcp.2015.05.017
Huang, C.-H., Chang, M.-T., Huang, M.-C., and Lee, F.-L. (2011). Application of the SNaPshot minisequencing assay to species identification in the Lactobacillus casei group. Mol. Cell. Probes 25, 153–157. doi: 10.1016/j.mcp.2011.03.002
Huang, C.-H., and Huang, L. (2018). Rapid species- and subspecies-specific level classification and identification of Lactobacillus casei group members using MALDI Biotyper combined with ClinProTools. J. Dairy Sci. 101, 979–991. doi: 10.3168/jds.2017-13642
Huang, C.-H., and Lee, F.-L. (2009). Development of novel species-specific primers for species identification of the Lactobacillus casei group based on RAPD fingerprints. J. Sci. Food Agric. 89, 1831–1837. doi: 10.1002/jsfa.3658
Huang, C.-H., and Lee, F.-L. (2011). The dnaK gene as a molecular marker for the classification and discrimination of the Lactobacillus casei group. Antonie Van Leeuwenhoek 99, 319–327. doi: 10.1007/s10482-010-9493-6
Huang, C.-H., Lee, F.-L., and Liou, J.-S. (2010). Rapid discrimination and classification of the Lactobacillus plantarum group based on a partial dnaK sequence and DNA fingerprinting techniques. Antonie Van Leeuwenhoek 97, 289–296. doi: 10.1007/s10482-009-9409-5
Huang, C.-H., Liou, J.-S., Lee, A.-Y., Tseng, M., Miyashita, M., Huang, L., et al. (2018). Polyphasic characterization of a novel species in the Lactobacillus casei group from cow manure of Taiwan: description of L. chiayiensis sp. nov. Syst. Appl. Microbiol. 41, 270–278. doi: 10.1016/j.syapm.2018.01.008
Huys, G., Vancanneyt, M., D’Haene, K., Vankerckhoven, V., Goossens, H., and Swings, J. (2006). Accuracy of species identity of commercial bacterial cultures intended for probiotic or nutritional use. Res. Microbiol. 157, 803–810. doi: 10.1016/j.resmic.2006.06.006
Killer, J., Pechar, R., Švec, P., Salmonová, H., Švejstil, R., Geigerová, M., et al. (2017). Lactobacillus caviae sp. nov., an obligately heterofermentative bacterium isolated from the oral cavity of a guinea pig (Cavia aperea f. porcellus). Int. J. Syst. Evol. Microbiol. 67, 2903–2909. doi: 10.1099/ijsem.0.002044
Killer, J., Votavova, A., Valterova, I., Vlkova, E., Rada, V., and Hroncova, Z. (2014). Lactobacillus bombi sp. nov., from the digestive tract of laboratory-reared bumblebee queens (Bombus terrestris). Int. J. Syst. Evol. Microbiol. 64, 2611–2617. doi: 10.1099/ijs.0.063602-0
Kim, E., Cho, Y., Lee, Y., Han, S.-K., Kim, C.-G., Choo, D.-W., et al. (2017). A proteomic approach for rapid identification of Weissella species isolated from Korean fermented foods on MALDI-TOF MS supplemented with an in-house database. Int. J. Food Microbiol. 243, 9–15. doi: 10.1016/j.ijfoodmicro.2016.11.027
Kim, M., Oh, H. S., Park, S. C., and Chun, J. (2014). Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 64, 346–351. doi: 10.1099/ijs.0.059774-0
Koort, J., Vandamme, P., Schillinger, U., Holzapfel, W., and Bjorkroth, J. (2004). Lactobacillus curvatus subsp. melibiosus is a later synonym of Lactobacillus sakei subsp. carnosus. Int. J. Syst. Evol. Microbiol. 54, 1621–1626. doi: 10.1099/ijs.0.63164-0
