- 1Department of Pathology, University of Texas Medical Branch at Galveston, Galveston, TX, United States
- 2Department of Microbiology, Tokushima University Graduate School of Medical Science, Tokushima, Japan
The family Arenaviridae is divided into three genera: Mammarenavirus, Reptarenavirus, and Hartmanivirus. The Mammarenaviruses contain viruses responsible for causing human hemorrhagic fever diseases including New World viruses Junin, Machupo, Guanarito, Sabia, and Chapare virus and Old World viruses Lassa, and Lujo virus. These two groups of arenaviruses share the same genome organization composed of two ambisense RNA segments. These segments contain four open reading frames that encode for four proteins: the nucleoprotein, glycoprotein precursor, L protein, and Z. Despite their genome similarities, these groups exhibit marked differences in their replication life cycles. This includes differences in attachment, entry, and immune evasion. By understanding the intricacy of replication in each of these viral species we can work to develop counter measures against human diseases. This includes the development of vaccines and antivirals for these emerging viral threats. Currently only the vaccine against Junin virus, Candid#1, is in use as well as Ribavirin for treatment of Lassa Fever. In addition, small molecule inhibitors can be developed to target various aspects of the virus life cycle. In these ways an understanding of the arenavirus replication cycle can be used to alleviate the mortality and morbidity of these infections worldwide.
Family Arenaviridae
The family Arenaviridae is a viral family encompassing three newly separated genera: Mammarenavirus, Reptarenavirus, and Hartmanivirus. As noted in the names, genus Mammarenavirus encompasses those viruses that infect mammalian hosts whereas the reptarenaviruses and hartmaniviruses infect reptilian hosts. The genus Mammarenavirus is further sub-divided into the Old World and New World groups. Currently, the International Committee for Taxonomy of Viruses (ICTV) recognizes 41 distinct viral species in the genus Mammarenavirus (Maes et al., 2018). All members of the family have a negative sense, bi-segmented genome consisting of a large (L) and small (S) segment. While varying between species on exact length, on average the L segment of arenaviruses is 7.2 kb long whereas the S segment is 3.4 kb (Buchmeier et al., 2007). Each segment contains two open reading frames (ORFs), utilizing an ambisense coding strategy, separated by an intergenic region (IGR). This coding strategy allows for the expression of early and late genes regulated by coding orientation. The L segment encodes the multi-functional matrix protein (Z) and the large RNA-dependent RNA polymerase (RdRp) complex (LP). The S segment encodes the viral nucleoprotein (NP) and glycoprotein precursor complex (GPC) that is later cleaved into a stable signal peptide (SSP), GP1, and GP2. Utilizing only a limited number of proteins, arenaviruses are still able to infect, replicate, and modulate host responses in complex and elegant fashions.
As stated previously, the genus Mammarenavirus is separated into two main groups, Old World and New World, based on their genetic, geographic, and epidemiological relationship (Wolff et al., 1978; Charrel et al., 2008). The natural host for Old World arenaviruses is the sub-family Murinae of the Muridae family of mice. Old World arenavirus species include prototypic members Lymphocytic choriomeningitis virus (LCMV) and Lassa virus (LASV) as well as newly emerged Lujo virus. The Old World viruses are generally geographically confined to the African continent with the exception of LCMV that circulates globally in the Mus musculus host. LASV strains were previously subdivided into 4 distinct lineages based on genetic relationship (Bowen et al., 2000; Manning et al., 2015). However, recent phylogenetic evaluation postulates the emergence of a fifth and sixth lineage (Manning et al., 2015; Whitmer et al., 2018). While the first 3 lineages cluster together geographically in Nigeria, lineage 4 LASV strains are found across several countries of West Africa.
The New World arenaviruses are further divided into four clades: A, B, C, and A/Rec (Clade D). The human hemorrhagic pathogens Junin virus (JUNV), Machupo virus (MACV), Guanarito virus (GTOV), Sabia virus (SABV), and Chapare virus, cluster in clade B together with the prototypic Tacaribe virus (TCRV). JUNV, MACV, GTOV, and SABV are the causative agents of Argentine Hemorrhagic Fever (AHF), Bolivian Hemorrhagic Fever (BHF), Venezuelan Hemorrhagic Fever (VHF), and Brazilian Hemorrhagic Fever (BzHF), respectively. Pichinde virus (PICV) is a prototypic clade A virus that is not pathogenic to humans (Buchmeier et al., 1974). To date, it has been widely utilized as a surrogate for LASV pathogenesis, mimicking several disease manifestations in the guinea pig model (Jahrling et al., 1981). Clade A/Rec (Clade D) is hypothesized to be a recombination of clades A and B (Charrel et al., 2001, 2002; Fulhorst et al., 2001; Archer and Rico-Hesse, 2002). This clade includes North American viruses Whitewater Arroyo virus (WWAV) and Bear Canyon virus (BCNV). The New World arenaviruses are geographically located on the South and North American continents. The natural host for the New World viruses is the Sigmodontinae sub-family of Muridae-family mice (Arenaviridae CDC, 2013), with the exception of TCRV, which is found in Artibeus bats (Salazar-Bravo et al., 2002). Recently, TCRV was isolated from Amblyomma americanum ticks in Florida opening the possibility of viral evolution aimed at adaptation to arthropod hosts (Sayler et al., 2014). No further studies have been undertaken to quantify the presence of other New World arenaviruses in arthropods.
Recent surveillance has uncovered the circulation of novel arenaviruses in Asia. Wenzhou Virus (WENV) was isolated in China and is phylogenetically related to the Old World viruses (Li et al., 2015). Further studies indicate that a Cambodian strain of this virus can cause human infection (Blasdell, 2016). These viruses have been found in several rodent vectors (Li et al., 2015), leading to the possibility of a large natural host range.
Both Old World and New World viruses cause significant disease burden in their endemic areas of circulation. According to the latest (Centers for Disease Control and Prevention [CDC], 2013, 2015) reports. LASV causes approximately 100,000–300,000 clinical cases of Lassa fever (LF) a year and results in an estimated 5,000 deaths (Lassa Fever CDC, 2015). These cases are primarily localized in the Western African nations of Sierra Leon, Liberia, Guinea, and Nigeria. In South America, JUNV, MACV, GTOV, and Sabia cause fewer total numbers of disease cases but human infections result in higher case fatality rates. According to the most recent reports AHF, BHF, and VHF result in 15–20, 35, and 33% case fatality rates, respectively (de Manzione et al., 1998; Enria et al., 2004; Patterson et al., 2014).
