- 1College of Food Science, South China Agricultural University, Guangzhou, China
- 2Guangdong Open Laboratory of Applied Microbiology, Guangdong Provincial Key Laboratory of Microbial Culture Collection and Application, State Key Laboratory of Applied Microbiology Southern China, Guangdong Institute of Microbiology, Guangzhou, China
- 3Institute of Microbiology, College of Life Sciences, Zhejiang University, Hangzhou, China
Heat-shock transcription factors (HSFs) with a HSF domain are regulators of fungal heat-shock protein (HSP) genes and many others vectoring heat-shock elements, to which the domain binds in response to heat shock and other stress cues. The fungal insect pathogen Beauveria bassiana harbors three HSF domain-containing orthologous to Hsf1, Sfl1, and Skn7 in many fungi. Here, we show that the three proteins are interrelated at transcription level, play overlapping or opposite roles in activating different families of 28 HSP genes and mediate differential expression of some genes required for asexual developmental and intracellular Na+ homeostasis. Expression levels of skn7 and sfl1 largely increased in Δhsf1, which is evidently lethal in some other fungi. Hsf1 was distinct from Sfl1 and Skn7 in activating most HSP genes under normal and heat-shocked conditions. Sfl1 and Skn7 played overlapping roles in activating more than half of the HSP genes under heat shock. Each protein also activated a few HSP genes not targeted by two others under certain conditions. Deletion of sfl1 resulted in most severe growth defects on rich medium and several minimal media at optimal 25°C while such growth defects were less severe in Δhsf1 and minor in Δskn7. Conidiation level was lowered by 76% in Δskn7, 62% in Δsfl1, and 39% in Δhsf1. These deletion mutants also showed differential changes in cell wall integrity, antioxidant activity, virulence and cellular tolerance to osmotic salt, heat shock, and UV-B irradiation. These results provide a global insight into vital roles of Hsf1, Sfl1, and Skn7 in B. bassiana adaptation to environment and host.
Introduction
Fungal heat-shock proteins (HSPs) classified to different families by molecular sizes enable to protect cells from stress damages (Hjorth-Sørensen et al., 2001). Their coding genes bear a typical heat-shock element (HSE), i.e., a tandem array of three oppositely oriented DNA consensus motifs (nGAAn), and can be activated by heat-shock transcription factors (HSFs) that bind specifically to the HSE in response to external stress cues (Lindquist, 1986; Lindquist and Craig, 1988; Morimoto et al., 1992; Thompson et al., 2008). In Saccharomyces cerevisiae, Hsf1 is an HSF required for not only expression of basal HSP genes, such as SSA1 and SSA4 in the Hsp70 family, in response to stress cues but also a variety of cellular events (Sorger and Pelham, 1988; Smith and Yaffe, 1991; Gallo et al., 1993; Andrew et al., 1995). In Candida albicans, Hsf1 was shown to regulate expression of hsp104, hsp90, and hsp70 genes under normal and stressful conditions, and its mutants lacking the negative regulatory domain CE2 (residues 535–550) became thermosensitive and less virulent (Nicholls et al., 2009, 2011). Inactivation of Hsf1 in Coniothyrium minitans resulted in reduced conidiation and decreased tolerance to thermal and osmotic stresses (Hamid et al., 2013).
Aside from Hsf1, Sfl1 also bears a DNA-binding domain (i.e., HSF domain) in yeast (Fujita et al., 1989) and has been identified as a key activator of hsp30 genes under basal and stressful conditions (Galeote et al., 2007). The HSF domain is crucial for Sfl1 to bind specifically to HSE with an inverted DNA repeat region (AGAA-n-TTCT) (Conlan and Tzamarias, 2001). Deletion of sfl1 in C. albicans resulted in flocculated clumps in cells, enhanced filamentous growth in several media, and depressed expression of hsp30 and hsp90 genes under a heat shock (Bauer and Wendland, 2007; Li et al., 2007; Zhang et al., 2008). Deletion of sfl1 in Magnaporthe oryzae led to suppressed expression of hsp30 and hsp98 genes, invasive growth, elevated thermosensitivity, and attenuated virulence (Li et al., 2011). Hsf1 and Sfl1 (Hsf2) were proven to be essential for hyphal growth and asexual development in Neurospora crassa (Thompson et al., 2008).
Skn7 is another stress-responsive transcription factor that contains the HSF domain and is highly conserved in fungi (Brown et al., 1993). In yeast, Skn7 and Hsf1 may co-activate hsp12, hsp26, hsp70, hsp82, and hsp104 genes in response to oxidative stress so that deletion of skn7 caused increased sensitivity to oxidative and heat stresses (Morgan et al., 1997; Lee et al., 1999; Raitt et al., 2000). In other fungi, Skn7 is vital for cellular responses to oxidative, osmotic, and cell wall perturbing stresses (Fassler and West, 2011; Hussain et al., 2016) but not necessarily important for pathogenicity. For instance, Skn7 was evidently required for the virulence of Alternaria alternata (Chen et al., 2012) and Metarhizium robertsii (Shang et al., 2015) but not for the virulence of M. oryzae (Motoyama et al., 2008), Batrytis cinerea (Yang et al., 2015), and Fusarium graminearum (Jiang et al., 2015).
Above all, Hsf1 and Sfl1 mainly regulated the fungal growth, conidiation, thermotolence, and virulence, while Skn7 played important roles in the adaptation to various stress conditions, including oxidative, osmotic stresses, cell wall damage agents, and/or virulence and all of them may take part in controlling the expression of several heat shock proteins in previous studied (Table 1). Meanwhile, fungal growth, development, stress tolerance, and virulence regulated by Hsf1, Sfl1, and Skn7 are phenotypes crucial for the pest control potential of filamentous fungal insect pathogens, such as Beauveria bassiana widely applied in biological control programs of arthropod pests (Wang and Feng, 2014; Ortiz-Urquiza et al., 2015). The genomic database of B. bassiana (Xiao et al., 2012) has Hsf1, Sfl1, and Skn7 orthologs and 28 HSPs, which fall into the small HSP, Hsp40, Hsp60, Hsp70, Hsp90, and Hsp100 families with each containing 1–15 members. However, either three HSF or most HSP genes remain functionally unexplored because only two hsp40 genes, i.e., mas5 and mdj1, have been characterized in B. bassiana (Wang et al., 2016, 2017). It is unclear whether Hsf1, Sfl1, and Skn7 activate different families of HSP genes in an independent or cooperative manner. This study seeks to explore transcriptional linkages of Hsf1, Sfl1, and Skn7 with all 28 HSP genes in B. bassiana and to elucidate their functions by multi-phenotypic analyses of deletion/complement mutants. Our results provide a global insight into differential roles for Hsf1, Sfl1, and Skn7 in activating different families of HSP genes and sustaining asexual cycle, virulence, and multiple stress tolerance in B. bassiana.
Materials and Methods
Microbial Strains and Culture Conditions
The wild-type strain B. bassiana ARSEF 2860 (designated WT herein) and its mutants were cultivated in Sabouraud dextrose agar [SDAY (4% glucose, 1% peptone, and 1.5% agar) plus 1% yeast extract] at 25°C in a light/dark cycle of 12:12 h for normal growth and conidiation and in Czapek-Dox agar (CZA; 3% sucrose, 0.3% NaNO3, 0.1% K2HPO4, 0.05% KCl, 0.05% MgSO4, and 0.001% FeSO4 plus 1.5% agar) or 1/4 SDAY (amended with 1/4 of each SDAY nutrient) for their responses to nutritional and chemical stresses at the same regime. Escherichia coli DH5α from Invitrogen (Shanghai, China) for plasmid propagation was grown in Luria-Bertani medium at 37°C.
Bioinformatic Analysis of Three HSF Domain-Containing Proteins in B. bassiana
The Hsf1, Sfl1, and Skn7 sequences of S. cerevisiae were used as queries to search through in the B. bassiana genome database under the NCBI accession NZ_ADAH00000000 (Xiao et al., 2012). The located three orthologs were structurally compared through online blast analysis at https://blast.ncbi.nlm.nih.gov/Blast.cgi. Their sequences were aligned with the counterparts of other representative filamentous fungi for phynogenetic analysis with a neighbor-jointing method in MEGA7 software at http://www.megasoftware.net.
Generation of hsf1, sfl1, and skn7 Mutants
The same strategy for the deletion of mas5 or mdj1 and associated backbone plasmids (Wang et al., 2016, 2017) were used to delete hsf1, sfl1, and skn7 (Gene ID: 19887609, 19887228, and 19884300, respectively) from the wild-type strain B. bassiana ARSEF2860 (designated WT herein). Briefly, the 5′ and 3′ coding/flanking fragments of each gene were amplified from the WT via PCR with paired primers (Supplementary Table S1) under the action of LaTaq DNA polymerase (Promega, Madison, MI, United States) and inserted into p0380-bar. The resultant plasmid p0380-5′x-bar-3′x (x = hsf1, sfl1, or skn7) was transformed into the WT for targeted gene deletion by homogeneous recombination of the 5′ and 3′ fragments separated by bar marker through Agrobacterium-mediated transformation. Subsequently, the full-length coding sequence of each gene with flanking regions was cloned from the WT and ligated into p0380-sur-gateway to exchange for the gateway fragment. The new plasmid p0380-sur-x was ectopically integrated into the corresponding deletion mutant via the same transformation system. Putative deletion or complemented mutant colonies were screened in terms of the bar resistance to phosphinothricin (200 μg/ml) or the sur resistance to chlorimuron ethyl (10 μg/ml) on a selective medium and identified through PCR, quantitative real-time PCR (qRT-PCR) and Southern blotting analyses with paired primers and amplified probes (Supplementary Table S1). Positive deletion mutants were evaluated in parallel with the WT and complemented strains (control strains) in the following experiments of three replicates (cultures or samples from the cultures).
