- 1Laboratório de Genética Celular e Molecular, Departamento de Biologia Geral, Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais, Belo Horizonte, Brazil
- 2Kroton Educacional, Faculdade Pitágoras, Contagem, Brazil
- 3Centro Federal de Educação Tecnológica de Minas Gerais, Coordenação de Ciências, Belo Horizonte, Brazil
Lactococcus lactis has been used historically in fermentation and food preservation processes as it is considered safe for human consumption (GRAS—Generally Recognized As Safe). Nowadays, in addition to its wide use in the food industry, L. lactis has been used as a bioreactor for the production of molecules of medical interest, as well as vectors for DNA delivery. These applications are possible due to the development of promising genetic tools over the past few decades, such as gene expression, protein targeting systems, and vaccine plasmids. Thus, this review presents some of these genetic tools and their evolution, which allow us to envision new biotechnological and therapeutic uses of L. lactis. Constitutive and inductive expression systems will be discussed, many of which have been used successfully for heterologous production of different proteins, tested on animal models. In addition, advances in the construction of new plasmids to be used as potential DNA vaccines, delivered by this microorganism, will also be viewed. Finally, we will focus on the scene of gene expression systems known as “food-grade systems” based on inducing compounds and safe selection markers, which eliminate the need for the use of compounds harmful to humans or animal health and potential future prospects for their applications.
Lactococcus lactis: the Model Lactic Acid Bacteria
Lactic acid bacteria (LAB) constitute a diverse group of Gram-positive microorganisms, including species of genera Lactobacillus, Lactococcus, Streptococcus, Pediococcus Leuconostoc, and Oenococcus, which, among other shared characteristics, have the capacity to convert sugars into lactic acid (Makarova and Koonin, 2007). Because they have long been used in fermentation and food preservation processes, most of these bacteria are considered safe for human consumption, possessing the GRAS (Generally Recognized As Safe) status (van de Guchte et al., 2006). Among the representatives of this group, L. lactis is the best characterized species and figures as the model organism for its study. This, in turn, is due not only to its evident economic importance, but also to the fact that this bacterium is a microorganism of easy manipulation and has its genome sequenced. A great number of genetic tools have been developed. Thus, L. lactis has also been widely used in the field of biotechnology for the production and delivery of antigens and cytokines to mucosal surfaces (Wells, 2011), and more recently as a vehicle for the delivery of DNA vaccines (Pereira et al., 2014, 2017; Pontes et al., 2014; Zurita-Turk et al., 2014; Souza et al., 2016; Mancha-Agresti et al., 2017).
This microorganism has several characteristics that make it an interesting vector for the production and delivery of biomolecules, especially for oral route, such as resisting stomach acidic environment and being able to survive in the gastrointestinal tract. Another attractive property of L. lactis is that it lacks lipopolysaccharides in its cell wall, which eliminates the risk of endotoxin shock. Finally, this bacterium is poorly immunogenic, and for this reason, it can be continuously used in immunization programs (Mercenier et al., 2000). Thus, L. lactis offers great versatility, and several vectors to be used in this microorganism have already been constructed, allowing a wide range of biotechnological applications, besides its use in the food industry.
Lactococcus lactis as a Cell Factory—Proteins of Biotechnological Interest
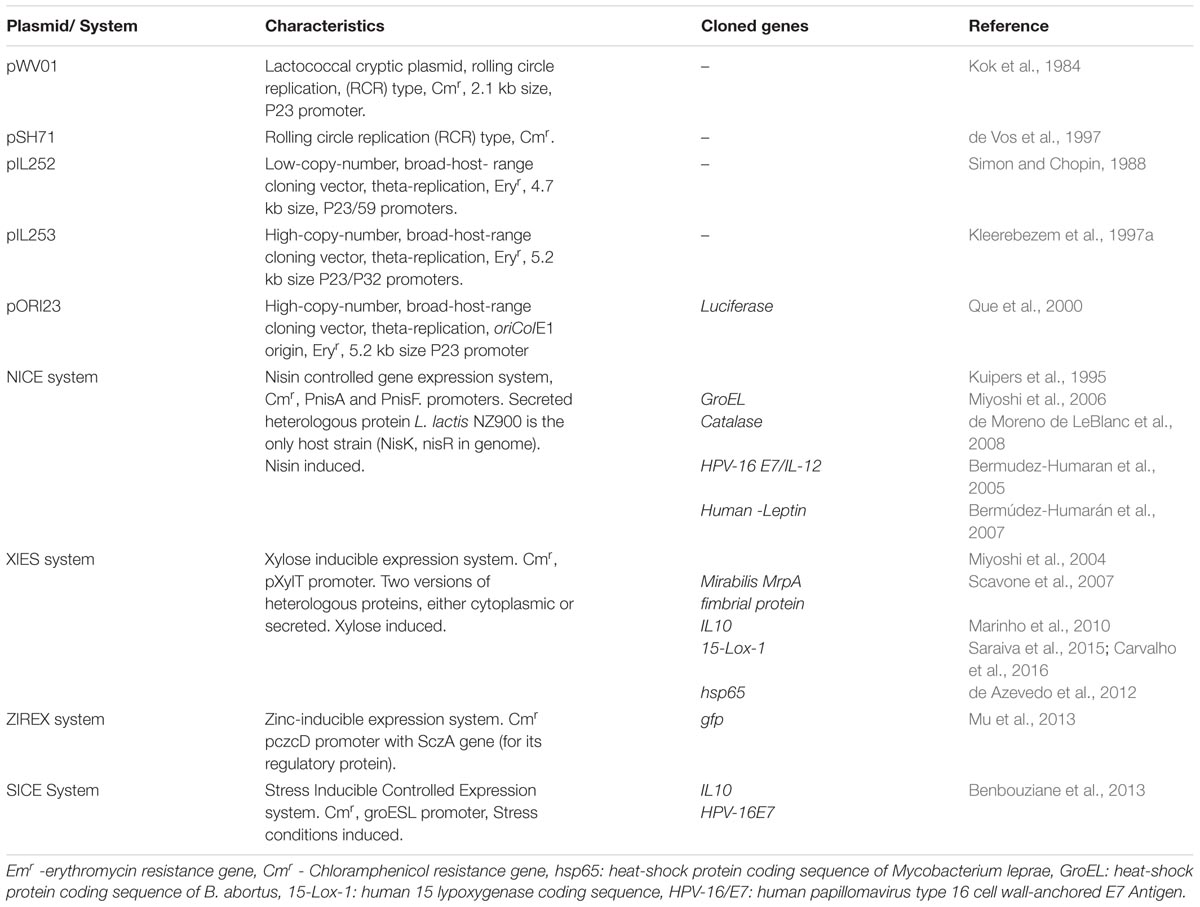
The use of L. lactis as a cell factory for high-level protein production was developed using constitutive and inducible promoters. The history regarding the use of vectors for cloning exogenous genes in the LAB started with two cryptic broad host-range plasmids: pWV01 (Kok et al., 1984) and pSH71 (de Vos et al., 1997). The replicons of these plasmids have been the base for creating useful cloning vectors being able to replicate in a broad range of genera, such as L. lactis and also in Escherichia coli (recA+strain) (De Vos and Simons, 1994). In general, these small plasmids undergo a rolling-circle replication with chloramphenicol resistance. Another family of vectors derived from pAMbeta1 (from Enterococcus faecalis), which can only replicate in Gram-positive host strains, was constructed, pIL252 and pIL253, showing low and high copy plasmids, respectively, as having erythromycin resistance (Simon and Chopin, 1988; Kleerebezem et al., 1997b). These large plasmids replicate via “theta” replication and are equipped with E. coli replicons, generating pOri23 (Que et al., 2000), allowing efficient shuttling between Gram-positive and Gram-negative bacteria (O’Sullivan and Klaenhammer, 1993).
