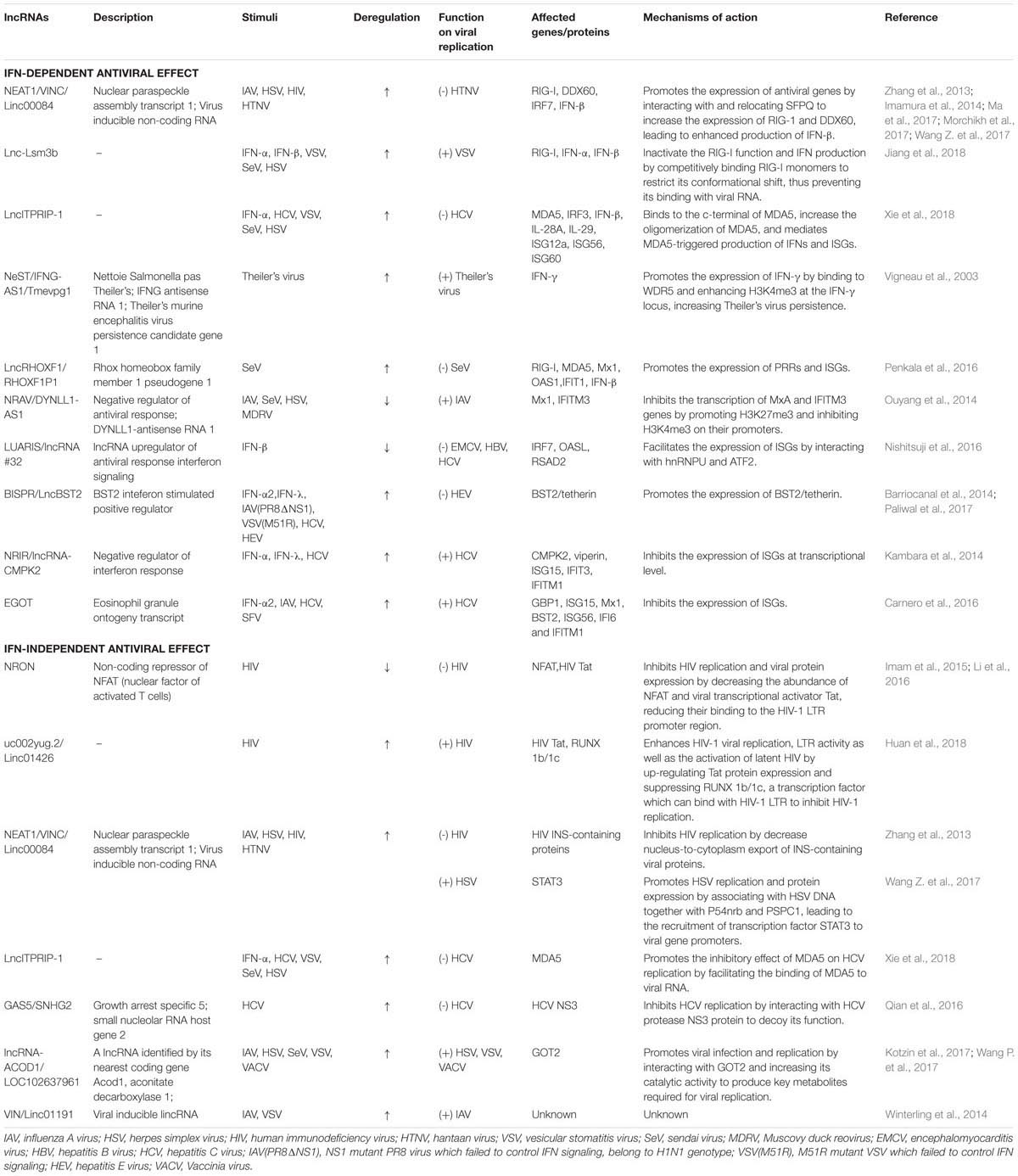
- Institute of Life Sciences, Jiangsu University, Zhenjiang, China
Interferons (IFNs) are a family of cytokines providing a robust first line of host innate defense against pathogenic infection, and have now been part of the standard treatment for viral infection. However, IFN based therapy can best be described as modestly effective. Long non-coding RNAs (lncRNAs) are a novel class of non-protein-coding RNAs that are capable of regulating gene expression at different levels, including chromatin, transcription, post-transcription, and translation. Recently, lncRNAs are found to be deregulated upon viral infection or IFN treatment, and some of them can modulate viral infection in an IFN-dependent or -independent manner. Due to the crucial roles of lncRNAs in viral infection and the IFN antiviral response, the modulation of specific lncRNAs may be involved to increase the IFN antiviral response and improve the clinical result of IFN-based therapy. In this review, we summarize lncRNAs that are deregulated by viral infection, with special focus on the functions and underlying mechanisms of some essential lncRNAs, and discuss their roles in viral infection and the antiviral response of IFN.
Introduction
Interferons (IFNs) are a family of cytokines providing a robust first line of host innate defense against pathogenic infection. Upon viral infection, IFNs are actively transcribed, which then induces the expression of various interferon stimulated genes (ISGs), establishing an antiviral state in the target cells (Borden et al., 2007). Currently, IFNs are attractive therapeutic options to control chronic virus infections. They are classified into three types: type I (IFN-α,-β, -𝜀, -κ, and -ω), II (IFN-γ), and III (IFN-λ1/IL-29, -λ2/IL-28A, -λ3/IL-28B). Type I IFNs, predominantly interferon-α (IFN-α) and IFN-β, have been part of the standard treatment for hepatitis B virus (HBV) and hepatitis C virus (HCV) infection, and play important roles in the initial stages of viral infection (Borden et al., 2007; Lin and Young, 2014). However, IFN based therapy can best be described as modestly effective. In a study of HBV infection, IFN based therapy only reached 33% HBV e Antigen (HBeAg) seroconversion [from HBeAg to HBV e antibody (anti-HBeAg)], with 25% of HBeAg positive patients achieving undetectable HBV DNA (Yuen and Lai, 2011). Therefore, great effort is needed to improve the clinical result of IFN-based therapy.
Long non-coding RNAs (lncRNAs), a novel class of non-protein-coding RNAs exceeding 200 nucleotides in length, are capable of regulating gene expression at different levels, including chromatin, transcription, post-transcription, and translation, and thus likely to be involved in innate immunity and viral replication (Imamura et al., 2014; Ouyang et al., 2014, 2016; Fortes and Morris, 2016; Valadkhan and Gunawardane, 2016). Recent studies demonstrated that, in response to viral infection or IFN, many lncRNAs were deregulated, and some of them impact on viral replication in an IFN-dependent or -independent manner; some viruses may hijack host lncRNAs to facilitate their replication and latency (Li et al., 2016; Ma et al., 2017; Wang P. et al., 2017). Due to the crucial roles of lncRNAs in viral infection and the IFN antiviral response, the modulation of specific lncRNAs may be involved to increase the antiviral response of IFN and improve the clinical result of IFN-based therapy. In this review, we summarize lncRNAs that are deregulated by viral infection and IFN, with special focus on the functions and underlying mechanisms of some essential lncRNAs, and discuss their roles in viral infection and the antiviral response of IFN.
IFN Induction
Upon viral infection, viral features or pathogen-associated molecular patterns (PAMPs) are sensed by pattern recognition receptors (PRRs), such as retinoid acid inducible gene I (RIG)-I-like receptors (RLRs), Toll-like receptors (TLRs), and cyclic guanosine-monophosphate adenosine-monophosphate (cGAMP) synthase (cGAS). Subsequently, adaptor proteins, such as mitochondrial activator of virus signaling [MAVS, IFN-β promoter stimulator 1 (IPS-1), or Cardif], Toll-IL-1 receptor (TIR)-domain-containing adaptor inducing IFN-β (TRIF), myeloid differentiation primary response 88 (MyD88), and stimulator of IFN genes (STING), are activated, which then phosphorylate kinases NF-κB activator (TANK)-binding kinase-1 (TBK1) and inhibitor of κB (IκB) kinase (IKK) 𝜀, or the IKKα and IKKβ kinases, leading to the activation of interferon response factors (IRF) 3/7 or NF-κB. Phosphorylated dimers of IRF3/7 or NF-κB then translocate to the nucleus, bind to the promoters of target genes, and trigger the expression of IFNs (Figure 1; Schneider et al., 2014; Makris et al., 2017).
