- 1Center for Biomedical Research, Northwest Minzu University, Lanzhou, China
- 2State Key Laboratory of Veterinary Etiological Biology, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Lanzhou, China
Viral infections trigger the innate immune system, serving as the first line of defense, and are characterized by the production of type I interferon (IFN). Type I IFN is expressed in a broad spectrum of cells and tissues in the host and includes various subtypes (IFN-α, IFN-β, IFN-δ, IFN-ε, IFN-κ, IFN-τ, IFN-ω, IFN-ν, and IFN-ζ). Since the discovery of type I IFN, our knowledge of the biology of type I IFN has accumulated immensely, and we now have a substantial amount of information on the molecular mechanisms of the response and induction of type I IFN, as well as the strategies utilized by viruses to evade the type I IFN response. Foot-and-mouth disease virus (FMDV) can selectively alter cellular pathways to promote viral replication and evade antiviral immune activation of type I IFN. RNA molecules generated by FMDV are sensed by the cellular receptor for pathogen-associated molecular patterns (PAMPs). FMDV preferentially activates different sensor molecules and various signal transduction pathways. Based on knowledge of the virus or RNA pathogen specificity as well as the function-structure relationship of RNA sensing, it is necessary to summarize numerous signaling adaptors that are reported to participate in the regulation of IFN gene activation.
Introduction
Foot-and-mouth disease virus (FMDV) belongs to the Aphthovirus genus in the Picornaviridae family, and a highly infection disease caused by FMDV is regarded as an important concern for animal health (Knight-Jones et al., 2016). During FMDV evolutionary process, high mutation rates of the viral genome and quasispecies dynamics are considered major genetic factors (Shih et al., 1992). Thus, a series of studies were conducted to examine the relationship between genetic changes of the viral genome and viral fitness and different host/viral pathogenicities. Except for positive/negative selection and the random drift of the genome (Domingo et al., 2003), synonymous codon usage patterns of the FMDV genome also dominate its host ranges and viral proteins with normal biological functions (Zhou et al., 2010a,b, 2011, 2013a,b,c; Ahn et al., 2011; Ma et al., 2013; Ma X.X. et al., 2016; Gao et al., 2014). Due to the high genetic diversity of FMDV, the measures for controlling this disease need to be developed comprehensively, including killing infected and in-contact animals, the limitation of animal movement and vaccination based on conventional vaccines or new typical ones (Robinson et al., 2016). To further improve measures involved in antiviral treatments and novel vaccines for controlling rapid FMDV spread, it is important to obtain a deep understanding of the interaction between the host and FMDV. The antiviral immune response is the major focus on resisting FMDV infection, including innate/adaptive immune activations (Golde et al., 2008; Toka and Golde, 2013). The innate immune system serves as the first line of defense for resisting viral infections. The rapid induction of type I interferon (IFN) and other antiviral cytokines at the site of infection are part of the defense involved in antiviral immunity. The type I IFN family of placental mammals comprises 9 recognized classes identified to date: IFN-α, IFN-β, IFN-δ, IFN-ε, IFN-κ, IFN-τ, IFN-ω, IFN-ν, and IFN-ζ (Krause and Pestka, 2005; Detournay et al., 2013).
Type I IFNs exhibit direct antiviral activities by inhibiting viral replication and mediating the cellular immune functions of both the innate and adaptive immune system, resulting in both early limitation of the virus and long-term immunity. However, viruses are capable of selecting various strategies to evade the host immune system and thus contributing to viral pathogenicity (Schulz and Mossman, 2004; Jackson et al., 2017; Sumner et al., 2017). For FMDV infection, type I IFNs also play important roles in counteracting viral infection represent a potent biotherapeutic method against FMDV (Rodríguezpulido et al., 2011; Borrego et al., 2017). This minireview summarizes the current knowledge on how type I IFN is resistant to FMDV infection and how FMDV counteracts type I IFN induction and signaling transduction to evade the type I IFN system of host.
Recognition of Viral Genome for IFN Production
Once viral infection occurs, cells of the infected host can trigger a series of activations of cytokines, including type I and type III IFNs. These IFNs can perform multiple biological functions related to antiviral, antiproliferative and immunomodulatory activations and trigger various interferon stimulated genes (ISGs), thereby contributing to the establishment of the antiviral state in which various steps of viral replication are restricted (Sen and Sarkar, 2007; Fensterl et al., 2015). Notably, both type I and type III IFNs represent similar patterns of expression and mechanisms of induction (Uzé and Monneron, 2007). A pivotal feature of IFN expression is the requirement for detection of the invading pathogens by pathogen-associated receptors. Generally, the innate immune system relies on germ-line-encoded pattern recognition receptors (PRRs) to recognize non-self RNA (viral RNA) which is one of the pathogen-associated molecular patterns (PAMPs) (Akira et al., 2006). For recognizing the viral RNA genome by the PRRs, there are two major classes of PRRs: Toll-like receptor (TLRs) at the cell surface or in endosomes, and retinoic acid-inducible gene-I (RIG-I) like receptors (RLRs) (Akira et al., 2006; Bruns et al., 2014). Among TLRs related to antiviral immune response, TLR 3 can recognize double-stranded RNA (ds RNA) and TLRs 7/8 can recognize single-stranded RNA (Alexopoulou et al., 2001; Diebold et al., 2004; Heil et al., 2004). However, TLR3 can sense ssRNA of poliovirus in some conditions (Tatematsu et al., 2013). Turning to RIG-I like receptor, which is involved in the antiviral immune response (Figure 1), melanoma differentiation-associated gene 5 (MDA5), laboratory of genetics and physiology 2 (LGP2) and RIG-I, which are ubiquitous cytosolic RNA helicases, play a pivotal role in recognizing viral RNA fragments (Yoneyama et al., 2005). It has been accepted that RIG-I can preferentially sense short dsRNAs, while MDA5 can recognize long dsRNA (Kato et al., 2008; Berke et al., 2013; Liu et al., 2016). Compared with the physical organizations and biological functions of MDA5 and RIG-I, LGP2 loses its CARD domain and displays a regulatory role that works as a concentration dependent biphasic status to mediate activations of MDA5 (Bruns et al., 2013, 2014; Uchikawa et al., 2016). Interestingly, RIG-I deficiency does not inhibit the replication of Picornaviridae, while MDA5 displays a remarkable role in resisting these viruses (Kato et al., 2006). Depending on the mice model with LGP2 deletion, dendritic cells (DCs) derived from these mice fail to generate IFN production upon infection by encephalomyocarditis virus (EMCV), vesicular stomatitis virus (VSV), and Newcastle disease virus (NDV) (Venkataraman et al., 2007; Satoh et al., 2010), suggesting that LGP2 might play a positive role in vitro by recognize some viral infections to promote IFN production.
