- 1Research Service, Atlanta Veterans Affairs Medical Center, Decatur, GA, United States
- 2Research Service, Louis Stokes Cleveland Department of Veterans Affairs Medical Center, Cleveland, OH, United States
- 3Department of Medicine, Pharmacology and Molecular Biology and Microbiology, Case Western Reserve University School of Medicine, Cleveland, OH, United States
- 4Case Western Reserve University Veterans Affairs Center for Antimicrobial Resistance (Case-VA CARES), Cleveland, OH, United States
- 5Department of Microbiology and Immunology, Emory University School of Medicine, Atlanta, GA, United States
- 6Emory Antibiotic Resistance Center, Emory University School of Medicine, Atlanta, GA, United States
Acinetobacter nosocomialis is a member of the Acinetobacter calcoaceticus-Acinetobacter baumannii (ACB) complex. Increasingly, reports are emerging of the pathogenic profile and multidrug resistance (MDR) phenotype of this species. To define novel therapies to overcome resistance, we queried the role of the major efflux pumps in A. nosocomialis strain M2 on antimicrobial susceptibility profiles. A. nosocomialis strains with the following mutations were engineered by allelic replacement; ΔadeB, ΔadeJ, and ΔadeB/adeJ. In these isogenic strains, we show that the ΔadeJ mutation increased susceptibility to beta-lactams, beta-lactam/beta-lactamase inhibitors, chloramphenicol, monobactam, tigecycline, and trimethoprim. The ΔadeB mutation had a minor effect on resistance to certain beta-lactams, rifampicin and tigecycline. In addition, the ΔadeJ mutation resulted in a significant decrease in surface motility and a minor decrease in biofilm formation. Our results indicate that the efflux pump, AdeIJK, has additional roles outside of antibiotic resistance in A. nosocomialis.
Introduction
Acinetobacter nosocomialis is a Gram-negative opportunistic pathogen that is grouped into the Acinetobacter calcoaceticus-Acinetobacter baumannii (ACB) complex (Nemec et al., 2011; Visca et al., 2011). The ability of A. nosocomialis to cause disease in humans is well-recognized (Wisplinghoff et al., 2012; Chusri et al., 2014; Huang et al., 2014), although studies suggest the virulence of this bacterium may be lower than the closely related bacterium Acinetobacter baumannii (Peleg et al., 2012; Lee et al., 2013; Yang et al., 2013; Fitzpatrick et al., 2015). Many potential virulence factors have been identified in A. nosocomialis and include a CTFR inhibitory factor (Cif), a protein O-glycosylation system, a type-I secretion system, a type-II secretion system, secretion of outer membrane vesicles, the OmpA protein, the CpaA protease, and quorum sensing (Niu et al., 2008; Bahl et al., 2014; Harding et al., 2015, 2016, 2017; Nho et al., 2015; Weber et al., 2015; Kim et al., 2016; Kinsella et al., 2017).
A. nosocomialis strain M2 was isolated in 1996 from a hip infection and has been extensively studied, particularly with respect to the virulence factors described above. M2 was formerly classified as A. baumannii, but whole genome sequencing resulted in its reclassification (Carruthers et al., 2013). While A. nosocomialis can be highly resistant to antibiotics, the role of RND-type efflux pumps in this process has not been investigated in this bacterium. Two primary efflux systems in the closely related A. baumannii are the AdeABC and AdeIJK efflux systems (Magnet et al., 2001; Damier-Piolle et al., 2008). Each efflux system is composed of an outer membrane channel (AdeC, AdeK), a membrane fusion protein (AdeA, AdeI) and an inner membrane transporter (AdeB, AdeJ). In addition to the efflux of antimicrobials, these systems can impact additional phenotypes in the cell, such as surface motility, biofilm formation, and virulence (Yoon et al., 2015; Richmond et al., 2016).
