- 1State Key Laboratory of Food Science and Technology, Jiangnan University, Wuxi, China
- 2School of Food Science and Technology, Jiangnan University, Wuxi, China
- 3National Engineering Research Center for Functional Food, Jiangnan University, Wuxi, China
- 4Department of Cancer Biology, Wake Forest School of Medicine, Winston-Salem, NC, United States
- 5Beijing Innovation Centre of Food Nutrition and Human Health, Beijing Technology and Business University, Beijing, China
Omega-3 long-chain polyunsaturated fatty acids (LC-PUFAs) have important therapeutic and nutritional benefits in humans. In the biosynthesis pathways of these LC-PUFAs, omega-3 desaturase plays a critical role. In this study, we report a new omega-3 desaturase (PPD17) from Phytophthora parasitica. This desaturase shares high similarities with the known omega-3 desaturases and was expressed in Saccharomyces cerevisiae for the activity and substrate specificity research. The desaturase has a wide omega-6 fatty acid substrate, containing both 18C and 20C fatty acids, and exhibits a strong activity of delta-17 desaturase but a weak activity of delta-15 desaturase. The new desaturase converted the omega-6 arachidonic acid (AA, C20:4) to EPA (an omega-3 LC-PUFA, C20:5) with a substrate conversion rate of 70%. To obtain a high EPA-producing strain, we transformed PPD17 into Mortierella alpina, an AA-producing filamentous fungus. The EPA content of the total fatty acids in reconstruction strains reached 31.5% and was followed by the fermentation optimization of the EPA yield of up to 1.9 g/L. This research characterized a new omega-3 desaturase and provides a possibility of industrially producing EPA using M. alpina.
Introduction
Omega-3 long-chain polyunsaturated fatty acids (LC-PUFAs), particularly eicosapentaenoic acid (EPA, C20:5) and docosahexaenoic acid (DHA, C22:6), are critical for human health (Xue et al., 2013a). Mammals cannot de novo synthesize EPA and DHA, and have to acquire by diet. However, marine microorganisms and phytoplankton can de novo synthesize EPA and DHA, and these fatty acids are accumulated in marine fishes through the food chain (Xie et al., 2017). Natural marine fish oil is typically the EPA and DHA source for human consumption, but due to human population growth and oceanic pollution, the current practice of harvesting fish for EPA and DHA is unsustainable (Ji et al., 2015; Okuda et al., 2015).
Many plants, algae and fungi have been investigated as potential hosts to facilitate sustainable commercial EPA or DHA production (Xue et al., 2013a; Sun et al., 2014; Ruiz-Lopez et al., 2015). DHA has been commercially produced in large-scale fermentation processes by using the microalgae (Kyle, 2001), but for a long time, EPA production has not achieved sufficiently high yields. Recently, however, DuPont assembled 30 copies of 9 different heterologous genes in Yarrowia lipolytica and the recombination strain accumulated 30% (w/w) lipids in biomass and EPA accounted for 56.6% (w/w) of the total fatty acids (TFAs; Xue et al., 2013b). In contrast to the reconstitution of the EPA biosynthetic pathway in Y. lipolytica through the co-expression of a large number of heterologous genes, single-gene modification for EPA production in the microorganisms containing omega-3 LC-PUFA pathways could be simpler and more stable.
Mortierella alpina, an oleaginous filamentous fungus producing commercial arachidonic acid (AA, C20:4) used as an additive in infant formulas, has been recognized as a potential strain to commercially produce EPA from AA via an omega-3 desaturase (Sakuradani et al., 2013; Ji et al., 2014). There has been researcher overexpressed an endogenous omega-3 desaturase in M. alpina 1S-4, resulting in an increase in the EPA proportion in the TFA content and the maximum value achieved was up to 40% (w/w). However, it had to be grown at a low temperature (12°C) (Ando et al., 2009). Our previous study investigated the omega-3 fatty acid desaturase of M. alpina ATCC 32222 and homologously expressed it in M. alpina, but the content of EPA was no more than 5% in the TFA. The little EPA production could be attributed to the low activity of the endogenous omega-3 desaturase gene at room temperature. Thus, we need to use high-activity heterologous omega-3 desaturase.
Considerable effort has been made to investigate different omega-3 desaturases from a variety of sources. Researches had shown that the omega-3 desaturases from plants (e.g., Sapium sebiferum and Helianthus annuus) exclusively desaturated 18C omega-6 PUFAs, and that from Caenorhabditis elegans preferentially desaturated 18C PUFAs compared with the 20C substrates (Meesapyodsuk et al., 2000; Venegas-Caleron et al., 2006; Niu et al., 2008). However, some omega-3 desaturases with high activity of delta-17 desaturase in preference to C20 substrates have been discovered recently. The omega-3 desaturase (SDD17) from Saprolegnia diclina was found to exclusively desaturate 20C PUFAs with a remarkable preference for AA (Pereira et al., 2004; Okuda et al., 2015). Another omega-3 desaturase (OPIN17) from Phytophthora infestans also could convert 31% AA into EPA (Fu et al., 2013). In addition, Xue et al. (2013a) found that three omega-3 desaturases (PaD17 from Pythium aphanidermatum, PsD17 from Phytophthora sojae, PrD17 from Phytophthora ramorum) had strong delta-17 desaturase activity and PaD17 had the highest conversion rate from AA to EPA. Indeed, we had transformed the PaD17 into M. alpina and increased the conversion rate of AA to EPA (49.7%) in previous study (Ge et al., 2017).
For commercial EPA production by M. alpina, a high-efficiency, ordinary-temperature, and preferential AA substrate omega-3 desaturase is very important. Thus, in this study, we used sequence comparison and structure prediction to study desaturases and found a new omega-3 desaturase defined as PPD17 preferentially converting AA to EPA from Phytophthora parasitica. Then, we characterized this new omega-3 desaturase by expressing it in S. cerevisiae and transformed it into M. alpina. Furthermore, we optimized the culture medium and the conditions of EPA-producing M. alpina transformation.
Materials and Methods
Sequence Comparison and Structure Prediction
Sequence homology was analyzed by the BLAST tools at NCBI. Sequence alignment was done with the Clustal W software. Initial prediction of the transmembrane (TM) domain was performed through the programs TMHMM, HMMTOP, and TOP-PRED.
Strains and Plasmid
Mortierella alpina CCFM 501 was uracil-auxotrophic through being modified from M. alpina ATCC 32222 (Hao et al., 2014). Agrobacterium tumefaciens C58C1, which was provided by Yasuyuki Kubo, is a transfer DNA donor for M. alpina transformation. Escherichia coli Top 10 was used to construct the plasmid. Both the INVSc1 yeast strain and the pYES2/NT C plasmid were purchased from Invitrogen, but the pBIG2-ura5s-ITs plasmid was constructed in our laboratory (Hao et al., 2014).
Media and Culture
Escherichia coli Top 10 was cultivated at 37°C on LB medium and S. cerevisiae was grown on YPD medium at 28°C. M. alpina strains were grown on the GY medium and 0.05 g/L uracil were added when culturing uracil auxotroph, and A. tumefaciens C58C1 was cultured in a YEP medium (Hao et al., 2014). SC-U was the synthetic minimal medium for yeast to induce the recombinant protein expression; its composition is described in our previous work (Shi et al., 2015). The transformation media, including SC medium, MM and IM, were as previously described (Takeno et al., 2004; Ando et al., 2009). When analysing the fatty acid of the M. alpina transformants, this fungus was grown for 7 days at 28°C in a modified broth medium (50 g/L glucose, 10 g/L KNO3, 5 g/L yeast extract, 1 g/L KH2PO4⋅H2O, 0.25 g/L MgSO4⋅7H2O).
