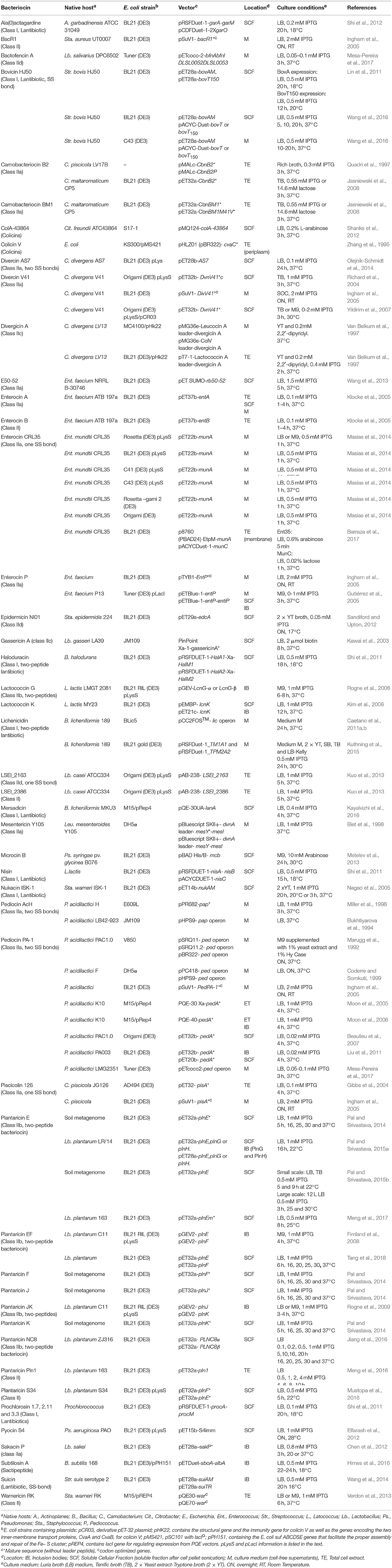
- 1Teagasc Food Research Centre, Teagasc Moorepark, Fermoy, Cork, Ireland
- 2APC Microbiome Ireland, University College Cork, Cork, Ireland
- 3School of Microbiology, University College Cork, Cork, Ireland
- 4College of Science Engineering and Food Science, University College Cork, Cork, Ireland
Bacteriocins, a heterogenous group of antibacterial ribosomally synthesized peptides, have potential as bio-preservatives in in a wide range of foods and as future therapeutics for the inhibition of antibiotic-resistant bacteria. While many bacteriocins have been characterized, several factors limit their production in large quantities, a requirement to make them commercially viable for food or pharma applications. The identification of new bacteriocins by database mining has been promising, but their potential is difficult to evaluate in the absence of suitable expression systems. E. coli has been used as a heterologous host to produce recombinant proteins for decades and has an extensive set of expression vectors and strains available. Here, we review the different expression systems for bacteriocin production using this host and identify the most important features to guarantee successful production of a range of bacteriocins.
Introduction
Bacteriocins are antimicrobial peptides produced by Gram-negative and Gram-positive bacteria. These molecules have attracted considerable interest, especially those produced by GRAS (Generally Recognized As Safe) microorganisms, as natural food preservatives in the food industry. They also represent potential alternatives to traditional antibiotics in the treatment of infections in humans and food-producing animals (Cotter et al., 2005; Desriac et al., 2010; Svetoch and Stern, 2010) and, in some cases, show promise as drugs for cancer treatment (reviewed in Kaur and Kaur, 2015).
Since the identification of colicin in 1925 (Gratia, 1925), many bacteriocins have been described (BAGEL4 database, http://bagel4.molgenrug.nl). However, only nisin (e.g., Nisaplin®) and pediocin PA-1 (e.g., ALTA®-2431) have been commercialized to any significant extent and these mainly as food biopreservatives. This may be explained by the multitude of studies that are required before a bacteriocin can be considered commercially viable, including characterization, potent antimicrobial activity, product stability, mechanism of action, mode of delivery, toxicity and assessment of their applications and industrial-scale production (Ingham and Moore, 2007).
The most important prerequisite for these studies is the production of high levels of biologically active bacteriocin. Although these peptides can be purified from their native producing strains, the process is time-consuming and laborious, and bacteriocin yields are often low (Rodríguez et al., 2003). Synthetic production is one alternative in some cases, but the complexity of some of the bacteriocins and the cost of the process limit the synthesis of large quantities (Chen et al., 2012). Therefore, attempts to increase the production of bacteriocins using alternative hosts such as Lactococcus lactis and other lactic acid bacteria (LAB) have been investigated (reviewed in Cintas et al., 2011). These strains, which are food-grade organisms, offer a safer choice for industrial food products, and provide the genetic and secretory machinery for efficient LAB bacteriocin production. However, the expression strains available are highly specific and the results in terms of yield are still disappointing at industrial level, restricting the variety and quantity of bacteriocin produced (Rodríguez et al., 2003). Given these limitations E. coli, the most commonly used organism for heterologous protein production, is an attractive option for the heterologous bacteriocin expression due to its rapid growth on inexpensive media, its extensive genetic characterization and the availability of versatile cloning tools, expression systems and strains (Mergulhao et al., 2004; Rosano and Ceccarelli, 2014; Jia and Jeon, 2016). This could facilitate the functional characterization and establishment of a production process in bacteriocins from sources that are difficult to cultivate, in addition to those bacteriocins discovered by data mining from sequenced bacterial genomes (Kuo et al., 2013), increasing their potential for manufacture and commerialization by food and pharmaceutical industries (Ongey and Neubauer, 2016). However, this approach is not without obstacles which may arise during the expression, secretion or processing of these peptides in E. coli (Choi and Lee, 2004).
In order to provide a guide for a design of a successful expression system for bacteriocin production in this host, the present review focuses on the different systems currently available for bacteriocin production in E. coli.
General Considerations Before the Expression System Selection
Bacteriocin
The choice of an expression system is not simple. Firstly, the characteristics of the bacteriocin should be taken into account, such as the presence of post-translational modifications and disulfide bonds, because these may affect its heterologous production.
According to Cotter et al. (2012), LAB bacteriocins are classified into those which are either post-translationally modified (class I) and unmodified or minimally modified peptides (class II). Class I can be subdivided into lantibiotics (with lanthionine bridges), linaridins, proteusins, linear azole-or azoline-containing peptides, cyanobactins, thiopeptides, lasso peptides, sactibiotics (contain sulfur-α-carbon linkages), bottromycins, glycocins, and modified microcins that do not belong to other subgroups. Class II is further divided into class IIa (the pediocin-like bacteriocins), IIb (the two-peptide bacteriocins), IIc (circular bacteriocins), IId (unmodified, linear, non-pediocin-like, single-peptide bacteriocins that do not belong to other subclasses), and IIe (the microcin E492-like bacteriocins). In this classification, the large (>10 kDa), heat-labile antimicrobial peptides bacteriolysins (formerly class III bacteriocins) were removed from the bacteriocin category.
Similarly, bacteriocins from Gram-negative organisms can be divided into small peptides, such as microcins [class I (presence of modifications) or II (unmodified)], and large peptides such as colicins (Drider and Rebuffat, 2011).