In this study, we investigated the role of AdeABC and AdeIJK orthologs in A. nosocomialis. Similar to what is observed in A. baumannii, loss of AdeIJK had a major impact on antibiotic susceptibility profiles. In contrast, the loss of AdeABC had a minimal impact on susceptibility. Interestingly, the loss of AdeIJK reduced surface motility, indicating additional roles for this RND-type efflux system in A. nosocomialis.
Materials and Methods
Bacterial Growth Conditions, Strains, and Plasmids
A. nosocomialis strain M2 was used for all studies and has been described previously (Carruthers et al., 2013). E. coli strains EC100D and CC118 were used for general cloning. E. coli strain SM10 was used for bacterial conjugations. Growth media consisted of 10 g tryptone, 5 g yeast extract, and 5 g NaCl per liter. Agar was added at 15 g per liter. For sucrose counter-selections, media was prepared as described above, but without NaCl and containing 10% sucrose. Cloning vectors were pBC.SK- (Agilent) and pKNG101 (Kaniga et al., 1991).
Construction of adeB and adeJ Mutations
Internal fragments of the adeB and adeJ genes were obtained by PCR amplification of M2 genomic DNA using the following primers. peg93.for 5′- TTGCTAAGTATTCCTAAATTAC-3′ and peg93.rev 5′- TTAGGAAGAGATTTTTTTC−3′ for adeB, and peg1681.for 5′- ATGGCACAATTTTTTATTCATC−3′ and peg1681.rev 5′- TCACGATTTATGCTCCTGAG-3′ for adeJ. The resulting PCR generated fragments were cloned into the pBC.SK digested plasmid with SmaI, creating padeB and padeJ. The padeB plasmid was then digested with NarI, which digests once in the middle of the adeB gene and treated with T4 DNA polymerase to create blunt ends. This was then re-ligated to create a frameshift mutation in adeB. The plasmid padeJ was digested with SphI, which cuts once in the middle of adeJ, treated with T4 DNA polymerase to create blunt ends and re-ligated to create an adeJ frameshift mutation. The mutated adeB and adeJ genes were then excised as an XbaI-SalI fragment and cloned into the suicide vector pKNG101 digested with XbaI and SalI. Each plasmid was transformed into E. coli SM10 and then introduced into the A. nosocomialis M2 chromosome by conjugation. Exconjugants were grown for 10 generations in LB broth without antibiotic and dilutions were plated on lysogeny broth (LB) plates without sodium chloride and containing 10% sucrose. Colonies containing the adeB or adeJ frameshift mutations were identified by PCR amplifying each gene and the digesting the resulting PCR products with either NarI for adeB or SphI for adeJ. The presence of each chromosomal mutation was indicated by the failure of each enzyme to digest the fragment and each mutation was verified by DNA sequence analysis. To create an adeB, adeJ double mutant, the adeB mutant was used as the parent and the adeJ mutation was crossed into the chromosome as described above. To create an adeB::Km mutation, an EZ-Tn5 <Kan-2> insertion centrally located in the adeB gene present in pKNG101 was recombined into the chromosomal copy of adeB as described above.
Antimicrobial Susceptibility Testing
A. nosocomialis strain M2 and its isogenic derivatives were subject to antimicrobial susceptibility testing using E-Test Strips, Trek, and MicroScan platforms. Additionally, disk diffusion assays were performed using Mueller Hinton agar for several antibiotics alone and in combination with boronic acid transition state inhibitor (BATSI) compounds SM23 and S02030 (Powers et al., 2014; Nguyen et al., 2016). For TREK, strains were tested once. For the disc diffusion and Etest assays, strains were tested in duplicate.
Motility Assays
The base media for motility assays consisted of 10 g tryptone, 5 g yeast extract, and 5 g NaCl per liter. Media was solidified using 0.35% Eiken agar (Eiken Chemical Ltd. Tokyo, Japan). Plates were used the same day they were prepared. For testing the motility of the M2 strain and various mutants, cultures were grown up to early log phase, adjusted to the same optical density of A600 = 0.15 by the addition of sterile LB broth and a 1 μl drop was placed on the center of the plate. Plates were incubated at 30°C and motility was measured after 14 h. Statistical analysis was done using the Student's T-test.
