
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 17 August 2018
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.01937
 Yuan-Yuan Zheng1,2†
Yuan-Yuan Zheng1,2† Ruo-Lan Du2†
Ruo-Lan Du2† Sen-Yuan Cai1
Sen-Yuan Cai1 Zhi-Hua Liu3
Zhi-Hua Liu3 Zhi-Yuan Fang3
Zhi-Yuan Fang3 Ting Liu3
Ting Liu3 Lok-Yan So2
Lok-Yan So2 Yu-Jing Lu1,4*
Yu-Jing Lu1,4* Ning Sun2,3,5*
Ning Sun2,3,5* Kwok-Yin Wong2*
Kwok-Yin Wong2*New generation of antibacterial agents are urgently needed in order to fight the emergence of multidrug-resistant bacteria. FtsZ is currently identified as a promising target for new types of antimicrobial compounds development because of its conservative characteristics and its essential role played in bacterial cell division. In the present study, the antibacterial activity of a series of benzofuroquinolinium derivatives was investigated. The results show that the compounds possess potent antibacterial activity against drug resistant pathogens including MRSA, VREF and NDM-1 Escherichia coli. Biological studies reveal that the compound is an effective inhibitor that is able to suppress FtsZ polymerization and GTPase activity and thus stopping the cell division and causing cell death. More importantly, this series of compounds shows low cytotoxicity on mammalian cells and therefore they could be new chemotypes for the development of new antibacterial agents targeting the cell-division protein FtsZ.
The use of antibiotics to treat infectious diseases is a common and very effective practice in the past decades (Brown, 2015). However, because of our past misuse and abuse of antibiotics, multi-drug resistant bacteria, such as methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus faecium (VREF), pose the greatest threat to human health (Levy and Marshall, 2004; Fischbach and Walsh, 2009; Willyard, 2017). The World Health Organization (WHO) reported that antibacterial resistance is one of the biggest health problems and the world is running out of antibiotics (WHO, 2017). Bacteria can be intrinsically resistant to antibiotics or develop resistance rapidly after exposure to antibiotics (Blair et al., 2015). The mechanisms of antibiotic resistance are generally developed through the following pathways: (i) bacteria producing specific enzyme to inactivate antibacterial drugs (Lin et al., 2015); (ii) modifying drug targets to render antibiotics ineffective (Walsh, 2003; Mi et al., 2016); (iii) changing the permeability of the outer membrane to prevent drugs entering the cell body, affecting the efflux system to pump out the drugs (Walsh, 2003; Nikaido and Pagès, 2012; Wang et al., 2018). Therefore, new generation of antibacterial agents that act through novel mechanisms and/or on novel targets are needed (Payne, 2008; Rice, 2008).
It has been recognized that new drug targets of antibacterial agents is possibly obtained through the study of bacterial cell division machinery (Margalit et al., 2004). While in the bacterial cell-division cycle, filamenting temperature-sensitive mutant Z (FtsZ) is targeted as one of the most important proteins. Because it undergoes GTP-dependent polymerization at mid-cell and recruits the other cell division proteins to complete the division process (Bi and Lutkenhaus, 1991; Lutkenhaus, 1993). In addition, the characteristics of FtsZ proteins are conserved in most bacteria (Lock and Harry, 2008; Kapoor and Panda, 2009).
Over the past few years, some FtsZ-targeting compounds were reported to inhibit bacterial cell division through disrupting GTPase activity and polymerization of FtsZ (Domadia et al., 2007; Haydon et al., 2008; Ruiz-Avila et al., 2013; Stokes et al., 2014; Haranahalli et al., 2016; Liu et al., 2018). These studies suggested that the molecules impair bacterial growth through disrupting the dynamic assembly and GTP hydrolysis of FtsZ. These inhibitors can be divided into natural products and chemically synthesized small molecules. Natural products mainly include curcumin, berberine, totarol and cinnamaldehyde (Domadia et al., 2007; Jaiswal et al., 2007; Domadia et al., 2008; Kaur et al., 2010). The synthesized small molecules mainly include 3-MBA derivatives, rhodanine derivatives and taxane derivatives (Haydon et al., 2008; Singh et al., 2012, 2014; Bi et al., 2018). Recently, we successfully screened some FtsZ inhibitors with strong antibacterial effects including berberine derivatives (Sun et al., 2014), quindolinium derivatives (Sun et al., 2017a), pyrimidine derivatives (Chan et al., 2017), and tiplaxtinin (Sun et al., 2017c), and found that molecular structures bearing a benzothiazole or quinolinium scaffold is commonly able to inhibit the bacterial cell division and disrupt FtsZ function. In this study, on the basis of our understanding on the benzothiazole and quinolinium molecular frameworks, we attempted to further study benzothiazole-substituted benzofuroquinolinium derivatives 1–5 (Figure 1) to search for FstZ inhibitors with potent antimicrobial activity. These compounds were published as G-quadruplex fluorescent probes (Lu et al., 2015), but their binding affinities were low. However, our previous reports about FtsZ-targeting quinolinium derivatives (Sun et al., 2017a, 2018) encourage us to investigate the antibacterial activities and their mode of action of these benzofuroquinolinium derivatives 1–5. The results obtained reveal that these derivatives can effectively inhibit the GTPase activity and polymerization of FtsZ. The compounds also exhibit cell division inhibition phenomenon with a promising antibacterial activity against bacterial strains including the drug-resistant strains.
The minimum inhibitory concentrations (MICs) of the test compounds were determined using a broth microdilution method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (Wikler et al., 2009). A single colony of test bacterial strain was inoculated from TSB agar plate to 5 mL Mueller Hinton broth (MHB) or Cation-adjusted Mueller hinton broth (CA-MHB) and then overnight incubated at 37°C. Cells were diluted to a final concentration of approximately 5 × 105 CFU/mL in a 96-well microtiter plate. The compounds were then added at a series concentration (0.125, 0.25, 0.5, 1, 2, 4, 8, 16, 32, 64 μg/mL) and the plate was incubated at 37°C for 18 h. The MICs are determined as the minimum concentration of the visually clear wells. 10 μl of broth cultures from the MIC assays were incubated overnight on the MH agar at 37°C. The minimum bactericidal concentration (MBC) was reported as the lowest concentration at which colonies were not observed (Wikler et al., 2009). Three independent assays were performed for all the tests.
Staphylococcus aureus ATCC 29213 and Escherichia coli ATCC 25922 were diluted to approximately 1 × 105 CFU.mL-1 with MHB, various concentrations of compounds were added, and the cultures were incubated at 37°C (Wikler et al., 2009; Sun et al., 2017a). Hundred microliter samples were removed and diluted appropriate times and then spread on to MH agar at 1, 2, 4, 6, 8, 24 h. Agar plates were incubated at 37°C for 24 h to count the cell colonies (CFU.ml-1). Three independent assays were performed for all the tests.
Synergistic effect of these compounds with methicillin against S. aureus ATCC BAA41 (MRSA) was tested using a checkerboard method by following the previous reported procedures (Pillai et al., 2005). In addition, we also detected the growing curve of S. aureus ATCC BAA41 cultured with or without our compounds with methicillin. Cells were cultured overnight at LB broth and then diluted to 1 × 105 CFU/mL with CA-MHB (Liu et al., 2000; Wang et al., 2017). Compounds and methicillin at a different combination concentration were added and cells were then incubated at 37°C. OD600 of the mixtures were measured using a UV-visible spectrophotometer at 1, 2, 4, 6, 8, 24 h. FIC index = MICA/A + MICB/B, A and B stand for MIC when acting alone. FIC index: <0.5, two drugs for synergy; 0.5–1 for the additive effect; 1–2 is irrelevant; >2 for the antagonistic effect. Three independent assays were performed for all the tests.
The B. subtilis 168 cells or E. coli 25922 were overnight incubated at 37°C in 5 mL Luria-Bertani (LB) broth (Sun et al., 2017b). Then the cells were diluted to 1 × 105 CFU/mL, treated with different concentrations of compounds at 37°C for 4 h. For membrane staining, the B. subtilis cells were further incubated with 1.6 μM of FM 4–64 for an additional 30 min at 37°C without shaking before harvested. The bacterial cells were then harvested and re-suspended in PBS containing 0.25% agarose. Then the sample mixtures were placed on a microscopic slide pre-treated with 0.1% (w/v) poly-L-lysine. The morphology of the bacterial cells was observed using an Olympus FSX100 microscope at 400× magnification.