Genes Required for Bacteriocin Expression
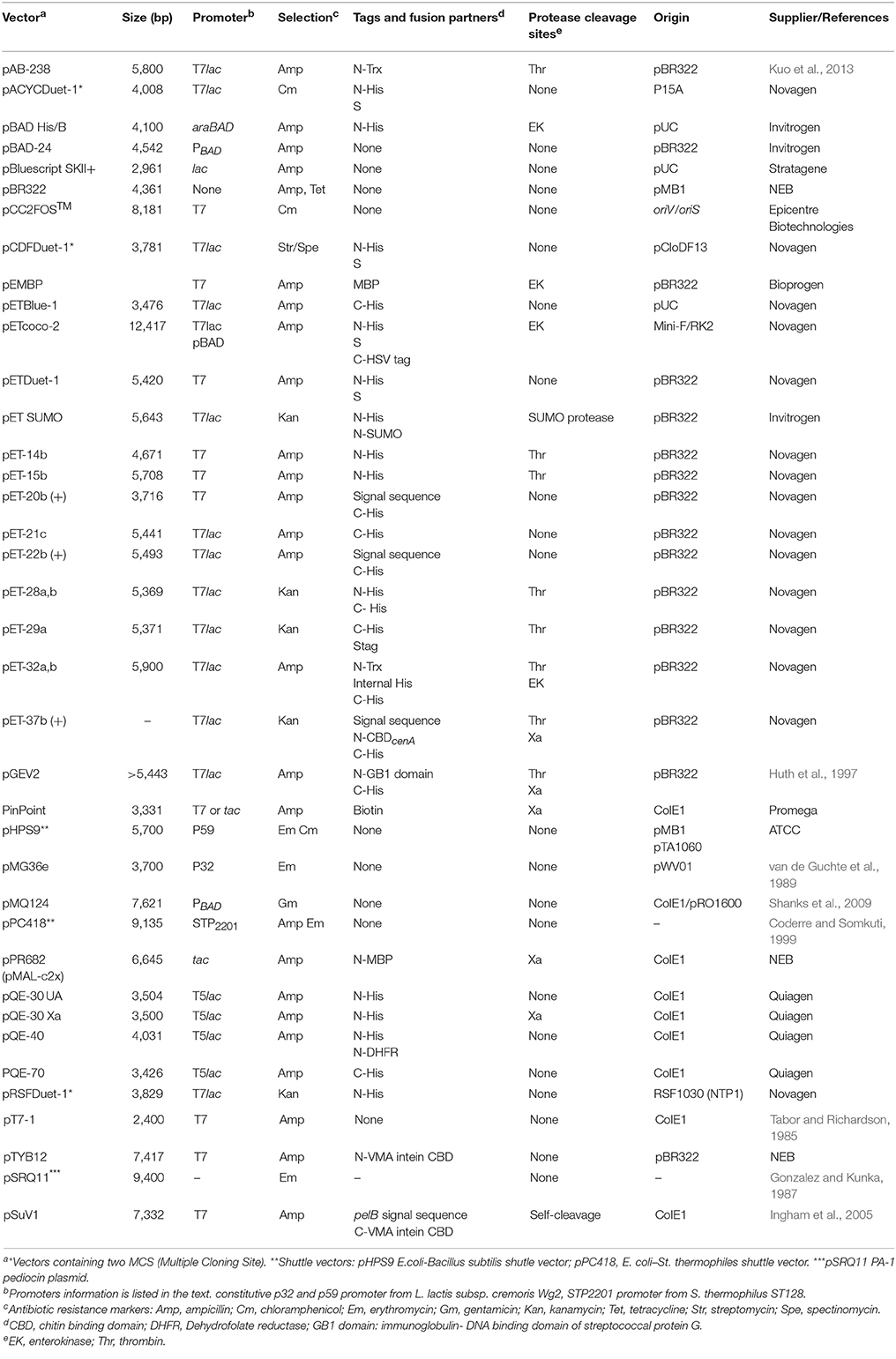
In general, the production of bacteriocins in the native host requires several genes including a structural gene that encodes the prepeptide (or two structural genes for the two peptide bacteriocins). Other genes encode an immunity protein, specialized secretion machinery and in many cases proteins capable of performing modifications and regulatory sequences (Nes et al., 1996). Therefore, different strategies are required depending on the characteristics of each bacteriocin to ensure production (see Table 1).
In most cases, the expression of the structural gene or its mature sequence is enough to produce the active bacteriocin. Some examples include carnobacteriocin B2 (Jasniewski et al., 2008), divercin AS7 and V41(Richard et al., 2004; Ingham et al., 2005; Yildirim et al., 2007; Olejnik-Schmidt et al., 2014), epidermicin NI01 (Sandiford and Upton, 2012), gassericin A (Kawai et al., 2003), or sakacin P (Chen et al., 2012) (see Table 1). However, the transporter gene is also necessary for the synthesis of some bacteriocins, for example in the production of pediocin PA-1 and bactofencin A (Bukhtiyarova et al., 1994; Mesa-Pereira et al., 2017). In other cases, the co-expression of the structural gene with the genes involved in post-translational modifications on the same or different plasmids are required for the heterologous expression of lantibiotics such as lichenicidin (Caetano et al., 2011a,b; Kuthning et al., 2015), nukacin ISK-1 (Nagao et al., 2005), prochlorosin, haloduracin, nisin (Shi et al., 2011), suicin (Wang et al., 2014), and the sactibiotic subtilosin A (Himes et al., 2016), amongst others.
Toxicity
The potential toxicity to E. coli due to the overexpression of the mature peptides or components of the secretion machinery and other bacterial integral membrane proteins (Fath and Kolter, 1993; Miller et al., 1993) must also be considered as these could interfere with the growth and viability of E. coli, limiting bacteriocin production (Bentley et al., 1990; Mccormick et al., 1996; Biet et al., 1998; Gutiérrez et al., 2005; Ingham et al., 2005; Moon et al., 2005; Masias et al., 2014; Mesa-Pereira et al., 2017).
Plasmids for Bacteriocin Expression in E. coli
General Features
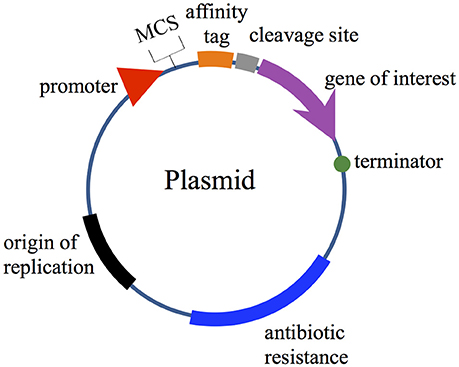
Expression vectors require various components to carry out their functions, including; (i) an origin of replication; (ii) a selection marker (generally genes encoding resistance to antibiotics); (iii) a promoter region for gene transcription initiation; and (iv) multiple unique restriction enzyme sites arranged in a polylinker region after the promoter to facilitate the cloning (referred to as multiple cloning sites; MCS). In some cases, two or more MCS are available in commercial plasmids (i.e., Duet vectors and pRSFDuet™-1) for cloning several genes of interest without the need to use multiple plasmids. In addition, the MCS can additionally provide fusion tags to facilitate the purification of the expressed bacteriocins. Plasmids must also contain one or several terminators to ensure an efficient transcriptional termination and prevent the transcription downstream of the coding sequence of interest (Figure 1). In terms of translational features, the plasmid must include a ribosome binding site (RBS) with a Shine-Dalgarno sequence (UAAGGAGG) located 5–13 bases upstream of the start codon for the interaction with the 3′ end of rRNA during translation initiation (reviewed in Mergulhao et al., 2004; Terpe, 2006; Durani et al., 2012; Rosano and Ceccarelli, 2014).
Plasmid Copy Number
A replicon consists of one origin of replication with associated control elements and is involved in plasmid replication and copy number control (del Solar and Espinosa, 2000). The number of copies per cell can vary between one and approximately one hundred. Theoretically, the higher the copy number of a plasmid, the higher the expression of the gene of interest. However, this often results in aggregation, misfolding or protein degradation in E. coli (Tolia and Joshua-Tor, 2006), and could also cause cellular toxicity when the overexpression of secretion machinery and other integral membrane proteins are required for bacteriocin production. For this reason, low and middle copy vectors (15–20 copies per cell) based on ColE1, pMB1 replicons, including the pBR322 and pET vector systems and pACYC plasmids with an origin replication derived from p15A, have been successfully used for bacteriocin expression (Table 2). In addition, the use of two plasmids with compatible origins (e.g., ColE1 with p15A, pMB1 with p15A) has allowed for dual expression of proteins for functional characterization (Table 1). In this regard, the expression vector pETcoco-2 could be a useful tool for studying whole operons since it enables the control of copy number by arabinose induction, facilitating the optimization of bacteriocin expression (Sektas and Szybalski, 2002; Mesa-Pereira et al., 2017).