Phenotypic Experiments
Aliquots of 1 μL 106 conidia/mL suspension were spotted centrally onto the plates (9 cm diameter) of rich Sabouraud dextrose agar, minimal CZA, and modified CZA media with different carbon or nitrogen sources and availability. The modified media were prepared by deleting 3% sucrose (carbon starvation) or 0.3% NaNO3 (nitrogen starvation) from the standard CZA, replacing 3% sucrose with 3% of glucose, galactose, glycerol, fructose, trehalose, maltose, or acetate (NaAc) as sole carbon source, and replacing 0.3% NaNO3 with 0.3% of NaNO2 or NH4Cl as sole nitrogen source, respectively. After 8-day incubation at 25°C and 12:12 h, the mean diameter of each colony was estimated as a growth index of each strain on a given medium using two measurements taken perpendicular to each other across the center of the colony. Additionally, SDAY colonies initiated as above were incubated at 34°C and 12:12 h for 8 days, followed by measuring colony diameters as a growth index of each strain at the high temperature.
SDAY cultures for quantification of conidiation capacity were initiated by spreading aliquots of 100 μL 107 conidia/mL suspension. From day 4 onwards during 7-day incubation at the same regime, three culture plugs (5 mm diameter) were taken daily from each plate using a cork borer. Conidia on each plug were released into 1 mL of 0.02% Tween 80 via thorough vibration. The concentration of conidia in the suspension was assessed with a hemocytometer and converted to the number of conidia per cm2 plate culture.
The viability of the conidia from the SDAY cultures was assessed as median germination time (GT50) for each strain to reach 50% germination at the same regime. Conidial thermotolerance and UV-B resistance were quantified as median lethal time (LT50; min) after exposure to a wet heat at 45°C for 0–120 min and median lethal dose (LD50; J/cm2) after exposure to UV-B irradiation (weighted 312 nm) at 0-0.8 J/cm2, as described previously (Wang J. et al., 2013). Conidial virulence was bioassayed on Galleria mellonella larvae by immersing cohorts of ∼35 larvae for 10 s in 30 ml aliquots of a 107 conidia/mL suspension for normal cuticle infection and injecting 5 μL of a 105 conidia/mL suspension was injected into the hemocoel of each larva in each cohort for cuticle-bypassing infection as described previously (Wang et al., 2016). All treated cohorts were maintained in Petri dishes for up to 10 days at 25°C and monitored daily for mortality records, followed by probit analyses of time-mortality trends for the estimates of median lethal time (LT50) for each strain against the model insect.
To assess hyphal sensitivity to cell wall perturbation, hyphal mass disks (5 mm diameter) were taken from the 3-days-old cellophane-overlaid SDAY cultures initiated by spreading 100 μL of a 107 conidia/mL suspension per plate and attached centrally to the plates (9 cm diameter) of 1/4 SDAY (amended with 1/4 of each SDAY nutrient) alone (control) or supplemented with a gradient of Congo red (0.5–3 mg/mL), followed by 6-day incubation at 25°C. An effective concentration (EC50) for Congo red to suppress 50% of colony growth was estimated by modeling analysis of relative growth indices of each strain over the chemical gradient. The relative growth index was computed as a colony diameter ratio of each chemical concentration over the control (Wang J. et al., 2013). Conidial sensitivity of each strain to a sensitive concentration of Congo red (1 mg/mL) was assessed as the ratio of its germination percentage at the concentration over that in the control after 24-h incubation at 25°C.
Cell wall fragility of hyphae was assessed using a method of cell wall degradation (Zeng et al., 2012). Aliquots of ∼100 mg hyphal cells (fresh weight) were collected from the 2-days-old liquid cultures in SDB (agar-free SDAY) containing 106 conidia/mL, washed twice with PBS (pH 7.0) and resuspended in 2 mL aliquots of 0.8 M sucrose containing 10 mg/mL of snailase and lysing enzymes (Sigma). After 6-h incubation at 37°C, the cell suspensions were kept in ice for termination of cell wall lysing. The concentration of protoplasts released from the hyphal cells of each suspension was quantified as an index of the cell wall fragility using a hemocycometer.
Carbohydrate epitopes on the surfaces of conidia were probed with the Alexa fluor 488-labeled lectins concanavalin A [ConA specific to α-glucose and α-N-acetylglucosamine (α-GlcNAc)], Galanthus nivalis (GNL lectin specific to mannose residues), and wheat germ agglutinin (WGA specific to β-GlcNAc and sialic acids) from Molecular Probes-Invitrogen and Vector Laboratories, respectively. Briefly, conidia were fixed in 3% formaldehyde for overnight at 4°C, washed three times in PBS, and resuspended in the buffer, followed by centrifugation. The pretreated conidia were then suspended in each lectin-binding buffer for 1 h labeling in darkness following the user’s guide. Unbound lectin was removed by washing repeatedly with the binding buffer. Fluorescent intensity in every 2 × 104 labeled conidia was quantified on the flow cytometer FC 500 MCL (Beckman Coulter, California, United States) using an argon laser at the excitation/emission wavelengths of 488/530 nm. Each lectin assay included three conidial samples as replicates.
Hyphal sensitivities to osmotic and oxidative agents were assayed by incubating the hyphal mass disks on the plates of 1/4 SDAY alone (control) or supplemented with the gradients of NaCl (0.4–2 M), menadione (2–8 mM), and H2O2 (20–80 mM) and quantified as an EC50 for each chemical to suppress 50% colony growth, as described for Congo red.
Transcriptional Profiling of Phenotype-Related Genes
For each strain, 100 μL aliquots of a 107 conidia/ml suspension were spread onto cellophane-overlaid plates of SDAY alone or 1/4 SDAY supplemented with NaCl (0.8 M), followed by 3-day incubation at 25°C. For a heat shock treatment, the 3-days-old hyphal cultures were exposed to 40°C for 1 h. Total RNA was extracted from each of the cultures under the action of an RNAisoTM Plus Reagent (TaKaRa, Dalian, China) and reversely transcribed into cDNA with PrimeScript® RT reagent kit (TaKaRa), respectively. The experiments of quantitative real-time PCR (qRT-PCR) with paired primers (Supplementary Table S2) were performed under the action of SYBR® Premix Ex TaqTM (TaKaRa) to assess the transcripts of: (1) three HSF domain-containing genes, several conidiation-related genes and 28 HSP genes in three cDNA samples (10-fold dilution, the same below) derived from the normal SDAY cultures; (2) three HSF domain-containing genes and 28 HSP-coding genes in the cDNA samples derived from the heat-shocked SDAY cultures; and (3) five Na+-ATPase genes in the cDNA samples derived from the 1/4 SDAY cultures co-cultivated with NaCl. The transcript of the fungal γ-actin gene in each treatment was used as internal standard. Relative transcript level of each gene was calculated as the ratio of its transcript in the cDNA of each mutant over that in the cDNA of the WT strain using the 2-ΔΔCt method (Livak and Schmittgen, 2001).
Quantification of Antioxidant Enzyme Activities
Aliquots of 0.5 g hyphal mass from the 3-days-old SDAY cultures of each strain grown at 25°C were ground in liquid nitrogen, suspended in 50 mM phosphate buffer (pH 7.4), and centrifuged for 20 min by 16 000 ×g at 4°C. The protein concentration (mg/mL) in the supernatant was determined using a bicinchoninic acid (BCA) Protein Assay Kit (KeyGen, Nanjing, China). Total activity (U/mg) of superoxide dismutases (SODs) in the extract was assayed based on the inhibition of spontaneous autoxidation of pyrogallol. One unit of SOD activity was defined as the SOD amount required to inhibit 50% pyrogallol autoxidation rate. To assay total activity of catalases (CAT), the homogenized hyphal samples were suspended in 20 mM sodium phosphate buffer (pH 7.3), centrifuged as above, and filtered through a 0.22 mm Millex HPF PVDF membrane (GE Healthcare, Shanghai, China), followed by assessing the protein concentration with the BCA kit. Total catalase activity (U/mg) in the supernatant was assayed by reading the OD240 value indicative of H2O2 decomposition. One unit of CAT activity was defined as 1 μM H2O2 consumed per min.
Statistic Analysis
All data from the phenotypic experiments of three replicates were subjected to one-factor (strain) analysis of variance, followed by Tukey’s honestly significant difference (HSD) test for the difference of each phenotype between each deletion mutant and two control strains.
Results
Bioinformatic Features of Three HSF Domain-Containing Orthologs in B. bassiana
Orthologs of Hsf1, Sfl1, and Skn7 (NCBI accession codes: EJP66657, EJP66923, and EJP69323 respectively) located in the genomic database of B. bassiana (Xiao et al., 2012) consist of 724, 539, and 585 amino acids with molecular masses of 78.8, 59.1, and 63.7 kDa, respectively, and share a conserved HSF domain at N-termini (Supplementary Figure S1a). Additionally, Hsf1 has a central Tmemb_cc2 domain typical for transmembrane and coiled-coil-2 proteins (Zhang et al., 2014) and a C-terminal SOG2 domain typical for the proteins of RAM signaling pathway involved in cell separation and cytokinesis (Nelson et al., 2003). Skn7 possesses an RR (response regulator) domain. Sfl1 has a unique mitogen-activated protein kinase (MAPK) docking site (KRGDIAGR at residues 119–126). In phylogeny, the sequences of Hsf1, Sfl1, and Skn7 in B. bassiana share an identity of 26–49% with one another and are much more identical to the counterparts of Cordyceps than those of other filamentous fungi and yeasts (Supplementary Figure S1b).
