- 1Gut Microbes and Health Programme, Quadram Institute Bioscience, Norwich Research Park, Norwich, United Kingdom
- 2Norwich Medical School, University of East Anglia, Norwich Research Park, Norwich, United Kingdom
by Gohari, I. M., and Prescott, J. F. (2018). Front. Microbiol. 9:1856. doi: 10.3389/fmicb.2018.01856
We firstly would like to express our appreciation for the comments and efforts made by Gohari and Prescott regarding their detailed commentary of our Clostridium perfringens Whole Genome Sequencing (WGS) based genomic and phylogenetic study we published in December 2017. We are particularly pleased to note that the authors acknowledge that our “work has contributed significantly to understanding of genomic diversity of this bacterium.”
The aim of our study was to conduct a robust large-scale WGS study on this important pathogen, and to highlight genomic insights to the global scientific community. We believe that WGS could prove extremely useful in exploring new traits in C. perfringens, as has been performed in many different bacterial pathogens. We agree with the authors, and indeed highlight this throughout our original study, that alongside genomic-based studies, clinical metadata, epidemiological studies, and phenotypic testing, will be central to determine the impact of genetic variation in C. perfringens in the context of human and animal health.
We would first like to respond to the authors commentary suggesting a “clear mistake” or “incorrect conclusion” in that netB gene (a gene that encode NetB toxin, which is associated with avian Necrotising Enteritis) has no role in canine haemorrhagic gastroenteritis and foal necrotising enteritis (Keyburn et al., 2008, 2010; Rood et al., 2016). We appreciate the authors have a track record of working in this particular area of C. perfringens virulence, as highlighted by citation of their own work throughout the commentary. They highlight that netB and netE genes (which encodes proteins NetB and NetE respectively) share high sequence identity of 78% amino acid (according to their published work in 2015, 79% amino acid sequence identity; Mehdizadeh Gohari et al., 2015), which may have potentially contributed to our “understandable misinterpretation” of the data. Based on our informatics filtering parameters, we also determined that netB and netE are highly identical and not distinctive at nucleotide sequence level. This was based on strict double-filtering strategy at 80% identity and E-value of 10−20, (routinely applied in WGS studies to infer identical genes; Pearson, 2013; Kiu et al., 2017), and was used as we were undertaking a global in silico-based approach to explore a significant number of C. perfringens genomes for virulence-associated traits (not solely focused on these two toxin genes). Consequently, we have reanalysed the data and determined that a higher sequence identity threshold at 90% (BLASTn) confirms an absence of the netB gene in these NetF-associated genomes. We thank the authors for highlighting the high sequence similarly of these toxin genes, and will factor these parameters in for future studies (Camacho et al., 2009). As we carried out a purely bioinformatic-based study, and are aware of the sensitivity/specificity of different computational pipelines and parameters, we were careful not to come to any definitive “conclusions,” and “suggested” that NetB toxin “might be involved,” thus clarifying our discussion points. Notably, with the advancement in bioinformatics tools, differentiation between highly similar genes may be possible using a “best-match” approach to avoid inaccurate annotations.
To address the question as to whether or not NCTC8503 is an “NE isolate,” we have re-traced the source of this isolate and its sequence data. NCTC8503 is a C. perfringens strain isolated by Bennetts (1930) according to the history record shown on the Public Health England NCTC official website1 Nairn and Bamford (1967) state “Bennetts (1930) had earlier reported the isolation of Bacillus welchii from a bowel lesion in a Black Orpington pullet with intestinal coccidiosis, and he considered that an enterotoxaemia had contributed to the bird's death”; B. welchii was later renamed as C. perfringens. Furthermore, Williams et al. states “A frequent, although sporadic, poultry clostridiosis (necrotic enteritis [NE]) was first recorded by Bennetts (1930) in Australia,” thus these descriptions indicated a likely link to strain NCTC8503, a C. perfringens strain isolated in 1930. We agree that no type D C. perfringens strains have been linked with poultry NE. Unfortunately, as NCTC8503 is a historical isolate with no detailed source information recorded, we have been unable to definitively confirm the isolate's origin (Bennetts, 1930; Williams, 2005). However, we can reaffirm that this isolate has not lost the epsilon toxin plasmid during laboratory passage as confirmed by both WGS (Figure 1), and multiplex PCR toxinotyping (Baums et al., 2004; Kiu et al., 2017).