Biofilm Analysis
Cells for biofilm analysis were taken directly from freezer stocks and grown in 2 ml 0.5X LB without shaking at room temperature to an optical density A600 of 0.1. Each tube was then used to inoculate wells of a 96 well microtiter plate with 150 μl of culture. Plates were incubated stationary at 30 or 37°C for 24 h. The optical density of each well was read at A600 for cell growth and the planktonic cells were removed. To stain biofilms, 250 μl of 10% crystal violet was added to each well for 30 min. The crystal violet was gently decanted and each well was gently washed three times with distilled water. Three hundred microliters of 33% acetic acid was added to each well to solubilize the crystal violet and the absorbance of a 1/10 dilution was read at A585. Statistical analysis was done using the Student's T-test.
Results
Analysis of AdeABC and AdeIJK RND-Efflux Systems in A. nosocomialis
A. nosocomialis strain M2 contains orthologs of AdeA, AdeB and AdeC that share 94, 98, and 92 percent amino acid identify, respectively, to the corresponding proteins in A. baumannii strain AB5075.UW. In addition, orthologs of the AdeIJK proteins were found with 97, 99, and 98 percent identity to the corresponding proteins in A. baumannii AB5075.UW. To investigate the function of each RND-type efflux system, null alleles in the adeB and adeJ genes, encoding the inner membrane transporter for each system were constructed by introducing frameshift mutations in each gene into the chromosome by allelic replacement (section Materials and Methods).
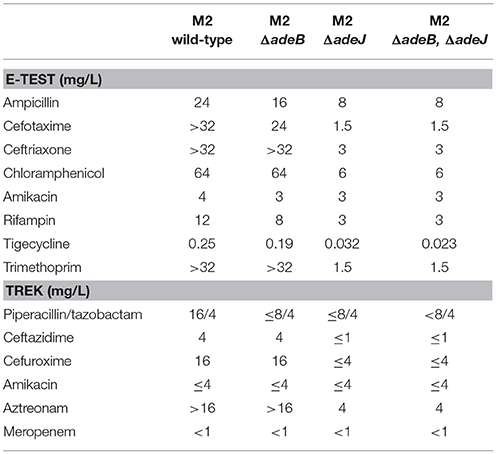
The antibiotic susceptibility profile of each mutant was then determined for a panel of antibiotics representing different classes (Table 1). The loss of adeB had a minimal effect on the overall levels of resistance and a slight increase in susceptibility was observed for ampicillin, cefotaxime, amikacin, rifampin, and tigecyline (Table 1). This result was surprising as the AdeABC system has a prominent role in antibiotic resistance in A. baumannii (Magnet et al., 2001). To determine if this adeB frameshift mutation was somehow being suppressed or was not a null allele, we constructed an adeB::Km mutation, where the adeB gene was disrupted in the middle of the coding region. However, this adeB::km mutant displayed the same level of resistance to ampicillin (128 μg/ml), tetracycline (2 μg/ml), and ciprofloxacin (0.38 μg/ml) as wild-type, indicating that the previously isolated frameshift mutation in adeB was non-functional.
The effect of a mutation in adeJ on antibiotic susceptibility was far more pronounced, where cells became more susceptible to the following antibiotics; ampicillin (3-fold), cefotaxime (>15-fold), ceftriaxone (>10-fold), chloramphenicol (>10-fold), rifampin (4-fold), tigecycline (8-fold), and trimethoprim (>20-fold) (Table 1). The antibiotic susceptibility profiles were also examined for an adeB/adeJ double mutant to determine if the loss of both efflux systems had additional effects. However, the adeB/adeJ double mutant essentially phenocopied the adeJ single mutant (Table 1).
