- 1Department of Natural Resource Sciences, McGill University, Sainte-Anne-de-Bellevue, QC, Canada
- 2Biotechnology Research Institute, National Research Council of Canada, Montreal, QC, Canada
Permafrost subzero environments harbor diverse, active communities of microorganisms. However, our understanding of the subzero growth, metabolisms, and adaptive properties of these microbes remains very limited. We performed transcriptomic analyses on two subzero-growing permafrost isolates with different growth profiles in order to characterize and compare their cold temperature growth and cold-adaptive strategies. The two organisms, Rhodococcus sp. JG3 (-5 to 30°C) and Polaromonas sp. Eur3 1.2.1 (-5 to 22°C), shared several common responses during low temperature growth, including induction of translation and ribosomal processes, upregulation of nutrient transport, increased oxidative and osmotic stress responses, and stimulation of polysaccharide capsule synthesis. Recombination appeared to be an important adaptive strategy for both isolates at low temperatures, likely as a mechanism to increase genetic diversity and the potential for survival in cold systems. While Rhodococcus sp. JG3 favored upregulating iron and amino acid transport, sustaining redox potential, and modulating fatty acid synthesis and composition during growth at -5°C compared to 25°C, Polaromonas sp. Eur3 1.2.1 increased the relative abundance of transcripts involved in primary energy metabolism and the electron transport chain, in addition to signal transduction and peptidoglycan synthesis at 0°C compared to 20°C. The increase in energy metabolism may explain why Polaromonas sp. Eur3 1.2.1 is able to sustain growth rates at 0°C comparable to those at higher temperatures. For Rhodococcus sp. JG3, flexibility in use of carbon sources, iron acquisition, control of membrane fatty acid composition, and modulating redox and co-factor potential may be ways in which this organism is able to sustain growth over a wider range of temperatures. Increasing our understanding of the microbes in these habitats helps us better understand active pathways and metabolisms in extreme environments. Identifying novel, thermolabile, and cold-active enzymes from studies such as this is also of great interest to the biotechnology and food industries.
Introduction
The discovery of active, growing communities in polar habitats has given rise to much interest in studying the microbes that inhabit these regions and obtaining a greater understanding of the mechanisms and adaptive features that allow them to grow in cryoenvironments, generally defined as environments that exist continuously and predominantly at subzero temperatures. Microorganisms living in cryoenvironments must adapt to overcome numerous physiological and kinetic challenges, including decreased membrane fluidity, reduced diffusion and reaction rates, lower enzyme activity, stable secondary inhibitory DNA/RNA structures, and protein misfolding (Bakermans et al., 2012; De Maayer et al., 2014). Subzero environments, including permafrost and ice, are characterized by limited water availability, and liquid water in these habitats is believed to exist primarily in salty brine veins where increased solute and salt concentrations prevent freezing (Gilichinsky et al., 2005). As such, mechanisms must also exist to overcome osmotic stress and high salt concentrations.
Organisms isolated from these environments and capable of growth at low (≤0°C) temperatures have generally been classified as eurypsychrophiles (formerly psychrotrophs/psychrotolerant), as those organisms possess a broader growth range (Tmax > 20°C), and while capable of growth at very low temperatures (-15°C), they usually retain optimum growth rates at temperatures above 20°C (Feller and Gerday, 2003; Cavicchioli, 2006; Bakermans et al., 2012). In comparison, stenopsychrophiles (formerly “true psychrophiles”) exhibit narrower growth ranges (Tmax ∼20°C), and usually grow optimally at temperatures ranging between 5 and 15°C. We will use the term cold-adapted bacteria or psychrophile here to refer to both steno- and eury-psychrophiles. While it is true that eurypsychrophiles generally grow optimally at >20°C, they often exhibit growth over a wide range of temperatures and their low temperature minimum is often lower than stenopsychrophiles; indeed, almost all of the organisms reported to date capable of growth at -5°C and below are eurypsychrophiles (Bakermans et al., 2007; Rodrigues et al., 2008; Bergholz et al., 2009; Mykytczuk et al., 2013; Koh et al., 2017). This is an interesting observation and raises questions about the cold-adaptive properties and limitations of both types of cold-adapted bacteria.
The vast majority of studies into cold adaptation and growth, especially proteomes and transcriptomes, have focused heavily on eurypsychrophiles, and most of these, on above zero growth conditions (reviewed extensively in Kawamoto et al., 2017 and Raymond-Bouchard and Whyte, 2017). Comparatively little is known about stenopsychrophiles, especially in bacteria, whose members are more limited, and include Colwellia psychrerythraea and Desulfotalea psychrophila (Rabus et al., 2004; Methé et al., 2005). One characteristic of stenopsychrophiles that may allow for faster growth at lower temperatures is their ability to increase energy and carbon metabolism at these temperatures, when compared to higher suboptimal temperatures (Mock and Hoch, 2005; Mock et al., 2005; Hwang et al., 2008; Chong et al., 2011). These mechanisms are often downregulated in eurypsychrophiles at lower temperatures (Bergholz et al., 2009; Mykytczuk et al., 2013; Tribelli et al., 2015).
While both types of organisms have been studied for their cold-adaptive properties, almost no studies have directly compared the two types of psychrophiles (eury- and steno-psychrophiles) to elucidate their cold-adapted features and compare and contrast the different strategies that characterize their varied growth capabilities and optimums. We aim to increase our knowledge of cold and subzero growth in permafrost isolated eury- and steno-psychrophiles and gain a better understanding of their cold-adaptive strategies for growth at these temperatures. The eurypsychrophile Rhodococcus sp. JG3, isolated from permafrost (Goordial et al., 2016) in the McMurdo Dry Valley, Antarctica, is capable of growth from at least -5 to 30°C, and grows optimally at 20–25°C. Polaromonas sp. Eur3 1.2.1, isolated from permafrost in Eureka (Steven et al., 2008), Ellesmere Island, Nunavut, is a stenopsychrophile capable of growth from -5 up to 22°C and exhibits a faster generation time (g) at 10°C (3.1 days) than 22°C (3.4 days), and only slightly lower at 0°C (3.6 days) (Raymond-Bouchard et al., 2018). Polaromonas sp. Eur3 1.2.1 is a slow-growing, fastidious organism, and while growth down to -5°C has been detected on ½ R2A agar supplemented with 7% sucrose, so far no liquid media adequate for subzero growth has been found. In this study, our goal was to perform transcriptomic analyses of both organisms at their lower temperature limits of growth, 0 and -5°C, in addition to higher ≥20°C temperatures, in order to characterize and compare their low temperature growth and cold-adaptive strategies.
We chose to focus on potentially relevant transcriptomic changes in each organism during cold growth, as determined using RNA-seq, since our goal was to highlight cold-active pathways and genes of interest, which could be targets of future studies. We acknowledge that these organisms are evolutionarily distant. Our goal was to observe cold-adaptive changes and trends between evolutionarily different organisms. While very similar organisms (i.e., same genus) would allow us to narrow down very specific cold adaptations or changes in specific strains, our interest lies in beginning to elucidate adaptations on a larger scale, such as those that may be universal, compared to those that may be more specific to certain groups. Using evolutionarily different organisms allow us to investigate these potential differences by looking at each organism individually, in parallel, from a lower to higher temperatures and then looking for relative changes or trends between them.
Materials and Methods
Culturing and Growth Conditions
Polaromonas sp. Eur3 1.2.1 and Rhodococcus sp. JG3 were cultured on ½ R2A and R2A agar (BD Difco, Franklin Lakes, NJ, United States), respectively. For RNA extraction, liquid cultures of each strain were grown in biological triplicate to mid-late exponential phase on ½ R2A at 0 and 20°C (OD600 0.165) for Polaromonas sp. Eur3 1.2.1, and in tryptic soy broth (TSB; BD Difco) supplemented with 5% salt and 5% sucrose at -5°C (OD600 1.0) and 25°C (OD600 2.0) for Rhodococcus sp. JG3. Polaromonas sp. Eur3 1.2.1 is a slow-growing organism with low cell densities, and as a result, several cultures had to be combined into one for each triplicate condition to compensate for the reduced amount of biomass obtained. It is challenging to identify adequate low temperature growth media for psychrophilic organisms. Ideally, we would have liked to grow both organisms on the same media. However, Polaromonas sp. Eur3 1.2.1 is a very fastidious organism and we have only been successful in growing the organism on ½ R2A. Rhodococcus sp. JG3, on the other hand grows much better on TSB at subzero temperatures and exhibits very slow growth rates on ½ R2A such that it was not feasible to perform this experiment on ½ R2A with Rhodococcus sp. JG3 at -5°C. We understand that some of the differences in transcriptional responses may be a result of differential growth media and have attempted to take this under consideration in our discussion. However, it is important to state that we are primarily looking at changes in one organism during growth at higher vs. lower temperatures. Subsequently, we make comparisons between the two organisms when general trends of interest can be seen.
RNA Extraction and Library Preparation
RNA extraction was performed using the Direct-zol RNA MiniPrep Plus Kit (#R2070) from Zymo Research (Irvine, CA, United States) according to manufacturer’s instructions. RNA quantity and purity were checked using the Qubit 3.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, United States) and 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States). Sequencing libraries were prepared using the Illumina (San Diego, CA, United States) TruSeq Stranded mRNA LT Sample Prep Kit as outlined in the manufacturer’s guide.
Illumina Sequencing and Bioinformatics Analysis
Sequencing was performed by RNAseq on the Illumina MiSeq system using a Reagent Kit v3 150 cycles with a 2 × 75 bp paired-end configuration. Sequencing raw data (728 mega-bases) were processed through our metatranscriptomics bioinformatic pipelines. Read count summaries are provided for each sequencing library in Supplementary Table S1. Sequencing adapters were removed from each read and bases at the end of reads having a quality score less than 30 were cutoff (Trimmomatic v0.32) (Bolger et al., 2014) to generate quality controlled (QC) reads. QC-passed reads were mapped (BWA mem v0.7.10)1 (unpublished) against Polaromonas sp. Eur3 1.2.1 or Rhodococcus sp. JG3 genome references to obtain contig abundance profiles. Polaromonas sp. Eur3 1.2.1 and Rhodococcus sp. JG3 genomes were obtained from the Joint Genome Institute2, ID 2619618817 and 2529292502, respectively. Alignment files in bam format were sorted by read coordinates using samtools v1.1 (Li et al., 2009) and only properly aligned read pairs were kept for downstream steps. Each bam file (containing properly aligned paired-reads only) was analyzed for coverage of genes and contigs using bedtools (v2.17.0) (Quinlan and Hall, 2010) using corresponding gene models (gene coordinates on each contig for both genome references). Only paired-reads both overlapping their contigs or genes were considered for gene counts. Coverage profiles of each sample were merged to generate an abundance matrix (rows = contig, columns = samples) for which a corresponding counts per million abundance matrix (edgeR v3.10.2) (Robinson et al., 2009) was generated. According to our experimental design, differentially expressed genes were assessed with edgeR (v3.10.2) using its generalized linear model approach detailed by the authors (see section 3.2.3, page 29)3 with transcriptomics raw counts matrices as input. Genes having a log fold-change (logFC) ratio ≥| 1.5| and false discovery rate (FDR) <0.05 were considered as differentially expressed between the two temperatures in each organism. We did not directly compare Rhodococcus sp. JG3 and Polaromonas sp. Eur3 1.2.1 counts, rather we observed them individually in parallel and looked for relative differences at each organism’s reaction to cold-growth conditions. Metadata for all samples reported in this study are available in Supplementary Table S1. Raw sequence reads of the shotgun transcriptomics data were submitted to the Sequence Read Archive under BioProject ID PRJNA4225964.