Afterward, several vectors containing constitutive or inducible promoters were developed, and they represent the basis of all expression systems in L. lactis and other LAB (Pontes et al., 2011). Although the high production of heterologous proteins in L. lactis has been achieved using constitutive promoters (De Vos, 1999), this continuous production of high-level protein could generate intracellular accumulation, which could lead to degradation in the cytoplasm and sometimes could be toxic to the cell. This drawback, together with the fact that inducible promoters offer better handling, makes them the vectors of choice. In 1995, the most promising and frequently used system based on genes involved in biosynthesis and regulation of nisin, an antimicrobial peptide whose biosynthesis is encoded by a cluster of 11 genes, was developed (Kuipers et al., 1995). The NICE system (nisin-controlled expression), as it is called, comprises NIS operon regulatory elements, NisR and NisK code, for the regulator two-component system. The histidine-protein kinase NisK located in the cytoplasmic membrane binds the nisin molecule and activates NisR via phosphorylation, which in turn induces transcription of two promoters in the nisin gene cluster: PnisA and PnisF. In fact, this system has been demonstrated to have high protein production and is also easy to use as it has been extensively used in the industry. However, nisin has a high cost limiting its use. Due to nisin, researchers developed another system that is inducible by xylose sugar (Miyoshi et al., 2004). The xylose-inducible expression system (XIES) has the PxylT promoter, and in the presence of other sugars such as glucose, fructose, and/or mannose, it is tightly repressed. Thus, in the presence of xylose, PxylT is transcriptionally activated (Lokman et al., 1994). Another important characteristic of this system is its capacity to produce cytoplasmic or secreted proteins, and it can be easily switched on or off by adding either xylose or glucose, respectively (Miyoshi et al., 2004). As a sugar is used to operate this system, it is considered cheap and very useful for implementation in biotechnological processes. The secretion of heterologous proteins has advantages over intracellular-expressed ones, in the way in which only a simple purification step is necessary, higher yields are reached, and better target interactions are achieved (Le Loir et al., 2005).
Two other inducible systems for L. lactis were described recently. One is the zinc-inducible expression system, called ZIREX. This system allows induction of genes regulated by zinc ions and the pneumococcal repressor SczA (Mu et al., 2013). The other inducible system for L. lactis is stress-inducible controlled expression system (SICE). This system uses a promoter of the L. lactis groESL operon, whose expression is induced under stress conditions, which does not need exogenous induction, nor the presence of regulatory genes (Benbouziane et al., 2013). Characteristics of all these vectors are summarized in Table 1. Many of these vector applications have been described with satisfactory results. A recent report showed the gastrointestinal passage time of bacteria and their spatial localization in the gut. To this end, the authors used a plasmid from the NICE system (pNZ8148) in which IRFP reporters (pNZIRFP713 and pNZ-IRFP682) were cloned, resulting in near-infrared fluorescent proteins being expressed in the LAB (Berlec et al., 2015).
Concerning the XIES system, some therapeutic molecules such as 15-lipoxygenase were cloned in it. L. lactis (pXIES:CYT:15lox-1)-fermented milk was effective in the prevention of intestinal damage associated with inflammatory bowel disease (IBD) in a trinitrobenzenesulfonic acid-induced IBD mouse model (Saraiva et al., 2015) and, in addition, a decrease in IFN-γ and IL-4 was also observed. Also, an increase in IL-10 was observed in a dextran sodium sulfate-induced (DSS-induced) IBD mouse model, where mice were orally administrated with the culture of this strain (Carvalho et al., 2016). L. lactis (pXIES:hsp65) expressing the 65-kDa heat shock protein from Mycobacterium leprae (de Azevedo et al., 2012) was able to prevent the development of experimental autoimmune encephalomyelitis in C57BL/6 mice which received this bacterium by oral administration. The reduction of IL-17 and the increase of IL-10 in mesenteric lymph node and spleen cell cultures were observed (Rezende et al., 2013). The same recombinant strain was tested in the DSS-induced colitis mouse model. In colonic tissue levels, proinflammatory cytokines (IFN-γ, IL-6, and TNF-α) were reduced by increasing IL-10 production. An expansion of regulatory T cells (CD4+Foxp3+ and CD4+LAP+) was also observed in the spleen, as well as in mesenteric lymph nodes. Thus, these data indicate that oral pretreatment with genetically modified L. lactis Hsp65 protein production is able to prevent DSS-induced colitis in C57BL/6 mice (Gomes-Santos et al., 2017).
Dna Vaccine Vectors
Since the early 1990s, several studies have been conducted to test the efficiency of DNA vaccines in the prevention or treatment of different diseases (Huang and Gorman, 1990; Galvin et al., 2000). Modern vaccinology employs plasmids obtained from bacteria that encode antigen polypeptide sequences. These constructions are made using classical molecular biology techniques and more recently molecular techniques such as synthetic biology. There are many advantages of using bacterial plasmids as a vaccine strategy, and some of them are related to their stability at room temperature and reduction of production costs once the purification of the protein in interest is not required, which increases safety in administration (Suschak et al., 2017). Since these vaccine techniques began to be developed, many studies have been conducted aiming at empowering the delivery of these plasmids to host cells, and among these different strategies, we can cite, for example, the different routes of administration and use of adjuvants. However, much emphasis has been placed on the design of vectors that will be used as a vaccine platform (Sørensen et al., 2005; Oliveira and Mairhofer, 2013; Williams, 2014; Suschak et al., 2017).
