- 1College of Science and Engineering, James Cook University, Townsville, QLD, Australia
- 2College of Public Health, Medical and Veterinary Sciences, James Cook University, Townsville, QLD, Australia
Symbiotic bacterial communities resident on amphibian skin can benefit their hosts. For example, antibiotic production by community members can control the pathogen Batrachochytrium dendrobatidis (Bd) and it is possible for these community members to be used as probiotics to reduce infection levels. In the early 1990s, the emergence of Bd caused declines and disappearances of frogs in the Australian Wet Tropics; the severity of its effects varied among species and sites. Some species have since recolonized despite enzootic Bd within their populations. This variation in history among species and sites provided an opportunity to investigate the role of anti-fungal cutaneous bacteria in protecting frogs against Bd infection. We collected cutaneous swab samples from three species of frogs at two upland and two lowland sites in the Wet Tropics, and used in vitro challenge assays to identify culturable Bd-inhibitory bacterial isolates for further analysis. We sequenced DNA from cultured inhibitory isolates to identify taxa, resulting in the classification of 16 Bd-inhibitory OTUs, and determined whether inhibitory taxa were associated with frog species, site, or intensity of infection. We present preliminary results showing that the upper limit of Bd infection intensity was negatively correlated with number of inhibitory OTUs present per frog indicating that increased numbers of Bd-inhibiting taxa may play a role in reducing the intensity of Bd infections, facilitating frog coexistence with enzootic Bd. One upland site had a significantly lower prevalence of Bd infection, a significantly higher proportion of frogs with one or more culturable Bd-inhibitory OTUs, a greater number of inhibitory bacterial genera present per frog, and statistically significant clustering of individual frogs with similar Bd-inhibitory signatures when compared to all other sites. This suggests that Bd-inhibitory taxa are likely to be particularly important to frogs at this site and may have played a role in their ability to recolonize following population declines. Our findings suggest that the use of multi-taxon Bd-inhibitory probiotics to support at-risk amphibian populations may be more effective than single-taxon alternatives.
Introduction
Antibiotic-producing bacterial symbionts can protect their hosts from disease (Currie et al., 1999; Haas and Défago, 2005; Scott et al., 2008; Mao-Jones et al., 2010; Mattoso et al., 2012). For example, a high proportion of bacteria isolated from healthy coral can produce antibiotic compounds active against coral pathogens (Ritchie, 2006; Mao-Jones et al., 2010; Zhang et al., 2013). However, the best-studied mutualisms exist between insects and their bacterial symbionts. Both fungus-farming ants and pine beetles house antibiotic-producing bacteria in specialized cuticular compartments to control pathogens that threaten their food supply (Currie et al., 1999, 2006; Scott et al., 2008). In addition, solitary digger wasps use antibiotic-producing bacteria to protect their cocooned larvae against pathogens (Kroiss et al., 2010), and fungus-farming ants maintain a bacterial biofilm on their cuticles to control a fungal pathogen (Mattoso et al., 2012). Variation in the bacterial strain present can cause differential morbidity in fungus-farming ants (Poulsen et al., 2010), demonstrating that antibiotic production by symbionts is important for host protection.
Antibiotic production by symbiotic bacteria is also important in vertebrates. In two bird species, the European Hoopoe and the Green Woodhoopoe, nestlings harbor antibiotic-producing bacteria in their uropygial glands to control feather-degrading pathogens (Soler et al., 2008; Martín-Vivaldi et al., 2010). Symbiotic bacterial communities resident on amphibian skin can also produce, antibiotic compounds that contribute to the control of Batrachochytrium dendrobatidis (Bd; Becker et al., 2009, 2015; Harris et al., 2009a,b; Lam et al., 2010; Kueneman et al., 2016), a pathogenic fungus responsible for worldwide amphibian declines (Berger et al., 1998; Stuart et al., 2004; Skerratt et al., 2007; Wake and Vredenburg, 2008).
One aspect of community ecology theory predicts that complex communities, with higher numbers of species and hence more potential interactions, are generally more resistant to invasion than simple communities with fewer species (Robinson and Valentine, 1979). This has been demonstrated in soil bacterial communities (Matos et al., 2005; van Elsas et al., 2012; Hol et al., 2015; Hu et al., 2016), and also in the locust gut (Dillon et al., 2005) where higher numbers of inoculated bacterial isolates have been associated with greater resistance against pathogens.
Studies of amphibian skin microbiota have produced similar results. In vitro studies have demonstrated the potential importance of multi-species bacterial communities in resistance to Bd (Loudon et al., 2014; Piovia-Scott et al., 2017). Co-cultured synthetic multi-species communities were more effective against Bd than single species alone (Piovia-Scott et al., 2017). While there are several possible explanations for this effect, the presence of greater numbers of Bd-inhibitory metabolites following co-culture of bacterial isolates can have a synergistic effect (Loudon et al., 2014). While no field studies have reported a reduction of Bd load with increased Bd-inhibitory isolate richness, increased Proteobacteria phylotype richness on the skin of Eleutherodactylus coqui frogs was correlated with reduced Bd infection loads (Longo et al., 2015). A large portion of Bd-inhibitory isolates described to date fall within this phylum (Woodhams et al., 2007; Lam et al., 2010; Walke et al., 2011; Flechas et al., 2012; Bell et al., 2013; Daskin et al., 2014; Becker et al., 2015; Holden et al., 2015; Madison et al., 2017).
Patterns of frog decline and recovery in the Australian Wet Tropics following the arrival of the pathogen Bd in the late 1980s (Laurance et al., 1996; Berger et al., 1999; McDonald and Alford, 1999) differed among species and sites. Frog populations of all species at lowland sites (below 400 m) did not appear to experience declines, while several species, including Litoria nannotis (waterfall frog) and Litoria rheocola (common mist frog), suffered either declines or local extirpation at all upland rainforest sites (above 400 m; Richards et al., 1993; McDonald and Alford, 1999). Some populations of these species have subsequently reappeared at some but not all sites from which they disappeared (McDonald et al., 2005; Woodhams and Alford, 2005; Skerratt et al., 2010; McKnight et al., 2017). This suggests multiple, potentially independent appearances of resistance to Bd, or loss of virulence by the pathogen (McKnight et al., 2017). Another species, Litoria serrata (green-eyed tree frog) appeared to suffer temporary population declines at upland sites, followed by recovery within a few years (Richards et al., 1993; McDonald and Alford, 1999; Richards and Alford, 2005).
The mechanisms allowing population recovery or recolonization are likely to vary among species and sites (McKnight et al., 2017). There is substantial genetic separation among Wet Tropics frog populations (Schneider et al., 1998; Cunningham, 2001; Hoskin et al., 2005; Richards et al., 2010; Bell et al., 2012); variation among populations in both innate and acquired immune defenses due to selection pressures imposed by the pathogen may affect host resistance to Bd and ultimately survival (Rollins-Smith et al., 2002, 2011; Woodhams et al., 2010; Savage and Zamudio, 2011; Fites et al., 2012, 2014; Bataille et al., 2015; McKnight et al., 2017; Voyles et al., 2018). Pathogen virulence may also vary among regions (Berger et al., 2005; Farrer et al., 2011; Becker et al., 2017). Therefore, resistance to Bd infection is likely to have been acquired independently at these sites.
Symbiotic cutaneous bacteria are considered to be an important component of the amphibian innate immune defense system (Woodhams et al., 2007, 2014; Harris et al., 2009a). It is likely that the suite of cutaneous bacteria present on amphibian skin can evolve broad antimicrobial activity against invading pathogens much faster than adaptation of the host’s innate and acquired defenses (Rosenberg et al., 2007; King et al., 2016; Rosenberg and Zilber-Rosenberg, 2016). Therefore it is possible that at least some of the recolonized frogs in the Wet Tropics uplands have developed resistance to Bd through acquisition or evolution of more effective cutaneous bacterial symbionts.
The patterns of frog reappearances in the Wet Tropics uplands provide an opportunity to investigate whether the anti-fungal cutaneous bacterial microbiota have played a role in the ability of frogs to recolonize upland sites. We describe the culturable anti-Bd cutaneous bacterial microbiota of three species of rainforest frogs from four sites in northern Queensland, Australia, and assess how its composition may contribute to the health and population recovery of frogs at sites with different histories of pathogen-related population declines.
Materials and Methods
Field Sites and Species
We collected cutaneous swab samples from the stream-dwelling rainforest frogs L. serrata, L. nannotis, and L. rheocola at four rainforest field sites in the Wet Tropics bioregion where enzootic Bd infection exists, with one upland and one lowland site in each of two latitudinally separated national parks (Table 1). L. nannotis are the most, and L. serrata the least, strongly associated with water, and L. serrata are the least susceptible to population fluctuations as a result of Bd infection (McDonald and Alford, 1999; Woodhams and Alford, 2005). The upland sites were subject to extensive frog population declines in the early 1990s, while frog populations at the lowland sites persisted. Sampling was conducted in winter, when Bd infection is typically more prevalent (Berger et al., 2004; Woodhams and Alford, 2005; Kriger and Hero, 2007; Sapsford et al., 2013), to maximize chances of including Bd-infected frogs in the study. This study was conducted in compliance with the National Health and Medical Research Council’s Australian Code of Practice for the Care and Use of Animals for Scientific Purposes, 7th Edition, 2004, the Queensland Animal Care and Protection Act, 2001 and the Queensland Nature Conservation Act, 1992. Approval was granted from James Cook University Animal Ethics Committee (A1316 and A1420) and the Queensland government (scientific purposes permit number WITK05922209).
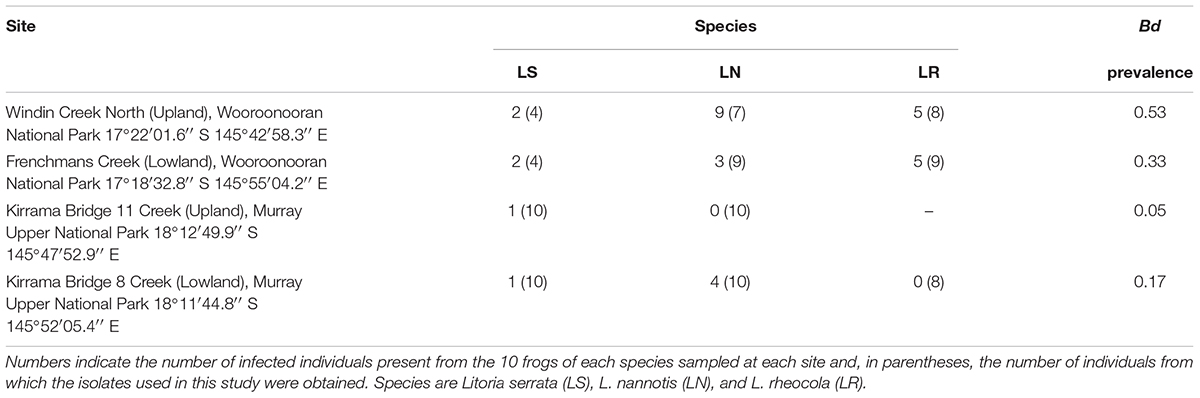
TABLE 1. Locations, species and Batrachochytrium dendrobatidis infection status of frogs sampled for each survey site, and overall Bd infection prevalence by site.
Collection of Bacterial Samples
We hand-captured frogs from vegetation and rocks bordering the creeks, in the winters of 2009 and 2010. All handling was carried out with a new plastic bag and pair of vinyl gloves for each animal to prevent contact with bacteria from our skin, and to preclude the transfer of pathogens or symbionts between animals. We gently restrained each animal by hand and rinsed it twice with a stream of sterile distilled water from a wash bottle to remove transient bacteria, which can differ from the resident microbiota (Lauer et al., 2007). We then swabbed over the dorsal and ventral skin from knee to neck five times using sterile rayon swabs (MW112, MW&E, Bath UK) moistened with sterile distilled water, and immediately transferred the swab’s contents to a low-nutrient agar plate (R2A, BD, Franklin Lakes, NJ, United States) by rotating the swab on the plate surface. Agar plates were sealed with parafilm (Parafilm “M,” Pechiney Plastic Packaging, Inc., Chicago, IL, United States), held at ambient temperature (10–25°C) and returned to the laboratory within 72 h. We collected an additional swab sample (MW100, MW&E, Bath UK) by rotating the swab over the abdomen, hands, feet, and thighs twice for analysis of Bd infection status.