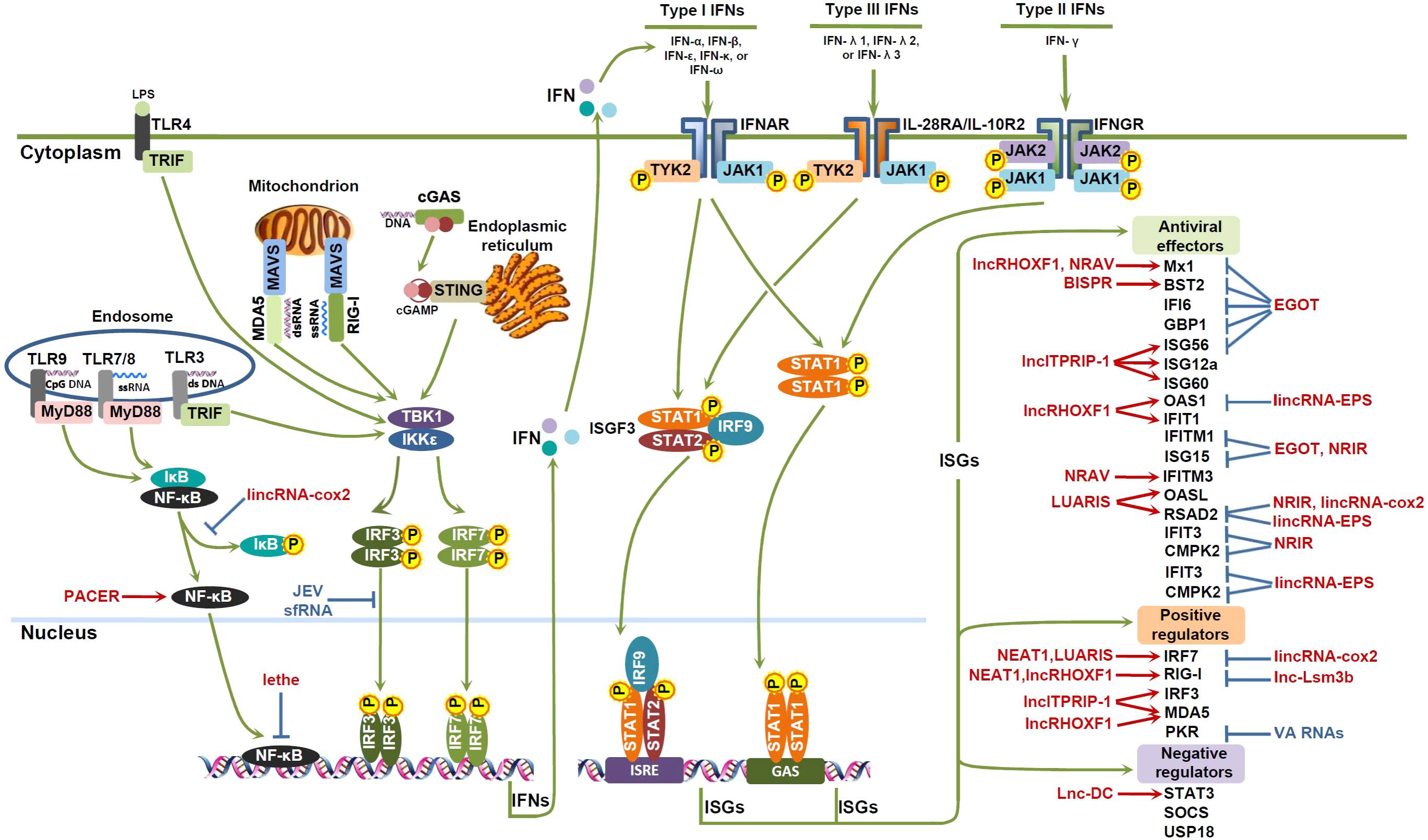
FIGURE 1. lncRNAs involved in the IFN antiviral response. Viral DNAs or RNAs are recognized by pattern recognition receptors (PRRs), such as retinoid acid inducible gene I (RIG)-I-like receptors (RLRs), Toll-like receptors (TLRs), and cyclic guanosine-monophosphate adenosine-monophosphate (cGAMP) synthase (cGAS). Subsequently, adaptor proteins, such as mitochondrial activator of virus signaling (MAVS), Toll-IL-1 receptor (TIR)-domain-containing adaptor inducing IFN-β (TRIF), myeloid differentiation primary response 88 (MyD88), and stimulator of IFN genes (STING), are activated, leading to the activation of interferon response factors (IRF) 3/7 or NF-κB. Phosphorylated dimers of IRF3/7 or NF-κB then translocate to the nucleus, bind to the promoters of target genes, and trigger the expression of IFNs. IFNs exerts their antiviral effects by binding to their corresponding receptors, and signaling through Janus kinase (JAK)-signal transducer and activator of transcription (STAT) pathway to induce the expression of IFN-stimulated genes (ISGs), which can function as antiviral effectors, positive regulators or negative regulators of the IFN pathway. Some host lncRNAs (red color) and viral lncRNAs (blue color) can modulate the activities of transcription factors, or the expression of ISGs, and thus impact on the IFN antiviral response.
RLR family consists of three members, RIG-I (also known as DDX58), melanoma differentiation-associated gene 5 (MDA5, IFIH1), and laboratory of genetics and physiology 2 (LGP2), all of which are expressed in cytoplasm and specifically recognize viral RNA (Bruns and Horvath, 2015). Although they share similar structures, LGP2 lacks caspase activation and recruitment domain (CARD), and could not directly initiate downstream signaling like RIG-I and MDA5, while it was shown to participate in MDA5-mediated signaling (Bruns and Horvath, 2015). Upon binding of viral RNA, both of RIG-I and MDA5 were transported to the mitochondria, where they interact with the CARD of an adaptor protein MAVS and trigger the expression of type I IFNs (Liu S. et al., 2015). Viral nucleic acids can also be recognized by endosomal TLRs, including TLR-3 (ds RNA), TLR7/8 (ssRNA), and TLR-9 (unmethylated CpG DNA) (Lester and Li, 2014). Whist TLR3 interacts with TRIF and initiates TRIF-dependent signaling cascade, TLR7/8 and TLR-9 activate MyD88-dependent pathways (Lester and Li, 2014; Makris et al., 2017). Another TLR, cell surface expressed TLR4, may sense lipopolysaccharides (LPS) and induce the production of IFN-β through TRIF-mediated signaling (Fitzgerald et al., 2003). cGAS recognizes viral DNAs, and catalyzes the synthesis of the second messenger cGAMP, which binds to endoplasmic-reticulum-resident protein STING, resulting in the activation of IRF3 and induction of type I IFN (Ablasser et al., 2013; Sun et al., 2013; Wu et al., 2013). Other potential DNA sensors include DNA-dependent protein kinase (DNA-PK), IFN-γ-inducible protein 16 (IFI16, also known as AIM2, absent in melanoma 2), DNA-dependent activator of IFN-regulatory factors (DAI), and DExD/H-box helicase (DDX) 41, etc. (Mansur et al., 2014).
IFN Signaling
Generally, IFNs may bind to their receptors, and exert antiviral effects in an autocrine/paracrine manner by signaling through Janus kinase (JAK)-signal transducer and activator of transcription (STAT) pathway to induce the expression of ISGs, establishing an antiviral state in the target cells (Figure 1; Borden et al., 2007). Except for the JAK-STAT pathway, IFNs also function by activating other STAT-independent pathways, such as mitogen-activated protein kinases (MAPKs) p38 and extracellular signal regulated kinases (ERKs), as well as the phosphatidylinositol 3-kinase (PI3K) pathway (Wang et al., 2004; Platanias, 2005).
Type I IFNs bind to a heterodimeric transmembrane receptor consisting of IFN-α receptor 1/2 (IFNAR1/IFNAR2), which then activating IFNAR1/IFNAR2-associated tyrosine kinase 2 (TYK2) and JAK1, resulting in the phosphorylation of STAT1 and STAT2. STAT1/STAT2, together with IRF9, forms the IFN-stimulated gene factor 3 (ISGF3), which translocates to the nucleus and binds to IFN-stimulated response elements (ISREs) in the promoters of ISGs to initiate their transcription. In addition to ISGF3, type I IFN also induce the formation of STAT1 homodimers, which directly translocate to the nucleus without assembly with IRF9, then bind to gamma-activated sequences (GASs) in the promoter of ISGs to stimulate gene transcription (Schneider et al., 2014). Type II IFNs bind to IFN-γ receptors 1/2 (IFNGR1/IFNGR2), which then activates their associated tyrosine kinases JAK1 and JAK2, leading to the phosphorylation of only STAT1 (Schroder et al., 2004). Phosphorylated STAT1 can form homodimers, which then translocate to the nucleus and bind to GAS in the promoter of ISGs to promote their transcription (Schroder et al., 2004). Type III IFNs, in contrast to type I IFNs, bind to a distinct receptor complex composed of IL-28RA and IL-10R2, while it triggers the same JAK-STAT signaling transduction cascades to activate gene transcription (Lopusna et al., 2013).