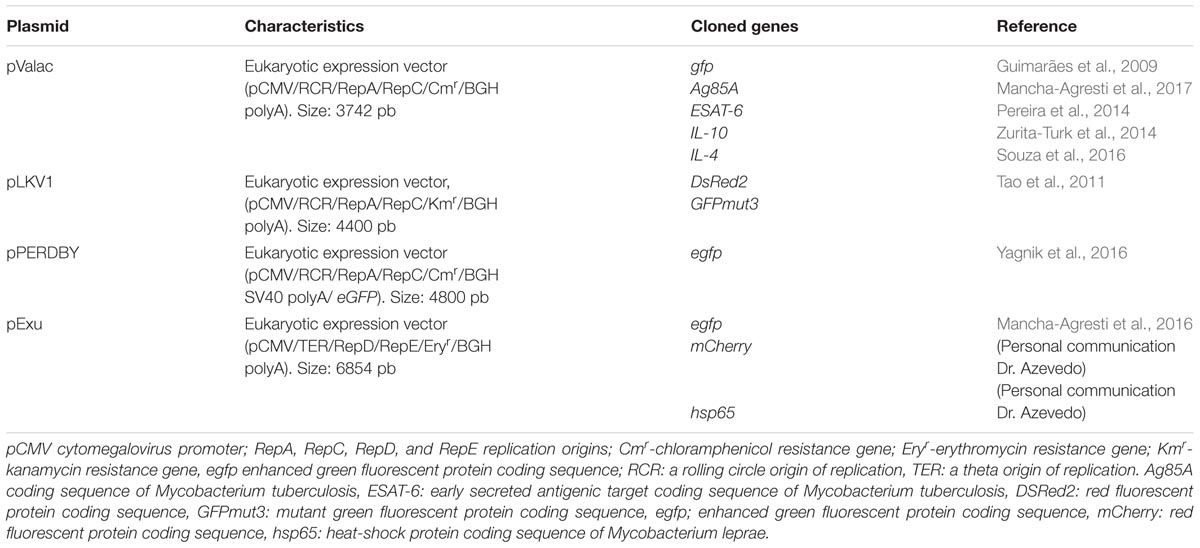
Currently, there are very few groups worldwide working on the development of vectors for DNA vaccines using L. lactis as delivery vehicles, and in recent years, only a few papers have been published in the scientific literature (Guimarães et al., 2009; Tao et al., 2011; Mancha-Agresti et al., 2016; Yagnik et al., 2016, 2017). Vectors generally exhibit some common features such as the presence of a strong eukaryotic promoter, such as cytomegalovirus, multiple cloning sites, a polyadenylation signal in addition to a prokaryotic region that includes a selection marker, usually an antibiotic resistance gene, as well as a prokaryotic origin of replication (Kutzler and Weiner, 2008). In 2009, our research group published a new vector for DNA delivery, pValac (vaccination using lactic acid bacteria), using lactococci. This vector exhibits the characteristics cited above including bovine growth hormone (BGH polyA) polyadenylation sequence, prokaryotic region that allows its replication in both E. coli and L. lactis strains, and chloramphenicol resistance gene as a selection marker, in addition to presenting a rolling circle origin of replication, which contributes to the relatively small size of the plasmid (3742 bp). L. lactis carrying pValac was able to efficiently deliver the vector to eukaryotic cells, allowing expression of green fluorescent protein (GFP) that could be observed by fluorescence microscopy and flow cytometry (Guimarães et al., 2009). Tao et al. (2011) published a paper describing the construction of the pLKV1 vector (4400 bp), which is quite similar to pValac. However, its selection marker is the kanamycin resistance gene, which is seen as an advantage due to the Food and Drug Administration recommending its use in DNA vaccines. The DsRed2 gene, which encodes a red fluorescent protein, was cloned in it. The functionality of this plasmid was verified by visualizing the expression of this protein in epifluorescent microscopy 48 h after the transfection of Caco-2 cells (Tao et al., 2011). However, there are no reports in the literature about in vivo studies using this plasmid. More recently, Yagnik et al. (2016) published the construction of the pPERDBY vector (4800 bp), which, in addition to sharing features common to other vectors already mentioned above, shows the polyadenylation signal of SV40 and the enhanced green fluorescent protein, eGFP, linked to the multiple cloning sites. This feature was designed to facilitate other protein expression, since the eGFP is expressed fused to the protein of interest where the presence of immunostimulatory CpG motifs, which could act as adjuvants, has been described in it. In the Chinese hamster ovary cells (CHO) cell transfection assay, it was possible to observe eGFP expression, demonstrating the functionality of the vector. However, in the flow cytometry analysis, the percentage of cells expressing the reporter protein was extremely low, showing only 0.53% of positive cells (Yagnik et al., 2016). In 2016, our research group published a study relating to the construction of the pExu vector (extra chromosomal unit) with 6854 bp, which in comparison with the described vectors presents the “theta” type origin of replication, which gives it both greater structural and segregational stability. Eukaryotic cells were transfected with pExu:egfp plasmid and analyzed by confocal microscopy and flow cytometry 48 h after transfection. The flow cytometric analysis showed that 18.7% cells were positive for eGFP expression. In addition, in vivo expression kinetics were performed on mice that had received the L. lactis (pExu:egfp) strain by oral administration. The results demonstrated that intestinal cells, specifically in the duodenal region of the bowel, were able to produce the protein of interest from 12 h to 72 h after treatment (Mancha-Agresti et al., 2016) in high amounts. These interesting results would be beneficial from a therapeutic standpoint because it reduces the need for multiple doses in a short period of time. Table 2 summarizes some characteristics of major plasmids constructed for DNA vaccine.
Food-Grade Vectors for Lactococcus lactis
The use of L. lactis for the production of heterologous proteins, as well as for the delivery of DNA vaccines, is currently a promising reality. However, for further clinical trials, the assessment and minimization of risks to human health will be necessary. One of the major concerns related to the use of recombinant bacteria in clinical trials is related to the safety of their use by the host. Historically, cloning and expression vectors are designed using antibiotic resistance genes as marker selection; therefore, this feature may make it unfeasible to use extra-laboratories of L. lactis in the food and pharmaceutical industry.