Promoter Region
A careful balance of promoter strength and gene copy number is necessary for the optimization of the bacteriocin expression level. Since bacteriocins could be toxic for the host, the promoter strength should be adequate in order to minimize the metabolic burden on E. coli prior to the production phase and prevent the saturation of the host transport machinery (Rosenberg, 1998; Mergulhão et al., 2003; Mergulhao et al., 2004). This might explain the failure to generate successful expression plasmids under the control of constitutive promoters (Mccormick et al., 1998; McCormick et al., 1999), while the use of inducible promoters enables stable bacteriocin expression (Gutiérrez et al., 2005). Thus, an appropriate promoter must be strong, have a low basal expression level (i.e., be highly repressible) and a cost-effective induction system.
A large number of promoter systems have been described for protein production in E. coli (reviewed in Terpe, 2006; Durani et al., 2012; Rosano and Ceccarelli, 2014). In terms of bacteriocin expression, the L-arabinose inducible araBAD (PBAD) promoter has been used in some studies (Shanks et al., 2012; Metelev et al., 2013; Barraza et al., 2017), while most bacteriocins have been expressed using lac-derived promoters inducible by lactose or its non-hydrolyzable analog isopropyl β-D-thiogalactopyranoside (IPTG), including tac promoter (Miller et al., 1998; Kawai et al., 2003), T5lac (Moon et al., 2005, 2006; Verdon et al., 2013; Kayalvizhi et al., 2016) and the most widely used T7 and T7lac promoters (Table 2).
In T7 promoter systems, the gene of interest is cloned behind a promoter recognized by the phage T7 RNA polymerase that is provided by another plasmid or in the bacterial genome in a prophage (λDE3). T7 RNA polymerase is under the transcriptional control of a lacUV5 promoter inducible by lactose or IPTG. Basal expression can be controlled by the introduction of a mutated promoter of the lacI gene, called lacIQ, that increases the expression of the lac promoter repressor LacI. Additionally, the co-expression of T7 lysozyme provided in a compatible plasmid (pLysS or pLysE) can inhibit the transcription of T7 RNA polymerase. In the case of the T7lac promoter, this also includes a lacO operator downstream of the promoter that avoids basal expression (reviewed in Rosano and Ceccarelli, 2014).
Protein Targeting-Signal Secretion Sequences
Since E. coli is a Gram-negative bacterium, bacteriocin production can be confined to the cytoplasm where it can accumulate in a soluble form or aggregate in insoluble inclusion bodies, or can also be secreted into the periplasmic space or into the culture medium. Several factors including protein size, amino acid composition, and the type of leader peptide can affect bacteriocin translocation to these locations (reviewed in Mergulhao et al., 2004).
Most bacteriocins use a dedicated secretion machinery in their natural hosts to export the bacteriocin to the extracellular media. However, the production of active bacteriocins directly to the culture medium by cloning their whole operons in E. coli has been reported on a limited number of occasions (Figure 2) (Caetano et al., 2011a; Mesa-Pereira et al., 2017). Generally, the overexpression of cloned native genes leads to bacteriocin accumulation in inclusion bodies in the cytoplasm. At first glance, the formation of inclusion bodies could be advantageous as the expressed bacteriocins are inactive and protected against host proteases, facilitating their purification and a high protein yield. However, this aggregation can affect the host metabolism and additional steps (inclusion body isolation, solubilization of the aggregates, and protein refolding) are required for their purification, limiting their large-scale production (Mergulhao et al., 2004).
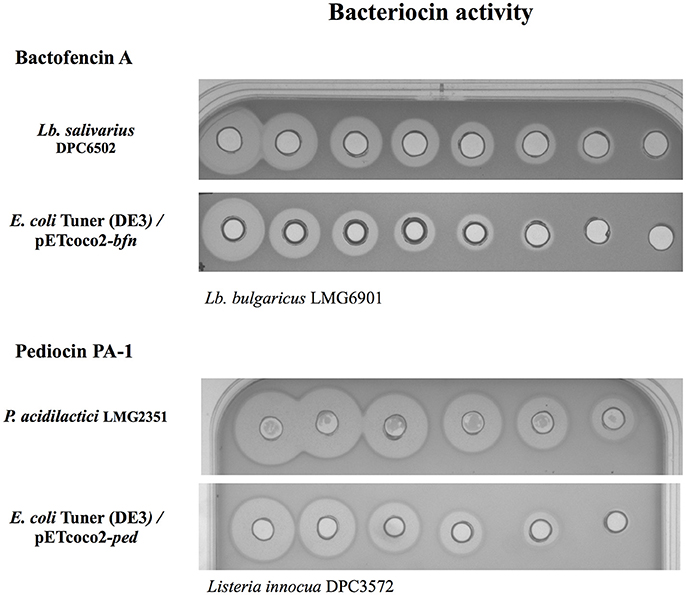
Figure 2. Bacteriocin activity of 2-fold serial dilutions of cell-free supernatants. Antimicrobial activity of bactofencin A against Lb. bulgaricus LMG 6901 produced by Lb. salivarius DPC6502 (natural producer) and E. coli Tuner (DE3) carrying the vector expressing the whole bactofencin A operon (bfn) and pediocin PA-1 against Listeria innocua DPC3572 produced by P. acidilactici LMG2351 (natural producer) or by E. coli Tuner (DE3) carrying the plasmid with the pediocin PA-1 operon (ped).
The processing and the correct folding of bacteriocins as well as their recovery can be simplified when the peptide is secreted into the E. coli periplasm or into the culture medium. To achieve that, signal sequences of proteins recognized by a general protein secretory pathway (sec pathway), such as the maltose-binding protein malE (Miller et al., 1998), the pectase lyase secretion signal pelB (Ingham et al., 2005), and the outer membrane protein ompA (Zhang et al., 1995), have been fused to the bacteriocin sequence for targeting their secretion to the periplasm. In addition, the bacteriocin divergicin A signal peptide has been used to direct the expression of mesentericin Y105 in E. coli in the absence of their dedicated secretion machinery (Biet et al., 1998). This relies on the fact that divergicin A, as well as other bacteriocins such as hiracin JM79 (Sánchez et al., 2007), acidocin B (Leer et al., 1995), and enterocin P (Cintas et al., 1997; Gutiérrez et al., 2005), can be exported by the E. coli sec pathway. Subsequently, the peptides can be secreted into the culture medium by osmotic shock or cell wall permeabilization or, alternatively, be released directly into the medium using a periplasmic leaky E. coli host (E609L, Miller et al., 1998).
It is important to mention that bacteriocins containing disulfide bonds are normally accumulated in the periplasm where disulfide binding proteins catalyze the oxidation process (Miller et al., 1998) or alternatively overexpressed in the cytoplasm by thioreductase-deficient (trxB) and glutathione reductase (gor) deficient strains (Terpe, 2006).
Affinity Tags and Other Fusion Partners
Given the small size of bacteriocins, the incorporation of affinity tags, such as poly-His-tags, facilitates their detection and allows for one-step affinity purification. In addition, the use of fusion protein partners can increase the expression level, enhance protein solubility and assist in correct folding and disulfide bond formation (Ingham and Moore, 2007). Common fusion partners used for bacteriocin production include the cellulose binding domain (CBDcenA) (Klocke et al., 2005), maltose-binding protein (MBP) (Quadri et al., 1997; Kim et al., 2006), thiorredoxin (Trx) (Gibbs et al., 2004; Richard et al., 2004; Beaulieu et al., 2007; Yildirim et al., 2007; Jasniewski et al., 2008; Caetano et al., 2011a; Liu et al., 2011; Pal and Srivastava, 2014, 2015a,b; Jiang et al., 2016; Mustopa et al., 2016; Meng et al., 2017; Tang et al., 2018) and the small ubiquitin-related modifier SUMO (Wang et al., 2013).