Data Analysis
Predicted COG and KEGG assignments for Rhodococcus sp. JG3 (ID 2529292502) and Polaromonas sp. Eur3 1.2.1 (ID 2619618817) were downloaded from the JGI IMG/ER website5, and used to obtain preliminary function and pathway assignments. These were then manually cross-referenced against available literature for the proteins discussed in the text to ensure function and pathway assignments are as accurate as possible. Focus was given to those genes with logFC ≥ 1.5 (FDR < 0.05) for which functional and pathway assignments could best be predicted based on homology assignments, conserved domains, and literature data. The volcano plot was created in R (v3.4.0) (R Core Team, 2017).
Results and Discussion
Using RNAseq we carried out transcriptomic analyses on (triplicate) cultures of Rhodococcus sp. JG3 grown at -5 and 25°C and Polaromonas sp. Eur3 1.2.1 grown at 0 and 20°C. Overall, 515 and 359 transcripts were found to be differentially expressed for Rhodococcus sp. JG3 and Polaromonas sp. Eur3 1.2.1, respectively, between their respective higher and lower temperatures (Table 1 and Supplementary Figure S1). Of these about 66 and 45% could be assigned to COG and KEGG categories for both organisms. The largest shifts in abundance of transcripts observed for Rhodococcus sp. JG3 between -5 and 25°C, according to COG assignments, were in translation and ribosomal proteins, amino acid and inorganic ion transport and metabolism, lipid transport and metabolism, and energy production and conversion (Figure 1). Translation and amino acid and ion transport and metabolism processes saw significant upshifts in transcripts at -5°C, while transcripts involved in energy production and conversion, and lipid transport and metabolism were downregulated. Like Rhodococcus sp. JG3, Polaromonas sp. Eur3 1.2.1 also showed upregulation of transcripts involved in translation and ribosomal proteins at low temperature (0°C), compared to 20°C (Figure 1). However, unlike Rhodococcus sp. JG3, Polaromonas sp. Eur3 1.2.1 upregulated genes involved in energy production at low temperature and downregulated gene expression of amino acid transport and metabolism. In addition, noticeable increases in abundance of transcripts involved in signal transduction and carbohydrate transport and metabolism were also detected, along with a decrease in coenzyme transport and metabolism. Interestingly, at both 0 and 20°C Polaromonas sp. Eur3 1.2.1 upregulated a unique subset of transposon elements (discussed further below).
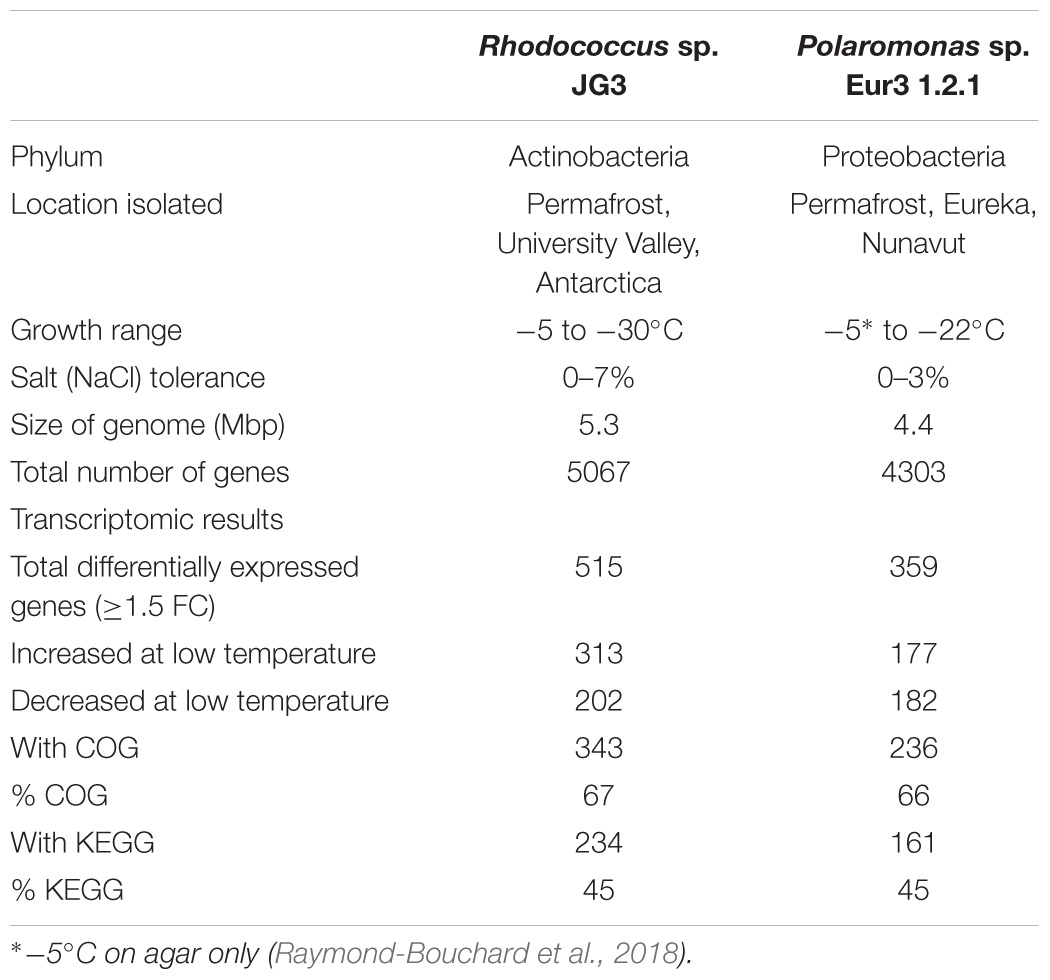
TABLE 1. Strain information and summary of transcriptomic results for Rhodococcus sp. JG3 and Polaromonas sp. Eur3 1.2.1.
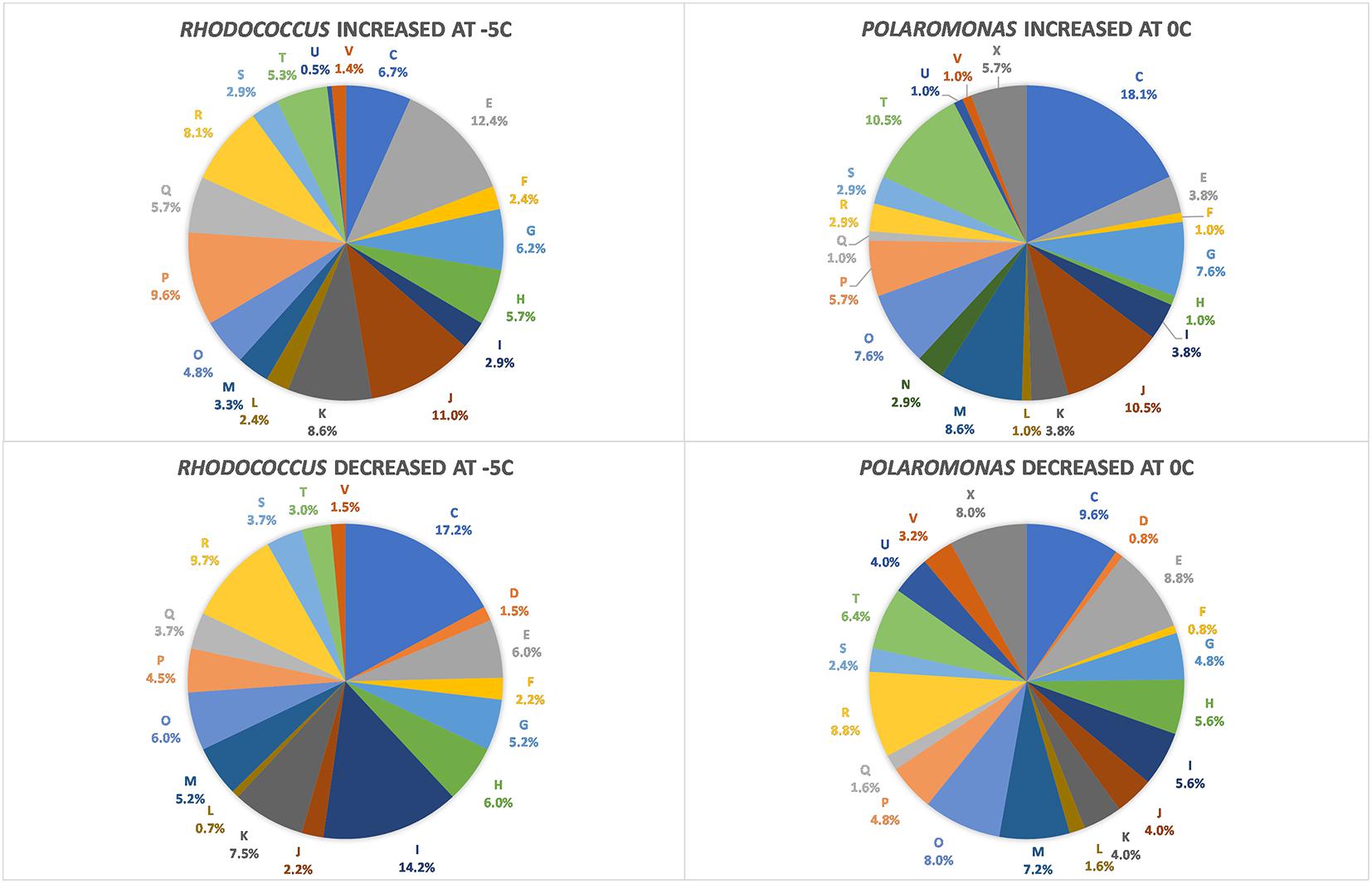
FIGURE 1. Diagram of COG categories with differentially expressed genes in Polaromonas sp. Eur3 1.2.1 at 0°C and Rhodococcus sp. JG3 at –5°C. The number of differentially expressed genes in each category is expressed as a percent of total. Letters correspond to COG categories: C, energy production and conversion; D, cell cycle control, cell division, and chromosome partitioning; E, amino acid metabolism and transport; F, nucleotide metabolism and transport; G, carbohydrate metabolism and transport; H, coenzyme metabolism and transport; I, lipid metabolism and transport; J, translation, ribosome structure, and biogenesis; K, transcription; L, replication, recombination, and repair; M, cell wall/membrane/envelope biogenesis; N, cell motility and secretion; O, post-translational modification, protein turnover, and chaperone functions; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport, and catabolism; T, signal transduction; V, defense mechanisms; and R, general functional prediction only.