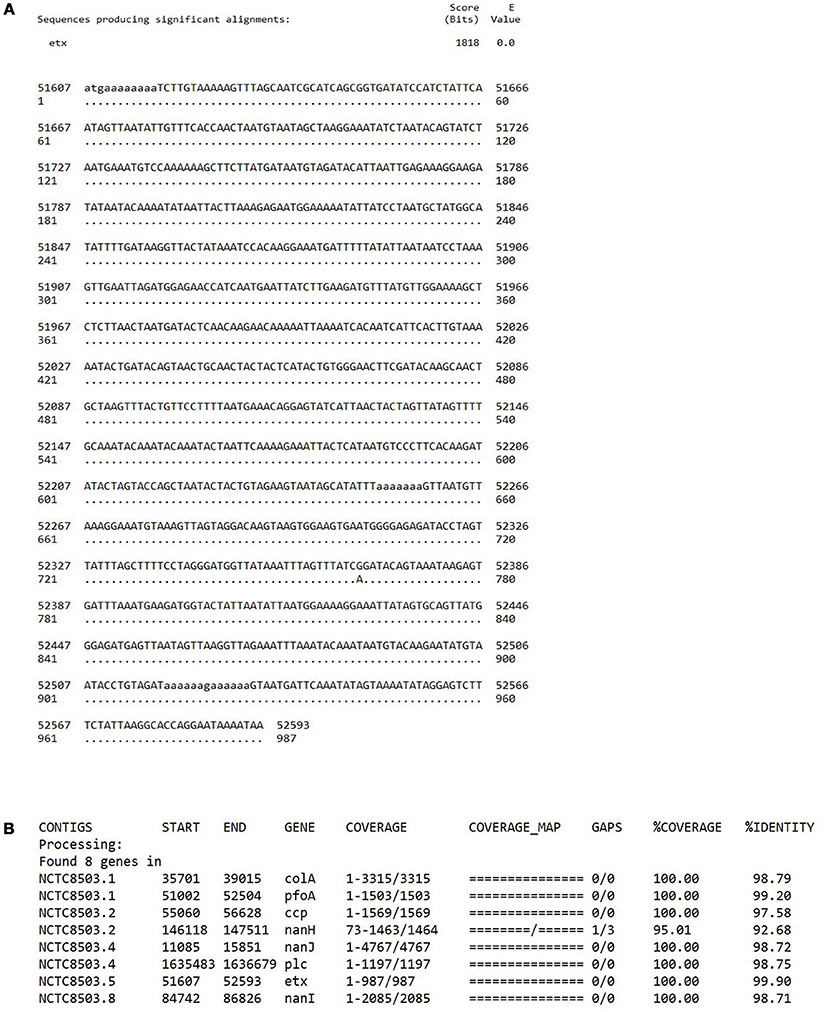
Figure 1. (A) A BLASTn sequence alignment performed on a local machine that aligns epsilon toxin gene etx (NCBI accession: M95206.1) with PacBio-sequenced NCTC8503 previously assembled genome (ENA accession: SAMEA3879480; contig 5: nucleotide position 51607-52593). Only a point mutation at position 762 is detected with 100% coverage and > 99% sequence identity. (B) Toxin profile of NCTC8503 isolate, performed via sequence similarity search (“best-match” approach) pipeline ABRicate (https://github.com/tseemann/abricate; BLASTn-based tool) that confirmed the presence of etx gene in NCTC8503 genome with 99.90% identity (at 100% coverage).