The assay using a strain of B. subtilis 168 containing an IPTG-inducible plasmid, it can produce the green fluorescence protein (GFP)-tagged FtsZ to make the Z-ring visible (Sun et al., 2014). A Single colonies of the B. subtilis was overnight incubated at 37°C in 5 mL LB broth, and then were diluted to 1 × 105 CFU/mL with MHB containing 40 μM IPTG (Isopropyl β-D-1-thiogalactopyranoside). The cells were treated with the test compounds at the MIC concentration for 4 h at 37°C. After that they were centrifuged and re-suspended in PBS and then observed using the Nikon N-SIM Super-resolution Imaging System.
Escherichia coli FtsZ was cloned, overexpressed, and purified as previous report (Li and Ma, 2015). The GTPase activity of E. coli FtsZ was measured by using an ATPase/GTPase Activity Assay Kit (SIGMA MAK113) according to an optimized protocol and the Kit instructions (Sun et al., 2014). The E. coli FtsZ protein was diluted at a final concentration of 10 μm in MOPS-KOH (50 mM MOPS, 50 mM KCl, 5 mM MgCl2, pH 6.5). The Assay Reaction Mixes contains of 20 μL Assay Buffer, 5 μL protein and 5 μL DMSO/compounds (0.5, 1, 2, 4, 8, 16, 32, 64 μg/mL). Mix Assay Buffer with 4 mM GTP in a 2:1 ratio to set up the Reaction Mix. Add 30 μL of Reaction Mix to each sample, background blank, and negative control well. Incubate the reaction for 30 min at room temperature. Then, 200 μL of reagents were added to each well and the plates were incubated for an additional 30 min at room temperature to terminate the enzyme reaction and generate the colorimetric product. The absorbance was read at 620nm for all samples, standards, blanks, and controls. The phosphorus standard curve has been measured and shown at Supplementary Table S2, the standard curve equation is y = 0.0092x + 0.289 and R2 = 0.996. Each assay was carried out in triplicates.
Escherichia coli FtsZ protein was diluted at a final concentration of 10 μm in MOPS-KOH (50 mM MOPS, 50 mM KCl, 5 mM MgCl2, pH 6.5). Compound 5 was added to the protein at a series of concentrations (8, 16, 32, 64 μg/mL) and DMSO was used as negative control. Then the reaction was initiated by adding 1mM GTP and the mixtures were incubated at 25°C for 1 h. Then the samples were centrifuged at 14,000x rpm for 60 min, pellets were re-suspended in MOPS-KOH and then analyzed by 12% SDS-PAGE gel. Gels were stained with Instant Blue and the protein content of binding bands were measured by densitometric quantification using Image J software (Mukherjee et al., 2001; Margalit et al., 2004; Andreu et al., 2010). Each assay was carried out in triplicates.
The drug resistance study of compound 1–5 was performed by following the protocol of previous study (Lin et al., 2017). The initial MIC values of Compound 1–5 and methicillin against S. aureus ATCC 29213 were determined according to method described above. The bacteria in the 0.5× MIC wells from the above experiment were diluted to approximately 1× 105 CFU.mL-1 with MHB for the next MIC test. Various concentrations of compounds were added to the corresponding bacteria, the MICs are determined after incubated 24 h at 37°C. The process was repeated continuously for 25 days, assays were carried out with biological replicates.
Molecular modeling was carried out by CDOCKER of Discovery Studio 2016, the X-ray crystal structure of the S. aureus FtsZ bound with PC190723 (PDB code: 4DXD) and GDP was downloaded from the RCSB Protein Data Bank1. The ligands and water molecules were removed, and the protein was prepared for docking using an automated protein preparation protocol.
Compounds 1–5 were tested with Mouse fibroblasts cells (L929), renal epithelial cells (HK-2) and human hepatocellular liver carcinoma cells (HepG2) by MTT method to determine the cytotoxicity of eukaryotic mammalian cells. Cells are resuscitated to log phase, then collected and diluted with cell culture medium to about 8000 cells per mL. For the MTT assay, cells were 100 μL per well seeded in a 96-wells microplate and incubated at 37°C about 16 h until cells adhere at the bottom of wells. Then cells were treated with compounds for 24 h, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) were added into cells at a final concentration of 0.5 mg/mL for another 4 h to form formazan, then the formazan were dissolved with DMSO and the concentration was measured by reading OD490 with a multi-function microporous plate detector (Sun et al., 2017b). Each assay was carried out in triplicates.
Compounds 1–5 were tested with Human erythrocytes (Hongquan bio-tel, GZ) at a concentration of 2% as previous report described (Zheng et al., 2017). The assay volumes are 200 μL. Different concentrations of compounds were added 100 μL per well to the 96-well plate, then each well added 100 μL of human blood. After incubated 1 h at 37°C, the plate was centrifuged at 500 × g for 5 min, then 100 μL of the supernatant were to determine the absorbance at 540 nm. Each assay was carried out in triplicates.
The test compounds were synthesized according to the previous reported procedures (Lu et al., 2015). Their structures and purity were confirmed by NMR and ESI-MS (refer to Supplementary Materials).
The antibacterial activity of the compounds was evaluated, and the results show that all compounds possess potent antibacterial activity against the tested strains (Table 1). For Gram-positive bacteria, the compounds inhibited the growth of antibiotic-susceptible S. aureus and methicillin-resistant S. aureus (MRSA) with minimum inhibitory concentrations (MIC) in the range of 0.25–8.0 μg/mL (0.48–16.17 μM). Among the compounds, 5 containing an extended N-isopropyl-N-propylpropan-1-amine group from the benzothiazole moiety was the most effective one against MRSA with MIC values ranged from 0.5–1.0 μg/mL (0.96–1.91 μM). Its antibacterial activity is over 200-fold better than methicillin. In addition, the growth of vancomycin-resistant E. faecium (VRE) was inhibited by 5 with an MIC value of 1 μg/mL (1.91 μM). The antibacterial effects of 5 on the VRE was much more potent than that of vancomycin (MIC > 64 μg/mL) (Sun et al., 2017a). For Gram-negative bacteria, the compounds can inhibit the growth of E. coli and its antibiotics resistant mutant with MIC values of 1–8 μg/mL (1.91–15.73 μM). When tested with NDM-1 E. coli, methicillin shows just little effect even at the concentration MIC > 64 μg/mL (>159.05 μM). It is noteworthy that 5 can effectively inhibit NDM-1 producing E. coli with a much lower MIC value of 1 μg/mL (1.91 μM). In addition, Acinetobacter baumannii, Pseudomonas aeruginosa, and Klebsiella pneumoniae, which are resistant to most of the clinical antibiotics can be inhibited by our compounds with MIC values of 4–64 μg/mL (7.65–125.82 μM). Compound 5 can act effectively against these multi-drug resistant Gram-negative strains with MIC values of 4–8 μg/mL (7.65–15.30 μM) (Table 1). The minimum bactericidal concentration (MBC) of 5 is 1 μg/mL (1.91 μM) against E. coli ATCC 25922 and S. aureus ATCC 29213, suggesting that the antibacterial activity of our compounds with the same mode of bactericidal effect.
Time killing curves were determined by counting bacterial colonies at different time points to determine whether the compounds were bactericidal or not. The results of time killing curves from 5 against E. coli ATCC 25922 and S. aureus ATCC 29213 are shown in Figure 2. The results show that the growth of E. coli ATCC 25922 and S. aureus ATCC 29213 can be completely inhibited at 1x MIC and at 4x MIC respectively.