Isolation, Purification, and Storage of Microbial Cultures
Inverted R2A agar plates were incubated for 48–72 h at ambient temperature (22–25°C) in the laboratory until we observed microbial culture growth. We examined plates daily for 5 days using a dissecting microscope and selected all colonies with different morphological characteristics for isolation to pure (axenic) culture using standard microbiological techniques (Salle, 1961). When initial agar plates generated from swabs plated in the field did not yield any viable bacterial colonies, possibly due to media choice or the presence of potent antimicrobial peptides on the frogs, we excluded samples from those frogs from further analysis. Of a total of nine initial plates that produced no viable cultures, five were from L. rheocola, and four were from L. nannotis. In contrast, R2A agar plates originating from L. serrata at the upland and lowland sites in Wooroonooran National Park produced very large numbers of viable bacterial colonies making isolation difficult and time-consuming; all of these plates appeared visually very similar. Because time and available effort constrained the number of these plates we could fully sample, we selected agar plates haphazardly from only four L. serrata at each of the Wooroonooran sites (Table 1). For all other combinations of site and species, all plates with bacterial colonies present were included.
Diagnosis of Batrachochytrium dendrobatidis Infection
DNA was extracted from skin swabs with PrepManTM Ultra (Applied Biosystems, Scoresby, VIC, Australia) and quantitative PCR (Real-time TaqMan® assay) used to diagnose Bd infection status as per Boyle et al. (2004) with the addition of BSA (Garland et al., 2010). We considered frogs to be infected with Bd when at least two of the three technical PCR replicates were positive. Bd intensity was calculated as the mean of the technical replicates.
Challenge Assays
We conducted challenge assays on cell free supernatants from axenic isolates, and categorized them as totally inhibitory, partially inhibitory, or non-inhibitory according to their effects on Bd growth as described by Bell et al. (2013). Briefly, cell-free supernatants for each isolate were obtained by centrifuging and filtering bacterial cultures incubated in TGhL medium (8 g tryptone, 1 g hydrolysed gelatin, 2 g lactose, to 1 L deionised water, autoclaved) at 23°C for 48 h. Bd isolate “Gibbo River, L. Les, 06-LB-1” was incubated on TGhL agar plates (as above recipe with 10 g bacteriological agar) at 23°C for 3 days. Zoospores were flushed from the plate with sterile TGhL medium, filtered to remove sporangia and resuspended in sterile TGhL at 2 × 106 zoospores ml-1. We conducted challenge assays in 96-well microplates (Costar 3595, Corning) with five replicates of each sample (50 μl cell-free supernatant and 50 μl of Bd zoospores), positive, negative, and medium only controls. Plates were incubated at 23°C for approximately 7 days until maximum growth in the positive controls was observed. Daily absorbance readings were collected at 492 nm using a spectrophotometer (Multiskan Ascent, Thermo Scientific) and from these we calculated the proportion of inhibition observed in each sample on its maximum growth day relative to the positive control. Bacterial isolates that produced cell-free supernatants exhibiting total inhibition against Bd were selected for taxonomic identification by DNA sequencing. Due to financial constraints it was not possible to sequence other isolates.
Bacterial DNA Extraction
We used a sterile toothpick to inoculate each axenic isolate into a 1.7 ml sterile microtube containing 400 μl molecular grade water. Samples were vortexed to create a cell suspension and subjected to three freeze-thaw cycles (70°C/-80°C; 10 min each), and then centrifuged at 7500 ×g for 5 min to pellet cell debris. We used the supernatant directly as template in the DNA amplification reaction, but if this was unsuccessful, DNA was extracted using a Qiagen DNeasy Blood and Tissue Kit (Qiagen, Doncaster, VIC, Australia) as per the manufacturer’s protocol.
Amplification of 16S rRNA Gene
We amplified the 16S rRNA gene from pure bacterial isolates by PCR on Bio-Rad C1000/S1000 thermal cyclers (Bio-Rad, Hercules, CA, United States) with the bacteria-specific primer 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and the universal primer 1492R (5′-GGTTACCTTGTTACGACTT-3′) (Lane, 1991). The PCR reaction mix contained 0.2 μM of each primer, 0.2 mM dNTPs, 3 mM MgCl2, 0.2 mg ml-1 BSA, 1.25 U HotStar Taq polymerase (Qiagen, Doncaster, VIC, Australia) with 1× buffer and <1 ug template DNA. The thermocycling parameters were: 95°C for 15 min followed by 35 cycles of 94°C for 1 min, 48°C for 1 min, 72°C for 1.5 min, and a final extension for 10 min at 72°C.
DNA Sequencing and Analysis
PCR product purification and Sanger DNA sequencing were conducted by Macrogen, Inc. (South Korea) using both forward and reverse primers described above. We aligned forward and reverse nucleotide sequences in Geneious Pro (Biomatters, Ltd.; Drummond et al., 2010) to create a consensus sequence of approximately 1400 bp. Following alignment of sequences, we compiled a FASTA file containing a single representative of each unique (one or more bp difference from any other sequence) consensus sequence and defined operational taxonomic units (OTUs) using mothur (v1.36.1; Schloss et al., 2009). Sequences were clustered at 96% sequence similarity to create genus-level OTUs and taxonomy was assigned using the SILVA reference database v128 (Quast et al., 2013; Yilmaz et al., 2014). When three or more sequences were present in a given OTU, the sequence used to assign taxonomy was the sequence that was the minimum distance to the other sequences in the OTU. Clustering was based on 96% sequence similarity to group the isolates into unique genera because our goal was to examine the diversity of cultured taxonomic groups likely to have different mechanisms of inhibiting the growth of Bd. Although some genera could comprise multiple OTUs that each produce different antifungal compounds, it is likely that some subgeneric OTUs would have the same mechanisms. For example, multiple Pseudomonas species can produce the antifungal compound 2,4-diacetylphloroglucinol (Gutierrez-Garcia et al., 2017). Splitting genera into multiple OTUs may overestimate the diversity of inhibitory mechanisms, so we chose the more conservative approach of leaving genera intact. We examined how well our sequenced data were likely to reflect the total number of culturable inhibitory bacterial OTUs present across all frog taxa at all of our sampling sites by constructing a species accumulation curve using the program EstimateS (Colwell, 2013). Because our study was primarily aimed at examining how the composition of culturable bacterial assemblages on individual frogs affected their Bd infection status, we also examined whether the isolates we cultured from each individual were likely to have captured all culturable totally inhibitory OTUs present on their skins. We did this by plotting number of totally inhibitory OTUs found on each frog against the total number of cultured isolates (inhibitory and non-inhibitory) obtained from each frog, and fitting a species accumulation curve to this relationship.
We added additional 16S sequences from closely related bacterial type strains (SILVA reference database v123; Quast et al., 2013; Yilmaz et al., 2014) to our FASTA file sequences and conducted a SINA alignment (Pruesse et al., 2012). From this alignment, we constructed a neighbor-joining phylogenetic tree in Geneious Pro (Biomatters, Ltd.; Drummond et al., 2010) and visualized this in Figtree v1.4.3 (Rambaut, 2016).
Data Manipulation and Analysis
As data were not normally distributed, and attempts at normalization via transformation were unsuccessful, we used non-parametric statistical tests where applicable. All statistical analyses were carried out using R (v3.2.4)1 and figures produced using ggplot2 (Wickham and Chang, 2016). We excluded L. rheocola from most of the analyses to balance species across sites, but included them in the graphics when appropriate. By including only the two species that were present at all sites, we minimized the possibility that site effects could reflect differences between species.
We used a generalized linear model (GLM) with a binomial response to compare prevalence of infection in L. serrata and L. nannotis (the species that occurred at all sites) across sites. We used Tukey’s post hoc pairwise comparisons (Tukey, 1949) to examine differences among pairs of sites using the function “glht” in the package multcomp (Hothorn et al., 2017), and adjusted p-values using the false discovery rate method (Benjamini and Hochberg, 1995). We assessed significance of the effects in this and other GLMs using the “ANOVA” function from the package car (Fox et al., 2017). To determine whether the ability of bacterial cell-free supernatants to inhibit Bd differed with infection status, species and site (L. serrata and L. nannotis only), we used permutational multivariate analysis of variance (PERMANOVA) on the proportions of isolates from each frog (L. serrata and L. nannotis only) that were in each of two challenge assay result categories; totally inhibitory and partially inhibitory. For this, we used the function “adonis2” with marginal terms (variables were assessed in the model together to show their impact when added last) in the community ecology package vegan (Oksanen et al., 2017). Tests were performed with 10000 permutations using Euclidean distance as the distance measure. We used the function “pairwise.adonis” (Martinez Arbizu, 2016) with Benjamini and Hochberg corrected p-values to examine differences among site pairs.
Following OTU classification, when totally inhibitory isolates were present on Bd-infected frogs, we used quantile regression at the 90th percentile level to examine the relationship between the intensity of Bd infection and the number of OTUs present on each frog using the package quantreg (Koenker et al., 2017) with 1000 bootstrap replications, and calculated pseudo-R2 (Koenker and Machado, 1999). We used quantile regression because it is recommended for use when examining relationships in which the independent variable sets a boundary for the dependent variable, but the value the dependent variable takes within that boundary is affected by many other factors (Cade and Noon, 2003). This is the case for Bd infection intensity, which can be affected by time since exposure, recent history of body temperature, and effects of innate and inducible immune responses not related to bacteria, among other things. We used the 90th percentile because we were interested in whether inhibitory bacteria set an upper boundary, and the 90th percentile was the highest we could practically examine given the number of observations in our data set. Frogs that did not carry any totally inhibitory isolates were excluded from our quantile regressions because the goal of these analyses was to examine how the upper limit of intensity of infection responds as number of inhibitory taxa increases. Frogs with no inhibitory OTUs will necessarily have no upper limit set by their microbiota, and are thus in a separate class, not comparable for our purpose to frogs with one or more inhibitory OTUs.
If inhibitory bacterial assemblages have evolved in response to Bd, they might differ across sites due to the differential availability of candidate sets of bacteria. Similarly, each frog species is likely to provide a different substrate for microbes to colonize. However, we would expect them to be similar across individuals within sites and species. Therefore, to determine if individual frogs clustered according to the taxonomy of their Bd-inhibitory bacterial microbiota within sites, frog species or with their interaction, we used PERMANOVA as described above with the Bray–Curtis distance measure (Bray and Curtis, 1957) with 10000 permutations. We excluded L. rheocola to ensure that species were balanced across sites. When the interaction term was not significant we repeated the analysis using only the two main effects. We checked for heterogeneity of multivariate dispersion using function “betadisper” in package vegan (Oksanen et al., 2017) with 10000 permutations. We used the function “pairwise.adonis” (Martinez Arbizu, 2016) with Benjamini and Hochberg corrected p-values to examine differences among pairs of sites. When clustering was observed, we visualized results with non-metric multidimensional scaling (nMDS) using the function “metaMDS.” When the nMDS failed to converge, we identified samples that were disconnected using the function “disconnected.” As a result, we removed three samples that each had just one unique Bd-inhibitory OTU from the nMDS analyses.
We used a GLM, specifying that the response came from a Poisson distribution, to examine species and site-specific differences in the number of Bd-inhibitory cutaneous bacterial OTUs present on frogs. We used Tukey’s post hoc pairwise comparisons (Tukey, 1949) to examine differences among site pairs as above with p-values adjusted using the false discovery rate method (Benjamini and Hochberg, 1995). For this, we excluded all data from L. rheocola to ensure that species were balanced across sites, and analyzed data as species-site pairs to test whether one species was responsible for any site-specific effects observed. Finally, we used a Fisher’s Exact Test to examine the association between the proportion of frogs with Bd-inhibitory OTUs and field site; we did not use a GLM to examine the effects of site and species simultaneously because numbers of individuals from which any OTUs were isolated was as low as 4 for some species-site combinations, so that the statistical power of a full analysis would have been near zero. Because this analysis excluded “species” as a factor, any species effects could have been confounded with site effects, since the numbers of each species from which OTUs were isolated differed among sites. To examine this possibility, we conducted a separate Fisher’s Exact Test to determine whether there were across-site effects of species (L. nannotis and L. serrata only, as these were the species in the site analysis) on the proportion of frogs with Bd-inhibitory OTUs.