Interrelationships of Hsf1, Sfl1, and Skn7 and Their Contributions to Heat Tolerance
The expected recombinant events for the deletion and complementation of hsf1, sfl1, or skn7 were confirmed by PCR and Southern blotting analyses (Supplementary Figure S2). The transcript of each deleted gene was undetectable in either normal or heat-shocked cultures of each deletion mutant in qRT-PCR, providing further evidence for the success of each deletion.
Intriguingly, deletion of one HSF domain-containing gene altered drastically transcript levels of two others in the 3-days-old SDAY cultures with respect to the WT standard under normal and heat-shocked conditions, as illustrated in Figure 1A. Deletion of hsf1 resulted in an upregulation of sfl1 and skn7 by 1.1- and 8.5-fold under normal conditions, respectively, despite no impact on expression of either gene in response to a 1-h heat shock at 40°C. Transcript levels of hsf1 and skn7 in the Δsfl1 cultures were reduced respectively by 74 and 93% in response to the heat shock but not affected under normal culture conditions. In Δskn7, expression of hsf1 and sfl1 was depressed respectively by 79 and 75% under normal conditions and 94 and 92% under the heat shock. These data indicated that the three HSF genes were conditionally interrelated at transcriptional level in B. bassiana. Particularly, the heat shock stimulated expression of skn7 and sfl1 in the absence of hsf1 and depressed expression of hsf1 and skn7 in the absence of sfl1. Either hsf1 or sfl1 was highly sensitive to the absence of skn7 irrespective of being exposed to the heat shock or not.
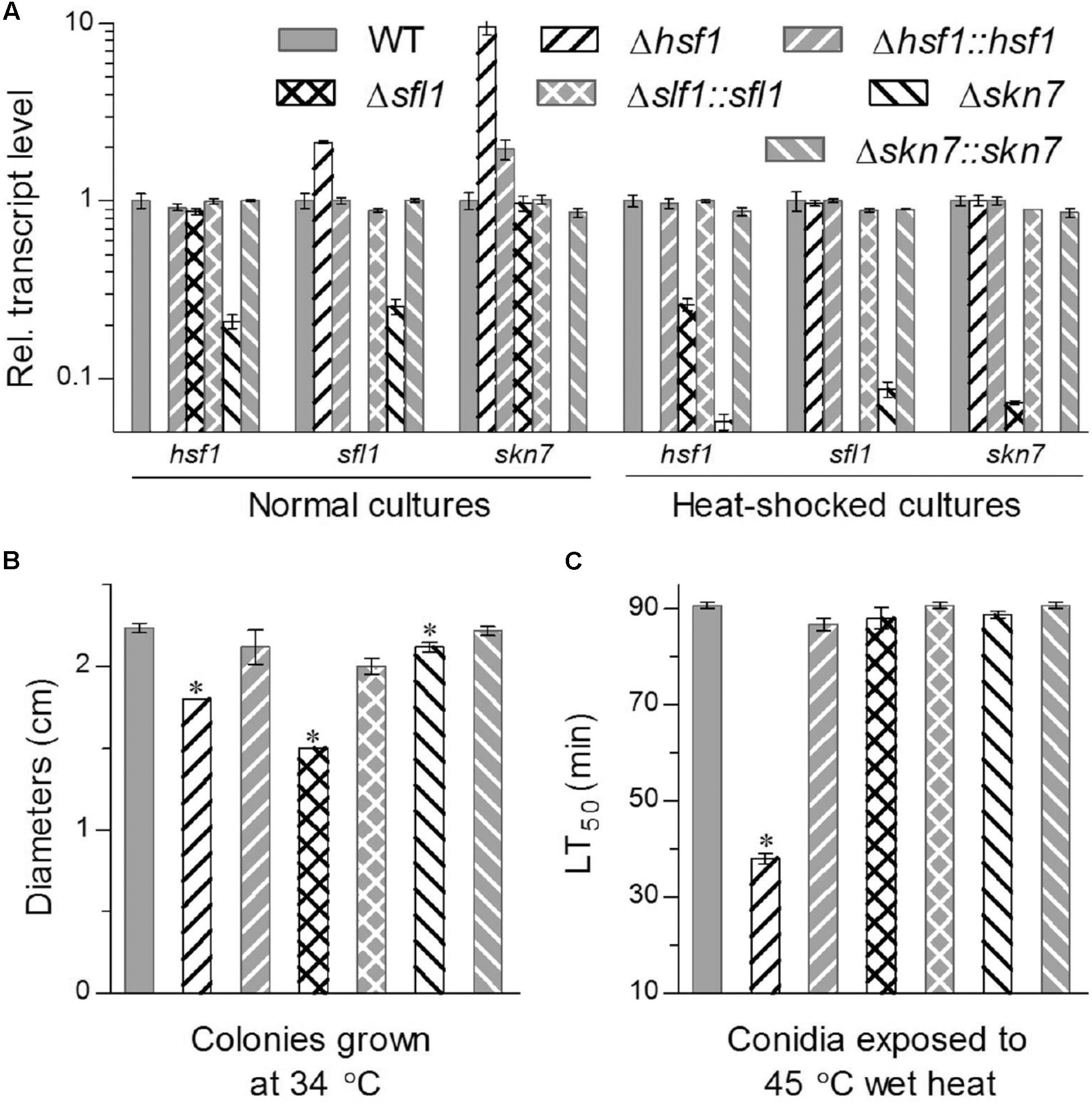
FIGURE 1. Deletion of hsf1, sfl1 or skn7 altered transcriptional profiles of two other genes and response to high temperature in B. bassiana. (A) Relative transcript levels of HSF domain-containing genes in the 3-day-old SDAY cultures of each mutant exposed or not exposed for 1 h to a heat shock at 40°C with respect to the WT standard. (B) Diameters of fungal colonies after 8 days of cultivation in SDAY at 34°C. Each colony was initiated by spotting 1 μL of conidial suspension. (C) LT50 (min) for conidial tolerance to a wet-heat stress at 45°C. Asterisked bars in each bar group differ significantly from those unmarked (Tukey’s HSD, P < 0.05). Error bars: SD from three replicates.
The three deletion mutants were differentially responsive to thermal stress. They grew significantly slower than paired control strains during 8-day incubation at the high temperature of 34°C, resulting in a decrease of mean colony size by 33% in Δsfl1, 19% in Δhsf1, and 5% in Δskn7 (Figure 1B). LT50 for conidial tolerance to a wet-heat stress at 45°C (Figure 1C) was shortened by 58% in Δhsf1 but did not differ significantly between two other mutants and their control strains (Tukey’s HSD, P > 0.05).
Roles of Hsf1, Sfl1, and Skn7 in Activating HSP Genes of Different Families
Transcript levels of all 28 HSP genes in B. bassiana were quantified in the 3-days-old SDAY cultures of each mutant versus the WT strain triggered with or without 1 h of heat shock at 40°C. As shown in Supplementary Table S3, significant changes were marked with red for the HSP genes upregulated (↑) by ≥ twofold or with green for those downregulated (↓) by ≥ 50% in each deletion mutant. The Δhsf1 mutant showed transcript levels of 13 HSP genes increased by 3- to 35-fold and of seven genes decreased by 50–94% under normal conditions while most of the upregulated genes were expressed at approximate WT levels or depressed significantly in response to the heat shock. Those showing remarked transcript changes in Δhsf1 under normal and heat-shocked conditions were hsp20 (25% ↑ vs. 90% ↓), hsp30a (50% ↑ vs. 60% ↓), hsp40b (90% ↓ vs. 20% ↑), hsp40c (5-fold ↑ vs. 70% ↓), hsp70b (89% ↓ vs. 1.6-fold ↑), hsp70g (94% ↓ vs. 1.3-fold ↑), hsp70h (53% ↓ vs. 22% ↑), hsp70k (55% ↓ vs. 48% ↑), and hsp104 (1.7-fold ↑ vs. 58% ↓). The normal and heat-shocked Δsfl1 cultures contained 10 and 23 HSP genes depressed by 50–88% and 50–98%, respectively. Striking transcript changes in the two types of Δsfl1 cultures were present in hsp20 (37% ↑ vs. 89% ↓), hsp40b (32% ↑ vs. 89% ↓), hsp40d–f (WT level vs. 76–86% ↓), hsp70a/c/d/j/k/m–o (0.3- to 1.3-fold ↑ vs. 50–98% ↓), and hsp104 (37% ↓ vs. 2.6-fold ↑). In Δskn7, transcript levels of 18 HSP genes were reduced by 50–89% without the heat shock while 15 depressed (50–95%) HSP genes concurred with two significantly upregulated genes under the heat shock. The differentially expressed HSP genes in Δskn7 included hsp30b (75% ↓ vs. WT level), hsp40a (38% ↓ vs. 2.3-fold ↑), hsp70b (1.6-fold ↑ vs. 94% ↓), hsp70g/h/k (1.3- to 1.8-fold ↑ vs. 50–57% ↓), hsp90 (50% ↓ vs. WT level), and hsp78 (80% ↓ vs. 2.7-fold ↑). All of these transcript changes were restored to the WT levels by targeted gene complementation irrespective of being exposed or not exposed to the heat shock. These data implicated that, in B. bassiana, Hsf1 was distinct from Sfl1 and Skn7 in activating most HSP genes under normal and heat-shocked conditions and that Sfl1 and Skn7 played overlapping roles in activating many HSP genes under heat shock although each activated preferentially a few HSP genes not targeted by two other HSFs.