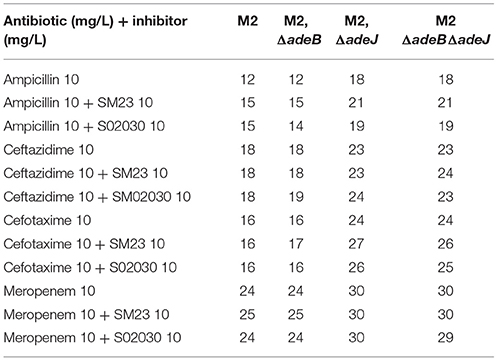
We next assayed the role of AdeB and AdeJ efflux pumps in the handling of the boronic acid transition state inhibitors (BATSIs) SM23 and S02030 (Powers et al., 2014; Nguyen et al., 2016). These BATSIs either mimic the acylation or deacylation transition state. Paired with a penicillin (ampicillin), carbapenem (meropenem), or cephalosporin (ceftazidimne or cefepime) as performed herein, the BATSI can act to inhibit serine based beta-lactamases in-vitro. As a result of this mechanism of action, class C cephalosporinases possess the greatest affinity for these compounds (e.g., ADC cephalosporinase in A. nosocomialis). Our results indicate that the BATSI studied are substrates for the AdeIJK efflux pump in A. nosocomialis (Table 2). In particular, the susceptibility of wild-type M2 to cefotaxime is unaffected by these inhibitors, but in the presence of the adeJ mutation, these inhibitors now increase susceptibility to cefotaxime (Table 2).
Role of AdeABC and AdeIJK in Motility
A. nosocomialis strain M2 is capable of rapidly translocating across soft agar surfaces (Clemmer et al., 2011). Although the mechanism responsible for this motility is unclear, a number of genes have been identified that reduce motility including mutations in the abaI autoinducer synthase responsible for quorum sensing signal production (Clemmer et al., 2011). We tested the wild-type M2 parent and the isogenic adeB and adeJ mutants for their motility phenotypes at 30 degrees. The adeB mutation did not significantly alter surface motility (Figures 1A,B). In contrast, the adeJ mutation had a pronounced effect on surface motility, with a greater than 50% reduction relative to the wild-type M2 parent (Figure 1). Interestingly, this motility defect was temperature dependent, at 37 degrees the adeJ mutant exhibited a similar level of motility as wild-type (Figure 1C).
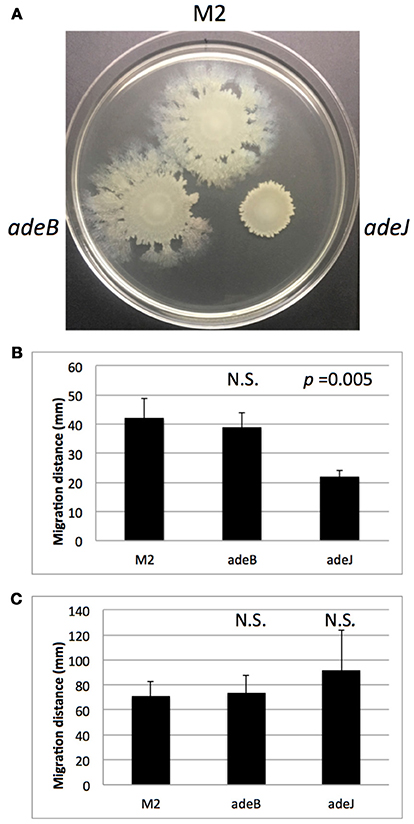
Figure 1. Surface motility of wild-type M2 and efflux mutants. Wild-type M2 and the isogenic adeB and adeJ mutants were assayed for surface motility as described in the Materials and Methods. In (A), motility of the indicated strains is shown after 14 h at 30°C. Motility was quantitated from 4 separate experiments at 30°C (B) and 37°C (C). Error bars represent standard deviation of the mean. N.S. indicates a p-value > 0.05.
In a previous study, the motility of A. nosocomialis M2 was shown to be dependent on production of the quorum sensing signal 3-OH C12-HSL (Clemmer et al., 2011). To investigate the possibility that the motility defect in the adeJ mutant was due to the failure to export 3-OH-C12-HSL, an Agrobacterium tumefaciens traG-lacZ biosensor strain was used to assay signal production in the adeJ mutant and wild-type M2 (Niu et al., 2008). However, no significant differences in signal production were observed between these strains (Supplementary Figure 1).