Results
Infection Status of Frogs
Thirty-two of the 110 frogs (29%) sampled were infected with Bd (Table 1). Our GLM on data for the two species (L. nannotis and L. serrata) that were present at all sites indicated that the prevalence of infection differed significantly between those species (ANOVA: χ2 = 7.77, df = 1, p = 0.005) and among sites (ANOVA; χ2 = 14.98, df = 3, p = 0.0002); the interaction effect was not significant (ANOVA; χ2 = 7.50, df = 3, p = 0.058). Post hoc comparisons using Benjamini and Hochberg adjusted p-values showed that the Wooroonooran upland and Kirrama upland sites differed significantly (p = 0.019). Two of the five remaining pairwise comparisons were suggestive (Wooroonooran upland vs. Kirrama lowland and Wooroonooran upland vs. Wooroonooran lowland, p = 0.0892 for both cases). The remaining pairwise comparisons were not significantly different (p > 0.100 in all cases). Prevalence in L. nannotis and L. serrata combined (the species common to all sites) was 5% at Kirrama Uplands, 25% at Kirrama Lowlands, 25% at Wooroonooran Lowlands, and 55% at Wooroonooran Uplands.
We could not statistically compare intensity of infection among species within each site due to the low numbers of infected frogs. However, of the 29 infected frogs, 13 had infections of 10 zoospore equivalents or less indicating relatively low infection levels. Higher infection intensities occurred almost exclusively in L. rheocola at the Wooroonooran sites.
Culture and Challenge Assays
We sampled bacterial isolates from a total of 89 frogs (Table 1). We obtained a total of 1005 bacterial isolates across all frog species and sites, with a mean of 11.3 cultured bacterial isolates per frog (Range = 1–36, SD = 8.3). This is broadly in line with the number of isolates reported in other studies (Woodhams et al., 2007; Walke et al., 2011, 2015a; Becker et al., 2015). Challenge assay results showed that 68.5% of frogs from which isolates were obtained had one or more isolates with some activity against Bd; 55% had totally inhibitory isolates and 58% had partially inhibitory isolates [following the definition of Bell et al. (2013); Figure 1].
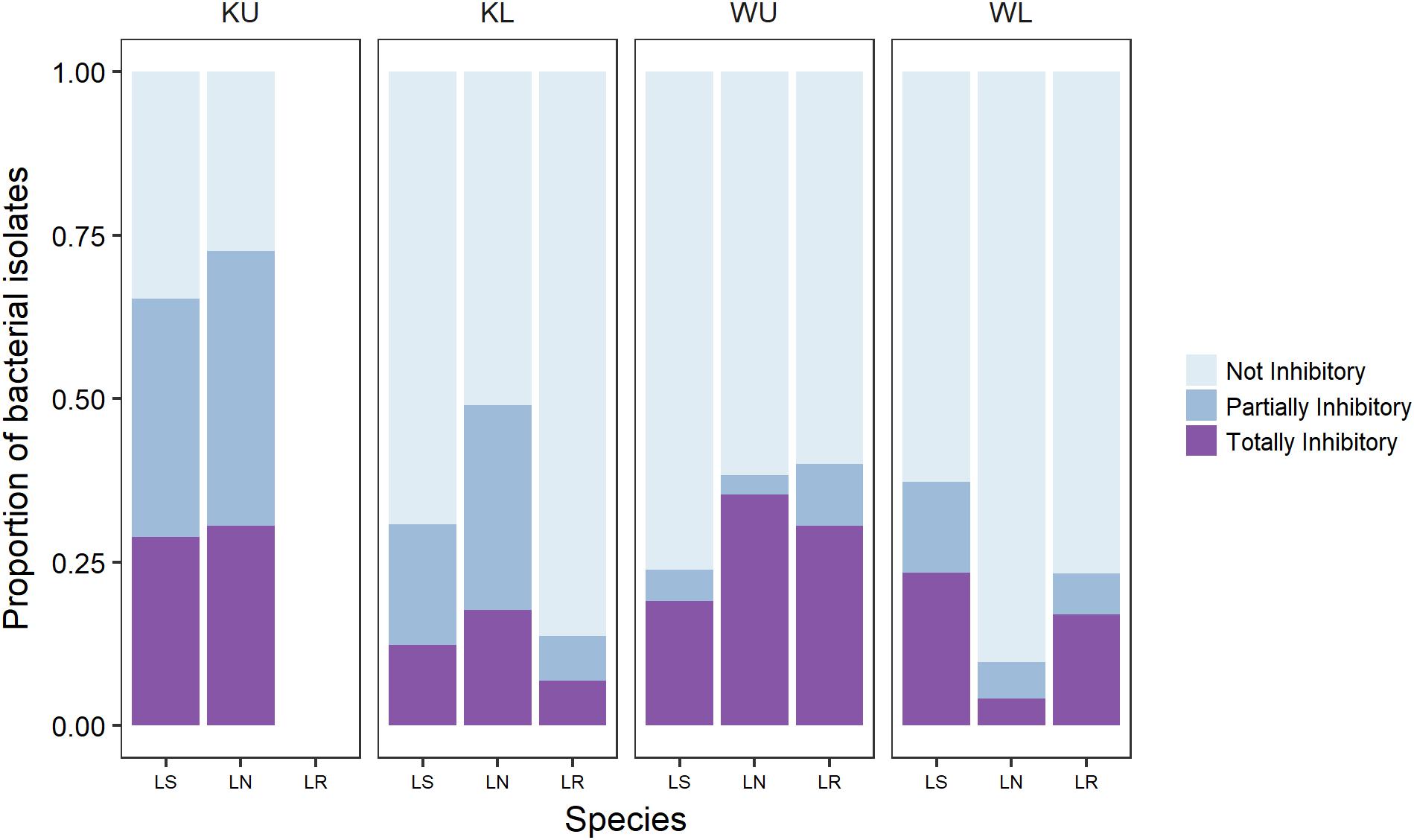
FIGURE 1. The proportion of culturable isolates as categorized by challenge assay results for each frog species at each site. Category definitions follow Bell et al. (2013). Species are Litoria serrata (LS), L. nannotis (LN), and L. rheocola (LR). Sites are Kirrama upland (KU), Kirrama lowland (KL), Wooroonooran upland (WU), and Wooroonooran lowland (WL).
Our PERMANOVA analysis of data for L. nannotis and L. serrata, the species that occurred at all sites, showed that the proportion of isolates in each category of inhibition (totally inhibitory and partially inhibitory) did not depend significantly upon infection status or species (PERMANOVA: infection status, pseudo-F1,58 = 0.478, p = 0.628, species, pseudo-F1,58 = 0.880, p = 0.422), but differed significantly among sites (Figure 1; PERMANOVA; pseudo-F3,58 = 6.189, p < 0.001). Post hoc comparisons among sites using Benjamini and Hochberg adjusted p-values showed that frogs at the Kirrama upland site had a significantly greater proportion of Bd-inhibitory isolates than those at all other sites (p < 0.01 in all cases). Frogs at the Wooroonooran upland and Kirrama lowland sites also differed significantly (p = 0.0285). The remaining site-pairs were not significantly different from each other (p > 0.100 for both cases).
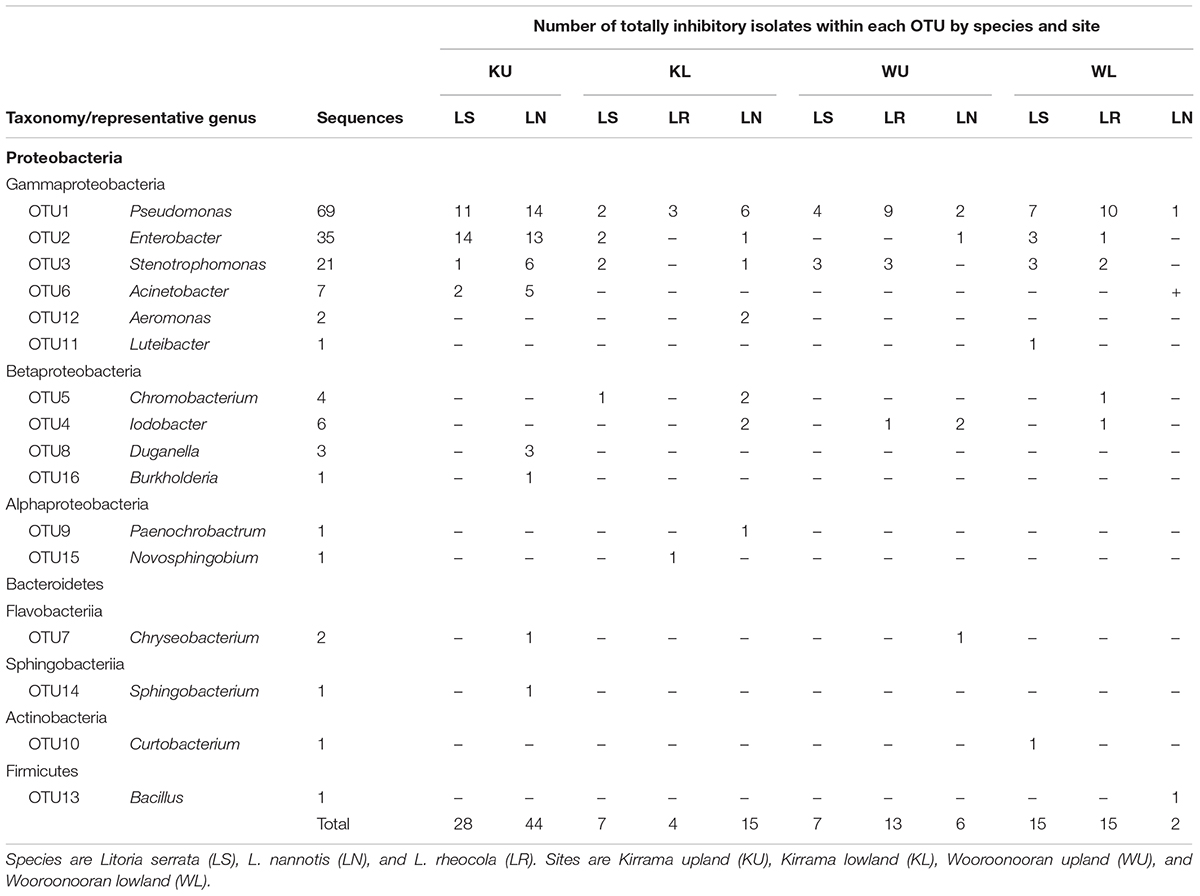
Identification of Bacterial Isolates
From a total of 156 totally inhibitory isolates sequenced, we identified 105 unique 16S rRNA sequences. These comprised 16 OTUs at the 96% sequence similarity level, each representative of one genus (Figure 2 and Table 2). 75% of OTUs fell within the Proteobacteria phylum with small contributions from the phyla Bacteriodetes, Actinobacteria, and Firmicutes, which is in line with the findings of other studies (Harris et al., 2006; Woodhams et al., 2007; Lam et al., 2010; Walke et al., 2011, 2015a; Flechas et al., 2012; Bell et al., 2013; Daskin et al., 2014; Becker et al., 2015; Holden et al., 2015; Madison et al., 2017). 16S rRNA gene sequences have been lodged in Genbank with accession numbers KJ191368–KJ191378, KJ191380–KJ191390, and MG491528–MG491661.