Since secretion in the native hosts involves the cleavage of the signal sequence, such fusions often lack antimicrobial activity until chemical (e.g., cyanogen bromide cleaves proteins on the C-terminal side of methionine residues) or enymatic cleavage occurs. The cyanogen bromide (CNBr) chemical cleavage strategy has been used to release the mature PlnE and PlnF (Fimland et al., 2008) and PlnJ and PlnK (Rogne et al., 2009), as has also been described for production of carnobacteriocin B2 and BM1 (Jasniewski et al., 2008) and piscicolin 126 (Gibbs et al., 2004). However, the most common approach to release recombinant bacteriocins is to include a sequence, between the signal peptide and the bacteriocin, recognized by Factor Xa (Quadri et al., 1997; Kawai et al., 2003; Klocke et al., 2005; Moon et al., 2006; Ingham and Moore, 2007; Rogne et al., 2008; Shi et al., 2011), trypsin (Shi et al., 2012; Himes et al., 2016), thrombin (Klocke et al., 2005), enterokinases (Beaulieu et al., 2007; Jasniewski et al., 2008; Liu et al., 2011; Pal and Srivastava, 2014, 2015a,b; Jiang et al., 2016; Meng et al., 2016; Tang et al., 2018), and SUMO proteases (Wang et al., 2013). Alternatively, the use of intein fusions has been described for the cloning and expression of self-cleaving fusion forms of unmodified bacteriocins under appropriate buffer conditions (Ingham et al., 2005).
E. coli Strains for Bacteriocin Expression
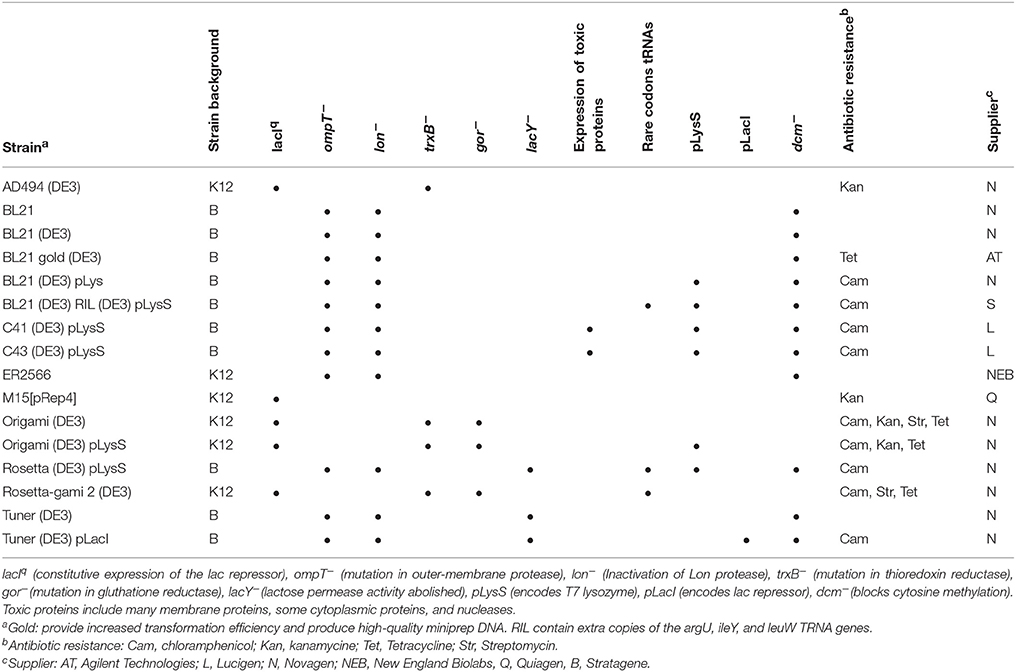
Since bacteriocins can easily be degraded in the expression strain (Chen et al., 2012), E. coli BL21 (DE3) and its derivates are most frequently used for bacteriocin expression (Tables 1, 3) as they are deficient in the Lon protease and the outer membrane protease OmpT (Gottesman, 1996). Tuner™ (DE3) strains (Novagen) are lacZY deletion mutants of BL21. The lac permease mutation (lacY) allows uniform entry of IPTG into all cells in the population, which enables the regulation of the levels of protein expression by adjusting the concentration of the inductor IPTG. Other B related strains such as C41 (DE3) and C43 (DE3), described specially for the production of toxic proteins (Terpe, 2006), have been used successfully for the expression of the bacteriocin bovicin HJ50 while the use of E. coli BL21 (DE3) did not produce this peptide (Wang et al., 2016).
The designation DE3 indicates that the host is a lysogen of λDE3 which carries a chromosomal copy of the T7 RNA polymerase under the lacUV5 promoter required for the expression of genes under a T7 promoter (Rosano and Ceccarelli, 2014). To suppress the basal expression of T7 RNA polymerase prior to induction, especially important for toxic protein expression that affect cell growth and viability of the host, pLysS hosts carry a plasmid that encodes the T7 lysozyme, an inhibitor of T7 RNA polymerase. The pLacI designation is given to hosts bearing a plasmid that encodes the lac repressor.
K-12 derivatives have also been used for bacteriocin expression. E. coli JM109, which is lon protease deficient, has been used for gassericin A expression (Kawai et al., 2003). Divercin V41, enterocin CRL35, LSE_2163 and LSE_2386 and pediocin PA-1 that require disulphide bond formation for proper folding have been expressed in Origami™ (DE3) strains (Beaulieu et al., 2007; Yildirim et al., 2007; Kuo et al., 2013; Masias et al., 2014), which carry a double mutation in thioredoxin reductase (trxB) and glutathione reductase (gor) genes with an oxidative cytoplasmic environment that allows disulfide bond formation. The same trxB/gor mutations are carried by a derived Tuner™ strain, Origami B, combining the characteristics of BL21 and Origami hosts in one strain (Novagen). Other combinations, including Rosetta-gami™ (K-12 derivative) and Rosetta-gami™ B (BL21 derivative), facilitate the expression of eukaryotic proteins containing rare codons and cytoplasmic disulphide bond formation at the same time. Table 3 summarizes the features of commercial E. coli strains most used for bacteriocin production.
Codon Bias
Major differences between the codon usage of E. coli and the overexpressed protein may be an obstacle for bacteriocin production, especially in the case of rare codons. Rare codons are defined as codons used by E. coli at a frequency <1% (Kane, 1995), which means the tRNA for these codons are rare or lacking in the expression host. To overcome this problem, two strategies have been used: codon optimization of the mature sequences of bacteriocins (Richard et al., 2004; Ingham et al., 2005; Verdon et al., 2013) or increasing the availability of underrepresented tRNAs by supplying pRIL or pRARE plasmids. pRIL vector provides extra genes for the tRNAs for Arg (AGG/AGA), Ile (AUA), and Leu (CUA) while pRARE encodes tRNA genes for all the above-mentioned codons plus Gly (GGA) and Pro (CCC). pRIL or pRARE plasmids are supplied in the BL21 derivatives, BL21 (DE3) Codon Plus strain (Stratagene), and Rosetta (DE3) strains (Novagen), respectively. However, it is important to mention that although these strains can improve the level of production, they sometimes can cause a decrease in protein solubility (Rosano and Ceccarelli, 2014) or even decrease the growth rate of E. coli significantly (Masias et al., 2014).
Selecting a Suitable E. coli Strain
Given that the expression level of bacteriocins can vary in different E. coli strains (Figure 3), it is recommended to test different strains, chosen to reflect the properties of the bacteriocin (e.g., disulfide bonds, rare codons, etc.) to select the best host for the heterologous expression. A clear example is described by Masias et al. (2014), who showed the expression of enterocin CRL35 in E. coli BL21, C41, C43, Origami, and Rosetta-gami 2. In this study, the expression of enterocin CRL35 was lower in E. coli C41 and C43 than in other E. coli strains. The best strains for enterocin CRL35 expression were E. coli Rosetta and E. coli Rosetta-gami 2 since they are able to synthesize proteins despite the presence of rare codons. In addition, this study showed that E. coli Rosetta produced an additional enterocin CRL35 variant. Therefore, the correct choice of the strain is critical for bacteriocin expression.