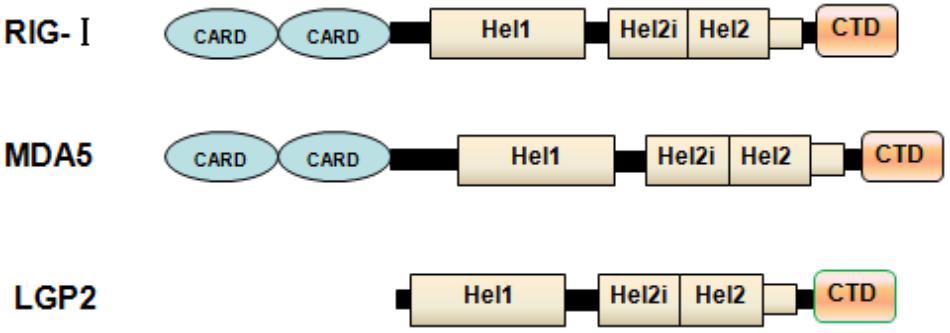
FIGURE 1. Physical organization of RLs. Schematic representation of RLR domains. The caspase activation and recruitment domain (CARD), helicase domain and RNA recognition domain are shown. LGP2 lacks the N-terminal CARD domain used by MDA5 and RIG-I for signal transduction.
Recognition of FMDV RNA by TLRS and RLRS
The differential viral recognition by RIG-I and MDA5 is partly due to the differential recognition of distinct types of RNA patterns. It has been reported that MDA5 can recognize ssRNA involved in EMCV, Theiler’s virus and Mengo virus to activate IFN systems (Kato et al., 2006). Like those (+) ssRNA viruses mentioned above, FMDV RNA was recognized by MDA5 rather than by TLR3 or RIG-I in porcine epithelial cells, based on the result of the porcine kidney (PK-15) cells with MDA5 knockdown by RNA interference (Hüsser et al., 2011). According to the current knowledge on the length of RNA sensed by RIG-I and MDA5 (Kato et al., 2008), the FMDV genomes longer than 8000 nt are proposed to be sensed by MDA5 but not by RIG-I. Even though FMDV breaks into target cells and inhibits host translational systems by generation of Lpro and 3Cpro to evade the innate immune response against viral infection, MDA5 can first initiate IFN-β expression regardless of impairment of host translational systems. In addition, EMCV can produce high molecular weight RNA with a single- and double-stranded structure, and this RNA structure is capable of triggering MDA5 (Pichlmair et al., 2009). Notably, similar RNA structure exists in the 5′untranslation region of the FMDV genome (Carrillo et al., 2005), hence contributing to the activation of MDA5 to some degree. Most recently, it has been reported that the S fragment in the 5′untranslation region (UTR) of the FMDV genome is required for the replication and modulation of the innate immune response in host cells (Kloc et al., 2017). In human HEK 293 cell line, the S fragment and internal ribosomal entry site of 5′UTR and 3′UTR in the FMDV genome can trigger IFN-β promoter activation (Borrego et al., 2015). IFN-α/β can trigger double-stranded RNA-dependent protein kinase R (PKR) in the direct inhibition of FMDV replication in vitro (Chinsangaram et al., 2001; De et al., 2006). Turning to FMDV infection recognized by TLRs, the expression of TLR-4 in nasal-associated lymphoid tissue (NALT) during the acute stage of FMDV infection is higher than that of uninfected cattle, but non-infected and infected cattle do not differ regarding the transcription levels between TLR3 and TLR4 in NALT (Zhang et al., 2006). In addition, the FMDV genome can be recognized by TLR7 and TLR9 in plasmacytoid dendritic cells (pDCs) (Guzylack-Piriou et al., 2006; Lannes et al., 2012).
FMDV-Induced Signaling Cascade and IFN Gene Activation
After RIG-I or MDA5 bind to viral RNA, the tandem CARDs are unmasked and interact with MAVS (also termed as Cardif, IPS-1 or VISA), which contains one CARD, by CARD-CARD interaction (Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005; Xu et al., 2005). MAVS are located on the outer membrane of the mitochondria and this specific localization is pivotal for signal transduction, because the formation of the RLR-MAVS complex contributes to the recruitment of numerous signaling adaptors (Seth et al., 2005). Recently, Sun Hur and her colleagues have postulated a novel mechanism showing that two CARD tetramer formation is essential for triggering the MAVS prion-like structure, leading to type I interferon production (Wu and Hur, 2015). Notably, the Atg5-Atg12 conjugate, an essential regulator of the autophagic machinery, directly associates with the CARD of the RIG-I and IFN-β promoter (also called IPS-1 or MAVS) and further impair the interaction between RIG-I and MAVS, hence contributing to RNA virus replication in host cells (Jounai et al., 2007). Interestingly, FMDV 3Cpro can degrade the Atg5-Atg12 complex to suppress autophagy and antiviral responses mediated by the NF-κB pathway (Fan et al., 2017). However, it needs to be noted that FMDV Lpro can degrade NF-κB as well and decrease IRF3/7 expression to suppress dsRNA-induced type I IFN production, thereby impairing the expression of IFNs (Wang et al., 2010, 2012). Taken together, FMDV can adopt multiple measures to go against the antiviral immune response. IFNs are induced through transcriptional mechanisms involving the transcription factors nuclear factor (NF)-κB and the IFN regulatory factors (IRFs). The RLR-MAVS complex can recruit TRAF (tumor necrosis factor receptor-associated factor) family members and transmit signals to downstream protein kinases (inhibitor of NF-κB (IκB) kinase (IKK) family members, which are pivotal for activating the transcription factors IRF-3, IRF-7, and NF-κB (Karin and Benneriah, 2000; Xu et al., 2005), thereby contributing to IFNs expression. As an ovarian tumor domain (OUT)-containing enzyme, de-ubiquitinating enzyme (DUB) A interacts directly with TRAF3 and catalyzes the cleavage of Lys-63-linked ubiquitin chains of TRAF3, thereby contributing to the dissociation of TBK1 from TRAF3 and blocking signal transductions mediated by RLRs (Kayagaki et al., 2007). During FMDV evolution, viral Lpro evolved a specific function domain which is similar to the cellular DUBs and possesses DUB activity to impair signal transduction related to type I IFN production (Wang et al., 2011b). The canonical IKK complex, which is composed of IKK-α, IKK-β and the regulatory subunit NF-κB essential modulator (NEMO, or called IKK-γ), phosphorylates IκB, and subsequent proteasome-dependent degradation of IκB results in functional NF-κB moving into the nucleus (Karin and Benneriah, 2000). In contrast, non-canonical IKKs, TRAF family associated NF-κB activator (TANK)-binding kinase 1 (TBK1), and IKK-iii, activate the signal-dependent phosphorylation of IRF-3 and IRF-7 (Fitzgerald et al., 2003; Hemmi et al., 2004; Mcwhirter et al., 2004; Perry et al., 2004). Interaction between MAVS and TRAF3 is pivotal for the recruitment of both IKK complexes, while TRAF2 and TRAF6 are likely to be responsible for NF-κB activation (Xu et al., 2005; Khandekar et al., 2006). As a regulatory component, TANK and NAK-associated protein 1 (NAP1) are commonly involved in the TBK-1/IKK- iii-mediated activation of IRFs (Sasai et al., 2006; Guo and Cheng, 2007). IKK-γ (NEMO) serves as a regulatory subunit for the canonical IKK complex, which plays a role in the TBK-1/IKK- iii-mediated activation of IRFs (Zhao et al., 2007). However, FMDV 3Cpro can also target IKK-γ at the Gln383 residue and cleave off the C-terminal Zinc finger domain from IKK-γ, thereby disrupting the RIG-I/MDA5 pathway and contributing to the inhibition of IFN production (Wang et al., 2012). Most recently, crosstalk between the canonical IKK complex and non-canonical IKK complex exists in RLR-mediated signal transduction to a perform antiviral response (Fang et al., 2017). Collectively, these studies might imply that the immune systems have evolved a comprehensive network where various cytokines and signal molecules can perform cross-talk among different signal pathways to counteract the evasion of antiviral immune responses induced by viruses.