Lassa virus and JUNV cause the prototypic human disease manifestations for the Old World and New World viruses, respectively. Lassa fever is a zoonotic disease usually resulting from contact with infected murine feces or urine, however, human-to-human transmission is possible. Lassa virus infections have an average incubation time of 10 days and are usually characterized by general flu-like symptoms: fever, malaise, and headache. Patients that progress to severe cases of LF can then develop hemorrhaging and/or neurologic involvement that may be fatal (Frame et al., 1970; Monath et al., 1974; Monson et al., 1984, 1987; McCormick et al., 1987; Hirabayashi et al., 1988; Frame, 1989; Bausch et al., 2001; Enria et al., 2004). In LF survivors, high rates of neurological sequelae have been observed, with nearly 25% of survivors developing deafness (Liao et al., 1992; WHO, 2005; Okokhere et al., 2009). JUNV infection also occurs from exposure to infected murine feces and urine, mostly associated with agricultural activities, and may develop into AHF with an incubation time of 6–14 days. Like LF, AHF initially presents with flu-like symptoms including fever, malaise, anorexia, and headache. Disease progression can result in neurologic symptoms, leukopenia, and thrombocytopenia. One hallmark of AHF as well as other New World virus infections is the large spike of interferon production during infection that is not seen in Old World virus infections (Enria et al., 2004).
Viral Life Cycle
Virus Entry
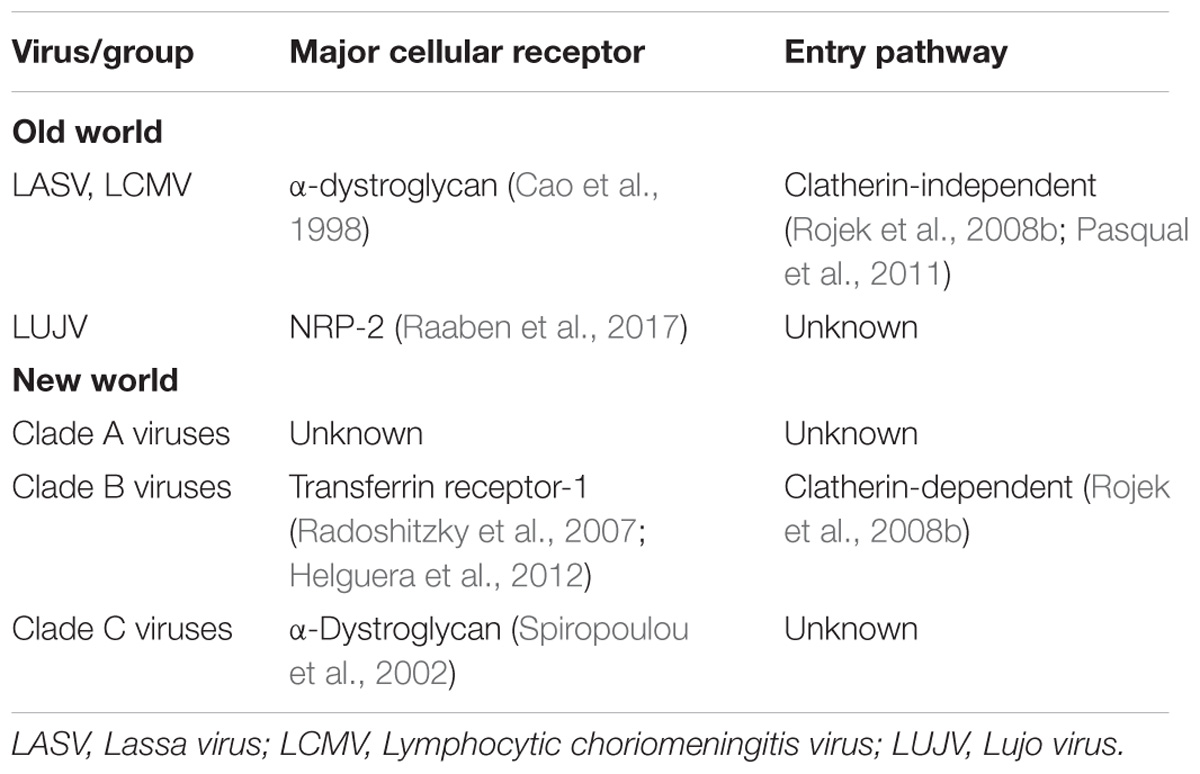
Virus particles initially enter the host through inhalation of aerosolized particles. It is hypothesized that alveolar macrophages are the first cell types infected due to the route of infection and virus present in macrophages early in infection (Gonzalez et al., 1980). While arenaviruses and antigen have been detected in the pulmonary epithelium of infected humans and animals, it is unclear whether these cells are the initial targets (Walker et al., 1982; Hall et al., 1996). These initial infected macrophages can then move to the draining lymph node, resulting in migration and spread of the virus to various tissues (Gonzalez et al., 1980; Buchmeier et al., 2013). Aside from the different cellular receptors, Old World and New World viruses also utilize different entry pathways into the cell. The process of entry is initiated when the GP1 attaches to the corresponding cellular receptor Old World viruses, such as LASV and LCMV as well as Clade C New World viruses utilize α-dystroglycan, an extracellular matrix protein commonly found in the basement membrane (Cao et al., 1998; Spiropoulou et al., 2002), for entry. LUJV was recently discovered to utilize the neurophin (NRP)-2 receptor to mediate entry (Raaben et al., 2017). Recently, TAM family, C-type lectins, and AxI have also been implicated as candidate receptors for LASV and LCMV (Shimojima and Kawaoka, 2012; Shimojima et al., 2012; Fedeli et al., 2017). This raises the possibility of multiple receptors and entry methods for Old World arenaviruses. For Clade B New World arenaviruses, GP1 attaches to transferrin receptor-1 (Radoshitzky et al., 2007; Helguera et al., 2012). TIM family proteins have also been shown to enhance New World arenavirus infectivity (Jemielity et al., 2013). Additionally, siRNA screens indicate that voltage-gated calcium channel (VGCC) subunits play a role in JUNV cell fusion and cellular entry (Lavanya et al., 2013), indicating possible co-receptors used in New World arenavirus entry. To date, the cellular receptor utilized by Clade A New World arenaviruses has yet to be established. After GP1 attaches to the cellular receptor, the virion is endocytosed. New World viruses, such as JUNV, are endocytosed through a clatherin-dependent pathway, whereas the Old World viruses proceed through an unknown clatherin-independent pathway (Rojek et al., 2008b; Pasqual et al., 2011). This entry indicates the role of Endosomal Sorting Complex Required for Transport (ESCRT) proteins for successful cellular entry (Pasqual et al., 2011). Recent studies additionally indicate a possible role of macropinocytosis pathways in Old World viral entry (Oppliger et al., 2016) (see Table 1 for summary of arenavirus entry). Following endocytosis and acidification of the endosome, GP2, a class I fusion protein, facilitates membrane fusion and the release of the viral genome and replication complexes into the cell (Eschli et al., 2006). Additional evidence has shown that in the process of LASV entry into the cell acidification of the endosome results in an altered G1 receptor binding domain. This results in GP1 binding Lysosomal-associated membrane protein 1 (LAMP1) and potentially assisting in the fusion activity of GP2 (Cohen-Dvashi et al., 2016). LUJV has been shown to utilize the same strategy but binding to CD63 instead of LAMP1 to mediate fusion and escape from the endosome (Raaben et al., 2017).