Overall, the response in Polaromonas sp. Eur3 1.2.1 was relatively more attenuated than in Rhodococcus sp. JG3. Fewer genes were found to be differentially regulated, with less than 40 genes upregulated >threefold change (logFC) at 0°C, and no transcripts with logFC > 5.8, as compared to Rhodococcus sp. JG3 where ∼150 genes were upregulated >threefold and up to 8.7 logFC (Table 1 and Supplementary Figure S1). It is possible, given the reduced response and the fairly consistent generation times (3.6 days at 0°C, 3.1 at 10°C, and 3.4 days at 22°C) observed for Polaromonas sp. Eur3 1.2.1 over its temperature range that it remains somewhat constitutively adapted to the lower temperatures and does not need to shift processes to the extent that Rhodococcus sp. JG3 does at lower temperatures.
Regardless of the influence of different media used for the two organisms, differences between the cold and the optimal temperatures give us key insight into global transcriptional changes that can occur in an organism in response to cold. Our main aim was to compare the transcriptional differences in each organism from their higher to lower growth temperatures and to highlight potentially interesting changes that may be relevant to cold adaptation in each organism. Following this, in cases where differences could be observed between the organisms, we believed there was value in discussing these, especially since some of the differences observed are consistent with previous studies on steno- and eury-psychrophiles.
Translation and Ribosomal Proteins
The importance of sustaining appropriate and necessary translation and ribosomal processes at low temperatures is apparent in the responses of both Rhodococcus sp. JG3 and Polaromonas sp. Eur3 1.2.1 at -5°C and 0°C, respectively. Rhodococcus sp. JG3 shows upregulation of at least 24 genes with logFC > 1.5 involved in translation and ribosomal processes, including numerous ribosomal proteins, a GTPase Era, the methylase RsmH, and the translation elongation factor EF-Ts (Supplementary Table S2) compared to the higher temperature 25°C. A queuine tRNA-ribosyltransferase and an O-acetyl-ADP-ribose deacetylase, which downregulates RNase III activity, as well as the ribosome binding factor A (rbfA) and a superfamily II DNA and RNA helicase (includes DEAD/DEAH-box family proteins) were upregulated >3 logFC. Helicases of this family are a common feature of cold growth in numerous cold-adapted bacteria, owing to their importance in stabilizing translation processes (Piette et al., 2011b). RbfA, involved in ribosome maturation and translation initiation, binds to the 30S subunit and is important for cold tolerance in Escherichia coli, where overexpression of this protein at cold temperatures (15°C) suppresses a cold-sensitive mutation and increases protein synthesis and growth (Dammel and Noller, 1995; Jones and Inouye, 1996). Upregulation of RbfA has also been reported for the psychrophiles Exiguobacterium sibiricum at -2.5°C (Rodrigues et al., 2008) and Psychrobacter arcticus 273-4 at -6°C (Bergholz et al., 2009). A fivefold upregulation of ADP-ribosylglycohydrolase, responsible for removal of ADP-ribose from ADP-ribosylated proteins was also observed in Rhodococcus sp. JG3 and may be important in the regulation of many cellular processes, including DNA/protein repair, DNA recombination, signal transduction, and gene transcription (Liu and Yu, 2015). Conversely, very few decreases were observed in this category (Supplementary Tables S2, S4) and included mostly translation inhibitors, such as the gene for an mRNA degradation ribonuclease and the ribosome-associated translation inhibitor RaiA, indicating the importance of keeping translation processes active at low temperatures.
Similarly, Polaromonas sp. Eur3 1.2.1 increased expression of transcripts for a number of ribosomal proteins, a superfamily II helicase, and the translation initiation factor IF-1, with concurrently small fold decreases in transcripts in this category (Supplementary Tables S3, S5) at 0°C compared to 20°C. Two of the most upregulated genes in Polaromonas sp. Eur3 1.2.1 were molecular chaperones, a disulfide isomerase subunit, dsbG, of the thiol:disulfide interchange protein, and a second thiol:disulfide isomerase. Another thiol:disulfide interchange subunit, dsbD, was also increased. The thiol:disulfide isomerases are required for proper folding of proteins through isomerization of incorrect disulfide bonds to the correct pattern (Shao et al., 2000; Gleiter and Bardwell, 2008). They prevent misfolding and aggregation of proteins, which is believed to be more prevalent at lower temperatures (Doyle et al., 2012). Polaromonas sp. Eur3 1.2.1 also downshifts expression of the Ribonuclease E in favor of its homolog Ribonuclease G, an enzyme with more limited enzymatic capabilities than RNase E (Deana and Belasco, 2004). The importance of sustaining translation at low temperatures has been highlighted in studies of both eury- and steno-psychrophiles (Goodchild et al., 2004; Mock et al., 2005; Nunn et al., 2015; Koh et al., 2017) and has been postulated to be the rate-limiting step for protein synthesis in certain cold communities (Toseland et al., 2013). Shifts in ribosomal and translation proteins ultimately allow the ribosome to become “cold-adapted” and preserve translation at cold temperatures, thus maintaining growth and metabolic activity.
Transposons, DNA Recombination, and Genomic Redundancy
Polaromonas sp. Eur3 1.2.1 appears to express transcripts for a number of transposase elements, and at 0°C, shifts expression in favor of IS5 family transposases (Supplementary Table S3) compared to 20°C. IS5 can mediate directed mutations and causes gene activation or inactivation through insertion upstream of targeted genes near the promoter (Zhang and Saier, 2012). Transcripts for the competent protein, ComEA, important for free DNA uptake and horizontal gene transfer, was also increased slightly. While Rhodococcus sp. JG3 does not appear to differentially express transposon elements from -5 to 25°C, several genes with roles in DNA recombination, including a single strand DNA binding protein, the Holliday junction helicase (RuvA), and the alpha subunit (RecD) of exonuclease V were all increased ∼twofold at -5°C relative to 25°C (Supplementary Table S2). Two transposons and several recombination proteins were expressed in the psychrophilic archaeon Methanococcoides burtonii at 4°C (Goodchild et al., 2004) and several psychrophilic genomes, including the genome of subzero-growing P. arcticus 273-4, have been shown to possess large numbers of elements that contribute to genome plasticity, such as plasmids, transposons, and other mobile elements (Allen et al., 2009; Ayala-del-Río et al., 2010; Math et al., 2012). Recombination, both from transposition and site-specific recombination, may serve as a general adaptive strategy to increase genetic diversity and the potential for survival and growth in cold-temperature systems.
Indeed, genomic redundancy and isozyme exchange has been shown to be a cold-adaptive strategy in certain cold-adapted bacteria, including E. sibiricum, P. arcticus, Planococcus halocryophilus, and Psychrobacter sp. PAMC 21119, with different copies of genes being expressed at different temperatures (Rodrigues et al., 2008; Bergholz et al., 2009; Mykytczuk et al., 2013; Koh et al., 2017). Both Rhodococcus sp. JG3 and Polaromonas sp. Eur3 1.2.1 show evidence of genomic redundancy with high copy numbers of certain genes known to have roles during cold-temperature growth (Goordial et al., 2016; Raymond-Bouchard et al., 2018). In this study, differential expression was observed for several gene copies including catalase, the iron uptake regulator fur, sigma-70 polymerase, universal stress protein (uspA), TRAP dicarboxylate permease, and ABC-type polar amino acid transporter (Table 2) when comparing the higher and lower growth temperatures. This may confer an advantage during cold growth whereby cold-adapted versions of a given gene product would be expressed and likely function better under that condition.

TABLE 2. Genomically redundant differentially expressed genes in Rhodococcus sp. JG-3 and Polaromonas sp. Eur3 1.2.1.
Iron Acquisition
Fourteen genes with roles in iron acquisition and transport were found to be upregulated greater than 1.5-fold at -5°C in Rhodococcus sp. JG3 compared to 25°C, with 1/2 of these increased >5-fold (Supplementary Table S2). This response may be one way to counteract the reduced solubility of iron at cold temperatures (Lide, 2005). The most highly upregulated were those involved in the biosynthesis and transport of siderophores, including mycobactin (mtbA, mtbB, and mtbI) and hydroxamate siderophores (pvdA), as well as a ferric siderophore reductase likely involved in reduction-mediated release of the iron from the siderophore. Siderophores are low molecular weight molecules with very high affinity for ferric iron (Fe3+). The gene for the transcriptional repressor Fur, which binds ferrous iron (Fe2+) and negatively regulates iron uptake genes, was downregulated over fourfold (Supplementary Table S2). Fe3+ is a crucial co-factor in numerous cellular processes and the concurrently large increases in transcripts for genes containing or synthesizing porphyrin and Fe–S cluster containing molecules, such as protoporphyrinogen IX oxidase, catalases, selenocysteine dehydrogenase, and flavoproteins, explains the greater need for iron in Rhodococcus sp. JG3. Genes for iron uptake were not differentially expressed in Polaromonas sp. Eur3 1.2.1 at 0°C relative to 20°C, though a few genes for Fe–S cluster formation were slightly increased (1.5–2 logFC) (Supplementary Table S3). As with Rhodococcus sp. JG3, Fe–S clusters are important co-factors for a number of processes that are increased at cold temperature in Polaromonas sp. Eur3 1.2.1 including tetrathionate reductase and cytochromes important for energy generation.
Large increases in iron acquisition mechanisms are not usually observed in cold-adapted bacteria during cold-temperature growth, and there is evidence that iron uptake genes and iron-associated proteins are suppressed in cold-adapted bacteria (Piette et al., 2011a; Ronholm et al., 2015; Tribelli et al., 2015). This suppression is theorized to perhaps contribute to alleviating oxidative stress by iron (Fenton reaction). However, as with Polaromonas sp. Eur3 1.2.1, increases in Fe–S cluster generating proteins is one exception that has been reported (Goodchild et al., 2005; Bergholz et al., 2009). While this strategy of reliance on additional iron during cold growth in Rhodococcus sp. JG3 can be partly explained within the context of its natural environment, the McMurdo Dry Valleys, which have relatively high soluble (bioavailable) iron content and noticeable ferric oxides (Tamppari et al., 2012; Bhattachan et al., 2015), it is intriguing and warrants further investigation.