Although binding to different receptors, three types of IFNs signaling through similar JAK-STAT pathway. Types I and III IFNs activate the transcription of overlapping ISGs, while type II IFNs induces overlapping but distinct set of ISGs (Hertzog et al., 2011; Lopusna et al., 2013). These ISGs may function as antiviral factors, positive regulators or negative regulators of the IFN pathway (Figure 1). Antiviral factors impact on the different stages of the viral cycle. Among them, Myxovirus resistance (Mx) family of guanosine triphosphate (GTP)ases, IFN-inducible transmembrane (IFITM) and the tripartite motif (TRIM) family of proteins influence viral entry; IFN-induced protein with tetratricopeptide repeats (IFIT) family, the 2′–5′ oligoadenylate synthetase (OAS)-directed RNaseL pathway family proteins, protein kinase R (PKR), guanylate-binding proteins (GBPs), ISG15, ISG12a (IFI27), and G1P3 (IFI6, or 6–16) impact on viral replication, transcription, and translation; while radical s-adenosyl methionine domain-containing protein 2 (RSAD2, viperin) and bone marrow stromal cell antigen (BST)-2 (also known as tetherin) regulate viral assembly and release (Zhu and Liu, 2003; Zhu et al., 2003; Cheriyath et al., 2011; Sun et al., 2013; Liu et al., 2014; Yang et al., 2014; Xue et al., 2016; Chen et al., 2017). Positive regulators of IFN pathway include PRRs and signal transducing proteins, such as IRF3/7/9 and STAT1/2, and can be enhanced by IFN and reinforce the IFN response (Schroder et al., 2004). However, negative regulators, such as suppressor of cytokine signaling (SOCS), ubiquitin specific peptidase (USP) 18 and STAT3, may help IFN-induced cells to return to cellular homeostasis (Schroder et al., 2004; Wang et al., 2011).
Deregulation of lncRNA Upon Viral Infection
Using lncRNA array and next-generation sequencing, aberrantly expressed lncRNAs in viral infection have been progressively unveiled. Various viruses, including influenza A virus (IAV), HCV, human immunodeficiency virus (HIV), herpes simplex virus (HSV), etc. have been reported to modulate the expression of multiple lncRNAs in host cells. One specific virus can result in the deregulation of multiple lncRNAs. For example, the infection of wild type or mutant IAV lacking NS1 (PR8ΔNS1, which failed to control IFN in infected cells) induces the expression of lncRNA EGOT (eosinophil granule ontogeny transcript), NEAT1 (Nuclear paraspeckle assembly transcript 1), BISPR (BST2 IFN-stimulated positive regulator, also known as LncBST2), VIN (virus inducible lincRNA), ISR (IFN-stimulated lncRNA)2 and ISR8, while it inhibits the expression of lncRNA NRAV (negative regulator of antiviral response, also known as DYNLL1AS1) (Barriocanal et al., 2014; Carnero et al., 2014, 2016; Imamura et al., 2014; Ouyang et al., 2014; Winterling et al., 2014). HCV infection enhances the expression of lncRNA BISPR, NRIR (negative regulator of interferon response), EGOT, ISR2, ISR8, GAS5 (growth arrest-specific 5), and lncITPRIP-1, etc. (Barriocanal et al., 2014; Carnero et al., 2014, 2016; Kambara et al., 2014; Qian et al., 2016; Xie et al., 2018). HIV infection promotes the expression of ISR2 and NEAT1, while it inhibits the expression of NRON [non-coding repressor of Nuclear Factor of Activated T cells (NFAT)] (Zhang et al., 2013; Carnero et al., 2014; Imam et al., 2015). Likely, one specific lncRNA can also be deregulated by the infection of different viruses. For instance, NEAT1 was reported to be up-regulated by IAV, HIV, HSV, and Hantaan virus (HTNV); ACOD1, a lncRNA identified by its nearest coding gene aconitate decarboxylase 1 (Acod1), can be increased by IAV, HSV and vascular stomatitis virus (VSV); while NRAV can be inhibited by the infection of IAV, HSV and Sendai virus (SeV) (Zhang et al., 2013; Imamura et al., 2014; Ouyang et al., 2014; Ma et al., 2017; Wang P. et al., 2017; Wang Z. et al., 2017). These results indicate that lncRNAs were differentially expressed in viral infection, and may be involved in viral replication and pathogenesis.
Host lncRNAs as Pivotal Regulators of IFN Antiviral Response
Host lncRNAs Modulate IFN Induction
NEAT1, an essential component for paraspeckle formation, may interact with mammalian Drosophila melanogaster behavior and human splicing (DBHS) proteins, including splicing factor proline- and glutamine-rich (SFPQ) protein, paraspeckle component 1 (PSPC1), and a non-POU domain containing, octamer-binding NONO/p54nrb protein (Naganuma and Hirose, 2013). Recently, it was reported to be involved in cancer progression and the infection of several viruses, including HTNV, HSV-1, IAV, HIV, and KSHV (Zhang et al., 2013; Imamura et al., 2014; Idogawa et al., 2017; Ma et al., 2017; Wang Y. et al., 2017; Wang Z. et al., 2017; Chen et al., 2018). Among these viruses, NEAT1 modulates HTNV through impacting on IFN induction (Ma et al., 2017). In HTNV infected human umbilical vein endothelial cells (HUVECs), silencing NEAT1 by siRNA significantly accelerated, while over-expression of NEAT1 by NEAT1-expressing plasmid effectively inhibited, HTNV replication, viral-specific mRNA and nucleocapsid protein (NP) expression (Ma et al., 2017). Results from in vivo studies demonstrated similar inhibitory effect of NEAT1 on HTNV virus titers and NP expression, and also found decreased IFN-β production in serum and remarkably reduced body weight (Ma et al., 2017). Further mechanism studies revealed that NEAT1 may promote IFN responses by acting as a positive feedback for RIG-I signaling (Ma et al., 2017). By interacting with and relocating SFPQ from the promoter regions of RIG-I and DDX60 to paraspeckles, NEAT1 removed the transcriptional inhibitory effects of SFPQ on RIG-I and DDX60, resulting in increased expression of transcriptional factor IRF7, which in turn induced the expression of IFN and NEAT1 (Ma et al., 2017). Except for this, NEAT1 also forms a multi-subunit complex with HEXIM1, which may interact with cGAS sensor and its partner polyglutamine binding protein 1 (PQBP1), resulting in the release of paraspeckle proteins, recruitment of STING, and activation of IRF3, initiating the activation pathway leading to the production of type 1 IFN (Morchikh et al., 2017). These results indicate that NEAT1 has a critical role in the antiviral response of IFN through RIG-I signaling and cGAS-STING-IRF3 pathway.
Most recently, lnc-Lsm3b and lncITPRIP-1, two IFN-inducible lncRNAs, were reported to impact on viral infection and IFN production by modulating PRRs RIG-I and MDA5, respectively (Jiang et al., 2018; Xie et al., 2018). Lnc-Lsm3b, which is transcribed from Lsm3 loci, could be up-regulated upon the infection of RNA viruses, such as VSV or SeV, as well as DNA virus HSV-1, and could also be stimulated by high concentration of type I IFNs and various TLR ligands (Jiang et al., 2018). Knocking down of lnc-Lsm3b using siRNA significantly increased the production of IFN-α and -β, resulting in decreased replication of VSV (Jiang et al., 2018). Mechanism studies revealed that in viral infected cells, lnc-Lsm3b may competitively bind with RIG-I monomer to restrict its conformational shift, thus preventing its binding with viral RNA, resulting the inactivation of RIG-I downstream signaling and termination of IFNs production (Jiang et al., 2018). By contrast, lncITPRIP-1, the genomic loci of which is near to inositol 1,4,5-trisphosphate receptor interacting protein (ITPRIP, also known as DNACR), was reported to be induced by multiple viruses, including HCV, VSV, SeV, and HSV, while over-expression of this lncRNA significantly inhibited HCV replication (Xie et al., 2018). Further studies revealed that lncITPRIP-1 can bind to the c-terminal of MDA5, increase the oligomerization of MDA5, and mediate MDA5-triggered production of IFNs (IFN-β, IL-28A, and IL-29) and ISGs (ISG12a, ISG56, and ISG60), leading to the suppression of HCV replication (Xie et al., 2018).