Roles of Hsf1, Sfl1, and Skn7 in Vegetative Growth and Asexual Development
Compared to WT, the mutants Δhsf1, Δsfl1, and Δskn7 displayed differential defects in vegetative growth, conidiation capacity and conidial germination. Most severe growth defects on rich SDAY, minimal CZA, and 11 CZA media modified with different carbon/nitrogen sources and availability were observed in Δsfl1, whose colony sizes diminished by 21–51% after 8-day incubation on the media at the optimum 25°C (Figure 2A). Significant, but less severe, growth defects with no more than 18% decrease in colony size occurred in Δhsf1 grown on most media excluding NH4+ as sole nitrogen source and maltose as sole carbon source. The sizes of Δskn7 colonies diminished by no more than 12% on the nitrogen sources of NO2- and NH4+ and the carbon sources of glycerol, glucose, fructose, galactose and maltose but were not significantly affected on other tested media (Tukey’s HSD, P > 0.05).
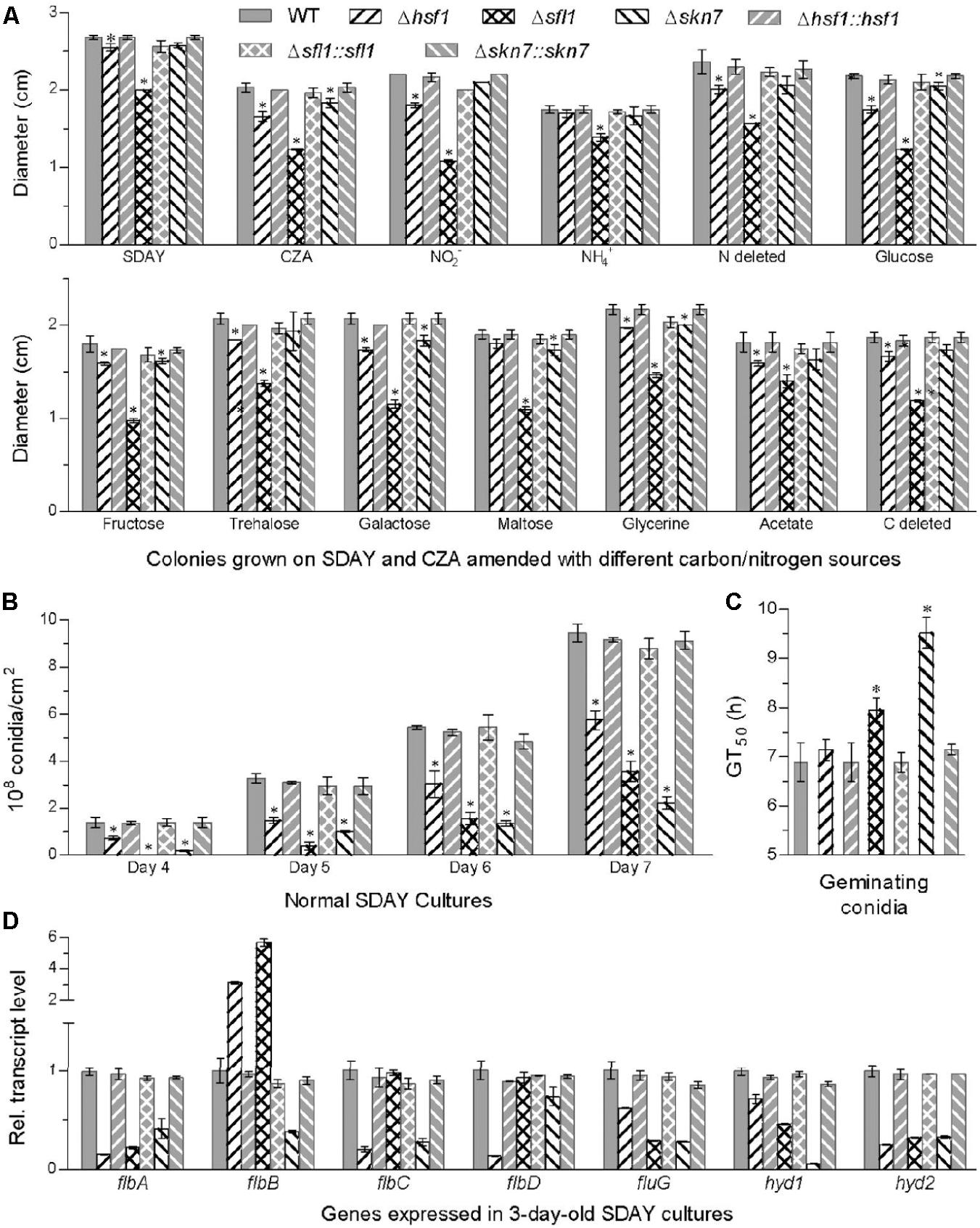
FIGURE 2. Hsf1, Sfl1, and Skn7 play differential roles for in the asexual cycle of B. bassiana. (A) Diameters of fungal colonies after 8 days of cultivation at 25°C on rich SDAY, minimal CZA, and modified CZA media with different carbon/nitrogen sources and rich SDAY. Each colony was initiated with the spotting method. (B) Conidial yields quantified over the days of normal cultivation on SDAY plates spread with 100 μL aliquots of conidial suspension for culture initiation. (C) Median germination time (GT50) as an index of conidial germination rate. (D) Relative transcript levels of conidiation-required genes in the 3-day SDAY cultures of each mutant with respect to the WT standard. Asterisked bars in each bar group differ significantly from those unmarked (Tukey’s HSD, P < 0.05). Error bars: SD from three replicates.
Conidial yields quantified from the 7-days-old SDAY cultures were averagely reduced by 76% in Δskn7, followed by 62% in Δsfl1 and 39% in Δhsf1, as compared with the WT yield (Figure 2B). Meanwhile, all the deletion mutants showed much fewer conidiophores and conidia than the control strains in microscopic examination of samples taken from colonies on day 3 (Supplementary Figure S3). Additionally, Δskn7 and Δsfl1 required longer time to achieve 50% of conidial germination at 25°C than the WT strain, but this difference disappeared in Δhsf1, although germination percentages within 16 h were not affected significantly in all the mutants (Figure 2C and Supplementary Figure S3).
Intriguingly, transcript levels of several conidiation-required genes decreased drastically in the deletion mutants, including those encoding five transcription factors (FlbA–D and FluG) essential for activation of fungal development pathway (Etxebeste et al., 2010; Park and Yu, 2012) and two hydrophobins (Hyd1/2) crucial for conidial structure and hydrophobicity in B. bassiana (Zhang et al., 2011). As shown in Figure 2D, most of these gene transcripts decreased by 75–87% in Δhsf1 (flbA/C/D and hyd2), 54–77% in Δsfl1 (flbA, fluG and hyd1/2), and 58–94% in Δskn7 (all except flbD). The depressed expression of these genes in the deletion mutants could be causative of their conidiation defects, suggesting important roles for Hsf1, Sfl1, and Skn7 in the asexual development of B. bassiana.
Roles of Hsf1, Sfl1, and Skn7 in Cell Wall Integrity
Cell wall integrity was differentially impaired by the deletions of hsf1, sfl1, and skn7 in B. bassiana. First, an EC50 for the cell wall stressor Congo red to suppress 50% hyphal growth was lowered by 78% in Δhsf1, 69% in Δskn7, and 43% in Δsfl1 as compared with an estimate of 1.86 mg/mL from the WT (Figure 3A). Adding Congo red (1 mg/mL) to a medium suppressed conidial germination of the deletion mutants 10–42% more than that observed in the WT (Figure 3B). Second, more protoplasts were released from the hyphal cells of each deletion mutant than of the WT after 6 h cell lysing with enzymes (Figure 3C). The released protoplasts increased by 3.1-fold in Δhsf1, followed by 1.8-fold in Δskn7 and 65% in Δsfl1. These data indicated that hyphal cell walls became much more fragile in the absence of hsf1 or skn7 and of sfl1. Third, the deletion of each gene altered cell wall composition of conidia, as indicated by the data from the fluorescent lectin-binding assays (Figure 3D). The content of α-GlcNAc increased by 79% on the surfaces of ConA-labeled Δsfl1 conidia. The content of GNL-labeled mannose residues increased by 25, 9, and 6% on the surfaces of Δhsf1, Δsfl1, and Δskn7 conidia, respectively. The content of WGA-labeled β-GlcNAc increased by 59 and 32% on the surfaces of Δhsf1 and Δskn7 conidia, respectively. Finally, examination by transmission electron microscopy (TEM) revealed that an outermost layer of conidial wall in the deletion mutants was less distinctly outlined compared with the WT counterpart, making the mutant conidia readily deformed in the pretreatment for TEM (Supplementary Figure S4).
