- Institute of Animal Husbandry and Veterinary Medicine, Beijing Academy of Agriculture and Forestry Sciences, Beijing, China
Weaning disturbs the intestinal barrier function and increases the risk of infection in piglets. Probiotics exert beneficial health effects, mainly by reinforcing the intestinal epithelium and modulating the gut microbiota. However, the mechanisms of action, and especially, the specific regulatory effects of modulated microbiota by probiotics on the intestinal epithelium have not yet been elucidated. The present study aimed to decipher the protective effects of the probiotic Lactobacillus plantarum strain ZLP001 on the intestinal epithelium and microbiota as well as the effects of modulated microbiota on epithelial function. Paracellular permeability was measured with fluorescein isothiocyanate-dextran (FD-4). Gene and protein expression levels of tight junction (TJ) proteins, proinflammatory cytokines, and host defense peptides were determined by RT-qPCR, ELISA, and western blot analysis. Short-chain fatty acid (SCFA) concentrations were measured by ion chromatography. Fecal microbiota composition was assessed by high-throughput sequencing. The results showed that pretreatment with 108 colony forming units (CFU) mL−1 of L. plantarum ZLP001 significantly counteracted the increase in gut permeability to FD-4 induced by 106 CFU mL−1 enterotoxigenic Escherichia coli (ETEC). In addition, L. plantarum ZLP001 pretreatment alleviated the reduction in TJ proteins (claudin-1, occludin, and ZO-1) and downregulated proinflammatory cytokines IL-6 and IL-8, and TNFα expression and secretion caused by ETEC. L. plantarum ZLP001 also significantly increased the expression of the host defense peptides pBD2 and PG1-5 and pBD2 secretion relative to the control. Furthermore, L. plantarum ZLP001 treatment affected piglet fecal microbiota. The abundance of butyrate-producing bacteria Anaerotruncus and Faecalibacterium was significantly increased in L. plantarum ZLP001-treated piglets, and showed a positive correlation with fecal butyric and acetic acid concentrations. In addition, the cell density of Clostridium sensu stricto 1, which may cause epithelial inflammation, was decreased after L. plantarum ZLP001 administration, while the beneficial Lactobacillus was significantly increased. Our findings suggest that L. plantarum ZLP001 fortifies the intestinal barrier by strengthening epithelial defense functions and modulating gut microbiota.
Introduction
The sudden changes in diet and the physical and social environment associated with weaning are significant piglet stressors. Elevated plasma cortisol and corticotropin-releasing factor levels are indices of weaning stress (Van der Meulen et al., 2010). The feed intake of most piglets after weaning is relatively low because of the dietary change from liquid milk to solid feed. Decreased feed and water intake cause small intestinal villous atrophy (Lallès et al., 2004), which in turn results in diminished digestive and absorptive capacities and reduced growth rate. Maternal separation and changes in environment are social and environmental stresses that cause tension in piglets and weaken their immune system. Furthermore, the dietary and environmental changes associated with weaning are associated with a substantial modification of the intestinal microbiota and may cause post-weaning diarrhea and enteric infection (Lallès et al., 2007). Perturbations of the intestinal epithelium, weakened immune system, and modified intestinal microbiota induced by weaning stress can profoundly impact piglet health and growth performance and may, in some cases, lead to mortality (Campbell et al., 2013).
The intestine plays a critical role in the defense against harmful external factors. Poor intestinal defense renders piglets more susceptible to weaning stress, leading to infection and disease. In post-weaning piglets, transepithelial electrical resistance (TER) significantly decreases while paracellular permeability increases (Hu et al., 2013). The expression of the tight junction (TJ) proteins occludin, claudin-1, and ZO-1 decreases during weaning, and as a result, barrier integrity is impaired. This facilitates pathogen penetration and permits bacteriotoxins to enter the body. Weaned piglets also exhibit elevated expression of the proinflammatory cytokines TNF-α and IL-6 (Hu et al., 2013), which is associated with weak epithelium and inflammatory disease. Furthermore, weaning lowers intestinal microbial diversity and alters the microbiota composition in that the abundance of obligate anaerobic bacteria decreases and that of facultative anaerobic bacteria increases (Winter et al., 2013), which weakens intestinal function. For instance, Lactobacillus spp. and other beneficial bacteria play an important role in protecting against intestinal pathogens, and the reduction in their abundance after weaning enhances disease risk (Konstantinov et al., 2006). S. enterica and E. coli are two major pathogens infecting piglets. An increased abundance of these pathogens in the intestine often results in severe infection.
Evidence indicates that the consumption of probiotic bacteria contributes to intestinal function by maintaining paracellular permeability, enhancing the physical mucous layer, stimulating the immune system, and modulating resident microbiota composition and activity (Boirivant and Strober, 2007). The regulatory effects of probiotics on human, pig, and chicken intestinal homeostasis have been studied extensively (Van Baarlen et al., 2013; Cisek and Binek, 2014; Gresse et al., 2017), and interactions between probiotic and commensal bacteria and the epithelial barrier are thought to be the main underlying mechanism. Lactobacillus is a predominant indigenous bacterial genus found in the human and animal gastrointestinal tract, and species of this genus are commonly used as probiotics. In vitro and in vivo studies in various cell lines and animal models have demonstrated that L. plantarum MB452, L. casei, L. rhamnosus GG, and L. reuteri I5007 affect TER and epithelial permeability and modulate TJ protein expression and distribution (Anderson et al., 2010; Eun et al., 2011; Patel et al., 2012; Yang et al., 2015). Lactobacillus spp. boost the immune system by promoting the expression of anti-inflammatory cytokines, such as IL-10 and IFN-γ (Lactobacillus GG, Kopp et al., 2008; L. rhamnosus CRL1505, Villena et al., 2014), or by inhibiting that of pro-inflammatory cytokines, such as IL-6, IL-8, and TNF-α (L. reuteri LR1, Wang et al., 2016; L. plantarum 2142, Farkas et al., 2014). L. reuteri I5007 and L. plantarum DSMZ 12028 can modulate the synthesis of antimicrobials by the intestinal epithelium (Paolillo et al., 2009; Liu et al., 2017). Multiple studies have confirmed that Lactobacillus spp., such as L. salivarius UCC118 and L. acidophilus, significantly modulate resident intestinal microbiota composition and activity (Riboulet-Bisson et al., 2012; Li et al., 2017). Thus, through enhancing intestinal epithelial function, probiotics improve host health. However, the effects of probiotics on intestinal barrier function are strain-dependent and not ubiquitous. The unique effects of specific strains on the intestinal epithelium and microbiota, and whether the modulated microbiota affect intestinal epithelial function remain unclear.
The results of our previous studies indicated that dietary supplementation with Lactobacillus plantarum ZLP001 isolated from healthy piglet intestinal tract (Wang et al., 2011) improves growth performance and antioxidant status in post-weaning piglets (Wang et al., 2012). However, its impact on intestinal barrier function and microbiota, and the interaction between barrier function and microbiota after L. plantarum ZLP001 treatment remained to be investigated. In this study, the impact of L. plantarum ZLP001 on intestinal epithelial function was evaluated by measuring gut permeability and the expression of TJ proteins, inflammatory cytokines, and host defense peptides (HDP). Further, we evaluated the ability of this strain to regulate microbiota composition and community structure. The regulatory effects of the microbiota modulated by the probiotic strain on intestinal epithelium function were also analyzed.
Materials and Methods
Bacteria and Culture Conditions
L. plantarum ZLP001 was isolated in our laboratory from the intestine of a healthy piglet. It was identified by the China Center of Industrial Culture Collection (Beijing, China) and preserved in the China General Microbiological Culture Collection Center (CGMCC No. 7370). It was grown in improved De Man, Rogosa, and Sharpe liquid medium (10 g peptone, 5 g yeast powder, 20 g glucose, 10 g beef extract, 5 g sodium acetate, 2 g ammonium citrate dibasic, 2 g dipotassium phosphate, 0.58 g magnesium sulfate, 0.19 g manganese sulfate, 1 mL of Tween 80, and water to 1,000 mL; pH 6.5) at 37°C under anaerobic conditions.
Enteropathic E. coli strain expressing F4 (F4+ ETEC), serotype O149:K91, K88ac, was obtained from the China Veterinary Culture Collection Center. It was grown in Luria-Bertani medium (Oxoid, Basingstoke, United Kingdom) at 37°C.
Cell Line and Culture Conditions
The porcine intestinal epithelial cell line (IPEC-J2) used in this study was purchased from JENNIO Biological Technology (Guangzhou, China). It was originally derived from the jejuna of neonatal piglets. The cells were cultured in DMEM/F12 (Dulbecco’s modified Eagle’s medium/nutrient mixture F-12, a 1:1 mixture of DMEM and Ham’s F-12; Invitrogen, Carlsbad, CA, United States) supplemented with 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, United States), streptomycin (100 μg mL−1), and amphotericin B (0.5 μg mL−1). IPEC-J2 cells were cultured at 37°C in a 5% CO2/95% air atmosphere and 90% relative humidity. Cells were separated at each passage with 0.25% w/v trypsin (Invitrogen, Carlsbad, CA, United States) and replenished with fresh media every 2–3 days.
Paracellular Permeability Determination
Changes in paracellular permeability after L. plantarum ZLP001 treatment were determined with fluorescein isothiocyanate-dextrans (FD-4; average molecular mass, 4.4 kDa; Sigma-Aldrich Corp., St. Louis, MO, United States) according to the method reported by Wang et al. (2016) with some modifications. IPEC-2 cells were seeded into 6-well Transwell insert chambers (0.4 μm pore size; Corning, Inc., Corning, NY, United States) at a density of 2.5 × 105 cells per well and were cultured to form differentiated monolayers. The cells were pretreated or not with L. plantarum ZLP001 (LP, 108 CFU mL−1) for 6 h and then challenged or not with 106 CFU mL−1 ETEC for 3 h. FD-4 was added to the apical sides of the IPEC-J2 cell monolayers at a final concentration of 1 mg mL−1. After incubation, medium (100 μL) was sampled from the basolateral chambers, and the FD-4 concentration was quantified using a fluorescence microplate reader (FLx800; BioTek, Winooski, VT, United States). Calibration curves were plotted with an FD-4 gradient series. All experiments were carried out in triplicate.
IPEC-J2 Cells Treatment With L. plantarum ZLP001 and ETEC
IPEC-J2 cells were seeded into 6-well plates (Corning, Inc., Corning, NY, United States) at a density of 2.5 × 105 cells per well. At 80% confluence, the cells were pretreated or not with L. plantarum ZLP001 (LP, 108 CFU mL−1) at 37°C for 6 h. Then, the cells were washed three times with PBS and the supernatant was removed. The cells were challenged or not with 106 CFU mL−1 ETEC at 37°C for 3 h. Fresh medium containing bacteria was prepared by resuspending and diluting collected bacteria in DMEM/F12 without FBS or streptomycin/penicillin. After incubation, the cells were rinsed with PBS three times and collected for subsequent assays. Culture medium supernatants were collected simultaneously. All experiments were carried out in triplicate.