Cell Envelope and Extracellular Polysaccharides
Some psychrophiles, such as P. halocryophilus and Colwellia psychrerythraea, create unique extracellular cell envelope structures at low temperatures, believed to offer some level of cryoprotection and antifreeze properties (Mykytczuk et al., 2013, 2015; De Maayer et al., 2014; Carillo et al., 2015). In C. psychrerythraea, this structure is a polysaccharide capsule with similarities to antifreeze proteins and glycoproteins. Both Polaromonas sp. Eur3 1.2.1 and Rhodococcus sp. JG3 increase the abundance of transcripts predicted to be in involved in exopolysaccharide (EPS) and capsular polysaccharide biosynthesis at their respective low temperatures compared to their respective higher temperatures. These included colanic acid biosynthesis, UDP-glucose lipid carrier transferase, cellulose synthase, EPS biosynthesis proteins, mannose-6-phosphate isomerase, and heparin binding hemagglutinin (Supplementary Tables S2, S3). The EPS biosynthesis protein is part of the EPS system in Bacillus subtilis (Roux et al., 2015), and heparin binding hemagglutinin induces aggregation of cells in mycobacteria (Menozzi et al., 1996). Mutational analyses to study impaired cold-temperature growth in the deep-sea bacterium Photobacterium profundum, found the largest fraction of genes associated with temperature sensitivity to be involved in the cell envelope formation, specifically extracellular polysaccharide biosynthesis (Lauro et al., 2008), implying that some level of EPS synthesis is especially relevant at cold temperatures. In addition, the capsule from C. psychrerythraea has been theorized to function as a protective mechanism at subzero temperatures (Carillo et al., 2015).
Cell Wall/Membrane
A common feature of many cold-adapted microorganisms is the presence of a system to increase or modulate membrane flexibility at lower temperatures (De Maayer et al., 2014). There is evidence for membrane modifications in Rhodococcus sp. JG3 at -5°C compared to 25°C with significant upregulation of an acyl-CoA thioesterase, an important regulator of lipid metabolism involved in both synthesis and degradation of fatty acids (Hunt and Alexson, 2002), as well as an acyl dehydratase and a putative flippase (Supplementary Table S2). Flippases are important for maintaining asymmetric distributions of phospholipids, while acyl dehydratases can create double bonds in fatty acids. Increasing content of unsaturated fatty acids in the membrane is a common mechanism to increase flexibility in cold-adapted organisms (De Maayer et al., 2014). Similar increases were not observed in Polaromonas sp. Eur3 1.2.1 However, emphasis on increased peptidoglycan synthesis and remodeling at 0°C compared to 20°C was apparent in Polaromonas sp. Eur3 1.2.1 with a ∼twofold upregulation of transcripts for three enzymes, undecaprenyl diphosphate synthase and UDP-N-acetylmuramyl tripeptide synthase (murE), vital for peptidoglycan synthesis, and membrane-bound lytic murein transglycosylase B implicated in cell wall remodeling and cell growth (Hunt and Alexson, 2002). P. halocryophilus also increases synthesis of peptidoglycans at cold temperatures, and forms a protective envelope on its surface made up partly of peptidoglycan (Mykytczuk et al., 2013, 2015; Raymond-Bouchard et al., 2017).
Transporters
Overall, numerous transcripts for transporters were strongly increased in both Rhodococcus sp. JG3 and Polaromonas sp. Eur3 1.2.1 at their colder temperatures compared to the warmer temperatures. In Rhodococcus sp. JG3, these were primarily branched-chain amino acid and peptide transporters, while Polaromonas sp. Eur3 1.2.1 increased a polar amino acid transport system. Both organisms upregulated sulfate transport and MFS family efflux pumps, including multidrug transporters in Rhodococcus sp. JG3, which can recognize a variety of substrates and may also be important for toxin removal. A dicarboxylate symporter was strongly induced in Rhodococcus sp. JG3 at -5°C compared to 25°C (>sevenfold; Supplementary Table S2), while in Polaromonas sp. Eur3 1.2.1 two tripartite tricarboxylate transporter receptor components, tctC, and a TRAP-type dicarboxylate transport system were increased two- to fivefold (Supplementary Table S3) at 0°C compared to the 20°C condition. Substrates for these transporters can include a variety of carboxylate containing molecules, including malate, fumarate, 2-oxobutyrate, and pyruvate, as well as the compatible solutes ectoine (Mulligan et al., 2011), which may be relevant for osmoregulation (discussed below). The most upregulated (5.6 logFC) transporter transcripts in Polaromonas sp. Eur3 1.2.1 at 0°C relative to 20°C were from an ompA-ompF type porin, which forms a channel for uptake of a variety of hydrophilic solutes (Confer and Ayalew, 2013). These types of porins were also increased at 4°C in the Antarctic bacterium Shewanella livingstonensis (Kawamoto et al., 2007). Overall, increasing transporters at low temperatures is an important strategy for eury- and steno-psychrophiles (Mock et al., 2005; Campanaro et al., 2011; Koh et al., 2017), who must overcome reduced diffusion rates and ensure efficient nutrient uptake (Nedwell, 1999).
Amino Acid Metabolism
In addition to the large increases in expression of amino acid transporters (discussed above), Rhodococcus sp. JG3 strongly upregulated transcripts for two of the enzymes involved in methionine biosynthesis at -5°C compared to 25°C, MetE and MetF, by four- and fivefold, respectfully (Supplementary Table S2), likely due to this amino acid’s importance in protein biosynthesis. In addition, methionine is a precursor to the major cellular methyl donor in methylation reactions, S-adenosyl methionine (Figge, 2006), and is consistent with increases in several methyltransferases during growth at -5°C (Supplementary Tables S2, S4). Smaller increases (<threefold) in expression of transcripts for enzymes involved in the synthesis of glutamine, cysteine, and branched-chain amino acids were also observed. Increases in amino acid synthesis was an important part of the response to cold growth in the marine bacterium Sphingopyxis alaskensis (Ting et al., 2010). Also of note is the induced presence of transcripts for two enzymes involved in the Kynurenine pathway, KynA and KynU, leading to the production of the co-enzyme nicotinamide adenine dinucleotide (NAD+) from the catabolism of tryptophan. NAD+ is an important co-factor in numerous cellular processes.
Carbon, Energy, and Co-enzyme Metabolism
Transcripts for numerous enzymes involved in glycolysis, D-mannose metabolism, the electron transport chain (ETC) and oxidative phosphorylation were increased in Polaromonas sp. Eur3 1.2.1 at 0°C compared to 20°C, including all subunits of NADH-quinone oxidoreductase, ATP synthase, cytochrome c553, triosephosphate isomerase, and mannose-6-phosphate isomerase (Supplementary Table S3). The increased abundance of all subunits of tetrathionate reductase highlights the potential for Polaromonas sp. Eur3 1.2.1 to utilize tetrathionate as a terminal electron acceptor and may be an important strategy for growth in permafrost at subzero temperatures by providing respiratory flexibility, including anaerobic respiration, for energy generation. The ability of stenopsychrophiles to have higher growth rates at cold temperatures has been attributed, at least in part, to the ability to increase energy acquisition. Photosynthesis in the stenopsychrophiles Fragilariopsis cylindrus and Chlorella sp. UMACC 234 was increased at -1 and 4°C, respectively (Mock and Hoch, 2005; Mock et al., 2005; Chong et al., 2011). The increase in expression of genes involved in oxidative phosphorylation may explain how Polaromonas sp. Eur3 1.2.1 is able to maintain growth rates at 0°C (0.19 d-1) that are very similar to >20°C (.21 d-1), by keeping energy metabolism at a constant rate and offsetting reduced reaction rates at low temperatures.
These same increases were not seen in Rhodococcus sp. JG3 for ETC and glycolysis, and in fact multiple cytochromes were decreased at -5°C compared to 25°C (Supplementary Tables S2, S4). However, there was a sevenfold upregulation of L-lactate dehydrogenase, responsible for either the conversion of L-lactate to pyruvate or the reverse reaction. There was also significant upregulation of genes involved in co-factor biosynthesis, biotin, NAD, porphyrin, and Fe–S clusters, and for maintaining redox potential. Amongst those most induced was six- and sevenfold upregulation of two enzymes for protoporphyrinogen IX oxidase, menaquinone-dependent, required for porphyrin synthesis (Supplementary Table S2). A pimeloyl-ACP methyl ester carboxylesterase, almost fivefold upregulated, functions as a gatekeeper to remove the methyl group from pimeloyl-ACP methyl ester producing pimeloyl-ACP which stops further elongation of fatty acid synthesis and shuttles pimeloyl-ACP into the biosynthesis pathway for the co-factor biotin (Agarwal et al., 2012). Iron–sulfur cluster biosynthesis was also increased (discussed above). Maintenance of redox potential could be observed at -5°C in Rhodococcus sp. JG3 through the increased presence of numerous dehydrogenases and oxidoreductases (Supplementary Tables S2, S4) when compared to 25°C. Overall, these results indicate that Rhodococcus sp. JG3 places emphasis on preserving cellular activity through redox homeostasis and co-factor synthesis at subzero temperatures.
The ability of Rhodococcus sp. JG3 to catabolize alcohols under subzero conditions was observed with large fold-change increases in homologs of a propanol-preferring alcohol dehydrogenase (7.21 logFC) and a secondary alcohol dehydrogenase (5.5 logFC). The large subunit of ethanolamine ammonia-lyase (eutB) was increased 6.5-fold, and is responsible, along with smaller subunit eutC, for cleaving ethanolamine to ammonia and ethanol, and subsequently, acetaldehyde (Tsoy et al., 2009). This suggests the potential for secondary alcohols, aromatic alcohols, and ethanolamine to serve as carbon sources and, in the case of ethanolamine, a nitrogen source as well (Roof and Roth, 1988). Numerous members of the genus Rhodococcus are capable of breaking down secondary and aromatic alcohols (Ludwig et al., 1995; Peng et al., 2006).
Finally, while strongly suppressed at -5°C relative to 25°C, all of the genes involved in the four steps of the phenylacetic acid catabolism pathway for the breakdown of aromatic compounds (paaA, paaB, paaC, paaD, paaE, paaG, paaI, paaZ, and paaH) were expressed in Rhodococcus sp. (Supplementary Table S2). While this pathway does not appear to be important for growth of Rhodococcus sp. at -5°C, where it may be preferable to focus available energy on streamlining primary metabolism and downregulate secondary metabolic processes, it is nevertheless worthwhile to note the catabolic potential of this organism.