Kudo, Y., Oki, K., and Watanabe, K. (2012). Lactobacillus delbrueckii subsp. sunkii subsp. nov., isolated from sunki, a traditional Japanese pickle. Int. J. Syst. Evol. Microbiol. 62, 2643–2649. doi: 10.1099/ijs.0.037051-0
Kwon, H.-S., Yang, E.-H., Yeon, S.-W., Kang, B.-H., and Kim, T.-Y. (2004). Rapid identification of probiotic Lactobacillus species by multiplex PCR using species-specific primers based on the region extending from 16S rRNA through 23S rRNA. FEMS Microbiol. Lett. 239, 267–275. doi: 10.1016/j.femsle.2004.08.049
Lai, Q., Liu, Y., Yuan, J., Du, J., Wang, L., Sun, F., et al. (2014). Multilocus sequence analysis for assessment of phylogenetic diversity and biogeography in Thalassospira bacteria from diverse marine environments. PLoS One 9:e106353. doi: 10.1371/journal.pone.0106353
Lee, I., Kim, Y. O., Park, S.-C., and Chun, J. (2016). OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Li, C., Lai, Q., Li, G., Liu, Y., Sun, F., and Shao, Z. (2014). Multilocus sequence analysis for the assessment of phylogenetic diversity and biogeography in Hyphomonas bacteria from diverse marine environments. PLoS One 9:e101394. doi: 10.1371/journal.pone.0101394
Li, L., Wieme, A., Spitaels, F., Balzarini, T., Nunes, O. C., Manaia, C. M., et al. (2014). Acetobacter sicerae sp. nov., isolated from cider and kefir, and identification of species of the genus Acetobacter by dnaK, groEL and rpoB sequence analysis. Int. J. Syst. Evol. Microbiol. 64, 2407–2415. doi: 10.1099/ijs.0.058354-0
Liu, Y., Lai, Q., and Shao, Z. (2017). A multilocus sequence analysis scheme for phylogeny of Thioclava bacteria and proposal of two novel species. Front. Microbiol. 8:1321. doi: 10.3389/fmicb.2017.01321
Macheras, E., Roux, A. L., Bastian, S., Leao, S. C., Palaci, M., Sivadon-Tardy, V., et al. (2011). Multilocus sequence analysis and rpoB sequencing of Mycobacterium abscessus (Sensu Lato) strains. J. Clin. Microbiol. 49, 491–499. doi: 10.1128/JCM.01274-10
Mattarelli, P., Holzapfel, W., Franz, C. M., Endo, A., Felis, G. E., Hammes, W., et al. (2014). Recommended minimal standards for description of new taxa of the genera Bifidobacterium, Lactobacillus and related genera. Int. J. Syst. Evol. Microbiol. 64, 1434–1451. doi: 10.1099/ijs.0.060046-0
Mehlen, A., Goeldner, M., Ried, S., Stindl, S., Ludwig, W., and Schleifer, K.-H. (2004). Development of a fast DNA–DNA hybridization method based on melting profiles in microplates. Syst. Appl. Microbiol. 27, 689–695. doi: 10.1078/0723202042369875
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H.-P., and Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60
Meier-Kolthoff, J. P., Klenk, H.-P., and Göker, M. (2014). Taxonomic use of DNA G+C content and DNA–DNA hybridization in the genomic age. Int. J. Syst. Evol. Microbiol. 64, 352–356. doi: 10.1099/ijs.0.056994-0
Modesto, M., Michelini, S., Oki, K., Biavati, B., Watanabe, K., and Mattarelli, P. (2018). Bifidobacterium catulorum sp. nov. a novel taxa from the faeces of the baby common marmoset (Callithrix jacchus). Int. J. Syst. Evol. Microbiol. 68, 575–581. doi: 10.1099/ijsem.0.002545
Mulet, M., Lalucat, J., and Garcia-Valdes, E. (2010). DNA sequence-based analysis of the Pseudomonas species. Environ. Microbiol. 12, 1513–1530.