Replication
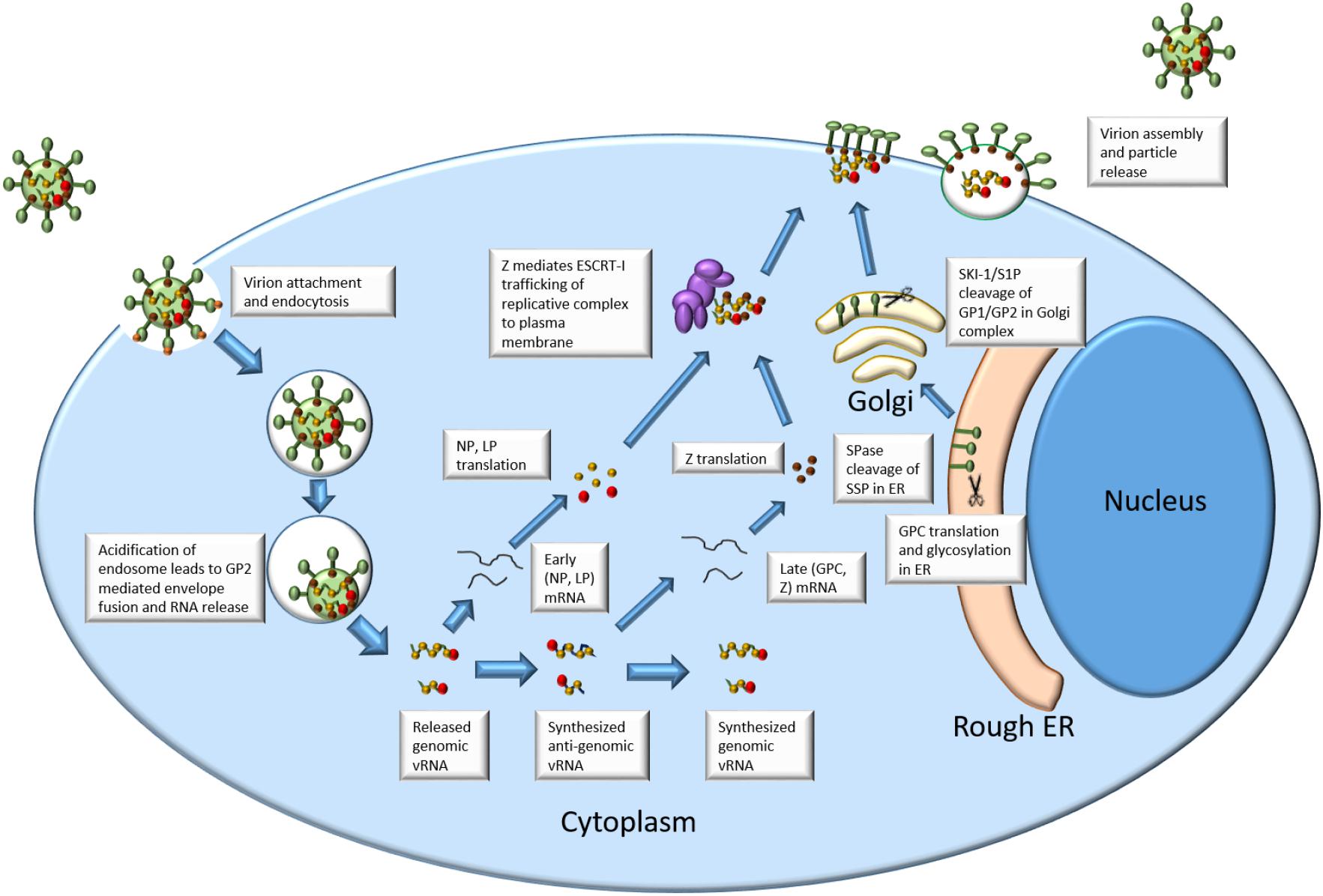
See Figure 1 for an outline of arenavirus replication.
Once the replication complex has been released into the cytoplasm, the viral LP, in conjunction with NP, begins replication of the viral genome. The association of LP, NP, and the viral genome in the cell is considered to be the minimum requirement for initiation of replication (Lee et al., 2000; López et al., 2001; Hass et al., 2004). The LP contains 4 proposed domains (Brunotte et al., 2011). Domain 1 contains the endonuclease domain hypothesized to facilitate acquisition of 5′ mRNA caps critical to transcription during the replication cycle. The 5′ mRNA cap is an m7G structure that is found on the 5′ end of eukaryotic mRNA. This cap serves the purposes enhancing mRNA translation and stability (Both et al., 1975; Furuichi et al., 1977). This LP function is due to the domain’s sequence and structure homology with influenza cap-acquisition protein PA as well as enzymatic studies utilizing LCMV Domain 1 expression (Morin et al., 2010). Domain 3 of the LP contains conserved sequences identified as elements of an RdRp (Vieth et al., 2004). Domains 2 and 4 have as yet unknown functions that may relate to stability, regulation, or other protein-protein interactions (Brunotte et al., 2011). Replication is initiated by LP and NP acquiring 5′ caps from host mRNA. These caps prime the initiation of early gene transcription: NP and LP ORFs on the S and L segments, respectively. Since the arenavirus RNA is negative sense, it is unable to be directly translated in its genomic form. This necessitates nascent RNA-dependent RNA transcription that is produced 5′–3′. The NP and LP ORFs are found in this first transcribed region leading to their initial production as early products (Meyer et al., 2002). Transcription is terminated through the utilization of intergenic hairpin structures found in both the L and S segment IGR (Pinschewer et al., 2005). Translated NP binds to viral RNA to facilitate further transcription, replication, and eventually assembly of progeny virus. Crystal structure analysis of NP has shown that the N terminal domain may accommodate the “stolen” host mRNA cap facilitating further transcription (Qi et al., 2010).
The transition from early gene products to late gene products is facilitated through the production of anti-genomic RNA (Meyer et al., 2002). This is produced when the polymerase reads through the IGR hairpins and makes a complimentary RNA genome. These anti-genomic RNAs are then utilized as the template for transcribing mRNAs from the Z and GPC ORFs. Since they require the production of antigenomic RNA and later genomic mRNA (Meyer et al., 2002), Z and GPC constitute the late gene products in the viral replication cycle. GPC is translated in the endoplasmic reticulum where it undergoes N-linked glycosylation and cleavage of the SSP from GP1/GP2 by SPase (Eichler et al., 2003). The majority of type 1 fusion proteins are cleaved by the host furin like proteases, but arenavirus GPC utilizes a divergent pathway (Harrison, 2015). Precleaved GPC is additionally cleaved between GP1 and GP2 by Subtilisin Kexin Isozyme 1/Site 1 Protease (SKI-1/S1P) into the final SSP, GP1, and GP2 in the trans-Golgi network (Lenz et al., 2001; Rojek et al., 2008a). These three cleaved products stay associated and form a tripartite glycoprotein spike in the cellular plasma membrane. This again is unique in the fact that the signal peptide is incorporated into the final virion instead of being degraded following final protein localization. The Z protein is a short peptide from 90 to 99 aa in length depending on viral species (Capul et al., 2011). This protein contains three domains that contribute to its multifunctional nature. The N-terminal domain contains a myristylation site that allows the protein to localize and anchor itself to the plasma membrane (Perez et al., 2004). The central domain contains a Really Interesting New Gene (RING) domain that chelates Zinc ions. This domain has been shown to be critical for Z-NP and Z-LP interaction but not for Z-GPC interaction (Wilda et al., 2008; Capul et al., 2011; Loureiro et al., 2011). Finally, the C-terminal domain of Z contains proline-rich motifs known to interact with the ESCRT machinery. This interaction is due to interactions of the late-domain of Z with the ESCRT proteins (Perez et al., 2003). Within arenaviruses, there are many different late domains that fall into several large groups. In general, Old World viruses contain the PTAP and PPPY amino acid domains whereas the New World viruses generally contain the PT/SAP amino acid domain (Urata and de la Torre, 2011).