Oxidative and Universal Stress Responses
There is some evidence that oxidative stress is higher at colder temperatures due to increased gas solubility and increases in rates of enzyme activity to adapt to reduced catalytic rates at lower temperatures (Chattopadhyay et al., 2011; De Maayer et al., 2014). This appears to be the case in Rhodococcus sp. JG3 and Polaromonas sp. Eur3 1.2.1 given the increase in transcripts associated with iron acquisition and redox potential observed at -5°C compared to 25°C in Rhodococcus sp. JG3, and the emphasis on formation and activity of Fe–S cofactors by both Rhodococcus sp. JG3 and Polaromonas sp. Eur3 1.2.1 at their respective colder temperatures when compared to their respective higher temperatures. Iron is involved in the production of oxygen radicals via the Fenton reaction (Fenton, 1894; Troxell and Hassan, 2013). To relieve oxidative stress, two catalases and a cytochrome peroxidase were increased almost twofold in Polaromonas sp. Eur3 1.2.1 (Supplementary Table S3). In Rhodococcus sp. JG3, a specific catalase gene was upregulated ∼threefold, replacing three other catalase genes, which were downregulated (Supplementary Table S2). A superoxide dismutase, responsible for converting the superoxide radical to oxygen or hydrogen peroxide, was also increased in Rhodococcus sp.
Rhodococcus sp. and Polaromonas sp. Eur3 1.2.1 possess 16 and 13 copies, respectively, of the uspA in their genome (Goordial et al., 2016; Raymond-Bouchard et al., 2018). Four of these were strongly (3.5- to 6.5-fold) induced during subzero growth in Rhodococcus sp. JG3 (Supplementary Table S2) compared to 25°C, while one copy of was upregulated about 3.5-fold in Polaromonas sp. Eur3 1.2.1, and two copies were decreased (Supplementary Table S3) from the higher to lower temperature. While the exact function of this family of genes remains poorly elucidated, they are induced by a variety of environmental stressors, such as nutrient starvation, extreme temperatures, high salinity, and drought. They are thought to protect the cell from stress and damage and in some cases are linked to protection against DNA-damaging agents (Kvint et al., 2003). Further studies looking into the function of uspA genes at low temperatures in Rhodococcus sp. JG3 and Polaromonas sp. Eur3 1.2.1 would be interesting, given their selective upregulation and differential expression patterns.
Compatible Solutes and Osmoregulation
Compatible solutes, small water soluble organic compounds, are accumulated by cold-adapted microbes and play an important role in resisting osmotic pressure caused by high salinity and low water activity associated with cryoenvironments, such as the brine veins in permafrost and ice (Doyle et al., 2012). In addition, compatible solutes also play a role as cryoprotectants and increase the stability of macromolecules, membranes, and proteins, as well as enhancing folding and ligand binding in the latter (Thomas et al., 2001; Yancey, 2005). Genes for the biosynthesis or transport of compatible solutes are induced at cold temperatures in several cold-adapted strains, including E. sibiricum, M. burtonii, and P. arcticus (Rodrigues et al., 2008; Bergholz et al., 2009; Campanaro et al., 2011), as well as this study. The choline transporter betT was increased by threefold, and components of the opuABCD system for uptake of the compatible solute glycine betaine were over fivefold increased at -5°C in Rhodococcus sp. JG3, as was a putative osmotically induced protein (osmC) (Supplementary Table S2) when compared to 25°C. OsmC in Mycobacterium spp. functions as a hydroperoxide reductase and may therefore have a role as an antioxidant in Rhodococcus sp. JG3 (Saikolappan et al., 2011). However, betT was increased, and genes involved in the synthesis of glycine betaine, betA and betB, were more than six- and eightfold decreased in Rhodococcus sp. JG3. This might indicate preference for the uptake of available solutes from the environment, rather than energy-demanding de novo synthesis of solutes. It is also possible that the synthesis genes were induced during the initial acclimation period and were subsequently downregulated once optimal concentrations of compatible solutes had been achieved, and thus were not detected in these cultures which were harvested at mid-late exponential phase. A similar theory has been proposed for P. halocryophilus (Mykytczuk et al., 2013).
Polaromonas sp. Eur3 1.2.1 does not appear to upregulate transport of compatible solutes to the extent that Rhodococcus sp. JG3 does, though this may be explained by the fact that the media used to culture Polaromonas sp. Eur3 1.2.1 did not contain additional NaCl beyond what was present in the growth medium (R2A). Nevertheless, Polaromonas sp. Eur3 1.2.1 also increases the abundance of transcripts for an osmotically inducible protein (osmB) by almost fourfold (Supplementary Table S3). A >threefold increase in a TRAP system capable of transporting ectoine suggests the potential for accumulation of compatible solutes. Even under low salt conditions, cold growth appears to be linked at least partially with osmoregulation systems.
Transcription, Signaling, and Motility
Numerous transcription factors (TFs) were strongly upregulated during cold-temperature growth in Rhodoccocus sp. and in Polaromonas sp. Eur3 1.2.1, though slightly less so. The predicted regulatory functions of these TFs are consistent with many transcripts increased in abundance in both organisms. Ten transcriptional regulators were found to be >threefold upregulated in Rhodoccocus sp. at -5°C compared to 25°C, with 9 more than fivefold upregulated (Supplementary Tables S2, S4), including two acrR family regulators, known to be involved in modulating responses to osmotic stress, modification and elimination of toxic substances, and lipid metabolism (Deng et al., 2013), the primary sigma-70 factor required for transcription initiation (Paget and Helmann, 2003), and the whiB7 transcriptional regulator, which is highly conserved in actinomycetes, with characterized roles in redox homeostasis, cell metabolism, and antibiotic resistance (Burian et al., 2012).
NsrR is a nitric oxide (NO) sensitive transcription repressor which can upregulate the ResDE two-component system required for induction of nitrate respiration genes and NO detoxifying enzymes in the presence of NO (Yukl et al., 2008). The elevated transcription of nsrR and the flavohemoglobin NO-detoxifying enzyme hmp suggests that Rhodococcus sp. JG3 may be under heightened NO stress at subzero temperatures when compared to higher temperatures. One of the genes involved in nitrate reduction, nitrite reductase, was detected, suggesting that Rhodococcus sp. JG3 may be capable of denitrification but it was >4.5-fold downregulated at -5°C relative to 25°C. MarR (∼sixfold increase) is important in the stress response and modification and export of toxic compounds, partly through the induction of efflux pumps (Grove, 2013), and ytrA (5.5-fold increased) is involved in regulation of ATP-binding cassette transporters (Suvorova et al., 2015). These TFs may have roles in the induction of the MFS pumps and ABC transporters observed in Rhodococcus sp. JG3 at -5°C (discussed above).
At cold temperatures both Rhodococcus sp. JG3 and Polaromonas sp. Eur3 1.2.1 show increased levels of transcription factors of the narL/fixJ/luxR and iclR families (Supplementary Tables S2, S3). In addition, Polaromonas sp. Eur3 1.2.1 induces 2 lysR family TFs and a regulator of the gntR family, the same family as ytrA in Rhodococcus sp. JG3 during 0°C growth as compared to 20°C. Members of the iclR family are transcriptional activators and repressors often involved in regulating carbon metabolism, such as the glyoxylate bypass operon, degradation of aromatic compounds, quorum-sensing, and multidrug resistance (Sunnarborg et al., 1990; Chao and Zhou, 2013). narL/fixJ/luxR comprises a large family of TFs and, therefore, the exact role of these in Polaromonas sp. Eur3 1.2.1 and Rhodococcus sp. JG3 is hard to pinpoint but potential roles could include activating the nitrate reductase operon and nitrogen-fixation genes and uptake of the compatible solute ectoine (Rabin and Stewart, 1993; Galiniers et al., 1994; Rodríguez-Moya et al., 2010). The presence of the nac/lysR regulator, or nitrogen assimilation control protein, in Polaromonas sp. Eur3 1.2.1 would suggest that at cold temperatures Polaromonas sp. Eur3 1.2.1 experiences nitrogen limitation (Bender, 2010). Signaling systems associated with these transcription factors were induced about two- to threefold at 0°C in Polaromonas sp. Eur3 1.2.1 (Supplementary Tables S3, S5) including the two-component system of fixJ/luxR, adenylate cyclase, and components of the cAMP receptor protein, a global transcriptional activator that regulates transcription of many genes, including energy metabolism, consistent with increases in energy metabolism observed in the bacterium at 0°C. Overall, the transcription factors upregulated at cold temperatures suggest the induction of numerous pathways important during cold growth, including lipid metabolism, redox homeostasis, ABC transporters, and osmoregulation in Rhodococcus sp. JG3, and ABC transporters, carbon and energy metabolism, and export of toxic compounds in Polaromonas sp. Eur3 1.2.1.
Conclusion
Overall, the transcriptomic responses to cold growth in Rhodococcus sp. and Polaromonas sp. Eur3 1.2.1, shared many cold-adaptive features (Figure 2). Common responses included induction of translation and ribosomal processes, which we hypothesize results in translationally active cold-adapted ribosomes, upregulation of nutrient transport, increased oxidative and osmotic stress responses, modulating cell wall/membrane features, induction of EPS synthesis, and accumulation of compatible solutes, though this last item was much more pronounced in Rhodococcus sp. Recombination and genomic redundancy also appeared to be a shared strategy at low temperatures, though the mechanism used to achieve this was different in each organism. Polaromonas sp. Eur3 1.2.1 utilized specific transposases, while Rhodococcus sp. induced recombination proteins. The presence of the above properties in most psychrophiles, whether eury- or steno-, suggests that these may be conserved adaptive features, necessary for growth at low temperatures in most organisms.