Determination of mRNA Expression
The mRNA expression levels of TJ proteins, cytokines, and HDPs were determined by quantitative real-time PCR (RT-qPCR). IPEC-J2 cells collected after incubation were lysed with RNAzol (MRC, Cincinnati, OH, United States). Total RNA was extracted following the manufacturer’s instructions. RNA concentrations were determined with a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States) and purity was verified by A260:A280 and A260:A230 absorbance ratios. The RNA was reverse transcribed with an iScript cDNA Synthesis Kit (Bio-Rad Laboratories Ltd., Hercules, CA, United States) according to the manufacturer’s instructions. qPCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad Laboratories Ltd., Hercules, CA, United States) on a QuantStudio 3 real-time PCR system (Thermo Fisher Scientific, Waltham, MA, United States). Porcine-specific primers are listed in Supplementary Table S1. The expression of each gene was normalized to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) to yield a relative transcript level. PCR conditions were 95°C for 10 min followed by 40 amplification cycles (95°C for 30 s, 60°C for 30 s, and 72°C for 20 s). Relative gene expression was calculated by the 2−ΔΔCT method.
Protein Extraction and Immunoblotting
Total protein from IPEC-J2 cells was extracted after the various treatments using a lysis buffer containing 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris–HCl adjusted to pH 7.4 and supplemented with a protease inhibitor cocktail (Applygene, Beijing, China). The IPEC-J2 cells were collected into precooled lysis buffer and kept on ice for 30 min. The lysed samples were centrifuged at 4°C and 12,000 × g for 5 min to collect the supernatants. Protein concentrations were determined with a Bicinchoninic Acid Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, United States). After separation on 10% SDS polyacrylamide gel, proteins were electrophoretically transferred to polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, United States). The membranes were blocked with 5% skim milk and then incubated with primary antibodies overnight (∼12–16 h) at 4°C. They were then incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h at 20–25°C. The antibodies used are listed in Supplementary Table S2. Immunoreactive proteins were detected on a ChemiDoc XRS imaging system (Bio-Rad Laboratories Ltd., Hercules, CA, United States) using Western Blotting Luminol Reagent (Santa Cruz Biotechnology, Dallas, TX, United States). Band densities were analyzed with ImageJ (National Institutes of Health, Bethesda, MD, United States). Results were calculated and recorded as the protein abundance relative to β-actin.
Proinflammatory Cytokine and Porcine β-Defensin 2 (pBD2) Measurement
Proinflammatory cytokines and porcine β-defensin 2 were measured by an enzyme linked immunosorbent assay (ELISA). Cell culture medium supernatant (500 μL) was centrifuged at 4,000 × g for 10 min and then passed through a 0.25-μm pore diameter filter (Corning Inc., Corning, NY, United States). The concentrations of interleukin 6 (IL-6), interleukin 8 (IL-8), tumor necrosis factor α (TNF-α), and pBD2 were determined with porcine-specific ELISA Kits (Abcam, Cambridge, United Kingdom), according to the manufacturer’s instructions.
Animal Groups and Diets
The experimental protocol was reviewed and approved by the Ethics Committee of the Institute of Animal Husbandry and Veterinary Medicine, Beijing Academy of Agriculture and Forestry Sciences, Beijing, PRC. Humane animal care was practiced throughout the trial.
Ten post-weaning piglets (siblings; Large White × Landrace; 8.54 ± 0.58 kg) were assigned to L. plantarum ZLP001 treatment or placebo control groups. Each group consisted of two males and three females. Animals were raised at the Beijing Xiqingminfeng Farm (Beijing, China) in a separate room decontaminated prior to the study and were housed at 25–28°C. Each piglet was kept in an individual 1.28-m2 pen with a mesh floor. Each pen contained a feeder and a water nipple. Free access to feed and water was provided throughout the 30-day trial. Piglets received a complete feed specially formulated according to the NRC (2012) and the Feeding Standard of Swine (2004). Detailed information about the diet is shown in Supplementary Table S3. The control group received a basal diet supplemented with placebo (2 g kg−1 feed). The treatment group was administered the basal diet supplemented with freeze-dried L. plantarum ZLP001 (5.0 × 109 CFU g−1, 2 g kg−1 feed).
Fecal Sample Collection and Microbiota Analysis
Fresh fecal samples were individually collected from piglet recta at the end of the feeding experiment. The samples were immediately transferred to the laboratory and processed for genomic DNA extraction with an E.Z.N.A. Stool DNA Kit (Omega Bio-Tek, Norcross, GA, United States) according to the manufacturer’s instructions. V3+V4 hypervariable sequences of 16S rDNA were amplified by PCR with TransStart FastPfu DNA Polymerase (TransGen Biotech Ltd., Beijing, China) with barcode-modified universal primers (forward: 338F, 5′-ACTCCTACGGGAGGCAGCA-3′; reverse: 806R, 5′-GGACTACHVGGGTWTCTAAT-3′). Amplified products were separated on 2% agarose gels and extracted and purified with an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, United States). Barcoded V3+V4 amplicons were sequenced by the paired-end method with Illumina MiSeq at Shanghai Majorbio Bio-pharm Technology Co. (Shanghai, China). Raw sequences were denoised using Trimmomatic and FLASH software and filtered according to their barcodes and primer sequences with QIIME v. 1.5.0. Chimeras were identified and excluded using the UCHIME algorithm v. 4.2.40. Optimized, high-quality sequences were clustered into operational taxonomic units (OTUs) at 97% sequence identity against a subset of the Silva 16S sequence database (Release 1191). Taxon-dependent analysis was carried out using the Ribosomal Database Project (RDP) naive Bayesian classifier, with an 80% bootstrap cutoff. Alpha diversity (Shannon and Simpson indices), abundance (Chao1 and ACE indices), and Good’s coverage and rarefaction were analyzed with mothur v. 1.31.2. Principle coordinates analysis (PCoA) was conducted to visualize differences in fecal community composition. PCoA plots were generated on the basis of Bray–Curtis indices. The linear discriminant analysis effect size (LEfSe) algorithm was used to identify the taxa responsible for the differences between the treatment and control groups. The biomarkers used in the present study had an effect-size threshold of two.
Determination of Fecal Short-Chain Fatty Acid (SCFA) Concentrations
Fecal SCFA concentrations were determined following modified procedures of Qiu and Jin (2002). Half-gram fecal samples were homogenized in 10 mL of double-distilled water. After centrifugation at 12,000 × g for 10 min, the supernatants were removed and filtered through 0.25-μm-pore filters (Corning Inc., Corning, NY, United States). Acetic, propionic, and butyric acids were measured with an ion chromatography system (Dionex Corp., Sunnyvale, CA, United States).
Statistical Analysis
SPSS v. 22.0 (IBM Corp., Armonk, NY, United States) was used for statistical analysis. The FD-4 concentration and mRNA expression levels were analyzed by one-way ANOVA. Fecal SCFA concentrations were analyzed with unpaired Student’s two-tailed t-tests. The results are expressed as the mean ± standard error of the mean (SEM). The significance level was P < 0.05. Correlations between fecal SCFA concentration and intestinal-associated microbiota were examined with Spearman’s rank-order correlation test in R v. 3.2.1.
Results
Effects of L. plantarum ZLP001 on Epithelial Permeability
FD-4 diffusion is a good indicator of paracellular permeability. Therefore, FD-4 transport was measured in this study to evaluate the protective effect of L. plantarum ZLP001 on epithelial integrity (Figure 1). FD-4 concentrations in the L. plantarum ZLP001-treated group were not significantly different from those in the untreated control. When IPEC-J2 cells were exposed to 106 CFU mL−1 ETEC alone, FD-4 permeation was significantly increased relative to that of the control group (P < 0.05). Pretreatment with L. plantarum ZLP001 (108 CFU mL−1) significantly counteracted this permeation.
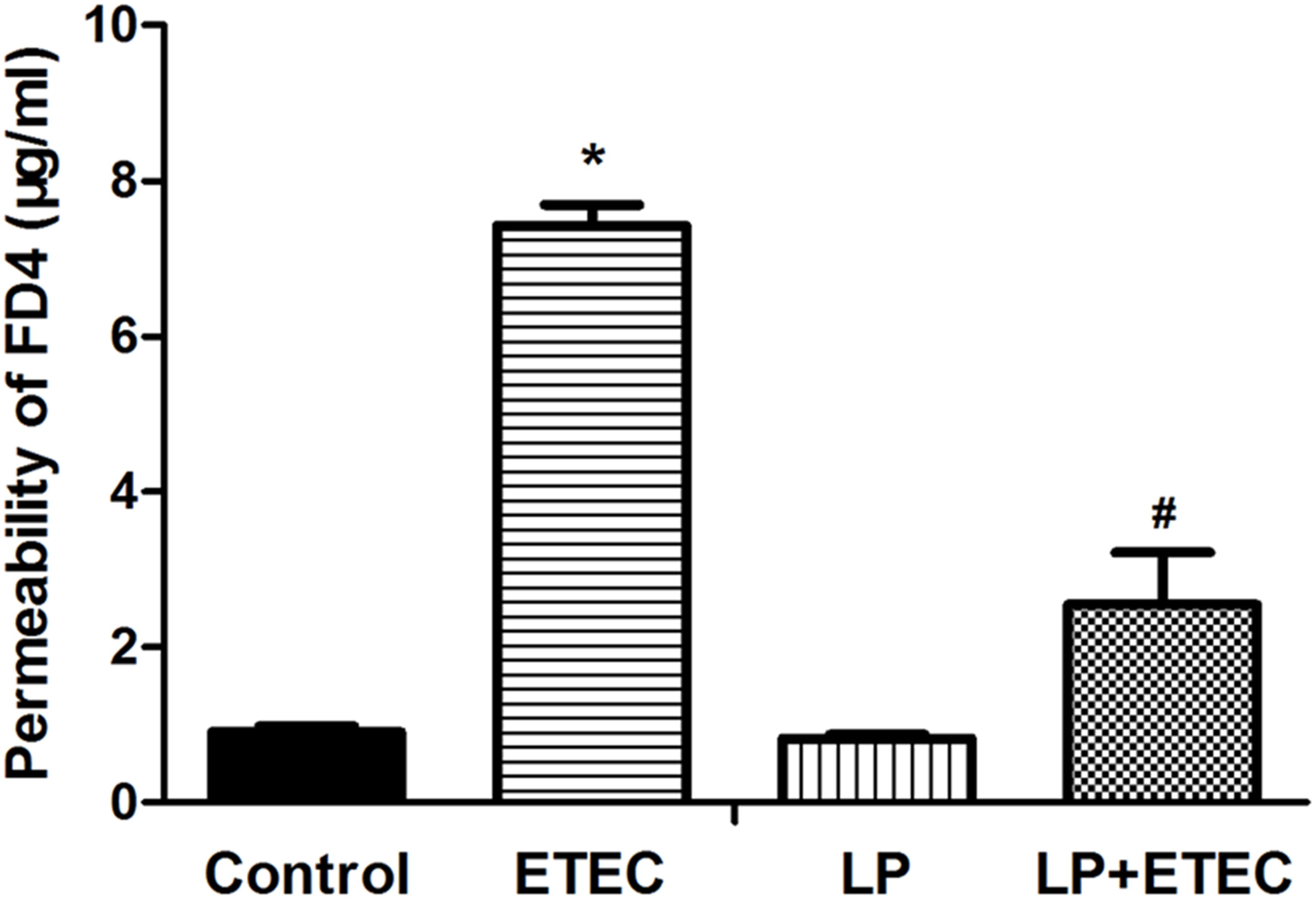
FIGURE 1. FD-4 diffusion in L. plantarum ZLP001-treated IPEC-J2 cells. Cells were left untreated or pretreated with L. plantarum ZLP001 (LP, 108 CFU mL−1) for 6 h on the apical sides of the cell monolayers and then challenged or not with 106 CFU mL−1 ETEC for 3 h. FD-4 concentrations in the basal compartments were measured. Values are shown as the means ± SE of three independent experiments. ∗P < 0.05 vs. non-treated controls; #P < 0.05 vs. ETEC alone.