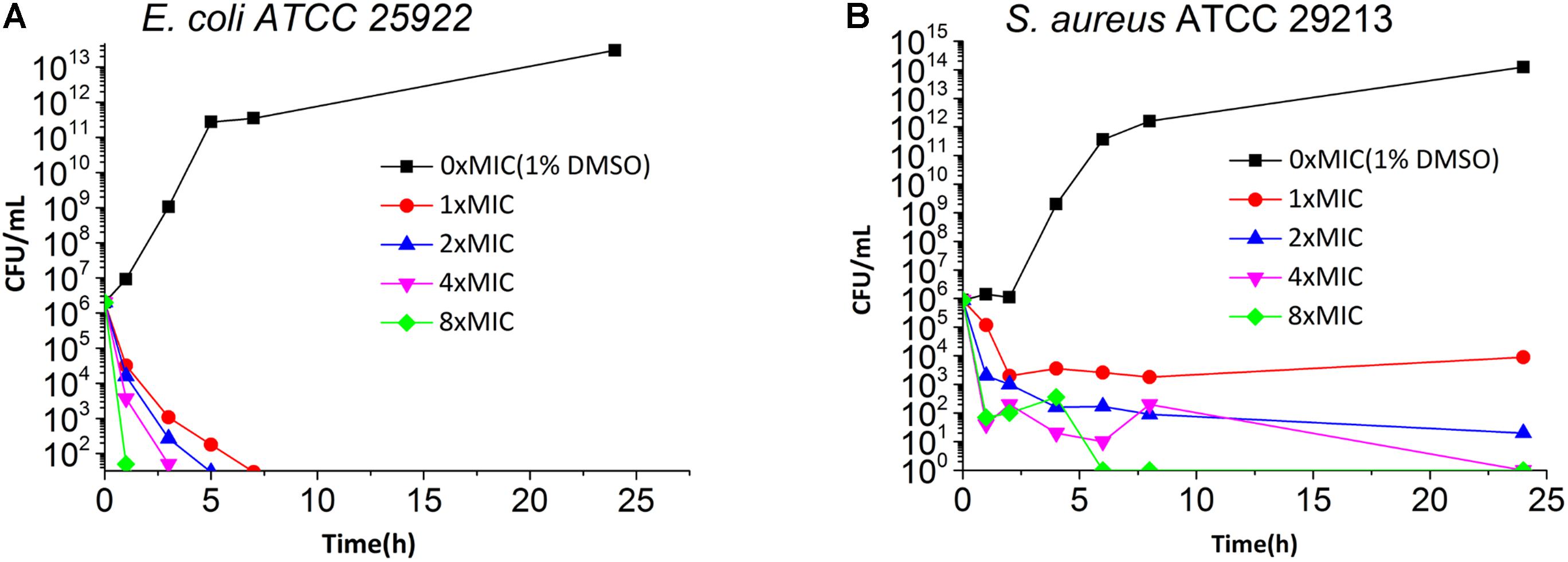
FIGURE 2. Time Killing Curve of 5 against E. coli ATCC 25922 (A) and S. aureus ATCC 29213 (B). The different concentration of 5 was represented by different colors, 0× MIC (1% DMSO) (black), 1× MIC (red), 2× MIC (blue), 4× MIC (pink) and 8× MIC (green).
To observe whether the compounds can restore the antibacterial activity of methicillin, an MRSA strain (S. aureus ATCC BAA41) was treated with methicillin combined with compounds 1–5. The MIC of methicillin against BAA41 is 1024 μg/mL (2544.75 μM), but in the combination test with 5 at the concentration of 25% MIC (0.25 μg/mL) value or with other compounds at the concentration of 50% MIC value, it can be reduced to 256 μg/mL (1272.37 μM) (Figure 3, Supplementary Figure S1, and Supplementary Table S2). The results of checkerboard test (Supplementary Table S2) and growing curve (Figure 3 and Supplementary Figure S1) suggest that the combination of the compounds with methicillin displays synergistic or partial synergistic effect against MRSA, since the FIC index is 0.5 for 5 with methicillin, and FIC index is 1 for other compounds with methicillin (Sung and Lee, 2008).
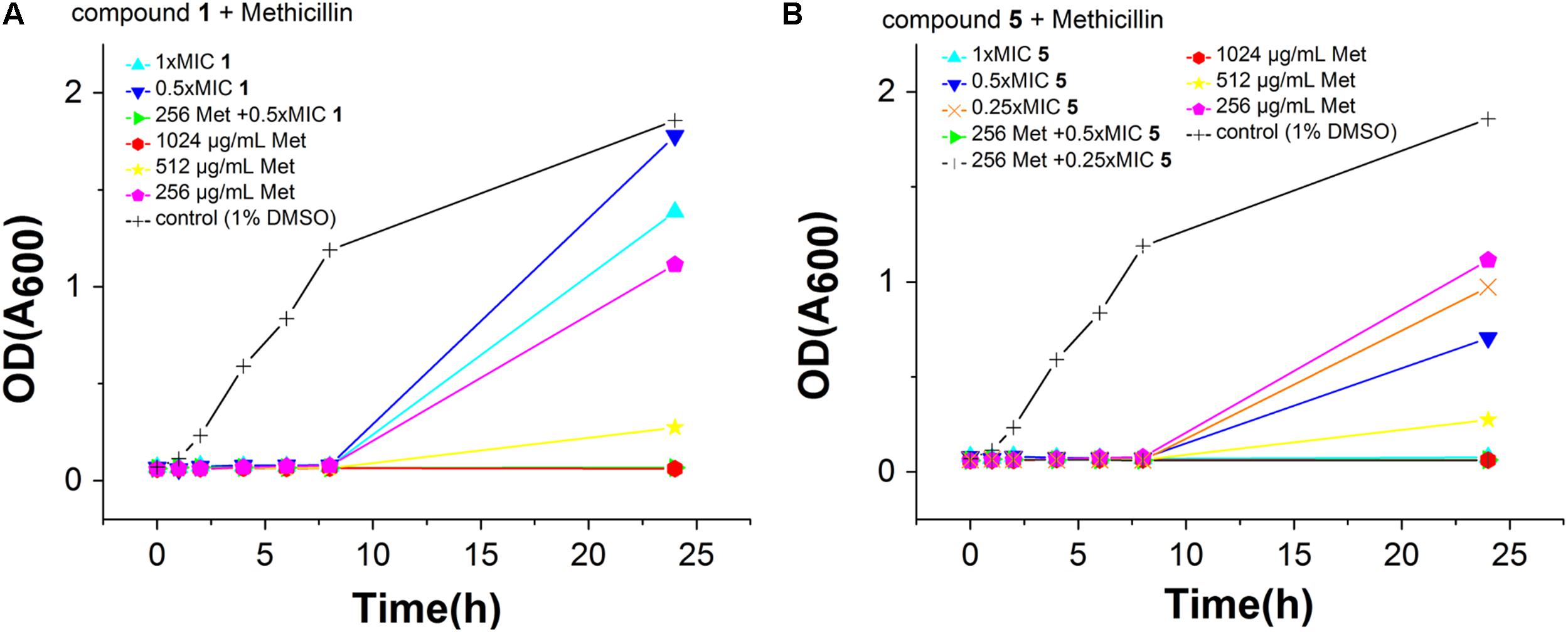
FIGURE 3. Effects of Compound 2 (A) and 5 (B) combination with methicillin against S. aureus ATCC BAA41. The different concentration of compounds was represented by different colors, control (1% DMSO) (black), 1× MIC (cyan), 0.5× MIC (blue) and 0.25× MIC (orange). The different concentration of methicillin was represented by different colors, 1024 μg/mL (red), 512 μg/mL (yellow), and 256 μg/mL (pink). In addition, green and dark gray represent the effect of methicillin with 0.5 or 0.25× MIC of compound, respectively.
FtsZ targeting compounds can disturb FtsZ function and lead to bacterial cell division inhibition. In order to further investigate the antibacterial mechanism of our compounds, we observed the cell morphological effect of the compounds against B. subtilis 168 and E. coli 25922 cells. The results show that compounds 1–5 can both significantly elongate B. subtilis and E. coli 25922 cells compared to the control (Figure 4 and Supplementary Figures S2, S3). For example, the normal cell length of B. subtilis cells is around 5–10 μm (Figure 4B). After incubation with 5 at the MIC concentration, the cell length increased was longer than 20 μm (Figure 4A). A similar situation happened to E. coli 25922, the normal cell length is only 2–4 μm while the treated cells are longer than 12 μm. The cell length increased about 4–6 times compared to the control cells. These phenomena suggest that the benzofuroquinolinium derivatives inhibit the growth of bacteria through a mechanism of cell division inhibition. The histogram of cell length refers to the Supplementary Figure S4. Since perturbation of membrane structure can also lead to bacterial cell death, the effect of benzofuroquinolinium derivatives on the bacterial cell membrane was further investigated using a red fluorescent dye FM4-64 on B. subtilis cells. In spite of an increased length of B. subtilis cells (Supplementary Figure S5), these compounds did not induce any detectable perturbation of the cell membrane, as compared to untreated cells (Supplementary Figure S5). These results suggest that these benzofuroquinolinium derivatives inhibit bacterial proliferation by inducing cell elongation without perturbing the bacterial membrane.