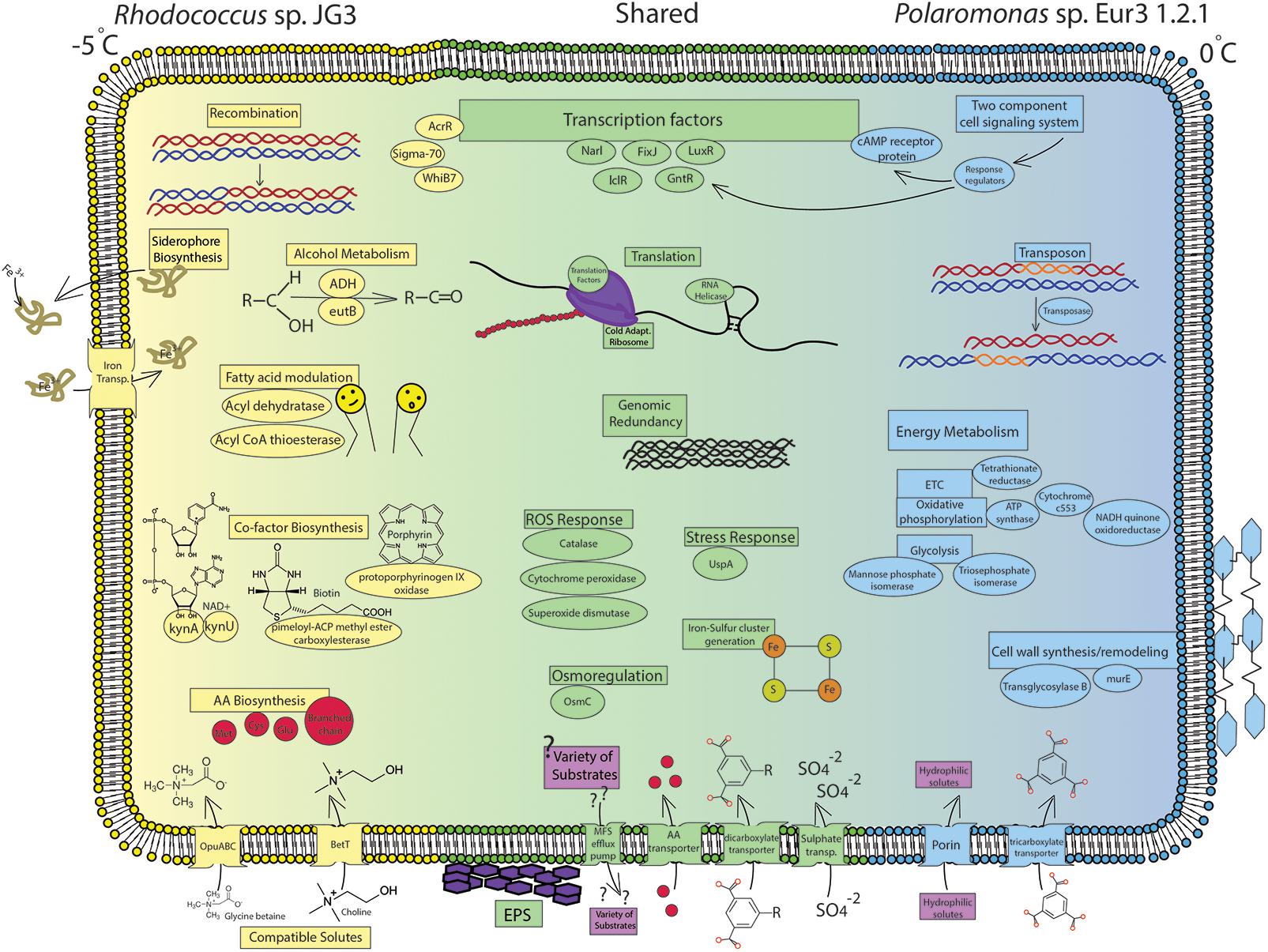
FIGURE 2. Cell diagram showing important processses and pathways increased during cold growth. Blue labels represent processes increased in Polamonas sp. Eur3 1.2.1 at 0°C, yellow labels represent processes increased in Rhodococcus sp. JG3 at –5°C, and green labels represent shared processes increased in both organisms during cold growth. Full gene descriptions corresponding to the short protein names used in the figure can be found in Supplementary Tables S2–S5.
In Rhodococcus sp. JG3, the marked differences during cold growth featured increased abundance of transcripts involved in iron transport, amino acid transport and metabolism, modulating fatty acid synthesis and composition, catabolism of alcohols/ethanolamine, and sustaining redox potential (Figure 2). Conversely, Polaromonas sp. Eur3 1.2.1 was found to induce energy metabolism relating to the ETC, oxidative phosphorylation, and glycolysis, as well as global signal transduction mechanisms and transport and metabolism of carboxylates (Figure 2). In addition, enzymes vital to peptidoglycan synthesis and modulation were increased. Finally, Polaromonas sp. Eur3 1.2.1 differentially up/downregulated a specific subset of transposases at the two growth temperatures.
As mentioned previously, the two organisms used in this study are from two different phyla. As such, we were able to investigate relative changes that may be largely conserved cold adaptations vs. ones that may be specific to certain groups. Though we have yet to identify such candidates from permafrost, in the future, comparing a steno-and a eury-psychrophile from the same genus would also be worthwhile to narrow down more specific adaptive features. It is also important to note that some functions involved in cold adaptation may not be regulated at the transcriptional level and it is likely that some regulation occurs at the posttranscriptional level as well. Overall, however, this study does provide us with valuable insight into the cold-adaptive strategies of two different cold-adapted organisms. We also recognize that we focused exclusively on RNA-seq data. Our goal was to identify genes and mechanisms of interest that may be important during cold growth to provide a basis for future studies.
Increased activity of primary energy production and the ETC in Polaromonas sp. Eur3 1.2.1 and other stenopsychrophiles, is likely to be one major mechanism by which these organisms are able to sustain optimal growth at colder temperatures or consistent growth rates over their temperature range. The more attenuated transcriptomic response in Polaromonas sp. Eur3 1.2.1 also suggests that the organism may be, at least partly, constitutively adapted to colder temperatures or for growth over its temperature range and does not need to upregulate its response to the same levels as Rhodococcus sp. JG3 at lower temperatures. However, more research will need to be done to determine if this is true and to identify those features which are unique to these types of organisms, and differ from eurypsychrophiles. Certainly, it may simply be that stenopsychrophiles are adapted to grow at a narrow range and temperature changes, both above and below this range, are difficult for these organisms. In the same vein, it is difficult to pinpoint the mechanisms that favor eurypsychrophilic growth at subzero temperatures from this study alone, though in the case of Rhodococcus sp. JG3 increased co-factor formation, flexibility in use of carbon sources, control of fatty acid composition in the membrane, iron acquisition, and sustaining redox potential appear to be important factors. These may represent adaptive strategies that allow this organism to grow at subzero temperatures, in addition to sustaining growth over a wide range of temperatures (-5 to 30°C). It is hoped that further studies will better elucidate the role that these mechanisms may play in cold growth in both eury- and steno-psychrophiles.
Author Contributions
IR-B wrote the manuscript, performed all laboratory experiments, and all downstream analyses with the results of the pipeline data. JT performed bioinformatic analysis of the raw RNAseq data using the transcriptomic pipeline, provided experimental guidance, and manuscript feedback and editing. IA helped with creation of the cell figure, assisted with library preparation, and participated in manuscript editing and discussion. CG and LW provided guidance, and critical feedback and editing of the manuscript.
Funding
Funding for this research was provided by the Natural Sciences and Engineering Research Council of Canada (NSERC) through an NSERC CREATE Ph.D. fellowship and an NSERC Canadian Graduate Scholarship to IR-B, and through NSERC Discovery and Northern Research Supplement grants and a Polar Continental Shelf Project grant to LW.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the U.S. Department of Energy Joint Genome Institute (JGI) who sequenced both genomes under a Community Sequencing Program Project (Quartely; ID 1015712). We also wish to acknowledge Compute Canada for access to the McGill University High Performance Computing (HPC) infrastructure (Guillimin system).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01565/full#supplementary-material
Footnotes
- ^ http://bio-bwa.sourceforge.net
- ^ https://img.jgi.doe.gov/cgi-bin/mer/main.cgi
- ^ https://www.bioconductor.org/packages/3.3/bioc/vignettes/edgeR/inst/doc/edgeRUsersGuide.pdf
- ^ http://www.ncbi.nlm.nih.gov/bioproject/422596
- ^ https://img.jgi.doe.gov/cgi-bin/mer/main.cgi
References
Agarwal, V., Lin, S., Lukk, T., Nair, S. K., and Cronan, J. E. (2012). Structure of the enzyme-acyl carrier protein (ACP) substrate gatekeeper complex required for biotin synthesis. Proc. Natl. Acad. Sci. U.S.A. 109, 17406–17411. doi: 10.1073/pnas.1207028109
Allen, M. A., Lauro, F. M., Williams, T. J., Burg, D., Siddiqui, K. S., De Francisci, D., et al. (2009). The genome sequence of the psychrophilic archaeon, Methanococcoides burtonii: the role of genome evolution in cold adaptation. ISME J. 3, 1012–1035. doi: 10.1038/ismej.2009.45
Ayala-del-Río, H. L., Chain, P. S., Grzymski, J. J., Ponder, M. A., Ivanova, N., Bergholz, P. W., et al. (2010). The genome sequence of Psychrobacter arcticus 273-4, a psychroactive Siberian permafrost bacterium, reveals mechanisms for adaptation to low-temperature growth. Appl. Environ. Microbiol. 76, 2304–2312. doi: 10.1128/AEM.02101-09
Bakermans, C., Bergholz, P. W., Rodrigues, D. F., Vishnivetskaya, T. A., Ayala-Del-Rio, H., and Tiedje, J. M. (2012). “Genomic and expression analyses of cold-adapted microorganisms,” in Polar Microbiology: Life in a Deep Freeze, eds R. V. Miller and L. G. Whyte (Washington, DC: ASM Press), 126–155. doi: 10.1128/9781555817183.ch6
Bakermans, C., Tollaksen, S. L., Giometti, C. S., Wilkerson, C., Tiedje, J. M., and Thomashow, M. F. (2007). Proteomic analysis of Psychrobacter cryohalolentis K5 during growth at subzero temperatures. Extremophiles 11, 343–354. doi: 10.1007/s00792-006-0042-1
Bender, R. A. (2010). A NAC for regulating metabolism: the nitrogen assimilation control protein (NAC) from Klebsiella pneumoniae. J. Bacteriol. 192, 4801–4811. doi: 10.1128/JB.00266-10
Bergholz, P. W., Bakermans, C., and Tiedje, J. M. (2009). Psychrobacter arcticus 273-4 uses resource efficiency and molecular motion adaptations for subzero temperature growth. J. Bacteriol. 191, 2340–2352. doi: 10.1128/JB.01377-08
Bhattachan, A., Wang, L., Miller, M. F., Licht, K. J., and D’Odorico, P. (2015). Antarctica’s dry valleys: A potential source of soluble iron to the southern ocean? Geophys. Res. Lett. 42, 1912–1918. doi: 10.1002/2015GL063419
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Burian, J., Ramón-García, S., Sweet, G., Gómez-Velasco, A., Av-Gay, Y., and Thompson, C. J. (2012). The mycobacterial transcriptional regulator whiB7 gene links redox homeostasis and intrinsic antibiotic resistance. J. Biol. Chem. 287, 299–310. doi: 10.1074/jbc.M111.302588
Campanaro, S., Williams, T. J., Burg, D. W., De Francisci, D., Treu, L., Lauro, F. M., et al. (2011). Temperature-dependent global gene expression in the Antarctic archaeon Methanococcoides burtonii. Environ. Microbiol. 13, 2018–2038. doi: 10.1111/j.1462-2920.2010.02367.x
Carillo, S., Casillo, A., Pieretti, G., Parrilli, E., Sannino, F., Bayer-Giraldi, M., et al. (2015). A unique capsular polysaccharide structure from the psychrophilic marine bacterium Colwellia psychrerythraea 34H that mimics antifreeze (glyco)proteins. J. Am. Chem. Soc. 137, 179–189. doi: 10.1021/ja5075954
Cavicchioli, R. (2006). Cold-adapted archaea. Nat. Rev. Microbiol. 4, 331–343. doi: 10.1038/nrmicro1390
Chao, H., and Zhou, N. Y. (2013). GenR, an IclR-type regulator, activates and represses the transcription of gen genes involved in 3-hydroxybenzoate and gentisate catabolism in Corynebacterium glutamicum. J. Bacteriol 195, 1598–1609. doi: 10.1128/JB.02216-12
Chattopadhyay, M. K., Raghu, G., Sharma, Y. V., Biju, A. R., Rajasekharan, M. V., and Shivaji, S. (2011). Increase in oxidative stress at low temperature in an antarctic Bacterium. Curr. Microbiol. 62, 544–546. doi: 10.1007/s00284-010-9742-y
Chong, G.-L., Chu, W.-L., Othman, R. Y., and Phang, S.-M. (2011). Differential gene expression of an Antarctic Chlorella in response to temperature stress. Polar Biol. 34, 637–645. doi: 10.1007/s00300-010-0918-5
Confer, A. W., and Ayalew, S. (2013). The OmpA family of proteins: roles in bacterial pathogenesis and immunity. Vet. Microbiol. 163, 207–222. doi: 10.1016/j.vetmic.2012.08.019
Dammel, C. S., and Noller, H. F. (1995). Suppression of a cold-sensitive mutation in 16s rRNA by overexpression of a novel ribosome-binding factor, RbfA. Genes Dev. 9, 626–637. doi: 10.1101/gad.9.5.626
De Maayer, P., Anderson, D., Cary, C., and Cowan, D. A. (2014). Some like it cold: understanding the survival strategies of psychrophiles. EMBO Rep. 15, 508–517. doi: 10.1002/embr.201338170
Deana, A., and Belasco, J. G. (2004). The function of RNase G in Escherichia coli is constrained by its amino and carboxyl termini. Mol. Microbiol. 51, 1205–1217. doi: 10.1046/j.1365-2958.2003.03905.x
Deng, W., Li, C., and Xie, J. (2013). The underling mechanism of bacterial TetR/AcrR family transcriptional repressors. Cell. Signal. 25, 1608–1613. doi: 10.1016/j.cellsig.2013.04.003
Doyle, S., Dieser, M., Broemsen, E., and Christner, B. (2012). “General characteristics of cold-adapted microorganisms,” in Polar Microbiology: Life in a Deep Freeze, eds R. V. Miller and L. G. Whyte (Washington, DC: ASM Press), 103–125.