Many questions are raised, not only about this specific issue, but also about the long-term adverse effects, as well as questions about possible integration into genome, dissemination and toxicity of these plasmids during human trials. To minimize these concerns, the development of new expression systems composed of safer elements, called food-grade systems, that allow selection, induction, and maintenance in the host is necessary to overcome the use of conventional gene expression systems and preserve the GRAS status of this bacterium (Cotter et al., 2003; Landete, 2017). Thus, some tools can be used, such as optimization of vector design, allowing the construction of food grade plasmids, without antibiotic resistance markers, for example, which could increase the safety in use. In addition, bioinformatics tools can aid in this process, allowing the construction of more robust vectors without, however, compromising the safety in the use to treat diseases in humans (Cho et al., 2018).
Among the main mechanisms of resistance and induction used for the development of food-grade vectors such as auxotrophic complementation, chromosome integration, use of sugars, resistance to heavy metals, and more recently, the use of CRISPR/Cas technology can be mentioned (Berlec and Strukelj, 2009; Landete, 2017; Berlec et al., 2018). As an example of auxotrophic complementation, MacCormick and coworkers developed a food-grade system based on lactose metabolism of the L. lactis MG5267 strain. This strain, which has lac operon in its chromosome, was subjected to deletion of the lacF gene, and when this gene was expressed in a plasmid, its ability to metabolize lactose was restored, presenting potential for industrial use (MacCormick et al., 1995). Another selection system based on threonine complementation was described. For this reason, researchers constructed the pJAG5 vector containing the gene that encodes homoserine dehydrogenase-homoserine kinase protein as a selective marker, which catalyzes two steps of the conversion of the aspartate into threonine. This vector, when used in an L. lactis MG1363 strain that presents deletions in two genes encoding threonine biosynthetic enzymes, proved to be quite stable and was able to express green fluorescent protein in eukaryotic cells (Glenting et al., 2002). In another study, a purine auxotrophic L. lactis DN209 strain (Dickely et al., 1995) was transformed with replicative food-grade plasmid pFG1 coding for genes producing peptidases from Lactobacillus helveticus. This strain was able to reduce the ripening period in cheese manufacturing, as well as producing special varieties of cheese (Joutsjoki et al., 2002). Besides auxotrophic complementation, resistance to heavy metals can also be explored with the aim of developing food-grade expression systems for L. lactis (Landete, 2017). The pND302 vector identified in L. lactis M71 strain, for example, offers resistance to cadmium (Liu et al., 1996). Even though it seems paradoxical, the use of these markers is safe, because this vector uses a theta origin of replication, thus being stable even in the absence of selective pressure (Emond et al., 2001).
However, it is important to note that some food-grade vectors present structural instability, and to circumvent this problem, some systems based on chromosomal integration have been developed (Wegmann et al., 1999; Gosalbes et al., 2000; Sheng et al., 2015). Perhaps the most promising example of this technique is the work of Steidler et al. (2003). These researchers replaced thymidylate synthase gene (thyA) of L. lactis with a synthetic human IL10 gene by double homologous crossover (Steidler et al., 2003). This strain contains no antibiotic resistance markers, and because of its thymidine auxotrophy, it cannot spread in the environment, making it one of the safest genetically modified organisms ever designed. The efficacy of this strain was assessed in a first phase 1 clinical trial in a patient with Crohn’s disease, and the results from this study revealed that the use of genetically modified L. lactis to deliver therapeutic molecules, such as IL-10, at the mucosal level is a viable strategy in humans with chronic intestinal inflammation (Braat et al., 2006). Another elegant report showed a recombinant strain of LAB, capable to both produce and secrete the mucosal protectant human trefoil factor 1 at the site of the targeted oral mucosa [L. lactis (AGO13)]. This strain was engineered based on the same strategy (deficient in the gene encoding thymidylate synthase), being a food-grade strain. Results in patients with oral mucositis (phase 1 clinical trial) demonstrated that this strain was able to ameliorate the symptoms of this disease (Limaye et al., 2013).
Although research works with recombinant LAB show promising results in animal models of human disease, these particular ones were the only studies in which recombinant bacteria were used in human clinical trials, demonstrating its potential as a biotechnological tool for the treatment of diseases, but still remaining as a proof of concept. Concerning the regulation of gene expression in food-grade vectors, sugar, temperature, and pH-induced systems are highlighted. The promoter P170 present in L. lactis is inducible by the accumulation of lactic acid at pH 6.0–6.5, when the culture is reaching the stationary phase (Madsen et al., 1999). Therefore, this self-inducible promoter could be considered for the construction of new food-grade expression systems.
P1 and P2 are classic examples of temperature-induced promoters. The repressor of the P2 promoter is inactivated when the medium temperature reaches 40°C. Thus, shifting growth temperature to 24–42°C results in an increase of gene expression controlled by this promoter (Sanders et al., 1998). Sugars can be a safe and inexpensive way to induce gene expression in food-grade systems. Payne and colleagues developed a food-grade expression system induced by lactose; they used the previously mentioned L. lactis strain MG5267, which possesses the lactose operon integrated in the bacterial chromosome. After the lysin gene from Listeria monocytogenes bacteriophage LM-4 was integrated into the chromosome, the chromosomal lacG gene, which encodes phospho-beta-galactosidase, was inactivated by a double cross-over event. Thus, the expression of the lysin gene was shown to be regulated by growth in the presence of lactose, proving to be an important strategy for controlled protein expression in L. lactis (Payne et al., 1996).
Perspectives for Development of Next Generation Vectors
Lactococcus lactis, the model lactic acid bacteria, has enormous potential to be used as a biofactory for the production of numerous proteins of medical and industrial interests, as well as a carrier vehicle for DNA vaccines. Nowadays, this is a reality on the laboratory scale. As new information concerning this microorganism emerges, new possibilities for its use are being contemplated. Constant discoveries in the areas of genomics, transcriptomics, and proteomics of lactic acid bacteria provide us information for new approaches for the construction of the next generation of vectors. These should include the investigation of L. lactis cryptic plasmids, as well as exploration of new selection markers that exclude any type of resistance to antibiotics. In addition, features such as host range, stability, and copy number of plasmids must be considered and also the type of replication origin to be used in vector construction. The theta-type replication mechanism is indicated for reasons already mentioned in previous sections. Moreover, food-grade systems can also be developed by integrating vectors into the host’s chromosomal DNA allowing more stability. Finally, mechanisms of gene expression induction that include the use of cheap and non-harmful compounds such as sugar, pH, or temperature should be considered for future biomedical, biotechnological, and industrial applications. Furthermore, the most promising technique in the field of genetic studies is the CRISPR Cas9 (clustered regularly interspaced short palindromic repeats) system. CRISPR is known as the prokaryote adaptive immune system that provides resistance against foreign DNA, such as plasmids and bacteriophage (Bolotin et al., 2005; Deveau et al., 2008). This system has been studied for the last 30 years (Ishino et al., 1987), but only in 2013 the first experiments were conducted, demonstrating its functionality as an editing genome tool (Cong et al., 2013; Mali et al., 2013). This system has been used in different studies to help understand both disease models and therapeutic schemes for several diseases (Jiang et al., 2013; Makarova et al., 2015; Di Bella et al., 2016).