Yeast Transformation
The sequence of PPD17 gene was optimized on the basis of the codon preference of S. cerevisiae, and the codon-optimized gene, called oPPD17, was synthesized and cloned into the pUCsimple vector. The primer pair oPPD17 F/oPPD17 R (Supplementary Table 1) was used to amplify oPPD17, and the amplification products were ligated into pYES2/NT C to obtain the plasmid pYES2/NT C-oPPD17. Then, the obtained plasmid was transformed into INVSc1 through lithium acetate transformation method (Gietz et al., 1992).
Omega-6 Fatty Acid Feeding
INVSc1 containing pYES2/NT C-oPPD17 were cultivated on SC-U medium for 48 h and diluted to OD600 of 0.4, followed by aliquoting into 20 mL cultures containing 0.2 mM linoleic acid (LA, C18:2), 0.2 mM gamma-linolenic acid (GLA, C18:3), 0.2 mM dihomo-gamma-linolenic acid (DGLA, C20:3) and 0.2 mM AA. When grown for another 48 h, the cultures were collected and the pellets were used for further analysis.
T-DNA Binary Vector Construction and Agrobacterium tumefaciens-Mediated Transformation (ATMT)
The PPD17 gene was codon-optimized on the basis of the codon preference of M. alpina, called oPpFADS17 (GenBank accession No. KT372001) and then, synthesized and cloned into the pUCsimple vector. The primer pair oPpFADS17 F/oPpFADS17 R (Supplementary Table 1) was used to amplify oPpFADS17, and the oPpFADS17 gene was cloned into pBIG2-ura5s-Its to acquire pBIG2-ura5s-oPpFADS17. The vector carrying the oPpFADS17 gene was used for ATMT as previously described (Hao et al., 2014).
RT-qPCR Analysis
The total RNA of the transformants was isolated with TRIzol (Invitrogen, United States) and then converted to cDNA by the PrimeScript RT reagent kit (Takara, Japan). The quantitative PCR was carried out through CFX Connect RealTime System (Bio-Rad, United States). The PCR amplification involved 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 58°C for 30 s. The primer pairs are showed in Supplementary Table 1, and the 18S rRNA is the internal control in the PCR amplification. The relative transcription levels were calculated using the 2-ΔΔCt method (Shi et al., 2016). The introduced gene is exogenous and it is absent in control strains, thus one of the transformants was selected as standard for relative transcription level analysis.
Western Blot Analysis
Western blot analysis was carried out as described in our previous study (Chen et al., 2013). Briefly, 100 μg of cellular protein was loaded onto SDS-PAGE (12% gel), and PVDF membranes (Millipore, United States) were used for the analysis. The Anti-His antibodies (Nanjing GenScript Biotechnology Corporation, China) were used to detect the recombinant protein.
Cell Dry Weight (CDW) and Fatty Acid Methyl Ester (FAME) Analysis
The biomass was collected through filtration and washed, followed by being freeze-dried; then, CDW was determined gravimetrically. Extraction and methyl esterification of TFAs were carried out as described previously (Wang et al., 2011). Fatty acid methyl ester (FAME) analysis was used GCMS-QP2010 Ultra (Shimadzu Co., Japan) and the temperature program was described in our previous work (Hao et al., 2016).
Results
Bioinformatics Analysis of the New Sequence PPD17 From Phytophthora parasitica
The sequence named PPD17 from P. parasitica searched from GenBank which is 1,086 bp and encodes a peptide of 361 amino acids was aligned with several known omega-3 desaturases using Clustal W (Figure 1A). The sequence shares high similarities (>60%) with known desaturases. Conserved motifs were identified and all of these conserved motifs included the three histidine boxes that are known to be required for desaturase activity. The structure arrangements of all these desaturases are very similar and consist of four membrane-spanning domains and one catalytic domain on the cytosolic side of the membrane (Figure 1B). The predicted topology model was in accordance with other membrane-bound enzymes. The protein also contains KTKST, which was the C terminal motif, proposing to be a retention signal for a number of TM proteins in ER (endoplasmic reticulum).
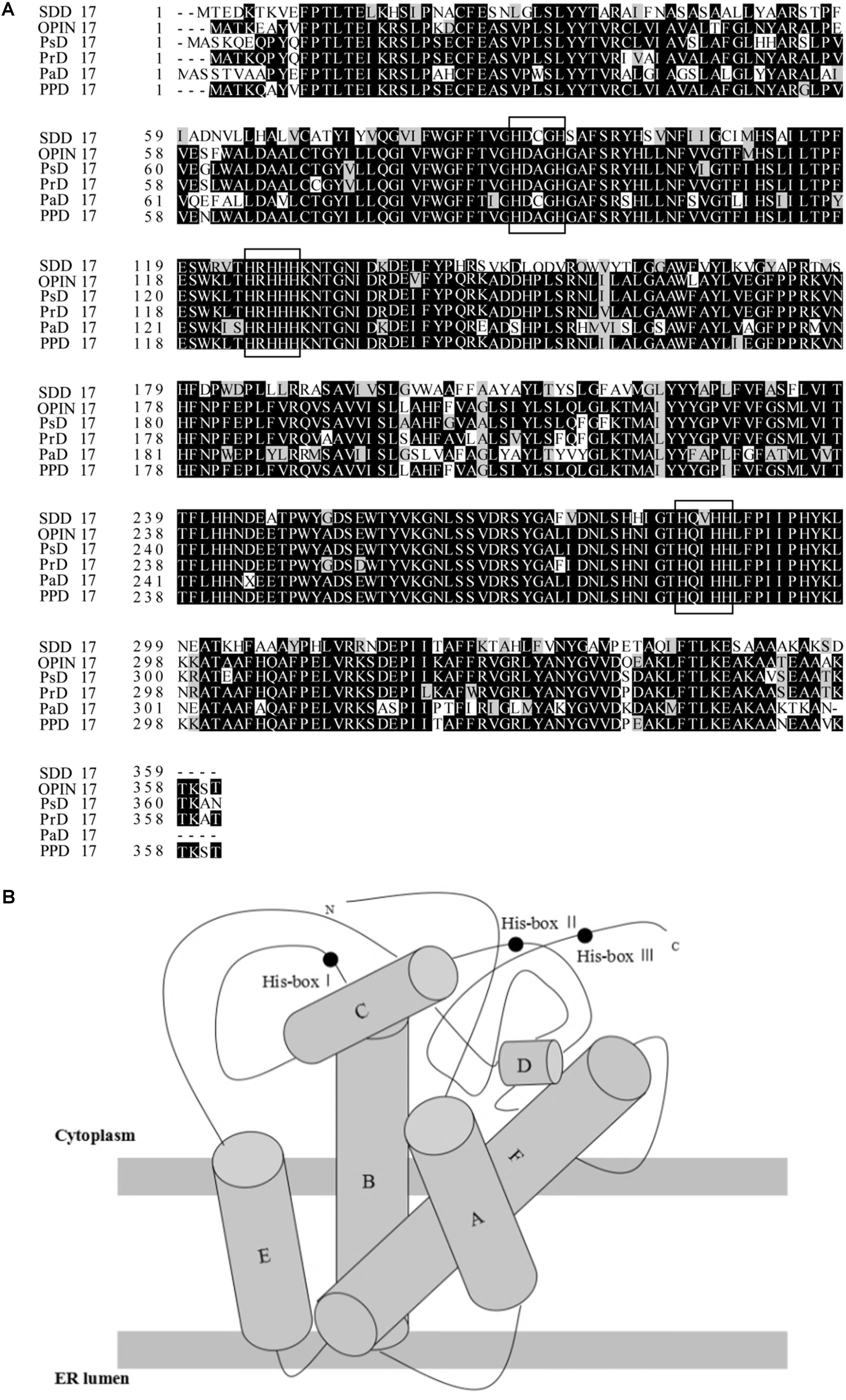
FIGURE 1. (A) Sequence comparison of selected omega-3 desaturases using the Clustal W (SDD17, Saprolegnia diclina delta-17 desaturase; OPIN17, Phytophthora infestans delta-17 desaturase; PrD17, Phytophthora ramorum delta-17 desaturase; PsD17, Phytophthora sojae delta-17 desaturase; PaD17, Pythium aphanidermatum delta-17 desaturase; PPD17, a sequence from Phytophthora parasitica). (B) Topology model of omega-3 desaturase PPD17.