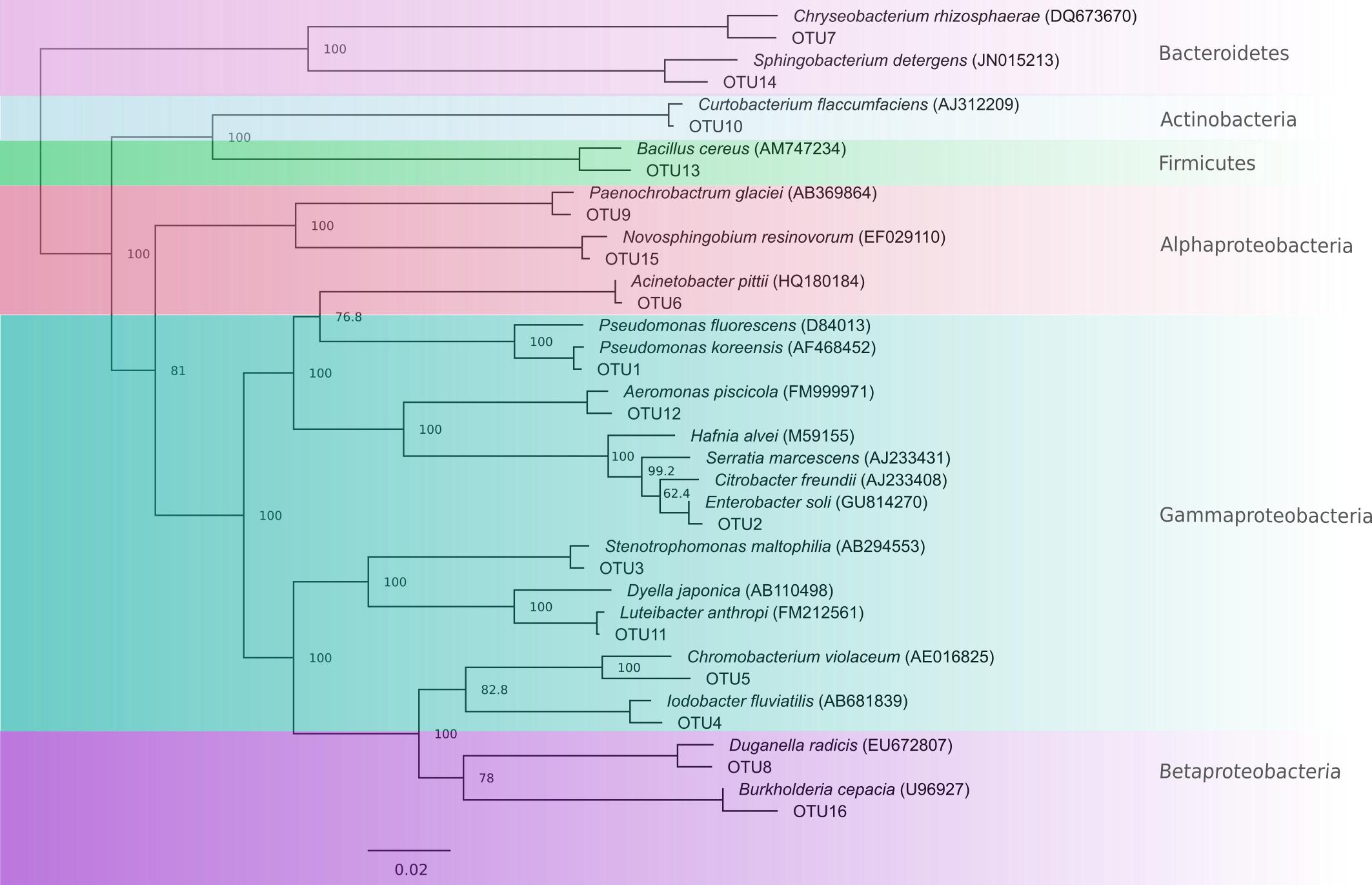
FIGURE 2. Neighbor-joining tree showing OTUs with representative type species for comparison. Nodes are labeled with percentage bootstrap support values. Labels with 100% bootstrap support were removed from some nodes to improve visualization.
Analysis of OTU Diversity Across All Sites and Species and Upon Individual Frogs
Figure 3A summarizes the relationship between total number of cultured inhibitory isolates sequenced and expected number of totally inhibitory OTUs obtained for our data aggregated across all species and sites. It shows that our sampling effort is not ideal because we typically sampled less than 30 isolates from any individual (Figure 3B). However, as we often sampled the majority of colonies that grew on the plates and in all cases we sampled all morphologically different isolates present, it is a limitation of culturing studies in general that it is impossible to attain adequate sampling effort. Figure 3B summarizes our analysis of the relationship between the total number of isolates we found on each individual frog and the number of unique cultured totally inhibitory OTUs included in those isolates. It provides additional information as it includes bacterial isolates that were sampled from agar plates that were not inhibitory. However, as our sampling effort is less than ideal due to the nature of culturing studies, our OTU-based analyses and results should be considered as preliminary findings.
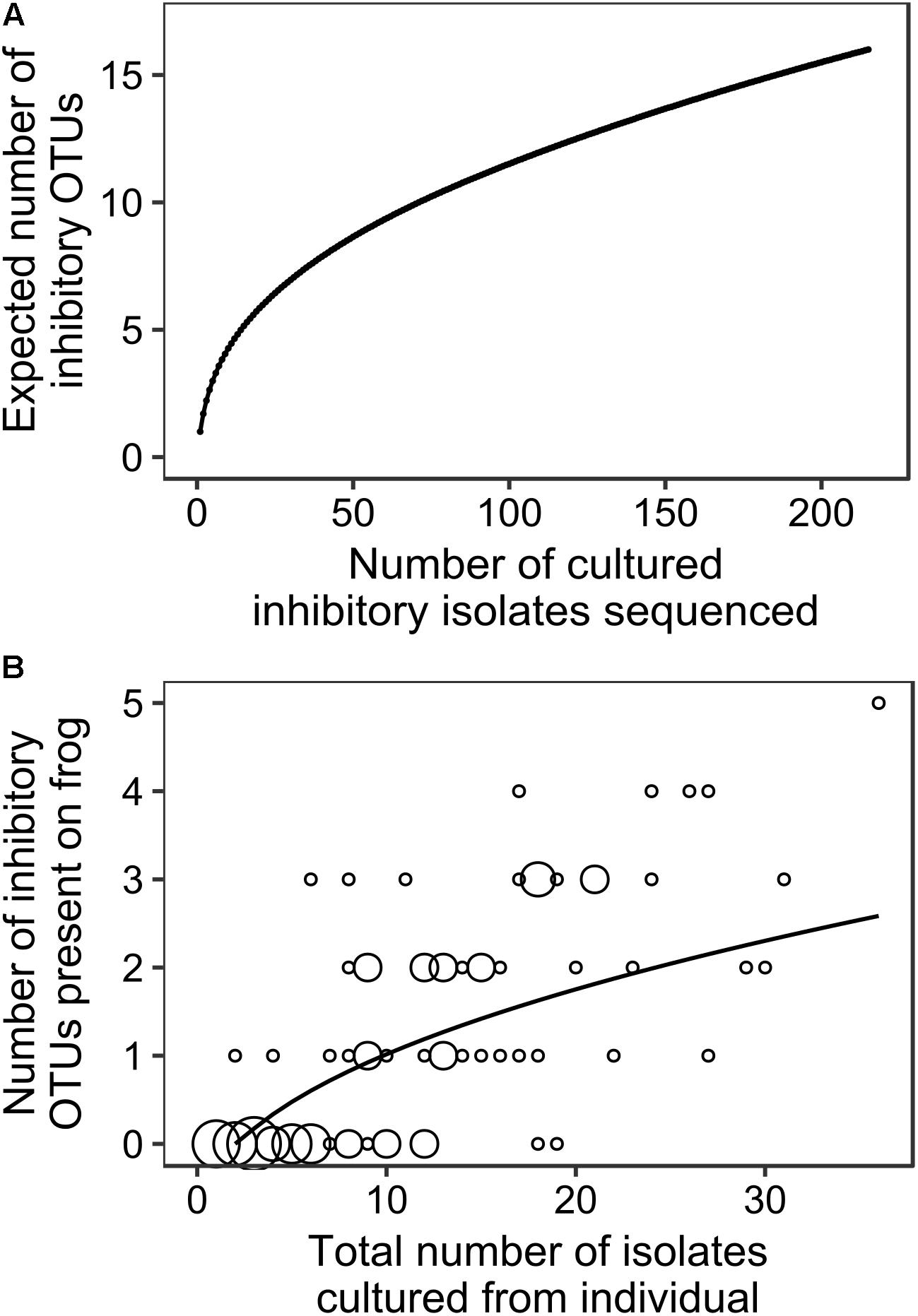
FIGURE 3. The relationships between (A) the number of totally inhibitory bacterial isolates identified and sequenced and the expected number of totally inhibitory OTUs (EstimateS; Colwell, 2013), and (B) the total number of isolates obtained from each individual frog and the number of totally inhibitory OTUs detected. The size of circles corresponds to the number of frogs at each combination of number of isolates and number of OTUs, with the smallest indicating one individual and the largest indicating eight. The best fit accumulation curve is a power curve fitted to the number of OTUs plus 1 (y = 0.717x0.449 with R2 = 0.505, p = 0.001).
Number of Totally Inhibitory OTUs and Bd Infection Intensity
Quantile regression at the 90th percentile level demonstrated a preliminary significant negative relationship between the number of Bd-inhibitory OTUs present per frog and the upper limits of Bd infection intensity (Figure 4; n = 14, t = -2.818, pseudo-R2 = 0.369, p = 0.0155). No frogs with highly intense Bd infections and high numbers of inhibitory bacteria were found during this study.
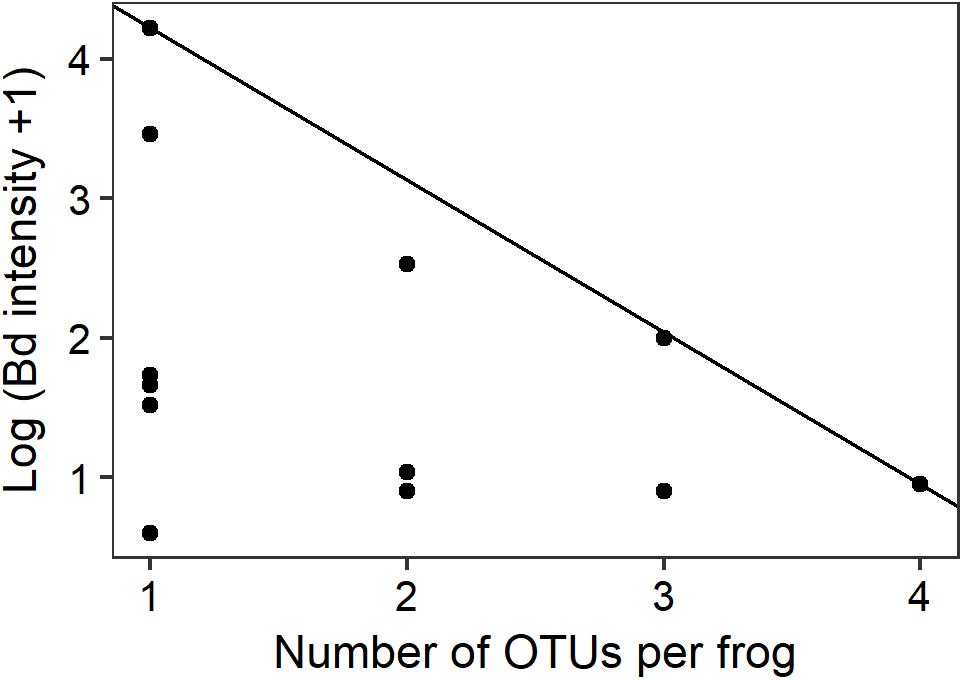
FIGURE 4. Intensity of Bd infection decreased as the number of inhibitory OTUs increased. The ninetieth percentile regression line shows the negative relationship between the number of OTUs per infected frog and the upper limits of Bd infection intensity (n = 14, t = -2.818, p = 0.0155).
Preliminary Species and Site-Specific Analyses
Our PERMANOVA examining the effects of species and site on the composition of Bd-inhibitory cutaneous bacterial OTUs showed no significant interaction between species and site (pseudo-F3,31 = 1.237, p = 0.221) and the effect of species was also not significant (pseudo-F1,34 = 0.661, p = 0.721), however, the effect of site was significant (pseudo-F3,34 = 1.780, p = 0.024). The multivariate dispersion of data using the Bray–Curtis distances used by the PERMANOVA was not significantly heterogeneous among sites (ANOVA; F3,35 = 2.541, p = 0.072). Inhibitory taxa present on L. nannotis and L. serrata from the Kirrama uplands clustered more tightly than those on frogs at any other site (Figure 5; post hoc Benjamini and Hochberg adjusted p-values: Kirrama upland – Kirrama lowland, p = 0.024: Kirrama upland – Wooroonooran upland, p = 0.046 and Kirrama upland – Wooroonooran lowland, p = 0.046, all other pairwise comparisons, p > 0.05).