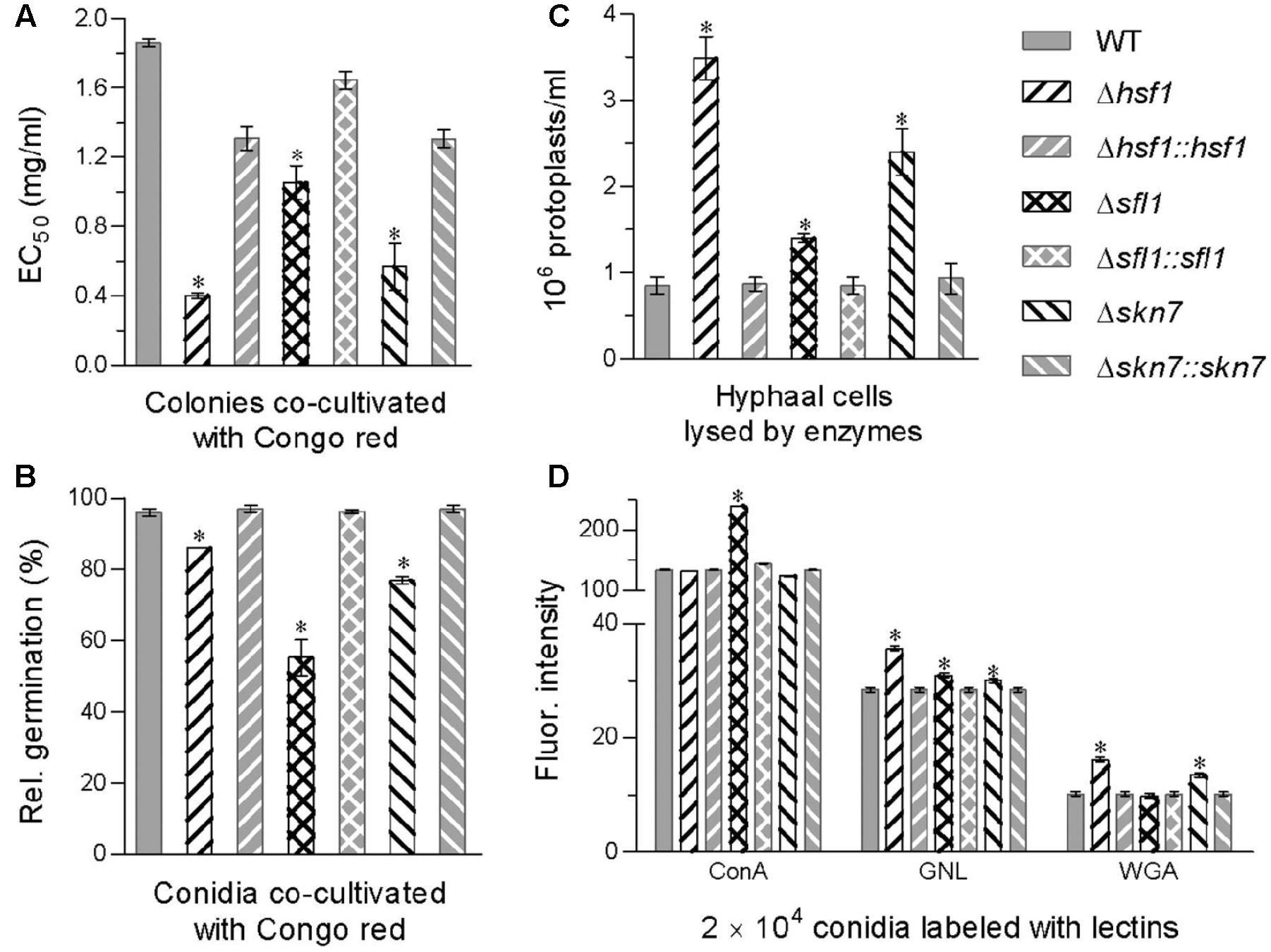
FIGURE 3. Contributions of Hsf1, Sfl1, and Skn7 to cell wall integrity of B. bassiana. (A) EC50 for Congo red to suppress 50% colony growth of each strain after 6-day incubation at 25°C on 1/4 SDAY plates, to which small hyphal disks were attached for colony initiation. (B) Relative germination percentages of conidia after 24-h incubation at 25°C in a medium alone (control) or supplemented with Congo red (1 mg/mL). (C) Concentrations of protoplasts released from the hyphal cells after 6 h treatment with cell wall lysing enzymes in osmotic solution of 0.8 M sucrose. (D) Fluorescence intensity as an index of cell wall composition from flow cytometry of 2 × 104 conidia labeled with the fluorescent lectins ConA, GNL, and WGA. Asterisked bars in each bar group differ significantly from those unmarked (Tukey’s HSD, P < 0.05). Error bar: SD from three replicates.
The increased cell wall sensitivity to Congo red, the increased cell wall fragility, the altered cell wall composition, and the less distinctly defined cell walls were largely or well restored by targeted gene complementation, indicating vital roles for Hsf1, Sfl1, and Skn7 in sustaining the fungal cell wall integrity.
Roles of Hsf1, Sfl1, and Skn7 in Stress Tolerance and Virulence
Compared to the WT strain, all deletion mutants became significantly more sensitive to two oxidants during colony growth (Tukey’s HSD, P < 0.05). The EC50 estimates for H2O2 and menadione to suppress 50% hyphal growth were lowered by 39 and 26% in Δhsf1, 35 and 20% in Δsfl1, and 24 and 13% in Δskn7, respectively (Figure 4A). Their reduced antioxidant capabilities were consistent with reduced activities of SODs and CATs, which were quantified with the protein extracts from 3-days-old SDAY cultures. The SOD and CAT activity were reduced by 35 and 61% in Δhsf1, 54 and 77% in Δsfl1, and 48 and 74% in Δskn7, respectively (Figure 4B). Interestingly, the EC50 values of menadione and H2O2 against all tested strains were linearly correlated with the total activities of their SODs (r2 = 0.71, F1,5 = 12.4, P = 0.017) and catalases (r2 = 0.72, F1,5 = 13.0, P = 0.016), respectively.
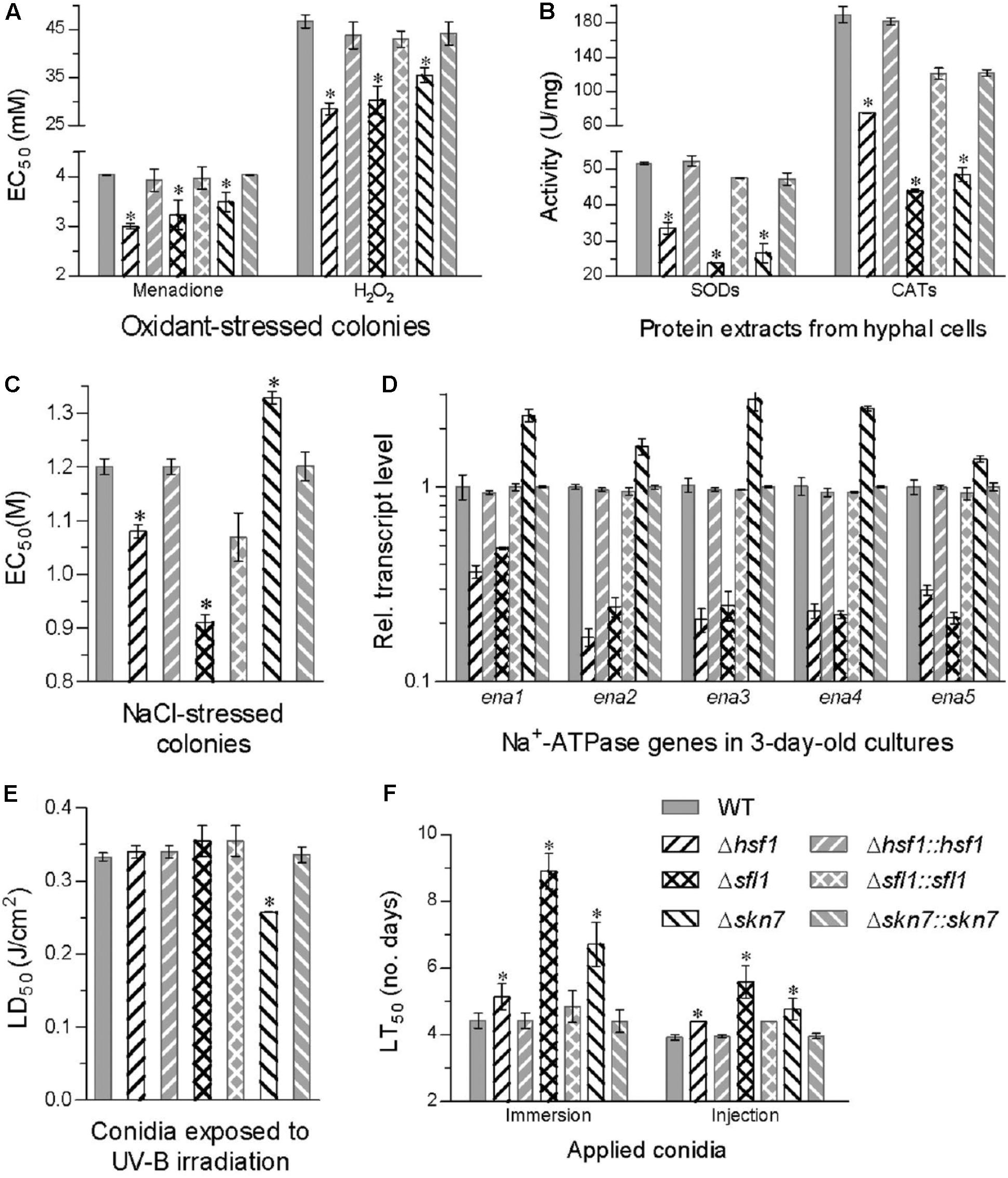
FIGURE 4. Deletion of each HSF domain-containing gene altered cellular sensitivity to oxidative, osmotic, and stresses, UV-B radiation and virulence. (A,B) EC50 estimates for menadione and H2O2 to suppress 50% colony growth after 6-day incubation at 25°C on 1/4 SDAY plates and total activities of SODs and CATs quantified in the protein extracts from 3-day-old SDAY cultures, respectively. (C,D) EC50 estimates for NaCl to suppress 50% colony growth on 1/4 SDAY and relative transcript levels of five Na+ ATPase genes in the 3-day-old 1/4 SDAY cultures of each mutant versus the WT strain co-cultivated with 0.8 M NaCl, respectively. (E) LD50 (J/cm2) estimates for conidial resistance to UV-B irradiation. (F) LT50 (D) for the virulence of each strain against G. mellonella larvae infected by topical application (immersion) or intrahemocoel injection of a standardized conidial suspension. Asterisked bars in each bar group differ significantly from those unmarked (Tukey’s HSD, P < 0.05). Error bar: SD from three replicates.