Characterization of Substrate Specificity of PPD17 in S. cerevisiae
To identify the activity and characterize the substrate specificity of PPD17, the desaturase was codon optimized and expressed in S. cerevisiae. The results of DNA agarose electrophoresis, RT-qPCR and Weston bolt analysis showed that oPPD17 was successfully expressed, transcribed and translated in the recombinant strains INVSc1 (Figure 2). To acquire insight into substrate requirements of PPD17, different fatty acid substrates were supplied in the media. The transformant INVSc1 containing pYES2/NT C-oPPD17 grown on the media supplemented with LA (converting into α-linolenic acid, ALA, C18:3), GLA (converting into stearidonic acid, SDA, C18:4), DGLA (converting into eicosatetraenoic acid, ETA, C20:4) and AA (converting into EPA), respectively, at 12°C°C and 28°C (Figure 3). The results indicated PPD17 convert 18C-PUFAs at a low efficiency (<10%) while the efficiency on 20C-PUFAs is much higher, especially AA, which almost reached up to 50%. In addition, compared with low temperature (12°C), the conversion rate was high at ordinary temperature (28°C), which was beneficial for the application of this enzyme. Furthermore, the effects of different AA-added concentration on the conversion rate of the yeast transformants was analyzed, it was found that AA conversion rate decreased as AA-added concentration increasing and the highest conversion rate (∼70%) was achieved when added 0.05 mM AA (Figure 4).
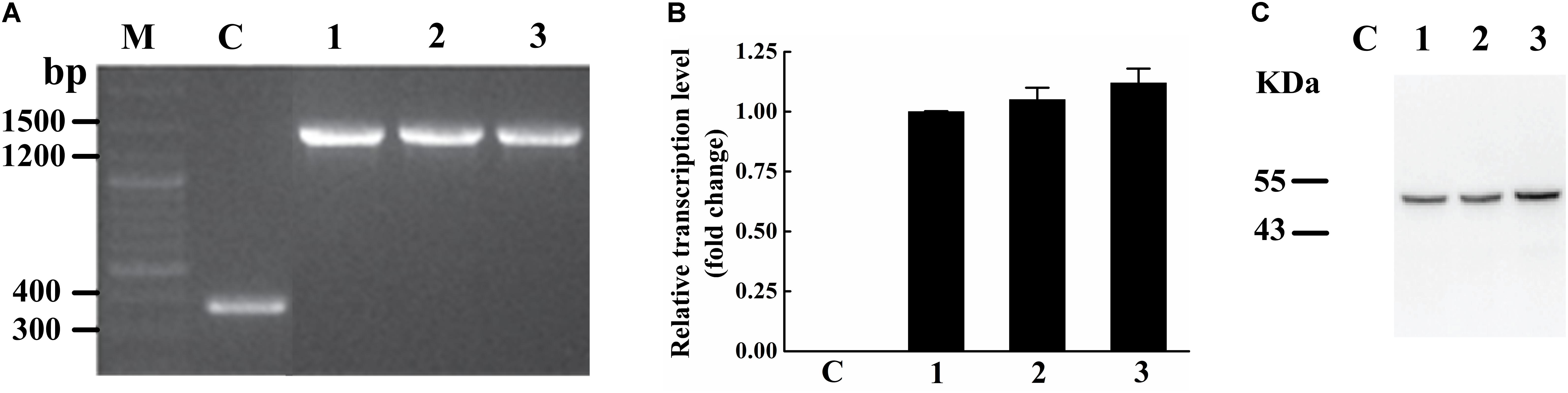
FIGURE 2. Verification of transformants expressing PPD17. (A) Map of fragment bands in the genome. (B) PPD17 relative transcription level. (C) PPD17 translation level. M, nucleic acid marker; C, INVSc1-pYES2/NT control strains; 1–3, INVSc1 (pYES2/NT C-oPPD17-1/2/3).