Assembly and Budding
Interaction with the ESCRT machinery allows for transport of Z to the membrane to support virion budding. Specifically, the late domain of Z has been show to interact with Tumor susceptibility gene 101 (Tsg-101) of the ESCRT pathway (Wilda et al., 2008). Recently, it has been suggested that the late domain motif of LCMV drives the production of defective interfering particles (Ziegler et al., 2016). This is thought to be mediated by the PPPY late domain found in some Old World viruses but not in New World (Ziegler et al., 2016). Accumulation of Z protein performs many late-replication functions in the cell. First, the Z protein interacts with LP and inhibits polymerase function (Kranzusch and Whelan, 2011). In TACV, the LP-Z interaction has been mapped to residues in Domain 1 and Domain 3 of the L polymerase (Perez et al., 2003). This interaction, along with NP-Z interactions, facilitates translocation of the replication complex to the cell membrane in preparation for virion budding that occurs through the interaction of the Z late domain with Tsg-101. Finally, accumulation of the Z protein at the membrane results in viral budding and the production of new, infectious virions. In the absence of other viral proteins and genomic RNA, Z has been shown to initiate self-budding resulting in production of virus-like particles which is why it is considered to be the minimal budding factor for the virus (Hoenen et al., 2007; Ziegler et al., 2016).
Immune Evasion
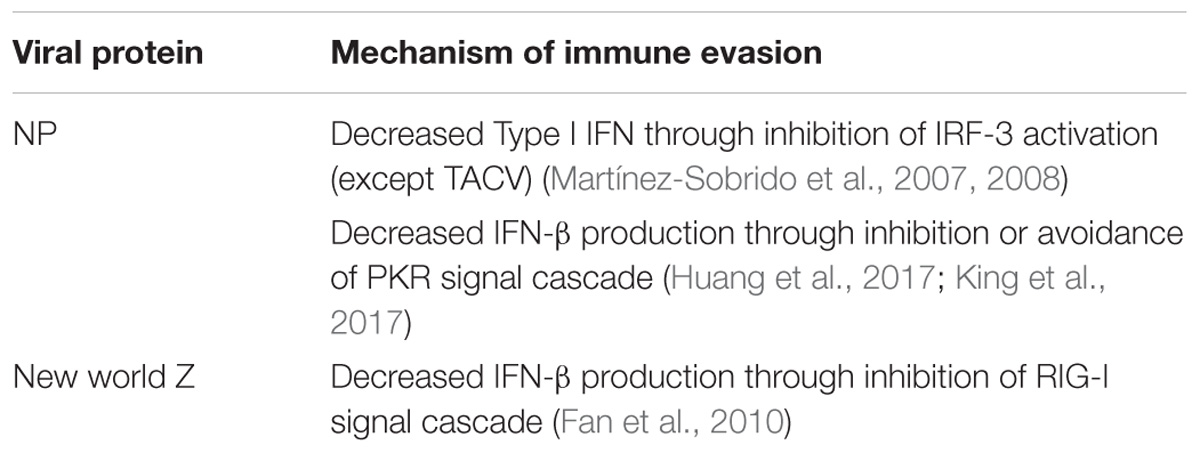
During replication, viral evasion from the immune system is critical for productive replication and dissemination in the host. Arenaviruses have been shown to accomplish this in many ways. First, the NP protein has been shown to have strong anti-immune functions in the cell. Interferon regulatory factor 3 (IRF-3) is an important component of the type 1 interferon response. Studies have shown that NP inhibits IRF-3 activation of downstream genes resulting in a limited interferon response (Martínez-Sobrido et al., 2007, 2008). Interestingly, TCRV does not exhibit this same function in the NP perhaps suggesting differential adaptation to an alternative host. The downregulation of interferon-beta in infected cells has been linked to the exonuclease function of the C terminal domain of NP (Qi et al., 2010). This results in the degradation of small dsRNAs that may trigger host signal peptides. One of these potent cellular defenses against viral infection is the dsRNA activated protein kinase (PKR). PKR is able to recognize dsRNA in the cell and become phosphorylated (García et al., 2006). Phosphorylated PKR triggers phosphorylation of other signal proteins as well as the phosphorylation of transcription factors needed to halt viral replication. While some studies have shown that the NP of JUNV is able to stop phosphorylated PKR from resulting in downstream signally and phosphorylation of translation factor eIF2α (King et al., 2017) others indicate that JUNV and MACV infections still produce phosphorylated eIF2α (Huang et al., 2017). Recent studies have demonstrated that while JUNV and MACV activate PKR, LASV infection does not (Huang et al., 2017). It was also shown that this was not due to a prevention of PKR activation, but likely an avoidance of detection. In this way LASV may avoid activation of immune responses in infected cells.
In New World viruses, the Z protein has been shown to also play a role in immune evasion. The Z protein binds to the retinoic acid-inducible gene I (RIG-I) and inhibits interferon-beta production (Fan et al., 2010). RIG-I normally acts as a cellular sensor for double stranded RNA and activates interferon production through a signal cascade (Loo and Gale, 2011). The binding with New World viral Z prevents RIG-I from binding mitochondrial antiviral signaling protein (MAVS), the next protein in the signaling cascade. This halts interferon-beta production and allows the virus to replicate without the presence of a robust immune response (see Table 2 for a summary of arenavirus immune evasion).
Another way in which arenaviruses may activate the innate immune system and promote a Type I interferon response is through the activation of various Toll-like receptors (TLRs) (Vidya et al., 2018). JUNV infection has been shown to induce a Type I response mediated through activation of TLR 2 (Cuevas et al., 2011; Cuevas and Ross, 2014). However, induction of this response did not result in decreased viral replication in brain cells. Old World LCMV has been shown to produce a TLR 7 and TLR 9 mediated Type I response (Lee et al., 2009; Macal et al., 2012; Walsh et al., 2012; Buechler et al., 2015). A productive TLR 7 mediated response was also shown to be necessary for robust adaptive immune responses as well as monocyte recruitment (Macal et al., 2012; Walsh et al., 2012). Interesting, chronic LCMV infection of dendritic cells inhibited the TLR 7 and 9 responses of cells treated with each respective agonist (Cuevas and Ross, 2014) in an ex vivo model. When these agonist were used to treat in vitro cells chronically infected with LCMV, there was no significant difference in the TLR 7 and 9 response. Further studies are necessary to determine the exact contributions of each of these signaling pathways and the viral impact on their activation.