Evasion of Antiviral Response of Type I IFN Mediated by FMDV
Upon viral infection, innate immune responses serving as the first line of defense against viruses can induce signal transduction involving IFN and finally the expression of type I IFN, which then contributes to an antiviral stage in the cells. However, many viruses have evolved strategies to evade the antiviral immune response and prevent IFN production. Picornavirus-induced host translation system shut-off has been known to result in IFNs and other cytokine suppression (Buenz and Howe, 2006). Due to these RNA viruses being armed with some viral proteases (i.e., Lpro, 2Apro, and 3Cpro), these proteases can target different signal transduction nodes mediated by IFN (Porter et al., 2006; Yang et al., 2007; Papon et al., 2009). FMDV is no exception and has evolved to use its production to evade the antiviral immune response against FMDV (Figure 2) and maximize viral replication and dissemination. In addition to promoting the generation of viral products with biological functions, FMDV Lpro and 3Cpro plays pivotal roles in disrupting the translational system of the host. For FMDV Lpro, this protease is associated with translocation to the nucleus and cleavage of the p65 subunit of NF-κB (de Los Santos et al., 2007; Zhu et al., 2010). Although FMDV Lpro can inhibit IRF3/7 to promote the interferon-stimulated response element (ISRE) (Wang et al., 2011a,c), IRF9 can assist type I and III IFNs to perform antiviral response against FMDV infection (Stark et al., 2010; Perezmartin et al., 2012), implying that the host has evolved multiple immune pathways to meet with the evasion immune response induced by FMDV. Most recently, it has been reported that FMDV Lpro can bind to the host transcription factor ADNP (activity dependent neuroprotective protein) to suppress IFN and ISG transcription and enhance FMDV replication (Medina et al., 2017). FMDV 3Cpro specializes in cleaving FMDV polyprotein into viral proteins with biological functions and ruins eIF4G, eIF4A and histone H3 to block the translation system of the host (Falk et al., 1990; Li et al., 2001). In addition to inhibition of cell protein generation by 3Cpro, it also disrupts the transcriptional levels of ISGs, blocks the translocation of STAT1-STAT2 complex to the nucleus and suppresses the ISKE promoter (Du et al., 2014). These biological functions of FMDV 3Cpro seem to ruin the IFN system against viral infection in the broad spectrum. Like the role of EMCV 3Cpro in impairing the interaction TANK and TRAF6-mediated NF-κB signaling (Huang et al., 2015), FMDV 3Cpro also cleaves TANK to block the non-canonical IKK complex signaling pathway, rather than FMDV Lpro only disrupting TANK binding activity. In general, protease encoded by FMDV can negatively regulate innate immune signaling by degradation of essential molecules in different pathway.
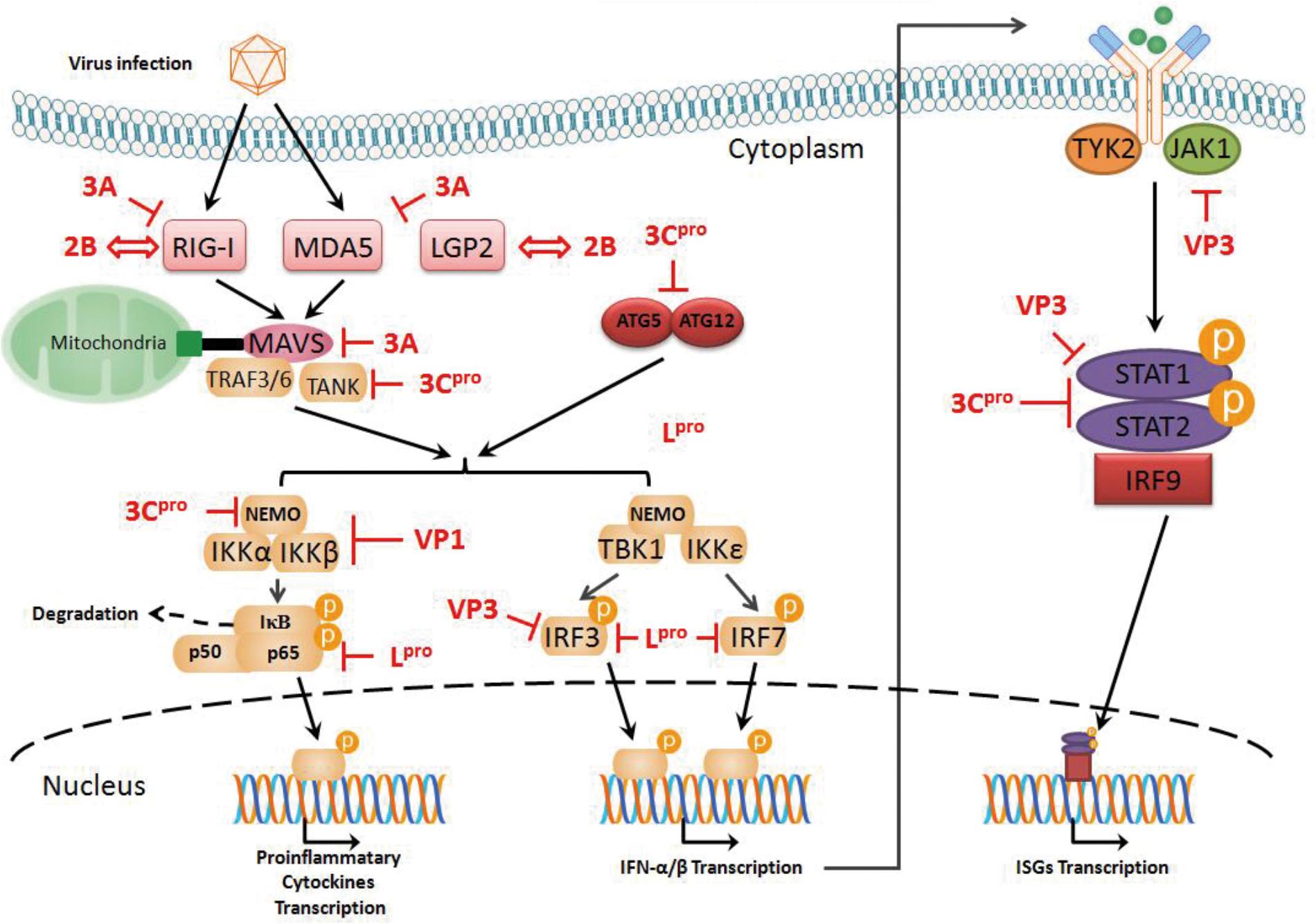
FIGURE 2. Evasion of antiviral response of type I IFN mediated by FMDV. FMDV Lpro can cleave the p65 subunit of NF-κB and inhibit IRF3/7, which promote the interferon-stimulated response element (ISRE). FMDV 3Cpro blocks the translocation of STAT1-STAT2 complex to the nucleus and suppresses the ISKE promoter. Recombinant VP1 can disrupt the formation of the canonical IKK complex to block NF-κB activity. VP3 can disrupt the assembly of the Jak1/STAT1 complex and inhibit IRF3 phosphorylation and dimerization. FMDV 2B can bind to RIG-I or LGP2 to impair related signal transduction and enhance viral replication. FMDV 3A protein can disrupt the transcriptional levels involved in RIG-I, MDA5, and MAVS and block the interaction between RIG-I/MDA5 and MAVS via its N-terminal region.