Effects of L. plantarum ZLP001 on TJ Expression
Abundances of claudin-1 (Figure 2A), occludin (Figure 2B), and ZO-1 (Figure 2C) transcripts in IPEC-J2 cells after bacterial treatments were examined by RT-qPCR (Figure 2). After IPEC-J2 cells were incubated with ETEC alone, the mRNA expression levels of these genes were significantly decreased (P < 0.05) relative to those in the untreated control. L. plantarum ZLP001 treatment alone had no significant influence on TJ mRNA expression as compared to the untreated control. L. plantarum ZLP001 pretreatment significantly (P < 0.05) abrogated the decreases in TJ-related mRNA expression caused by ETEC infection.
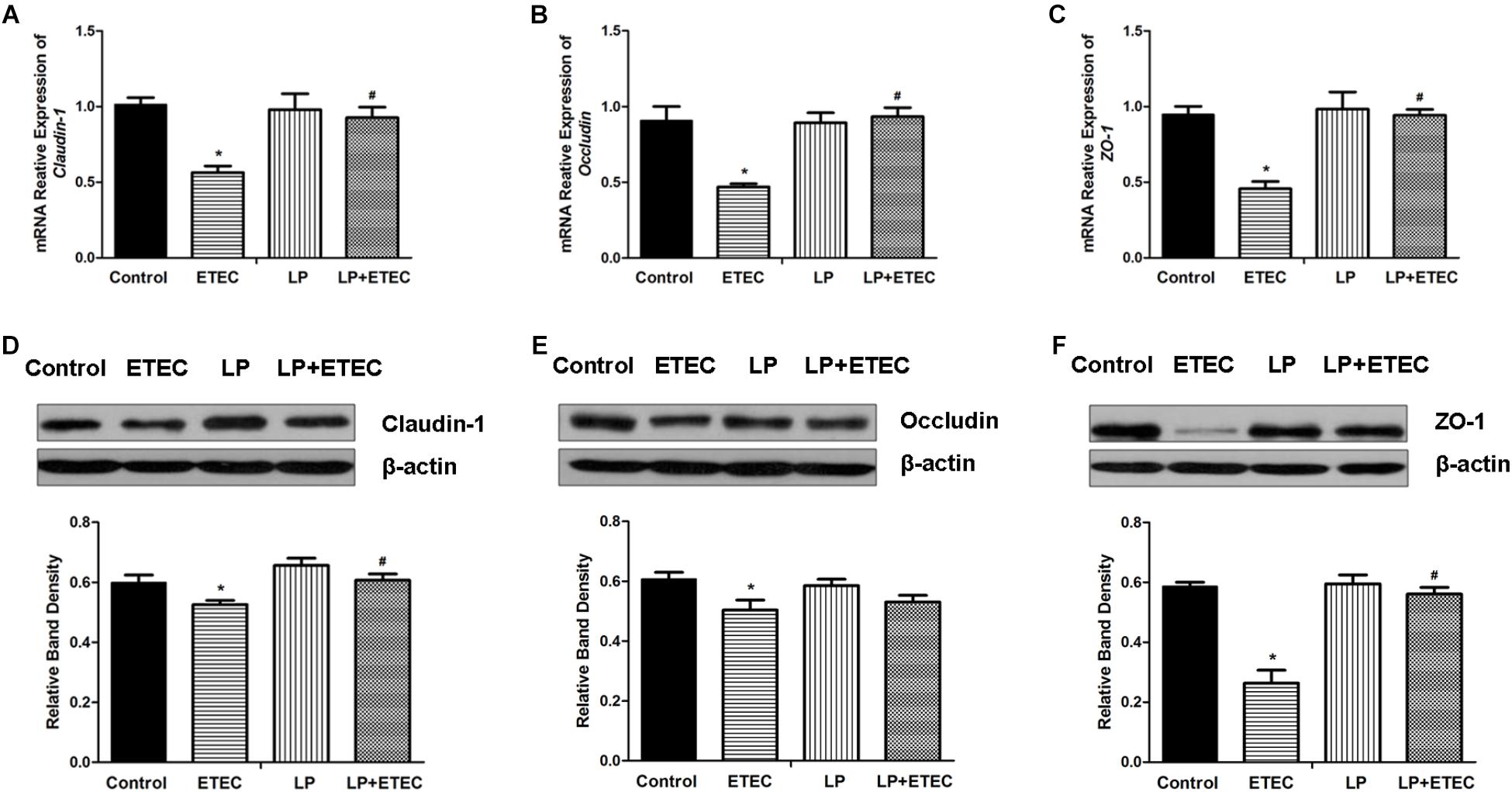
FIGURE 2. Relative mRNA transcript and protein levels of TJ proteins in IPEC-J2 cells left untreated or pretreated with L. plantarum ZLP001 (LP, 108 CFU mL−1) for 6 h, and then challenged or not with 106 CFU mL−1 ETEC for 3 h. mRNA levels of claudin-1 (A), occludin (B), and ZO-1 (C) were standardized to that of GAPDH. Expression levels relative to non-treated controls were calculated by the 2−ΔΔCT method. TJ protein levels were assessed by immunoblotting. Data are western blotting results and gradation analysis of claudin-1 (D), occludin (E), ZO-1 (F). Values are shown as the means ± SE of three independent experiments. ∗P < 0.05 vs. non-treated controls; #P < 0.05 vs. ETEC alone.
Differences in TJ protein expression after the bacterial treatments were examined by western blotting. Expression levels of claudin-1 (Figure 2D), occludin (Figure 2E), and ZO-1 (Figure 2F) were significantly lower (P < 0.05) in cells exposed to ETEC than in the untreated controls. These results were consistent with those for mRNA expression. L. plantarum ZLP001 treatment alone did not significantly affect protein expression relative to the untreated control. L. plantarum ZLP001 pretreatment negated the reduction in claudin-1 (Figure 2D) and ZO-1 (Figure 2F) abundance caused by ETEC treatment.
Effects of L. plantarum ZLP001 on the Epithelial Immunological Barrier
Proinflammatory cytokines in IPEC-J2 cells were quantified after the treatments (Figures 3A–C). Incubation with ETEC alone significantly upregulated IL-6, IL-8, and TNFα transcripts. Treatment with L. plantarum ZLP001 alone had no significant effect on cytokine expression. However, pretreatment with L. plantarum ZLP001 prior to the ETEC challenge reduced cytokine expression in the IPEC-J2 cells to levels lower than those observed after ETEC treatment alone (P < 0.05). Therefore, L. plantarum ZLP001 reduced the ETEC-induced upregulation of proinflammatory cytokines.
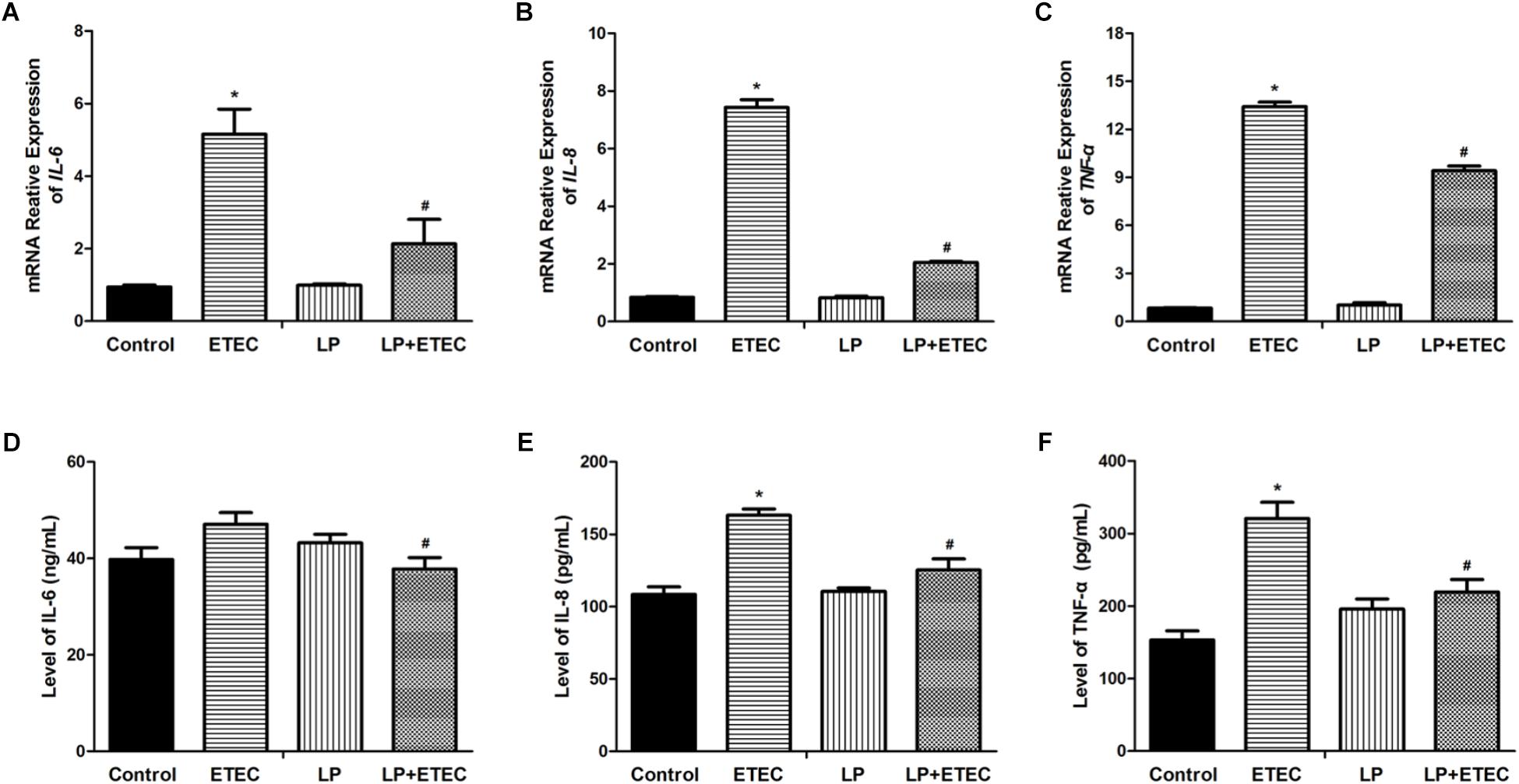
FIGURE 3. Relative mRNA transcript levels and concentrations of proinflammatory cytokines in the culture supernatant of IPEC-J2 cells left untreated or pretreated with L. plantarum ZLP001 (LP, 108 CFU mL−1) for 6 h then either unchallenged or challenged with 106 CFU mL−1 ETEC for 3 h. mRNA levels of IL-6 (A), IL-8 (B), and TNF-α (C) were standardized to that of GAPDH. Expression levels relative to non-treated controls were calculated by the 2−ΔΔCT method. Protein expression of IL-6 (D), IL-8 (E), and TNF-α (F) was assessed by ELISA. Values are shown as the means ± SE of three independent experiments. ∗P < 0.05 vs. non-treated controls; #P < 0.05 vs. ETEC alone.