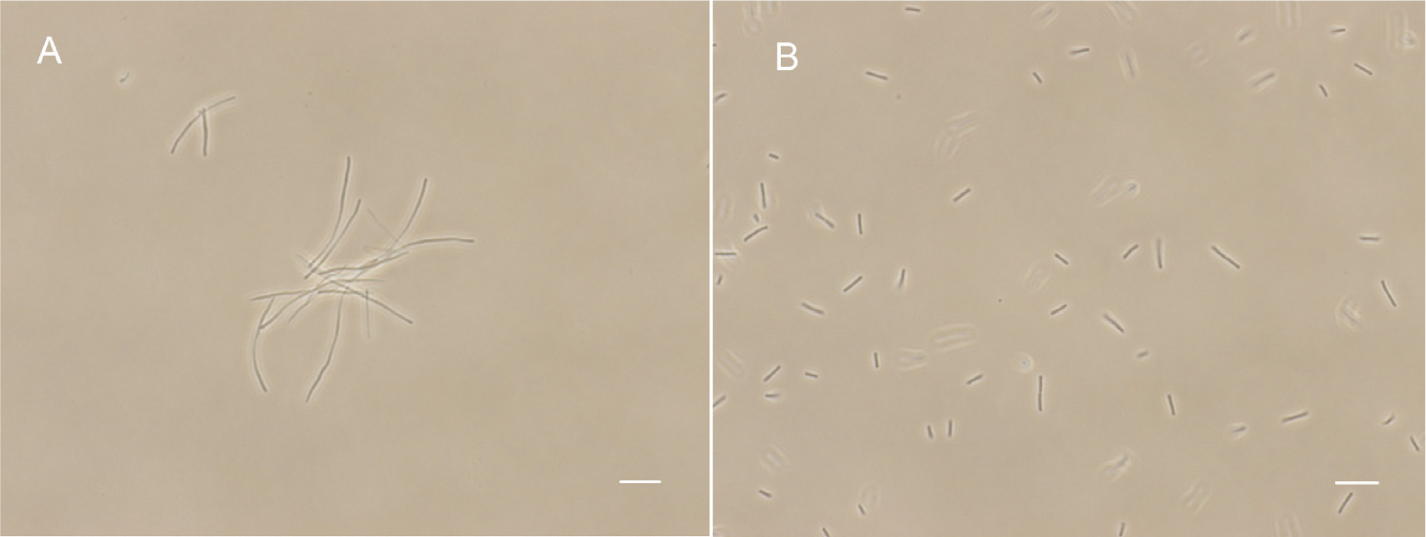
FIGURE 4. The effect of 5 on B. subtilis 168 morphology. Cells of B. subtilis 168 were grown with 0.25 μg/mL of compound 5 (A), and without compound (B). The scale bar is 10 μm.
Previous studies have shown that some FtsZ inhibitors can perturb FtsZ function to inhibit the formation of Z-ring (Domadia et al., 2007; Haydon et al., 2008; Keffer et al., 2013). The results show that there is a green fluorescent band at the middle of the cell and it becomes some green points at the present of test compounds (Figure 5). Both compound 2 and 5 can affect the Z-ring assembly to inhibit cell division and result in cell elongation, probably indicating FtsZ is a target of the benzofuroquinolinium derivatives.
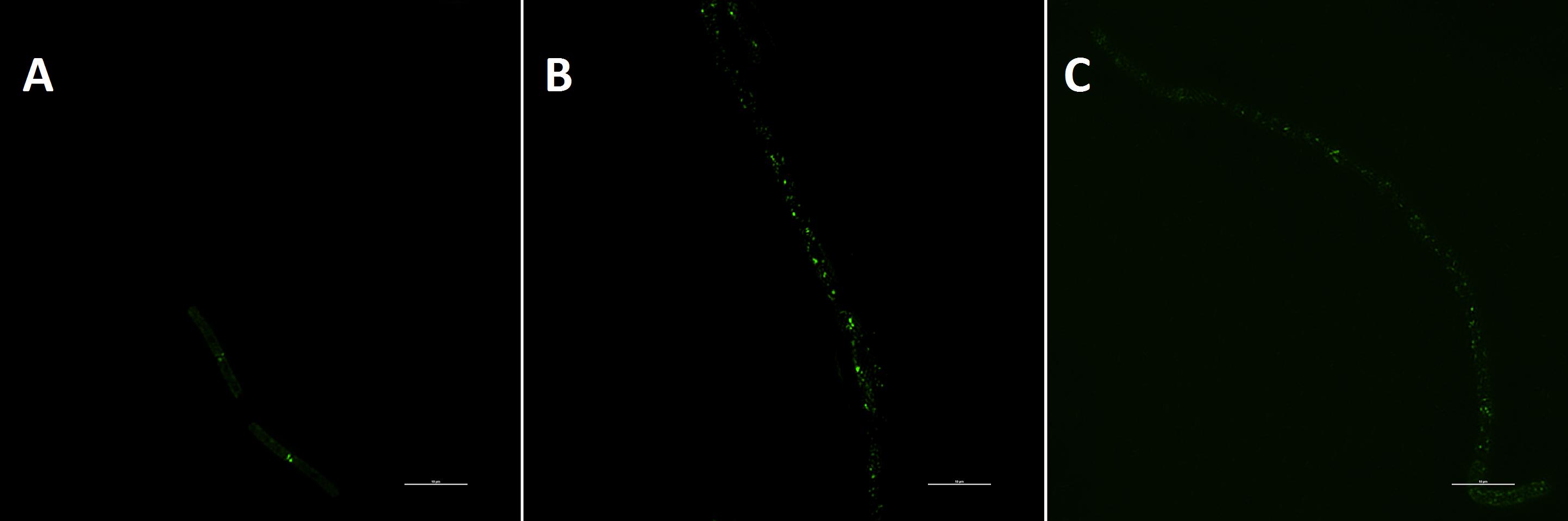
FIGURE 5. The visualization of Z-ring in bacterial cells. Cells of B. subtilis 168 were respectively grown with 1% DMSO (A), 1 μg/mL of compound 2 (B) and 0.25 μg/mL of compound 5 (C). The scale bar is 10 μm.
To study the interaction between FtsZ and the compounds, E. coli FtsZ was cloned, overexpressed, and purified as previous report (Li and Ma, 2015). A GTPase activity assay kit was used to assess the effects of our compounds on the GTPase activity of E. coli FtsZ protein. The results showed that all compounds inhibited the GTPase activity of FtsZ protein in a dose dependent manner (Figure 6A and Supplementary Figure S6). Among the compounds tested for GTPase inhibitory efficacy comparison, 5 is the most effective one. Typically, 5 at a concentration of 64 μg/mL was able to achieve 70% inhibition while other compounds were showing 60% inhibition or less.