Apart from viral proteases of FMDV, other viral proteins play roles in viral immune evasion. VP1 and VP3 serve as viral structural proteins, which are the main components of the viral capsid (Mason et al., 2003). When the vp1 gene was over-expressed in the human HEK293 cell line, VP1 and sorcin can reduce NF-κB at the transcriptional levels and weaken type I IFN activity; in addition, recombinant VP1 can disrupt the formation of the canonical IKK complex to block NF-κB activity (Li et al., 2013; Ho et al., 2014). When the vp3 gene was over-expressed in the human HEK293T cell line, VP3 disrupted the assembly of the Jak1/STAT1 complex, suggesting FMDV VP3 impairs the type II IFN signaling pathway, inducing Jak1 degradation via a lysosomal pathway; Moreover, FMDV VP3 can inhibit IRF3 phosphorylation and dimerization, thereby contributing to the impairment of signal transduction involved in the type I IFN (Li et al., 2016a,b).
Turning to some non-structural proteins of FMDV, the 2B protein is capable of improving membrane permeability and carrying out cellular protein secretory pathway shut-off (Moffat et al., 2007). Recently, it has been reported that FMDV 2B can bind to RIG-I or LGP2 to impair related signal transduction and enhance viral replication, but the detailed mechanisms have not been established yet (Zhu et al., 2016, 2017). Enterovirus 71 2C proteins can bind to RelA (p65) and suppress IKKβ phosphorylation to disrupt the formation of the canonical IKK complex and impair NF-κB activation (Du et al., 2015; Li et al., 2016c). Since the 2C protein of picornaviruses are highly conserved (Gorbalenya et al., 1989), it is assumed that FMDV 2C should possess most of these activities involved in immune evasion. The FMDV 3A protein is a multifunctional non-structural protein in viral replication, virulence and host-specific genetic features (Mason et al., 2003; Ma X. et al., 2016; Bhatt et al., 2017; Bohórquez et al., 2017; Lotufo et al., 2017). For the role of FMDV 3A in the antiviral immune response, when 3A gene was over-expressed in the PK-15 cell line (swine) and the HEK293 cell line (human), 3A protein can disrupt the transcriptional levels involved in RIG-I, MDA5, and MAVS and block the interaction between RIG-I/MDA5 and MAVS via its N-terminal region (Li et al., 2016d). Although some studies involved in the structural and non-structural proteins of FMDV have noted that these viral proteins take part in immune evasion by disrupting various nodes along signaling transduction mediated by IFNs, the over-expressed individual protein with in vitro physical structure and biological function may not reflect the real situation in infected cells by FMDV.
Conclusion
Based on recent knowledge on IFNs, it is now well established that these cytokines function in the innate immune response against viral infection. Collectively, even though a series of recent reports strongly indicate that the host has evolved a complex network related to signal transduction induced by IFN against FMDV infection and FMDV relies on its viral proteins to suppress or ruin antiviral immune responses induced by IFN systems, the detailed and precise regulatory mechanisms need to be elucidated. This field is thus at a stage where there is an urgent need for the better understanding of both the basic biology and therapeutic antiviral activity of IFNs, and for deep investigations of how those proteins, without protease activities, derived from FMDV infection can influence and control the IFN signaling transduction in vivo.
Author Contributions
X-XM, L-NM, Q-YC, and L-JL wrote the draft manuscript. PM and Y-YW produced the figures. Z-RM and XC gave instruction and proof-read the article.
Funding
This work was supported by the Program for Changjiang Scholars and the Innovative Research Team in University (IRT13091), the National Natural Science foundation of China (No. 31160033) and the Central Universities foundation derived from the Northwest University for Nationalities (31920150077 and 31920170158), Innovative Research Team in University (IRT_17R88); Gansu Provincial Science and Technology Grant (1504WKCA094); the Open Fund of Ministry of Education Key Laboratory of Molecular Microbiology and Technology, Nankai University; Characteristic discipline of bioengineering construction for the special guide project of the “world-class universities and world-class disciplines” of Northwest Minzu University (10018703) and Ministry of Science and Technology Assistance Project Grant (KY201501005).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ahn, I., Bae, S. E., and Son, H. S. (2011). Comparative study of codon substitution patterns in foot-and-mouth disease virus (serotype o). Exp. Mol. Med. 43, 587–595. doi: 10.3858/emm.2011.43.10.066
Akira, S., Uematsu, S., and Takeuchi, O. (2006). Pathogen recognition and innate immunity. Cell 124, 783–801. doi: 10.1016/j.cell.2006.02.015
Alexopoulou, L., Holt, A. C., Medzhitov, R., and Flavell, R. A. (2001). Recognition of double-stranded RNA and activation of NF-kappab by toll-like receptor 3. Nature 413, 732–738. doi: 10.1038/35099560
Berke, I. C., Li, Y., and Modis, Y. (2013). Structural basis of innate immune recognition of viral RNA. Cell. Microbiol. 15, 386–394. doi: 10.1111/cmi.12061
Bhatt, M., Mohapatra, J. K., Pandey, L. K., Mohanty, N. N., Das, B., Prusty, B. R., et al. (2017). Mutational analysis of foot and mouth disease virus nonstructural polyprotein 3ab-coding region to design a negative marker virus. Virus Res. 243, 36–43. doi: 10.1016/j.virusres.2017.10.010
Bohórquez, J. A., Defaus, S., Muñozgonzález, S., Perezsimó, M., Rosell, R., Fraile, L., et al. (2017). A bivalent dendrimeric peptide bearing a T-cell epitope from foot-and-mouth disease virus protein 3a improves humoral response against classical swine fever virus. Virus Res. 238, 8–12. doi: 10.1016/j.virusres.2017.05.020