ELISA was used to verify the protective effect of L. plantarum ZLP001 on epithelial immunological function at the protein level after ETEC challenge (Figures 3D–F). The results confirmed that, while ETEC did not significantly induced IL-6, L. plantarum ZLP001 pretreatment suppressed the increases in IL-6, IL-8, and TNFα secretion in IPEC-J2 cells challenged with ETEC relative to the levels observed in cells incubated with ETEC alone.
Effects of L. plantarum ZLP001 on HDP Production
The modulatory effect of L. plantarum ZLP001 on the innate immune response was evaluated by measuring porcine HDP mRNA expression (Figures 4A,B). Cathelicidins and β-defensins are the two main mammalian HDP families. We selected pBD2 (a β-defensin, Figure 4A) and PG1-5 (a cathelicidin, Figure 4B) as target genes in this study. The results showed that treatment with L. plantarum ZLP001 significantly induced mRNA expression of both HDPs in IPEC-J2 cells (P < 0.05). ETEC exposure had no significant effect on pBD2 expression, but significantly induced PG1-5 expression. Challenge with ETEC 3 h after L. plantarum ZLP001 pretreatment had no significant effect on the HDP expression levels observed after incubation with L. plantarum ZLP001 alone.
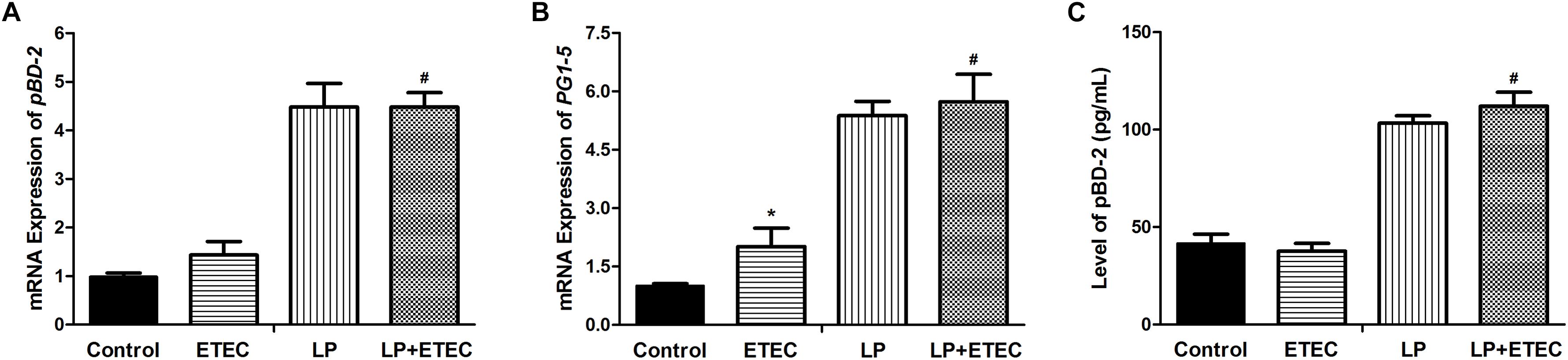
FIGURE 4. Relative mRNA transcript levels of pBD2 (A) and PG1-5 (B) and concentrations of pBD2 (C) in the culture supernatant of IPEC-J2 cells left untreated or pretreated with L. plantarum ZLP001 (LP, 108 CFU mL−1) for 6 h and then challenged or not with 106 CFU mL−1 ETEC for 3 h. mRNA expression levels were standardized to that of GAPDH. Expression levels relative to non-treated controls were calculated by the 2−ΔΔCT method. The pBD2 concentration was assessed by ELISA. Values are shown as the means ± SE of three independent experiments. ∗P < 0.05 vs. non-treated controls; #P < 0.05 vs. ETEC alone.
Effects of bacterial treatment on pBD2 secretion were evaluated by ELISA (Figure 4C). The result was consistent with pBD2 mRNA expression. Exposure to L. plantarum ZLP001 significantly induced pBD2 secretion in IPEC-J2 cells. In contrast, ETEC treatment had no significant effect on pBD2 secretion. Challenge with ETEC 3 h after L. plantarum ZLP001 pretreatment had no significant influence on pBD2 secretion by L. plantarum ZLP001 treatment alone.
Sequencing Results
Sequencing of the amplified 16S rRNA genes produced 373,846 reads after quality checks. An average of 37,385 ± 4,742 reads were obtained for each sample. Among the high-quality sequences, >99% were >400 bp. The average read length for each sample was 435 bp. Reads were clustered into 3,746 OTUs using a 97% similarity cut-off. We obtained 329–404 OTUs per sample. Based on rarefaction analysis, the sequencing depth adequately reflected species richness, suggested that the Illumina MiSeq sequencing system detected most of the fecal bacterial diversity in our study.
Effects of L. plantarum ZLP001 on Alpha Diversity of Fecal Microbiota
We used the Chao1, ACE, Shannon, and Simpson indices to estimate fecal microbiome taxon abundance and diversity (Table 1). L. plantarum ZLP001-treated groups exhibited higher diversity than the control group according to the Shannon and Simpson indices. Nevertheless, the difference was not significant (P > 0.05). L. plantarum ZLP001 treatment had no significant effect on fecal microbiota abundance according to the Chao1 and ACE indices. Good’s coverage was >99.6% for all samples. Thus, the dominant bacterial phylotypes present in the feces were captured by this analysis.
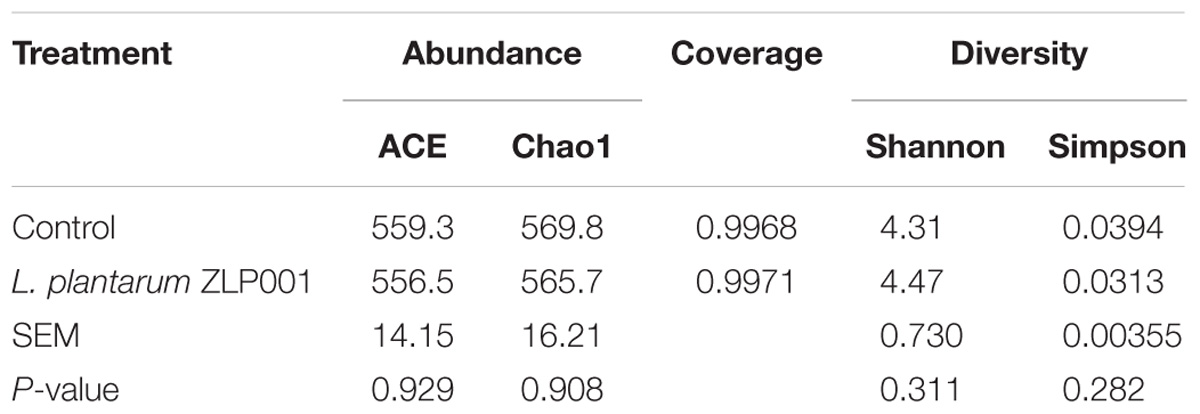
TABLE 1. Effects of L. plantarum ZLP001 treatment on average richness and diversity of bacterial community in piglet feces.
Effects of L. plantarum ZLP001 on Fecal Microbiota Composition
Taxon-dependent analysis was used to compare microbiota compositions of the feces from piglets treated with L. plantarum ZLP001 and those receiving the placebo (Figure 5). Firmicutes and Bacteroidetes were the most abundant phyla in both groups and accounted for >97% of the total sequences on average. Firmicutes was the dominant phylum and constituted 58.1% in all treatments. Bacteroidetes accounted for 39.6%. Other phyla were present at lower frequencies. Figure 5B shows a hierarchically clustered heatmap of the fecal microbiota composition at the genus level. Prevotella was the most abundant; it accounted for an average of 21.5% of the sequences in all treatments by the end of the experiment. Clostridium sensu stricto 1 (14.2%) was identified in the control piglets. Lactobacillus (12.8%) was detected in the L. plantarum ZLP001-treated piglets. LEfSe analysis indicated no significant differences between the placebo- and L. plantarum-treated piglets at the phylum level in terms of relative OTU abundance. Significant differences were observed between groups at several other taxa (Figure 6). The probiotic-treated group was enriched in Bacilli at the class level, Lactobacillales at the order level, Lactobacillaceae and Ruminococcaceae at the family level, and Alloprevotella, Anaerotruncus, Faecalibacterium, Lactobacillus, Subdoligranulum, unclassified Lachnospiraceae, and no-rank Ruminococcaceae at the genus level. However, it was depleted in Clostridiaceae_1 and Peptostreptococcaceae at the family level and Clostridium sensu stricto 1, Terrisporobacter, Ruminococcaceae_UCG_007, Ruminococcaceae_UCG_004, and Ruminococcaceae_UCG_009 at the genus level. Fecal microbiota composition PCoA revealed that L. plantarum ZLP001 treatment significantly affected overall fecal microbiota composition. The microbiota communities in the piglets treated with L. plantarum ZLP001 were clustered together and were distinctly separated from those of the control pigs (Supplementary Figure S1).
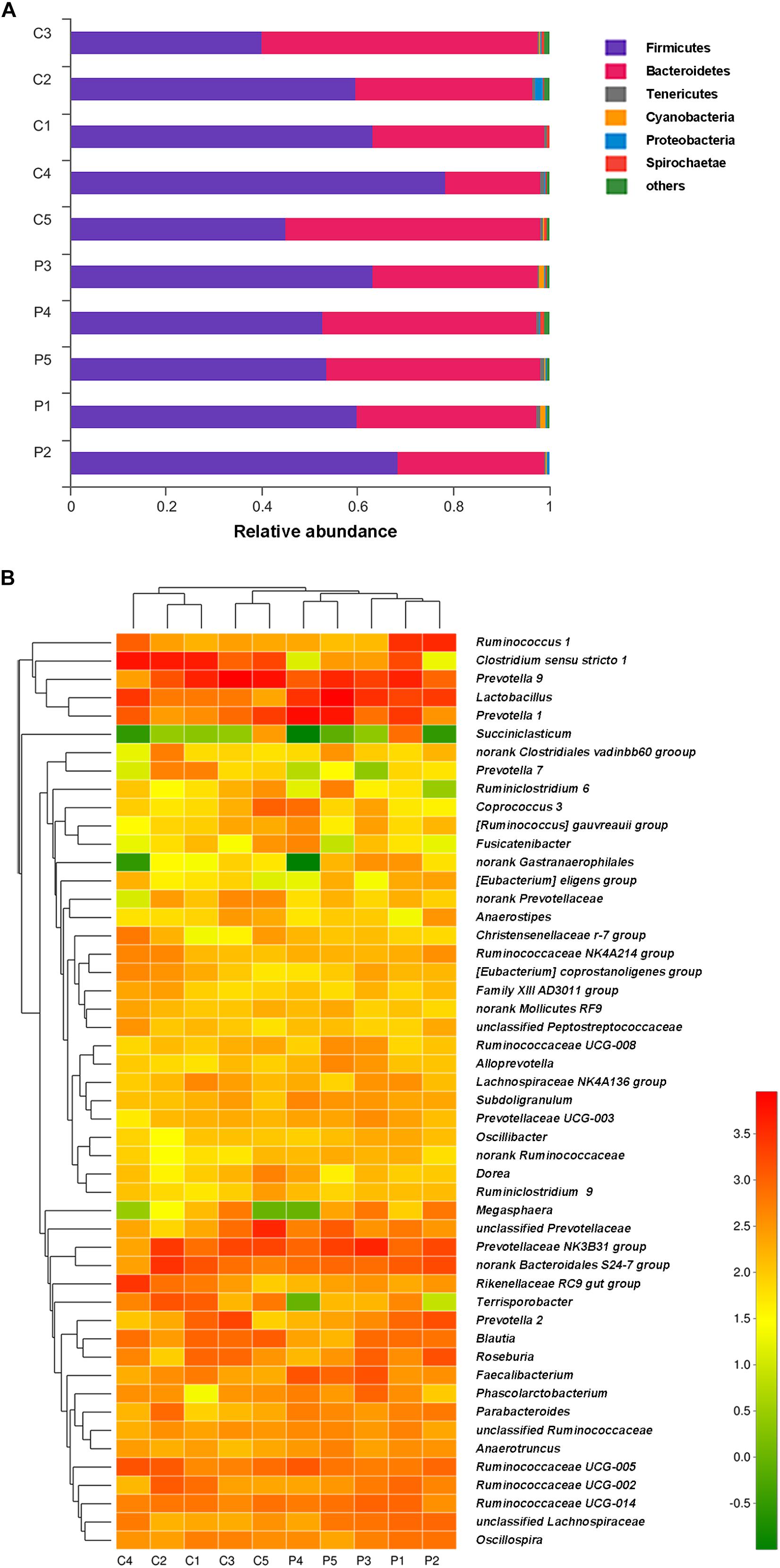
FIGURE 5. Phylum-level microbiota profile of L. plantarum ZLP001-treated piglets as compared to that of placebo-treated control piglets (A). Stacked column chart showing the relative phylum-level bacterial abundance per fecal sample. Genus-level microbiota profile of L. plantarum ZLP001-treated piglets as compared to that of placebo-treated control piglets (B). The heatmap shows genera whose relative abundance was >0.1%. Relative abundance is indicated by a color gradient from green to red, with green representing low abundance and red representing high abundance. C and P represent the control and the probiotic-treated group, respectively. Numbers represent individual animals.