Hyphal sensitivity to NaCl increased by 24% in Δsfl1 and 10% in Δhsf1 but decreased by 11% in Δskn7 as compared with a mean EC50 of 1.2 M from the WT response to the salt (Figure 4C). The salt tolerance was closely linked to transcriptional changes of five Na+ ATPase genes (ena1–5) in the 3-days-old cultures co-cultivated with NaCl (0.8 M). Transcript levels of all five genes decreased by 64–83% in Δhsf1 and 52–79% in Δsfl1 but increased by 0.4- to 1.8-fold in Δskn7 with respect to the WT standard (Figure 4D). Additionally, only Δskn7 showed a remarked reduction (23%) in conidial tolerance to UV-B irradiation, as indicated by LD50 estimates from all tested strains (Figure 4E).
In standardized bioassays, a mean LT50 for the WT strain to kill 50% of G. mellonella larvae was 4.4 ± 0.2 days through topical application of 107 conidia/mL suspension for normal cuticle infection and 3.9 ± 0.1 days through intrahemocoel injection for cuticle-bypassing infection (Figure 4F). Compared to the two means, the lethal action was significantly delayed in three deletion mutants irrespective of the infection through cuticular penetration or bypassing the insect cuticle (Tukey’s HSD, P < 0.05). The lethal action through normal infection was most delayed by 4.1 days in Δsfl1, followed by a delay of 2.3 days in Δskn7 and 0.7 days in Δhsf1. The delay of the lethal action through the cuticle-passing infection decreased to only 0.5–1.6 days in the three deletion mutants.
All of these changes were restored by targeted gene complementation, indicating that Hsf1, Sfl1, and Skn7 play differential, but important, roles in sustaining antioxidant activity, salt tolerance, and virulence of B. bassiana.
Discussion
Our study unveils that three HSF domain-containing genes are transcriptionally interrelated in B. bassiana. The drastic depressions of hsf1 and sfl1 in Δskn7 and of hsf1 and skn7 in Δsfl1 indicate that skn7 or sfl1 regulates expression of two other HSF genes positively under normal conditions and/or heat shock. In contrast, hsf1 acts as a negative mediator of sfl1 and skn7 under normal conditions since its absence resulted in remarked upregulation of the two genes. The compensation of increased skn7 and sfl1 expression levels for the absence of hsf1 provides an interpretation on the viability of Δhsf1 in B. bassiana, which might be inferred according to the observations in S. cerevisiae where deletion of skn7 exacerbated the growth defect of the hsf1 temperature-sensitive allele (hsf1ts) strain and high-copy expression of skn7 rescued the growth defect of the hsf1ts strain at 35°C (Raitt et al., 2000). This is very different from an indispensability of hsf1 for S. cerevisiae and N. crassa, in which deletion of hsf1 caused lethality (Wiederrecht et al., 1988; Thompson et al., 2008). Except for heat shock, Skn7 and Hsf1 might in some way cooperate in the response to free radical stress, which demonstrated again that there is clearly some overlap between the functions of skn7 and hsf1 in S. cerevisiae (Raitt et al., 2000).
Meanwhile, Hsf1, Sfl1, and Skn7 played distinctive or overlapping roles not only in activating HSP genes in response to heat shock but also in regulating expression of some genes crucial for asexual development and intracellular Na+ homeostasis and perhaps many more associated with multi-phenotypic alterations but not examined in this study. Function loss of each resulted in increased sensitivity to 34°C during colony growth, largely in agreement with increased thermosensitivity in the absence of other fungal hsf1 (Nicholls et al., 2011; Hamid et al., 2013), sfl1 (Li et al., 2011), or skn7 (Shang et al., 2015; Hussain et al., 2016). In C. albicans, Hsf1 can activate hsp104, hsp90, and hsp70 under normal and stressful conditions (Nicholls et al., 2009, 2011). Sfl1 also mediates expression of hsp30, hsp90, and hsp98 in S. cerevisiae, C. albicans, and M. oryzae under normal and/or stressful conditions (Galeote et al., 2007; Li et al., 2007, 2011; Zhang et al., 2008). The yeast Skn7 and Hsf1 can co-mediate expression of hsp12, hsp26, several hsp70, hsp82, and hsp104 under oxidative stress (Lee et al., 1999; Raitt et al., 2000). In this study, each HSF domain-containing gene could specifically activate some HSP genes under normal or heat-shocked conditions. We found overlapping roles of Sfl1 and Skn7 in activating hsp30b, hsp70f/e, hsp90, and hsp78 and of Sfl1 and Hsf1 in activating hsp70i/l but opposite roles of Hsf1 and Skn7 in activating hsp20, four hsp40, hsp60, and five hsp70 genes in B. bassiana under normal conditions. In response to heat shock, Sfl1 cooperated with Skn7 for positive mediation of hsp20, hsp40d–f and nine hsp70 genes or with Hsf1 for positive mediation of hsp30b, hsp40f, hsp70a, and hsp90 and opposite mediation of hsp104; Skn7 cooperated with Hsf1 for positive mediation of hsp40c/f. Regardless of more or less involvement in heat tolerance and regulating HSP genes alone or cooperatively, three HSF domain-containing genes were involved in physiological and cellular processes relevant to the fungal adaptation to environment and host, as discussed below.
First, three HSF domain-containing genes play important, but differential roles in sustaining the asexual cycle of B. bassiana. The growth defects of their deletion mutants on different media indicate that Sfl1 is more important than Hsf1 for the fungal pathogen to make use of carbon/nitrogen sources for vegetative growth. The importance of Sfl1 for the fungal growth is different from what have been learned from some other fungi, in which Δsfl1 mutants could grow as well in rich medium as wild-type strains (Li et al., 2007, 2011; Thompson et al., 2008). The limited role of skn7 in the fungal growth is similar to minor growth defect due to skn7 deletion in F. graminearum (Jiang et al., 2015) but somewhat different from no obvious role of the same gene in other fungal growth (Viefhues et al., 2015; Hussain et al., 2016; Zhang et al., 2016). The role of Hsf1 is dispensable for B. bassiana, unlike its indispensability for S. cerevisiae and N. crassa (Wiederrecht et al., 1988; Thompson et al., 2008). Aside from differential roles in nutritional exploitation and vegetative growth, three HSF domain-containing genes are all required for full conidiation in B. bassiana despite different contributions. Particularly, Skn7 is most important for not only conidiation but also conidial viability, followed by Sfl1 and Hsf1. Their roles in conidiation are consistent with those of Hsf1 in Coniothyrium minitans (Hamid et al., 2013), Sfl1 in M. oryzae (Li et al., 2011), and N. crassa (Thompson et al., 2008) and Skn7 in other filamentous fungi (Jiang et al., 2015; Yang et al., 2015; Hussain et al., 2016; Zhang et al., 2016). Our transcriptional analyses indicate prominent links of three HSF domain-containing genes to most transcription factors, which are required for the activation of central developmental pathway to govern asexual development (Etxebeste et al., 2010; Park and Yu, 2012), and hydrophobins that ensure conidial surface structure and hydrophobicity (Zhang et al., 2011). We speculate that the regulatory roles of three HSF domain-containing genes in the fungal asexual cycle are likely associated with expression levels of those phenotype-related genes.
Moreover, various cellular events rely upon cell wall integrity in fungi, including glycosylation of proteins, biosynthesis of glycosyl- phoshatidylinositol (GPI) anchors, quality control of secretory proteins, and delivery of cell wall components for assembly (Lesage and Bussey, 2006; Scrimale et al., 2009). In this study, Hsf1, Sfl1, and Skn7 were vital for the cell wall composition and integrity of B. bassiana due to altered cell wall components (α-GlcNAc, β-GlcNAc, and mannose residues), increased cell wall fragility, and higher sensitivity to cell wall perturbation in the absence of each. Our results are well in accordance with increased sensitivities of Δskn7 mutants to cell wall stressors, such as Congo red and/or Calcofluor white, in other filamentous fungi (Shang et al., 2015; Viefhues et al., 2015; Zhang et al., 2016) but are different from null response of Δskn7 mutants to cell wall stress in two yeasts (Singh et al., 2004; Fassler and West, 2011).
Furthermore, three HSF domain-containing genes also function in sustaining antioxidant activity of B. bassiana. This is indicated by reduced activities of intracellular SODs and CATs and increased cellular sensitivities to two typical oxidants in Δhsf1, Δsfl1, and Δskn7. Total activities of SODs and CATs are crucial for B. bassiana responses to menadione and H2O2, respectively (Xie et al., 2012; Wang Z.L. et al., 2013; Li et al., 2015) and were linearly correlated to antioxidant responses of all tested strains in this study. This implicates that maintenance of antioxidant activity by the three HSF domain-containing genes relies upon their involvements in mediating expression of antioxidant enzymes. Previously, increased sensitivity to oxidative stress was also caused by deletion of skn7 in yeast (Lee et al., 1999) or filamentous fungi (Lamarre et al., 2007; Viefhues et al., 2015; Yang et al., 2015; Hussain et al., 2016; Zhang et al., 2016) but not in M. robertsii, another fungal insect pathogen (Shang et al., 2015). In addition, transcriptional changes of five Na+ ATPase genes coincide well with reduced or increased tolerance of each deletion to NaCl in this study. Particularly, the increased tolerance of Δskn7 to NaCl is very different from null response or increased sensitivity of Δskn7 to osmotic salts in other fungi (Chen et al., 2012; Shang et al., 2015; Yang et al., 2015). Conidial thermotolerance and UV-B resistance crucial for B. bassiana survival in the field were also lowered in Δhsf1 and Δskn7, respectively. Altogether, Hsf1, Sfl1, and Skn7 play differential roles in the fungal adaptation to oxidative, ion osmotic, thermal, and UV-B irradiative stresses.