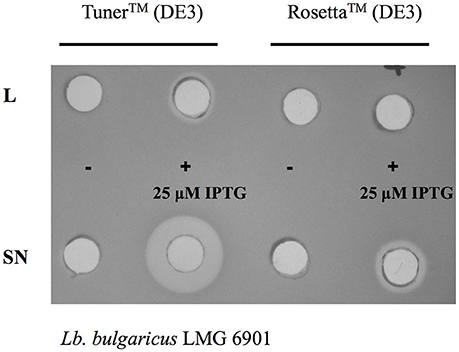
Figure 3. Differences in antimicrobial activity of cell-free supernatants (SN) and lysates (L) of E. coli Tuner™ (DE3) and Rosetta™ (DE3) cells carrying the plasmid encoding the whole operon of pediocin PA-1 without induction (−) and after 3 h of 25 μM IPTG induction (+).
Culture Conditions for Bacteriocin Expression
Both culture media composition and culture conditions are important for optimizing the heterologous expression of bacteriocins and must be optimized for each bacteriocin in each expression system as reviewed in Table 1. Even two peptides of the same bacteriocin might require different conditions, for example the maximum soluble fraction of PLNC8α was observed under 0.5 mM IPTG induction for 16 h at 20°C, while for PLNC8β it was 0.2 mM IPTG for 20 h at 16°C (Jiang et al., 2016). Bacteriocin production can be also increased with different production strategies such as batch and fed-batch cultivation (Gibbs et al., 2004; Yildirim et al., 2007). Although it is hard to generalize, there are some observations that can be taken into account in terms of culture conditions to facilitate this trial-and-error process.
Growth Media
Luria Broth (LB) is the most commonly used medium for culturing E. coli for bacteriocin expression as it is easy to make and it is nutritionally rich. However, the cell density obtained with this medium is low, affecting bacteriocin yield. To overcome this problem, there are media superior to LB available for reaching higher cell densities such as 2 × yeast extract tryptone (YT), Terrific Broth (TB), and Super Broth (SB) (Rosano and Ceccarelli, 2014). 2 × YT medium has been used for expression of nukacin ISK-1 (Nagao et al., 2005) and epidermicin NI01 (Sandiford and Upton, 2012), while TB has been used for divercin V41 (Richard et al., 2004), carnobacteriocin Cbn1 and CbnB2 production (Jasniewski et al., 2008). Other bacteriocins have been produced using minimal medium (Rogne et al., 2008; Metelev et al., 2013). Therefore, there are no general rules. Pal and Srivastava (2015b) found higher Plantaricin E yield in LB than in TB, while Kuthning et al. (2015) found the best-producing conditions for Bliα and Bliβ lichenicidin peptides was medium M compared to 2 × YT, SB, TB, and LB-Kelly.
Alternatively, M9 medium supplementation with 10 mM of EDTA and 0.05% Tween 20 at the time of induction could also increase the final yield (Masias et al., 2014). In addition, the pH of the culturing media has also an impact on peptide yields either promoting bacteriocin expression or increasing peptide stability as described for lichenicidin expression which increased at pH 6.5 compared to pH 8 (Kuthning et al., 2015).
Inducer Agents
Bacteriocin expression levels can be tuned by varying the inducer concentration (Gutiérrez et al., 2005; Yildirim et al., 2007; Masias et al., 2014; Jiang et al., 2016), but use of a high concentration to fully induce the promoter does not necessarily lead to maximal expression due to the metabolic burden and toxicity of the inducer to the cells (Glick, 1995).
Although the IPTG inducible lac expression system of E. coli is the most used for bacteriocin expression, IPTG is expensive and toxic and therefore not suitable for large-scale production. This problem could be solved by replacing IPTG by lactose, which is not toxic, and has resulted in an increase in the production yield of some bacteriocins such as the carnobacteriocins Cbn BM1 and Cbn B2 (Jasniewski et al., 2008).
Temperature
Temperature is one of the most important factors for the expression of functional proteins (Sambrook and Russell, 2001). Chen et al. (2012) showed that the expression level of sakacin P was higher when E. coli BL21 (DE3) carrying pET28a-sakP was induced at 20°C than at 37°C. Similar results were observed when the expression of plantaricin E was induced at 25°C rather than at 37°C (Pal and Srivastava, 2015b). Although the optimum growth temperature for E. coli is around 37°C, the cell growth and the protein synthesis are slowed down at lower temperatures, which provides the peptides the time and optimal environment to fold into their native conformation (Sambrook and Russell, 2001; Peng et al., 2004), decreasing the aggregation and increasing the expression of soluble protein. In addition, lowering the temperature, in combination with the time after induction, might shift the codon usage bias in E. coli sufficiently to solve some codon-usage based expression problems (Terpe, 2006). Therefore, it is essential to determine the optimal induction temperature in each case to improve the amount of the soluble fraction. When inclusion bodies formation is a problem, it is recommended to express the protein in the range of 15–25°C (Rosano and Ceccarelli, 2014).
Conclusion
The development of heterologous expression systems to improve bacteriocin yield may facilitate their characterization and broaden their applications in food and pharmaceutical industries. Currently E. coli is the most popular recombinant protein expression platform. However, choosing the perfect combination of expression vector and strain for bacteriocin production in E. coli is not possible a priori due to the many variables that can affect bacteriocin production. This review covers different strategies used for the bacteriocin expression in E. coli to help the process of choosing the best expression system and the conditions for any particular bacteriocin with a view to producing bacteriocins economically for both food and pharmaceutical applications.
Author Contributions
BM-P, MR, PC, CH, and RR wrote the manuscript and approved its final version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank all of the members of the laboratory for their insightful comments on the manuscript, and their helpful suggestions to improve this work. This work was supported by Science Foundation Ireland (SFI) under grant number SFI/12/RC/2273.
References
Barraza, D. E., Ríos Colombo, N. S., Galván, A. E., Acuña, L., Minahk, C. J., Bellomio, A., et al. (2017). New insights into enterocin CRL35: mechanism of action and immunity revealed by heterologous expression in Escherichia coli. Mol. Microbiol. 105, 922–933. doi: 10.1111/mmi.13746
Beaulieu, L., Tolkatchev, D., Jette, J. F., Groleau, D., and Subirade, M. (2007). Production of active pediocin PA-1 in Escherichia coli using a thioredoxin gene fusion expression approach: cloning, expression, purification, and characterization. Can. J. Microbiol. 53, 1246–1258. doi: 10.1139/w07-089
Bentley, W. E., Mirjalili, N., Andersen, D. C., Davis, R. H., and Kompala, D. S. (1990). Plasmid-encoded protein: the principal factor in the “metabolic burden” associated with recombinant bacteria. Biotechnol. Bioeng. 35, 668–681.
Biet, F., Berjeaud, J. M., Worobo, R. W., Cenatiempo, Y., and Fremaux, C. (1998). Heterologous expression of the bacteriocin mesentericin Y105 using the dedicated transport system and the general secretion pathway. Microbiology 144, 2845–2854. doi: 10.1099/00221287-144-10-2845
Bukhtiyarova, M., Yang, R., and Ray, B. (1994). Analysis of the pediocin AcH gene cluster from plasmid pSMB74 and its expression in a pediocin-negative Pediococcus acidilactici strain. Appl. Environ. Microbiol. 60, 3405–3408.
Caetano, T., Krawczyk, J. M., Mösker, E., Süssmuth, R. D., and Mendo, S. (2011a). Heterologous expression, biosynthesis, and mutagenesis of type II lantibiotics from Bacillus licheniformis in Escherichia coli. Chem. Biol. 18, 90–100. doi: 10.1016/j.chembiol.2010.11.010
Caetano, T., Krawczyk, J. M., Mösker, E., Süssmuth, R. D., and Mendo, S. (2011b). Lichenicidin biosynthesis in Escherichia coli: licFGEHI immunity genes are not essential for lantibiotic production or self-protection. Appl. Environ. Microbiol. 77, 5023–5026. doi: 10.1128/AEM.00270-11
Chen, H., Tian, F., Li, S., Xie, Y., Zhang, H., and Chen, W. (2012). Cloning and heterologous expression of a bacteriocin sakacin P from Lactobacillus sakei in Escherichia coli. Appl. Microbiol. Biotechnol. 94, 1061–1068. doi: 10.1007/s00253-012-3872-z
Choi, J. H., and Lee, S. Y. (2004). Secretory and extracellular production of recombinant proteins using Escherichia coli. Appl. Microbiol. Biotechnol. 64, 625–635. doi: 10.1007/s00253-004-1559-9
Cintas, L. M., Casaus, P., Håvarstein, L. S., Hernández, P. E., and Nes, I. F. (1997). Biochemical and genetic characterization of enterocin P, a novel sec- dependent bacteriocin from Enterococcus faecium P13 with a broad antimicrobial spectrum. Appl. Environ. Microbiol. 63, 4321–4330.