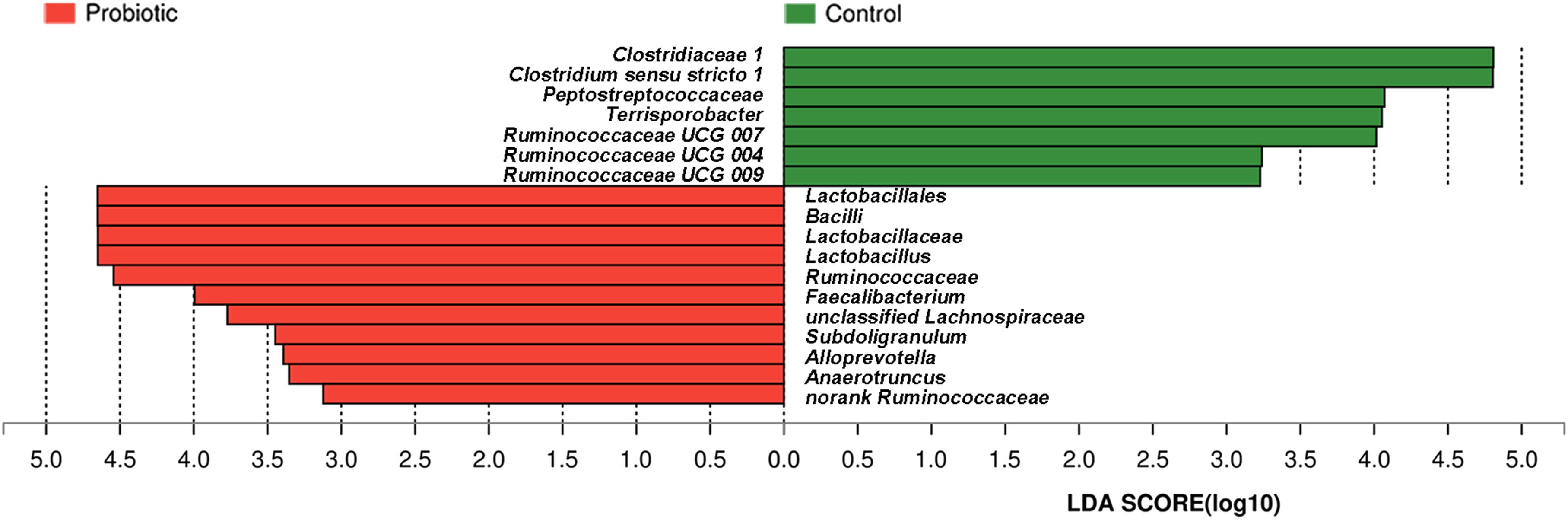
FIGURE 6. Differences between bacterial taxa in L. plantarum ZLP001-treated piglets and placebo-treated control piglets. The histogram shows differentially abundant bacteria in the treatment and control groups ranked by linear discriminant analysis (LDA) scores. Red bars (negative LDA scores) represent bacteria that are more abundant in probiotic-treated fecal samples than in controls. Green bars (positive LDA scores) represent bacteria that are more abundant in placebo-treated fecal samples than in probiotic-treated fecal samples.
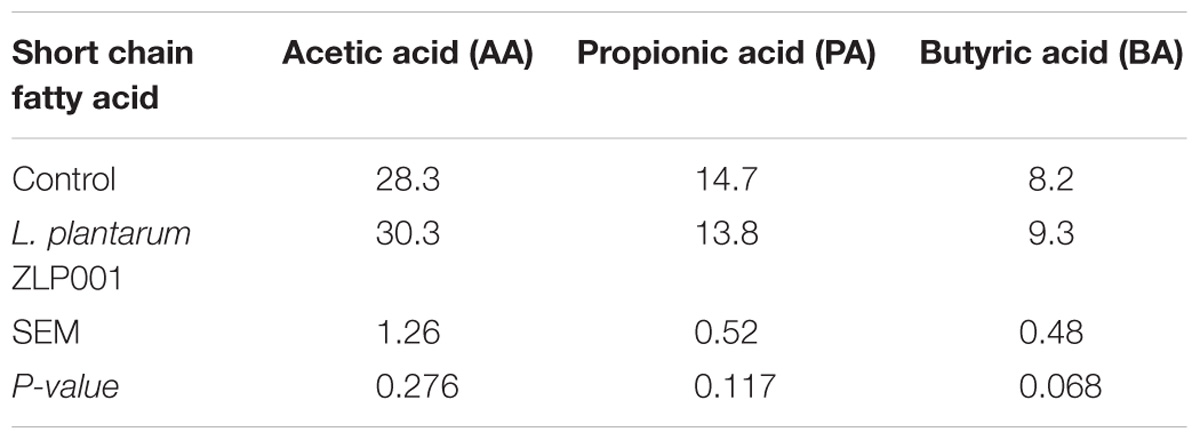
Effects of L. plantarum ZLP001 on Fecal SCFA Concentrations
Table 2 shows the SCFA concentrations in piglet feces. L. plantarum ZLP001 treatment increased butyric acid concentrations relative to those in the controls (P = 0.068). Acetic and propionic acid concentrations did not significantly differ between the placebo and L. plantarum ZLP001 treatments (P > 0.05).
Correlation Between SCFA Concentrations and Fecal Microbiota
Correlations between SCFA concentration and fecal bacterial abundance (relative abundance of the top 30 genera) are shown in Figure 7. The results demonstrated positive associations between butyric acid concentration and Anaerotruncus (r = 0.721, P = 0.019) and unclassified_f_Lachnospiraceae (r = 0.758, P = 0.011) abundance. Acetic acid concentration was positively correlated with Faecalibacterium (r = 0.879, P = 0.001), Subdoligranulum (r = 0.721, P = 0.019), and Prevotellaceae_NK3B31_group (r = 0.648, P = 0.043) abundance. It was negatively correlated with Clostridium sensu stricto 1 abundance (r = −0.661, P = 0.038). Propionic acid concentration was negatively correlated with Phascolarctobacterium abundance (r = −0.697, P = 0.025).
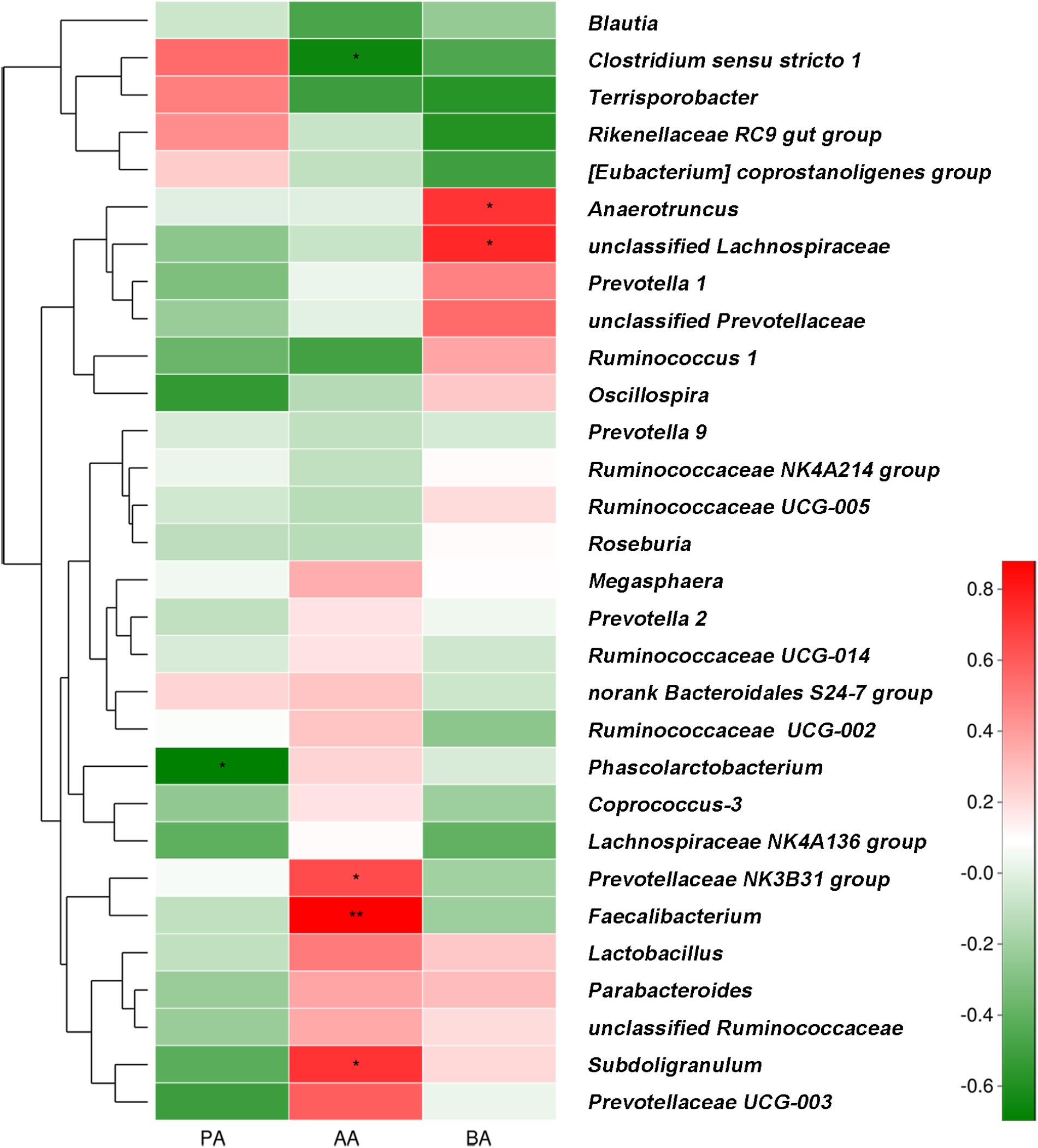
FIGURE 7. Correlations between relative generic abundance and SCFA concentrations in feces obtained from post-weaning piglets. Only the relative abundances of the top 30 bacterial genera are shown. Correlation is indicated by a color gradient from green to red based on Spearman’s correlation coefficients. Asterisks in red cells represent significant positive correlations (∗P < 0.05; ∗∗P < 0.01). Asterisks in green cells represent significant negative correlations (P < 0.05). AA, acetic acid; PA, propionic acid; BA, butyric acid.