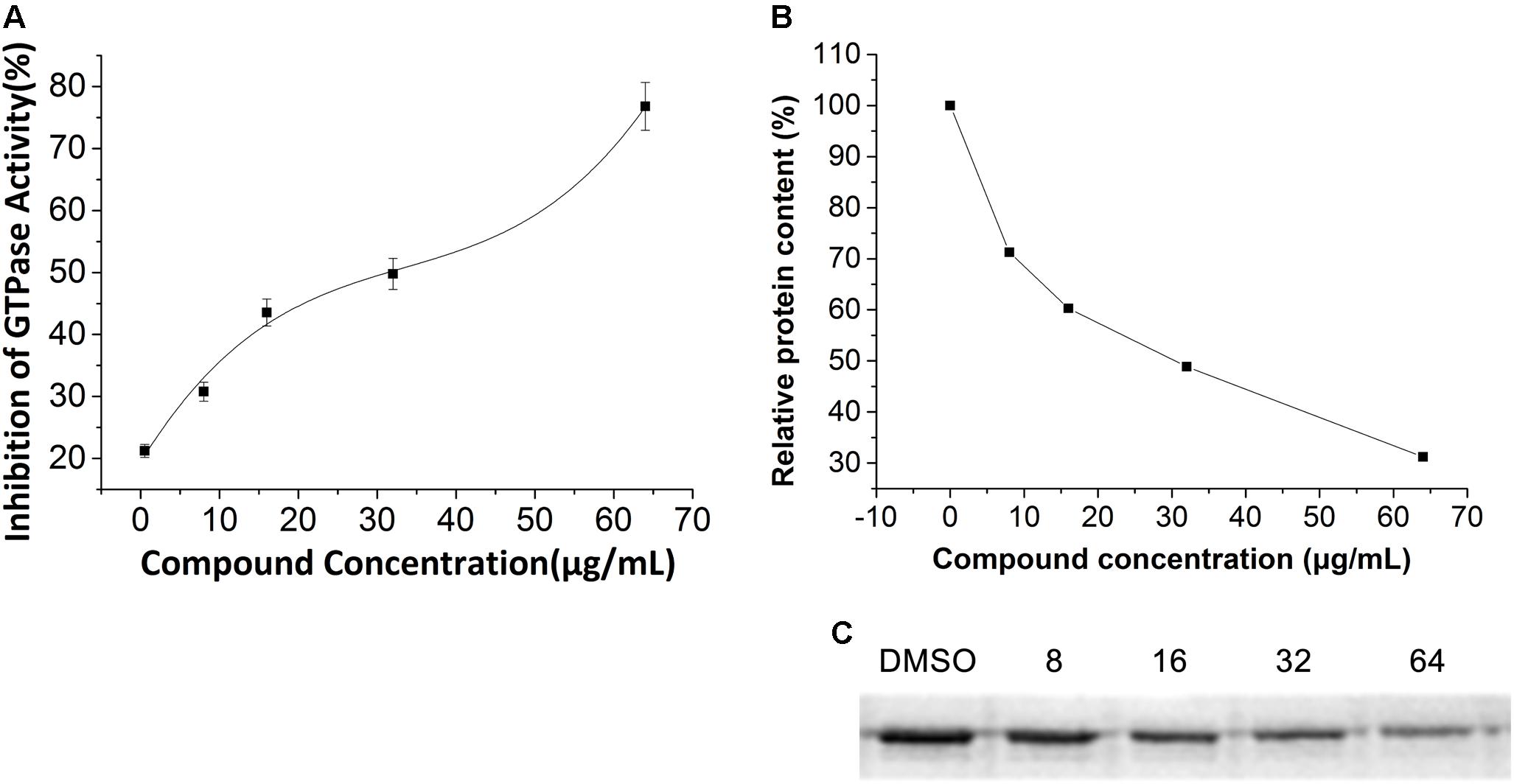
FIGURE 6. Effect of 5 on polymerization and GTPase Activity of E. coli FtsZ. (A) Protein were treated with compound 5 at a series concentration of 1, 8, 16, 32, 64 μg/mL. (B) Relative protein content of the pellet of the polymerization assay. Optical density analysis relative protein precipitation results by Image J. (C) SDS-PAGE of protein pellet after treated with vehicle (5% DMSO) or 8, 16, 32, 64 μg/mL compound 5. The concentration of protein is 0.4 mg/mL.
The influence of 5 on the polymerization of the protein was visualized by 12% SDS-PAGE. As shown in Figures 6B,C, the precipitation of E. coli FtsZ decreased when increasing the concentration of 5, meaning that the inhibition of protein polymerization is in a dose dependent manner.
The results of compounds 1–5 on drug resistance study suggested that these compounds have a low spontaneous frequency of resistance on S. aureus ATCC 29213. As shown in Figure 7, S. aureus ATCC 29213 did not produce resistant mutants at sublethal concentrations of 5 over a period of 25 days, and the MIC values of 5 did not increase more than 4-fold after 25 passages. Similar results can be found on the compounds 1–4 (data no shown). By contrast, the MIC value of Methicillin against S. aureus ATCC 29213 increased 8-fold after 11 passages, indicating the development of resistant mutants. These results indicate that compound 1–5 can kill bacteria effectively and avoid the development of drug resistance. So far, no drug-resistant mutants have been reported in the literature about the FtsZ inhibitors which targeting the GTP binding site, such as chrysophaentin A and pyrimidine derivatives (Plaza et al., 2010; Chan et al., 2017). The possible reason may be due to the importance for recognizing the GTP molecules of this binding site. Mutation of amino acid at this binding site may lead to improper recognition of GTP and hamper normal FtsZ polymerization. Based on these results, we therefore select GTP binding site to predict the possible binding poses of the compounds in the FtsZ in the next test.
There are two binding site in the FtsZ protein. One is located in the C-terminal interdomain cleft. Most of FtsZ inhibitors, such as PC 190723 and benzamides derivatives, which were found to stablize FtsZ polymerization, are reported to interact with FtsZ via binding into this area. Another pocket is GTP-binding site, compounds predicted to bind into this pocket were found to inhibit the GTPase activity and FtsZ polymerization. Since the features are in accordance with our benzofuroquinolinium derivatives, we carried out the molecular modeling studyusing GTP-binding site of FtsZ. The docking process generated several docking poses. The results revealed that compounds 4 and 5 which containing linear terminal amine group on their side chains have higher scores than that of compounds containing cyclic amine group (Supplementary Table S4). On the other hand, the top-score docking poses of 4 and 5 revealed that 5 can found more interactions with FtsZ protein (Figure 8 and Supplementary Figure S7), which are in accordance with the biological actives of these compounds. The top-score docking pose indicates that 5 binds to the FtsZ via interacting with several residues around the GTP-binding pocket (Figure 8). The figure shows that there are 8 amino acids involved in the main interactions with 5. Ala 26, Leu 169, Phe 183 and Ala 186 of FtsZ protein are interact with dipropyl group of 5 through alkyl or Pi-alkyl hydrophobic interactions. Gly 22 can found a conventional hydrogen bond with dipropylamine group of 5. In addition, Gly 21 and Asp 46 can found non-classical hydrogen bonds with benzofuroquinolinium core of 5. Moreover, Gly 21 can also interact with 5 through Pi-sigma and amide-Pi stacked interactions. On the other hand, the cationic quinolinium of 5 is predicted to participate in the charge interaction with the negative charged side chain of Asp 46. Last but not least, Glu 139 can interact with the benzene ring of 5 through a Pi-anion interaction (Figure 8B).
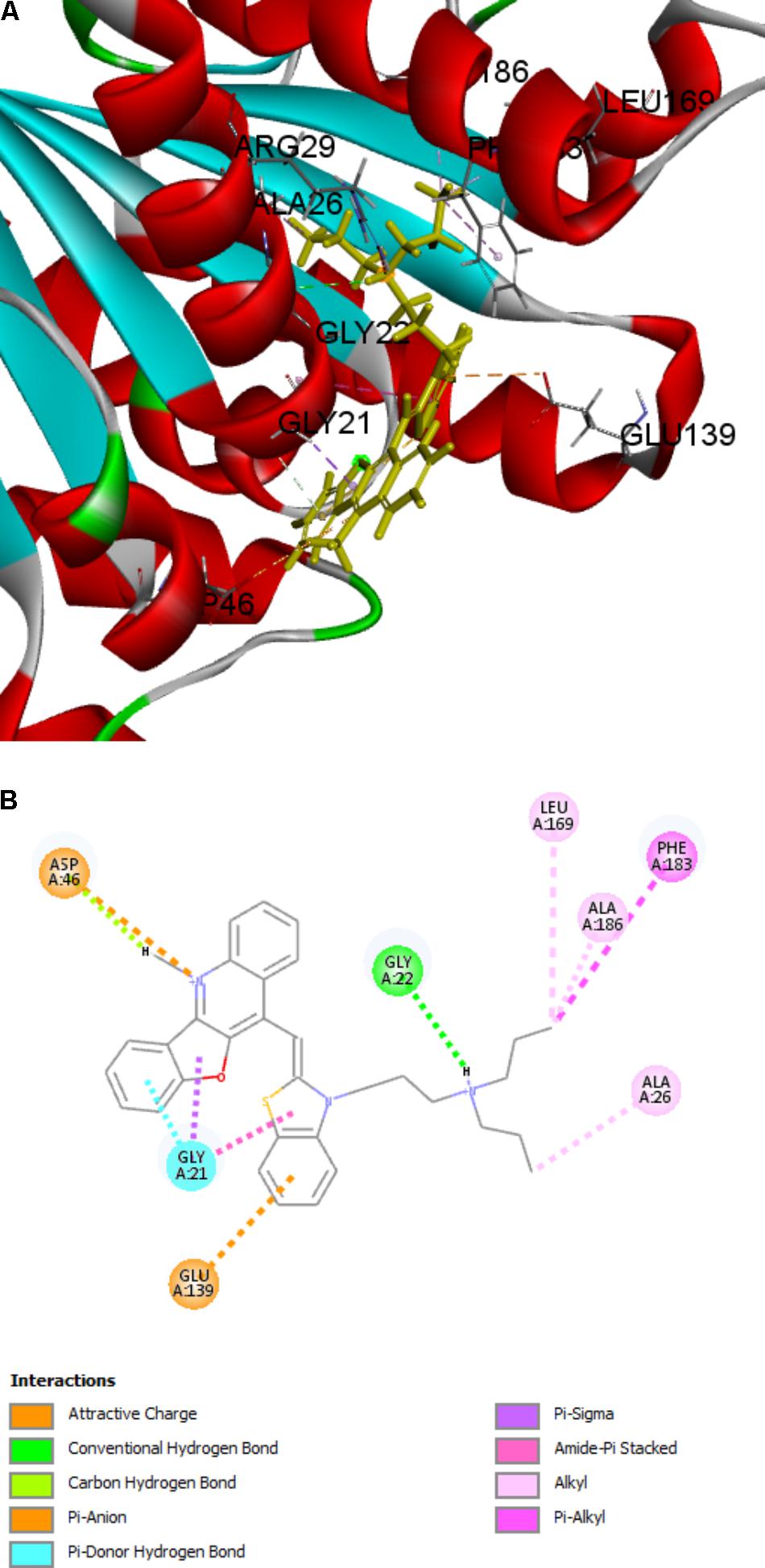
FIGURE 8. Predicted binding modes of compound 5 bound to FtsZ. (A) Compound 5 bound to the GTP-binding site of FtsZ (PDB: 4DXD); (B) Predicted interaction between compound 5 and amino acids of FtsZ.