NeST (IFNG-AS1, or Tmevpg1), the genomic loci of which is located adjacent to the IFN-γ gene in both humans and mice, was reported to promote IFN-γ expression, and is associated with host’s persistence from Theiler’s virus (Vigneau et al., 2003; Gomez et al., 2013). Mechanism study revealed that NeST can bind to WD repeat-containing protein 5 (WDR5), a component of histone H3 lysine 4 (H3K4) methyltransferase complexes, resulting in enhanced H3K4 trimethylation (H3K4me3) at the IFN-γ locus, and leading to enhanced transcription of IFN-γ (Gomez et al., 2013).
Another lncRNA, lncRHOXF1, is specifically expressed in human trophectoderm cells. It was found to be significantly increased by the infection of SeV, while knocking down of this lncRNA greatly reduced the production of viral mRNAs and decreased expression of ISGs Mx1 and IFIH (Penkala et al., 2016). Further studies demonstrated that in human trophectoderm progenitor cells, depleting lncRHOXF1 using siRNAs resulted in significant increase of proteins in IFN induction pathway and ISGs, such as PRR MDA5 and RIG-I, IFN-β, as well as ISGs Mx1, OAS1, and IFIT1 (Penkala et al., 2016). These results indicate that lncRHOXF1 play a pivotal role in controlling SeV infection by modulating the IFN antiviral response.
Host lncRNAs Interfere With ISGs Expression
NRAV, located on chromosome 12q24, was primarily found to be down-regulated in IAV-infected A549 cells, and could significantly affect the replication of IAV (Ouyang et al., 2014). Over-expression of NRAV by retroviral vectors significantly enhanced, whereas knocking down of this lncRNA by shRNA-based lentivectors greatly reduced, IAV replication (Ouyang et al., 2014). In in vivo studies, transgenic mice expressing human NRAV demonstrated greater replication of IAV and more severe inflammation in lungs compared with wide-type mice (Ouyang et al., 2014). By cDNA array screening and qRT-PCR validation, differentially expressed genes in NRAV over-expressing A549 cells infected with IAV were detected. The results showed that the expression of several critical ISGs, including IFIT2, IFIT3, IFITM3, OASL, and Mx1, were greatly reduced in NRAV over-expressing cells; while the expression of these ISGs were significantly increased in NRAV knocking down cells (Ouyang et al., 2014). Similar results were obtained in IAV infected NRAV transgenic mice (Ouyang et al., 2014). However, forced expression of these antiviral ISGs in NRAV over-expressing cells greatly reversed the effect of NRAV on the replication of IAV, suggesting that NRAV may function by regulating the expression of these ISGs during viral infection (Ouyang et al., 2014). Further mechanism studies revealed that NRAV over-expression significantly increased transcription activation marker histone 3 lysine 27 trimethylation (H3K27me3) enrichment at mxA gene locus, whereas NRAV knockdown greatly promoted transcription inhibition marker H3K4me3 enrichment and inhibited H3K27me3 enrichments at mxA and ifitm3 transcription start sites (Ouyang et al., 2014). These data suggest that NRAV promotes viral replication by inhibiting the expression of ISGs such as MxA and IFITM3 via promoting H3K27me3 and reducing of H3K4me3.
LUARIS (lncRNA upregulator of antiviral response interferon signaling), also known as lncRNA #32, is an IFN down-regulated lncRNA widely expressed in various human tissues, and has been reported to inhibit the replication of EMCV, HBV, and HCV (Nishitsuji et al., 2016). In immortalized human hepatocytes (HuS cells), over-expressing of LUARIS greatly inhibited EMCV levels, while silencing of this lncRNA significantly enhanced viral titers (Nishitsuji et al., 2016). Results from cDNA microarray demonstrated that, at the absence of LUARIS, treatment of IFN-β in HuS cells significantly reduced the expression of many known ISGs and chemokines, including IRF7, OASL (2′–5′-oligoadenylate synthetase-like protein), RSAD2, CCL5 (Chemokine C-C motif ligand 5), CXCL (C-X-C motif chemokine ligand) 11/ITAC (interferon gamma-inducible T-cell alpha chemoattractant), and CXCL10/IP-10; while in LUARIS over-expressing cells, many of them were greatly increased (Nishitsuji et al., 2016). Similar results were found for anti-HBV and anti-HCV activities of IFN-β in human primary hepatocytes, indicating the inhibitory role of LUARIS on viral replication may associate with its regulation of ISGs and chemokines (Nishitsuji et al., 2016). Mechanism studies revealed that the function of LUARIS may associated with heterogeneous nuclear ribonucleoprotein (hnRNP) U and activating transcription factor 2 (ATF2) (Nishitsuji et al., 2016). HnRNPU, a protein previously reported to inhibit the replication of RNA viruses, was found to improve the stabilization of LUARIS, and thus impacted on the expression of IP-10 and RSAD2 (Pichlmair et al., 2012; Nishitsuji et al., 2016). In contrast, ATF2 may bind to the promoter region of ISGs and regulate their expression with the help of LUARIS (Nishitsuji et al., 2016). These data implied that LUARIS may be negatively regulated by IFN to prevent the possible inflammatory damage induced by excessive IFN response.
BISPR, the genomic loci of which near to BST2, was identified to be highly up-regulated by IFN-α2 or IFN-λ in a dose- and time-dependent manner, and can be significantly increased upon the infection of mutant viruses IAV (PR8ΔNS1) and VSVM51R, two of which failed to control IFN in infected cells (Garcia-Sastre et al., 1998; Terstegen et al., 2001; Garcia-Sastre, 2011; Barriocanal et al., 2014). Similar results were shown in HCV or HEV infected Huh7 cells and the liver of HCV-infected patients (Barriocanal et al., 2014; Paliwal et al., 2017). Bioinformatics analysis suggested that BISPR could share the same promoter with BST2 (Barriocanal et al., 2014). Furthermore, the expression of both BISPR and BST2 was STAT-dependent, and BISPR could positively regulate the expression of BST2 (Barriocanal et al., 2014). Based on the inhibitory effect of BST2 on virion secretion, BISPR may be involved in regulating viral infection partially by increasing the expression of antiviral protein BST2.
NRIR, also known as lncRNA-CMPK2, is located near the genomic loci ISG CMPK2 (cytodine/uridine monophosphate kinase 2) and RASD2 (Kambara et al., 2014). It was found to be stimulated by IFN-α or IFN-γ (Kambara et al., 2014). However, when the JAK-STAT pathway was suppressed by JAK inhibitor ruxolitinib or depleting of STAT2, the up-regulation of CMPK2 was abrogated, suggesting the direct stimulating role of IFN on its transcription (Kambara et al., 2014). Furthermore, in IFN-stimulated hepatocytes, knocking down of NRIR resulted in the transcriptional up-regulation of many ISGs, including CMPK2, RASD2, ISG15, CXCL10, IFIT3, and IFITM1, and also led to remarkable suppression of HCV replication (Kambara et al., 2014). Clinical results demonstrated that the expression of NRIR was up-regulated in liver tissues of chronic HCV infected patients which have active IFN response (Kambara et al., 2014). These data suggest that NRIR promotes HCV replication by negatively regulate the IFN antiviral response.
EGOT is another lncRNA that can be stimulated by IFN-α (Carnero et al., 2016). It was found to modulate the infection of different viruses, including HCV, IAV and Semliki forest virus (SFV), and also play important roles in several cancers, such as gastric cancer, glioma and renal cell carcinoma (Xu et al., 2015; Carnero et al., 2016; Jin et al., 2017; Peng et al., 2017; Wu et al., 2017). Upon HCV infection, viral RNA was sensed by RIG-I and PKR, then NF-κB was activated and bound to the promoter region of EGOT, leading to increased expression of EGOT (Carnero et al., 2016). In HCV-infected Huh7 cells, depletion of EGOT using gapmers greatly reduced HCV genomes, titer, core and NS3 proteins (Carnero et al., 2016). Similar results were also observed in SFV-infected Huh7 cells (Carnero et al., 2016). Mechanism studies revealed that in EGOT depleting cells with or without HCV infection, the expression of several ISGs, including GBP1, ISG15, Mx1, BST2, ISG56, IFI6, and IFITM1, was observed to be significantly increased, indicating that EGOT may promote viral replication by blocking the IFN antiviral response (Carnero et al., 2016).