Discussion
Using porcine IPEC-2 cells as a model, we demonstrated that L. plantarum ZLP001 plays multiple protective roles in epithelial barrier regulation. The present study showed that ETEC treatment significantly increased gut permeability to FD-4, whereas treatment with probiotic L. plantarum ZLP001 alone had no significant effect on gut permeability. These findings corroborate those of a previous study, in which probiotic L. reuteri did not significantly change FD-4 fluorescence intensity in IPEC-J1 cells (Wang et al., 2016). On the other hand, L. plantarum ZLP001 pretreatment significantly suppressed the increase in gut permeability caused by ETEC infection, suggesting that L. plantarum ZLP001 can alleviate epithelial damage caused by ETEC. This result was consistent with that of a previous study in which permeability to FD-4 indicated that L. reuteri treatment maintains the barrier integrity of IPEC-J1 cells exposed to ETEC (Wang et al., 2016).
TJ proteins play crucial roles in maintaining barrier integrity and function. They include transmembrane proteins such as claudins and occludins, and cytoplasmic scaffolding proteins, such as the ZO family, which have linking and sealing effects (Suzuki, 2013). In this study, relative TJ transcript and protein abundances were significantly reduced after ETEC infection. However, these reductions were abrogated by pretreatment with L. plantarum ZLP001. Previous studies using various probiotic strains reported similar results in vivo and in vitro (Yang et al., 2014; Wu et al., 2016). Therefore, probiotic L. plantarum ZLP001 may fortify intestinal epithelial resistance to pathogens by maintaining TJ protein abundance.
Cytokines play significant regulatory roles in the intestinal inflammatory response. Several studies have demonstrated the effects of probiotics on cytokine expression. Nevertheless, this regulatory action varies with strain. L. reuteri ACTT 6475 shows immunosuppressive action by inhibiting TNFα overexpression in LPS-activated human monocytoid THP1 cells (Lin et al., 2008). However, L. reuteri ACTT 55730 significantly stimulates TNF-α production as an immunostimulatory action (Jones and Versalovic, 2009). In the present study, L. plantarum ZLP001 per se did not influence the expression of proinflammatory cytokines, but inhibited their ETEC-induced overexpression, thus exerting an immunosuppressive action. Certain pro-inflammatory cytokines reportedly are associated with pathogen-induced TJ protein changes (Otte and Podolsky, 2004). ETEC K88 substantially increases IL-8 and disrupts the membrane barrier; however, this disruption can be alleviated by L. plantarum pretreatment (Wu et al., 2016). We obtained similar results in the present study. ETEC-induced increases in IL-6-, IL-8-, and TNF-α expression were effectively counteracted by L. plantarum ZLP001 pretreatment. This observation was consistent with the FD-4 assay results and TJ protein expression levels. Similar findings indicated that L. reuteri inhibits TNF-α expression and may protect TJ proteins (Yang et al., 2015). High-throughput sequencing analysis of post-weaning piglet feces after L. plantarum ZLP001 or control treatment revealed relatively low abundances of certain bacterial genera in the probiotic-treated group. Some of these are associated with various pathological conditions, e.g., Peptostreptococcaceae incertae sedi, which is dominant in viral diarrhea (Ma et al., 2011). Wang et al. (2017) reported that IL-1β and TNF-α transcript levels were positively correlated with Clostridium sensu stricto 1 enrichment in the sheep colon. In the present study, Clostridium sensu stricto 1 was significantly less abundant in the probiotic-treated than in the control group. Certain Clostridium spp. are harmful to host health. Epithelial inflammation observed in weaned piglets may be correlated with Clostridium sensu stricto 1 enrichment in their intestinal mucosa (Wang et al., 2017). The protective effect of probiotics in terms of epithelial immunity may be partially explained by microbiota modulation. One limitation of the present study was that we did not evaluate proinflammatory cytokines in the piglet intestinal tissue and thus, we could not analyze the correlation with microbiota abundance. Such relationships merit further investigation.
The secretion of HDPs, which exert both antimicrobial and immunomodulatory activities, is an epithelial innate immunity mechanism (Bevins et al., 1999; Zhang et al., 2000). Enhancing endogenous HDP synthesis improves the early response to bacterial infection and inflammation (Veldhuizen et al., 2008). Nutrients, such as VD3, butyrate, and zinc induce HDP secretion (Talukder et al., 2011; Zeng et al., 2013; Merriman et al., 2015). Probiotics can also stimulate HDP expression (Schlee et al., 2008; Liu et al., 2017). In the present study, increased pBD2 and PG1-5 expression and pBD2 secretion were observed after L. plantarum ZLP001 treatment, suggesting that this strain can induce HDPs, to protect against bacterial infection. Similar results have been reported for the probiotic strain L. reuteri I5007 (Liu et al., 2017). The administration of synthetic HDPs reportedly can improve weaned piglet growth performance, nutrient digestion and assimilation, intestinal health, and antioxidant capacity (Xiao et al., 2013; Yoon et al., 2013; Yu et al., 2017). Therefore, the induction of HDP expression by L. plantarum ZLP001 may be correlated with the improved growth and reduced risk of diarrhea reported in our previous studies. SCFAs, especially butyrate, induce HDPs (Zeng et al., 2013). Butyrate production by enteric microbiota is the only microbial stimulus capable of inducing HDP expression (Schauber et al., 2003). Most butyrate-producing bacteria belong to Clostridium clusters IV and XIVa. Butyrate metabolism has been observed in species of Faecalibacterium and Anaerotruncus, e.g., Faecalibacterium prausnitzii and Anaerotruncus colihominis are butyrate producers (El Aidy et al., 2013; Wrzosek et al., 2013). A. colihominis has been shown to specifically colonize the lumen whereas F. prausnitzii is enriched in the mucus (Van den Abbeele et al., 2013). In the present study, Faecalibacterium spp. and Anaerotruncus spp. were significantly abundant in L. plantarum ZLP001-treated piglets. Therefore, increasing the abundances of these genera may elevate butyrate levels and epithelial HDP expression. The present study confirmed a positive correlation between fecal butyric acid and Anaerotruncus spp. abundance after L. plantarum ZLP001 treatment. In addition, Faecalibacterium spp. abundance showed a positive correlation with acetic acid concentration. Acetic acid can be converted to butyrate by Eubacterium rectale (Duncan and Flint, 2008), which may enhance HDP expression. The modulation of butyrate producers with probiotics to generate butyrate to stimulate HDP levels in the epithelium may thus be a meaningful approach for future interventions that aim to improve intestinal balance. Few studies have focused on the association between HDP production and probiotic function. Future studies involving probiotical and the non-pathogenic enteric bacterial regulation of antimicrobial peptides may elucidate the beneficial effects of probiotics against pathogen infection.
The piglet intestinal microbiota undergoes substantial dynamic changes after weaning, and this alteration can be associated with severe disorders and bowel disease. In the present study, fecal bacterial communities were dominated by Firmicutes and Bacteroidetes, regardless of treatment. This result was expected because the colon is a strictly anaerobic environment and most of the species within these phyla are anaerobic. Similar results were reported in previous pig studies (Konstantinov et al., 2004; Kim and Isaacson, 2015). Prevotella, which is associated with hemicelluloses degradation, reportedly is the predominant genus in piglets at nursery stage (Konstantinov et al., 2004). A high Prevotella spp. abundance may be essential for post-weaning piglets to be able to digest plant-based diets. Post-weaning increases in the proportions of Lactobacillus spp. are desirable and beneficial. Previous studies have shown that oral administration of lactic acid bacteria enhances the relative abundance of intestinal Lactobacillus spp. in weaned piglets (Hu et al., 2015; Zhang et al., 2016). In the present study, dietary L. plantarum ZLP001 supplementation significantly increased Lactobacillus spp. abundance in the post-weaning piglet intestine. L. plantarum ZLP001 may produce molecules that stimulate Lactobacillus spp. growth in the piglet intestine (Ohashi et al., 2001). Alternatively, the observed increase in Lactobacillus abundance may have resulted from the proliferation of the administered probiotic strain (Takahashi et al., 2007). Certain lactobacilli, such as L. rhamnosus, L. reuteri, and L. plantarum, protect TJ proteins after stress and infection and may, therefore, contribute to TJ integrity and intestinal barrier function (Yang et al., 2015; Blackwood et al., 2017). Further studies are needed to investigate specific species and their effects on piglet epithelial TJ proteins. The effects on gut microbiota composition and community observed after L. plantarum ZLP001 treatment in this study suggest that the modulation of the intestinal flora by L. plantarum ZLP001 helps to maintain a well-balanced gut microbiota, thereby improving the health and growth of pigs.
Conclusion
Our IPEC-J2 model demonstrated that L. plantarum ZLP001 enhances intestinal barrier function by (1) maintaining epithelial integrity and preventing ETEC-induced gut permeability, (2) forming TJs and reducing ETEC-induced TJ damage, (3) modulating immune function and repressing the ETEC-induced immune response, and (4) inducing the secretion of antimicrobial peptides to protect against pathogens. Our study also indicated that L. plantarum ZLP001 supplementation improved gut bacterial ecology and barrier function in weaned piglets by (1) reducing the abundance of certain bacterial species correlated with proinflammatory cytokine expression, (2) modulating butyrate-producing enteric microbiota to induce epithelial HDP expression, and (3) enhancing intestinal Lactobacillus abundance to improve the gut microbiota composition and reinforce TJs (Figure 8). Elucidation of the mechanisms by which probiotics act on the intestinal barrier will promote their use in livestock production. In addition, further animal studies are required to determine how L. plantarum ZLP001 protects the piglet intestinal barrier both before and after pathogenesis.