Since FtsZ is a homologue of eukaryotic tubulin, we further tested the toxicity of the compounds to mammalian cells. Two types of normal cells (L929 and HK-2) and one cancer cell line (HepG2) were examined and the inhibition rates are shown in Table 2. We treated the cells with 50 μg/mL (>20-fold of MIC against MRSA) of our compounds and found that the cell inhibition rates are about 50–65%, indicating that compounds may have no significant cytotoxicity on mammalian cells.
To investigate the toxicity of the compounds to human erythrocytes, we tested the hemolytic activity of compounds 1–5 using a human red cell lysis assay. The results showed that red blood cells treated with the detergent Triton X-100 were almost completely hemolyzed. On the other hand, 64 μg/mL of our compounds did not show significant hemolysis (HC50 > 64 μg/mL) (Figure 9). When compared with antibacterial activity of these compounds against MRSA, all the selectivity indices (HC50/MIC) are higher than 32 (Table 3), suggesting that our compounds do not have toxicity to human erythrocytes.
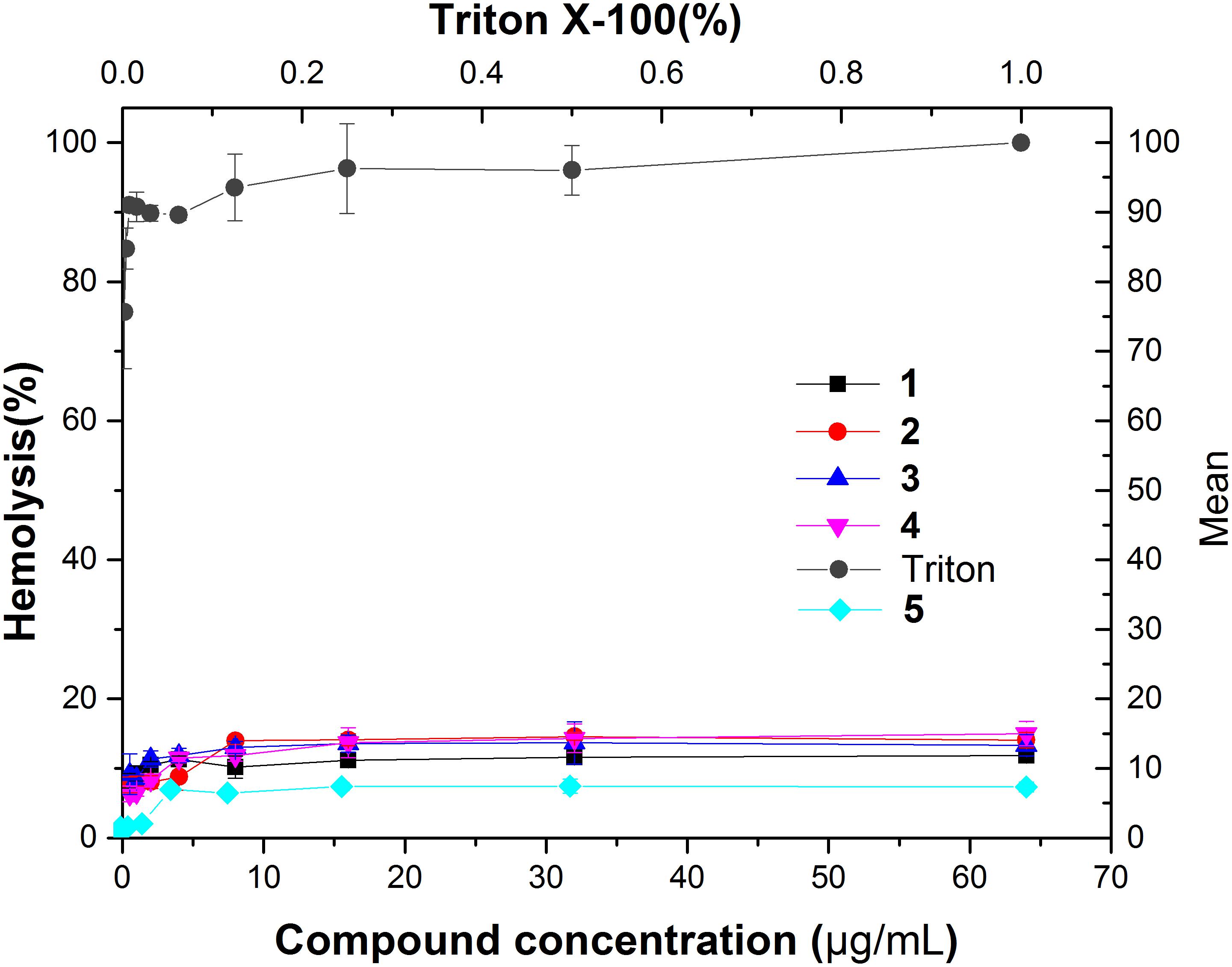
FIGURE 9. Hemolytic activity of compound 1–5. Human erythrocytes were treated with serial dilutions of Triton X-100 (0.002–1%) or compounds 1–5 (0.125–64 g/ml).
From the above studies, compounds 1–5 showed different degree of potency and a broad spectrum antibacterial activity. The bacterial experiments showed that the compounds restored the antibacterial activity of methicillin and elongate the B. subtilis 168 by inhibiting the cell division process. In the previous study, some FtsZ inhibitors such as quinoline derivatives, benzamide derivatives and 9-phenoxyalkylberberine derivatives also gave similar cell elongation effects. These phenomena encouraged us to further investigate the mode of action of benzofuroquinolinium derivatives targeting FtsZ. It was found that these compounds can inhibit the polymerization and decreased the GTPase activity of FtsZ in vitro. Moreover, the cytotoxicity data of 1–5 against two normal mammalian cells revealed that these compounds were hypotoxicity.
Among these tested compounds, 5 shows remarkable inhibition against Gram-positive strains (including MRSA and VRE) with MIC values from 0.25–4 μg/mL (0.48–7.65 μM). It is as good as or even better than some reported FtsZ inhibitors with high potency. For example, the MIC of 9-phenoxyalkylberberine derivative 2 against MRSA and VREF is from 2 to 8 μg/mL (Sun et al., 2014). The cinnamaldehyde derivatives exhibited MIC values of 4 to 64 μg/mL against MRSA (Li and Ma, 2015). Besides, 5 can strongly inhibit the Gram-negative strains with the MIC values from 1 to 8 μg/mL (1.91–15.30 μM). On the other hand, compounds 9-phenoxyalkylberberine derivative 2 and quinuclidine 1 were reported as FtsZ inhibitors with the MIC value of 32 μg/mL against E. coli., and cinnamaldehyde derivatives inhibited the growth of E. coli with MIC value of 128 μg/mL (Li and Ma, 2015). The antibacterial activities of 5 is much stronger compared to 9-phenoxyalkylberberine derivatives, quinuclidine derivative, and cinamaldehyde derivatives, suggesting that 5 possesses a better ability to pass the outer membrane of Gram-negative bacteria. On the other hand, it was reported that GTPase activity of FtsZ can affect its assembly and most of the FtsZ inhibitors can restraint the GTPase activity (Margalit et al., 2004; Margolin, 2005; Chan et al., 2013; Sun et al., 2014; Hurley et al., 2016). Compound 5 can inhibit 50% inhibition of GTP hydrolysis at the concentration of 32 μg/mL (∼61.22 μM). From the above biological assays, 5 is targeting FtsZ with a strong antibacterial activity.