Miscellaneous Host lncRNAs Participate in the IFN Antiviral Response
Four lncRNAs, including IFN-stimulated lncRNA (ISR) 2, 8, 12 and lncISG15, can be stimulated by IFN and different viruses, and their genomic loci are near to ISGs GBP1, IRF1, IL-6, and ISG15, respectively (Barriocanal et al., 2014; Carnero et al., 2014). Upon the treatment of IFN, the expression of ISR2 (GBP1 pseudogene 1), ISR8 (AC116366.6), and ISR12 (LOC100506178) was significantly stimulated at early (6–12 h, ISR2, and 8) or later times (48–72 h, ISR12), mimicking that of their neighboring genes GBP1, IRF1, and IL-6 (Barriocanal et al., 2014). Bioinformatics analysis showed that STAT1 and 2 as well as IRF1 and 2 may bind to the promoter of ISR8 and impact on its transcription, while lncISG15 may share the same promoter with ISG15 (Barriocanal et al., 2014; Carnero et al., 2014). Results from virus infected cells and clinical samples showed that both ISR2 and ISR8 can be greatly up-regulated by HCV infection, while lncISG15 can be significantly enhanced by the infection of mutant IAV (PR8ΔNS1) and VSV (M51R), indicating their possible roles in viral infection and IFN response (Barriocanal et al., 2014; Carnero et al., 2014).
Two lncRNAs, lincRNA-cox-2 and lincRNA-EPS, can be modulated by TLR ligands, and the deregulation of these two lncRNAs influence the expression of some ISGs. LincRNA-cox2, also known as Ptgs2os2 [prostaglandin-endoperoxide synthase (PTGS) 2, opposite strand 2], can be induced by TLR ligands in a MyD88 and NF-κB dependent manner, while the up-regulation of this lncRNA significantly reduced the expression of some ISGs, such as irf7 and Rasd2 (Carpenter et al., 2013). Mechanism studies suggested that lincRNA-Cox-2 may interact with HnRNP-A/B and hnRNP-A2/B1 to regulate gene expression (Carpenter et al., 2013). On the contrary, lincRNA-EPS (erythroid prosurvival), can be suppressed by TLR ligands, and the reduction of lincRNA-EPS significantly increased the expression of ISGs ifit2, Rasd2, Oas1 and gbp5 by recruiting hnRNPL (Atianand et al., 2016).
Several lncRNAs, including PACER, NKILA, and lethe, may modulate the activity of NF-κB (Rapicavoli et al., 2013; Krawczyk and Emerson, 2014; Liu B. et al., 2015). PACER (p50-associated Cox2 extragenic RNA), also known as PTGS2-AS1, is located near the genomic loci of COX-2 gene, and functions as a positive regulator of NF-κB by binding to the repressive NF-KB subunit p50 and promoting the formation of RelA-p50 heterodimer; NKILA (NF-κB-interacting lncRNA) inhibits NF-κB activity by binding to NF-κB/IκB and blocking the phosphorylation sites of IκB from IκB kinase (IKK) and thus preventing the degradation of IκB; while lethe suppress NF-κB activity by blocking the binding site of RelA to target genes and subsequent transcription (Rapicavoli et al., 2013; Krawczyk and Emerson, 2014; Liu B. et al., 2015).
Besides, lnc-DC (lnc-Dendritic cell lncRNA), also known as WFDC21P (WAP four-disulfide core domain 21, pseudogene), may impact on IFN response by modulating STAT3 (Wang et al., 2014). It can directly bind to STAT3 and prevent its dephosphorylation by SHP1, and thus activate STAT3-dependent transcription. In another report, lncRNA lethe (after the “river of forgetfulness” in Greek mythology) is reported to be stimulated by STAT3, and promote HCV infection and inhibit the expression of some ISGs, including PKR, OAS, and IRF1 (Xiong et al., 2015). However, in this report, incorrect primers for lncRNA lethe were used, so it is still arguable whether this lncRNA play a role in HCV infection.
Virus-Encoded lncRNAs Impact on the IFN Antiviral Response
To counteract the IFN antiviral response, viruses have evolved different strategies to minimize IFN production and the activation of IFN signaling (Devasthanam, 2014; Chan and Gack, 2016; Schulz and Mossman, 2016). Interestingly, recent findings found that virus encoded lncRNAs may also participate in the antiviral response of IFN. For example, PAN RNA (polyadenylated nuclear RNA) from KSHV interacts with several virus- and host-encoded factors, including IRF4. During the lytic phase of KSHV infection, the expression of PAN RNA reduces the expression of IFNα, IFNγ, and ISG RNaseL; sfRNAs (subgenomic flavivirus RNAs) from several viruses, including Japanese encephalitis virus (JEV), dengue virus, and West Nile Virus (WNV), may antagonize the antiviral response of IFN by inhibiting the IFN signaling, the expression of IFN-β or specific ISGs; adenovirus virus-associated RNA (VA) target the ISG PKR to regulate the expression of ISGs (Rossetto and Pari, 2011; Schuessler et al., 2012; Chang et al., 2013; Bidet et al., 2014; Kondo et al., 2014). Interestingly, a chimeric lncRNA HBx-LINE1, which is generated by the integration of HBV into host cell genome, may attenuate the IFN antiviral response by inhibiting microRNA (miRNA)-122, a negative regulator of HBV replication which can be suppressed by IFN (Qiu et al., 2010; Chen et al., 2011; Wang et al., 2012; Hao et al., 2013; Lau et al., 2014).
Host lncRNAs Participate in IFN-Independent Antiviral Response
Except for functioning IFN antiviral response by regulating IFN induction and ISG expression, lncRNAs can also regulate viral infection and replication in an IFN-independent manner, impacting on the transcription of viral genes, the translocation of viral transcripts, the function of viral proteins, even host cell metabolism. For example, NRON, which has been reported to enhance HIV gene expression in primary CD4 T cells, decreases the binding of NFAT and viral transcriptional activator Tat to the HIV-1 long terminal repeat (LTR) promoter region, resulting in reduced HIV-1 replication and viral protein expression; whereas lncRNA uc002yug.2 (linc01426) enhances HIV-1 viral replication, LTR activity, and the activation of latent HIV by up-regulating HIV Tat and suppressing RUNX 1b/1c, a transcription factor which can bind with HIV-1 LTR to inhibit HIV-1 replication (Cron et al., 2000; Willingham et al., 2005; Imam et al., 2015; Li et al., 2016; Mousseau and Valente, 2017; Huan et al., 2018). NEAT1 modulates the replication of different viruses by multiple mechanisms. Except for impacting on RIG-I signaling and cGAS-STING-IRF3 pathway, NEAT1 also influence the transcription of viral genes or translocation of viral transcripts. In response to HSV infection, NEAT1 and two other paraspeckle protein components, P54nrb and PSPC1, may associate with HSV DNA. By doing so, PSPC1 may recruit STAT3 to paraspeckles and viral gene promoters, and thus facilitate HSV replication and protein expression (Wang Z. et al., 2017). In contrast, upon HIV infection, NEAT1 reduces nucleus-to-cytoplasm export of HIV-1 transcripts which containing cis-acting instability elements (INS), such as gag/pol and env RNAs, resulting in reduced HIV-1 replication (Zolotukhin et al., 2003; Kula et al., 2011; Yedavalli and Jeang, 2011; Zhang et al., 2013). Similarly, lncITPRIP-1 regulates HCV replication through both IFN-dependent and IFN-independent manners. Except for mediating MDA5-triggered production of IFNs and ISGs, lncITPRIP-1 also enhances the inhibitory effect of MDA5 on HCV replication by facilitating the binding of MDA5 to viral RNA (Xie et al., 2018). Another lncRNA, GAS5, reduces HCV replication by interacting with HCV protease NS3 protein to decoy its function (Qian et al., 2016). Interestingly, lncRNA-ACOD1 can promotes viral infection by modulating host cell metabolism. It directly interacts with metabolic enzyme glutamic-oxaloacetic transaminease (GOT2) in cytoplasm, resulting in increased catalytic activity of this enzyme, leading to enhanced production of key metabolites required for viral replication (Kotzin et al., 2017; Wang P. et al., 2017). Besides these, other lncRNAs, such as VIN (viral inducible lincRNA), also reported to modulate viral replication, while it did not change type I IFN response, the underlying mechanism of which is still unclear (Winterling et al., 2014).
Conclusion and Future Perspectives
IFN-mediated innate antiviral response is the first line of immune defense against viral infection. In the past decade, lncRNAs have been demonstrated to control fundamental biological processes at the epigenetic, transcription and post-transcriptional levels, and the deregulation of lncRNAs contributes to immune response, including IFN-mediated antiviral response. In this review, we summarized the deregulated lncRNAs upon viral infection, with special focus on the functions and underlying mechanisms of some important lncRNAs, and discussed their roles in the antiviral response of IFN. The functions and mechanisms of action of some essential lncRNAs in viral infection and IFN antiviral response are summarized in Figure 1 and Table 1.