Nacef, M., Chevalier, M., Chollet, S., Drider, D., and Flahaut, C. (2017). MALDI-TOF mass spectrometry for the identification of lactic acid bacteria isolated from a French cheese: the Maroilles. Int. J. Food Microbiol. 247, 2–8. doi: 10.1016/j.ijfoodmicro.2016.07.005
Naser, S. M., Dawyndt, P., Hoste, B., Gevers, D., Vandemeulebroecke, K., Cleenwerck, I., et al. (2007). Identification of lactobacilli by pheS and rpoA gene sequence analyses. Int. J. Syst. Evol. Microbiol. 57, 2777–2789. doi: 10.1099/ijs.0.64711-0
Naser, S. M., Thompson, F. L., Hoste, B., Gevers, D., Dawyndt, P., Vancanneyt, M., et al. (2005). Application of multilocus sequence analysis (MLSA) for rapid identification of Enterococcus species based on rpoA and pheS genes. Microbiology 151, 2141–2150. doi: 10.1099/mic.0.27840-0
Neville, S. A., LeCordier, A., Ziochos, H., Chater, M. J., Gosbell, I. B., Maley, M. W., et al. (2011). Utility of matrix-assisted laser desorption ionization-time of flight mass spectrometry following introduction for routine laboratory bacterial identification. J. Clin. Microbiol. 49, 2980–2984. doi: 10.1128/JCM.00431-11
Nguyen, D. T. L., Van Hoorde, K., Cnockaert, M., De Brandt, E., De Bruyne, K., Le, B. T., et al. (2013). A culture-dependent and -independent approach for the identification of lactic acid bacteria associated with the production of nem chua, a Vietnamese fermented meat product. Food Res. Int. 50, 232–240. doi: 10.1016/j.foodres.2012.09.029
Nyanzi, R., Jooste, P. J., Cameron, M., and Witthuhn, C. (2013). Comparison of rpoA and pheS gene sequencing to 16S rRNA gene sequencing in identification and phylogenetic analysis of LAB from probiotic food products and supplements. Food Biotechnol. 27, 303–327. doi: 10.1080/08905436.2013.838783
Oki, K., Kudo, Y., and Watanabe, K. (2012). Lactobacillus saniviri sp. nov. and Lactobacillus senioris sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 62, 601–607. doi: 10.1099/ijs.0.031658-0
Orlando, A., Linsalata, M., and Russo, F. (2016). Antiproliferative effects on colon adenocarcinoma cells induced by co-administration of vitamin K1 and Lactobacillus rhamnosus GG. Int. J. Oncol. 48, 2629–2638. doi: 10.3892/ijo.2016.3463
Pavlovic, M., Huber, I., Konrad, R., and Busch, U. (2013). Application of MALDI-TOF MS for the identification of food borne bacteria. Open Microbiol. J. 7, 135–141. doi: 10.2174/1874285801307010135
Peeters, C., Meier-Kolthoff, J. P., Verheyde, B., De Brandt, E., Cooper, V. S., and Vandamme, P. (2016). Phylogenomic study of Burkholderia glathei-like organisms, proposal of 13 novel Burkholderia species and emended descriptions of Burkholderia sordidicola, Burkholderia zhejiangensis, and Burkholderia grimmiae. Front. Microbiol. 7:877. doi: 10.3389/fmicb.2016.00877
Petti, C. A. (2007). Detection and identification of microorganisms by gene amplification and sequencing. Clin. Infect. Dis. 44, 1108–1114. doi: 10.1086/512818
Pontes, D. S., Lima-Bittencourt, C. I., Chartone-Souza, E., and Nascimento, A. M. A. (2007). Molecular approaches: advantages and artifacts in assessing bacterial diversity. J. Ind. Microbiol. Biotechnol. 34, 463–473. doi: 10.1007/s10295-007-0219-3
Pot, B., Felis, G. E., Bruyne, K. D., Tsakalidou, E., Papadimitriou, K., Leisner, J., et al. (2014). “The genus Lactobacillus,” in Lactic Acid Bacteria: Biodiversity and Taxonomy, eds W. H. Holzapfel and B. J. B. Wood (West Sussex: John Wiley & Sons, Ltd), 249–353. doi: 10.1002/9781118655252.ch19
Praet, J., Meeus, I., Cnockaert, M., Houf, K., Smagghe, G., and Vandamme, P. (2015). Novel lactic acid bacteria isolated from the bumble bee gut: Convivina intestini gen. nov., sp. nov., Lactobacillus bombicola sp. nov., and Weissella bombi sp. nov. Antonie Van Leeuwenhoek 107, 1337–1349. doi: 10.1007/s10482-015-0429-z
Reid, G. (2015). The growth potential for dairy probiotics. Int. Dairy J. 49, 16–22. doi: 10.1016/j.idairyj.2015.04.004
Ribeiro, T. G., Gonçalves, B. R., da Silva, M. S., Novais,Â, Machado, E., Carriço, J. A., et al. (2017). Citrobacter portucalensis sp. nov., isolated from an aquatic sample. Int. J. Syst. Evol. Microbiol. 67, 3513–3517. doi: 10.1099/ijsem.0.002154