In conclusion, we demonstrated that the class of benzofuroquinolinium derivatives exhibit potent antibacterial activities against Gram-positive, Gram-negative, and even drug resistant bacteria. Furthermore, the compounds can restore the antibacterial activity of methicillin and perturb the Z-ring assembly. The mode of action of the compound was also studied through biochemical evaluations. The results indicate that compound 5 can effectively inhibit the polymerization and GTPase activity of FtsZ in a dose-dependent manner. In addition, the compound probably interacts with the GTP-binding site and disrupts the GTPase activity and polymerization of FtsZ, which cause abnormal bacterial cell division resulting cell death. More importantly, the benzothiazole-substituted benzofuroquinolinium derivatives were shwoing low cytotoxicity on mammalian cells. Therefore, the class of compounds could be developed as the potent antibacterial agents against drug resistant bacteria.
NS, YJ-L, and KY-W conceived and designed the experiments. NS, Y-YZ, RL-D, SY-C, and Z-YF performed the experiments. NS, Y-JL, Y-YZ, and R-LD analyzed the data. L-YS, TL, and Z-HL contributed reagents, materials, and analysis tools. NS, Y-JL, and K-YW wrote the paper.
We acknowledge the support from the National Natural Science Foundation of China (81473082 and 81703333), Natural Science Foundation of Guangdong Province, China (2017A030313078), and Science and Technology Program of Guangdong Province (2016A020209009, 2016B020211002, and 510093637143). We are also grateful to the support from Nanshan Scholar Program of Guangzhou Medical University (B185006006008) and Innovation and Technology Commission of Hong Kong, The Hong Kong Polytechnic University. This study was also supported by the Open Project of Guangdong Provincial Key Laboratory of New Drug Screening (GDKLNDS-2018OF005).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Y. F. Yuen and the University Research Facility in Life Sciences, The Hong Kong Polytechnic University provide the technical support on Nikon N-SIM Super-Resolution Imaging System.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01937/full#supplementary-material
Andreu, J. M., Schaffner-Barbero, C., Huecas, S., Alonso, D., Lopez-Rodriguez, M. L., Ruiz-Avila, L. B., et al. (2010). The antibacterial cell division inhibitor PC190723 is an FtsZ polymer-stabilizing agent that induces filament assembly and condensation. J. Biol. Chem. 285, 14239–14246. doi: 10.1074/jbc.M109.094722
Bi, E., and Lutkenhaus, J. (1991). FtsZ ring structure associated with division in Escherichia coli. Nature 354, 161–164. doi: 10.1038/354161a0
Bi, F., Ji, S., Venter, H., Liu, J., Semple, S. J., and Ma, S. (2018). Substitution of terminal amide with 1H-1,2,3-triazole: identification of unexpected class of potent antibacterial agents. Bioorg. Med. Chem. Lett. 28, 884–891. doi: 10.1016/j.bmcl.2018.02.001
Blair, J. M., Webber, M. A., Baylay, A. J., Ogbolu, D. O., and Piddock, L. J. (2015). Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51. doi: 10.1038/nrmicro3380
Brown, D. (2015). Antibiotic resistance breakers: can repurposed drugs fill the antibiotic discovery void? Nat. Rev. Drug Discov. 14, 821–832. doi: 10.1038/nrd4675
Chan, F.-Y., Sun, N., Neves, M. A., Lam, P. C.-H., Chung, W.-H., Wong, L.-K., et al. (2013). Identification of a new class of FtsZ inhibitors by structure-based design and in vitro screening. J. Chem. Inf. Model. 53, 2131–2140. doi: 10.1021/ci400203f
Chan, K. F., Sun, N., Yan, S. C., Wong, I. L. K., Lui, H. K., Cheung, K. C., et al. (2017). Efficient synthesis of amine-linked 2,4,6-trisubstituted pyrimidines as a new class of bacterial FtsZ inhibitors. ACS Omega 2, 7281–7292. doi: 10.1021/acsomega.7b00701
Domadia, P., Swarup, S., Bhunia, A., Sivaraman, J., and Dasgupta, D. (2007). Inhibition of bacterial cell division protein FtsZ by cinnamaldehyde. Biochem. Pharmacol. 74, 831–840. doi: 10.1016/j.bcp.2007.06.029
Domadia, P. N., Bhunia, A., Sivaraman, J., Swarup, S., and Dasgupta, D. (2008). Berberine targets assembly of Escherichia coli cell division protein FtsZ. Biochemistry 47, 3225–3234. doi: 10.1021/bi7018546
Fischbach, M. A., and Walsh, C. T. (2009). Antibiotics for emerging pathogens. Science 325, 1089–1093. doi: 10.1126/science.1176667
Haranahalli, K., Tong, S., and Ojima, I. (2016). Recent advances in the discovery and development of antibacterial agents targeting the cell-division protein FtsZ. Bioorg. Med. Chem. 24, 6354–6369. doi: 10.1016/j.bmc.2016.05.003
Haydon, D. J., Stokes, N. R., Ure, R., Galbraith, G., Bennett, J. M., Brown, D. R., et al. (2008). An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science 321, 1673–1675. doi: 10.1126/science.1159961
Hurley, K. A., Santos, T. M., Nepomuceno, G. M., Huynh, V., Shaw, J. T., and Weibel, D. B. (2016). Targeting the bacterial division protein FtsZ. J. Med. Chem. 59, 6975–6998. doi: 10.1021/acs.jmedchem.5b01098
Jaiswal, R., Beuria, T. K., Mohan, R., Mahajan, S. K., and Panda, D. (2007). Totarol inhibits bacterial cytokinesis by perturbing the assembly dynamics of FtsZ. Biochemistry 46, 4211–4220. doi: 10.1021/bi602573e
Kapoor, S., and Panda, D. (2009). Targeting FtsZ for antibacterial therapy: a promising avenue. Expert Opin. Ther. Targets 13, 1037–1051. doi: 10.1517/14728220903173257
Kaur, S., Modi, N. H., Panda, D., and Roy, N. (2010). Probing the binding site of curcumin in Escherichia coli and Bacillus subtilis FtsZ–a structural insight to unveil antibacterial activity of curcumin. Eur. J. Med. Chem. 45, 4209–4214. doi: 10.1016/j.ejmech.2010.06.015
Keffer, J. L., Huecas, S., Hammill, J. T., Wipf, P., Andreu, J. M., and Bewley, C. A. (2013). Chrysophaentins are competitive inhibitors of FtsZ and inhibit Z-ring formation in live bacteria. Bioorg. Med. Chem. 21, 5673–5678. doi: 10.1016/j.bmc.2013.07.033
Levy, S. B., and Marshall, B. (2004). Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10, S122–S129. doi: 10.1038/nm1145
Li, X., and Ma, S. (2015). Advances in the discovery of novel antimicrobials targeting the assembly of bacterial cell division protein FtsZ. Eur. J. Med. Chem. 95, 1–15. doi: 10.1016/j.ejmech.2015.03.026
Lin, J., Nishino, K., Roberts, M. C., Tolmasky, M., Aminov, R. I., and Zhang, L. (2015). Mechanisms of antibiotic resistance. Front. Microbiol. 6:34. doi: 10.3389/fmicb.2015.00034
Lin, S., Koh, J. J., Aung, T. T., Sin, W. L. W., Lim, F., Wang, L., et al. (2017). Semisynthetic flavone-derived antimicrobials with therapeutic potential against methicillin-resistant Staphylococcus aureus (MRSA). J. Med. Chem. 60, 6152–6165. doi: 10.1021/acs.jmedchem.7b00380
Liu, I. X., Durham, D. G., and Richards, R. M. E. (2000). Baicalin synergy with β-lactam antibiotics against methicillin-resistant Staphylococcus aureus and other β-lactam-resistant strains of S. aureus. J. Pharm. Pharmacol. 52, 361–366. doi: 10.1211/0022357001773922
Liu, J., Ma, R., Bi, F., Zhang, F., Hu, C., Venter, H., et al. (2018). Novel 5-methyl-2-phenylphenanthridium derivatives as FtsZ-targeting antibacterial agents from structural simplification of natural product sanguinarine. Bioorg. Med. Chem. Lett. 28, 1825–1831. doi: 10.1016/j.bmcl.2018.04.015
Lock, R. L., and Harry, E. J. (2008). Cell-division inhibitors: new insights for future antibiotics. Nat. Rev. Drug Discov. 7, 324–338. doi: 10.1038/nrd2510
Lu, Y.-J., Wang, Z.-Y., Hu, D.-P., Deng, Q., Huang, B.-H., Fang, Y.-X., et al. (2015). Benzothiazole-substituted benzofuroquinolinium dyes as new fluorescent probes for G-quadruplex DNA. Dyes Pigm. 122, 94–102. doi: 10.1039/c1cc00020a
Lutkenhaus, J. (1993). FtsZ ring in bacterial cytokinesis. Mol. Microbiol. 9, 403–409. doi: 10.1111/j.1365-2958.1993.tb01701.x
Margalit, D. N., Romberg, L., Mets, R. B., Hebert, A. M., Mitchison, T. J., Kirschner, M. W., et al. (2004). Targeting cell division: small-molecule inhibitors of FtsZ GTPase perturb cytokinetic ring assembly and induce bacterial lethality. Proc. Natl. Acad. Sci. U.S.A. 101, 11821–11826. doi: 10.1073/pnas.0404439101
Margolin, W. (2005). FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell Biol. 6, 862–871. doi: 10.1038/nrm1745
Mi, Y., Ma, R., Li, J., Venter, H., and Ma, S. (2016). Synthesis and antibacterial activity of novel 3- O -descladinosylazithromycin derivatives. Eur. J. Med. Chem. 127, 874–884. doi: 10.1016/j.ejmech.2016.10.065
Mukherjee, A., Saez, C., and Lutkenhaus, J. (2001). Assembly of an FtsZ mutant deficient in GTPase activity has implications for FtsZ assembly and the role of the Z ring in cell division. J. Bacteriol. 183, 7190–7197. doi: 10.1128/JB.183.24.7190-7197.2001
Nikaido, H., and Pagès, J. (2012). Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol. Rev. 36, 340–363. doi: 10.1111/j.1574-6976.2011.00290.x
Payne, D. J. (2008). Desperately seeking new antibiotics. Science 321, 1644–1645. doi: 10.1126/science.1164586
Pillai, S. K., Moellering, R. C., and Eliopoulos, G. M. (2005). “Antimicrobial combinations,” in Antibiotics in Laboratory Medicine, 5th Edn, ed. V. Lorian (Philadelphia, PA: The Lippincott Williams & Wilkins Co), 365–440.