Role of AdeABC and AdeIJK in Biofilm Formation
The role of AdeABC and AdeIJK in biofilm formation was also examined. When biofilms were formed on the surface of polystyrene microtiter wells, biofilm formation by the adeB and adeJ mutants were similar to wild-type M2 after 24 h of growth at 30°C (Figure 2). At 37°C, only the adeJ mutant showed a statistically significant reduction in biofilm formation, with a 24% decrease relative to wild-type (Figure 2).
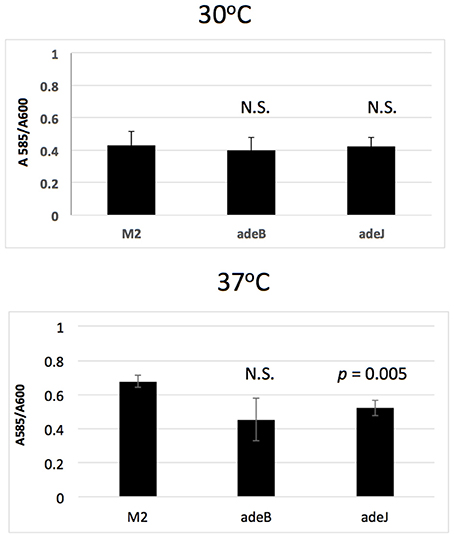
Figure 2. Biofilm formation. Wild-type M2 and the isogenic adeB and adeJ mutants were assayed for biofilm formation in microtiter wells grown at 30 or 37°C for 24 h. Values represent crystal violet staining/cell density (A585 / A600) ratio and error bars represent standard deviation of the mean. N.S. indicates a p-value > 0.05.
Discussion
In this study, the roles of AdeABC and AdeIJK orthologs in A. nosocomialis were addressed. Both a frameshift allele in the adeB gene and an adeB::Km disruption did not result in a major change in antibiotic resistance profiles, which is in contrast to that observed in A. baumannii (Magnet et al., 2001). Several possibilities can account for these differences. First, the adeABC genes may be expressed at very low levels in A. nosocomialis M2, therefore, the loss of this efflux system would have a minimal impact. In A. baumanii, the AdeABC system is typically expressed at low levels and inactivation of these genes in some strains does not produce a phenotype (Yoon et al., 2015; Leus et al., 2018). Increased expression can result from mutations in the AdeRS two-component system. In A. nosocomialis M2, the AdeR and AdeS proteins did not contain amino acid substitutions previously associated with increased AdeABC expression (Marchand et al., 2004; Yoon et al., 2013; Gerson et al., 2018). Based on this information, we propose that the AdeABC genes are tightly regulated by AdeRS and the levels of expression in the M2 strain do not contribute to intrinsic resistance. We also tested the role of AdeABC in both surface motility and biofilm formation and no significant changes were observed in the adeB mutant relative to wild-type (Figures 1, 2).
In contrast, the AdeIJK efflux system was shown to play a significant role in antibiotic efflux, where a mutation inactivating this system had a pronounced effect on antibiotic susceptibility (Table 1). This observation is consistent with previous studies in A. baumannii demonstrating that efflux mediated by AdeIJK contributes substantially to antibiotic resistance. In addition, the loss of AdeIJK strongly reduced surface motility with a greater than 50% reduction compared to wild-type (Figure 1). The loss of AdeIJK resulted in a modest (24%) reduction in biofilm formation, which is also consistent with previous studies in A. baumannii, where the loss of AdeIJK resulted in a 20% reduction in biofilm formation (Yoon et al., 2015). The decreased surface motility and biofilm formation in the adeJ mutant were not the result of decreased production of the quorum sensing signal 3-OH C12-HSL, which has been shown to be important for both surface motility and biofilm formation in A. nosocomialis (Niu et al., 2008; Clemmer et al., 2011).