Although several studies have been carried out with bacterial species using this strategy for genomic editing, only few studies are reported to be using LAB (Oh and van Pijkeren, 2014; Sanozky-Dawes et al., 2015; Song et al., 2017; Berlec et al., 2018). In this way, this technique can be explored for the development of food-grade expression systems and opens perspectives for the use of L. lactis in clinical routine in the near future.
Author Contributions
CP, MD, and PM-A conceptualized the study. CP, MD, PM-A, VB, and AN wrote the original draft of the paper. CP, MD, PM-A, and VA reviewed and edited the manuscript. VA acquired funding and supervised the study.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors want to thank the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG).
References
Benbouziane, B., Ribelles, P., Aubry, C., Martin, R., Kharrat, P., Riazi, A., et al. (2013). Development of a Stress-Inducible Controlled Expression (SICE) system in Lactococcus lactis for the production and delivery of therapeutic molecules at mucosal surfaces. J. Biotechnol. 168, 120–129. doi: 10.1016/j.jbiotec.2013.04.019
Berlec, A., Škrlec, K., Kocjan, J., Olenic, M., and Štrukelj, B. (2018). Single plasmid systems for inducible dual protein expression and for CRISPR-Cas9/CRISPRi gene regulation in lactic acid bacterium Lactococcus lactis. Sci. Rep. 8:1009. doi: 10.1038/s41598-018-19402-1
Berlec, A., and Strukelj, B. (2009). Novel applications of recombinant lactic acid bacteria in therapy and in metabolic engineering. Recent Pat. Biotechnol. 3, 77–87. doi: 10.2174/187220809788700175
Berlec, A., Završnik, J., Butinar, M., Turk, B., and Štrukelj, B. (2015). In vivo imaging of Lactococcus lactis, Lactobacillus plantarum and Escherichia coli expressing infrared fluorescent protein in mice. Microb. Cell Fact. 14:181. doi: 10.1186/s12934-015-0376-4
Bermudez-Humaran, L. G., Cortes-Perez, N. G., Lefevre, F., Guimaraes, V., Rabot, S., Alcocer-Gonzalez, J. M., et al. (2005). A novel mucosal vaccine based on live Lactococci expressing E7 antigen and IL-12 induces systemic and mucosal immune responses and protects mice against human papillomavirus type 16-induced tumors. J. Immunol. 175, 7297–7302. doi: 10.4049/jimmunol.175.11.7297
Bermúdez-Humarán, L. G., Nouaille, S., Zilberfarb, V., Corthier, G., Gruss, A., Langella, P., et al. (2007). Effects of intranasal administration of a leptin-secreting Lactococcus lactis recombinant on food intake, body weight, and immune response of mice. Appl. Environ. Microbiol. 73, 5300–5307. doi: 10.1128/AEM.00295-07
Bolotin, A., Quinquis, B., Sorokin, A., and Ehrlich, S. D. (2005). Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151, 2551–2561. doi: 10.1099/mic.0.28048-0
Braat, H., Rottiers, P., Hommes, D. W., Huyghebaert, N., Remaut, E., Remon, J.-P., et al. (2006). A phase I trial with transgenic bacteria expressing interleukin-10 in Crohn’s disease. Clin. Gastroenterol. Hepatol. 4, 754–759. doi: 10.1016/j.cgh.2006.03.028
Carvalho, R. D. O., Morais, K., Pereira, V. B., Gomes-Santos, A. C., Luerce, T. D., de Azevedo, M. S., et al. (2016). Oral administration of Lactococcus lactis expressing recombinant 15-lipoxygenase-1 (15 LOX-1) modulates chemically induced colitis in mice. Med. Res. Arch. 4:1. doi: 10.18103/mra.v4i7.612
Cho, S., Shin, J., and Cho, B.-K. (2018). Applications of CRISPR/cas system to bacterial metabolic engineering. Int. J. Mol. Sci. 19:E1089. doi: 10.3390/ijms19041089
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Cotter, P. D., Hill, C., and Ross, R. P. (2003). A food-grade approach for functional analysis and modification of native plasmids in Lactococcus lactis. Appl. Environ. Microbiol. 69, 702–706. doi: 10.1128/AEM.69.1.702-706.2003
de Azevedo, M. S. P., Rocha, C. S., Electo, N., Pontes, D. S., Molfetta, J. B., Gonçalves, E. D. C., et al. (2012). Cytoplasmic and extracellular expression of pharmaceutical-grade mycobacterial 65-kDa heat shock protein in Lactococcus lactis. Genet. Mol. Res. 11, 1146–1157. doi: 10.4238/2012.April.27.14
de Moreno de LeBlanc, A., LeBlanc, J. G., Perdigón, G., Miyoshi, A., Langella, P., Azevedo, V., et al. (2008). Oral administration of a catalase-producing Lactococcus lactis can prevent a chemically induced colon cancer in mice. J. Med. Microbiol. 57, 100–105. doi: 10.1099/jmm.0.47403-0
De Vos, W. M. (1999). Gene expression systems for lactic acid bacteria. Curr. Opin. Microbiol. 2, 289–295. doi: 10.1016/S1369-5274(99)80050-2
de Vos, W. M., Kleerebezem, M., and Kuipers, O. P. (1997). Expression systems for industrial gram-positive bacteria with low guanine and cytosine content. Curr. Opin. Biotechnol. 8, 547–553. doi: 10.1016/S0958-1669(97)80027-4
De Vos, W. M., and Simons, G. F. M. (1994). “Gene cloning and expression systems in Lactococci,” in Genetics and Biotechnology of Lactic Acid Bacteria, eds M. J. Gasson and W. de Vos (Dordrecht: Springer), 52–105. doi: 10.1007/978-94-011-1340-3_2
Deveau, H., Barrangou, R., Garneau, J. E., Labonté, J., Fremaux, C., Boyaval, P., et al. (2008). Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190, 1390–1400. doi: 10.1128/JB.01412-07
Di Bella, S., Ascenzi, P., Siarakas, S., Petrosillo, N., and di Masi, A. (2016). Clostridium difficile toxins A and B: insights into pathogenic properties and extraintestinal effects. Toxins 8:E134. doi: 10.3390/toxins8050134