Finally, Old World and New World viruses appear to exhibit differences in terms of immune protection against fatal infection. In New World JUNV, passive antibody transfer from immune sera has been shown to dramatically improve survival of infection in humans (Maiztegui et al., 1979; Ruggiero et al., 1986). Old World LASV, however, does not show any improved survival upon treatment with immune sera (McCormick et al., 1986). It is important to note that some patients treated with the JUNV immune plasma did develop late neurological complications. These results indicate an importance of a B cell mediated response in JUNV infection and a likely T cell mediated response in LASV infection.
Conclusion
The family Arenaviridae encompasses a large variety of viral species found in a variety of animal hosts. Within the genus Mammarenavirus, great diversity is still seen in terms of cellular entry strategies, and host immune evasion. Old World and New World viruses demonstrate a marked divergence in these pathways that makes an understanding of the varied mechanisms crucial to the study of their replication and pathogenesis. Within these broader groups there is still an observed divergence in cellular receptor usage and natural host range adding additional complexity to their life cycles. As arenavirus research continues, similarities and differences between the varied species will further allow for the development of pathogenesis models and treatments that are encompassing for species that share those similarities, but not be mistakenly utilized on divergent members. This refinement of arenavirus biology is crucial for all future research and development utilizing these agents.
Author Contributions
SH compiled and wrote the manuscript. TK, JM, and SP were involved with reviewing and editing.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Archer, A. M., and Rico-Hesse, R. (2002). High genetic divergence and recombination in Arenaviruses from the Americas. Virol. J. 304, 274–281. doi: 10.1006/viro.2002.1695
Bausch, D. G., Demby, A. H., Coulibaly, M., Kanu, J., Goba, A., Bah, A., et al. (2001). Lassa fever in Guinea: I. Epidemiology of human disease and clinical observations. Vector Borne Zoonotic Dis. 1, 269–281. doi: 10.1089/15303660160025903
Blasdell, K. R. (2016). Evidence of human infection by a new mammarenavirus endemic to Southeastern Asia. eLife 5:e13135. doi: 10.7554/eLife.13135
Both, G. W., Banerjee, A. K., and Shatkin, A. J. (1975). Methylation-dependent translation of viral messenger RNAs in vitro. Proc. Natl. Acad. Sci. U.S.A. 72, 1189–1193. doi: 10.1073/pnas.72.3.1189
Bowen, M. D., Rollin, P. E., Ksiazek, T. G., Hustad, H. L., Bausch, D. G., Demby, A. H., et al. (2000). Genetic diversity among Lassa virus strains. J. Virol. 74, 6992–7004. doi: 10.1128/JVI.74.15.6992-7004.2000
Brunotte, L., Lelke, M., Hass, M., Kleinsteuber, K., Becker-Ziaja, B., and Günther, S. (2011). Domain structure of Lassa virus L protein. J. Virol. 85, 324–333. doi: 10.1128/JVI.00721-10
Buchmeier, M., Adam, E., and Rawls, W. E. (1974). Serological evidence of infection by Pichinde virus among laboratory workers. Infect. Immun. 9, 821–823.
Buchmeier, M. J., de la Torre, J., and Peters, C. J. (2007). “Arenaviridae: the viruses and their replication,” in Fields Virology, 4th Edn, eds D. L. Knipe and P. M. Howley (Philadelphia, PA: Lippincott-Raven).
Buchmeier, M. J., de La Torre, J. C., Peters, C. J., et al. (2013). “Arenaviridae,” in Fields Virology, 6th Edn, eds D. M. Knipe and P. M. Holey (Philadelphia, PA: Lippincott Williams & Wilkins), 1283–1303.
Buechler, M. B., Gessay, G. M., Srivastava, S., Campbell, D. J., Hamerman, J. A., et al. (2015). Hematopoietic and non-hematopoietic cells promote type I interferon- and toll-like receptor 7-dependent monocytosis during low-dose Lymphocytic choriomeningitis virus infection. Eur. J. Immunol. 45, 3064–3072. doi: 10.1002/eji.201445331
Cao, W., Henry, M. D., Borrow, P., Yamada, H., Elder, J. H., Ravkov, E. V., et al. (1998). Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science 282, 2079–2081. doi: 10.1126/science.282.5396.2079
Capul, A. A., de la Torre, J. C., and Buchmeier, M. J. (2011). Conserved residues in Lassa fever virus Z protein modulate viral infectivity at the level of the ribonucleoprotein. J. Virol. 85, 3172–3178. doi: 10.1128/JVI.02081-10
Centers for Disease Control and Prevention [CDC] (2013). Arenaviridae. Available at: https://www.cdc.gov/vhf/virus-families/arenaviridae.html [accessed July 2, 2018].
Centers for Disease Control and Prevention [CDC] (2015). Lassa Fever. Available at: https://www.cdgov/vhf/lassa/ [accessed June 28, 2018].
Charrel, R. N., de Lamballerie, X., and Fulhorst, C. F. (2001). The Whitewater Arroyo virus: natural evidence for genetic recombination among tacaribe serocomplex viruses (family Arenaviridae). Virol. J. 283, 161–166. doi: 10.1006/viro.2001.0874
Charrel, R. N., de Lamballerie, X., and Emonet, S. (2008). Phylogeny of the genus Arenavirus. Curr. Opin. Microbiol. 11, 362–368. doi: 10.1016/j.mib.2008.06.001
Charrel, R. N., Feldmann, H., Fulhorst, C. F., Khelifa, R., de Chesse, R., and de Lamballerie, X. (2002). Phylogeny of New World arenaviruses based on the complete coding sequences of the small genomic segment identified an evolutionary lineage produced by intrasegmental recombination. Biochem. Biophys. Res. Commun. 296, 1118–1124. doi: 10.1016/S0006-291X(02)02053-3
Cohen-Dvashi, H., Israeli, H., Shani, O., Katz, A., and Diskin, R. (2016). Role of LAMP1 binding and pH sensing by the spike complex of Lassa virus. J. Virol. 90, 10329–10338. doi: 10.1128/JVI.01624-16
Cuevas, C. D., Lavanya, M., Wang, E., and Ross, S. R. (2011). Junin virus infects mouse cells and induces innate immune responses. J. Virol. 85, 11058–11068. doi: 10.1128/JVI.05304-11
Cuevas, C. D., and Ross, S. R. (2014). Toll-like receptor 2-mediated innate immune responses against Junin virus in mice lead to antiviral adaptive immune responses during systemic infection and do not affect viral replication in the brain. J. Virol. 88, 7703–7714. doi: 10.1128/JVI.00050-14
de Manzione, N., Salas, R. A., Paredes, H., Godoy, O., Rojas, L., Araoz, F., et al. (1998). Venezuelan hemorrhagic fever: clinical and epidemiological studies of 165 cases. Clin. Infect. Dis. 26, 308–313. doi: 10.1086/516299
Eichler, R., Lenz, O., Strecker, T., and Garten, W. (2003). Signal peptide of Lassa virus glycoprotein GP-C exhibits an unusual length. FEBS Lett. 538, 203–206. doi: 10.1016/S0014-5793(03)00160-1
Enria, D., Bowen, M. D., Mills, J. N., Shieh, W. J., Bausch, D., et al. (2004). “Arenavirus infections,” in Tropical Infectious Diseases: Principles, Pathogens, and Practice, eds R. L. Guerrant, D. H. Walker, P. F. Weller, and W. B. Saunders (Amarillo, TX: Saunders), 1191–1212.