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106,19126–19131. doi: 10.1073/pnas.0906412106
Richter, M., Rosselló-Móra, R., Glöckner, F. O., and Peplies, J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931. doi: 10.1093/bioinformatics/btv681
Ricker, N., Qian, H., and Fulthorpe, R. R. (2012). The limitations of draft assemblies for understanding prokaryotic adaptation and evolution. Genomics 100,167–175. doi: 10.1016/j.ygeno.2012.06.009
Rosselló-Móra, R., and Amann, R. (2015). Past and future species definitions for Bacteria and Archaea. Syst. Appl. Microbiol. 38, 209–216. doi: 10.1016/j.syapm.2015.02.001
Ruiz-Moyano, S., Tao, N., Underwood, M. A., and Mills, D. A. (2012). Rapid discrimination of Bifidobacterium animalis subspecies by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Food Microbiol. 30, 432–437. doi: 10.1016/j.fm.2011.12.012
Salvetti, E., Torriani, S., and Felis, G. E. (2012). The genus Lactobacillus: a taxonomic update. Probiotics Antimicrob. Proteins 4, 217–226. doi: 10.1007/s12602-012-9117-8
Samb-Ba, B., Mazenot, C., Gassama-Sow, A., Dubourg, G., Richet, H., Hugon, P., et al. (2014). MALDI-TOF identification of the human gut microbiome in people with and without diarrhea in Senegal. PLoS One 9:e87419. doi: 10.1371/journal.pone.0087419
Sandrin, T. R., Goldstein, J. E., and Schumaker, S. (2013). MALDI TOF MS profiling of bacteria at the strain level: a review. Mass Spectrom. Rev. 32, 188–217. doi: 10.1002/mas.21359
Santos, I. C., Hildenbrand, Z. L., and Schug, K. A. (2016). Applications of MALDI-TOF MS in environmental microbiology. Analyst 141, 2827–2837. doi: 10.1039/C6AN00131A
Sardaro, M. L. S., Levante, A., Bernini, V., Gatti, M., Neviani, E., and Lazzi, C. (2016). The spxB gene as a target to identify Lactobacillus casei group species in cheese. Food Microbiol. 59, 57–65. doi: 10.1016/j.fm.2016.05.004
Sato, H., Teramoto, K., Ishii, Y., Watanabe, K., and Benno, Y. (2011). Ribosomal protein profiling by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for phylogenety-based subspecies resolution of Bifidobacterium longum. Syst. Appl. Microbiol. 34, 76–80. doi: 10.1016/j.syapm.2010.07.003
Sato, H., Torimura, M., Kitahara, M., Ohkuma, M., Hotta, Y., and Tamura, H. (2012). Characterization of the Lactobacillus casei group based on the profiling of ribosomal proteins coded in S10-spc-alpha operons as observed by MALDI-TOF MS. Syst. Appl. Microbiol. 35, 447–454. doi: 10.1016/j.syapm.2012.08.008
Šedo, O., Vávrová, A., Vad’urová, M., Tvrzová, L., and Zdráhal, Z. (2013). The influence of growth conditions on strain differentiation within the Lactobacillus acidophilus group using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry profiling. Rapid Commun. Mass Spectrom. 27,2729–2736. doi: 10.1002/rcm.6741
Seegers, J. F. (2002). Lactobacilli as live vaccine delivery vectors: progress and prospects. Trends Biotechnol. 20, 508–515. doi: 10.1016/S0167-7799(02)02075-9
Shida, K., and Nomoto, K. (2013). Probiotics as efficient immunopotentiators: translational role in cancer prevention. Indian J. Med. Res. 138, 808–814.
Skerman, V. B. D., McGowan, V., and Sneath, P. H. A. (1980). Approved lists of bacterial names. Int. J. Syst. Bacteriol. 30, 225–420. doi: 10.1099/00207713-30-1-225
Song, Y.-L., Kato, N., Liu, C.-X., Matsumiya, Y., Kato, H., and Watanabe, K. (2000). Rapid identification of 11 human intestinal Lactobacillus species by multiplex PCR assays using group- and species-specific primers derived from the 16S–23S rRNA intergenic spacer region and its flanking 23S rRNA. FEMS Microbiol. Lett. 187, 167–173.
Spinali, S., van Belkum, A., Goering, R. V., Girard, V., Welker, M., Van Nuenen, M., et al. (2015). Microbial typing by matrix-assisted laser desorption ionization–time of flight mass spectrometry: Do we need guidance for data interpretation? J. Clin. Microbiol. 53, 760–765. doi: 10.1128/JCM.01635-14
Stackebrandt, E., and Ebers, J. (2006). Taxonomic parameters revisited: tarnished gold standards. Microbiol. Today 6, 152–155.