Dickely, F., Nilsson, D., Hansen, E. B., and Johansen, E. (1995). Isolation of Lactococcus lactis nonsense suppressors and construction of a food-grade cloning vector. Mol. Microbiol. 15, 839–847. doi: 10.1111/j.1365-2958.1995.tb02354.x
Emond, E., Lavallée, R., Drolet, G., Moineau, S., and LaPointe, G. (2001). Molecular characterization of a theta replication plasmid and its use for development of a two-component food-grade cloning system for Lactococcus lactis. Appl. Environ. Microbiol. 67, 1700–1709. doi: 10.1128/AEM.67.4.1700-1709.2001
Galvin, T. A., Muller, J., and Khan, A. S. (2000). Effect of different promoters on immune responses elicited by HIV-1 gag/env multigenic DNA vaccine in Macaca mulatta and Macaca nemestrina. Vaccine 18, 2566–2583. doi: 10.1016/S0264-410X(99)00569-1
Glenting, J., Madsen, S. M., Vrang, A., Fomsgaard, A., and Israelsen, H. (2002). ). A plasmid selection system in Lactococcus lactis and its use for gene expression in L. lactis and human kidney fibroblasts. Appl. Environ. Microbiol. 68, 5051–5056. doi: 10.1128/AEM.68.10.5051-5056.2002
Gomes-Santos, A. C., de Oliveira, R. P., Moreira, T. G., Castro-Junior, A. B., Horta, B. C., Lemos, L., et al. (2017). Hsp65-producing Lactococcus lactis prevents inflammatory intestinal disease in mice by IL-10- and TLR2-dependent pathways. Front. Immunol. 8:30. doi: 10.3389/fimmu.2017.00030
Gosalbes, M. J., Esteban, C. D., Galán, J. L., and Pérez-Martínez, G. (2000). Integrative food-grade expression system based on the lactose regulon of Lactobacillus casei. Appl. Environ. Microbiol. 66, 4822–4828. doi: 10.1128/AEM.66.11.4822-4828.2000
Guimarães, V., Innocentin, S., Chatel, J.-M., Lefèvre, F., Langella, P., Azevedo, V., et al. (2009). A new plasmid vector for DNA delivery using Lactococci. Genet. Vaccines Ther. 7:4. doi: 10.1186/1479-0556-7-4
Huang, M. T., and Gorman, C. M. (1990). Intervening sequences increase efficiency of RNA 3′ processing and accumulation of cytoplasmic RNA. Nucleic Acids Res. 18, 937–947. doi: 10.1093/nar/18.4.937
Ishino, Y., Shinagawa, H., Makino, K., Amemura, M., and Nakata, A. (1987). Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169, 5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987
Jiang, W., Bikard, D., Cox, D., Zhang, F., and Marraffini, L. A. (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31, 233–239. doi: 10.1038/nbt.2508
Joutsjoki, V., Luoma, S., Tamminen, M., Kilpi, M., Johansen, E., and Palva, A. (2002). Recombinant Lactococcus starters as a potential source of additional peptidolytic activity in cheese ripening. J. Appl. Microbiol. 92, 1159–1166. doi: 10.1046/j.1365-2672.2002.01652.x
Kleerebezem, M., Beerthuyzen, M. M., Vaughan, E. E., De Vos, W. M., and Kuipers, O. P. (1997a). Controlled gene expression systems for lactic acid bacteria: transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl. Environ. Microbiol. 63, 4581–4584.
Kleerebezem, M., Quadri, L. E. N., Kuipers, O. P., and de Vos, W. M. (1997b). Quorum sensing by peptide pheromones and two-component signal-transduction systems in gram-positive bacteria. Mol. Microbiol. 24, 895–904. doi: 10.1046/j.1365-2958.1997.4251782.x
Kok, J., Van Der Vossen, J. M., and Venema, G. (1984). Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl. Environ. Microbiol. 48, 726–731.
Kuipers, O. P., Beerthuyzen, M. M., de Ruyter, P. G., Luesink, E. J., and de Vos, W. M. (1995). Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J. Biol. Chem. 270, 27299–27304. doi: 10.1074/jbc.270.45.27299
Kutzler, M. A., and Weiner, D. B. (2008). DNA vaccines: ready for prime time? Nat. Rev. Genet. 9, 776–788. doi: 10.1038/nrg2432
Landete, J. M. (2017). A review of food-grade vectors in lactic acid bacteria: from the laboratory to their application. Crit. Rev. Biotechnol. 37, 296–308. doi: 10.3109/07388551.2016.1144044
Le Loir, Y., Azevedo, V., Oliveira, S. C., Freitas, D. A., Miyoshi, A., Bermúdez-Humarán, L. G., et al. (2005). Protein secretion in Lactococcus lactis: an efficient way to increase the overall heterologous protein production. Microb. Cell Fact. 4:2. doi: 10.1186/1475-2859-4-2
Limaye, S. A., Haddad, R. I., Cilli, F., Sonis, S. T., Colevas, A. D., Brennan, M. T., et al. (2013). Phase 1b, multicenter, single blinded, placebo-controlled, sequential dose escalation study to assess the safety and tolerability of topically applied AG013 in subjects with locally advanced head and neck cancer receiving induction chemotherapy. Cancer 119, 4268–4276. doi: 10.1002/cncr.28365
Liu, C. Q., Leelawatcharamas, V., Harvey, M. L., and Dunn, N. W. (1996). Cloning vectors for Lactococci based on a plasmid encoding resistance to cadmium. Curr. Microbiol. 33, 35–39. doi: 10.1007/s002849900070
Lokman, B. C., Leer, R. J., van Sorge, R., and Pouwels, P. H. (1994). Promoter analysis and transcriptional regulation of Lactobacillus pentosus genes involved in xylose catabolism. Mol. Gen. Genet. 245, 117–125. doi: 10.1007/BF00279757
MacCormick, C. A., Griffin, H. G., and Gasson, M. J. (1995). Construction of a food-grade host/vector system for Lactococcus lactis based on the lactose operon. FEMS Microbiol. Lett. 127, 105–109. doi: 10.1111/j.1574-6968.1995.tb07457.x