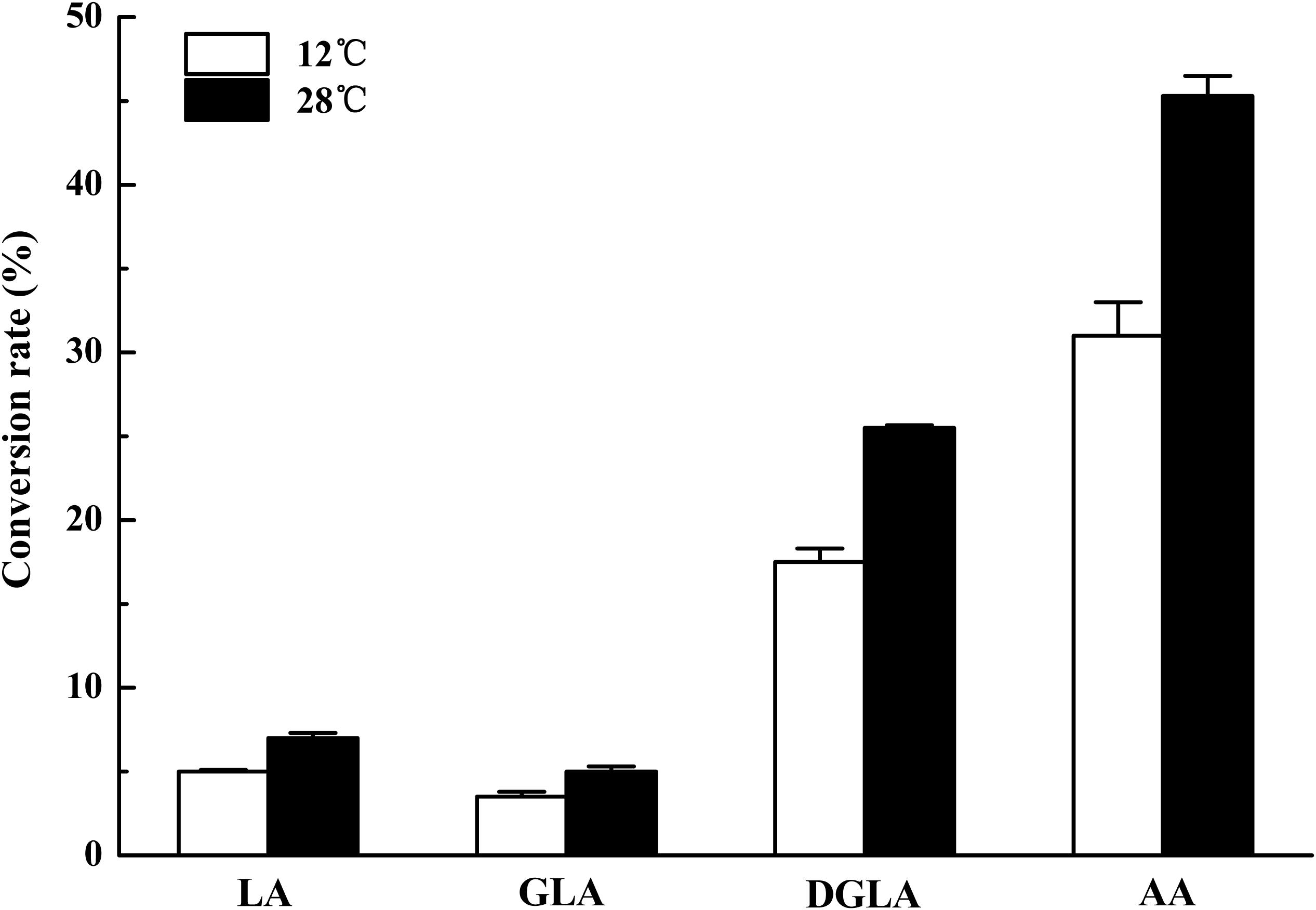
FIGURE 3. Conversion rate of yeast transformants on different fatty acids at different temperature. The transformants were cultivated in 0.2 mM LA, GLA, DGLA, and AA. Conversion rate is calculated as 100% × Product/(Product + Substrate).
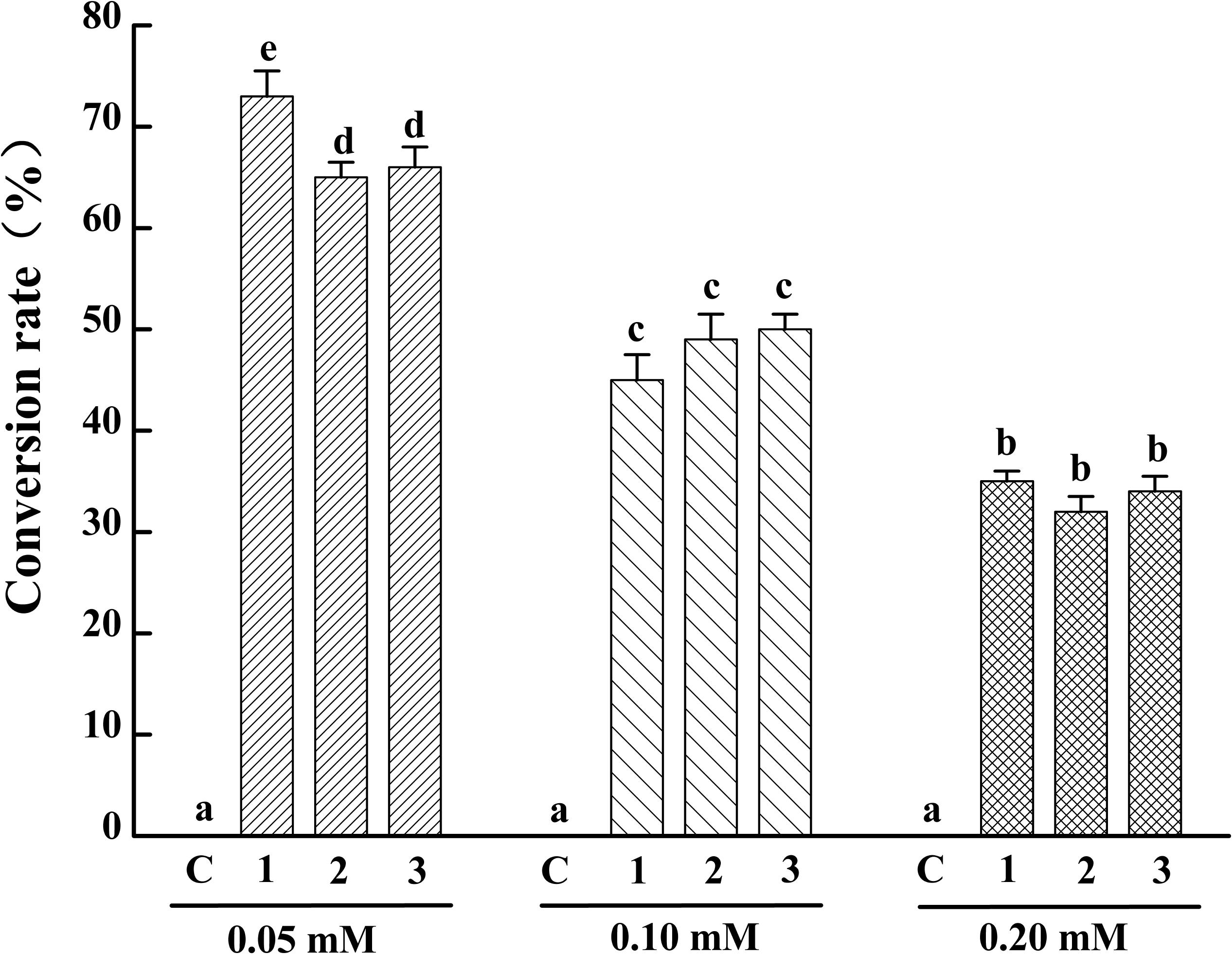
FIGURE 4. Conversion rate of yeast transformants with different concentration AA adding. C, Control, INVSc1 (pYES2/NT C); 1–3, yeast transformants harboring pYES2/NT C-oPPD17-1/2/3. Values share no common superscripts were significantly different (P < 0.05).
Transformation of PPD17 in M. alpina and Fatty Acid Analysis
Mortierella alpina can accumulated a large number of AA (more than 50% of total fatty acids) (Wang et al., 2011) and PPD17 has a great capability of converting AA to EPA, therefore we codon optimized PPD17 gene (oPaFADS17) and heterologously expressed it in M. alpina auxotrophic strain for EPA production. In this study, five stable transformants were acquired. The ura5 (818 bp) and oPpFADS17 (1,250 bp) fragments indicate that the oPpFADS17 were successful inserted into the M. alpina transformants’ genome (Figure 5A). The RT-qPCR analysis showed that oPpFADS17 was successfully transcribed in all of the five transformants (Figure 5B).