Finally, Sfl1 contributes much more to the virulence of B. bassiana than Skn7 and Hsf1 based on delayed lethal action in the absence of each. The attenuated virulence of their deletion mutants is similar to what was caused by the deletion of hsf1 (Nicholls et al., 2011; Hamid et al., 2013), sfl1 (Li et al., 2011), or skn7 (Singh et al., 2004; Wormley et al., 2005; Chen et al., 2012; Shang et al., 2015) in other fungal pathogens. However, these observations are different from little change in the Δskn7 virulence of Aspergillus fumigatus (Lamarre et al., 2007), B. cinerea (Yang et al., 2015), F. graminearum (Jiang et al., 2015), and M. oryzae (Motoyama et al., 2008). For the deletion mutants in this study, the degrees of attenuated virulence to the tested model insect via normal cuticle infection are largely attributable to partial losses of their ability to grow on scant insect integument for hyphal penetration through the host cuticle, as shown with their growth defects on minimal media, and reduced antioxidant activity. The latter phenotype is important for fungal ability to resist an oxidative stress generated from the host immunity defense and linearly correlated with virulence in B. bassiana (Xie et al., 2012, 2013).
Conclusion
There exists three HSF domain-containing genes transcriptionally interrelated in B. bassiana, which are respectively homologous to hsf1, sfl1, and skn7 in yeast and other fungi. Here, our results indicate that three HSF-containing genes play vital but differential roles in sustaining the asexual cycle, virulence, multiple stress tolerance, and activating different families of HSP genes in B. bassiana, which reveals possible means to improving field persistence and efficacy of a fungal formulation by manipulating the HSF domain-containing genes of a candidate strain. Additionally, further investigations will be needed to have a deeper insight into the interrelationships of three HSF domain-containing genes which might be not limited in the transcriptional level.
Ethics Statement
This study does not involved any experiment with human participants or animal performed.
Author Contributions
GZ and JW designed and performed the experiments, analyzed the data, and prepared the manuscript. S-HY and XF contributed to the manuscript revision. YH performed the experiments. M-GF contributed to the manuscript revision and overall support of this study.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos: 31600060, 31572054, 31500036, 31770091, and 31671855).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01677/full#supplementary-material
References
Andrew, K. S., Fumihiko, Y., Wei, Y., Terumi, M., Tersuo, M., and Dennis, R. W. (1995). Mutated yeast heat shock transcription factor exhibits elevated basal transcriptional activation and confers metal resistance. J. Biol. Chem. 270, 25079–25086. doi: 10.1074/jbc.270.42.25079
Bauer, J., and Wendland, J. (2007). Candida albicans Sfl1 suppresses flocculation and filamentation. Eukaryot. Cell 6, 1736–1744. doi: 10.1128/EC.00236-07
Brown, J., North, S., and Bussey, H. (1993). SKN7, a yeast multicopy suppressor of a mutation affecting cell wall beta-glucan assembly, encodes a product with domains homologous to prokaryotic two-component regulators and to heat shock transcription factors. J. Bacteriol. 175, 6908–6915. doi: 10.1128/jb.175.21.6908-6915.1993
Chen, L. H., Lin, C. H., and Chung, K. R. (2012). Roles for SKN7 response regulator in stress resistance, conidiation and virulence in the citrus pathogen Alternaria alternata. Fungal Genet. Biol. 49, 802–813. doi: 10.1016/j.fgb.2012.07.006
Conlan, R. S., and Tzamarias, D. (2001). Sfl1 functions via the co-repressor Ssn6-Tup1 and the cAMP-dependent protein kinase Tpk2. J. Mol. Biol. 309, 1007–1015. doi: 10.1006/jmbi.2001.4742
Etxebeste, O., Garzia, A., Espeso, E. A., and Ugalde, U. (2010). Aspergillus nidulans asexual development: making the most of cellular modules. Trends Microbiol. 18, 569–576. doi: 10.1016/j.tim.2010.09.007
Fassler, J. S., and West, A. H. (2011). Fungal Skn7 stress responses and their relationship to virulence. Eukaryot. Cell 10, 156–167. doi: 10.1128/EC.00245-10
Fujita, A., Kikuchi, Y., Kuhara, S., Misumi, Y., Matsumoto, S., and Kobayashi, H. (1989). Domains of the SFL1 protein of yeasts are homologous to Myc oncoproteins or yeast heat-shock transcription factor. Gene 85, 321–328. doi: 10.1016/0378-1119(89)90424-1
Galeote, V. A., Alexandre, H., Bach, B., Delobel, P., Dequin, S., and Blondin, B. (2007). Sfl1p acts as an activator of the HSP30 gene in Saccharomyces cerevisiae. Curr. Genet. 52, 55–63. doi: 10.1007/s00294-007-0136-z
Gallo, G. J., Prentice, H., and Kingston, R. E. (1993). Heat shock factor is required for growth at normal temperatures in the fission yeast Schizosaccharomyces pombe. Mol. Cell. Biol. 13, 749–761. doi: 10.1128/MCB.13.2.749
Hamid, M. I., Zeng, F. Y., Cheng, J. S., Jiang, D. H., and Fu, Y. P. (2013). Disruption of heat shock factor 1 reduces the formation of conidia and thermotolerance in the mycoparasitic fungus Coniothyrium minitans. Fungal Genet. Biol. 53, 42–49. doi: 10.1016/j.fgb.2012.12.002
Hjorth-Sørensen, B., Hoffmann, E. R., Lissin, N. M., Sewell, A. K., and Jakobsen, B. K. (2001). Activation of heat shock transcription factor in yeast is not influenced by the levels of expression of heat shock proteins. Mol. Microbiol. 39, 914–923. doi: 10.1046/j.1365-2958.2001.02279.x
Hussain, M., Hamid, M. I., Wang, N. N., Bin, L., Xiang, M. C., and Liu, X. Z. (2016). The transcription factor SKN7 regulates conidiation, thermotolerance, apoptotic-like cell death and parasitism in the nematode endoparasitic fungus Hirsutella minnesotensis. Sci. Rep. 6:30047. doi: 10.1038/srep30047
Jiang, C., Zhang, S. J., Zhang, Q., Tao, Y., Wang, C. F., and Xu, J. R. (2015). FgSkn7 and FgATF1 have overlapping functions in ascosporogenesis, pathogenesis and stress responses in Fusarium graminearum. Environ. Microbiol. 17, 1245–1260. doi: 10.1111/1462-2920.12561
Lamarre, C., Ibrahim-Granet, O., Du, C., Calderone, R., and Latge, J. P. (2007). Characterization of the SKN7 ortholog of Aspergillus fumigatus. Fungal Genet. Biol. 44, 682–690. doi: 10.1016/j.fgb.2007.01.009
Lee, J., Godon, C., Lagniel, G., Spector, D., Garin, J., Labarre, J., et al. (1999). Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 274, 16040–16048. doi: 10.1074/jbc.274.23.16040
Lesage, G., and Bussey, H. (2006). Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70, 317–343. doi: 10.1128/MMBR.00038-05
Li, F., Shi, H. Q., Ying, S. H., and Feng, M. G. (2015). Distinct contributions of one Fe- and two Cu/Zn-cofactored superoxide dismutases to antioxidation, UV tolerance and virulence of Beauveria bassiana. Fungal Genet. Biol. 81, 160–171. doi: 10.1016/j.fgb.2014.09.006
Li, G. T., Zhou, X. Y., Kong, L. G., Wang, Y. L., Zhang, H., Zhu, H., et al. (2011). MoSfl1 is important for virulence and heat tolerance in Magnaporthe oryzae. PLoS One 6:e19951. doi: 10.1371/journal.pone.0019951
Li, Y., Su, C., Mao, X., Cao, F., and Chen, J. (2007). Roles of Candida albicans Sfl1 in hyphal development. Eukaryot. Cell 6, 2112–2121. doi: 10.1128/EC.00199-07
Lindquist, S. (1986). The heat shock response. Annu. Rev. Biochem. 55, 1151–1191. doi: 10.1146/annurev.bi.55.070186.005443
Lindquist, S., and Craig, E. A. (1988). The heat-shock proteins. Annu. Rev. Genet. 22, 631–677. doi: 10.1146/annurev.ge.22.120188.003215
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Morgan, B. A., Banks, G. R., Toone, W. M., Raitt, D., Kuge, S., and Johnston, L. H. (1997). The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 16, 1035–1044. doi: 10.1093/emboj/16.5.1035
Morimoto, R. I., Sarge, K. D., and Abravaya, K. (1992). Transcriptional regulation of heat shock genes, A paradigm for inducible genomic responses. J. Biol. Chem. 267, 21987–21990.