Borrego, B., Blanco, E., Pulido, M. R., Mateos, F., Lorenzo, G., Cardillo, S., et al. (2017). Combined administration of synthetic RNA and a conventional vaccine improves immune responses and protection against foot-and-mouth disease virus in swine. Antiviral Res. 142, 30–36. doi: 10.1016/j.antiviral.2017.03.009
Borrego, B., Rodríguezpulido, M., Revilla, C., Álvarez, B., Sobrino, F., Domínguez, J., et al. (2015). Synthetic RNAs mimicking structural domains in the foot-and-mouth disease virus genome elicit a broad innate immune response in porcine cells triggered by RIG-I and TLR activation. Viruses 7, 3954–3973. doi: 10.3390/v7072807
Bruns, A., Leser, G., Lamb, R., and Horvath, C. (2014). The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5-RNA interaction and filament assembly. Mol. Cell 55, 771–781. doi: 10.1016/j.molcel.2014.07.003
Bruns, A. M., Pollpeter, D., Hadizadeh, N., Myong, S., Marko, J. F., and Horvath, C. M. (2013). ATP hydrolysis enhances RNA recognition and antiviral signal transduction by the innate immune sensor, laboratory of genetics and physiology 2 (lgp2). J. Biol. Chem. 288, 938–946. doi: 10.1074/jbc.M112.424416
Buenz, E. J., and Howe, C. L. (2006). Picornaviruses and cell death. Trends Microbiol. 14, 28–36. doi: 10.1016/j.tim.2005.11.003
Carrillo, C., Tulman, E. R., Delhon, G., Lu, Z., Carreno, A., Vagnozzi, A., et al. (2005). Comparative genomics of foot-and-mouth disease virus. J. Virol. 79, 6487–6504. doi: 10.1128/JVI.79.10.6487-6504.2005
Chinsangaram, J., Koster, M., and Grubman, M. J. (2001). Inhibition of l-deleted foot-and-mouth disease virus replication by alpha/beta interferon involves double-stranded RNA-dependent protein kinase. J. Virol. 75, 5498–5503. doi: 10.1128/JVI.75.12.5498-5503.2001
De, L. S. T., De, A. B. S., Weiblen, R., and Grubman, M. J. (2006). The leader proteinase of foot-and-mouth disease virus inhibits the induction of beta interferon mRNA and blocks the host innate immune response. J. Virol. 80, 1906–1914. doi: 10.1128/JVI.80.4.1906-1914.2006
de Los Santos, T., Diaz-San Segundo, F., and Grubman, M. J. (2007). Degradation of nuclear factor kappa B during foot-and-mouth disease virus infection. J. Virol. 81, 12803–12815. doi: 10.1128/JVI.01467-07
Detournay, O., Morrison, D. A., Wagner, B., Zarnegar, B., and Wattrang, E. (2013). Genomic analysis and mRNA expression of equine type I interferon genes. J. Interferon Cytokine Res. 33, 746–759. doi: 10.1089/jir.2012.0130
Diebold, S. S., Kaisho, T., Hemmi, H., Akira, S., and Sousa, C. R. E. (2004). Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303, 1529–1531. doi: 10.1126/science.1093616
Domingo, E., Escarmís, C., Baranowski, E., Ruizjarabo, C. M., Carrillo, E., Núñez, J. I., et al. (2003). Evolution of foot-and-mouth disease virus. Virus Res. 91, 47–63. doi: 10.1016/S0168-1702(02)00259-9
Du, H., Yin, P., Yang, X., Zhang, L., Jin, Q., and Zhu, G. (2015). Enterovirus 71 2c protein inhibits NF-κB activation by binding to Rela(p65). Sci. Rep. 5:14302. doi: 10.1038/srep14302
Du, Y., Bi, J., Liu, J., Liu, X., Wu, X., Jiang, P., et al. (2014). 3Cpro of foot-and-mouth disease virus antagonizes the interferon signaling pathway by blocking STAT1/STAT2 nuclear translocation. J. Virol. 88, 4908–4920. doi: 10.1128/JVI.03668-13
Falk, M. M., Grigera, P. R., Bergmann, I. E., Zibert, A., Multhaup, G., and Beck, E. (1990). Foot-and-mouth disease virus protease 3C induces specific proteolytic cleavage of host cell histone H3. J. Virol. 64, 748–756.
Fan, X., Han, S., Yan, D., Gao, Y., Wei, Y., Liu, X., et al. (2017). Foot-and-mouth disease virus infection suppresses autophagy and NF- κb antiviral responses via degradation of ATG5-ATG12 by 3Cpro. Cell Death Dis. 8:e2561. doi: 10.1038/cddis.2016.489
Fang, R., Jiang, Q., Zhou, X., Wang, C., Guan, Y., Tao, J., et al. (2017). MAVS activates TBK1 and IKKε through TRAFs in NEMO dependent and independent manner. PLoS Pathog. 13:e1006720. doi: 10.1371/journal.ppat.1006720
Fensterl, V., Chattopadhyay, S., and Sen, G. C. (2015). No love lost between viruses and interferons. Annu. Rev. Virol. 2, 549–572. doi: 10.1146/annurev-virology-100114-055249
Fitzgerald, K. A., Mcwhirter, S. M., Faia, K. L., Rowe, D. C., Latz, E., Golenbock, D. T., et al. (2003). Ikkepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4, 491–496. doi: 10.1038/ni921
Gao, Z. L., Zhou, J. H., Zhang, J., Ding, Y. Z., and Liu, Y. S. (2014). The silent point mutations at the cleavage site of 2A/2B have no effect on the self-cleavage activity of 2A of foot-and-mouth disease virus. Infect. Genet. Evol. 28, 101–106. doi: 10.1016/j.meegid.2014.08.006
Golde, W. T., Nfon, C. K., and Toka, F. N. (2008). Immune evasion during foot-and-mouth disease virus infection of swine. Immunol. Rev. 225, 85–95. doi: 10.1111/j.1600-065X.2008.00672.x
Gorbalenya, A. E., Blinov, V. M., Donchenko, A. P., and Koonin, E. V. (1989). An NTP-binding motif is the most conserved sequence in a highly diverged monophyletic group of proteins involved in positive strand RNA viral replication. J. Mol. Evol. 28, l256–l268. doi: 10.1007/BF02102483
Guo, B., and Cheng, G. (2007). Modulation of the interferon antiviral response by the TBK1/IKKi adaptor protein tank. J. Biol. Chem. 282, 11817–11826. doi: 10.1074/jbc.M700017200
Guzylack-Piriou, L., Bergamin, F., Gerber, M., Mccullough, K. C., and Summerfield, A. (2006). Plasmacytoid dendritic cell activation by foot-and-mouth disease virus requires immune complexes. Eur. J. Immunol. 36, 1674–1683. doi: 10.1002/eji.200635866
Heil, F., Hemmi, H., Hochrein, H., Ampenberger, F., Kirschning, C., Akira, S., et al. (2004). Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303, 1526–1529. doi: 10.1126/science.1093620
Hemmi, H., Takeuchi, O., Sato, S., Yamamoto, M., Kaisho, T., Sanjo, H., et al. (2004). The roles of two IκB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J. Exp. Med. 199, 1641–1650. doi: 10.1084/jem.20040520