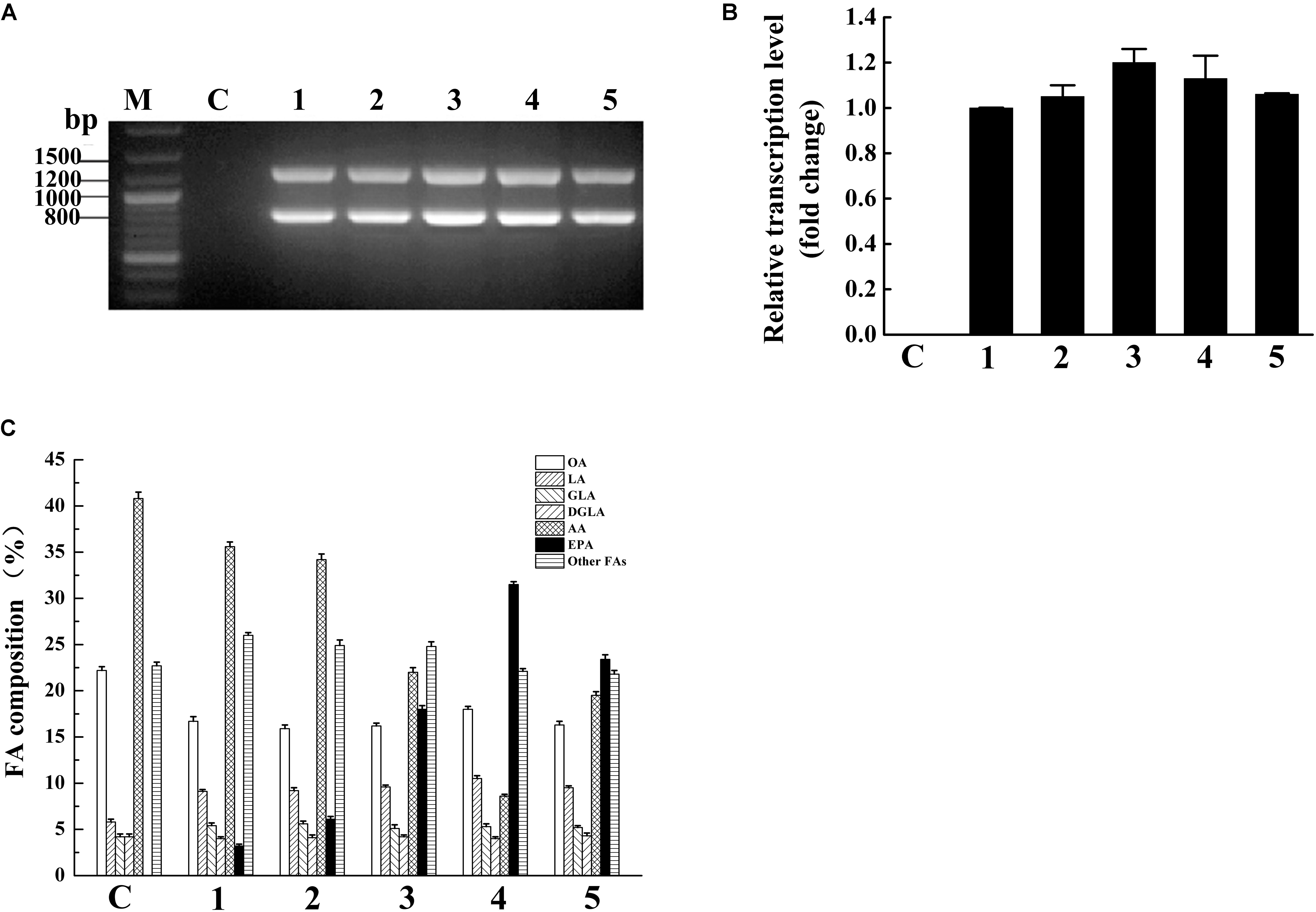
FIGURE 5. Verification of oPpFADS17 fragments in the genome (A), and relative transcription level (B), and fatty acid composition (C) in M. alpina expressing oPpFADS17. M, marker; C, M. alpina (control); 1–5, M. alpina-oPpFADS17-(1–5).
Fatty acid methyl ester analysis showed that the transformants and wild-type M. alpina had a similar fatty acid composition except for AA and EPA (Figure 5C). The wild-type M. alpina had no significant EPA accumulation. Although all of the five transformants could accumulate EPA, their EPA accumulation capacity were obviously different due to random integration based on ATMT method (Ando et al., 2009), which resulting in different effect on EPA production. Among all of the transformants, MA-oPpFADS17-4 (named M. alpina CCFM 698) exhibited the strongest capacity to accumulate EPA, and the EPA content in the TFA increased up to 31.5%. In this transformant, the accumulated AA decreased to 8.6% and the conversion rate of AA to EPA reached 78.6%.
Effects of Carbon/Nitrogen Source on EPA Production
The carbon substrates are important to cell growth and lipid accumulation in oleaginous microorganisms, and M. alpina can use a variety of inexpensive starch-based carbon sources for lipid production (Zhu et al., 2003; Nisha and Venkateswaran, 2011). To enhance EPA production and reduce the fermentation cost, the effects of different carbon sources on CDW, TFA, and EPA production in the recombinant M. alpina CCFM 698 were investigated (Figure 6A). Compared to other carbon sources, corn starch stimulated the growth of this fungus and the CDW reached up to 12.5 g/L, which was 24% high than that of glucose (10.1 g/L). However, the CDW grown on soluble starch and potato starch were no more than glucose. The TFA and EPA yield revealed corn starch and glucose are beneficial to TFA and EPA accumulation. The EPA production grown on corn starch and glucose were up to 1.2 g/L and 1.0 g/L, respectively, which were significantly higher than that grown on soluble starch and potato starch. Thus, corn starch and glucose are favorable carbon sources for EPA production in M. alpina CCFM 698.