Motoyama, T., Ochiai, N., Morita, M., Iida, Y., Usami, R., and Kudo, T. (2008). Involvement of putative response regulator genes of the rive blast fungus Magnaporthe oryzae in osmotic stress response, fungicide action, and pathogenicity. Curr. Genet. 54, 185–195. doi: 10.1007/s00294-008-0211-0
Nelson, B., Kurischko, C., Horecka, J., Mody, M., Nair, P., Pratt, L., et al. (2003). RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol. Biol. Cell 14, 3782–3803. doi: 10.1091/mbc.e03-01-0018
Nicholls, S., MacCallum, D. M., Kaffarnik, F. A. R., Selway, L., Peck, S. C., and Brown, A. J. P. (2011). Activation of the heat shock transcription factor Hsf1 is essential for the full virulence of the fungal pathogen Candida albicans. Fungal Genet. Biol. 48, 297–305. doi: 10.1016/j.fgb.2010.08.010
Nicholls, S., Michelle, D. L., Claire, L. P., and Alistair, J. P. B. (2009). Role of the heat shock transcription factor, Hsf1, in a major fungal pathogen that is obligately associated with warm-blooded animals. Mol. Microbiol. 74, 844–861. doi: 10.1111/j.1365-2958.2009.06883.x
Ortiz-Urquiza, A., Luo, Z. B., and Keyhani, N. O. (2015). Improving mycoinsecticides for insect biological control. Appl. Microbiol. Biotechnol. 99, 1057–1068. doi: 10.1007/s00253-014-6270-x
Park, H. S., and Yu, J. H. (2012). Genetic control of asexual sporulation in filamentous fungi. Curr. Opin. Microbiol. 15, 669–677. doi: 10.1016/j.mib.2012.09.006
Raitt, D. C., Johnson, A. L., Erkine, A. M., Makino, K., Morgan, B., Gross, D. S., et al. (2000). The Skn7 response regulator of Saccharomyces cerevisiae interacts with Hsf1 in vivo and is required for the induction of heat shock genes by oxidative stress. Mol. Biol. Cell 11, 2335–2347. doi: 10.1091/mbc.11.7.2335
Scrimale, T., Didone, L., de Mesy Bentley, K. L., and Krysan, D. J. (2009). The unfolded protein response is induced by the cell wall integrity mitogen-activated protein kinase signalling cascade and is required for cell wall integrity in Saccharomyces cerevisiae. Mol. Biol. Cell 20, 164–175. doi: 10.1091/mbc.e08-08-0809
Shang, Y. F., Chen, P. L., Chen, Y. X., Lu, Y. Z., and Wang, C. S. (2015). MrSkn7 controls sporulation, cell wall integrity, autolysis and virulence in Metarhizium robertsii. Eukaryot. Cell 14, 396–405. doi: 10.1128/EC.00266-14
Singh, P., Chauhan, N., Ghosh, A., Dixon, F., and Calderone, R. (2004). SKN7 of Candida albicans: mutant construction and phenotype analysis. Infect. Immun. 72, 2390–2394. doi: 10.1128/IAI.72.4.2390-2394.2004
Smith, B. J., and Yaffe, M. P. (1991). Uncoupling thermotolerance from the induction of heat shock proteins. Proc. Natl. Acad. Sci. U.S.A. 88, 11091–11094. doi: 10.1073/pnas.88.24.11091
Sorger, P. K., and Pelham, H. R. B. (1988). Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell 54, 855–864. doi: 10.1016/S0092-8674(88)91219-6
Thompson, S., Croft, N. J., Sotiriou, A., Piggins, H. D., and Crosthwaite, S. K. (2008). Neurospora crassa heat shock factor 1 is an essential gene; a second heat shock factor-like gene, hsf2, is required for asexual spore formation. Eukaryot. Cell 7, 1573–1581. doi: 10.1128/EC.00427-07
Viefhues, A., Schlathoelter, I., Simon, A., Viaud, M., and Tudzynski, P. (2015). Unraveling the function of the response regulator BcSkn7 in the stress signaling network of Botrytis cinerea. Eukaryot. Cell 14, 636–651. doi: 10.1128/EC.00043-15
Wang, C. S., and Feng, M. G. (2014). Advances in fundamental and applied studies in China of fungal biocontrol agents for use against arthropod pests. Biol. Control 68, 129–135. doi: 10.1016/j.biocontrol.2013.06.017
Wang, J., Ying, S. H., Hu, Y., and Feng, M. G. (2016). Mas5, a homologue of bacterial DnaJ, is indispensable for the host infection and environmental adaptation of a filamentous fungal insect pathogen. Environ. Microbiol. 18, 1037–1047. doi: 10.1111/1462-2920.13197
Wang, J., Ying, S. H., Hu, Y., and Feng, M. G. (2017). Vital role for the J-domain protein Mdj1 in asexual development, multiple stress tolerance, and virulence of Beauveria bassiana. Appl. Microbiol. Biotechnol. 101, 185–195. doi: 10.1007/s00253-016-7757-4
Wang, J., Zhou, G., Ying, S. H., and Feng, M. G. (2013). P-type calcium ATPase functions as a core regulator of Beauveria bassiana growth, conidiation and responses to multiple stressful stimuli through cross-talk with signalling networks. Environ. Microbiol. 15, 967–979. doi: 10.1111/1462-2920.12044
Wang, Z. L., Zhang, L. B., Ying, S. H., and Feng, M. G. (2013). Catalases play differentiated roles in the adaptation of a fungal entomopathogen to environmental stresses. Environ. Microbiol. 15, 409–418. doi: 10.1111/j.1462-2920.2012.02848.x
Wiederrecht, G., Seto, D., and Parker, C. S. (1988). Isolation of the gene encoding the S. cerevisiae heat shock transcription factor. Cell 54, 841–853. doi: 10.1016/S0092-8674(88)91197-X
Wormley, F. L. Jr., Heinrich, G., Miller, J. L., Perfect, J. R., and Cox, G. M. (2005). Identification and characterization of an SKN7 homologue in Cyptococus neoformans. Infect. Immun. 73, 5022–5030. doi: 10.1128/IAI.73.8.5022-5030.2005
Xiao, G., Ying, S. H., Zheng, P., Wang, Z. L., Zhang, S., Xie, X. Q., et al. (2012). Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci. Rep. 2:483. doi: 10.1038/srep00483
Xie, X. Q., Guan, Y., Ying, S. H., and Feng, M. G. (2013). Differentiated functions of Ras1 and Ras2 proteins in regulating the germination, growth, conidiation, multi-stress tolerance and virulence of Beauveria bassiana. Environ. Microbiol. 15, 447–462. doi: 10.1111/j.1462-2920.2012.02871.x
Xie, X. Q., Li, F., Ying, S. H., and Feng, M. G. (2012). Additive contributions of two manganese-cored superoxide dismutases (MnSODs) to antioxidation, UV tolerance and virulence of Beauveria bassiana. PLoS One 7:e30298. doi: 10.1371/journal.pone.0030298
Yang, Q. Q., Yin, D. F., Yin, Y. N., Cao, Y., and Ma, Z. H. (2015). The response regulator BcSkn7 is required for vegetative differentiation and adaptation to oxidative and osmotic stresses in Botrytis cinerea. Mol. Plant Pathol. 16, 276–287. doi: 10.1111/mpp.12181
Zeng, F. Y., Gong, X. Y., Hamid, M. I., Fu, Y. P., Xie, J. T., Cheng, J. S., et al. (2012). A fungal cell wall integrity-associated MAP kinase cascade in Coniothyrium minitans is required for conidiation and mycoparasitism. Fungal Genet. Biol. 49, 347–357. doi: 10.1016/j.fgb.2012.02.008
Zhang, C., Kho, Y., Wang, Z., Chiang, Y. T., Ng, G. K. H., Shaw, P., et al. (2014). Transmembrane and coiled-coil domain family 1 is a novel protein of the endoplasmic reticulum. PLoS One 9:e85206. doi: 10.1371/journal.pone.0085206
Zhang, F., Xu, G. P., Geng, L. P., Lu, X. Y., Yang, K. L., Yuan, J., et al. (2016). The stress response regulator AflSkn7 influences morphological development, stress response, and pathogenicity in the fungus Aspergillus flavus. Toxins 8, 202–215. doi: 10.3390/toxins8070202
Zhang, S. Z., Xia, Y. X., Kim, B., and Keyhani, N. O. (2011). Two hydrophobins are involved in fungal spore coat rodlet layer assembly and each play distinct roles in surface interactions, development and pathogenesis in the entomopathogenic fungus, Beauveria bassiana. Mol. Microbiol. 80, 811–826. doi: 10.1111/j.1365-2958.2011.07613.x
Keywords: entomopathogenic fungi, heat shock transcription factor, heat shock proteins, gene expression and regulation, asexual development, multiple stress responses, virulence
Citation: Zhou G, Ying S-H, Hu Y, Fang X, Feng M-G and Wang J (2018) Roles of Three HSF Domain-Containing Proteins in Mediating Heat-Shock Protein Genes and Sustaining Asexual Cycle, Stress Tolerance, and Virulence in Beauveria bassiana. Front. Microbiol. 9:1677. doi: 10.3389/fmicb.2018.01677
Received: 19 May 2018; Accepted: 04 July 2018;
Published: 25 July 2018.
Edited by:
Hector Mora Montes, Universidad de Guanajuato, MexicoReviewed by:
Chengshu Wang, Institute of Plant Physiology and Ecology (SIBS-CAS), ChinaZheng Wang, Yale University, United States
Copyright © 2018 Zhou, Ying, Hu, Fang, Feng and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiang Fang, ZnhpYW5nQHNjYXUuZWR1LmNu Ming-Guang Feng, bWdmZW5nQHpqdS5lZHUuY24= Jie Wang, d2FuZ2ppZWxhbmdqaW5nQDEyNi5jb20=
 Gang Zhou
Gang Zhou Sheng-Hua Ying
Sheng-Hua Ying Yue Hu3
Yue Hu3 Ming-Guang Feng
Ming-Guang Feng Jie Wang
Jie Wang