Madsen, S. M., Arnau, J., Vrang, A., Givskov, M., and Israelsen, H. (1999). Molecular characterization of the pH-inducible and growth phase-dependent promoter P170 of Lactococcus lactis. Mol. Microbiol. 32, 75–87. doi: 10.1046/j.1365-2958.1999.01326.x
Makarova, K. S., and Koonin, E. V. (2007). Evolutionary genomics of lactic acid bacteria. J. Bacteriol. 189, 1199–1208. doi: 10.1128/JB.01351-06
Makarova, K. S., Wolf, Y. I., Alkhnbashi, O. S., Costa, F., Shah, S. A., Saunders, S. J., et al. (2015). An updated evolutionary classification of CRISPR-cas systems. Nat. Rev. Microbiol. 13, 722–736. doi: 10.1038/nrmicro3569
Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., DiCarlo, J. E., et al. (2013). RNA-guided human genome engineering via Cas9. Science 339, 823–826. doi: 10.1126/science.1232033
Mancha-Agresti, P., de Castro, C. P., dos Santos, J. S. C., Araujo, M. A., Pereira, V. B., LeBlanc, J. G., et al. (2017). Recombinant invasive Lactococcus lactis carrying a DNA vaccine coding the Ag85A antigen increases INF-γ, IL-6, and TNF-α cytokines after intranasal immunization. Front. Microbiol. 8:1263. doi: 10.3389/fmicb.2017.01263
Mancha-Agresti, P., Drumond, M. M., Rosa, do Carmo, F. L., Santos, M. M., Coelho, et al. (2016). A new broad range plasmid for DNA delivery in eukaryotic cells using lactic acid bacteria: in vitro and in vivo assays. Mol. Ther. Methods Clin. Dev. 4, 83–91. doi: 10.1016/j.omtm.2016.12.005
Marinho, F. A. V., Pacífico, L. G. G., Miyoshi, A., Azevedo, V., Le Loir, Y., Guimarães, V. D., et al. (2010). An intranasal administration of Lactococcus lactis strains expressing recombinant interleukin-10 modulates acute allergic airway inflammation in a murine model. Clin. Exp. Allergy 40, 1541–1551. doi: 10.1111/j.1365-2222.2010.03502.x
Mercenier, A., Müller-Alouf, H., and Grangette, C. (2000). Lactic acid bacteria as live vaccines. Curr. Issues Mol. Biol. 2, 17–25.
Miyoshi, A., Bermúdez-Humarán, L. G., Ribeiro, L. A., Le Loir, Y., Oliveira, S. C., Langella, P., et al. (2006). Heterologous expression of Brucella abortus GroEL heat-shock protein in Lactococcus lactis. Microb. Cell Fact. 5:14. doi: 10.1186/1475-2859-5-14
Miyoshi, A., Jamet, E., Commissaire, J., Renault, P., Langella, P., and Azevedo, V. (2004). A xylose-inducible expression system for Lactococcus lactis. FEMS Microbiol. Lett. 239, 205–212. doi: 10.1016/j.femsle.2004.08.018
Mu, D., Montalbán-López, M., Masuda, Y., and Kuipers, O. P. (2013). Zirex: a novel zinc-regulated expression system for Lactococcus lactis. Appl. Environ. Microbiol. 79, 4503–4508. doi: 10.1128/AEM.00866-13
Oh, J.-H., and van Pijkeren, J.-P. (2014). CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res. 42:e131. doi: 10.1093/nar/gku623
Oliveira, P. H., and Mairhofer, J. (2013). Marker-free plasmids for biotechnological applications? implications and perspectives. Trends Biotechnol. 31, 539–547. doi: 10.1016/j.tibtech.2013.06.001
O’Sullivan, D. J., and Klaenhammer, T. R. (1993). High- and low-copy-number Lactococcus shuttle cloning vectors with features for clone screening. Gene 137, 227–231. doi: 10.1016/0378-1119(93)90011-Q
Payne, J., MacCormick, C. A., Griffin, H. G., and Gasson, M. J. (1996). Exploitation of a chromosomally integrated lactose operon for controlled gene expression in Lactococcus lactis. FEMS Microbiol. Lett. 136, 19–24. doi: 10.1111/j.1574-6968.1996.tb08019.x
Pereira, V. B., da Cunha, V. P., Preisser, T. M., Souza, B. M., Turk, M. Z., De Castro, C. P., et al. (2017). Lactococcus lactis carrying a DNA vaccine coding for the ESAT-6 antigen increases IL-17 cytokine secretion and boosts the BCG vaccine immune response. J. Appl. Microbiol. 122, 1657–1662. doi: 10.1111/jam.13449
Pereira, V. B., Saraiva, T. D. L., Souza, B. M., Zurita-Turk, M., Azevedo, M. S. P., De Castro, C. P., et al. (2014). Development of a new DNA vaccine based on mycobacterial ESAT-6 antigen delivered by recombinant invasive Lactococcus lactis FnBPA+. Appl. Microbiol. Biotechnol. 99, 1817–1826. doi: 10.1007/s00253-014-6285-3
Pontes, D., Azevedo, M., Innocentin, S., Blugeon, S., Lefévre, F., Azevedo, V., et al. (2014). Immune response elicited by DNA vaccination using Lactococcus lactis is modified by the production of surface exposed pathogenic protein. PLoS One 9:e84509. doi: 10.1371/journal.pone.0084509
Pontes, D. S., de Azevedo, M. S. P., Chatel, J.-M., Langella, P., Azevedo, V., and Miyoshi, A. (2011). Lactococcus lactis as a live vector: heterologous protein production and DNA delivery systems. Protein Expr. Purif. 79, 165–175. doi: 10.1016/j.pep.2011.06.005
Que, Y. A., Haefliger, J. A., Francioli, P., and Moreillon, P. (2000). Expression of Staphylococcus aureus clumping factor a in Lactococcus lactis subsp. cremoris using a new shuttle vector. Infect. Immun. 68, 3516–3522. doi: 10.1128/IAI.68.6.3516-3522.2000
Rezende, R. M., Oliveira, R. P., Medeiros, S. R., Gomes-Santos, A. C., Alves, A. C., Loli, F. G., et al. (2013). Hsp65-producing Lactococcus lactis prevents experimental autoimmune encephalomyelitis in mice by inducing CD4+LAP+ regulatory T cells. J. Autoimmun. 40, 45–57. doi: 10.1016/j.jaut.2012.07.012
Sanders, J. W., Leenhouts, K., Burghoorn, J., Brands, J. R., Venema, G., and Kok, J. (1998). A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol. Microbiol. 27, 299–310. doi: 10.1046/j.1365-2958.1998.00676.x