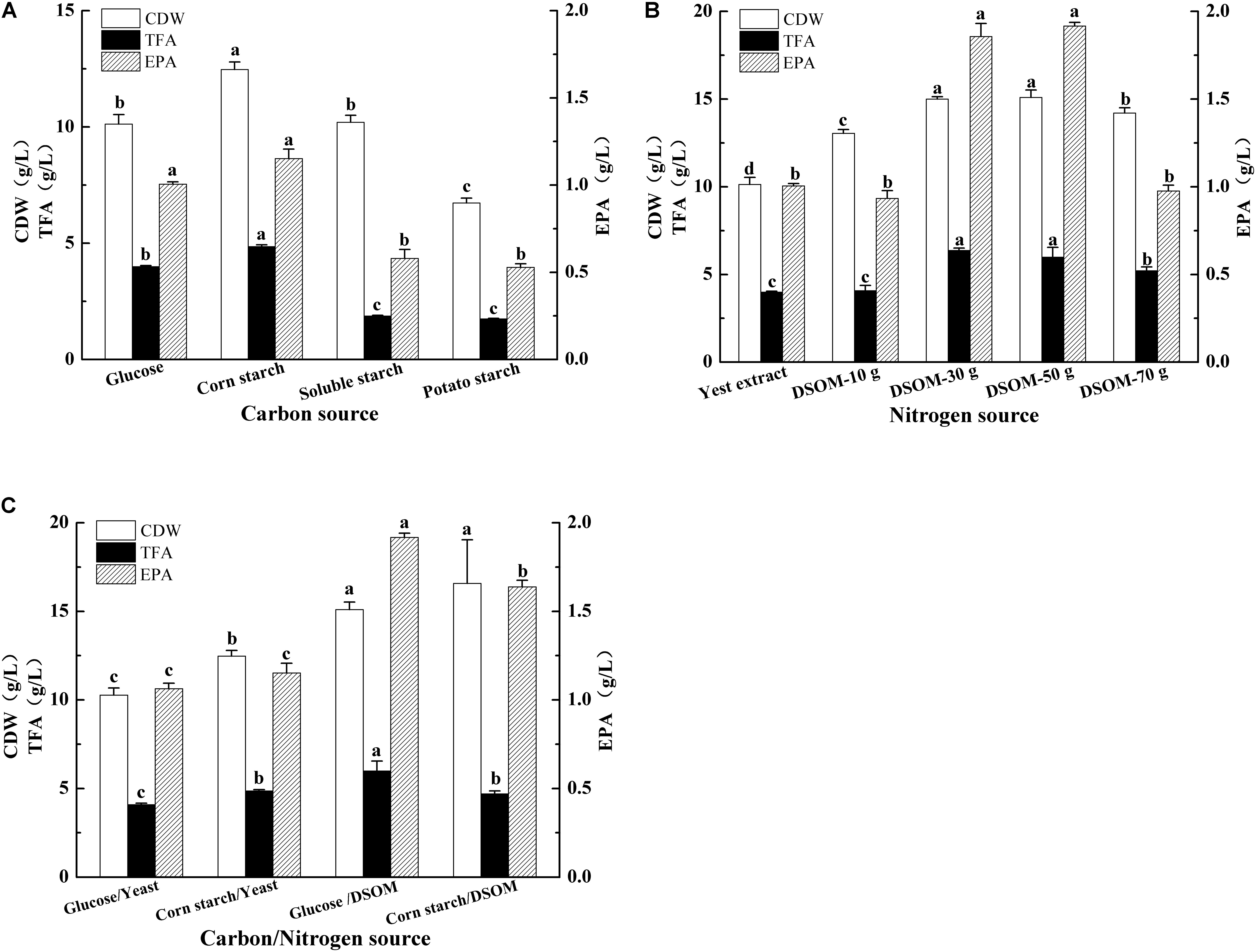
FIGURE 6. Effects of different carbon (A), nitrogen (B), combination (C) sources on cell dry weight (CDW), total fatty acids (TFAs), and EPA production of the recombinant M. alpina CCFM 698.
Nitrogen are key components in the growth media for microorganisms and the inexpensive nitrogen source soybean meal had been reported to enhance the biomass and fatty acid production in M. alpina (Cao et al., 2015). As the mycelia and soybean meal are hard to be separated during biomass collection, M. alpina CCFM 698 was grown on defatted soybean meal (DSOM) in this study. As shown in Figure 6B, compared to yeast extract, the high EPA-produced strain grown on 30 g/L and 50 g/L DSOM achieved higher CDW, TFA, and EPA yield. When M. alpina CCFM 698 was grown on 50 g/L DSOM, the EPA yield reach the highest value 1.9 g/L.
These above results suggested corn starch and glucose are favorable carbon sources, and 50 g/L DSOM is appropriate nitrogen source for EPA production in the high EPA-produced strain M. alpina CCFM 698. To obtain the optimal carbon/nitrogen source recipe, this strain was grown on glucose/DMSO and corn starch/DMSO, respectively, and using the yeast extract nitrogen source as control (Figure 6C). Compared with corn starch/DSOM, glucose/DMSO gave higher TFA and EPA yield although their CDW had no significant difference, this may due to the glucose is more beneficial to lipid accumulation. Thus, in this study, the highest EPA production was obtained when grown in glucose and DSOM, and the highest EPA yield was up to 1.9 g/L.
Discussion
Phytophthora is known to produce LC-PUFAs (e.g., AA and EPA), and synthesize these PUFAs via desaturase/elongase pathway rather than polyketide pathway (Fu et al., 2013; Xue et al., 2013a). Thus, it is highly possible to screen new desaturase or elongase genes for EPA pathway in these microorganisms. In this study, a new omega-3 desaturase in P. parasitica was identified and named PPD17. The new sequence PPD17 has high similarities with the published desaturases (Pereira et al., 2004; Fu et al., 2013; Xue et al., 2013a; Okuda et al., 2015), and contains three typical histidine motifs HDAGH, HRHHH, HQIHH, suggesting it owns the characteristic of membrane-bound desaturase. These motifs were proposed to be related to the iron atoms ligation in the active-site. Meanwhile, PPD17 contains four high hydrophobic regions clusters, which are the putative membrane spanning helices.
Omega-3 desaturase (e.g., delta-17 desaturase) is necessary for conversion of omega-6 PUFAs into their omega-3 counterparts and play a vital role in EPA synthesis from AA (Xue et al., 2013a; Xie et al., 2015). The activity and substrate preference of PPD17 are analyzed by expressing the protein in S. cerevisiae. Fatty acid feeding experiments showed that PPD17 had stronger substrate preference for C20-PUFA substrate AA and DGLA, especially AA, and lower preference for C18-PUFA substrate LA and GLA, which was in according with the identified delta-17 desaturase PaD17, PrD17, and PsD17 (Xue et al., 2013a). However, the other delta-17 desaturase SDD17 was found to be an exclusive desaturase for C20-PUFA and had no activity on C18 substrates (Pereira et al., 2004). In contrast to SSD17, the PPD17 had the activity of delta-15 desaturase and can convert LA and GLA to ALA and SDA, respectively. When LA and AA were fed simultaneously, the conversation rate of AA was much higher than LA, further indicating that PPD17 is much more preference AA. The highest conversion rate to AA can reach 70% when 0.05 mM AA was added at ordinary temperature 28°C, suggesting that PPD17 has great potential to produce EPA from AA.
Oleaginous fungus M. alpina can accumulate 50% AA of total fatty acid composed by different 18C- and 20C-PUFAs (Wang et al., 2011). Furthermore, M. alpina also can accumulate little EPA at low temperature (Shimizu et al., 1989; Sakuradani et al., 2005). There has been researcher attempted to enhance EPA production in M. alpina through overexpressing endogenous omega-3 desaturase, but the EPA production was no more than 0.7 g/L and needed to be cultured at 12°C rather than ordinary temperature (Takeno et al., 2004). For engineering an organism with high EPA production, the conversion efficiency of the omega-3 desaturase used is one of the important factors, and the high activity of the newly identified PPD17 render its excellent choices for genetic engineering in the EPA production. In the present study, the codon optimized PPD17 gene was successful heterologously expressed in M. alpina using the A. tumefaciens transformation method and the conversion rate of AA to EPA in a transformant MA-oPpFADS17-4 reached up to 78.6%, which were higher than that in engineered EPA-produced Y. lipolytica strains which harbored the omega-3 desaturase PaD17, PrD17, or PsD17 (Xue et al., 2013a). Furthermore, the conversion rate of AA to EPA of PPD17 in this strain was also much higher than PaD17 (49.7%) from our previous study (Ge et al., 2017). The EPA content of the TFA in PPD17-harboring M. alpina transformant increased up to 31.5%, which was higher than PaD17-harboring M. alpina transformant (18.7%) (Ge et al., 2017), but less than that of recombination Y. lipolytica (56.6%) which assembled 30 copies of nine different heterologous genes (Xue et al., 2013b; Xie et al., 2015). However, M. alpina, as an oleaginous filamentous fungus producing commercial AA, was introduced a single-copy of PPD17 coding gene and was able to convert nearly 80% AA to EPA. M. alpina transformant integrated PPD17 may have great potential for future commercial EPA production.