Eschli, B., Quirin, K., Wepf, A., Weber, J., Zinkernagel, R., and Hengartner, H. (2006). Identification of an N-terminal trimeric coiled-coil core within Arenavirus glycoprotein 2 permits assignment to class I viral fusion proteins. J. Virol. 80, 5897–5907. doi: 10.1128/JVI.00008-06
Fan, L., Briese, T., and Lipkin, W. I. (2010). Z proteins of New World arenaviruses bind RIG-I and interfere with type I interferon induction. J. Virol. 84, 1785–1791. doi: 10.1128/JVI.01362-09
Fedeli, C., Torriani, G., Galan-Navarro, C., Moraz, M. L., Moreno, H., Gerold, G., et al. (2017). Axl can serve as entry factor for Lassa virus depending on the functional glycosylation of dystroglycan. J. Virol. 92:e01613-17. doi: 10.1128/JVI.01613-17
Frame, J. D. (1989). Clinical features of Lassa fever in Liberia. Rev. Infect. Dis. 11(Suppl. 4), S783–S789. doi: 10.1093/clinids/11.Supplement_4.S783
Frame, J. D., Baldwin, J. M. Jr., Gocke, D. J., and Troup, J. M. (1970). Lassa fever, a new virus disease of man from West Africa. I. Clinical description and pathological findings. Am. J. Trop. Med. Hyg. 19, 670–676. doi: 10.4269/ajtmh.1970.19.670
Fulhorst, C. F., Charrel, R. N., Weaver, S. C., Ksiazek, T. G., Bradley, R. D., Milazzo, M. L., et al. (2001). Geographic distribution and genetic diversity of Whitewater Arroyo virus in the southwestern United States. Emerg. Infect. Dis. 7, 403–407. doi: 10.3201/eid0703.017306
Furuichi, Y., LaFiandra, A., and Shatkin, A. J. (1977). 5′-Terminal structure and mRNA stability. Nature 266, 235–239. doi: 10.1038/266235a0
García, M. A., Gil, J., Ventoso, I., Guerra, S., Domingo, E., Rivas, C., et al. (2006). Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 70, 1032–1060. doi: 10.1128/MMBR.00027-06
Gonzalez, P. H., Cossio, P. M., Arana, R., Maiztegui, J. I., and Laguens, R. P. (1980). Lymphatic tissue in Argentine hemorrhagic fever. Pathologic features. Arch. Pathol. Lab. Med. 104, 250–254.
Hall, W. C., Geisbert, T. W., Huggins, J. W., and Jahrling, P. B. (1996). Experimental infection of guinea pigs with Venezuelan hemorrhagic fever (Guanarito): a model of human disease. Am. J. Trop. Med. Hyg. 55, 81–88. doi: 10.4269/ajtmh.1996.55.81
Harrison, S. C. (2015). Viral membrane fusion. Virology 479–480, 498–507. doi: 10.1016/j.virol.2015.03.043
Hass, M., Gölnitz, U., Müller, S., Becker-Ziaja, B., and Günther, S. (2004). Replicon system for Lassa virus. J. Virol. 78, 13793–13803. doi: 10.1128/JVI.78.24.13793-13803.2004
Helguera, G., Jemielity, S., Abraham, J., Cordo, S. M., Martinez, M. G., Rodríguez, J. A., et al. (2012). An antibody recognizing the apical domain of human transferrin receptor 1 efficiently inhibits the entry of all New World hemorrhagic fever arenaviruses. J. Virol. 86, 4024–4028. doi: 10.1128/JVI.06397-11
Hirabayashi, Y., Oka, S., Goto, H., Shimada, K., Kurata, T., Fisher-Hoch, S. P., et al. (1988). An imported case of Lassa fever with late appearance of polyserositis. J. Infect. Dis. 158, 872–875. doi: 10.1093/infdis/158.4.872
Hoenen, T., Kolesnikova, L., and Becker, S. (2007). Recent advances in filovirus- and arenavirus-like particles. Future Virol. 2, 193–203. doi: 10.2217/17460794.2.2.193
Huang, C., Kolokoltsova, O. A., Mateer, E. J., Koma, T., and Paessler, S. (2017). Highly pathogenic New World arenavirus infection activates the pattern recognition receptor Protein Kinase R without attenuating virus replication in human cells. J. Virol. 91:e01090-17. doi: 10.1128/JVI.01090-17
Jahrling, P. B., Hesse, R. A., Rhoderick, J. B., Elwell, M. A., and Moe, J. B. (1981). Pathogenesis of a pichinde virus strain adapted to produce lethal infections in guinea pigs. Infect. Immun. 32, 872–880.