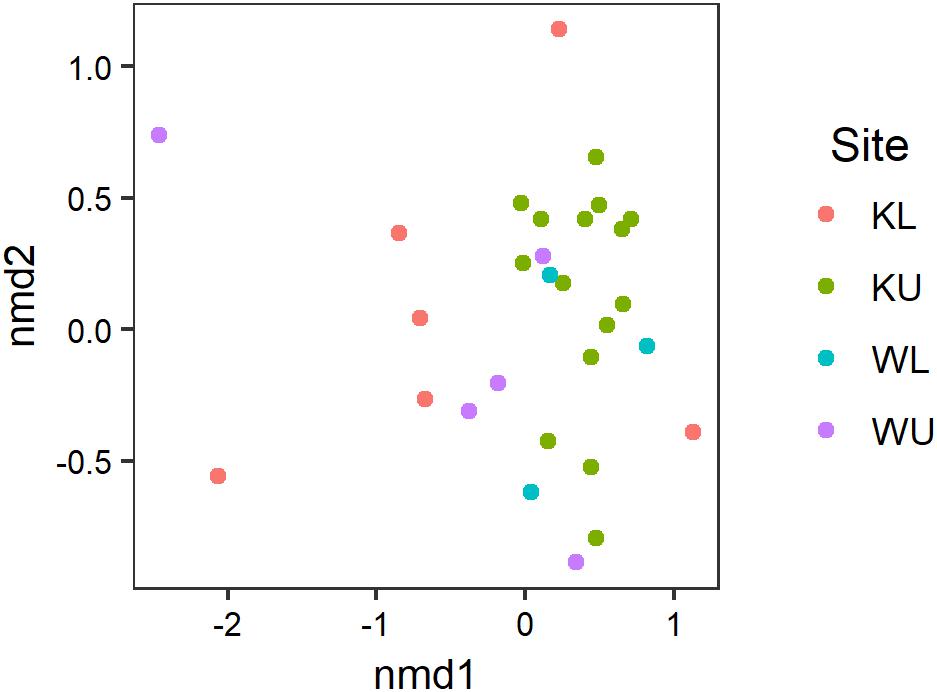
FIGURE 5. Results of an nMDS illustrating how the OTU signatures of frogs cluster by site. Stress = 0.124. Sites are KL, Kirrama lowland; KU, Kirrama upland; WL, Wooroonooran lowland; WU, Wooroonooran upland.
The outcome of our GLM examining the number of Bd-inhibitory OTUs present on frogs as a Poisson-distributed response showed a significant interaction effect, indicating that site and species had effects that were not independent (ANOVA; χ2 = 44.938, df = 7, p < 0.0001). We therefore performed a post hoc analysis comparing 28 pairs of combinations of species and site (Tukey, 1949), using Benjamini and Hochberg adjusted p-values. This revealed that the differences among species by site were largely driven by L. nannotis at the Kirrama upland site. L. nannotis from the Kirrama upland site had significantly greater numbers of Bd-inhibitory bacterial OTUs than L. nannotis at all three other sites (for each site, p < 0.05) and significantly greater numbers of Bd-inhibitory bacterial OTUs than L. serrata at Kirrama lowland and upland sites (p < 0.05). There was also a suggestion of greater OTU numbers in L. nannotis from the Kirrama upland site compared with Wooroonooran upland L. serrata (p = 0.061). A full set of pairwise comparisons are provided as Supplementary Table 1. Bd-inhibitory members of the genera Acinetobacter and Duganella were unique to frogs at the Kirrama upland site (Figure 6).
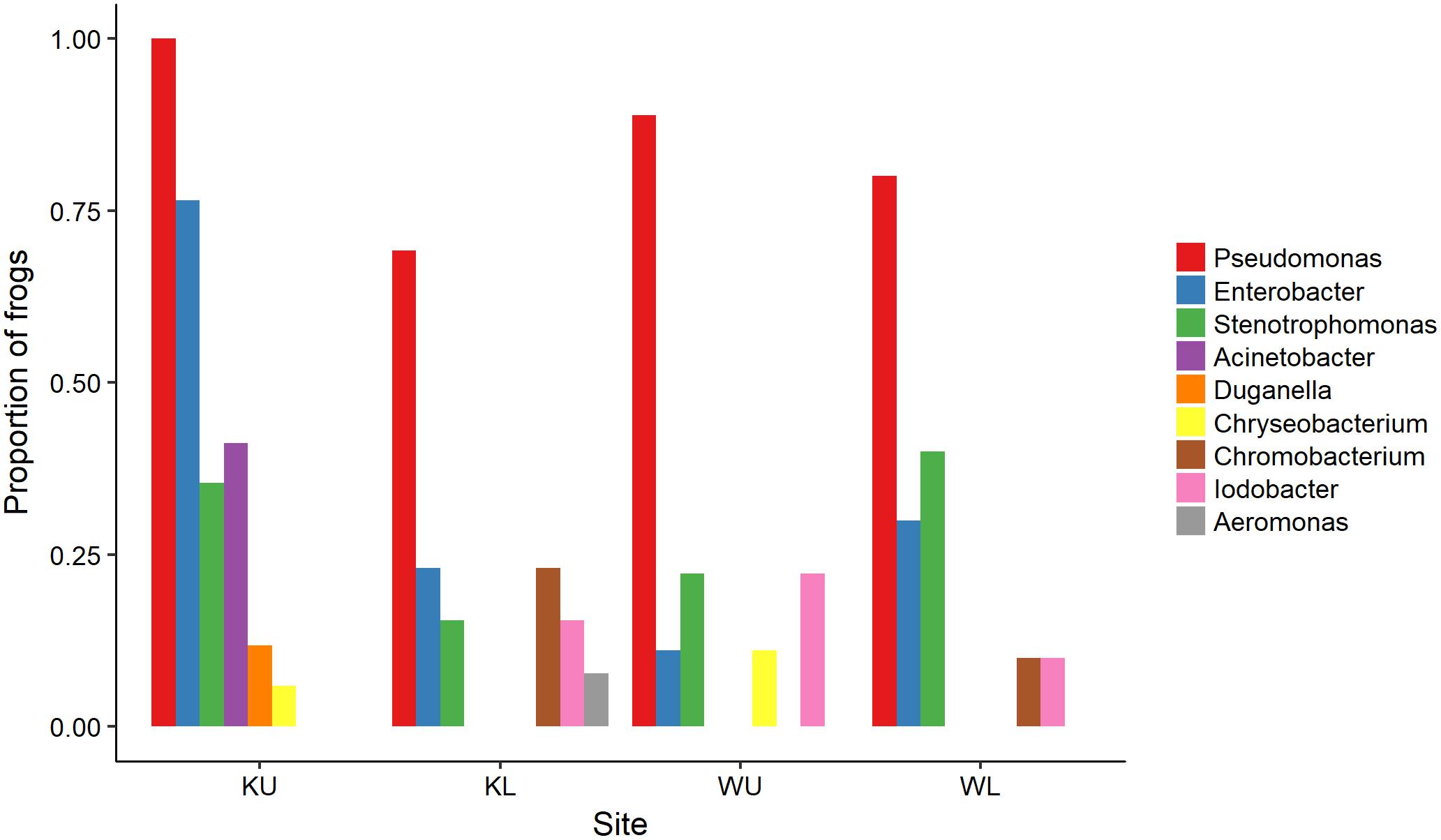
FIGURE 6. Proportion of frogs with isolates in each OTU by site. For ease of visualization, OTUs that occurred on fewer than two frogs at each site have been excluded. KU, Kirrama upland; KL, Kirrama lowland; WU, Wooroonooran upland; WL, Wooroonooran lowland.
As well as having significantly more Bd-inhibitory bacterial OTUs on L. nannotis, the Kirrama upland site also had a significantly higher proportion of frogs with one or more Bd-inhibitory OTUs than other sites (Figure 7; Fisher’s Exact Test; p = 0.0497; L. nannotis and L. serrata only due to the absence of L. rheocola from the Kirrama upland site). When the Kirrama upland site was removed from the analyses, all other sites did not differ significantly from each other (Fisher’s Exact Test; p = 0.861). The proportion of frogs with Bd-inhibitory OTUs did not differ significantly between species (Fisher’s Exact Test; p = 0.797; L. nannotis and L. serrata only). This suggests that the site effect we detected was not a confounded effect of species.
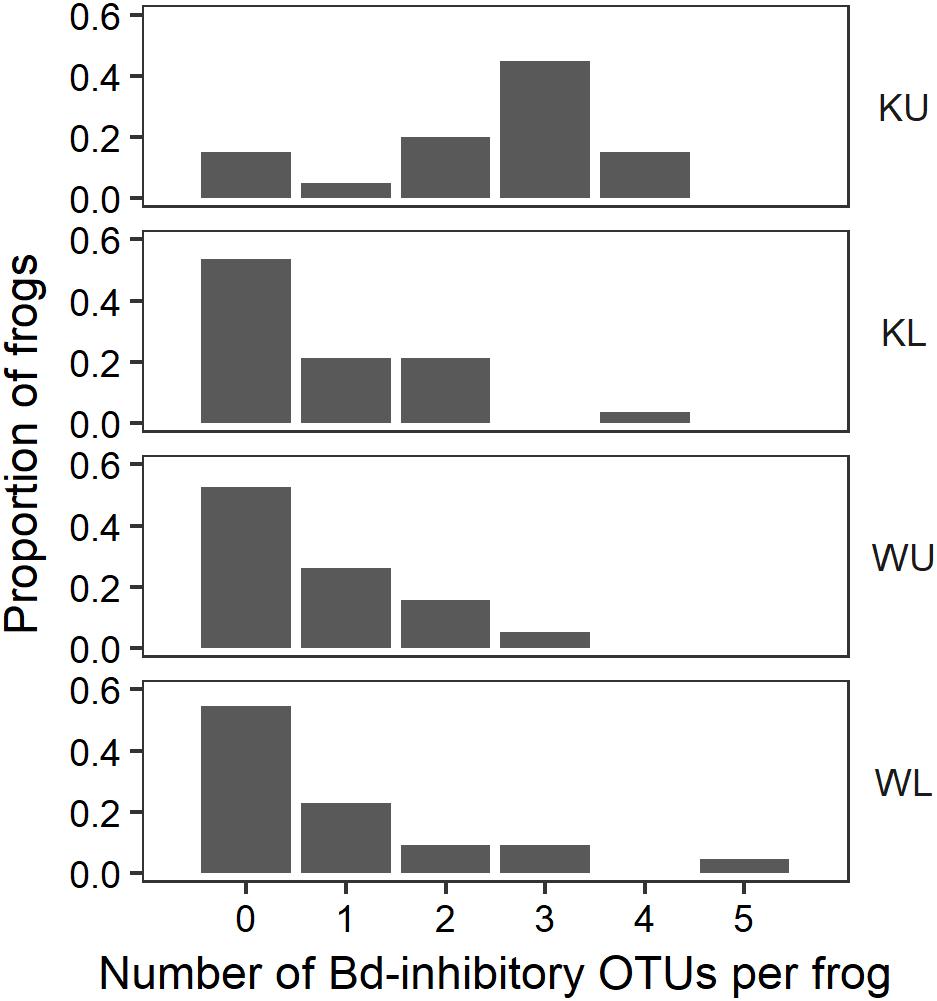
FIGURE 7. The proportion of frogs with Bd-inhibitory OTUs for each field site. KU, Kirrama upland; KL, Kirrama lowland; WU, Wooroonooran upland; WL, Wooroonooran lowland.
Discussion
We investigated the potential role that Bd-inhibitory cutaneous bacterial microbiota of Australian Wet Tropics frogs may have played in the reappearance of upland frog populations following population declines in the early 1990s and found that frogs from the Kirrama upland site differed from frogs at other sites in the presence and nature of their Bd-inhibitory bacteria. There was also a significant negative relationship between the upper limit of Bd infection intensity and the number of totally inhibitory bacterial genera on infected frogs. This suggests that greater inhibitory potential is linked to a reduction in the intensity of Bd infection, and that it therefore is possible that greater numbers of Bd-inhibitory bacterial genera may contribute to tolerance of Bd infection.
While our sampling effort was not ideal, it is often impossible for studies based on cultured isolates to achieve adequate sampling effort. For example, Figure 3A suggests that approximately 30 cultured totally inhibitory isolates per frog would be adequate. On average, our study found that 22% of the cultured isolates that we tested were totally inhibitory to Bd. Therefore, this would have required us to sample and test about 130 isolates from each frog/agar plate (more than 11,500 isolates from the 89 frogs in this study) in order to find 22% inhibitory. In almost all cases, there were considerably less colonies present on the agar plates than this and in many cases we sampled the majority of colonies that grew on the plate. In every case, we sampled all colony morphotypes present on the agar plates in an attempt to capture the diversity present. From our sequencing results, we often obtained identical sequences from individual frogs indicating that we probably over-sampled our agar plates, rather than under-sampled them. For these reasons, it is highly likely that we have captured the majority of cultured inhibitory OTUs present. The use of multiple culture media types may have resulted in the discovery of more OTUs, but it is likely that only a small fraction of all bacteria present on the frogs could ever be captured by culturing.