Carbon substrates and nitrogen sources are very vital chemical compounds in microorganism cultivation. M. alpina can use various carbon and nitrogen sources for growth and lipid accumulation (Nisha and Venkateswaran, 2011; Yu et al., 2011). Our results demonstrated the inexpensive carbon source corn starch enhanced the biomass and TFA production in the recombinant M. alpina CCFM 698. For the development of PUFAs production by large-scale fermentation process, it is necessary to utilize inexpensive medium components to reduce the cost (Zhu et al., 2003). Thus, the corn starch has a potential of industrialize applications as carbon source for EPA production in M. alpina. For nitrogen source, organic nitrogen compounds are in favor of cell growth and lipid accumulation compared with inorganic nitrogen sources (Lu et al., 2011). Compared with the common organic nitrogen yeast extract, the DSOM obviously increased the biomass, TFA, and EPA yield in M. alpina CCFM 698, which is in accordance with the recent finding that the inexpensive nitrogen source soybean meal could increase the biomass and fatty acid production in M. alpina (Cao et al., 2015). Indeed, the highest EPA yield reached up to 1.9 g/L when the recombinant grown on 50 g/L DSOM at ordinary temperature in this study. This EPA yield was much higher than that (0.6 g/L) in our previous engineered strain which was heterogeneous expressed an ALA-preferring delta-6 desaturase (Shi et al., 2016). Nevertheless, there has been other study reported that EPA production was up to 1.8 g/L through expressing the S. diclina delta-17 desaturase, which was little less than our result (Okuda et al., 2015). Furthermore, the EPA yield of PPD17-harboring M. alpina transformant in this study was also higher than PaD17-harboring transformant which yield 1.7 g/L EPA after a series of fermentation optimization (Ge et al., 2017). PPD17 was first discovered and characterized in this study and the conversion rate of AA to EPA with PPD17 was much higher than PaD17.
In general, this recombinant M. alpina CCFM 698 has great potential for EPA industrialized production. Furthermore, although optimal carbon/nitrogen source recipe indicated that the glucose/DMSO gave higher EPA yield than corn starch/DSOM, the low cost of corn starch might make this carbon source own a broad application prospect.
Conclusion
In conclusion, a new omega-3 desaturase PPD17 from Phytophthora parasitica was reported in this study. This new omega-3 desaturase shares high similarities with known omega-3 desaturases and has a wide omega-6 fatty acid substrate preference including both 18-C and 20-C omega-6 fatty acids with strong delta-17 desaturase activity but weaker delta-15 desaturase activity. The conversion rate from AA to EPA was up to 70% when this desaturase was expressed in S cerevisiae. For obtaining a high EPA-producing strain, this desaturase was transformed into Mortierella alpina, an AA-producing filamentous fungus. The EPA content of total fatty acid in reconstruction strains reached 31.5% and followed by fermentation optimization the EPA yield was up to 1.9 g/L.
Author Contributions
XT, TM, and CG carried out the experiments and drafted the manuscript. HC and ZG analyzed the data and helped to draft the manuscript. HC, HZ, YC, and WC conceived and designed the study and revised the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (31722041 and 31530056), the Fundamental Research Funds for the Central Universities (JUSRP51702A), the Natural Science Foundation of Jiangsu Province (BK20160172), the Project funded by China Postdoctoral Science Foundation (2017M611701), the Postdoctoral Science Foundation of Jiangsu Province (1701061C), and the Jiangsu Province “Collaborative Innovation Center for Food Safety and Quality Control.”
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer GP and handling Editor declared their shared affiliation at time of review.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01878/full#supplementary-material
References
Ando, A., Sumida, Y., Negoro, H., Suroto, D. A., Ogawa, J., Sakuradani, E., et al. (2009). Establishment of Agrobacterium tumefaciens-mediated transformation of an oleaginous fungus, Mortierella alpina 1S-4, and its application for eicosapentaenoic acid producer breeding. Appl. Environ. Microbiol. 75, 5529–5535. doi: 10.1128/AEM.00648-09
Cao, G., Guan, Z., Liao, X., and Cai, Y. (2015). Arachidonic acid production by Mortierella alpina using raw crop materials. Acta Sci. Pol. Technol. Aliment. 14, 133–143. doi: 10.17306/J.AFS.2015.2.15
Chen, H., Gu, Z., Zhang, H., Wang, M., Chen, W., Lowther, W. T., et al. (2013). Expression and purification of integral membrane fatty acid desaturases. PLoS One 8:e58139. doi: 10.1371/journal.pone.0058139
Fu, Y., Fan, X., Li, X., Wang, H., and Chen, H. (2013). The desaturase OPIN17 from Phytophthora infestans converts arachidonic acid to eicosapentaenoic acid in CHO cells. Appl. Biochem. Biotechnol. 171, 975–988. doi: 10.1007/s12010-013-0332-x
Ge, C., Chen, H., Mei, T., Tang, X., Chang, L., Gu, Z., et al. (2017). Application of a ω-3 desaturase with an arachidonic acid preference to eicosapentaenoic acid production in Mortierella alpina. Front. Bioeng. Biotechnol. 5:89. doi: 10.3389/fbioe.2017.00089
Gietz, D., St Jean, A., Woods, R. A., and Schiestl, R. H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. doi: 10.1093/nar/20.6.1425
Hao, G., Chen, H., Gu, Z., Zhang, H., Chen, W., and Chen, Y. Q. (2016). Metabolic engineering of Mortierella alpina for enhanced arachidonic acid production through the NADPH supplying strategy. Appl. Environ. Microbiol. 82, 3280–3288. doi: 10.1128/AEM.00572-16
Hao, G., Chen, H., Wang, L., Gu, Z., Song, Y., Zhang, H., et al. (2014). Role of malic enzyme during fatty acid synthesis in the oleaginous fungus Mortierella alpina. Appl. Environ. Microbiol. 80, 2672–2678. doi: 10.1128/AEM.00140-14
Ji, X., Ren, L., and Huang, H. (2015). Omega-3 biotechnology: a green and sustainable process for omega-3 fatty acids production. Front. Bioeng. Biotechnol. 3:158. doi: 10.3389/fbioe.2015.00158
Ji, X., Ren, L., Nie, Z., Huang, H., and Ouyang, P. (2014). Fungal arachidonic acid-rich oil: research, development and industrialization. Crit. Rev. Biotechnol. 34, 197–214. doi: 10.3109/07388551.2013.778229
Kyle, D. J. (2001). “The large-scale production and use of a single-cell oil highly enriched in docosahexaenoic acid,” in Omega-3 Fatty Acids, eds F. Shahidi and J. W. Finley (New York, NY: ACS Publications), 92–107.