The “extreme” level of pangenome (12.6% core genes) in C. perfringens has not been widely reported to date (McInerney et al., 2017). However, as stated by the authors a previous study carried out in 2010 in Escherichia coli, which analysed 53 genomes, indicated 11% core genes (Lukjancenko et al., 2010). Notably, although these 2 bacteria can colonise the GI tract, they are fundamentally different regarding their oxygen sensitivity and ability to form spores. As described by Lukjanenko et al., analysis of 53 genomes (not 61 genomes as claimed) identified 1,472 core genes out of 13,296 genes in the pangenome (11%), which is lower than the 12.6% reported in our genomic C. perfringens analysis, although we note some differences in approaches used. More recently a pangenome study based on 228 E. coli genomes identified 23.8% core genes (2,722 core genes in 11,401 gene families) (McNally et al., 2016), and using web-based tool panX pangenome analysis2 and databases (Ding et al., 2018), based on the same cutoff as our study indicated 13% of core genes, based on analysis of 307 E. coli genomes (as of May 2018). We reiterate that our “extreme” observation for spore-forming Gram-positive C. perfringens is a rare trait, and is therefore of interest to the wider research community. We agree with their statement “describing a species as having extreme variation depends very much to what it is being compared,” and furthermore we would emphasise that prediction of genetic diversity will likely vary due to different factors including sampling bias, number of strains selected, and parameters used during informatics analysis, thus impacting diversity measurements. In this study we compared C. perfringens with Clostridium difficile (30.3%, which has been reclassified as Clostridioides difficile), a closer relative of C. perfringens, and Streptococcus pneumoniae (46.5%), Salmonella enterica (16%), and Klebsiella pneumoniae (26%) (Lawson et al., 2016; Kiu et al., 2017).
We agree that there is currently no definitive pathogenicity link with C. perfringens prophages, although previous studies have indicated they enhance sporulation, which would be expected to enhance transmission, and can be viewed as a virulence trait (Stewart and Johnson, 1977). Notably, in the closely related pathogen C. difficile, bacteriophages are linked to toxin-secretion (Goh et al., 2005). Consequently, we speculate that C. perfringens phages contribute to their virulence, which could be confirmed in future studies.
We hope that our commentary provides clarification and context and we look forward to wider discussion with all investigators in the C. perfringens field.
Author Contributions
RK and LH co-wrote the manuscript. RK performed the bioinformatics analysis.
Funding
Funding by a Wellcome Trust Investigator Award (100974/C/13/Z), and the Biotechnology and Biological Sciences Research Council (BBSRC); Institute Strategic Programme Gut Microbes and Health BB/R012490/1, and Institute Strategic Programme Gut Health and Food Safety BB/J004529/1 to LH. RK is a recipient of University of East Anglia International Bursary (Faculty of Medical and Health Sciences).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Baums, C. G., Schotte, U., Amtsberg, G., and Goethe, R. (2004). Diagnostic multiplex PCR for toxin genotyping of Clostridium perfringens isolates. Vet. Microbiol. 100, 11–16. doi: 10.1016/S0378-1135(03)00126-3
Bennetts, H. W. (1930). Bacillus welchii and bowel lesions. With special reference to a case of coccidiosis. Aust. Vet. J. 6, 153–154.