Many recent studies have used next generation sequencing techniques to investigate the diversity of amphibian cutaneous bacteria (reviewed by Jimenez and Sommer, 2017), but there is generally little evidence for a negative relationship between uncultured bacterial species richness and Bd infection load. However, uncultured proteobacteria phylotype richness was negatively correlated with Bd load in E. coqui frogs in Puerto Rico (Longo et al., 2015) and the relative abundance of certain taxa was negatively correlated with Bd infection load in Rana sierrae in California (Jani and Briggs, 2014). There are two possible reasons for negative relationships between Bd infection intensity and Bd-inhibitory OTU numbers. Frogs with naturally lower numbers of inhibitory bacterial taxa could be more susceptible to Bd or conversely, more intense Bd infection may cause a reduction in the numbers of inhibitory bacterial taxa.
Bd exposure can change the microbiome’s composition (Jani and Briggs, 2014; Walke et al., 2015b; Bataille et al., 2016; Jani et al., 2017). Studies have demonstrated that some OTUs can respond positively or negatively to Bd infection (Jani and Briggs, 2014; Familiar López et al., 2017; Longo and Zamudio, 2017) and that higher microbiome richness is associated with host persistence against Bd (Jani et al., 2017). However, increasing Bd load has not been demonstrated to lead to decreases in overall bacterial diversity or in the diversity of Bd-inhibitory bacteria on hosts. Laboratory experiments in vivo are therefore needed to ascertain whether a progressing Bd infection can cause a reduction in the richness of symbiotic bacterial communities, especially those inhibitory to Bd.
In vitro challenge assays have shown that combinations of multiple cultured bacterial strains are more effective at inhibiting Bd than are individual strains alone through additive and synergistic effects (Loudon et al., 2014; Piovia-Scott et al., 2017). This provides strong supporting evidence to our observations that increased numbers of inhibitory genera are associated with reduced susceptibility to Bd. Similar additive and synergistic effects have led to stronger fungal inhibition in hydra (Fraune et al., 2015), and soil Pseudomonads (Jousset et al., 2014). Three principal Bd-inhibitory genera (Pseudomonas, Enterobacter, and Stenotrophomonas) were present at all of our sites. The diverse secondary metabolites that can be produced by these principal genera might perform complementary roles. For example, one compound may act on the zoospore cell membrane while another interferes with transcription of DNA and therefore cell replication (Ghannoum and Rice, 1999; Kathiravan et al., 2012). It therefore seems possible that frogs in our study with greater numbers of Bd-inhibitory genera had more synergistic interactions among bacterial community members, leading to the production of additional antimicrobial metabolites and a reduction in either or both the probability of infection by Bd and the upper limits of infection load if infected (Figure 4).
Ideally, laboratory inoculation-infection experiments are needed to determine whether artificially increasing the number of Bd-inhibitory bacterial taxa on frog skin alters susceptibility to, or intensity of Bd infections. Similar experiments have been conducted on both plants and locusts and have demonstrated reduced pathogen density when a greater number of bacterial symbionts was present (Dillon et al., 2005; Matos et al., 2005; Hol et al., 2015) indicating the protective effect of a more diverse microbiome.
Our results from the Kirrama upland site showed that frogs at this site differed from all other sites in a number of ways, and together, provide support for protection against Bd infection by increased number of Bd-inhibitory bacterial genera. Frogs at this site had a significantly lower prevalence of infection than those at the Wooroonooran upland site, a significantly higher proportion of frogs with one or more Bd-inhibitory OTUs than other sites (though this could in part have been influenced by species effects), a higher proportion of inhibitory isolates than their conspecifics at other sites, significantly more Bd-inhibitory bacterial OTUs per frog in L. nannotis (suggesting increased synergistic anti-Bd potential), and statistically significant clustering of individual frogs with similar Bd-inhibitory taxonomic signatures (suggesting local selective pressure on skin microbiota). Some of the patterns we observed at the Kirrama upland site were clearly driven by L. nannotis. However, given that L. serrata only suffered temporary upland declines, it seems likely that the acquisition of greater numbers of Bd-inhibitory genera by L. nannotis at the Kirrama upland site may have facilitated their recolonization. A higher proportion of Kirrama upland frogs had Bd-inhibitory bacteria from the genera Pseudomonas and Enterobacter than at any other site, and the genera Acinetobacter and Duganella were unique to frogs from this site. Together, these differences suggest that Kirrama upland frogs, under pressure from enzootic Bd infection, have actively or passively acquired a greater number of Bd-inhibitory genera than frogs elsewhere, and that this may facilitate a reduction in the prevalence of Bd at this site.
All three species of frogs in this study have recolonized the Wooroonooran upland site despite having a lower proportion of Bd-inhibitory isolates than frogs at the Kirrama upland site and a lower proportion of frogs with Bd-inhibitory bacteria. Given the substantial genetic separation between frogs in the two regions (Schneider et al., 1998; Cunningham, 2001; Hoskin et al., 2005; Richards et al., 2010; Bell et al., 2012), it is likely that acquisition of resistance to Bd infection has evolved independently at the two upland sites. Therefore, factors other than symbiotic bacteria have probably played a part in aiding recolonization of the Wooroonooran upland site. These could include host immune defense, behavior, and environmental factors (Rollins-Smith et al., 2011; Savage and Zamudio, 2011; Rowley and Alford, 2013; Jani et al., 2017; McKnight et al., 2017; Voyles et al., 2018).
Bacteria that can inhibit the growth of Bd have received much attention in recent years, as it is possible that they could be cultured and used as probiotics to support at-risk populations of frogs through disease outbreaks (Becker et al., 2012; Bletz et al., 2013; Küng et al., 2014). Therefore the need to culture bacterial isolates to assess inhibitory potential against Bd remains essential to facilitate probiotic development, because it is not yet possible to accurately predict the functional capability of a community from 16S amplicon sequence data alone (Bletz et al., 2013; Becker et al., 2015). Fortunately, many of the more common OTUs described in 16S amplicon studies also appear to be culturable (Walke et al., 2015a) giving confidence that the functional capacity of the dominant community members was likely to be captured in our study. Future use of shotgun metagenomics, metatranscriptomics and metabolomics will enable insight into the functional capacity of bacterial communities without the need for culture (Rebollar et al., 2016) and help to elucidate the mechanisms by which individuals acquire and maintain communities of cutaneous bacteria that are inhibitory to Bd.
To our knowledge, we provide the first evidence that a greater number of Bd-inhibitory genera is correlated with lower Bd infection loads in wild frogs. This highlights the necessity for the creation of appropriate multiple-isolate combinations to promote potential synergistic interactions when developing probiotic candidates for supplementation of at-risk amphibian populations.
Author Contributions
SB and RA designed the study, conducted the data analysis, and produced the first draft of the manuscript. SB completed the fieldwork. SB and SG processed samples in the laboratory. All authors edited the manuscript.
Funding
Research funding was made available through funding from the Australian Research Council Discovery grant “Understanding and Managing Resistance to the Amphibian Chytrid Fungus Batrachochytrium dendrobatidis in Australian Tropical Rainforest Frogs” (DP0986537; 2009–2011) to RA and James Cook University’s Internal Graduate Research Scheme funding to SB.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful for field assistance provided by Betsy Roznik, David Pike, and Kiyomi Yasumiba, and laboratory assistance provided by Kiyomi Yasumiba, Josh Daskin, and Martha Silva Velasco. We thank three reviewers for their thorough comments, which helped us to improve the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01604/full#supplementary-material
Footnotes
References
Bataille, A., Cashins, S. D., Grogan, L., Skerratt, L. F., Hunter, D., Mcfadden, M., et al. (2015). Susceptibility of amphibians to chytridiomycosis is associated with MHC class II conformation. Proc. R. Soc. B Biol. Sci. 282:20143127. doi: 10.1098/rspb.2014.3127
Bataille, A., Lee-Cruz, L., Tripathi, B., Kim, H., and Waldman, B. (2016). Microbiome variation across amphibian skin regions: implications for chytridiomycosis mitigation efforts. Microb. Ecol. 71, 221–232. doi: 10.1007/s00248-015-0653-0
Becker, C. G., Greenspan, S. E., Tracy, K. E., Dash, J. A., Lambertini, C., Jenkinson, T. S., et al. (2017). Variation in phenotype and virulence among enzootic and panzootic amphibian chytrid lineages. Fungal Ecol. 26, 45–50. doi: 10.1016/j.funeco.2016.11.007
Becker, M. H., Brucker, R. M., Schwantes, C. R., Harris, R. N., and Minbiole, K. P. C. (2009). The bacterially produced metabolite violacein is associated with survival of amphibians infected with a lethal fungus. Appl. Environ. Microbiol. 75, 6635–6638. doi: 10.1128/AEM.01294-09
Becker, M. H., Harris, R. N., Minbiole, K. P. C., Schwantes, C. R., Rollins-Smith, L. A., Reinert, L. K., et al. (2012). Towards a better understanding of the use of probiotics for preventing chytridiomycosis in Panamanian golden frogs. Ecohealth 8, 501–506. doi: 10.1007/s10393-012-0743-0
Becker, M. H., Walke, J. B., Murrill, L., Woodhams, D. C., Reinert, L. K., Rollins-Smith, L. A., et al. (2015). Phylogenetic distribution of symbiotic bacteria from Panamanian amphibians that inhibit growth of the lethal fungal pathogen Batrachochytrium dendrobatidis. Mol. Ecol. 24, 1628–1641. doi: 10.1111/mec.13135
Bell, R. C., Mackenzie, J. B., Hickerson, M. J., Chavarria, K. L., Cunningham, M., Williams, S., et al. (2012). Comparative multi-locus phylogeography confirms multiple vicariance events in co-distributed rainforest frogs. Proc. R. Soc. B Biol. Sci. 279, 991–999. doi: 10.1098/rspb.2011.1229
Bell, S. C., Alford, R. A., Garland, S., Padilla, G., and Thomas, A. D. (2013). Screening bacterial metabolites for inhibitory effects against Batrachochytrium dendrobatidis using a spectrophotometric assay. Dis. Aquat. Organ. 103, 77–85. doi: 10.3354/dao02560
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate - A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300.
Berger, L., Marantelli, G., Skerratt, L. L., and Speare, R. (2005). Virulence of the amphibian chytrid fungus Batrachochytrium dendrobatidis varies with the strain. Dis. Aquat. Organ. 68, 47–50. doi: 10.3354/dao068047
Berger, L., Speare, R., Daszak, P., Green, D. E., Cunningham, A. A., Goggin, C. L., et al. (1998). Chytridiomycosis causes amphibian mortality associated with population declines in the rain forests of Australia and Central America. Proc. Natl. Acad. Sci. U.S.A. 95, 9031–9036. doi: 10.1073/pnas.95.15.9031
Berger, L., Speare, R., Hines, H. B., Marantelli, G., Hyatt, A. D., McDonald, K. R., et al. (2004). Effect of season and temperature on mortality in amphibians due to chytridiomycosis. Aust. Vet. J. 82, 434–439. doi: 10.1111/j.1751-0813.2004.tb11137.x
Berger, L., Speare, R., and Hyatt, A. (1999). “Chytrid fungi and amphibian declines: overview, implications and future directions,” in Declines and Disappearances of Australian Frogs, ed. A. Campbell (Canberra: Biodiversity Group Environment Australia), 23–33.
Bletz, M. C., Loudon, A. H., Becker, M. H., Bell, S. C., Woodhams, D. C., Minbiole, K. P. C., et al. (2013). Mitigating amphibian chytridiomycosis with bioaugmentation: characteristics of effective probiotics and strategies for their selection and use. Ecol. Lett. 16, 807–820. doi: 10.1111/ele.12099
Boyle, D. G., Boyle, D. B., Olsen, V., Morgan, J. A., and Hyatt, A. D. (2004). Rapid quantitative detection of chytridiomycosis (Batrachochytrium dendrobatidis) in amphibian samples using real-time Taqman PCR assay. Dis. Aquat. Organ. 60, 141–148. doi: 10.3354/dao060141
Bray, J. R., and Curtis, J. T. (1957). An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27, 325–349. doi: 10.2307/1942268
Cade, B. S., and Noon, B. R. (2003). A gentle introduction to quantile regression for ecologists. Front. Ecol. Environ. 1, 412–420. doi: 10.1890/1540-92952003001[0412:AGITQR]2.0.CO;2
Colwell, R. K. (2013). EstimateS: Statistical Estimation of Species Richness and Shared Species from Samples, Version 9.1.0. Available at: http://purl.oclc.org/estimates
Cunningham, M. (2001). Vicariance, Speciation and Diversity in Australopapuan Rainforest Frogs. Ph.D. thesis, University of Queensland, Brisbane.