The mechanism that results in loss of motility when the AdeIJK system in inactivated is unknown, but may indicate a role for AdeIJK in secretion of a lipopeptide surfactant that is required for motilty (Clemmer et al., 2011; Rumbo-Feal et al., 2017) or in the secretion of 1,3-diaminopropane, also required for motility (Skiebe et al., 2012). This also indicates that in addition to antibiotic efflux, there are cellular functions mediated by AdeIJK, indicating a general role for this RND-type efflux system in general physiology of A. nosocomialis.
Author Contributions
DK, SR, and PR conducted experiments. PR and RB wrote the manuscript. SR, RB, and PR edited the manuscript.
Funding
This work was supported by grant R01 AI072219 from the National Institutes of Health to PR and RB. PR is also supported by grants from the Merit Review program I01BX001725 and a Research Career Scientist Award IK6BX004470, both from Department of Veterans Affairs.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Katy Clemmer for assistance with strain constructions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01902/full#supplementary-material
References
Bahl, C. D., Hvorecny, K. L., Bridges, A. A., Ballok, A. E., Bomberger, J. M., Cady, K. C., et al. (2014). Signature motifs identify an Acinetobacter Cif virulence factor with epoxide hydrolase activity. J. Biol. Chem. 289, 7460–7469. doi: 10.1074/jbc.M113.518092
Carruthers, M. D., Harding, C. M., Baker, B. D., Bonomo, R. A., Hujer, K. M., Rather, P. N., et al. (2013). Draft genome sequence of the clinical isolate Acinetobacter nosocomialis strain M2. Genome Announc. 1:e00906-13 doi: 10.1128/genomeA.00906-13
Chusri, S., Chongsuvivatwong, V., Rivera, J. I., Silpapojakul, K., Singkhamanan, K., McNeil, E., et al. (2014). Clinical outcomes of hospital-acquired infection with Acinetobacter nosocomialis and Acinetobacter pittii. Antimicrob. Agents Chemother. 58, 4172–4179. doi: 10.1128/AAC.02992-14
Clemmer, K. M., Bonomo, R. A., and Rather, P. N. (2011). Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology 157, 2534–2544. doi: 10.1099/mic.0.049791-0
Damier-Piolle, L., Magnet, S., Bremont, S., Lambert, T., and Courvalin, P. (2008). AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob. Agents Chemother. 52, 557–562. doi: 10.1128/AAC.00732-07
Fitzpatrick, M. A., Ozer, E., Bolon, M. K., and Hauser, A. R. (2015). Influence of ACB complex genospecies on clinical outcomes in a U.S. hospital with high rates of multidrug resistance. J. Infect. 70, 144–152. doi: 10.1016/j.jinf.2014.09.004
Gerson, S., Nowak, J., Zander, E., Ertel, J., Wen, Y., Krut, O., et al. (2018). Diversity of mutations in regulatory genes of resistance-nodulation-cell division efflux pumps in association with tigecycline resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 73, 1501–1508. doi: 10.1093/jac/dky083
Harding, C. M., Kinsella, R. L., Palmer, L. D., Skaar, E. P., and Feldman, M. F. (2016). Medically relevant acinetobacter species require a type II secretion system and specific membrane-associated chaperones for the export of multiple substrates and full virulence. PLoS Pathog. 12:e1005391. doi: 10.1371/journal.ppat.1005391
Harding, C. M., Nasr, M. A., Kinsella, R. L., Scott, N. E., Foster, L. J., Weber, B. S., et al. (2015). Acinetobacter strains carry two functional oligosaccharyltransferases, one devoted exclusively to type IV pilin, and the other one dedicated to O-glycosylation of multiple proteins. Mol. Microbiol. 96, 1023–1041. doi: 10.1111/mmi.12986