Stackebrandt, E., Garrity, G. M., Trüper, H. G., Whitman, W. B., Grimont, P. A. D., Nesme, X., et al. (2002). Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 52, 1043–1047.
Stackebrandt, E., and Goebel, B. M. (1994). Taxonomic note: a place for DNA–DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 44, 846–849. doi: 10.1099/00207713-44-4-846
Tamang, J. P., Watanabe, K., and Holzapfel, W. H. (2015). Review: diversity of microorganisms in global fermented foods and beverages. Front. Microbiol. 7:377. doi: 10.3389/fmicb.2016.00377
Tanigawa, K., Kawabata, H., and Watanabe, K. (2010). Identification and typing of Lactococcus lactis by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 76, 4055–4062. doi: 10.1128/AEM.02698-09
Teeling, H., Waldmann, J., Lombardot, T., Bauer, M., and Glöckner, F. (2004). TETRA: a web-service and a stand-alone program for the analysis and comparison of tetranucleotide usage patterns in DNA sequences. BMC Bioinformatics 5:163. doi: 10.1186/1471-2105-5-163
Timilsina, S., Jibrin, M. O., Potnis, N., Minsavage, G. V., Kebede, M., Schwartz, A., et al. (2015). Multilocus sequence analysis of xanthomonads causing bacterial spot of tomato and pepper plants reveals strains generated by recombination among species and recent global spread of Xanthomonas gardneri. Appl. Environ. Microbiol. 81, 1520–1529. doi: 10.1128/AEM.03000-14
Tindall, B. J. (2008). The type strain of Lactobacillus casei is ATCC 393, ATCC 334 cannot serve as the type because it represents a different taxon, the name Lactobacillus paracasei and its subspecies names are not rejected and the revival of the name ‘Lactobacillus zeae’ contravenes Rules 51b (1) and (2) of the International Code of Nomenclature of Bacteria. Opinion 82. Int. J. Syst. Evol. Microbiol. 58, 1764–1765. doi: 10.1099/ijs.0.2008/005330-0
Torriani, S., Clementi, F., Vancanneyt, M., Hoste, B., Dellaglio, F., and Kersters, K. (2001). Differentiation of Lactobacillus plantarum, L. pentosus and L. paraplantarum species by RAPD-PCR and AFLP. Syst. Appl. Microbiol. 24, 554–560. doi: 10.1078/0723-2020-00071
Tran, A., Alby, K., Kerr, A., Jones, M., and Gilligan, P. H. (2015). Cost savings realized by implementation of routine microbiological identification by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 53, 2473–2479. doi: 10.1128/JCM.00833-15
Van de Peer, Y., Chapelle, S., and De Wachter, R. (1996). A quantitative map of nucleotide substitution rates in bacterial rRNA. Nucleic Acids Res. 24,3381–3391. doi: 10.1093/nar/24.17.3381
Vandamme, P., and Peeters, C. (2014). Time to revisit polyphasic taxonomy. Antonie Van Leeuwenhoek 106, 57–65. doi: 10.1007/s10482-014-0148-x
Vásquez, A., Ahrne, S., Pettersson, B., and Molin, G. (2001). Temporal temperature gradient gel electrophoresis (TTGE) as a tool for identification of Lactobacillus casei, Lactobacillus paracasei, Lactobacillus zeae and Lactobacillus rhamnosus. Lett. Appl. Microbiol. 32, 215–219. doi: 10.1046/j.1472-765X.2001.00901.x
Ventura, M., Canchaya, C., Meylan, V., Klaenhammer, T. R., and Zink, R. (2003). Analysis, characterization, and loci of the tuf genes in Lactobacillus and Bifidobacterium species and their direct application for species identification. Appl. Environ. Microbiol. 69, 6908–6922. doi: 10.1128/AEM.69.11.6908-6922.2003
Ward, L. J., and Timmins, M. J. (1999). Differentiation of Lactobacillus casei, Lactobacillus paracasei and Lactobacillus rhamnosus by polymerase chain reaction. Lett. Appl. Microbiol. 29, 90–92. doi: 10.1046/j.1365-2672.1999.00586.x