Lu, J., Peng, C., Ji, X., You, J., Cong, L., Ouyang, P., et al. (2011). Fermentation characteristics of Mortierella alpina in response to different nitrogen sources. Appl. Biochem. Biotechnol. 164, 979–990. doi: 10.1007/s12010-011-9189-z
Meesapyodsuk, D., Reed, D. W., Savile, C. K., Buist, P. H., Ambrose, S. J., and Covello, P. S. (2000). Characterization of the regiochemistry and cryptoregiochemistry of a Caenorhabditis elegans fatty acid desaturase (FAT-1) expressed in Saccharomyces cerevisiae. Biochemistry 39, 11948–11954. doi: 10.1021/bi000756a
Nisha, A., and Venkateswaran, G. (2011). Effect of culture variables on mycelial arachidonic acid production by Mortierella alpina. Food Bioprocess Technol. 4, 232–240. doi: 10.1007/s11947-008-0146-y
Niu, B., Guo, L., Zhao, M., Luo, T., Zhang, R., Zhang, F., et al. (2008). Molecular cloning, characterization, and expression of an omega-3 fatty acid desaturase gene from Sapium sebiferum. J. Biosci. Bioeng. 106, 375–380. doi: 10.1263/jbb.106.375
Okuda, T., Ando, A., Negoro, H., Muratsubaki, T., Kikukawa, H., Sakamoto, T., et al. (2015). Eicosapentaenoic acid (EPA) production by an oleaginous fungus Mortierella alpina expressing heterologous the Δ17-desaturase gene under ordinary temperature. Eur. J. Lipid Sci. Technol. 117, 1919–1927. doi: 10.1002/ejlt.201400657
Pereira, S. L., Huang, Y.-S., Bobik, E. G., Kinney, A. J., Stecca, K. L., Packer, J. C., et al. (2004). A novel omega3-fatty acid desaturase involved in the biosynthesis of eicosapentaenoic acid. Biochem. J. 378, 665–671. doi: 10.1042/bj20031319
Ruiz-Lopez, N., Usher, S., Sayanova, O. V., Napier, J. A., and Haslam, R. P. (2015). Modifying the lipid content and composition of plant seeds: engineering the production of LC-PUFA. Appl. Microbiol. Biotechnol. 99, 143–154. doi: 10.1007/s00253-014-6217-2
Sakuradani, E., Abe, T., Iguchi, K., and Shimizu, S. (2005). A novel fungal ω3-desaturase with wide substrate specificity from arachidonic acid-producing Mortierella alpina 1S-4. Appl. Microbiol. Biotechnol. 66, 648–654. doi: 10.1007/s00253-004-1760-x
Sakuradani, E., Ando, A., Shimizu, S., and Ogawa, J. (2013). Metabolic engineering for the production of polyunsaturated fatty acids by oleaginous fungus Mortierella alpina 1S-4. J. Biosci. Bioeng. 116, 417–422. doi: 10.1016/j.jbiosc.2013.04.008
Shi, H., Chen, H., Gu, Z., Song, Y., Zhang, H., Chen, W., et al. (2015). Molecular mechanism of substrate specificity for delta 6 desaturase from Mortierella alpina and Micromonas pusilla. J. Lipid Res. 56, 2309–2321. doi: 10.1194/jlr.M062158
Shi, H., Chen, H., Gu, Z., Zhang, H., Chen, W., and Chen, Y. Q. (2016). Application of a delta-6 desaturase with α-linolenic acid preference on eicosapentaenoic acid production in Mortierella alpina. Microb. Cell Fact. 15:117. doi: 10.1186/s12934-016-0516-5
Shimizu, S., Kawashima, H., Akimoto, K., Shinmen, Y., and Yamada, H. (1989). Conversion of linseed oil to an eicosapentaenoic acid-containing oil by Mortierella alpina 1S-4 at low temperature. Appl. Microbiol. Biotechnol. 32, 1–4. doi: 10.1007/BF00164813
Sun, L., Ren, L., Zhuang, X., Ji, X., Yan, J., and Huang, H. (2014). Differential effects of nutrient limitations on biochemical constituents and docosahexaenoic acid production of Schizochytrium sp. Bioresour. Technol. 159, 199–206. doi: 10.1016/j.biortech.2014.02.106
Takeno, S., Sakuradani, E., Murata, S., Inohara-Ochiai, M., Kawashima, H., Ashikari, T., et al. (2004). Establishment of an overall transformation system for an oil-producing filamentous fungus, Mortierella alpina 1S-4. Appl. Microbiol. Biotechnol. 65, 419–425. doi: 10.1007/s00253-004-1622-6
Venegas-Caleron, M., Muro-Pastor, A. M., Garces, R., and Martinez-Force, E. (2006). Functional characterization of a plastidial omega-3 desaturase from sunflower (Helianthus annuus) in cyanobacteria. Plant Physiol. Biochem. 44, 517–525. doi: 10.1016/j.plaphy.2006.09.005
Wang, L., Chen, W., Feng, Y., Ren, Y., Gu, Z., Chen, H., et al. (2011). Genome characterization of the oleaginous fungus Mortierella alpina. PLoS One 6:e28319. doi: 10.1371/journal.pone.0028319
Xie, D., Jackson, E. N., and Zhu, Q. (2015). Sustainable source of omega-3 eicosapentaenoic acid from metabolically engineered Yarrowia lipolytica: from fundamental research to commercial production. Appl. Microbiol. Biotechnol. 99, 1599–1610. doi: 10.1007/s00253-014-6318-y
Xie, D., Miller, E., Sharpe, P., Jackson, E., and Zhu, Q. (2017). Omega-3 production by fermentation of Yarrowia lipolytica: from fed-batch to continuous. Biotechnol. Bioeng. 114, 798–812. doi: 10.1002/bit.26216
Xue, Z., He, H., Hollerbach, D., Macool, D. J., Yadav, N. S., Zhang, H., et al. (2013a). Identification and characterization of new Δ-17 fatty acid desaturases. Appl. Microbiol. Biotechnol. 97, 1973–1985. doi: 10.1007/s00253-012-4068-2
Xue, Z., Sharpe, P. L., Hong, S.-P., Yadav, N. S., Xie, D., Short, D. R., et al. (2013b). Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica. Nat. Biotechnol. 31, 734–740. doi: 10.1038/nbt.2622
Yu, A. Q., Zhu, J. C., Zhang, B., Xing, L. J., and Li, M. (2011). Effects of different carbon sources on the growth, fatty acids production, and expression of three desaturase genes of Mortierella alpina ATCC 16266. Curr. Microbiol. 62, 1617–1622. doi: 10.1007/s00284-011-9902-8
Keywords: omega-3 desaturase, PPD17, Mortierella alpina, EPA, carbon/nitrogen source
Citation: Tang X, Chen H, Mei T, Ge C, Gu Z, Zhang H, Chen YQ and Chen W (2018) Characterization of an Omega-3 Desaturase From Phytophthora parasitica and Application for Eicosapentaenoic Acid Production in Mortierella alpina. Front. Microbiol. 9:1878. doi: 10.3389/fmicb.2018.01878
Received: 14 March 2018; Accepted: 26 July 2018;
Published: 14 August 2018.
Edited by:
Giovanna Suzzi, Università degli Studi di Teramo, ItalyReviewed by:
Xiao-Jun Ji, Nanjing Tech University, ChinaGiorgia Perpetuini, Università degli Studi di Teramo, Italy
Copyright © 2018 Tang, Chen, Mei, Ge, Gu, Zhang, Chen and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiqin Chen, aGFpcWluY2hlbkBqaWFuZ25hbi5lZHUuY24=
 Xin Tang1,2
Xin Tang1,2 Haiqin Chen
Haiqin Chen Zhennan Gu
Zhennan Gu Yong Q. Chen
Yong Q. Chen