Ho, M. Y., Hung, S. W., Liang, C. M., and Liang, S. M. (2014). Recombinant viral capsid protein VP1 suppresses lung cancer metastasis by inhibiting COX-2/PGE2 and MIG-7. Oncotarget 5, 3931–3943. doi: 10.18632/oncotarget.2040
Huang, L., Liu, Q., Zhang, L., Zhang, Q., Hu, L., Li, C., et al. (2015). Encephalomyocarditis virus 3C protease relieves traf family member-associated NF-κB activator (tank) inhibitory effect on traf6-mediated NF-κB signaling through cleavage of tank. J. Biol. Chem. 290, 27618–27632. doi: 10.1074/jbc.M115.660761
Hüsser, L., Alves, M. P., Ruggli, N., and Summerfield, A. (2011). Identification of the role of RIG-I, MDA5 and TLR3 in sensing RNA viruses in porcine epithelial cells using Lentivirus-driven RNA interference. Virus Res. 159, 9–16. doi: 10.1016/j.virusres.2011.04.005
Jackson, S. E., Redeker, A., Arens, R., Van, B. D., Sph, V. D. B., Benedict, C. A., et al. (2017). CMV immune evasion and manipulation of the immune system with aging. Geroscience 39(Suppl. 3), 1–19. doi: 10.1007/s11357-017-9986-6
Jounai, N., Takeshita, F., Kobiyama, K., Sawano, A., Miyawaki, A., Xin, K. Q., et al. (2007). The ATG5-ATG12 conjugate associates with innate antiviral immune responses. Proc. Natl Acad. Sci. U.S.A. 104, 14050–14055. doi: 10.1073/pnas.0704014104
Karin, M., and Benneriah, Y. (2000). Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu. Rev. Immunol. 18, 621–663. doi: 10.1146/annurev.immunol.18.1.621
Kato, H., Takeuchi, O., Mikamo-Satoh, E., Hirai, R., Kawai, T., Matsushita, K., et al. (2008). Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid–inducible gene-1 and melanoma differentiation–associated gene 5. J. Exp. Med. 205, 1601–1610. doi: 10.1084/jem.20080091
Kato, H., Takeuchi, O., Sato, S., Yoneyama, M., Yamamoto, M., Matsui, K., et al. (2006). Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441, 101–105. doi: 10.1038/nature04734
Kawai, T., Takahashi, K., Sato, S., Coban, C., Kumar, H., Kato, H., et al. (2005). IPS-1, an adaptor triggering RIG-I and MDA5-mediated type I interferon induction. Nat. Immunol. 6, 981–988. doi: 10.1038/ni1243
Kayagaki, N., Phung, Q., Chan, S., Chaudhari, R., Quan, C., O’Rourke, K. M., et al. (2007). DUBA: a deubiquitinase that regulates type I interferon production. Science 318, 1628–1632. doi: 10.1126/science.1145918
Khandekar, S. S., Brauer, P. P., Naylor, J. W., Chang, H. C., Kern, P., Newcomb, J. R., et al. (2006). Regulation of antiviral responses by a direct and specific interaction between TRAF3 and cardif. EMBO J. 25, 3257–3263. doi: 10.1038/sj.emboj.7601220
Kloc, A., Diazsan, F. S., Schafer, E. A., Rai, D. K., Kenney, M., De, T. L. S., et al. (2017). Foot-and-mouth disease virus 5′-terminal S fragment is required for replication and modulation of the innate immune response in host cells. Virology 512, 132–143. doi: 10.1016/j.virol.2017.08.036
Knight-Jones, T. J. D., Robinson, L., Charleston, B., Rodriguez, L. L., Gay, C. G., Sumption, K. J., et al. (2016). Global foot-and-mouth disease research update and gap analysis: 1 - overview of global status and research needs. Transbound. Emerg. Dis. 63(Suppl. S1), 3–13. doi: 10.1111/tbed.12522
Krause, C. D., and Pestka, S. (2005). Evolution of the class 2 cytokines and receptors, and discovery of new friends and relatives. Pharmacol. Ther. 106, 299–346. doi: 10.1016/j.pharmthera.2004.12.002
Lannes, N., Python, S., and Summerfield, A. (2012). Interplay of foot-and-mouth disease virus, antibodies and plasmacytoid dendritic cells: virus opsonization under non-neutralizing conditions results in enhanced interferon-alpha responses. Vet. Res. 43:64. doi: 10.1186/1297-9716-43-64
Li, D., Wei, J., Yang, F., Liu, H. N., Zhu, Z. X., Cao, W. J., et al. (2016a). Foot-and-mouth disease virus structural protein VP3 degrades janus kinase 1 to inhibit IFN-γ signal transduction pathways. Cell Cycle 15, 850–860. doi: 10.1080/15384101.2016.1151584
Li, D., Yang, W., Yang, F., Liu, H., Zhu, Z., Lian, K., et al. (2016b). The VP3 structural protein of foot-and-mouth disease virus inhibits the IFN-β signaling pathway. FASEB J. 30, 1757–1766. doi: 10.1096/fj.15-281410
Li, Q., Zheng, Z., Liu, Y., Zhang, Z., Liu, Q., Meng, J., et al. (2016c). 2C proteins of enteroviruses suppress IKKβ phosphorylation by recruiting protein phosphatase1. J. Virol. 90, 5141–5151. doi: 10.1128/JVI.03021-15
Li, D., Lei, C., Xu, Z., Yang, F., Liu, H., Zhu, Z., et al. (2016d). Foot-and-mouth disease virus non-structural protein 3A inhibits the interferon-β signaling pathway. Sci. Rep. 6:21888. doi: 10.1038/srep21888
Li, W., Rosssmith, N., Proud, C. G., and Belsham, G. J. (2001). Cleavage of translation initiation factor 4ai (eif4ai) but not eif4aii by foot-and-mouth disease virus 3C protease: identification of the eif4ai cleavage site. FEBS Lett. 507, 1–5. doi: 10.1016/S0014-5793(01)02885-X
Li, X., Wang, J., Liu, J., Li, Z., Wang, Y., Xue, Y., et al. (2013). Engagement of soluble resistance-related calcium binding protein (SORCIN) with foot-and-mouth disease virus (FMDV) VP1 inhibits type I interferon response in cells. Vet. Microbiol. 166, 35–46. doi: 10.1016/j.vetmic.2013.04.028
Liu, Y., Olagnier, D., and Lin, R. (2016). Host and viral modulation of RIG-I-mediated antiviral immunity. Front. Immunol. 7:662. doi: 10.3389/fimmu.2016.00662
Lotufo, C. M., Bergmann, I. E., Mattion, N. M., Wilda, M., and Grigera, P. R. (2017). Recombinant foot-and-mouth disease virus (FMDV) non-structural protein 3A fused to enhanced green fluorescent protein (EGFP) as a candidate probe to identify FMDV-infected cattle in serosurveys. Arch. Virol. 162, 2279–2286. doi: 10.1007/s00705-017-3359-4
Ma, X., Li, P., Sun, P., Lu, Z., Bao, H., Bai, X., et al. (2016). Genome sequence of foot-and-mouth disease virus outside the 3A region is also responsible for virus replication in bovine cells. Virus Res. 220, 64–69. doi: 10.1016/j.virusres.2016.04.011