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Ding, W., Baumdicker, F., and Neher, R. A. (2018). panX: pan-genome analysis and exploration. Nucleic Acids Res. 46, e5. doi: 10.1093/nar/gkx977
Goh, S., Chang, B. J., and Riley, T. V. (2005). Effect of phage infection on toxin production by Clostridium difficile. J. Med. Microbiol. 54(Pt 2), 129–135. doi: 10.1099/jmm.0.45821-0
Keyburn, A. L., Bannam, T. L., Moore, R. J., and Rood, J. I. (2010). NetB, a pore-forming toxin from necrotic enteritis strains of Clostridium perfringens. Toxins 2, 1913–1927. doi: 10.3390/toxins2071913
Keyburn, A. L., Boyce, J. D., Vaz, P., Bannam, T. L., Ford, M. E., Parker, D., et al. (2008). NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathog. 4:e26. doi: 10.1371/journal.ppat.0040026
Kiu, R., Caim, S., Alexander, S., Pachori, P., and Hall, L. J. (2017). Probing genomic aspects of the multi-host pathogen Clostridium perfringens reveals significant pangenome diversity, and a diverse array of virulence factors. Front. Microbiol. 8:2485. doi: 10.3389/fmicb.2017.02485
Lawson, P. A., Citron, D. M., Tyrrell, K. L., and Finegold, S. M. (2016). Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O'Toole 1935) Prevot 1938. Anaerobe 40, 95–99. doi: 10.1016/j.anaerobe.2016.06.008
Lukjancenko, O., Wassenaar, T. M., and Ussery, D. W. (2010). Comparison of 61 sequenced Escherichia coli genomes. Microb. Ecol. 60, 708–720. doi: 10.1007/s00248-010-9717-3
McInerney, J. O., McNally, A., and O'Connell, M. J. (2017). Why prokaryotes have pangenomes. Nat. Microbiol. 2:17040. doi: 10.1038/nmicrobiol.2017.40
McNally, A., Oren, Y., Kelly, D., Pascoe, B., Dunn, S., Sreecharan, T., et al. (2016). Combined analysis of variation in core, accessory and regulatory genome regions provides a super-resolution view into the evolution of bacterial populations. PLoS Genet. 12:e1006280. doi: 10.1371/journal.pgen.1006280
Mehdizadeh Gohari, I., Parreira, V. R., Nowell, V. J., Nicholson, V. M., Oliphant, K., and Prescott, J. F. (2015). A novel pore-forming toxin in type A Clostridium perfringens is associated with both fatal canine hemorrhagic gastroenteritis and fatal foal necrotizing enterocolitis. PLoS ONE 10:e0122684. doi: 10.1371/journal.pone.0122684
Nairn, M. E., and Bamford, V. W. (1967). Necrotic enteritis of broiler chickens in western Australia. Aust. Vet. J. 43, 49–54.
Pearson, W. R. (2013). An introduction to sequence similarity (“homology”) searching. Curr. Protoc. Bioinformatics Chapter 3, Unit3 1. doi: 10.1002/0471250953.bi0301s42
Rood, J. I., Keyburn, A. L., and Moore, R. J. (2016). NetB and necrotic enteritis: the hole movable story. Avian Pathol. 45, 295–301. doi: 10.1080/03079457.2016.1158781
Stewart, A. W., and Johnson, M. G. (1977). Increased numbers of heat-resistant spores produced by two strains of Clostridium perfringens bearing temperate phage s9. J. Gen. Microbiol. 103, 45–50. doi: 10.1099/00221287-103-1-45
Keywords: pangenome, antimicrobial resistance, Clostridium perfringens, genomics, whole genome sequencing, Clostridial infection, exotoxins, pathogen
Citation: Kiu R and Hall LJ (2018) Response: Commentary: Probing Genomic Aspects of the Multi-Host Pathogen Clostridium perfringens Reveals Significant Pangenome Diversity, and a Diverse Array of Virulence Factors. Front. Microbiol. 9:1857. doi: 10.3389/fmicb.2018.01857
Received: 02 June 2018; Accepted: 24 July 2018;
Published: 16 August 2018.
Edited by:
John R. Battista, Louisiana State University, United StatesReviewed by:
Karl Hassan, University of Newcastle, AustraliaCopyright © 2018 Kiu and Hall. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lindsay J. Hall, TGluZHNheS5IYWxsQHF1YWRyYW0uYWMudWs=
 Raymond Kiu
Raymond Kiu Lindsay J. Hall
Lindsay J. Hall