Cintas, L. M., Herranz, C., and Hernández, P. E. (2011). “Natural and heterologous production of bacteriocins,” in Prokaryotic Antimicrobial Peptides: From Genes to Applications, eds D. Drider and S. Rebuffat (New York, NY: Springer), 115–143.
Coderre, P. E., and Somkuti, G. A. (1999). Cloning and expression of the pediocin operon in Streptococcus thermophilus and other lactic fermentation bacteria. Curr. Microbiol. 39, 295–301. doi: 10.1007/s002849900462
Cotter, P. D., Hill, C., and Ross, R. P. (2005). Food microbiology: bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3, 777–788. doi: 10.1038/nrmicro1273
Cotter, P. D., Ross, R. P., and Hill, C. (2012). Bacteriocins — a viable alternative to antibiotics? Nat. Rev. Microbiol. 11, 95–105. doi: 10.1038/nrmicro2937
del Solar, G., and Espinosa, M. (2000). Plasmid copy number control: an ever-growing story. Mol. Microbiol. 37, 492–500. doi: 10.1046/j.1365-2958.2000.02005.x
Desriac, F., Defer, D., Bourgougnon, N., Brillet, B., Le Chevalier, P., and Fleury, Y. (2010). Bacteriocin as weapons in the marine animal-associated bacteria warfare: inventory and potential applications as an aquaculture probiotic. Mar. Drugs 8, 1153–1177. doi: 10.3390/md8041153
Drider, D., and Rebuffat, S. (2011). Prokaryotic Antimicrobial Pep-tides: From Genes to Applications. (New York, NY: Springer).
Durani, V., Sullivan, B. J., and Magliery, T. J. (2012). Simplifying protein expression with ligation-free, traceless and tag-switching plasmids. Protein Expr. Purif. 85, 9–12. doi: 10.1016/j.pep.2012.06.007
Elfarash, A., Wei, Q., and Cornelis, P. (2012). The soluble pyocins S2 and S4 from Pseudomonas aeruginosa bind to the same FpvAI receptor. Microbiologyopen 1, 268–275. doi: 10.1002/mbo3.27
Fath, M. J., and Kolter, R. (1993). ABC transporters: bacterial exporters. Microbiol. Rev. 57, 995–1017.
Fimland, N., Rogne, P., Fimland, G., Nissen-Meyer, J., and Kristiansen, P. E. (2008). Three-dimensional structure of the two peptides that constitute the two-peptide bacteriocin plantaricin EF. Biochim. Biophys. Acta Proteins Proteomics 1784, 1711–1719. doi: 10.1016/j.bbapap.2008.05.003
Gibbs, G. M., Davidson, B. E., and Hillier, A. J. (2004). Novel expression system for large-scale production and purification of recombinant class IIa bacteriocins and its application to piscicolin 126. Appl. Environ. Microbiol. 70, 3292–3297. doi: 10.1128/AEM.70.6.3292-3297.2004
Glick, B. R. (1995). Metabolic load and heterologous gene expression. Biotechnol. Adv. 13, 247–261. doi: 10.1016/0734-9750(95)00004-A
Gonzalez, C. F., and Kunka, B. S. (1987). Plasmid-Associated Bacteriocin Production and Sucrose Fermentation in Pediococcus acidilactici. Appl. Environ. Microbiol. 53, 2534–2538.
Gottesman, S. (1996). Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30, 465–506. doi: 10.1146/annurev.genet.30.1.465
Gratia, A. (1925). Sur un rémarquable example d' antagonisme entre deux souches de colibacille. Biochimie 84, 1040–1041.
Gutiérrez, J., Criado, R., Citti, R., Martín, M., Herranz, C., Nes, I. F., et al. (2005). Cloning, production and functional expression of enterocin P, a sec-dependent bacteriocin produced by Enterococcus faecium P13, in Escherichia coli. Int. J. Food Microbiol. 103, 239–250. doi: 10.1016/j.ijfoodmicro.2004.11.035
Himes, P. M., Allen, S. E., Hwang, S., and Bowers, A. A. (2016). Production of Sactipeptides in Escherichia coli: probing the substrate promiscuity of subtilosin A biosynthesis. ACS Chem. Biol. 11, 1737–1744. doi: 10.1021/acschembio.6b00042
Huth, J. R., Bewley, C. A., Jackson, B. M., Hinnebusch, A. G., Clore, G. M., and Gronenborn, A. M. (1997). Design of an expression system for detecting folded protein domains and mapping macromolecular interactions by NMR. Protein Sci. 6, 2359–2364. doi: 10.1002/pro.5560061109
Ingham, A. B., and Moore, R. J. (2007). Recombinant production of antimicrobial peptides in heterologous microbial systems. Biotechnol. Appl. Biochem. 47(Pt 1), 1–9. doi: 10.1042/BA20060207
Ingham, A. B., Sproat, K. W., Tizard, M. L. V., and Moore, R. J. (2005). A versatile system for the expression of nonmodified bacteriocins in Escherichia coli. J. Appl. Microbiol. 98, 676–683. doi: 10.1111/j.1365-2672.2004.02502.x
Jasniewski, J., Cailliez-Grimal, C., Gelhaye, E., and Revol-Junelles, A. M. (2008). Optimization of the production and purification processes of carnobacteriocins Cbn BM1 and Cbn B2 from Carnobacterium maltaromaticum CP5 by heterologous expression in Escherichia coli. J. Microbiol. Methods 73, 41–48. doi: 10.1016/j.mimet.2008.01.008
Jia, B., and Jeon, C. O. (2016). High-throughput recombinant protein expression in Escherichia coli: current status and future perspectives. Open Biol. 6:160196. doi: 10.1098/rsob.160196
Jiang, H., Li, P., and Gu, Q. (2016). Heterologous expression and purification of plantaricin NC8, a two-peptide bacteriocin against Salmonella spp. from Lactobacillus plantarum ZJ316. Protein Expr. Purif. 127, 28–34. doi: 10.1016/j.pep.2016.06.013
Kane, J. F. (1995). Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr. Opin. Biotechnol. 6, 494–500. doi: 10.1016/0958-1669(95)80082-4
Kaur, S., and Kaur, S. (2015). Bacteriocins as potential anticancer agents. Front. Pharmacol. 6:272. doi: 10.3389/fphar.2015.00272
Kawai, Y., Arakawa, K., Itoh, A., Saitoh, B., Ishii, Y., Nishimura, J., et al. (2003). Heterologous expression of gassericin A, a bacteriocin produced by Lactobacillus gasseri LA39. Anim. Sci. J. 74, 45–51. doi: 10.1046/j.1344-3941.2003.00085.x
Kayalvizhi, N., Rameshkumar, N., and Gunasekaran, P. (2016). Cloning and characterization of mersacidin like bacteriocin from Bacillus licheniformis MKU3 in Escherichia coli. J. Food Sci. Technol. 53, 2298–2306. doi: 10.1007/s13197-016-2195-y
Kim, Y. S., Kim, M. J., Kim, P., and Kim, J. H. (2006). Cloning and production of a novel bacteriocin, lactococcin K, from Lactococcus lactis subsp. lactis MY23. Biotechnol. Lett. 28, 357–362. doi: 10.1007/s10529-005-5935-z
Klocke, M., Mundt, K., Idler, F., Jung, S., and Backhausen, J. E. (2005). Heterologous expression of enterocin A, a bacteriocin from Enterococcus faecium, fused to a cellulose-binding domain in Escherichia coli results in a functional protein with inhibitory activity against Listeria. Appl. Microbiol. Biotechnol. 67, 532–538. doi: 10.1007/s00253-004-1838-5
Kuo, Y. C., Liu, C. F., Lin, J. F., Li, A. C., Lo, T. C., and Lin, T. H. (2013). Characterization of putative class II bacteriocins identified from a non-bacteriocin-producing strain Lactobacillus casei ATCC 334. Appl. Microbiol. Biotechnol. 97, 237–246. doi: 10.1007/s00253-012-4149-2
Kuthning, A., Mösker, E., and Süssmuth, R. D. (2015). Engineering the heterologous expression of lanthipeptides in Escherichia coli by multigene assembly. Appl. Microbiol. Biotechnol. 99, 6351–6361. doi: 10.1007/s00253-015-6557-6
Leer, R. J., van der Vossen, J. M., van Giezen, M., van Noort, J. M., and Pouwels, P. H. (1995). Genetic analysis of acidocin B, a novel bacteriocin produced by Lactobacillus acidophilus. Microbiology 141(Pt 7), 1629–1635. doi: 10.1099/13500872-141-7-1629
Lin, Y., Teng, K., Huan, L., and Zhong, J. (2011). Dissection of the bridging pattern of bovicin HJ50, a lantibiotic containing a characteristic disulfide bridge. Microbiol. Res. 166, 146–154. doi: 10.1016/j.micres.2010.05.001
Liu, S., Han, Y., and Zhou, Z. (2011). Fusion expression of pedA gene to obtain biologically active pediocin PA-1 in Escherichia coli. J. Zhejiang Univ. Sci. B 12, 65–71. doi: 10.1631/jzus.B1000152
Marugg, J. D., Gonzalez, C. F., Kunka, B. S., Ledeboer, A. M., Pucci, M. J., Toonen, M. Y., et al. (1992). Cloning, expression, and nucleotide sequence of genes involved in production of pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0. Appl. Environ. Microbiol. 58, 2360–2367.