Except for impacting on IFN-mediated antiviral response, lncRNAs also modulate viral infection or replication by other mechanisms, such as regulating viral gene transcription, viral RNA translocation, viral protein function, and host cell metabolism. Interestingly, one lncRNA, NEAT1, has been reported to exert different effects on different viruses: it inhibits HTNV or HIV replication by activating IFN signaling or improving the translocation of INS-containing viral RNAs, while enhances HSV replication by facilitating the binding of STAT3 on the viral gene promoters. These indicate that different therapeutic strategies should be used to control different viruses. When host lncRNAs exert modulation on immune response, viruses have evolved to facilitate their survival and replication by regulating the expression of lncRNAs to influence different host pathways, suggesting the pivotal regulatory roles of lncRNAs in the interplay between host and viruses. Until now, only a small part of lncRNAs have been identified and characterized to participate in the IFN antiviral response, while the vast majority of uncharacterized lncRNAs remain to be further explored. More extensive studies are required to describe the precise profile of virus regulated lncRNAs and their functions in viral infection. Furthermore, the regulatory mechanisms of these lncRNAs by different viruses, as well as the underlying mechanisms of these lncRNAs in viral infection need to be fully elucidated. The investigation of the interaction between lncRNA and the IFN antiviral response may not only deepen our understanding of antiviral response, but also provide novel applications for better prognosis and antiviral therapy.
Author Contributions
LQ, TW, QT, GL, PW, and KC conceived and wrote the manuscript. LQ did the figure.
Funding
This work was supported by the National Natural Science Foundation of China (81402840, 31402016, and 31572467); the Natural Science Foundation of Jiangsu Province, China (BK20130495); the Training Project of Young Backbone Teachers of Jiangsu University.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ablasser, A., Goldeck, M., Cavlar, T., Deimling, T., Witte, G., Rohl, I., et al. (2013). CGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498, 380–384. doi: 10.1038/nature12306
Atianand, M. K., Hu, W., Satpathy, A. T., Shen, Y., Ricci, E. P., Alvarez-Dominguez, J. R., et al. (2016). A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell 165, 1672–1685. doi: 10.1016/j.cell.2016.05.075
Barriocanal, M., Carnero, E., Segura, V., and Fortes, P. (2014). Long non-coding RNA BST2/BISPR is induced by ifn and regulates the expression of the antiviral factor tetherin. Front. Immunol. 5:655. doi: 10.3389/fimmu.2014.00655
Bidet, K., Dadlani, D., and Garcia-Blanco, M. A. (2014). G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog. 10:e1004242. doi: 10.1371/journal.ppat.1004242
Borden, E. C., Sen, G. C., Uze, G., Silverman, R. H., Ransohoff, R. M., Foster, G. R., et al. (2007). Interferons at age 50: past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 6, 975–990. doi: 10.1038/nrd2422
Bruns, A. M., and Horvath, C. M. (2015). LGP2 synergy with MDA5 in RLR-mediated RNA recognition and antiviral signaling. Cytokine 74, 198–206. doi: 10.1016/j.cyto.2015.02.010
Carnero, E., Barriocanal, M., Prior, C., Pablo Unfried, J., Segura, V., Guruceaga, E., et al. (2016). Long noncoding RNA EGOT negatively affects the antiviral response and favors HCV replication. EMBO Rep. 17, 1013–1028. doi: 10.15252/embr.201541763
Carnero, E., Barriocanal, M., Segura, V., Guruceaga, E., Prior, C., Borner, K., et al. (2014). Type I interferon regulates the expression of long non-coding RNAs. Front. Immunol. 5:548. doi: 10.3389/fimmu.2014.00548
Carpenter, S., Aiello, D., Atianand, M. K., Ricci, E. P., Gandhi, P., Hall, L. L., et al. (2013). A long noncoding RNA mediates both activation and repression of immune response genes. Science 341, 789–792. doi: 10.1126/science.1240925
Chan, Y. K., and Gack, M. U. (2016). Viral evasion of intracellular DNA and RNA sensing. Nat. Rev. Microbiol. 14, 360–373. doi: 10.1038/nrmicro.2016.45
Chang, R. Y., Hsu, T. W., Chen, Y. L., Liu, S. F., Tsai, Y. J., Lin, Y. T., et al. (2013). Japanese encephalitis virus non-coding RNA inhibits activation of interferon by blocking nuclear translocation of interferon regulatory factor 3. Vet. Microbiol. 166, 11–21. doi: 10.1016/j.vetmic.2013.04.026
Chen, Q., Cai, J., Wang, Q., Wang, Y., Liu, M., Yang, J., et al. (2018). Long noncoding RNA NEAT1, regulated by the EGFR pathway, contributes to glioblastoma progression through the WNT/beta-catenin pathway by scaffolding EZH2. Clin. Cancer Res. 24, 684–695. doi: 10.1158/1078-0432.CCR-17-0605
Chen, Y., Jiao, B., Yao, M., Shi, X., Zheng, Z., Li, S., et al. (2017). ISG12a inhibits HCV replication and potentiates the anti-HCV activity of IFN-alpha through activation of the Jak/STAT signaling pathway independent of autophagy and apoptosis. Virus Res. 227, 231–239. doi: 10.1016/j.virusres.2016.10.013
Chen, Y., Shen, A., Rider, P. J., Yu, Y., Wu, K., Mu, Y., et al. (2011). A liver-specific microRNA binds to a highly conserved RNA sequence of hepatitis B virus and negatively regulates viral gene expression and replication. FASEB J. 25, 4511–4521. doi: 10.1096/fj.11-187781
Cheriyath, V., Leaman, D. W., and Borden, E. C. (2011). Emerging roles of FAM14 family members (G1P3/ISG 6-16 and ISG12/IFI27) in innate immunity and cancer. J. Interferon Cytokine Res. 31, 173–181. doi: 10.1089/jir.2010.0105
Cron, R. Q., Bartz, S. R., Clausell, A., Bort, S. J., Klebanoff, S. J., and Lewis, D. B. (2000). NFAT1 enhances HIV-1 gene expression in primary human CD4 T cells. Clin. Immunol. 94, 179–191. doi: 10.1006/clim.1999.4831
Devasthanam, A. S. (2014). Mechanisms underlying the inhibition of interferon signaling by viruses. Virulence 5, 270–277. doi: 10.4161/viru.27902
Fitzgerald, K. A., Rowe, D. C., Barnes, B. J., Caffrey, D. R., Visintin, A., Latz, E., et al. (2003). LPS-TLR4 signaling to IRF-3/7 and NF-kappaB involves the toll adapters TRAM and TRIF. J. Exp. Med. 198, 1043–1055. doi: 10.1084/jem.20031023
Fortes, P., and Morris, K. V. (2016). Long noncoding RNAs in viral infections. Virus Res. 212, 1–11. doi: 10.1016/j.virusres.2015.10.002
Garcia-Sastre, A. (2011). Induction and evasion of type I interferon responses by influenza viruses. Virus Res. 162, 12–18. doi: 10.1016/j.virusres.2011.10.017
Garcia-Sastre, A., Egorov, A., Matassov, D., Brandt, S., Levy, D. E., Durbin, J. E., et al. (1998). Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252, 324–330. doi: 10.1006/viro.1998.9508