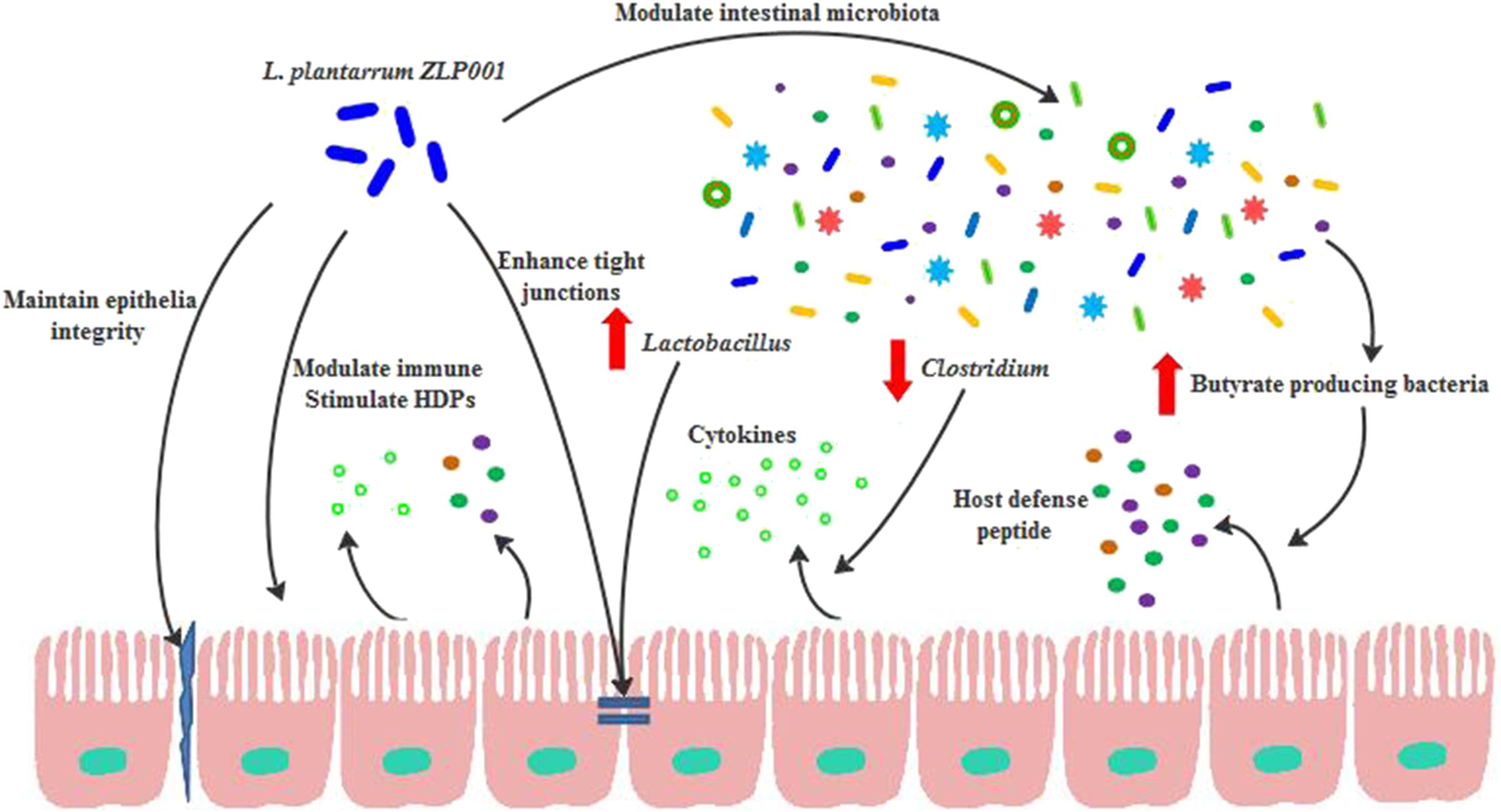
FIGURE 8. Suggested mechanism by which L. plantarum ZLP001 protects intestinal barrier function from weaning stress. L. plantarum ZLP001 functions not only through maintaining epithelial integrity, improving TJs, regulating the immune response, and stimulating HDPs, but also via modulating the intestinal indigenous microbiota. Modulated microbiota and alterations in certain bacterial taxa in turn enhance epithelial function.
Author Contributions
JW and HJ conceived and designed the experiments. JW, SW, HL, and DZ performed the experiments. JW and WZ analyzed the data. YW contributed reagents and materials. JW and HJ helped to draft the manuscript. All authors read and approved the final manuscript.
Funding
This study was financially supported by the Youth Fund of the Beijing Academy of Agriculture and Forestry Sciences (Grant No. QNJJ201607), the Beijing Innovation Consortium of Agriculture Research System (Grant No. BAIC02-2017), the Special Program on Science and Technology Innovation Capacity Building of BAAFS (Grant Nos. KJCX20180109 and KJCX20161503), and the International Scientific and Technological Cooperation Funding (Grant No. GJHZ2018-06).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01953/full#supplementary-material
FIGURE S1 | Effects of L. plantarum ZLP001 on microbial community structure in piglet feces based on principal coordinate analysis (PCoA). PCoA plot showing microbiota clustering in various treatments. Each dot represents an individual sample. Red and green indicate control and probiotic-treated samples, respectively. C and P represent control and probiotic-treated groups, respectively. Numbers represent individual animals.
TABLE S1 | Primers used in this study.
TABLE S2 | Antibodies used in this study.
TABLE S3 | Ingredients and chemical composition of the basal diet.
Footnotes
References
Anderson, R. C., Cookson, A., McNabb, W., Park, Z., McCann, M., Kelly, W., et al. (2010). Lactobacillus plantarum MB452 enhances the function of the intestinal barrier by increasing the expression levels of genes involved in tight junction formation. BMC Microbiol. 10:316. doi: 10.1186/1471-2180-10-316
Bevins, C. L., Martin-Porter, E., and Ganz, T. (1999). Defensins and innate host defence of the gastrointestinal tract. Gut 45, 911–915. doi: 10.1136/gut.45.6.911
Blackwood, B. P., Yuan, C. Y., Wood, D. R., Nicolas, J. D., Grothaus, J. S., and Hunter, C. J. (2017). Probiotic Lactobacillus species strengthen intestinal barrier function and tight junction integrity in experimental necrotizing enterocolitis. J. Probiotics Health 5:159. doi: 10.4172/2329-8901.1000159
Boirivant, M., and Strober, W. (2007). The mechanism of action of probiotics. Curr. Opin. Gastroenterol. 23, 679–692. doi: 10.1097/MOG.0b013e3282f0cffc
Campbell, J. M., Crenshaw, J. D., and Polo, J. (2013). The biological stress of early weaned piglets. J. Anim. Sci. Biotechnol. 4:19. doi: 10.1186/2049-1891-4-19
Cisek, A. A., and Binek, M. (2014). Chicken intestinal microbiota function with a special emphasis on the role of probiotic bacteria. Pol. J. Vet. Sci. 17, 385–394. doi: 10.2478/pjvs-2014-0057
Duncan, S. H., and Flint, H. J. (2008). Proposal of a neotype strain (A1–86) for Eubacterium rectale. Request for an opinion. Int. J. Syst. Evol. Microbiol. 58, 1735–1736. doi: 10.1099/ijs.0.2008/004580-0
El Aidy, S., Van den Abbeele, P., Van de Wiele, T., Louis, P., and Kleerebezem, M. (2013). Intestinal colonization: how key microbial players become established in this dynamic process. Bioessays 35, 913–923. doi: 10.1002/bies.201300073
Eun, C. S., Kim, Y. S., Han, D. S., Choi, J. H., Lee, A. R., and Park, Y. K. (2011). Lactobacillus casei prevents impaired barrier function in intestinal epithelial cells. APMIS 119, 49–56. doi: 10.1111/j.1600-0463.2010.02691.x
Farkas, O., Mátis, G., Pászti-Gere, E., Palócz, O., Kulcsár, A., Petrilla, J., et al. (2014). Effects of Lactobacillus plantarum 2142 and sodium n-butyrate in lipopolysaccharide-triggered inflammation: comparison of a porcine intestinal epithelial cell line and primary hepatocyte monocultures with a porcine enterohepatic co-culture system. J. Anim. Sci. 92, 3835–3845. doi: 10.2527/jas.2013-7453
Gresse, R., Chaucheyras-Durand, F., Fleury, M. A., Van de Wiele, T., Forano, E., and Blanquet-Diot, S. (2017). Gut microbiota dysbiosis in postweaning piglets: understanding the keys to health. Trends Microbiol. 25, 851–873. doi: 10.1016/j.tim.2017.05.004
Hu, C. H., Xiao, K., Luan, Z. S., and Song, J. (2013). Early weaning increases intestinal permeability, alters expression of cytokine and tight junction proteins, and activates mitogen-activated protein kinases in pigs. Anim. Sci. 91,1094–1101. doi: 10.2527/jas2012-5796
Hu, Y., Dun, Y., Li, S., Zhang, D., Peng, N., Zhao, S., et al. (2015). Dietary Enterococcus faecalis LAB 31 improves growth performance, reduces diarrhea, and increases fecal Lactobacillus number of weaned piglets. PLoS One 10:e0116635. doi: 10.1371/journal.pone.0116635
Jones, S. E., and Versalovic, J. (2009). Probiotic Lactobacillus reuteri biofilms produce antimicrobial and anti-inflammatory factors. BMC Microbiol. 9:35. doi: 10.1186/1471-2180-9-35
Kim, H. B., and Isaacson, R. E. (2015). The pig gut microbial diversity: understanding the pig gut microbial ecology through the next generation high throughput sequencing. Vet. Microbiol. 177, 242–251. doi: 10.1016/j.vetmic.2015.03.014
Konstantinov, S. R., Awati, A. A., Williams, B. A., Miller, B. G., Jones, P., Stokes, C. R., et al. (2006). Post-natal development of the porcine microbiota composition and activities. Environ. Microbiol. 8, 1191–1199. doi: 10.1111/j.1462-2920.2006.01009.x
Konstantinov, S. R., Favier, C. F., Zhu, W. Y., Williams, B. A., Klüß, J., Souffrant, W. B., et al. (2004). Microbial diversity studies of the porcine gastrointestinal ecosystem during weaning transition. Anim. Res. 53, 317–324. doi: 10.1051/animres:2004019
Kopp, M. V., Goldstein, M., Dietschek, A., Sofke, J., Heinzmann, A., and Urbanek, R. (2008). Lactobacillus GG has in vitro effects on enhanced interleukin- 10 and interferon-g release of mononuclear cells but no in vivo effects in supplemented mothers and their neonates. Clin. Exp. Allergy 38, 602–610. doi: 10.1111/j.1365-2222.2007.02911.x
Lallès, J. P., Bosi, P., Smidt, H., and Stokes, C. R. (2007). Weaning – A challenge to gut physiologists. Livest. Sci. 108, 82–93. doi: 10.1016/j.livsci.2007.01.091
Lallès, J. P., Boudry, G., Favier, C., Floc’h, N. L., Luron, I., Montagne, L., et al. (2004). Gut function and dysfunction in young pigs: physiology. Anim. Res. 53, 301–316. doi: 10.1051/animres:2004018
Li, Z., Wang, W., Liu, D., and Guo, Y. (2017). Effects of Lactobacillus acidophilus on gut microbiota composition in broilers challenged with Clostridium perfringens. PLoS One 12:e0188634. doi: 10.1371/journal.pone.0188634
Lin, Y. P., Thibodeaux, C. H., Pena, J. A., Ferry, G. D., and Versalovic, J. (2008). Probiotic Lactobacillus reuteri suppress proinflammatory cytokines via c-Jun. Inflamm. Bowel Dis. 14, 1068–1083. doi: 10.1002/ibd.20448
Liu, H., Hou, C., Wang, G., Jia, H., Yu, H., Zeng, X., et al. (2017). Lactobacillus reuteri I5007 modulates intestinal host defense peptide expression in the model of IPEC-J2 cells and neonatal piglets. Nutrients 9:559. doi: 10.3390/nu9060559
Ma, C., Wu, X., Nawaz, M., Li, J., Yu, P., Moore, J. E., et al. (2011). Molecular characterization of fecal microbiota in patients with viral diarrhea. Curr. Microbiol. 63, 259–266. doi: 10.1007/s00284-011-9972-7
Merriman, K. E., Kweh, M. F., Powell, J. L., Lippolis, J. D., and Nelson, C. D. (2015). Multiple beta defensin genes are upregulated by the vitamin D pathway in cattle. J. Steroid. Biochem. Mol. Biol. 154, 120–129. doi: 10.1016/j.jsbmb.2015.08.002
NRC (2012). Nutrient Requirements of Swine, 11th revised Edn. Washington, DC: National Academy Press.