Jemielity, S., Wang, J. J., Chan, Y. K., Ahmed, A. A., Li, W., Monahan, S., et al. (2013). TIM-family proteins promote infection of multiple enveloped viruses through virion-associated phosphatidylserine. PLoS Pathog. 9:e1003232. doi: 10.1371/journal.ppat.1003232
King, B. R., Hershkowitz, D., Eisenhauer, P. L., Weir, M. E., Ziegler, C. M., Russo, J., et al. (2017). A map of the arenavirus nucleoprotein-host protein interactome reveals that Junin virus selectively impairs the antiviral activity of PKR. J. Virol. 91:e01090-17. doi: 10.1128/JVI.00763-17
Kranzusch, P. J., and Whelan, S. P. (2011). Arenavirus Z protein controls viral RNA synthesis by locking a polymerase-promoter complex. Proc. Natl. Acad. Sci. U.S.A. 108, 19743–19748. doi: 10.1073/pnas.1112742108
Lavanya, M., Cuevas, C. D., Thomas, M., Cherry, S., and Ross, S. R. (2013). siRNA screen for genes that affect Junin virus entry uncovers voltage-gated calcium channels as a therapeutic target. Sci. Transl. Med. 5:204ra131. doi: 10.1126/scitranslmed.3006827
Lee, K. J., Novella, I. S., Teng, M. N., Oldstone, M. B., and de La Torre, J. C. (2000). NP and L proteins of lymphocytic choriomeningitis virus (LCMV)are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J. Virol. 74, 3470–3477. doi: 10.1128/JVI.74.8.3470-3477.2000
Lee, L. N., Burke, S., Montoya, M., and Borrow, P. (2009). Multiple mechanisms contribute to impairment of type 1 interferon production during chronic lymphocytic choriomeningitis virus infections of mice. J. Immunol. 182, 7178–7189. doi: 10.4049/jimmunol.0802526
Lenz, O., ter Meulen, J., Klenk, H. D., Seidah, N. G., and Garten, W. (2001). The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc. Natl. Acad. Sci. U.S.A. 98, 12701–12705. doi: 10.1073/pnas.221447598
Li, K., Lin, X. D., Wang, W., Shi, M., Guo, W. P., Zhang, X. H., et al. (2015). Isolation and characterization of a novel Arenavirus harbored by Rodents and Shrews in Zhejiang province, China. Virology 476, 37–42. doi: 10.1016/j.virol.2014.11.026
Liao, B. S., Byl, F. M., and Adour, K. K. (1992). Audiometric comparison of Lassa fever hearing loss and idiopathic sudden hearing loss: evidence for viral cause. Otolaryngol. Head Neck Surg. 106, 226–229. doi: 10.1177/019459989210600303
Loo, Y. M., and Gale, M. Jr. (2011). Immune signaling by RIG-I-like receptors. Immunity 34, 680–692. doi: 10.1016/j.immuni.2011.05.003
López, N., Jácamo, R., and Franze-Fernández, M. T. (2001). Transcription and RNA replication of tacaribe virus genome and antigenome analogs require N and L proteins: Z protein is an inhibitor of these processes. J. Virol. 75, 12241–12251. doi: 10.1128/JVI.75.24.12241-12251.2001
Loureiro, M. E., Wilda, M., Levingston Macleod, J. M., D’Antuono, A., Foscaldi, S., Marino Buslje, C., et al. (2011). Molecular determinants of arenavirus Z protein homo-oligomerization and L polymerase binding. J. Virol. 85, 12304–12314. doi: 10.1128/JVI.05691-11
Macal, M., Lewis, G. M., Kunz, S., Flavell, R., Harker, J. A., and Zúñiga, E. I. (2012). Plasmacytoid dendritic cells are productively infected and activated through TLR-7 early after Arenavirus infection. Cell Host Microbe 11, 617–630. doi: 10.1016/j.chom.2012.04.017
Maes, P., Alkhovsky, S. V., Bào, Y., Beer, M., Birkhead, M., Briese, T., et al. (2018). Taxonomy of the family Arenaviridae and the order Bunyavirales: update 2018. Arch. Virol. doi: 10.1007/s00705-018-3843-5 [Epub ahead of print].
Maiztegui, J. I., Fernandez, N. J., and de Damilano, A. J. (1979). Efficacy of immune plasma in treatment of Argentine haemorrhagic fever and association between treatment and a late neurological syndrome. Lancet 2, 1216–1217. doi: 10.1016/S0140-6736(79)92335-3
Manning, J. T., Forrester, N., and Paessler, S. (2015). Lassa virus isolates from Mali and the Ivory Coast represent an emerging fifth lineage. Front. Microbiol. 6:1037. doi: 10.3389/fmicb.2015.01037
Martínez-Sobrido, L., Giannakas, P., Cubitt, B., García-Sastre, A., and de la Torre, J. C. (2007). Differential inhibition of type 1 interferon induction by arenavirus nucleoproteins. J. Virol. 81, 12696–12703. doi: 10.1128/JVI.00882-07
Martinez-Sobrido, L., Zúñiga, E. I., Rosario, D., García-Sastre, A., and de la Torre, J. C. (2008). Inhibition of the type I interferon response by the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J. Virol. 80, 9192–9199. doi: 10.1128/JVI.00555-06
McCormick, J. B., King, I. J., Webb, P. A., Johnson, K. M., O’Sullivan, R., Smith, E. S., et al. (1987). A case-control study of the clinical diagnosis and course of Lassa fever. J. Infect. Dis. 155, 445–455. doi: 10.1093/infdis/155.3.445
McCormick, J. B., King, I. J., Webb, P. A., Scribner, C. L., Craven, R. B., Johnson, K. M., et al. (1986). Lassa fever. Effective therapy with ribavirin. N. Engl. J. Med. 314, 20–26. doi: 10.1056/NEJM198601023140104
Meyer, B. J., de la Torre, J. C, and Southern, P. J. (2002). “Arenaviruses: genomic RNAs, transcription, and replication,” in Arenaviruses I, ed. M. B. Oldstone (Berlin: Springer-Verlag), 139–149. doi: 10.1007/978-3-642-56029-3_6
Monath, T. P., Maher, M., Casals, J., Kissling, R. E., and Cacciapuoti, A. (1974). Lassa fever in the Eastern Province of Sierra Leone, 1970–1972. II. Clinical observations and virological studies on selected hospital cases. Am. J. Trop. Med. Hyg. 23, 1140–1149. doi: 10.4269/ajtmh.1974.23.1140
Monson, M. H., Cole, A. K., Frame, J. D., Serwint, J. R., Alexander, S., and Jahrling, P. B. (1987). Pediatric Lassa fever: a review of 33 Liberian cases. Am. J. Trop. Med. Hyg. 36, 408–415. doi: 10.4269/ajtmh.1987.36.408
Monson, M. H., Frame, J. D., Jahrling, P. B., and Alexander, K. (1984). Endemic Lassa fever in Liberia. I. Clinical and epidemiological aspects at Curran Lutheran Hospital, Zorzor, Liberia. Trans. R. Soc. Trop. Med. Hyg. 78, 549–553. doi: 10.1016/0035-9203(84)90082-8
Morin, B., Coutard, B., Lelke, M., Ferron, F., Kerber, R., Jamal, S., et al. (2010). The N-terminal domain of the arenavirus L protein is an RNA endonuclease essential in mRNA transcription. PLoS Pathog. 6:e1001038. doi: 10.1371/journal.ppat.1001038
Okokhere, P. O., Ibekwe, T. S., and Akpede, G. O. (2009). Sensorineural hearing loss in Lassa fever: two case reports. J. Med. Case Rep. 3:36. doi: 10.1186/1752-1947-3-36