Gomez, J. A., Wapinski, O. L., Yang, Y. W., Bureau, J. F., Gopinath, S., Monack, D. M., et al. (2013). The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell 152, 743–754. doi: 10.1016/j.cell.2013.01.015
Hao, J., Jin, W., Li, X., Wang, S., Zhang, X., Fan, H., et al. (2013). Inhibition of alpha interferon (IFN-alpha)-induced microRNA-122 negatively affects the anti-hepatitis B virus efficiency of IFN-alpha. J. Virol. 87, 137–147. doi: 10.1128/JVI.01710-12
Hertzog, P., Forster, S., and Samarajiwa, S. (2011). Systems biology of interferon responses. J. Interferon Cytokine Res. 31, 5–11. doi: 10.1089/jir.2010.0126
Huan, C., Li, Z., Ning, S., Wang, H., Yu, X. F., and Zhang, W. (2018). Long noncoding RNA uc002yug.2 Activates HIV-1 latency through regulation of mRNA levels of various RUNX1 isoforms and increased tat expression. J. Virol. 92:e01844-17. doi: 10.1128/JVI.01844-17
Idogawa, M., Ohashi, T., Sasaki, Y., Nakase, H., and Tokino, T. (2017). Long non-coding RNA NEAT1 is a transcriptional target of p53 and modulates p53-induced transactivation and tumor-suppressor function. Int. J. Cancer 140, 2785–2791. doi: 10.1002/ijc.30689
Imam, H., Bano, A. S., Patel, P., Holla, P., and Jameel, S. (2015). The lncRNA NRON modulates HIV-1 replication in a NFAT-dependent manner and is differentially regulated by early and late viral proteins. Sci. Rep. 5:8639. doi: 10.1038/srep08639
Imamura, K., Imamachi, N., Akizuki, G., Kumakura, M., Kawaguchi, A., Nagata, K., et al. (2014). Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell 53, 393–406. doi: 10.1016/j.molcel.2014.01.009
Jiang, M., Zhang, S., Yang, Z., Lin, H., Zhu, J., Liu, L., et al. (2018). Self-recognition of an inducible host lncRNA by RIG-I feedback restricts innate immune response. Cell 173, 906.e13–919.e13. doi: 10.1016/j.cell.2018.03.064
Jin, L., Quan, J., Pan, X., He, T., Hu, J., Li, Y., et al. (2017). Identification of lncRNA EGOT as a tumor suppressor in renal cell carcinoma. Mol. Med. Rep. 16, 7072–7079. doi: 10.3892/mmr.2017.7470
Kambara, H., Niazi, F., Kostadinova, L., Moonka, D. K., Siegel, C. T., Post, A. B., et al. (2014). Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucleic Acids Res. 42, 10668–10680. doi: 10.1093/nar/gku713
Kondo, S., Yoshida, K., Suzuki, M., Saito, I., and Kanegae, Y. (2014). Adenovirus-encoding virus-associated RNAs suppress HDGF gene expression to support efficient viral replication. PLoS One 9:e108627. doi: 10.1371/journal.pone.0108627
Kotzin, J. J., Mowel, W. K., and Henao-Mejia, J. (2017). Viruses hijack a host lncRNA to replicate. Science 358, 993–994. doi: 10.1126/science.aar2300
Krawczyk, M., and Emerson, B. M. (2014). P50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-kappaB complexes. eLife 3:e01776. doi: 10.7554/eLife.01776
Kula, A., Guerra, J., Knezevich, A., Kleva, D., Myers, M. P., and Marcello, A. (2011). Characterization of the HIV-1 RNA associated proteome identifies matrin 3 as a nuclear cofactor of rev function. Retrovirology 8:60. doi: 10.1186/1742-4690-8-60
Lau, C. C., Sun, T., Ching, A. K., He, M., Li, J. W., Wong, A. M., et al. (2014). Viral-human chimeric transcript predisposes risk to liver cancer development and progression. Cancer Cell 25, 335–349. doi: 10.1016/j.ccr.2014.01.030
Lester, S. N., and Li, K. (2014). Toll-like receptors in antiviral innate immunity. J. Mol. Biol. 426, 1246–1264. doi: 10.1016/j.jmb.2013.11.024
Li, J., Chen, C., Ma, X., Geng, G., Liu, B., Zhang, Y., et al. (2016). Long noncoding RNA NRON contributes to HIV-1 latency by specifically inducing tat protein degradation. Nat. Commun. 7:11730. doi: 10.1038/ncomms11730
Lin, F. C., and Young, H. A. (2014). Interferons: success in anti-viral immunotherapy. Cytokine Growth Factor Rev. 25, 369–376. doi: 10.1016/j.cytogfr.2014.07.015
Liu, B., Sun, L., Liu, Q., Gong, C., Yao, Y., Lv, X., et al. (2015). A cytoplasmic NF-kappaB interacting long noncoding RNA blocks ikappab phosphorylation and suppresses breast cancer metastasis. Cancer Cell 27, 370–381. doi: 10.1016/j.ccell.2015.02.004
Liu, N., Long, Y., Liu, B., Yang, D., Li, C., Chen, T., et al. (2014). ISG12a mediates cell response to newcastle disease viral infection. Virology 462–463, 283–294. doi: 10.1016/j.virol.2014.06.014
Liu, S., Cai, X., Wu, J., Cong, Q., Chen, X., Li, T., et al. (2015). Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347:aaa2630. doi: 10.1126/science.aaa2630
Lopusna, K., Rezuchova, I., Betakova, T., Skovranova, L., Tomaskova, J., Lukacikova, L., et al. (2013). Interferons lambda, new cytokines with antiviral activity. Acta Virol. 57, 171–179. doi: 10.4149/av-2013-02-171
Ma, H., Han, P., Ye, W., Chen, H., Zheng, X., Cheng, L., et al. (2017). The long noncoding RNA NEAT1 exerts antihantaviral effects by acting as positive feedback for RIG-I signaling. J. Virol. 91:e02250-16. doi: 10.1128/JVI.02250-16
Makris, S., Paulsen, M., and Johansson, C. (2017). Type I interferons as regulators of lung inflammation. Front. Immunol. 8:259. doi: 10.3389/fimmu.2017.00259
Mansur, D. S., Smith, G. L., and Ferguson, B. J. (2014). Intracellular sensing of viral DNA by the innate immune system. Microbes Infect. 16, 1002–1012. doi: 10.1016/j.micinf.2014.09.010
Morchikh, M., Cribier, A., Raffel, R., Amraoui, S., Cau, J., Severac, D., et al. (2017). HEXIM1 and NEAT1 long non-coding RNA form a multi-subunit complex that regulates DNA-mediated innate immune response. Mol. Cell 67, 387.e5–399.e5. doi: 10.1016/j.molcel.2017.06.020
Mousseau, G., and Valente, S. T. (2017). Role of host factors on the regulation of tat-mediated HIV-1 transcription. Curr. Pharm. Des. 23, 4079–4090. doi: 10.2174/1381612823666170622104355
Naganuma, T., and Hirose, T. (2013). Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol. 10, 456–461. doi: 10.4161/rna.23547
Nishitsuji, H., Ujino, S., Yoshio, S., Sugiyama, M., Mizokami, M., Kanto, T., et al. (2016). Long noncoding RNA #32 contributes to antiviral responses by controlling interferon-stimulated gene expression. Proc. Natl. Acad. Sci. U.S.A. 113, 10388–10393. doi: 10.1073/pnas.1525022113
Ouyang, J., Hu, J., and Chen, J. L. (2016). lncRNAs regulate the innate immune response to viral infection. Wiley Interdiscip. Rev. RNA 7, 129–143. doi: 10.1002/wrna.1321
Ouyang, J., Zhu, X., Chen, Y., Wei, H., Chen, Q., Chi, X., et al. (2014). NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe 16, 616–626. doi: 10.1016/j.chom.2014.10.001
Paliwal, D., Joshi, P., and Panda, S. K. (2017). Hepatitis E Virus (HEV) egress: role of BST2 (Tetherin) and interferon induced long non- coding RNA (lncRNA) BISPR. PLoS One 12:e0187334. doi: 10.1371/journal.pone.0187334
Peng, W., Wu, J., Fan, H., Lu, J., and Feng, J. (2017). LncRNA EGOT promotes tumorigenesis via hedgehog pathway in gastric cancer. Pathol. Oncol. Res. doi: 10.1007/s12253-017-0367-3 [Epub ahead of print].