Feller, G., and Gerday, C. (2003). Psychrophilic enzymes: hot topics in cold adaptation. Nat. Rev. Microbiol. 1, 200–208. doi: 10.1038/nrmicro773
Fenton, H. J. H. (1894). Oxidation of tartaric acid in the presence of iron. J. Chem. Soc 65, 899–910. doi: 10.1039/ct8946500899
Figge, R. M. (2006). “Methionine biosynthesis in Escherichia coli and Corynebacterium glutamicum,” in Amino Acid Biosynthesis, ed. V. F. Wendisch (Berlin: Springer-Verlag), 163–193. doi: 10.1007/7171
Galiniers, A., Garnerone, A., Kahn, D., and Batutn, J. (1994). Phosphorylation of the Rhizobium meliloti FixJ protein induces its binding to a compound regulatory region at the fixK promoter∗. J. Biol. Chem. 269, 23784–23789.
Gilichinsky, D., Rivkina, E., Bakermans, C., Shcherbakova, V., Petrovskaya, L., Ozerskaya, S., et al. (2005). Biodiversity of cryopegs in permafrost. FEMS Microbiol. Ecol. 53, 117–128. doi: 10.1016/j.femsec.2005.02.003
Gleiter, S., and Bardwell, J. C. A. (2008). Disulfide bond isomerization in prokaryotes Stefan. Biochim. Biophys. Acta 1783, 530–534. doi: 10.1016/j.bbamcr.2008.02.009
Goodchild, A., Raftery, M., Saunders, N. F. W., Guilhaus, M., and Cavicchioli, R. (2004). Biology of the cold-adapted archaeon, Methanococcoides burtonii, determined by proteomics using liquid chromatography-tandem mass spectrometry. J. Proteome Res. 3, 1164–1176. doi: 10.1021/pr0498988
Goodchild, A., Raftery, M., Saunders, N. F. W., Guilhaus, M., and Cavicchioli, R. (2005). Cold adaptation of the Antarctic archaeon, Methanococcoides burtonii assessed by proteomics using ICAT. J. Proteome Res. 4, 473–480. doi: 10.1021/pr049760p
Goordial, J., Raymond-Bouchard, I., Zolotarov, Y., de Bethencourt, L., Ronholm, J., Shapiro, N., et al. (2016). Cold adaptive traits revealed by comparative genomic analysis of the eurypsychrophile Rhodococcus sp. JG3 isolated from high elevation McMurdo Dry Valley permafrost, Antarctica. FEMS Microbiol. Ecol. 92:fiv154. doi: 10.1093/femsec/fiv154
Grove, A. (2013). MarR family transcription factors. Curr. Biol. 23, R142–R143. doi: 10.1016/j.cub.2013.01.013
Hunt, M. C., and Alexson, S. E. H. (2002). The role Acyl-CoA thioesterases play in mediating intracellular lipid metabolism. Prog. Lipid Res. 41, 99–130. doi: 10.1016/S0163-7827(01)00017-0
Hwang, Y., Jung, G., and Jin, E. (2008). Transcriptome analysis of acclimatory responses to thermal stress in Antarctic algae. Biochem. Biophys. Res. Commun. 367, 635–641. doi: 10.1016/j.bbrc.2007.12.176
Jones, P. G., and Inouye, M. (1996). RbfA, a 30S ribosomal binding factor, is a cold-shock protein whose absence triggers the cold-shock response. Mol. Microbiol. 21, 1207–1218. doi: 10.1111/j.1365-2958.1996.tb02582.x
Kawamoto, J., Kurihara, T., and Esaki, N. (2017). “Proteomic insights of psychrophiles,” in Psychrophiles: From Biodiversity to Biotechnology, ed. R. Margesin (Cham: Springer), 423–435. doi: 10.1007/978-3-319-57057-0_17
Kawamoto, J., Kurihara, T., Kitagawa, M., Kato, I., and Esaki, N. (2007). Proteomic studies of an Antarctic cold-adapted bacterium, Shewanella livingstonensis Ac 10, for global identification of cold-inducible proteins. Extremophiles 11, 819–826. doi: 10.1007/s00792-007-0098-6
Koh, H. Y., Park, H., Lee, J. H., Han, S. J., Sohn, Y. C., and Lee, S. G. (2017). Proteomic and transcriptomic investigations on cold-responsive properties of the psychrophilic Antarctic bacterium Psychrobacter sp. PAMC 21119 at subzero temperatures. Environ. Microbiol. 19, 628–644. doi: 10.1111/1462-2920.13578
Kvint, K., Nachin, L., Diez, A., and Nyström, T. (2003). The bacterial universal stress protein: function and regulation. Curr. Opin. Microbiol. 6, 140–145. doi: 10.1016/S1369-5274(03)00025-0
Lauro, F. M., Tran, K., Vezzi, A., Vitulo, N., Valle, G., and Bartlett, D. H. (2008). Large-scale transposon mutagenesis of Photobacterium profundum SS9 reveals new genetic loci important for growth at low temperature and high pressure. J. Bacteriol. 190, 1699–1709. doi: 10.1128/JB.01176-07
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Lide, D. (2005). “Aqueous solubility of inorganic compounds at various temperatures,” in CRC Handbook of Chemistry and Physics ed. D. R. Lide (Boca Raton, FL: CRC Press), 1–6.
Liu, C., and Yu, X. (2015). ADP-ribosyltransferases and poly ADP-ribosylation. Curr. Protein Pept. Sci. 16, 491–501. doi: 10.2174/1389203716666150504122435
Ludwig, B., Akundi, A., Kendall, K., Ludwig, B., Akundi, A., and Kendall, K. (1995). A long-chain secondary alcohol dehydrogenase from Rhodococcus erythropolis ATCC 4277. Appl. Environ. Microbiol. 61, 3729–3733.