Harding, C. M., Pulido, M. R., Di Venanzio, G., Kinsella, R. L., Webb, A. I., Scott, N. E., et al. (2017). Pathogenic Acinetobacter species have a functional type I secretion system and contact-dependent inhibition systems. J. Biol. Chem. 292, 9075–9087. doi: 10.1074/jbc.M117.781575
Huang, L., Chen, T. L., Lee, Y. T., Lee, M. H., Kuo, S. C., Yu, K. W. P., et al. (2014). Risk factors for imipenem-nonsusceptible Acinetobacter nosocomialis bloodstream infection. J. Microbiol. Immunol. Infect. 47, 311–317. doi: 10.1016/j.jmii.2013.02.002
Kaniga, K., Delor, I., and Cornelis, G. R. (1991). A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109, 137–141. doi: 10.1016/0378-1119(91)90599-7
Kim, S. W., Oh, M. H., Jun, S. H., Jeon, H., Kim, S. I., Kim, K., et al. (2016). Outer membrane protein A plays a role in pathogenesis of Acinetobacter nosocomialis. Virulence 7, 413–426. doi: 10.1080/21505594.2016.1140298
Kinsella, R. L., Lopez, J., Palmer, L. D., Salinas, N. D., Skaar, E. P., Tolia, N. H., et al. (2017). Defining the interaction of the protease CpaA with its type II secretion chaperone CpaB and its contribution to virulence in Acinetobacter species. J. Biol. Chem. 292, 19628–19638. doi: 10.1074/jbc.M117.808394
Lee, Y. T., Kuo, S. C., Yang, S. P., Lin, Y. T., Chiang, D. H., Tseng, F. C., et al. (2013). Bacteremic nosocomial pneumonia caused by Acinetobacter baumannii and Acinetobacter nosocomialis: a single or two distinct clinical entities? Clin. Microbiol. Infect. 19, 640–645. doi: 10.1111/j.1469-0691.2012.03988.x
Leus, I. V., Weeks, J. W., Bonifay, V., Smith, L., Richardson, S., and Zgurskaya, H. I. (2018). Substrate specificities and efflux efficiencies of RND efflux pumps of Acinetobacter baumannii. J. Bacteriol. 200:JB.00049–18. doi: 10.1128/JB.00049-18
Magnet, S., Courvalin, P., and Lambert, T. (2001). Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob. Agents Chemother. 45, 3375–3380. doi: 10.1128/AAC.45.12.3375-3380.2001
Marchand, I., Damier-Piolle, L., Courvalin, P., and Lambert, T. (2004). Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob. Agents Chemother. 48, 3298–3304. doi: 10.1128/AAC.48.9.3298-3304.2004
Nemec, A., Krizova, L., Maixnerova, M., van der Reijden, T. J., Deschaght, P., Passet, V., et al. (2011). Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex with the proposal of Acinetobacter pittii sp. nov. (formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov. (formerly Acinetobacter genomic species 13TU). Res. Microbiol. 162, 393–404. doi: 10.1016/j.resmic.2011.02.006
Nguyen, N. Q., Krishnan, N. P., Rojas, L. J., Prati, F., Caselli, E., Romagnoli, C., et al. (2016). Crystal structures of KPC-2 and SHV-1 beta-lactamases in complex with the boronic acid transition state analog S02030. Antimicrob. Agents Chemother. 60, 1760–1766. doi: 10.1128/AAC.02643-15
Nho, J. S., Jun, S. H., Oh, M. H., Park, T. I., Choi, C. W., Kim, S. I., et al. (2015). Acinetobacter nosocomialis secretes outer membrane vesicles that induce epithelial cell death and host inflammatory responses. Microb. Pathog. 81, 39–45. doi: 10.1016/j.micpath.2015.03.012
Niu, C., Clemmer, K. M., Bonomo, R. A., and Rather, P. N. (2008). Isolation and characterization of an autoinducer synthase from Acinetobacter baumannii. J. Bacteriol. 190, 3386–3392. doi: 10.1128/JB.01929-07