Masias, E., Picariello, G., Acuña, L., Chalon, M., Sesma, F., Morero, R., et al. (2014). Co-expression and characterization of enterocin CRL35 and its mutant in Escherichia coli Rosetta. Peptidomics 1, 30–42. doi: 10.2478/ped-2014-0003
McCormick, J. K., Klaenhammer, T. R., and Stiles, M. E. (1999). Colicin V can be produced by lactic acid bacteria. Lett. Appl. Microbiol. 29, 37–41. doi: 10.1046/j.1365-2672.1999.00571.x
Mccormick, J. K., Poon, A., Sailer, M., Gao, Y., Roy, K. L., Mcmullen, L. M., et al. (1998). Genetic characterization and heterologous expression of brochocin-C, an antibotulinal, two-peptide bacteriocin produced by Brochothrix campestris ATCC 43754. Appl. Environ. Microbiol. 64, 4757–4766.
Mccormick, J. K., Worobo, R. W., and Stiles, M. E. (1996). Expression of the antimicrobial peptide carnobacteriocin B2 by a signal peptide-dependent general secretory pathway. Appl. Environ. Microbiol. 62, 4095–4099.
Meng, F., Zhao, H., Zhang, C., Lu, F., Bie, X., and Lu, Z. (2016). Expression of a novel bacteriocin—the plantaricin Pln1—in Escherichia coli and its functional analysis. Protein Expr. Purif. 119, 85–93. doi: 10.1016/j.pep.2015.11.008
Meng, F., Zhu, X., Lu, F., Bie, X., and Lu, Z. (2017). Functional analysis of plantaricin E and its mutant by heterologous expression in Escherichia coli. Appl. Biochem. Biotechnol. 182, 311–323. doi: 10.1007/s12010-016-2328-9
Mergulhao, F. J. M., Monteiro, G. A., Cabral, J. M. S., and Taipa, M. A. (2004). Design of bacterial vector systems for the production of recombinant proteins in Escherichia coli. J. Microbiol. Biotechnol. 14, 1–14.
Mergulhão, F. J. M., Monteiro, G. A., Larsson, G., Bostrom, M., Farewell, A., Nyström, T., et al. (2003). Evaluation of inducible promoters on the secretion of a ZZ-proinsulin fusion protein in Escherichia coli. Biotechnol. Appl. Biochem. 38, 87–93. doi: 10.1042/BA20030043
Mesa-Pereira, B., O'Connor, P. M., Rea, M. C., Cotter, P. D., Hill, C., and Ross, R. P. (2017). Controlled functional expression of the bacteriocins pediocin PA-1 and bactofencin A in Escherichia coli. Sci. Rep. 7:3069. doi: 10.1038/s41598-017-02868-w
Metelev, M., Serebryakov, M., Ghilarov, D., Zhao, Y., and Severinov, K. (2013). Structure of microcin B-like compounds produced by Pseudomonas syringae and species specificity of their antibacterial action. J. Bacteriol. 195, 4129–4137. doi: 10.1128/JB.00665-13
Miller, K. W., Konen, P. L., Olson, J., and Ratanavanich, K. M. (1993). Membrane protein topology determination by proteolysis of maltose binding protein fusions. Anal. Biochem. 215, 118–128. doi: 10.1006/abio.1993.1563
Miller, K. W., Schamber, R., Chen, Y., and Ray, B. (1998). Production of active chimeric pediocin AcH in Escherichia coli in the absence of processing and secretion genes from the Pediococcus pap operon. Appl. Environ. Microbiol. 64, 14–20.
Moon, G. S., Pyun, Y. R., and Kim, W. J. (2005). Characterization of the pediocin operon of Pediococcus acidilactici K10 and expression of his-tagged recombinant pediocin PA-1 in Escherichia coli. J. Microbiol. Biotechnol. 15, 403–411.
Moon, G. S., Pyun, Y. R., and Kim, W. J. (2006). Expression and purification of a fusion-typed pediocin PA-1 in Escherichia coli and recovery of biologically active pediocin PA-1. Int. J. Food Microbiol. 108, 136–140. doi: 10.1016/j.ijfoodmicro.2005.10.019
Mustopa, A. Z., Kusdianawati Fatimah, Umami, R. N., Budiarto, R. B., and Danuri, H. (2016). Cloning and expression of plantaricin E and F genes of Lactobacillus Plantarum S34 isolated from Indonesia traditional-fermented meat (Bekasam). Int. Food Res. J. 23, 762–769.
Nagao, J. I., Harada, Y., Shioya, K., Aso, Y., Zendo, T., Nakayama, J., et al. (2005). Lanthionine introduction into nukacin ISK-1 prepeptide by co-expression with modification enzyme NukM in Escherichia coli. Biochem. Biophys. Res. Commun. 336, 507–513. doi: 10.1016/j.bbrc.2005.08.125
Nes, I. F., Diep, D. B., Håvarstein, L. S., Brurberg, M. B., Eijsink, V., and Holo, H. (1996). Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Van Leeuwenhoek 70, 113–128.