Oppliger, J., Torriani, G., Herrador, A., and Kunz, S. (2016). Lassa virus cell entry via dystroglycan involves an unusual pathway of macropinocytosis. J. Virol. 90, 6412–6429. doi: 10.1128/JVI.00257-16
Pasqual, G., Rojek, J. M., Masin, M., Chatton, J. Y., and Kunz, S. (2011). Old world arenaviruses enter the host cell via the multivesicular body and depend on the endosomal sorting complex required for transport. PLoS Pathog. 7:e1002232. doi: 10.1371/journal.ppat.1002232
Patterson, M., Grant, A., and Paessler, S. (2014). Epidemiology and pathogenesis of bolivian hemorrhagic fever. Curr. Opin. Virol. 5, 82–90. doi: 10.1016/j.coviro.2014.02.007
Perez, M., Craven, R. C., and de la Torre, J. C. (2003). The small ring finger protein z drives arenavirus budding: implications for antiviral strategies. Proc. Natl. Acad. Sci. U.S.A. 100, 12978–12983. doi: 10.1073/pnas.2133782100
Perez, M., Greenwald, D. L., and de la Torre, J. C. (2004). Myristoylation of the RING finger Z protein is essential for arenavirus budding. J. Virol. 78, 11443–11448. doi: 10.1128/JVI.78.20.11443-11448.2004
Pinschewer, D. D., Perez, M., and de la Torre, J. C. (2005). Dual role of the lymphocytic choriomeningitis virus intergenic region in transcription termination and virus propagation. J. Virol. 79, 4519–4526. doi: 10.1128/JVI.79.7.4519-4526.2005
Qi, X., Lan, S., Wang, W., Schelde, L. M., Dong, H., Wallat, G. D., et al. (2010). Cap binding and immune evasion revealed for Lassa nucleoprotein structure. Nature 468, 779–783. doi: 10.1038/nature09605
Raaben, M., Jae, L. T., Herbert, A. S., Kuehne, A. I., Stubbs, S. H., Chou, Y. Y., et al. (2017). NRP2 and CD63 are host factors for Lujo virus cell entry. Cell Host Microbe 22, 688.e5–696.e5. doi: 10.1016/j.chom.2017.10.002
Radoshitzky, S. R., Abraham, J., Spiropoulou, C. F., Kuhn, J. H., Nguyen, D., Li, W., et al. (2007). Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature 446, 92–96. doi: 10.1038/nature05539
Rojek, J. M., Lee, A. M., Nguyen, N., Spiropoulou, C. F., and Kunz, S. (2008a). Site 1 protease is required for proteolytic processing of the glycoproteins of the South American hemorrhagic fever viruses Junin. Machupo, and Guanarito. J. Virol. 82, 6045–6051. doi: 10.1128/JVI.02392-07
Rojek, J. M., Sanchez, A. B., Nguyen, N. T., de la Torre, J. C., and Kunz, S. (2008b). Different mechanisms of cell entry by human-pathogenic Old World and New World arenaviruses. J. Virol. 82, 7677–7687. doi: 10.1128/JVI.00560-08
Ruggiero, H. A., Pérez Isquierdo, F., Milani, H. A., Barri, A., Val, A., and Maglio, F. (1986). Treatment of Argentine hemorrhagic fever with convalescent’s plasma. 4433 cases. Presse Med. 15, 2239–2242.
Salazar-Bravo, J., Ruedas, L. A., and Yates, T. L. (2002). Mammalian reservoirs of arenaviruses. Curr. Top. Microbiol. Immunol. 262, 25–63. doi: 10.1007/978-3-642-56029-3_2
Sayler, K. A., Barbet, A. F., Chamberlain, C., Clapp, W. L., Alleman, R., Loeb, J. C., et al. (2014). Isolation of Tacaribe virus, a Caribbean arenavirus, from host-seeking Amblyomma americanum ticks in Florida. PLoS One 9:e115769. doi: 10.1371/journal.pone.0115769
Shimojima, M., and Kawaoka, Y. (2012). Cell surface molecules involved in infection mediated by lymphocytic choriomeningitis virus glycoprotein. J. Vet. Med. Sci. 74, 1363–1366. doi: 10.1292/jvms.12-0176
Shimojima, M., Ströher, U., Ebihara, H., Feldmann, H., and Kawaoka, Y. (2012). Identification of cell surface molecules involved in dystroglycan-independent Lassa virus cell entry. J. Virol. 86, 2067–2078. doi: 10.1128/JVI.06451-11
Spiropoulou, C. F., Kunz, S., Rollin, P. E., Campbell, K. P., and Oldstone, M. B. (2002). New World arenavirus clade C, but not clade A and B viruses, utilizes α-dystroglycan as its major receptor. J. Virol. 76, 5140–5146. doi: 10.1128/JVI.76.10.5140-5146.2002
Urata, S., and de la Torre, J. C. (2011). Arenavirus budding. Adv. Virol. 2011:180326. doi: 10.1155/2011/180326
Vidya, M. K., Kumar, V. G., Sejian, V., Bagath, M., Krishnan, G., and Bhatta, R. (2018). Toll-like receptors: significance, ligands, signaling pathways, and functions in mammals. Int. Rev. Immunol. 37, 20–36. doi: 10.1080/08830185.2017.1380200
Vieth, S., Torda, A. E., Asper, M., Schmitz, H., and Günther, S. (2004). Sequence analysis of L RNA of Lassa virus. Virology 318, 153–168. doi: 10.1016/j.virol.2003.09.009
Walker, D. H., McCormick, J. B., Johnson, K. M., Webb, P. A., Komba-Kono, G., Elliott, L. H., et al. (1982). Pathologic and virologic study of fatal Lassa Fever in Man. Am. J. Pathol. 107, 349–356.
Walsh, K. B., Teijaro, J. R., Zuniga, E. I., Welch, M. J., Fremgen, D. M., Blackburn, S. D., et al. (2012). Toll-like receptor 7 is required for effective adaptive immune responses that prevent persistent virus infection. Cell Host Microbe 11, 643–653. doi: 10.1016/j.chom.2012.04.016
Whitmer, S. L. M., Strecker, T., Cadar, D., Dienes, H. P., Faber, K., Patel, K., et al. (2018). New lineage of Lassa virus, Togo, 2016. Emerg. Infect. Dis. 24, 599–602. doi: 10.3201/eid2403.171905
WHO (2005). Lassa Fever Fact Sheet N°179. Available at: http://www.who.int/mediacentre/factsheets/fs179/en/
Wilda, M., Lopez, N., Casabona, J. C., and Franze-Fernandez, M. T. (2008). Mapping of the tacaribe arenavirus Z-protein binding sites on the L protein identified both amino acids within the putative polymerase domain and a region at the N terminus of L that are critically involved in binding. J. Virol. 82, 11454–11460. doi: 10.1128/JVI.01533-08
Wolff, H., Lange, J. V., and Webb, P. A. (1978). Interrelationships among Arenaviruses measured by indirect immunofluorescence. Intervirology 9, 344–350. doi: 10.1159/000148956
Keywords: arenavirus, virus replication, lassa virus, junin virus, viral entry, virus immune evasion
Citation: Hallam SJ, Koma T, Maruyama J and Paessler S (2018) Review of Mammarenavirus Biology and Replication. Front. Microbiol. 9:1751. doi: 10.3389/fmicb.2018.01751
Received: 19 March 2018; Accepted: 13 July 2018;
Published: 03 August 2018.
Edited by:
Lisa Sedger, University of Technology Sydney, AustraliaReviewed by:
Susan R. Ross, University of Illinois at Chicago, United StatesIgor S. Lukashevich, University of Louisville, United States
Copyright © 2018 Hallam, Koma, Maruyama and Paessler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Slobodan Paessler, c2xwYWVzc2xAdXRtYi5lZHU=
 Steven J. Hallam
Steven J. Hallam Takaaki Koma
Takaaki Koma Junki Maruyama
Junki Maruyama Slobodan Paessler
Slobodan Paessler