Peleg, A. Y., de Breij, A., Adams, M. D., Cerqueira, G. M., Mocali, S., Galardini, M., et al. (2012). The success of acinetobacter species; genetic, metabolic and virulence attributes. PLoS ONE 7:e46984. doi: 10.1371/journal.pone.0046984
Powers, R. A., Swanson, H. C., Taracila, M. A., Florek, N. W., Romagnoli, C., Caselli, E., et al. (2014). Biochemical and structural analysis of inhibitors targeting the ADC-7 cephalosporinase of Acinetobacter baumannii. Biochemistry 53, 7670–7679. doi: 10.1021/bi500887n
Richmond, G. E., Evans, L. P., Anderson, M. J., Wand, M. E., Bonney, L. C., Ivens, A., et al. (2016). The Acinetobacter baumannii two-component system AdeRS regulates genes required for multidrug efflux, biofilm formation, and virulence in a strain-specific manner. MBio 7:e00430-16. doi: 10.1128/mBio.00430-16
Rumbo-Feal, S., Perez, A., Ramelot, T. A., Alvarez-Fraga, L., Vallejo, J. A., Beceiro, A., et al. (2017). Contribution of the A. baumannii A1S_0114 gene to the interaction with eukaryotic cells and virulence. Front. Cell. Infect. Microbiol. 7:108. doi: 10.3389/fcimb.2017.00108
Skiebe, E., de Berardinis, V., Morczinek, P., Kerrinnes, T., Faber, F., Lepka, D., et al. (2012). Surface-associated motility, a common trait of clinical isolates of Acinetobacter baumannii, depends on 1,3-diaminopropane. Int. J. Med. Microbiol. 302, 117–128. doi: 10.1016/j.ijmm.2012.03.003
Visca, P., Seifert, H., and Towner, K. J. (2011). Acinetobacter infection–an emerging threat to human health. IUBMB Life 63, 1048–1054. doi: 10.1002/iub.534
Weber, B. S., Harding, C. M., and Feldman, M. F. (2015). Pathogenic acinetobacter: from the cell surface to infinity and beyond. J. Bacteriol. 198, 880–887. doi: 10.1128/JB.00906-15
Wisplinghoff, H., Paulus, T., Lugenheim, M., Stefanik, D., Higgins, P. G., Edmond, M. B., et al. (2012). Nosocomial bloodstream infections due to Acinetobacter baumannii, Acinetobacter pittii and Acinetobacter nosocomialis in the United States. J. Infect. 64, 282–290. doi: 10.1016/j.jinf.2011.12.008
Yang, Y. S., Lee, Y. T., Tsai, W. C., Kuo, S. C., Sun, J. R., Yang, C. H., et al. (2013). Comparison between bacteremia caused by carbapenem resistant Acinetobacter baumannii and Acinetobacter nosocomialis. BMC Infect. Dis. 13:311. doi: 10.1186/1471-2334-13-311
Yoon, E. J., Chabane, Y. N., Goussard, S., Snesrud, E., Courvalin, P., De, E., et al. (2015). Contribution of resistance-nodulation-cell division efflux systems to antibiotic resistance and biofilm formation in Acinetobacter baumannii. mBio 6:e00309-15. doi: 10.1128/mBio.00309-15
Keywords: Acinetobacter, RND-efflux, motility, biofilm, antimicrobial resistance
Citation: Knight DB, Rudin SD, Bonomo RA and Rather PN (2018) Acinetobacter nosocomialis: Defining the Role of Efflux Pumps in Resistance to Antimicrobial Therapy, Surface Motility, and Biofilm Formation. Front. Microbiol. 9:1902. doi: 10.3389/fmicb.2018.01902
Received: 25 May 2018; Accepted: 27 July 2018;
Published: 21 August 2018.
Edited by:
Gilberto Igrejas, University of Trás-os-Montes and Alto Douro, PortugalReviewed by:
Bart Antonie Eijkelkamp, University of Adelaide, AustraliaKarl Hassan, University of Newcastle, Australia
Ayush Kumar, University of Manitoba, Canada
Copyright © 2018 Knight, Rudin, Bonomo and Rather. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Philip N. Rather, cHJhdGhlckBlbW9yeS5lZHU=
 Daniel B. Knight1
Daniel B. Knight1 Philip N. Rather
Philip N. Rather