Olejnik-Schmidt, A. K., Schmidt, M. T., Sip, A., Szablewski, T., and Grajek, W. (2014). Expression of bacteriocin divercin AS7 in Escherichia coli and its functional analysis. Ann. Microbiol. 64, 1197–1202. doi: 10.1007/s13213-013-0759-x
Ongey, E. L., and Neubauer, P. (2016). Lanthipeptides: chemical synthesis versus in vivo biosynthesis as tools for pharmaceutical production. Microb. Cell Fact. 15:97. doi: 10.1186/s12934-016-0502-y
Pal, G., and Srivastava, S. (2014). Cloning and heterologous expression of plnE, -F, -J and -K genes derived from soil metagenome and purification of active plantaricin peptides. Appl. Microbiol. Biotechnol. 98, 1441–1447. doi: 10.1007/s00253-013-5097-1
Pal, G., and Srivastava, S. (2015a). In vitro activity of a recombinant ABC transporter protein in the processing of plantaricin E pre-peptide. Arch. Microbiol. 197, 843–849. doi: 10.1007/s00203-015-1120-5
Pal, G., and Srivastava, S. (2015b). Scaling up the production of recombinant antimicrobial plantaricin E from a Heterologous Host, Escherichia coli. Probiotics Antimicrob. Proteins 7, 216–221. doi: 10.1007/s12602-015-9193-7
Peng, L., Xu, Z., Fang, X., Wang, F., and Cen, P. (2004). High-level expression of soluble human β-defensin-2 in Escherichia coli. Process Biochem. 39, 2199–2205. doi: 10.1016/j.procbio.2003.11.011
Quadri, L. E. N., Yan, L. Z., Stiles, M. E., and Vederas, J. C. (1997). Effect of amino acid substitutions on the activity of carnobacteriocin B2. Overproduction of the antimicrobial peptide, its engineered variants, and its precursor in Escherichia coli. J. Biol. Chem. 272, 3384–3388. doi: 10.1074/jbc.272.6.3384
Richard, C., Drider, D., Elmorjani, K., Marion, D., and Prévost, H. (2004). Heterologous expression and purification of active divercin V41, a class IIa bacteriocin encoded by a synthetic gene in Escherichia coli. J. Bacteriol. 186, 4276–4284. doi: 10.1128/JB.186.13.4276-4284.2004
Rodríguez, J. M., Martínez, M. I., Horn, N., and Dodd, H. M. (2003). Heterologous production of bacteriocins by lactic acid bacteria. Int. J. Food Microbiol. 80, 101–116. doi: 10.1016/S0168-1605(02)00153-8
Rogne, P., Fimland, G., Nissen-Meyer, J., and Kristiansen, P. E. (2008). Three-dimensional structure of the two peptides that constitute the two-peptide bacteriocin lactococcin G. Biochim. Biophys. Acta 1784, 543–554. doi: 10.1016/j.bbapap.2007.12.002
Rogne, P., Haugen, C., Fimland, G., Nissen-Meyer, J., and Kristiansen, P. E. (2009). Three-dimensional structure of the two-peptide bacteriocin plantaricin JK. Peptides 30, 1613–1621. doi: 10.1016/j.peptides.2009.06.010
Rosano, G. L., and Ceccarelli, E. A. (2014). Recombinant protein expression in Escherichia coli: advances and challenges. Front. Microbiol. 5:172. doi: 10.3389/fmicb.2014.00172
Rosenberg, H. F. (1998). Isolation of recombinant secretory proteins by limited induction and quantitative harvest. BioTechniques 24, 188–192.
Sambrook, J., and Russell, W. D. (2001). Molecular Cloning: A Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory Press.
Sánchez, J., Diep, D. B., Herranz, C., Nes, I. F., Cintas, L. M., and Hernández, P. E. (2007). Amino acid and nucleotide sequence, adjacent genes, and heterologous expression of hiracin JM79, a sec-dependent bacteriocin produced by Enterococcus hirae DCH5, isolated from Mallard ducks (Anas platyrhynchos). FEMS Microbiol. Lett. 270, 227–236. doi: 10.1111/j.1574-6968.2007.00673.x
Sandiford, S., and Upton, M. (2012). Identification, characterization, and recombinant expression of epidermicin NI01, a novel unmodified bacteriocin produced by Staphylococcus epidermidis that displays potent activity against staphylococci. Antimicrob. Agents Chemother. 56, 1539–1547. doi: 10.1128/AAC.05397-11
Sektas, M., and Szybalski, W. (2002). Novel single-copy pETcoco™ vector with dual controls for amplification and expression. Innovations 14, 6.
Shanks, R. M. Q., Dashiff, A., Alster, J. S., and Kadouri, D. E. (2012). Isolation and identification of a bacteriocin with antibacterial and antibiofilm activity from Citrobacter freundii. Arch. Microbiol. 194, 575–587. doi: 10.1007/s00203-012-0793-2
Shanks, R. M. Q., Kadouri, D. E., MacEachran, D. P., and O'Toole, G. A. (2009). New yeast recombineering tools for bacteria. Plasmid 62, 88–97. doi: 10.1016/j.plasmid.2009.05.002
Shi, Y., Bueno, A., and van der Donk, W. A. (2012). Heterologous production of the lantibiotic Ala(0)actagardine in Escherichia coli. Chem. Commun. 48:10966. doi: 10.1039/c2cc36336d
Shi, Y., Yang, X., Garg, N., and Van Der Donk, W. A. (2011). Production of lantipeptides in Escherichia coli. J. Am. Chem. Soc. 133, 2338–2341. doi: 10.1021/ja109044r
Svetoch, E. A., and Stern, N. J. (2010). Bacteriocins to control Campylobacter spp. in poultry-A review. Poult. Sci. 89, 1763–1768. doi: 10.3382/ps.2010-00659
Tabor, S., and Richardson, C. C. (1985). A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. U.S.A. 82, 1074–1078. doi: 10.1073/pnas.82.4.1074
Tang, X., Wu, S., Wang, X., Gu, Q., and Li, P. (2018). Antimicrobial activity and preliminary mode of action of PlnEF expressed in Escherichia coli against Staphylococci. Protein Expr. Purif. 143, 28–33. doi: 10.1016/j.pep.2017.10.008
Terpe, K. (2006). Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 72, 211–222. doi: 10.1007/s00253-006-0465-8
Tolia, N. H., and Joshua-Tor, L. (2006). Strategies for protein coexpression in Escherichia coli. Nat. Methods 3, 55–64. doi: 10.1038/nmeth0106-55
Van Belkum, M. J., Worobo, R. W., and Stiles, M. E. (1997). Double-glycine-type leader peptides direct secretion of bacteriocins by ABC transporters: colicin V secretion in Lactococcus lactis. Mol. Microbiol. 23, 1293–1301. doi: 10.1046/j.1365-2958.1997.3111677.x
van de Guchte, M., van der Vossen, J. M., Kok, J., and Venema, G. (1989). Construction of a lactococcal expression vector: expression of hen egg white lysozyme in Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 55, 224–228.
Verdon, J., Girardin, N., Marchand, A., Héchard, Y., and Berjeaud, J. M. (2013). Purification and antibacterial activity of recombinant warnericin RK expressed in Escherichia coli. Appl. Microbiol. Biotechnol. 97, 5401–5412. doi: 10.1007/s00253-012-4417-1
Wang, J., Gao, Y., Teng, K., Zhang, J., Sun, S., and Zhong, J. (2014). Restoration of bioactive lantibiotic suicin from a remnant lan locus of pathogenic Streptococcus suis serotype 2. Appl. Environ. Microbiol. 80, 1062–1071. doi: 10.1128/AEM.03213-13
Wang, J., Ge, X., Zhang, L., Teng, K., and Zhong, J. (2016). One-pot synthesis of class II lanthipeptide bovicin HJ50 via an engineered lanthipeptide synthetase. Sci. Rep. 6:38630. doi: 10.1038/srep38630
Wang, Q., Fu, W., Ma, Q., Yu, Z., and Zhang, R. (2013). Production of bacteriocin E50-52 by small ubiquitin-related modifier fusion in Escherichia coli. Polish J. Microbiol. 62, 345–350.
Yildirim, S., Konrad, D., Calvez, S., Drider, D., Prévost, H., and Lacroix, C. (2007). Production of recombinant bacteriocin divercin V41 by high cell density Escherichia coli batch and fed-batch cultures. Appl. Microbiol. Biotechnol. 77, 525–531. doi: 10.1007/s00253-007-1188-1
Keywords: bacteriocins, heterologous expression, E. coli, vectors, strains
Citation: Mesa-Pereira B, Rea MC, Cotter PD, Hill C and Ross RP (2018) Heterologous Expression of Biopreservative Bacteriocins With a View to Low Cost Production. Front. Microbiol. 9:1654. doi: 10.3389/fmicb.2018.01654
Received: 27 March 2018; Accepted: 02 July 2018;
Published: 26 July 2018.
Edited by:
Fliss Ismail, Laval University, CanadaReviewed by:
Giuseppe Spano, University of Foggia, ItalyJosé María Landete, Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA), Spain
Copyright © 2018 Mesa-Pereira, Rea, Cotter, Hill and Ross. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: R. Paul Ross, cC5yb3NzQHVjYy5pZQ==
 Beatriz Mesa-Pereira
Beatriz Mesa-Pereira Mary C. Rea
Mary C. Rea Paul D. Cotter1,2
Paul D. Cotter1,2 Colin Hill
Colin Hill R. Paul Ross
R. Paul Ross