Watanabe, K., Fujimoto, J., Sasamoto, M., Dugersuren, J., Tumursuh, T., and Demberel, S. (2008). Diversity of lactic acid bacteria and yeasts in Airag and Tarag, traditional fermented milk products of Mongolia. World J. Microbiol. Biotechnol. 24, 1313–1325. doi: 10.1007/s11274-007-9604-3
Watanabe, K., Fujimoto, J., Tomii, Y., Sasamoto, M., Makino, H., Kudo, Y., et al. (2009). Lactobacillus kisonensis sp. nov., Lactobacillus otakiensis sp. nov. Lactobacillus rapi sp. nov. and Lactobacillus sunkii sp. nov., four novel heterofermentative species isolated from sunki, a Japanese traditional pickle. Int. J. Syst. Evol. Microbiol. 59, 754–760. doi: 10.1099/ijs.0.004689-0
Wayne, L. G., Moore, W. E. C., Stackebrandt, E., Kandler, O., Colwell, R. R., Krichevsky, M. I., et al. (1987). Report of the Ad Hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Evol. Microbiol. 37, 463–464. doi: 10.1099/00207713-37-4-463
Welker, M., and Moore, E. R. B. (2011). Applications of whole-cell matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry in systematic microbiology. Syst. Appl. Microbiol. 34, 2–11. doi: 10.1016/j.syapm.2010.11.013
Whitman, W. B. (2016). Modest proposals to expand the type material for naming of prokaryotes. Int. J. Syst. Evol. Microbiol. 66, 2108–2112. doi: 10.1099/ijsem.0.000980
Wieme, A., Cleenwerck, I., Van Landschoot, A., and Vandamme, P. (2012). Pediococcus lolii DSM 19927T and JCM 15055T are strains of Pediococcus acidilactici. Int. J. Syst. Evol. Microbiol. 62, 3105–3108. doi: 10.1099/ijs.0.046201-0
Wieme, A. D., Spitaels, F., Aerts, M., De Bruyne, K., Van Landschoot, A., and Vandamme, P. (2014). Identification of beer-spoilage bacteria using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Int. J. Food Microbiol. 185, 41–50. doi: 10.1016/j.ijfoodmicro.2014.05.003
Wuyts, S., Wittouck, S., De Boeck, I., Allonsius, C. N., Pasolli, E., Segata, N., et al. (2017). Large-scale phylogenomics of the Lactobacillus casei group highlights taxonomic inconsistencies and reveals novel clade-associated features. mSystems 2, e00061-e17. doi: 10.1128/mSystems.00061-17
Yanokura, E., Oki, K., Makino, H., Modesto, M., Pot, B., Mattarelli, P., et al. (2015). Subspeciation of Bifidobacterium longum by multilocus approaches and amplified fragment length polymorphism: description of B. longum subsp. suillum subsp. nov., isolated from the faeces of piglets. System. Appl. Microbiol. 38, 305–314. doi: 10.1016/j.syapm.2015.05.001
Yu, J., Sun, Z., Liu, W., Bao, Q., Zhang, J., and Zhang, H. (2014). Phylogenetic study of Lactobacillus acidophilus group, L. casei group and L. plantarum group based on partial hsp60, pheS and tuf gene sequences. Eur. Food Res. Technol. 234, 927–934. doi: 10.1007/s00217-012-1712-0
Keywords: Lactobacillus casei group, MALDI-TOF MS, MLSA, species-specific PCR, whole genome sequence
Citation: Huang C-H, Li S-W, Huang L and Watanabe K (2018) Identification and Classification for the Lactobacillus casei Group. Front. Microbiol. 9:1974. doi: 10.3389/fmicb.2018.01974
Received: 20 June 2018; Accepted: 06 August 2018;
Published: 22 August 2018.
Edited by:
Teresa Zotta, Consiglio Nazionale delle Ricerche (CNR), ItalyReviewed by:
Marta Ewa Dec, University of Life Sciences in Lublin, PolandAnnette Carola Anderson, Universitätsklinikum Freiburg, Germany
Copyright © 2018 Huang, Li, Huang and Watanabe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Koichi Watanabe, a29pY2hpX3d0bmJAeWFob28uY28uanA=
 Chien-Hsun Huang
Chien-Hsun Huang Shiao-Wen Li
Shiao-Wen Li Lina Huang1
Lina Huang1 Koichi Watanabe
Koichi Watanabe