Penkala, I., Wang, J., Syrett, C. M., Goetzl, L., Lopez, C. B., and Anguera, M. C. (2016). lncRHOXF1, a long noncoding RNA from the X chromosome that suppresses viral response genes during development of the early human placenta. Mol. Cell. Biol. 36, 1764–1775. doi: 10.1128/MCB.01098-15
Pichlmair, A., Kandasamy, K., Alvisi, G., Mulhern, O., Sacco, R., Habjan, M., et al. (2012). Viral immune modulators perturb the human molecular network by common and unique strategies. Nature 487, 486–490. doi: 10.1038/nature11289
Platanias, L. C. (2005). Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5, 375–386. doi: 10.1038/ni1604
Qian, X., Xu, C., Zhao, P., and Qi, Z. (2016). Long non-coding RNA GAS5 inhibited hepatitis C virus replication by binding viral NS3 protein. Virology 492, 155–165. doi: 10.1016/j.virol.2016.02.020
Qiu, L., Fan, H., Jin, W., Zhao, B., Wang, Y., Ju, Y., et al. (2010). miR-122-induced down-regulation of HO-1 negatively affects miR-122-mediated suppression of HBV. Biochem. Biophys. Res. Commun. 398, 771–777. doi: 10.1016/j.bbrc.2010.07.021
Rapicavoli, N. A., Qu, K., Zhang, J., Mikhail, M., Laberge, R. M., and Chang, H. Y. (2013). A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. eLife 2:e00762. doi: 10.7554/eLife.00762
Rossetto, C. C., and Pari, G. S. (2011). Kaposi’s sarcoma-associated herpesvirus noncoding polyadenylated nuclear RNA interacts with virus- and host cell-encoded proteins and suppresses expression of genes involved in immune modulation. J. Virol. 85, 13290–13297. doi: 10.1128/JVI.05886-11
Schneider, W. M., Chevillotte, M. D., and Rice, C. M. (2014). Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 32, 513–545. doi: 10.1146/annurev-immunol-032713-120231
Schroder, K., Hertzog, P. J., Ravasi, T., and Hume, D. A. (2004). Interferon-gamma: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 75, 163–189. doi: 10.1189/jlb.0603252
Schuessler, A., Funk, A., Lazear, H. M., Cooper, D. A., Torres, S., Daffis, S., et al. (2012). West Nile virus noncoding subgenomic RNA contributes to viral evasion of the type I interferon-mediated antiviral response. J. Virol. 86, 5708–5718. doi: 10.1128/JVI.00207-12
Schulz, K. S., and Mossman, K. L. (2016). Viral evasion strategies in type I IFN signaling - a summary of recent developments. Front. Immunol. 7:498. doi: 10.3389/fimmu.2016.00498
Sun, L., Wu, J., Du, F., Chen, X., and Chen, Z. J. (2013). Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791. doi: 10.1126/science.1232458
Terstegen, L., Gatsios, P., Ludwig, S., Pleschka, S., Jahnen-Dechent, W., Heinrich, P. C., et al. (2001). The vesicular stomatitis virus matrix protein inhibits glycoprotein 130-dependent STAT activation. J. Immunol. 167, 5209–5216. doi: 10.4049/jimmunol.167.9.5209
Valadkhan, S., and Gunawardane, L. S. (2016). lncRNA-mediated regulation of the interferon response. Virus Res. 212, 127–136. doi: 10.1016/j.virusres.2015.09.023
Vigneau, S., Rohrlich, P. S., Brahic, M., and Bureau, J. F. (2003). Tmevpg1, a candidate gene for the control of theiler’s virus persistence, could be implicated in the regulation of gamma interferon. J. Virol. 77, 5632–5638. doi: 10.1128/JVI.77.10.5632-5638.2003
Wang, F., Ma, Y., Barrett, J. W., Gao, X., Loh, J., Barton, E., et al. (2004). Disruption of Erk-dependent type I interferon induction breaks the myxoma virus species barrier. Nat. Immunol. 5, 1266–1274. doi: 10.1038/ni1132
Wang, P., Xu, J., Wang, Y., and Cao, X. (2017). An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science 358, 1051–1055. doi: 10.1126/science.aao0409
Wang, P., Xue, Y., Han, Y., Lin, L., Wu, C., Xu, S., et al. (2014). The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 344, 310–313. doi: 10.1126/science.1251456
Wang, S., Qiu, L., Yan, X., Jin, W., Wang, Y., Chen, L., et al. (2012). Loss of microRNA 122 expression in patients with hepatitis B enhances hepatitis B virus replication through cyclin G(1) -modulated P53 activity. Hepatology 55, 730–741. doi: 10.1002/hep.24809
Wang, W. B., Levy, D. E., and Lee, C. K. (2011). STAT3 negatively regulates type I IFN-mediated antiviral response. J. Immunol. 187, 2578–2585. doi: 10.4049/jimmunol
Wang, Y., Wang, C., Chen, C., Wu, F., Shen, P., Zhang, P., et al. (2017). Long non-coding RNA NEAT1 regulates epithelial membrane protein 2 expression to repress nasopharyngeal carcinoma migration and irradiation-resistance through miR-101-3p as a competing endogenous RNA mechanism. Oncotarget 8, 70156–70171. doi: 10.18632/oncotarget.19596
Wang, Z., Fan, P., Zhao, Y., Zhang, S., Lu, J., Xie, W., et al. (2017). Neat1 modulates herpes simplex virus-1 replication by regulating viral gene transcription. Cell. Mol. Life Sci. 74, 1117–1131. doi: 10.1007/s00018-016-2398-4
Willingham, A. T., Orth, A. P., Batalov, S., Peters, E. C., Wen, B. G., Aza-Blanc, P., et al. (2005). A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 309, 1570–1573. doi: 10.1126/science.1115901
Winterling, C., Koch, M., Koeppel, M., Garcia-Alcalde, F., Karlas, A., and Meyer, T. F. (2014). Evidence for a crucial role of a host non-coding RNA in influenza A virus replication. RNA Biol. 11, 66–75. doi: 10.4161/rna.27504
Wu, J., Sun, L., Chen, X., Du, F., Shi, H., Chen, C., et al. (2013). Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830. doi: 10.1126/science.1229963
Wu, Y., Liang, S., Xu, B., Zhang, R., Zhu, M., Zhou, W., et al. (2017). Long noncoding RNA eosinophil granule ontogeny transcript inhibits cell proliferation and migration and promotes cell apoptosis in human glioma. Exp. Ther. Med. 14, 3817–3823. doi: 10.3892/etm.2017.4949
Xie, Q., Chen, S., Tian, R., Huang, X., Deng, R., Xue, B., et al. (2018). LncITPRIP-1 positively regulates innate immune response through promoting oligomerization and activation of MDA5. J. Virol. doi: 10.1128/JVI.00507-18 [Epub ahead of print].
Xiong, Y., Yuan, J., Zhang, C., Zhu, Y., Kuang, X., Lan, L., et al. (2015). The STAT3-regulated long non-coding RNA Lethe promote the HCV replication. Biomed. Pharmacother. 72, 165–171. doi: 10.1016/j.biopha.2015.04.019
Xu, S. P., Zhang, J. F., Sui, S. Y., Bai, N. X., Gao, S., Zhang, G. W., et al. (2015). Downregulation of the long noncoding RNA EGOT correlates with malignant status and poor prognosis in breast cancer. Tumour Biol. 36, 9807–9812. doi: 10.1007/s13277-015-3746-y
Xue, B., Yang, D., Wang, J., Xu, Y., Wang, X., Qin, Y., et al. (2016). ISG12a restricts hepatitis c virus infection through the ubiquitination-dependent degradation pathway. J. Virol. 90, 6832–6845. doi: 10.1128/JVI.00352-16
Yang, D., Meng, X., Xue, B., Liu, N., Wang, X., and Zhu, H. (2014). MiR-942 mediates hepatitis C virus-induced apoptosis via regulation of ISG12a. PLoS One 9:e94501. doi: 10.1371/journal.pone.0094501
Yedavalli, V. S., and Jeang, K. T. (2011). Matrin 3 is a co-factor for HIV-1 rev in regulating post-transcriptional viral gene expression. Retrovirology 8:61. doi: 10.1186/1742-4690-8-61
Yuen, M. F., and Lai, C. L. (2011). Treatment of chronic hepatitis B: evolution over two decades. J. Gastroenterol. Hepatol. 26(Suppl. 1), 138–143. doi: 10.1111/j.1440-1746.2010.06545.x
Zhang, Q., Chen, C. Y., Yedavalli, V. S., and Jeang, K. T. (2013). Neat1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. mBio 4:e00596-12. doi: 10.1128/mBio.00596-12
Zhu, H., and Liu, C. (2003). Interleukin-1 inhibits hepatitis C virus subgenomic RNA replication by activation of extracellular regulated kinase pathway. J. Virol. 77, 5493–5498. doi: 10.1128/JVI.77.9.5493-5498.2003
Zhu, H., Zhao, H., Collins, C. D., Eckenrode, S. E., Run, Q., McIndoe, R. A., et al. (2003). Gene expression associated with interferon alfa antiviral activity in an HCV replicon cell line. Hepatology 37, 1180–1188. doi: 10.1053/jhep.2003.50184
Keywords: long non-coding RNA, interferon, antiviral response, viral infection, interferon-stimulated genes
Citation: Qiu L, Wang T, Tang Q, Li G, Wu P and Chen K (2018) Long Non-coding RNAs: Regulators of Viral Infection and the Interferon Antiviral Response. Front. Microbiol. 9:1621. doi: 10.3389/fmicb.2018.01621
Received: 15 May 2018; Accepted: 28 June 2018;
Published: 19 July 2018.
Edited by:
Akihide Ryo, Yokohama City University, JapanReviewed by:
Masahiro Shuda, University of Pittsburgh, United StatesTakayuki Hishiki, Kanagawa Prefectural Institute of Public Health, Japan
Haizhen Zhu, Hunan University, China
Copyright © 2018 Qiu, Wang, Tang, Li, Wu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lipeng Qiu, dHVsaXBfbGlwZW5nQDE2My5jb20= Keping Chen, a3BjaGVuQHVqcy5lZHUuY24=
 Lipeng Qiu
Lipeng Qiu Tao Wang
Tao Wang Qi Tang
Qi Tang Guohui Li
Guohui Li Peng Wu
Peng Wu Keping Chen
Keping Chen