Math, R. K., Jin, H. M., Kim, J. M., Hahn, Y., Park, W., Madsen, E. L., et al. (2012). Comparative genomics reveals adaptation by Alteromonas sp. SN2 to marine tidal-flat conditions: cold tolerance and aromatic hydrocarbon metabolism. PLoS One 7:e35784. doi: 10.1371/journal.pone.0035784
Menozzi, F. D., Rouse, J. H., Alavi, M., Laude-Sharp, M., Muller, J., Bischoff, R., et al. (1996). Identification of a heparin-binding hemagglutinin present in mycobacteria. J. Exp. Med. 184, 993–1001. doi: 10.1084/jem.184.3.993
Methé, B. A., Nelson, K. E., Deming, J. W., Momen, B., Melamud, E., Zhang, X., et al. (2005). The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc. Natl. Acad. Sci. U.S.A. 102, 10913–10918. doi: 10.1073/pnas.0504766102
Mock, T., and Hoch, N. (2005). Long-term temperature acclimation of photosynthesis in steady-state cultures of the polar diatom Fragilariopsis cylindrus. Photosynth. Res. 85, 307–317. doi: 10.1007/s11120-005-5668-9
Mock, T., Krell, A., Valentin, K., Glockner, G., Kolukisaoglu, U., and Valentin, K. (2005). Analysis of expressed sequence tags (ESTs) from the polar diatom Fragilariopsis cylindrus. J. Phycol. 42, 78–85. doi: 10.1111/j.1529-8817.2005.00164.x
Mulligan, C., Fischer, M., and Thomas, G. H. (2011). Tripartite ATP-independent periplasmic (TRAP) transporters in bacteria and archaea. FEMS Microbiol. Rev. 35, 68–86. doi: 10.1111/j.1574-6976.2010.00236.x
Mykytczuk, N. C., Foote, S. J., Omelon, C. R., Southam, G., Greer, C. W., and Whyte, L. G. (2013). Bacterial growth at -15 °C; molecular insights from the permafrost bacterium Planococcus halocryophilus Or1. ISME J. 7, 1211–1226. doi: 10.1038/ismej.2013.8
Mykytczuk, N. C. S., Lawrence, J. R., Omelon, C. R., Southam, G., and Whyte, L. G. (2015). Microscopic characterization of the bacterial cell envelope of Planococcus halocryophilus Or1 during subzero growth at -15 °C. Polar Biol. 39, 701–712. doi: 10.1007/s00300-015-1826-5
Nedwell, D. D. (1999). Effect of low temperature on microbial growth: lowered affinity for substrates limits growth at low temperature. FEMS Microbiol. Ecol. 30, 101–111. doi: 10.1111/j.1574-6941.1999.tb00639.x
Nunn, B. L., Slattery, K. V., Cameron, K. A., Timmins-Schiffman, E., and Junge, K. (2015). Proteomics of Colwellia psychrerythraea at subzero temperatures - a life with limited movement, flexible membranes and vital DNA repair. Environ. Microbiol. 17, 2319–2335. doi: 10.1111/1462-2920.12691
Paget, M. S. B., and Helmann, J. D. (2003). The sigma70 family of sigma factors. Genome Biol. 4:203. doi: 10.1186/gb-2003-4-1-203
Peng, X., Taki, H., Komukai, S., Sekine, M., Kanoh, K., Kasai, H., et al. (2006). Characterization of four Rhodococcus alcohol dehydrogenase genes responsible for the oxidation of aromatic alcohols. Appl. Microbiol. Biotechnol. 71, 824–832. doi: 10.1007/s00253-005-0204-6
Piette, F., D’Amico, S., Mazzucchelli, G., Danchin, A., Leprince, P., and Feller, G. (2011a). Life in the cold: a proteomic study of cold-repressed proteins in the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Appl. Environ. Microbiol. 77, 3881–3883. doi: 10.1128/AEM.02757-10
Piette, F., Struvay, C., and Feller, G. (2011b). The protein folding challenge in psychrophiles: facts and current issues. Environ. Microbiol. 13, 1924–1933. doi: 10.1111/j.1462-2920.2011.02436.x
Quinlan, A. R., and Hall, I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. doi: 10.1093/bioinformatics/btq033
R Core Team (2017). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Rabin, R. S., and Stewart, V. (1993). Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coli K-12. J. Bacteriol. 175, 3259–3268. doi: 10.1128/jb.175.11.3259-3268.1993
Rabus, R., Ruepp, A., Frickey, T., Rattei, T., Fartmann, B., Stark, M., et al. (2004). The genome of Desulfotalea psychrophila, a sulfate-reducing bacterium from permanently cold Arctic sediments. Environ. Microbiol. 6, 887–902. doi: 10.1111/j.1462-2920.2004.00665.x
Raymond-Bouchard, I., Chourey, K., Altshuler, I., Iyer, R., Hettich, R. L., and Whyte, L. G. (2017). Mechanisms of subzero growth in the cryophile Planococcus halocryophilus determined through proteomic analysis. Environ. Microbiol. 19, 4460–4479. doi: 10.1111/1462-2920.13893
Raymond-Bouchard, I., Goordial, J., Zolotarov, Y., Ronholm, J., Stromvik, M., Bakermans, C., et al. (2018). Conserved genomic and amino acid traits of cold adaptation in subzero-growing Arctic permafrost bacteria. FEMS Microbiol. Ecol. 94, 1–13. doi: 10.1093/femsec/fiy023
Raymond-Bouchard, I., and Whyte, L. G. (2017). “From transcriptomes to metatranscriptomes: cold adaptation and active metabolisms of psychrophiles from cold environments,” in Psychrophiles: From Biodiversity to Biotechnology, ed. R. Margesin (Berlin: Springer), 437–457. doi: 10.1007/978-3-319-57057-0
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2009). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rodrigues, D. F., Ivanova, N., He, Z., Huebner, M., Zhou, J., and Tiedje, J. M. (2008). Architecture of thermal adaptation in an Exiguobacterium sibiricum strain isolated from 3 million year old permafrost: a genome and transcriptome approach. BMC Genomics 9:547. doi: 10.1186/1471-2164-9-547
Rodríguez-Moya, J., Argandoña, M., Reina-Bueno, M., Nieto, J. J., Iglesias-Guerra, F., Jebbar, M., et al. (2010). Involvement of EupR, a response regulator of the NarL/FixJ family, in the control of the uptake of the compatible solutes ectoines by the halophilic bacterium Chromohalobacter salexigens. BMC Microbiol. 10:256. doi: 10.1186/1471-2180-10-256
Ronholm, J., Raymond-Bouchard, I., Creskey, M., Cyr, T., Cloutis, E. A., and Whyte, L. G. (2015). Characterizing the surface-exposed proteome of Planococcus halocryophilus during cryophilic growth. Extremophiles 19, 619–629. doi: 10.1007/s00792-015-0743-4
Roof, D. M., and Roth, J. R. (1988). Ethanolamine utilization in Salmonella typhimurium. J. Bacteriol. 170, 3855–3863. doi: 10.1128/jb.170.9.3855-3863.1988
Roux, D., Cywes-Bentley, C., Zhang, Y. F., Pons, S., Konkol, M., Kearns, D. B., et al. (2015). Identification of Poly-N-acetylglucosamine as a major polysaccharide component of the bacillus subtilis biofilm matrix. J. Biol. Chem. 290, 19261–19272. doi: 10.1074/jbc.M115.648709
Saikolappan, S., Das, K., Sasindran, S. J., Jagannath, C., and Dhandayuthapani, S. (2011). OsmC proteins of Mycobacterium tuberculosis and Mycobacterium smegmatis protect against organic hydroperoxide stress. Tuberculosis 91, 1–22. doi: 10.1016/j.tube.2011.10.021
Shao, F., Bader, M. W., Jakob, U., and Bardwell, J. C. A. (2000). DsbG, a protein disulfide isomerase with chaperone activity. J. Biol. Chem. 275, 13349–13352. doi: 10.1074/jbc.275.18.13349
Steven, B., Pollard, W. H., Greer, C. W., and Whyte, L. G. (2008). Microbial diversity and activity through a permafrost/ground ice core profile from the Canadian high Arctic. Environ. Microbiol. 10, 3388–3403. doi: 10.1111/j.1462-2920.2008.01746.x
Sunnarborg, A., Klumpp, D., Chung, T., and LaPorte, D. C. (1990). Regulation of the glyoxylate bypass operon: Cloning and characterization of iclR. J. Bacteriol. 172, 2642–2649. doi: 10.1128/jb.172.5.2642-2649.1990
Suvorova, I. A., Korostelev, Y. D., and Gelfand, M. S. (2015). GntR family of bacterial transcription factors and their DNA binding motifs: structure, positioning and co-evolution. PLoS One 10:e0132618. doi: 10.1371/journal.pone.0132618
Tamppari, L. K., Anderson, R. M., Archer, P. D., Douglas, S., Kounaves, S. P., Mckay, C. P., et al. (2012). Effects of extreme cold and aridity on soils and habitability: McMurdo Dry Valleys as an analogue for the Mars Phoenix landing site. Antarct. Sci. 24, 211–228. doi: 10.1017/S0954102011000800
Thomas, T., Kumar, N., and Cavicchioli, R. (2001). Effects of ribosomes and intracellular solutes on activities and stabilities of elongation factor 2 proteins from psychrotolerant and thermophilic Methanogens. J. Bacteriol. 183, 1974–1982. doi: 10.1128/JB.183.6.1974
Ting, L., Williams, T. J., Cowley, M. J., Lauro, F. M., Guilhaus, M., Raftery, M. J., et al. (2010). Cold adaptation in the marine bacterium, Sphingopyxis alaskensis, assessed using quantitative proteomics. Environ. Microbiol. 12, 2658–2676. doi: 10.1111/j.1462-2920.2010.02235.x
Toseland, A., Daines, S. J., Clark, J. R., Kirkham, A., Strauss, J., Uhlig, C., et al. (2013). The impact of temperature on marine phytoplankton resource allocation and metabolism. Nat. Clim. Chang. 3, 979–984. doi: 10.1038/nclimate1989
Tribelli, P. M., Venero, E. C. S., Ricardi, M. M., Gómez-Lozano, M., Iustman, L. J. R., Molin, S., et al. (2015). Novel essential role of ethanol oxidation genes at low temperature revealed by transcriptome analysis in the Antarctic bacterium Pseudomonas extremaustralis. PLoS One 10:e0145353. doi: 10.1371/journal.pone.0145353
Troxell, B., and Hassan, H. M. (2013). Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria. Front. Cell. Infect. Microbiol. 3:59. doi: 10.3389/fcimb.2013.00059
Tsoy, O., Ravcheev, D., and Mushegian, A. (2009). Comparative genomics of ethanolamine utilization. J. Bacteriol 191, 7157–7164. doi: 10.1128/JB.00838-09
Yancey, P. H. (2005). Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J. Exp. Biol. 208, 2819–2830. doi: 10.1242/jeb.01730
Yukl, E., Elbaz, M., Nakano, M., and Moënne-Loccoz, P. (2008). Transcription factor NsrR from Bacillus subtilis senses nitric oxide with a 4Fe- 4S Cluster†. Biochemistry 47, 13084–13092. doi: 10.1021/bi801342x.Transcription
Keywords: eurypsychrophile, stenopsychrophile, transcriptomics, cold growth, cold adaptation, permafrost, Polaromonas, Rhodococcus
Citation: Raymond-Bouchard I, Tremblay J, Altshuler I, Greer CW and Whyte LG (2018) Comparative Transcriptomics of Cold Growth and Adaptive Features of a Eury- and Steno-Psychrophile. Front. Microbiol. 9:1565. doi: 10.3389/fmicb.2018.01565
Received: 09 April 2018; Accepted: 25 June 2018;
Published: 31 July 2018.
Edited by:
Masahiro Ito, Toyo University, JapanReviewed by:
Jun Kawamoto, Kyoto University, JapanDoug Bartlett, University of California, San Diego, United States
Copyright © 2018 Raymond-Bouchard, Tremblay, Altshuler, Greer and Whyte. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lyle G. Whyte, bHlsZS53aHl0ZUBtY2dpbGwuY2E=
 Isabelle Raymond-Bouchard
Isabelle Raymond-Bouchard Julien Tremblay
Julien Tremblay Ianina Altshuler
Ianina Altshuler Charles W. Greer
Charles W. Greer Lyle G. Whyte
Lyle G. Whyte