Ma, X. X., Feng, Y. P., Gu, Y. X., Zhou, J. H., and Ma, Z. R. (2016). Effect of the nucleotides surrounding the start codon on the translation of foot-and-mouth disease virus RNA. Acta Virol. 60, 151–155. doi: 10.4149/av_2016_02_151
Ma, X. X., Feng, Y. P., Liu, J. L., Ma, B., Chen, L., Zhao, Y. Q., et al. (2013). The effects of the codon usage and translation speed on protein folding of 3D(pol) of foot-and-mouth disease virus. Vet. Res. Commun. 37, 243–250. doi: 10.1007/s11259-013-9564-z
Mason, P. W., Grubman, M. J., and Baxt, B. (2003). Molecular basis of pathogenesis of FMDV. Virus Res. 91, 9–32. doi: 10.1016/S0168-1702(02)00257-5
Mcwhirter, S. M., Fitzgerald, K. A., Rosains, J., Rowe, D. C., Golenbock, D. T., and Maniatis, T. (2004). IFN-regulatory factor 3-dependent gene expression is defective in TBK1-deficient mouse embryonic fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 101, 233–238. doi: 10.1073/pnas.2237236100
Medina, G. N., Knudsen, G. M., Greninger, A. L., Kloc, A., Segundo, D. S., Rieder, E., et al. (2017). Interaction between FMDVLpro, and transcription factor ADNP is required for optimal viral replication. Virology 505, 12–22. doi: 10.1016/j.virol.2017.02.010
Meylan, E., Curran, J., and Hofmann, K. (2005). Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437, 1167–1172. doi: 10.1038/nature04193
Moffat, K., Knox, C., Howell, G., Clark, S. J., Yang, H., Belsham, G. J., et al. (2007). Inhibition of the secretory pathway by foot-and-mouth disease virus 2BC protein is reproduced by coexpression of 2B with 2C, and the site of inhibition is determined by the subcellular location of 2C. J. Virol. 81, 1129–1139. doi: 10.1128/JVI.00393-06
Papon, L., Oteiza, A., Imaizumi, T., Kato, H., Brocchi, E., Lawson, T. G., et al. (2009). The viral RNA recognition sensor RIG-I is degraded during encephalomyocarditis virus (EMCV) infection. Virology 393, 311–318. doi: 10.1016/j.virol.2009.08.009
Perezmartin, E., Weiss, M., Diazsan, S. F., Pacheco, J. M., Arzt, J., Grubman, M. J., et al. (2012). Bovine type III interferon significantly delays and reduces the severity of foot-and-mouth disease in cattle. J. Virol. 86, 4477–4487. doi: 10.1128/JVI.06683-11
Perry, A. K., Chow, E. K., Goodnough, J. B., Yeh, W. C., and Cheng, G. (2004). Differential requirement for tank-binding kinase-1 in type I interferon responses to Toll-like receptor activation and viral infection. J. Exp. Med. 199, 1651–1658. doi: 10.1084/jem.20040528
Pichlmair, A., Schulz, O., Tan, C. P., Rehwinkel, J., Kato, H., Takeuchi, O., et al. (2009). Activation of MDA5 requires higher-order RNA structures generated during virus infection. J. Virol. 83, 10761–10769. doi: 10.1128/JVI.00770-09
Porter, F. W., Bochkov, Y. A., Albee, A. J., Wiese, C., and Palmenberg, A. C. (2006). A picornavirus protein interacts with RAN-GTPase and disrupts nucleocytoplasmic transport. Proc. Natl. Acad. Sci. U.S.A. 103, 12417–12422. doi: 10.1073/pnas.0605375103
Robinson, L., Knight-Jones, T. J., Charleston, B., Rodriguez, L. L., Gay, C. G., Sumption, K. J., et al. (2016). Global foot-and-mouth disease research update and gap analysis: 3 - vaccines. Transbound. Emerg. Dis. 63(Suppl. S1), 30–41. doi: 10.1111/tbed.12521
Rodríguezpulido, M., Sobrino, F., Borrego, B., and Sáiz, M. (2011). Inoculation of newborn mice with non-coding regions of foot-and-mouth disease virus RNA can induce a rapid, solid and wide-range protection against viral infection. Antiviral Res. 92, 500–504. doi: 10.1016/j.antiviral.2011.10.005
Sasai, M., Shingai, M., Funami, K., Yoneyama, M., Fujita, T., Matsumoto, M., et al. (2006). NAK-associated protein 1 participates in both the TLR3 and the cytoplasmic pathways in type IIFN induction. J. Immunol. 177, 8676–8683. doi: 10.4049/jimmunol.177.12.8676
Satoh, T., Kato, H., Kumagai, Y., Yoneyama, M., Sato, S., Matsushita, K., et al. (2010). Lgp2 is a positive regulator of RIG-I– and MDA5-mediated antiviral responses. Proc. Natl. Acad. Sci. U.S.A. 107, 1512–1517. doi: 10.1073/pnas.0912986107
Schulz, K. S., and Mossman, K. L. (2004). Viral evasion strategies in type IIFN signaling – a summary of recent developments. Evol. Ecol. 18, 85–105. doi: 10.3389/fimmu.2016.00498
Sen, G. C., and Sarkar, S. N. (2007). The interferon-stimulated genes: targets of direct signaling by interferons, double-stranded RNA, and viruses. Curr. Top. Microbiol. 316, 233–250. doi: 10.1007/978-3-540-71329-6_12
Seth, R. B., Sun, L., Ea, C. K., and Chen, Z. J. (2005). Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF3. Cell 122, 669–682. doi: 10.1038/nature04193
Shih, J. P., Zeng, H. Q., and Ogawa, M. (1992). Enrichment of murine marrow cells for progenitors of multilineage hemopoietic colonies. Leukemia 6, 193–198. doi: 10.1016/j.meegid.2014.08.006
Stark, G. R., Kerr, I. M., Williams, B. R. G., Silverman, R. H., and Schreiber, R. D. (2010). How cells respond to interferons. J. Clin. Oncol. 28, 227–264. doi: 10.1146/annurev.biochem.67.1.227
Sumner, R. P., Thorne, L. G., Fink, D. L., Khan, H., Milne, R. S., and Towers, G. J. (2017). Are evolution and the intracellular innate immune system key determinants in HIV transmission? Front. Immunol. 8:1246. doi: 10.3389/fimmu.2017.01246
Tatematsu, M., Nishikawa, F., Seya, T., and Matsumoto, M. (2013). Toll-like receptor 3 recognizes incomplete stem structures in single-stranded viral RNA. Nat. Commun. 4:1833. doi: 10.1038/ncomms2857
Toka, F. N., and Golde, W. T. (2013). Cell mediated innate responses of cattle and swine are diverse during foot-and-mouth disease virus (FMDV) infection: a unique landscape of innate immunity. Immunol. Lett. 152, 135–143. doi: 10.1016/j.imlet.2013.05.007
Uchikawa, E., Lethier, M., Malet, H., Brunel, J., Gerlier, D., and Cusack, S. (2016). Structural analysis of dsRNA binding to anti-viral pattern recognition receptors LGP2 and MDA5. Mol. Cell 62, 586–602. doi: 10.1016/j.molcel.2016.04.021