Ohashi, Y., Inoue, R., Tanaka, K., Matsuki, T., Umesaki, Y., and Ushida, K. (2001). Lactobacillus casei strain Shirota-fermented milk stimulates indigenous lactobacilli in the pig intestine. J. Nutr Sci. Vitaminol. 47, 172–176. doi: 10.3177/jnsv.47.172
Otte, J. M., and Podolsky, D. K. (2004). Functional modulation of enterocytes by gram-positive and gram-negative microorganisms. Am. J. Physiol. Gastrointest. Liver Physiol. 286, G613–G626. doi: 10.1152/ajpgi.00341.2003
Paolillo, R., Romano Carratelli, C., Sorrentino, S., Mazzola, N., and Rizzo, A. (2009). Immunomodulatory effects of Lactobacillus plantarum on human colon cancer cells. Int. Immunopharmacol. 9, 1265–1271. doi: 10.1016/j.intimp.2009.07.008
Patel, R. M., Myers, L. S., Kurundkar, A. R., Maheshwari, A., Nusrat, A., and Lin, P. W. (2012). Probiotic bacteria induce maturation of intestinal claudin 3 expression and barrier function. Am. J. Pathol. 180, 626–635. doi: 10.1016/j.ajpath.2011.10.025
Qiu, J., and Jin, X. (2002). Development and optimization of organic acid analysis in tobacco with ion chromatography and suppressed conductivity detection. J. Chromatogr. A 950, 81–88. doi: 10.1016/S0021-9673(02)00034-1
Riboulet-Bisson, E., Sturme, M. H. J., Jeffery, I. B., O’Donnell, M. M., Neville, B. A., Forde, B. M., et al. (2012). Effect of Lactobacillus salivarius Bacteriocin Abp118 on the mouse and pig intestinal microbiota. PLoS One 7:e31113. doi: 10.1371/journal.pone.0031113
Schauber, J., Svanholm, C., Termén, S., Iffland, K., Menzel, T., Scheppach, W., et al. (2003). Expression of the cathelicidin LL-37 is modulated by short chain fatty acids in colonocytes: relevance of signalling pathways. Gut 52, 735–741. doi: 10.1136/gut.52.5.735
Schlee, M., Harder, J., Koten, B., Stange, E. F., Wehkamp, J., and Fellermann, K. (2008). Probiotic lactobacilli and VSL#3 induce enterocyte β-defensin 2. Clin. Exp. Immunol. 151, 528–535. doi: 10.1111/j.1365-2249.2007.03587.x
Suzuki, T. (2013). Regulation of intestinal epithelial permeability by tight junctions. Cell. Mol. Life Sci. 70, 631–659. doi: 10.1007/s00018-012-1070-x
Takahashi, S., Egawa, Y., Simojo, N., Tsukahara, T., and Ushida, K. (2007). Oral administration of Lactobacillus plantarum strain Lq80 to weaning piglets stimulates the growth of indigenous lactobacilli to modify the lactobacillal population. J. Gen. Appl. Microbiol. 53, 325–332. doi: 10.2323/jgam.53.325
Talukder, P., Satho, T., Irie, K., Sharmin, T., Hamady, D., Nakashima, Y., et al. (2011). Trace metal zinc stimulates secretion of antimicrobial peptide LL-37 from Caco-2 cells through ERK and p38 MAP kinase. Int. Immunopharmacol. 11, 141–144. doi: 10.1016/j.intimp.2010.10.010
Van Baarlen, P., Wells, J. M., and Kleerebezem, M. (2013). Regulation of intestinal homeostasis and immunity with probiotic lactobacilli. Trends Immunol. 34, 208–215. doi: 10.1016/j.it.2013.01.005
Van den Abbeele, P., Belzer, C., Goossens, M., Kleerebezem, M., De Vos, W. M., Thas, O., et al. (2013). Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 7, 949–961. doi: 10.1038/ismej.2012.158
Van der Meulen, J., Koopmans, S. J., Dekker, R. A., and Hoogendoorn, A. (2010). Increasing weaning age of piglets from 4 to 7 weeks reduces stress, increases post-weaning feed intake but does not improve intestinal functionality. Animal 4, 1653–1661. doi: 10.1017/S1751731110001011
Veldhuizen, E. J., Rijnders, M., Claassen, E. A., Van Dijk, A., and Haagsman, H. P. (2008). Porcine beta-defensin 2 displays broad antimicrobial activity against pathogenic intestinal bacteria. Mol. Immunol. 45, 386–394. doi: 10.1016/j.molimm.2007.06.001
Villena, J., Chiba, E., Vizoso-Pinto, M. G., Tomosada, Y., Takahashi, T., Ishizuka, T., et al. (2014). Immunobiotic Lactobacillus rhamnosus strains differentially modulate antiviral immune response in porcine intestinal epithelial and antigen presenting cells. BMC Microbiol. 14:126. doi: 10.1186/1471-2180-14-126
Wang, J., Ji, H., Wang, S., Zhang, D., Liu, H., Shan, D., et al. (2012). Lactobacillus plantarum ZLP001: in vitro assessment of antioxidant capacity and effect on growth performance and antioxidant status in weaning piglets. Asian Austral. J. Anim. Sci. 25, 1153–1158. doi: 10.5713/ajas.2012.12079
Wang, J., Ji, H., Zhang, D., Liu, H., Wang, S., Shan, D., et al. (2011). Assessment of probiotic properties of Lactobacillus plantarum ZLP001 isolated from gastrointestinal tract of weaning pigs. Afr. J. Biotechnol. 10, 11303–11308. doi: 10.5897/AJB11.255
Wang, Y., Xu, L., Liu, J., Zhu, W., and Mao, S. (2017). A high grain diet dynamically shifted the composition of mucosa-associated microbiota and induced mucosal injuries in the colon of sheep. Front. Microbiol. 8:2080. doi: 10.3389/fmicb.2017.02080
Wang, Z., Wang, L., Chen, Z., Ma, X., Yang, X., Zhang, J., et al. (2016). In Vitro evaluation of swine-derived Lactobacillus reuteri: probiotic properties and effects on intestinal porcine epithelial cells challenged with enterotoxigenic Escherichia coli K88. J. Microbiol. Biotechnol. 26, 1018–1025. doi: 10.4014/jmb.1510.10089
Winter, S. E., Winter, M. G., Xavier, M. N., Thiennimitr, P., Poon, V., Keestra, A. M., et al. (2013). Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339, 708–711. doi: 10.1126/science.1232467
Wrzosek, L., Miquel, S., Noordine, M. L., Bouet, S., Joncquel Chevalier-Curt, M., Robert, V., et al. (2013). Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol. 11:61. doi: 10.1186/1741-7007-11-61
Wu, Y., Zhu, C., Chen, Z., Chen, Z., Zhang, W., Ma, X., et al. (2016). Protective effects of Lactobacillus plantarum on epithelial barrier disruption caused by enterotoxigenic Escherichia coli in intestinal porcine epithelial cells. Vet. Immunol. Immunopathol. 172, 55–63. doi: 10.1016/j.vetimm.2016.03.005
Xiao, H., Wu, M. M., Tan, B. E., Yin, Y. L., Li, T. J., Xiao, D. F., et al. (2013). Effects of composite antimicrobial peptides in weanling piglets challenged with deoxynivalenol: I. Growth performance, immune function, and antioxidation capacity. J. Anim. Sci. 91, 4772–4780. doi: 10.2527/jas.2013-6426
Yang, F., Wang, A., Zeng, X., Hou, C., Liu, H., and Qiao, S. (2015). Lactobacillus reuteri I5007 modulates tight junction protein expression in IPEC-J2 cells with LPS stimulation and in newborn piglets under normal conditions. BMC Microbiol. 15:32. doi: 10.1186/s12866-015-0372-1
Yang, K. M., Jiang, Z. Y., Zheng, C. T., Wang, L., and Yang, X. F. (2014). Effect of Lactobacillus plantarum on diarrhea and intestinal barrier function of young piglets challenged with enterotoxigenic Escherichia coli K88. J. Anim. Sci. 92, 1496–1503. doi: 10.2527/jas.2013-6619
Yoon, J. H., Ingale, S. L., Kim, J. S., Kim, K. H., Lohakare, J., Park, Y. K., et al. (2013). Effects of dietary supplementation with antimicrobial peptide-P5 on growth performance, apparent total tract digestibility, faecal and intestinal microflora and intestinal morphology of weanling pigs. J. Sci. Food Agric. 93, 587–592. doi: 10.1002/jsfa.5840
Yu, H. T., Ding, X. L., Li, N., Zhang, X. Y., Zeng, X. F., Wang, S., et al. (2017). Dietary supplemented antimicrobial peptide microcin J25 improves the growth performance, apparent total tract digestibility, fecal microbiota, and intestinal barrier function of weaned pigs. J. Anim. Sci. 95, 5064–5076. doi: 10.2527/jas2017.1494
Zeng, X., Sunkara, L. T., Jiang, W., Bible, M., Carter, S., Ma, X., et al. (2013). Induction of porcine host defense peptide gene expression by short-chain fatty acids and their analogs. PLoS One 8:e72922. doi: 10.1371/journal.pone.0072922
Zhang, G., Ross, C. R., and Blecha, F. (2000). Porcine antimicrobial peptides: new prospects for ancient molecules of host defense. Vet. Res. 31, 277–296. doi: 10.1051/vetres:2000121
Zhang, W., Zhu, Y. H., Zhou, D., Wu, Q., Song, D., Dicksved, J., et al. (2016). Oral administration of a select mixture of Bacillus probiotics affects the gut microbiota and goblet cell function following Escherichia coli challenge in newly weaned pigs of genotype MUC4 that are supposed to be enterotoxigenic E. coli F4ab/ac receptor negative. Appl. Environ. Microbiol. 83:e02747-16. doi: 10.1128/AEM.02747-16
Keywords: Lactobacillus plantarum, permeability, tight junction, immune response, host defense, microbiota
Citation: Wang J, Ji H, Wang S, Liu H, Zhang W, Zhang D and Wang Y (2018) Probiotic Lactobacillus plantarum Promotes Intestinal Barrier Function by Strengthening the Epithelium and Modulating Gut Microbiota. Front. Microbiol. 9:1953. doi: 10.3389/fmicb.2018.01953
Received: 09 April 2018; Accepted: 02 August 2018;
Published: 24 August 2018.
Edited by:
Maria Olivia Pereira, University of Minho, PortugalReviewed by:
M. Andrea Azcarate-Peril, University of North Carolina at Chapel Hill, United StatesMaryam Dadar, Razi Vaccine and Serum Research Institute, Iran
Copyright © 2018 Wang, Ji, Wang, Liu, Zhang, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haifeng Ji, amhmMjA3QDEyNi5jb20=
 Jing Wang
Jing Wang Haifeng Ji*
Haifeng Ji* Wei Zhang
Wei Zhang Dongyan Zhang
Dongyan Zhang