Plaza, A., Keffer, J. L., Bifulco, G., Lloyd, J. R., and Bewley, C. A. (2010). Chrysophaentins A-H, antibacterial bisdiarylbutene macrocycles that inhibit the bacterial cell division protein FtsZ. J. Am. Chem. Soc. 132, 9069–9077. doi: 10.1021/ja102100h
Rice, L. B. (2008). Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: no ESKAPE. Chicago, IL: The University of Chicago Press.
Ruiz-Avila, L. B., Huecas, S., Artola, M., VergonϸOìS, A., Ramírez-Aportela, E., Cercenado, E., et al. (2013). Synthetic inhibitors of bacterial cell division targeting the GTP-binding site of FtsZ. ACS Chem. Biol. 8, 2072–2083. doi: 10.1021/cb400208z
Singh, D., Bhattacharya, A., Rai, A., Dhaked, H. P., Awasthi, D., Ojima, I., et al. (2014). SB-RA-2001 inhibits bacterial proliferation by targeting FtsZ assembly. Biochemistry 53, 2979–2992. doi: 10.1021/bi401356y
Singh, P., Jindal, B., Surolia, A., and Panda, D. (2012). A rhodanine derivative CCR-11 inhibits bacterial proliferation by inhibiting the assembly and GTPase activity of FtsZ. Biochemistry 51, 5434–5442. doi: 10.1021/bi201813u
Stokes, N. R., Baker, N., Bennett, J. M., Chauhan, P. K., Collins, I., Davies, D. T., et al. (2014). Design, synthesis and structure–activity relationships of substituted oxazole–benzamide antibacterial inhibitors of FtsZ. Bioorg. Med. Chem. Lett. 24, 353–359. doi: 10.1016/j.bmcl.2013.11.002
Sun, N., Chan, F.-Y., Lu, Y.-J., Neves, M. A., Lui, H.-K., Wang, Y., et al. (2014). Rational design of berberine-based FtsZ inhibitors with broad-spectrum antibacterial activity. PLoS One 9:e97514. doi: 10.1371/journal.pone.0097514
Sun, N., Du, R. L., Zheng, Y. Y., Guo, Q., Cai, S. Y., Liu, Z. H., et al. (2018). Antibacterial activity of 3-methylbenzo[d]thiazol-methylquinolinium derivatives and study of their action mechanism. J. Enzyme. Inhib. Med. Chem. 33, 879–889. doi: 10.1080/14756366.2018.1465055
Sun, N., Du, R.-L., Zheng, Y.-Y., Huang, B.-H., Guo, Q., Zhang, R.-F., et al. (2017a). Antibacterial activity of N-methylbenzofuro [3, 2-b] quinoline and N-methylbenzoindolo [3, 2-b]-quinoline derivatives and study of their mode of action. Eur. J. Med. Chem. 135, 1–11. doi: 10.1016/j.ejmech.2017.04.018
Sun, N., Lu, Y.-J., Chan, F.-Y., Du, R.-L., Zheng, Y.-Y., Zhang, K., et al. (2017b). A thiazole orange derivative targeting the bacterial protein FtsZ shows potent antibacterial activity. Front. Microbiol. 8:855. doi: 10.3389/fmicb.2017.00855
Sun, N., Zheng, Y.-Y., Du, R.-L., Cai, S.-Y., Zhang, K., So, L.-Y., et al. (2017c). New application of tiplaxtinin as an effective FtsZ-targeting chemotype for an antimicrobial study. MedChemComm 8, 1909–1913. doi: 10.1039/C7MD00387K
Sung, W. S., and Lee, D. G. (2008). Mechanism of decreased susceptibility for Gram-negative bacteria and synergistic effect with ampicillin of indole-3-carbinol. Biol. Pharm. Bull. 31, 1798–1801. doi: 10.1248/bpb.31.1798
Walsh, C. (2003). Antibiotics: Actions, Origins, Resistance. Washington, DC: American Society for Microbiology. doi: 10.1128/9781555817886
Wang, R., Hou, S., Dong, X., Chen, D., Shao, L., Qian, L., et al. (2017). Synergism of fused bicyclic 2-aminothiazolyl compounds with polymyxin B against Klebsiella pneumoniae. MedChemComm 8, 2060–2066. doi: 10.1039/C7MD00354D
Wang, Y., Mowla, R., Ji, S., Guo, L., De Barros Lopes, M. A., Jin, C., et al. (2018). Design, synthesis and biological activity evaluation of novel 4-subtituted 2-naphthamide derivatives as AcrB inhibitors. Eur. J. Med. Chem. 143, 699–709. doi: 10.1016/j.ejmech.2017.11.102
WHO (2017). Critically Important Antimicrobials for Human Medicine: Ranking of Antimicrobial Agents for Risk Management of Antimicrobial Resistance due to Non-Human Use. Geneva: World Health Organization.
Wikler, M. A., Hindler, J. F., Cookerill, F. R., Patel, J. B., Bush, K., and Powell, M. (2009). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that grow Aerobically. Wayne, PA: Clinical and Laboratory, M07–M08.
Willyard, C. (2017). The drug-resistant bacteria that pose the greatest health threats. Nature 543:15. doi: 10.1038/nature.2017.21550
Keywords: bacterial resistance, antibacterial activity, cell division, FtsZ inhibitor, FtsZ polymerization
Citation: Zheng Y-Y, Du R-L, Cai S-Y, Liu Z-H, Fang Z-Y, Liu T, So L-Y, Lu Y-J, Sun N and Wong K-Y (2018) Study of Benzofuroquinolinium Derivatives as a New Class of Potent Antibacterial Agent and the Mode of Inhibition Targeting FtsZ. Front. Microbiol. 9:1937. doi: 10.3389/fmicb.2018.01937
Received: 13 March 2018; Accepted: 31 July 2018;
Published: 17 August 2018.
Edited by:
Henrietta Venter, University of South Australia, AustraliaReviewed by:
Nagendran Tharmalingam, Alpert Medical School, United StatesCopyright © 2018 Zheng, Du, Cai, Liu, Fang, Liu, So, Lu, Sun and Wong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu-Jing Lu, bHV5akBnZHV0LmVkdS5jbg== Ning Sun, bmluZy5zdW5AY29ubmVjdC5wb2x5dS5oaw== Kwok-Yin Wong, a3dvay15aW4ud29uZ0Bwb2x5dS5lZHUuaGs=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.