Uzé, G., and Monneron, D. (2007). IL-28 and IL-29: newcomers to the interferon family. Biochimie 89, 729–734. doi: 10.1016/j.biochi.2007.01.008
Venkataraman, T., Valdes, M., Elsby, R., Kakuta, S., Caceres, G., Saijo, S., et al. (2007). Loss of DEXD/HBOXRNA helicase LGP2 manifests disparate antiviral responses. J. Immunol. 178, 6444–6455. doi: 10.4049/jimmunol.178.10.6444
Wang, D., Fang, L., Bi, J., Chen, Q., Cao, L., Luo, R., et al. (2011a). Foot-and-mouth disease virus leader proteinase inhibits dsRNA-induced rantes transcription in pk-15 cells. Virus Genes 42, 388–393. doi: 10.1007/s11262-011-0590-z
Wang, D., Fang, L., Li, P., Sun, L., Fan, J., Zhang, Q., et al. (2011b). The leader proteinase of foot-and-mouth disease virus negatively regulates the type I interferon pathway by acting as a viral deubiquitinase. J. Virol. 85, 3758–3766. doi: 10.1128/JVI.02589-10
Wang, D., Fang, L., Liu, L., Zhong, H., Chen, Q., Luo, R., et al. (2011c). Foot-and-mouth disease virus (FMDV) leader proteinase negatively regulates the porcine interferon-jjj1 pathway. Mol. Immunol. 49, 407–412. doi: 10.1016/j.molimm.2011.09.009
Wang, D., Fang, L., Li, K., Zhong, H., Fan, J., Ouyang, C., et al. (2012). Foot-and-mouth disease virus 3C protease cleaves NEMO to impair innate immune signaling. J. Virol. 86, 9311–9322. doi: 10.1128/JVI.00722-12
Wang, D., Fang, L., Luo, R., Ye, R., Fang, Y., Xie, L., et al. (2010). Foot-and-mouth disease virus leader proteinase inhibits dsRNA-induced type I interferon transcription by decreasing interferon regulatory factor 3/7 in protein levels. Biochem. Biophys. Res. Commun. 399, 72–78. doi: 10.1016/j.bbrc.2010.07.044
Wu, B., and Hur, S. (2015). How RIG-I like receptors activate MAVS. Curr. Opin. Virol. 12, 91–98. doi: 10.1016/j.coviro.2015.04.004
Xu, L. G., Wang, Y. Y., Han, K. J., Li, L. Y., Zhai, Z., and Shu, H. B. (2005). VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19, 727–740. doi: 10.1016/j.molcel.2005.08.014
Yang, Y., Liang, Y., Qu, L., Chen, Z., Yi, M., Li, K., et al. (2007). Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc. Natl. Acad. Sci. U.S.A. 104, 7253–7258. doi: 10.1073/pnas.0611506104
Yoneyama, M., Kikuchi, M., Matsumoto, K., Imaizumi, T., Miyagishi, M., Taira, K., et al. (2005). Shared and unique functions of the DEXD/H-BOX helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175, 2851–2858. doi: 10.4049/jimmunol.175.5.2851
Zhang, Z., Bashiruddin, J. B., Doel, C., Horsington, J., Durand, S., and Alexandersen, S. (2006). Cytokine and Toll-like receptor mRNAs in the nasal-associated lymphoid tissues of cattle during foot-and-mouth disease virus infection. J. Comp. Pathol. 134, 56–62. doi: 10.1016/j.jcpa.2005.06.011
Zhao, T., Yang, L., Sun, Q., Arguello, M., Ballard, D. W., Hiscott, J., et al. (2007). The NEMO adaptor bridges the nuclear factor-kappab and interferon regulatory factor signaling pathways. Nat. Immunol. 8, 592–600. doi: 10.1038/ni1465
Zhou, J. H., Gao, Z. L., Zhang, J., Ding, Y. Z., Stipkovits, L., Szathmary, S., et al. (2013a). The analysis of codon bias of foot-and-mouth disease virus and the adaptation of this virus to the hosts. Infect. Genet. Evol. 14, 105–110. doi: 10.1016/j.meegid.2012.09.020
Zhou, J. H., You, Y. N., Chen, H. T., Zhang, J., Ma, L. N., Ding, Y. Z., et al. (2013b). The effects of the synonymous codon usage and tRNA abundance on protein folding of the 3C protease of foot-and-mouth disease virus. Infect. Genet. Evol. 16, 270–274. doi: 10.1016/j.meegid.2013.02.017
Zhou, J. H., Zhang, J., Sun, D. J., Ma, Q., Ma, B., Pejsak, Z., et al. (2013c). Potential roles of synonymous codon usage and tRNA concentration in hosts on the two initiation regions of foot-and-mouth disease virus RNA. Virus Res. 176, 298–302. doi: 10.1016/j.virusres.2013.06.006
Zhou, J. H., Jie, Z., Chen, H. T., Ma, L. N., and Liu, Y. S. (2010a). Analysis of synonymous codon usage in foot-and-mouth disease virus. Vet. Res. Commun. 34, 393–404. doi: 10.1007/s11259-010-9359-4
Zhou, J. H., Zhang, J., Ding, Y. Z., Chen, H. T., Ma, L. N., and Liu, Y. S. (2010b). Characteristics of codon usage bias in two regions downstream of the initiation codons of foot-and-mouth disease virus. Biosystems 101, 20–28. doi: 10.1016/j.biosystems.2010.04.001
Zhou, J. H., Zhang, J., Chen, H. T., Ma, L. N., Ding, Y. Z., Pejsak, Z., et al. (2011). The codon usage model of the context flanking each cleavage site in the polyprotein of foot-and-mouth disease virus. Infect. Genet. Evol. 11, 1815–1819. doi: 10.1016/j.meegid.2011.07.014
Zhu, J., Weiss, M., Grubman, M. J., and de los Santos, T. (2010). Differential gene expression in bovine cells infected with wild type and leaderless foot-and-mouth disease virus. Virology 404, 32–40. doi: 10.1016/j.virol.2010.04.021
Zhu, Z., Li, C., Du, X., Wang, G., Cao, W., Yang, F., et al. (2017). Foot-and-mouth disease virus infection inhibits LGP2 protein expression to exaggerate inflammatory response and promote viral replication. Cell Death Dis. 8:e2747. doi: 10.1038/cddis.2017.170
Keywords: type I interferon, Picornaviridae, FMDV, RIG-I, MDA5
Citation: Ma X-X, Ma L-N, Chang Q-Y, Ma P, Li L-J, Wang Y-Y, Ma Z-R and Cao X (2018) Type I Interferon Induced and Antagonized by Foot-and-Mouth Disease Virus. Front. Microbiol. 9:1862. doi: 10.3389/fmicb.2018.01862
Received: 16 April 2018; Accepted: 24 July 2018;
Published: 13 August 2018.
Edited by:
Penghua Wang, University of Connecticut Health Center, United StatesReviewed by:
Xin Zhao, Institute of Microbiology (CAS), ChinaJianwei Wang, Chinese Academy of Medical Sciences, China
Copyright © 2018 Ma, Ma, Chang, Ma, Li, Wang, Ma and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhong-ren Ma, bXpyQHhibXUuZWR1LmNu Xin Cao, Y2FveGluQHhibXUuZWR1LmNu
†These authors have contributed equally to this work.
 Xiao-xia Ma1†
Xiao-xia Ma1† Xin Cao
Xin Cao