Sanozky-Dawes, R., Selle, K., O’Flaherty, S., Klaenhammer, T., and Barrangou, R. (2015). Occurrence and activity of a type II CRISPR-cas system in Lactobacillus gasseri. Microbiology 161, 1752–1761. doi: 10.1099/mic.0.000129
Saraiva, T. D. L., Morais, K., Pereira, V. B., de Azevedo, M., Rocha, C. S., Prosperi, C. C., et al. (2015). Milk fermented with a 15-lipoxygenase-1-producing Lactococcus lactis alleviates symptoms of colitis in a murine model. Curr. Pharm. Biotechnol. 16, 424–429. doi: 10.2174/1389201015666141113123502
Scavone, P., Miyoshi, A., Rial, A., Chabalgoity, A., Langella, P., Azevedo, V., et al. (2007). Intranasal immunisation with recombinant Lactococcus lactis displaying either anchored or secreted forms of Proteus mirabilis MrpA fimbrial protein confers specific immune response and induces a significant reduction of kidney bacterial colonisation in mi. Microbes Infect. 9, 821–828. doi: 10.1016/j.micinf.2007.02.023
Sheng, J., Ling, P., and Wang, F. (2015). Constructing a recombinant hyaluronic acid biosynthesis operon and producing food-grade hyaluronic acid in Lactococcus lactis. J. Ind. Microbiol. Biotechnol. 42, 197–206. doi: 10.1007/s10295-014-1555-8
Simon, D., and Chopin, A. (1988). Construction of a vector plasnfid family and its use for molecular cloning in Streptococcus lactis. Biochimie 70, 559–566. doi: 10.1016/0300-9084(88)90093-4
Song, X., Huang, H., Xiong, Z., Ai, L., and Yang, S. (2017). CRISPR-Cas9D10A nickase-assisted genome editing in Lactobacillus casei. Appl. Environ. Microbiol. 83:e1259-17. doi: 10.1128/AEM.01259-17
Sørensen, S. J., Bailey, M., Hansen, L. H., Kroer, N., and Wuertz, S. (2005). Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol. 3, 700–710. doi: 10.1038/nrmicro1232
Souza, B. M., Preisser, T. M., Pereira, V. B., Zurita-Turk, M., de Castro, C. P., da Cunha, V. P., et al. (2016). Lactococcus lactis carrying the pValac eukaryotic expression vector coding for IL-4 reduces chemically-induced intestinal inflammation by increasing the levels of IL-10-producing regulatory cells. Microb. Cell Fact. 15, 150. doi: 10.1186/s12934-016-0548-x
Steidler, L., Neirynck, S., Huyghebaert, N., Snoeck, V., Vermeire, A., Goddeeris, B., et al. (2003). Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nat. Biotechnol. 21, 785–789. doi: 10.1038/nbt840
Suschak, J. J., Williams, J. A., and Schmaljohn, C. S. (2017). Advancements in DNA vaccine vectors, non-mechanical delivery methods, and molecular adjuvants to increase immunogenicity. Hum. Vaccin. Immunother. 13, 2837–2848. doi: 10.1080/21645515.2017.1330236
Tao, L., Pavlova, S. I., Ji, X., Jin, L., and Spear, G. (2011). A novel plasmid for delivering genes into mammalian cells with noninvasive food and communal lactic acid bacteria. Plasmid 65, 8–14. doi: 10.1016/j.plasmid.2010.09.001
van de Guchte, M., Penaud, S., Grimaldi, C., Barbe, V., Bryson, K., Nicolas, P., et al. (2006). The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc. Natl. Acad. Sci. U.S.A. 103, 9274–9279. doi: 10.1073/pnas.0603024103
Wegmann, U., Klein, J. R., Drumm, I., Kuipers, O. P., and Henrich, B. (1999). Introduction of peptidase genes from Lactobacillus delbrueckii subsp. lactis into Lactococcus lactis and controlled expression. Appl. Environ. Microbiol. 65, 4729–4733.
Wells, J. (2011). Mucosal vaccination and therapy with genetically modified lactic acid bacteria. Annu. Rev. Food Sci. Technol. 2, 423–445. doi: 10.1146/annurev-food-022510-133640
Williams, J. A. (2014). Improving DNA vaccine performance through vector design. Curr. Gene Ther. 14, 170–189. doi: 10.2174/156652321403140819122538
Yagnik, B., Padh, H., and Desai, P. (2016). Construction of a new shuttle vector for DNA delivery into mammalian cells using non-invasive Lactococcus lactis. Microbes Infect. 18, 237–244. doi: 10.1016/j.micinf.2015.11.006
Yagnik, B., Sharma, D., Padh, H., and Desai, P. (2017). Dual recombinant Lactococcus lactis for enhanced delivery of DNA vaccine reporter plasmid pPERDBY. Microbiol. Immunol. 61, 123–129. doi: 10.1111/1348-0421.12473
Zurita-Turk, M., Del Carmen, S., Santos, A. C. G., Pereira, V. B., Cara, D. C., Leclercq, S. Y., et al. (2014). Lactococcus lactis carrying the pValac DNA expression vector coding for IL-10 reduces inflammation in a murine model of experimental colitis. BMC Biotechnol. 14:73. doi: 10.1186/1472-6750-14-73
Keywords: lactic acid bacteria, recombinant L. lactis, expression systems, DNA vaccine, food grade vectors
Citation: de Castro CP, Drumond MM, Batista VL, Nunes A, Mancha-Agresti P and Azevedo V (2018) Vector Development Timeline for Mucosal Vaccination and Treatment of Disease Using Lactococcus lactis and Design Approaches of Next Generation Food Grade Plasmids. Front. Microbiol. 9:1805. doi: 10.3389/fmicb.2018.01805
Received: 09 April 2018; Accepted: 18 July 2018;
Published: 14 August 2018.
Edited by:
Aleš Berlec, Jožef Stefan Institute (IJS), SloveniaReviewed by:
Priti Desai, Institute of Advanced Research, IndiaCatherine Daniel, Institut Pasteur de Lille, France
Copyright © 2018 de Castro, Drumond, Batista, Nunes, Mancha-Agresti and Azevedo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vasco Azevedo, dmFzY29AaWNiLnVmbWcuYnI=
 Camila Prosperi de Castro
Camila Prosperi de Castro Mariana M. Drumond
Mariana M. Drumond Viviane L. Batista
Viviane L. Batista Amanda Nunes
Amanda Nunes Pamela Mancha-Agresti
Pamela Mancha-Agresti Vasco Azevedo
Vasco Azevedo