Currie, C. R., Poulsen, M., Mendenhall, J., Boomsma, J. J., and Billen, J. (2006). Coevolved crypts and exocrine glands support mutualistic bacteria in fungus-growing ants. Science 311, 81–83. doi: 10.1126/science.1119744
Currie, C. R., Scott, J. A., Summerbell, R. C., and Malloch, D. (1999). Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature 398, 701–704. doi: 10.1073/pnas.0812082106
Daskin, J. H., Bell, S. C., Schwarzkopf, L., and Alford, R. A. (2014). Cool temperatures reduce antifungal activity of symbiotic bacteria of threatened amphibians - implications for disease management and patterns of decline. PLoS One 9:e100378. doi: 10.1371/journal.pone.0100378
Dillon, R. J., Vennard, C. T., Buckling, A., and Charnley, A. K. (2005). Diversity of locust gut bacteria protects against pathogen invasion. Ecol. Lett. 8, 1291–1298. doi: 10.1111/j.1461-0248.2005.00828.x
Drummond, A. J., Ashton, B., Buxton, S., Cheung, M., Cooper, A., Heled, J., et al. (2010). Geneious v5.3. Biomatters Ltd. Available at: http://www.geneious.com
Familiar López, M., Rebollar, E. A., Harris, R. N., Vredenburg, V. T., and Hero, J. M. (2017). Temporal variation of the skin bacterial community and Batrachochytrium dendrobatidis infection in the terrestrial cryptic frog Philoria loveridgei. Front. Microbiol. 8:2535. doi: 10.3389/fmicb.2017.02535
Farrer, R. A., Weinert, L. A., Bielby, J., Garner, T. W. J., Balloux, F., Clare, F., et al. (2011). Multiple emergences of genetically diverse amphibian-infecting chytrids include a globalized hypervirulent recombinant lineage. Proc. Natl. Acad. Sci. U.S.A. 108, 18732–18736. doi: 10.1073/pnas.1111915108
Fites, J. S., Ramsey, J. P., and Rollins-Smith, L. A. (2012). Paralysis of amphibian lymphocyte functions by products of the chytrid fungus, Batrachocytrium dendrobatidis. Integr. Comp. Biol. 52:E58.
Fites, J. S., Reinert, L. K., Chappell, T. M., and Rollins-Smith, L. A. (2014). Inhibition of local immune responses by the frog-killing fungus Batrachochytrium dendrobatidis. Infect. Immun. 82, 4698–4706. doi: 10.1128/IAI.02231-14
Flechas, S. V., Sarmiento, C., Cardenas, M. E., Medina, E. M., Restrepo, S., and Amezquita, A. (2012). Surviving chytridiomycosis: differential anti-Batrachochytrium dendrobatidis activity in bacterial isolates from three lowland species of Atelopus. PLoS One 7:e44832. doi: 10.1371/journal.pone.0044832
Fox, J., Weisberg, S., Adler, D., Bates, D., Baud-Bovy, G., Ellison, S., et al. (2017). Companion to Applied Regression, Package ‘car’. Version 2.1-6. Available at: https://cran.r-project.org/web/packages/car/car.pdf
Fraune, S., Anton-Erxleben, F., Augustin, R., Franzenburg, S., Knop, M., Schröder, K., et al. (2015). Bacteria–bacteria interactions within the microbiota of the ancestral metazoan Hydra contribute to fungal resistance. ISME J. 9, 1543–1556. doi: 10.1038/ismej.2014.239
Garland, S., Baker, A., Phillott, A. D., and Skerratt, L. F. (2010). BSA reduces inhibition in a TaqMan (R) assay for the detection of Batrachochytrium dendrobatidis. Dis. Aquat. Organ. 92, 113–116. doi: 10.3354/dao02053
Ghannoum, M. A., and Rice, L. B. (1999). Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin. Microbiol. Rev. 12, 501–517.
Gutierrez-Garcia, K., Neira-González, A., Pérez-Gutiérrez, R. M., Granados-Ramírez, G., Zarraga, R., Wrobel, K., et al. (2017). Phylogenomics of 2,4-diacetylphloroglucinol-producing Pseudomonas and novel antiglycation endophytes from Piper auritum. J. Nat. Prod. 80, 1955–1963. doi: 10.1021/acs.jnatprod.6b00823
Haas, D., and Défago, G. (2005). Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3, 307–319. doi: 10.1038/nrmicro1129
Harris, R. N., Brucker, R. M., Walke, J. B., Becker, M. H., Schwantes, C. R., Flaherty, D. C., et al. (2009a). Skin microbes on frogs prevent morbidity and mortality caused by a lethal skin fungus. ISME J. 3, 818–824. doi: 10.1038/ismej.2009.27
Harris, R. N., James, T. Y., Lauer, A., Simon, M. A., and Patel, A. (2006). Amphibian pathogen Batrachochytrium dendrobatidis is inhibited by the cutaneous bacteria of Amphibian species. Ecohealth 3, 53–56. doi: 10.1007/s10393-005-0009-1
Harris, R. N., Lauer, A., Simon, M. A., Banning, J. L., and Alford, R. A. (2009b). Addition of antifungal skin bacteria to salamanders ameliorates the effects of chytridiomycosis. Dis. Aquat. Organ. 83, 11–16. doi: 10.3354/dao02004
Hol, W. H. G., Garbeva, P., Hordijk, C., Hundscheid, M. P. J., Gunnewiek, P., Van Agtmaal, M., et al. (2015). Non-random species loss in bacterial communities reduces antifungal volatile production. Ecology 96, 2042–2048. doi: 10.1890/14-2359.1
Holden, W. M., Hanlon, S. M., Woodhams, D. C., Chappell, T. M., Wells, H. L., Glisson, S. M., et al. (2015). Skin bacteria provide early protection for newly metamorphosed southern leopard frogs (Rana sphenocephala) against the frog-killing fungus. Batrachochytrium dendrobatidis. Biological Conservation 187, 91–102. doi: 10.1016/j.biocon.2015.04.007
Hoskin, C. J., Higgie, M., McDonald, K. R., and Moritz, C. (2005). Reinforcement drives rapid allopatric speciation. Nature 437, 1353–1356. doi: 10.1038/nature04004
Hothorn, T., Bretz, F., Westfall, P., Heiberger, R. M., Schuetzenmeister, A., and Scheibe, S. (2017). Silmultaneous Inference in General Parametric Models Package ’multcomp’. Version 2.4-5. Available at: https://cran.r-project.org/web/packages/multcomp/multcomp.pdf
Hu, J., Wei, Z., Friman, V. P., Gu, S. H., Wang, X. F., Eisenhauer, N., et al. (2016). Probiotic diversity enhances rhizosphere microbiome function and plant disease suppression. mBio 7:e01790-16. doi: 10.1128/mBio.01790-16
Jani, A. J., and Briggs, C. J. (2014). The pathogen Batrachochytrium dendrobatidis disturbs the frog skin microbiome during a natural epidemic and experimental infection. Proc. Natl. Acad. Sci. U.S.A. 111, E5049–E5058. doi: 10.1073/pnas.1412752111
Jani, A. J., Knapp, R. A., and Briggs, C. J. (2017). Epidemic and endemic pathogen dynamics correspond to distinct host population microbiomes at a landscape scale. Proc. R. Soc. B Biol. Sci. 284:20170944. doi: 10.1098/rspb.2017.0944
Jimenez, R. R., and Sommer, S. (2017). The amphibian microbiome: natural range of variation, pathogenic dysbiosis, and role in conservation. Biodivers. Conserv. 26, 763–786. doi: 10.1007/s10531-016-1272-x
Jousset, A., Becker, J., Chatterjee, S., Karlovsky, P., Scheu, S., and Eisenhauer, N. (2014). Biodiversity and species identity shape the antifungal activity of bacterial communities. Ecology 95, 1184–1190. doi: 10.1890/13-1215.1
Kathiravan, M. K., Salake, A. B., Chothe, A. S., Dudhe, P. B., Watode, R. P., Mukta, M. S., et al. (2012). The biology and chemistry of antifungal agents: a review. Bioorg. Med. Chem. 20, 5678–5698. doi: 10.1016/j.ymeth.2016.11.014
King, K. C., Brockhurst, M. A., Vasieva, O., Paterson, S., Betts, A., Ford, S. A., et al. (2016). Rapid evolution of microbe-mediated protection against pathogens in a worm host. ISME J. 10, 1915–1924. doi: 10.1038/ismej.2015.259
Koenker, R., and Machado, J. A. F. (1999). Goodness of fit and related inference processes for quantile regression. J. Am. Stat. Assoc. 94, 1296–1310. doi: 10.1080/01621459.1999.10473882
Koenker, R., Portnoy, S., Ng, P. T., Zeileis, A., Grosjean, P., and Ripley, B. D. (2017). Quantile Regression Package ‘quantreg’. Version 5.33. Available at: https://cran.r-project.org/web/packages/quantreg/quantreg.pdf
Kriger, K. M., and Hero, J. M. (2007). Large-scale seasonal variation in the prevalence and severity of chytridiomycosis. J. Zool. 271, 352–359.
Kroiss, J., Kaltenpoth, M., Schneider, B., Schwinger, M.-G., Hertweck, C., Maddula, R. K., et al. (2010). Symbiotic streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nat. Chem. Biol. 6, 261–263. doi: 10.1038/nchembio.331
Kueneman, J. G., Woodhams, D. C., Van Treuren, W., Archer, H. M., Knight, R., and Mckenzie, V. J. (2016). Inhibitory bacteria reduce fungi on early life stages of endangered Colorado boreal toads (Anaxyrus boreas). ISME J. 10, 934–944. doi: 10.1038/ismej.2015.168
Küng, D., Bigler, L., Davis, L. R., Gratwicke, B., Griffith, E., and Woodhams, D. C. (2014). Stability of microbiota facilitated by host immune regulation: informing probiotic strategies to manage amphibian disease. PLoS One 9:e87101. doi: 10.1371/journal.pone.0087101
Lam, B. A., Walke, J. B., Vredenburg, V. T., and Harris, R. N. (2010). Proportion of individuals with anti-Batrachochytrium dendrobatidis skin bacteria is associated with population persistence in the frog Rana muscosa. Biol. Conserv. 143, 529–531. doi: 10.1016/j.biocon.2009.11.015
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (Chichester: John Wiley & Sons), 115–175.
Lauer, A., Simon, M. A., Banning, J. L., Andre, E., Duncan, K., and Harris, R. N. (2007). Common cutaneous bacteria from the eastern red-backed salamander can inhibit pathogenic fungi. Copeia 3, 630–640. doi: 10.1643/0045-8511 (2007)2007[630:CCBFTE]2.0.CO;2
Laurance, W. F., McDonald, K. R., and Speare, R. (1996). Epidemic disease and the catastrophic decline of Australian rain forest frogs. Conserv. Biol. 10, 406–413. doi: 10.1046/j.1523-1739.1996.10020406.x
Longo, A. V., Savage, A. E., Hewson, I., and Zamudio, K. R. (2015). Seasonal and ontogenetic variation of skin microbial communities and relationships to natural disease dynamics in declining amphibians. R. Soc. Open Sci. 2:140377. doi: 10.1098/rsos.140377
Longo, A. V., and Zamudio, K. R. (2017). Environmental fluctuations and host skin bacteria shift survival advantage between frogs and their fungal pathogen. ISME J. 11, 349–361. doi: 10.1038/ismej.2016.138
Loudon, A. H., Holland, J. A., Umile, T. P., Burzynski, E. A., Minbiole, K. P., and Harris, R. N. (2014). Interactions between amphibians’ symbiotic bacteria cause the production of emergent anti-fungal metabolites. Front. Microbiol. 5:441. doi: 10.3389/fmicb.2014.00441
Madison, J. D., Berg, E. A., Abarca, J. G., Whitfield, S. M., Gorbatenko, O., Pinto, A., et al. (2017). Characterization of Batrachochytrium dendrobatidis inhibiting bacteria from amphibian populations in Costa Rica. Front. Microbiol. 8:290. doi: 10.3389/fmicb.2017.00290
Mao-Jones, J., Ritchie, K. B., Jones, L. E., and Ellner, S. P. (2010). How microbial community composition regulates coral disease development. PLoS Biol. 8:e1000345. doi: 10.1371/journal.pbio.1000345
Martinez Arbizu, P. (2016). “How can I do perMANOVA Pairwise Contrasts in R? Response to the Question Posted. Available at: www.researchgate.net
Martín-Vivaldi, M., Peña, A., Peralta-Sánchez, J. M., Sánchez, L., Ananou, S., Ruiz-Rodríguez, M., et al. (2010). Antimicrobial chemicals in hoopoe preen secretions are produced by symbiotic bacteria. Proc. R. Soc. B Biol. Sci. 277, 123–130. doi: 10.1098/rspb.2009.1377
Matos, A., Kerkhof, L., and Garland, J. L. (2005). Effects of microbial community diversity on the survival of Pseudomonas aeruginosa in the wheat rhizosphere. Microb. Ecol. 49, 257–264. doi: 10.1007/s00248-004-0179-3
Mattoso, T. C., Moreira, D. D., and Samuels, R. I. (2012). Symbiotic bacteria on the cuticle of the leaf-cutting ant Acromyrmex subterraneus subterraneus protect workers from attack by entomopathogenic fungi. Biol. Lett. 8, 461–464. doi: 10.1098/rsbl.2011.0963
McDonald, K., and Alford, R. (1999). “A review of Declining Frogs in Northern Queensland,” in Declines and Disappearances of Australian Frogs, ed. A. Campbell (Canberra: Biodiversity Group Environment Australia), 14–22.
McDonald, K. R., Méndez, D., Müller, R., Freeman, A. B., and Speare, R. (2005). Decline in the prevalence of chytridiomycosis in upland frog populations in North Queensland, Australia. Pac. Conserv. Biol. 11, 114–129. doi: 10.1071/PC050114
McKnight, D. T., Alford, R. A., Hoskin, C. J., Schwarzkopf, L., Greenspan, S. E., Zenger, K. R., et al. (2017). Fighting an uphill battle: the recovery of frogs in Australia’s Wet Tropics. Ecology 98, 3221–3223. doi: 10.1002/ecy.2019
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., Mcglinn, D., et al. (2017). Community Ecology Package ‘vegan’. Version 2.4-4. Available at: https://cran.r-project.org/web/packages/vegan/vegan.pdf
Piovia-Scott, J., Rejmanek, D., Woodhams, D. C., Worth, S. J., Kenny, H., Mckenzie, V., et al. (2017). Greater species richness of bacterial skin symbionts better suppresses the amphibian fungal pathogen Batrachochytrium dendrobatidis. Microb. Ecol. 74, 217–226. doi: 10.1007/s00248-016-0916-4
Poulsen, M., Cafaro, M. J., Erhardt, D. P., Little, A. E., Gerardo, N. M., Tebbets, B., et al. (2010). Variation in Pseudonocardia antibiotic defence helps govern parasite-induced morbidity in Acromyrmex leaf-cutting ants. Environ. Microbiol. Rep. 2, 534–540. doi: 10.1111/j.1758-2229.2009.00098.x
Pruesse, E., Peplies, J., and Glöckner, F. O. (2012). SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28, 1823–1829. doi: 10.1093/bioinformatics/bts252
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rambaut, A. (2016). FigTree. Version 1.4.3. Available at: http://tree.bio.ed.ac.uk/software/figtree/
Rebollar, E. A., Antwis, R. E., Becker, M. H., Belden, L. K., Bletz, M. C., Brucker, R. M., et al. (2016). Using “omics” and integrated multi-omics approaches to guide probiotic selection to mitigate chytridiomycosis and other emerging infectious diseases. Front. Microbiol. 7:68. doi: 10.3389/fmicb.2016.00068
Richards, S. J., and Alford, R. A. (2005). Structure and dynamics of a rainforest frog (Litoria genimaculata) population in northern Queensland. Aust. J. Zool. 53, 229–236. doi: 10.1071/ZO03036
Richards, S. J., Hoskin, C. J., Cunningham, M. J., McDonald, K., and Donnellan, S. C. (2010). Taxonomic re-assessment of the Australian and New Guinean green-eyed treefrogs Litoria eucnemis, L. genimaculata and L. serrata (Anura: Hylidae). Zootaxa 2391, 33–46.
Richards, S. J., McDonald, K. R., and Alford, R. A. (1993). Declines in populations of Australia’s endemic tropical rainforest frogs. Pac. Conserv. Biol. 1, 66–77. doi: 10.1071/PC930066
Ritchie, K. B. (2006). Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 322, 1–14. doi: 10.3354/meps322001
Robinson, J. V., and Valentine, W. D. (1979). The concepts of elasticity, invulnerability and invadability. J. Theor. Biol. 81, 91–104. doi: 10.1016/0022-5193(79)90083-3
Rollins-Smith, L. A., Carey, C., Longcore, J. E., Doersam, J. K., Reinert, L. K., Boutte, A., et al. (2002). Antimicrobial peptide defenses against Batrachochytrium dendrobatidis, the chytrid fungus associated with global amphibian declines. FASEB J. 16, A291–A291.
Rollins-Smith, L. A., Ramsey, J. P., Pask, J. D., Reinert, L. K., and Woodhams, D. C. (2011). Amphibian immune defenses against chytridiomycosis: Impacts of changing environments. Integr. Comp. Biol. 51, 552–562. doi: 10.1093/icb/icr095
Rosenberg, E., Koren, O., Reshef, L., Efrony, R., and Zilber-Rosenberg, I. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362. doi: 10.1038/nrmicro1635
Rosenberg, E., and Zilber-Rosenberg, I. (2016). Microbes drive evolution of animals and plants: the hologenome concept. mBio 7:e01395-15. doi: 10.1128/mBio.01395-15
Rowley, J. J. L., and Alford, R. A. (2013). Hot bodies protect amphibians against chytrid infection in nature. Sci. Rep. 3:1515. doi: 10.1038/srep01515
Salle, A. J. (1961). Laboratory Manual on Fundamental Principles of Bacteriology. London: McGraw-Hill.
Sapsford, S. J., Alford, R. A., and Schwarzkopf, L. (2013). Elevation, temperature, and aquatic connectivity all influence the infection dynamics of the amphibian chytrid fungus in adult frogs. PLoS One 8:e82425. doi: 10.1371/journal.pone.0082425
Savage, A. E., and Zamudio, K. R. (2011). MHC genotypes associate with resistance to a frog-killing fungus. Proc. Natl. Acad. Sci. U.S.A. 108, 16705–16710. doi: 10.1073/pnas.1106893108
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Schneider, C. J., Cunningham, M., and Moritz, C. (1998). Comparative phylogeography and the history of endemic vertebrates in the Wet Tropics rainforests of Australia. Mol. Ecol. 7, 487–498. doi: 10.1016/j.ympev.2011.10.010
Scott, J. J., Oh, D. C., Yuceer, M. C., Klepzig, K. D., Clardy, J., and Currie, C. R. (2008). Bacterial protection of beetle-fungus mutualism. Science 322:63. doi: 10.1126/science.1160423
Skerratt, L. F., Berger, L., Speare, R., Cashins, S., McDonald, K. R., Phillott, A. D., et al. (2007). Spread of chytridiomycosis has caused the rapid global decline and extinction of frogs. Ecohealth 4, 125–134. doi: 10.1007/s10393-007-0093-5
Skerratt, L. F., McDonald, K. R., Hines, H. B., Berger, L., Mendez, D., Phillott, A. D., et al. (2010). Application of the survey protocol for chytridiomycosis to Queensland, Australia. Dis. Aquat. Organ. 92, 117–129. doi: 10.3354/dao02272
Soler, J. J., Martín-Vivaldi, M., Ruiz-Rodríguez, M., Valdivia, E., Martín-Platero, A. M., Martínez-Bueno, M., et al. (2008). Symbiotic association between hoopoes and antibiotic-producing bacteria that live in their uropygial gland. Funct. Ecol. 22, 864–871. doi: 10.1111/j.1365-2435.2008.01448.x
Stuart, S. N., Chanson, J. S., Cox, N. A., Young, B. E., Rodrigues, A. S., Fischman, D. L., et al. (2004). Status and trends of amphibian declines and extinctions worldwide. Science 306, 1783–1786. doi: 10.1126/science.1103538
Tukey, J. (1949). Comparing individual means in the analysis of variance. Biometrics 5, 99–114. doi: 10.2307/3001913
van Elsas, J. D., Chiurazzi, M., Mallon, C. A., Elhottovā, D., Krištůfek, V., and Salles, J. F. (2012). Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc. Natl. Acad. Sci. U.S.A. 109, 1159–1164. doi: 10.1073/pnas.1109326109
Voyles, J., Woodhams, D. C., Saenz, V., Byrne, A. Q., Perez, R., Rios-Sotelo, G., et al. (2018). Shifts in disease dynamics in a tropical amphibian assemblage are not due to pathogen attenuation. Science 359, 1517–1519. doi: 10.1126/science.aao4806
Wake, D. B., and Vredenburg, V. T. (2008). Are we in the midst of the sixth mass extinction? A view from the world of amphibians. Proc. Natl. Acad. Sci. U.S.A. 105, 11466–11473. doi: 10.1073/pnas.0801921105
Walke, J. B., Becker, M. H., Hughey, M. C., Swartwout, M. C., Jensen, R. V., and Belden, L. K. (2015a). Most of the dominant members of amphibian skin bacterial communities can be readily cultured. Appl. Environ. Microbiol. 81, 6589–6600. doi: 10.1128/AEM.01486-15
Walke, J. B., Becker, M. H., Loftus, S. C., House, L. L., Teotonio, T. L., Minbiole, K. P., et al. (2015b). Community structure and function of amphibian skin microbes: an experiment with bullfrogs exposed to a chytrid fungus. PLoS One 10:e0139848. doi: 10.1371/journal.pone.0139848
Walke, J. B., Harris, R. N., Reinert, L. K., Rollins-Smith, L. A., and Woodhams, D. C. (2011). Social immunity in amphibians: evidence for vertical transmission of innate defenses. Biotropica 43, 396–400. doi: 10.1111/j.1744-7429.2011.00787.x
Wickham, H., and Chang, W. (2016). Create Elegant Data Visualisations Using the Grammar of Graphics Package ‘ggplot2’. Version 2.2.1. Available at: https://cran.r-project.org/web/packages/ggplot2/ggplot2.pdf
Woodhams, D. C., and Alford, R. A. (2005). Ecology of chytridiomycosis in rainforest stream frog assemblages of tropical Queensland. Conserv. Biol. 19, 1449–1459. doi: 10.1111/j.1523-1739.2005.004403.x
Woodhams, D. C., Brandt, H., Baumgartner, S., Kielgast, J., Kupfer, E., Tobler, U., et al. (2014). Interacting symbionts and immunity in the amphibian skin mucosome predict disease risk and probiotic effectiveness. PLoS One 9:e96375. doi: 10.1371/journal.pone.0096375
Woodhams, D. C., Kenyon, N., Bell, S. C., Alford, R. A., Chen, S., Billheimer, D., et al. (2010). Adaptations of skin peptide defences and possible response to the amphibian chytrid fungus in populations of Australian green-eyed treefrogs, Litoria genimaculata. Divers. Distrib. 16, 703–712. doi: 10.1111/j.1472-4642.2010.00666.x
Woodhams, D. C., Vredenburg, V. T., Simon, M. A., Billheimer, D., Shakhtour, B., Shyr, Y., et al. (2007). Symbiotic bacteria contribute to innate immune defenses of the threatened mountain yellow-legged frog, Rana muscosa. Biol. Conserv. 138, 390–398. doi: 10.1016/j.biocon.2007.05.004
Yilmaz, P., Parfrey, L. W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., et al. (2014). The SILVA and “All-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42, D643–D648. doi: 10.1093/nar/gkt1209
Keywords: amphibian, cutaneous bacteria, Batrachochytrium dendrobatidis, microbiota, cell-free supernatant, inhibitory bacteria, chytridiomycosis, disease mitigation
Citation: Bell SC, Garland S and Alford RA (2018) Increased Numbers of Culturable Inhibitory Bacterial Taxa May Mitigate the Effects of Batrachochytrium dendrobatidis in Australian Wet Tropics Frogs. Front. Microbiol. 9:1604. doi: 10.3389/fmicb.2018.01604
Received: 31 October 2017; Accepted: 27 June 2018;
Published: 18 July 2018.
Edited by:
Eria Alaide Rebollar, National Autonomous University of Mexico (Morelos), MexicoReviewed by:
Molly Bletz, Technische Universitat Braunschweig, GermanyMatthew Henry Becker, Smithsonian Institution, United States
Copyright © 2018 Bell, Garland and Alford. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sara C. Bell, c2FyYWNiZWxsQGdtYWlsLmNvbQ==
†Present address: Sara C. Bell, Australian Institute of Marine Science, Townsville, QLD, Australia
 Sara C. Bell
Sara C. Bell Stephen Garland
Stephen Garland Ross A. Alford
Ross A. Alford